Programmed cell death preventing heterokaryon formation in fungi has been proposed to represent a defense mechanism against conspecific genome exploitation and mycoviruses. Here, Daskalov et al. identified a novel genetic determinant of allorecognition...
Keywords: Neurospora, programmed cell death, allorecognition, heterokaryon incompatibility, cell fusion
Abstract
Nonself recognition following cell fusion between genetically distinct individuals of the same species in filamentous fungi often results in a programmed cell death (PCD) reaction, where the heterokaryotic fusion cell is compartmentalized and rapidly killed. The allorecognition process plays a key role as a defense mechanism that restricts genome exploitation, resource plundering, and the spread of deleterious senescence plasmids and mycoviruses. Although a number of incompatibility systems have been described that function in mature hyphae, less is known about the PCD pathways in asexual spores, which represent the main infectious unit in various human and plant fungal pathogens. Here, we report the identification of regulator of cell death-1 (rcd-1), a novel allorecognition gene, controlling PCD in germinating asexual spores of Neurospora crassa; rcd-1 is one of the most polymorphic genes in the genomes of wild N. crassa isolates. The coexpression of two antagonistic rcd-1-1 and rcd-1-2 alleles was necessary and sufficient to trigger cell death in fused germlings and in hyphae. Based on analysis of wild populations of N. crassa and N. discreta, rcd-1 alleles appeared to be under balancing selection and associated with trans-species polymorphisms. We shed light on genomic rearrangements that could have led to the emergence of the incompatibility system in Neurospora and show that rcd-1 belongs to a much larger gene family in fungi. Overall, our work contributes toward a better understanding of allorecognition and PCD in an underexplored developmental stage of filamentous fungi.
ALLORECOGNITION or discrimination against conspecific but genetically distinct nonself is a phenomenon that is important in various colonial organisms that limits genome exploitation and resource plundering (Debets and Griffiths 1998; Laird et al. 2005; Ho et al. 2013; Bastiaans et al. 2016). The phenomenon has been described in early diverging invertebrate metazoans, including the ascidian Botryllus schlosseri, the cnidarian Hydractinia symbiolongicarpus, and in the social amoebae Dictyostelium discoideum, where it restricts colonial chimerism to closely related individuals (Laird et al. 2005; Lakkis et al. 2008; Rosengarten and Nicotra 2011). In filamentous fungi, where cell fusion (anastomosis) between genetically different fungal colonies is often abortive, allorecognition mechanisms play a similar role by triggering a programmed cell death (PCD) reaction termed vegetative or heterokaryon incompatibility (HI) (Glass and Dementhon 2006; Paoletti 2016; Daskalov et al. 2017). The cell death reaction is spatially and temporally restricted to the heterokaryotic fusion cells in the contact zone between two genetically distinct individuals, and is often associated with the formation of a pigmented demarcation line or barrage between incompatible strains (Perkins et al. 2001). Fusion cells undergo extensive vacuolization, hyper-septation, ROS production, lipid droplet accumulation, and plasma membrane disruption (Saupe et al. 2000; Glass and Kaneko 2003). HI has been described in dozens of filamentous ascomycete species, and the populations of numerous phytopathogenic species have been characterized in terms of vegetative compatibility groups (VCGs). Strains belonging to the same VCG are able to form heterokaryons and transmit genetic content horizontally and often exhibit similar virulence and host specificity, while strains of different VCGs are incompatible and do not from viable heterokaryons (Zeise and Von Tiedemann 2002; Fan et al. 2018). By restricting chimerism between individuals, HI limits the horizontal transmission of deleterious cytoplasmic elements (i.e., mycoviruses, senescence plasmids), thus functioning as a fungal defense mechanism (Debets et al. 1994; Zhang et al. 2014). In Cryphonectria parasitica, the causal agent of chestnut blight disease, systematic disruption of allorecognition genes that prevented heterokaryon formation between strains of different VCG resulted in the creation of a super mycovirus donor strain (Zhang and Nuss 2016). The “super mycovirus donor strain” carries a hypo-virulence plasmid, and, hence, can be an effective biocontrol agent (Zhang and Nuss 2016). Similar approaches underscore the importance of genetically identifying and characterizing novel allorecognition loci in fungi.
Neurospora crassa is a model filamentous ascomycete species for studying cell–cell communication, anastomosis formation, and HI (Herzog et al. 2015; Zhao et al. 2015; Daskalov et al. 2017). At least 11 loci control HI in N. crassa (Mylyk 1975). Several het genes (for HI) have been molecularly identified and shown to induce cell death when coexpressed with incompatible alleles of the same gene (allelic HI systems) or with incompatible alleles of a different gene (nonallelic HI systems) (Kaneko et al. 2006; Paoletti 2016). The identified het genes are often highly polymorphic, with several different alleles segregating in near equal frequencies in wild populations of N. crassa (Wu et al. 1998; Hall et al. 2010). Such a distribution pattern of antagonistic alleles in wild isolates is an indication that a particular locus is under balancing selection. Balancing selection has been described to operate on the self-incompatibility locus (SI) in flowering plants, the major histocompatibility complex (MHC) in mammals, and has been associated with allorecognition loci in other animals (Richman 2000; Nydam et al. 2017). Alleles of a gene under balancing selection can be maintained in populations undergoing speciation events, which results in a trans-species segregation of multiple allelic haplogroups (Klein et al. 1998). Evolutionary hallmarks of long-term balancing selection and trans-species polymorphism have been recently used in a population genomics analysis of N. crassa wild isolates to successfully identify novel het genes (Zhao et al. 2015). However, in spite of the frequently shared molecular hallmarks of evolution, het gene function seems poorly conserved between species (Paoletti 2016).
The products of several het genes belong to a large family of fungal proteins, analogous to NOD-like receptors (NLRs) (Dyrka et al. 2014). NLR proteins (also known as nucleotide-binding site and leucine-rich repeats containing proteins or NBs-LRR) are intracellular innate immune receptors controlling cell death in plants and animals in response to immunogenic cues (Duxbury et al. 2016; Jones et al. 2016). The similarities between het determinants and genes involved in innate immunity in other eukaryotic lineages have prompted the hypothesis that HI may have originated from signaling pathways mediating interspecific biotic interactions, possibly in the context of an uncharacterized fungal innate immunity system (Paoletti and Saupe 2009; Dyrka et al. 2014; Uehling et al. 2017). Thus, the molecular identification and characterization of novel allorecognition factors in fungi could help us elucidate the evolutionary history of nonself recognition processes in Eukaryotes.
Previously identified HI-inducing systems are suppressed during the Neurospora sexual cycle, and also in germinating conidia (germlings), which play a fundamental role for vegetative propagation (Shiu and Glass 1999; Ishikawa et al. 2012). However, recently Heller et al. (2018) identified a gene pair controlling PCD and allorecognition in germlings of N. crassa. Germling-regulated death (GRD) is triggered when germlings of incompatible haplotypes undergo cellular fusions during early colony establishment. The cell death reaction occurs very rapidly (∼20 min) with the appearance of large vacuoles in the cytoplasm of fused incompatible germlings preceding cellular lysis and uptake of vital dyes (Heller et al. 2018). The two genes controlling GRD are linked in the genome of Neurospora. One gene (plp-1) encodes an NLR-like protein containing a phospholipase domain, and the other (sec-9) encodes a homolog of SEC9 from yeast, a protein involved in plasma membrane fusion and exocytosis (Brennwald et al. 1994; Heller et al. 2018).
The discovery of GRD represents an exciting possibility to study fungal PCD in a well-controlled biological system and during a developmental stage of the fungal life cycle (asexual spores) of great importance for clinical and agricultural applications, because it constitutes the infectious stage of many human and plant fungal pathogens (Brown et al. 2012). Here, we further investigated the molecular basis of GRD, and identified a novel genetic determinant of allorecognition in N. crassa. By using bulked segregant analysis (BSA) of GRD-specific DNA pools and population genomics data, the NCU05712 locus was identified as a second GRD-controlling allorecognition factor, termed regulator of cell death-1 (rcd-1). Viable germling fusion of cells containing incompatible alleles of rcd-1 (rcd-1-1 and rcd-1-2) was prevented by PCD induction. The rcd-1 alleles are highly polymorphic in N. crassa populations and show signs of balancing selection. Furthermore, we identified genomic rearrangements in the rcd-1 locus, which could be at the origin of this allorecognition system in Neurospora. Such genomic rearrangements could represent a general evolutionary mode for fungal incompatibility systems. Moreover, we show that that rcd-1 belongs to a large gene family in filamentous ascomycete species. The latter observation suggests that rcd-1 might be an important component of cell death pathways in fungi with broader role in nonself recognition.
Materials and Methods
Strains, growth media, and molecular constructs
Strains were grown using standard procedures and protocols that can be found on the Neurospora homepage at FGSC (http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm). Vogel’s minimal media (VMM) (with supplements, if required) was used to culture all strains, except when specified otherwise (Vogel 1956). Crosses were performed on Westergaard’s synthetic cross medium (Westergaard and Mitchell 1947). For flow cytometry experiments, thermo-reversible solid Vogel’s medium was obtained substituting the agar with 20% Pluronic F-127 (Sigma-Aldrich).
All wild N. crassa strains in the present study were isolated from Louisiana, have been described previously, and are available at the FGSC (Dettman et al. 2003; Palma-Guerrero et al. 2013; Zhao et al. 2015; Corcoran et al. 2016) (Supplemental Material, Table S1). All engineered strains were from the genetic background of the wild-type laboratory strain (FGSC2489; OR74A). The ΔNCU05712 deletion strain was obtained from the single gene deletion collection of N. crassa strains at the FGSC (Colot et al. 2006).
The rcd-1-1 and rcd-1-2 alleles with native promoters were cloned in the pMF272 vector (Freitag et al. 2004) using the restrictions enzymes NotI and ApaI and introduced in the his-3 locus of a Δrcd-1 strain. Molecular fusions of rcd-1 with fluorescent proteins (GFP or mCherry) were produced by cloning the rcd-1 alleles in pMF272-derived vector using MscI/PacI (for the rcd-1-1 allele) and XbaI/PacI (for the rcd-1-2 allele) under the regulation of the tef-1 promoter. Positive transformants were backcrossed with an Δrcd-1 strain to construct homokaryotic strains that were subsequently verified by PCR.
Bulked segregant analyses and genome resequencing
Bulked segregant analysis was performed as described (Heller et al. 2018). The GRD phenotype of progeny from a cross between FGSC2489 and a wild isolate JW258 (FGSC 10679) was determined by pairing germlings with each parental strain and assessing PCD. Two GRD-specific DNA pools were produced (60 ng from 50 progeny strains in each DNA pool) for library preparation and sequencing. All paired-end libraries were sequenced on a HiSeq2000 sequencing platform using standard Illumina operating procedures (Vincent J. Coates Genomics Sequencing Laboratory, University of California, Berkeley). The mapped reads for each group of 50 pooled segregants are available at the Sequence Read Archive (PRJNA556444; https://www.ncbi.nlm.nih.gov/sra/PRJNA556444).
Flow cytometry
Flow cytometry was performed according to Heller et al. (2018). We recorded 20,000 events per sample for each experiment. Experiments were performed at least three times. Data were analyzed with custom MATLAB (MathWorks) script with ungerminated conidia excluded from the analyses (Gonçalves et al. 2019). Cell death is shown as the average percentage of fluorescent events from all experiments.
Evolutionary analysis of DNA and protein sequences
We characterized homologs of NCU05712 in a collection of 194 publicly available Sordariales genomes representing 16 Neurospora species (Lasiosphaeriaceae II) (Kruys et al. 2015), one non-Neurospora Lasiosphaeriaceae II species, two species in Lasiosphaeriaceae IV, one species in Lasiosphaeriaceae I, and 14 species of thermophilic subfamily Chaetomiaceae (Table S1). The dataset included resequencing data for 27 N. crassa isolates from Louisiana (Galagan et al. 2003; Zhao et al. 2015); 92 N. tetrasperma strains from North America, Europe, and Oceania (Ellison et al. 2011; Corcoran et al. 2016); and 43 N. discreta PS4 from North America, Europe, and Asia (Gladieux et al. 2015). We used publicly available genome assemblies and predicted genes where possible. Other genomes were assembled using ABySS (Simpson et al. 2009), and annotated using Augustus, with N. crassa as the reference species and intron locations obtained by mapping other species proteins onto each focal genome using Exonerate (Slater and Birney 2005).
We identified NCU05712 homologs in Neurospora genomes using BlastP against predicted proteins and keeping hits with an e-value <1e-5 and score >300. The resulting set of protein sequences was filtered from proteins whose length was >270 aa, aligned using Clustalo (Sievers and Higgins 2018), and used to build a hidden Markov model (HMM) profile using hmmbuild (Finn et al. 2011). We then searched for homologs in non-Neurospora genomes using hmmsearch (Finn et al. 2011), keeping only proteins in the inclusion threshold (Table S2). We removed from the dataset homologs identified in Neurospora tetrasperma reference genome, because gene models predicted premature stops codons. N. tetrasperma rcd-1 ORF sequences were aligned using BLAST Global align (at NCBI), and sequences with SNPs or micro indels (up to six nucleotides) leading to predicted stop codons in the N. tetrasperma rcd-1 ORFs were removed.
For each taxon with resequencing data, genomic variation at rcd-1 was compared to a set of orthologous genes. Orthologous relationships were analyzed using Orthofinder (Emms and Kelly 2015) using Diamond (Buchfink et al. 2015) as the sequence aligner. We generated sets of single-copy orthologs present in 95% of isolates, and removed coding sequences not starting with “ATG” or having >10 Ns. TranslatorX (Abascal et al. 2010) and aligned coding sequences, while keeping the coding frame (protein sequence aligner: Muscle; sequence cleaning: GBlocks with parameters -g “-b2 8 -b3 3 -b4 3 -b5 n”). The number of protein variants was estimated based on nonsynonymous substitutions using Biopython, Ksmax (the maximum number of synonymous substitutions between sequences pairs) using yn00 in PAML (Yang 2007), and π (the average number of nucleotide differences between sequence pairs), s (number of segregating sites standardized by sequence length), and Tajima’s D (a measure of skewness of the allele frequency spectrum) using Egglib v3 (De Mita and Siol 2012). Gene genealogy of rcd-1 was inferred using the GTRGAMMA model in RAxML v8.2.10 (Stamatakis 2014). The genome genealogy within Sordariaceae used the same approach and program by concatenating coding sequences at 874 single copy orthologs present in at least 90% of isolates.
Long-term positive selection was assessed using a maximum likelihood framework for parameter inference and codon-based substitution models that allow the dN/dS ratio ω to vary among sites and among branches, as implemented in PAML’s codeml program. For the site-specific analysis, we compared three codon substitution models: M7 which is a model of purifying selection that allows individual sites to evolve under differing levels of constraint and assumes a beta distribution of negatively selected (0 < ω < 1), M8a which is a model of purifying selection that adds neutral sites ω = 1 to M7, and M8, which is a model of positive selection that adds positively selected sites (ω > 1) to M7. For the branch-site analysis, assuming variable selective pressures among branches and sites, we specified rcd-1-2 as the foreground branch and rcd-1-1 as the background branch. We compared model A1, which is a model of purifying selection (0 < ω ≤ 1) with model A, which adds positively selected sites along the foreground branch (ω > 1) to model A1. Nested models were compared using likelihood ratio tests.
To gather rcd-1 homologs outside the Sordariales, we used the HMMER web server (Finn et al. 2011). The initial query sequence was rcd-1-1 from N. crassa strain FGSC2489. Three iterative searches were performed to retrieve as many rcd-1 homologs as possible (Table S2). Significant hits were aligned with Clustal, sorted by size, and sequences <230 aa and >350 were discarded. Sequences were assigned to taxonomic groups (Order) using NCBI Taxonomy, followed by manual curation of “Unclassified” taxa using Index Fungorum. Pseudogymnoascus destructans accounted for most of the sequences left “Unclassified” after manual curation. Protein sequences were aligned using Clustalo, and a protein genealogy was inferred using model PROTGAMMAJTTF in RAxML v8.2.10.
Microscopy
Microscopy was performed on Zeiss Axioskop two equipped with a Q Imaging Retiga-2000R camera (Surrey), using a 40×/1.30 Plan-Neofluar oil immersion objective and the iVision Mac4.5 software. Agar squares (∼1 cm2) were excised from plates with growing germlings and observed under the microscope after 4–5 hr of growth (Heller et al. 2016).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Mapped sequencing reads for BSA analysis of pooled segregant strains are available at the Sequence Read Archive (PRJNA556444). Files used for evolutionary analysis of rcd-1 homologs are deposited at figshare and are publically accessible with the following link https://doi.org/10.6084/m9.figshare.10298045. The deposited datasets are as follows – File 1: DNA sequences of rcd-1 homologs in Sordariaceae used for phylogeny in Figure 5 and positive selection analyses. File 2: Protein sequences of rcd-1 homologs in fungi and bacteria used for phylogeny presented in Figure 6. File 3: DNA sequences of genes representing the genomic background in N. crassa used for analyses of balancing selection at rcd-1 in N. crassa. File 4: DNA sequences of genes representing the genomic background in N. discreta used for analyses of balancing selection at rcd-1 in N. discreta. File 5: DNA sequences of genes representing the genomic background in N. tetrasperma used for analyses of balancing selection at rcd-1 in N. tetrasperma. Raw flow cytometry data files are available upon demand. Supplementary Tables and Figures are available at figshare: https://doi.org/10.25386/genetics.10162688.
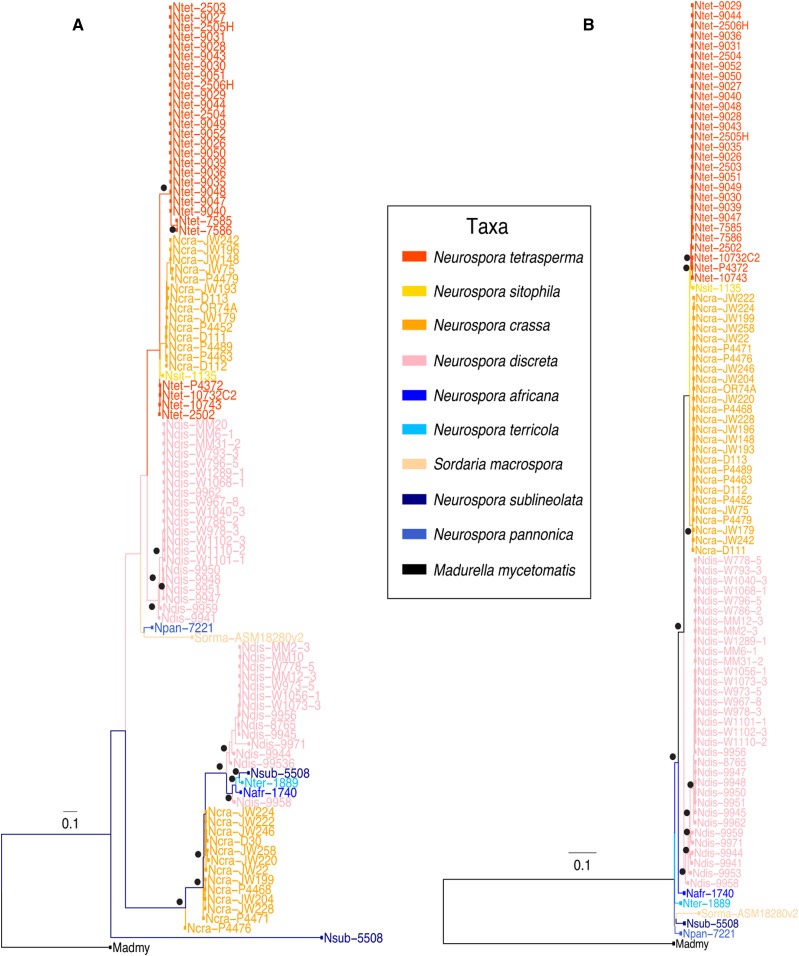
Figure 5.
Phylogenetic distribution of rcd-1 in Sordariaceae (A) as compared with species phylogeny (B). Species are color-coded and nodes with bootstrap values above 0.7 are indicated with black dots. Detailed species and strain information is available in Table S1.
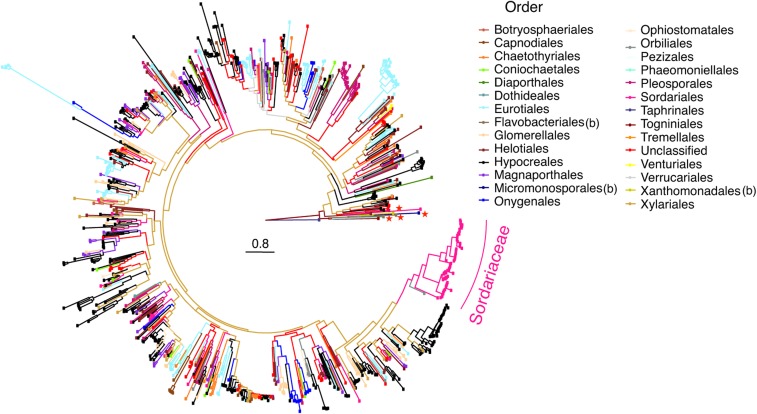
Figure 6.
Phylogenetic distribution of rcd-1 in fungi and bacteria. Maximum likelihood tree of ∼940 rcd-1 homologs. Branches of the phylogenetic tree are color-coded in accordance with the taxonomic rank at order level for the species from which the various rcd-1 sequences have been retrieved. Order names are indicated in the panel next to the phylogenetic tree. The gene family expansion in Sordariaceae family (order Sordariales), which display the various rcd-1-1 and rcd-1-2 Neurospora alleles from this study, is highlighted. Branches corresponding to the four bacterial sequences are indicated with red stars. Bacterial phylogenetic orders are indicated with the letter (b) in the legend of the figure. Sequences from species with no clearly established phylogeny at order level, are grouped under “unclassified”.
Results
BSA and population genomics identify NCU05712 as a highly polymorphic candidate gene controlling GRD
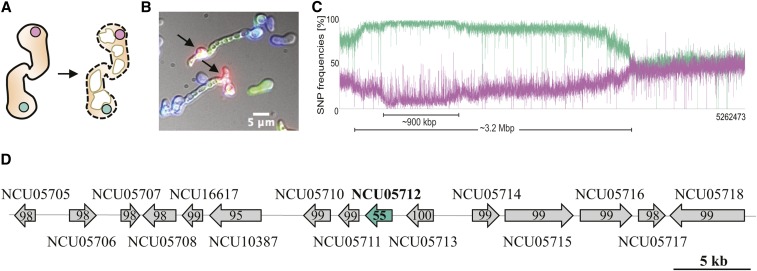
To identify novel GRD-inducing genes, we used progeny from a cross between two wild isolates, the laboratory reference strain FGSC2489 and JW258 (FGSC10679). Genomic analyses of the progeny from this cross (FGSC2489 x JW258) were used previously to identify the kind of recognition determinants that act during chemotropic interactions between germlings and two linked GRD-inducing genes, plp-1 and sec-9 (Heller et al. 2016, 2018). The GRD phenotype (Figure 1, A and B) segregated 3:1 in progeny of this cross, suggesting that two independent loci regulated the GRD phenotype (sec-9/plp-1 and a second unidentified locus). To identify the second GRD locus, we performed a BSA with progeny from an F2 backcross between FGSC2489 and an F1 progeny (Segregant 121) that showed GRD with FGSC2489 in spite of being isogenic to FGSC2489 at sec-9/plp-1 (GRD due to second locus). More than 100 F2 progeny strains were phenotyped for GRD in cocultures with germlings of FGSC2489. Approximately half of the progeny formed viable heterokaryons with FGSC2489 germlings, while the other half showed rapid death of germlings ∼20 min postfusion (Video 1). We extracted DNA from F2 progeny strains and sequenced two GRD-specific DNA pools (compatible with FGSC2489 and incompatible with FGSC2489) for BSA analysis. A random SNP distribution of ∼50% was observed for the two groups on all chromosomes with the exception of a large region (∼900 kbp) on chromosome III, where SNPs segregated at ∼100% frequency between GRD groups (Figure 1C). Within this interval, we searched for highly polymorphic genes, hypothesizing that strongly divergent alleles might represent good candidates for being involved in allorecognition. While most genes in this region showed high identity (>95%) between the two parental strains (FGSC2489 and JW258), NCU05712 was highly polymorphic and parental alleles showed only 55% identity in the ORF region (Figure 1D). The high degree of sequence divergence between the two allelic haplogroups of NCU05712 was not observed at any of the adjacent genes, which showed identity percentage ranging from 95 to 100%.
Figure 1.
Bulked segregant analysis (BSA) and comparative genomics identifies GRD-controlling candidate gene. (A) Cartoon of germling-regulated death (GRD). Shown are two genetically incompatible germlings, represented by nuclei (circles) in pink and green, undergoing cellular fusion (left). The GRD reaction (right) manifests with extensive vacuolization of the germlings (white shapes) and cell lysis (shown with dashed line). (B) GRD reaction occurring between genetically incompatible asexual spores of two different strains as revealed by the uptake of the vital dye propidium iodide (PI, black arrows) at fusion points between incompatible germlings. One strain is marked with calcofluor-white (blue) and the other strain expresses cytoplasmic GFP (green). Bar, 5 µm. (C) SNP segregation on linkage group III after sequencing of two genomic DNA pools of 50 segregant strains each from the FGSC2489 X JW258 cross. Purple line: SNP frequencies of pooled DNA of segregant strains showing GRD with FGSC2489. Green line: SNP frequencies of pooled DNA of segregant strains compatible with FGSC2489. (D) Genomic organization of the locus containing the GRD candidate gene NCU05712 or regulator of cell death-1. Percentage of identity between the parental strains (FGSC2489 and JW258) is shown for each ORF in the region.
We asked how the observed level of polymorphism in NCU05712 compared to a set of reference genes representing the broader gene polymorphism found in the genomes of N. crassa isolates from Louisiana (the population of origin of the laboratory reference strain FGSC2489). We computed five summary statistics of genetic variation for the set of reference genes, and compared the scores obtained for each statistic, representing the overall polymorpism level in the genome, to scores obtained with NCU05712 for the same five statistics (Table 1). Our analysis ranked the GRD candidate gene NCU05712 in the top 1% of most polymorphic genes in the N. crassa genome for all five statistics (Table 1). Alleles at NCU05712 were in the 99.9% percentile on three of the computed five statistics—the number of amino acid sequences based on nonsynonymous substitutions (Pvar), the average number of nucleotide difference between sequences (π), and Tajima’s neutrality statistic D (Table 1). NCU05712 featured equally among the most polymorphic genes in Neurospora discreta, but not in Neurospora tetrasperma, consistent with the fact that we could retrieve only one of the NCU05712 allelic lines in the latter species (Table 1). These results indicate that NCU05712 is one of the most polymorphic genes in the genomes of at least two Neurospora species. Based on our analysis, we hypothesized that NCU05712 was the likeliest candidate gene to control GRD in the segregating region.
Table 1. Average summary statistics (and 95% percentile) for rcd-1 and reference genes representing the genomic background in three Neurospora species.
| Species/gene set | Sa | πb | Dc | KSmaxd | Pvare |
|---|---|---|---|---|---|
| Neurospora crassa | |||||
| rcd-1 | 0.451** | 0.220*** | 3.491*** | 1.762** | 22.0*** |
| Reference genes (n = 5632) | 0.043 (0.136) | 0.011 (0.031) | −0.490 (1.072) | 0.261 (0.248) | 8.0 (18.0) |
| Neurospora discreta | |||||
| rcd-1 | 0.491** | 0.202*** | 2.637** | 2.318** | 16.0** |
| Reference genes (n = 5265) | 0.032 (0.085) | 0.008 (0.019) | 0.244 (1.580) | 0.324 (0.149) | 5.7 (12.0) |
| Neurospora tetrasperma | |||||
| rcd-1 | 0.116 | 0.023 | −0.908 | 0.218 | 6.0 |
| Reference genes (n = 4698) | 0.054 (0.154) | 0.011 (0.032) | −0.0367 (2.190) | 0.480 (0.319) | 7.735 (17.0) |
Number of polymorphic sites per base pair.
Average number of nucleotide differences between sequence pairs.
Tajima’s neutrality statistic (Tajima 1989).
Maximum number of synonymous substitutions between sequences pairs.
Number of amino-acid sequences based on nonsynonymous substitutions (*** in 99.9% percentile, ** in 99% percentile, * in 95% percentile).
NCU05712 is necessary and sufficient for GRD
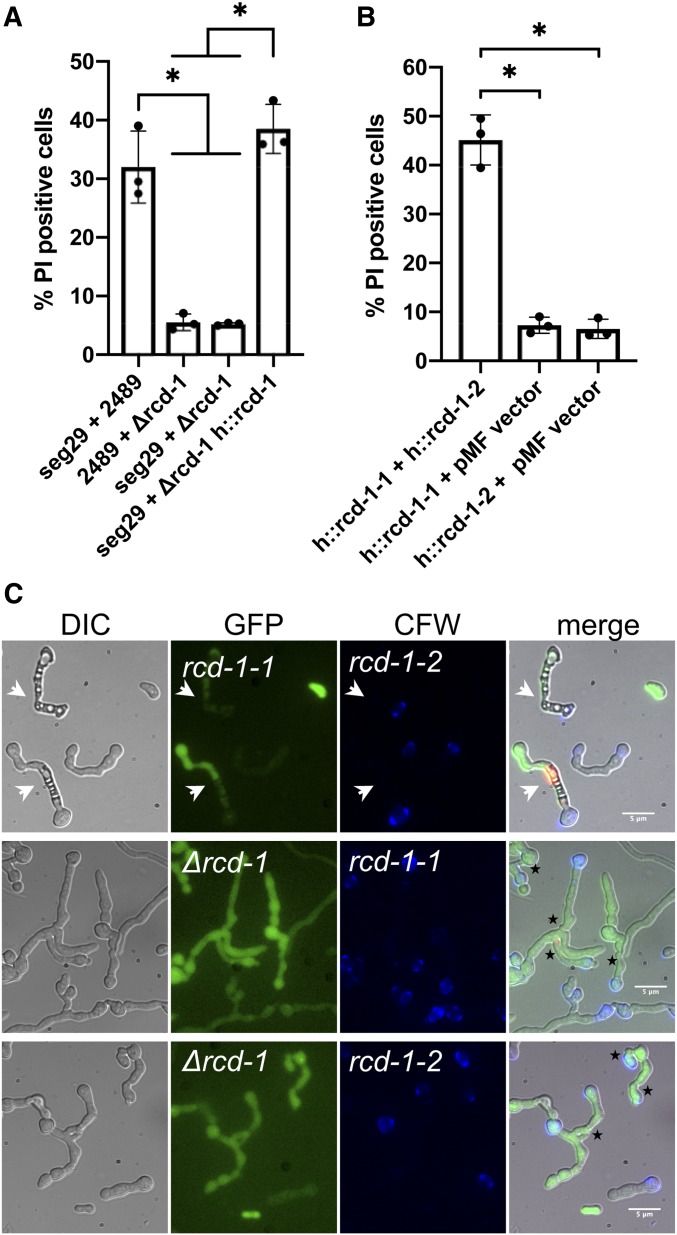
To test the hypothesis that NCU05712 controls GRD, a NCU05712 deletion strain from the Neurospora deletion collection (Colot et al. 2006; Dunlap et al. 2007) was characterized. First, we showed that the ΔNCU05712 mutant was not affected in growth or conidiation as compared to the parental strain (FGSC2489) (Figure S1). We then compared the frequency of GRD in pairings between FGSC2489 germlings with an incompatible segregant strain (seg29) carrying the JW258 allele of NCU05712 or in pairs of ΔNCU05712 germlings with seg29 using flow cytometry and the vital dye propidium iodide (PI) (Figure 2A; Video 1). When germlings of FGSC2489 were mixed in 1:1 ratio with germlings from seg29, we observed ∼30% of pairs that exhibited PI uptake (a proxy for GRD). This frequency of GRD was significantly higher than that observed for mixtures of ΔNCU05712 and seg29 germlings, where GRD was ∼5% (Figure 2A); ∼5% cell death was also observed in germlings undergoing self-fusion in a single strain (Figure S2). The strong reduction in GRD in ΔNCU05712 + seg29 pairings segregated with the hygromycin resistance marker used to construct the deletion strain (Figure S3). Furthermore, the GRD phenotype was restored when NCU05712 was reintroduced into the ΔNCU05712 strain (Figure 2A). We concluded that NCU05712 was necessary for the allorecognition reaction, and named the locus regulator of cell death or rcd-1. We termed the two allelic haplogroups as rcd-1-1 (as represented by FGSC2489) and rcd-1-2 (as represented by JW258).
Figure 2.
NCU05712 (rcd-1) is necessary and sufficient for GRD induction in N. crassa. (A) A flow cytometry-based quantification of GRD using the vital dye PI (Heller et al. 2018) shows that rcd-1 is necessary for the GRD phenotype. (B) GRD quantification with isogenic strains of N. crassa carrying different rcd-1 alleles. Strains carrying ectopic rcd-1 allele at the his-3 locus are indicated with (h::) preceding the allele. Experiments were performed in triplicate, with 20,000 events counted per experiment. *P < 0.0006, one-way ANOVA with Tukey’s multiple comparisons test. (C) Reconstitution of rcd-1-1/rcd-1-2 GRD phenotype in isogenic N. crassa strains. Cells undergoing GRD (white arrows) show strong vacuolization and uptake of PI. Deletion of rcd-1 abolishes GRD and genetically distinct strains can undergo cell fusion to form viable cell networks. Fusion points between genetically distinct germlings are shown with black asterisks. Bar, 5 µm.
To test if polymorphism at the rcd-1 locus was sufficient to trigger cell death, we targeted an rcd-1-1 allele or an rcd-1-2 allele to the his-3 locus of a Δrcd-1 strain. Paired germlings of engineered rcd-1-1 and rcd-1-2 isogenic strains, expressing the antagonistic rcd-1 alleles in otherwise isogenic backgrounds, produced a strong GRD reaction with a ∼45% cell death frequency (Figure 2B). Strains expressing just rcd-1-1 or rcd-1-2 did not show GRD above the background level when grown by themselves or in cocultivations with a Δrcd-1 strain transformed by an empty control vector (pMF274) (Figure 2, B and C and Figure S3). In addition, we observed an incompatible reaction in ascospore progeny bearing both rcd-1-1 and rcd-1-2 (Figure S4), while heterokaryotic fusion cells bearing fluorescently tagged RCD-1-1 and RCD-1-2 showed strong vacuolization and compartmentation of fused cells. These results indicated that allelic differences at the rcd-1 locus were necessary and sufficient to trigger cell death in fused germling pairs and also in hyphae in N. crassa, and established rcd-1 as a second allorecognition system controlling GRD in N. crassa.
rcd-1 does not act downstream of plp-1 in the induction of GRD
The similar GRD phenotype between the two loci that regulate death upon fusion of germlings, sec-9/plp-1 and rcd-1, suggested that a functional relationship might exist in the induction of GRD by allelic differences at these loci. Therefore, we assessed whether the GRD phenotype induced in germlings with different sec-9/plp-1 allelic specificities was dependent on rcd-1, by assessing sec-9/plp-1-induced GRD in a Δrcd-1 mutant. Genetic differences at sec-9/plp-1 were sufficient to induce GRD in a Δrcd-1 mutant background, with a frequency of cell death and cellular phenotype that was indistinguishable from strains inducing sec-9/plp-1-triggered GRD in a wild-type genetic background (Figure S5). Thus, RCD-1 does not function downstream of SEC-9/PLP-1 in the execution of cell death, suggesting that these two GRD pathways act independently.
Genomic rearrangements at the rcd-1 locus in sordariaceae
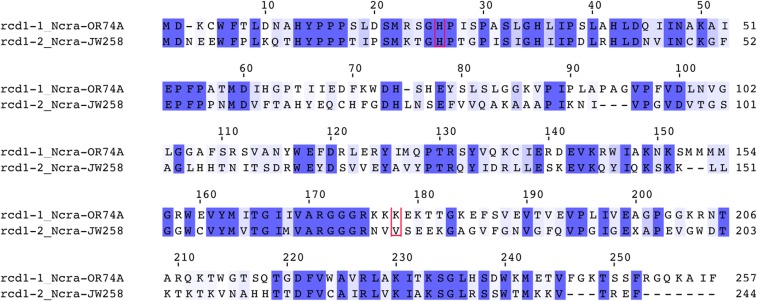
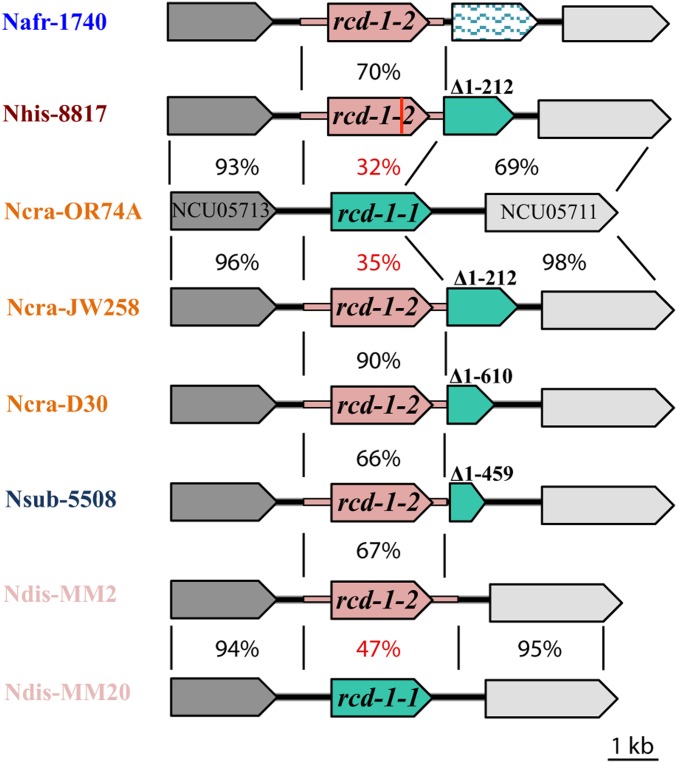
The rcd-1-1 and rcd-1-2 alleles encode proteins of unknown function of 257 and 244 amino acids, respectively. The two allelic variants showed 38% identity (54% similarity) on the protein level (Figure 3). The coding region of both rcd-1 alleles consisted of two exons, as confirmed by sequencing rcd-1-1 cDNA from FGSC2489. The analysis of rcd-1 gene structure showed a partial duplication of rcd-1-1 in the 3′ end of the rcd-1-2 allele in the JW258 strain. The duplicated rcd-1-1 in the rcd-1-2 background lacked 212 bp from the 5′ end of the ORF, including the start codon of the gene and the promoter (Figure 4). We examined the rcd-1 locus of other rcd-1-2-encoding N. crassa strains, and found that all sequenced wild isolates of the rcd-1-2 genotype carried a partial duplication of the rcd-1-1 allele at the 3′ end of rcd-1-2. However, unlike JW258, the other N. crassa rcd-1-2 strains have a shorter partial duplication of the rcd-1-1 allele (rcd-1-1Δ1−610), comprising mostly the second exon (Figure 4). These data suggest that an rcd-1-2 allele inserted into an rcd-1-1 allele, thereby disrupting the gene structure of the latter.
Figure 3.
Alignment of GRD-inducing allelic variants RCD-1-1 and RCD-1-2 from the wild-type reference strain FGSC2489 and the wild isolate JW258. Identity between the two proteins is shown in dark blue, similarity in light blue. Amino acids residues found to be under positive selection are boxed in red.
Figure 4.
Genetic rearrangements in the rcd-1 locus in Neurospora. rcd-1-1 (green) and rcd-1-2 (pink) alleles from different species are shown as cartoons in the rcd-1 locus. The extended region in pink in rcd-1-2 represents the sequence with sharp decrease in homology (or regions with lack of homology) at nucleotide level between strains of the rcd-1-1 and rcd-1-2 genotype. Percentage identities between two different genomic regions are shown between black lines delimiting the comparisons. Numbers over the rcd-1-1 allele indicate the limits of the truncated ORF in the genome of different Neurospora species and strains. Red line in the rcd-1-2 allele of Neurospora hispaniola (Nhis-8817) represents a premature stop codon. Species names are abbreviated as follows; N. africana (Nafr), N. hispaniola (Nhis), N. crassa (Ncra), N. sublineolata (Nsub), N. discreta (Ndis).
Sequenced N. crassa strains carrying the rcd-1-2 allele had a partially duplicated rcd-1-1Δ1−610 or rcd-1-1Δ1−212 copy, with the vast majority of strains carrying rcd-1-1Δ1−610 and only JW258 presenting the longer duplication (Figure S6). Yet, the insertion site in the intergenic region upstream of the rcd-1-2 locus in sequenced rcd-1-2 N. crassa strains was the same, strongly suggesting that one ancestral insertion event was the origin of the rcd-1-1 gene disruption in N. crassa and that the partially duplicated rcd-1-1Δ1−610 and rcd-1-1Δ1−212 alleles evolved during a subsequent independent event (Figure S7). The molecular signature of the rcd-1 gene disruption event was also found in the genomes of other related Neurospora species. For example, in the genome of N. sublineolata (FGSC5508), downstream of the rcd-1-2 allele was a partially duplicated rcd-1-1Δ1−459 sequence, while in N. hispaniola (FGSC8817), the rcd-1-2 allele had the same insertion sites as in N. crassa JW258 strain and thus carried a partially duplicated rcd-1-1Δ1−212 allele (Figure 4 and Figure S6). This observation suggested that the genomic rearrangement was a relatively ancient event, and likely predates speciation. Although, we did not find a partially duplicated rcd-1-1 ORF in N. discreta rcd-1-2 strains, we identified clear breakpoints in the sequence homology of the rcd-1 locus between N. discreta strains carrying rcd-1-1 and rcd-1-2 alleles (Figure S8). As the sudden decrease in sequence similarity was in the upstream/downstream region of the rcd-1-2 gene, similarly to the N. crassa strains, we concluded that, in N. discreta, the insertion of rcd-1-2 led to the complete loss of the rcd-1-1 ORF. Likely, taking into account the observed degeneration of the pseudogenic partial rcd-1-1 alleles in the rcd-1-2 strains of N. crassa and N. hispaniola, the rcd-1-2 locus in N. discreta strains has resulted from the same initial ancestral event of an rcd-1-2 allele disrupting an rcd-1-1 locus. These data suggest that the partial rcd-1-1 allele present in the rcd-1-2 strain was not sufficient to induce GRD. Consistent with this hypothesis, we quantified the amount of cell death in mixtures of germlings expressing the rcd-1-1 allele, and germlings expressing the rcd-1-2 allele lacking the partial duplication of rcd-1-1 (rcd-1-2-NPD) or rcd-1-2 in the presence of partially duplicated rcd-1-1Δ1−610. A significant difference in the frequency of cell death between these two germlings mixtures was not observed (Figure S9).
Evolutionary signatures of balanced and long-term positive selection associated with rcd-1
As rcd-1 is a novel allorecognition determinant, we investigated the evolutionary signatures associated with the gene. Natural selection has been shown to operate on allorecognition loci via a mechanism of negative frequency dependent selection that favors rare alleles and results in the maintenance of multiple alleles at relatively high frequencies (Wu et al. 1998; Hall et al. 2010; Zhao et al. 2015). The high positive Tajima’s D scores calculated for rcd-1 in N. crassa (D = 3.491) and N. discreta (D = 2.637) indicated an excess of intermediate frequency alleles at rcd-1 in the two Neurospora populations (Table 1). To deepen our analysis of the allelic distribution of rcd-1, we investigated the phylogenetic history of this locus in members of the Sordariaceae. We analyzed 194 fungal genomes from 26 species and identified 113 rcd-1 homologs in 104 of the sequenced strains (Table S1). Two Chaetomiaceae species (Chaetomium cochliodes and Thielavia hyrcaniae) and one Lasiosphaeriaceae II had two copies of rcd-1, and one Lasiosphaeriaceae IV (Cladorrhinum bulbillosum) had seven copies. However, rcd-1 homologs were not identified in 90 genomes, representing 12 species of Chaetomiaceae and Lasiosphaeriaceae I and II. We found that 81 of these genomes belonged to N. tetrasperma (65) and N. discreta (16) strains, where gene models encoding longer versions (>500 aa) of rcd-1 were predicted in different lineages, likely corresponding to annotation errors. When examined manually, rcd-1 alleles from N. tetrasperma strains (FGSC7586, FGSC9035) carried premature stop codons, similar to an rcd-1 allele from N. hispaniola (FGSC8817).
Phylogenetic analysis (maximum-likelihood) of rcd-1 in 27 wild isolates of N. crassa from Louisiana revealed a nearly equal distribution of two main polyphyletic clades (14 rcd-1-1-like and 13 rcd-1-2-like homologs in each clade) (Figure 5A and Table S1). Furthermore, in N. discreta, a relatively equal distribution (60–40%) between the two clades for the 35 analyzed rcd-1 homologs was also identified. The lack of monophyletic segregation between the N. crassa and N. discreta alleles indicates that the allele-specific polymorphisms predate the speciation event, and represent an example of trans-species polymorphism (Figure 5). Other Neurospora species carrying rcd-1 alleles also split between the two clades with N. pannonica and N. sitophila rcd-1 alleles being closer to rcd-1-1 alleles, while N. terricola, N. africana, and N. sublineolata carried sequences situated on the rcd-1-2 branch. The allele-specific polymorphisms were distributed over the entire length of the allelic variants (Figure S10). We did not find marks of trans-species polymorphism on the adjacent genes (NCU05713 and NCU05711) to rcd-1 (NCU05712), which are situated at ∼1.3 kb and ∼0.9 kb, respectively, from NCU05712 (Figure S11). The equal distribution of allelic frequencies in the wild populations of Neurospora and the observed trans-species polymorphism suggest that the rcd-1 locus is under long-term balancing selection.
Positive diversifying selection has been reported previously to operate on different allorecognition genes in Neurospora and Podospora (Bastiaans et al. 2014; Zhao et al. 2015). Sites under positive selection could be of importance for subsequent functional analyses of the rcd-1 allorecognition process. We thus tested for long-term positive selection by comparing, with likelihood-ratio tests, models assuming positive selection with null models in PAML’s codeml program. For site-specific analyses, positive selection model M8 (which assumes that one class of sites has ω > 1) provided a better fit to the data than purifying selection models M7 and M8a (which assumes 0 < ω < 1 and 0 < ω ≤ 1, respectively; likelihood ratio test: P = 0.0397 for M7 vs. M8, P = 0.0374 for M8a vs. M8) (Table 2). Under M8a, a single amino acid had a >0.95 posterior Bayesian probability of being under positive selection using the Bayes empirical Bayes method. For branch-site analyses, positive selection model A (which assumes positive selection ω > 1 along foreground branch rcd-1-2) provided a better fit to the data than purifying selection model A (which assumes 0 < ω ≤ 1 along all branches; likelihood ratio test: P = 0.0003). Under model M8 and A, respectively, one and two amino acids (plus the stop codon) had a >0.95 posterior Bayesian probability of being under positive selection using the empirical Bayes method. Both models found the position 173 (V173 in RCD-1-2; boxed in red, Figure 3) as being under positive selection, while model A identified H26 (RCD-1-2) (a histidine residue present in same position in the RCD-1-1 variant from FGSC2489 and the RCD-1-2 variant of JW258, but with a greater sequence diversity outside of the two parental strains) (boxed in red, Figure 3 and Figure S9).
Table 2. Parameter estimates and likelihood scores of negative selection (A1, M7, and M8a) and positive selection models (A, M8).
| Model | dN/dS | Parameter estimates | PSS | Likelihood |
|---|---|---|---|---|
| M7 | 0.364 | p = 1.06499, q = 1.81851 | NA | −7758.9 |
| M8 | 0.365 | p0 = 0.99506 P = 1.13305 q = 2.02613 (p1 = 0.00494) ω = 2.65855 | 1 (1) | −7755.6 |
| M8a | 0.359 | p0 = 0.96236 P = 1.21055 q = 2.35599 (p1 = 0.03764) ω = 1.00000 | NA | −7757.8 |
| A1 | Site class 0 1 2a 2b | NA | −7774.8 | |
| Proportion 0.75587 0.20934 0.02724 0.00754 | ||||
| Background w 0.24450 1.00000 0.24450 1.00000 | ||||
| Foreground w 0.24450 1.00000 1.00000 1.00000 | ||||
| A | Site class 0 1 2a 2b | 4 (3) | −7764.5 | |
| Proportion 0.76963 0.21413 0.01270 0.00353 | ||||
| Background w 0.25112 1.00000 0.25112 1.00000 | ||||
| Foreground w 0.25112 1.00000 15.33251 15.33251 |
The dN/dS ratio is an average across all sites categories. PSS, number of positively selected sites (number of sites with posterior probability cutoff P > 95%).
Overall, the evolutionary analysis of rcd-1 uncovered molecular signatures typically associated with allorecognition genes. These results suggest that the rcd-1 allorecognition process operates in the wild.
Distribution of rcd-1 homologs in Ascomycota
Functional allorecognition genes are generally poorly conserved between different fungal species (Fedorova et al. 2005; Paoletti 2016; Daskalov et al. 2017). However, genes encoding proteins carrying predicted domains associated with allorecognition processes fall in several large gene families and are distributed throughout fungi (Van der Nest et al. 2014; Zhao et al. 2015; Paoletti 2016). As rcd-1 is not related to any currently known fungal cell death-inducing determinant, we decided to explore the broader distribution of rcd-1 homologs. BLAST searches with either of the allelic variants of RCD-1 retrieved ∼260 significant hits (E value <0.05) from the nonredundant proteins database (NCBI). All identified sequences were encoded in the genomes of filamentous ascomycetes (Pezizomycotina), predominantly in species in the Sordariomycetes. The number of rcd-1 homologs increased to ∼940 sequences when we performed a profile HMM search using HMMER with RCD-1-1 as initial query sequence (Finn et al. 2011; Potter et al. 2018); the HMM search was performed until no new sequences were retrieved, which occurred after the third iteration. Almost all significant hits (>99.9% of hits with E value <0.005) were distributed in 24 fungal orders (23 in Ascomycota and 1 in Basidiomycota) and three bacterial orders containing only four sequences (Figure 6 and Table S2). The unique rcd-1 homolog found in Basidiomycota was from Naematelia encephala (Tremellales), while the prokaryotic sequences were distributed in three Gram-negative species (Flavobacteriales and Xanthomonadales) and one rcd-1 homolog in the filamentous actinomycete Actinoplanes missouriensis (Micromonosporales) (Table S2). The four bacterial sequences grouped near the root of the tree in two different nodes with other fungal rcd-1 homologs (Figure 6). Two fungal orders—Hypocreales (324) and Eurotiales (112)—contained approximately half of the analyzed rcd-1 homologs. Yet, rcd-1 genealogy did not follow Ascomycota phylogeny, because a large number of nodes grouped rcd-1 homologs from phylogenetically distant taxa (Figure 6) https://figshare.com/articles/Programmed_Cell_Death_in_Neurospora_crassa_Is_Controlled_by_the_Allorecognition_Determinant_rcd-1/10298045. For example, rcd-1 homologs from Hypocreales (black in Figure 6) are interspersed among homologs from other orders of Ascomycota and are found on most branches of the tree. A similar distribution is observed for homologs belonging to Eurotiales (sky blue), Magnaporthales (purple), and, in general, most orders with high number of rcd-1 homologs (Figure 6). Sequences from Sordariales (pink) were scattered in distant nodes of the tree, with previously analyzed rcd-1 homologs from the Sordariaceae (mainly Neurospora species) (Figure 5A)—where the incompatible rcd-1-1 and rcd-1-2 alleles could be found—grouped together in a separate branch of the tree. The latter finding was not surprising as the sequences situated on the Sordariaceae branch are alleles of rcd-1 from one species (N. crassa, N. discreta, and N. tetrasperma) and/or rcd-1 orthologs from closely related species.
At least one rcd-1 homolog was present in ∼180 fungal species, with a mean of five and median of three rcd-1 homologs per strains (sequenced genome). The genomes of more than a dozen ascomycete taxa contained >10 rcd-1 homologs. Among the species with higher numbers of rcd-1 homologs were several Trichoderma species, where individual strains carried up to 23 rcd-1 genes in their genome (T. atroviride ATCC 20476). Remarkably, the genome of the closely related species T. reesei QM6A contained only two rcd-1 genes. We concluded from these results that the allorecognition determinant rcd-1 identified in N. crassa is a member of a large gene family, widespread in the Ascomycota. The phylogeny of rcd-1 in fungi suggests that the gene has experienced lineage-specific gene expansions and was subject to horizontal transfers between species. Similar distribution patterns have also been reported for other families of nonself recognition determinants in fungi (Dyrka et al. 2014; Zhao et al. 2015).
Discussion
Here, we investigated the molecular basis of programmed cell-death occurring during nonself recognition in germinating conidia of N. crassa, and identified NCU05712 as a novel allorecognition determinant, which we named regulator of cell death-1 or rcd-1. Two incompatible alleles, rcd-1-1 and rcd-1-2, were identified at the rcd-1 locus and the coexpression of these alleles in fused germlings and hyphae was necessary and sufficient to trigger massive vacuolization and cell death. The two alleles showed evidence of balancing-selection and were associated with trans-species polymorphism, two molecular hallmarks associated with genes involved in the control of nonself discrimination (Richman 2000; Hall et al. 2010; Milgroom et al. 2018). Furthermore, close examination of the rcd-1 locus revealed that an rcd-1-2 allele inserted in an rcd-1-1 allele, disrupting the gene structure of the later. We find this observation particularly intriguing as it underscores a trend of genomic rearrangements frequently associated with nonself recognition loci. For example, several distinct GRD-specific haplotypes are present at the recently identified plp-1 locus in wild populations of Neurospora and Podospora (Heller et al. 2018). Multiple rearranged haplotypes were also found at the doc-1 (determinant of communication-1) locus, which defines nonself recognition at distance (precontact) in germlings (Heller et al. 2016), while, in Cryphonectria parasitica, the incompatibility locus vic4 contains two idiomorphic genes (Choi et al. 2012). Population genomics data and evolutionary signatures of rcd-1 suggest that the genomic rearrangements may have initiated events that led to the emergence of this incompatibility system in at least some Neurospora species. This hypothesis is based on the following observations: (i) rcd-1 alleles appear to be under balancing selection, evenly distributed in the populations of N. crassa and N. discreta, which suggests that natural selection is acting on the rcd-1-1/rcd-1-2 incompatibility; and (ii) all currently sequenced rcd-1-2 strains from N. crassa and N. discreta carry molecular evidence of a rearrangement in the rcd-1 locus. If the rcd-1-1/rcd-1-2 incompatibility system predates the rearrangements in the rcd-1 locus, and, assuming that the adaptive value of a hypothetical wild-type rcd-1-2 and the “inserted rcd-1-2” is equivalent, one could ask why the locus carrying the insertion is so strongly over-represented in N. crassa and N. discreta. A plausible explanation is that the rcd-1-2 allele has been horizontally transferred or introgressed; this event could have been possible only if it disrupted the rcd-1-1 allele (coexpression of both alleles is lethal).
Interspecies transfer of HI genes and mating-related genes has been previously documented in Ophiostoma novo-ulmi (Paoletti et al. 2006). In Neurospora, introgression has been shown to play a role in shaping mating-type chromosomes, and, possibly, the evolution of the rsk (resistant to spore killer) locus (Sun et al. 2012; Svedberg et al. 2018). Further phylogenetic analyses are needed to explore the evolutionary origin of rcd-1 incompatibility, notably sequencing of additional Neurospora strains and/or species closely related to Neurospora within the Sordariales. The accumulation of such genomic data could identify more rcd-1 haplotypes from species with population samples, and offer a better view of the allelic diversity of rcd-1 in Sordariales, which could reveal a more precise hypothesis regarding the emergence of rcd-1-1/rcd-1-2 incompatibility. The discovery of the rcd-1 incompatibility system represents an exciting opportunity to investigate the evolution of allorecognition genes and how particular genomic rearrangements impact their distribution in fungi.
Several key contributions have been made recently toward a better understanding of the molecular basis of allorecognition in N. crassa, with three key steps or checkpoints that regulate early colony establishment (Dyer 2019). The first checkpoint regulates chemotropic interactions between germlings, with Neurospora populations showing five communication groups, and which is regulated by allelic specificity at the determinate of communication or doc loci. Isolates from the same communication group (and with identical doc allelic specificity) show robust chemotropic interactions, while isolates from different communication groups (with different doc allelic specificity) show significantly reduced communication, and, thus, cell fusion (Heller et al. 2016). The second checkpoint, controlled by the cwr-1 and cwr-2 (cell wall remodeling) loci, regulates whether cell wall dissolution and cytoplasmic mixing occurs between adhered germlings (Gonçalves et al. 2019). Cells with identical allelic specificity at cwr-1 and cwr-2 undergo cell fusion and cytoplasmic mixing, while adhered germlings with alternate cwr-1 cwr-2 allelic specificity are blocked. If cells have identity at the doc and cwr loci, somatic cell fusion proceeds, but the viability and fitness of fused germling are dependent upon the GRD loci – plp-1/sec-9 (Heller et al. 2018) and rcd-1. These loci also control allorecognition in mature hyphae and thus could play a critical role in fungal interindividual relations beyond germling interactions and colony establishment.
In future work, investigations into the molecular mechanisms of rcd-1-induced cell death are of major interest as the gene is active in a developmental stage (asexual spores) frequently associated with threats to human and plant health. Targeting of endogenous cell-death pathways in fungi could represent a viable strategy in the fight against various fungal pathogens; it was recently proposed that mice clear inhaled spores from their lungs by targeting fungal PCD (Shlezinger et al. 2017; Kulkarni et al. 2019). The identification of rcd-1 diversifies the inventory of allorecognition genes identified in fungi. Considering the widespread nature and abundance of rcd-1 homologs, especially in the Pezizomycotina, we hypothesize that conspecific nonself discrimination is unlikely to be the sole role performed by rcd-1 in all of these species. This hypothesis is especially intriguing in light of recent reports positioning various allorecognition determinants inside broader gene families, which control innate immunity in animals and plants (Paoletti and Saupe 2009; Dyrka et al. 2014; Gonçalves et al. 2017). Future work on the molecular mechanism of recognition and induction of cell death mediated by RCD-1 will test its role in the paradigm of allorecognition and potentially provide clues to alternative functions of this gene family in the Pezizomycotina.
Acknowledgments
The authors wish to thank Amy Powell and Don Natvig for the kind permission to use Thielavia genome sequences for analysis of rcd-1 homology. We thank the Berkeley Flow Cytometry Facility and the College of Natural Resources (CNR) Biological Imaging Facility for their technical support. This work used the Vincent J. Coates Genomics Sequencing Laboratory (University of California, Berkeley), supported by National Institutes of Health (NIH) S10 Instrumentation Grants S10RR029668 and S10RR027303. This work was supported by a Laboratory Directed Research and Development Program of Lawrence Berkeley National Laboratory under United States Department of Energy Contract No. DE-AC02-05CH11231 to N.L.G.
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.10298045; https://doi.org/10.25386/genetics.10162688.
Present address: CNRS, UMR 5248, European Institute of Chemistry and Biology, University of Bordeaux, Pessac, 33607, France.
Communicating editor: M. Freitag
Literature Cited
- Abascal F., Zardoya R., and Telford M. J., 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38: W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaans E., Debets A. J. M., Aanen D. K., van Diepeningen A. D., Saupe S. J. et al. , 2014. Natural variation of heterokaryon incompatibility gene het-c in Podospora anserina reveals diversifying selection. Mol. Biol. Evol. 31: 962–974. 10.1093/molbev/msu047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaans E., Debets A. J. M., and Aanen D. K., 2016. Experimental evolution reveals that high relatedness protects multicellular cooperation from cheaters. Nat. Commun. 7: 11435 10.1038/ncomms11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Kearns B., Champion K., Keränen S., Bankaitis V. et al. , 1994. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 79: 245–258. 10.1016/0092-8674(94)90194-5 [DOI] [PubMed] [Google Scholar]
- Brown G. D., Denning D. W., Gow N. A. R., Levitz S. M., Netea M. G. et al. , 2012. Hidden killers: human fungal infections. Sci. Transl. Med. 4: 165rv13 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., and Huson D. H., 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12: 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Choi G. H., Dawe A. L., Churbanov A., Smith M. L., Milgroom M. G. et al. , 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190: 113–127. 10.1534/genetics.111.133983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M. et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 (erratum: Proc. Natl. Acad. Sci. USA 103: 16614). 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran P., Anderson J. L., Jacobson D. J., Sun Y., Ni P. et al. , 2016. Introgression maintains the genetic integrity of the mating-type determining chromosome of the fungus Neurospora tetrasperma. Genome Res. 26: 486–498. 10.1101/gr.197244.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A., Heller J., Herzog S., Fleißner A., and Glass N. L., 2017. Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol. Spectr. 5 10.1128/microbiolspec.FUNK-0015-2016 [DOI] [PubMed] [Google Scholar]
- Debets A. J. M., and Griffiths A. J. F., 1998. Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycol. Res. 102: 1343–1349. 10.1017/S095375629800639X [DOI] [Google Scholar]
- Debets F., Yang X., and Griffiths A. J. F., 1994. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr. Genet. 26: 113–119. 10.1007/BF00313797 [DOI] [PubMed] [Google Scholar]
- De Mita S., and Siol M., 2012. EggLib: processing, analysis and simulation tools for population genetics and genomics. BMC Genet. 13: 27 10.1186/1471-2156-13-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman J. R., Jacobson D. J., and Taylor J. W., 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. 10.1111/j.0014-3820.2003.tb01514.x [DOI] [PubMed] [Google Scholar]
- Dunlap J. C., Borkovich K. A., Henn M. R., Turner G. E., Sachs M. S. et al. , 2007. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 57: 49–96. 10.1016/S0065-2660(06)57002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury Z., Ma Y., Furzer O. J., Huh S. U., Cevik V. et al. , 2016. Pathogen perception by NLRs in plants and animals: parallel worlds. BioEssays 38: 769–781. 10.1002/bies.201600046 [DOI] [PubMed] [Google Scholar]
- Dyer P. S., 2019. Self/Non-self recognition: microbes playing hard to get. Curr. Biol. 29: R866–R868. 10.1016/j.cub.2019.08.001 [DOI] [PubMed] [Google Scholar]
- Dyrka W., Lamacchia M., Durrens P., Kobe B., Daskalov A. et al. , 2014. Diversity and variability of NOD-like receptors in fungi. Genome Biol. Evol. 6: 3137–3158. 10.1093/gbe/evu251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison C. E., Stajich J. E., Jacobson D. J., Natvig D. O., Lapidus A. et al. , 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189: 55–69. 10.1534/genetics.111.130690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R., Cockerton H. M., Armitage A. D., Bates H., Cascant-Lopez E. et al. , 2018. Vegetative compatibility groups partition variation in the virulence of Verticillium dahliae on strawberry. PLoS One 13: e0191824 10.1371/journal.pone.0191824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N. D., Badger J. H., Robson G. D., Wortman J. R., and Nierman W. C., 2005. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics 6: 177 10.1186/1471-2164-6-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., and Eddy S. R., 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39: W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Raju N. B., Selker E. U., and Read N. D., 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41: 897–910. 10.1016/j.fgb.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D. et al. , 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. 10.1038/nature01554 [DOI] [PubMed] [Google Scholar]
- Gladieux P., Wilson B. A., Perraudeau F., Montoya L. A., Kowbel D. et al. , 2015. Genomic sequencing reveals historical, demographic and selective factors associated with the diversification of the fire-associated fungus Neurospora discreta. Mol. Ecol. 24: 5657–5675. 10.1111/mec.13417 [DOI] [PubMed] [Google Scholar]
- Glass N. L., and Kaneko I., 2003. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell 2: 1–8. 10.1128/EC.2.1.1-8.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., and Dementhon K., 2006. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 9: 553–558. 10.1016/j.mib.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Gonçalves A. P., Heller J., Daskalov A., Videira A., and Glass N. L., 2017. Regulated forms of cell death in fungi. Front. Microbiol. 8: 1837 10.3389/fmicb.2017.01837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves A. P., Heller J., Span E. A., Rosenfield G., Do H. P. et al. , 2019. Allorecognition upon fungal cell-cell contact determines social cooperation and impacts the acquisition of multicellularity. Curr. Biol. 29: 3006–3017.e3. 10.1016/j.cub.2019.07.060 [DOI] [PubMed] [Google Scholar]
- Hall C., Welch J., Kowbel D. J., and Glass N. L., 2010. Evolution and diversity of a fungal self/nonself recognition locus. PLoS One 5: e14055 10.1371/journal.pone.0014055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Zhao J., Rosenfield G., Kowbel D. J., Gladieux P. et al. , 2016. Characterization of greenbeard genes involved in long-distance kind discrimination in a microbial eukaryote. PLoS Biol. 14: e1002431 10.1371/journal.pbio.1002431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Clavé C., Gladieux P., Saupe S. J., and Glass N. L., 2018. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl. Acad. Sci. USA 115: E2292–E2301. 10.1073/pnas.1719705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S., Schumann M. R., and Fleißner A., 2015. Cell fusion in Neurospora crassa. Curr. Opin. Microbiol. 28: 53–59. 10.1016/j.mib.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Ho H.-I., Hirose S., Kuspa A., and Shaulsky G., 2013. Kin recognition protects cooperators against cheaters. Curr. Biol. 23: 1590–1595. 10.1016/j.cub.2013.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F. H., Souza E. A., Shoji J.-Y., Connolly L., Freitag M. et al. , 2012. Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PLoS One 7: e31175 10.1371/journal.pone.0031175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Vance R. E., and Dangl J. L., 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354: pii: aaf6395. 10.1126/science.aaf6395 [DOI] [PubMed] [Google Scholar]
- Kaneko I., Dementhon K., Xiang Q., and Glass N. L., 2006. Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172: 1545–1555. 10.1534/genetics.105.051490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J., Sato A., Nagl S., and O’hUigín C., 1998. Molecular trans-species polymorphism. Annu. Rev. Ecol. Syst. 29: 1–21. 10.1146/annurev.ecolsys.29.1.1 [DOI] [Google Scholar]
- Kruys Å., Huhndorf S. M., and Miller A. N., 2015. Coprophilous contributions to the phylogeny of Lasiosphaeriaceae and allied taxa within Sordariales (Ascomycota, fungi). Fungal Divers. 70: 101–113. 10.1007/s13225-014-0296-3 [DOI] [Google Scholar]
- Kulkarni M., Stolp Z. D., and Hardwick J. M., 2019. Targeting intrinsic cell death pathways to control fungal pathogens. Biochem. Pharmacol. 162: 71–78. 10.1016/j.bcp.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird D. J., De Tomaso A. W., and Weissman I. L., 2005. Stem cells are units of natural selection in a colonial ascidian. Cell 123: 1351–1360. 10.1016/j.cell.2005.10.026 [DOI] [PubMed] [Google Scholar]
- Lakkis F. G., Dellaporta S. L., and Buss L. W., 2008. Allorecognition and chimerism in an invertebrate model organism. Organogenesis 4: 236–240. 10.4161/org.4.4.7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgroom M. G., Smith M. L., Drott M. T., and Nuss D. L., 2018. Balancing selection at nonself recognition loci in the chestnut blight fungus, Cryphonectria parasitica, demonstrated by trans-species polymorphisms, positive selection, and even allele frequencies. Heredity 121: 511–523. 10.1038/s41437-018-0060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylyk O. M., 1975. Heterokaryon incompatibility genes in Neurospora crassa detected using duplication-producing chromosome rearrangements. Genetics 80: 107–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydam M. L., Stephenson E. E., Waldman C. E., and De Tomaso A. W., 2017. Balancing selection on allorecognition genes in the colonial ascidian Botryllus schlosseri. Dev. Comp. Immunol. 69: 60–74. 10.1016/j.dci.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Palma-Guerrero J., Hall C. R., Kowbel D., Welch J., Taylor J. W. et al. , 2013. Genome wide association identifies novel loci involved in fungal communication. PLoS Genet. 9: e1003669 10.1371/journal.pgen.1003669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M., 2016. Vegetative incompatibility in fungi: from recognition to cell death, whatever does the trick. Fungal Biol. Rev. 30: 152–162. 10.1016/j.fbr.2016.08.002 [DOI] [Google Scholar]
- Paoletti M., and Saupe S. J., 2009. Fungal incompatibility: evolutionary origin in pathogen defense? BioEssays 31: 1201–1210. 10.1002/bies.200900085 [DOI] [PubMed] [Google Scholar]
- Paoletti M., Buck K. W., and Brasier C. M., 2006. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol. Ecol. 15: 249–262. 10.1111/j.1365-294X.2005.02728.x [DOI] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., and Sachs M. S., 2001. The Neurospora Compendium: chromosomal loci, Academic Press, Cambridge, MA. [Google Scholar]
- Potter S. C., Luciani A., Eddy S. R., Park Y., Lopez R. et al. , 2018. HMMER web server: 2018 update. Nucleic Acids Res. 46: W200–W204. 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman A., 2000. Evolution of balanced genetic polymorphism. Mol. Ecol. 9: 1953–1963. 10.1046/j.1365-294X.2000.01125.x [DOI] [PubMed] [Google Scholar]
- Rosengarten R. D., and Nicotra M. L., 2011. Model systems of invertebrate allorecognition. Curr. Biol. 21: R82–R92. 10.1016/j.cub.2010.11.061 [DOI] [PubMed] [Google Scholar]
- Saupe S. J., Clavé C., and Bégueret J., 2000. Vegetative incompatibility in filamentous fungi: Podospora and Neurospora provide some clues. Curr. Opin. Microbiol. 3: 608–612. 10.1016/S1369-5274(00)00148-X [DOI] [PubMed] [Google Scholar]
- Shiu P. K., and Glass N. L., 1999. Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics 151: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlezinger N., Irmer H., Dhingra S., Beattie S. R., Cramer R. A. et al. , 2017. Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death. Science 357: 1037–1041. 10.1126/science.aan0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., and Higgins D. G., 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27: 135–145. 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. M. et al. , 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19: 1117–1123. 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G. S. C., and Birney E., 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Corcoran P., Menkis A., Whittle C. A., Andersson S. G. E. et al. , 2012. Large-scale introgression shapes the evolution of the mating-type chromosomes of the filamentous ascomycete Neurospora tetrasperma. PLoS Genet. 8: e1002820 10.1371/journal.pgen.1002820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg J., Hosseini S., Chen J., Vogan A. A., Mozgova I. et al. , 2018. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat. Commun. 9: 4242 10.1038/s41467-018-06562-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima D. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehling J., Deveau A., and Paoletti M., 2017. Do fungi have an innate immune response? An NLR-based comparison to plant and animal immune systems. PLoS Pathog. 13: e1006578 10.1371/journal.ppat.1006578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Nest M. A., Olson A., Lind M., Vélëz H., Dalman K. et al. , 2014. Distribution and evolution of het gene homologs in the basidiomycota. Fungal Genet. Biol. 64: 45–57. 10.1016/j.fgb.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora crassa. Microbial Genet. Bull. 13: 42–46. [Google Scholar]
- Westergaard M., and Mitchell H. K., 1947. NEUROSPORA V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. 10.1002/j.1537-2197.1947.tb13032.x [DOI] [Google Scholar]
- Wu J., Saupe S. J., and Glass N. L., 1998. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl. Acad. Sci. USA 95: 12398–12403. 10.1073/pnas.95.21.12398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zeise K., and Von Tiedemann A., 2002. Host specialization among vegetative compatibility groups of Verticillium dahliae in relation to Verticillium longisporum. J. Phytopathol. 150: 112–119. 10.1046/j.1439-0434.2002.00730.x [DOI] [Google Scholar]
- Zhang D.-X., and Nuss D. L., 2016. Engineering super mycovirus donor strains of chestnut blight fungus by systematic disruption of multilocus vic genes. Proc. Natl. Acad. Sci. USA 113: 2062–2067. 10.1073/pnas.1522219113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.-X., Spiering M. J., Dawe A. L., and Nuss D. L., 2014. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics 197: 701–714. 10.1534/genetics.114.164574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Gladieux P., Hutchison E., Bueche J., Hall C. et al. , 2015. Identification of allorecognition loci in Neurospora crassa by genomics and evolutionary approaches. Mol. Biol. Evol. 32: 2417–2432. 10.1093/molbev/msv125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Mapped sequencing reads for BSA analysis of pooled segregant strains are available at the Sequence Read Archive (PRJNA556444). Files used for evolutionary analysis of rcd-1 homologs are deposited at figshare and are publically accessible with the following link https://doi.org/10.6084/m9.figshare.10298045. The deposited datasets are as follows – File 1: DNA sequences of rcd-1 homologs in Sordariaceae used for phylogeny in Figure 5 and positive selection analyses. File 2: Protein sequences of rcd-1 homologs in fungi and bacteria used for phylogeny presented in Figure 6. File 3: DNA sequences of genes representing the genomic background in N. crassa used for analyses of balancing selection at rcd-1 in N. crassa. File 4: DNA sequences of genes representing the genomic background in N. discreta used for analyses of balancing selection at rcd-1 in N. discreta. File 5: DNA sequences of genes representing the genomic background in N. tetrasperma used for analyses of balancing selection at rcd-1 in N. tetrasperma. Raw flow cytometry data files are available upon demand. Supplementary Tables and Figures are available at figshare: https://doi.org/10.25386/genetics.10162688.
Figure 5.
Phylogenetic distribution of rcd-1 in Sordariaceae (A) as compared with species phylogeny (B). Species are color-coded and nodes with bootstrap values above 0.7 are indicated with black dots. Detailed species and strain information is available in Table S1.
Figure 6.
Phylogenetic distribution of rcd-1 in fungi and bacteria. Maximum likelihood tree of ∼940 rcd-1 homologs. Branches of the phylogenetic tree are color-coded in accordance with the taxonomic rank at order level for the species from which the various rcd-1 sequences have been retrieved. Order names are indicated in the panel next to the phylogenetic tree. The gene family expansion in Sordariaceae family (order Sordariales), which display the various rcd-1-1 and rcd-1-2 Neurospora alleles from this study, is highlighted. Branches corresponding to the four bacterial sequences are indicated with red stars. Bacterial phylogenetic orders are indicated with the letter (b) in the legend of the figure. Sequences from species with no clearly established phylogeny at order level, are grouped under “unclassified”.