Abstract
When the soil bacterium Pseudomonas putida grows in a complete medium, it prioritizes the assimilation of preferred carbon sources, optimizing its metabolism and growth. This regulatory process is orchestrated by the Crc and Hfq proteins. The present work examines the changes that occur in metabolic fluxes when the crc gene is inactivated and cells grow exponentially in LB complete medium. Analyses were performed at three different moments during exponential growth, examining the assimilation rates for the compounds present in LB, changes in the proteome, and the changes in metabolic fluxes predicted by the iJN1411 metabolic model for P. putida KT2440. During the early exponential phase, consumption rates for sugars, many organic acids and most amino acids were higher in a Crc‐null strain than in the wild type, leading to an overflow of the metabolic pathways and the leakage of pyruvate and acetate. These accelerated consumption rates decreased during the mid‐exponential phase, when cells mostly used sugars and alanine. At later times, pyruvate was recovered from the medium and utilized. The higher consumption rates of the Crc‐null strain reduced the growth rate. The lack of the Crc/Hfq regulatory system thus led to unbalanced metabolism with poorly optimized metabolic fluxes.
Introduction
Pseudomonas putida KT2440, a non‐pathogenic soil bacterium, has a versatile and robust metabolism and is very resistant to harsh conditions and energetic stress. It is widely recognized as a suitable host organism for bioprocessing and for developing next generation synthetic organisms (Ebert et al., 2011; Poblete‐Castro et al., 2012; Nikel et al., 2014; Adams, 2016; Belda et al., 2016; Kampers et al., 2019). However, its sophisticated regulatory systems complicate any modification of the metabolic fluxes towards the synthesis or degradation of target compounds, particularly when cultivating cells in complex growth media containing mixtures of carbon sources.
When pseudomonads grow in complex media, the assimilation of the available resources is coordinated by a regulatory process termed catabolite repression, which ensures the sequential and hierarchical use of the carbon sources present (reviewed by Rojo, 2010). Catabolite repression not only prevents the assimilation of non‐preferred compounds until preferred ones have been consumed but also coordinates the simultaneous assimilation of two or more substrates (del Castillo and Ramos, 2007; Moreno et al., 2009; La Rosa et al., 2016). In these bacteria, catabolite repression relies mainly on the Crc and Hfq regulatory proteins, the activities of which are controlled by small RNAs of the CrcZ family (Sonnleitner et al., 2009; Moreno et al., 2012; Moreno et al., 2015; Sonnleitner et al., 2018). The Crc and Hfq proteins can form complexes with mRNAs containing A‐rich target sequences, inhibiting their translation (Madhushani et al., 2015; Moreno et al., 2015; Sonnleitner et al., 2018; Pei et al., 2019). This regulatory phenomenon is very prominent when cells grow in complex media such as lysogeny broth (LB), a commonly used medium because it allows for high growth rates.
LB medium contains free amino acids, peptides of different length, sugars, organic acids, (poly)nucleotides, vitamins and micronutrients (Sezonov et al., 2007; La Rosa et al., 2016; Molina et al., 2019). Pseudomonas putida catabolizes amino acids in a hierarchical manner using different pathways that, depending on the amino acid in question, eventually converge into pyruvate, acetyl‐CoA or different intermediates of the tricarboxylic acid (TCA) cycle (Moreno et al., 2009; La Rosa et al., 2016, and references therein). The sugars in LB are mostly polysaccharides derived from the yeast extract used to prepare the medium. A recent study showed that although P. putida KT2440 cannot use polysaccharides such as glycogen or starch as the sole carbon source, it can profit from them when other carbon sources are also present, as occurs in LB medium (Molina et al., 2019). Other polysaccharides present in the LB, such as mannan, are not assimilated by this bacterial strain. Interestingly, the glucose polymers were found to be used differently depending on whether cells were in the early, mid or late exponential phase of growth. During the early phase, sugars were utilized mainly as an energy source, but not as a carbon source, although at later times they were used both as energy and carbon sources. This is a consequence of the particular way in which pseudomonads consume glucose. Glucose enters the periplasm through the OprB‐I/OprB‐II porins. It is then either transported to the cytoplasm through an ABC transport system or oxidized to gluconate in the periplasm by periplasmic glucose dehydrogenase (Gcd). Gluconate can be transported into the cell or be oxidized to 2‐ketogluconate in the periplasm by gluconate dehydrogenase (Gad). The electrons detached from the glucose during its oxidation in the periplasm are transferred to the ubiquinones of the cytoplasmic membrane, providing reducing power to the electron transport chain (Matsushita et al., 1979; van Schie et al., 1985). Gluconate and 2‐keto‐gluconate can either be transported to the cytoplasm or be expelled to the medium. In the latter event, the cells would be using glucose as an energy source without processing its carbon skeleton. If they are transported to the cytoplasm, however, gluconate and 2‐keto‐gluconate, as well as glucose, converge into 6‐phosphogluconate (6PG), which is catabolized to pyruvate and glyceraldehyde‐3‐phosphate (G3P) through a cycle formed by enzymes of the Entner–Doudoroff, Embden–Meyerhof–Parnas and pentose phosphate pathways (del Castillo et al., 2007; del Castillo et al., 2008; Nikel et al., 2015).
A recent report showed that the sequential assimilation of LB medium components is paralleled by successive changes in the metabolic fluxes of the cells, which allow their metabolism to be adapted as preferred compounds are consumed and new ones start to be used (Molina et al., 2019). These analyses were performed at three points during exponential growth (early, mid and late), predicting the metabolic fluxes that might account for the observed consumption rates with the help of the iJN1411 metabolic model for P. putida strain KT2440 (Nogales et al., 2017). The changes observed in the proteome at these same times served to validate the flux predictions made. In the present work, the same approach was used to analyse the changes that occur in the metabolic fluxes of a Crc‐null derivative of strain KT2440. The results obtained show that the Crc protein has a key role in the coordination and distribution of the metabolic fluxes as cells sequentially use the compounds in LB medium. A lack of Crc led to a disordered assimilation of the compounds available, which resulted in metabolic overflow and a significant secretion of pyruvate and acetate, thus leading to inefficient metabolism and growth.
Results
Influence of the Crc regulator on the uptake rates of the nutrients present in LB
Pseudomonas putida KTCRC, a Crc‐null derivative of strain KT2440, was inoculated at a turbidity (A600) of 0.05 into a flask containing LB medium and allowed to grow under vigorous aeration. The composition of the medium was analysed when the cells were at the initial stage of exponential growth (A600 of 0.2 after 2 h of growth), at mid exponential growth (A600 of 0.6 after 3 h of growth), and at the end of exponential growth (A600 of 2 after 7 h of growth). The results are shown in the Supporting Information Table S1, and a comparison with results previously reported for the wild‐type strain in similar assays is presented in Figures 1 and 2.
Figure 1.
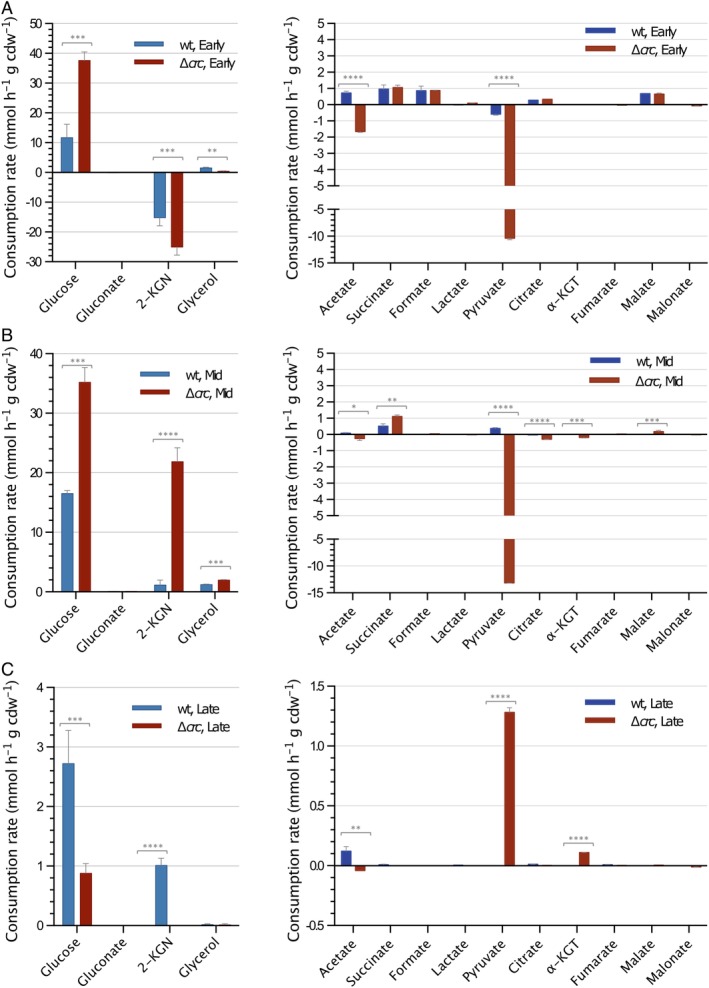
Consumption rates for the sugars and organic acids present in LB. The values observed for P. putida strains KT2440 (wild type; blue bars) and KTCRC (∆crc; red bars) at different times of their exponential growth are indicated.A. Early phase of growth. B. mid‐exponential phase. C. Late exponential phase. Error bars indicate the standard deviation of three biological replicates. The scale is not the same in all panels. 2‐KGN; 2‐ketogluconate; α‐KGT, α‐ketoglutarate. The statistical significance of the differences for the values of the wild type and the ∆ crc mutant was determined by one‐way ANOVA (* p < 0.1; **p < 0.01; ***p < 0.001; ****p < 0.0001).
Figure 2.
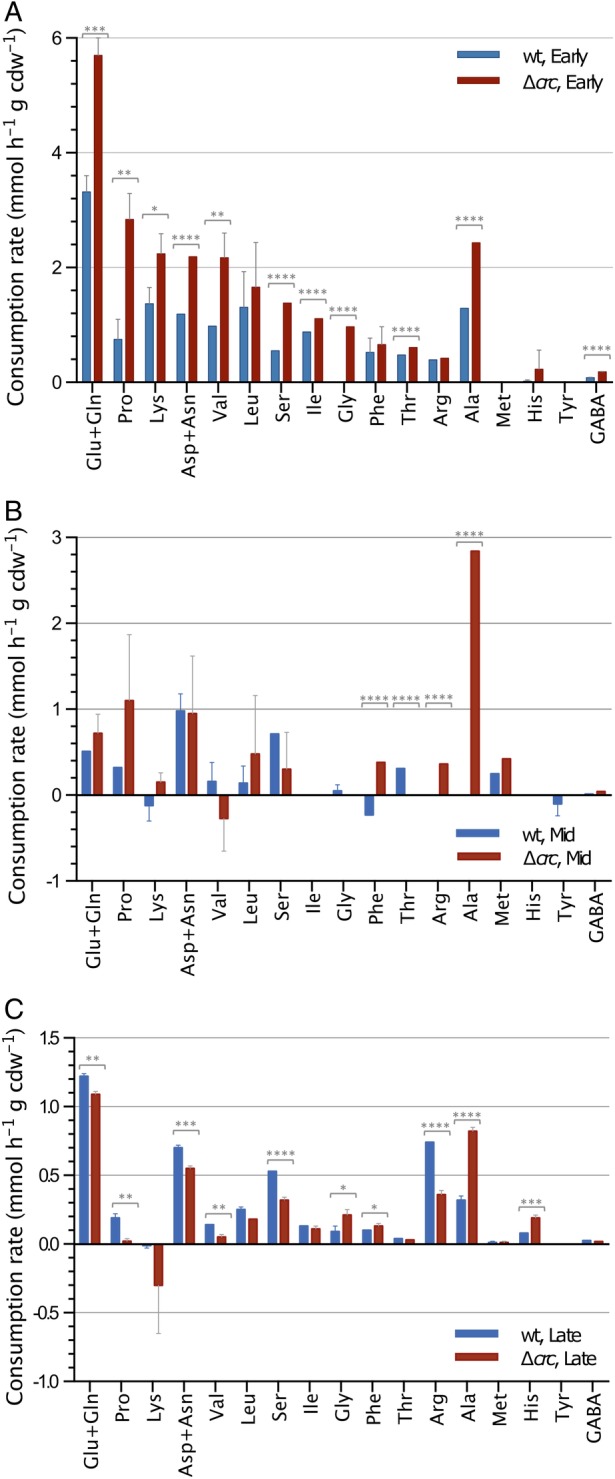
Consumption rates for the amino acids present in LB. The values observed for P. putida strains KT2440 (wild type; blue bars) and KTCRC (∆crc; red bars) at different times of their exponential growth are indicated.A. Early phase of growth. B. Mid‐exponential phase. C. Late exponential phase. Error bars indicate the standard deviation of three biological replicates. The scale is not the same in all panels. The statistical significance of the differences in the values of the wild type and the ∆ crc mutant was determined by one‐way ANOVA (* p < 0.1; **p < 0.01; ***p < 0.001; ****p < 0.0001).
The Crc‐null strain began growth by consuming sugars (free glucose and glucose polysaccharides, labelled as “total glucose” in the Supporting Information Table S1), glycerol, succinate, formate, malate and several amino acids, with glutamate/glutamine the most consumed. The consumption rate of glucose during the first 2 h of growth was more than three times that reported for the wild‐type strain cultivated under the same conditions (Molina et al., 2019) (Fig. 1A). Glucose consumption was paralleled by the secretion of 2‐ketogluconate, likely derived from glucose oxidation in the periplasm (Fig. 1A). However, while in the wild‐type strain, nearly all the glucose consumed during the first 2 h of growth was released into the medium as 2‐ketogluconate, in the Crc‐null strain the 2‐ketogluconate detected in the medium represented only 65% of the glucose consumed, the rest presumably being transported to the cytoplasm (either as glucose, gluconate or 2‐ketogluconate; Fig. 1A). During the early phase, the consumption rate of several amino acids was higher in the Crc‐null strain than in the wild type (Fig. 2A), which was reflected in a strong release of ammonia (Supporting Information Table S1) that was almost six times that reported for the wild type (Molina et al., 2019). Finally, the early phase of growth saw the substantial secretion of pyruvate and, to a lesser extent, acetate (Fig. 1A), as previously reported (La Rosa et al., 2016).
During the mid‐exponential phase, the consumption of glucose continued at a high rate, but the fate of gluconate and 2‐ketogluconate changed. Their concentration in the medium decreased substantially, indicating them to have been recovered from the medium and catabolized (Fig. 1B). The consumption of amino acids decreased substantially with the exception of alanine, which continued at a high rate in the Crc‐null strain, but not in the wild type (Fig. 2B). Pyruvate secretion by the mutant strain continued at a rate similar to that observed during the previous phase; TCA intermediates such as citrate and α‐ketoglutarate were also secreted during this growth phase (Fig. 1B).
In the late phase of growth, the glucose consumption rate decreased substantially (by more than 35‐fold in the crc mutant strain), and pyruvate was recovered from the medium and consumed (Fig. 1C). The consumption of amino acids was low in both the wild‐type and Crc‐null strains, and the use of glutamate and glutamine was partially regained compared to the previous phase (Fig. 2C).
Construction of condition‐specific metabolic models and proteomic analyses of the Crc‐null strain at different moments of the growth phase
To visualize the changes in the metabolic fluxes derived from the inactivation of the crc gene, the iJN1411 metabolic model developed for P. putida KT2440 was used. This model includes 2826 metabolic and transport reactions and 1411 genes and offers a solid representation of P. putida metabolism (Nogales et al., 2008; Nogales et al., 2017). Condition‐specific models for the metabolic status of cells in the early, mid and late exponential phases of growth were constructed using the experimentally obtained data for the consumption or expulsion of metabolites during these growth phases (indicated in the Supporting Information Table S1). The solution space for each growth phase‐specific model was then examined by flux‐balance analysis using growth rate as the objective function. The flux distributions between the different condition‐specific models were then systematically compared. The objective function used was the maximization of the growth rate. The specific growth rates (μ) for each time point estimated by the flux model were very similar to those determined experimentally on the basis of the increase in biomass (Fig. 3). This strongly supports the idea that the flux model correctly contemplates the uptake rates and assimilation of the nutrients by the cells.
Figure 3.
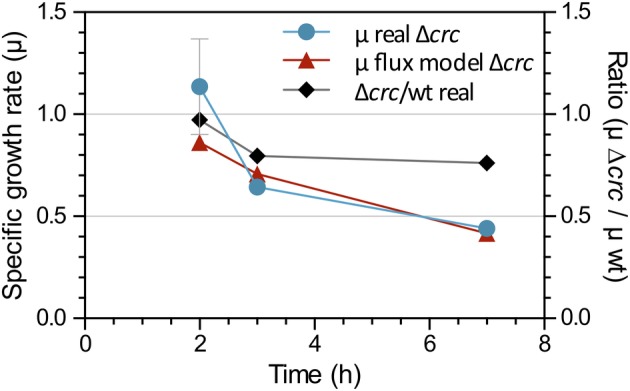
Growth rate (μ) of cells during the early‐, mid‐ and late‐exponential phases of growth. Blue, values determined experimentally for the Crc‐null strain measuring the increase in cell dry weight (error bars indicate the standard deviation of three biological replicates). Red, values predicted by the flux model for the Crc‐null strain. The ratio of the experimentally determined growth rates for the Crc‐null versus the wild‐type strain at each growth phase is indicated in black.
In addition, the proteomes of the cells collected at the early, mid and late exponential phase were analysed to confirm that the changes in the predicted metabolic fluxes agreed with the increase or reduction in the proteins involved in the corresponding metabolic pathways. More than 1730 proteins were detected. The transition from the early to the mid‐exponential phase of growth was paralleled by a change in the abundance of 8.2% of the proteins detected, most of them being downregulated (summarized in the Supporting Information Table S2; see Table S3 in the Supporting Information for a complete list of the differentially expressed proteins). However, this picture changed as cells entered the late exponential phase of growth, during which the abundance of 16.6% of the proteins changed relative to the early phase, most of them being upregulated. A visual summary of the changes detected is presented in the Supporting Information Figure S1.
The absence of Crc changes the configuration of the metabolic fluxes during the early exponential phase
Earlier work performed with the wild type strain KT2440 using the same experimental design as used in the present work indicated that, during the early exponential phase of growth in LB medium, the cells use different organic acids (acetate, citrate, malate and succinate) as carbon sources, plus a selected set of amino acids, mostly glutamate/glutamine (Molina et al., 2019). Sugars (glucose monomers and glucose polymers derived from yeast extract) were also consumed during this early stage, but rather than being used as a carbon source they were mostly utilized to supply electrons to the electron transport chain. The oxidized sugar products were mostly released into the medium. In addition, the predicted carbon fluxes suggested that the TCA cycle operated exclusively through the glyoxylate shunt, and the metabolic fluxes towards glucose‐6‐P indicated that the cells were functioning in a gluconeogenic configuration.
In the case of the Crc‐null strain, the metabolic fluxes predicted using the model for the early growth phase showed clear differences to those of the wild‐type strain. Figure 4 shows a brief summary of the fluxes and of their modification throughout growth, while Figures [Link], [Link] (Supporting Information) provide a more detailed description. In addition, a comparison of the fluxes of the Crc‐null and wild‐type strains at each of the growth phases analysed can be found in Figure 5 and in more detail in the Supporting Information Figures [Link], [Link]. According to the model predictions, and contrary to that which occurred in the wild‐type strain, the Crc‐null strain allowed the entry of some of the glucose and gluconate/ketogluconate into the cytoplasm, which were then converted to pyruvate. As mentioned earlier, 65% of the glucose consumed was recovered as gluconate or ketogluconate in the medium (Supporting Information Table S1), indicating that about 35% of the glucose/gluconate/ketogluconate molecules that entered the cells were metabolized, feeding the pyruvate pool. Pyruvate and acetate were produced not only from glucose but from lactate, alanine, serine, leucine, isoleucine, malate or fatty acids. The flux model indicated that about 80% of the pyruvate produced was expelled to the medium, while only a small fraction was converted to acetyl‐CoA. The secretion of pyruvate during the early growth phase was about 18 times that observed earlier for the wild‐type strain (La Rosa et al., 2016; Molina et al., 2019).
Figure 4.
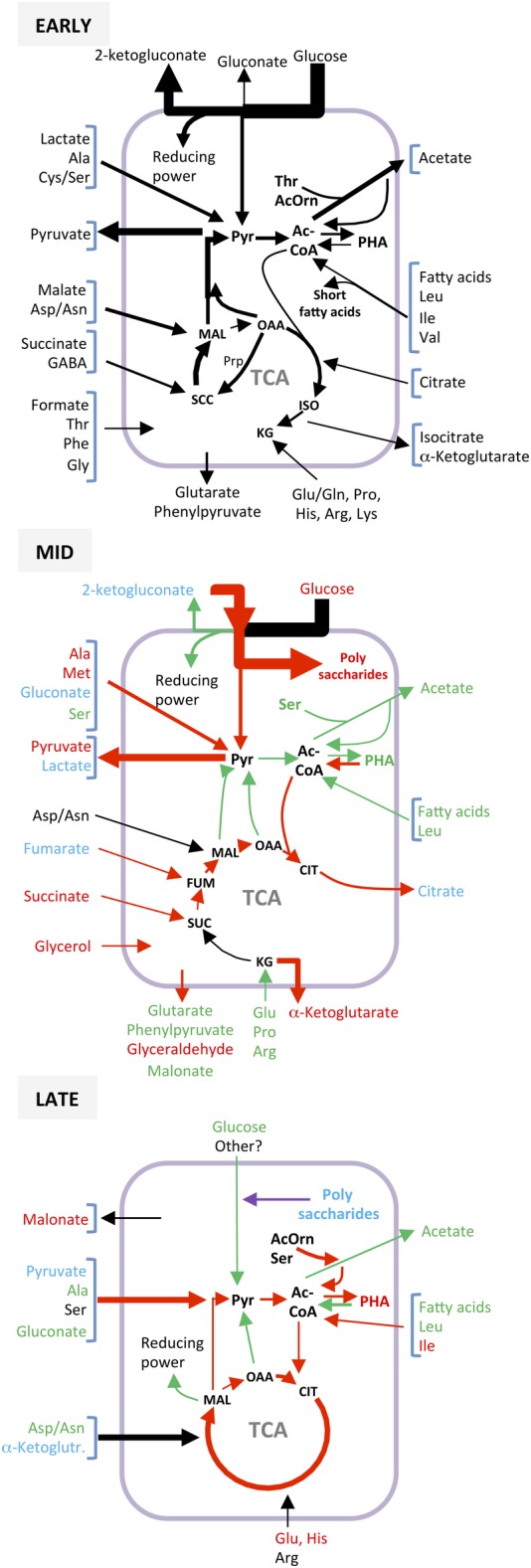
Configuration of the metabolite fluxes related to central carbon metabolism in the Crc‐null strain during early, mid and late exponential growth. Summary of the metabolite fluxes as predicted by the iJN1411 metabolic model constrained by the consumption data indicated in the Supporting Information Table S1. The fluxes corresponding to the early exponential phase are indicated in black. Fluxes that change at later times of growth are indicated in red (increased flux) or green (decreased flux). Compounds that are released to the medium and later recycled are indicated in blue. A more detailed distribution of the fluxes is given in the Supporting Information Figures S2 (early exponential phase), S3 (mid exponential phase) and S5 (late exponential phase). TCA, tricarboxylic acids cycle; G6P, glucose‐6‐phosphate; Ac‐CoA, acetyl‐CoA; PHA, polyhydroxyalkanoates; OAA, oxaloacetate; SUC, succinate; ISO, isocitrate, MAL, malate; CIT, citrate, Pyr, pyruvate, KG, α‐ketoglutarate.
Figure 5.
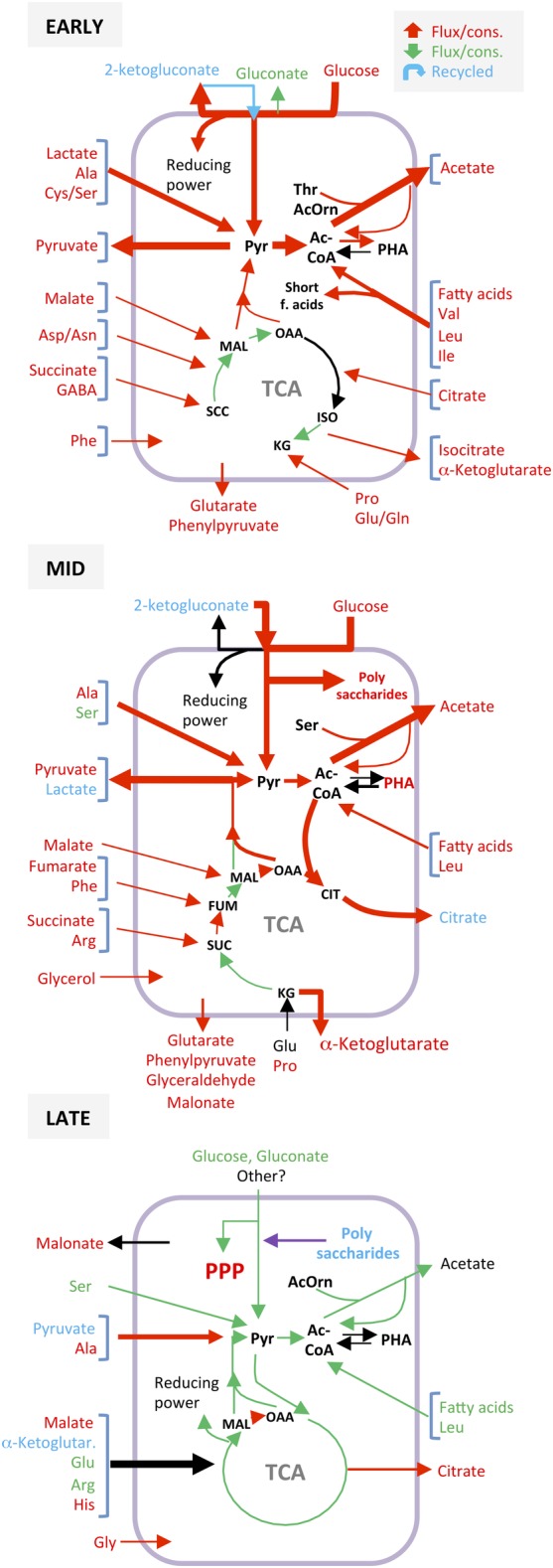
Effect of inactivating the crc gene on the configuration of the metabolite fluxes related to central carbon metabolism during early, mid and late exponential growth. The fluxes that increased (in red), decreased (in green), or remained unchanged (in black) in the Crc‐null strain as compared to the wild type, are highlighted. Compounds that were released to the medium and later recycled are indicated in blue. A more detailed description is given in the Supporting Information Figures S7 (early exponential phase), S9 (mid‐exponential phase) and S11 (late exponential phase). For abbreviations, see Fig. 4.
The most notable modification to the TCA cycle derived from the absence of the Crc protein came via the drastic reduction in the flow of metabolites through the glyoxylate shunt. In the wild type, the glyoxylate shunt played an important role during this phase, bypassing the oxidative steps of the TCA cycle and promoting the production of the C4 metabolites that drive gluconeogenesis (Molina et al., 2019). The absence of the glyoxylate shunt in the Crc‐null strain impaired the funnelling of carbon skeletons towards gluconeogenesis and therefore the flux from succinate to oxaloacetate decreased significantly (Fig. 5). In contrast, the methyl‐citrate cycle (Prp proteins) was activated, which allowed an important fraction of the oxaloacetate to be converted into succinate and pyruvate, alleviating the accumulation of TCA intermediates derived from the interruption of the cycle at the level of ketoglutarate (Fig. 4 and Supporting Information Fig. S2). This reconfiguration of the TCA cycle was not completely efficient, however, and a large amount of carbon was predicted to be secreted in the form of ketoglutarate. Finally, part of the carbon overflow towards pyruvate was directed towards the synthesis of polyhydroxyalkanoates (PHAs), a process predicted to be more active at this stage in the Crc mutant than in the wild‐type strain.
Overall, the fluxes through the TCA cycle during the early growth phase were smaller in the Crc mutant than in the wild‐type strain, which reduced the amount of NADH generated in this cycle. This reduction was compensated by the NADH produced via the pentose phosphate pathway (PPP) through the oxidation of glucose and glutamate.
The consumption of amino acids during the early growth phase was greater in the Crc‐null strain than in the wild type (Fig. 2) and was paralleled by an increased abundance of proteins involved in the uptake and assimilation of amino acids such as glutamate/aspartate (ABC‐transport proteins GltL, GltK, GltI and GltJ, AnsB glutaminase‐asparaginase and GdhA glutamate dehydrogenase), proline (PutA and PutP proteins), histidine (histidine‐specific outer membrane protein OpdC and the His/Lys/Arg permease HisQ), and aromatic amino acids (AroP‐II transporter and the PhhAB and Hpd proteins). Proteins involved in the uptake of dipeptides (DppA‐I, DppA‐II, DppA‐III, DppB, DppD, OpdP) were also more abundant in the Crc‐null strain than in the wild type (Supporting Information Fig. S8, red arrows, and Supporting Information Table S3). The greater consumption of amino acids fed the pools of pyruvate, acetyl‐CoA and intermediates of the TCA cycle (Fig 6), supporting the idea that the internal pools of pyruvate and acetyl‐CoA were beyond cellular demand, resulting in their secretion.
Figure 6.
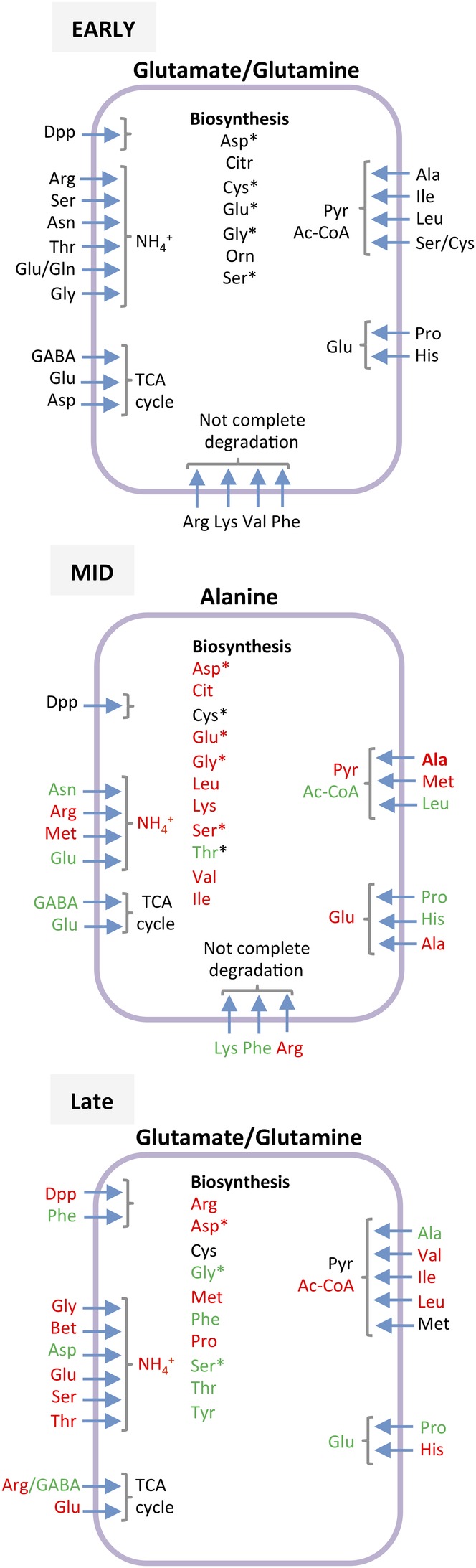
Metabolism of amino acids in the Crc‐null strain during early, mid and late exponential growth. Summary of the fate of the amino acids used as predicted by the iJN1411 metabolic model constrained by the consumption data indicated in the Supporting Information Table S1. Those used during the early exponential phase are indicated in black. Amino acids for which consumption increased (red) or decreased (green) relative to early exponential growth are highlighted. Those that, after their uptake, are biodegraded within cells are indicated with an asterisk. A more detailed distribution of the fluxes is given in the Supporting Information Figures S2 (early exponential phase), S4 (mid‐exponential phase) and S6 (late exponential phase). *Amino acids that were biosynthesized and later transformed, according to the flux model.
Influence of Crc on the configuration of the metabolic fluxes during the mid‐exponential phase
During the mid‐exponential phase, the consumption of glucose by the Crc‐null strain remained nearly as high as in the previous phase, although it was not paralleled by an accumulation of gluconate or ketogluconate in the growth medium. On the contrary, these two compounds were recovered from the medium and metabolized to the point that the 2‐ketogluconate accumulated in the previous phase (3.36 mM) now disappeared from the medium (Supporting Information Table S1 and Fig. 4). In comparison, in the wild‐type strain, glucose consumption during this phase was about half that of the Crc‐null strain, and the concentration of 2‐ketogluconate in the medium remained high; apparently only part of it was transported to the cytoplasm and consumed (Molina et al., 2019). According to the flux model, in the Crc‐null strain, about half of the sugar metabolization products made during this growth phase were funnelled towards pyruvate, while a large part was predicted to be transformed into polysaccharides (Figs. 4 and 5).
The pyruvate pool was also fed by the metabolization of alanine, the uptake of which was significantly greater in the Crc‐null strain than in the wild type (Figs. 2, 3, 4, 5). Since, according to the model, the production of AcCoA and PHAs from pyruvate decreased, it is not surprising that the secretion of pyruvate should increase in this growth phase relative to the previous (Supporting Information Table S1). Acetate secretion was also evident; this compound was produced both from pyruvate and from the assimilation of fatty acids and leucine.
During mid exponential growth, the flux model indicated that the TCA cycle to operate from α‐ketoglutarate towards citrate, but citrate was not further transformed. This possibly explains the observed secretion of citrate into the medium (Supporting Information Table S1 and Fig. 4), something that barely occurred in the wild‐type strain. This configuration of the TCA cycle allowed the assimilation of compounds such as succinate, fumarate, malate, glycerol, glutamate, aspartate/asparagine and proline, although the scant activity of ketoglutarate dehydrogenase led to a secretion of ketoglutarate, which accumulated in the growth medium.
The consumption of amino acids by the Crc‐null strain during the mid‐exponential growth decreased significantly compared to the previous phase, with the exception of alanine, the consumption of which remained high (Fig. 2). Wild‐type cells, however, did not consume alanine during this phase (La Rosa et al., 2016; Molina et al., 2019). The flux model predicted the activation of several amino acid biosynthesis pathways during this phase to compensate for their lower intake (Fig. 6).
Configuration of metabolism during the late exponential phase
Seven hours after inoculation, the cells were in the late exponential phase, still growing albeit at a lower rate (Fig. 3). At this time, the consumption pattern of the Crc‐null strain and the ensuing metabolic fluxes again differed from those of the preceding phase: the assimilation of sugars ceased (the glucose concentration had decreased to a 10th of its initial value) and the cells now used the pyruvate excreted to the medium in the preceding phases, along with some amino acids, mostly glutamate/glutamine, alanine, aspartate/asparagine, serine and arginine (Supporting Information Table S1 and Figs. 4 and 6). Acetate was not recovered; in fact, some secretion was still detected (Supporting Information Table S1). The TCA cycle, which in the previous phase operated in two divergent branches, now functioned in a fully cyclical and oxidative manner, fed by amino acids such as glutamate/glutamine, aspartate/asparagine and arginine. This allowed pyruvate and NADH/NADPH to be made, compensating for the strong reduction in the production of these compounds derived from the oxidation of sugars, which was now much less than in the previous phases (Supporting Information Table S1). The production of PHAs was predicted to increase relative to the previous phase and to be greater than in the wild‐type strain, which agrees with earlier experimental evidence (La Rosa et al., 2014). Several amino acids in short supply in the medium were predicted to be synthesized (Fig. 7). Compared to the wild type, many central metabolic pathways were less active (Fig. 5); metabolism appeared to be driven by a mixture of carbon sources, among which glutamate/glutamine, aspartate/asparagine and alanine stood out.
Figure 7.
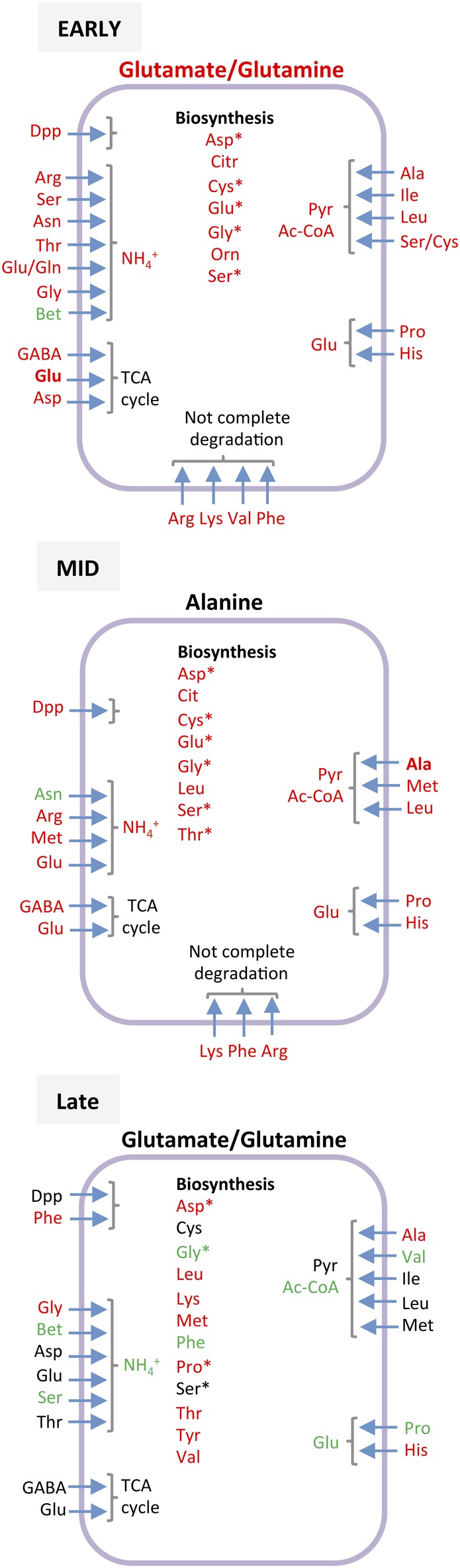
Effect of inactivating the crc gene on the metabolism of amino acids during early, mid and late exponential growth. The amino acids whose consumption increases (in red), decreases (in green), or remains unchanged (in black) in the Crc‐null strain compared to the wild type, are highlighted. A more detailed description is given in the Supporting Information Figures S8 (early exponential phase), S10 (mid‐exponential phase) and S12 (late exponential phase). *Amino acids that were biosynthesized and later transformed, according to the flux model.
Discussion
Overall, the present results indicate that the Crc‐null strain consumed nutrients much faster than the wild strain during the early exponential phase of growth. The consumption rate of glucose during this phase more than tripled, and that of most amino acids was also higher (about two to three times that for many of those showing the highest consumption rates, such as Glu + Gln, Pro, Lys, Asp/Asn, Ala, Val or Ser). The consumption rates for other amino acids was, however, similar in both strains (Phe, Thr, Arg, Ile). While the wild‐type strain obtained energy (electrons) but not carbon skeletons from glucose at this time, the Crc‐null strain obtained both. In other words, while the wild type strain restricted the entry of glucose and the glucose‐oxidation products generated at the periplasm during early growth, the Crc‐null strain allowed the entry of about 35% of these compounds into the cytoplasm, which were then converted into pyruvate. As a result, the configuration of the main metabolic pathways in the Crc‐null strain differed in several aspects from that of the wild type. The flow of the TCA cycle intermediates towards pyruvate added to the pyruvate generated from glucose and amino acids such as Ala or Ser. The cells were apparently unable to make full use of the pyruvate produced, or to direct it towards the formation of storage products, and thus it was expelled at a rate almost 18 times that detected in the wild type strain during this phase of growth. Acetate was also expelled by the mutant strain, albeit at a lower rate. In spite of the greater consumption of nutrients, the growth rate of the Crc‐null strain during the early exponential phase was very similar to that of the wild type, indicating that the increase in nutrient use did not translate into more efficient growth.
Proteomic analyses detected several proteins involved in the transport and assimilation of sugars and amino acids that were more abundant in the Crc‐null strain than in the wild type (Supporting Information Table S3), which might explain the more rapid assimilation of these compounds by the mutant strain. Such is the case of the GtsD protein (which forms part of the GtsABCD glucose transport system), the GntT gluconate transporter, and different proteins involved in the uptake of dipeptides and the assimilation of amino acids, including Glu, Asp, Lys, Arg, Ala, and aromatic and branched‐chain amino acids (see Supporting Information Table S3). Many of these are encoded by genes the mRNAs of which have a target site for the Crc/Hfq proteins (see Supporting Information Table S4). In addition, genes that are regulated by transcriptional activators that include Crc/Hfq targets can also be strongly – albeit indirectly – regulated by Crc/Hfq. For example, the expression of the GtsABCD‐OprB‐1 transport system involved in glucose uptake is activated by the GltR‐2 transcriptional regulator (del Castillo et al., 2008; Daddaoua et al., 2014). The gltR‐2 gene, as well as gtsA and gtsB (included in the gtsABCD‐oprB‐1 polycistronic mRNA), contain putative Crc/Hfq targets. As predicted, the abundance of the GtsA and GtsD proteins was greater in the Crc‐null strain than in the wild‐type strain during the exponential phase of growth.
Glucose consumption during the mid‐exponential phase remained high – in the Crc mutant it was double that noted for the wild‐type strain – and most of glucose and glucose oxidation products entered the cell in both strains. Unlike that seen for the early growth phase, the model now predicted that a substantial part of the glucose oxidation products would be converted into polysaccharides (storage products), while only a small fraction of the flux would continue towards pyruvate. This organic acid was produced in significant amounts, as well from other sources such as Gln/Glu, Pro, Asp/Asn, Phe and, most notably Ala. The final result was that the pyruvate pool remained too large for cellular demands, and its secretion increased by 26% compared with the previous phase. The flow of metabolites through most of the main central metabolic pathways was stronger in the Crc‐null strain than in the wild type. Despite this, the growth rate of the Crc‐null strain decreased compared to the previous phase and was only about 80% that of the wild type. This again indicates that cells could not profit from the increased consumption of resources. It would therefore seem likely that Crc regulates the entry and consumption of compounds present in the medium to meet the demands and metabolic capacity of the cell, avoiding the excessive entry of assimilable compounds when these are in high concentration.
During the late phase of growth, the metabolic configuration of the Crc‐null strain suffered a notable change. The consumption rate of glucose decreased by about 40‐fold and fell to a third of that recorded for the wild type. Pyruvate secretion ceased; indeed, this compound was now recovered from the medium and used as a carbon source. The consumption of amino acids was low and included mainly Gln/Glu, Asp/Asn and Ala. The TCA cycle now operated in a fully oxidative configuration, as in the wild type, but its activity was less than in the wild type. Part of the resources were diverted towards the production of PHAs, as previously observed (La Rosa et al., 2014). The growth rate did not recover but continued at values not exceeding 80% of that observed for the wild‐type strain. The overall picture was that of a strain with an unbalanced metabolism.
In summary, the present results show the Crc/Hfq system to be a key regulator of carbon metabolism that controls the uptake and assimilation of carbon sources. This helps keep the flow of metabolites through the corresponding pathways at below overflow levels, avoiding the accumulation and secretion of metabolites such as pyruvate and acetate. Crc affected and coordinated tasks such as nutrient uptake, central metabolism and the generation of energy. Although the role of Crc/Hfq system as a regulator of the expression of particular genes has been well documented, its overall influence on the flow of metabolites through the main metabolic pathways was poorly known. The present results provide a clear indication of the key role of Crc/Hfq regulatory system in the optimization of metabolism when cells grow in the presence of an ample supply of carbon sources. This is key for the fitness of bacteria that are metabolically versatile and live in environments in which the supply of nutrients can suffer frequent and significant changes (Chubukov et al., 2014). The signals that drive the regulation of the metabolic fluxes in P. putida are unclear. It has been proposed that the concentration and/or the flow of certain ‘flux‐signalling’ metabolites through particular enzymes may serve as signals to control metabolic fluxes in microbes (for a review, see Litsios et al., 2018). The activity of the Crc/Hfq system can change not only according to nutrient availability and internal signals (Monteagudo‐Cascales et al., 2019), but to temperature as well, which has important consequences for cell physiology (Fonseca et al., 2011, 2013). Thus, the Crc/Hfq system seems to act as an information hub that allows cells to adapt their metabolic fluxes to the prevailing conditions, thereby optimizing growth.
Experimental Procedures
Bacterial strains and culture media
Pseudomonas putida KTCRC, a KT2440 derivative containing an inactivated crc allele, has been described elsewhere (Hernández‐Arranz et al., 2013). Cells were cultured at 30°C in LB medium (10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl). Tryptone and yeast extract were obtained from Conda (Spain).
Determination of chemical compounds in LB
Twenty milliliters of fresh LB were inoculated with an overnight culture of P. putida KTCRC at a turbidity (A600) of 0.05. The cells were incubated with vigorous aeration at 30°C to an A600 of 0.2 (early exponential growth), 0.6 (mid‐exponential growth), or 2 (late exponential growth). At these points, cells were centrifuged at 7500 rpm for 10 min and the supernatants obtained filtered through 0.22 μm pore filters (Millipore‐Merck) to eliminate any remaining cells. They were then frozen at −80°C until analysis. To determine the total content of glucose in the samples, glucose polysaccharides were hydrolysed with 2 M trifluoroacetic acid at 120°C for 1 h. Glucose was then detected by HPLC as previously described (Molina et al., 2019). The concentration of 2‐ketogluconate, gluconate, lactate, fatty acids, nucleotides, α‐ketoglutarate, citrate, GABA, isocitrate and malate in the growth medium was performed as described elsewhere (Molina et al., 2019). In all cases, three independent assays (biological replicates) were performed.
The total amino acid content of the culture samples (free amino acids plus those forming polypeptides) was determined by Alphalyse (Denmark) after acid hydrolysis of the samples for 20 h at 110°C in 6 M HCl, 0.1% phenol and 0.1% thioglycolic acid under reduced pressure in an argon atmosphere. The 20 common amino acids, except for tryptophan and cysteine, were all determined in two biological replicates.
To measure the redox potential of the cells, these were collected from 1.5 ml of culture and treated with 2,3,4‐triphenyl‐2H‐tetrazolium chloride as previously described (Defez et al., 2017). pH was measured using a pH‐meter. To determine the dry weight of culture samples, 50 ml of culture were centrifuged at 7000 rpm for 10 min. The resulting pellets were dried for 24 h at 60°C and weighed using an analytical balance.
Constraint‐based analyses of metabolic fluxes
Metabolic flux estimations were undertaken using the iJN1411 genome‐scale metabolic model for P. putida KT2440 (Nogales et al., 2017). This model includes 2826 metabolic and transport reactions, 2087 metabolites and 1411 genes, and provides the best available representation of P. putida metabolism. The model accounts for reactions mimicking the exchange of up to 337 different metabolites with the environment. It can contemplate 220 metabolites as the sole carbon and energy source, and 105 metabolites as the sole nitrogen source, and can explain growth in rich media, such as LB (Nogales et al., 2017). Growth rate predictions and initial flux distributions were performed using flux balance analysis (FBA) (Varma and Palsson, 1995, Orth et al., 2010), which is based on solving linear optimization problems by maximizing or minimizing a given objective function ‘Z’ subject to a set of constraints. The constraint ‘S·v = 0’ corresponds to a situation of steady‐state mass conservation where the change in concentration of the metabolites over time is zero. ‘S’ is an m × n matrix containing all the stoichiometric coefficients in the model of m metabolites and n reactions, and the vector ‘v’ has n elements that represent the individual flux values for each reaction. These fluxes are additionally constrained by the imposed lower and upper limits (bounds) ‘vl’ and ‘vu’. The output is a flux distribution that maximizes or minimizes a given objective function. The growth rate was used routinely as the objective function. All computational simulations were performed using the COBRA toolbox (Schellenberger et al., 2011) in the MATLAB environment (The MathWorks Inc.). Linear optimization problems were solved using the GNU Linear Programming Kit (GLPK) (http://www.gnu.org/software/glpk).
Construction of growth‐phase condition specific models
To simulate bacterial growth in LB, an initial in silico medium was defined based on the composition of LB reported in the literature (Oh et al., 2007) and gene analysis of strain KT2440 (Nogales et al., 2017; Molina‐Henares et al., 2010). The initial in silico LB composition was then completed with the metabolites detected in the present analyses (Supporting Information Table S1). The default growth conditions included the free uptake of the metabolites present in the medium while allowing for the free exchange of CO2, H2O, H+, HCO3 −, Na+, NH4 +, Pi, Fe2+ and SO4 2−. Constraint‐based metabolic models calculate intracellular flux distributions that satisfy three types of constraints: steady‐state mass‐balance, reaction reversibility and flux capacities. For the latter, the flux measured for a given reaction, including nutrient uptake rates, can be imposed on the model, constraining the solution space. The growth phase‐specific models were constructed by constraining to experimental values the bounds of the exchange reactions corresponding to the different compounds for which uptake and/or secretion rates could be determined (sugars, organic acids, amino acids, lipids, nucleotides and ammonium) at each time point (Supporting Information Table S1). This approach allowed specific models reproducing the growth rate and nutrient exchange during early, mid and late exponential growth to be constructed.
Quantitative analysis of proteomes by mass spectrometry using isobaric tagging relative and absolute quantitation
Cells were collected, disrupted and processed as described earlier (Molina et al., 2019). Proteomic analyses were performed at the CNB‐CSIC proteomics facility (Madrid, Spain; http://proteo.cnb.csic.es/proteomica/) using iTRAQ, as described elsewhere (Molina et al., 2019). Protein identification and the analysis of protein differential expression were performed by Proteobotics S.L. (Madrid, Spain), also as described earlier (Molina et al., 2019). Differential regulation was measured using linear models (López‐Serra et al., 2014). Statistical significance was measured using q‐values (FDR). All analyses were conducted using software from Proteobotics. Proteins showing an abundance change of ≥1.4‐fold (log2 fold change of 0.49), and a q‐value of ≤0.05, were considered differentially expressed (Koul et al., 2014).
Supporting information
Table S1. Composition of LB before and after inoculation with P. putida KT2440. The indicated concentrations correspond to those observed before inoculation (labelled as “initial”), or when cells were at the early (A600=0.2), mid (A600=0.6) or late (A600=2) exponential phase of growth. The consumption rates for each compound during these three growth periods are indicated; the highest value for each compound is highlighted in grey. The values underlined were determined following predictions made by the flux model. Glucose concentration refers to free glucose plus that present in polysaccharides (indicated as mM of free glucose monomers). N.d., not determined. (*) Data from reference (La Rosa et al., 2016); (**) TTC reduction assay (A510 nm mg−1 of cell dry weight (CDW)).
Table S2. Proteins detected as differentially expressed (q‐value<0.05, fold‐change >1.4) in the three growth phases for strains KT2440 (indicated as “wt”) and KTCRC (indicated as “crc”), and for KTCRC in each phase compared to the wild type strain (indicated as “crc/wt”). A visual representation is provided in Figure S1.
Table S3. Proteins showing increased or reduced abundance (change ≥1.4 fold; log2 of 0.49), and a q‐value of ≤0.05, when comparing the following conditions: wild type or Crc‐null strains in mid exponential growth relative to early exponential growth (Excel sheets labelled “Wt_0.6_vs_Wt_0.2” or “CRC_0.6_vs_CRC_0.2”, respectively), or in late exponential growth relative to mid exponential growth (Excel sheets labelled “Wt_2.0_vs_Wt_0.6” or “CRC_2.0_vs_CRC_0.6”), or the Crc‐null strain versus the wild type strain under the three growth conditions compared (Excel sheets labelled “CRC_0.2_vs_Wt_0.2”, “CRC_0.6_vs_Wt_0.6”, or “CRC_2.0_vs_Wt_2.0”).
Table S4. Proteins showing increased or reduced abundance (change <1.4 fold; log2 of 0.49; values for which q <0.05 are in bold face), and that are produced from mRNAs that include a putative Hfq/Crc binding site close to or overlapping the translation initiation codon. Upregulated proteins are indicated in yellow, downregulated proteins in green.
Figure S1. Comparison of the proteome of cells collected at early, mid and late exponential growth. Volcano plots of the proteins showing increased or reduced abundance during early, mid and late exponential growth. Those showing a fold‐change >1.4 (q‐value<0.05) are highlighted in red (up‐regulated) or green (down‐regulated). A detailed list of the differentially expressed proteins is provided in Table S3 (Supplementary Material).
Figure S2. Metabolite fluxes predicted for central carbon metabolism (A) and for the metabolism of amino acids (B) in the Crc‐null strain KTCRC during early exponential growth. Fluxes were predicted by the iJN1411 metabolic model constrained by the consumption data in Table S1. Fluxes >0.1 mmol/g cell dry weight/h are indicated in parentheses. Compounds predicted to be released to the medium are boxed in yellow.
Figure S3. Modifications observed in the central carbon metabolism of the Crc‐null strain KTCRC at mid exponential growth relative to early exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. Arrows indicate fluxes (A) or proteins (B) that are more abundant (red), or less abundant (green), or did not change (black) relative to the previous phase. Blue, reactions that did not occur in the previous phase, or that change direction. Arrow thickness is roughly proportional to the fluxes/changes observed. Dashed green lines indicate that some enzymes in the pathway are repressed. Numbers in parenthesis indicate the fold change in the corresponding fluxes (panel A), or in protein levels (panel B). Compounds predicted to be expelled are boxed in yellow; those that accumulate are boxed in blue. Reactions inverted relative to the previous phase (*), or not observed in the previous phase (**), are indicated.
Figure S4. Modifications observed in the metabolism of amino acids by the Crc‐null strain KTCRC in mid exponential growth relative to early exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. For details, see legend to Figure S3.
Figure S5. Modifications observed in the central carbon metabolism of the Crc‐null strain KTCRC at late exponential growth relative to mid exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. For details, see legend to Figure S3.
Figure S6. Modifications observed in the metabolism of amino acids by the Crc‐null strain KTCRC in late exponential growth relative to mid exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. For details, see legend to Figure S3.
Figure S7. Consequences of the inactivation of the crc gene on the metabolite fluxes for central carbon metabolism during early exponential growth in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S2). Arrows indicate fluxes (A) or proteins (B) that are more abundant (red), or less abundant (green), or do not change (black) in the Crc‐null strain relative to the wild type strain. Blue, reactions that do not occur in the wild type strain, or that change direction. Arrow thickness is roughly proportional to the fluxes/changes observed. Dashed green lines indicate that some enzymes in the pathway are repressed. Numbers in parenthesis indicate the fold change in the corresponding fluxes (panel A), or in protein levels (panel B). Compounds predicted to be expelled are boxed in yellow; those that accumulate are boxed in blue. Reactions inverted (*) or nor observed (**) relative to the previous phase are indicated. Fluxes for the Crc‐null strain were predicted by the iJN1411 metabolic model constrained by the consumption data in Table S1; those for the wild type were predicted using the same procedure, and have been reported earlier (Molina et al., 2019).
Figure S8. Effects of the inactivation of the crc gene on the metabolite fluxes for the metabolism of amino acids during the early exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) the proteomic analysis reported in Table S3. For details, see legend to Figure S7.
Figure S9. Effects of the inactivation of the crc gene on the metabolite fluxes for central carbon metabolism during the mid‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S3). For details, see legend to Figure S7.
Figure S10. Effects of the inactivation of the crc gene on the metabolite fluxes for the metabolism of amino acids during the mid‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S3). For details, see legend to Figure S7.
Figure S11. Effects of the inactivation of the crc gene on the metabolite fluxes for central carbon metabolism during the late‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) the proteomic analysis presented in Table S3. For details, see legend to Figure S7.
Figure S12. Effects of the inactivation of the crc gene on the metabolite fluxes for the metabolism of amino acids during the late‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S3). For details, see legend to Figure S7.
Acknowledgements
We are grateful to L. Yuste for excellent technical assistance. This work was funded by grant PGC2018‐094143‐B‐I00 to FR from the Ministerio de Ciencia, Innovación y Universidades, Spain (AEI/FEDER, EU).
References
- Adams, B.L. (2016) The Next Generation of Synthetic Biology Chassis: Moving Synthetic Biology from the Laboratory to the Field. ACS Synth Biol 5: 1328–1330. [DOI] [PubMed] [Google Scholar]
- Belda, E. , van Heck, R.G. , José López‐Sánchez, M. , Cruveiller, S. , Barbe, V. , Fraser, C. , et al (2016) The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18: 3403–3424. [DOI] [PubMed] [Google Scholar]
- Chubukov, V. , Gerosa, L. , Kochanowski, K. , and Sauer, U. (2014) Coordination of microbial metabolism. Nat Rev Microbiol 12: 327–340. [DOI] [PubMed] [Google Scholar]
- Daddaoua, A. , Molina‐Santiago, C. , de la Torre, J. , Krell, T. , and Ramos, J.L. (2014) GtrS and GltR form a two‐component system: the central role of 2‐ketogluconate in the expression of exotoxin A and glucose catabolic enzymes in Pseudomonas aeruginosa . Nucleic Acids Res 42: 7654–7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defez, R. , Andreozzi, A. , and Bianco, C. (2017) Quantification of triphenyl‐2H‐tetrazoliumchloride reduction activity in bacterial cells. Bio‐protocol 7: e2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo, T. , and Ramos, J.L. (2007) Simultaneous catabolite repression between glucose and toluene metabolism in Pseudomonas putida is channeled through different signaling pathways. J Bacteriol 189: 6602–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo, T. , Ramos, J.L. , Rodríguez‐Hervá, J.J. , Fuhrer, T. , Sauer, U. , and Duque, E. (2007) Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J Bacteriol 189: 5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo, T. , Duque, E. , and Ramos, J.L. (2008) A set of activators and repressors control peripheral glucose pathways in Pseudomonas putida to yield a common central intermediate. J Bacteriol 190: 2331–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, B.E. , Kurth, F. , Grund, M. , Blank, L.M. , and Schmid, A. (2011) Response of Pseudomonas putida KT2440 to increased NADH and ATP demand. Appl Environ Microbiol 77: 6597–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, P. , Moreno, R. , and Rojo, F. (2011) Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ Microbiol Rep 3: 329–339. [DOI] [PubMed] [Google Scholar]
- Fonseca, P. , Moreno, R. , and Rojo, F. (2013) Pseudomonas putida growing at low temperature shows increased levels of CrcZ and CrcY sRNAs, leading to reduced Crc‐dependent catabolite repression. Environ Microbiol 15: 24–35. [DOI] [PubMed] [Google Scholar]
- Hernández‐Arranz, S. , Moreno, R. , and Rojo, F. (2013) The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi‐tier regulation strategy. Environ Microbiol 15: 227–241. [DOI] [PubMed] [Google Scholar]
- Kampers, L.F.C. , Volkers, R.J.M. , and Martins Dos Santos, V.A.P. (2019) Pseudomonas putida KT2440 is HV1 certified, not GRAS. Microb Biotechnol 12: 845–848. 10.1111/1751-7915.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul, A. , Vranckx, L. , Dhar, N. , Gohlmann, H.W. , Ozdemir, E. , Neefs, J.M. , et al (2014) Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun 5: 3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, R. , de la Peña, F. , Prieto, M.A. , and Rojo, F. (2014) The Crc protein inhibits the production of polyhydroxyalkanoates in Pseudomonas putida under balanced carbon/nitrogen growth conditions. Environ Microbiol 16: 278–290. [DOI] [PubMed] [Google Scholar]
- La Rosa, R. , Behrends, V. , Williams, H.D. , Bundy, J.G. , and Rojo, F. (2016) Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas . Environ Microbiol 18: 807–818. [DOI] [PubMed] [Google Scholar]
- Litsios, A. , Ortega, A.D. , Wit, E.C. , and Heinemann, M. (2018) Metabolic‐flux dependent regulation of microbial physiology. Curr Opin Microbiol 42: 71–78. [DOI] [PubMed] [Google Scholar]
- López‐Serra, P. , Marcilla, M. , Villanueva, A. , Ramos‐Fernández, A. , Palau, A. , Leal, L. , et al (2014) A DERL3‐associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat Commun 5: 3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhushani, A. , Del Peso‐Santos, T. , Moreno, R. , Rojo, F. , and Shingler, V. (2015) Transcriptional and translational control through the 5'‐leader region of the dmpR master regulatory gene of phenol metabolism. Environ Microbiol 17: 119–133. [DOI] [PubMed] [Google Scholar]
- Matsushita, K. , Shinagawa, E. , Adachi, O. , and Ameyama, M. (1979) Membrane‐bound D‐gluconate dehydrogenase from Pseudomonas aeruginosa. Its kinetic properties and a reconstitution of gluconate oxidase. J Biochem 86: 249–256. [PubMed] [Google Scholar]
- Molina, L. , Rosa, R. , Nogales, J. , and Rojo, F. (2019) Pseudomonas putida KT2440 metabolism undergoes sequential modifications during exponential growth in a complete medium as compounds are gradually consumed. Environ Microbiol 21: 2375–2390. 10.1111/1462-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina‐Henares, M.A. , de la Torre, J. , García‐Salamanca, A. , Molina‐Henares, A.J. , Herrera, M.C. , Ramos, J.L. , and Duque, E. (2010) Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome‐wide mutant library. Environ Microbiol 12: 1468–1485. [DOI] [PubMed] [Google Scholar]
- Monteagudo‐Cascales, E. , García‐Mauriño, S.M. , Santero, E. , and Canosa, I. (2019) Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two‐component system in Pseudomonas putida . Sci Rep 9: 9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, R. , Martínez‐Gomariz, M. , Yuste, L. , Gil, C. , and Rojo, F. (2009) The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9: 2910–2928. [DOI] [PubMed] [Google Scholar]
- Moreno, R. , Fonseca, P. , and Rojo, F. (2012) Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol 83: 24–40. [DOI] [PubMed] [Google Scholar]
- Moreno, R. , Hernández‐Arranz, S. , La Rosa, R. , Yuste, L. , Madhushani, A. , Shingler, V. , and Rojo, F. (2015) The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol 17: 105–118. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Martínez‐García, E. , and de Lorenzo, V. (2014) Biotechnological domestication of pseudomonads using synthetic biology. Nat Rev Microbiol 12: 368–379. [DOI] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarria, M. , Fuhrer, T. , Sauer, U. , and de Lorenzo, V. (2015) Pseudomonas putida KT2440 Strain metabolizes glucose through a cycle formed by enzymes of the Entner‐Doudoroff, Embden‐Meyerhof‐Parnas, and pentose phosphate pathways. J Biol Chem 290: 25920–25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, J. , Palsson, B.O. , and Thiele, I. (2008) A genome‐scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst Biol 2: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, J. , Gudmundsson, S. , Duque, E. , Ramos, J.L. , and Palsson, B.O. (2017) Expanding the computable reactome in Pseudomonas putida reveals metabolic cycles providing robustness. bioRxiv 139121: 10.1101/139121. [DOI]
- Oh, Y.K. , Palsson, B.O. , Park, S.M. , Schilling, C.H. , and Mahadevan, R. (2007) Genome‐scale reconstruction of metabolic network in Bacillus subtilis based on high‐throughput phenotyping and gene essentiality data. J Biol Chem 282: 28791–28799. [DOI] [PubMed] [Google Scholar]
- Orth, J.D. , Thiele, I. , and Palsson, B.O. (2010) What is flux balance analysis? Nat Biotechnol 28: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, X.Y. , Dendooven, T. , Sonnleitner, E. , Chen, S. , Blasi, U. , and Luisi, B.F. (2019) Architectural principles for Hfq/Crc‐mediated regulation of gene expression. Elife 8:e43158 10.7554/eLife.43158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poblete‐Castro, I. , Becker, J. , Dohnt, K. , Dos Santos, V.M. , and Wittmann, C. (2012) Industrial biotechnology of Pseudomonas putida and related species. Appl Microbiol Biotechnol 93: 2279–2290. [DOI] [PubMed] [Google Scholar]
- Rojo, F. (2010) Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34: 658–684. [DOI] [PubMed] [Google Scholar]
- Schellenberger, J. , Que, R. , Fleming, R.M. , Thiele, I. , Orth, J.D. , Feist, A.M. , et al (2011) Quantitative prediction of cellular metabolism with constraint‐based models: the COBRA Toolbox v2.0. Nat Protocol 6: 1290–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezonov, G. , Joseleau‐Petit, D. , and D'Ari, R. (2007) Escherichia coli physiology in Luria‐Bertani broth. J Bacteriol 189: 8746–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner, E. , Abdou, L. , and Haas, D. (2009) Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 106: 21866–21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner, E. , Wulf, A. , Campagne, S. , Pei, X.Y. , Wolfinger, M.T. , Forlani, G. , et al (2018) Interplay between the catabolite repression control protein Crc, Hfq and RNA in Hfq‐dependent translational regulation in Pseudomonas aeruginosa . Nucleic Acids Res 46: 1470–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie, B.J. , Hellingwerf, K.J. , van Dijken, J.P. , Elferink, M.G. , van Dijl, J.M. , Kuenen, J.G. , and Konings, W.N. (1985) Energy transduction by electron transfer via a pyrrolo‐quinoline quinone‐dependent glucose dehydrogenase in Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter calcoaceticus (var. lwoffi). J Bacteriol 163: 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, A. , and Palsson, B.O. (1995) Parametric sensitivity of stoichiometric flux balance models applied to wild‐type Escherichia coli metabolism. Biotechnol Bioeng 45: 69–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Composition of LB before and after inoculation with P. putida KT2440. The indicated concentrations correspond to those observed before inoculation (labelled as “initial”), or when cells were at the early (A600=0.2), mid (A600=0.6) or late (A600=2) exponential phase of growth. The consumption rates for each compound during these three growth periods are indicated; the highest value for each compound is highlighted in grey. The values underlined were determined following predictions made by the flux model. Glucose concentration refers to free glucose plus that present in polysaccharides (indicated as mM of free glucose monomers). N.d., not determined. (*) Data from reference (La Rosa et al., 2016); (**) TTC reduction assay (A510 nm mg−1 of cell dry weight (CDW)).
Table S2. Proteins detected as differentially expressed (q‐value<0.05, fold‐change >1.4) in the three growth phases for strains KT2440 (indicated as “wt”) and KTCRC (indicated as “crc”), and for KTCRC in each phase compared to the wild type strain (indicated as “crc/wt”). A visual representation is provided in Figure S1.
Table S3. Proteins showing increased or reduced abundance (change ≥1.4 fold; log2 of 0.49), and a q‐value of ≤0.05, when comparing the following conditions: wild type or Crc‐null strains in mid exponential growth relative to early exponential growth (Excel sheets labelled “Wt_0.6_vs_Wt_0.2” or “CRC_0.6_vs_CRC_0.2”, respectively), or in late exponential growth relative to mid exponential growth (Excel sheets labelled “Wt_2.0_vs_Wt_0.6” or “CRC_2.0_vs_CRC_0.6”), or the Crc‐null strain versus the wild type strain under the three growth conditions compared (Excel sheets labelled “CRC_0.2_vs_Wt_0.2”, “CRC_0.6_vs_Wt_0.6”, or “CRC_2.0_vs_Wt_2.0”).
Table S4. Proteins showing increased or reduced abundance (change <1.4 fold; log2 of 0.49; values for which q <0.05 are in bold face), and that are produced from mRNAs that include a putative Hfq/Crc binding site close to or overlapping the translation initiation codon. Upregulated proteins are indicated in yellow, downregulated proteins in green.
Figure S1. Comparison of the proteome of cells collected at early, mid and late exponential growth. Volcano plots of the proteins showing increased or reduced abundance during early, mid and late exponential growth. Those showing a fold‐change >1.4 (q‐value<0.05) are highlighted in red (up‐regulated) or green (down‐regulated). A detailed list of the differentially expressed proteins is provided in Table S3 (Supplementary Material).
Figure S2. Metabolite fluxes predicted for central carbon metabolism (A) and for the metabolism of amino acids (B) in the Crc‐null strain KTCRC during early exponential growth. Fluxes were predicted by the iJN1411 metabolic model constrained by the consumption data in Table S1. Fluxes >0.1 mmol/g cell dry weight/h are indicated in parentheses. Compounds predicted to be released to the medium are boxed in yellow.
Figure S3. Modifications observed in the central carbon metabolism of the Crc‐null strain KTCRC at mid exponential growth relative to early exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. Arrows indicate fluxes (A) or proteins (B) that are more abundant (red), or less abundant (green), or did not change (black) relative to the previous phase. Blue, reactions that did not occur in the previous phase, or that change direction. Arrow thickness is roughly proportional to the fluxes/changes observed. Dashed green lines indicate that some enzymes in the pathway are repressed. Numbers in parenthesis indicate the fold change in the corresponding fluxes (panel A), or in protein levels (panel B). Compounds predicted to be expelled are boxed in yellow; those that accumulate are boxed in blue. Reactions inverted relative to the previous phase (*), or not observed in the previous phase (**), are indicated.
Figure S4. Modifications observed in the metabolism of amino acids by the Crc‐null strain KTCRC in mid exponential growth relative to early exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. For details, see legend to Figure S3.
Figure S5. Modifications observed in the central carbon metabolism of the Crc‐null strain KTCRC at late exponential growth relative to mid exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. For details, see legend to Figure S3.
Figure S6. Modifications observed in the metabolism of amino acids by the Crc‐null strain KTCRC in late exponential growth relative to mid exponential growth according to (A) the metabolic flux model, and (B) proteomic analysis. For details, see legend to Figure S3.
Figure S7. Consequences of the inactivation of the crc gene on the metabolite fluxes for central carbon metabolism during early exponential growth in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S2). Arrows indicate fluxes (A) or proteins (B) that are more abundant (red), or less abundant (green), or do not change (black) in the Crc‐null strain relative to the wild type strain. Blue, reactions that do not occur in the wild type strain, or that change direction. Arrow thickness is roughly proportional to the fluxes/changes observed. Dashed green lines indicate that some enzymes in the pathway are repressed. Numbers in parenthesis indicate the fold change in the corresponding fluxes (panel A), or in protein levels (panel B). Compounds predicted to be expelled are boxed in yellow; those that accumulate are boxed in blue. Reactions inverted (*) or nor observed (**) relative to the previous phase are indicated. Fluxes for the Crc‐null strain were predicted by the iJN1411 metabolic model constrained by the consumption data in Table S1; those for the wild type were predicted using the same procedure, and have been reported earlier (Molina et al., 2019).
Figure S8. Effects of the inactivation of the crc gene on the metabolite fluxes for the metabolism of amino acids during the early exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) the proteomic analysis reported in Table S3. For details, see legend to Figure S7.
Figure S9. Effects of the inactivation of the crc gene on the metabolite fluxes for central carbon metabolism during the mid‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S3). For details, see legend to Figure S7.
Figure S10. Effects of the inactivation of the crc gene on the metabolite fluxes for the metabolism of amino acids during the mid‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S3). For details, see legend to Figure S7.
Figure S11. Effects of the inactivation of the crc gene on the metabolite fluxes for central carbon metabolism during the late‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) the proteomic analysis presented in Table S3. For details, see legend to Figure S7.
Figure S12. Effects of the inactivation of the crc gene on the metabolite fluxes for the metabolism of amino acids during the late‐exponential growth phase in LB medium according to (A) the iJN1411 metabolic flux model, and (B) proteomic analysis (see Table S3). For details, see legend to Figure S7.


