Abstract
Background.
Real-time RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) is considered the gold standard for Zika virus (ZIKV) infection diagnosis, despite its low sensitivity. Diagnosis using recommended serologic cutoffs in co-circulating Flaviviruses areas maybe inadequate due to in-vitro cross-reactivities of Flaviviruses-specific antibodies. We evaluated Zika diagnosis in symptomatic patients using serial RT-PCR and develop a classification model using serial Dengue virus (DENV) and ZIKV serologies.
Methods.
A prospective longitudinal multicentric study in Southern Mexico () enrolled symptomatic and non-symptomatic participants. In the classification model, true positives were symptomatic (using a modified World Health Organization /Pan American Health Organization definition) with RT-PCR positive for ZIKV or DENV. True negatives were non-symptomatic with negative RT-PCR. Serial serology measurements were used to predict disease status.
Results.
Analyzing ZIKV and DENV RT-PCR at 3 timepoints between days 3 and 13 of symptom onset detected 25% more cases than a single PCR analysis between day 0 and 6. When considering sensitivity and specificity together, the serial serology model predicted all categories of disease and negatives better than manufactures cutoffs. Their cutoffs optimized sensitivity or specificity but not both.
Conclusions.
We demonstrated the importance of serial RT-PCR and antibody measurements to diagnose arbovirus infection in symptomatic patients living in regions with co-circulating flaviviruses.
Keywords: Zika Virus, Dengue Virus, Reverse Transcriptase Polymerase Chain Reaction, Antibodies, Kinetics, Viremia
1. Background
Zika virus (ZIKV) was declared a public health emergency by the World Health Organization (WHO) on February 2016, in response to the large spread of the virus in the Americas, the increasing evidence of congenital malformations in neonates from mothers that were infected with ZIKV during pregnancy, and because of neurological disorders found in infected adults [1–3]. ZIKV, along with the Dengue virus (DENV), belongs to the genus Flavivirus of the Flaviviridae family. Both are arthropod-borne viruses (arboviruses), possessing a single-stranded, positive-sense Ribonucleic acid (RNA) genome of approximately 11kb that codes for a polyprotein containing 3 structural proteins, among which is the envelope protein (E) [4]. The primary epitopes targeted by human antibodies during a flavivirus infection are found in the E protein, which is structurally conserved among flaviviruses. Immune mediated enhancement with closely related flavivirus, through the antibodies produced during a primary infection, remains controversial [5–8]. Those antigenic similarities have microbiological consequences [9,10], such as the in-vitro cross-reaction of serologic tests against close flaviviruses [11].
ZIKV and DENV transmission to humans occurs either through the bites of infected mosquitoes (Aedes aegypti and Aedes albopictus [12]) or, for ZIKV, through sexual, mother-to-fetus or blood transfusion transmissions [13]. Most arboviral infections are non-symptomatic or only cause influenza-like illness [13]. DENV and ZIKV infections display the same phenotype during minor forms, characterized by moderate fever, arthralgia, myalgia, leading to possible misdiagnosis in areas with co-circulation of the two viruses. During the 2015–2017 ZIKV epidemic in the Americas, DENV and ZIKV were both prevalent in Mexico with 130,069 DENV cases [14] and 7,320 ZIKV cases [15] reported in 2016 by the Pan American Health Organization (PAHO).
The Centers for Disease Control and Prevention (CDC) defines ZIKV infection based on viremia in blood and urine in non-pregnant symptomatic individuals with symptom onset within 14 days who have a possible exposure to ZIKV [16]. If onset of symptoms is after 14 days or if viremia is negative, serologic studies of specific Immunoglobulin M (IgM) and Immunoglobulin G (IgG) are recommended. Based on this testing algorithm, many symptomatic patients are left without any confirmed diagnosis. Indeed, the peak of plasma ZIKV viremia has been reported to be lower and of shorter duration than other flaviviruses, raising issues about ability of RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) to diagnose all cases and thus avoid false negatives [17]. Because of the high degree of cross-reactivity within specific anti-flaviviruses antibodies, establishing a definitive diagnosis is challenging, especially in an area with co-circulation of different flaviviruses.
In this study, we used a cohort study performed in Tapachula City, State of Chiapas, Mexico, which includes true cases of “Definitive Zika”, “Definitive Dengue”, “Household” (healthy relative controls) and symptomatic patients with negative real-time RT-PCR tests. The primary goal of this paper was to evaluate the potential benefit of serial viremia (serum and urine) to diagnose ZIKV and DENV infection in symptomatic patients in areas with virus co-circulation. We also evaluated the utility of combining serial serologic evaluations and the current serologic assay cutpoints through a tree model.
2. Methods
2.1. Study population and study design
A prospective observational longitudinal multicentric study in the Tapachula City area, State of Chiapas Mexico was conducted by the Mexican Emerging Infectious Disease Clinical Research Network (LaRED). Patients were enrolled from 5 clinical care centers: Clínica Hospital Dr. Roberto Nettel Flores, Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado; Hospital Regional de Alta Especialidad de Tapachula Ciudad Salud, Hospital General de Tapachula, and Unidad de Medicina Familiar No.11, Instituto Mexicano del Seguro Social. The first cohort included consenting adults with febrile rash, who met the modified WHO/PAHO probable Zika case definition [1] and consulted within 7 days after symptom onset, referred to as “symptomatic cohort”. In brief, suspected ZIKV patients were defined as patients with one or more of the following symptoms (not explained by other medical conditions): rash, elevated body temperature (> 37.2 °C), arthralgia, myalgia, non-purulent conjunctivitis or conjunctival hyperemia, headache, malaise. The second cohort enrolled household members of those participants in the symptomatic cohort, but who did not themselves display any symptoms that matched the WHO/PAHO criteria (referred to as the “household cohort”). If they had symptoms at screening or during follow-up, they were excluded from the household cohort and could be evaluated for inclusion in the symptomatic cohort. The study didn’t have any exclusion criteria.
2.2. Ethical considerations and regulatory approval
The study protocol was approved by an Institution Review Board in two participating institutions (“Comité de Ética en Investigación del Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán” - Registration number: CONBIOETICA −09CEI-011–20160627 and “Comité de Investigación del Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán” - Registration number: 18 CI 09 012 005). The study was recorded on Clinical.Trials.gov (). It was supported by the Mexico Ministry of Health and US National Institute of Allergy and Infectious Diseases (NIAID) and sponsored by the Division of Clinical Research of NIAID. Signed informed consent was obtained from all subjects or from a legal representative if the subject was not able to sign the consent form.
2.3. Study planning and clinical evaluation
All participants enrolled in the symptomatic cohort were evaluated on Study-visit (SV) days 0, 3, 7 and 28. During each SV, real-time RT-PCR was run on serum and urine samples. Serologies (IgM and IgG) for ZIKV, DENV and Chikungunya Virus (CHIKV)were also run on serum samples at each SV. Plaque Reduction Neutralization Test (PRNT) was not performed on the positive serologic samples. At SV0 and 28, cell blood count and other blood chemistries were also performed. The baseline clinical evaluation collected data on demographic characteristics, medical history and exposure history. All SV collected: extensive clinical data, symptoms from a checklist, potential pregnancy, neurocognitive and hearing function assessment, disability using WHODAS 12 (WHO Disability Assessment Schedule), any complications, and assessment of Guillain-Barre syndrome, with dedicated tests if present [18]. All subjects enrolled in the household cohort were evaluated on SV0 and 28, with the same evaluations performed as for the symptomatic cohort.
2.4. Laboratory procedures
Serologies of the three arboviruses were measured using commercial Enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s instructions: anti-ZIKV specific-IgM (ZIKV-sIgM) and IgG (ZIKV-sIgG) ELISA (EuroImmun®, Lübeck, Germany), anti-CHIKV specific-IgM (CHIKV-sIgM) and IgG (CHIKV-sIgG) ELISA (EuroImmun®, Lübeck, Germany), anti-DENV specific-IgG (Panbio Dengue IgG Capture ELISA - panbio diagnostics®, Republic of Korea) and anti-DENV specific-IgG (Panbio Dengue IgM Capture ELISA - panbio diagnostics®, Republic of Korea). Four real-time RT-PCR assays were performed on the same sample, evaluating ZIKV [19], DENV [20], CHIKV [21] and panflavivirus [22], as previously described. Briefly, for RNA extraction, total nucleic acids from 500 μl of serum and urine were extracted using the NucliSENS® easyMAG® system (bioMerieux®, Netherlands) and eluted in 55 μl, according to manufacturer instructions. For amplification, the oligonucleotide primers and dual labeled 5’-fluorescent probes used in the assay are listed in the Supplemental Table 1 [19–22]. The amplification of the human RNaseP (RP) gene was carried out for each sample as an internal control to demonstrate the presence of RNA and the validation of the extraction process. The amplification of the NS5 gene was also carried out for the generic detection of Flavivirus as another control of ZIKV and DENV and to determine the possible presence of other flaviviruses in the sample. Amplifications were performed in singleplex (each virus detected in a separate reaction) by one-step RT-PCR reaction in 25 μl with SuperScript III Platinum One-Step quantitative RT-PCR System (Invitrogen®, ThermoFisher Scientific®, Waltham, MA, USA) and 5 μl of sample. Cycle sequencing was: retrotranscription at 50°C for 30 minutes, initial PCR denaturation at 94°C for 2 minutes followed by 45 cycles of denaturation at 94°C for 15 seconds and annealing and extension at 60°C for 1 minute in the Applied Biosystems 7500 Fast Real Time PCR System (Applied Biosystems, ThermoFisher Scientific®, Waltham, MA, USA).
2.5. Cases definitions
In adherence with the CDC guidance for ZIKV infection [16], patients had their first sample within 7 days after symptoms onset (SV0). Real-time RT-PCR was performed on urine and serum at SV0, 3, and 7. As in previous studies [23], patients were classified either as “Definitive Zika”, “Definitive Dengue” or “Definitive Chikungunya” when real-time RT-PCR was positive for one of these viruses at any of the three SV samples. In this study the maximum possible delay for real-time RT-PCR testing was 13 days (i.e. testing at SV7 with a symptom onset of 6 days at SV0). The study team considered this delay to be acceptable because of the known persistence of ZIKV and DENV viremia [24]. When real-time RT-PCR was negative for all three arboviruses, patients of the symptomatic cohort were considered as “undefined fever events” (UFE) and patients of the household cohort as true negative. CHIKV infections were included in the original protocol because of its large prevalence in 2015. However, because the number of cases in this study was below 10, we did not include them in this analysis.
2.6. Statistical analyses
Numerous classification methods were considered as candidates to model the relationship between serial serology and disease category. We chose to work with a decision tree paradigm due to its relative simplicity of interpretation and its ability to easily handle a response variable with more than two categories. We used the R statistical software [25] with the rpart package [26] to build a classification tree model using serial serology measurements to predict disease status. The classification tree procedure picks important variables and cutoffs to use to predict disease status. We examined the following variables in the classification tree procedure: ZIKV-sIgG, ZIKV-sIgM, DENV-sIgG, and DENV-sIgM at SV0, SV28, and change in those variables from SV0 to SV28. To maintain interpretability and parsimony, we constrained the classification tree to not exceed a depth of 4 (maximum number of decision criteria that can be used to assign categories) and required a minimum node size of 5 subjects (at least 5 subjects must be assigned to a category based on each decision criterion). After the classification tree procedure chose the best model given the above constraints, we examined the quality of the model by performing 10-fold stratified cross-validation and calculating out-of-fold error for each of the three groups. Calculating out-of-fold error provided an estimate of the performance of the model as if we had had a new independent set of data. The model performance was evaluated using sensitivity and specificity. After fitting the model and obtaining measures of model quality, we wished to compare the performance of the classification tree model to the performance of the cutoffs recommended by the manufacturers of the commercial serology kits. We applied the recommended cutoffs to the group of known subjects and observed the resulting sensitivity and specificity. Finally, we took the classification tree model result from the fit on the known subjects and applied the prediction model and the manufacturers recommended cutoffs to the UFE subjects.
3. Results
3.1. Patients’ inclusion and clinical presentation
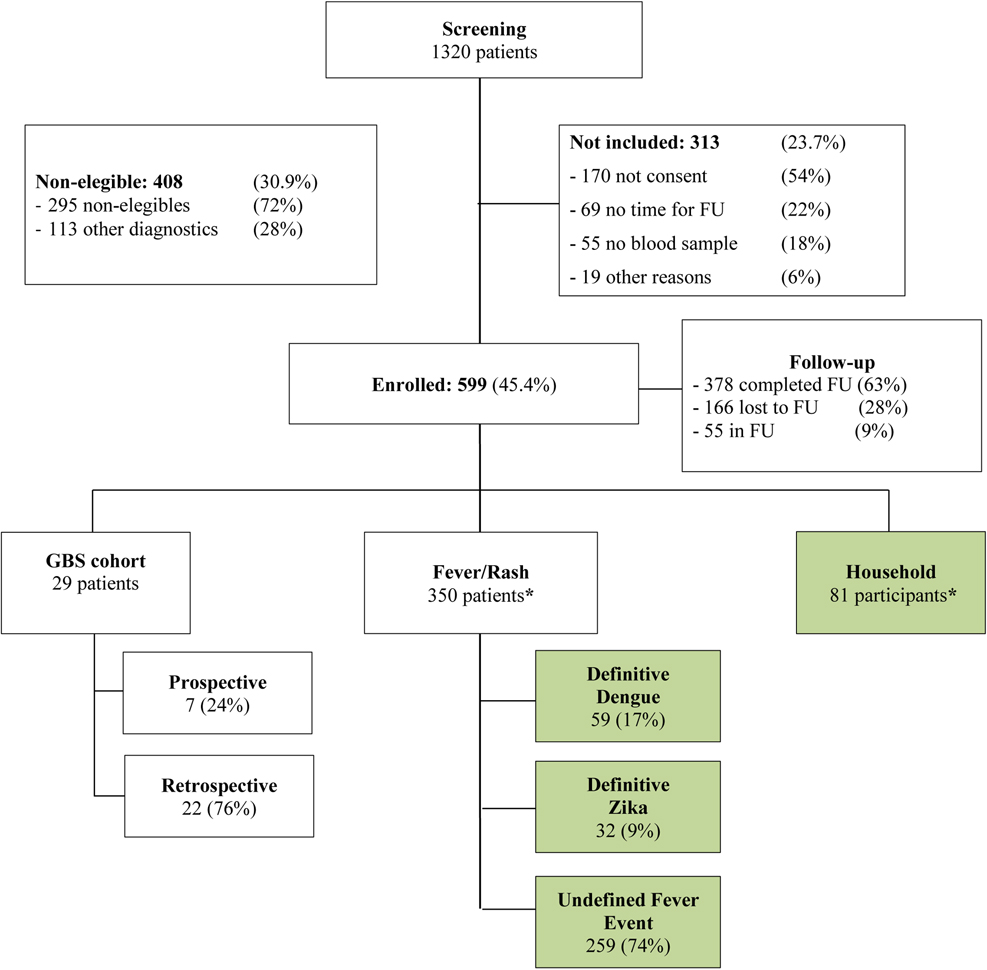
We prospectively enrolled 467 symptomatic participants between June 20, 2016 and August 31, 2018. Among those, 350 symptomatic participants were included in this analysis (see Flowchart in Figure 1) and 81 Household participants. The median age at enrollment was 32 years old (Interquartile Range (IQR) 23.5) and 61% of them were women (Table 1). Patients’ inclusion criteria symptoms at SV0 are presented in Table 1. Among the symptomatic patients, 32 (9%) had positive ZIKV real-time RT-PCR and 59 (17%) positive DENV real-time RT-PCR on SV0, 3 or 7, with 259 (74%) classified as UFE. Among the 81 households’ subjects enrolled, all their indexrelated cases were classified among the UFE participants. They did not present with any symptoms.
Figure 1.
Consort diagram. Green boxes represent groups included in our analysis.
* : only included those subjects who have no missing serology data on both SV0 and 28, and at most one missing PCR measurement.
FU = Follow-Up. GBS = Guillain Barre Syndrom.
Table 1.
Table of Demographics for patients in this analysis
| Variable | ZIKV Patients (n=32) | DENV Patients (n=59) | Household Patients (n=81) | Undefined fever events (n=259) |
|---|---|---|---|---|
| Median age (IQR) | 33.5 (12.2) | 26 (22) | 42 (27) | 31 (22.5) |
| Gender female, n (%) | 19 (59%) | 34 (58%) | 45 (56%) | 165 (64%) |
| Days after symptoms onset at enrolment | ||||
| 0 day | 1 (3%) | 0 (0%) | N/A | 2 (1%) |
| 1 days | 2 (6%) | 2 (3%) | N/A | 30 (12%) |
| 2 days | 10 (31%) | 5 (8%) | N/A | 47 (18%) |
| 3 days | 8 (25%) | 8 (14%) | N/A | 46 (18%) |
| 4 days | 5 (16%) | 10 (17%) | N/A | 48 (19%) |
| 5 days | 4 (13%) | 12 (20%) | N/A | 41 (16%) |
| 6 days | 2 (6%) | 12 (20%) | N/A | 37 (14%) |
| 7 days | 0 (0%) | 10 (17%) | N/A | 8 (3%) |
| Clinical symptoms, n (%) | ||||
| Fever | 22 (69%) | 57 (97%) | 0 (0%) | 228 (88%) |
| Non-purulent conjunctivitis or conjunctival hyperemia | 26 (81%) | 29 (49%) | 0 (0%) | 149 (58%) |
| Rash | 24 (75%) | 51 (86%) | 8 (10%) | 127 (49%) |
| Pruritus | 25 (78%) | 50 (85%) | 19 (24%) | 150 (58%) |
| Arthralgia | 26 (81%) | 54 (92%) | 21 (26%) | 243 (94%) |
| Myalgia | 32 (100%) | 56 (94%) | 35 (43%) | 248 (96%) |
| Headache | 28 (88%) | 59 (100%) | 49 (61%) | 250 (97%) |
| Diarrhea | 12 (38%) | 36 (61%) | 16 (20%) | 127 (49%) |
| Malaise | 31 (97%) | 59 (100%) | 81 (100%) | 258 (99%) |
| Eye pain | 25 (78%) | 45 (76%) | 0 (0%) | 205 (79%) |
| Median number of symptoms (IQR) | 8 (2) | 9 (1) | 2 (2) | 8 (2) |
3.2. RT-PCR and serologies measurements
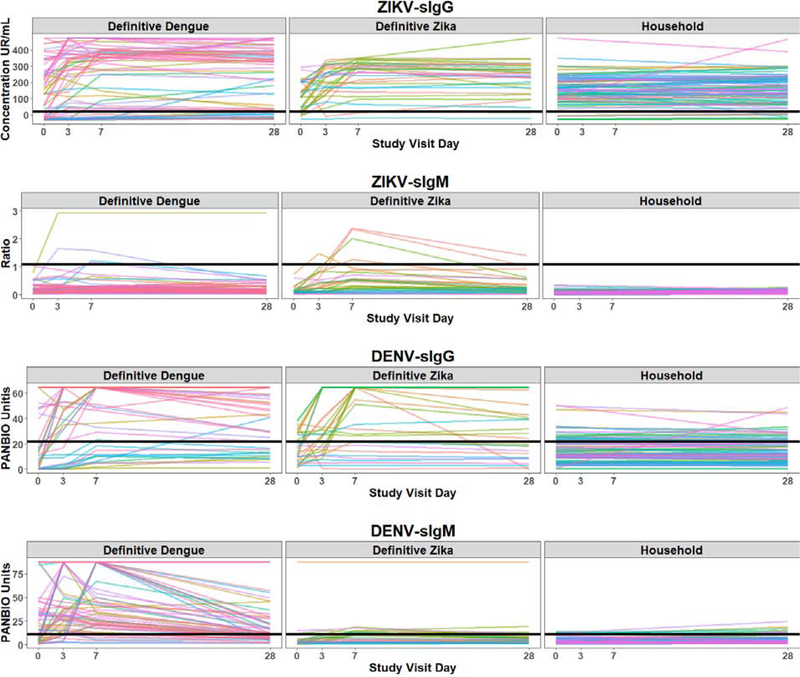
Among the 32 definitive Zika patients, 8 (25%) had their first viremia or viruria detected on SV3 or 7, corresponding to a delay after symptoms onset ranging from 3 to 13 days. As illustrated in Figure 2, 90% of the households’ subjects had a ZIKV-sIgG above the recommended cutoffs at SV0 and 28, with a flat slope. For definitive Zika patients, 78% had an increase from SV0 to SV28 in ZIKV-sIgG and 66% in DENV-sIgG, with a positive slope, underlying the in-vitro sIgG cross-reactivities (same for definitive Dengue patients and evolution of DENV-sIgG and ZIKV-sIgG). When analysing the performance of the manufacturer recommended IgG cutoffs, there was a tradeoff between sensitivity and specificity with only of the two measures falling in the acceptable range (for example above 80%) (Table 2).
Figure 2.
Spaghetti plots for ZIKV and DENV sIgG and sIgM among the three cohorts: ZIKV-sIgG (row 1), ZIKV-sIgM (row 2), DENV-sIgG (row 3) and DENV-sIgM (row 4) by study visit for each cohort: Definitive Dengue, Definitive Zika and Household. Each colored line represents the trajectory of one subject in the study. The horizontal black line represents the manufacturer-recommended cutoff for defining seropositivity.
Table 2.
Sensitivity and Specificity of decision criteria. These are the results of applying both our decision tree model and the manufacturer recommended cutoffs to those subjects with known disease status.
| Zika PCR+ | Dengue PCR+ | Household | ||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Performance of Tree Model1 | 13/32 = 41% |
137/140 = 98% |
50/59 = 85% |
103/113 = 91% |
75/81 = 93% |
70/91 = 77% |
|
Manufacturer Cutoffs: IgM Day 0 |
0/32 = 0% |
140/140 = 100% |
36/59 = 61% |
104/113 = 92% |
75/81 = 93% |
39/91 = 43% |
|
Manufacturer Cutoffs: IgM Day 28 |
1/32 = 3% |
139/140 = 99% |
33/59 = 56% |
99/113 = 88% |
72/81 = 89% |
39/91 = 43% |
|
Manufacturer Cutoffs: IgG Day 0 |
30/32 = 94% |
20/140 = 14% |
33/59 = 56% |
80/113 = 71% |
7/81 = 9% |
69/91 = 76% |
|
Manufacturer Cutoffs: IgG Day 28 |
30/32 = 94% |
16/100 = 16% |
46/59 = 78% |
73/113 = 65% |
8/81 = 10% |
81/91 = 89% |
Calculated on out-of-fold predictions with 10-fold cross-validation
3.3. Statistical decision tree model
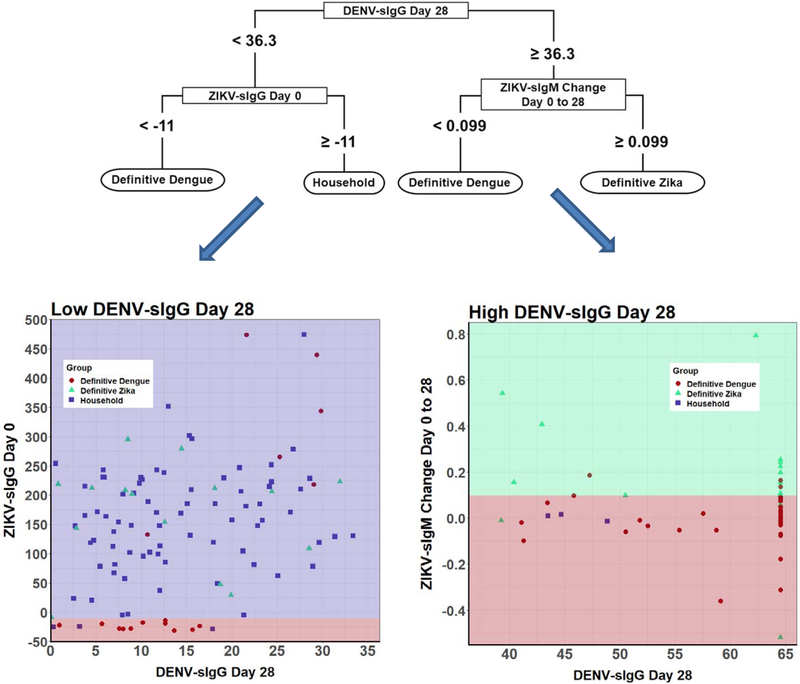
The variables that were important in predicting infection were: DENV-sIgG at SV28, change in ZIKV IgM, and ZIKV IgG at SV0 (Figure 3). The variables picked by the model do not per se reflect the ones with the most biological/clinical sense, but the ones with the best predictive capacity. The classification model picked the cutoffs that determine whether a serological measurement at a specific timepoint or a change in measurements between timepoints is “small” or “large”. These cutoffs do not discriminate perfectly as seen by the sensitivity and specificity values less than 100%. When looking at the sensitivity and specificity in unison, the classification model performed better for all three groups. (Table 2). The manufactures cutoffs performed well on one measure (either sensitivity or specificity) at the expense of the other. In addition, when the decision criteria are applied to the UFE population, the manufacturer cutoffs would classify a large number of subjects as both ZIKV and DENV, which is unlikely to be an accurate assertion given our knowledge of the properties of flavivirus infection (Table 3).
Figure 3:
Decision Tree results. In the flow diagram each level shows the decision criteria and variable to use to predict the disease group. The colored figures show the criteria diagrammatically. The left figure shows the cut points for the initial cutpoint of low DENV-sIgG SV 28. The purple area shows prediction of household patients with ZIKV-sIgG high and the red area shows the prediction of DENV infected patients. The dots show the true disease status of the people who would fall in the purple and red areas. Red dots are true DENV infected patients, green triangles are true ZIKV infected patients and purple squares are Household subjects. The figure on the right is for the initial cutpoint of high DENV-sIgG SV 28. The green area shows prediction of ZIKV infected patients with ZIKV-sIgM change high and the red area shows the prediction of DENV infected patients.
Table 3.
Apply decision criteria to undefined fever events patients as an example. These are the results of applying both our decision tree model and the manufacturer recommended cutoffs to those subjects with unknown disease status.
| %ZIKV alone | %DENV alone | %(ZIKV and DENV) | %Undefined fever events | |
|---|---|---|---|---|
| Tree Model | 4/259 = 2% |
43/259 = 17% |
0/259 = 0% |
212/259 = 82% |
|
Manufacturer Cutoffs: IgM Day 0 |
1/259 = <1% |
25/259 = 10% |
0/259 = 0% |
233/259 = 90% |
|
Manufacturer Cutoffs: IgM Day 28 |
0/259 = 0% |
27/259 = 10% |
1/259 = <1% |
231/259 = 89% |
|
Manufacturer Cutoffs: IgG Day 0 |
142/259 = 55% |
5/259 = 2% |
79/259 = 31% |
33/259 = 13% |
|
Manufacturer Cutoffs: IgG Day 28 |
149/259 = 58% |
5/259 = 2% |
68/259 = 27% |
37/259 = 14% |
4. Discussion
Establishing a final arbovirus diagnosis in a co-circulation endemic area is challenging. Currently, recommendations for ZIKV and DENV diagnosis are based on virus detection and/or specific-IgM detection according to the delay within symptoms onset. To the best of our knowledge, most assay recommendations assign a cut-point using a single evaluation with the exception of WHO interim guidance suggesting analysis of paired serum specimens collected 2–3 weeks apart [27]. So far, no benefit of serial measurements has been reported in the literature. In this study, we established the value of serial real-time RT-PCR measurements of ZIKV and DENV during the symptomatic phase of the flavivirus infection to help physicians provide a diagnosis. Our tree model demonstrates the importance of looking at the change in DENV/ZIKV-sIgM and DENV/ZIKV-sIgG to distinguish ZIKV infected patients from DENV infected patients. Our model shows that due to the cross reactivity of flaviviruses, a single threshold for a single antibody cannot adequately diagnose ZIKA or DENV infection in co-circulating areas, especially if the cutoffs recommended were validated in a traveler population.
Clinical distinction of ZIKV suspected cases in arboviruses co-circulating areas is particularly difficult because symptoms are not pathognomonic, with failure to develop a clinical score with both high sensitivity and specificity in this context [23]. Therefore, confirmation of ZIKV infection requires a biological evaluation, by identification either of the virus or of specific antibodies against ZIKV. In our study, among the 350 symptomatic subjects, 32 were diagnosed “definitive Zika”, 59 “definitive Dengue”, leaving 74% of patients with unknown diagnosis, underlying the importance of diagnosis improvement. Limitations of ZIKV diagnosis tools are well reported. For viremia, in an external quality assessment among European labs in 2016 [28] (i.e. countries testing predominantly travelers returning from tropical regions) and in Brazil in 2017 [29] (i.e. resource-limited settings with multiple co-circulating endemic arboviruses), 40% and 27% were respectively correctly able to perform accurate ZIKV molecular diagnostics on all provided samples. In our study, 24% of the patients would not have been identified neither with a single real-time RT-PCR evaluation at SV0 nor with evaluation of ZIKV-sIgM that was largely below the recommended threshold. Interestingly, the delay between consultation and symptoms onset of these patients ranged from 0 to 7 days, thus not explaining that lack of diagnosis. The peak of ZIKV viremia has been reported to be lower and of shorter duration compared to others flaviviruses. But based on our results, a second later evaluation of viremia would improve ZIKV and DENV infection diagnosis and should be considered (Supplemental Table 2). Neither blood nor urine testing alone were able to define the whole set of ZIKV and DENV positive patients, no matter which time point and site were considered. A single evaluation at SV0 would have missed 25% and 20% of the ZIKV and DENV infected patients respectively [30].
The second step in the recommended CDC algorithm is based on sIgM evaluation. One major issue of serological evaluation of flaviviruses is the antibodies cross-reactivity [6,31]. To be able to assess antibodies specificity, PRNT or other techniques have been suggested. However, they are either expensive or time consuming, with almost a week being required before interpretation of the results; thus they are not routinely performed. To the best of our knowledge, most studies evaluating the cross-reactivity within DENV and ZIKV antibodies with the EuroImmun® serologies kits were performed among infected travelers returning from an endemic area [32,33]. Even if the manufacturer reports a low cross-reactivity within DENV and ZIKV antibodies, patients living in an endemic area with high DENV prevalence are distinct from naïve flavivirus European patients. Indeed, ZIKV patients have been reported to have two serologic profiles during the active phase of infection according to their origin. Whereas most travelers display high IgM ratio values and moderate IgG ratios, the endemic-area residents had infections with very high IgG ratios with negative or low IgM ratios [32]. Plus, sensitivity of the ZIKV-sIgM EuroImmun® kit has been reported to be as low as 27% in an endemic population [34]. We could thus assume that both the recommended cutoff and the sensitivity/specificity reported in a non-endemic population would differ in an endemic area population. Because of the lack of sensitivity and specificity, other studies have evaluated a combination of different assays to improve ZIKV diagnosis, without answering the question of DENV misdiagnosis [35–37].
Establishing the exact onset of disease is not easy in an endemic area considering the non-specificity of the symptoms. That date is always established retrospectively and may thus not be accurate. Precise timing of specific IgM onset has not been fully established yet, but based on related flaviviruses knowledge, it is assumed that ZIKV-sIgM appear 4–5 days after symptoms onset and last up to 12 weeks [38]. However, that assumption is made on a flavivirus-naïve patient, and both the value and the kinetics may be different as previously mentioned (60% of the Mexican population have already been exposed to DENV [39,40]).
Based on the antibody-dependent enhancement hypothesis [6,41], after a previous DENV infection in an individual, a ZIKV infection may be considered by the host as a second infection and lead to an early elevated ZIKV-sIgG response with a low or absent ZIKV-sIgM. In our study, based on the recommended cutoff for IgM and IgG, results are in agreement with that assumption with no more than 20% of the ZIKV infected patients with positive ZIKV-sIgM (whatever timepoint considered) and 97% of all the cohort (infected and household patients) with positive ZIKV-sIgG. Determining different cutoffs and a ZIKV diagnosis algorithm according to the origin area of the population studied is thus essential to help physicians to target the population at risk. Flaviviruses cross-reactivity influences also the results of DENV tests, with the Panbio® DENV-sIgG and DENV-sIgM kits [9]. Thus, the presence of both flaviviruses serologies in our tree model reflects the complexity of the in-vitro testing in our population.
Our study has several limitations. First, because 70–80% of ZIKV infected patients are supposed to be asymptomatic [42] and because of the low sensitivity of the real-time RT-PCR, the true negativity of the household cohort could be argued. However, panflavivirus real-time RT-PCR of the households were negative, their ZIKV-sIgG slope was flat and all the index case patients associated with each household member were classified among the UFE patients. Some of those patients from the household cohort had minor symptoms, such as previously reported among 19% of the non-infected household members of a ZIKV-infected case patients during the Yap Island outbreak [42]. Thus, we believe that this population is a suitable matched negative population because they shared the same endemic origin. Second, our tree strategy won’t be suitable neither for patients presenting with arbovirus co-infections, as previously reported with DENV and CHIKV [43–45] or with DENV and ZIKV [46–48], nor across different populations because of the different background flavivirus seropositivity. However, the relevance of both DENV and ZIKV serial serology testing is of particular interest for (potential) pregnant women. The tree also emphasizes the complexity of a final diagnosis because of the antibodies’ cross-reactivities. Third, although days since symptom onset is an important variable to consider, we did not adjust for it in the tree model, because this data was non-existent for the household cohort. Because of the known rise and fall of the serologic curves, it is likely that the part of the reason for misclassifications is due to people seeking care at different times. But since in practice people will seek care at different times, this model reflects clinical practice. Finally, we do not claim that the cutpoints found in this analysis will work for all kits or even are the best cutpoints for these kits.
5. Conclusion
Establishing a final flaviviruses diagnosis is challenging considering the pitfalls of the existing tools. Based on our study, a significant proportion of patients could benefit from a serial ZIKV viremia and viruria evaluation because of the low sensitivity of that test. Considering that some of the serologic tests were validated in traveler cohorts, analyzing serologic kinetics could help to target population at higher risk for ZIKV infection, especially for pregnant women or for men having sex with pregnant women.
Supplementary Material
Highlights.
Serial RT-PCR evaluation until day 13 increases by 25% the identification of patients infected by Zika Virus.
Cutoffs for Zika and Dengue virus serologic tests validated in traveler cohorts do not apply on endemic population.
Zika virus serologic kinetics could help to target population that should benefit from a closer monitoring.
Acknowledgments
Dr. Francisco Camas-Durán, Dr. Zoila Priego-Smith (Clínica Hospital Dr. Roberto Nettel Flores, Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado); Dr. Karla Reyna Navarro Fuentes (Unidad de Medicina Familiar No.11, Instituto Mexicano del Seguro Social); Dr. Paola Alcalá, Dr. Alfredo Vera-Maloof, Dr. Luis Rojas, Dr. Cielo Mayorga, Dr.Benjamin Hidalgo, NP Erika Estudillo(LaRED) participated in data collection. Alexander López (Hospital Regional de Alta Especialidad Ciudad Salud), Nora Mora Suárez, Mónica Reyes Romero, Julia Marinez López, Gustavo Pedraza (Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, INCMNSZ) participated in sample collection, management and laboratory procedures. Víctor Hugo López (LaRED), Gustavo Rosales, Hugo Arroyo, and Dr. Lourdes Guerrero (INCMNSZ) participated in implementation of laboratory process and data quality management and control.
Funding
Funded through the Mexico Emerging Infectious Diseases Clinical Research Network (La Red). La Red is collaboration between the Mexico Ministry of Health and the U.S. National Institute of Allergy and Infectious Diseases. This study has been funded in part by CONACYT (Fondo Sectorial SSA/IMSS/ISSSTE, Projects No. 71260 and No. 127088). This project was funded in whole or in part with federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- CDC
Centers for Disease Control and Prevention
- CHIKV
Chikungunya Virus
- DENV
Dengue Virus
- ELISA
Enzyme-linked immunosorbent assay
- IgM
Immunoglobulin M
- IgG
Immunoglobulin G
- IQR
Interquartile Range
- PAHO
National Institute of Allergy and Infectious Diseases (NIAID Pan American Health Organization
- PRNT
Plaque Reduction Neutralization Test
- RT-PCR
Reverse Transcriptase Polymerase Chain Reaction
- RNA
Ribonucleic acid
- SV
Study-visit
- UFE
Undefined Fever Events
- WHODAS
WHO Disability Assessment Schedule
- WHO
World Health Organization
- ZIKV
Zika Virus
Footnotes
Conflicts of interest:
Pablo F Belaunzarán-Zamudio has received non-financial support as consultant for Sanofi-Pasteur and funding for projects not related to this work. The other authors, Aurelie Gouel-Cheron, Keith Lumbard, Sally Hunsberger, Fernando J. Arteaga-Cabello, John Beigel, Pablo F. Belaunzarán-Zamudio, Sandra Caballero-Sosa, Kenia Escobedo-López, Violeta Ibarra-González, Gabriel Nájera-Cancino, Héctor Rincón-León, Emilia Ruiz-Hernández, Karina Trujillo-Murillo, have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6 References
- 1.PAHO/WHO, 1 December 2015. Epidemiological Alert Neurological syndrome, congenital malformations, and Zika virus infection. Implications for public health in the Americas. (http://www.paho.org/hq/index.php?option=com_docman&task=doc_download&Itemid=&gid=32405&lang=en). 2015;. [Google Scholar]
- 2.Cao-Lormeau V-M, Blake A, Mons S, et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016; 387:1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects--Reviewing the Evidence for Causality. N Engl J Med. 2016; 374:1981–7. [DOI] [PubMed] [Google Scholar]
- 4.Hasan SS, Sevvana M, Kuhn RJ, Rossmann MG. Structural biology of Zika virus and other flaviviruses. Nat Struct Mol Biol. 2018; 25:13–20. [DOI] [PubMed] [Google Scholar]
- 5.Beltramello M, Williams KL, Simmons CP, et al. The Human Immune Response to Dengue Virus Is Dominated by Highly Cross-Reactive Antibodies Endowed with Neutralizing and Enhancing Activity. Cell Host Microbe. 2010; 8:271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dejnirattisai W, Supasa P, Wongwiwat W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol. 2016; 17:1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castanha PMS, Nascimento EJM, Braga C, et al. Dengue Virus–Specific Antibodies Enhance Brazilian Zika Virus Infection. J Infect Dis. 2017; 215:781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardina SV, Bunduc P, Tripathi S, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017; 356:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felix AC, Souza NCS, Figueiredo WM, et al. Cross reactivity of commercial anti-dengue immunoassays in patients with acute Zika virus infection. J Med Virol. 2017; 89(8):1477–1479. [DOI] [PubMed] [Google Scholar]
- 10.L’Huillier AG, Hamid-Allie A, Kristjanson E, et al. Evaluation of Euroimmun Anti-Zika virus ELISAs (IgM & IgG) for Zika virus serologic testing. J Clin Microbiol. 2017; 55(8):2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langerak T, Mumtaz N, Tolk VI, et al. The possible role of cross-reactive dengue virus antibodies in Zika virus pathogenesis. PLoS Pathog. 2019; 15(4):e1007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasilakis N, Weaver SC. Flavivirus transmission focusing on Zika. Curr Opin Virol. 2017; 22:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musso D, Gubler DJ. Zika Virus. Clin Microbiol Rev. 2016; 29:487–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PAHO. Number of Reported Cases of Dengue and Severe Dengue (SD) in the Americas, by Country. Figures for 2016 (to week noted by each country) (http://www.paho.org/hq/index.php?option=com_docman&task=doc_download&Itemid=270&gid=37782&lang=en). 2016;. [Google Scholar]
- 15.PAHO. Zika Cumulative Cases - 29 December 2016 (http://www.paho.org/hq/index.php?option=com_docman&task=doc_view&Itemid=270&gid=37582&lang=en). 2016;. [Google Scholar]
- 16.CDC. Guidance for US Laboratories Testing for Zika Virus Infection. July 24, 2017. (https://www.cdc.gov/zika/laboratories/lab-guidance.html). 2017;. [Google Scholar]
- 17.Paz-Bailey G, Rosenberg ES, Doyle K, et al. Persistence of Zika Virus in Body Fluids — Final Report. N Engl J Med. 2018; 379(13):1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuki N, Hartung H-P. Guillain–Barré Syndrome. N Engl J Med. 2012; 366:2294–2304. [DOI] [PubMed] [Google Scholar]
- 19.Lanciotti RS, Kosoy OL, Laven JJ, et al. Genetic and Serologic Properties of Zika Virus Associated with an Epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008; 14:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waggoner JJ, Abeynayake J, Sahoo MK, et al. Development of an Internally Controlled Real-Time Reverse Transcriptase PCR Assay for Pan-Dengue Virus Detection and Comparison of Four Molecular Dengue Virus Detection Assays. J Clin Microbiol. 2013; 51:2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carletti F, Bordi L, Chiappini R, et al. Rapid detection and quantification of Chikungunya virus by a one-step reverse transcription polymerase chain reaction real-time assay. Am J Trop Med Hyg. 2007; 77:521–4. [PubMed] [Google Scholar]
- 22.Patel P, Landt O, Kaiser M, et al. Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol J. 2013; 10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braga JU, Bressan C, Dalvi APR, et al. Accuracy of Zika virus disease case definition during simultaneous Dengue and Chikungunya epidemics. PLoS One. 2017; 12:e0179725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lessler J, Ott CT, Carcelen AC, et al. Times to key events in Zika virus infection and implications for blood donation: a systematic review. Bull World Health Organ. 2016; 94:841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Team. R: a language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2017. Available from: https://www.R-project.org/ [Google Scholar]
- 26.Therneau T, Atkinson B. rpart: Recursive Partitioning and Regression Trees. R package version 4.1–13. [Internet]. 2018. Available from: https://CRAN.R-project.org/package=rpart [Google Scholar]
- 27.WHO. Laboratory testing for Zika virus infection. Interim guidance. 23 March 2016. (http://apps.who.int/iris/bitstream/handle/10665/204671/WHO_ZIKV_LAB_16.1_eng.pdf?sequence=1). 2016;. [Google Scholar]
- 28.Charrel R, Mögling R, Pas S, et al. Variable Sensitivity in Molecular Detection of Zika Virus in European Expert Laboratories: External Quality Assessment, November 2016. J Clin Microbiol. 2017; 55:3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer C, Pedroso C, Mendrone A, et al. External Quality Assessment for Zika Virus Molecular Diagnostic Testing, Brazil. Emerg Infect Dis. 2018; 24(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh RK, Dhama K, Karthik K, et al. Advances in Diagnosis, Surveillance, and Monitoring of Zika Virus: An Update. Front Microbiol. 2017; 8:2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stettler K, Beltramello M, Espinosa DA, et al. Specificity, cross-reactivity and function of antibodies elicited by Zika virus infection. Science. 2016; 353(6301):823–6. [DOI] [PubMed] [Google Scholar]
- 32.Steinhagen K, Probst C, Radzimski C, et al. Serodiagnosis of Zika virus (ZIKV) infections by a novel NS1-based ELISA devoid of cross-reactivity with dengue virus antibodies: a multicohort study of assay performance, 2015 to 2016. Euro Surveill. 2016; 21:30426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huzly D, Hanselmann I, Schmidt-Chanasit J, Panning M. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro Surveill. 2016; 21(16). [DOI] [PubMed] [Google Scholar]
- 34.EuroImmun. Medizinische Labordiagnostika G. Zika virus infections. EUROIMMUN test systems for the diagnosis of Zika virus infections (https://www.euroimmun.com/documents/Indications/Infections/Zika-virus/HI_2668_I_UK_B.pdf). 2018;. [Google Scholar]
- 35.Mendoza EJ, Makowski K, Barairo N, et al. Establishment of a comprehensive and high throughput serological algorithm for Zika virus diagnostic testing. Diagn Microbiol Infect Dis. 2019;. [DOI] [PubMed] [Google Scholar]
- 36.Michelson Y, Lustig Y, Avivi S, Schwartz E, Danielli A. Highly Sensitive and Specific Zika Virus Serological Assays Using a Magnetic Modulation Biosensing System. J Infect Dis. 2019; 219(7):1035–1043. [DOI] [PubMed] [Google Scholar]
- 37.De Ory F, Sánchez-Seco MP, Vázquez A, et al. Comparative Evaluation of Indirect Immunofluorescence and NS-1-Based ELISA to Determine Zika Virus-Specific IgM. Viruses. 2018; 10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika Virus. N Engl J Med. 2016; 374:1552–1563. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez Rodríguez D, Garza Rodríguez M, Chavarria AM, et al. Dengue virus antibodies in blood donors from an endemic area. Transfus Med. 2009; 19:125–131. [DOI] [PubMed] [Google Scholar]
- 40.Arellanos-Soto D, B. d l Cruz V, Mendoza-Tavera N, et al. Constant risk of dengue virus infection by blood transfusion in an endemic area in Mexico. Transfus Med. 2015; 25:122–4. [DOI] [PubMed] [Google Scholar]
- 41.Priyamvada L, Suthar MS, Ahmed R, Wrammert J. Humoral Immune Responses Against Zika Virus Infection and the Importance of Preexisting Flavivirus Immunity. J Infect Dis. 2017; 216:S906–S911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duffy MR, Chen T-H, Hancock WT, et al. Zika Virus Outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009; 360:2536–2543. [DOI] [PubMed] [Google Scholar]
- 43.Ruckert C, Weger-Lucarelli J, Garcia-Luna SM, et al. Impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nat Commun. 2017; 8:15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caron M, Paupy C, Grard G, et al. Recent Introduction and Rapid Dissemination of Chikungunya Virus and Dengue Virus Serotype 2 Associated With Human and Mosquito Coinfections in Gabon, Central Africa. Clin Infect Dis. 2012; 55:e45–e53. [DOI] [PubMed] [Google Scholar]
- 45.Furuya-Kanamori L, Liang S, Milinovich G, et al. Co-distribution and co-infection of chikungunya and dengue viruses. BMC Infect Dis. 2016; 16:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waggoner JJ, Gresh L, Vargas MJ, et al. Viremia and Clinical Presentation in Nicaraguan Patients Infected With Zika Virus, Chikungunya Virus, and Dengue Virus. Clin Infect Dis. 2016; 63:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villamil-Gómez WE, González-Camargo O, Rodriguez-Ayubi J, Zapata-Serpa D, Rodriguez-Morales AJ. Dengue, chikungunya and Zika co-infection in a patient from Colombia. J Infect Public Health. 2016; 9:684–686. [DOI] [PubMed] [Google Scholar]
- 48.Chia PY, Yew HS, Ho H, et al. Clinical features of patients with Zika and dengue virus co-infection in Singapore. J Infect. 2017; 74:611–615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.