Abstract
Connectivity modeling in functional neuroimaging has become widely used method of analysis for understanding functional architecture. One method for deriving directed connectivity models is Group Iterative Multiple Model Estimation (GIMME; Gates & Molenaar, 2012). GIMME looks for commonalities across the sample to detect signal from noise and arrive at edges that exist across the majority in the group (“group-level edges”) and individual-level edges. In this way, GIMME obtains generalizable results via the group-level edges while also allowing for between subject heterogeneity in connectivity, moving the field closer to obtaining reliable personalized connectivity maps. In this article, we present a novel extension of GIMME, confirmatory subgrouping GIMME, which estimates subgroup-level edges for a priori known groups (e.g. typically developing controls vs. clinical group). Detecting edges that consistently exist for individuals within predefined subgroups aids in interpretation of the heterogeneity in connectivity maps and allows for subgroup-specific inferences. We describe this algorithm, as well as several methods to examine the results. We present an empirical example that finds similarities and differences in resting state functional connectivity among four groups of children: typically developing controls (TDC), children with autism spectrum disorder (ASD), children with Inattentive (ADHD-I) and Combined (ADHD-C) Type ADHD. Findings from this study suggest common involvement of the left Broca’s area in all the clinical groups, as well as several unique patterns of functional connectivity specific to a given disorder. Overall, the current approach and proof of principle findings highlight a novel and reliable tool for capturing heterogeneity in complex mental health disorders.
Introduction
In recent decades, functional neuroimaging has become an increasingly widely used tool for investigating a variety of cognitive behaviors as well as both medical and psychological disorders. One specific use of functional imaging is in determining differences in brain processes between groups of subjects, such as comparing children diagnosed with autism to typically developing controls with regard to functional activation or connectivity.
Functional connectivity approaches have emerged as powerful tools for studying group differences in functional organization. However, within the functional connectivity framework there are many different methodologies for analyzing group differences, each of which have advantages and disadvantages. Many times, investigators interested in assessing functional connectivity begin by starting with a graph, or a matrix form that indicates how brain regions relate to each other across time. Using a correlation matrix of contemporaneous (i.e., lag-0) relations between regions of interest (ROIs) represents the most common approach for arriving at such graphs. Here, the correlation coefficients are considered “edges,” where the brain regions themselves are considered nodes. The use of correlation matrices for group comparisons has limitations – primarily, each paired relationship does not take into account the potential influence of other regions (Marrelec et al., 2006; Varoquaux & Craddock, 2013). Methods which do consider indirect effects such as partial correlations, have been considered as ways to handle this issue (Smith et al., 2011); however, the use of partial correlations must also be cautioned since each estimated coefficient between two given regions has arbitrarily controlled for every other region’s potential influence. This heavy-handed approach could cause true edges among regions to be missed due to suppression effects (Kutner, Nachtsheim, Neter, & Li, 2005).
A different set of methods, causal search algorithms, provides a means to both account for indirect effects, as well as induce sparsity. These algorithms take the BOLD time series of the ROIs of interest and return a sparse, directed weighted functional connectivity1 matrix that contains only edges that most parsimoniously describe the overall pattern of functional connectivity for a subject. A number of desirable qualities emerge. By being directed, the algorithms test whether a putative causal relationship exists between two given regions after controlling for other relevant brain regions. Additionally, these algorithms provide weighted edges, enabling inferences as to whether a given region inhibits (i.e., is negative) or excites (positive) another region. The sparsity in these connectivity patterns differs from the sparsity induced in correlation and partial correlation approaches in important ways. For one, the sparsity is arrived at without the need to arbitrarily threshold at a given value as is often done in correlation-based approaches. When using causal search algorithms, sparsity is identified through a data-driven approach, rather than specified a priori or inferred post hoc. Two, rather than control for all possible influences from other brain regions (as in partial correlation) or none of them (as in correlation), estimates obtained from causal search algorithms control for only those regions that have been found to have an effect on the target variable. In this way, bias in the estimate is reduced by considering third-party variables but suppression is less likely to happen. Three, rather than being symmetric and providing the presence of a bidirectional relation, these algorithms ascertain which brain region explains a statistically significant amount of variability in a given brain region (controlling for other possible regions). Together, these benefits attend more closely to the underlying hypotheses of interest in connectivity analysis by detecting relations among brain regions in ways that decrease the likelihood of spurious edges and false negatives (Mumford & Ramsey, 2014).
There are a variety of causal search algorithms used in fMRI research (for review see: Henry & Gates, 2017), and here we use a novel variant of one of the algorithms found to be most reliable: Group Iterated Multiple Model Estimation (GIMME; Gates and Molenaar, 2012). We term the variant Confirmatory Subgrouping GIMME (CS-GIMME) since predefined classifications of individuals will be taken into account during the model search procedure. Prior to the inception of GIMME, Smith and colleagues (2011) revealed that most analytic approaches for arriving at directed patterns of connectivity perform poorly when individual-level analysis is conducted. GIMME performed as well as the best approaches in the simulations of Smith and colleagues (2011) in terms of determining the presence of a connection, with the added benefit of being uniquely able to detect the directionality of effects (Gates & Molenaar, 2012).
Additionally, GIMME is one of few techniques that do not assume this homogeneity in brain processes. Said differently, GIMME does not presuppose that brain processes are ergodic when looking across individuals (see Molenaar, 2004). The algorithm achieves this by not forcing models to be similar across individuals. It does look for similarities, should they exist, in patterns of edges across individuals in order to detect signal from noise at the start of the algorithm. Looking for edges that replicate across the majority of individuals improves the reliability of the results for the search for individual-level, or unique, edges (Gates & Molenaar, 2012). By contrast, methods that aggregate individuals that are heterogeneous in their dynamic processes lead to spurious results that may fail to describe any one individual in the sample (Molenaar & Campbell, 2009). Because of this quality, reliable group- and individual-level edges are obtained from GIMME at rates higher than most competing approaches. GIMME has been highlighted as one of the best options available for recovering the presence of individual-level edges (Mumford & Ramsey, 2014), due to its high rate of recovery in both the presence of edges and the direction of those edges, and has been used in fMRI studies with focuses ranging from language processing (Yang, Gates, Molenaar, & Li, 2015) to substance use (Beltz et al., 2013; Zelle, Gates, Fiez, Sayette, & Wilson, 2017) and clinical diagnoses (Gates, Molenaar, Hillary, Ram, & Rovine, 2010; Price et al., 2017).
The current manuscript extends GIMME by also searching for subgroup-level edges that exist for predefined (i.e., user-specified) subsets of individuals. Following the heuristic for the group-level edge search, the subgroup-level search identifies patterns of edges that exist for the majority of individuals in each subgroup. Prior work has suggested that searching for similarities across smaller subsets of individuals using the same algorithmic approach within GIMME further improves recovery of edges (Gates, Lane, Varangis, Giovanello, & Guiskewicz, 2017; Lane, Gates, Pike, Beltz, & Wright, In Press). A critical benefit of this approach is that it is possible to have no subgroup-level edges - the algorithm only returns them if they truly exist. At the end of the model searches, all individuals have unique estimates for the group- and subgroup-level edges, in addition to unique edges if needed to explain variance in the brain regions. Simulated data studies have demonstrated that the algorithm within GIMME for arriving at subgroup-level edges can recover them at very high rates (Gates et al., 2017; Lane et al, In press). A drawback of these previously investigations is that the user was not able to a priori define the subgroups. With CS-GIMME, a novel extension to GIMME developed for use in the present paper, the researcher has control over how the participants are organized into subsets for comparisons.
CS-GIMME provides several additional advantages over other network neuroscience methods of analysis for examining group differences. As it provides both whole sample level edges (i.e., common between groups), as well as subgroup level edges (i.e., unique to a given group), researchers can describe differences in the strength of common connections in addition to the presence or absence of edges. It must be stressed that searches for edges that exist on the group or subgroup levels do not assume that the individuals share commonalities. Rather than force them to be estimated for all individuals (as is done in multilevel approaches and concatenation), CS-GIMME simply detects them if they are there by using an effective method for detecting signal from noise. Furthermore, CS-GIMME allows for the analysis of an arbitrary number of groups, pooling information from all groups to discover common functional connectivity. This allows for a principled positive control analysis, where for example, in addition to analyzing differences from typically developing patients, one can compare between clinical groups as well in the same analysis while also assessing similarities across all groups.
This article is structured as follows; first we describe CS-GIMME in detail, and describe various tuning parameters that govern its behavior. Second, we apply CS-GIMME to an empirical dataset of resting state scans of typically developing controls (TDC), children with attention deficit hyperactivity disorder combined subtype (ADHD-C), children with attention deficit hyperactivity disorder inattentive subtype (ADHD-I) and children with autism spectrum disorder (ASD). In this study, we expand on the base output of CS-GIMME and describe several inferential techniques to better evaluate specific group differences. Finally, we summarize the findings from the empirical example, and discuss other use cases for CS-GIMME, as well as several limitations.
Confirmatory subgroup GIMME:
CS-GIMME extends the original GIMME (Gates & Molenaar, 2012) and is implemented in the R package gimme, (Lane et al., 2018). The overarching modeling framework is the unified Structural Equation Model (uSEM; Gates et al., 2010; Kim et al., 2007), a model that incorporates both lagged and contemporaneous directed edges among brain regions. Modeling contemporaneous edges is crucial for fMRI studies due to the low temporal resolution. The heavy reliance of contemporaneous edges in functional connectivity approaches (e.g., correlational and ICA) plus results from simulated data studies (Smith et al., 2011) support this notion. However, including lagged edges is also important. For one, the measurements used from fMRI studies are based on processes that are known to be autocorrelated due to the hemodynamic response to neuronal activity (Logothetis, 2008), so omitting these known influences on the signal likely would result in biased estimates. Two, including autoregressive (AR) relations enables Granger causality testing (Granger, 1969). Briefly, if a brain region Y is said to “Granger cause” another region Z, that means that region Y can explain variability in Z after taking into account the AR effect for region Z. By including the AR effects via lagged edges early in the model building process GIMME detects directionality at higher rates than if they were omitted (Lane et al., In Press).
The general uSEM may be formally defined as:
where A is a p × p matrix containing the directed contemporaneous edges among p brain regions (with a zero diagonal to prevent contemporaneous self-prediction), ϕ is a p × p matrix containing the lagged edges among p brain regions with AR effect estimates on the diagonal, η is the p × 1 observed time series of brain activity at time t, and the p × 1 vector ζ contains residuals with a mean of zero and diagonal covariance matrix. The residuals are assumed to be white noise processes and thus contain no temporal dependencies. The intercept is omitted here for clarity in presentation but would exist if the data are not mean-centered.
These edges can be decomposed for each individual into group-, subgroup-, and individual-level relations. That is, certain edges exist for the entire sample; certain edges exist within a given subgroup k; and certain additional edges exist for a given individual i. This decomposition can be expressed as:
Here, A, ϕ, and ζ are defined as before, where the superscripts s and g indicate that these edges exist at the subgroup- and group-level, respectively. Importantly, should no subgroup division exist (i.e., all individuals are in one “subgroup”), this matrix will only contain zeros for individuals in that subgroup. Parameter matrices A and ϕ that lack a superscript denote matrices containing only estimates for the individual-level pattern of edges. Finally, the subscript i on all matrices indicates that each edge is estimated at the individual-level, even in the cases where there are group- and subgroup-level patterns of edges.
Group-level search.
The ultimate goal of CS-GIMME is to find patterns of edges that tend to exist for the entire sample, patterns of edges that exist for previously defined subgroups of individuals, and additional edges that may exist for particular individuals. In this way, signal is detected from noise and the models are not driven entirely by individual nuances. If one were to approach the data-driven search the opposite way by first arriving at individual-level models and then assessing similarities for the sample and the subgroups, the inferences may be driven by noise due to nuances in the individual-level data and potential for any algorithm to model noise. By looking for consistencies across individuals in early steps of the data-driven search CS-GIMME detects signal from noise – first at the sample (or group) level, then at the subgroup level. The reliably-obtained group-level and subgroup-level edge patterns in turn greatly aid in accurately recovering individual-level edges (Lane et al., In Press). Full details of the model selection procedure can be found in Gates and colleagues (2017). Here, we briefly describe the relevant steps. The model selection procedure is implemented in the freely distributed R package, gimme (Lane et al., 2018), which now allows for both traditional GIMME and CS-GIMME to be implemented.
The group-level search is guided by the use of modification indices (MIs), related to Lagrange multipliers (Engle, 1984), which are scores that indicate the extent to which the addition of a potential edge will improve the overall model fit for that individual (Sörbom, 1989). As MIs are asymptotically chi-square distributed, significance can be directly tested for each MI. It has previously been suggested that models built using MIs need to be replicated to demonstrate consistency of relations (MacCallum, 1986). As such, GIMME only includes edges at the group level that exist across the majority of individuals. The GIMME algorithm begins by counting, for each edge, the number of individuals whose models would significantly improve should that edge be freely estimated. This results in a count matrix, and the element from the constrained set that the edge with the highest count is selected. Due to the testing of MIs across all individuals, the criterion for significance uses a strict Bonferroni correction of, α = .05/N, where N = the number of individuals. This starkly contrasts methods that identify edges to include in the group model by looking at the average of edge weights, as the GIMME approach cannot be influenced by outlier cases and is impervious to sign differences (such as large absolute values for all individuals that are negative for some individuals and positive for others). In fact, information regarding the sign of the weight is not used in the group-level search—here, only the absolute magnitude is considered. Should there be a tie in the count of significant MIs then the algorithm selects the element with the highest sum of MIs taken across all individuals. The selection of group-level edges terminates when no edge is significant for a prespecified proportion of individuals that is considered the majority (see below for a discussion on this in the “Tuning Parameters” subsection). By requiring the edges be significant for a majority of individuals GIMME ensures the final group-level edges truly apply to the individuals in the sample. Other aggregation approaches may lead group-level relations that do not describe a given individual in the sample (Molenaar, 2004).
Confirmatory Subgroup-level search.
Following the identification of the group-level edges (i.e., edges that replicate for the majority of the sample), CS-GIMME conducts the search for edges that exist for the majority of individuals in each respective subgroup. CS-GIMME searches for subgroup-level edges in a similar manner to the group-level search by using MIs to guide the addition of edges. Beginning with the group-level edges as the new null model from which to search, CS- GIMME identifies the edge that, if estimated for everyone in that subgroup, would improve the greatest number of individuals’ model fits. The addition of the edge must improve the majority of individuals’ model fits as indicated by Bonferroni corrected p-values obtained from the MI tests. Again, the threshold for what constitutes the “majority” for the subgroup can be defined by the researcher. Once identified, this edge is then estimated for everyone in the subgroup, with each edge parameter estimated uniquely for each. The procedure stops adding edges to the models once there are none that will improve the model for the majority of individuals in that subgroup. Finally, using the group- and subgroup-level edges as null models for individual-level searches, CS-GIMME searches for any additional edges that are needed to best explain each individual’s functional connectivity. Here, MIs are again used and edges with significant MIs are added in a feed-forward iterative fashion. This method for arriving at subgroup-specific paths has been extensively evaluated using simulation studies (Gates et al., 2017; Lane et al., In Press) but has yet to be available for predefined subgroup classifications. Nonetheless, as CS-GIMME is an extension of the GIMME algorithm, it inherits GIMME’s robustness. A comprehensive independent review of causal search methods concluded that GIMME performs as well as competing algorithms out there, and in some cases offers advantages (see Mumford & Ramsey, 2014). The search algorithm used for subgroup-level paths is the same as that used at the group level.
Discussion of tuning parameters for group and subgroup level edges.
The group and subgroup-level edge selection processes both require setting thresholds for what is considered the “majority”. Informed by prior research and simulations, the default cutoff for the majority threshold for the group-level search is 75% since this value resulted in exceptionally good recovery of the data-generating models (Gates & Molenaar, 2012). Smith et al. (2011) note that the ability to detect signal from noise in data simulated to emulate fMRI data varies according to the length of the series. They found in their simulations that the best method can be expected to detect the presence of edges 95% of the time when block length is 10 minutes, 77% of the time when it is 5 minutes, and 59% of the time when the block length is 2.5 minutes. Of course, these rough guidelines will be influenced by the repetition time for scans since the power to detect effects relies on the length of the time series. In the GIMME function available in R, the researcher can easily adjust this value if a more or less strict criterion is desired by adjusting the “groupcutoff =” argument. Historically, the subgroup-level edges have used a looser criterion for the majority threshold. The rationale here is that the subgroups may be smaller, and reaching a majority threshold as high as 75% may require that almost all the of the individuals in the subgroup has the edge (e.g., 6 out 7 individuals would have to have an edge for it to be above this threshold). While the default is that over 50% of the individuals must have a significant edge (as suggested by the corresponding MI) for it to be added to the subgroup-level model, this can be adjusted by the researcher by changing the “subcutoff =” argument in the gimme function. We suggest a stricter threshold for the use of CS-GIMME, such as .75, as in this case the groups are known a priori, and recovery of consistent subgroup level paths is the goal of the analysis and the researcher likely has adequately-sized subgroups based on their study design.
Discussion of optimal sample sizes for CS-GIMME.
For functional neuroimaging studies both the number of time points and the number of individuals must be considered. In terms of the number of time points, GIMME has been previously shown to have excellent path presence recovery (92–100%) in simulated data with numbers of time points as low as 50 (2.5 minutes of data at TR of 3 seconds, see Smith et al., 2011) while GIMME’s ability to distinguish the directionality of paths begins to suffer by being no better than chance (Gates & Molenaar, 2012). Accurate recovery of both the presence and direction of edges occurs consistently with 200 time points (using simulated data from Smith et al., 2011 and in Gates et al., 2017). Regarding the number of participants, the algorithm for detecting signal from noise in the edges performs well with as few as 10 individuals (Gates & Molenaar, 2012). Given these results, we suggest that the minimum sample size requirements to use CS-GIMME are as follows: at least 10 subjects per confirmatory group, with at least 200 time points (e.g., 400 seconds with a repetition time of 2 seconds) remaining after motion correction. We stress that these are minimal suggested requirements, with accuracy and the power to detect relations improving with larger numbers of individuals and time points.
Comparing TDC, ASD, ADHD-I and ADHD-C
In this section we apply CS-GIMME to a dataset consisting of resting state scans for age, gender and IQ matched typically developing children and children with ASD, ADHD-C or ADHD-I. This dataset provides an ideal case to demonstrate the utility of the CS-GIMME approach for analyzing differences (and similarities) in directed functional connectivity between a variety of groups. First, we present a brief overview of the disorders, along with a rationale for the following analysis.
ASD and ADHD:
ASD and ADHD are neurodevelopmental disorders. In terms of prototypical symptomology, ASD and ADHD exhibit different profiles, with ASD being characterized by delays in social communication/language along with restricted interests and/or repetitive behaviors (Constantino & Charman, 2016), while ADHD manifests as solely inattentive/disorganized or both inattentive/disorganized and hyperactive/impulsive behaviors (American Psychiatric Association, 2013; Matthews, Nigg, & Fair, 2013). Controversy has led to efforts to identify a specifically inattentive-only group of youth with ADHD (ADHD-I), such that new brain imaging data have been called for (Willcutt et al., 2012).
Despite these differences in symptomology ASD and ADHD presentations show some overlaps in neurobiological and cognitive findings such as executive functioning (Gardiner & Iarocci, 2017; Semrud-Clikeman, Walkowiak, Wilkinson, & Minne, 2010), although the ASD effects may be attributable to comorbid ADHD (Karalunas et al., 2018). Given both their unique and overlapping properties, ASD and ADHD are well suited to act as positive controls to one another, as examining directed functional connectivity differences between disordered groups (and compared to typically developing children) can provide insight into how neural correlates of behavior are dissociated between the disorders. In the following analysis we focus on three functional subnetworks that have been previously related to either executive functioning in general, or the developmental disorders: the Default Mode network (DMN), the Salience Network and the Ventral Attention Network.
The DMN has been widely studied for both its overall role in executive functioning, as well as its relation with developmental disorders. Previous work has shown atypical organization for both ASD and ADHD samples (For reviews see: Henry & Cohen, In Press; Konrad & Eickhoff, 2010; Padmanabhan, Lynch, Schaer, & Menon, 2017; Posner, Park, & Wang, 2014), making the DMN an ideal choice of a functional subnetwork to examine. The Salience network has been shown to be involved in task maintenance (Menon & Uddin, 2010; Seeley et al., 2007) and has been implicated in ASD (Di Martino et al., 2009; Supekar et al., 2013; Uddin et al., 2013; Uddin & Menon, 2009). Finally, the Ventral Attention network is vital for response inhibition (Zhang, Geng, & Lee, 2017), which shows deficits in ADHD (Booth, Burman, & Meyer, 2005).
Methods
Sample:
Families were recruited from the community and a tertiary ASD clinic. A sample of 152 children are included here, assigned as ASD (n=39, mean age=11.79), ADHD DSM-IV combined type (n=38, mean age=11.17), ADHD DSM-IV primarily inattentive type (n=38, mean age=10.83), and typically developing controls (n=37, mean age=11.04), with an overall age range of 7–15 years and a mean estimated IQ of 108. Groups were comparable on age, gender, and estimated IQ, and no significant differences on these variables were found (all p > 0.05). After preprocessing (see below), the final sample used in this analysis consisted of 133 children (ASD = 31, ADHD-C = 31, ADHD-I = 34, TDC = 37). Diagnosis was carefully characterized with a multi-method, multi-informant research assessment protocol based on DSM-V classification. This included parent and teacher standardized ratings, a Kiddie Schedule for Affective Disorders and Schizophrenia (Orvaschel, 1994), and, for ASD, an Autism Diagnostic Observation Schedule (Lord et al., 1989) confirmed by a consensus of research-reliable clinicians using a best-estimate review of all available information. ASD symptomotology was measured using the Social Responsiveness Scale (SRS; Constantino & Gruber, 2005). Children in the ASD group were permitted to have a co-morbid diagnosis of ADHD, although other major psychicatric diagnoses, neurological conditions and non-stimulant psychoactive medications were ruled out for all groups. IQ was estimated with three subtests of the WISC-IV (Wechsler, 2003), and children with an estimated IQ < 70 were excluded. All children on stimulant medication completed a minimum washout of 5 half lives (24–48 hours) prior to the MRI. Descriptives of age, gender, symptomology, comorbidity status and in-scanner motion are provided in Table 1 below:
Table 1:
Demographics. Mean (SD). % M is percent males in group. H-Score is total hyperactivity symptom score. I-Score is total inattentive symptom score. SRS is total SRS score.
| Group | N | % M | Age | IQ | H-Score | I-Score | SRS |
|---|---|---|---|---|---|---|---|
| TDC | 37 | 62 | 11.1 (1.42) | 111.86 (13.83) | 2.78 (3.85) | 3.78 (4.23) | NA |
| ADHD-I | 34 | 76 | 11.3 (1.64) | 108.42 (13.23) | 5.74 (5.46) | 14.09 (5.59) | NA |
| ADHD-C | 31 | 80 | 11.25 (1.42) | 105.64 (16.27) | 14.39 (4.94) | 17.06 (5.35) | NA |
| ASD | 31 | 83 | 11.59 (2.39) | 102.21 (17.56) | 5.22 (4.05) | 10.67 (7.1) | 89.89 (29.83) |
Of the 31 children with ASD, 16 reached criteria for co-morbid diagnosis of ADHD-C. Implications of this comorbidity is discussed in the Discussion section.
Rs-fcMRI:
Acquired functional imaging series underwent well described and widely used preprocessing steps converted to a Nipype (Gorgolewski et al., 2011) workflow to minimize artifacts and spurious noise (Fair et al., 2007, 2009, 2012; Mills et al., 2012; Costa Dias et al. 2013). Three resting-state functional MRI scans were attempted (5 minutes each for a maximum of 15 minutes of resting state data) for each subject using standard acquisition techniques (EPI sequence: TR=2500ms, TE=30ms, FA=90°, 3.8mm3 voxels, 36 slices, FOV=240×240mm). Of the 133 subjects, 87 had a full 15 minutes of resting state data (360 TRs), and 46 had 10 minutes of resting state data (240 TRs). Standard preprocessing methods included slice time correction, debanding, rigid body head motion correction, and within-series signal intensity normalization to a whole-brain mode value of 1000. Anatomical T1-weighted MPRAGE images were resampled and transformed to standard Talairach atlas space (Talairach & Tournoux, 1988) and used for co-registration of the functional series. Connectivity preprocessing included detrending the functional signal, removal of nuisance regressors (Friston, Williams, Howard, Frackowiak, & Turner, 1996; Power et al., 2013), and (iii) temporal low-pass filtering (f > 0.1Hz) via second order Butterworth filter. Nuisance regressors applied in step (ii) of connectivity preprocessing consisted of: (a) movement from 3 rotational and 3 translational parameters obtained in the previous rigid body head motion step, (b) the global whole-brain functional signal, (c) the averaged ventricular signal, and (d) first-order whole-brain, ventricular, and white matter derivatives (Fair et al., 2013; Power et al., 2013). Additionally, all imaging data were visually inspected upon preprocessing completion to exclude series with unsatisfactory co-registration or significant blood-oxygen-level dependent (BOLD) signal dropout. Additional movement correction was applied utilizing a framewise-displacement (FD) threshold of 0.3mm, following methods described by Power and colleagues (Power et al., 2013; Power, Barnes, Snyder, Schlaggar, & Petersen, 2011). Observations identified as having high motion were replaced with a “missing” placeholder so that temporal ordering was maintained. Children who had at least 60% of TRs remaining after scrubbing were retained in this analysis (resulting in a minimum of 216 TRs for a 15 minute session, and 158 TRs for a 10 minute session). Before scrubbing the diagnostic subgroups were marginally different in their frame displacement (F(3, 129) = 2.23, p = .08). After scrubbing at .3, there were no differences between subgroups in terms of frame displacement (F(3, 129) = .6, p = .61). Additionally, there was no correlation between age and frame displacement, either before or after scrubbing (r = −.04, .008,p = .58, .92). Table 2 below shows motion and TR related information.
Table 2:
Motion information. Mean (SD). FD is framewise displacement.
| Group | FD | FD After Scrub | Remaining TRs |
|---|---|---|---|
| TDC | 0.14 (0.05) | 0.11 (0.03) | 284.11 (59.29) |
| ADHD-I | 0.17 (0.07) | 0.12 (0.02) | 270.44 (61.1) |
| ADHD-C | 0.17 (0.09) | 0.12 (0.03) | 267.35 (58.75) |
| ASD | 0.15 (0.06) | 0.12 (0.03) | 281.55 (55.74) |
Parcellations and functional networks:
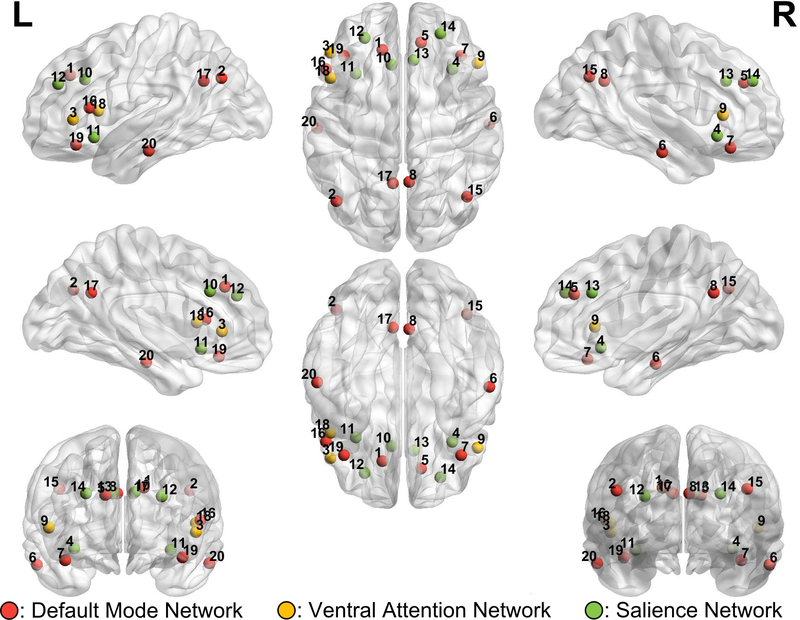
Timecourses were calculated as the average signal within the ROIs defined by the parcellation schemas by Cary and colleagues (2017). This parcellation schema defines 79 ROIs using the InfoMap community detection algorithm (Rosvall & Bergstrom, 2008) applied to the voxelwise functional connectivity matrix. These 79 ROIs are divided into 19 functional subnetworks, labeled as follows: Cingulo-Opercular, Default Mode, Cuneus-Midlingual, Somatosensory-motor, Dorsal Attention, Lateral Occ and Fusiform, Frontoparietal Task Control, Hippocampus, Salience, Temporal Pole, Superior Temporal, Ventral Attention, PCC Borders, Head. Hippocampus, Temporal Occipital Junction, OFC, Frontal Pole, Post OFC and Ventral Lateral Prefrontal. The ROIs are irregularly shaped but non-overlapping and only contain grey matter voxels. For visualizations of all ROIs used, see Supplementary Materials. In the present analysis we selected only ROIs that were members of the Default Mode Network (DMN), Salience and Ventral Attention. The total number of ROIs used is 20, and MNI coordinates for the centroid of the ROI along with anatomical labels are provided in Table 1. ROI AAL label and Brodmann area are classified based on centroid location. A voxelwise visualization of the parcellation is provided in the Supplementary Materials.
CS-GIMME:
CS-GIMME was run on the dataset using a .75 threshold for both group and subgroup level edges. Subgroup labeling corresponded to clinical group, resulting in four total subgroups with nearly identical sample sizes both in terms of number of TRs and participants. CS-GIMME produces primarily two sets of applicable results for inquiries regarding subgroup differences. The first are group level edge coefficients, which are estimated for each subject and allow comparisons of connectivity strength of group edges between all subgroups. The second set of results are in the subgroup level edges. These edges are estimated for all subjects within a particular subgroup. It is important to note that these edges are not necessarily exclusive to a subgroup, as several subgroups might have several of the same subgroup edges. Furthermore, subgroup edges can be estimated as individual level edges for specific participants. CS-GIMME as an extension of GIMME also produces several other sets of output that are not examined in this study. For example, in addition to group and subgroup level edges, individual level edges are estimated, allowing good model fits to be obtained for every subject. These edges can be examined to see if there are additional (but less common) commonalities within subgroup, but this strategy was not pursued here. CS-GIMME additionally produces participant level model fit information, as well as plots of connectivity patterns for the convenience of the analyst. Figures in this study were produced using BrainNet Viewer (Xia, Wang, & He, 2013). Note, ROIs are represented visually as small spheres at the central point of the ROI to provide better visualization of the edges.
Group-Level Edge Analysis:
As group level edges were estimated for all subjects, the distribution of estimates will be approximately normal. Hence the strength of these edges can be compared between subgroups. In this study, simple linear regression models were fit to each edge, comparing the edge strength of each clinical subgroup (ADHD-C, ADHD-I, ASD) to the TDC subgroup. The Benjamani-Hochberg correction for multiple comparisons (Benjamini & Hochberg, 1995) was used to adjust each subgroup comparison across all edges tested. This type of analysis can be extended to account for subject level demographics or any other subject level variable of interest, as the edge strengths are calculated on a per subject basis. Group-level analysis is performed only on the contemporaneous paths. As the data is being sampled at a rate slower than the actual underlying phenomena, the relations between the ROI timeseries may be best captured in the contemporaneous edges, once the lagged relations are taken into account (Granger, 1969). Furthermore, previous work examining the performance of causal search algorithms suggest that only methods that used contemporaneous connections recovered the correct data generating models (Smith et al., 2011). As such, we restrict our group and subgroup level analyses to contemporaneous edges.
Subgroup-Level Edge Analysis:
Subgroup level edges are estimated for all subjects within a particular subgroup, which makes comparison between subgroups less useful as some subgroups might not contain that edge resulting in zero-inflated distributions of estimates. Instead we can examine the presence of common subgroup level edges between subgroups (such as shared edges between ADHD-I and ADHD-C), as well as if edges are within a functional network or between a functional network.
Additionally, we perform a permutation analysis to assess the probability that a given subgroup edge is due to chance. We randomly permute the subgroup labels, which leads to a new label set with the same proportion of subjects in a given subgroup as the original subgroup label set. We then apply CS-GIMME to the data using the permuted label set and extract any subgroups edges that are estimated. By examining the number of times a subgroup edge that was detected in the true label set was also present as a subgroup edge in the permuted label set, we obtain a measure of how likely a given subgroup edge is simply due to chance. A higher rate of occurrence suggests that a given path is not truly specific to a subgroup but is more likely a group level path that was erroneously classified as a subgroup level path. A low, or zero, rate of occurrence suggests that a given subgroup level path is specific to the subgroup in question. We perform this permutation analysis 1000 times.
Results
Group level edges:
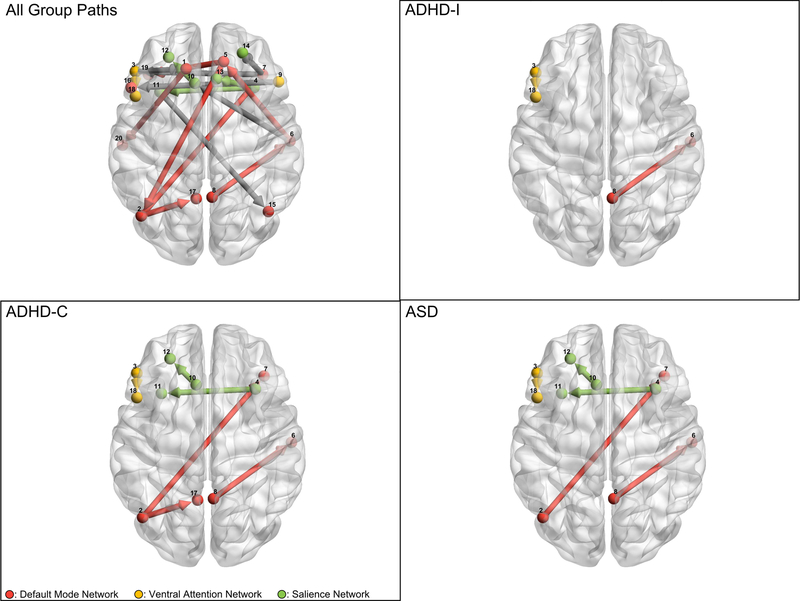
Figure 2 shows the common group level edges, as well as the group level edges with weights that are significantly different than TDC weights for each clinical group. Table 4 lists all group level paths. Table 5 contains information about the significantly different group edges (comparing each clinical subgroup to TDCs).
Figure 2:
Comparison of clinical groups to TDC with regard to group level edge strength. Top Left Panel shows all contemporaneous group level edges and directions. Other panels show significantly different (after BH correction) group level edges for a given clinical group compared with the TDC group. All significantly different edges were stronger in clinical groups. Grey arrows represent between functional subnetwork edges, colored arrows represent within functional subgroup edges. Numerical differences are presented in Table 5.
Table 4:
Group Level Paths
| From | To | Edge Type |
|---|---|---|
| 1 | 20 | Within DMN |
| 2 | 17 | Within DMN |
| 3 | 18 | Within Ventral Attention |
| 4 | 13 | Within Salience |
| 4 | 11 | Within Salience |
| 5 | 2 | Within DMN |
| 2 | 7 | Within DMN |
| 3 | 1 | Ventral Attention to DMN |
| 6 | 5 | Within DMN |
| 7 | 14 | DMN to Salience |
| 8 | 6 | Within DMN |
| 5 | 19 | Within DMN |
| 9 | 16 | Ventral Attention to DMN |
| 10 | 12 | Within Salience |
| 7 | 3 | DMN to Ventral Attention |
| 6 | 10 | DMN to Salience |
| 11 | 15 | Salience to DMN |
Table 5:
Significantly different (from TDC) group edges.
| From | To | Edge Type | Subgroup | Difference (Subgroup - TDC) | Corrected p |
|---|---|---|---|---|---|
| 3 | 18 | Within VA | ADHD-I | 0.091186 | 0.003878 |
| 8 | 6 | Within DMN | ADHD-I | 0.081496 | 0.029656 |
| 3 | 18 | Within VA | ADHD-C | 0.087411 | 0.000831 |
| 2 | 17 | Within DMN | ADHD-C | 0.047516 | 0.009066 |
| 2 | 7 | Within DMN | ADHD-C | 0.101253 | 0.000217 |
| 8 | 6 | Within DMN | ADHD-C | 0.078122 | 0.000188 |
| 10 | 12 | Within Salience | ADHD-C | 0.042201 | 0.044368 |
| 4 | 11 | Within Salience | ADHD-C | 0.048907 | 0.000135 |
| 3 | 18 | Within VA | ASD | 0.088543 | 0.00128 |
| 2 | 7 | Within DMN | ASD | 0.102563 | 0.000357 |
| 8 | 6 | Within DMN | ASD | 0.079133 | 0.002375 |
| 10 | 12 | Within Salience | ASD | 0.042747 | 0.009066 |
| 4 | 11 | Within Salience | ASD | 0.04954 | 0.011747 |
There are several notable results apparent from examining subgroup differences in group level edge magnitudes. The first is the smaller number of significantly different group edges for individuals with ADHD-I compared to individuals with ADHD-C, consistent with concerns that ADHD-I may capture essentially a clinically mild version of ADHD compared to ADHD-C (Willcutt et al., 2012). Individuals with ADHD-I exhibited increased connectivity from Region 3, corresponding to BA45 to region 18, corresponding to BA44. These two regions correspond to the left Broca’s area. Additionally, individuals with ADHD-I showed increased connectivity between Region 8, contained in right precuneus, and Region 6, contained in the middle temporal cortex. Notably, individuals with ADHD-C show the same two significant connections, as well as additional greater connectivity between Regions 2 and 7, and Regions 2 and 17, both within the DMN, as well as between Regions 10 and 12, and Regions 4 and 11, both within Salience network connections.
Individuals with ASD showed identical patterns of increased functional connectivity as individuals with ADHD-C, with the exception of the edge between Regions 2 and 17, which is only significant for individuals with ADHD-C. Finally, across all disorders, the only edges that were significantly different from typically developing controls were within network edges, and the difference in connectivity was only ever positive, with individuals in the clinical subgroups only showing increased connectivity on edges relative to TDCs. This suggests that aberrant functional connectivity within functional networks is not only a difference in the structure but can also be conceptualized as a difference in the strength of the connections.
Subgroup Level Edges:
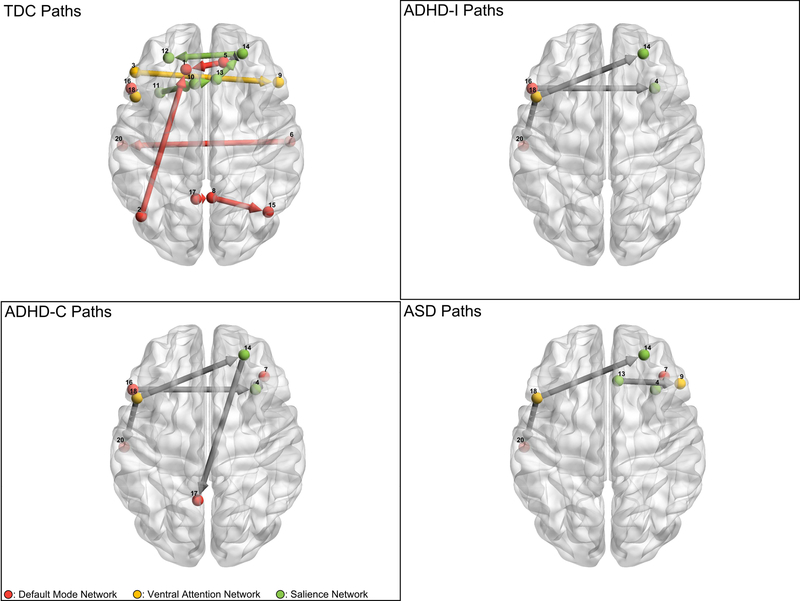
Figure 3 below shows the subgroup level edges estimated with CS-GIMME for all sample groups. Table 6 provides average weights and average standard errors for these edges.
Figure 3:
Subgroup specific edges. Each panel shows edges that are specific (though not necessarily unique) to each group. Grey arrows represent between functional subnetwork edges, while colored edges represent within functional subnetwork edges. Note that TDC specific subgroup edges were primarily within functional subnetwork, while clinical groups exhibit subgroup paths between functional subnetworks.
Table 6:
Subgroup Level Edges
| From | To | Edge Type | Subgroup | Mean Beta | Mean SE | Permute % |
|---|---|---|---|---|---|---|
| 18 | 16 | VA to DMN | TDC | 0.605 | 0.062 | 0 |
| 3 | 9 | Within VA | TDC | 0.383 | 0.054 | 7 |
| 2 | 1 | Within DMN | TDC | 0.354 | 0.039 | 0 |
| 17 | 8 | Within DMN | TDC | 0.866 | 0.043 | 0 |
| 5 | 1 | Within DMN | TDC | 0.554 | 0.043 | 0 |
| 8 | 15 | Within DMN | TDC | 0.610 | 0.056 | .3 % |
| 6 | 20 | Within DMN | TDC | 0.470 | 0.045 | 0 |
| 11 | 10 | Within Sal | TDC | 0.311 | 0.049 | 0 |
| 10 | 13 | Within Sal | TDC | 0.578 | 0.055 | 0 |
| 14 | 5 | Sal to DMN | TDC | 0.292 | 0.042 | 16 % |
| 14 | 12 | Within Sal | TDC | 0.470 | 0.052 | .02 % |
| 13 | 14 | Within Sal | TDC | 0.342 | 0.053 | 3.6 % |
| 18 | 20 | VA to DMN | ADHD-I | 0.218 | 0.031 | 0 |
| 18 | 14 | VA to Sal | ADHD-I | 0.313 | 0.035 | 0 |
| 16 | 4 | DMN to Sal | ADHD-I | 0.400 | 0.068 | 0 |
| 18 | 20 | VA to DMN | ADHD-C | 0.275 | 0.034 | 0 |
| 18 | 14 | VA to Sal | ADHD-C | 0.302 | 0.035 | 0 |
| 16 | 4 | DMN to Sal | ADHD-C | 0.279 | 0.045 | 0 |
| 14 | 17 | Sal to DMN | ADHD-C | 0.320 | 0.053 | 0 |
| 4 | 7 | Sal to DMN | ADHD-C | 0.192 | 0.03 | 0 |
| 18 | 20 | VA to DMN | ASD | 0.176 | 0.03 | 0 |
| 18 | 14 | VA to Sal | ASD | 0.291 | 0.037 | 0 |
| 4 | 7 | Sal to DMN | ASD | 0.227 | 0.04 | 0 |
| 13 | 9 | Sal to VA | ASD | 0.427 | 0.076 | .09 % |
There are several interesting findings from the subgroup edge detection. To begin, one notable difference between the TDC group and the clinical groups is that the unique edges to the TDC group are for the most part within network edges, while the unique edges for the clinical groups are entirely between network edges. This suggests that functional connectivity associated with the disordered groups is characterized as differences in the structure of connections between functional networks in addition to differences in strengths of common connections. Similarly to the group edge difference results, individuals with ADHD-I exhibit a subset of the edges that individuals with ADHD-C exhibit, where ADHD-C shows involvement with more default mode regions, specifically Region 17, in the left precuneus, and Region 7 in the inferior orbital frontal cortex. Individuals with ASD show subgroup edges primarily localized to areas near the right insula, specifically the edge between Region 4, contained in the right insula, and Region 7, contained in BA47.
Consistent with emerging evidence of a common liability factor shared across many domains of psychopathology (Pettersson, Lahey, Larsson, & Lichtenstein, 2018) and neurodevelopment including ASD and ADHD, two subgroup edges that appear in all three disorder subgroups, the edge from Region 18 to Region 20, and the edge between Region 18 and Region 14. Region 18, again, is contained in the left BA44, part of Broca’s area, while Region 20 encompasses the left middle temporal cortex, while Region 14 is contained in the right middle frontal cortex.
The permutation analysis suggests that subgroup level paths indicated for clinical subgroups are specific to the given subgroup labeling, as they did not appear as subgroup level paths when the subgroup labeling was permuted randomly. One paths that was indicated for the TDC subgroup appears to be less robust, specifically the path from ROI 14 to 5. This suggests that this path was close to being considered a group level path and is not specific to the TDC group.
Discussion:
GIMME is a search algorithm that consistently and reliably recovers data-generating edges in benchmark simulated data that emulates functional connectivity maps (e.g., Smith et al., 2011). It does so by detecting signal from noise in a manner that does not assume homogeneity across individuals. Prior work had arrived at data-driven subgroups within the GIMME framework (termed, S-GIMME) and found that adding edges from using the S-GIMME approach worked well even on these smaller sets of individuals (Gates et al., 2017; Lane et al., In Press). However, until researchers could not specify a priori which subgroup each individual was in. This proved to be a hindrance to researchers with targeted areas of foci who wanted to compare specific groups. Hitherto, researchers would have to run GIMME separately on these subgroups of interest to obtain the edges specific to that subgroup. Now, with CS-GIMME, researchers can capitalize on the similarities that may exist for the sample at large by obtaining group-level edges for the whole while also obtaining subgroup-specific edges should they exist.
Broadly speaking, CS-GIMME has a series of advantages over more general methods for analyzing functional connectivity. As it is a variant of the GIMME algorithm, it brings the advantages of appropriately analyzing individual heterogeneity, while extracting group level directed functional connectivity edges common across the sample. Furthermore, the confirmatory aspect of the method allows researchers to analyze specific functional hypotheses, involving a priori defined groups. As was shown in the empirical example, the results gleaned from the CS-GIMME method can be analyzed post hoc, allowing for researchers to examine differences and similarities among groups in both strength of functional connectivity for group level edges, and presence or absence of subgroup level edges. Finally, though not performed in this study, CS-GIMME produces individual, sparse functional connectivity networks suitable for further network analysis, such as with a network neuroscience framework (Bassett & Sporns, 2017). As CS-GIMME explicitly models subgroup differences, network analysis based on these networks will reflect subgroup differences in overall topology more so than a more uninformed network construction method.
The importance of generating functional connectivity patterns that truly capture individuals’ brain processes is an important step to understanding human cognition and emotions. Recently, obtaining subject-specific functional connectivity maps has been highlighted as a major goal in the field, with efforts such as “fingerprinting” originally defined by our group (Miranda-Dominguez et al., 2014; Miranda-Domínguez et al., 2017), amongst others (Finn et al., 2015), and other analytic methods for individual-level analysis being at the forefront (Gordon et al., 2017; Laumann et al., 2015; Ramsey, Hanson, & Glymour, 2011; Smith et al., 2011; Smith, 2012). Subject-specific functional connectivity maps are particularly critical for moving towards the use of MRI and related methods in personalized medicine. Still, among the nuances seen when looking across individuals there typically are also some similarities. This may be partly due to physiological constraints of the brain, and thus there may be some edges that exist for most people regardless of their classifications. CS-GIMME provides a reliable method for investigating connectivity across and within subgroups in a manner that attends to individual nuances.
Empirical findings unique and interesting to ADHD:
These findings both support findings from previous literature and suggest several new productive avenues of research. The first finding of note is that ADHD-I appears to involve a subset of the disrupted functional connectivity seen in individuals with ADHD-C, and specifically does not show the same involvement of more midbrain DMN regions, such as the precuneus, as evidenced by different subgroup edges. This is consistent with previous findings suggesting that ADHD-C shows more atypical connectivity in midline default mode network regions compared to ADHD-I (Fair et al., 2013). It is also consistent with concerns that ADHD-I as defined in the DSM largely captures a milder subset of ADHD than ADHD-C (Willcutt et al., 2012). Furthermore, the increased connectivity from the left BA45 (Region 16) to the right insula (Region 4) is consistent with previous findings of disrupted functional connectivity in the insula found in individuals with ADHD (Zhao et al., 2017), however no research to date has specifically examined subtype differences in insular functional connectivity.
Empirical findings unique and interesting to ASD:
In this sample, individuals with ASD were distinguished from TDCs and both ADHD subtypes by unique subgroup edges primarily localized around the right insula, with increased connectivity between the right insula (Region 4) and right BA47 (Region 7), as well as increased connectivity between the right anterior cingulate (Region 13) and the right BA45 (Region 9). Previous research has implicated both the insula and the ACC, in their roles as components of the salience network, in ASD (Di Martino et al., 2009; Uddin & Menon, 2009), and these findings are consistent with that. Recent research has found that functional connectivity of language processing regions is disrupted in individuals with ASD (Lee, Park, James, Kim, & Park, 2017; Verly et al., 2014), and the current study suggests that there is increased connectivity between the salience network and language processing regions. We did not separately consider ASD with and without ADHD for several reasons. The first is that splitting the ASD group in two leads to a corresponding reduction in power and increase in complexity. The second is that splitting the ASD group leads to unbalanced group sizes, which changes the interpretability of the subgroup path threshold parameter. For more on this, see the Using CS-GIMME section later in the discussion. Prior work suggests that a separate analysis of ASD with and without comorbid ADHD will clarify which features of overlap are attributable to co-occurring ADHD in the ASD group (Karalunas et al., 2018) and we encourage future research to examine this.
Commonalities across disorders:
Finally, CS-GIMME allows for the comparison of the similarities between clinical subgroups as well as TDCs. For one, the group-level edges suggest that those connections may be generalizable to the population as the majority of individuals, regardless of subgroup, had those edges in their models. In addition to these, a number of similar patterns emerged among the clinical subgroups. For instance, ADHD-C and ASD showed similar patterns of increased connectivity in group edges and were primarily distinguished by different subgroup level edges. The commonalities between the group edge differences is likely due to the presence of comorbid ADHD in the ASD group, which makes the unique edges presented by the ASD group particularly salient. Additionally, all three clinical subgroups shared the same two edges from Region 18 to Region 20 and Region 14. The centroid for Region 18 is in left BA44 and can be alternately classified as part of the left inferior frontal gyrus, while Region 20 encompasses a large part of the left temporal lobe, with the centroid contained in the left BA21, Region 14 is contained primarily in right BA9 and more generally the frontal middle gyrus. While left BA44 is a component of Broca’s area, and is related to semantic processing, the left BA21 plays a role in auditory processing of language, while right BA9 is related to attention, particularly related to auditory attention. This pattern of findings hints at a shared disruption in auditory and language processing across all three disorders, suggesting that a further investigation of both functional and structural aberration of these regions might further clarify their relation to these disorders. Such findings could provide clues to general liability factors for frequently-comorbid neurodevelopmental disorders.
Directionality of paths:
CS-GIMME estimates directed functional connectivity, and the directionality of these paths can be used to understand how ROIs and functional subnetworks interact with one another. Previous work using Granger causality has shown that in healthy adults the default mode network exerts greater influence on various task positive networks, rather than the other way around (Uddin, Kelly, Biswal, Castellanos, & Milham, 2009), while elements of the salience network, specifically the right anterior insula, appears to have a causal role in switching between the central executive network and the default mode network (Menon & Uddin, 2010). There has been no work we are aware of that examines differences in the directionality of the DMN to other network relation in individuals with ADHD or ASD, and there has only been a few recent articles examining directional connectivity differences in ASD. Notably, Bernas et al., (2018) showed that the causal connectivity from the ventral attention network to salience and executive network regions is weaker in males with high functioning autism.
The directionality of the paths in the empirical analysis suggest the influence of the default mode network on the ventral attention network and salience network is disrupted in both ADHD and ASD. Specifically, the majority of between network subgroup paths originated from the VAN or Salience network, rather than the DMN, and many of those paths were to the DMN. This is contrasted to the estimated group level paths, many of which were from the DMN to the VAN or Salience networks. This increase in bi-directional between network relations in the clinical subgroups suggests that while between network connectivity is increased in clinical subgroups, this increase is explicitly direction, e.g. we cannot say that one network is exerting more influence over another for individuals with ADHD. Instead, the directionality of between network connections is less well structured in our clinical subgroups.
An analysis of the directionality of paths is best suited for a more restricted ROI set where researchers have a priori hypotheses regarding differences in direction and strength of edges between ROIs. For example, given previous work on ADHD, a natural application of CS-GIMME would be to examine differences in DMN to task positive network path directionality and strength for individuals with ADHD vs. healthy controls. As previous literature suggests that the regulatory influence of the DMN is reduced in children with ADHD, it would be interesting to examine if there is any change in the directionality of between network edges, and which ROIs those are specifically associated with.
Using CS-GIMME:
To provide guidance to researchers interested in using CS-GIMME, we want to summarize several important decision points here. The first is determining if one’s own dataset is appropriate to use with CS-GIMME. An important assumption of the confirmatory subgrouping component of CS-GIMME is that within a subgroup, all subjects have the same set of subgroup level paths. What counts as a subgroup level path is determined by the subgroup threshold tuning parameter. Researchers can use this parameter to reflect the level of heterogeneity they are willing to allow in their subgroup paths. For example, when analyzing a demographically matched group of clinical patients who are fairly homogeneous in their symptom expression, the use of a high subgroup path threshold can lead to the estimation of a consistent set of subgroup paths. On the other hand, if one’s sample consists of a highly heterogenous set of subjects, a high subgroup path threshold could lead to very few subgroup paths being estimated. Another consideration in determining the subgroup path threshold is the size of the subgroups. In small subgroup sizes, a low subgroup threshold could lead to results that reflect sampling variability and would not reflect the groups under study. This is a particular concern for unbalanced group sizes. A general rule of thumb is that the subgroup path threshold should increase with both increasing group size as well as increasing theoretical homogeneity within the groups. In all cases, researchers should report and justify the subgroup path threshold value. Model based methods of selecting the subgroup path threshold value are an active area of methodological research.
A second aspect of CS-GIMME that researchers should consider is the ability to include a priori paths. While our empirical example does not explore CS-GIMME’s (or more generally GIMMEs) ability to test a priori hypotheses, researchers can propose that certain paths exist at the group or subgroup level before the algorithm is applied. For example, if one’s interest is in testing between network connectivity, it would be reasonable to propose that all ROIs within a functional subnetwork are connected. We propose researchers with strong a priori hypotheses fit CS-GIMME models with and without the a priori paths and compare the relative performance of these models subject by subject using BIC values. Expanding GIMME’s a priori hypothesis testing capabilities is also an active area of research.
Finally, we have included example code in the Supplementary Material for researchers to use or adapt to perform the group level significance tests.
Limitations and Future Directions:
There are, of course, several limitations to both the empirical example presented here, as well as CS-GIMME more broadly. The specific parcellation and choice of the default mode network, salience network and ventral attention network was informed by prior literature, but a different choice of functional networks would lead to different findings. Specifically, given findings suggesting that the interactions between the DMN and executive control networks (ECN; Seeley et al., 2007) are altered in ADHD (for review see: Rubia, 2018), we suggest that researchers include ROIs in both the DMN and ECNs in future applications of CS-GIMME on individuals with ADHD. CS-GIMME is not a whole brain algorithm due to computational constraints and requires researchers to choose specific regions of interest to analyze. The choice of these regions will impact results. This is due to CS-GIMME, and GIMME more generally modeling functional connectivity of the network as a complete whole, and so analysis using CS-GIMME needs to consider each ROI set used as a distinct analysis. Finally, CS-GIMME is computationally intensive, with rapid increase in the time taken to analyze a sample when both the number of ROIs and the number of subjects increases. This however can, and will, be alleviated with further methodological developments.
Confirmatory Subgrouping GIMME is a powerful new tool for researchers to use to examine differences in directed functional connectivity between a priori defined subgroups, and one that is grounded in the well validated GIMME algorithm. Further extensions of GIMME such as latent variable modeling of functional networks, improvements in estimation methods, implementation of HRF functions and dynamic modeling of functional connectivity will all further improve the capabilities of CS-GIMME in the near future and continue to improve its usefulness in studying directed functional connectivity differences in both clinical disorders, as well as subgroups of healthy controls.
Supplementary Material
Figure 1:
ROI Centroid Coordinates. Color corresponds to functional network, with red being the default mode network, yellow being the ventral attention network and green being the salience network.
Table 3:
ROI Centroid Locations and Labels
| Node Label | Network | X Coord | Y Coord | Z Coord | AAL Label | Brodmann Area |
|---|---|---|---|---|---|---|
| 1 | DMN | −14 | 35 | 33 | Frontal_Sup_L | Left BA9 |
| 2 | DMN | −45 | −67 | 31 | Angular_L | Left BA39 |
| 3 | Ventral Attention | −49 | 33 | 3 | Frontal_Inf_Tri_L | Left BA45 |
| 4 | Salience | 33 | 22 | −8 | Insula_R | Right Insula |
| 5 | DMN | 12 | 40 | 28 | Frontal_Sup_R | Right BA9 |
| 6 | DMN | 58 | −15 | −19 | Temporal_Mid_R | Right BA21 |
| 7 | DMN | 39 | 31 | −16 | Frontal_Inf_Orb_R | Right BA47 |
| 8 | DMN | 4 | −54 | 29 | Precuneus_R | Right BA31 |
| 9 | Ventral Attention | 50 | 26 | 6 | Frontal_Inf_Tri_R | Right BA45 |
| 10 | Salience | −8 | 25 | 30 | Cingulum_Ant_L | Left BA32 |
| 11 | Salience | −32 | 19 | −9 | Insula_L | Left Insula |
| 12 | Salience | −26 | 43 | 27 | Frontal_Mid_L | Left BA10 |
| 13 | Salience | 7 | 28 | 29 | Cingulum_Ant_R | Right BA8 |
| 14 | Salience | 25 | 46 | 29 | Frontal_Mid_R | Right BA9 |
| 15 | DMN | 43 | −64 | 32 | Angular_R | Right BA39 |
| 16 | DMN | −52 | 22 | 11 | Frontal_Inf_Tri_L | Left BA45 |
| 17 | DMN | −7 | −55 | 29 | Precuneus_L | Left BA31 |
| 18 | Ventral Attention | −49 | 16 | 9 | Frontal_Inf_Oper_L | Left BA44 |
| 19 | DMN | −40 | 31 | −14 | Frontal_Inf_Orb_L | Left BA47 |
| 20 | DMN | −58 | −18 | −18 | Temporal_Mid_L | Left BA21 |
Funding Acknowledgement:
We gratefully acknowledge support from NIH National Institute of Biomedical Imaging and Bioengineering (Award Number: 1-R01-EB022904-01; PI Gates) and NIH National Institute of Mental Health (R01 MH115357 PIs: Fair, Nigg; R01 MH096773 PI: Fair; R01 MH086654 PI: Nigg).
Footnotes
In following convention and technical definitions, “directed functional connectivity” is used rather than “effective connectivity” since the latter requires modeling of causal influences which typically requires the probing changes in connectivity patterns in the presence of experimental manipulation (Friston, 2011).
References
- American Psychiatric Association. (2013). Diagnostic and Statistical Manual of Mental Disorders (5th ed.). Arlington, VA: American Psychiatric Publishing. [Google Scholar]
- Bassett DS, & Sporns O (2017). Network neuroscience. Nature Neuroscience, 20(3), 353–364. 10.1038/nn.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz AM, Gates KM, Engels AS, Molenaar PCM, Pulido C, Turrisi R, … Wilson SJ (2013). Changes in alcohol-related brain networks across the first year of college: A prospective pilot study using fMRI effective connectivity mapping. Addictive Behaviors, 38(4), 2052–2059. 10.1016/j.addbeh.2012.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B, 57. [Google Scholar]
- Bernas A, Barendse EM, Aldenkamp AP, Backes WH, Hofman PAM, Hendriks MPH, … Jansen JFA (2018). Brain resting-state networks in adolescents with high-functioning autism: Analysis of spatial connectivity and temporal neurodynamics. Brain and Behavior, 8(2). 10.1002/brb3.878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth J, Burman D, & Meyer J (2005). Larger deficits in brain networks for response inhibition than for visual selective attention in attention deficit hyperactivity disorder (ADHD). Journal of Child …, 46, 94–111. 10.1111/j.1469-7610.2004.00337.x [DOI] [PubMed] [Google Scholar]
- Cary RP, Ray S, Grayson DS, Painter J, Carpenter S, Maron L, … Fair DA (2017). Network structure among brain systems in adult ADHD is uniquely modified by stimulant administration. Cerebral Cortex, 27(8), 3970–3979. 10.1093/cercor/bhw209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, & Charman T (2016). Diagnosis of autism spectrum disorder: reconciling the syndrome, its diverse origins, and variation in expression. The Lancet Neurology, 15(3), 279–291. 10.1016/S1474-4422(15)00151-9 [DOI] [PubMed] [Google Scholar]
- Constantino JN, & Gruber CP (2005). Social Responsiveness Scale (SRS). Los Angeles, CA: Western Psychological Services. [Google Scholar]
- Di Martino A, Shehzad Z, Kelly C, Roy AK, Gee DG, Uddin LQ, … Milham MP (2009). Relationship between cingulo-insular functional connectivity and autistic traits in neurotypical adults. American Journal of Psychiatry, 166(8), 891–899. 10.1176/appi.ajp.2009.08121894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle RF (1984). Chapter 13 Wald, likelihood ratio, and Lagrange multiplier tests in econometrics. Handbook of Econometrics, 2, 775–826. 10.1016/S1573-4412(84)02005-5 [DOI] [Google Scholar]
- Fair DA, Nigg JT, Iyer S, Bathula D, Mills KL, Dosenbach NUF, … Milham MP (2013). Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Frontiers in Systems Neuroscience, 6(February), 80 10.3389/fnsys.2012.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, … Todd Constable R (2015). Functional connectome fingerprinting: Identifying individuals based on patterns of brain connectivity HHS Public Access. Nat Neurosci, 18(11), 1664–1671. 10.1038/nn.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ (2011). Functional and Effective Connectivity: A Review. Brain Connectivity, 1(1), 13–36. 10.1089/brain.2011.0008 [DOI] [PubMed] [Google Scholar]
- Friston KJ, Williams S, Howard R, Frackowiak RSJ, & Turner R (1996). Movement-related effects in fMRI time-series. Magnetic Resonance in Medicine, 35(3), 346–355. 10.1002/mrm.1910350312 [DOI] [PubMed] [Google Scholar]
- Gardiner E, & Iarocci G (2017). Everyday executive function predicts adaptive and internalizing behavior among children with and without autism spectrum disorder. Autism Research : Official Journal of the International Society for Autism Research, 11(2), 284–295. 10.1002/aur.1877 [DOI] [PubMed] [Google Scholar]
- Gates KM, Lane ST, Varangis E, Giovanello K, & Guiskewicz K (2017). Unsupervised Classification During Time-Series Model Building. Multivariate Behavioral Research, 52(2), 129–148. 10.1080/00273171.2016.1256187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates KM, & Molenaar PCM (2012). Group search algorithm recovers effective connectivity maps for individuals in homogeneous and heterogeneous samples. NeuroImage, 63(1), 310–319. 10.1016/j.neuroimage.2012.06.026 [DOI] [PubMed] [Google Scholar]
- Gates KM, Molenaar PCM, Hillary FG, Ram N, & Rovine MJ (2010). Automatic search for fMRI connectivity mapping: An alternative to Granger causality testing using formal equivalences among SEM path modeling, VAR, and unified SEM. NeuroImage, 50(3), 1118–1125. 10.1016/j.neuroimage.2009.12.117 [DOI] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, … Dosenbach NUF (2017). Precision Functional Mapping of Individual Human Brains. Neuron, 95(4), 791–807.e7. 10.1016/j.neuron.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, & Ghosh SS (2011). Nipype: A Flexible, Lightweight and Extensible Neuroimaging Data Processing Framework in Python. Frontiers in Neuroinformatics, 5(13). 10.3389/fninf.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CWJ (1969). Investigating Causal Relations by Econometric Models and Cross-spectral Methods. Econometrica, 37(3), 424 10.2307/1912791 [DOI] [Google Scholar]
- Henry T, & Gates K (2017). Causal search procedures for fMRI: review and suggestions. Behaviormetrika, 44(1), 193–225. 10.1007/s41237-016-0010-8 [DOI] [Google Scholar]
- Henry TR, & Cohen JR (n.d.). Dysfunctional brain network organization in neurodevelopmental disorders In Laurienti PJ, Munsell B, & Wu G (Eds.), CONNECTOMICS: METHODS, MATHEMATICAL MODELS AND APPLICATIONS. Elsevier. [Google Scholar]
- Karalunas SL, Hawkey E, Gustafsson H, Miller M, Langhorst M, Cordova M, … Nigg JT (2018). Overlapping and Distinct Cognitive Impairments in Attention-Deficit/Hyperactivity and Autism Spectrum Disorder without Intellectual Disability. Journal of Abnormal Child Psychology. 10.1007/s10802-017-0394-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zhu W, Chang L, Bentler PM, & Ernst T (2007). Unified structural equation modeling approach for the analysis of multisubject, multivariate functional MRI data. Human Brain Mapping, 28(2), 85–93. 10.1002/hbm.20259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad K, & Eickhoff SB (2010). Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Human Brain Mapping, 31(6), 904–916. 10.1002/hbm.21058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner MH, Nachtsheim C, Neter J, & Li W (2005). Applied Linear Statistical Models. 10.2307/2984653 [DOI] [Google Scholar]
- Lane ST, Gates Kathleen Marie, Molenaar PCM, Hallquist MN, Pike H, Fisher Z, … Henry TR (2018). gimme: Group Iterative Multiple Model Estimation. Retrieved from https://cran.r-project.org/package=gimme [Google Scholar]
- Lane ST, Gates KM, Pike H, Beltz AM, & Wright AG (n.d.). Uncovering general, shared, and unique temporal patterns in ambulatory assessment data. Psychological Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, … Petersen SE (2015). Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron, 87(3), 658–671. 10.1016/j.neuron.2015.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Park B, James O, Kim S, & Park H (2017). Autism Spectrum Disorder Related Functional Connectivity Changes in the Language Network in Children, Adolescents and Adults, 11(August), 1–11. 10.3389/fnhum.2017.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK (2008). What we can do and what we cannot do with fMRI. Nature, 453(7197), 869–878. 10.1038/nature06976 [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, & Schopler E (1989). Austism diagnostic observation schedule: A standardized observation of communicative and social behavior. Journal of Autism and Developmental Disorders, 19(2), 185–212. 10.1007/BF02211841 [DOI] [PubMed] [Google Scholar]
- MacCallum R (1986). Specification Searches in Covariance Structure Modeling. Psychological Bulletin, 100(1), 107–120. 10.1037/0033-2909.100.1.107 [DOI] [Google Scholar]
- Marrelec G, Krainik A, Duffau H, Peregrini-Issac M, Lehericy S, Doyon J, & Benali H (2006). Partial correlation for functional brain interactivity investigation in functional MRI. NeuroImage, 32, 228–237. [DOI] [PubMed] [Google Scholar]
- Matthews M, Nigg JT, & Fair DA (2013). Attention Deficit Hyperactivity Disorder. In Journal of Educational Measurement (Vol. 35, pp. 235–266). 10.1007/7854_2013_249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V, & Uddin LQ (2010). Saliency, switching, attention and control: a network model of insula function. Brain Structure and Function, 214(5–6), 655–667. 10.1007/s00429-010-0262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Domínguez Ó, Feczko E, Grayson DS, Walum H, Nigg JT, & Fair DA (2017). Heritability of the human connectome: a connectotyping study. Network Neuroscience, 1–48. 10.1162/NETN_a_00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Carpenter SD, Grant KA, Kroenke CD, Nigg JT, & Fair DA (2014). Connectotyping: Model based fingerprinting of the functional connectome. PLoS ONE, 9(11). 10.1371/journal.pone.0111048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar PCM (2004). A Manifesto on Psychology as Idiographic Science: Bringing the Person Back Into Scientific Psychology, This Time Forever. Measurement: Interdisciplinary Research & Perspective, 2(4), 201–218. 10.1207/s15366359mea0204_1 [DOI] [Google Scholar]
- Molenaar PCM, & Campbell CG (2009). The New Person-Specifc Paradigm in Psychology. Psychological Science, 18(2), 112–117. 10.1111/j.1467-8721.2009.01619.x [DOI] [Google Scholar]
- Mumford JA, & Ramsey JD (2014). Bayesian networks for fMRI: A primer. NeuroImage, 86, 573–582. 10.1016/j.neuroimage.2013.10.020 [DOI] [PubMed] [Google Scholar]
- Orvaschel H (1994). Schizophrenia and Affective Disorders Schedule for Children—Epidemiological Version (KSADS-E). Nova Southwestern University, Florida. [Google Scholar]
- Padmanabhan A, Lynch CJ, Schaer M, & Menon V (2017). The Default Mode Network in Autism. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 2(6), 476–486. 10.1016/j.bpsc.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson E, Lahey BB, Larsson H, & Lichtenstein P (2018). Criterion Validity and Utility of the General Factor of Psychopathology in Childhood: Predictive Associations With Independently Measured Severe Adverse Mental Health Outcomes in Adolescence. Journal of the American Academy of Child and Adolescent Psychiatry, 57(6), 372–383. 10.1016/j.jaac.2017.12.016 [DOI] [PubMed] [Google Scholar]
- Posner J, Park C, & Wang Z (2014). Connecting the dots: A review of resting connectivity MRI studies in attention-deficit/hyperactivity disorder. Neuropsychology Review, 24(1), 3–15. 10.1007/s11065-014-9251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, & Petersen SE (2011). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage, 59(3), 2142–2154. 10.1016/j.neuroimage.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, & Petersen SE (2013). Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage, 84, 320–341. 10.1016/j.neuroimage.2013.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RB, Lane S, Gates K, Kraynak TE, Horner MS, Thase ME, & Siegle GJ (2017). Parsing Heterogeneity in the Brain Connectivity of Depressed and Healthy Adults During Positive Mood. Biological Psychiatry, 81(4), 347–357. 10.1016/j.biopsych.2016.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JD, Hanson SJ, & Glymour C (2011). Multi-subject search correctly identifies causal connections and most causal directions in the DCM models of the Smith et al. simulation study. NeuroImage, 58(3), 838–848. 10.1016/j.neuroimage.2011.06.068 [DOI] [PubMed] [Google Scholar]
- Rosvall M, & Bergstrom CT (2008). Maps of random walks on complex networks reveal community structure. Proceedings of the National Academy of Sciences, 105(4), 1118–1123. 10.1073/pnas.0706851105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubia K (2018). Cognitive Neuroscience of Attention Deficit Hyperactivity Disorder (ADHD) and Its Clinical Translation. Frontiers in Human Neuroscience, 12 10.3389/fnhum.2018.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, … Greicius MD (2007). Dissociable Intrinsic Connectivity Networks for Salience Processing and Executive Control. Journal of Neuroscience, 27(9), 2349–2356. 10.1523/JNEUROSCI.5587-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrud-Clikeman M, Walkowiak J, Wilkinson A, & Minne E (2010). Direct and Indirect Measures of Social Perception, Behavior, and Emotional Functioning in Children with Asperger’s Disorder, Nonverbal Learning Disability, or ADHD. J Abnorm Child Psychol, 38(4), 509–519. 10.1007/s10802-009-9380-7 [DOI] [PubMed] [Google Scholar]
- Smith SM (2012). The future of FMRI connectivity. NeuroImage, 62(2), 1257–1266. 10.1016/j.neuroimage.2012.01.022 [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, … Woolrich MW (2011). Network modelling methods for FMRI. NeuroImage, 54(2), 875–891. 10.1016/j.neuroimage.2010.08.063 [DOI] [PubMed] [Google Scholar]
- Sörbom D (1989). Model modification. Psychometrika, 54(3), 371–384. 10.1007/BF02294623 [DOI] [Google Scholar]
- Supekar K, Uddin LQ, Khouzam A, Phillips J, Gaillard WD, Kenworthy LE, … Menon V (2013). Brain Hyperconnectivity in Children with Autism and its Links to Social Deficits. Cell Reports, 5(3), 738–747. 10.1016/j.celrep.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, & Tournoux P (1988). Co-planar stereotaxic atlas of the human brain. New York: Thieme Medical Publishers. [Google Scholar]
- Uddin LQ, Kelly AMC, Biswal BB, Castellanos FX, & Milham MP (2009). Functional Connectivity of Default Mode Network Components: Correlation, Anticorrelation, and Causality. Human Brain Mapping, 30(2), 625–637. 10.1002/hbm.20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, & Menon V (2009). The anterior insula in autism: Under-connected and under-examined. Neuroscience and Biobehavioral Reviews, 33(8), 1198–1203. 10.1016/j.neubiorev.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Lynch CJ, Khouzam A, Phillips J, Feinstein C, … Menon V (2013). Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry, 70(8), 869–879. 10.1001/jamapsychiatry.2013.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G, & Craddock RC (2013). Learning and comparing functional connectomes across subjects. NeuroImage, 80, 405–415. 10.1016/j.neuroimage.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Verly M, Verhoeven J, Zink I, Mantini D, Peeters R, Deprez S, … Sunaert S (2014). Altered functional connectivity of the language network in ASD: Role of classical language areas and cerebellum. NeuroImage: Clinical, 4, 374–382. 10.1016/j.nicl.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D (2003). Wechsler Intelligence Scale for Children - Fourth Edition (WISC-IV). London: Pearson. [Google Scholar]
- Willcutt EG, Nigg JT, Pennington BF, Solanto MV, Rohde LA, Tannock R, … Lahey BB (2012). Validity of DSM-IV attention deficit/hyperactivity disorder symptom dimensions and subtypes. Journal of Abnormal Psychology, 121(4), 991–1010. 10.1037/a0027347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, & He Y (2013). BrainNet Viewer: A Network Visualization Tool for Human Brain Connectomics. PLoS ONE, 8(7). 10.1371/journal.pone.0068910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gates KM, Molenaar P, & Li P (2015). Neural changes underlying successful second language word learning: An fMRI study. Journal of Neurolinguistics, 33, 29–49. 10.1016/j.jneuroling.2014.09.004 [DOI] [Google Scholar]
- Zelle SL, Gates KM, Fiez JA, Sayette MA, & Wilson SJ (2017). The first day is always the hardest: Functional connectivity during cue exposure and the ability to resist smoking in the initial hours of a quit attempt. NeuroImage, 151, 24–32. 10.1016/j.neuroimage.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Geng X, & Lee TMC (2017). Large-scale functional neural network correlates of response inhibition: an fMRI meta-analysis. Brain Structure and Function, 222(9), 3973–3990. 10.1007/s00429-017-1443-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Li H, Yu X, Huang F, Wang Y, Liu L, … Wang Y (2017). Abnormal resting-state functional connectivity of insular subregions and disrupted correlation with working memory in adults with attention deficit/hyperactivity disorder. Frontiers in Psychiatry, 8(OCT). 10.3389/fpsyt.2017.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.