Abstract
Alveolar macrophages (AMs) are the first cells to be infected during Mycobacterium tuberculosis (M.tb.) infection. Thus the AM response to infection is the first of many steps leading to initiation of the adaptive immune response, required for efficient control of infection. A hallmark of M.tb. infection is the slow initiation of the adaptive response, yet the mechanisms responsible for this are largely unknown. To study the initial AM response to infection, we developed a system to identify, sort and analyze M.tb.-infected AMs from the lung within the first 10 days of infection. In contrast to what has been previously described using in vitro systems, M.tb.-infected AMs up-regulate a cell-protective antioxidant transcriptional signature that is dependent on the lung environment but not bacterial virulence. Computational approaches including pathway analysis and transcription factor motif enrichment analysis identify NRF2 as a master regulator of the response. Using knock-out mouse models, we demonstrate that NRF2 drives expression of the cell protective signature in AMs and impairs the control of early bacterial growth. AMs up-regulate a significant pro-inflammatory response to M.tb. infection only 10 days following infection, yet comparisons with bystander AMs from the same infected animals demonstrate that M.tb.-infected AMs generate a less robust inflammatory response than the uninfected cells around them. Our findings demonstrate that the initial macrophage response to M.tb. in the lung is far less inflammatory than has previously been described by in vitro systems and may impede the overall host response to infection.
One Sentence Summary:
Induction of a NRF2-dependent cell protective signature impairs alveolar macrophages from controlling M.tb. infection in vivo.
Introduction
Infection with Mycobacterium tuberculosis (M.tb.) is now the leading cause of death by a single pathogen worldwide (1). Transmission of M.tb. generally occurs via aerosols expelled by cough from an infected person. Although inhaled M.tb. is rapidly engulfed by alveolar macrophages (AMs) in the lung (2), mobilization of a robust immune response is extremely slow and the lack of a speedy response is thought to contribute to disease progression (3). In animal models, recruitment of innate cells such as neutrophils and monocytes to the lung and T cell priming in the draining mediastinal lymph nodes do not occur until 11–14 days following infection, by which time the bacteria have replicated nearly 1,000-fold in the lung (2, 4–7). The mechanisms that restrain or subvert the host response in the lung immediately following infection remain largely unknown.
AMs are the first cells in the lung to be infected with M.tb. after aerosol transmission (2). AMs are the largest resident macrophage population in the lung and serve as pulmonary immune sentinels, constantly sampling the airway for foreign particles (8). In addition, AMs perform a critical homeostatic role clearing inhaled material from the airway and recycling pulmonary surfactant, which requires AMs to uncouple phagocytic function from inflammatory responses (9, 10). Due to their steady-state functions, AMs express unique transcriptional and epigenetic profiles that are highly distinct from those of other tissue-resident macrophages (11–13).
Similar to other macrophage populations that are constantly exposed to environmental stimuli, AMs express a number of inhibitory receptors that have been shown to dampen their responses. However, under conditions such as acute lung injury, AMs can become highly activated and release damaging levels of pro-inflammatory mediators that contribute to airway disease (8). During other pulmonary infections such as influenza, in which AMs serve as secondary or tertiary responders, AMs exhibit pro-inflammatory transcriptional responses and can serve as an immunotherapy target to limit inflammation (14, 15). However, during these infections the AMs are likely influenced by cytokines released into the pulmonary environment by other cell types.
Unlike most pulmonary infections, the initial infectious dose of M.tb. is very low and for the first several days AMs are the only cells productively infected with the bacteria (2). Therefore, M.tb. infection provides an opportunity to assess the ability of AMs to respond to direct intracellular infection rather than environmental cues. Results from two recent studies provide hints that the response of AMs to M.tb. might be suboptimal: At two weeks following infection, when M.tb. can be found in multiple cell populations, AMs are significantly less effective at controlling bacterial replication than interstitial macrophages, another macrophage subset found in the lung (16); and AMs were shown to facilitate relocalization of the bacteria from the airway into the lung interstitium (2).
Due to the paucity of M.tb.-infected AMs during the first few days following aerosol challenge, their presumed immediate response to the bacteria has been extrapolated from in vitro culture systems using models such as bone marrow-derived macrophages (BMDMs). These studies have identified numerous sensing pathways and cytokine responses including TNF, Type I IFN, and IL-1β (17–22) that have been shown to play crucial roles in host immunity in both animal and human studies (23). However, experiments using knock-out mice have demonstrated that these mediators do not affect the course of disease during the first week of infection in vivo (24–26), despite the fact that they are up-regulated within hours by M.tb.-infected macrophages in vitro.
In order to more precisely define the response of AMs to M.tb. in vivo, we developed an infection model and isolation procedure that yields sufficient numbers of infected AMs to perform global systems-level analyses at any point within the first 10 days following infection. Our results demonstrate that the rapid up-regulation of pro-inflammatory genes observed for macrophages responding to M.tb. in vitro is delayed in AMs infected in vivo, occurring days instead of hours following infection. Immediately following in vivo infection, AMs up-regulate a cell-protective antioxidant transcriptional program regulated by the transcription factor, NRF2. Activation of this program is dependent on the lung microenvironment. In the absence of NRF2, AMs have an enhanced ability to control early bacterial growth.
Results
Alveolar macrophages provide a replication niche for M.tb. through the first 10 days of infection
In a standard low-dose M.tb. aerosol challenge model, mice are infected with ~100 CFU, which, due to the low replication rate of the bacteria, does not allow for the isolation of sufficient numbers of M.tb.-infected cells from a reasonable number of animals for analysis. Therefore, we developed a high-dose model in which mice are infected with 2 – 4 × 103 CFU of an mEmerald-expressing H37Rv strain of M.tb. Using this model, we found that over the first two weeks the vast majority of infected cells early on were AMs with significant numbers of infected neutrophils (PMN) and monocytes-derived macrophages (MDMs) appearing between day 10 and 14 (Fig. 1A, fig. S1, S2, table S1). The localization of the bacteria and the timing of recruitment of innate immune cells in the high-dose model is consistent with other studies that have used a low-dose model (2, 5), suggesting that the higher dose does not alter the initial immune response in the lung. At this dose, ~0.5–2% of AMs were infected (Fig. 1B) and the total number of M.tb.-infected AMs did not change significantly over the first 2 weeks (Fig. 1C). By day 14, the numbers of infected neutrophils and AMs were equivalent.
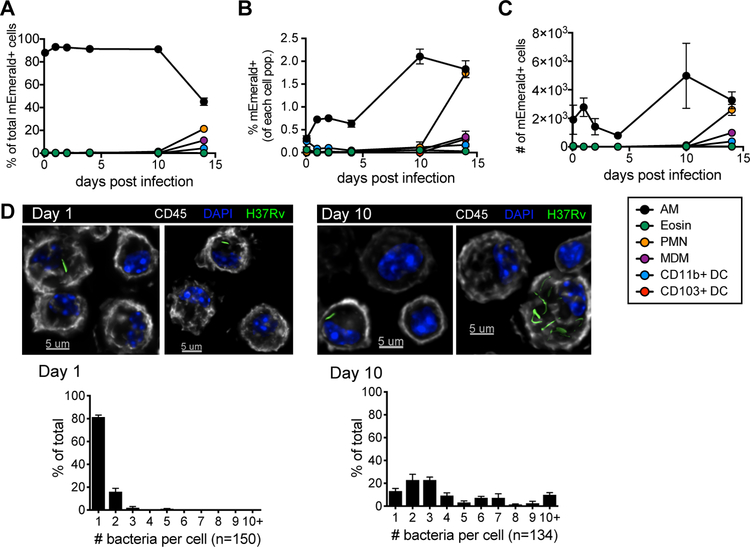
Figure 1: Alveolar macrophages provide a replication niche for M.tb. through the first 10 days of infection.
(A) % of total mEmerald+ cells, (B) % mEmerald+ for each cell population, and (C) total number of mEmerald+ cells in the lung between 2 hours and 14 days after high dose aerosol infection with mEmerald-tagged H37Rv (n = 3 mice/time point). (D) Microscopy of BAL samples 1 and 10 days after high dose aerosol infection with mEmerald-tagged H37Rv and quantitation of bacteria per AM (n = 3 replicates/time point, each replicate was pooled from 3 mice). Abbreviations: AM = alveolar macrophages, PMN = neutrophils, Eosin = eosinophils, MDM = myeloid-derived macrophages, DC = dendritic cells. Data are presented as mean ± SEM. Data is representative of 2 independent experiments.
We found that M.tb.-infected AMs were enriched within bronchoalveolar lavage (BAL) samples compared to the total lung (fig. S3) and used high-throughput microscopy to analyze BAL samples from mice following high-dose infection with mEmerald-H37Rv to quantify the number of bacteria in each infected cell. At 1 day post-infection, 81.3 ± 1.8% (mean ± SEM) of infected AMs contained a single bacillus; by day 10 only 13.2 ± 2.3% (mean ± SEM) of AMs contained a single bacillus, with 58.4 ± 6.1% of AMs containing 2–5 bacilli and 28.4 ± 3.8% containing more than 5 bacilli (Fig 1D). These data demonstrate that AM serve as a replication niche for M.tb. through the first 10 days infection, even in the setting of a high dose infection, an observation that has been similarly described at 1 and 2 weeks following infection in other studies (2, 16).
M.tb.-infected alveolar macrophages up-regulate an NRF2-associated cell protective response in vivo
To characterize the early macrophage transcriptional response to M.tb. infection, we infected mice with a high dose of mEmerald-H37Rv, and isolated AMs by expression of both Siglec F and CD64 (27, 28) from BAL by fluorescence activated cell sorting (FACS) 24 hours following infection (fig S3, S4). Isolating AMs by BAL allowed the samples to be kept on ice throughout processing and sorting, eliminating the need for lengthy and harsh digestion of lung tissue. Three populations were sorted for analysis by RNA-seq: M.tb.-infected AMs (mEmerald+), bystander AMs (mEmerald−), and AMs from naïve mice (Fig 2A).
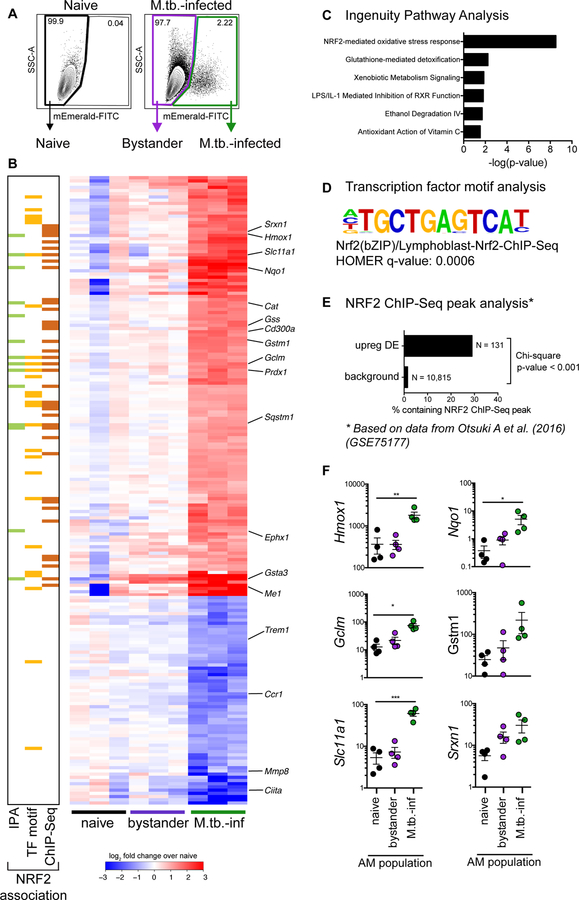
Figure 2: M.tb.-infected alveolar macrophages up-regulate an NRF2-associated antioxidant gene signature.
(A) Gating scheme to sort naïve, bystander, and M.tb.-infected AMs from bronchoalveolar lavage (BAL) samples after high dose aerosol infection with mEmerald-tagged H37Rv. (B) Heatmap of gene expression (log2 fold change over average of naïve AMs) for 196 differentially expressed genes between naïve and M.tb.-infected AMs (Filtering criteria: average CPM >1, |fold change| > 2 and FDR < 0.01, Benjamini-Hochberg calculated). Columns are independent experiments (pooled mice) and rows are genes. Genes called out are known NRF2 target genes of interest as well as downregulated pro-inflammatory genes. Colored bars to the left indicate NRF2 association as determined by 3 different methods: (C) Ingenuity Pathway Analysis, (D) transcription factor promoter binding motif enrichment analysis (HOMER), and (E) ChIP-seq peak analysis. (F) RT-qPCR validation of NRF2 associated genes for naïve, bystander and M.tb.-infected AMs 24 hours post-infection. Values are relative to Ef1a. Data is presented from 3 independent experiments with one-way ANOVA with Dunnett’s post-test *p < 0.05, **p < 0.01, ***p < 0.001.
At 24 hours post-infection, we identified 196 genes that were differentially expressed (DE) (average counts per million (CPM) > 1, |fold change| > 2, FDR < 0.01) between M.tb.-infected and naïve AMs (Fig. 2B, table S3). No genes were DE between bystander and naïve AMs at this stringency, indicating an absence of systemic changes to the lung 24 hours after infection.
To identify enriched pathways and potential transcriptional regulators, we analyzed the up-regulated (131) and down-regulated (65) genes independently using three complementary computational approaches. Ingenuity Pathway Analysis (IPA) revealed that the up-regulated genes were significantly enriched for the “NRF2-mediated oxidative stress response” pathway (p-value = 10−11.3) (Fig. 2B (green squares), Fig. 2C). This pathway includes genes involved in a number of cell protective functions including: antioxidant production (Nqo1, Cat, Prdx1, Txnrd1), iron metabolism (Hmox1), and glutathione metabolism (Gstm1, Gclm, Gsta3) (29). We also searched for enriched transcription factor promoter binding motifs using HOMER (Hypergeometric Optimization of Motif EnRichment) (30). The top four enriched transcription factor binding motifs in the 131 up-regulated genes (q-value = 0.0006) were motifs for either NRF2 or BACH1, which are known to compete for the same binding sites (31) (Fig. 2B (yellow squares), Fig. 2D). To validate these results, we used a published NRF2 ChIP-Seq analysis of peritoneal macrophages that were stimulated with the NRF2 agonist diethyl maleate (GSE75177) (32). 29% of the 131 up-regulated genes contained an NRF2 ChIP-Seq peak, compared to 1.5% of background genes. The difference in proportions is highly significant, χ2 (1, N = 10,946) = 531.21, p-value < 2.2e−16) (Fig. 2B, (orange squares), Fig. 2E). We validated the up-regulation of several of the NRF2-associated genes by qPCR (Fig. 2F). These results demonstrate that the transcription factor NRF2 is associated with the up-regulated transcriptional signature in M.tb.-infected AMs 24 hours after infection in vivo. A similar analysis of the down-regulated genes did not uncover any significantly enriched pathways or candidate transcriptional regulators.
M.tb.-infected alveolar macrophages do not immediately up-regulate classical pro-inflammatory genes
We examined whether, in addition to NRF2-associated genes, M.tb.-infected AMs up-regulated classical pro-inflammatory genes that have been shown to be expressed by various macrophage subsets in response to M.tb. including cytokines and chemokines (e.g. Tnf, Il1b, Cxcl9) (23), pro-inflammatory receptors, costimulatory molecules, and Fc receptors. Very few of these genes were significantly up-regulated by M.tb.-infected AMs (fig. S5). In contrast, several genes involved in glycolytic metabolism (Hif1a, Pkm, Aldoa) were significantly up-regulated by M.tb.-infected AMs (33). While previous studies have shown that at 2 weeks following infection M.tb.-infected AMs have relatively higher expression of genes involved in fatty acid oxidation and lower expression of genes involved in glycolysis compared to M.tb.-infected IMs (16), our data demonstrate that M.tb.-infected AMs do up-regulate genes involved in glycolysis relative to their naïve state.
Virulent M.tb. is not required to activate the NRF2-associated signature generated by alveolar macrophages
To determine whether the NRF2-signature was specifically induced by virulent M.tb., we repeated the high-dose infection using an mEmerald-expressing avirulent strain of M.tb. lacking the RD1 virulence locus (ΔRD1-H37Rv) (fig S6A). The ΔRD1 strain lacks a functional ESX-1 secretion system, which has been shown to contribute to pathogenicity in vivo (34–36). Macrophage in vitro studies have demonstrated that the RD1 region is required for M.tb.-mediated activation of host cytosolic receptors and production of type I interferon (17, 37) as well inflammasome activation and IL-1β production (38, 39). As an additional control, we exposed mice to fluorescent 1μm carboxylate-coated latex beads by aerosol treatment (fig S6B). To confirm the beads were not contaminated with TLR-ligands, BMDMs were treated with the beads for 8 hours and gene expression changes were measured. No significant transcriptional changes in pro-inflammatory or NRF2-associated genes were observed in vitro (fig S7).
Both the ΔRD1-infected and bead-positive AMs displayed up-regulation of NRF2-associated genes at 24 hours, although the magnitude of the fold change for some genes was smaller than for H37Rv-infected AMs (Fig. 3A, table S3). Unlike in vitro models that have shown a role for the ESX-1 locus, contained within RD1, in regulating the type I IFN responses of macrophages to M.tb. (17, 37, 40), there were very few significant differences between the transcriptional responses of H37Rv vs ΔRD1-infected AMs 24 hours after infection (Fig. 3B). The NRF2-mediated oxidative stress response was the most highly enriched pathway in ΔRD1-infected AMs by IPA (table S2). Based on the stringencies imposed on the 24 hour H37Rv-infected AMs, no genes were significantly changed in the bead-positive AMs and similarly no pathways were enriched by IPA (Fig. 3C, table S2). However, it is worth noting that within the most changed genes (with filtering criteria of FDR < 0.05, fold change > 1.5) NRF2-associated genes showed the greatest enrichment. Overall, these data demonstrate that AMs up-regulate the NRF2-associated signature as a response to virulent bacteria, avirulent bacteria, or inert beads, suggesting that it is a more general response by AMs to the uptake of particles. While it is not surprising that particle uptake leads to transcriptional changes, it is notable that infection with a virulent pathogen appears to induce no additional host response within the first day of infection, suggesting that classical pathogen sensing is deficient, inhibited, or delayed in AMs.
Figure 3: Up-regulation of the NRF2-associated signature does not require virulent M.tb. infection.
(A) Heatmap of log2 fold change gene expression over average of naïve AMs for H37Rv-infected, ΔRD1-infected, and bead-positive AMs 24 hours after treatment. Columns represent averages of 3 independent experiments. Rows represent 196 DE genes described in Figure 2. Colored bars to the left indicate NRF2 association as described in Figure 2. (B, C) Scatterplots comparing gene expression values (log2 fold change over average of naïve AMs) for H37Rv-infected versus ΔRD1-infected AMs (B) or H37Rv-infected versus bead-positive AMs (C) 24 hours post-infection with significant differentially expressed genes highlighted (average CPM >1, |fold change| > 2 and FDR < 0.01, Benjamini-Hochberg calculated). Data is presented from 3 independent experiments.
M.tb.-infected alveolar macrophages exhibit both NRF2-associated and pro-inflammatory responses 10 days after infection
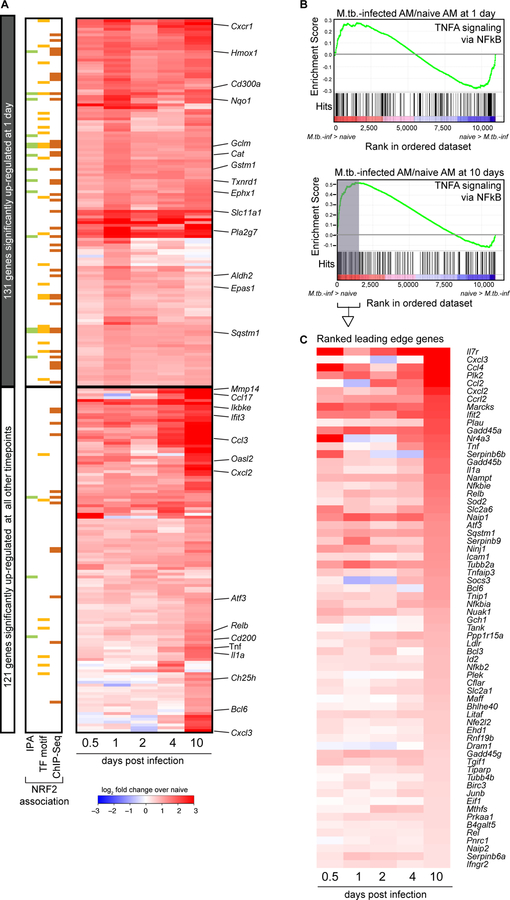
To characterize the kinetics of the AM response to M.tb. infection, we extended our transcriptional analysis to include 4 additional time points: 0.5 (12 hours), 2, 4, and 10 days post-infection. We identified 288 genes that were significantly up-regulated at one or more of these time points compared to naïve AMs (Fig. 4A, table S3). Many of the 131 genes up-regulated at 24 hours and associated with NRF2 by either IPA, transcription factor motif analysis or ChIP-Seq analysis showed sustained expression through 10 days of infection (Fig. 4A, top). Furthermore, by IPA, the “NRF2-mediated oxidative stress response” pathway was the most highly enriched pathway at all 5 timepoints (p-values: 10−5.3−10−10.1) (table S2). HOMER analysis also pinpointed a NRF2 motif as the most enriched transcription factor motif at 2 4, and 10 days, and identified no enriched motif at 0.5 days (table S2).
Figure 4: M.tb.-infected alveolar macrophages induce both NRF2-associated and pro-inflammatory responses 10 days after infection.
(A) Heatmap of gene expression (log2 fold change over naïve AMs) for 252 genes up-regulated in M.tb.-infected AMs compared to naïve AMs for at least one out of five time points (Filtering criteria: average CPM >1, |fold change| > 2 and FDR < 0.01, Benjamini-Hochberg calculated). Top 131 genes are significantly up-regulated in M.tb.-infected AMs at 1 day post-infection. Bottom 121 genes are not significantly up-regulated in M.tb.-infected AMs at 1 day post-infection. Colored bars indicate NRF2 association as described in Figure 2. (B, C) Gene set enrichment analysis and top 50 ranked leading edge genes in the “TNFA signaling via NFkB” pathway for M.tb.-infected AMs at 10 days post-infection. Data is presented from 3 independent experiments per time point.
The kinetic analysis also identified a late pro-inflammatory response up-regulated in M.tb.-infected AMs primarily at 10 days post-infection (Fig. 4A, bottom). At 10 days post-infection, M.tb.-infected AMs displayed significant up-regulation of genes in the TNFA signaling via NFκB Pathway as determined by Gene Set Enrichment Analysis (GSEA) compared to naïve AMs (NES = 1.8, FDR = 0.0013), a pathway not significantly enriched in M.tb.-infected AMs 1 day following infection (Fig. 4B). The ranked leading edge genes in this pathway reveal the changes in gene expression over time, including increases in expression of: Il1a, Tnf, Rel, Relb, NFkb2, Ccl2, Bhlhe40, and Nfe2l2 (Fig. 4C). A number of pro-inflammatory cytokine and chemokine genes (Tnf, Il1a, Cxcl2, Cxcl3, Ccl17) are significantly up-regulated in M.tb.-infected AMs only 10 days after infection (fig S8).
Bystander AMs express a pro-inflammatory transcriptional signature 10 days after infection
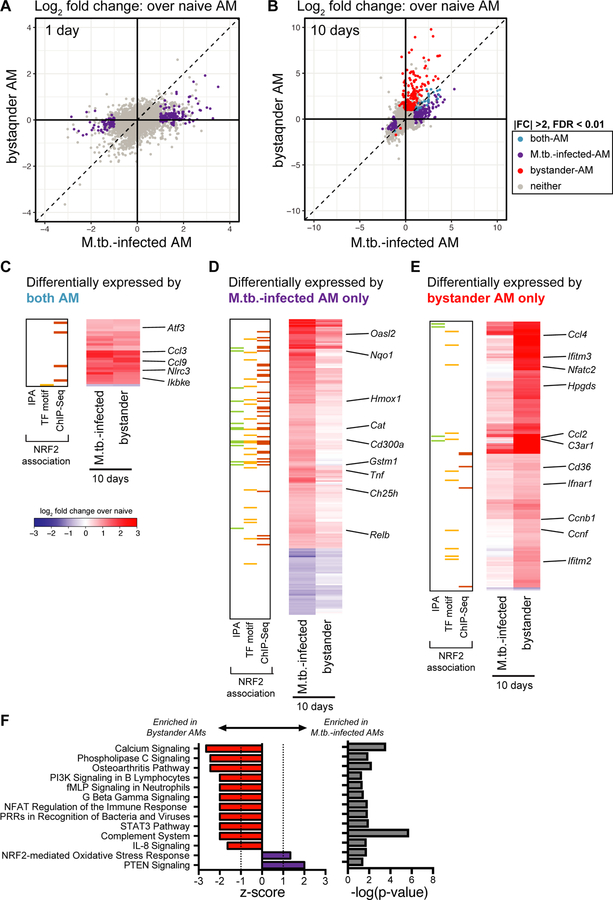
To disentangle the responses to intracellular infection from the responses to the inflammatory milieu in the lung, we analyzed the transcriptomes of the bystander AMs (uninfected cells from infected animals) at 1 and 10 days post-infection. As described above, bystander AMs displayed no significant gene expression changes compared to naïve AMs at 1 day after infection (Fig. 5A). In contrast, by 10 days after infection bystander AMs showed abundant changes in gene expression with a total of 205 significantly changed genes (Fig. 5B). 28 of these genes (highlighted in blue) were shared with the M.tb.-infected AMs (Fig. 5C), while 177 of them (highlighted in red) were uniquely changed in bystander but not in M.tb.-infected AMs (Fig. 5E). Another 200 genes (highlighted in purple) were differentially expressed only in M.tb.-infected AMs (Fig. 5D). Comparison between these sets of genes demonstrates that expression changes found only in M.tb.-infected AMs are enriched for NRF2-associated genes (Fig 5D), while expression changes found only in bystander AMs are enriched for inflammatory pathways (Fig 5E). These differences are confirmed by IPA. While NRF2-mediated oxidative stress response and PTEN signaling were more highly enriched in M.tb.-infected AMs compared to bystander AMs, bystander AMs showed differential expression for genes in a number of other pathways including calcium signaling, NFAT regulation of the immune response, role of pattern recognition receptors in recognition of bacteria and viruses, and STAT3 pathway (Fig. 5F). Enrichment of these pathways indicate that some of the inflammatory signals received by bystander cells are either blocked or integrated differently in infected cells 10 days after infection. Overall, these data suggest that over the first week and a half AMs respond directly to infection as well as to systemic changes in the lung environment and that these two signals may cross-regulate.
Figure 5: Bystander AMs express a unique transcriptional signature 10 days after infection.
(A, B) Scatterplots comparing gene expression values (log2 fold change over average of naïve AMs) for H37Rv-infected versus bystander AMs at 1 day (A) and 10 days (B) post-infection with significant differentially expressed genes highlighted (|fold change| > 2 and FDR < 0.01, Benjamini-Hochberg calculated). Data is presented from 3 independent experiments. (C, D, E) Heatmaps of log2 fold change gene expression at 10 days over average of naïve AMs. Colored bars indicate NRF2 association as described in Figure 2. (C) 28 genes differentially expressed by both bystander and M.tb.-infected AMs. (D) 200 genes differentially expressed only by M.tb.-infected AMs. (E) 177 genes differentially expressed only by bystander AMs. Columns represent the average of three independent experiments. Genes of interest noted to the right. (F) Ingenuity Pathway Analysis comparing gene expression from bystander AMs and M.tb.-infected AMs. Canonical pathways with |z-scores| >1 and p-values < 0.05 were reported.
Both cell-intrinsic and environmental factors shape the alveolar macrophage response to M.tb.
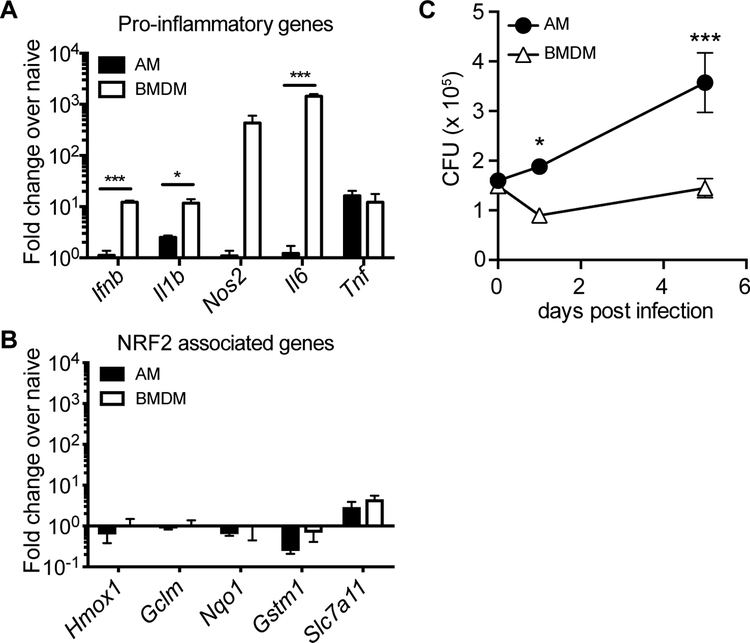
To determine whether the AM response to M.tb. is cell-intrinsic or environment-dependent, we compared the response of AMs infected in vitro to the in vivo measurements described above. We isolated AMs by BAL from naïve WT mice, allowed them to adhere for 18 hours, infected them with H37Rv, and measured their transcriptional response and ability to control bacterial growth. In parallel, we performed identical experiments with BMDMs, which have been used extensively, including by our group, to investigate how macrophages respond to M.tb. (41). Similar to their response to M.tb. infection in vivo, AMs displayed little to no increase in pro-inflammatory gene expression (including Il1b, Il6 and Nos2) after infection in vitro, while BMDMs robustly up-regulated these genes (Fig. 6A). One notable exception was Tnf, which was significantly up-regulated by AMs in response to H37Rv infection in vitro. In contrast to AMs infected in vivo, neither AMs nor BMDMs infected in vitro up-regulated NRF2-associated genes (Fig. 6B). Similar gene expression results were observed for AMs cultured with or without GM-CSF (fig. S9) and at 12 or 24 hour time points following infection, matching the time points measured in vivo (fig. S10). Overall, AMs were more permissive to bacterial growth than BMDMs, leading to a significant increase in bacterial burden as measured by CFU 5 days after infection (Fig. 6C). These results suggest that the inability of AMs to up-regulate pro-inflammatory genes in response to intracellular infection is cell-intrinsic, while the up-regulation of the NRF2-associated pathway is dependent on signals from the lung microenvironment.
Figure 6: The alveolar macrophage response to M.tb. is driven by both cell type and environment.
In vitro H37Rv infection of AMs and BMDMs. (A) RT-qPCR gene expression analysis of pro-inflammatory genes 8 hours post-infection. (B) RT-qPCR analysis of NRF2-associated genes 8 hours post-infection. (C) Colony forming unit (CFU) assay to measure bacterial burden in each cell type over 5 days. Data is representative of 3 independent experiments with three technical replicates each. Multiple t-tests with Holm-Sidak correction. *p<0.05, ***p < 0.001
Expression of NRF2 impairs the ability of AMs to control bacterial growth
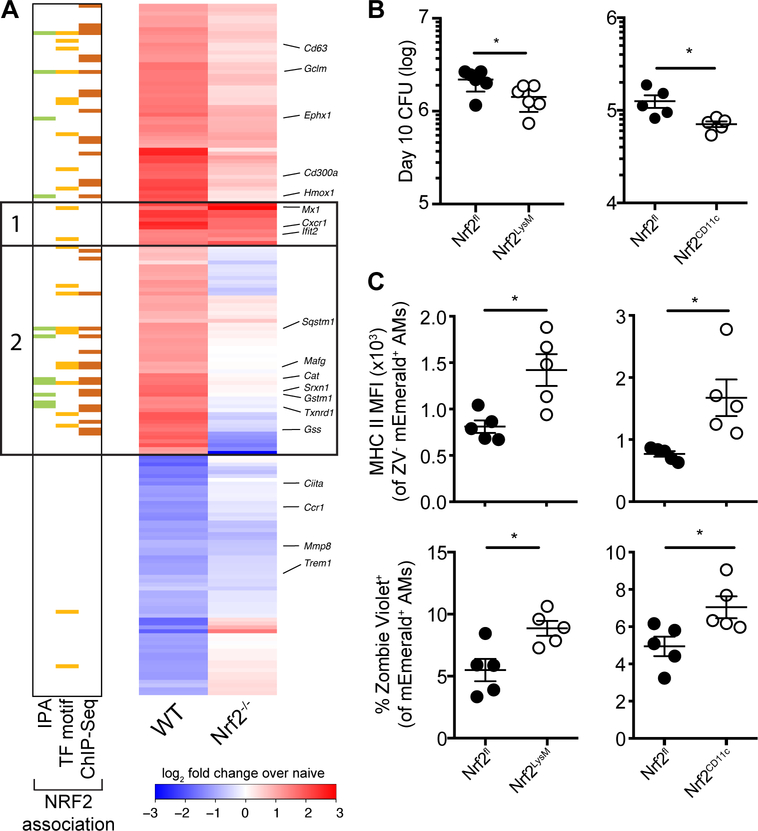
Our computational analyses identified NRF2 as a potential regulator of the in vivo AM response to M.tb.-infection. To test this prediction, we isolated naïve and M.tb.-infected AMs from Nrf2−/− mice 24 hours after high dose infection with mEmerald-H37Rv and performed RNA-sequencing. Deletion of NRF2 altered the expression of only a small number genes in the basal transcriptional profile of AMs (only 11 genes were DE (|fold change| > 2, FDR < 0.05) including Nfe2l2, the gene encoding NRF2, and several known NRF2 targets (Srxn1, Hmox1, Slc11a1)) (fig S11, table S4). In contrast, the response of Nrf2−/− AMs to M.tb. infection was strongly attenuated compared to that of WT AMs and many of the genes exhibiting altered responses were associated with NRF2 by IPA, transcription factor motif analysis, or ChIP-Seq analysis (Fig. 7A, group 2, table S4). The set of genes that responded to M.tb. infection in Nrf2−/− AM was completely contained within the set of 196 genes that responded to M.tb.-infection in WT AMs (|fold change| > 2, FDR < 0.01) suggesting that NRF2 does not act to restrain the transcriptional response in this setting, as has been reported previously (42). Notably, there was a small set of pro-inflammatory genes (i.e. Mx1, Cxcr1, Ifit2) that were up-regulated by both WT and Nrf2−/− AM (Fig. 7A, group 1).
Figure 7: Modulation of NRF2 activity alters macrophage response and control of M.tb.
(A) Heatmap of gene expression (log2 fold change over average of respective naïve AMs) for WT (first column) and Nrf2−/− (second column) M.tb.-infected AMs, averaged from at least two independent experiments. Rows depict all genes that are differentially expressed between naïve and M.tb. infected WT AMs, as shown in Fig 2B (181 genes expressed out of original 196 gene list). Groups 1 and 2 are defined in the text. Colored bars indicate NRF2 association as described in Figure 2. (B) Lung bacterial burden measured by CFU assay at 10 days post-infection with low dose H37Rv from Nrf2LysM, Nrf2CD11c and their respective Nrf2fl littermate controls. (C) MHC II MFI of Zombie Violet− M.tb.-infected AMs (top) and % dead (Zombie Violet+) of M.tb.-infected AMs (bottom) as measured by flow cytometry at 10 days post-infection with high dose mEmerald-H37Rv from Nrf2LysM, Nrf2CD11c and their respective Nrf2fl littermate controls. Data is presented from 2 independent experiments (A) or representative of 2 independent experiments with 5 mice/group (B-C). Two-tailed unpaired Student’s t-test * p< 0.05, ** p < 0.01.
NRF2 functions as a master regulator of an antioxidant stress response and likely plays critical roles in many cell types. Flow cytometric analysis of the lungs of naïve Nrf2−/− mice showed altered MHC II expression in AM and DCs, decreased numbers of MDMs, and an increased percentage of CD8+ T cells (fig S12); therefore, we chose to evaluate the functional role of NRF2 in AMs during M.tb. infection by generating NRF2 conditional knockout mice. While there is no model for AM-specific gene deletion, we utilized both LysMcre/+ and CD11ccre/+ strains that delete floxed genes in either macrophages and neutrophils or AMs and dendritic cells, respectively (43), in combination with an Nrf2floxed/floxed strain. These breedings generated the following conditional knockout models: Nrf2floxed/floxed; LysMcre/+ (Nrf2LysM) and Nrf2floxed/floxed; CD11ccre/+ (Nrf2CD11c). We confirmed the deletion of Nrf2 transcript and the absence of NRF2 protein in AMs in both strains by sequencing and Western Blot (fig S13), as previously described (44, 45). Analysis of myeloid cell number and activation, T cell frequencies and activation, and pro-inflammatory cytokine protein expression identified only one significant difference between naïve lungs of Nrf2CD11c and Nrf2fl controls: increased MHC II expression in AMs lacking NRF2 expression. There was a similar trend for AMs from Nrf2LysM mice compared Nrf2fl littermate controls, but it didn’t reach significance (fig S14). We repeated the transcriptional profiling on M.tb.-infected AMs from Nrf2LysM, Nrf2CD11c, and Nrf2fl littermate controls and found similar differences in gene expression compared to what had been observed between M.tb.-infected WT and Nrf2−/− AMs (fig S15, table S5). We examined the bacterial burden in these mice 10 days following infection, the latest time point when the vast majority of bacteria are still within AMs and found that both conditional knock-out models had lower bacterial burdens than their Nrf2fl littermate controls (Fig. 7B). M.tb.-infected AMs lacking NRF2 expression had higher levels of MHC II surface expression, reflecting the differences observed at baseline, and were more prone to cell death as measured by increased staining with viability dye Zombie Violet (Fig. 7C). The differences in cell viability were limited to M.tb.-infected cells and not observed in bystander cells (fig. S16). These results suggest that NRF2 may impede host control by limiting baseline AM activation and by blocking cell death of M.tb.-infected AMs, which could allow the bacteria more time to replicate in a permissive cell type and prevent M.tb. from being taken up by cells with more bactericidal capacity.
Discussion
Here we describe an NRF2-driven transcriptional signature generated by M.tb.-infected AMs in vivo. This innate response to M.tb. is very different from what has been previously observed in macrophage infection studies, including our own, that have shown M.tb.-infected macrophages in vitro up-regulate pro-inflammatory genes primarily driven by NF-κB and type I IFN signaling (17, 19, 37, 41). This dissimilarity is somewhat unexpected given that AMs express of almost all of the canonical pathogen-sensing machinery thought to be required to detect M.tb. infection (46). Our data suggest that the initial host response to M.tb. infection is relatively anti-inflammatory and may contribute to the extended time required for antigen transport to the draining lymph node and subsequent activation of the adaptive immune response. These results complement previous studies have that shown that the earliest events in the lung, particularly the interactions between macrophages and M.tb., have a major impact on subsequent disease outcome (2, 4, 16, 47–49). While NRF2 and several of its target genes, notably Hmox1, have been shown to be up-regulated during chronic M.tb. infection (50–52), our results suggest that the early NRF2 signature in AMs is a more general response to phagocytic activity rather than to bacterial infection. In the context of a meta-analysis of 32 studies covering 77 different host-pathogen interactions, this NRF2-driven transcriptional response falls far outside the norm (53). Our results highlight the importance of using in vivo systems to dissect lung-specific immunological events and suggest that established in vitro models do not adequately replicate the initial host response to M.tb. (54).
Studies using Nrf2−/− mice have demonstrated both protective (S. pneumoniae, S. aureus, RSV, and P. aeruginosa) and harmful (H. influenzae, Marburg virus) roles for NRF2 during infection (55–61). To evaluate the functional role of NRF2 in AMs during M.tb. infection, we chose to use two conditional knockout models (LysMcre and CD11ccre). One caveat is that cre expression by these promoters also deletes NRF2 in either neutrophils and MDMs (LysM) or dendritic cells (CD11c). During the first 10 days of infection when M.tb. almost exclusively infects AMs, this approach allows us to interrogate the direct role of NRF2 in M.tb.-infected AMs. Conversely, beyond 10 days, significant numbers of neutrophils, MDMs and dendritic cells begin to participate in the response making it impossible to link changes in the outcome of infection at later times to AM-specific NRF2 expression. In addition to infection, NRF2 can be activated by lung injury caused by noncommunicable conditions such as chronic obstructive pulmonary disease (COPD), cigarette smoking, cystic fibrosis, and exposure to air pollution (62, 63). The finding that NRF2 expression hinders AMs from controlling intracellular infection suggests an intriguing mechanism by which smoking and indoor air pollution may contribute to TB risk (64, 65).
The finding that AMs do not initially mount an inflammatory response to M.tb. infection is supported by bacterial transcriptional profiling we performed in collaboration on M.tb. residing within infected AMs in vivo using a novel bead-based method, Path-Seq, to enrich for bacterial transcripts (66). These studies by Peterson et al show that unlike infection of BMDMs in vitro, which causes M.tb. to dynamically regulate mycolic acid biosynthesis and increase expression of environmental stress regulators such as DosR, infection of AMs leads to fewer changes to the stress-related networks in the bacterial transcriptome. Targeting AMs may be one of M.tb.’s most important virulence strategies. To spread effectively following aerosol transmission of small numbers of bacteria, M.tb. targets a macrophage population that does not activate classical innate inflammatory pathways and instead responds by safeguarding against cellular damage. Our findings provide one explanation for the “macrophage paradox”, coined by Vance and Price, in which intracellular pathogens often infect the very cell type bestowed with effector functions to control their growth (67).
AMs express a number of inhibitory receptors at baseline that are thought to contribute to their immunoregulatory phenotype (8). We don’t yet understand how these other pathways might impact the AM response to M.tb. or provide redundancy with the NRF2 pathway. Our data suggest that M.tb.-infected AMs undergo a shift towards an increasingly pro-inflammatory phenotype over the first 10 days of infection, but the underlying mechanisms and the subsequent events remain unknown. Is this change cell-intrinsic or dependent on uptake and recognition of bacteria by other cell types following AM cell death? Similarly, we don’t yet understand why a portion of the pro-inflammatory program expressed by bystander AMs 10 days following infection is absent in infected AMs. Are there additional inhibitory mechanisms apart from NRF2 that specifically restrain M.tb.-infected AMs? Finally, we don’t yet know how human AMs respond to M.tb. upon initial infection in vivo. These studies were performed using mice living in specific-pathogen free facilities that are exposed to a very limited set of environmental factors, while humans are continuously exposed to a variety of airborne stimuli and pathogens that may alter the baseline state of AMs. How pre-exposure to different stimuli impacts human AM function, and therefore TB susceptibility, is not yet understood. Further investigation into the intervening events between initial AM infection and the adaptive T cell response as well as how the AM response to M.tb. can be modified by environmental exposure may facilitate a better understanding of the early events of TB infection and inform future TB vaccine strategies.
Study Design
The aim of this study was to measure the immune response of AMs to intracellular M.tb. infection in vivo. We characterized the transcriptional profile of murine M.tb.-infected alveolar macrophages after aerosol infection by sorting cells and performing low-input library amplification, followed by RNA-sequencing. To determine the effect of the transcription factor NRF2 on the response of AMs, we generated NRF2 conditional knock-out mouse models (Nrf2LysM and Nrf2CD11c) and tested their responses against Nrf2fl littermate controls. The number of replicates for each experiment is indicated in the figure caption. No animals were excluded in these studies.
Materials and Methods
Mice
C57BL/6 and Nrf2−/− (B6.129X1-Nfe2l2tm1Ywk/J) mice were purchased from Jackson Laboratories (Bar Harbor, ME). Nrf2floxed (C57BL/6-Nfe2l2tm1.1Sred/SbisJ), CD11ccre (B6.Cg-Tg(Itgax-cre)1–1Reiz/J) and LysMcre (B6.129P2-Lyz2tm1(cre)Ifo/J) were purchased from Jackson Laboratories and bred to generate Nrf2CD11c and Nrf2LysM mice. Mice were housed and maintained in specific pathogen-free conditions at Seattle Children’s Research Institute and experiments were performed in compliance with the Institutional Animal Care and Use Committee. 6–12 week old male and female mice were used for all experiments, except for RNA-sequencing, which used only female mice for uniformity. Mice infected with M.tb. were housed in a Biosafety Level 3 facility in an Animal Biohazard Containment Suite.
M. tuberculosis Aerosol Infections and Lung Mononuclear Cell Isolation
Most aerosol infections were performed with a stock of wildtype H37Rv transformed with an mEmerald reporter pMV261 plasmid, generously provided by Dr. Chris Sassetti and Christina Baer (University of Massachusetts Medical School, Worcester, MA). Some infections used an M.tb. strain with a deletion of the virulence determinant RD1 region (ΔRD1), provided by Dr. David Sherman (SCRI, Seattle, WA), and transformed with the same mEmerald expression plasmid. For both standard (~100 CFU) and high dose (~2,000–4,000 CFU) infections, mice were enclosed in an aerosol infection chamber (Glas-Col) and frozen stocks of bacteria were thawed and placed inside the associated nebulizer. To determine the infectious dose, three mice in each infection were sacrificed one day later and lung homogenates were plated onto 7H10 plates for CFU enumeration, as previously described (68).
Bead aerosolization
Carboxylate 1.0 μm fluorescent beads (ThermoFisher) were aerosolized using a LC Sprint Reusable Nebulizer (PARI) attached to a vacuum pump and an air flow regulator as described previously (69).
Lung Single Cell Suspensions
At each time point, lungs were removed and single-cell suspensions of lung mononuclear cells were prepared by Liberase Blendzyme 3 (70 ug/mL, Roche) digestion containing DNaseI (30 μg/ml; Sigma-Aldrich) for 30 mins at 37°C and mechanical disruption using a gentleMACS dissociator (Miltenyi Biotec), followed by filtering through a 70 μm cell strainer. Cells were resuspended in FACS buffer (PBS, 1% FBS, and 0.1% NaN3) prior to staining for flow cytometry.
Alveolar Macrophage Isolation
Bronchoalveolar lavage was performed by exposing the trachea of euthanized mice, puncturing the trachea with Vannas Micro Scissors (VWR) and injecting 1 mL PBS using a 20G-1” IV catheter (McKesson) connected to a 1 mL syringe. The PBS was flushed into the lung and then aspirated three times and the recovered fluid was placed on ice. The wash was repeated 3 additional times. Cells were filtered and spun down. For antibody staining, cells were suspended in FACS buffer. For cell culture, cells were plated at a density of 1 × 105 cells/well (96-well plate) in complete RPMI (RPMI plus FBS (10%, VWR), L-glutamine (2mM, Invitrogen), and Penicillin-Streptomycin (100 U/ml; Invitrogen) with or without GM-CSF (10 ng/ml, Peprotech). and allowed to adhere overnight in a 37°C humidified incubator (5% CO2). Media with antibiotics was washed out prior to infection.
Cell Sorting and Flow Cytometry
Fc receptors were blocked with anti-CD16/32 (2.4G2, BD Pharmingen). Cell viability was assessed using Zombie Violet dye (Biolegend). Cells were suspended in 1X PBS (pH 7.4) containing 0.01% NaN3 and 1% fetal bovine serum. Surface staining included antibodies specific for murine: Siglec F (E50–2440, BD Pharmingen), CD11b (M1/70), CD64 (X54–5/7.1), CD45 (104), CD3 (17A2, eBiosciences), CD4 (RM4–4), CD8 (53–6.7, BD Biosciences), CD19 (1D3, eBiosciences), CD11c (N418), CD103 (M290, BD Biosciences), (I-A/I-E (M5/114.15.2), and Ly6G (1A8) (reagents from Biolegend unless otherwise noted). Cell numbers were enumerated using Polybead Polystyrene 15.0 um Microspheres (Polysciences). Cell sorting was performed on a FACS Aria (BD Biosciences). Sorted cells were collected in complete media, spun down, resuspended in Trizol, and frozen at −80°C overnight prior to RNA isolation. Samples for flow cytometry were fixed in 2% paraformaldehyde solution in PBS and analyzed using a LSRII flow cytometer (BD Biosciences) and FlowJo software (Tree Star, Inc.).
RNA-sequencing and Analysis
RNA isolation was performed using TRIzol (Invitrogen), two sequential chloroform extractions, Glycoblue carrier (Thermo Fisher), isopropanol precipitation, and washes with 75% ethanol. RNA was quantified with the Bioanalyzer RNA 6000 Pico Kit (Agilent). Due to the low number of M.tb.-infected cells recovered (~2,000–4,000 cells total after pooling BAL from 10–12 mice), all cDNA libraries were constructed and amplified using the SMARTer Stranded Total RNA-Seq Kit (v1 or v2) - Pico Input Mammalian (Clontech) per the manufacturer’s instructions. Sequencing was performed on an Illumina NextSeq (2 × 75, paired-end). Stranded paired-end reads of length 76 were preprocessed: The first three nucleotides of R1 (v1 kit) or R2 (v2 kit) were removed as described in the SMARTer Stranded Total RNA-Seq Kit User Manual (v1: 112215, v2: 063017) and read ends consisting of 50 or more of the same nucleotide were removed. The remaining read pairs were aligned to the mouse (mm10) and M.tb. H37Rv genome using the gsnap aligner (v. 2016-08-24) allowing for novel splicing. Concordantly mapping read pairs (average 15-million / sample) that aligned uniquely were assigned to exons using the subRead program and gene definitions from Ensembl Mus_Musculus GRCm38.78 coding and non-coding genes. Only genes for which at least three samples had at least 10 counts and had an average CPM > 1.0 were retained, resulting in a total of 10,946 genes. Fold change comparisons between WT and NRF2 gene deletion strains were further filtered on genes with less than two samples with zero reads. Differential expression was calculated using the edgeR package from bioconductor.org. False discovery rate was computed with the Benjamini-Hochberg algorithm (table S3, S4, S5). Hierarchical clusterings were performed in R using ‘TSclust’ and ‘hclust’ libraries. Heat map and scatterplot visualizations were generated in R using the ‘heatmap.2’ and ‘ggplot2’ libraries, respectively.
Ingenuity Pathway Analysis (IPA)
IPA (QIAGEN) was used to identify enriched pathways for differentially expressed genes between naïve and M.tb.-infected or naïve and bystander AMs (cut-off values: FDR < 0.01, |FC| > 2). Canonical pathways with enrichment score p-value < 0.05 with greater than 10 gene members are reported. IPA was also used to identify differentially enriched pathways between bystander and M.tb.-infected AMs at 10 days post-infection (cut-off values: FDR < 0.05, |FC| > 2). Canonical pathways with |z-scores| >1 and p-values < 0.05 were reported.
Promoter Scanning (HOMER)
Promoter regions of genes that were up-regulated (|FC| > 2, FDR < 0.01, log2(average counts per million) > 1.0) were scanned for DNA protein-binding motif over-representation using the HOMER program (v4.9.1, homer.salk.edu) (30). Promoter regions were defined as 2000 nucleotides upstream of the gene start to 1000 nucleotides downstream. Background sequences were taken from the promoter regions of expressed genes defined by Ensembl Mus_Musculus GRCm38.78 (N=10,946). 402 known motifs were scanned and hypergeometric p-values computed.
NRF2 ChIP-seq Analysis
Fastq files from the GEO data set GSE75175 were downloaded for three NRF2 ChIP-seq experiments assaying peritoneal macrophages from wild-type mice including the corresponding sequencing of input DNA (32). For each sample, single ended reads of length 101 were filtered to remove those consisting of 50 or more of the same nucleotide or of low-quality base calls. Filtered reads were aligned against mm10 using gsnap (v. 2011-11-20) with no allowance for splicing. Uniquely mapped reads were filtered for duplicates based on alignment position. A total of 3,975 peaks were called using MACS2 (v.2.1.0) for the combined ChiP-seq samples using the input DNA libraries as the controls. Called peaks were annotated by checking for overlap with promoter regions as described above.
Gene Set Enrichment Analysis (GSEA)
Input data for GSEA consisted of lists, ranked by -log(p-value), comparing RNA-seq expression measures of target samples and naïve controls including directionality of fold-change. Mouse orthologs of human Hallmark genes were defined using a list provided by Molecular Signatures Database (MSigDB) (70). GSEA software was used to calculate enrichment of ranked lists in each of the respective hallmark gene lists, as described previously (71). A nominal p-value for each ES is calculated based on the null distribution of 1,000 random permutations. To correct for multiple hypothesis testing, a normalized enrichment score (NES) is calculated that corrects the ES based on the null distribution. A false-discovery rate (FDR) is calculated for each NES. Leading edge subsets are defined as the genes in a particular gene set that are part of the ranked list at or before the running sum reaches its maximum value.
BMDM Isolation and Culture
Bone marrow-derived macrophages (BMDMs) were cultured in complete RPMI with recombinant human M-CSF (50 ng/ml; PeproTech Inc.) for 6 days. Media with antibiotics were washed out prior to infection with H37Rv and replaced with complete RPMI without antibiotics. M-CSF was included in the BMDM media for the entire course of the experiments.
M.tb. In Vitro Culture and Infection
H37Rv was grown in 7H9 media at 37°C to O.D. of 0.1–0.3. The final concentration was calculated based on strain titer and bacteria was added to macrophages at an effective multiplicity of infection (MOI) of 0.5 for two hours. Cultures were then washed three times to remove extracellular bacteria. Infected macrophages were cultured for up to 5 days. For CFU measurement, cells were lysed with 1% Triton X-100/PBS and lysate from triplicate conditions were plated in serial dilutions on Middlebrook 7H10 agar plates (ThermoFisher Scientific) and cultured at 37°C for 21 days.
qRT-PCR
For gene expression analysis, 1 × 105 AMs or BMDMs were plated in 96-well plates overnight, followed by in vitro infection as described above. RNA isolation was performed as described above. Quantitative PCR reactions were carried out using TaqMan primer probes (ABI) and TaqMan Fast Universal PCR Master Mix (ThermoFisher) in a CFX384 Touch Real-Time PCR Detection System (BioRad). Data were normalized by the level of EF1a expression in individual samples. Fold induction was computed with respect to the normalized expression levels of respective macrophages under unstimulated conditions within the same experiment.
Microscopy
Cells acquired by BAL were spun down and plated on a Lab-Tek II 8-chamber glass-bottom slide (Nunc). The slides were kept for 2 hours in a 37°C humidified incubator (5% CO2) to allow adherence before media removal and fixation with 2% paraformaldehyde in PBS. Surface staining was performed with anti-CD45 (clone 30-F11; Biolegend). ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher) was used for mounting. Cells were imaged using a 100X objective (1.40 NA) on a DeltaVision Elite with the following excitation filter cubes: Cy5 (632/22), GFP (475/28), and DAPI (390/18) and emission cubes: Cy5(679/34), GFP (525/48), and DAPI (435/48). The entire cell volume was captured using a series of Z-stack images with a 0.2um step size. Number of Mycobacteria cells per macrophage were enumerated by manual counting using the Z-stack of images. Representative images were first deconvolved with a theoretical point spread function using SVI Huygens Essential before a maximum intensity project image was created in Imaris Image Analysis Software (Bitplane).
ELISA
Lung homogenate was digested in 1mL RIPA Lysis and Extraction Buffer (Thermo Scientific)) with Halt Protease Inhibitor (Thermo Scientific) followed by mechanical disruption using a gentleMACS dissociator (Miltenyi Biotec). Samples were double filtered through 0.2 μm filters to remove bacteria. Total protein concentration was measured by Pierce BCA Protein Assay (Thermo Scientific) and samples were normalized to 1.5 mg/mL total protein. Murine IL-6, IL-1β, and TNF DuoSet ELISAs were performed in accordance with the manufacturer’s instructions (R&D Systems). Absorbance was recorded at 450 nm with wavelength correction performed by subtracting 570 nm absorbance using the Spectramax M2 plate reader (Molecular Devices).
Western Blot
Naïve AMs were isolated by BAL and pooled from 5 mice, plated overnight for adherence and stimulated for 5 hours with the NRF2 agonist, dimethyl fumarate (3.65 μM; Sigma-Aldrich) prior to protein collection with RIPA buffer (Cell Signaling Technology) and HALT protease inhibitor (ThermoFisher). Western blotting analyses were performed using standard techniques and transblotted onto nitrocellulose membranes. Membranes were probed with relevant primary antibodies: rabbit anti-NRF2 (D1Z9C) (1:1000; Cell Signaling Technology), rabbit anti-mouse beta-actin1-HRP antibody (1:5000, Jackson Immunoresearch). Primary antibodies were detected by a secondary rat anti-rabbit-HRP antibody (1:2000, Jackson Immunoresearch).
Statistical Analyses
RNA-sequencing was analyzed using the edgeR package from Bioconductor.org and the false discovery rate was computed using the Benjamini-Hochberg algorithm. All other data are presented as mean ± SEM. For comparison of multiple conditions, data was analyzed by one-way analysis of variance (ANOVA) (95% confidence interval) with Holm-Sidak correction (for multiple comparisons) or Dunnett’s correction (for multiple comparisons to a single control). For comparison of two conditions, data was analyzed by two-tailed unpaired Student’s t-test (with Holm-Sidak correction for multiple testing). Statistical analysis and graphical representation of data was performed using either GraphPad Prism v6.0 software or R. At least 3–5 mice were used per group in each experiment and experiments were performed at least 2–3 times, as indicated in the figure legends. To denote significance, * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Material
Fig. S1: Lung flow cytometry gating scheme for myeloid cells
Fig. S2: Lung flow cytometry gating scheme for detection of M.tb. infected cells with overlay of mEmerald+ populations
Fig. S3: BAL and lung flow cytometry gating scheme for cell sorting
Fig. S4: Bacterial burden at the time of alveolar macrophage cell sorting
Fig. S5: Gene expression heatmaps of naïve, bystander and M.tb.-infected alveolar macrophages 24 hours following aerosol infection
Fig. S6: Gating strategy for sorting of ΔRD1-infected alveolar macrophages and bead-positive alveolar macrophages
Fig. S7: Gene expression of bone marrow-derived macrophages following treatment with fluorescent beads
Fig. S8: Gene expression heatmaps of M.tb.-infected alveolar macrophages 0.5, 1, 2, 4 and 10 days following aerosol infection
Fig. S9: Alveolar macrophage response to in vitro M.tb. infection with and without GM-CSF treatment
Fig. S10: Alveolar macrophage response to in vitro M.tb. infection at 8, 12 and 24 hours following infection
Fig. S11: Gene expression differences between naive WT and Nrf2−/− alveolar macrophages
Fig. S12: Baseline lung conditions of WT and Nrf2−/− mice
Fig. S13: Gene expression and protein confirmation of NRF2 knockout in alveolar macrophages from Nrf2LysM, Nrf2CD11c, Nrf2fl littermate controls, WT and Nrf2−/− mice
Fig. S14: Baseline lung conditions of Nrf2LysM, Nrf2CD11c, and Nrf2fl mice
Fig. S15: Gene expression heatmaps of M.tb.-infected Nrf2LysM, Nrf2CD11c, Nrf2fl, WT and Nrf2−/− alveolar macrophages 24 hours following aerosol infection
Fig. S16: Viability of bystander Nrf2LysM, Nrf2CD11c, and Nrf2fl alveolar macrophages 10 days following high dose aerosol infection
Table S1: Summary table accounting for M.tb.-infected cells in the lung 1–14 days following high dose mEmerald-H37Rv infection
Table S2: Ingenuity Pathway Analysis and HOMER results tables
Table S3: RNA-Sequencing data for WT alveolar macrophages
Table S4: RNA-Sequencing data for Nrf2−/− alveolar macrophages
Table S5: RNA-Sequencing data for Nrf2fl, Nrf2LysM, and Nrf2CD11c alveolar macrophages
Table S6: Raw data file
Acknowledgements:
We thank the staff at Seattle Children’s Research Institute vivarium for animal care, Pamela Troisch and the Next Gen Sequencing core at the Institute for Systems Biology, and Chris Sassetti and Christina Baer at the University of Massachusetts Medical School for the mEmerald bacterial strains. Members of the Urdahl lab provided helpful discussions.
Funding: This work was supported by National Institute of Allergy and Infectious Disease of the National Institute of Health under Awards U19AI106761, U19AI135976, U19AI100627, and R01AI032972 (A.A).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw and processed RNA-sequencing data can be accessed from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE125287.
References and Notes:
- 1.Reid MJA, Arinaminpathy N, Bloom A, Bloom BR, Boehme C, Chaisson R, Chin DP, Churchyard G, Cox H, Ditiu L, Dybul M, Farrar J, Fauci AS, Fekadu E, Fujiwara PI, Hallett TB, Hanson CL, Harrington M, Herbert N, Hopewell PC, Ikeda C, Jamison DT, Khan AJ, Koek I, Krishnan N, Motsoaledi A, Pai M, Raviglione MC, Sharman A, Small PM, Swaminathan S, Temesgen Z, Vassall A, Venkatesan N, van Weezenbeek K, Yamey G, Agins BD, Alexandru S, Andrews JR, Beyeler N, Bivol S, Brigden G, Cattamanchi A, Cazabon D, Crudu V, Daftary A, Dewan P, Doepel LK, Eisinger RW, Fan V, Fewer S, Furin J, Goldhaber-Fiebert JD, Gomez GB, Graham SM, Gupta D, Kamene M, Khaparde S, Mailu EW, Masini EO, McHugh L, Mitchell E, Moon S, Osberg M, Pande T, Prince L, Rade K, Rao R, Remme M, Seddon JA, Selwyn C, Shete P, Sachdeva KS, Stallworthy G, Vesga JF, Vilc V, Goosby EP, Building a tuberculosis-free world: The Lancet Commission on tuberculosis. Lancet 393, 1331–1384 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, Sherman DR, Gerner MY, Urdahl KB, Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe 24, 439–446 e434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urdahl KB, Understanding and overcoming the barriers to T cell-mediated immunity against tuberculosis. Semin Immunol 26, 578–587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf AJ, Desvignes L, Linas B, Banaiee N, Tamura T, Takatsu K, Ernst JD, Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med 205, 105–115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf AJ, Linas B, Trevejo-Nunez GJ, Kincaid E, Tamura T, Takatsu K, Ernst JD, Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol 179, 2509–2519 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Srivastava S, Ernst JD, Cell-to-cell transfer of M tuberculosis antigens optimizes CD4 T cell priming. Cell Host Microbe 15, 741–752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reiley WW, Calayag MD, Wittmer ST, Huntington JL, Pearl JE, Fountain JJ, Martino CA, Roberts AD, Cooper AM, Winslow GM, Woodland DL, ESAT-6-specific CD4 T cell responses to aerosol Mycobacterium tuberculosis infection are initiated in the mediastinal lymph nodes. Proc Natl Acad Sci U S A 105, 10961–10966 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussell T, Bell TJ, Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol 14, 81–93 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, Dickersin GR, Bachurski CJ, Mark EL, Whitsett JA, et al. , Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science 264, 713–716 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN, Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 210, 1977–1992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ, Immunological Genome C, Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I, Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK, Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snelgrove RJ, Goulding J, Didierlaurent AM, Lyonga D, Vekaria S, Edwards L, Gwyer E, Sedgwick JD, Barclay AN, Hussell T, A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat Immunol 9, 1074–1083 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Brandes M, Klauschen F, Kuchen S, Germain RN, A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 154, 197–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Nazarova EV, Tan S, Liu Y, Russell DG, Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med 215, 1135–1152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanley SA, Johndrow JE, Manzanillo P, Cox JS, The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol 178, 3143–3152 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, Kugler D, Hieny S, Caspar P, Nunez G, Schlueter D, Flavell RA, Sutterwala FS, Sher A, Caspase-1 independent IL-1beta production is critical for host resistance to mycobacterium tuberculosis and does not require TLR signaling in vivo. J Immunol 184, 3326–3330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNab FW, Ewbank J, Rajsbaum R, Stavropoulos E, Martirosyan A, Redford PS, Wu X, Graham CM, Saraiva M, Tsichlis P, Chaussabel D, Ley SC, O’Garra A, TPL-2-ERK1/2 signaling promotes host resistance against intracellular bacterial infection by negative regulation of type I IFN production. J Immunol 191, 1732–1743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS, The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe 17, 811–819 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, Shiloh MU, Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe 17, 820–828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H, Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun 65, 298–304 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP, The immune response in tuberculosis. Annu Rev Immunol 31, 475–527 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgwick JD, Britton WJ, Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol 162, 3504–3511 (1999). [PubMed] [Google Scholar]
- 25.Desvignes L, Wolf AJ, Ernst JD, Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. J Immunol 188, 6205–6215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fremond CM, Togbe D, Doz E, Rose S, Vasseur V, Maillet I, Jacobs M, Ryffel B, Quesniaux VF, IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J Immunol 179, 1178–1189 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H, Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 49, 503–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopf M, Schneider C, Nobs SP, The development and function of lung-resident macrophages and dendritic cells. Nat Immunol 16, 36–44 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Gorrini C, Harris IS, Mak TW, Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12, 931–947 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK, Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem 280, 16891–16900 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Otsuki A, Suzuki M, Katsuoka F, Tsuchida K, Suda H, Morita M, Shimizu R, Yamamoto M, Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic Biol Med 91, 45–57 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Braverman J, Sogi KM, Benjamin D, Nomura DK, Stanley SA, HIF-1alpha Is an Essential Mediator of IFN-gamma-Dependent Immunity to Mycobacterium tuberculosis. J Immunol 197, 1287–1297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley SA, Raghavan S, Hwang WW, Cox JS, Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 100, 13001–13006 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR, Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. The Journal of infectious diseases 187, 117–123 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell DG, The ins and outs of the Mycobacterium tuberculosis-containing vacuole. Cell Microbiol 18, 1065–1069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS, Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11, 469–480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koo IC, Wang C, Raghavan S, Morisaki JH, Cox JS, Brown EJ, ESX-1-dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol 10, 1866–1878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishra BB, Moura-Alves P, Sonawane A, Hacohen N, Griffiths G, Moita LF, Anes E, Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell Microbiol 12, 1046–1063 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, Ablasser A, Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host Microbe 17, 799–810 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Rothchild AC, Sissons JR, Shafiani S, Plaisier C, Min D, Mai D, Gilchrist M, Peschon J, Larson RP, Bergthaler A, Baliga NS, Urdahl KB, Aderem A, MiR-155-regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 113, E6172–E6181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, Tanaka N, Moriguchi T, Motohashi H, Nakayama K, Yamamoto M, Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun 7, 11624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abram CL, Roberge GL, Hu Y, Lowell CA, Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J Immunol Methods 408, 89–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan K, Lu R, Chang JC, Kan YW, NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 93, 13943–13948 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy NM, Potteti HR, Mariani TJ, Biswal S, Reddy SP, Conditional deletion of Nrf2 in airway epithelium exacerbates acute lung injury and impairs the resolution of inflammation. Am J Respir Cell Mol Biol 45, 1161–1168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamm CE, Collins AC, Shiloh MU, Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunol Rev 264, 204–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chackerian AA, Alt JM, Perera TV, Dascher CC, Behar SM, Dissemination of Mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun 70, 4501–4509 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Day J, Friedman A, Schlesinger LS, Modeling the immune rheostat of macrophages in the lung in response to infection. Proc Natl Acad Sci U S A 106, 11246–11251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marino S, Kirschner DE, The human immune response to Mycobacterium tuberculosis in lung and lymph node. J Theor Biol 227, 463–486 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Andrade BB, Pavan Kumar N, Amaral EP, Riteau N, Mayer-Barber KD, Tosh KW, Maier N, Conceicao EL, Kubler A, Sridhar R, Banurekha VV, Jawahar MS, Barbosa T, Manganiello VC, Moss J, Fontana JR, Marciano BE, Sampaio EP, Olivier KN, Holland SM, Jackson SH, Moayeri M, Leppla S, Sereti I, Barber DL, Nutman TB, Babu S, Sher A, Heme Oxygenase-1 Regulation of Matrix Metalloproteinase-1 Expression Underlies Distinct Disease Profiles in Tuberculosis. J Immunol 195, 2763–2773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chinta KC, Rahman MA, Saini V, Glasgow JN, Reddy VP, Lever JM, Nhamoyebonde S, Leslie A, Wells RM, Traylor A, Madansein R, Siegal GP, Antony VB, Deshane J, Wells G, Nargan K, George JF, Ramdial PK, Agarwal A, Steyn AJC, Microanatomic Distribution of Myeloid Heme Oxygenase-1 Protects against Free Radical-Mediated Immunopathology in Human Tuberculosis. Cell Rep 25, 1938–1952 e1935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scharn CR, Collins AC, Nair VR, Stamm CE, Marciano DK, Graviss EA, Shiloh MU, Heme Oxygenase-1 Regulates Inflammation and Mycobacterial Survival in Human Macrophages during Mycobacterium tuberculosis Infection. J Immunol 196, 4641–4649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenner RG, Young RA, Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol 3, 281–294 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Torrelles JB, Schlesinger LS, Integrating Lung Physiology, Immunology, and Tuberculosis. Trends Microbiol 25, 688–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez JC, Dang H, Martin JR, Doerschuk CM, Nrf2 Modulates Host Defense during Streptococcus pneumoniae Pneumonia in Mice. J Immunol 197, 2864–2879 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Athale J, Ulrich A, MacGarvey NC, Bartz RR, Welty-Wolf KE, Suliman HB, Piantadosi CA, Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice. Free Radic Biol Med 53, 1584–1594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reddy NM, Kleeberger SR, Kensler TW, Yamamoto M, Hassoun PM, Reddy SP, Disruption of Nrf2 impairs the resolution of hyperoxia-induced acute lung injury and inflammation in mice. J Immunol 182, 7264–7271 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Page A, Volchkova VA, Reid SP, Mateo M, Bagnaud-Baule A, Nemirov K, Shurtleff AC, Lawrence P, Reynard O, Ottmann M, Lotteau V, Biswal SS, Thimmulappa RK, Bavari S, Volchkov VE, Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep 6, 1026–1036 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF, The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep 6, 1017–1025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho HY, Imani F, Miller-DeGraff L, Walters D, Melendi GA, Yamamoto M, Polack FP, Kleeberger SR, Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am J Respir Crit Care Med 179, 138–150 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lugade AA, Vethanayagam RR, Nasirikenari M, Bogner PN, Segal BH, Thanavala Y, Nrf2 regulates chronic lung inflammation and B-cell responses to nontypeable Haemophilus influenzae. Am J Respir Cell Mol Biol 45, 557–565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boutten A, Goven D, Artaud-Macari E, Boczkowski J, Bonay M, NRF2 targeting: a promising therapeutic strategy in chronic obstructive pulmonary disease. Trends in molecular medicine 17, 363–371 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S, Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. The Journal of clinical investigation 114, 1248–1259 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shang S, Ordway D, Henao-Tamayo M, Bai X, Oberley-Deegan R, Shanley C, Orme IM, Case S, Minor M, Ackart D, Hascall-Dove L, Ovrutsky AR, Kandasamy P, Voelker DR, Lambert C, Freed BM, Iseman MD, Basaraba RJ, Chan ED, Cigarette smoke increases susceptibility to tuberculosis--evidence from in vivo and in vitro models. The Journal of infectious diseases 203, 1240–1248 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Gordon SB, Bruce NG, Grigg J, Hibberd PL, Kurmi OP, Lam KB, Mortimer K, Asante KP, Balakrishnan K, Balmes J, Bar-Zeev N, Bates MN, Breysse PN, Buist S, Chen Z, Havens D, Jack D, Jindal S, Kan H, Mehta S, Moschovis P, Naeher L, Patel A, Perez-Padilla R, Pope D, Rylance J, Semple S, Martin WJ, 2nd, Respiratory risks from household air pollution in low and middle income countries. The Lancet. Respiratory medicine 2, 823–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peterson EJ, Bailo R, Rothchild AC, Arrieta-Ortiz ML, Kaur A, Pan M, Mai D, Abidi AA, Cooper C, Aderem A, Bhatt A, Baliga NS, Path-seq identifies an essential mycolate remodeling program for mycobacterial host adaptation. Mol Syst Biol 15, e8584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price JV, Vance RE, The macrophage paradox. Immunity 41, 685–693 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Shafiani S, Tucker-Heard G, Kariyone A, Takatsu K, Urdahl KB, Pathogen-specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. J Exp Med 207, 1409–1420 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schroeder WG, Mitrescu LM, Hart ML, Unnithan R, Gilchrist JM, Smith EE, Shanley C, Benedict KM, Taraba L, Volckens J, Basaraba RJ, Schenkel AR , Flexible low-cost system for small animal aerosol inhalation exposure to drugs, proteins, inflammatory agents, and infectious agents. Biotechniques 46, Piii–Pviii (2009). [DOI] [PubMed] [Google Scholar]
- 70.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Lung flow cytometry gating scheme for myeloid cells
Fig. S2: Lung flow cytometry gating scheme for detection of M.tb. infected cells with overlay of mEmerald+ populations
Fig. S3: BAL and lung flow cytometry gating scheme for cell sorting
Fig. S4: Bacterial burden at the time of alveolar macrophage cell sorting
Fig. S5: Gene expression heatmaps of naïve, bystander and M.tb.-infected alveolar macrophages 24 hours following aerosol infection
Fig. S6: Gating strategy for sorting of ΔRD1-infected alveolar macrophages and bead-positive alveolar macrophages
Fig. S7: Gene expression of bone marrow-derived macrophages following treatment with fluorescent beads
Fig. S8: Gene expression heatmaps of M.tb.-infected alveolar macrophages 0.5, 1, 2, 4 and 10 days following aerosol infection
Fig. S9: Alveolar macrophage response to in vitro M.tb. infection with and without GM-CSF treatment
Fig. S10: Alveolar macrophage response to in vitro M.tb. infection at 8, 12 and 24 hours following infection
Fig. S11: Gene expression differences between naive WT and Nrf2−/− alveolar macrophages
Fig. S12: Baseline lung conditions of WT and Nrf2−/− mice
Fig. S13: Gene expression and protein confirmation of NRF2 knockout in alveolar macrophages from Nrf2LysM, Nrf2CD11c, Nrf2fl littermate controls, WT and Nrf2−/− mice
Fig. S14: Baseline lung conditions of Nrf2LysM, Nrf2CD11c, and Nrf2fl mice
Fig. S15: Gene expression heatmaps of M.tb.-infected Nrf2LysM, Nrf2CD11c, Nrf2fl, WT and Nrf2−/− alveolar macrophages 24 hours following aerosol infection
Fig. S16: Viability of bystander Nrf2LysM, Nrf2CD11c, and Nrf2fl alveolar macrophages 10 days following high dose aerosol infection
Table S1: Summary table accounting for M.tb.-infected cells in the lung 1–14 days following high dose mEmerald-H37Rv infection
Table S2: Ingenuity Pathway Analysis and HOMER results tables
Table S3: RNA-Sequencing data for WT alveolar macrophages
Table S4: RNA-Sequencing data for Nrf2−/− alveolar macrophages
Table S5: RNA-Sequencing data for Nrf2fl, Nrf2LysM, and Nrf2CD11c alveolar macrophages
Table S6: Raw data file