Significance
Seasonality is a widespread adaptation in nature, which is driven by changes in day length (i.e., photoperiod) in organisms living at temperate latitudes. In insects, despite decades of research, the molecular and genetic mechanisms by which photoperiodic changes are sensed and translated into seasonal changes in developmental, physiological, and behavioral processes have remained elusive. The present study supports a role for circadian clock genes in sensing photoperiodic signals and genetically demonstrates that the vitamin A in the insect brain, which is photoperiod- and clock-regulated, is necessary for regulating seasonal responses. These findings also establish the monarch butterfly as one of the few animal models in which to genetically dissect the molecular mechanisms that underlie photoperiodic responsiveness.
Keywords: circadian clocks, insect photoperiodism, CRISPR/Cas9, vitamin A, monarch butterfly
Abstract
Seasonal adaptation to changes in light:dark regimes (i.e., photoperiod) allows organisms living at temperate latitudes to anticipate environmental changes. In nearly all animals studied so far, the circadian system has been implicated in measurement and response to the photoperiod. In insects, genetic evidence further supports the involvement of several clock genes in photoperiodic responses. Yet, the key molecular pathways linking clock genes or the circadian clock to insect photoperiodic responses remain largely unknown. Here, we show that inactivating the clock in the North American monarch butterfly using loss-of-function mutants for the circadian activators CLOCK and BMAL1 and the circadian repressor CRYPTOCHROME 2 abolishes photoperiodic responses in reproductive output. Transcriptomic approaches in the brain of monarchs raised in long and short photoperiods, summer monarchs, and fall migrants revealed a molecular signature of seasonal-specific rhythmic gene expression that included several genes belonging to the vitamin A pathway. We found that the rhythmic expression of these genes was abolished in clock-deficient mutants, suggesting that the vitamin A pathway operates downstream of the circadian clock. Importantly, we showed that a CRISPR/Cas9-mediated loss-of-function mutation in the gene encoding the pathway’s rate-limiting enzyme, ninaB1, abolished photoperiod responsiveness independently of visual function in the compound eye and without affecting circadian rhythms. Together, these results provide genetic evidence that the clock-controlled vitamin A pathway mediates photoperiod responsiveness in an insect. Given previously reported seasonal changes associated with this pathway in the mammalian brain, our findings suggest an evolutionarily conserved function of vitamin A in animal photoperiodism.
Understanding how changes in photoperiod are sensed and translated into seasonal changes in animal developmental, physiological, and behavioral processes at the molecular level remains one of the fundamental yet poorly understood questions in the field of biological rhythms. Many classical resonance and night interruption experiments have suggested a circadian basis for photoperiodic responses in nearly all insects, birds, and seasonal-breeding mammals studied so far (1, 2). However, owing to the fact that most species with strong photoperiodic responses are nontraditional model organisms, a formal genetic demonstration that the circadian clock, an endogenous timekeeping mechanism that governs 24-h rhythms, plays a role in photoperiodic responsiveness is generally lacking. A few notable exceptions are in insects in which the use of genetic knockdowns or knockouts has demonstrated that at least some of the core clock genes are necessary for photoperiodic responses (3–7). Yet, because the impact of a given clock gene disruption on photoperiodic responses can be interpreted as a pleiotropic effect, the notion that the circadian clock acts as a functional system for photoperiod sensing and responsiveness remains debated (8). Perhaps for this reason, the key molecular pathways linking clock genes or the circadian clock to insect photoperiodic responses have remained virtually unexplored to date.
With a strong seasonal biology, the availability of a draft genome sequence (9) and functional genomic tools (10, 11), the eastern North American migratory monarch butterfly is well-suited not only to assess the effect of clock gene mutants on photoperiodic responses but also to probe the molecular bases underlying photoperiod responsiveness (12, 13). Each fall, coincident with the decreasing photoperiod, migrating monarchs undergo a switch in physiology and behavior, starting their epic southward migration from the northern United States to their Mexican overwintering sites in a state of reproductive dormancy (i.e., diapause) in which they remain until the spring. Although complete reproductive diapause appears to require more than a single diapause-inducing cue, subjecting laboratory-raised monarchs to a short photoperiod has been shown to robustly decrease the production of mature oocytes by females (14), making it possible to measure monarch photoperiodic responsiveness in laboratory conditions. Furthermore, the development of gene-editing technologies in this species has also permitted the generation of several clock gene loss-of-function mutants (10, 11, 15). The monarch circadian clock relies on transcriptional/translational feedback loops initiated by the heterodimeric transcription factor CLOCK (CLK):BMAL1 that drives the rhythmic transcription of the core clock genes period (Per), timeless (Tim), and cryptochrome 2 (Cry2). Once translated, PER, TIM, and CRY2 form a repressive complex that in turn rhythmically inhibits CLOCK:BMAL1-mediated transcription (16). Loss-of-function mutants for each of the circadian activators CLK and BMAL1, and for the main circadian repressor CRY2, which are available in our laboratory, can be used to genetically test if both positive and negative elements of the clock are required to elicit monarch photoperiodic responses.
In the work presented here, we demonstrate that both circadian activators and the main circadian repressor are necessary for monarch photoperiodic responses. In addition, RNA-sequencing (RNA-seq) over the course of the day in the brains of laboratory strains of monarchs raised in long and short photoperiods, as well as in the brains of summer monarchs and wild-caught fall migrants, reveals a seasonal signature of photoperiod-dependent rhythmic expression that includes several components of the vitamin A pathway. We further genetically demonstrate that the vitamin A pathway functions downstream of the brain circadian clock and that it is necessary for photoperiodic responses in the monarch butterfly. Together, these data provide a molecular link between circadian clock genes and insect photoperiodic responsiveness.
Results
Circadian Clock Activators and Repressors Are Required for Photoperiod Responsiveness.
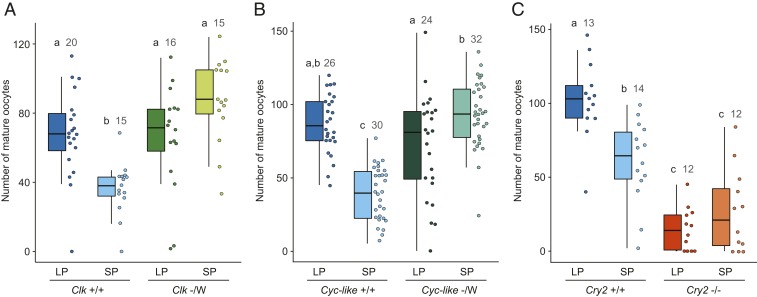
To genetically determine if circadian clock genes are required for monarch photoperiodic sensing and responses, we tested photoperiodic responsiveness of monarch loss-of-function mutant strains for the 2 circadian activators Clk and Bmal1 and for the main circadian repressor Cry2 (Fig. 1). All of these strains harbor dysfunctional molecular clocks and exhibit arrhythmic adult eclosion behavior (10, 15). All mutants were raised along with their respective wild-type siblings from eggs to adults under long-photoperiod (LP) conditions (15 h light:9 h dark) or short-photoperiod (SP) conditions (10 h light:14 h dark) at a constant temperature of 21 °C, and the number of mature oocytes produced by females was quantified 14 d post adult emergence (Fig. 1). The temperature of 21 °C was chosen because this temperature not only reflects the average day/night temperature that fall migratory monarchs experience across the eastern United States during their southward migration but also is permissive to female oocyte development in both LP and SP (14) (Fig. 1). Consistent with previous reports (14), we found that wild-type monarch females responded to the photoperiod by producing significantly more mature oocytes in LP than in SP (Fig. 1 A–C). In contrast, females of each of the 3 loss-of-function mutants tested lost the ability to respond to the photoperiod (Fig. 1 A–C). Loss-of-function mutant females for either Clk or Bmal1 [designated Cyc-like as this mutant lacks the C-terminal transactivation domain lost in Drosophila Cycle (15)] produced, both in LP and SP, significantly more mature oocytes than wild-type females raised in SP and as many as wild-type females raised in LP (Fig. 1 A and B). In contrast to monarchs lacking functional circadian activators, female monarchs lacking a functional CRY2 circadian repressor produced, regardless of the photoperiod during which they were raised, significantly fewer oocytes than did wild-type monarchs raised in SP (Fig. 1C). We verified that the low level of mature oocyte production in absence of a functional CRY2 was not due to sterility but rather to juvenile hormone (JH) deficiency. Topical application of methoprene, a JH analog, restored oocyte maturation in Cry2 homozygous mutant females to levels similar to those in treated wild-type females (SI Appendix, Fig. S1). Therefore, the lack of photoperiodic responses observed in Clk, Bmal1, and Cry2 loss-of-function mutants demonstrated that these 3 clock genes are necessary for photoperiodic responses in the monarch.
Fig. 1.
Circadian clock genes are required for photoperiodic responses. Number of mature oocytes produced 14 d post adult emergence in loss-of-function hemizygous Clock (Clk; A), hemizygous Bmal1 (Cyc-like; B), and homozygous Cryptochrome 2 (Cry2; C) mutant females, and in their respective wild-type siblings, all raised in LP and SP at 21 °C. Monarchs have a ZW sex-determination system; males are the homogametic sex (ZZ) while females are the heterogametic sex (ZW), and Clk and Bmal1 are located on the Z chromosome. For each condition, box plots and raw data points are shown. Error bars on box plots represent 1.5 times the interquartile range. LP: 15 h light, 9 h dark; SP: 10 h light, 14 h dark. Different letters over bars indicate groups that are statistically significant (interaction genotype × photoperiod, 2-way ANOVA, Tukey’s pairwise comparisons, P < 0.05), and numbers of dissected females are indicated.
Photoperiodic and Clock Regulation of the Vitamin A Pathway in the Brain.
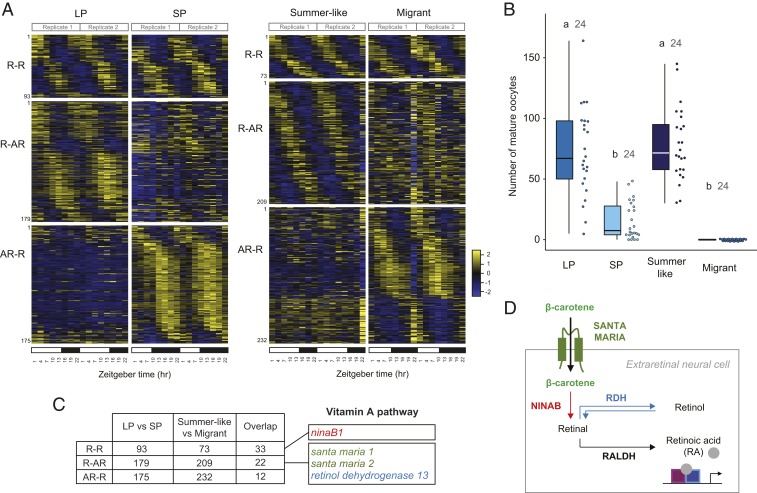
If circadian clock components are necessary for photoperiodic responses, we reasoned that molecular pathways mediating photoperiod responsiveness may be under rhythmic transcriptional regulation and may show seasonal-specific expression patterns of rhythmic expression in the brain, the organ known to function in photoperiodic reception in lepidopteran and some fly species (17, 18). To identify photoperiod-specific expression patterns of rhythmic expression in the monarch adult brain, we first carried out RNA-seq at 3-h intervals over the 24-h light:dark cycle in the brain of monarchs raised in LP and in SP at 21 °C (Fig. 2 A, Left). Expression levels for each gene were examined for rhythmic variation using RAIN (19) and MetaCycle (20), and genes with a maximum/minimum fold-change ≥1.3 and a P value corrected for multiple testing ≤0.05 were considered rhythmic. Using these criteria, we identified sets of genes with a seasonal molecular signature of rhythmic gene expression; while 93 genes were expressed rhythmically in both LP and SP conditions (rhythmic-rhythmic; R-R), we found 179 genes expressed rhythmically only in LP (rhythmic-arrhythmic; R-AR), and 175 genes expressed rhythmically only in SP (arrhythmic-rhythmic; AR-R) (Fig. 2 A, Left; SI Appendix, Table S1; and Dataset S1). To pinpoint which of these genes may be involved in photoperiodic responsiveness, we next sought to compare the differential rhythmic gene expression found between LP and SP monarchs to the one between different seasonal forms of wild monarchs exposed to summer and fall photoperiods. We predicted that key mediators of photoperiod-regulated responses should show similar patterns of differential expression in the brain of LP and SP monarchs and in the brain of summer monarchs and fall migrants. Unlike fall migrants, which migrate en masse and can therefore be caught in substantial numbers in a single round of collection, wild summer monarchs often move individually, making it challenging to catch the number necessary to perform RNA-seq around the clock at fine temporal resolution. Instead of wild-caught summer monarchs, we therefore used a laboratory strain of wild-type monarchs raised in a 15 h light:9 h dark summer-like long day (LD) and summer-like temperatures (25 °C). Importantly, summer-like monarchs and fall migrants exhibited similar photoperiodic reproductive responses to LP and SP monarchs raised at 21 °C, with summer-like females producing as many mature oocytes as LP females, and fall migrant females being in complete reproductive diapause (Fig. 2B). Comparable to results obtained with LP and SP monarchs, 3 classes of genes were identified by RNA-seq in the brains of summer-like monarchs and wild-caught fall migrants, with 73 genes expressed rhythmically in both seasonal forms (R-R), 209 genes expressed rhythmically only in summer-like monarchs (R-AR), and 232 genes expressed rhythmically only in fall migrants (AR-R) (Fig. 2 A, Right; SI Appendix, Table S2; and Dataset S2). As predicted, comparing the LP/SP and summer-like/migrant datasets revealed a number of genes expressed in a similar manner in both datasets; specifically, 33 genes were found to be rhythmic in all photoperiodic/seasonal forms (R-R), 22 genes were rhythmic only in LP and summer-like monarchs (R-AR), and 12 genes were rhythmic only in SP and wild-caught migrants (AR-R) (Fig. 2C and SI Appendix, Table S3).
Fig. 2.
Rhythmic gene expression analysis reveals photoperiodic regulation of the vitamin A pathway in the monarch brain. (A) Heatmaps of relative RNA levels for genes rhythmically expressed in brains of monarchs raised in either LP or SP at 21 °C (Left) and in brains of either summer-like or wild-caught migrant monarch (Right). R-R, genes rhythmic in both conditions; R-AR: genes rhythmic only in summer-like or LP; AR-R, genes rhythmic only in migrants or SP. Columns represent samples collected over a 24-h LD cycle. Transcripts are arranged by phase, and their order along the vertical axis is conserved in both conditions. White bars: light conditions; black bars: dark conditions. (B) Number of mature oocytes produced by females 9 to 15 d post adult emergence for summer-like monarchs and 7 to 11 d for LP/SP monarchs. Wild-caught migrants of unknown age were dissected 8 d after capture. Boxplots as in Fig. 1. One-way ANOVA, post hoc Tukey test, P < 0.05. (C) Table showing the number of genes found in any given category and the number of overlapping genes between comparisons. Genes involved in the vitamin A pathway are shown. The complete lists are shown in SI Appendix, Tables S1, S3, and S5. (D) Proposed model of the vitamin A pathway in insect brain cells. Beta-carotene is transported into extraretinal neural cells of the adult brain via santa maria and converted to retinal by ninaB. Retinal can either be interconverted into retinol by a retinol dehydrogenase (RDH) or converted into RA by a retinaldehyde dehydrogenase (RALDH). RA binds to retinoid receptors to regulate transcription of target genes.
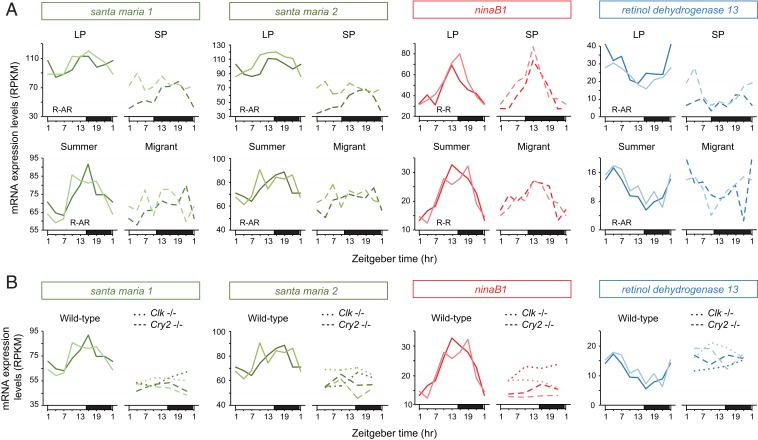
A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway-enrichment analysis identified circadian rhythm as an enriched term in the R-R category that contained the robustly cycling clock genes of the interlocked core and the stabilizing loops period, timeless, vrille, and clockwork orange (9, 21) (SI Appendix, Fig. S2). Although no other KEGG pathway was found enriched in genes rhythmic in LP and summer-like monarchs (R-AR) or in SP and wild-caught migrants (AR-R), 3 of 22 genes in the R-AR category stood out based on their known function in the vitamin A pathway in both Drosophila and mammals. Two of the 3 genes—scavenger receptor acting in neural tissue 1 and 2 (santa maria 1 and 2)—encode homologs of a transmembrane protein responsible for the uptake of beta-carotene into extraretinal neural cells in the Drosophila brain (22, 23), and the third gene, retinol dehydrogenase 13 (rdh 13), encodes an enzyme belonging to a family known to reversibly convert retinaldehyde (also called retinal) to retinol in mammals (24) (Figs. 2 C and D and 3A). Interestingly, a homolog of neither inactivation nor afterpotential B (ninaB1), the gene encoding the rate-limiting enzyme of this pathway that converts carotenoids into retinaldehyde (25) (Fig. 2D), was found expressed rhythmically in all groups (Figs. 2C and 3A). Together, these data suggested that the vitamin A pathway in the monarch brain could be involved in photoperiodic responsiveness. Because we demonstrated that both functional circadian activators CLK and BMAL1 and repressor CRY2 were necessary for photoperiodic responses, we then used already available RNA-seq datasets (21) to test whether ninaB1, santa maria 1, santa maria 2, and rdh13 rhythmic expression in brains of monarchs subjected to 15:9 LD could be Clk- and Cry2-dependent. Consistent with the idea that the vitamin A pathway acts downstream of the circadian clock or clock genes, we found that the rhythmic expression of all 4 genes was abolished in brains of Clk and Cry2 loss-of-function monarch mutants (Fig. 3B).
Fig. 3.
The vitamin A pathway is under both photoperiodic and clock regulation. (A) Temporal mRNA expression profiles of santa maria 1, santa maria 2, ninaB1, and retinol dehydrogenase 13 in brains of monarch raised in LP and SP (Top) and summer-like and migrant monarchs (Bottom). R-AR: rhythmic in LP and summer-like monarchs, arrhythmic in SP monarchs and migrants; R-R: rhythmic in LP, SP, summer-like monarchs, and migrants. The R-R and R-AR categories were defined based on the analysis of rhythmic gene expression reported in Fig. 2. (B) Temporal mRNA expression profiles of santa maria 1, santa maria 2, ninaB1, and retinol dehydrogenase 13 in brains of wild-type summer-like monarchs (plain lines), and Clk (dotted lines) and Cry2 (dashed lines) loss-of-function mutants raised in summer-like conditions. For each condition, 2 biological replicates are plotted.
Interestingly, we also identified collagen type IV subunit alpha 1 (α-1) among the genes expressed rhythmically only in the brain of SP monarchs and wild-caught migrants (SI Appendix, Table S3). This gene, which encodes one of 2 subunits of a central component of basement membranes (26), has previously been reported to show strong signatures of divergence in migratory and nonmigratory monarch populations (27). Based on differential expression in flight muscle of migratory and nonmigratory monarchs that were correlated with flight metabolic rates, collagen type IV α-1 has been proposed to be associated with the regulation of flight efficiency during long-distance migration (27). The differential rhythmic expression that we observed in the brain of monarch seasonal forms suggests that collagen type IV α-1 may serve an additional and/or different function with respect to seasonal migration.
A Functional Vitamin A Pathway Is Necessary for Photoperiodic Responses.
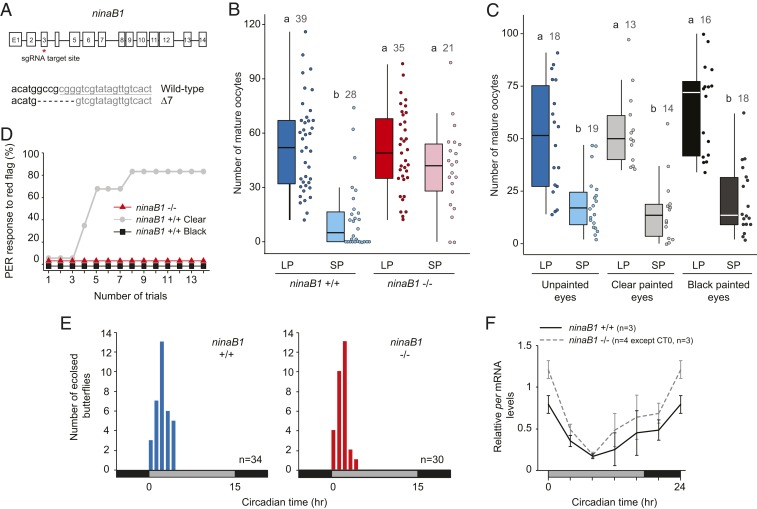
Given that several genes belonging to the vitamin A pathway were found differentially expressed in the brains of seasonal forms of monarchs and that seasonal changes associated with this pathway have been reported in the brain of mammals (28–31), we next focused on genetically testing if the vitamin A pathway was necessary for monarch photoperiodic responses. To this end, we generated a CRISPR/Cas9-mediated germline mutation in ninaB1, the gene encoding the rate-limiting enzyme responsible for the production of retinal. We selected a 7-bp deletion in the targeted third exon, resulting in a protein truncated by more than 80% to establish a loss-of-function mutant line (Fig. 4A) that we raised in both LP and SP conditions. We found that, similar to clock-deficient monarchs, homozygous mutant ninaB1 females lost the ability to respond to the photoperiod, producing in both LP and SP as many mature oocytes as their wild-type siblings raised in LP (Fig. 4B and SI Appendix, Fig. S3 A and B). The LP-like state of oocyte production observed in mutant ninaB1 females was surprising because ninaB1 was expressed at constitutively low levels in Cry2 loss-of-function mutants (Fig. 3B), which are themselves locked in a SP-like state of low oocyte production (Fig. 1C). The presence of paralogs that have some degree of functional redundancy for the production of retinal and that are expressed in the brain could explain this unexpected result. Blasting ninaB1 against protein sequences encoded in the monarch genome revealed the existence of 2 paralogs, ninaB2 and ninaB3, which showed ∼50% sequence conservation at the amino acid level with ninaB1 (SI Appendix, Fig. S3C). Only ninaB2 was found expressed at detectable levels by RNA-seq in the monarch brain (SI Appendix, Fig. S3D), and its product was also the only one of the 2 paralogs to share with ninaB1, a conserved residue known to be essential for the catalytic activity of Drosophila ninaB (25) (SI Appendix, Fig. S3C). Unlike ninaB1, ninaB2 was expressed at low levels and in an arrhythmic manner in the monarch brain (SI Appendix, Fig. S3 D and E). If monarch ninaB2 is indeed capable of converting carotenoids into retinal at a high-enough level to promote the production of a mature oocyte in a LP-like state, our data may indicate that the rhythmic production of retinal in the brain could be critical for photoperiodic responsiveness. If this is the case, the phase of ninaB1 expression, which is locked to “lights on” at the messenger RNA (mRNA) level with a peak of expression in the light phase in LP and in the dark phase in SP (Fig. 3A), might be important for the induction of a photoperiodic response.
Fig. 4.
Disruption of the vitamin A pathway abolishes photoperiodic responses. (A) Generation of a ninaB1 loss-of-function mutant using CRISPR/Cas9-mediated targeted mutagenesis. (Top) Genomic structure of ninaB1 in the monarch. Red star, position of the site targeted by sgRNA. (Bottom) Nucleotide sequence of the targeted site (underlined) and introduced frame-shifting mutation (7-bp deletion). (B) Number of mature oocytes produced 10 d post adult emergence in ninaB1 homozygous mutant and wild-type sibling females. Boxplots as in Fig. 1. Interaction genotype × photoperiod, 2-way ANOVA, Tukey’s pairwise comparisons, P < 0.05. (C) Number of mature oocytes produced 10 d post adult emergence in wild-type female monarchs with unpainted eyes or eyes covered with either clear or black paint at the day of emergence. Interaction genotype × painting condition, 2-way ANOVA, Tukey’s pairwise comparisons, P < 0.05. (D) Associative visual conditioning of the proboscis extension reflex (PER) of ninaB1 homozygous mutants (red line, n = 6) and wild-type monarchs with eyes painted clear (gray line, n = 6) or black (black line, n = 6) for 14 training trials on day 3 of training. (E) Profiles of adult eclosion in DD of wild-type (blue) and ninaB1 loss-of-function siblings (red). Gray bars: subjective day; black bars: subjective night. Kolmogorov–Smirnov test, P > 0.05. (F) Circadian expression of period in brains of wild-type (solid black lines) and ninaB1 loss-of-function (dashed gray lines) siblings. Values are mean ± SEM, and the numbers of animals for each genotype and time point are indicated. Interaction genotype × time, 2-way ANOVA, P > 0.05.
Retinal is most conventionally known to be essential for visual function, and Drosophila ninaB loss-of-function mutants are blind (22, 25, 32). Because our monarch ninaB1 loss-of-function mutant was not brain-specific, we sought to verify that the observed loss of photoperiodic responses in this mutant did not result from an inability to perceive light through the compound eyes. To this end, we raised wild-type monarchs in LP and SP conditions and covered the adult compound eyes on the day of adult emergence with either an enamel-based black paint or clear paint (for controls), respectively blocking or allowing light input to monarch tissues (33). Importantly, we found that the photoperiodic response of monarch females with black painted eyes was indistinguishable from that of control females with either unpainted or clear painted eyes (Fig. 4C). We further excluded the possibility that the black paint was not fully blocking eye visual function by showing that, in contrast to wild-type monarchs with clear painted eyes, monarchs with black painted eyes trained to associate a colored flag with a sugar reward were not able to learn the association (Fig. 4D), likely because their vision was impaired. Consistent with the defects in vision caused by mutations in Drosophila ninaB (25), we also found that monarch homozygous ninaB1 mutants were unable to associate a color with the reward (Fig. 4D). Using similar painting experiments, we also excluded the possibility that the antennae, which harbor the navigational clocks for monarch migration, were necessary for monarch photoperiodic responses (SI Appendix, Fig. S4). These results strongly suggest that the loss of photoperiodic responsiveness in monarch ninaB1 loss-of-function mutants results from disruption of the vitamin A pathway in the brain and not in the compound eyes. Finally, we show that the lack of photoperiodic responses in homozygous ninaB1 monarch mutants did not result from a disrupted circadian clock, as both behavioral rhythms of adult eclosion and molecular rhythms of Per in the brain were intact in this mutant (Fig. 4 E and F). Together, our findings provide a functional demonstration that the vitamin A pathway in the brain, which acts downstream of the circadian clock and is seasonally regulated, is necessary to regulate photoperiodic responsiveness in the monarch.
Discussion
For decades, progress in understanding the molecular and genetic bases of photoperiodism in insects has been limited by the fact that the key genetically tractable model organism Drosophila melanogaster lacks a pronounced diapause, the prominent photoperiodic response exhibited by insects (34). The advent of molecular tools and their applicability to nontraditional model insects with a more dramatic photoperiodic response has recently changed this trend. While photoperiodic effects on the oscillation patterns of some circadian clock components provided molecular evidence that clock genes may be involved in sensing and transmitting the photoperiodic information to output cascades (35–38), recent analysis using gene knockdowns and knockouts in the bean bug Riptortus pedestris and the mosquito Culex pipiens have unambiguously demonstrated the functional involvement of core clock genes in photoperiodism (3–6). Whether clock gene products act pleiotropically or as part of a functional circadian system remains debatable as the use of individual clock gene knockouts that render the clock dysfunctional cannot formally distinguish between these 2 possibilities. However, our findings that monarchs bearing nonfunctional circadian activators and repressors display opposite egg maturation phenotypes mirror previous findings obtained in the bean bug (3, 5). Together, these data indicate that circadian activators and repressors may work in a coordinated fashion to sense photoperiod, likely through a feedback loop. As previously demonstrated in other insects (39), photoperiodic induction of reproductive diapause is induced by a reduction of JH synthesis and/or secretion by the corpora allata, a pair of neurohemal organs located behind the brain. Being able to restore the production of mature oocytes to high levels in Cry2 knockout females by application of a JH analog suggests not only that JH titers are likely low in this mutant but also that Cry2 is not involved in the cascade of events downstream of JH synthesis and/or secretion. Similarly, while knocking out Clk and Bmal1 disrupts photoperiodic responses, this treatment does not affect the levels of mature oocytes produced by mutant females, suggesting that JH titers may be similar between these mutants and wild-type siblings raised in LP. Together, these data indicate that circadian clock genes likely act upstream of JH synthesis and/or secretion, as previously demonstrated in the bean bug (5). Our discovery that the seasonally regulated vitamin A pathway is also under clock control in the monarch brain further adds support to the idea that a functional circadian system may be involved in photoperiodic responses. It also provides a molecular link in the insect brain between clock genes and the diapause response.
Interestingly, the involvement of vitamin A and carotenoids in insect photoperiodism is not without precedent. Classic dietary-deprivation experiments showed decades ago that many species of insects and mites reared on an artificial diet deficient in carotenoids lose their photoperiodic response (40–43), leading to the hypothesis that a yet-unknown vitamin A-based pigment, retinal-opsin, was the likely photoperiodic receptor. Here, we provide genetic evidence that regulation of the vitamin A pathway in the insect brain mediates photoperiodic responses. The findings that visual function from the compound eyes is dispensable for photoperiodic responses in the monarch is consistent with the major role that extraretinal deep-brain photoreceptors play in photoperiodic reception in other lepidopteran species (44). By knocking out the rate-limiting enzyme that converts carotenoids into retinal, we show that retinal conversion is necessary for photoperiodic responses in the monarch. As previously proposed, one possible function of retinal in the brain could be the production of a deep brain photoreceptor for photoperiodic induction that is different from Cryptochrome 1, the key photoreceptor used for circadian clock entrainment (16). Determining which of the 5 opsins present in the monarch genome are expressed in monarch brain and knocking them out could help functionally test this hypothesis and identify the potential relevant opsin.
Alternatively, similar to that described in mammals (28–31), retinoic acid (RA), a product of retinal, could be responsible for the seasonal regulation of the transcriptional program and/or the neuronal plasticity in the brain that regulates photoperiodic responses. In rats, concentration and signaling of RA, which regulates gene transcription through the activation of nuclear receptors, are under photoperiod regulation and reduced in SP relative to LP (28, 29, 31). Until recently, the RA signaling pathway was believed to occur only in vertebrates. However, reports that not only RA but also orthologs of the retinoic acid receptors are expressed in insects (45) suggest that a similar pathway may also operate in these species. While we found that genes involved in beta-carotene uptake (santa-maria 1 and 2) and conversion of vitamin A to retinal (rdh13) were under photoperiodic regulation and that their rhythms of expression were dampened in SP relative to LP, the gene encoding the enzyme catalyzing the conversion of retinal to RA (retinaldehyde dehydrogenase) was not found under photoperiodic regulation in the monarch brain. However, this does not exclude the possibility of a photoperiodic regulation of concentration and signaling of RA in the monarch brain. Measuring the levels of RA in the brain of monarchs subjected to LP and SP will be necessary to ultimately implicate RA signaling in insect photoperiod responses. In mammals, RA is produced in ependymal tanycytes, a glial cell type lining the third ventricle adjacent to the hypothalamus that controls the season-dependent production of the active form of the thyroid hormone involved in seasonal reproduction (28). Interestingly, tanycytes have recently been shown to undergo a seasonal change in morphology in hamster and sheep (30, 46), where they enclose the synapses of neurons producing the reproductive hormone gonadotropin-releasing hormone (GRH) during the nonbreeding season, presumably blocking the release of GRH (30). A similar role of RA-positive cells in brain neuronal plasticity for driving monarch seasonal reproduction is an intriguing possibility. Identifying the cell type in which the vitamin A operates and developing knock-in approaches to mark cells with membrane-tagged fluorescent proteins will be necessary to test whether vitamin A-positive cells also undergo a seasonal remodeling in the monarch brain.
In conclusion, our work in the monarch provides genetic evidence that the vitamin A pathway, which is under seasonal and clock regulation in the brain, mediates photoperiodic responses in an insect. Given the parallel in seasonal changes associated with this pathway in the brain of monarchs and mammals, it is tantalizing to speculate that the function of vitamin A in animal photoperiodism may be evolutionarily conserved. The continued molecular and genetic dissection of this pathway in the monarch and its involvement in brain photoreception, seasonal regulation of transcription, or seasonal neuronal plasticity should shed light on the mechanisms of action of vitamin A in insect photoperiodic responsiveness and ultimately reveal the degree of conservation with its role in mammalian seasonal biology.
Materials and Methods
Animal Husbandry.
For photoperiodic experiments, laboratory stocks of wild-type and mutant monarchs were raised from eggs to adults in Percival incubators under 15-h light:9-h dark (LP) or 10-h light:14-h dark (SP) at a constant temperature of 21 °C with 70% humidity. For RNA-seq experiments, fall migratory monarch butterflies were captured on October 15, 2014, as they migrated through College Station, Texas (latitude 30°37′ N, longitude 96°20′ W). After capture, migrants were housed in a Percival incubator under fall-like conditions with a LD cycle set to prevailing light conditions (11 h:13 h LD, 0730 to 1830 Central standard time), at a constant temperature of 21 °C with 70% humidity. They were manually fed a 25% honey solution every other day and dissected 8 d after capture. Monarchs used for the LP and SP datasets were raised in the same conditions as for the photoperiodic experiments and were dissected 7 to 11 d post eclosion. For detailed descriptions, see SI Appendix, Materials and Methods.
Evaluation of Female Reproductive Status.
Female abdomens were transected dorsally, and mature oocytes were individually counted from each of the ovarioles in both ovaries under a dissecting microscope. Oocytes were considered mature when chorionated, i.e., when the presence of vertical ridges on the chorion was visible.
Compound Eye Painting and Antennae Removal.
Compound eyes were covered on the day of eclosion with enamel-based clear paint (Model master clear top coat; Testors no. 2736) or black paint (Glossy black; Testors no. 1147) under a dissecting microscope. The completeness of painting was verified the next day, and touch ups were performed if needed. Antennae were removed by clipping them with scissors at the base of the flagellum as previously described (33). Details are available in SI Appendix, Materials and Methods.
RNA-Seq Experiments.
Brains of monarchs raised indoors in LP and SP and of wild-caught migrants were dissected in 0.5× RNA later (Invitrogen) and stored at −80 °C until use. For each seasonal phenotype/photoperiodic condition, 3 pooled brains were collected in 2 replicates at Zeitgeber time (ZT)1, ZT4, ZT7, ZT10, ZT13, ZT16, ZT19, and ZT22. Total RNA was extracted using an RNeasy Mini kit (Qiagen) and polyA+ RNA was isolated from total RNA with the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). For samples from wild-caught migrants, multiplexed libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina and NEBNext Multiplex Oligos (New England Biolabs), following the manufacturer’s recommendations. For samples from monarchs raised in LP and SP, multiplexed libraries were prepared by the Texas A&M AgriLife Genomics and Bioinformatics Facility using the TruSeq Stranded mRNA Library Prep Kit for Illumina, following the manufacturer’s recommendations. Libraries were sequenced on a Hi-sEq. 2500 (Illumina) using 50-bp single-end reads. RNA-seq datasets from brains of summer-like monarchs and clock-deficient mutants used in this study were already available (21). For detailed descriptions, see SI Appendix, Materials and Methods.
RNA-Seq Data Processing, Mapping, and Identification of Cycling Transcripts.
After quality control and demultiplexing, reads were mapped to the monarch genome [assembly v3 (47)] using TopHat2 (48), averaging 91.5% of reads mapped across LP, SP, summer-like and migrant monarchs (SI Appendix, Table S4). Expression levels of transcripts were quantified using Cufflinks (49, 50), and cycling transcripts were identified using RAIN (19) and MetaCycle (20). Only genes with a minimum of 3 reads per kilobase per million mapped reads in at least one time point were considered as expressed. Rhythmically expressed genes were determined based on fold-change and P values adjusted for multiple testing using the Benjamini–Hochberg procedure to control for false discovery rate. Transcripts were considered rhythmically expressed when meeting the following criteria: 1) adjusted P values ≤ 0.05 for both RAIN and MetaCycle and 2) fold-change (maximal/minimal experimental values within a time series) ≥1.3. Percentages of genes determined as rhythmically expressed are shown in SI Appendix, Table S5. For comparisons of rhythmic expression between photoperiod conditions and seasonal forms, genes were sorted based on their rhythmic expression and classified in 3 groups: 1) rhythmic in both LP and SP or summer-like and migrant monarchs, 2) rhythmic only in LP or summer-like monarchs, and 3) rhythmic only in SP or migrant monarchs. Heatmaps were produced using heatmap.2 in R. Enriched gene ontology of biological processes and the KEGG pathway were determined using Metascape (metascape.org). Details are available in SI Appendix, Materials and Methods.
CRISPR/Cas9-Mediated Targeted Mutagenesis of Monarch ninaB1.
The guide RNA (gRNA) site was selected within exon 3 of the 14 exon-containing ninaB1 gene using CHOPCHOP (51, 52). The gRNA expression vector was constructed by inserting annealed synthetic oligomers into the DR274 plasmid from Addgene (53) at the BsaI cleavage site using the following primers (F, forward; R, reverse): F, 5′-TAGGAGTGACAACTATACGACCCG-3′ and R, 5′-AAACCGGGTCGTATAGTTGTCACT-3′. In vitro transcription of the single guide RNA (sgRNA) was performed using T7 RNA polymerase (Promega) from purified PCR products amplified from the DR274 vectors, as previously described (15). In vitro transcription of Streptococcus pyogenes Cas9 mRNA was performed using the mMessage mMachine T3 transcription kit (Invitrogen) and the pCS2-nCas9n expression plasmid from Addgene (54), as previously described (15). Cas9 mRNAs and the sgRNA were quantitated by spectrophotometry (NanoDrop 1000), respectively, diluted in RNase-free water to final concentrations of 0.5 μg/μL and 0.25 μg/μL and microinjected into monarch eggs, as previously described (10). For screening for the presence of CRISPR/Cas9-induced mutations, PCR fragments flanking the targeted region were amplified from genomic DNA extracted from larval sensors of potential founders with the following primers: ninaB1F, 5′-GTTTCACTTGTACCGTGACTTC-3′ and ninaB1R, 5′-GGATACTGTTTAGCCAGGTACC-3. Purified PCR products (250 to 350 ng) were subjected to Cas9-based cleavage assays using a recombinant Cas9 protein (750 ng) and the sgRNA (400 to 600 ng), as previously described (10). Surviving adults of opposite sexes presenting the highest level of somaticism, estimated based on the relative abundance of uncleaved fragments, were hand-paired to establish a mutant line. The progeny were screened for the presence of mutated alleles as described above, and uncleaved fragments were sequenced. A 7-bp deletion causing a frameshift and the introduction of a premature stop codon was selected to establish a mutant line. For detailed procedures, see SI Appendix, Materials and Methods.
Proboscis Extension Reflex Assay.
For proboscis extension tests, individual butterflies were harnessed in 15-mL polypropylene conical tubes, fed daily with 150 µL of a 25% honey solution, and starved for 24 h the day prior to stimulus conditioning, as previously described (33). Wild-type monarchs with eyes covered with either clear or black paint and ninaB1−/− monarchs were conditioned to a colored stimulus (red flag; conditioned stimulus, CS) by presenting the stimulus for 5 s (CS only), contacting the middle legs with a cotton-tipped applicator soaked in 50% sucrose solution for 15 s (CS+US), and removing the stimulus after 5 s (US only). Individuals were then held for 5 min and US-CS pairing was repeated for 13 to 14 trials a day for 3 consecutive days until the individual extended its proboscis upon the initial presentation of the CS. Monarchs were considered to have a positive proboscis extension reflex (PER) (conditioned response) upon full extension of their proboscis in response to the CS. Details are available in SI Appendix, Materials and Methods.
Eclosion Behavior Assay.
Eclosion behavior was monitored during the first day of constant darkness (DD) as previously described (10) from butterflies raised in LD 15:9 in a Percival incubator at 25 °C and 70% humidity. Eclosion data were analyzed and plotted as 1-h bins.
Real-Time qPCR.
Brains of adult wild-type and ninaB1 homozygous mutant monarchs entrained to 7 15:9 LD cycles were dissected in 0.5× RNA later (Invitrogen) under red light on the first day of transfer to DD at circadian time (CT)0, CT4, CT8, CT12, CT16, and CT20. Total RNA was extracted using a RNeasy Mini kit (Qiagen) and reverse-transcribed with SuperScript II Reverse Transcriptase (Thermo Scientific) and random hexamers, following the manufacturers’ instructions. Quantifications of gene expression were performed on a QuantStudio 6 Flex Real-Time PCR System (Thermo Scientific) using iTaq Universal SYBR Green Supermix (Bio-Rad) and monarch Per and control rp49 primers, as previously described (10). The data were normalized to rp49 as an internal control and normalized to the mean of one sample within a set for statistics. See SI Appendix, Materials and Methods, for details.
Statistical Analysis.
P values were calculated using Student’s t-tests, 1-way and 2-way ANOVAs followed by post hoc analyses, and Kolmogorov–Smirnov tests using online calculators at www.wessa.net/rwasp_Two%20Factor%20ANOVA.wasp (55), http://www.physics.csbsju.edu/stats/KS-test.n.plot_form.html, and https://www.graphpad.com/quickcalcs/posttest1.cfm.
Data Availability.
The RNA-seq datasets have been deposited in the Gene Expression Omnibus repository under accession number GSE126336.
Supplementary Material
Acknowledgments
We thank Matthew Markert and Justin Vann for assistance with the wild-caught migrant collection; Sarah Kenny, Kendall Bowen, Catherine Bogdan, and Jason Park for assistance with husbandry; Jerome Menet and Ben Greenwell for help with bioinformatic analyses; the Texas A&M AgriLife Genomics and Bioinformatics facility for sequencing services; and Jerome Menet and Paul Hardin for comments on the manuscript. We also thank Steven M. Reppert for use of the data presented in SI Appendix, Fig. S1, which were generated by C.M. as a postdoctoral fellow in his laboratory. This work was supported by funds from Texas A&M University and NSF Grant IOS-1456985 (to C.M.). C.M. was also supported by a Klingenstein-Simons award in the neurosciences.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing datasets have been deposited in the Gene Expression Omnibus repository (accession no. GSE126336).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913915116/-/DCSupplemental.
References
- 1.Ikegami K., Yoshimura T., Seasonal time measurement during reproduction. J. Reprod. Dev. 59, 327–333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders D. S., Bertossa R. C., Deciphering time measurement: The role of circadian ‘clock’ genes and formal experimentation in insect photoperiodism. J. Insect Physiol. 57, 557–566 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ikeno T., Numata H., Goto S. G., Photoperiodic response requires mammalian-type cryptochrome in the bean bug Riptortus pedestris. Biochem. Biophys. Res. Commun. 410, 394–397 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Ikeno T., Numata H., Goto S. G., Circadian clock genes period and cycle regulate photoperiodic diapause in the bean bug Riptortus pedestris males. J. Insect Physiol. 57, 935–938 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Ikeno T., Tanaka S. I., Numata H., Goto S. G., Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 8, 116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meuti M. E., Stone M., Ikeno T., Denlinger D. L., Functional circadian clock genes are essential for the overwintering diapause of the Northern house mosquito, Culex pipiens. J. Exp. Biol. 218, 412–422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pegoraro M., Gesto J. S., Kyriacou C. P., Tauber E., Role for circadian clock genes in seasonal timing: Testing the Bünning hypothesis. PLoS Genet. 10, e1004603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meuti M. E., Denlinger D. L., Evolutionary links between circadian clocks and photoperiodic diapause in insects. Integr. Comp. Biol. 53, 131–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan S., Merlin C., Boore J. L., Reppert S. M., The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markert M. J., et al. , Genomic access to monarch migration using TALEN and CRISPR/Cas9-mediated targeted mutagenesis. G3 (Bethesda) 6, 905–915 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merlin C., Beaver L. E., Taylor O. R., Wolfe S. A., Reppert S. M., Efficient targeted mutagenesis in the monarch butterfly using zinc-finger nucleases. Genome Res. 23, 159–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denlinger D. L., Hahn D. A., Merlin C., Holzapfel C. M., Bradshaw W. E., Keeping time without a spine: What can the insect clock teach us about seasonal adaptation? Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reppert S. M., Guerra P. A., Merlin C., Neurobiology of monarch butterfly migration. Annu. Rev. Entomol. 61, 25–42 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Goehring L., Oberhauser K. S., Effects of photoperiod, temperature, and host plant age on induction of reproductive diapause and development time in Danaus plexippus. Ecol. Entomol. 27, 674–685 (2002). [Google Scholar]
- 15.Zhang Y., Markert M. J., Groves S. C., Hardin P. E., Merlin C., Vertebrate-like CRYPTOCHROME 2 from monarch regulates circadian transcription via independent repression of CLOCK and BMAL1 activity. Proc. Natl. Acad. Sci. U.S.A. 114, E7516–E7525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H., et al. , Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 6, e4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen M. F., Saunders D. S., Bollenbacher W. E., Gilbert L. I., In vitro reprogramming of the photoperiodic clock in an insect brain-retrocerebral complex. Proc. Natl. Acad. Sci. U.S.A. 81, 5881–5884 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders D. S., Insect photoperiodism: Measuring the night. J. Insect Physiol. 59, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Thaben P. F., Westermark P. O., Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu G., Anafi R. C., Hughes M. E., Kornacker K., Hogenesch J. B., MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics 32, 3351–3353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lugena A. B., Zhang Y., Menet J. S., Merlin C., Genome-wide discovery of the daily transcriptome, DNA regulatory elements and transcription factor occupancy in the monarch butterfly brain. PLoS Genet. 15, e1008265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T., Jiao Y., Montell C., Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J. Cell Biol. 177, 305–316 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., O’Tousa J. E., Cellular sites of Drosophila NinaB and NinaD activity in vitamin A metabolism. Mol. Cell. Neurosci. 35, 49–56 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Duester G., Families of retinoid dehydrogenases regulating vitamin A function: Production of visual pigment and retinoic acid. Eur. J. Biochem. 267, 4315–4324 (2000). [DOI] [PubMed] [Google Scholar]
- 25.von Lintig J., Dreher A., Kiefer C., Wernet M. F., Vogt K., Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation invivo. Proc. Natl. Acad. Sci. U.S.A. 98, 1130–1135 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnorrer F., et al. , Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464, 287–291 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Zhan S., et al. , The genetics of monarch butterfly migration and warning colouration. Nature 514, 317–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shearer K. D., et al. , Photoperiodic regulation of retinoic acid signaling in the hypothalamus. J. Neurochem. 112, 246–257 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Shearer K. D., et al. , Photoperiodic expression of two RALDH enzymes and the regulation of cell proliferation by retinoic acid in the rat hypothalamus. J. Neurochem. 122, 789–799 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Wood S. H., et al. , Binary switching of calendar cells in the pituitary defines the phase of the circannual cycle in mammals. Curr. Biol. 25, 2651–2662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helfer G., et al. , Photoperiod regulates vitamin A and Wnt/β-catenin signaling in F344 rats. Endocrinology 153, 815–824 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Voolstra O., et al. , NinaB is essential for Drosophila vision but induces retinal degeneration in opsin-deficient photoreceptors. J. Biol. Chem. 285, 2130–2139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merlin C., Gegear R. J., Reppert S. M., Antennal circadian clocks coordinate sun compass orientation in migratory monarch butterflies. Science 325, 1700–1704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tauber E., Kyriacou B. P., Insect photoperiodism and circadian clocks: Models and mechanisms. J. Biol. Rhythms 16, 381–390 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Goto S. G., Denlinger D. L., Short-day and long-day expression patterns of genes involved in the flesh fly clock mechanism: Period, timeless, cycle and cryptochrome. J. Insect Physiol. 48, 803–816 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Iwai S., Fukui Y., Fujiwara Y., Takeda M., Structure and expressions of two circadian clock genes, period and timeless in the commercial silkmoth, Bombyx mori. J. Insect Physiol. 52, 625–637 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Majercak J., Sidote D., Hardin P. E., Edery I., How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Qiu J., Hardin P. E., per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol. Cell. Biol. 16, 4182–4188 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denlinger D. L., Yocum G. D., Rinehart J. P., “Hormonal control of diapause” in Insect Endocrinology, Li G., Ed. (Academic Press, Cambridge, MA, 2011), pp. 430–463. [Google Scholar]
- 40.Veerman A., et al. , Vitamin-A is essential for photoperiodic induction of diapause in an eyeless mite. Nature 302, 248–249 (1983). [Google Scholar]
- 41.Veerman A., Slagt M. E., Alderlieste M. F. J., Veenendaal R. L., Photoperiodic induction of diapause in an insect is vitamin-A dependent. Experientia 41, 1194–1195 (1985). [Google Scholar]
- 42.Claret J., Volkoff N., Vitamin A is essential for two processes involved in the photoperiodic reaction in Pieris brassicae. J. Insect Physiol. 38, 569–574 (1992). [Google Scholar]
- 43.Saunders D. S., Insect Clocks (Elsevier, Amsterdam, 2002). [Google Scholar]
- 44.Truman J. W., Physiology of insect rhythms. II. The silkmoth brain as the location of the biological clock controlling eclosion. J. Comp. Physiol. 81, 99–114 (1972). [Google Scholar]
- 45.Bui-Göbbels K., Quintela R. M., Bräunig P., Mey J., Is retinoic acid a signal for nerve regeneration in insects? Neural Regen. Res. 10, 901–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolborea M., et al. , Melatonin controls photoperiodic changes in tanycyte vimentin and neural cell adhesion molecule expression in the Djungarian hamster (Phodopus sungorus). Endocrinology 152, 3871–3883 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Zhan S., Reppert S. M., MonarchBase: The monarch butterfly genome database. Nucleic Acids Res. 41, D758–D763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D., et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C., et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C., et al. , Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labun K., Montague T. G., Gagnon J. A., Thyme S. B., Valen E., CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., Valen E., CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang W. Y., et al. , Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jao L. E., Wente S. R., Chen W., Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holliday I. E., Two-Way ANOVA (v1.0.6) in Free Statistics Software (v1.2.1) (Office for Research Development and Education, 2019). https://www.wessa.net/rwasp_Two%20Factor%20ANOVA.wasp/. Accessed 21 November 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets have been deposited in the Gene Expression Omnibus repository under accession number GSE126336.