Short abstract
The transforming growth factor beta (TGFβ) signaling pathway orchestrates a wide breadth of biological processes, ranging from bone development to reproduction. Given this, there has been a surge of interest from the drug development industry to modulate the pathway – at several points. This review discusses and provides additional context for several layers of the TGFβ signaling pathway from a structural biology viewpoint. The combination of structural techniques coupled with biophysical studies has provided a foundational knowledge of the molecular mechanisms governing this high impact, ubiquitous pathway, underlying many of the current therapeutic pursuits. This work seeks to consolidate TGFβ-related structural knowledge and educate other researchers of the apparent gaps that still prove elusive. We aim to highlight the importance of these structures and provide the contextual information to understand the contribution to the field, with the hope of advancing the discussion and exploration of the TGFβ signaling pathway.
Impact statement
The transforming growth factor beta (TGFβ) signaling pathway is a multifacetted and highly regulated pathway, forming the underpinnings of a large range of biological processes. Here, we review and consolidate the key steps in TGFβ signaling using literature rooted in structural and biophysical techniques, with a focus on molecular mechanisms and gaps in knowledge. From extracellular regulation to ligand–receptor interactions and intracellular activation cascades, we hope to provide an introductory base for understanding the TGFβ pathway as a whole.
Keywords: Structural biology, X-ray crystallography, protein–protein interactions, NMR, kinase, structure function
Introduction
The transforming growth factor beta (TGFβ) superfamily controls several intricate signaling mechanisms that underlie every biological process from development and homeostasis to reproduction and wound-repair. Overall, the TGFβ family consists of over 33 unique ligands synthesized as a precursor that contains a large prodomain (∼200–300 residues) and a mature, signaling domain (∼115 residues). Each monomer has a growth-factor like fold with a pair of reciprocating β-strands divided by a central α-helix (Figure 1(a)). Once processed, ligands bind and assemble two signaling receptors, termed type I and type II (Figure 1(b)). Both type I and type II receptors have a single extracellular binding domain and an intracellular kinase domain. Upon assembly, the constitutively active type II receptor kinase phosphorylates the type I receptor kinase, initiating a downstream signaling cascade following phosphorylation of SMAD proteins. To handle the large array of ligands, only five different type II receptors (TβRII, ActRIIA, ActRIIB, BMPRII and AMHRII) and seven type I receptors (Alk1-7) are available where the combination of type I and type II receptors are ligand dependent. Along this, the ligands are classically organized into three classes: the TGFβs, for which the family is named, the bone morphogenetic proteins (BMPs), named for their roles in bone development, growth and homeostasis, and the activins, which play deterministic roles in reproduction and muscle development. Both TGFβ and activin class ligands canonically activate SMAD2/3 signaling, while the BMP ligands diverge, activating SMAD1/5/8. Additionally, distinct extra- and intracellular mechanisms have evolved to regulate each class at several levels providing exquisite control over structurally related signaling molecules. Throughout this review, we highlight our current structural understanding of these mechanisms including ligand–receptor interactions, extracellular antagonism, prodomain–ligand interactions, and recent insight into the intracellular kinase domains and SMAD proteins.
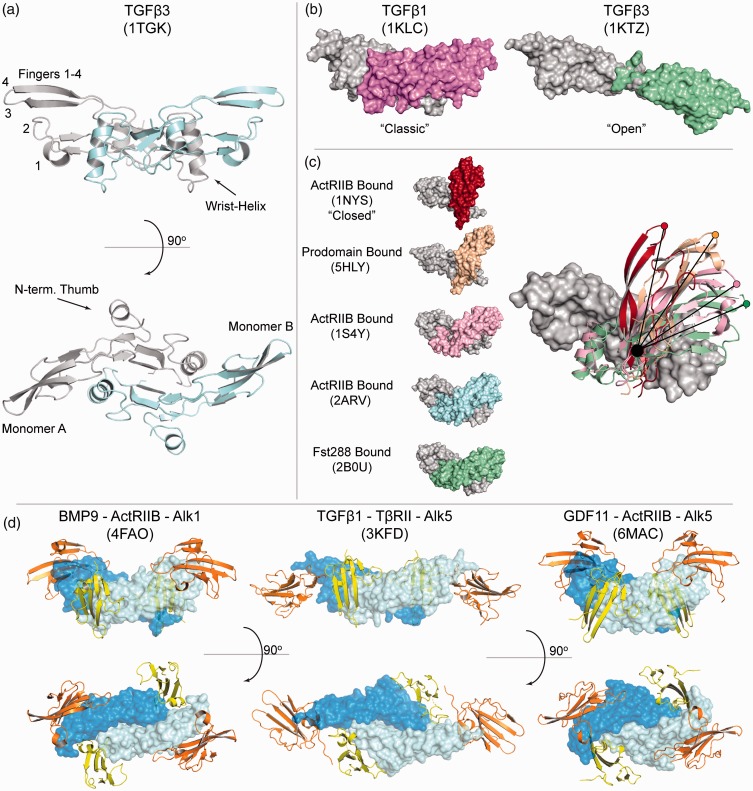
Figure 1.
TGFβ family ligand structure, flexibility, and receptor interactions. (a) ‘Classic’ butterfly and propeller structure of TGFβ3 displaying ‘hand-like’ labels: Wrist, Fingers, and Thumb. Monomer A is represented in gray, while monomer B is represented in cyan. (b) Structures of TGFβ1 and an altered TGFβ3 crystal form representing the classic and ‘open’ conformations, respectively. Monomer A (gray) is aligned and monomer B is shown in altered colors. (c) Structures of ActA displaying an altered position of monomer B and the flexible nature of the activin dimer interface. (d) Characteristic signaling complexes across the TGFβ family displaying complete extracellular assemblies. Ligands are in blue, type II receptors in orange, and type I receptors are colored, yellow. (A color version of this figure is available in the online journal.)
Receptor–ligand interactions across the TGFβ family
Ligand structures
Three-dimensional structures have been resolved for over 15 TGFβ family members either in complex with receptors, antagonists, their prodomains or unbound, in an apo-state. Representative structures are available for all three classes (TGFβ, BMP and activin) and coalesce into an archetypal structure described as a hand, with an α-helix (wrist) and two sets of antiparallel β-strands (fingers; Figure 1(a)).1 Despite having a similar overall architecture, TGFβ and activin members contain an N-terminal segment that folds back onto finger 1 and is connected by an additional disulfide bond. Structures of BMP ligands lack this N-terminal motif and subsequently, the additional disulfide bond.2 However, all family members contain a centralized cystine knot, a hallmark of other growth factors including nerve growth factor, platelet-derived growth factor, and vascular endothelial growth factor. The knot is constructed by two disulfide bonds between fingers 2 and 4, which form a ring that is pierced by a third disulfide between fingers 1 and 3. This knot contributes significantly to the folding and stability of the ligand, which generally lacks a hydrophobic core. The dimer is formed by an additional interchain disulfide bond, packing the two wrist helices into the opposing monomer’s palm to form a butterfly-like shape (Figure 1(a)). Within the palm of the ligand on finger 1 are two conserved tryptophan residues, part of a conserved WXXW motif, that are surface exposed and positioned on a helical turn, pointing into the concave pocket of the dimer interface. Previous studies have implicated that these residues are important for folding and play key roles in providing hydrophobic contact points with ligand binding partners, particularly the extracellular domains (ECD) of the corresponding receptors.3–13
Throughout the family, differences can be seen in residues that are not critical to the overall growth-factor fold. While these differences play an important role in defining which receptors and proteins they interact with, they can also have other functional roles. In particular, sequence variation can result in significant differences in the overall charge of the ligands. For instance, BMP ligands are positively charged leading to their ability to interact directly with heparin or heparan sulfate proteoglycans.2,14 Binding to heparin restricts their mobility, causing them to signal in a paracrine or autocrine manner. In contrast, some ligands, such as Activin A (ActA), are negatively charged at neutral pH and diffuse more readily, while others, such as GDF8, are polar where one half is positive and the other half is negative.15,16 Thus, the overall charge of the ligands is variable and can impact protein interactions, as well as their mobility postsecretion.
Ligands can also diverge heavily when comparing the overall dimer shape, specifically the relative position of each monomer. Structures of BMP2, 3, 6, 7, and 9 all display the ‘classic’ butterfly-like morphology where the dimer interface, including the wrist helix are well-ordered, implying rigidity of the dimer shape (Figure 1(a) and (b)).2–5,8,9,12,17–23 In contrast, several other ligands have displayed variation in the dimer shape, brought about by a flexible wrist region which permits free-rotation around the intermolecular disulfide bond. For example, TGFβ3 was crystallized in two different conformations: one, adopting the more classic configuration and a second receptor-bound structure, where the dimer is flattened, with an elongated or ‘open’ structure (Figure 1(b)).1,24 Interestingly, differences in conformational flexibility have been suggested to play a role in the differential activities of closely related isoforms.25
Similar to the TGFβ class, ActA, GDF8, and GDF11 of the activin class have been captured in multiple conformations including: an open state with an unresolved wrist helix (Apo-GDF11, antibody bound-GDF8), multiple structures in the classic state (Apo-GDF11, Fs288:GDF11, ActRIIB:Alk5:GDF11, Fs288/FSTL3:GDF8, Fs288/Fs315/FSTL3:ActA, and ActRIIB:ActA), and several structures displaying a unique ‘closed’state (prodomain:GDF8, Apo-GDF8, prodomain:ActA, and ActRIIB:ActA).13,15,16,23,26–35 A comparison of these different states describing the flexibility of ActA is highlighted in Figure 1(c). Together, these structures suggest that activin class ligands are highly flexible, with a large degree of variation in dimer conformations and an unstable wrist helix. Interestingly, occupancy of the concave binding interface by either an antagonist or the type I receptor seemingly favors the classic state, which may stabilize the complex.13,15,23,26,29,30,33 However, prodomain bound ActA and GDF8 prefer a more closed state.27,32 While these studies indicate that certain ligands are flexible, solution studies have been lacking that can quantify the degree of flexibility for each ligand.
Ligand–receptor complex structures
As mentioned, TGFβ family ligands initiate signaling through the assembly of a receptor complex consisting of a ligand, two type I, and two type II receptors. Each receptor ECD consists of ∼100 residues exhibiting a three-finger toxin fold of three β-strands. Both type I and type II receptors display a high degree of homology, except for the extended loops that flank the core β-sheet. Four of the five type II and four of the seven type I receptor ECDs have been structurally characterized either alone (type II: ActRIIA, TBRII, BMPII/type I: Alk1, Alk3, Alk5) or in a ligand–receptor complex (type II: ActRIIA, ActRIIB, TBRII, BMPII/type I: Alk1, Alk3, Alk5, Alk6).3–13,16,20,24,28,36–42 These structures have revealed a common set of four disulfide bonds that stabilize the β-sheet structure in each receptor. Additionally, ActRIIA, ActRIIB, BMPRII, and the type I receptors contain an additional disulfide bond to stabilize the β4-β5 loop and preventing the occlusion of the surface of the receptor, a crucial region for maintaining ligand binding, particularly for the type II receptors. TβRII is unique in having two additional disulfide bonds to tether the β1–β2 loop in a position for ligand interaction.
During evolution the ligands have expanded significantly faster than the receptors, and thus there are a far greater number of ligands than type I and type II receptors. As a consequence, receptors can interact with multiple ligands and vice versa, where ligands will typically interact with multiple receptors. It is important to note that the extent of ligand–receptor promiscuity is critical for defining signaling events.26,43,44 Along this, studies have only scratched the surface of the consequences of signaling when multiple ligands are present. Over the years, structural and biochemical studies have provided insight into the molecular basis of receptor specificity to define what ligand features are important for receptor specificity and what receptor features are used to engage certain ligands, while avoiding others.
Type II receptor–ligand structures
Of the five type II receptors, BMPRII, ActRIIA, and ActRIIB interact with several ligands whereas TβRII and AMHRII have a limited ligand binding repertoire.45,46 TβRII is highly specific for binding to the TGFβ class (TGFβ1–3), while AMHRII is reserved exclusively for the ligand, antiMullerian hormone (AMH). While AMHRII has not been structurally characterized, the limited specificity of TβRII for the TGFβ class is well understood.10 During complex formation with TGFβ, TβRII binds at the distal fingertips of the ligand, where the receptor β-sheet is perpendicular to the β-strands of the fingers (Figure 1(d), center, orange). Specifically, β2 of TβRII forms significant contacts and binds within a cleft formed from positively charged residues (Arg303 and Arg372 in TGFβ1) – a feature shared by the TGFβ ligands.11 These residues account for TGFβ ligand specificity for TβRII and are not present in BMP or activin class ligands.47,48 This interaction is further stabilized by an extension in the β1–β2 loop that is limited to TβRII and AMHRII.
Type II receptors, ActRIIA and ActRIIB interact with activin class ligands, as well as BMP ligands.46 BMPRII typically acts as a BMP class type II receptor, although in gonadotropic cells, activin has been shown to signal using BMPRII.49 While ligands can sometimes signal using multiple type II receptors, ligands can have differences in binding affinity, indicating that they exhibit preferences for specific type II receptors. Recombinant BMP 2/4/5/6/7/8 and GDF5/6/7 bind all three type II receptors with generally equal affinity, while GDF1, 3, 10, BMP3, GDF8, and GDF11 prefer ActRIIB.13,50–52 BMP9, 15, and GDF9 bind BMPRII with higher affinity, with BMP9 extending to ActRIIA.43 In general, activin class ligands bind type II receptors with higher affinity than BMP class ligands, suggesting a competitive environment may be present where limited amounts of ActRIIA or ActRIIB are present, pushing BMP ligands to bind BMPRII.13,53 For each ligand–receptor complex, the binding location of ActRIIA, ActRIIB and BMPRII is very similar where the receptor interacts using the concave β-sheet surface to compliment the convex, knuckle interface of the ligand (Figure 1(d), left and right, orange). This positions the β-sheets of the receptor to be parallel to the β-strands of the fingers. The location and orientation of ActRIIA and ActRIIB binding is completely different than that observed for TβRII.
For ActRIIA and ActRIIB, a conserved set of aromatic residues in the receptor core constitute the bulk of the interactions by contacting small hydrophobic residues present in the knuckle region of the activin and BMP ligands.16,20 Within finger 1 a trio of residues (Ile-Ala-Pro) are conserved in ActA, ActB, GDF8, and GDF11, which is similar in BMP2 (Val-Ala-Pro). Mutation of this central alanine in BMP2 attenuated type II receptor binding, confirming that small hydrophobic residues are crucial for type II binding.3 In addition to these central hydrophobic interactions, ActA and GDF11 form several electrostatic interactions that flank the ligand receptor interface.13,16 These interactions, along with slight alterations in the hydrophobic core likely account for the differences in affinity exhibited for BMP–type II interactions. Furthermore, TGFβ isoforms have a histidine and glutamate in the corresponding position of the IAP motif which prevents binding to ActRIIA and ActRIIB.3 Thus, two type II receptor binding modes have emerged where the placement and position of the type II receptor is different depending on the ligand. Since the specificity of AMHRII is more restricted, similar to TBRII, it will be interesting to determine if AMHRII binds at the convex knuckle region or the ligand fingertips.
Type I receptor–ligand structures
Structures of type I receptors in complex with various ligands have also illuminated the mechanisms of receptor assembly for each class. Ternary structures of TGFβ, BMP, and GDF11 have revealed a similar positioning of the type I receptor: in the concave interface formed between the ligand fingertips and the wrist helix, including significant contacts with the loop leading into the wrist helix, termed the prehelix loop (Figure 1(d), yellow).8–11,13 Multiple structures, including (BMP2:Alk3, BMP2:ActRIIA:Alk3, BMP2:Alk3/Alk6 chimera, GDF5:Alk3/Alk6) have shown that BMP type I receptors, Alk3 and Alk6, which bind with nM affinity, contact each ligand monomer extensively to form a composite interface.3–9,54 Central to this interaction is a phenylalanine residue of the receptor that projects into a hydrophobic pocket formed on the underside of the ligand fingers, termed a ‘knob-in-hole’. Another BMP type I receptor, Alk1, which is primarily activated through interactions with BMP9 and BMP10, was shown in the ternary structure of Alk1:BMP9:ActRIIB to have a slightly different placement and is tilted so that the β-sheet is pivoted away from the prehelix loop of BMP9 and towards the posthelix loop.12,55 This shift is caused by a two amino acid truncation in the β4–β5 variable loop, causing the loop to be more rigid and closer to the β-sheet, forcing an altered orientation. Modeling Alk1 onto BMP9 in a similar binding position to that seen in the Alk3:BMP2 structure induces severe steric clashes.
Alk5 is the canonical type I receptor for the TGFβ class.46 Structures of TGFβ in complex with Alk5 showed that, as compared to the BMP type I receptors, the type I position is shifted away from the wrist helix and toward the fingertip region, allowing for direct interaction with the long N-terminal portion of TβRII. In fact, inter-receptor interactions constitute a hydrophobic, cooperative interface that enhances binding of the type I receptor, which has extremely low μM affinity for the ligand that is not already bound to TβRII.10 Thus, the organization of TGFβ class receptor interactions is much different from the BMP class.
Interestingly, Alk5 also serves as a low affinity type I receptor for several activin class members, such as GDF8 and GDF11.54 In the recent structure of Alk5:GDF11:ActRIIB, Alk5 is positioned uniquely compared to the BMP and TGFβ type I receptors.13 Here, Alk5 utilizes a phenylalanine to form a knob-in-hole with GDF11 similar to Alk3 and Alk6 binding to BMP ligands, but in contrast, is shifted away from the dimer interface to primarily interact with the tips of the ligand fingers (Figure 1(d)). The fingertips act in a manner similar to TβRII and bind to Alk5 at the β4-β5 variable loop, stabilizing the receptor between the fingertips and the prehelix region. However, no direct inter-receptor interactions are observed, precluding a TGFβ-like cooperative mechanism to account for low type I affinity. This results in two classes of ligands that interact uniquely with the same type I receptor. In fact, Goebel et al. showed that a single point mutation in Alk5 (Phe 84) could render the receptor insensitive to GDF11 activation while maintaining its ability to be activated by TGFβ1.13
While Alk5 is the primary receptor for TGFβ, the activin class ligands are more promiscuous and various ligands can signal using multiple type I receptors, including Alk4, Alk5, and Alk7.26,46 Recently, it was shown that specific residues at the ligand fingertip play important roles in defining which receptors the different activin class ligands can signal through.13 Importantly, differences within the activin class have to be compatible with differences in the β4–β5 loop of the receptors for Alk4/5/7. In addition to the fingertip, the prehelix loop has been shown to be important for receptor specificity.15 While it is possible that the prehelix loop differentially engages ligands, it is likely that differences in the prehelix loop impact ligand flexibility, which has a direct effect on type I specificity. Thus, a combination of flexibility differences and amino acid differences at the ligand fingertip determines which receptors activin class ligands can utilize. This type of binding mode is compatible with a conformational selection model where the ligand can adopt distinct conformations that are compatible with certain receptors, and is distinct from the TGFβ and BMP paradigms.
Thus, is appears that ligands of the TGFβ, the BMP, and activin classes have evolved distinct mechanisms of receptor assembly, underpinned by differences in receptor affinity, receptor positioning, and degrees of ligand flexibility. These differences govern receptor utilization and signaling across the family. While only the extracellular components of a signaling complex have been resolved, a full-length ligand–receptor complex is currently lacking and would set the stage for linking intracellular and extracellular signaling mechanisms.
Extracellular regulation across the TGFβ family
TGFβ prodomain complexes
TGFβ ligands are synthesized as larger precursor proteins that include an N-terminal prodomain and a C-terminal mature signaling growth factor domain (Figure 2(a)).56 The prodomain facilitates proper folding of the ligands and is subsequently cleaved from the mature domain by furin proteases.56–58 In most cases, the prodomain remains non-covalently bound to the growth factor in what is typically referred to as a procomplex.27,32,57,59 The functional significance of the procomplex appears to be highly variable for different family members. Commonly, the procomplex can still signal where the prodomain is believed to be readily displaced by higher-affinity interactions with the receptors.27,59 Whether the prodomain is fully displaced or remains partially bound during signaling remains to be determined for individual family members.
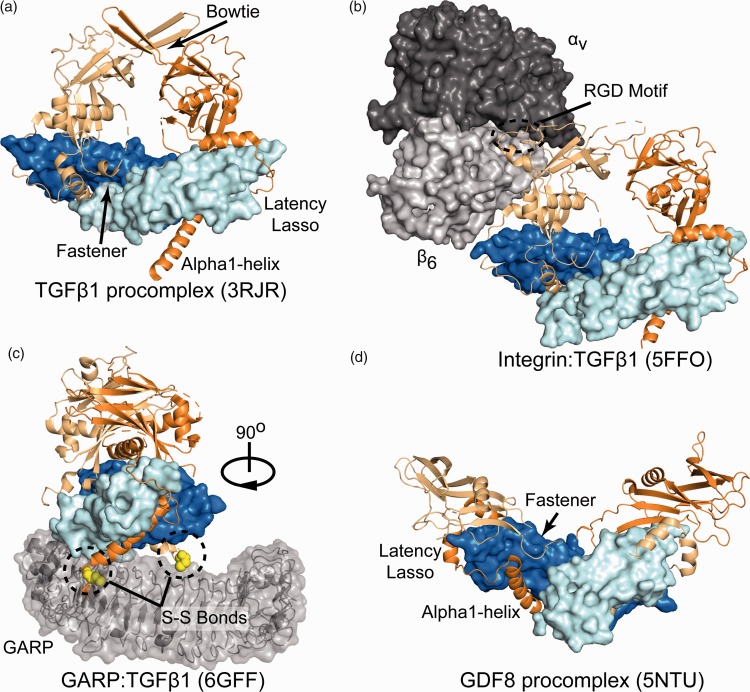
Figure 2.
TGFβ procomplex structures. (a) TGFβ1 procomplex in a closed conformation mediated by the bowtie. Ligand monomers are pale-cyan and blue, and prodomain monomers are orange and light orange with inhibitory elements labeled. (b) The TGFβ1 procomplex bound to αvβ6 integrin with the RGD motif of the prodomain circled. (c) TGFβ1 procomplex bound to GARP rotated 90° to reveal both disulfide bonds anchoring the two proteins. (d) The structure of GDF8 bound to its prodomain revealing the same inhibitory elements as TGFβ1 but adopting an open conformation. (A color version of this figure is available in the online journal.)
In specific cases, the prodomain can render a ligand inactive, keeping it trapped in a latent procomplex. Prodomain-mediated latency is observed within the TGFβ class and the activin class members, GDF8 and GDF11.58,60 For these ligands to signal, different mechanisms are used to liberate the ligand from the latent procomplex. For TGFβ1 and 3, the prodomain interacts with αvβ6 integrin and either latent TGFβ binding proteins (LTBPs) or glycoprotein-A repetitions predominant protein (GARP; Figure 2(b) and (c)).57,61–65 Interactions with LTBP and GARP involve a direct disulfide bond, which anchors the procomplex to the extracellular matrix or the cell surface, respectively.64–66 These larger latent complexes can interact with αvβ6 integrin, which can initiate a tensile force across the complex, releasing the ligand for signaling.57,62,64,67 In contrast, GDF8 and GDF11 are activated by an additional proteolytic event mediated by the tolloid family of metalloproteases.60,68,69 Processing of the prodomain by tolloid, fragments the inhibitory prodomain, weakening its interactions with the growth factor.32,60,70 Recently, structures of the TGFβ1 and GDF8 procomplexes, including higher order complexes of the TGFβ1 procomplex bound to integrin and GARP have provided a better understanding of prodomain-mediated latency and activation.32,57,64,67
The first structure of latent TGFβ1 (prodomain bound to the growth factor) was solved in 2011 and revealed key features of the prodomain that contributed to latency such as the α1-helix, latency lasso, and fastener region.57 These elements inhibit TGFβ signaling by blocking both the type I and type II receptor binding sites. The α1-helix of the prodomain lies perpendicular to the fingers of the ligand within the concave palm of the type I receptor interface (Figure 2(a)). While only a handful of TGFβ ligands contain inhibitory prodomains, the majority of prodomains are predicted to contain an α1-helix, suggesting that this element plays a major role in procomplex formation across the family.53 From the C-terminus of the α1-helix, the prodomain wraps around the fingertips of the ligand to block TGFβ type II receptor binding, in what is referred to as the latency lasso. Following the latency lasso, the prodomain traverses over the knuckles and back toward the C-terminus of the α1-helix. Here, the prodomain loops back to interact with α1-helix, forming the ‘fastener’, which includes key hydrogen bonds between distant prodomain segments that appear to pinch the base of the fingers. Interestingly, mutation of fastener residues has been shown to remove prodomain-mediated latency.32,57,60 Collectively, these inhibitory elements make up the ‘straitjacket’ of the prodomain. In addition, the two prodomain chains continue away from the ligand in the same direction joining together through an interprodomain disulfide bond. The top of this structure is termed the ‘bowtie’ with the prodomains forming either a compact or closed conformation (Figure 2(a)). The structure of the procomplex revealed that the consensus integrin binding motif, RGD, is surface exposed at the base of the bowtie far from the inhibitory straitjacket components, indicating that long distance interactions are important for triggering TGFβ activation.
In 2017, the structure of the TGFβ1 procomplex bound to the αvβ6 integrin head was solved (Figure 2(b)). In the lattice, only one chain of the TGFβ1 prodomain was bound to αvβ6 integrin, leaving one RGD motif unoccupied.67 In the structure, the integrin head is positioned such that the β6 chain is proximal to the growth factor with the αv chain positioned more distally with both the αv and β6 chains interacting with the RGD motif. Comparison to the unbound structure showed that significant structural differences occurred in the conformation of the RGD motif and surrounding area. Interestingly, conformational differences were also observed in the unbound prodomain, implying that integrin binding might have long-range effects that prime the procomplex for force-dependent release of the TGFβ1 ligand. Molecular dynamic simulations showed that a force applied through the β6 subunit of integrin, while artificially anchoring the N-terminus of the prodomains, can disrupt the ligand–straightjacket interactions.67 This study concluded that since the prodomains are covalently linked through a disulfide bond, a force applied through one prodomain monomer is sufficient to fully activate TGFβ1.
To get a better idea for how TGFβ is anchored at the N-terminus of the prodomain, a recent structure of TGFβ1 was solved in complex with GARP with the aid of a stabilizing antibody MHG-8FAB (Figure 2(c)).64 In the immune system, the immunosuppressive activity of Tregs is enhanced by TGFβ1 and has become a recent therapeutic target for cancer immunotherapy.71–73 GARP is a transmembrane protein expressed on the surface of Tregs and forms a disulfide bond with each TGFβ1 prodomain chain, tethering latent TGFβ1 to the cell surface.74–76 GARP contains leucine rich repeats that adopts a horseshoe-like structure similar to toll-like receptors. Within GARP, two unpaired cystines, C211 and C350, are involved in linking both prodomain chains of latent TGFβ1 procomplex through the N-terminus of the α1-helix.64
Together, these studies highlight a TGFβ activation mechanism that requires both chains of the prodomain to be tethered to GARP, while a force is generated by a single molecule of integrin.64,65,67 While GARP is bound to cell surface for TGFβ activation, TGFβ is also found tethered to the ECM through LTBPs.62,65 Since LTBPs are structurally distinct from GARP, containing multiple domains, future studies are needed to determine if LTBPs have a similar mechanism of activation for the latent TGFβ procomplexes.
Recently, the structure of the latent GDF8 procomplex was solved and was found to contain the same inhibitory elements described from TGFβ1 (Figure 2(d)).32 Similarly, mutations of the fastener residues resulted in a decrease in prodomain-mediated latency.60 Interestingly, mutation of hydrophobic residues, specifically I56 and I53 of GDF8, within the α1-helix also removed latency, but were shown to be active without the need for tolloid proteolysis.32,60 However, despite sharing similar inhibitory elements, the TGFβ1 and GDF8 procomplexes form distinct conformations. Unlike TGFβ, the prodomain of GDF8 lacks an interdisulfide bond, which allows for a more open V-like conformation as compared to the closed conformation of TGFβ1 (Figure 2(d)). Interestingly, this open conformation exposes the tolloid cleavage site required for latent GDF8 activation. Further studies using hydrogen–deuterium exchange revealed that processing by tolloid primes the prodomain for dissociation by weakening its interaction with the ligand.70 These proteolytic fragments are easily displaced by the receptors allowing for GDF8 to signal. Currently, the mechanism of latent GDF8 activation by tolloid, including recognition of the procomplex by tolloid and the spatial and temporal regulation of activation in vivo remains under investigation.
Interestingly, the structure of both the TGFβ1 and GDF8 procomplexes revealed a ‘cross-over’ mechanism, where the prodomain translated with one monomer forms interactions with the opposite monomer.32,77 It is interesting to speculate that a cross-over mechanism is important for defining which ligands can hetero- and homo-dimerize. Thus, the prodomain must be ‘compatible’ with the growth factor domain of the dimerization partner. In addition, prodomain–prodomain interactions might also be important as both the TGFβ1 prodomains and GDF8 prodomains directly contact each other. However, more research needs to be conducted to ascertain the role of the prodomain in hetero- and homodimer formation.
Besides serving to help properly fold the growth factor, these procomplexes could serve additional roles. As discussed, TGFβ superfamily ligands are highly hydrophobic and utilize their hydrophobicity to interact with cell surface receptors. Due to this characteristic, it is unlikely that ligands in serum are in an apo- or unbound state. Thus, it is likely that the prodomain serves to increase their half-life in serum or to protect the ligand from the variety of extracellular antagonists found within serum.78 Complexes of BMP9 and ActA procomplexes have been solved and represent non-inhibitory procomplexes.27,59 Both complexes were solved in an open configuration; however, they exhibited very different interactions with their prodomains. For ActA, the α1-helix was positioned in the palm of the dimer interface, while BMP9 adopted a completely different complex utilizing an α5-helix positioned within the palm. Thus, within the family prodomain–ligand interactions have evolved a variety of functions that provide ligand-specific regulation.
TGFβ antagonist complexes
TGFβ family ligands are also regulated by a wide array of structurally diverse extracellular protein antagonists (Figure 3). Protein antagonists consist of varying domain architectures and inhibit ligands through different binding mechanisms.53 The majority function by blocking both the two type I and two type II receptor binding sites to prevent receptor assembly and signaling. Often antagonists accomplish this by binding in a 2:1 stoichiometric ratio (2 antagonists, 1 ligand dimer) where one antagonist molecule will block a single type I and a single type II site. However, antagonists such as chordin and WFIKKN2 provide exceptions where each bind in an asymmetric 1:1 ratio.79–81 In this section, we will highlight the structural studies that have revealed different mechanisms for ligand antagonism.
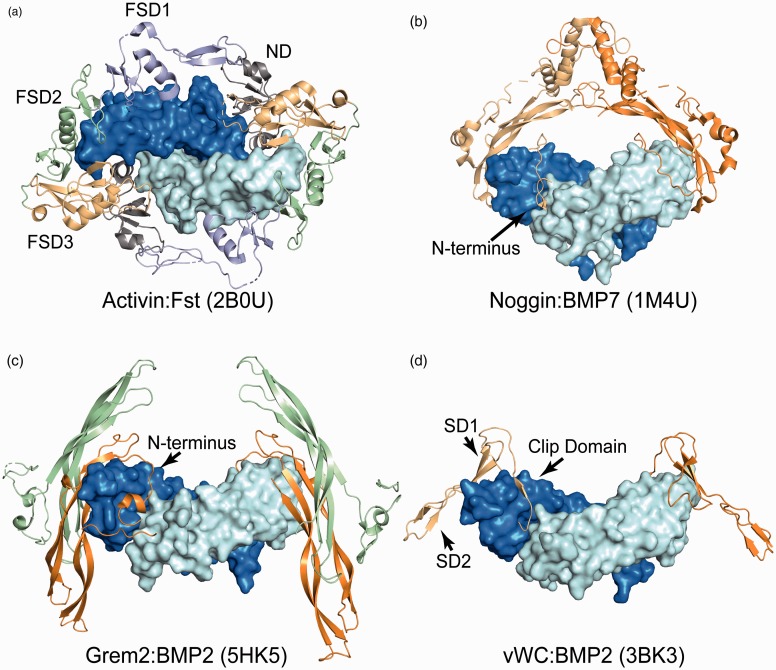
Figure 3.
TGFβ antagonist structures. (a) Top down view of an activin dimer, surface representation with monomers in pale-cyan and blue, bound to two monomers of follistatin (cartoon). Domains of one follistatin monomer are labeled, ND: N-terminal domain (gray), FSD1–3: follistatin domains 1–3, light blue, green and light orange. (b) Surface representation of BMP7 bound to two monomers of Noggin (cartoon). The N-terminus of one monomer can be seen threaded through the type I receptor site. (c) Structure of BMP2 (surface) bound to two Grem2 dimers, one monomer in orange and the other in green. The N-terminal extension occupying the type I receptor site is labeled. (d) BMP2 bound to two vWC domains of CV-2. The clip domain is within the type I receptor binding site, SD1 is bound to the type II receptor site and SD2 is not contacting the ligand. (A color version of this figure is available in the online journal.)
The first structure of an antagonist was solved in 2002 and revealed how the antagonist, Noggin, binds and inhibits BMP7 (Figure 3(b)).82 Interestingly, Noggin is single-domain protein secreted as a disulfide-linked homodimer, containing a cystine-knot motif and sharing remarkable homology to TGFβ ligands. The Noggin dimer binds symmetrically to the ligand dimer and is positioned similar to where the TGFβ prodomain is located. The structure revealed that each chain of Noggin sequesters a type I and type II receptor binding site. To interfere with type I receptor binding, Noggin binds BMP7 using an unstructured N-terminus that threads into the type I site similar to the strategy used by the α1-helix of the prodomains. A number of residues within the N-terminus were shown to be important for proper antagonism and mutations within this region cause a number of diseases such as symphalangism and tarsal/carpal coalition syndrome.83–85 On the outside of the ligand, the type II receptor sites are blocked by the ‘fingertips’ of Noggin somewhat similar to a caliper that pinches the distal ends of the ligand. Since the Noggin chains are disulfide-linked, the nM binding affinity is a result of avidity and binding of a single noggin dimer blocks all four receptor sites of BMP7.82
The next antagonist–ligand complex was solved in 2005 between Follistatin (Fst) and ActA (Figure 3(a)).30 Unlike Noggin, Fst is a multidomain protein containing an N-terminal Domain (ND) and three tandem follistatin domains (FSDs). Two molecules of Fst bind the ActA dimer in a ring-like structure. The ND of each Fst occupies the type I receptor binding site with FSD1 and FSD2 covering the type II receptor site. The two Fst molecules interact in a head-to-tail mechanism where the ND of one Fst interacts with the FSD3 of the neighboring Fst. Several structures of Fst in complex with various ligands have highlighted a conformational selection model where both the antagonist and ligand adopt a preferred conformation for binding.15,30,33,86 In particular, the ND of Fst can adopt different conformations to accommodate the wrist region of different ligands. This mechanism provides the basis for Fst’s ability to be promiscuous and target multiple ligands, including activin (ActA/B, GDF8/11) and BMP (BMP2/4/6/7) class ligands. Similarly, Follistatin-Like 3 (Fstl3), which lacks the third FSD, was solved in complex with both ActA and GDF8.29,33 Here, the ND ligand interactions are less flexible, partially explaining why Fstl3 antagonizes fewer family members. Collectively, these structures have been important for understanding how an antagonist binds different ligands, and also, how a particular ligand is neutralized by different antagonists.
Members for the differential screening-selected gene in neuroblastoma (DAN) family (Gremlin-1, Gremlin-2, Dan, Cerberus, Coco, USAG-1 and SOST) have been shown to bind and neutralize BMP ligands with varying potencies.53,87 Certain members, such as SOST, do not inhibit BMP, but instead interfere with Wnt signaling by interacting with the Wnt coreceptor, LRP5/6.88,89 The NMR structures of SOST revealed a growth factor-like fold, strikingly similar to TGFβ ligands, with a cystine rich domain and a set of β-stranded ‘fingers’.90,91 Subsequently, the crystal structures of Gremlin-2, DAN, and Gremlin-1 all revealed a unique head-to-tail dimeric structure where the dimer interfaces were stabilized by a strong antiparallel β-strand interaction between the two chains (Figure 3(c)).92–94 Unlike Noggin, the dimer was not disulfide-linked even though a number of DAN family members contain an unpaired cysteine at the dimer interface. In 2016, the structure of Gremlin-2 was solved in complex with GDF5 making it the only antagonist to date where the full-length structure was solved in a bound and unbound state.93,94 The structure of Gremlin-2:GDF5 resembles an H shape, with two Gremlin-2 dimers binding perpendicular at each end of the ligand dimer (Figure 3). Interestingly, within the crystal lattice each Gremlin-2 monomer is bound to a single chain of GDF5 creating a long daisy chain of alternating Gremlin-2 and GDF5 ligand dimers. A comparison of the two Gremlin-2 structures reveals that, like Noggin, a long, flexible, N-terminus binds to the type I receptor binding site of GDF5. However, having both the bound and unbound structures of Gremlin-2 provides insight into the molecular transitions that occur upon binding ligand. In the unbound state the N-terminus of Gremlin-2 is helical and sits on top of the dimer, shielding a major hydrophobic cleft. Upon ligand binding, the helix unwinds, forming a β-sheet interaction with the fingers of the ligand while threading the N-terminus into the palm of the ligand. This exposes a number of hydrophobic residues in Gremlin-2 that engage the ligand at the type II receptor binding location. Thus, the N-terminus appears to shield hydrophobic residues prior to ligand binding, offering protection from non-specific interactions.
WFIKKN1 and WFIKKN2, named for its conserved domain architecture, a whey acidic protein, FSD, immunoglobulin domain, 2 tandem Kunitz domains and a netrin domain, is a specific antagonist for GDF8 and GDF11.95–97 Limited structural information is available for how WFIKKN1/2 bind and antagonize ligands. Currently, the Kunitz 2 domain of WFIKKN1 and the FSD of WFIKKN2 have been solved by X-ray crystallography in the absence of ligand.98,99 The Kunitz 2 domain resembles a typical Kunitz protease inhibitor, containing two antiparallel β-strands, an adjacent α-helix and a flexible loop that is predicted to insert into target proteases, namely trypsin.98 The structure of WFIKKN2 FSD revealed a similar two domain architecture with an epidermal growth factor (EGF) and Kazal subdomain that is conserved in other FSDs. However, the orientation of the WFIKKN2 FSD subdomains is unique from Fst FSDs 1-3, including FSD1 of Fst unbound to ligand.99 Currently, the molecular mechanisms utilized by the multiple domains of WFIKKN to inhibit GDF8 and GDF11 is unknown, as no structure of the domains or full-length WFIKKN has been solved.
Like WFIKKN, the antagonist Chordin also contains a number of domains within the full-length protein, one C-terminal Von Willebrand factor C (vWC) domain, four tandem chordin specific domains, and three N-terminal vWC domains.81,100–102 No structure of mammalian chordin has been solved; however, insight into how vWC domains bind BMP ligands was revealed by the structure of the C-terminal vWC of drosophila crossveinless-2 (CV-2) in complex with BMP2 (Figure 3(d)).100 The vWC structure resembles a hook, with a clip domain (the hook) and two subdomains SD1 and SD2 that form a stalk. The vWC hooks over the top of the BMP2 knuckles and the clip is inserted into the concave palm of the type I receptor site with the SD1 pressed against the type II receptor site. Two other vWC structures not bound to a ligand from collagen 2a and CCN3, both known to interact with BMPs, were solved by X-ray crystallography.102 Interestingly, a structural comparison revealed that the other vWCs bind BMPs using a different molecular mechanism as they lack the N-terminal clip and SD1 epitopes found in CV-2 vWC. Although the vWC of a chordin homolog has been solved, more work is required to identify how mammalian chordin functions and what role the other domains serve during antagonism.
In general, TGFβ ligands are highly regulated by both the prodomains they are synthesized with and a variety of extracellular protein antagonists. Over the years, structural work has highlighted major differences in the mechanism of binding. However, many questions remain, including how higher-order complexes with the extracellular matrix impact ligand activity. Further, only four procomplexes of family members are available, each describing vastly different structures. Certainly, additional structural data are needed to define these mechanisms for individual ligands, and further to understand how large multidomain extracellular antagonists, such as WFIKKN and chordin neutralize ligands. These studies will help support the growing need to develop therapeutics that modulate TGFβ ligands.
Intracellular regulation across the TGFβ family
Structures of kinase domains and SMAD proteins
The type I and type II TGFβ receptors contain an intracellular serine/threonine kinase domain. Similar to other kinases, the domain is comprised of a small (∼70 residues) N-terminal lobe (N lobe), a hinge region linker, and a larger (∼250 residues) C-terminal lobe (C lobe). Type I receptors differ from the type II receptors by the presence of an N-terminal glycine-serine rich (GS) domain (∼30 residues) which regulates kinase activity and Smad binding. When not active, a small 12 kDa immunophilin protein termed FK506-binding protein 1A (FKBP12) engages the GS domain keeping the type I receptor in an inactive state.103–106 Activation of the type I receptor occurs upon assembly of the receptor–ligand complex, where the constitutively active type II receptor phosphorylates serine and threonine residues within the GS domain of the type I receptor, causing a conformational change to the GS domain.107 This conformational change dissociates FKBP12, liberating the active site of the GS domain for Smad binding.104 Subsequently, the active type I receptor will phosphorylate an S-X-S motif on the distal C-terminus of Smad proteins causing them to accumulate in the nucleus and to interact with transcriptional machinery.108,109 The intracellular components of the TGFβ family have been reviewed by many others including Chaikuad and Bullock,110 Massagué,111 Heldin and Moustakas,112 Hill,113 and Hata and Chen.114 The purpose of this review is to not only summarize other reviews, but to also focus on the information gained from structures of the intracellular molecules of the TGFβ family. This section will describe the structural and molecular features of the intracellular type I receptors, type II receptors, and Smad proteins that contribute to TGFβ signaling outcomes, and highlight certain gaps in our current understanding of TGFβ signal transduction.
Type I receptor kinase domain structures
The first solved kinase domain structure within the TGFβ family was the type I receptor, Alk5, in complex with FKBP12 by Huse et al.103 This structure revealed that overall type I receptors adopt a canonical protein kinase fold similar to other protein kinases such as, cAMP-dependent protein kinase PKA115 (Figure 4). As such, Alk5 contains an N lobe consisting of a twisted five-stranded β-sheet and a single α helix termed αC. Importantly, the loop between β4 and β5 (the L45 loop) contains sequence specific elements that are important for dictating specificity for Smad2/3 or Smad1/5/8.116,117 The GS domain, which is positioned in between the cell membrane and the N-lobe, is phosphorylated by the type II receptor and provides a critical switch mechanism regulating type I kinase activity. The GS domain adopts a helix–loop–helix segment, N-terminal to the β-sheet. In contrast to the N lobe, the C lobe of the receptor is mostly α-helical with a catalytic loop containing a His-Arg-Asp (HRD) motif and an activation loop initiated by an Asp-Phe/Leu-Gly (DF/LG) motif. The activation loop motif binds to ATP and a magnesium ion when the receptor is in the active conformation. However, in the structure of Alk5, the activation loop forms a β-hairpin supported by a one and half turn extension of an α-helix. This interacts with the GS region and the αC helix to restrict the motion of the helix and hold the receptor in its unphosphorylated, inactive state. Phosphorylation of the GS domain is thought to activate the kinase by causing a conformational change of the αC and activation loop providing substrate (Smad) access. When phosphorylated, the GS domain is thought to interact directly with the Smad substrate providing an additional binding epitope for Smad molecules.
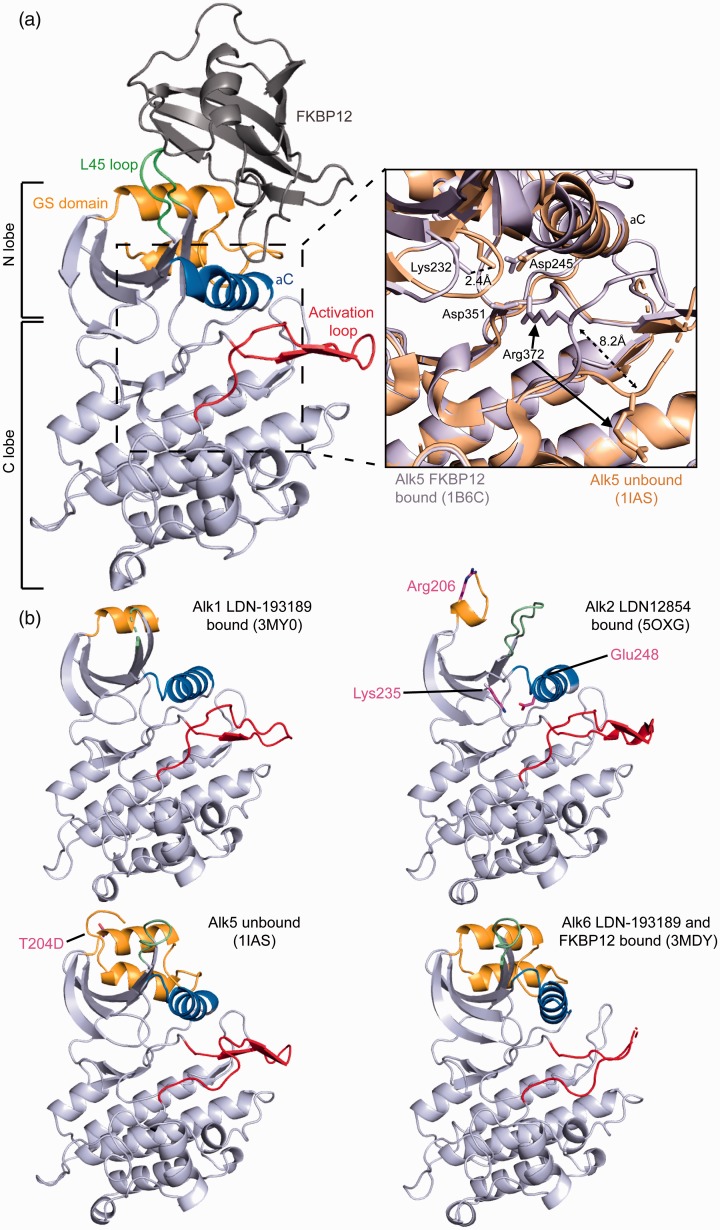
Figure 4.
TGFβ receptor kinase domain. Structure of Alk5 kinase domain in complex with FKBP12. Key structural features are colored and labeled, L45 loop (green), GS domain (orange), aC helix (blue), activation loop (red), FKBP12 (dark gray). N-terminal (N-lobe), and C-terminal (C-lobe) lobes are labeled to the left. Inlet shows the conformational changes made to two salt bridges in Alk5 either bound (light gray) or unbound (tan) to FKBP12. (A color version of this figure is available in the online journal.)
In 2001, the structure of Alk5 lacking FKBP12 was solved.104 This new structure provided the first insights into the dynamics of these receptors. By aligning the two structures and observing the differences in the FKBP12 bound and unbound receptors, it became clear that FKBP12 was in fact playing a crucial role in holding the receptor in an inactive state to inhibit ligand-independent Smad signaling. The structure revealed that a catalytic salt bridge formed between Glu245 on the αC helix and Lys232 on β3 is broken when FKBP12 is bound, while a second salt bridge is created between Arg372 and Asp351 that blocks the ATP binding site within the activation loop (Figure 4).103,104,110 These two structures set the framework for understanding the intracellular receptors at the molecular level mechanistically and providing a structural basis for FKBP12 regulation within the TGFβ family. Presently, crystal structures are available for four of the seven type I receptor kinase domains (Alk1, Alk2, Alk5, and Alk6103,104,118–138).
These receptor structures have allowed us to relate a number of mutations that occur in the kinase domain with their potential impact on kinase activity. For instance, a mutation, T204D, in Alk5 causes constitutive activation, resulting in signaling independent of ligand binding and phosphorylation by the type II receptor. Interestingly, this activity occurs because these mutations prevent the binding of FKBP12 to Alk5 and cause irregular signaling of the receptor. Furthermore, this ligand-independent activation of Alk5 has been shown to result in the development of ovarian sex cord-stromal tumors.139 The crystal structure of the Alk5 T204D kinase domain depicts that changes occur in the αC helix and L45 loop which could also alter Smad binding.129 However, how the T204D mutant of Alk5 is able to enhance autophosphorylation of the receptor is unclear. Further understanding of receptor activation would be greatly enhanced from structures of these receptors in a phosphorylated state or bound to a Smad protein.
Additionally, a mutation, R206H, in the GS region of the type I receptor Alk2 has been associated with the skeletal malformation disorder, fibrodysplasia ossificans progressive (FOP), a rare disorder characterized by the replacement of skeletal muscle and connective tissues by bone. The structure of Alk2 with this mutation shows that the salt bridge normally formed by the binding of FKBP12 that normally holds the receptor in an inactive state is disrupted, thus making the receptor constitutively active.124 While many studies have suggested this to be the basis for the disease, recent studies have shown that this mutation actually alters ligand specificity. Besides BMP ligands, Alk2 can also interact with ActA and for wild-type Alk2, ActA binding does not typically produce a signal, and can instead act as a competitive inhibitor to BMP. However, with the R260H mutation, ActA can now signal through Alk2 leading to aberrant activation of Smad 1/5/8.140 How this intracellular mutation leads to an alteration in ligand specificity outside of the cell remains unknown and emphasizes that receptor activation is more complicated than merely a co-localization of type I and type II receptors.
Two recently published structures of the kinase domain of Alk2 in complex with a small molecule inhibitor have helped to identify important residue contacts between the receptor and inhibitor. In the structure solved by Williams and Bullock in 2018, it was shown that important interactions between residues Lys235 and Glu248 in Alk2 play a crucial role in water-mediated hydrogen bonding with the small molecule inhibitor, LDN-212854.119 With their work, they were able to explain the specificity of this inhibitor for Alk2 where these interactions are not capable of forming in Alk5 and account for the increased potency when compared to other commonly studied Alk2 inhibitors, such as LDN-193189.120 By using the structure of Alk2 FOP mutant, R206H, in complex with a small molecule inhibitor, RK-59638, recent work has been able to identify potent inhibitors of Alk2 R206H with increased oral bioavailability.124 Studies such as the two described here, open the doors for better therapeutic design by being able to specifically target interactions to lock the receptor in non-signaling configuration.
Type II receptor kinase domain structures
The first structure of a type II receptor kinase domain was ActRIIB, which was solved in 2007.141 Much like their ECD, these Ser/Thr kinase receptors all show a high level of structural homology. The type II receptor structure revealed the same overall kinase fold seen in all other kinases. However, two major differences between the structure of ActRIIB, a type II receptor, and Alk5, a type I receptor, are the lack of a GS domain in ActRIIB and a difference in the activation loop of the C lobe (residues 339–368 in ActRIIB). As mentioned, in the Alk5 structure, this activation loop helps to hold Alk5 in an inactive state. However, in ActRIIB, this β-hairpin is absent and thus does not restrict the receptor in an inactive state. Interestingly, this region is sequentially variable between type I and type II receptors and shows a high level of flexibility.141 Additionally, the lack of the GS domain in the type II receptors limits FKBP12 to interactions with only the type I receptors. These differences in the type I and type II receptors are consistent with the fact that the type II receptors are constitutively active and not held inactive by FKBP12. To date, structures are available for four of the five type II receptor kinase domains (ActRIIA, ActRIIB, BMPRII, and TBRII121,129,137,141,142).
Smad protein structures
In addition to studying the receptor kinase domain structures and activation, structures of Smads and additional interacting proteins have been studied in order to better understand the mechanisms of signal transduction. As mentioned above, activated type I receptors will phosphorylate and activate receptor-specific Smad proteins or R-Smads. Upon phosphorylation, these R-Smad proteins will form heteromeric complexes with co-Smad4. Activated R-Smads, together with Smad4 will accumulate in the nucleus to regulate target gene expression. Smad proteins are composed of two globular domains termed Mad homology 1 (MH1) and MH2. The two domains are held together by a proline-rich linker. The N-terminal MH1 domain serves as the DNA-binding domain with a hairpin structure within this domain allowing it to bind DNA.111 The C-terminal MH2 domain serves as the binding site for the type I receptors which recognize a common Ser-X-Ser motif at the extreme C-terminus of R-Smad.143,144 Additionally, the MH2 domain is used for interactions with other Smad proteins, cofactors, co-activators, and co-repressors of this pathway.
The first crystal structure of a Smad protein was the structure of the co-Smad4 MH2 domain which showed a trimeric structure.145 Importantly, this structure showed that most tumor-derived missense mutations were located within the protein–protein interface that facilitated trimeric assembly, demonstrating that oligomerization is crucial for signaling.145 The MH2 domains contain a central β-sandwich core between conserved three-helix bundles on one side and a conserved loop-helix region on the other (Figure 5(a)). Following this structure, several other structures were solved, including the structures of unphosphorylated and phosphorylated Smad2 and the heterotrimeric complexes of R-Smad2 or R-Smad3 with co-Smad4.146–148
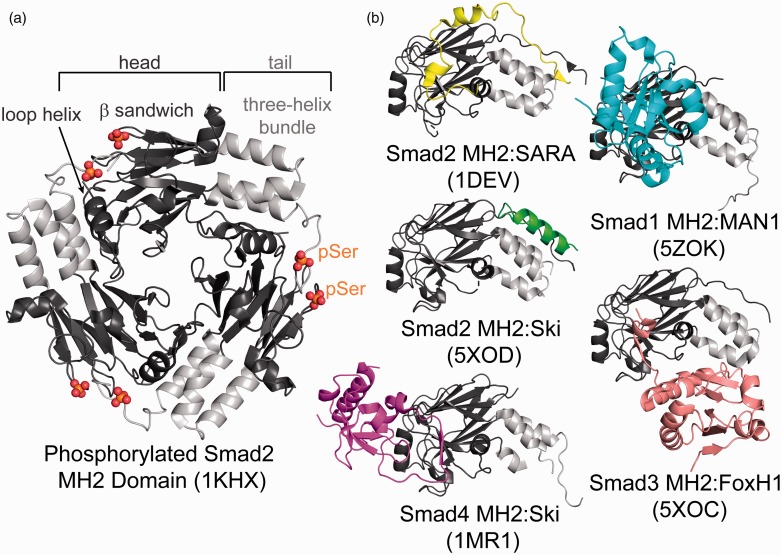
Figure 5.
Smad oligomerization and binding proteins. (a) Structure of the trimeric state of Smad 2 MH2 domain binding in a head (dark gray) to tail (light gray) mechanism. Phosphorylation sites on Ser residues are highlighted in red and orange. Structural features are labeled on top. (b) Structures of Smad MH2 domains (head: dark gray, tail: light gray) in complex with binding partners, SARA (yellow), MAN1 (blue), Ski (green and purple), FoxH1 (pink), showing differential binding sites on the Smad MH2 domain. (A color version of this figure is available in the online journal.)
Structures of the MH2 domain of R-Smad2, phosphorylated and unphosphorylated, have given insight into the tertiary structure of Smad molecules and how activation stabilizes the oligomeric state.146,147 The structure of phosphorylated R-Smad2 showed that there is a head-to-tail interaction where the phosphorylation at the C-terminus stabilized the oligomeric form through interactions with a basic patch on an adjacent Smad MH2 N-terminus147 (Figure 5(a)). Pivotal to understanding Smad–Smad interactions, in 2004 heterotrimeric complex structures of either two R-Smad2 or R-Smad3 MH2 domains in complex with the co-Smad4 MH2 domain were solved.148 The structure showed that unique electrostatic interactions within the heteromeric interfaces favor Smad assembly of two R-Smads and one co-Smad4 in TGFβ signaling.148
Over the years, it has been well established that MH2 domains form a primary hub for other Smad interaction proteins. The structure of the R-Smad2 MH2 domain in complex with the Smad binding domain (SBD) of Smad anchor for receptor activation (SARA) provided the first example for how proteins can interact with the Smad MH2 domains.146 This structure showed that the SBD peptide interactions with the MH2 domain were derived through binding hydrophobic residues across both the β-strand and three-helix bundle. Next, the complex structure between the co-Smad4 MH2 domain and the corepressor, Ski, shows that Ski binds within the binding surface on co-Smad4 required for R-Smad binding.149 Ski forms a compact structure containing a core with four β-strands which pack into three α-helices, a fourth large C-terminal α helix, and a fifth β-strand which binds co-Smad4. This β-strand is within a large interaction loop (I-loop) in the middle of Ski that forms hydrophobic interactions with the bottom of the β-sandwich of the co-Smad4 MH2 domain. Conversely, a recently solved structure of the complex between R-Smad2 and Ski shows a different binding mode, in which Ski only interacts with the three-helix bundle, making it a unique interaction compared to the structure with co-Smad4.150 Recently, the structure of MAN1, which terminates TGF-β signaling through Smad binding, was solved in complex with the MH2 domain of Smad1 and Smad2 and shows a different Smad binding mode. MAN1 engages the MH2 domain through interactions with α-helix 2 of the bundle motif and multiple β-strands in the β-sandwich including the L3 loop.151 Further diversity in Smad binding was shown by the same group, who published the structure of the Smad interacting domain of FoxH1 in complex with the MH2 domain of Smad3.150 These structures highlight that interactions can occur through different binding epitopes, demonstrating the complexity of these Smad protein mechanism (Figure 5(b)).
In 1998, the structure of the MH1 domain of Smad3 bound to DNA depicted the first structure of a MH1 domain.152 The MH1 domain consists of an N-terminus with three α-helices and a C-terminus with six β-strands which form two small β-sheets and one β-hairpin. In the middle of the MH1 domain is a hydrophobic core which contains a fourth α-helix. The β-hairpin interacts and binds the phosphodiester backbones of DNA by hydrogen bonding.152 Structures of the MH1 domains of Smad5 and co-Smad4 have also been resolved by X-ray crystallography.153,154
While the structures describe here have given us many tools for understanding the dynamic assemblies of intracellular proteins within the TGFβ family, many questions still exist within the field. In TGFβ signaling, it is generally thought that these kinase domains will interact with a number of proteins to affect downstream signaling. The structural basis of these interactions has not been resolved. In part, this could be because they are part of a higher order complex or because they are transient in nature, making them more difficult to study at the structural level. For example, a major area of uncertainty is the type I receptor and R-Smad interface. This complex structure will be crucial to finally understanding, at the molecular level, the innerworkings of how type I receptors interact with Smads and elude to preferential selection of different receptors for specific Smads. Another major gap is the molecular mechanism of how the type I and type II kinase domains interact. The interactions necessary for the type II receptors to phosphorylate the GS domain of the type I receptor are currently unknown. Interestingly, recent studies have suggested that type I receptors interact, independent of type II receptors, resulting in type I receptor cross-phosphorylation, which could be similar, in principle, to homodimeric activation of receptor tyrosine kinases.108 Furthermore, a structure of a full Smad complex (MH1 and MH2 domains) is lacking, partially because of the difficulties in making these proteins and the flexible linker between these MH domains, which is highly regulated by post-translational modifications. Certainly, the relative position of the intracellular kinase domains might be impacted by the spatial distribution of these receptors mediated through ligand assembly of the ECD, and thus there is a need to characterize the full-length receptor complex.
Concluding remarks
In summary, over the past two decades significant progress has been made understanding how TGF-β ligands signal and are regulated. However, given the complexity and diversity of the family significant gaps still exist for a number of ligands. In the coming decades, as structural techniques improve, methods such as cryo-EM will play an important role in imaging larger more complex structures.
AUTHOR CONTRIBUTIONS
All authors (EJG, KNH, JCM, and TBT) participated in the composing and writing of the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Thomas B Thompson is a consultant for Acceleron Pharma.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Foundation for the National Institutes of Health [grant number R01GM127726].
References
- 1.Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, Grütter MG. The crystal structure of TGF-beta 3 and comparison to TGF-beta 2: implications for receptor binding. Protein Sci 1996; 5:1261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheufler C, Sebald W, Hülsmeyer M. Crystal structure of human bone morphogenetic protein-2 at 2.7 a resolution. J Mol Biol 1999; 287:103–15 [DOI] [PubMed] [Google Scholar]
- 3.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol 2000; 7:492–6 [DOI] [PubMed] [Google Scholar]
- 4.Keller S, Nickel J, Zhang J-L, Sebald W, Mueller TD. Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol 2004; 11:481–8 [DOI] [PubMed] [Google Scholar]
- 5.Kotzsch A, Nickel J, Seher A, Heinecke K, van Geersdaele L, Herrmann T, Sebald W, Mueller TD. Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome. J Biol Chem 2008; 283:5876–87 [DOI] [PubMed] [Google Scholar]
- 6.Kotzsch A, Nickel J, Seher A, Sebald W, Müller TD. Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity. Embo J 2009; 28:937–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klammert U, Mueller TD, Hellmann TV, Wuerzler KK, Kotzsch A, Schliermann A, Schmitz W, Kuebler AC, Sebald W, Nickel J. GDF-5 can act as a context-dependent BMP-2 antagonist. BMC Biol 2015; 13:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci U S A 2006; 103:7643–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, Mueller TD. A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol 2007; 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 2008; 29:157–68 [DOI] [PubMed] [Google Scholar]
- 11.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem 2010; 285:14806–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Townson SA, Martinez-Hackert E, Greppi C, Lowden P, Sako D, Liu J, Ucran JA, Liharska K, Underwood KW, Seehra J, Kumar R, Grinberg AV. Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J Biol Chem 2012; 287:27313–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goebel EJ, Corpina RA, Hinck CS, Czepnik M, Castonguay R, Grenha R, Boisvert A, Miklossy G, Fullerton PT, Matzuk MM, Idone VJ, Economides AN, Kumar R, Hinck AP, Thompson TB. Structural characterization of an activin class ternary receptor complex reveals a third paradigm for receptor specificity. Proc Natl Acad Sci U S A 2015; 116:15505–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem 1996; 237:295–302 [DOI] [PubMed] [Google Scholar]
- 15.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: insights into receptor utilization and heparin binding. Embo J 2009; 28:2662–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActRIIB:activin a complex reveal a novel binding mode for TGF-beta ligand:receptor interactions. Embo J 2003; 22:1555–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allendorph GP, Isaacs MJ, Kawakami Y, Izpisua Belmonte JC, Choe S. BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors. Biochemistry 2007; 46:12238–47 [DOI] [PubMed] [Google Scholar]
- 18.Saremba S, Nickel J, Seher A, Kotzsch A, Sebald W, Mueller TD. Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J 2008; 275:172–83 [DOI] [PubMed] [Google Scholar]
- 19.Griffith DL, Keck PC, Sampath TK, Rueger DC, Carlson WD. Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily. Proc Natl Acad Sci U S A 1996; 93:878–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 2003; 11:605–17 [DOI] [PubMed] [Google Scholar]
- 21.Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, Roschke V, Sanyal I, Choe S. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem 2005; 280:25111–8 [DOI] [PubMed] [Google Scholar]
- 22.Wei Z, Salmon RM, Upton PD, Morrell NW, Li W. Regulation of bone morphogenetic protein 9 (BMP9) by redox-dependent proteolysis. J Biol Chem 2014; 289:31150–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S-I, Hyvönen M. Structural basis for the inhibition of activin signalling by follistatin. Embo J 2006; 25:1035–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TbetaR2 ectodomain–TGF-beta3 complex. Nat Struct Biol 2002; 9:203–8 [DOI] [PubMed] [Google Scholar]
- 25.Huang T, Schor SL, Hinck AP. Biological activity differences between TGF-β1 and TGF-β3 correlate with differences in the rigidity and arrangement of their component monomers. Biochemistry 2014; 53:5737–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker RG, Czepnik M, Goebel EJ, McCoy JC, Vujic A, Cho M, Oh J, Aykul S, Walton KL, Schang G, Bernard DJ, Hinck AP, Harrison CA, Martinez-Hackert E, Wagers AJ, Lee RT, Thompson TB. Structural basis for potency differences between GDF8 and GDF11. BMC Biol 2017; 15:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Fischer G, Hyvönen M. Structure and activation of pro-activin A. Nat Commun 2016; 7:12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwald J, Vega ME, Allendorph GP, Fischer WH, Vale W, Choe S. A flexible activin explains the membrane-dependent cooperative assembly of TGF-beta family receptors. Mol Cell 2004; 15:485–9 [DOI] [PubMed] [Google Scholar]
- 29.Stamler R, Keutmann HT, Sidis Y, Kattamuri C, Schneyer A, Thompson TB. The structure of FSTL3.activin a complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity. J Biol Chem 2008; 283:32831–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell 2005; 9:535–43 [DOI] [PubMed] [Google Scholar]
- 31.Lerch TF, Shimasaki S, Woodruff TK, Jardetzky TS. Structural and biophysical coupling of heparin and activin binding to follistatin isoform functions. J Biol Chem 2007; 282:15930–9 [DOI] [PubMed] [Google Scholar]
- 32.Cotton TR, Fischer G, Wang X, McCoy JC, Czepnik M, Thompson TB, Hyvönen M. Structure of the human myostatin precursor and determinants of growth factor latency. Embo J 2018; 37:367–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cash JN, Angerman EB, Kattamuri C, Nolan K, Zhao H, Sidis Y, Keutmann HT, Thompson TB. Structure of myostatin·follistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J Biol Chem 2012; 287:1043–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apgar JR, Mader M, Agostinelli R, Benard S, Bialek P, Johnson M, Gao Y, Krebs M, Owens J, Parris K, St Andre M, Svenson K, Morris C, Tchistiakova L. Beyond CDR-grafting: Structure-guided humanization of framework and CDR regions of an anti-myostatin antibody. MAbs 2016; 8:1302–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padyana AK, Vaidialingam B, Hayes DB, Gupta P, Franti M, Farrow NA. Crystal structure of human GDF11. Acta Crystallogr F Struct Biol Commun 2016; 72:160–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenwald J, Fischer WH, Vale WW, Choe S. Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase. Nat Struct Biol 1999; 6:18–22 [DOI] [PubMed] [Google Scholar]
- 37.Boesen CC, Radaev S, Motyka SA, Patamawenu A, Sun PD. The 1.1 a crystal structure of human TGF-beta type II receptor ligand binding domain. Structure 2002; 10:913–9 [DOI] [PubMed] [Google Scholar]
- 38.Marlow MS, Brown CB, Barnett JV, Krezel AM. Solution structure of the chick TGFbeta type II receptor ligand-binding domain. J Mol Biol 2003; 326:989–97 [DOI] [PubMed] [Google Scholar]
- 39.Mace PD, Cutfield JF, Cutfield SM. High resolution structures of the bone morphogenetic protein type II receptor in two crystal forms: implications for ligand binding. Biochem Biophys Res Commun 2006; 351:831–8 [DOI] [PubMed] [Google Scholar]
- 40.Mahlawat P, Ilangovan U, Biswas T, Sun L-Z, Hinck AP. Structure of the Alk1 extracellular domain and characterization of its bone morphogenetic protein (BMP) binding properties. Biochemistry 2012; 51:6328–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klages J, Kotzsch A, Coles M, Sebald W, Nickel J, Müller T, Kessler H. The solution structure of BMPR-IA reveals a local disorder-to-order transition upon BMP-2 binding. Biochemistry 2008; 47:11930–9 [DOI] [PubMed] [Google Scholar]
- 42.Zuniga JE, Ilangovan U, Mahlawat P, Hinck CS, Huang T, Groppe JC, McEwen DG, Hinck AP. The TβR-I pre-helix extension is structurally ordered in the unbound form and its flanking prolines are essential for binding. J Mol Biol 2011; 412:601–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadin D, Knaus P, Mueller TD. Structural insights into BMP receptors: specificity, activation and inhibition. Cytokine Growth Factor Rev 2016; 27:13–34 [DOI] [PubMed] [Google Scholar]
- 44.Derynck R, Budi EH. Specificity, versatility, and control of TGF-β family signaling. Sci Signal 2019; 12:eaav5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grootegoed JA, Baarends WM, Themmen AP. Welcome to the family: the anti-müllerian hormone receptor. Mol Cell Endocrinol 1994; 100:29–34 [DOI] [PubMed] [Google Scholar]
- 46.Liu F, Ventura F, Doody J, Massagué J. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol Cell Biol 1995; 15:3479–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Crescenzo G, Hinck CS, Shu Z, Zúñiga J, Yang J, Tang Y, Baardsnes J, Mendoza V, Sun L, López-Casillas F, O’Connor-McCourt M, Hinck AP. Three key residues underlie the differential affinity of the TGFbeta isoforms for the TGFbeta type II receptor. J Mol Biol 2006; 355:47–62 [DOI] [PubMed] [Google Scholar]
- 48.Baardsnes J, Hinck CS, Hinck AP, O’Connor-McCourt MD. TbetaR-II discriminates the high- and low-affinity TGF-beta isoforms via two hydrogen-bonded ion pairs. Biochemistry 2009; 48:2146–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rejon CA, Hancock MA, Li YN, Thompson TB, Hébert TE, Bernard DJ. Activins bind and signal via bone morphogenetic protein receptor type II (BMPR2) in immortalized gonadotrope-like cells. Cell Signal 2013; 25:2717–26 [DOI] [PubMed] [Google Scholar]
- 50.Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. Receptor oligomerization and beyond: a case study in bone morphogenetic proteins. BMC Biol 2009; 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci U S A 2001; 98:9306–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor β-like signaling pathway to block adipogenesis. Mol Cell Biol 2003; 23:7230–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinck AP, Mueller TD, Springer TA. Structural biology and evolution of the TGF-β family. Cold Spring Harb Perspect Biol 2016; 8:a022103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ten Dijke P, Yamashita H, Ichijo H, Franzén P, Laiho M, Miyazono K, Heldin CH. Characterization of type I receptors for transforming growth factor-beta and activin. Science 1994; 264:101–4 [DOI] [PubMed] [Google Scholar]
- 55.David L, Mallet C, Mazerbourg S, Feige J-J, Bailly S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 2007; 109:1953–61 [DOI] [PubMed] [Google Scholar]
- 56.Gray AM, Mason AJ. Requirement for activin a and transforming growth factor–beta 1 pro-regions in homodimer assembly. Science 1990; 247:1328–30 [DOI] [PubMed] [Google Scholar]
- 57.Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. Latent TGF-β structure and activation. Nature 2011; 474:343–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrison CA, Al-Musawi SL, Walton KL. Prodomains regulate the synthesis, extracellular localisation and activity of TGF-β superfamily ligands. Growth Factors 2011; 29:174–86 [DOI] [PubMed] [Google Scholar]
- 59.Mi L-Z, Brown CT, Gao Y, Tian Y, Le VQ, Walz T, Springer TA. Structure of bone morphogenetic protein 9 procomplex. Proc Natl Acad Sci U S A 2015; 112:3710–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker RG, McCoy JC, Czepnik M, Mills MJ, Hagg A, Walton KL, Cotton TR, Hyvönen M, Lee RT, Gregorevic P, Harrison CA, Thompson TB. Molecular characterization of latent GDF8 reveals mechanisms of activation. Proc Natl Acad Sci U S A 2018; 115:E866–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. J Cell Biol 2006; 175:111–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol 2004; 165:723–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarrazy V, Koehler A, Chow ML, Zimina E, Li CX, Kato H, Caldarone CA, Hinz B. Integrins αvβ5 and αvβ3 promote latent TGF-β1 activation by human cardiac fibroblast contraction. Cardiovasc Res 2014; 102:407–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liénart S, Merceron R, Vanderaa C, Lambert F, Colau D, Stockis J, van der Woning B, De Haard H, Saunders M, Coulie PG, Savvides SN, Lucas S. Structural basis of latent TGF-β1 presentation and activation by GARP on human regulatory T cells. Science 2018; 362:952–6 [DOI] [PubMed] [Google Scholar]
- 65.Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. Embo J 1991; 10:1091–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stockis J, Dedobbeleer O, Lucas S. Role of GARP in the activation of latent TGF-β1. Mol Biosyst 2017; 13:1925–35 [DOI] [PubMed] [Google Scholar]
- 67.Dong X, Zhao B, Iacob RE, Zhu J, Koksal AC, Lu C, Engen JR, Springer TA. Force interacts with macromolecular structure in activation of TGF-β. Nature 2017; 542:55–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/tolloid-like metalloproteinases. Matrix Biol 2007; 26:508–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge G, Hopkins DR, Ho W-B, Greenspan DS. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol 2005; 25:5846–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le VQ, Iacob RE, Tian Y, McConaughy W, Jackson J, Su Y, Zhao B, Engen JR, Pirruccello-Straub M, Springer TA. Tolloid cleavage activates latent GDF8 by priming the pro-complex for dissociation. Embo J 2018; 37:384–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stockis J, Liénart S, Colau D, Collignon A, Nishimura SL, Sheppard D, Coulie PG, Lucas S. Blocking immunosuppression by human Tregs in vivo with antibodies targeting integrin αVβ8. Proc Natl Acad Sci U S A 2017; 114:E10161–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuende J, Liénart S, Dedobbeleer O, van der Woning B, De Boeck G, Stockis J, Huygens C, Colau D, Somja J, Delvenne P, Hannon M, Baron F, Dumoutier L, Renauld J-C, De Haard H, Saunders M, Coulie PG, Lucas S. Monoclonal antibodies against GARP/TGF-β1 complexes inhibit the immunosuppressive activity of human regulatory T cells in vivo. Sci Transl Med 2015; 7 [DOI] [PubMed] [Google Scholar]
- 73.Manlove LS, Berquam-Vrieze KE, Pauken KE, Williams RT, Jenkins MK, Farrar MA. Adaptive immunity to leukemia is inhibited by cross-reactive induced regulatory T cells. J Immunol 2015; 195:4028–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A 2009; 106:13445–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang R, Zhu J, Dong X, Shi M, Lu C, Springer TA. GARP regulates the bioavailability and activation of TGFβ. Mol Biol Cell 2012; 23:1129–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stockis J, Colau D, Coulie PG, Lucas S. Membrane protein GARP is a receptor for latent TGF-beta on the surface of activated human Treg. Eur J Immunol 2009; 39:3315–22 [DOI] [PubMed] [Google Scholar]
- 77.Zhao B, Xu S, Dong X, Lu C, Springer TA. Prodomain-growth factor swapping in the structure of pro-TGF-β1. J Biol Chem 2018; 293:1579–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson KE, Makanji Y, Temple-Smith P, Kelly EK, Barton PA, Al-Musawi SL, Mueller TD, Walton KL, Harrison CA. Biological activity and in vivo half-life of pro-activin a in male rats. Mol Cell Endocrinol 2016; 422:84–92 [DOI] [PubMed] [Google Scholar]
- 79.Walker RG, Angerman EB, Kattamuri C, Lee Y-S, Lee S-J, Thompson TB. Alternative binding modes identified for growth and differentiation factor-associated serum protein (GASP) family antagonism of myostatin. J Biol Chem 2015; 290:7506–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Troilo H, Zuk AV, Tunnicliffe RB, Wohl AP, Berry R, Collins RF, Jowitt TA, Sengle G, Baldock C. Nanoscale structure of the BMP antagonist chordin supports cooperative BMP binding. Proc Natl Acad Sci U S A 2014; 111:13063–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang J-L, Huang Y, Qiu L-Y, Nickel J, Sebald W. Von willebrand factor type C domain-containing proteins regulate bone morphogenetic protein signaling through different recognition mechanisms. J Biol Chem 2007; 6:20002–14 [DOI] [PubMed] [Google Scholar]
- 82.Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Izpisua Belmonte JC, Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein noggin. Nature 2002; 420:636–42 [DOI] [PubMed] [Google Scholar]
- 83.Dixon ME, Armstrong P, Stevens DB, Bamshad M. Identical mutations in NOG can cause either tarsal/carpal coalition syndrome or proximal symphalangism. Genet Med 2001; 3:349–53 [DOI] [PubMed] [Google Scholar]
- 84.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 1998; 280:1455–7 [DOI] [PubMed] [Google Scholar]
- 85.Das Bhowmik A, Salem Ramakumaran V, Dalal A. Tarsal-carpal coalition syndrome: report of a novel missense mutation in NOG gene and phenotypic delineation. Am J Med Genet A 2018; 176:219–24 [DOI] [PubMed] [Google Scholar]
- 86.Innis CA, Hyvönen M. Crystal structures of the heparan sulfate-binding domain of follistatin. Insights into ligand binding. J Biol Chem 2003; 278:39969–77 [DOI] [PubMed] [Google Scholar]
- 87.Nolan K, Thompson TB. The DAN family: modulators of TGF-β signaling and beyond. Protein Sci 2014; 23:999–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical wnt signaling. J Biol Chem 2005; 280:19883–7 [DOI] [PubMed] [Google Scholar]
- 89.Li X, Ominsky MS, Niu Q-T, Sun N, Daugherty B, Agostin DD, Kurahara C, Gao Y, Cao J, Gong J, Asuncion F, Barrero M, Warmington K, Dwyer D, Stolina M, Morony S, Sarosi I, Kostenuik PJ, Lacey DL, Simonet WS, Ke HZ, Paszty C. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res 2008; 23:860–9 [DOI] [PubMed] [Google Scholar]
- 90.Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, Gong J, Qian X Paszty C, Taylor RJ, Robinson MK, Carr MD. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of wnt-mediated bone formation. J Biol Chem 2009; 284:10890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weidauer SE, Schmieder P, Beerbaum M, Schmitz W, Oschkinat H, Mueller TD. NMR structure of the wnt modulator protein sclerostin. Biochem Biophys Res Commun 2009; 380:160–5 [DOI] [PubMed] [Google Scholar]
- 92.Kišonaitė M, Wang X, Hyvönen M. Structure of gremlin-1 and analysis of its interaction with BMP-2. Biochem J 2016; 473:1593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nolan K, Kattamuri C, Luedeke DM, Deng X, Jagpal A, Zhang F, Linhardt RJ, Kenny AP, Zorn AM, Thompson TB. Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism. Structure 2013; 21:1417–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nolan K, Kattamuri C, Rankin SA, Read RJ, Zorn AM, Thompson TB. Structure of gremlin-2 in complex with GDF5 gives insight into DAN-family-mediated BMP antagonism. Cell Rep 2016; 16:2077–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Monestier O, Brun C, Cocquempot O, Petit D, Blanquet V. GASP/WFIKKN proteins: evolutionary aspects of their functions. PLoS One 2012; 7:e43710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kondás K, Szláma G, Trexler M, Patthy L. Both WFIKKN1 and WFIKKN2 have high affinity for growth and differentiation factors 8 and 11. J Biol Chem 2008; 283:23677–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hill JJ, Qiu Y, Hewick RM, Wolfman NM. Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol Endocrinol 2003; 17:1144–54 [DOI] [PubMed] [Google Scholar]
- 98.Liepinsh E, Nagy A, Trexler M, Patthy L, Otting G. Second kunitz-type protease inhibitor domain of the human WFIKKN1 protein. J Biomol NMR 2006; 35:73–8 [DOI] [PubMed] [Google Scholar]
- 99.McCoy JC, Walker RG, Murray NH, Thompson TB. Crystal structure of the WFIKKN2 follistatin domain reveals insight into how it inhibits growth differentiation factor 8 (GDF8) and GDF11. J Biol Chem 2019; 294:6333–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J-L, Qiu L-Y, Kotzsch A, Weidauer S, Patterson L, Hammerschmidt M, Sebald W, Mueller TD. Crystal structure analysis reveals how the chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev Cell 2008; 14:739–50 [DOI] [PubMed] [Google Scholar]
- 101.Fiebig JE, Weidauer SE, Qiu L-Y, Bauer M, Schmieder P, Beerbaum M, Zhang J-L, Oschkinat H, Sebald W, Mueller TD. The clip-segment of the von Willebrand domain 1 of the BMP modulator protein crossveinless 2 is preformed. Molecules 2013; 18:11658–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu E-R, Blythe EE, Fischer G, Hyvönen M. Structural analyses of von Willebrand factor C domains of collagen 2A and CCN3 reveal an alternative mode of binding to bone morphogenetic protein-2. J Biol Chem 2017; 292:12516–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huse M, Chen YG, Massagué J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell 1999; 96:425–36 [DOI] [PubMed] [Google Scholar]
- 104.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massagué J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell 2001; 8:671–82 [DOI] [PubMed] [Google Scholar]
- 105.Wang T, Li BY, Danielson PD, Shah PC, Rockwell S, Lechleider RJ, Martin J, Manganaro T, Donahoe PK. The immunophilin FKBP12 functions as a common inhibitor of the TGF beta family type I receptors. Cell 1996; 86:435–44 [DOI] [PubMed] [Google Scholar]
- 106.Chen YG, Liu F, Massague J. Mechanism of TGFbeta receptor inhibition by FKBP12. Embo J 1997; 16:3866–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature 1994; 370:341–7 [DOI] [PubMed] [Google Scholar]
- 108.Ramachandran A, Vizán P, Das D, Chakravarty P, Vogt J, Rogers KW, Müller P, Hinck AP, Sapkota GP, Hill CS. TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. Elife 2018; 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wrighton KH, Lin X, Feng X-H. Phospho-control of TGF-beta superfamily signaling. Cell Res 2009; 19:8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chaikuad A, Bullock AN. Structural basis of intracellular TGF-β signaling: receptors and smads. Cold Spring Harb Perspect Biol 2016; 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol 2012; 13:616–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heldin C-H, Moustakas A. Signaling receptors for TGF-β family members. Cold Spring Harb Perspect Biol 2016; 8:a022053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hill CS. Transcriptional control by the SMADs. Cold Spring Harb Perspect Biol 2016; 8:a022079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hata A, Chen Y-G. TGF-β signaling from receptors to smads. Cold Spring Harb Perspect Biol 2016; 8:a022061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Knighton DR, Zheng JH, Eyck LT, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991; 253:407–14 [DOI] [PubMed] [Google Scholar]
- 116.Feng X-H, Derynck R. A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling specificity. Embo J 1997; 16:3912–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massagué J. Determinants of specificity in TGF-beta signal transduction. Genes Dev 1998; 12:2144–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kerr G, Sheldon H, Chaikuad A, Alfano I, von Delft F, Bullock AN, Harris AL. A small molecule targeting ALK1 prevents notch cooperativity and inhibits functional angiogenesis. Angiogenesis 2015; 18:209–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hudson L, Mui J, Vázquez S, Carvalho DM, Williams E, Jones C, Bullock AN, Hoelder S. Novel quinazolinone inhibitors of ALK2 flip between alternate binding modes: structure–activity relationship, structural characterization, kinase profiling, and cellular proof of concept. J Med Chem 2018; 61:7261–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Williams E, Bullock AN. Structural basis for the potent and selective binding of LDN-212854 to the BMP receptor kinase ALK2. Bone 2018; 109:251–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chaikuad A, Alfano I, Kerr G, Sanvitale CE, Boergermann JH, Triffitt JT, von Delft F, Knapp S, Knaus P, Bullock AN. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J Biol Chem 2012; 287:36990–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mohedas AH, Wang Y, Sanvitale CE, Canning P, Choi S, Xing X, Bullock AN, Cuny GD, Yu PB. Structure–activity relationship of 3,5-diaryl-2-aminopyridine ALK2 inhibitors reveals unaltered binding affinity for fibrodysplasia ossificans progressiva causing mutants. J Med Chem 2014; 57:7900–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanvitale CE, Kerr G, Chaikuad A, Ramel M-C, Mohedas AH, Reichert S, Wang Y, Triffitt JT, Cuny GD, Yu PB, Hill CS, Bullock AN. A new class of small molecule inhibitor of BMP signaling. PLoS One 2013; 8:e62721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sekimata K, Sato T, Sakai N, Watanabe H, Mishima-Tsumagari C, Taguri T, Matsumoto T, Fujii Y, Handa N, Honma T, Tanaka A, Shirouzu M, Yokoyama S, Miyazono K, Hashizume Y, Koyama H. Bis-heteroaryl pyrazoles: identification of orally bioavailable inhibitors of activin receptor-like kinase-2 (R206H). Chem Pharm Bull 2019; 67:224–35 [DOI] [PubMed] [Google Scholar]
- 125.Gellibert F, Fouchet M-H, Nguyen V-L, Wang R, Krysa G, de Gouville A-C, Huet S, Dodic N. Design of novel quinazoline derivatives and related analogues as potent and selective ALK5 inhibitors. Bioorg Med Chem Lett 2009; 19:2277–81 [DOI] [PubMed] [Google Scholar]
- 126.Goldberg FW, Daunt P, Pearson SE, Greenwood R, Grist M, Debreczeni JÉ. Identification and optimisation of a series of N-(4-anilino-2-pyridyl)acetamide activin receptor-like kinase 1 (ALK1) inhibitors. Med Chem Commun 2016; 7:1204–8 [Google Scholar]
- 127.Goldberg FW, Ward RA, Powell SJ, Debreczeni JE, Norman RA, Roberts NJ, Dishington AP, Gingell HJ, Wickson KF, Roberts AL. Rapid generation of a high quality lead for transforming growth factor-beta (TGF-beta) type I receptor (ALK5). J Med Chem 2009; 52:7901–5 [DOI] [PubMed] [Google Scholar]
- 128.Sabat M, Wang H, Scorah N, Lawson JD, Atienza J, Kamran R, Hixon MS, Dougan DR. Design, synthesis and optimization of 7-substituted-pyrazolo[4,3-b]pyridine ALK5 (activin receptor-like kinase 5) inhibitors. Bioorg Med Chem Lett 2017; 27:1955–61 [DOI] [PubMed] [Google Scholar]
- 129.Tebben AJ, Ruzanov M, Gao M, Xie D, Kiefer SE, Yan C, Newitt JA, Zhang L, Kim K, Lu H, Kopcho LM, Sheriff S. Crystal structures of apo and inhibitor-bound TGFβR2 kinase domain: insights into TGFβR isoform selectivity. Acta Crystallogr D Struct Biol 2016; 72:658–74 [DOI] [PubMed] [Google Scholar]
- 130.Ogunjimi AA, Zeqiraj E, Ceccarelli DF, Sicheri F, Wrana JL, David L. Structural basis for specificity of TGFβ family receptor small molecule inhibitors. Cell Signal 2012; 24:476–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Czodrowski P, Hölzemann G, Barnickel G, Greiner H, Musil D. Selection of fragments for kinase inhibitor design: decoration is key. J Med Chem 2015; 58:457–65 [DOI] [PubMed] [Google Scholar]
- 132.Bonafoux D, Chuaqui C, Boriack-Sjodin PA, Fitch C, Hankins G, Josiah S, Black C, Hetu G, Ling L, Lee W-C. 2-Aminoimidazoles inhibitors of TGF-beta receptor 1. Bioorg Med Chem Lett 2009; 19:912–6 [DOI] [PubMed] [Google Scholar]
- 133.Guckian K, Carter MB, Lin EY-S, Choi M, Sun L, Boriack-Sjodin PA, Chuaqui C, Lane B, Cheung K, Ling L, Lee W-C. Pyrazolone based TGFbetaR1 kinase inhibitors. Bioorg Med Chem Lett 2010; 20:326–9 [DOI] [PubMed] [Google Scholar]
- 134.Sawyer JS, Beight DW, Britt KS, Anderson BD, Campbell RM, Goodson T, Herron DK, Li H-Y, McMillen WT, Mort N, Parsons S, Smith ECR, Wagner JR, Yan L, Zhang F, Yingling JM. Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-beta type I receptor kinase domain. Bioorg Med Chem Lett 2004; 14:3581–4 [DOI] [PubMed] [Google Scholar]
- 135.Roth GJ, Heckel A, Brandl T, Grauert M, Hoerer S, Kley JT, Schnapp G, Baum P, Mennerich D, Schnapp A, Park JE. Design, synthesis, and evaluation of indolinones as inhibitors of the transforming growth factor β receptor I (TGFβRI). J Med Chem 2010; 53:7287–95 [DOI] [PubMed] [Google Scholar]
- 136.Gellibert F, Woolven J, Fouchet M-H, Mathews N, Goodland H, Lovegrove V, Laroze A, Nguyen V-L, Sautet S, Wang R, Janson C, Smith W, Krysa G, Boullay V, De Gouville A-C, Huet S, Hartley D. Identification of 1,5-naphthyridine derivatives as a novel series of potent and selective TGF-beta type I receptor inhibitors. J Med Chem 2004; 47:4494–506 [DOI] [PubMed] [Google Scholar]
- 137.Zhang Y, Zhao Y, Tebben AJ, Sheriff S, Ruzanov M, Fereshteh MP, Fan Y, Lippy J, Swanson J, Ho C-P, Wautlet BS, Rose A, Parrish K, Yang Z, Donnell AF, Zhang L, Fink BE, Vite GD, Augustine-Rauch K, Fargnoli J, Borzilleri RM. Discovery of 4-Azaindole inhibitors of TGFβRI as immuno-oncology agents. ACS Med Chem Lett 2018; 9:1117–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harikrishnan LS, Warrier J, Tebben AJ, Tonukunuru G, Madduri SR, Baligar V, Mannoori R, Seshadri B, Rahaman H, Arunachalam PN, Dikundwar AG, Fink BE, Fargnoli J, Fereshteh M, Fan Y, Lippy J, Ho C-P, Wautlet B, Sheriff S, Ruzanov M, Borzilleri RM. Heterobicyclic inhibitors of transforming growth factor beta receptor I (TGFβRI). Bioorg Med Chem 2018; 26:1026–34 [DOI] [PubMed] [Google Scholar]
- 139.Gao Y, Vincent DF, Davis AJ, Sansom OJ, Bartholin L, Li Q. Constitutively active transforming growth factor β receptor 1 in the mouse ovary promotes tumorigenesis. Oncotarget 2016; 7:40904–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hatsell SJ, Idone V, Wolken DMA, Huang L, Kim HJ, Wang L, Wen X, Nannuru KC, Jimenez J, Xie L, Das N, Makhoul G, Chernomorsky R, D’Ambrosio D, Corpina RA, Schoenherr CJ, Feeley K, Yu PB, Yancopoulos GD, Murphy AJ, Economides AN. ACVR1 R206H receptor mutation causes fibrodysplasia ossificans progressiva by imparting responsiveness to activin A. Sci Transl Med 2015; 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Han S, Loulakis P, Griffor M, Xie Z. Crystal structure of activin receptor type IIB kinase domain from human at 2.0 angstrom resolution. Protein Sci 2007; 16:2272–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Horbelt D, Boergermann JH, Chaikuad A, Alfano I, Williams E, Lukonin I, Timmel T, Bullock AN, Knaus P. Small molecules dorsomorphin and LDN-193189 inhibit myostatin/GDF8 signaling and promote functional myoblast differentiation. J Biol Chem 2015; 290:3390–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Souchelnytskyi S, Tamaki K, Engström U, Wernstedt C, ten Dijke P, Heldin CH. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-beta signaling. J Biol Chem 1997; 272:28107–15 [DOI] [PubMed] [Google Scholar]
- 144.Abdollah S, Macías-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem 1997; 272:27678–85 [DOI] [PubMed] [Google Scholar]
- 145.Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 1997; 388:87. [DOI] [PubMed] [Google Scholar]
- 146.Wu G, Chen YG, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, Massagué J, Shi Y. Structural basis of Smad2 recognition by the smad anchor for receptor activation. Science 2000; 287:92–7 [DOI] [PubMed] [Google Scholar]
- 147.Wu J-W, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massagué J, Shi Y. Crystal structure of a phosphorylated Smad2: recognition of phosphoserine by the MH2 domain and insights on smad function in TGF-β signaling. Mol Cell 2011; 8:1277–89 [DOI] [PubMed] [Google Scholar]
- 148.Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell 2004; 15:813–23 [DOI] [PubMed] [Google Scholar]
- 149.Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein ski: insights on ski-mediated repression of TGF-beta signaling. Cell 2002; 111:357–67 [DOI] [PubMed] [Google Scholar]
- 150.Miyazono K-I, Moriwaki S, Ito T, Kurisaki A, Asashima M, Tanokura M. Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors. Sci Signal 2018; 11 [DOI] [PubMed] [Google Scholar]
- 151.Miyazono K-I, Ohno Y, Wada H, Ito T, Fukatsu Y, Kurisaki A, Asashima M, Tanokura M. Structural basis for receptor-regulated SMAD recognition by MAN1. Nucleic Acids Res 2018; 46:12139–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 1998; 94:585–94 [DOI] [PubMed] [Google Scholar]
- 153.Chai N, Li W-X, Wang J, Wang Z-X, Yang S-M, Wu J-W. Structural basis for the Smad5 MH1 domain to recognize different DNA sequences. Nucleic Acids Res 2015; 43:9051–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Martin-Malpartida P, Batet M, Kaczmarska Z, Freier R, Gomes T, Aragón E, Zou Y, Wang Q, Xi Q, Ruiz L, Vea A, Márquez JA, Massagué J, Macias MJ. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat Commun 2017; 8:2070. [DOI] [PMC free article] [PubMed] [Google Scholar]