To survive and replicate during infection, pathogens utilize different carbon and energy sources depending on the nutritional landscape of their host microenvironment. Salmonella enterica serovar Typhimurium is an intracellular bacterial pathogen that occupies diverse cellular niches. While it is clear that Salmonella Typhimurium requires access to glucose during systemic infection, data on the need for lipid metabolism are mixed.
KEYWORDS: Salmonella, lipid metabolism, macrophage
ABSTRACT
To survive and replicate during infection, pathogens utilize different carbon and energy sources depending on the nutritional landscape of their host microenvironment. Salmonella enterica serovar Typhimurium is an intracellular bacterial pathogen that occupies diverse cellular niches. While it is clear that Salmonella Typhimurium requires access to glucose during systemic infection, data on the need for lipid metabolism are mixed. We report that Salmonella Typhimurium strains lacking lipid metabolism genes were defective for systemic infection of mice. Bacterial lipid import, β-oxidation, and glyoxylate shunt genes were required for tissue colonization upon oral or intraperitoneal inoculation. In cultured macrophages, lipid import and β-oxidation genes were required for bacterial replication and/or survival only when the cell culture medium was supplemented with nonessential amino acids. Removal of glucose from tissue culture medium further enhanced these phenotypes and, in addition, conferred a requirement for glyoxylate shunt genes. We also observed that Salmonella Typhimurium needs lipid metabolism genes in proinflammatory but not anti-inflammatory macrophages. These results suggest that during systemic infection, the Salmonella Typhimurium that relies upon host lipids to replicate is within proinflammatory macrophages that have access to amino acids but not glucose. An improved understanding of the host microenvironments in which pathogens have specific metabolic requirements may facilitate the development of targeted approaches to treatment.
INTRODUCTION
Microbial pathogens encounter diverse niches throughout infection (1). Spatially, infection spreads to new tissues or cell types. Temporally, the innate immune response gives way to adaptive effectors, and infection may evolve into a chronic state. Within each unique pathogen niche, the availability of nutrients is influenced by factors such as the host metabolic state (2–4), the host inflammatory state (5–8), interactions with other microbes (9–11), and stochastic heterogeneity (12–14). Together, these factors define the nutritional microenvironment and influence pathogen replication, persistence, and transmission and the outcome of infection.
Salmonella enterica subspecies enterica serovar Typhimurium naturally infects mice and has been used to model the acute and chronic stages of human typhoid fever, which is caused by Salmonella enterica subspecies enterica serovar Typhi and is responsible for 200,000 deaths globally each year (15). Salmonella Typhimurium is acquired through contaminated food or water and reaches the anaerobic small intestine, where it traverses the epithelial cell layer (16). The bacteria are taken up by and replicate within professional phagocytes, which facilitate the colonization of tissues, including Peyer’s patches, mesenteric lymph nodes, spleen, and liver (17). Salmonella Typhimurium has been observed in humans and animals within lipid-rich macrophages that accumulate during systemic infection (18–21). In addition, the bacteria likely encounter many types of macrophages over the course of infection, including those that are polarized toward either a pro- or anti-inflammatory state (5, 22, 23). How the pathogen may exploit these different cellular environments is not clear.
Salmonella Typhimurium has been demonstrated to require access to glucose during systemic infection (24), but data on its use of lipids are mixed. Some studies of mutant strains lacking key enzymes in lipid metabolism report no in vivo defect (25–27), but others demonstrate attenuation (28, 29). While bulk expression profiling and proteomics do not point to a significant induction of lipid metabolism genes during infection of macrophages or mice (30–33), recent single-cell work with fluorescent reporters and metabolomics suggests some bacteria use lipids (34, 35). We examined the requirement for Salmonella Typhimurium lipid uptake, β-oxidation, and glyoxylate shunt genes in mice and in polarized macrophages. We found that the nutritional environment of the macrophage, specifically access to amino acids and glucose, dictates whether bacteria require lipid metabolism genes to replicate and/or survive. Moreover, we determined that that Salmonella Typhimurium uses lipid metabolism genes within proinflammatory, but not anti-inflammatory, macrophages.
RESULTS
Deletion of Salmonella Typhimurium lipid metabolism genes abrogates growth on lipids.
To verify the requirement for Salmonella Typhimurium lipid metabolism genes, we examined growth in broth (see Fig. S1 and S2 in the supplemental material) or on plates (see Table S1 in the supplemental material). Salmonella Typhimurium fatty acid degradation involves multiple steps, some of which can be executed by gene products encoded by either a canonical or a secondary lipid metabolism pathway (36–38). First, medium- and long-chain fatty acids are transported across the outer membrane and activated by the addition of coenzyme A (CoA) to produce acyl-CoA (Fig. 1A). We found that the fatty acid import genes fadL and fadD were required for growth on oleic acid as a sole carbon source, but the Salmonella Typhimurium paralog of fadD, denoted ydiD, was not. In Escherichia coli, ydiD contributes to anaerobic growth (39). However, our data showed that fadL and fadD, but not ydiD, were required for Salmonella Typhimurium anaerobic growth on lipids (Table S1). These defects were complemented by a plasmid containing the target gene (Fig. S1 and S2; Table S1).
FIG 1.
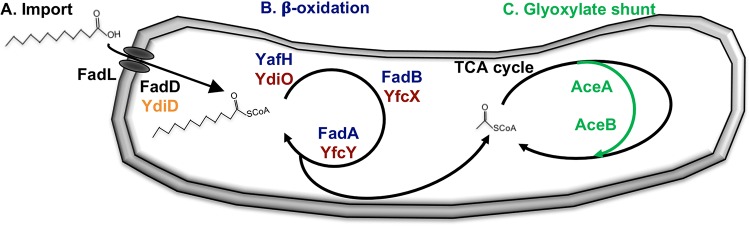
Schematic of lipid metabolism pathways in Salmonella Typhimurium. (A) Long-chain free fatty acids are imported via FadL (outer membrane translocase) and activated by FadD or YdiD (acyl-CoA synthetases). Canonical import genes are black; putative secondary import gene is orange. (B) Resulting acyl-thioesters are serially β-oxidized via YafH or YdiO (acyl-CoA dehydrogenases), FadB or YfcX (3-hydroxyacyl-CoA dehydrogenases), and FadA or YfcY (3-ketoacyl-CoA thiolases), producing acetyl-CoA, which is metabolized via the TCA cycle. Canonical β-oxidation genes are blue; putative secondary β-oxidation genes are red. (C) The glyoxylate shunt (green) consists of AceA (isocitrate lyase) and AceB (malate synthase), which bridge the TCA cycle to preserve carbon and cofactors during growth in the absence of glycolytic energy sources.
In the second step of lipid metabolism, acyl-CoA is oxidized by a series of enzymes to produce acetyl-CoA, which enters the tricarboxylic acid (TCA) cycle (37, 40) (Fig. 1B). As in E. coli (41, 42), Salmonella Typhimurium carries both canonical (yafH, fadB, and fadA) and secondary (ydiO, yfcX, and yfcY) β-oxidation pathways. For growth on lipids, Salmonella Typhimurium required the secondary pathway under anaerobic conditions. In the presence of oxygen, Salmonella Typhimurium relied largely upon the canonical pathway (Fig. S1; Table S1).
During the third step of lipid metabolism, the glyoxylate shunt pathway feeds the acetyl-CoA derived from β-oxidation into a truncated version of the TCA cycle (Fig. 1C). When lipids are the sole carbon source, the glyoxylate shunt is needed to preserve carbon for incorporation into biosynthetic pathways. The glyoxylate shunt consists of two enzymes encoded in an operon, isocitrate lyase (aceA) and malate synthase (aceB) (43). As anticipated, nonpolar deletions of either gene eliminated aerobic and anaerobic growth on oleic acid as a sole carbon source, and these defects were complemented by a plasmid containing the target gene (Fig. S1 and S2; Table S1).
Salmonella Typhimurium requires lipid degradation genes for orogastric infection of mice.
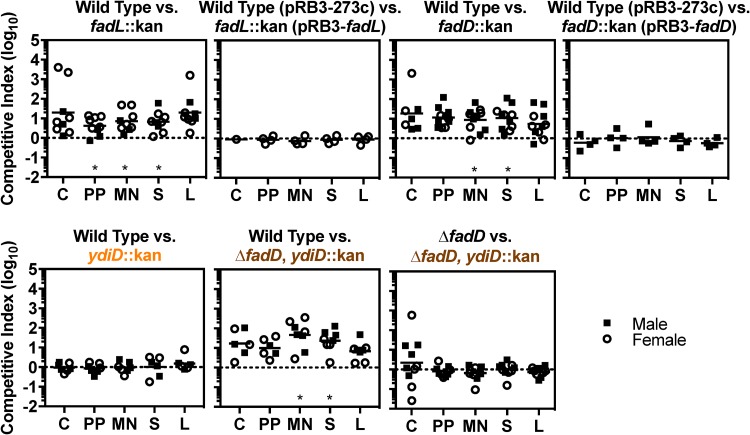
Several groups have found differing requirements for Salmonella Typhimurium lipid metabolism during infection. However, many of these studies were conducted in mouse strains that develop acute lethal infection due to a genetic lesion in innate immunity (25–29). To establish whether Salmonella Typhimurium requires lipid metabolism genes in immunocompetent mice, we performed mixed inoculations in Sv129S6/SvEvTac (Sv129) mice, which typically survive infection (44). Mice were orogastrically infected with equivalent numbers of wild-type and lipid-metabolism mutant bacteria and euthanized 2 weeks after infection, when bacteria have disseminated systemically and begun transitioning to a chronic infectious state (5, 19, 44). Tissues were harvested to enumerate CFU, and the competitive index was calculated as the ratio of recovered wild-type to mutant CFU, normalized to the ratio in the inoculum. Strains lacking the canonical import gene fadL or fadD were attenuated in competition with wild type (Fig. 2). Complementation of each of these strains with a plasmid containing the target gene restored colonization to wild-type levels. A strain lacking ydiD, the secondary paralog of fadD, competed equivalently with wild type, and loss of ydiD did not further attenuate a fadD mutant. These data indicate that ydiD is dispensable and that the canonical import genes fadL and fadD contribute to infection. Thus, Salmonella Typhimurium lipid uptake and activation are needed for tissue colonization in mice.
FIG 2.
Lipid import genes are needed for Salmonella Typhimurium to competitively colonize mouse tissues. Mice were orogastrically inoculated with equivalent numbers of wild-type or mutant bacteria, as determined by CFU (1 × 109 or 2 × 109 [ΔfadD versus ΔfadD, ydiD::kan experiment only]). After 2 weeks, the following tissues were harvested to enumerate CFU: cecum (C), Peyer’s patches (PP), mesenteric lymph nodes (MN), spleen (S), and liver (L). The competitive index was calculated as the ratio of the indicated strains at harvest divided by the ratio of the inoculum. Each symbol represents a tissue from one mouse. *, P < 0.05 by one-sample t test.
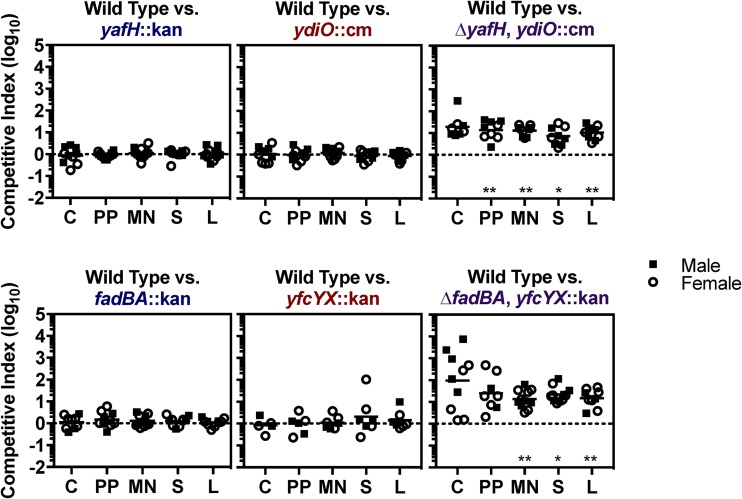
Having shown that lipid import is required for Salmonella Typhimurium colonization of tissues, we asked whether β-oxidation is required. In mixed infection experiments, strains lacking a single gene or operon from either the canonical or secondary β-oxidation pathway competed equivalently with wild type (Fig. 3). However, strains lacking genes encoding both cognate enzymes were attenuated. Thus, the canonical and secondary β-oxidation pathways are redundant during infection, and β-oxidation of lipids is necessary for tissue colonization.
FIG 3.
β-Oxidation genes are needed for Salmonella Typhimurium to competitively colonize mouse tissues. Mice were orogastrically inoculated with equivalent numbers of wild-type or mutant bacteria (1 × 109 CFU). After 2 weeks, the following tissues were harvested to enumerate CFU: cecum (C), Peyer’s patches (PP), mesenteric lymph nodes (MN), spleen (S), and liver (L). The competitive index was calculated as the ratio of the indicated strains at harvest divided by the ratio of the inoculum. Each symbol represents a tissue from one mouse. *, P < 0.05; **, P < 0.01 by one-sample t test.
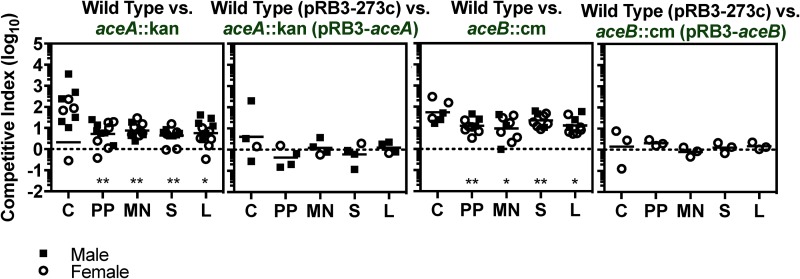
Since both lipid import and β-oxidation contribute to Salmonella Typhimurium colonization of tissues, we addressed whether the incorporation of acetyl-CoA into the TCA cycle via the glyoxylate shunt is also important. Mutant strains lacking either glyoxylate shunt pathway gene were outcompeted by wild type in multiple tissues, a phenotype complemented by the corresponding gene on a plasmid (Fig. 4). Therefore, in some situations, lipids may be the only carbon source available to Salmonella Typhimurium during infection.
FIG 4.
Glyoxylate shunt genes are needed for Salmonella Typhimurium to competitively colonize mouse tissues. Mice were orogastrically inoculated with equivalent numbers of wild-type or mutant bacteria (1 × 109 CFU). After 2 weeks, the following tissues were harvested to enumerate CFU: cecum (C), Peyer’s patches (PP), mesenteric lymph nodes (MN), spleen (S), and liver (L). The competitive index was calculated as the ratio of the indicated strains at harvest divided by the ratio of the inoculum. Each symbol represents a tissue from one mouse. *, P < 0.05; **, P < 0.01 by one-sample t test.
Lipid degradation genes are required for infection when Salmonella Typhimurium bypasses the gastrointestinal tract.
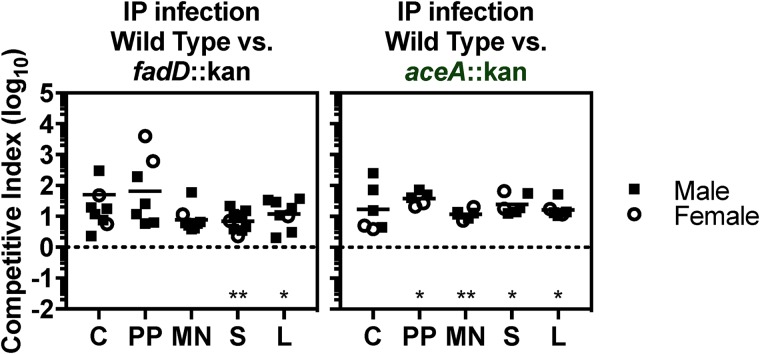
Prior to dissemination to the spleen and liver, orogastric inoculation exposes Salmonella Typhimurium to the intestinal tract. Fatty acids within the intestine alter the invasion of strains with defects in lipid metabolism and regulate the expression of Salmonella Typhimurium genes required for systemic infection (45–48). Therefore, we asked whether Salmonella Typhimurium requires lipid metabolism genes when mixed inoculations are delivered into the intraperitoneal cavity, a method that allows Salmonella Typhimurium to bypass the gastrointestinal tract and become rapidly engulfed by resident peritoneal macrophages (17). We found that mutant strains lacking fadD or aceA colonized tissues poorly compared with the wild-type (Fig. 5). These data indicate that Salmonella Typhimurium requires lipid metabolism genes to colonize tissues irrespective of passage through the gastrointestinal tract.
FIG 5.
Upon intraperitoneal inoculation, lipid metabolism genes are needed for Salmonella Typhimurium to competitively colonize mouse tissues. Mice were intraperitoneally inoculated with equivalent numbers of wild-type or mutant bacteria (1 × 103 CFU). After 2 weeks, the following tissues were harvested to enumerate CFU: cecum (C), Peyer’s patches (PP), mesenteric lymph nodes (MN), spleen (S), and liver (L). The competitive index was calculated as the ratio of the indicated strains at harvest divided by the ratio of the inoculum. Each symbol represents a tissue from one mouse. *, P < 0.05; **, P < 0.01 by one-sample t test.
In macrophages cultured with nonessential amino acids, Salmonella Typhimurium requires lipid import and beta oxidation to replicate.
A cell culture model of Salmonella Typhimurium lipid utilization would enable an examination of the intracellular niche(s) in which the bacterium uses lipids. Therefore, we infected bone marrow-derived macrophages (BMDMs) from Sv129 mice or RAW264.7 cell line macrophages and monitored bacterial load after 18 h. Two methods were used to quantify bacterial load, namely, standard plating for CFU and a flow cytometry-based fluorescence dilution assay (49). We also compared standard medium containing glucose (4.5 mg/ml) to glucose-free medium, with or without oleic acid, as bacteria may exploit the accumulated lipid droplets (50) (see Fig. S3 in the supplemental material). No differences in replication were observed between the wild type and mutant (see Fig. S4 in the supplemental material), indicating that under these conditions, Salmonella Typhimurium lipid metabolism genes are dispensable.
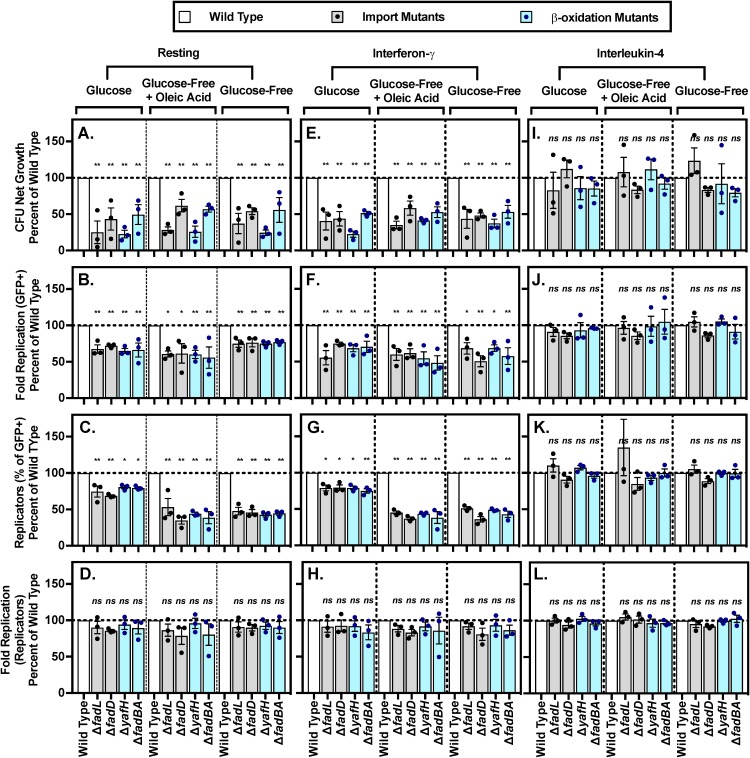
Since we observed a Salmonella Typhimurium need for lipid metabolism genes in mice but not macrophages, we hypothesized that a modified tissue culture medium may be required. A recent study demonstrated that RAW264.7 cells grown with nonessential amino acids (NEAAs) support Salmonella Typhimurium expression of the glyoxylate shunt operon, specifically, an aceBA-gfp reporter (34). We were not growing the macrophages under conditions similar to those in vivo. A recent study reported that RAW264.7 cells grown with NEAA restrict the replication of a Salmonella Typhimurium strain lacking a lipid uptake gene (fadD) (34). We found that primary and cell culture macrophages supplemented with NEAAs reduced the replication and/or survival of Salmonella Typhimurium lacking lipid import genes (fadL and fadD) or the canonical β-oxidation pathway (fadBA and yafH) (Fig. 6A and B; see Fig. S5A and B in the supplemental material). The presence of NEAAs also reduced the percentage of mutant bacteria that replicated (Fig. 6C; Fig. S5) but not the number of times replicating bacteria doubled (Fig. 6D; Fig. S5D). These results suggest that macrophage access to amino acids creates an intracellular environment in which a fraction of intracellular Salmonella Typhimurium import and oxidize lipids to replicate and/or survive. In the remainder of experiments, we supplemented the tissue culture media with NEAAs.
FIG 6.
Supplementation of macrophage media with NEAA reveals requirement for lipid import and β-oxidization genes in resting and interferon-γ-pretreated BMDMs. BMDMs were transferred into defined medium with the indicated carbon source for 18 to 24 h prior to infection with the indicated strains. Net growth was calculated as CFU at 18 h postinfection divided by CFU at 2 h postinfection (A, E, I). Fluorescence dilution was used to calculate fold replication for all GFP+ bacteria (B, F, J), percentage of GFP-low replicating bacteria (C, G, K), and fold replication of the GFP-low population of replicating bacteria (D, H, L). GFP-low replicating bacteria were gated based on the fluorescence of the inoculum. Data were normalized to wild type (100%). Average of triplicate samples from each of three independent biological replicates (circles) is superimposed on mean and SEM. *, P < 0.05; **, P < 0.01 by one-way ANOVA.
In macrophages cultured without glucose, Salmonella Typhimurium requires the glyoxylate shunt pathway.
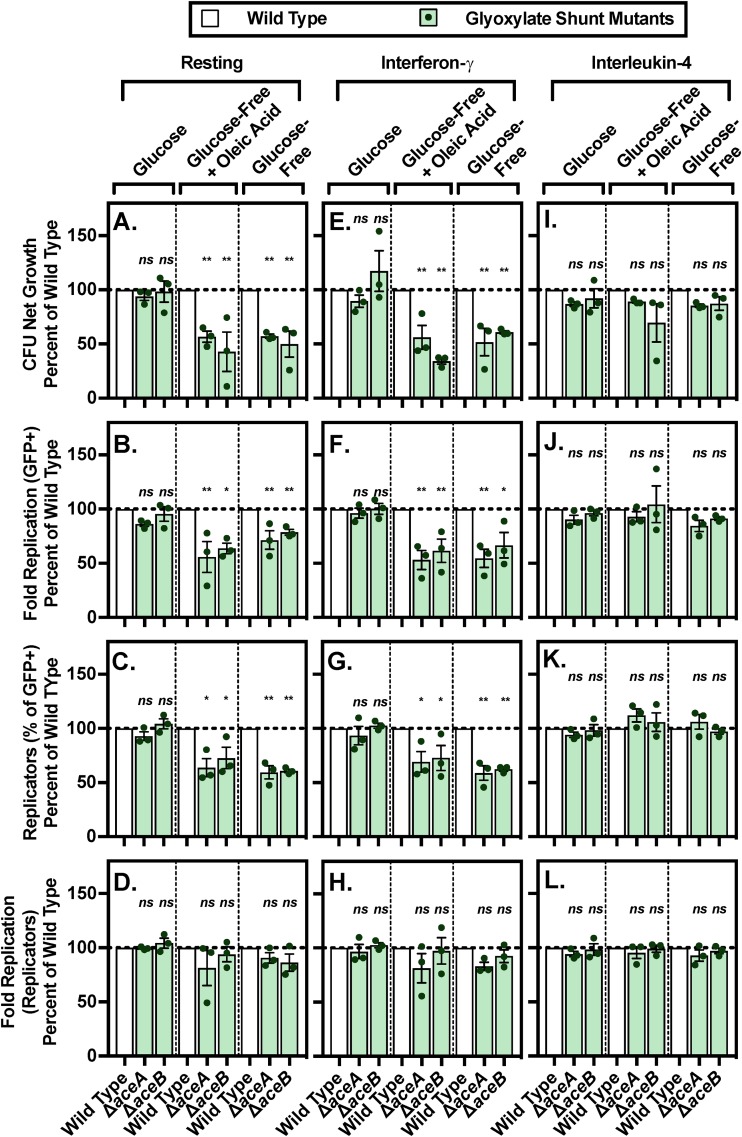
Having found macrophage growth conditions that require intracellular Salmonella Typhimurium to use the first two steps of lipid metabolism, we sought conditions that also required the glyoxylate shunt pathway (aceA and aceB). We found that in macrophages grown without glucose, Salmonella Typhimurium requires the glyoxylate shunt pathway to replicate and/or survive (Fig. 7A and B; Fig. S6A and B). Under these conditions, the percentage of mutant bacteria that replicated declined, but the number of times replicating bacteria doubled remained steady, compared with that in the presence of glucose (Fig. 7C and D; Fig. S6C and D). That glucose obviates the need for the glyoxylate shunt pathway is consistent with the role of this pathway in anaplerosis in the absence of glucose (43). Moreover, the data indicate that the presence of NEAAs within tissue culture media changes the metabolism of the macrophages and/or the bacteria to reflect or mimic conditions that the bacteria experience within an animal. That is, when NEAAs are supplied, a fraction of Salmonella Typhimurium use lipids to replenish the TCA cycle, consistent with the requirement for the glyoxylate shunt genes for the colonization of mice.
FIG 7.
A lack of glucose in the macrophage culture media uncovers a Salmonella Typhimurium requirement for the glyoxylate shunt genes in resting and interferon-γ-pretreated BMDMs. BMDMs were transferred into defined medium with the indicated carbon source for 18 to 24 h prior to infection with the indicated strains. Net growth was calculated as CFU at 18 h postinfection divided by CFU at 2 h postinfection (A, E, I). Fluorescence dilution was used to calculate fold replication for all GFP+ bacteria (B, F, J), percentage of GFP-low replicating bacteria (C, G, K), and fold replication of the GFP-low population of replicating bacteria (D, H, L). GFP-low replicating bacteria were gated based on the fluorescence of the inoculum. Data were normalized to wild type (100%). Average of triplicate samples from each of three independent biological replicates (circles) is superimposed on mean and SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA.
Salmonella Typhimurium requires lipid metabolism to replicate in proinflammatory but not anti-inflammatory macrophages.
In animals, Salmonella Typhimurium likely encounters many kinds of activated macrophages, particularly those polarized toward a classical proinflammatory (M1), microbiocidal state, or toward an alternative, anti-inflammatory (M2) phenotype (5, 7). The experiments described above were carried out with resting macrophages, meaning that they were not pretreated with cytokines. However, resting macrophages become proinflammatory upon exposure to lipopolysaccharide (LPS), which occurs upon infection with Salmonella Typhimurium (51). To clearly establish whether pro- or anti-inflammatory macrophages restrict the growth of Salmonella Typhimurium lacking lipid metabolism genes, we pretreated BMDMs or RAW264.7 cells with interferon gamma (IFN-γ) or interleukin-4 (IL-4), which polarize macrophages toward pro- and anti-inflammatory states, respectively (5, 7). We obtained similar results in resting and IFN-γ pretreated macrophages, namely, lipid import and β-oxidation contributed to replication whether glucose was present or not, and the glyoxylate shunt pathway was only needed in the absence of glucose (Fig. 6 and 7; Fig. S5 and S6 in the supplemental material). These observations are consistent with the proinflammatory activation state that resting macrophages acquire upon infection with Salmonella Typhimurium (51). IL-4-stimulated macrophages did not require any Salmonella Typhimurium lipid genes under any conditions tested. Thus, Salmonella Typhimurium lipid metabolism genes appear to be required for replication and/or survival in proinflammatory macrophages.
DISCUSSION
Diverse pathogens use fatty acids during infection (52–56), but little is known about the specific microenvironments that enable or necessitate lipid acquisition. Our study used Salmonella Typhimurium mutants lacking genes in every step of lipid metabolism, and we examined their phenotypes in microbiological media, in mice, and in cultured macrophages. Sv129 mice, which develop a systemic, nonfatal infection (44, 57), were used to test for lipid gene requirements. The glyoxylate shunt gene aceA is needed by Salmonella Typhimurium in Sv129 mice for the colonization of mesenteric lymph nodes, but this same study reported no phenotype for an aceA mutant strain in C3H/HeN mice, which develop fatal Salmonella Typhimurium infections (29). BALB/c mice infected with Salmonella Typhimurium also develop fatal infections, which do not require that Salmonella Typhimurium have lipid import genes (fadL [46] or fadD [25]), β-oxidation genes (yafH [27]), or the glyoxylate shunt gene aceA (25, 26). These data may suggest that lipids are dispensable in mouse models of fatal acute infection but contribute to long-term colonization of tissues.
We tested lipid mutants for growth in macrophages with the aim of modeling phenotypes observed in mice. Macrophages supplemented with NEAAs curbed the replication of Salmonella Typhimurium lacking genes for the first two lipid metabolism steps, uptake and β-oxidation. These results echo a report that in NEAA-grown RAW264.7 cells, a lipid uptake gene (fadD) is required for replication by the Salmonella Typhimurium strain NCTC 12023 (ATCC 14028) (34), which is genetically and phenotypically distinct from SL1344 (58, 59). We also observed a NEAA-dependent increase in Salmonella Typhimurium intracellular replication in primary and cell line macrophages from mice with different genetic backgrounds, namely, Sv129 and BALB/c, respectively (see Fig. S7 in the supplemental material). NEAAs may enable the observation of a macrophage phenotype for Salmonella Typhimurium with fatty acid metabolism gene mutations because it supports better bacterial growth. Alternatively, macrophage access to NEAAs may enable some bacteria to metabolize lipids based on access to lipids and/or changes in bacterial metabolic capabilities. The presence of NEAAs may also change the metabolism of the macrophage and/or of the bacteria such that a subset of bacteria now requires lipids for growth. Regardless, when macrophages have access to NEAAs, the nutritional microenvironment and/or capabilities of intracellular Salmonella Typhimurium change in a manner(s) that influences the outcome of infection. These observations highlight the fact that growing host cells under different conditions may enable in vivo phenotypes to be studied in cell culture.
We did not document any Salmonella Typhimurium requirement for glyoxylate shunt genes unless the culture media lacked glucose. These observations are consistent with the Salmonella Typhimurium requirement for glyoxylate shunt genes in microbiological media without glucose (43). These data suggest that macrophage access to NEAAs affects Salmonella Typhimurium access to and/or the ability to use glucose versus lipids as a carbon source. The Salmonella Typhimurium that requires the glyoxylate shunt to colonize mouse tissues may reside within macrophages that have little access to glucose but significant access to other nutrients. Thus, the host cell nutritional microenvironment has the potential to determine which intracellular pathogens replicate and contribute to infection.
Macrophage access to nutrients is not the only microenvironmental factor that affects the intracellular replication of Salmonella Typhimurium lipid metabolism mutants. Macrophage exposure to pro- or anti-inflammatory cytokines determines whether these cells rely upon glycolytic or oxidative metabolism, respectively (5, 7). We found that macrophages pretreated with pro- but not anti-inflammatory cytokines required Salmonella Typhimurium to have intact lipid metabolism genes to survive and/or replicate. Supplementation with NEAAs was essential to observe the effect of macrophage activation state on Salmonella Typhimurium replication, and all three steps of lipid metabolism, including the glyoxylate shunt, were needed in media lacking glucose. Thus, proinflammatory macrophages may provide Salmonella Typhimurium with access to lipids and/or enable, for instance, the expression of bacterial lipid metabolism genes. The bacteria within mice that need lipids to replicate may reside within proinflammatory macrophages.
One difference we observed between Salmonella Typhimurium lipid requirements in mice versus cell culture is that the canonical and secondary β-oxidation genes compensated for each other only in mice. In broth culture, anaerobiosis allows for the expression of the secondary pathway, suggesting Salmonella Typhimurium in mice may use lipids under anaerobic conditions, for example in granulomas.
Future efforts to define the specific tissue or cellular environment(s) in which Salmonella Typhimurium requires exogenous lipids to replicate may improve our understanding of the microenvironments in which the pathogen resides. It may also be of use to test whether Salmonella Typhimurium lipid metabolism genes are required in murine genetic and pharmacological models that respond to Salmonella Typhimurium infection with predominantly M1 or M2 macrophage production. This may be addressable by, for example, comparing the survival and cellular residency of wild-type and lipid-mutant Salmonella Typhimurium strains in C57BL/6 (M1) and BALB/c (M2) mice (60). However, interpretation of results is likely to be confounded by differences in the genetic backgrounds of these two strains (61, 62). Alternatively, mice can be pretreated with pharmacological agents that drive them to produce macrophages that are largely M1 (e.g., CpG oligodeoxynucleotides [63, 64]) or M2 (e.g., all-trans-retinoic acid [65] and melatonin [66]). Since these methods have primarily been examined in Nramp1−/− mouse backgrounds, their effect on Nramp1+/+ mice over time will need clarification prior to testing the hypothesis that lipid metabolism genes are needed for tissue colonization in animals with an M1 phenotype.
In conclusion, the entire lipid metabolism pathway is required for Salmonella Typhimurium replication in proinflammatory macrophages when glucose is limited and amino acids are abundant, consistent with bacterial requirements for tissue colonization in Sv129 mice. Thus, pathogen nutritional strategy can be strongly affected by the polarization state of the host cell type and by the availability of nutrients to that host cell.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Salmonella enterica serovar Typhimurium wild-type strain SL1344 (67) and derivatives were cultured in LB media containing 30 μg/ml streptomycin, 50 μg/ml ampicillin, 30 μg/ml kanamycin, or 34 μg/ml chloramphenicol. Liquid cultures were incubated overnight at 37°C with aeration. Deletion strains were marked at the indicated locus with kanamycin or chloramphenicol resistance cassettes (68, 69), which were P22 phages transduced into SL1344 (70). The resistance cassette was removed by the induction of FLP recombinase or I-SceI to yield an 84-bp scar or no scar, respectively. Mutant strains were validated by PCR and growth on oleic acid.
Complementation plasmids were derived from medium copy number plasmid pRB3-273c (71). The target gene and its promoter were amplified from Salmonella genomic DNA (350 bp upstream and 50 bp downstream of the gene) using PCR primers containing restriction digest sites for BamHI and HindIII (fadL, aceA, and aceB) or BamHI and KpnI (fadD). For aceA, the following two amplicons were initially generated: the aceBA promoter region with a downstream KpnI recognition sequence and the aceA gene with aceB Shine-Dalgarno sequence and KpnI recognition sequence engineered into the upstream PCR primer. The two amplicons were digested using KpnI, ligated, and amplified as a single fragment using PCR. For aceB, the entire aceBA operon was amplified, digested with HpaI to remove 1,157 bp of the aceA gene, ligated, and amplified as a single fragment using PCR. The fragment thus contained the promoter, aceB, the 31-bp intergenic region, the initial 117 nucleotides, and final 31 bp of aceA. Amplicons were digested with BamHI and HindIII or KpnI, gel purified, ligated into linearized pRB3-273c, and verified by sequencing of the entire insertion (Fig. S1; Table S1). This approach was used because the promoter-aceB amplicon only partially complemented mutants lacking aceB for unknown reasons.
For fluorescence dilution experiments, strains were transformed with pDiGi and chromosomally marked with green fluorescent protein (GFP) at the rpsM locus using P22 phage transduction (17). To induce the expression of dsRed prior to infection, strains were cultured overnight in media containing 170 mM morpholineethanesulfonic acid (MES) (pH 5.0), 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 10 mM MgCl2, 0.3% glycerol, 0.1% Casamino Acids, and 10 mM arabinose with appropriate antibiotics (49). We found that pDiGc, which encodes GFP under the rpsM promoter, significantly hindered Salmonella infection, presumably due to high GFP expression (data not shown).
Bacterial growth assays.
For broth growth experiments, overnight cultures were washed in phosphate-buffered saline (PBS) and diluted to an optical density at 600 nm (OD600) of 0.01 in 300 μl of LB or M9 minimal medium supplemented with 0.004% histidine, 1 mM MgSO4, 100 μM CaCl2, and either 0.4% dextrose, 0.4% glycerol, or 0.1% sodium oleate and 1% IGEPAL CA-630, which was necessary to solubilize oleate. Bacteria were grown in 96-well plates with shaking in an Eon or Synergy H1 microplate spectrophotometer (BioTek) at 37°C, and the OD600 was recorded every 20 min.
For growth on solid media, overnight cultures were washed in PBS and resuspended in PBS to an OD600 of 1.0, and 3 μl was spread onto M9 minimal agar plates supplemented with 1 mM MgSO4, 100 μM CaCl2, 0.004% histidine, 1.5% agar, 1% IGEPAL CA-630, and either 0.4% dextrose, 0.1% sodium oleate, 0.1% sodium decanoate, or 0.1% sodium octanoate. No growth was observed in the absence of carbon. For anaerobic growth, plates were supplemented with 25 mM nitrate as an alternative electron acceptor; no anaerobic growth was observed in the absence of nitrate. Anaerobic plates were grown at 37°C within GasPak EZ anaerobe pouches (BD).
Cell culture and bone marrow-derived macrophage generation.
Macrophages were routinely cultured in Dulbecco modified Eagle medium (DMEM) high glucose (Sigma) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, and 50 μM β-mercaptoethanol. To generate bone marrow-derived macrophages, femurs and tibias from 4- to 10-week-old wild-type Sv129S6/SvEvTac mice (Taconic Laboratories) were flushed with PBS to recover bone marrow, layered over Histopaque-1083 (Sigma), and centrifuged for 25 min at 500 × g. The mononuclear cell fraction was recovered and washed in complete media. Cells were seeded at 1 × 105 to 2 × 105 cells/ml in complete medium supplemented with 30% to 35% conditioned media from 3T3 cells expressing macrophage colony-stimulating factor (MCSF) and fed 3 to 4 days later. After 1 week in culture, BMDMs typically replicated 5- to 10-fold under these conditions. Twenty-four hours prior to infection, macrophages were transferred into defined glucose-free DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 10 mM HEPES, 50 μM β-mercaptoethanol, and 1× NEAA (alanine, asparagine, aspartic acid, glycine, serine, proline, and glutamic acid; Sigma). Importantly, we observed less replication of Salmonella Typhimurium within macrophages in the absence of NEAAs (Fig. S7). Glucose-free medium was supplemented with 4.5 g/liter glucose or 600 μM oleic acid (conjugated 6:1 with bovine serum albumin [BSA]; concentrated oleic acid dissolved in 0.1 M Na2CO3 at 55°C was added to BSA dissolved in PBS, stirred for 1 h at 37°C, filter sterilized, and stored at –20°C). Supplementation with BSA vehicle control yielded results identical to glucose-free medium. Macrophages were activated with 2 ng/ml recombinant murine IFN-γ (PeproTech) or 20 ng/ml recombinant murine IL-4 (PeproTech) for 18 to 24 h prior to infection.
Infection of cell culture macrophages.
For fluorescence dilution experiments, bacteria were added to BMDMs at a concentration of 3 × 107 CFU/ml in media. We found that this protocol reproducibly yielded infection of 70% to 80% of BMDMs with dsRed-expressing bacteria by flow cytometry (data not shown). After 45 min and 2 h, medium was exchanged for media containing 100 and 10 μg/ml gentamicin, respectively. At 2 and 18 h postinfection, parallel samples were washed three times with PBS and lysed with 0.1% Triton X-100. A portion of the lysate was diluted in PBS and plated to determine CFU. Net growth was calculated as the recovered CFU at 18 h divided by that at 2 h. The remainder of the lysate was centrifuged for 20 min at 2,500 × g at 10°C, fixed with 1.6% paraformaldehyde, and analyzed using a CyAn ADP cytometer (Beckman Coulter). A minimum of 30,000 GFP-positive events were collected for analysis. Data were analyzed with FlowJo. Samples were gated for GFP-positive bacteria. Bacterial fold replication was calculated as the dsRed geometric mean of the inoculum divided by that of the GFP-positive population at 18 h postinfection. Data were normalized to wild type (100%).
Mouse infections.
Experimental protocols were approved by the University of Colorado Institutional Committee for Animal Care and Use. Seven-week-old male and female Sv129S6/SvEvTac mice (Taconic Laboratories) were used for competitive infection studies. Mice were inoculated with a 1:1 mixture of two differentially marked strains. For orogastric infections, animals were fasted 8 to 12 h prior to oral gavage with 1 × 109 of each strain (2 × 109 total in 100 μl), except for the ΔfadD versus ΔfadD;ydiD coinfection, which was 2 × 109 each strain. For intraperitoneal infections, animals were inoculated with 1 × 103 of each strain (2 × 103 total in 100 μl). At 2 weeks postinfection, animals were euthanized by CO2 asphyxiation followed by cervical dislocation. Cecum, Peyer’s patches, mesenteric lymph nodes, spleen, and liver were harvested, homogenized in PBS, and serially diluted to enumerate CFU. The competitive index (CI) for each organ was calculated as (CFUstrain A/CFUstrain B) output/(CFUstrain A/CFUstrain B) input.
Statistics.
P values were calculated using one-way analysis of variance (ANOVA) or one-sample t test (GraphPad Prism) and considered significant if the P value was <0.05, as described in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the members of the Detweiler lab for helpful discussions over the course of this project and Paul Muhlrad for help with writing.
This work was supported by NIH grants AI121474 (T.A.N. and C.S.D.) and AI121365 (C.S.D.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. There are no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bumann D. 2015. Heterogeneous host-pathogen encounters: act locally, think globally. Cell Host Microbe 17:13–19. doi: 10.1016/j.chom.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Gutierrez E, Chidlaw AC, Le Gall G, Bowden SD, Tedin K, Kelly DJ, Thompson A. 2016. A comparison of the ATP generating pathways used by S. Typhimurium to fuel replication within human and murine macrophage and epithelial cell lines. PLoS One 11:e0150687. doi: 10.1371/journal.pone.0150687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Yu K, Zhou F, Ding T, Yang Y, Hu M, Liu X. 2017. Quantitative proteomics charts the landscape of Salmonella carbon metabolism within host epithelial cells. J Proteome Res 16:788–797. doi: 10.1021/acs.jproteome.6b00793. [DOI] [PubMed] [Google Scholar]
- 5.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM. 2013. Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14:171–182. doi: 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffé M, Emile J-F, Marchou B, Cardona P-J, de Chastellier C, Altare F. 2008. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog 4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saliba A-E, Li L, Westermann AJ, Appenzeller S, Stapels DAC, Schulte LN, Helaine S, Vogel J. 2016. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol 2:16206. doi: 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 8.Muraille E, Leo O, Moser M. 2014. Th1/Th2 paradigm extended: macrophage polarization as an unappreciated pathogen-driven escape mechanism? Front Immunol 5:603. doi: 10.3389/fimmu.2014.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammer ND, Cassat JE, Noto MJ, Lojek LJ, Chadha AD, Schmitz JE, Creech CB, Skaar EP. 2014. Inter- and intraspecies metabolite exchange promotes virulence of antibiotic-resistant Staphylococcus aureus. Cell Host Microbe 16:531–537. doi: 10.1016/j.chom.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Bäumler AJ. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Bäumler AJ. 2017. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13:e1006129. doi: 10.1371/journal.ppat.1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manina G, Dhar N, McKinney JD. 2015. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. 2008. Positive feedback and noise activate the stringent response regulator rel in Mycobacteria. PLoS One 3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull World Health Organ 82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 16.Jones BD, Ghori N, Falkow S. 1994. Salmonella Typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med 180:15. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 18.Nix RN, Altschuler SE, Henson PM, Detweiler CS. 2007. Hemophagocytic macrophages harbor Salmonella enterica during persistent infection. PLoS Pathog 3:e193. doi: 10.1371/journal.ppat.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown DE, McCoy MW, Pilonieta MC, Nix RN, Detweiler CS. 2010. Chronic murine typhoid fever is a natural model of secondary hemophagocytic lymphohistiocytosis. PLoS One 5:e9441. doi: 10.1371/journal.pone.0009441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh ZN, Rakheja D, Yadav TP, Shome DK. 2005. Infection-associated haemophagocytosis: the tropical spectrum. Clin Lab Haematol 27:312–315. doi: 10.1111/j.1365-2257.2005.00717.x. [DOI] [PubMed] [Google Scholar]
- 21.Shin B-M, Paik IK, Cho HI. 1994. Bone marrow pathology of culture proven typhoid fever. J Korean Med Sci 9:57–63. doi: 10.3346/jkms.1994.9.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jha AK, Huang S-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, Artyomov MN. 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Galván-Peña S, O'Neill LAJ. 2014. Metabolic reprograming in macrophage polarization. Front Immunol 5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden SD, Rowley G, Hinton JCD, Thompson A. 2009. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar Typhimurium. Infect Immun 77:3117–3126. doi: 10.1128/IAI.00093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchawa Yimga M, Leatham MP, Allen JH, Laux DC, Conway T, Cohen PS. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect Immun 74:1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YR, Brinsmade SR, Yang Z, Escalante-Semerena J, Fierer J. 2006. Mutation of phosphotransacetylase but not isocitrate lyase reduces the virulence of Salmonella enterica serovar Typhimurium in mice. Infect Immun 74:2498–2502. doi: 10.1128/IAI.74.4.2498-2502.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spector MP, DiRusso CC, Pallen MJ, Del Portillo FG, Dougan G, Finlay BB. 1999. The medium-/long-chain fatty acyl-CoA dehydrogenase (fadF) gene of Salmonella Typhimurium is a phase 1 starvation-stress response (SSR) locus. Microbiology 145:15–31. doi: 10.1099/13500872-145-1-15. [DOI] [PubMed] [Google Scholar]
- 28.Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, Farhan H, Mazé A, Bumann D. 2013. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 9:e1003301. doi: 10.1371/journal.ppat.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang FC, Libby SJ, Castor ME, Fung AM. 2005. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect Immun 73:2547–2549. doi: 10.1128/IAI.73.4.2547-2549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Liu Y, Fu J, Zhang B, Cheng S, Wu M, Wang Z, Jiang J, Chang C, Liu X. 2019. Salmonella proteomic profiling during infection distinguishes the intracellular environment of host cells. mSystems 4:e00314-18. doi: 10.1128/mSystems.00314-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker D, Selbach M, Rollenhagen C, Ballmaier M, Meyer TF, Mann M, Bumann D. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 33.Shi L, Adkins JN, Coleman JR, Schepmoes AA, Dohnkova A, Mottaz HM, Norbeck AD, Purvine SO, Manes NP, Smallwood HS, Wang H, Forbes J, Gros P, Uzzau S, Rodland KD, Heffron F, Smith RD, Squier TC. 2006. Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J Biol Chem 281:29131–29140. doi: 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- 34.Diacovich L, Lorenzi L, Tomassetti M, Méresse S, Gramajo H. 2017. The infectious intracellular lifestyle of Salmonella enterica relies on the adaptation to nutritional conditions within the Salmonella-containing vacuole. Virulence 8:975–992. doi: 10.1080/21505594.2016.1270493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Preciado-Llanes L, Aulicino A, Decker CM, Depke M, Gesell Salazar M, Schmidt F, Simmons A, Huang WE. 2019. Single-cell and time-resolved profiling of intracellular Salmonella metabolism in primary human cells. Anal Chem 91:7729–7737. doi: 10.1021/acs.analchem.9b01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heath RJ, Jackowski S, Rock CO. 2002. Fatty acid and phospholipid metabolism in prokaryotes, p 55–92. In Vance DE, Vance JE (ed), New comprehensive biochemistry, vol 36. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 37.Iram SH, Cronan JE. 2006. The β-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J Bacteriol 188:599–608. doi: 10.1128/JB.188.2.599-608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark D, Cronan J. 2005. Two-carbon compounds and fatty acids as carbon sources. EcoSal Plus 2005 doi: 10.1128/ecosalplus.3.4.4. [DOI] [PubMed] [Google Scholar]
- 39.Morgan-Kiss RM, Cronan JE. 2004. The Escherichia coli fadK (ydiD) gene encodes an anerobically regulated short chain acyl-CoA synthetase. J Biol Chem 279:37324–37333. doi: 10.1074/jbc.M405233200. [DOI] [PubMed] [Google Scholar]
- 40.Campbell JW, Cronan JE. 2002. The enigmatic Escherichia coli fadE gene is yafH. J Bacteriol 184:3759–3764. doi: 10.1128/jb.184.13.3759-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell JW, Morgan-Kiss RM, E Cronan J. 2003. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic β-oxidation pathway. Mol Microbiol 47:793–805. doi: 10.1046/j.1365-2958.2003.03341.x. [DOI] [PubMed] [Google Scholar]
- 42.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 43.Wilson RB, Maloy SR. 1987. Isolation and characterization of Salmonella Typhimurium glyoxylate shunt mutants. J Bacteriol 169:3029–3034. doi: 10.1128/jb.169.7.3029-3034.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monack DM, Bouley DM, Falkow S. 2004. Salmonella Typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNγ neutralization. J Exp Med 199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol 182:1872–1882. doi: 10.1128/JB.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. 2016. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio 7:e02170-15. doi: 10.1128/mBio.02170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viarengo G, Sciara MI, Salazar MO, Kieffer PM, Furlan RLE, Garcia VE. 2013. Unsaturated long-chain free fatty acids are input signals of the Salmonella enterica PhoP/PhoQ regulatory system. J Biol Chem 288:22346. doi: 10.1074/jbc.M113.472829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 49.Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. 2010. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci U S A 107:3746–3751. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. 2009. Autophagy regulates lipid metabolism. Nature 458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mosser DM, Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 53.Kumar Y, Cocchiaro J, Valdivia RH. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol 16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 54.Lorenz MC, Fink GR. 2001. The glyoxylate shunt is required for fungal virulence. Nature 412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 55.Wall DM, Duffy PS, DuPont C, Prescott JF, Meijer WG. 2005. Isocitrate lyase activity is required for virulence of the intracellular pathogen Rhodococcus equi. Infect Immun 73:6736. doi: 10.1128/IAI.73.10.6736-6741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindsey TL, Hagins JM, Sokol PA, Silo-Suh LA. 2008. Virulence determinants from a cystic fibrosis isolate of Pseudomonas aeruginosa include isocitrate lyase. Microbiology 154:1616–1627. doi: 10.1099/mic.0.2007/014506-0. [DOI] [PubMed] [Google Scholar]
- 57.Brown DE, Libby SJ, Moreland SM, McCoy MW, Brabb T, Stepanek A, Fang FC, Detweiler CS. 2013. Salmonella enterica causes more severe inflammatory disease in C57/BL6 Nramp1G169 mice than Sv129S6 mice. Vet Pathol 50:867–876. doi: 10.1177/0300985813478213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark L, Perrett CA, Malt L, Harward C, Humphrey S, Jepson KA, Martinez-Argudo I, Carney LJ, La Ragione RM, Humphrey TJ, Jepson MA. 2011. Differences in Salmonella enterica serovar Typhimurium strain invasiveness are associated with heterogeneity in SPI-1 gene expression. Microbiology 157:2072–2083. doi: 10.1099/mic.0.048496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Branchu P, Bawn M, Kingsley RA. 2018. Genome variation and molecular epidemiology of Salmonella enterica serovar Typhimurium pathovariants. Infect Immun 86:e00079-18. doi: 10.1128/IAI.00079-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. 2000. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 61.Mirkov I, Stojanovic I, Glamoclija J, Stosic-Grujicic S, Zolotarevski L, Kataranovski D, Kataranovski M. 2011. Differential mechanisms of resistance to sublethal systemic Aspergillus fumigatus infection in immunocompetent BALB/c and C57BL/6 mice. Immunobiology 216:234–242. doi: 10.1016/j.imbio.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 62.Mariman R, Tielen F, Koning F, Nagelkerken L. 2015. The probiotic mixture VSL#3 has differential effects on intestinal immune parameters in healthy female BALB/c and C57BL/6 mice. J Nutr 145:1354–1361. doi: 10.3945/jn.114.199729. [DOI] [PubMed] [Google Scholar]
- 63.Krieg AM. 1999. Mechanisms and applications of immune stimulatory CpG oligodeoxynucleotides. Biochim Biophys Acta 1489:107–116. doi: 10.1016/s0167-4781(99)00147-5. [DOI] [PubMed] [Google Scholar]
- 64.Chu RS, Targoni OS, Krieg AM, Lehmann PV, Harding CV. 1997. CpG oligodeoxynucleotides act as adjuvants that switch on T helper 1 (Th1) immunity. J Exp Med 186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vellozo NS, Pereira-Marques ST, Cabral-Piccin MP, Filardy AA, Ribeiro-Gomes FL, Rigoni TS, DosReis GA, Lopes MF. 2017. All-trans retinoic acid promotes an M1- to M2-phenotype shift and inhibits macrophage-mediated immunity to Leishmania major. Front Immunol 8:1560. doi: 10.3389/fimmu.2017.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi W-J, Kim TS. 2017. Melatonin protects mice against stress-induced inflammation through enhancement of M2 macrophage polarization. Int Immunopharmacol 48:146–158. doi: 10.1016/j.intimp.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 67.Merritt FF, Smith BP, Reina-Guerra M, Habasha F, Johnson E. 1984. Relationship of cutaneous delayed hypersensitivity to protection from challenge exposure with Salmonella typhimurium in calves. Am J Vet Res 45:1081–1085. [PubMed] [Google Scholar]
- 68.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, Webb AM, Kershner JP, Blaskowski S, Copley SD. 2014. A versatile and highly efficient method for scarless genome editing in Escherichia coli and Salmonella enterica. BMC Biotechnol 14:84. doi: 10.1186/1472-6750-14-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 71.Berggren RE, Wunderlich A, Ziegler E, Schleicher M, Duke RC, Looney D, Fang FC. 1995. HIV gp120-specific cell-mediated immune responses in mice after oral immunization with recombinant Salmonella. J Acquir Immune Defic Syndr Hum Retrovirol 10:489–495. doi: 10.1097/00042560-199510050-00001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.