Abstract
Prostate cancer (PCa) progression is characterized by increased expression and transcriptional activity of the androgen receptor (AR). In the advanced stages of prostate cancer, AR significantly upregulates the expression of genes involved in DNA repair. Upregulation of expression for base excision repair (BER) related genes is associated with poor patient survival. Thus, inhibition of the BER pathway may prove to be an effective therapy for prostate cancer. Using a high throughput BER capacity screening assay, we sought to identify BER inhibitors that can synergize with castration therapy. An FDA-approved drug library was screened to identify inhibitors of BER using a fluorescence-based assay suitable for HTS. A gel-based secondary assay confirmed the reduction of BER capacity by compounds identified in the primary screen. Five compounds were then selected for further testing in the independently derived, androgen-dependent prostate cancer cell lines, LNCaP and LAPC4, and in the nonmalignant prostate derived cell lines PNT1A and RWPE1. Further analysis led to the identification of a lead compound, natamycin, as an effective inhibitor of key BER enzymes DNA polymerase β (pol β) and DNA Ligase I (LIG I). Natamycin significantly inhibited proliferation of PCa cells in an androgen depleted environment at 1 μM concentration, however, growth inhibition did not occur with nonmalignant prostate cell lines, suggesting that BER inhibition may improve efficacy of the castration therapies.
Keywords: BER, prostate cancer, natamycin
1. Introduction
Prostate cancer is the second most frequently diagnosed cancer among men. When detected early, localized disease can be effectively treated with a prostatectomy. Advanced disease that has metastasized beyond the prostate is treated with castration therapies. While at first responsive to this therapy, the cancer invariably recurs as castration resistant prostate cancer (CRPC), for which there is no cure. High throughput genetic screening technologies, such as transcriptomics and next generation sequencing, have led to the stratification of prostate cancer into well-defined molecular subclasses with unique drivers [1–3], making it clear that changes to the DNA damage and repair pathways are an intrinsic part of prostate cancer progression to CRPC. An ETS gene fusion that is found in nearly 80% of advanced cancers induces DNA damage [1, 4, 5] and correlates with increased patient mortality [6]. DNA repair pathways are altered in 27% of metastatic prostate cancers [1]. The increased expression of DNA repair associated genes in general and base excision repair (BER) – associated genes in particular correlates with rapid recurrence, metastatic dissemination, and decreased patient survival [3]. These findings suggest that the DNA repair machinery may be a therapeutic target in most advanced prostate cancers. Indeed, inhibitors of PARP1, an enzyme required for efficient BER [7], are being tested in men with CRPC [8–10]. Moreover, preclinical studies indicate that a tumor suppressor CCDC6 is a biomarker for sensitivity of CRPC to PARP inhibitors [11, 12]. The PARP inhibitors rucaparib (, and ) and olaparib (, , , etc.) have been tested in clinical trials and were granted breakthrough designation by the FDA for metastatic CRPC.
BER is an essential DNA repair pathway in mammalian cells that removes a variety of DNA base lesions generated endogenously and exogenously by DNA damaging agents [13]. It also repairs single-strand DNA (ssDNA) breaks by coordinating PARP-1, X-ray cross-complementing protein 1 (XRCC1), and DNA polymerase β (pol β) [14, 15]. There are two BER sub-pathways: single-nucleotide (SN) and long-patch (LP) BER. SN-BER is initiated by the removal of a damaged DNA base by a damage-specific DNA glycosylase. This leaves an abasic site that is 5’-incised by AP endonuclease 1 (APE1) resulting in a 1 nucleotide-gapped intermediate with a 5’-deoxyribose phosphate (dRP) group. Subsequently, a native dRP group is directly removed by the dRP lyase activity of pol β [16] [17]. Pol β then fills in the gap with its polymerase activity leaving nicked DNA, which is sealed by DNA ligases, e.g. DNA Ligase I (LIG I) and LIG IIIα [18, 19]. In this scenario only one nucleotide is replaced. In LP-BER, a modified dRP group, such as an oxidized dRP, that is refractory to the dRP lyase activity of pol β, is removed by flap endonuclease 1 (FEN1). This leaves a one-nucleotide gap that is then filled in by pol β creating a nick sealed by LIG I. Alternatively, pol β can continuously perform the strand-displacement DNA synthesis to create a 5’-flap with a dRP group. FEN1 removes the flap, and LIG I seals the nick to accomplish the long-patch BER [19]. To facilitate the efficiency of BER, several co-factors, such as PARP1 and proliferating cell nuclear antigen (PCNA) coordinate and stimulate the activities of the core enzymes, pol β and FEN1 [20, 21]. It is proposed that PARP1 is activated in response to ssDNA breaks and catalyzes poly (ADP)ribosylation of XRCC1 [22, 23]. XRCC1-DNA ligase III (LIG III) complex is recruited to the strand breaks and facilitates the ligation of nicked DNA [23]. PARP1 can also coordinate with APE1 and FEN1 to modulate the efficiency of pol β-mediated LP-BER [15, 21]. These findings suggest a regulatory role for BER cofactors in the total capacity of BER in cells.
Rapid development of resistance to PARP inhibitors makes it is critically important to identify orthogonal suppressors of BER capacity. In this study we employed a high throughput BER capacity assay to screen a small molecule compound library of FDA approved drugs in prostate cancer cell extracts [24]. Following primary and secondary screens we identified 5 compounds that significantly reduced BER capacity. Using the independently derived androgen sensitive prostate cancer cell lines, LNCaP and LAPC4, and nonmalignant cell lines, PNT1A and RWPE1, we compared the inhibitory effect of these compounds on cellular proliferation and viability of prostate-derived cell lines. Natamycin showed a significant preference for inhibiting viability and proliferation of LNCaP and LAPC4, when compared to PNT1A and RWPE1 cell lines. Investigation of the mechanism of natamycin action showed that it inhibits key BER enzymes, pol β and LIG I. These findings support BER pathways as therapeutic targets and suggests that natamycin should be investigated as a potential treatment for advanced prostate cancer.
2. Materials & Methods:
2.1. Reagents
The SCREEN-WELL FDA approved drug library V2 was purchased from Enzo (Farmingdale, NY). All individual compounds were purchased from Sigma-Aldrich (St. Louis, MO). Nystatin (N4014), ceftazidime (CDS020667), calcipotriene (C4369) and prasugrel (SML0331) were all dissolved in sterile DMSO (MP Biomedicals, Solon, OH). Natamycin (P0440) is supplied as a 2.5% γ-irradiated saline solution. Oligonucleotides were from Integrated DNA Technologies (IDT, Coralville, IA). The oligonucleotide containing a BHQ-tagged T was from LGC Biosearch Technologies (Petaluma, CA). E. coli Exo III was from New England BioLabs (Ipswitch, MA). All chemical reagents were from Sigma-Aldrich (St Louis, MO) and Thermo Fisher Scientific Inc (Weston, FL).
2.2. High throughput screening assay for inhibitors of the BER pathway
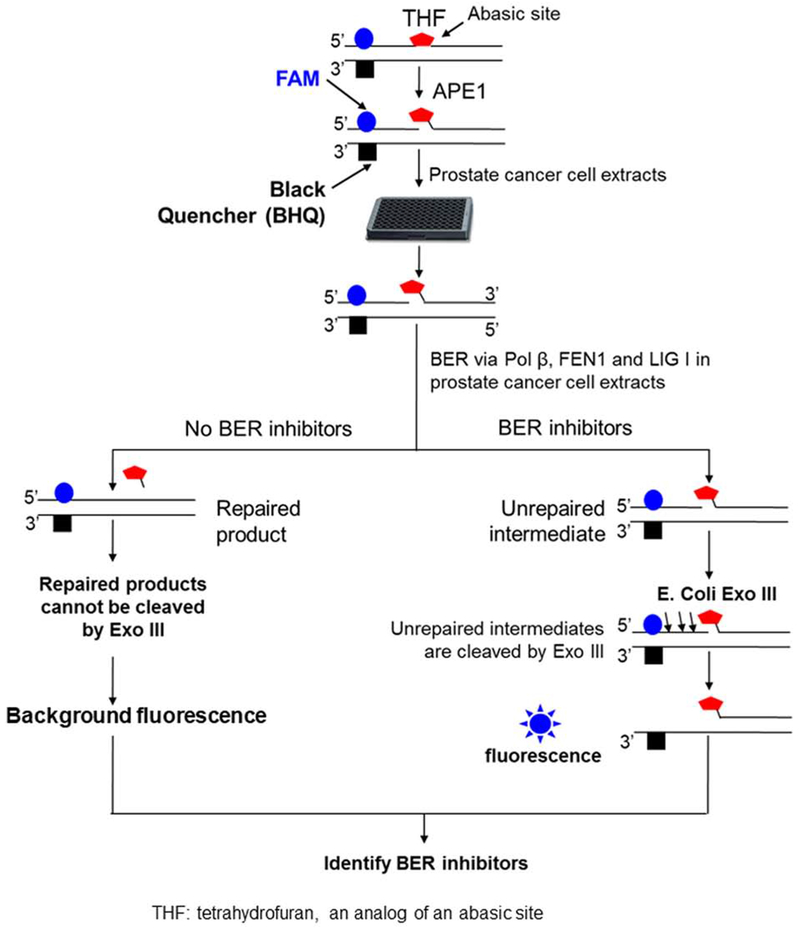
The high throughput screening assay was described in US Patent No. 9809843 B1. Briefly, a fluorescence-tagged oligonucleotide substrate that contains a synthesized abasic site, i.e., tetrahydrofuran (THF) was designed to determine the total capacity of BER in prostate cancer whole cell extracts. The sequence of the oligonucleotides for constructing the substrate is: 5’-CTGGA[FluorT]ACACGAACTTTAAGCATHFAGTCAATGAAGGACGCATATCAGTG-3’ (upper strand) and 5’-CACTGATATGCGTCCTTCATTGACTCTGCTTAAAGTTCG TG[T(BHQ-1)]ATCCAG-3 (bottom strand). A 6-carboxyfluorescein (6-FAM)-tagged-T is inserted upstream of the abasic site in the damaged strand and close to a black hole quencher (BHQ) tagged-T, which was inserted in the template strand (Figure 1). The substrate was constructed by annealing the damaged strand with the template strand at 1:1 ratio. The substrate (25 nM) was precut with 25 nM purified human AP endonuclease 1 (APE1) at 37 °C for 30 min. Subsequently, the substrate was incubated with 25 μg prostate cancer cell extracts (total volume of 10 μL) at 37 °C for 30 min allowing repair of the abasic site by BER. Unrepaired substrates were then subject to digestion by the 3′-5′ exonuclease activity of E. coli Exo III (0.5U) (New England BioLabs, Ipswitch, MA) at 37 °C for 10 min. This cleaved the upstream strand in the unrepaired substrates releasing the 6-FAM-tagged T and allowing the emission of fluorescence detected by a fluorescence plate reader at 528±20 nm (Biotek Instruments, Winoski, VT). Inhibition of BER enzymatic activity and/or the coordination among BER enzymes and their cofactors reduced the amount of repaired products and led to the accumulation of unrepaired substrates thereby significantly increasing the intensity of fluorescence signal. The approach was used with a 384-well platform in high throughput screening for inhibitors of the BER pathway.
Figure 1. The schematic diagram of the fluorescence-based high throughput screening of BER capacity inhibitors.
A fluorescence-tagged oligonucleotide substrate that contains the analog of an abasic lesion, tetrahydrofuran (THF) was employed to determine the inhibitory effects of 774 compounds from The Screen-Well® FDA Approved Drug Library V2 on the BER capacity of prostate cancer whole cell extracts. The procedure of the screening was conducted as descried in the “Materials and Methods”.
2.2.1. High Throughput Screening for BER inhibitors
The Screen-Well® FDA Approved Drug Library V2 with 774 compounds were purchased from Enzo. The 10 mM stock solutions in DMSO were diluted to 2 mM before 0.5 μL was added to 10 μL of each assay reaction mixture of 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 0.1 mM EDTA, 0.1 mg/ml bovine serum albumin, 0.01% Nonidet P-40, 25 nM APE1 pre-cut substrate and cancer cell extract (72 μg of LNCaP cell lysate per assay) in 384-well black plates (Corning 3821), for a final compound concentration of 100 μM. The control reaction also has 5% DMSO added. After mixing for 2 min and spinning at 200 g for 1 min, the plates were incubated at 37°C for 30 min. Freshly diluted Exo III (0.5 U, New England BioLabs) was then added for an additional incubation at 37°C for 10 min, followed by 30 min at 50°C. The reactions were terminated by adding 1 μL of 500 mM EDTA. Fluorescence signal (excitation wavelength of 485±20 nm and emission wavelength of 528±20 nm) were recorded with the Biotek Synergy HT Plate Reader. Compounds that showed a signal greater than DMSO control + 3 S.D for each plate were chosen as hits. Twenty-six hits were selected from 774 compounds (3.4%).
2.2.2. Secondary Assays of BER inhibitors
The hit compounds were subject to secondary assays to determine their ability to reduce the BER capacity when reconstituted with purified core BER enzymes, as well as their inhibitory effects on individual BER enzymes including pol β, FEN1 and LIG I. The effect of hit compounds on BER capacity and BER enzymes was examined using a denaturing sequencing gel-based assay. 10 μM hit compounds were incubated with 10 nM pol β, 10 nM FEN1 and 20 nM LIG I in BER reaction buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 0.1 mM EDTA, 0.1 mg/ml bovine serum albumin, and 0.01% Nonidet P-40 in the presence of 5 mM Mg2+ and 2 mM ATP. The compounds were initially preincubated with a BER enzyme at varying concentrations for 1 h with rotation. This was followed by the addition of 25 nM 32P-labeled substrate containing an abasic site, which was precut with 5 nM APE1. The reaction mixture (20 μL) was incubated at 37° C for 30 min, and the reaction was terminated by 50 mM EDTA. Substrates, products, and unrepaired BER intermediates were separated by 15% urea-denaturing sequencing gel and detected by a Pharos FX Plus PhosphorImager (Bio-Rad, Hercules, CA).
2.3. Cell Culture
LNCaP, LAPC4, PNT1A, and RWPE1 cell lines were purchased from ATCC (Manassas, VA) and used within 8 passages after plating. LNCaP and PNT1A and were maintained in RPMI-1640 supplemented with 10% FBS 1% PenStrep. LAPC4 were maintained in RPMI-1640 supplemented with 10% FBS, 1% PenStrep and 10−8M of R1881. RWPE1 was maintained in Keratinocyte-SFM supplemented with EGF and BPE, according to ATCC guidelines. All media and PenStrep were purchased from Gibco (Carlsbad, CA). FBS and charcoal stripped serum were purchased from Sigma-Aldrich (St. Louis, MO). Charcoal stripped Serum (CSS) was purchased from HyClone (Logan, UT). R1881 was purchased from Perkin Elmer (Waltham, MA).
2.4. Proliferation Assay
Proliferation assays were done using Roche DP Real Time Cell Analyzer (RTCA), as described by the manufacturer. Background impedance was established after incubating E-plates (Acea Biosciences, San Diego, CA) with 50 μL media at room temperature for 30 min and placed in RTCA. Cells were then seeded in 100 μL per well. Cells attached overnight and then treated. During attachment stage and after treatment impedance was measured every 30 min. Impedance is represented by cell index and is calculated as follows: CI= (Zi-Zo)/15Ω where Zi is impedance at an individual time point, and Zo is the background impedance. Average CI was calculated from four wells per treatment at each time point and normalized to the impedance immediately after compound addition. All data was normalized to the impedance at the time of treatment, which was assigned a value of 1.
2.5. Viability Assay
Cell viability was determined using the Cell Titer Glo Cell Viability Assay (Promega, Madison, WI), as recommended by manufacturer. All experiments were performed in quadruplicate.
2.6. Toxicity assay
Cellular toxicity was assessed using the MTT assay. Cells were seeded in a 96 well plate at 1x104 cells/well and allowed to adhere overnight in the presence of FBS or CSS. The following day treatment was added to the wells and the plates were incubated for 24 h or 48 h. After incubation, 50 μL of MTT (2 mg/mL) was added to each well and incubated in the dark for 4 h. After incubation 150 μL of the media/MTT solution was removed and 100 μL of DMSO was added to each well and incubated for 15 min. Absorbance at 570 nm was measured using a ClarioStar Plate Reader (BMG LabTech, Ortenburg, Germany).
2.7. Western Blotting
Whole cell lysate protein was extracted with protein extraction buffer [20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton-X] supplemented with protease inhibitors (GenDepot, Barker, TX). For each sample 20μg of protein was resolved on a 10% SDS-PAGE and transferred to a PVDF membrane (GE Healthcare, Germany). Immunoblotting was done using primary antibodies for Tubulin (1:5000; Millipore, Temecula, CA), γ-H2A.X [Ser139] (1:1000; Cell Signaling, Danvers, MA), and AR (1:1000; Santa Cruz, Dallas, TX). Chemiluminescent signal was captured using an ImageQuant LAS 500 (GE Healthcare, Uppsala, Sweden).
2.8. Measurement of the inhibitory effects of natamycin on pol β DNA synthesis and LIG I activity
The inhibitory activities of natamycin on pol β DNA synthesis were measured by incubating the substrates containing 1-nt gap (50 nM) with 5 nM pol β in the presence of 1, 2, 5 or 10 μM natamycin in BER reaction buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 0.1 mM EDTA, 0.1 mg/ml bovine serum albumin, and 0.01% Nonidet P-40) with 5 mM MgCl2 and 50 μM dNTPs. The inhibitory activities of natamycin on LIG I (5 nM) activity were tested on the substrate containing random DNA sequence with a nick in BER reaction buffer with 5 mM MgCl2 and 2 mM ATP in the presence of 0.1, 0.5, 1 or 5 μM natamycin. Reaction mixtures were assembled on ice and incubated at 37° C for 15 min. BER reactions were terminated by adding 2X stopping buffer containing 95% formamide and 10 mM EDTA. Reaction mixtures were then denatured at 95° C for 5 min and separated by 15% urea-denaturing polyacrylamide gel electrophoresis. Substrates and products were detected and analyzed using a Pharos FX Plus Phosphorlmager (Bio-Rad, Hercules, CA). For all the reactions, natamycin was preincubated with the BER enzymes for 2 h at 4° C with rotation.
3. Results
3.1. Primary screening of compounds that inhibit the BER pathway
To identify FDA approved compounds that can directly inhibit the activities of BER enzymes and co-factors we invented a high throughput, BER pathway – specific screening approach (Figure 1). This method was used to identify compounds that interfere with the interactions between BER enzymes and co-factors in the prostate cancer cell line LNCaP. This technique relies on the design of an oligonucleotide substrate with a fluorescent 6-FAM labeled abasic lesion located adjacent to a black quencher (BHQ) in the template strand. Efficient BER of the abasic lesion will lead to quenching of the 6-FAM fluorescent tag, whereas inhibition of BER will spatially separate 6-FAM and BHQ tags increasing fluorescent signal (Figure 1). Using this approach, we performed a high throughput screening of 774 compounds from the Screen-Well® FDA Approved Drug Library V2, in LNCaP cell lysates. The initial screen identified 26 compounds that exerted significant inhibition on BER as indicated by a more than 3-fold increase in fluorescent signal over the background.
3.2. Secondary screening of compounds that inhibit the activities of BER enzymes
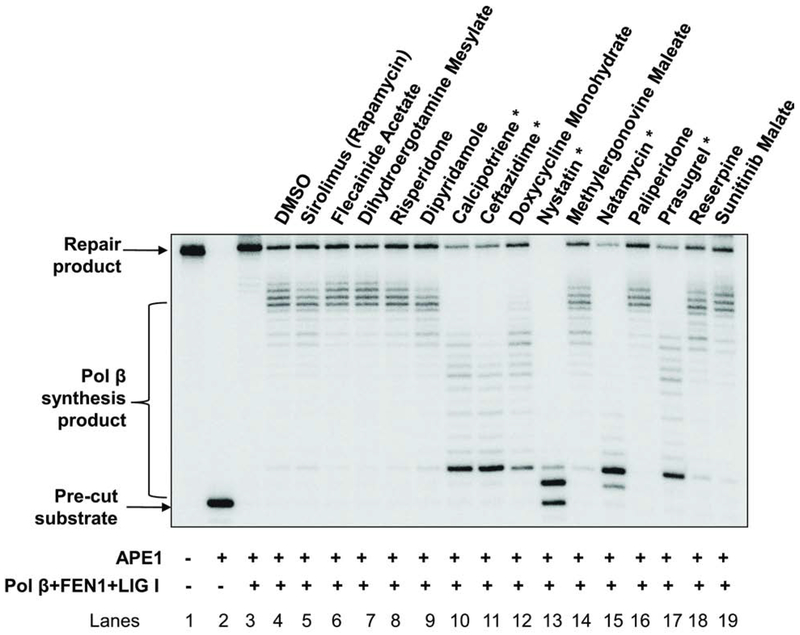
Among the 26 identified compounds, nine were DNA intercalators or inhibitors of human DNA topoisomerases and were excluded from further studies. Thus, fifteen FDA approved drugs were selected for further testing. To further confirm the inhibitory effects of the lead compounds on BER, we initially determined the effects of the compounds on BER reactions reconstituted with purified BER core enzymes. We reconstituted the substrate containing the abasic site analogue THF with purified Pol β, FEN1, and LIG I. The results demonstrated that 5 compounds, calcipotriene, ceftazidime, nystatin, natamycin and prasugrel (Figure 2, marked *) significantly reduced BER capacity and the production of the BER repair product suggesting that the compounds inhibited BER by inhibiting the activities of the BER core enzymes.
Figure 2. Secondary screen of BER inhibitors.
The secondary screening assay was performed as described in the “Materials and Methods”. Lane 1 represents the substrate only. Lane 2 indicates the reaction mixture with 5 nM APE1. Lanes 3 and 4 correspond to the reaction mixture with 5 nM APE1, 10 nM pol β, 10 nM FEN1 and 20 nM LIG I in the absence and presence of DMSO. Lanes 5 through 19 correspond to the reaction mixture with 5 nM APE1, 10 nM pol β, 10 nM FEN1 and 20 nM LIG I in the presence of indicated compounds. The experiments were repeated at least three times. A representative gel is illustrated. “*” denotes the hit compounds that exhibited significant BER capacity inhibitory effects in the secondary screening assay.
3.3. Validation of inhibitory effects of the lead compounds on the proliferation of malignant and non-malignant prostate cell lines.
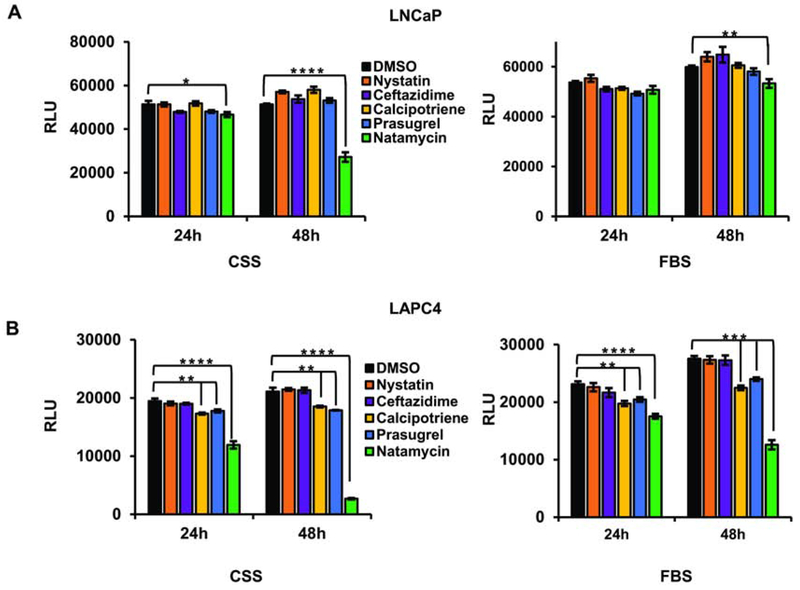
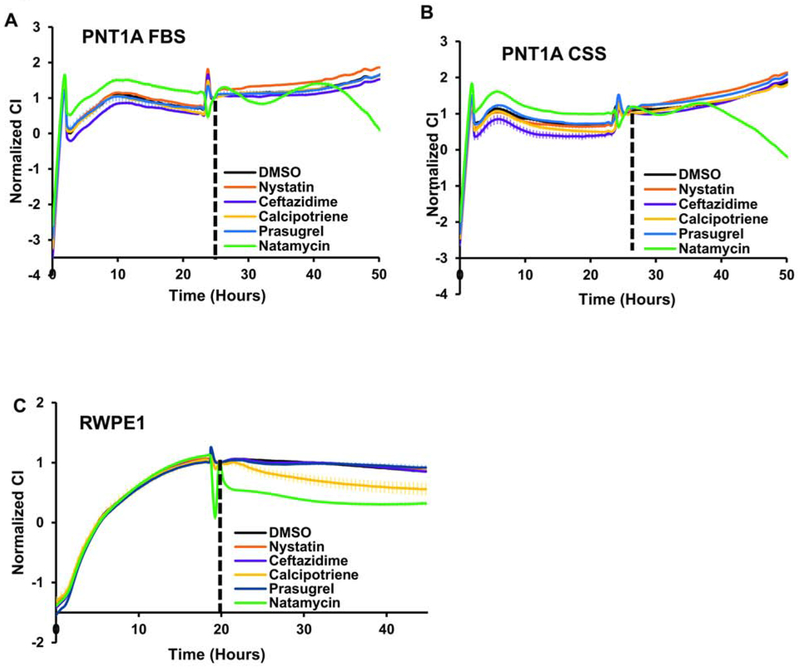
To determine whether lead BER inhibitors affect cell survival, we treated the independently derived androgen-dependent prostate cancer cells, LNCaP and LAPC4, with vehicle (DMSO) or 10 μM nystatin, ceftazidime, calcipotriene, prasugrel, or natamycin. The highest toxicity was observed in prostate cancer cells maintained in androgen depleted conditions treated with natamycin (Figure 3A–B).
Figure 3. Cellular toxicity of the lead compounds in androgen-dependent prostate cancer cell lines.
A. LNCaP cells were plated in a 96-well plate at 1x104 cells/well in the presence of FBS or CSS. Cells were allowed to attach overnight and were treated with either vehicle (DMSO) or 10 μM of indicated compound. At 24 and 48 h cellular viability was compared using the Cell Titer Glo Luminescent Cell Viability kit. B. LAPC4 cells were plated in a 96-well plate at a density of 1x104 cells/well in the presence of FBS or CSS. Cells were allowed to attach overnight and treated with 10 μM of indicated compound. At 24 h and 48 h cellular viability was compared as in A. (* signifies a difference between vehicle and treatment with *p≤0.05, ** p≤0.01, *** p≤0.001 **** p≤0.0001).
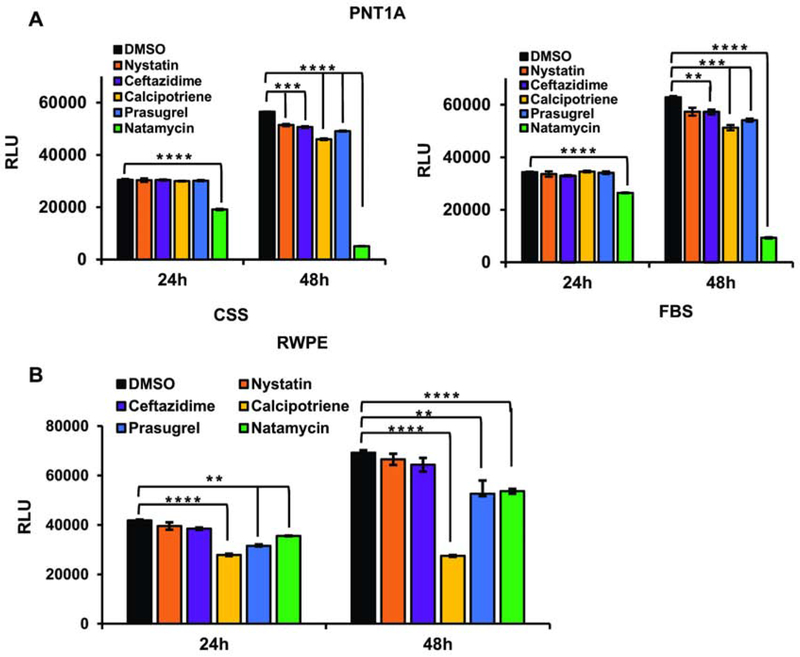
We next tested whether the same concentration of compounds decreases the viability of the AR negative and nonmalignant prostate derived cells, PNT1A and RWPE1. In PNT1A, natamycin displayed similar toxicity in both FBS and CSS supplemented medium (Figures 4A). Medium for RWPE1 cell lines does not include serum, rather it contains the growth hormone supplements, Epidermal Growth Factor (EGF) and Bovine Pituitary Extract (BPE). Calcipotriene, prasugrel, and natamycin significantly decreased RWPE1 cell viability at 10 μM after 24 h and 48 h (Figure 4B). Thus, androgen depletion significantly and specifically enhances natamycin toxicity in androgen dependent prostate cancer cell lines.
Figure 4. Cellular toxicity of lead compounds in nonmalignant androgen independent prostate-derived cell lines.
A. PNT1A cells were plated in a 96-well plate at 1x104 cell/well in the presence of FBS or CSS. Cells were allowed to attach overnight and were treated with either vehicle (DMSO) or 10 μM of indicated compound. At 24 and 48 h, cellular viability was compared using the Cell Titer Glo Luminescent Cell Viability kit. B. RWPE1 cells were plated in a 96-well plate at 1x104 cells/well using the specified serum free media. Cells were allowed to attach overnight and treated with 10 μM of indicated compound. At 24 h and 48 h, cellular viability was compared as in A. (* signifies a difference between vehicle and treatment with ** p≤0.01, *** p≤0.001, **** p≤0.0001).
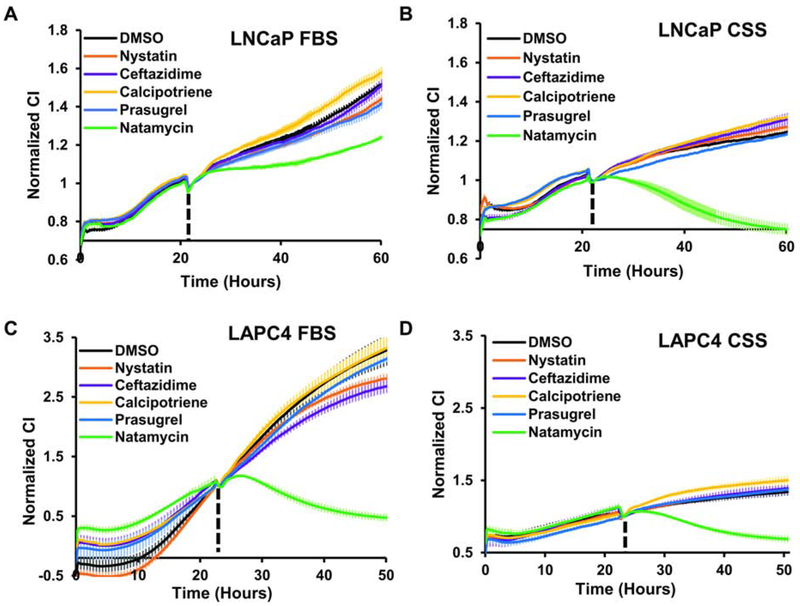
We next compared the effect of 10 μM nystatin, ceftazidime, calcipotriene, prasugrel, or natamycin on cellular proliferation in LNCaP, LAPC4, PNT1A, and RWPE1 cell lines. As seen in Figure 5A, in the presence of FBS LNCaP cells were able to proliferate in the presence of all inhibitors. However, natamycin treated cells had the slowest proliferation rate (Figure 5A). Consistent with previous reports, LNCaP cells proliferated more slowly in androgen depleted medium and addition of 10 μM natamycin abolished cellular impedance (Figure 5B) suggesting cell detachment. In LAPC4 cells, cellular proliferation and attachment was abolished after treatment with 10 μM natamycin in medium supplemented with both FBS (Figure 5C) and CSS (Figure 5D).
Figure 5. Natamycin suppresses cellular proliferation and adhesion in androgen dependent prostate cancer cell lines.
A-B. LNCaP cells were plated onto xCelligence E-plates at 104 cells per well in medium supplemented with either 10% FBS (A) or 10% CSS (B) serum. Cells attached overnight and were treated with vehicle (DMSO) or 10 μM of the indicated compounds. C-D. LAPC4 cells were plated at 5x104 cells per well onto xCelligence E-plates in media supplemented with FBS (C) or CSS (D) and allowed to attach overnight. Treatment was conducted as in (A-B) and impedance measured continuously in 30 min intervals. Impedance in all graphs was normalized to the time of treatment, which was assigned a value of 1.
The nonmalignant, AR negative cell line PNT1A proliferated in the presence of all inhibitors except natamycin, in both FBS (Figure 6A) and CSS supplemented media (Figure 6B). However, natamycin effect became evident only at 42 h of treatment. RWPE1 was slightly inhibited by 10 μM natamycin and calcipotriene (Figure 6C). Both cell lines remained attached after treatment with all inhibitors.
Figure 6. Androgen-independent prostate derived cell lines are sensitive to high concentrations of natamycin.
A-B. PNT1A cells were plated onto xCelligence E-plates at 5X104 cells per well in media supplemented with 10% FBS (A) or CSS (B). Cells were allowed to attach overnight and treated with vehicle (DMSO) or 10 μM of indicated compounds. Cellular impedance was monitored continuously with 30 min intervals. C. RWPE1 cells were plated at 5X104 cells per well in serum free medium supplemented with EGF and BGP onto E-plates and treated with 10 μM of indicated compounds as indicated. Impedance was continuously measured using RTCA every 30 min and was normalized to the time of treatment which was assigned a value of 1.
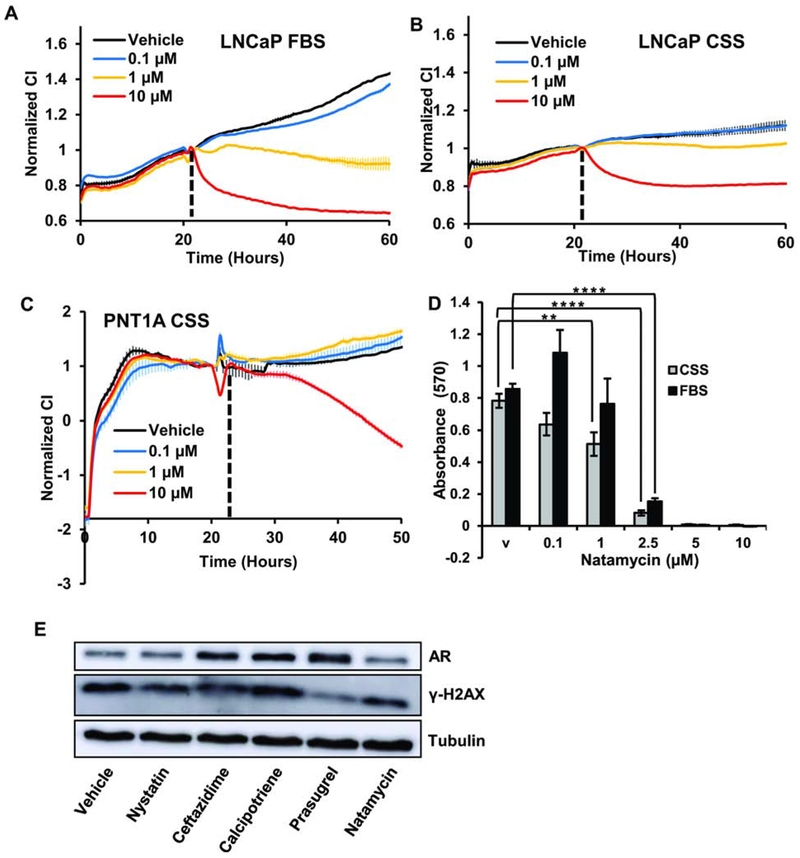
To determine whether AR positive and AR negative cell lines have different sensitivity to natamycin we treated LNCaP and PNT1A with 10 μM, 1 μM, and 0.1 μM of the compound. LNCaP cell proliferation was significantly inhibited by all concentrations in FBS medium (Figure 7A) and 10 μM and 1 μM were inhibitory in medium with CSS (Figure 7B). PNT1A cell proliferation was inhibited only with 10 μM natamycin in both CSS (Figure 7C) and FBS (not shown) medium. In LNCaP cells, natamycin reduced viability preferentially in androgen-depleted conditions. As seen in Figure 7D, natamycin reduced the viability of LNCaP cells in CSS medium at concentrations of 0.1 μM and higher, while treatment with 2.5 μM or higher concentration was required to reduce LNCaP viability in medium supplemented with FBS.
Figure 7. Androgen-dependent prostate cancer cell lines are more sensitive to natamycin treatment.
A. LNCaP cells were plated in 10 % FBS supplemented medium at 104 cells per well onto E-plates and treated with vehicle, 0.1 μM, 1 μM, or 10 μM of natamycin. B. LNCaP cells were plated in medium supplemented with 10% CSS at 104 cells/well and treated as in A. C. PNT1A cells were plated in medium supplemented with 10% CSS at 5x104 cells per well and treated as in A. D. LNCaP cells were plated in medium supplemented with 10% CSS or 10% FBS and cells were allowed to attach. Next day, cells were treated with vehicle, 0.1 μM, 1 μM, 2.5 μM, 5 μM, or 10 μM of natamycin. Forty eight hours later cellular viability was compared using MTT assay. E. LNCaP cells were grown in complete medium and treated for 48 h with 10 μM of indicated compounds. Protein was extracted and AR, γ-H2AX, and tubulin levels compared by Western blotting. Impedance in A-C was measured every 30 min and values were normalized to those at the time of treatment. (*signifies a difference between vehicle and treatment with ** p≤ 0.01, ****p≤ 0.0001).
The proliferation of androgen dependent cell lines is driven in part by AR signaling. To test whether these compounds affect AR signaling we compared AR protein levels after 48-hour treatment with 10 μM of each compound. As seen in Figure 7E, natamycin slightly decreased AR levels, while no decline was observed with other compounds. No significant increase in the marker of double-stranded breaks, γ-H2AX, was evident at 48 h after treatment with the indicated inhibitors.
3.4. The inhibitory effects of natamycin on BER enzymes
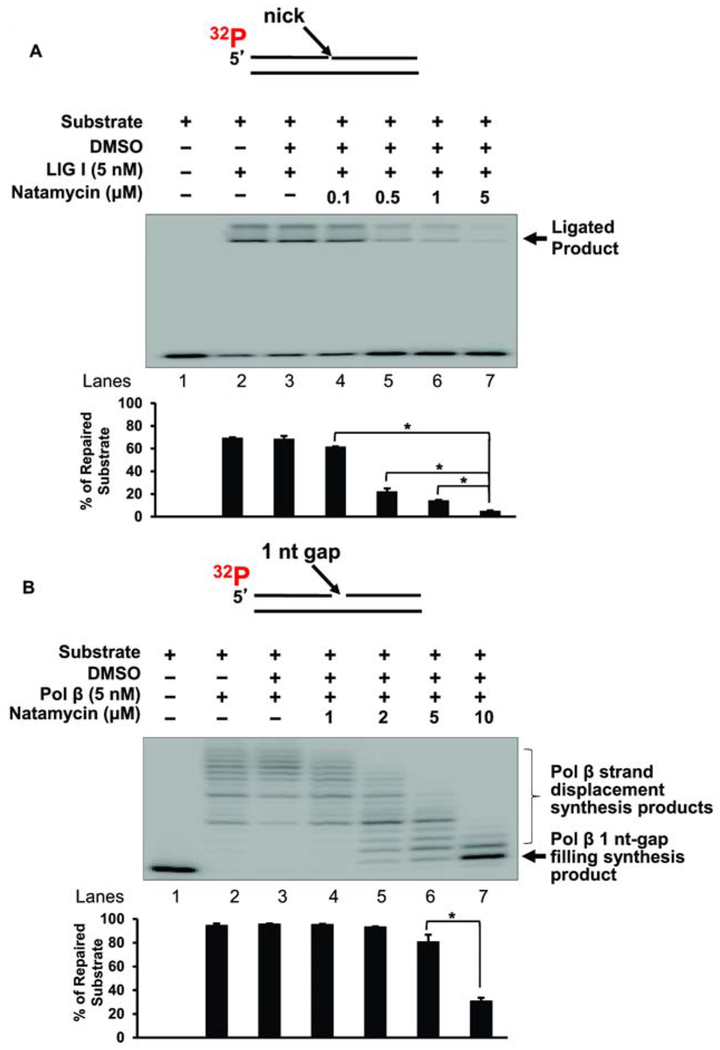
To further determine if natamycin reduced BER capacity by inhibiting a specific BER enzyme, we examined the inhibitory effects of natamycin on the activities of the core BER enzymes, LIG I, FEN1, and pol β. At 0.5 μM and 1 μM, natamycin reduced the activity of LIG I by 3.5-fold (Figure 8A). At 5 μM, it decreased the activity of LIG I by 14-fold (Figure 8A). Natamycin showed a significantly milder inhibitory effect on pol β at low concentrations. At 10 μM natamycin reduced pol β activity by 3-fold (Figure 8B). Moreover, our results showed that natamycin exhibited a significant inhibitory effect on pol β strand-displacement synthesis at 2 nM - 5 nM but did not inhibit pol β 1 nt gap-filling synthesis (Figure 8B, lanes 5-7) further indicating that natamycin specifically inhibited long-patch BER rather than SN-BER. No inhibitory effect of natamycin on FEN1 was detected (data not shown). The results indicate that natamycin moderately inhibited pol β DNA synthesis activity and exhibited a potent inhibition on LIG I activity. The results further indicate that natamycin reduced the BER capacity of prostate cancer cells primarily by inhibiting the activity of LIG I along with the moderate inhibition the strand-displacement synthesis of pol β DNA. This further suggests that natamycin can lead to the accumulation of single-strand DNA breaks in prostate cancer cells, impeding cell proliferation.
Figure 8. Natamycin inhibits BER enzymes pol β and LIG I.
A. The inhibitory effect of natamycin on LIG I activity was examined with the substrate containing a nick. Lane 1 represents the substrate only. Lanes 2 and 3 indicates the reaction mixture with 5 nM LIG I in the absence and presence of DMSO. Lanes 4-7 correspond to the reaction mixture with 5 nM LIG I in the presence of increasing concentrations of natamycin. The substrate was 32P-labeled at the 5’-end of the upstream strand and is illustrated above the gel. The experiments were repeated at least three times. Representative gel is illustrated. Quantification of the results is shown below the gel. Two-way ANOVA with Tukey’s multiple comparison posttests was used to determine statistically significant differences. * signifies p<0.05 compared to the control reaction with LIG I and DMSO. It should be noted that upper band is the smearing of the ligated products in the course of gel electrophoresis. B. The inhibitory effect of natamycin on pol β DNA synthesis was determined with the substrate containing a 1-nt gap as described in the “Materials and Methods”. Lane 1 represents the substrate only. Lanes 2 and 3 indicates the reaction mixture with 5 nM pol β in the absence and presence of DMSO. Lanes 4-7 correspond to the reaction mixture with 5 nM pol β in the presence of increasing concentrations of natamycin. The substrate was 32P-labeled at the 5’-end of the upstream strand and is illustrated above the gel. The experiments were repeated at least three times. Quantification of the results are shown below the gel. Two-way ANOVA with Tukey’s multiple comparison posttests was used to determine statistical significance. * signifies p< 0.05 compared to the control reaction with pol β and DMSO.
4. Discussion
Prostate cancer that has disseminated beyond the prostate capsule is an incurable disease. Castration therapies extend survival by a few months followed by progression to CRPC. Increasing evidence suggests that in prostate cancer the upregulation of multiple DNA repair pathways, including BER, helps to mitigate the inherent genomic instability of this disease [3]. To date, no attempts to target core enzymes of BER in prostate cancer have been made. We thought to test FDA approved compounds for their ability to inhibit BER and to evaluate BER inhibition as a potential therapeutic strategy for prostate cancer treatment. To conduct this screening, we developed a high throughput fluorescent assay that measures BER capacity. In this assay, we utilized a synthetic BER substrate containing both a fluorescent reporter and quencher; efficient BER keeps fluorescence at background levels due to proximity of reporter and quencher, while BER inhibition leads to degradation of the unrepaired substrate, liberating the fluorescent reporter from the quencher and increasing fluorescence. Using our innovative strategy, we screened 744 FDA approved compounds, of which 26 efficiently reduced fluorescence in the BER capacity assay. Some of these compounds were known DNA intercalators and topoisomerase inhibitors and were deemed nonspecific. However, nystatin, ceftazidime, calcipotriene, prasugrel, and natamycin, did not fall in this category, efficiently inhibited BER in a cell-free reaction, and were further evaluated for BER specificity. These five compounds included functionally disparate molecules with antifungal and anti-bacterial properties, vitamin D agonist, and an ADP receptor inhibitor [25–29]. Using cell-based assays we determined that the strongest inhibitor of viability and proliferation in two independently derived prostate cancer cells, LNCaP and LAPC4, was natamycin, a polyene macrolide antibiotic with broad antifungal activity and low toxicity against mammalian cells. Similar to most prostate cancers, both LNCaP and LAPC4 cells express androgen receptor and require androgens for optimal proliferation and viability. While natamycin efficiently inhibited both cell lines in complete medium, it essentially abolished proliferation in medium depleted of steroids. Remarkably, prostate derived non-malignant cell lines, PNT1A and RWPE, were significantly less sensitive to natamycin treatment in both proliferation and viability assays. Moreover, a 10 μM concentration was required to inhibit nonmalignant cells while 1 μM was sufficient for proliferation inhibition of LNCaP and LAPC4 cell lines, suggesting an increased sensitivity of cancer cells to BER inhibition. In previous reports, treatment of human lymphocytes with natamycin at concentrations ranging from 19.5 μM to 42.4 μM caused chromosomal abnormalities only at the highest concentration supporting the safety of 1 μM natamycin treatment for nonmalignant cells [30].
We next investigated which components of BER are inhibited by natamycin. Our results demonstrated that Natamycin exhibited potent inhibition on LIG I activity at 0.5 μM and pol β strand-displacement synthesis at 5 μM (Figure 8), suggesting that the inhibitory effects of natamycin result in the accumulation of ssDNA breaks leading to prostate cancer cell death. It has been found that the steady state levels of LIG I are elevated in cancer cell lines compared to normal cells, presumably due to the necessity of this enzyme for the aggressive proliferative activity of cancer cells [31]. Thus, the development of ligase inhibitors may potentiate the toxic effects of chemotherapeutic agents used for cancer treatment. As the central component of BER, pol β plays a significant role in the drug resistance of cancer therapy [32] due to its “translesion” DNA synthesis that can help cancer cells tolerate DNA damage caused by some anticancer therapies [33]. Pol β is mutated in approximately 30% of tumors leading to a reduction in pol β fidelity during DNA synthesis and promoting mutagenesis and survival of the tumor cells [34–36]. Thus, targeting pol β has been considered as a promising therapeutic strategy for the improvement of cancer treatment [37]. Inhibition of pol β DNA synthesis can result in the accumulation of gapped DNA intermediates that cannot be ligated by DNA ligases, suggesting that the synergy between the inhibition of both LIG I and pol β by natamycin may sensitize prostate cancer cells to endogenous cancer-specific DNA damage as well as DNA damaging and hormonal cancer therapies.
5. Conclusions
In this study, we show that FDA approved compounds contain molecules that specifically inhibit BER. We also show that androgen dependent prostate cancer cell lines require BER for optimal viability and proliferation. Therefore, BER inhibitors should be investigated further as a novel prostate cancer therapy.
Highlights.
A fluorescent high throughput assay to assess Base Excision Repair capacity was developed.
Compounds approved by U.S. Food and Drug Administration for human use contain long-patch Base Excision Repair (BER) inhibitors.
Prostate cancer cell lines are more sensitive to the BER inhibitors than nonmalignant prostate-derived epithelial cells.
Androgen deprivation and BER inhibition synergistically inhibit prostate cancer cell growth.
Acknowledgments
Funding
This research is supported the following Funds at the Community Foundation of Broward: Mary N. Porter Cancer Research Fund, Harold D. Franks Cancer Fund, Gene and Collette Herman Family Fund, Bank of American Unrestricted Fund. This research is also partially supported by NIH grant ES023569 (YLiu), and National Science Foundation of China grant No. 81773380 (ZZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Compliance with Ethical Standards
All studies in this manuscript were performed in accordance with the NIH Guiding Principles for Ethical Research.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Armenia J, et al. , The long tail of oncogenic drivers in prostate cancer. Nat Genet, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research, N., The Molecular Taxonomy of Primary Prostate Cancer. Cell, 2015. 163(4): p. 1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans JR, et al. , Patient-Level DNA Damage and Repair Pathway Profiles and Prognosis After Prostatectomy for High-Risk Prostate Cancer. JAMA Oncol, 2016. 2(4): p. 471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomlins SA, et al. , Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science, 2005. 310(5748): p. 644–8. [DOI] [PubMed] [Google Scholar]
- 5.Brenner JC, et al. , Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell, 2011. 19(5): p. 664–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demichelis F, et al. , TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene, 2007. 26(31): p. 4596–9. [DOI] [PubMed] [Google Scholar]
- 7.Dantzer F, et al. , Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry, 2000. 39(25): p. 7559–69. [DOI] [PubMed] [Google Scholar]
- 8.Mateo J, et al. , DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med, 2015. 373(18): p. 1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramakrishnan Geethakumari P, et al. , PARP Inhibitors in Prostate Cancer. Curr Treat Options Oncol, 2017. 18(6): p. 37. [DOI] [PubMed] [Google Scholar]
- 10.Athie A, et al. , Targeting DNA Repair Defects for Precision Medicine in Prostate Cancer. Curr Oncol Rep, 2019. 21(5): p. 42. [DOI] [PubMed] [Google Scholar]
- 11.Leone V, et al. , Ccdc6 knock-in mice develop thyroid hyperplasia associated to an enhanced CREB1 activity. Oncotarget, 2015. 6(17): p. 15628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morra F, et al. , The combined effect of USP7 inhibitors and PARP inhibitors in hormone-sensitive and castration-resistant prostate cancer cells. Oncotarget, 2017. 8(19): p. 31815–31829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace SS, Base excision repair: a critical player in many games. DNA Repair (Amst), 2014. 19: p. 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton JK, et al. , XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res, 2008. 18(1): p. 48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sukhanova MV, et al. , Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Res, 2005. 33(4): p. 1222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto Y and Kim K, Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science, 1995. 269(5224): p. 699–702. [DOI] [PubMed] [Google Scholar]
- 17.Prasad R, et al. , Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem, 1998. 273(24): p. 15263–70. [DOI] [PubMed] [Google Scholar]
- 18.Beard WA and Wilson SH, Structure and mechanism of DNA polymerase Beta. Chem Rev, 2006. 106(2): p. 361–82. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y and Wilson SH, DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem Sci, 2012. 37(4): p. 162–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, et al. , Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem, 1995. 270(38): p. 22109–12. [DOI] [PubMed] [Google Scholar]
- 21.Prasad R, et al. , DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J Biol Chem, 2001. 276(35): p. 32411–4. [DOI] [PubMed] [Google Scholar]
- 22.Hu LY, et al. , SUMOylation of XRCC1 activated by poly (ADP-ribosyl)ation regulates DNA repair. Hum Mol Genet, 2018. 27(13): p. 2306–2317. [DOI] [PubMed] [Google Scholar]
- 23.Caldecott KW, XRCC1 and DNA strand break repair. DNA Repair (Amst), 2003. 2(9): p. 955–69. [DOI] [PubMed] [Google Scholar]
- 24.Liu YL, Lei Y, R, High throughput measurement of DNA base lesion repair capacity, U. States, Editor. 2016, Florida International University: United States. [Google Scholar]
- 25.Ghannoum MA and Rice LB, Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev, 1999. 12(4): p. 501–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards DM and Brogden RN, Ceftazidime. A review of its antibacterial activity, pharmacokinetic properties and therapeutic use. Drugs, 1985. 29(2): p. 105–61. [DOI] [PubMed] [Google Scholar]
- 27.Angiolillo DJ, et al. , Prasugrel: a novel platelet ADP P2Y12 receptor antagonist. A review on its mechanism of action and clinical development. Expert Opin Pharmacother, 2008. 9(16): p. 2893–900. [DOI] [PubMed] [Google Scholar]
- 28.Binderup L and Bramm E, Effects of a novel vitamin D analogue MC903 on cell proliferation and differentiation in vitro and on calcium metabolism in vivo. Biochem Pharmacol, 1988. 37(5): p. 889–95. [DOI] [PubMed] [Google Scholar]
- 29.YASUNORI[JP], K.H.J.A.F.J.S.A.J.K.T.J.I.T.J.N.S.J.T., Tetrahydrothienopyridine derivatives, furo and pyrrolo analogs thereof and their preparation and uses for inhibiting blood platelet aggregation U. States, Editor. 1994, Ube Industries Limited, Ube; Sankyo Company, Limited,. p. 43. [Google Scholar]
- 30.Rencuzogullari E, et al. , Effects of natamycin on sister chromatid exchanges, chromosome aberrations and micronucleus in human lymphocytes. Drug Chem Toxicol, 2009. 32(1): p. 47–52. [DOI] [PubMed] [Google Scholar]
- 31.Sun D, et al. , Elevated expression of DNA ligase I in human cancers. Clin Cancer Res, 2001. 7(12): p. 4143–8. [PubMed] [Google Scholar]
- 32.Bergoglio V, et al. , Enhanced expression and activity of DNA polymerase beta in human ovarian tumor cells: impact on sensitivity towards antitumor agents. Oncogene, 2001. 20(43): p. 6181–7. [DOI] [PubMed] [Google Scholar]
- 33.Lange SS, Takata K, and Wood RD, DNA polymerases and cancer. Nat Rev Cancer, 2011. 11(2): p. 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starcevic D, Dalal S, and Sweasy JB, Is there a link between DNA polymerase beta and cancer? Cell Cycle, 2004. 3(8): p. 998–1001. [PubMed] [Google Scholar]
- 35.Chan K, et al. , Overexpression of DNA polymerase beta results in an increased rate of frameshift mutations during base excision repair. Mutagenesis, 2007. 22(3): p. 183–8. [DOI] [PubMed] [Google Scholar]
- 36.Albertella MR, Lau A, and O’Connor MJ, The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst), 2005. 4(5): p. 583–93. [DOI] [PubMed] [Google Scholar]
- 37.Wilson SH, et al. , Base excision repair and design of small molecule inhibitors of human DNA polymerase beta. Cell Mol Life Sci, 2010. 67(21): p. 3633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]