Abstract
Bacteria exhibit a myriad of different morphologies, through the synthesis and modification of their essential peptidoglycan (PG) cell wall. Our discovery of a fluorescent D-amino acid (FDAA)-based PG labeling approach provided a powerful method for observing how these morphological changes occur. Given that PG is unique to bacterial cells and a common target for antibiotics, understanding the precise mechanism(s) for incorporation of (F)DAA-based probes is a crucial determinant in understanding the role of PG synthesis in bacterial cell biology and could provide a valuable tool in the development of new antimicrobials to treat drug-resistant antibacterial infections. Here, we systematically investigate the mechanisms of FDAA probe incorporation into PG using two model organisms Escherichia coli (Gram-negative) and Bacillus subtilis (Gram-positive). Our in vitro and in vivo data unequivocally demonstrate that these bacteria incorporate FDAAs using two extracytoplasmic pathways: through activity of their D,D-transpeptidases, and, if present, by their L,D-transpeptidases and not via cytoplasmic incorporation into a D-Ala-D-Ala dipeptide precursor. Our data also revealed the unprecedented finding that the DAA-drug, D-cycloserine, can be incorporated into peptide stems by each of these transpeptidases, in addition to its known inhibitory activity against D-alanine racemase and D-Ala-D-Ala ligase. These mechanistic findings enabled development of a new, FDAA-based, in vitro labeling approach that reports on subcellular distribution of muropeptides, an especially important attribute to enable the study of bacteria with poorly defined growth modes. An improved understanding of the incorporation mechanisms utilized by DAA-based probes is essential when interpreting results from high resolution experiments and highlights the antimicrobial potential of synthetic DAAs.
Introduction
Of the known bacterial species, nearly all possess a peptidoglycan (PG) cell wall that surrounds the cell. PG is an essential cellular component that maintains the size and shape of the bacterial cell and helps protect the cell from its environment. Given its essential roles in cell growth and division, its relative accessibility when compared to potential cytoplasmic targets, and that it is unique to bacterial cells, some of the most successful antibiotics target PG biosynthesis.1
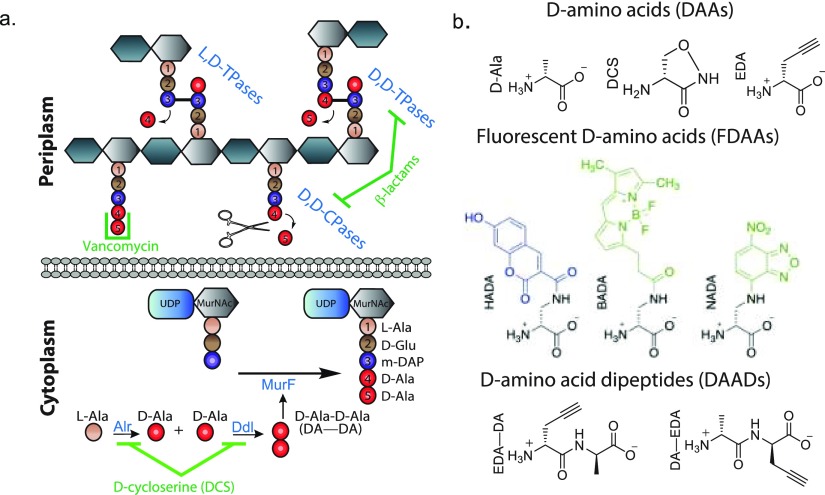
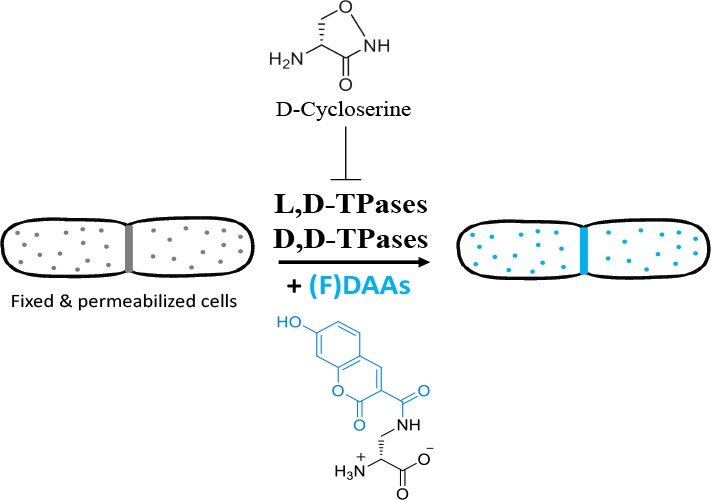
The PG cell wall is a macromolecule consisting of glycan strands cross-linked by short D-amino acid (DAA)-containing peptides. The PG biosynthetic pathway is promiscuous with respect to the assembly and/or utilization of the D-Ala-D-Ala (DA—DA) subunit at the terminus of the pentapeptide stem,2,3 and these steps are targets of many clinically important anti-PG drugs such as D-cycloserine (DCS), the β-lactams, and vancomycin4 (Figure 1a). Interestingly, DCS, a second-line drug currently used to treat multidrug-resistant strains of M. tuberculosis,4 and many β-lactams, are DAAs, while their mirror image counterparts (i.e., the L-enantiomers) show remarkably diminished antibacterial activities.5−7
Figure 1.
PG biosynthetic pathways are promiscuous and accept D-amino acid (DAA) based probes. (a) Simplified cartoon representation of the PG synthesis and modification pathways relevant to this work, along with representative PG synthesis inhibitors. (b) Representative DAA-based probes utilized in the experiments described in this manuscript.
The DA–DA subunit is assembled in the cytoplasm by D-Ala-D-Ala ligase (Ddl) and is appended to UDP-MurNAc-tripeptide by MurF to provide UDP-MurNAc pentapeptide (Figure 1a). After additional downstream processing into a C55-lipid-linked disaccharide pentapeptide (lipid II), this monomeric PG subunit is translocated across the cytoplasmic membrane and incorporated into elongating glycan strands in the periplasm. Subsequently, the DA–DA subunit of the stem peptide is a substrate for cross-linking reactions that are essential for providing the PG cell wall with the requisite load-bearing properties against the cell’s turgor pressure. These cross-links are installed by two distinct pathways: (1) the essential and ubiquitous D,D-transpeptidases (D,D-TPases or penicillin binding proteins, PBPs) that catalyze installation of 4–3 cross-links, or (2) the normally dispensable and more infrequent, L,D-transpeptidases (L,D-TPases) that catalyze installation of 3–3 cross-links (Figure 1a).8
Elucidating the molecular mechanisms regulating PG biosynthesis will provide valuable insights that can be applied to combat the alarming increase of multidrug resistance.9 Toward this goal, we and others have recently developed new classes of synthetic D-amino acid (DAA) based probes to visualize PG assembly and dynamics in virtually all PG-containing bacterial species.10−18 These probes, fluorescent D-amino acids (FDAAs), clickable DAAs, and D-amino acid dipeptides (DAADs), have advanced our fundamental knowledge of bacterial growth and division in diverse bacteria.10,14−53 In order to make maximal use of the data obtained from labeling experiments utilizing these probes, a detailed understanding of probe incorporation mechanisms is required.
Here we have systematically characterized the incorporation mechanisms of common DAA-based probes in two model organisms: Gram-positive B. subtilis and Gram-negative E. coli. We leveraged chemical genetics, genetics, microscopy, and in vitro approaches in combination with labeling experiments that utilized: EDA, a small clickable DAA; HADA, an FDAA; along with EDA–DA and DA–EDA, two clickable small DAADs (Figure 1b). Our data suggest that DAADs are incorporated using the cytoplasmic (lipid II) pathway, while single DAA-based probes (e.g., EDA, HADA) are incorporated outside the cytoplasm by the D,D-TPases, and, if present, the L,D-TPases. Our chemical genetics experiments revealed a previously unknown mode of action for DCS; specifically, that it is a substrate for both D,D-TPases and L,D-TPases and that it is readily incorporated into pentapeptide and tetrapeptide stems, respectively (Figure 1b), perhaps providing an important insight for further investigation of novel D-amino acid-based antibacterial agents. Finally, our improved mechanistic understanding helped us design a new, FDAA-based labeling approach that reports on subcellular distribution of muropeptides in bacterial cells.
Results
Chemical Genetics Approach Reveals Transpeptidase-Mediated Incorporation of DAAs in Bacillus subtilis
In order to probe the mechanism of FDAA incorporation in vivo, we first attempted a chemical genetics approach. Since bacterial species usually have multiple L,D-TPases and/or commonly essential D,D-TPases, deleting them all is not feasible. We first focused on vegetative B. subtilis cells that reportedly lacked 3,3-cross-links typical of L,D-TPase activity.54 This reduced the potential modes of FDAA installation to incorporation by the D,D-TPases or to cytoplasmic incorporation via the lipid II pathway. In order to verify the lack of L,D-TPase mediated incorporation, we prelabeled vegetative B. subtilis cells with HADA. This was followed by ethanol-fixing and permeabilization of the cells followed by treatment with an E. coliD,D-carboxypeptidase/endopeptidase (PBP4Ec, the product of the dacB gene), in vitro. While PBP4Ec did not remove HADA from tetrapeptides in E. coli cells (Figure SI1a), it completely removed the signal from vegetative ethanol-fixed pentapeptide-rich B. subtilis ΔdacA cells prelabeled with HADA (Figure SI1b). These results are in agreement with previous observations that vegetative B. subtilis cells incorporate HADA exclusively into their pentapeptides.15
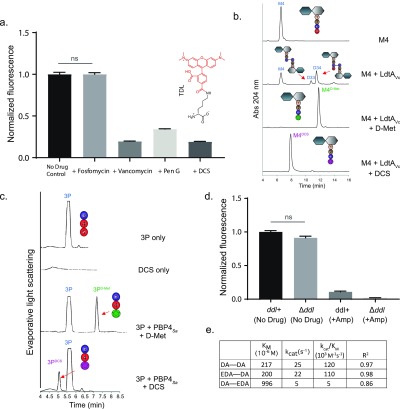
In the absence of L,D-TPase activity, chemical inhibition of extracellular D,D-TPases or inhibition of cytoplasmic D-Ala-D-Ala synthesis and incorporation could provide valuable information on the route of FDAA incorporation. We first confirmed that cells remained viable after such brief drug exposures (Figure SI1c), consistent with a recent report.55 Our initial experiments screening different FDAAs and drugs showed that both β-lactams and D-cycloserine (DCS) had the potential to significantly inhibit the incorporation of FDAAs in live B. subtilis in a drug- and FDAA-dependent manner (Figures 2a and SI1c). For example, the inhibition by DCS was particularly pronounced and comparable to penicillin G, a β-lactam, when the FDAA was TDL, a large red FDAA linked to D-lysine, (∼65–80% inhibition, Figure 2a). A similar effect was observed in E. coli wild-type cells. While D-cycloserine and ampicillin both inhibited HADA incorporation (by ∼30%), meropenem inhibited virtually all HADA incorporation (Figure SI1d).
Figure 2.
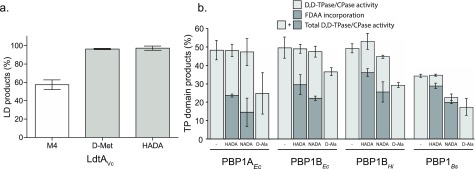
B. subtilis cells do not incorporate (F)DAAs cytoplasmically, but through reactions mediated by D,D-TPases, which can be inhibited by D-cycloserine. (a) A brief pretreatment of live B. subtilis cells with vancomycin, penicillin G (Pen G), and D-cycloserine (DCS), significantly inhibited FDAA incorporation. No inhibition of FDAA incorporation was obsereved with fosfomycin. (b) LdtAVc incorporated DCS into M4, comparable to other DAAs, e.g., D-Met, in vitro. (c) PBP4Sa incorporated DCS into a synthetic Nα, Nε-Diacetyl-L-Lys-D-Ala-D-Ala tripeptide (3P), in vitro. (d) Live B. subtilis wild-type and B. subtilis Δddl cells, grown in S750 minimal media supplemented with DA–DA, incorporated HADA comparably; HADA incorporation was significantly inhibited by ampicillin pretreatment. (e) DA–EDA was a poor substrate for MurFBs in vitro, but EDA–DA performed similarly well to the endogenous substrate, DA–DA. Column bar graphs represent mean relative signal ± SD quantified from at least N > 100 cells. Error bars are SEM.
Given the significant inhibition observed for both DCS and β-lactams—and the well-characterized inhibitory activity of the β-lactams toward the bacterial transpeptidases—we postulated that DCS, a cyclized D-serine analogue with a free amino group (Figure 1b), might behave as a competitive inhibitor of periplasmic L,D- and/or D,D-TPases. In support of this hypothesis, both LdtAVc, a representative L,D-TPase from Vibrio cholerae,56 and PBP4Sa, a soluble version of recombinant PBP4 from Staphylococcus aureus,17 incorporated DCS into appropriate soluble substrate analogues in vitro:13 LdtAVc incorporated DCS into tetrapeptides comparably to D-methionine, a DAA that is naturally produced by Vibrio cholerae cells2 (Figure 2b); the product of each reaction was confirmed by MS (Figure SI2a). PBP4Sa incorporated DCS into a synthetic Nα, Nε-diacetyl-L-Lys-D-Ala-D-Ala tripeptide (3P) albeit to a lower extent when compared to identical experiments utilizing D-methionine (Figure 2c). Moreover, excess DCS directly competed with incorporation of FDAAs into ethanol-fixed cells by LdtAVc in a FDAA-specific manner (Figure SI2b). These data suggest that, in addition to its known cytoplasmic targets (alanine racemase and D-Ala-D-Ala ligase),57 DCS, a clinically important antibiotic, has the previously uncharacterized potential to competitively inhibit L,D- and D,D-TPases, and therefore its utilization would not yield clear results probing (F)DAA incorporation mechanisms.
Vegetative B. subtilis Cells Incorporate FDAAs with D,D-TPases and Not Cytoplasmically
Given these difficulties, we turned to a conventional genetics approach for deletion of nonessential (or conditionally essential) pathways and interrogation of potential DAA incorporation mechanisms through comparison of FDAA accumulation between otherwise isogenic strains. Because vegetative B. subtilis cells lack L,D-TPase activity (Figure SI1b), knocking out its ability to form DA–DA would also eliminate its ability to incorporate DAAs cytoplasmically and leave D,D-transpeptidation as the only feasible route for DAA incorporation (Figure 1a). Deleting the single and essential DA–DA-ligase gene (ddl) in B. subtilis provided a strain that was auxotrophic for exogenously provided DA–DA. Under identical growth conditions, wild-type B. subtilis and B. subtilis Δddl cells accumulated a HADA signal to a comparable extent, while ampicillin treatment inhibited >80% of their initial signal (Figure 2d). This result strongly suggests that FDAAs are not incorporated cytoplasmically in B. subtilis.
Our in vitro findings with MurF from Bacillus subtilis (MurFBs), the next cytoplasmic enzyme downstream of Ddl, support the conclusion that FDAAs are not incorporated cytoplasmically in B. subtilis. These experiments revealed that MurFBs accepted EDA–DA, a clickable DAAD that eventually generates lipid II tagged in the fourth position of the pentapeptide stem,16 as well as its endogenous substrate, DA–DA both in terms of its affinity and turnover (Figure 2e). However, DA–EDA, a DAAD that generates a pentapeptide tagged in the fifth (i.e., terminal) position, is a significantly poorer (∼20-fold) substrate than DA–DA. This observation, along with the enzymatic activity of the endogenous carboxypeptidases (e.g., PBP5), may explain the differential DAAD labeling previously observed in live B. subtilis cells; specifically, EDA–DA labeled the PG of B. subtilis much more strongly than DA–EDA.16 The in vitro data also suggest that cytoplasmic DAA incorporation into the terminal position of the pentapeptide stem (e.g., with DA-EDA)—the pentapeptide terminus is the only site labeled by (F)DAAs in vegetative B. subtilis cells (Figure SI1b)—is particularly disfavored by MurF in B. subtilis (Figure 2e). Furthermore, excess DA–DA did not significantly compete with the FDAA label accumulation in live vegetative B. subtilis cells, but D-alanine and D-tyrosine, a DAA that is naturally produced by B. subtilis cells,2 (>50-fold decrease) did (Figure SI2c). Thus, we conclude that (F)DAAs are not incorporated cytoplasmically in vegetative B. subtilis cells. By the process of genetic elimination and in combination with the Supporting Information, our results strongly suggest that, in vegetative B. subtilis cells, FDAAs are incorporated predominantly (if not solely) through the activity of the periplasmic D,D-transpeptidases.
E. coli Cells Incorporate FDAAs by L,D-TPases in a Growth Independent Manner
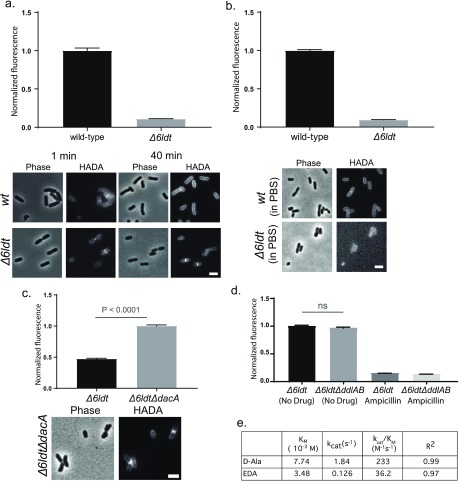
In contrast to B. subtilis, E. coli is rich in L,D-TPase activity.58 We recently reported on a mutant E. coli strain in which all the predicted L,D-TPases were deleted.36 Cells from this strain, E. coli BW255113Δ6LDT (hereafter referred to as Δ6LDT), revealed a ∼10-fold lower HADA signal compared to wild-type E. coli (Figure 3a). Among the six single L,D-TPase knockout strains, ΔldtD cells showed the least HADA incorporation (∼50%, Figure SI3a). Consistently, double, triple, and quadruple L,D-TPase mutants showed a significant decrease in label incorporation compared to wild-type as long as ldtD is one of the deleted genes (Figure SI3b). These results suggest that the major mode(s) of FDAA incorporation in actively growing E. coli is due to the L,D-TPases, primarily LdtD.41
Figure 3.
E. coli cells incorporate (F)DAAs through reactions mediated by L,D- and D,D-TPases, and not cytoplasmically. (a) HADA incorporation in E. coli BW25113Δ6LDT cells (Δ6LDT) was ∼10-fold lower than that observed in E. coli wild-type cells. Unlike wild-type cells, which reveal extensive accumulation of FDAA signal around the entire cell, FDAA labeling in the Δ6LDT strain was predominantly limited to the sites of new growth. (b) Live E. coli wild-type cells, but not Δ6LDT cells, incorporated FDAAs in a growth-independent manner. (c) Live E. coli Δ6LDTΔdacA cells accumulated significantly more HADA signal compared to Δ6LDT. (d) Live Δ6LDT and E. coli Δ6LDTΔddlAB cells, grown in M9 minimal media supplemented with DA–DA, incorporated HADA comparably; HADA incorporation was inhibited by ampicillin pretreatment. (e) Compared to D-Ala, EDA was a poor substrate for DdlBE. coliin vitro. Micrographs are adjusted for qualitative comparison only. Values in column bar graphs represent mean relative signal quantified from at least N > 100 cells. Scale bars, 2 μm.
Early labeling experiments revealed that wild-type E. coli cells begin to accumulate FDAA signal at sites of new growth (i.e., septum and side walls) eventually plateauing into uniformly labeled cells over 1–2 generations.15 In contrast, E. coli Δ6LDT cells accumulate FDAA signal to a lower extent and preserve the signal persistently at sites of new growth (Figure 3a). This uniform and peripheral accumulation of FDAA signal in wild-type E. coli cells suggested a growth-independent, L,D-Tpase-facilitated DAA incorporation.
Consistently, nutrient starved cells, which are kept in phosphate buffered saline (PBS) for 1 h at RT, of wild-type E. coli incorporated HADA strongly and uniformly, whereas E. coli Δ6LDT cells did not show any significant signal accumulation above background (Figure 3b). A complementation assay with nutrient starved E. coli cells suggested that this growth independent FDAA incorporation in buffer was predominantly due to overexpression of L,D-TPase genes known to generate 3–3 cross-links, ldtD and ldtE,58 and less so due to L,D-TPases that anchor Braun’s lipoprotein (ldtA, ldtB, or ldtC) or yafK (Figure SI3c). This property was not specific to E. coli. Diverse bacteria rich in predicted L,D-TPases, such as the deltaproteobacterium Bdellovibrio bacteriovorus(36) and the alphaproteobacterium Agrobacterium tumefaciens,27 also incorporated HADA in a growth-independent manner (Figure SI 3d).
Collectively, these results suggested that L,D-TPase mediated DAA incorporation in E. coli can occur independently of cell growth.59 On the other hand, the persistent signal observed in the absence of L,D-TPase activity (Figure 3a) points to the presence of an additional mode for DAA incorporation in E. coli.
D,D-TPases Incorporate DAAs into the Pentapeptides of PG in E. coli Cells
Pentapeptides are rare in E. coli PG.60 We hypothesized that the relatively faint FDAA signal localized at sites of new growth in cells lacking L,D-TPases represented probe incorporation into pentapeptides. If so, deleting the gene for one of the major E. coliD,D-CPases (dacA) in this background should increase the FDAA labeled pentapeptide pools in E. coli PG. Indeed, E. coli Δ6LDTΔdacA cells showed 2-fold greater incorporation of HADA signal than E. coli Δ6LDT (Figure 3c). We observed that HADA accumulation in E. coli Δ6LDTΔdacA cells could be eliminated by treatment with a carboxy/endopeptidase (PBP4Ec) in vitro (Figure SI4a). The labeling trends observed with these mutants and EDA, a small clickable DAA, were comparable (Figure SI4b). These results reveal previously underappreciated DAA incorporation into pentapeptides in E. coli.
This pentapeptide labeling, could be achieved via the cytoplasmic route (through Ddl/MurF), an exchange reaction mediated by the D,D-transpeptidases (i.e., the PBPs), or a combination of the two. Therefore, we deleted ddlA and ddlB on top of 6 L,D-TPases generating an E. coli Δ6LDTΔddlAB strain that is auxotrophic for DA–DA. This mutant strain lacks both L,D-TPase activity and the ability to incorporate DAAs cytoplasmically. Similar to the aforementioned case in B. subtilis, E. coli Δ6LDTΔddlAB cells accumulated HADA (Figure 3d) or EDA (Figure SI4c) signals comparably to identically treated E. coli Δ6LDT cells. Ampicillin treatment significantly inhibited HADA accumulation in both strains (Figure 3d). Moreover, DCS significantly inhibited (F)DAA signal incorporation even in the absence of Ddl, and in the presence of excess DA–DA, in both E. coli and B. subtilis (Figure SI4c-d), providing further support for the notion that DCS may also function as a DAA-like competitive inhibitor of D,D-TPases in both organisms. Collectively, these results suggest that the D,D-TPases, and not the cytoplasmic enzymes, are primarily responsible for incorporation of DAAs into pentapeptides in E. coli and in B. subtilis.
Ddl Disfavors the Cytoplasmic Incorporation of DAAs
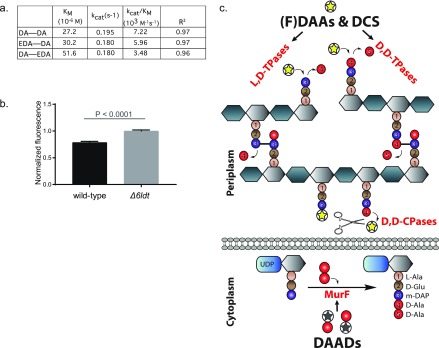
For cytoplasmic incorporation, following cellular import, a (F)DAA needs to be tolerated by Ddl and incorporated into a corresponding DAAD. To test our conclusion that (F)DAAs are not incorporated cytoplasmically, we assessed the ability of E. coli DdlB (DdlBEc) to ligate D-Ala to EDA, the small clickable DAA, forming DA–EDA, in vitro. Under previously published conditions, DdlBEc showed 2-fold higher affinity for EDA than for D-Ala; however, the apparent rate of the reaction with EDA was 10-fold slower than the reaction with D-Ala (Figure 3e). This suggests that cytoplasmic (F)DAA incorporation may be primarily rejected by Ddl, especially since the cytoplasmic steps lying downstream of Ddl seem to be significantly more tolerant.16 Indeed, MurF from Pseudomonas aeruginosa (MurFPa, an enzyme that shows high sequence and kinetic similarity to E. coli MurF61) incorporated both of the clickable DAADs, EDA–DA and DA–EDA, with comparable rates and affinities to the native substrate, DA–DA (Figure 4a). This tolerance of MurF (and presumably the downstream steps) could explain how small DAADs could be used as common and versatile metabolic PG labeling alternatives to FDAAs and why they can rescue DA–DA deficiencies in E. coli, B. subtilis, and Chlamydiae.16,37
Figure 4.
FDAAs are incorporated by periplasmic transpeptidases and DAADs cytoplasmically. (a) MurFPa incorporated DAADs as well as the native substrate (DA–DA), in vitro. (b) Live Δ6LDT cells incorporated a greater EDA—DA (1 mM for 1 h) signal than E. coli wild-type cells. Values in column bar graphs represent the mean relative signal quantified from at least N > 150 cells. (c) Cartoon representation depicting that in E. coli DAAs, including FDAAs and DCS, are substrates for periplasmic L,D-TPases, D,D-TPases, or D,D-CPases and that DAADs are substrates for cytoplasmic MurF.
Lastly, we wanted to test if E. coliL,D-TPases play a role in DAAD incorporation. Cells from E. coli Δ6LDT grown for 1–2 generations in the presence of EDA–DA incorporated approximately 20% more DAAD signal compared to wild-type E. coli cells (Figure 4b). These data provide strong evidence that L,D-TPases do not play a significant role in DAAD incorporation.
From our combined results, we conclude that the incorporation of FDAA and DAAD probes can be used to distinguish between two distinct biosynthetic pathways. FDAA probe incorporation reports on the activity of the extracytoplasmic L,D- and D,D-TPases, whereas DAAD probe incorporation reports on the cytoplasmic (lipid II) pathway, i.e., nascent PG synthesis (Figure 4c).
FDAAs Report on L,D- and D,D-TPase Activity and Muropeptide Distribution in Fixed Cells
Our results and previous in vitro work with D-amino acids (DAAs) revealed that both L,D-TPases and D,D-TPases of different bacterial origins can utilize DAAs as acyl-acceptor substrates in the presence of appropriate acyl-donors.17,56,62 Consistently, representative enzymes with known L,D-TPase or D,D-TPase activities could utilize larger fluorescent DAAs (FDAAs) as acyl-acceptor substrates in vitro. A representative L,D-TPase from Vibrio cholerae (LdtAVc) incorporated HADA into a soluble disaccharide tetrapeptide (Figure 5a) while four recombinant high molecular weight PBPs (E. coli, PBP1AEc and PBP1BEc; Haemophilus influenzae, PBP1AHi; and B. subtilis, PBP1Bs) facilitated incorporation of FDAAs during the synthesis of nascent PG from a lipid II precursor (Figure 5b). The FDAA incorporation activities were comparable to naturally produced D-methionine2 (Figure 5a) or were at the expense of their native cross-linking/carboxypeptidase activities (Figure 5b). This suggested that FDAAs did not significantly interfere with the native functions of these enzymes and could serve as proxy reporters for the DAA incorporation activities of L,D-TPases or for cross-linking/carboxypeptidase activities of D,D-TPases.
Figure 5.
FDAAs are efficiently incorporated into PG precursors. (a) LdtAVc incorporated HADA into the soluble muropeptide disaccharide tetrapeptide (M4) comparable to other DAAs, e.g., D-Met, in vitro. (b) Different high-molecular-weight D,D-TPases from diverse bacteria incorporated HADA and NADA during the in vitro synthesis of nascent PG from lipid-II without significantly changing their total D,D-TPase and dD,D-CPase product distribution. The values are the mean ± SD of three independent experiments.
Next, we tested whether these recombinant transpeptidases would incorporate FDAAs into isolated sacculi in vitro. Early attempts with isolated V. cholerae sacculi indicated high background/nonspecific labeling (Figure SI5a). This led us to utilize ethanol-fixed and permeabilized intact cells as transpeptidase substrates, a simple substrate preparation approach that we have previously employed.16 Treatment of bacterial cells with ice-cold ethanol inactivates virtually all cellular activity and permeabilizes membranes for downstream applications while keeping the cells intact and well separated for microscopy experiments.16 Indeed, recombinant LdtAV. cholerae incorporated HADA into ethanol-fixed V. cholerae cells in vitro (Figure 6a). Label incorporation was specific for PG (Figure SI5b); parallel experiments with the L-isomer, HALA, resulted in a 5-fold decrease in fluorescence incorporation (Figure SI5c). We similarly observed a minor, and V. cholerae-specific, HALA incorporation in live V. cholerae cells, but not live E. coli cells, albeit approximately 20-fold less efficiently than HADA (Figure SI 5d). LdtAVc incorporated HADA into ethanol-fixed cells from a variety of Gram-negative species (Figure SI6a). Similarly, a soluble version of recombinant PBP4 from S. aureus (PBP4Sa)17 incorporated HADA into ethanol-fixed Staphylococcus aureus cells (Figure 6b) and ethanol-fixed E. coli cells (Figure SI6b). Recombinant PBP4Sa also incorporated HALA into fixed E. coli cells, although it was incorporated approximately 20-fold less efficiently than HADA (Figure SI6b).
Figure 6.
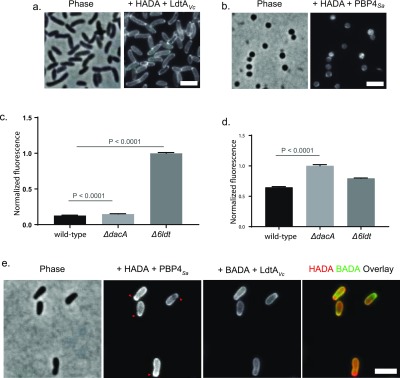
FDAAs report on the abundance and subcellular distribution of muropeptides in ethanol fixed and permeabilized bacterial cells in vitro. (a) Ethanol fixed V. cholerae cells are substrates for LdtAVc and FDAAs, e.g., HADA, in vitro. (b) Ethanol fixed S. aureus cells are substrates for PBP4Sa and FDAAs in vitro. (c–d) Ethanol fixed E. coli cells are substrates for LdtAVc or PBP4Sa and FDAAs in vitro. (c) FDAAs and LdtAVc can report on PG tetrapeptide abundance of the E. coli Δ6LDT and ΔdacA strains in vitro. (d) FDAAs and PBP4Sa can report on PG pentapeptide abundance of the E. coli Δ6LDT and ΔdacA strains in vitro. (e) Sequential PBP4Sa (with HADA) and LdtAVc (with BADA) labeling can report on differential subcellular muropeptide distribution of a strain, e.g., in ethanol fixed A. tumefaciens cells, red arrows. Column bar graphs represent mean relative signal quantified from at least N > 100 cells. Error bars are SEM. Scale bars, 2 μm.
The PG of E. coli is rich in tetrapeptides,58 but the abundance of pentapeptides increases in strains lacking D,D-carboxypeptidases60 (e.g., PBP5 encoded by dacA). Consistently, LdtAVc incorporated HADA into ethanol-fixed E. coli ΔdacA cells comparably to wild-type cells (Figure 6c), suggesting that the carboxypeptidase activity of DacA may not significantly impact overall tetrapeptide abundance. However, a 7-fold increase in HADA incorporation was observed using ethanol-fixed cells from an E. coli strain that lacked all of the six known or predicted genes encoding for L,D-TPases, Δ6LDT (Figure 6c). The enhanced HADA incorporation in the Δ6LDT strain relative to the wild-type strain suggests that LdtAVc, combined with the FDAAs, can report on the relative abundance of tetrapeptides in PG.
On the other hand, PBP4Sa incorporated HADA into fixed cells from an E. coli strain that lacked a major D,D-CPase, BW25133ΔdacA, significantly more than the wild-type, strongly suggesting that pentapeptides are the preferred substrate for PBP4Sa-mediated HADA incorporation (Figure 6d). Sequential enzymatic labeling of ethanol-fixed cells with differently colored FDAAs was possible. Treatment of ethanol-fixed Δ6LDTΔdacA cells with HADA and PBP4Sa, followed by BADA, a green, BODIPY-FL containing FDAA that is spectrally distinguishable from HADA, and LdtAVc resulted in superimposable labeling patterns (Figure SI6c). The same experimental approach with polarly growing Agrobacterium tumefaciens cells showed complementary labeling patterns particularly in young cells; a HADA signal (reporting on pentapeptides) appeared enriched at the very tip of the growing pole, followed by a band of BADA signal (reporting on tetrapeptides, Figure 6e). These results show that L,D- and D,D-TPases can incorporate FDAAs into ethanol fixed and permeabilized cells from diverse species in vitro while reporting on subcellular tetrapeptide/pentapeptide distribution of a strain. Moreover, these in vitro results support the direct role of these periplasmic L,D- and D,D-TPases in incorporating (F)DAAs in different bacteria.
Discussion
Bacteria have the evolutionarily preserved ability to produce DAAs, and their PG biosynthetic pathways are uniquely promiscuous for utilization of DAAs that provide them with a survival advantage in the wild.36,63 The design of multiple classes of DAA-based PG labeling probes that take advantage of this promiscuity are now widely used.10,14−18,29 While dissecting the routes for incorporation (and for turnover) of these PG probes has been challenging, without this knowledge, the interpretation of the data generated with these probes in a given species is limited. Here, we provide a roadmap to explore the mechanisms by which DAA-based probes are incorporated into the PG by combining genetics, chemical genetics, and in vitro approaches for two model organisms, E. coli and B. subtilis. We provide strong evidence that (F)DAAs, including the DAA drug DCS, are incorporated by extracytoplasmic (Gram-positive) or periplasmic (Gram-negative) L,D-TPases and D,D-TPases in contrast to DAADs, which are cytoplasmically incorporated (Figure 4c).
Prior to our discovery of the FDAAs, previous work with DAAs2,17,29,56,62 (e.g., D-methionine) and various DAADs,3,16,37,49,64 showed that incorporation of any DAA is tolerated by these D-Ala assembly/utilization steps that are common in PG containing bacteria. Previous work strongly suggested DAA probe incorporation was mediated by the bacterial transpeptidases; however, definitive experiments to rule out cytoplasmic incorporation had yet to be reported. The ability to distinguish these pathways has important ramifications with respect to interpretation of results from FDAA labeling experiments. For example, cytoplasmic incorporation of a DAA probe would report on nascent PG synthesis; the probe would be incorporated into the lipid II intermediate that would subsequently be detected upon polymerization into glycan strands. Alternatively, periplasmic incorporation would report on the localized activity of transpeptidases on the bacterial cell surface (Figure 1a). Therefore, any cytoplasmic/periplasmic cross-reactivity of a probe would significantly complicate either of these interpretations. The mechanistic picture is further complicated in experiments that are carried out in the presence of D-cycloserine (DCS). Since DCS is a well-established inhibitor of alanine racemase and D-Ala-D-Ala ligase (Ddl), enzymes that are critical for the cytoplasmic assembly of D-Ala-D-Ala, it is logical to conclude that diminished DAA probe incorporation in the presence of DCS is indicative of a cytoplasmic pathway for probe incorporation. However, our data have revealed that DCS has a previously unknown mode of action that also directly impacts TPase-mediated probe incorporation.
The mechanism question is convoluted partly because of the essentiality and/or redundancy of enzymes for a particular route of (F)DAA incorporation. For example, E. coli has 2 Ddls, 6 predicted L,D-TPases, and 12 D,D-TPases/CPases (i.e., penicillin-binding proteins, PBPs). Understandably, this creates an opportunity to utilize a chemical genetics approach where all enzymes in a pathway are inhibited simultaneously, as we and others have done previously.23,24,35,37,55,56 However, here we show that implementation of a chemical genetics approach, based on inhibition of probe incorporation by anti-PG antibiotics, was insufficient for multiple reasons. Even when the toxicity of these agents is controlled for, it is difficult to conclusively rule out indirect effects. For example, vancomycin inhibits FDAA incorporation in B. subtilis. However, this effect could arise directly, through inhibition of D,D-TPase-dependent FDAA incorporation, or indirectly, through inhibition of nascent PG synthesis that results in depletion of the substrate utilized in D,D-TPase-mediated FDAA incorporation. Interpretation of probe incorporation data obtained with antibiotics was further complicated by cross-reactivities of drugs like meropenem that inhibit FDAA incorporation by both the L,D-TPases and the D,D-TPases or, as demonstrated here, by DCS that inhibits each of the three possible routes for probe incorporation.
Since DCS is a well-established inhibitor of alanine racemase and D-Ala-D-Ala ligase (Ddl), enzymes that are critical for the cytoplasmic assembly of D-Ala-D-Ala, it was initially thought that diminished DAA probe incorporation in the presence of DCS was indicative of a cytoplasmic pathway for probe incorporation. However, our data have revealed that DCS can directly compete with transpeptidase-mediated (F)DAA incorporation. This unexpected behavior is likely because DCS is also a DAA and may point to a heretofore unidentified mode of action of DCS on TPase-mediated reactions. Indeed, a D,D-CPase mutation has been shown to confer resistance to DCS in Mycobacterium.65 Although this multiple target reactivity makes DCS a problematic reagent for investigation of FDAA incorporation mechanisms, such reactivity is highly desirable property for development of a potent drug.
The (F)DAA incorporation activities are driven by the exchange of the terminal D-Ala residues in tetrapeptides and pentapeptides catalyzed by L,D- and D,D-TPases (Figure 1a), respectively. We show that this differential activity can be utilized to probe the subcellular distribution of tetrapeptides and pentapeptides by the sequential incubation of recombinant L,D- and D,D-TPases and differently colored FDAAs with ethanol-fixed bacterial cells, in vitro. This sample preparation approach takes less than an hour while keeping the cells intact. Since the PG that is being labeled by this approach is made naturally by the strain of interest (i.e., it is label-free), this method also offers access to a native-like substrate scaffold rich in information regarding muropeptide spatial distribution. The visualization of FDAA labeling then decodes this distribution, the composition of which may vary with cells from different mutant strains. This method will be particularly useful for investigating the roles of relative distribution of PG tetrapeptides and pentapeptides in bacteria with poorly understood growth modes (e.g., polar growth).20 Indeed, the dual color labeling in ethanol-fixed Agrobacterium tumefaciens (Figure 6e) suggests that the polar growth of this bacterium might necessitate pentapeptide incorporation at the very tip of the growing cell.
Previous muropeptide analyses suggested that E. coli incorporate (F)DAAs predominantly into tetrapeptides.15 This label incorporation into tetrapeptides is L,D-TPase dependent; E. coli cells that lack all six L,D-TPases accumulate an order of magnitude less (F)DAA signal compared to the parent. Surprisingly, we observed that evolutionarily diverse and L,D-TPase-rich Gram negative-bacteria including A. tumefaciens (14 homologues), E. coli (six homologues), or B. bacteriovorus (19 homologues36) incorporate FDAAs under nutrient-starved conditions in a growth-independent manner. In E. coli, wild-type cells strongly incorporated FDAAs into PG under nutrient-starved conditions, but an isogenic L,D-TPase-null strain did not. This explains the loss of observable growth patterns during prolonged FDAA pulses in E. coli.15 Therefore, we caution that such growth-independent (F)DAA incorporation by L,D-TPases (or D,D-TPases39) can obscure the growth-specific FDAA labeling, especially in extended labeling experiments.
The signal in the L,D-TPase deficient strains represents exclusive pentapeptide labeling and a steady-state between (F)DAA incorporation and its removal by D,D-CPases (e.g.; PBP4Ec). This pentapeptide labeling could occur either cytoplasmically, via the combined activities of Ddl and MurF, or through the activity of the D,D-TPases. Since deletion (or inhibition) of every available D,D-TPase in a given bacterial species, without killing the cell, is virtually impossible, we elucidated their role(s) in incorporating FDAAs into PG of live E. coli and B. subtilis cells through the process of elimination. Ddl knockout strains in B. subtilis and E. coli that also lacked L,D-TPase activity showed (F)DAA labeling comparable to the parent strains with native Ddl activity. The observed labeling, in the absence of Ddl and L,D-TPase activity, provides compelling evidence that D,D-TPases incorporate (F)DAAs into pentapeptides in the PG of these two species in vivo. Consistently, β-lactams dramatically reduce in vivo (F)DAA incorporation into pentapeptides in these species. Our in vitro data also suggest that bacteria may have an intrinsic ability to limit utilization of DAAs other than D-Ala by Ddl and MurF. For example, DdlBEc has a ∼10-fold lower specific activity toward EDA relative to D-Ala, suggesting that the cytoplasmic incorporation route may be limited at this point even if (F)DAAs are taken up by the cell. Similarly, MurFBs rejects DA–EDA, the terminally tagged DAAD.
The incorporation mechanism for DAADs appears to be simpler. We, and others, have previously shown that DAADs could complement growth in E. coli, B. subtilis, Chlamydia trachomatis, Mycobacterium smegmatis, and even chloroplasts of the moss, Physcomitrella patens, when the cytoplasmic DA–DA pools were depleted by DCS and/or by the generation of auxotrophic Ddl mutants.16,64,66 Our data here are in agreement with this finding: DAADs perform similarly to the endogenous substrate (DA–DA) for dipeptide incorporation in vitro.
Collectively, our results imply that (F)DAAs report on the activities of periplasmic D,D-TPases and L,D-TPases, which explains why (F)DAAs label diverse bacteria so effectively. In addition, (F)DAAs do not report on any enzyme activity until the periplasmic biosynthesis of nascent PG is complete (Figure 1a). DAADs, along with a recently reported metabolic glycolabeling strategy, are complementary to FDAAs in that they report on the biosynthesis of nascent PG.16,49,67
Finally, we note that many clinically relevant antibiotics are DAAs (e.g., DCS, the β-lactams). Our data reveal that DCS, an important antituberculosis antibiotic, has a previously unrecognized mode of action in addition to its main inhibitory effect on cytoplasmic stages of PG synthesis. Similar to other DAAs, we show that DCS can target diverse TPases in evolutionarily diverse bacteria, including E. coli. Like DCS, many β-lactams—widely used and long-standing anti-D,D-TPase drugs—have terminal DAA moieties that confer them improved antibacterial activities relative to their L-amino acid counterparts.40,49 For example, D-ampicillin (or ampicillin) is up to 5-fold more potent than L-ampicillin, and a synergy between free DAAs and β-lactams has been shown.5,7,68 It is tantalizing to think that this synergy points to a DAA-dependent D,D-TPase “activation” leading to an enhanced sensitivity toward natural electrophilic β-lactams. If true, and given that (F)DAAs can selectively target bacteria in live animals,33 this may provide a promising new avenue for DAA-based antibacterial discovery.
Acknowledgments
We thank K. Hummels and D. Kearns for the B. subtilis Δddl strain; J.-E. Hugonnet for many of the E. coli ΔLdt strains; A. Randich for her help with protein purification. We also thank C. Dowson and A. Lloyd for support of the kinetics studies involving MurF and DdlB. We also thank P. T. van Bentum for support in the preparation of lipid II derivatives.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.9b00664.
A complete set of protocols, experimental details, and supplementary figures (PDF)
Author Contributions
E.K., A.R., X.M., A.E., G.B., and M.S.V. carried out (F)DAA labeling experiments, including quantitation; E.K. performed microscopy experiments; E.K., A.D.D., and R. R. performed in vitro enzyme experiments with (F)DAAs and DAADs; L.A. and F.C. carried out analysis of products deriving from in vitro (F)DAA exchange; E.B. provided samples of labeled lipid II for in vitro (F)DAA exchange experiments; E.K., D.R., F.C., W.V., Y.V.V., and M.S.V wrote the manuscript. All authors were involved in the design and execution of this work.
This work was supported by Life Sciences Research Foundation Fellowship awarded to E.K.. Work in the Vollmer laboratory was supported by a Wellcome Trust Senior Investigator Grant (101824/Z/13/Z). The Brun laboratory was supported by grants from the National Institutes of Health (GM113172, GM122556) and by a Canada 150 Research Chair in Bacterial Cell Biology to Y.B. Work in the VanNieuwenhze laboratory was supported by a Grant from the National Institutes of Health (GM113172).
The authors declare no competing financial interest.
Supplementary Material
References
- Koch A. L. (2003) Bacterial wall as target for attack: past, present, and future research. Clin. Microbiol. Rev. 16, 673–687. 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.; Oh D. C.; Cava F.; Takacs C. N.; Clardy J.; de Pedro M. A.; Waldor M. K. (2009) D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555. 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten J. A.; Bagga S.; Lloyd A. J.; de Pascale G.; Dowson C. G.; Roper D. I.; Bugg T. D. (2006) Fluorescent reagents for in vitro studies of lipid-linked steps of bacterial peptidoglycan biosynthesis: derivatives of UDPMurNAc-pentapeptide containing D-cysteine at position 4 or 5. Mol. BioSyst. 2, 484–491. 10.1039/b607908c. [DOI] [PubMed] [Google Scholar]
- Caminero J. A.; Sotgiu G.; Zumla A.; Migliori G. B. (2010) Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect. Dis. 10, 621–629. 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- Boman H. G.; Eriksson K. G. (1963) Penicillin-induced lysis in Escherichia coli. J. Gen. Microbiol. 31, 339–352. 10.1099/00221287-31-3-339. [DOI] [PubMed] [Google Scholar]
- Neuhaus F. C.d-Cycloserine and O-carbamyl-D-serine. In Mechanism of Action; Gottlieb D., and Shaw P. D., Eds.; Springer: Berlin, Heidelberg, 1967; pp 40–83. [Google Scholar]
- Tsuruoka T.; Tamura A.; Miyata A.; Takei T.; Inouye S.; Matsuhashi M. (1985) Second lytic target of beta-lactam compounds that have a terminal D-amino acid residue. Eur. J. Biochem. 151, 209–216. 10.1111/j.1432-1033.1985.tb09089.x. [DOI] [PubMed] [Google Scholar]
- de Pedro M. A.; Cava F. (2015) Structural constraints and dynamics of bacterial cell wall architecture. Front. Microbiol. 6, 449. 10.3389/fmicb.2015.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M. (2013) Antibiotic resistance: the last resort. Nature 499, 394–396. 10.1038/499394a. [DOI] [PubMed] [Google Scholar]
- Fura J. M.; Kearns D.; Pires M. M. (2015) D-Amino acid probes for penicillin binding protein-based bacterial surface labeling. J. Biol. Chem. 290, 30540–30550. 10.1074/jbc.M115.683342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-P.; Meng X.; VanNieuwenhze M. S. (2016) Methods for visualization of peptidoglycan biosynthesis. Methods Microbiol. 43, 3–48. 10.1016/bs.mim.2016.10.004. [DOI] [Google Scholar]
- Radkov A. D.; Hsu Y.-P.; Booher G.; VanNieuwenhze M. S. (2018) Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 87, 991–1014. 10.1146/annurev-biochem-062917-012921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-P.; Hall E.; Booher G.; Murphy B.; Radkov A. D.; Yablonowski J.; Mulcahey C.; Alvarez L.; Cava F.; Brun Y. V.; Kuru E.; VanNieuwenhze M. S. (2019) Fluorogenic D-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat. Chem. 11, 335–341. 10.1038/s41557-019-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-P.; Rittichier J.; Kuru E.; Yablonowski J.; Pasciak E.; Tekkam S.; Hall E.; Murphy B.; Lee T. K.; Garner E. C.; Huang K. C.; Brun Y. V.; VanNieuwenhze M. S. (2017) Full color palette of fluorescent D-amino acids for in situ labeling of bacterial cell walls. Chem. Sci. 8, 6313–6321. 10.1039/C7SC01800B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E.; Hughes H. V.; Brown P. J.; Hall E.; Tekkam S.; Cava F.; de Pedro M. A.; Brun Y. V.; VanNieuwenhze M. S. (2012) In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem., Int. Ed. 51, 12519–12523. 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti G. W.; Kuru E.; Hall E.; Kalinda A.; Brun Y. V.; VanNieuwenhze M.; Maurelli A. T. (2014) A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510. 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y.; Lebar M. D.; Schirner K.; Schaefer K.; Tsukamoto H.; Kahne D.; Walker S. (2014) Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc. 136, 14678–14681. 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M. S.; Whiteside S.; Jewett J. C.; Aditham A.; Cava F.; Bertozzi C. R. (2013) (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol. 8, 500–505. 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal S.; Giengkam S.; VanNieuwenhze M.; Salje J. (2016) Live imaging of the genetically intractable obligate intracellular bacteria Orientia tsutsugamushi using a panel of fluorescent dyes. J. Microbiol. Methods 130, 169–176. 10.1016/j.mimet.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski C.; Welsh M. A.; Sham L.-T.; Eskandarian H. A.; Lim H. C.; Kieser K. J.; Wagner J. C.; McKinney J. D.; Fantner G. E.; Ioerger T. R.; Walker S.; Bernhardt T. G.; Rubin E. J.; Rego E. H. (2018) Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife 7, e37516 10.7554/eLife.37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett T. M.; Bratton B. P.; Miguel A.; Duvshani A.; Sheng Y.; Martin N. R.; Nguyen J. P.; Persat A.; Desmarais S. M.; VanNieuwenhze M. S.; Huang K. C.; Zhu J.; Shaevitz J. W.; Gitai Z. (2017) A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168, 172–185. 10.1016/j.cell.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge M. J.; Mercy C.; Mortier-Barriere I.; VanNieuwenhze M. S.; Brun Y. V.; Grangeasse C.; Polard P.; Campo N. (2017) A programmed cell division delay preserves genome integrity during natural genetic transformation in Streptococcus pneumoniae. Nat. Commun. 8, 1621. 10.1038/s41467-017-01716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho A. W.; Hsu Y. P.; Squyres G. R.; Kuru E.; Wu F.; Jukes C.; Sun Y.; Dekker C.; Holden S.; VanNieuwenhze M. S.; Brun Y. V.; Garner E. C. (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743. 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma M. J.; Kuru E.; Rittichier J. T.; VanNieuwenhze M. S.; Brun Y. V.; Winkler M. E. (2015) Minimal peptidoglycan (PG) turnover in wild-type and PG Hydrolase and cell division mutants of Streptococcus pneumoniae D39 growing planktonically and in host-relevant biofilms. J. Bacteriol. 197, 3472–3485. 10.1128/JB.00541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J.; Durmort C.; Jacq M.; Mortier-Barriere I.; Campo N.; VanNieuwenhze M. S.; Brun Y. V.; Arthaud C.; Gallet B.; Moriscot C.; Morlot C.; Vernet T.; Di Guilmi A. M. (2017) Peptidoglycan O-acetylation is functionally related to cell wall biosynthesis and cell division in Streptococcus pneumoniae. Mol. Microbiol. 106, 832–846. 10.1111/mmi.13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella H.; Yang G.; Ouerfelli O.; Ehrt S.; Nathan C. F.; Vaubourgeix J. (2017) Distinct spatiotemporal dynamics of peptidoglycan synthesis between Mycobacterium smegmatis and Mycobacterium tuberculosis. mBio 8, 301183–17. 10.1128/mBio.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron T. A.; Anderson-Furgeson J.; Zupan J. R.; Zik J. J.; Zambryski P. C. (2014) Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 5, e01219–14 10.1128/mBio.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca M.; Pfister S. P.; Buschor S.; Bayramova F.; Hernandez S. B.; Cava F.; Kuru E.; Van Nieuwenhze M. S.; Brun Y. V.; Coelho F. M.; Hapfelmeier S. (2016) D-Alanine-controlled transient intestinal mono-colonization with non-laboratory-adapted commensal. PLoS One 11, e0151872 10.1371/journal.pone.0151872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro M. A.; Quintela J. C.; Holtje J. V.; Schwarz H. (1997) Murein segregation in Escherichia coli. J. Bacteriol. 179, 2823–2834. 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure L.; Fiche J.-B.; Espinosa L.; Ducret A.; Anantharaman V.; Luciano J.; Lhospice S.; Islam S.; Tréguier J.; Sotes M.; Kuru E.; VanNieuwenhze M.; Brun Y.; Theodoly O.; Aravind L.; Nollmann M.; Mignot T. (2016) The mechanism of force transmission at bacterial focal adhesion complexes. Nature 539, 530–535. 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A.; Lesterlin C.; Manuse S.; Zhao C.; Cluzel C.; Lavergne J.-P.; Franz-Wachtel M.; Macek B.; Combet C.; Kuru E.; VanNieuwenhze M. S.; Brun Y. V.; Sherratt D.; Grangeasse C. (2014) MapZ beacons the division sites and positions FtsZ-rings in Streptococcus pneumoniae. Nature 516, 259–262. 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie A.; Manuse S.; Zhao C.; Campo N.; Cluzel C.; Lavergne J.-P.; Freton C.; Combet C.; Guiral S.; Soufi B.; Macek B.; Kuru E.; VanNieuwenhze M. S.; Brun Y. V.; Di Guilmi A.-M.; Claverys J.-P.; Galinier A.; Grangeasse C. (2014) Interplay of the serine/threonine kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 10, e1004275 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak J. E.; Alvarez D.; Skelly A.; von Andrian U. H.; Kasper D. L. (2017) Illuminating vital surface molecules of symbionts in health and disease. Nat. Microbiol. 2, 17099. 10.1038/nmicrobiol.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J.-E.; Mengin-Lecreulx D.; Monton A.; den Blaauwen T.; Carbonnelle E.; Veckerle C.; Brun Y. V.; Van Nieuwenhze M.; Bouchier C.; Tu K.; Rice L. B; Arthur M. (2016) Factors essential for L, D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia col. eLife 5, 319469. 10.7554/eLife.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras B. L.; Scott M.; Parry B.; Biboy J.; Gray J.; Vollmer W.; Jacobs-Wagner C. (2016) Lyme disease and relapsing fever Borrelia elongate through zones of peptidoglycan synthesis that mark division sites of daughter cells. Proc. Natl. Acad. Sci. U. S. A. 113, 9162–9170. 10.1073/pnas.1610805113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E.; Lambert C.; Rittichier J.; Till R.; Ducret A.; Derouaux A.; Gray J.; Biboy J.; Vollmer W.; VanNieuwenhze M.; Brun Y. V.; Sockett R. E. (2017) Fluorescent D-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat. Microbiol. 2, 1648–1657. 10.1038/s41564-017-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti G.; Kuru E.; Packiam M.; Hsu Y. P.; Tekkam S.; Hall E.; Rittichier J. T.; VanNieuwenhze M.; Brun Y. V.; Maurelli A. T. (2016) Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog. 12, e1005590 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuse S.; Jean N. L.; Guinot M.; Lavergne J.-P.; Laguri C.; Bougault C. M.; VanNieuwenhze M. S.; Grangeasse C.; Simorre J.-P. (2016) Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ. Nat. Commun. 7, 12071. 10.1038/ncomms12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro J. M.; Fernandes P. B.; Vaz F.; Pereira A. R.; Tavares A. C.; Ferreira M. T.; Pereira P. M.; Veiga H.; Kuru E.; VanNieuwenhze M. S.; Brun Y. V.; Filipe S. R.; Pinho M. G. (2015) Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 6, 8055. 10.1038/ncomms9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro J. M.; Pereira A. R.; Reichmann N. T.; Saraiva B. M.; Fernandes P. B.; Veiga H.; Tavares A. C.; Santos M.; Ferreira M. T.; Macario V.; VanNieuwenhze M. S.; Filipe S. R.; Pinho M. G. (2018) Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532. 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montón Silva A.; Otten C.; Biboy J.; Breukink E.; VanNieuwenhze M.; Vollmer W.; den Blaauwen T. (2018) The fluorescent D-amino acid NADA as a tool to study the conditional activity of transpeptidases in Escherichia coli. Front. Microbiol. 9, 2101. 10.3389/fmicb.2018.02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq M.; Arthaud C.; Manuse S.; Mercy C.; Bellard L.; Peters K.; Gallet B.; Galindo J.; Doan T.; Vollmer W.; Brun Y. V.; VanNieuwenhze M. S.; Di Guilmi A. M.; Vernet T.; Grangeasse C.; Morlot C. (2018) The cell wall hydrolase Pmp23 safeguards the division ring in Streptococcus pneumoniae. Sci. Rep. 8, 7591. 10.1038/s41598-018-25882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos M.; Peters K.; Casanova M.; Palacios P.; VanNieuwenhze M. S.; Breukink E.; Vicente M.; Vollmer W. (2018) Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat. Commun. 9, 5090. 10.1038/s41467-018-07559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende N.; Wang J.; Weber P. M.; Verheul J.; Kuru E.; Rittmann S. K. R.; Leisch N.; VanNieuwenhze M. S.; Brun Y. V.; den Blaauwen T.; Bulgheresi S. (2018) Host-polarized cell growth in animal Ssymbionts. Curr. Biol. 28, 1039–1051. e5 10.1016/j.cub.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K.; Pazos M.; Edoo Z.; Hugonnet J.-E.; Martorana A. M.; Polissi A.; VanNieuwenhze M. S.; Arthur M.; Vollmer W. (2018) Copper inhibits peptidoglycan L, D-transpeptidase suppressing beta-lactam resistance due to by-pass of penicillin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 115, 10786–10791. 10.1073/pnas.1809285115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M.; Aistleitner K.; Biboy J.; Gray J.; Kuru E.; Hall E.; Brun Y. V.; VanNieuwenhze M. S.; Vollmer W.; Horn M.; Jensen G. J. (2013) Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 4, 2856. 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan Z. R.; Nairn B. L.; Wang J.; Hsu Y.-P.; Hesse L. E.; Beavers W. N.; Chazin W. J.; Trinidad J. C.; VanNieuwenhze M. S.; Giedroc D. P.; Skaar E. P. (2019) An Acinetobacter baumannii zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep. 26, P2009–2018. e6 10.1016/j.celrep.2019.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann N. T.; Tavares A. C.; Saraiva B. M.; Jousselin A.; Reed P.; Pereira A. R.; Monteiro J. M.; Sobral R. G.; VanNieuwenhze M. S.; Fernandes F.; Pinho M. G. (2019) SEDS-bPBP pairs direct lateral and septal peptidoglycan in Staphylococcus aureus. Nat. Microbiol. 4, 1368–1377. 10.1038/s41564-019-0437-2. [DOI] [PubMed] [Google Scholar]
- Sarkar S.; Libby E. A.; Pidgeon S. E.; Dworkin J.; Pires M. M. (2016) In vivo probe of lipid II-interacting proteins. Angew. Chem., Int. Ed. 55, 8401–8404. 10.1002/anie.201603441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva E. I.; López-Garrido J.; Hughes H. V.; Fredlund J.; Kuru E.; VanNieuwenhze M. S.; Brun Y. V.; Pogliano K.; Jensen G. J. (2013) Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol. 88, 673–686. 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.-C. T.; Boersma M. J.; Vella S. A.; Kocaoglu O.; Kuru E.; Peceny J. K.; Carlson E. E.; VanNieuwenhze M. S.; Brun Y. V.; Shaw S. L.; Winkler M. E. (2014) Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumonia D39. Mol. Microbiol. 94, 21–40. 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Lyu Z.; Miguel A.; McQuillen R.; Huang K. C.; Xiao J. (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747. 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A. R.; Hsin J.; Krol E.; Tavares A. C.; Flores P.; Hoiczyk E.; Ng N.; Dajkovic A.; Brun Y. V.; VanNieuwenhze M. S.; Roemer T.; Carballido-Lopez R.; Scheffers D.-J.; Huang K. C.; Pinho M. G. (2016) FtsZ-dependent elongation of a coccoid bacterium. mBio 7, e00908–16 10.1128/mBio.00908-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrih A.; Bacher G.; Allmaier G.; Williamson M. P.; Foster S. J. (1999) Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. Microbiology 181, 3956–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Angeles D.; Liu Y.; Hartman A. M.; Borisova M.; de Sousa Borges A.; de Kok N.; Beilharz K.; Veening J. W.; Mayer C.; Hirsch A. K.; Scheffers D. J. (2017) Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol. Microbiol. 104, 319–333. 10.1111/mmi.13629. [DOI] [PubMed] [Google Scholar]
- Cava F.; de Pedro M. A.; Lam H.; Davis B. M.; Waldor M. K. (2011) Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 30, 3442–3453. 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batson S.; de Chiara C.; Majce V.; Lloyd A. J.; Gobec S.; Rea D.; Fülöp V.; Thoroughgood C. W.; Simmons K. J.; Dowson C. G.; Fishwick C. W. G.; de Carvalho L. P. S.; Roper D. I. (2017) Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic d-cycloserine. Nat. Commun. 8, 1939. 10.1038/s41467-017-02118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S.; Dubost L.; Marie A.; Arthur M.; Gutmann L. (2008) Identification of the L, D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J. Bacteriol. 190, 4782–4785. 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparrós M.; Pisabarro A. G.; de Pedro M. A. (1992) Effect of dD-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol. 174, 5549–5559. 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K.; Kannan S.; Rao V. A.; Biboy J.; Vollmer D.; Erickson S. W.; Lewis R. J.; Young K. D.; Vollmer W. (2016) The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli. mBio 7, e00819–16 10.1128/mBio.00819-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolina B. A.; Yuan X.; Anderson M. S.; El-Sherbeini M. (2001) The cell wall and cell division gene cluster in the Mra operon of Pseudomonas aeruginosa: cloning, production, and purification of active enzymes. Protein Expression Purif. 21, 393–400. 10.1006/prep.2001.1390. [DOI] [PubMed] [Google Scholar]
- Lupoli T. J.; Tsukamoto H.; Doud E. H.; Wang T. S.; Walker S.; Kahne D. (2011) Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133, 10748–10751. 10.1021/ja2040656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F.; Lam H.; de Pedro M. A.; Waldor M. K. (2011) Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell. Mol. Life Sci. 68, 817–831. 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T.; Tanidokoro K.; Shimizu Y.; Kawarabayasi Y.; Ohshima T.; Sato M.; Tadano S.; Ishikawa H.; Takio S.; Takechi K.; Takano H. (2016) Moss chloroplasts are surrounded by a peptidoglycan wall containing D-amino acids. Plant Cell 28, 1521–1532. 10.1105/tpc.16.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteroy M.; Severin A.; Zhao F.; Rosner D.; Lopatin U.; Scherman H.; Belanger A.; Harvey B.; Hatfull G. F.; Brennan P. J.; Connell N. D. (2000) Characterization of a Mycobacterium smegmatis mutant that is simultaneously resistant to d-cycloserine and vancomycin. Antimicrob. Agents Chemother. 44, 1701–1704. 10.1128/AAC.44.6.1701-1704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Heredia A.; Pohane A. A.; Melzer E. S.; Carr C. R.; Fiolek T. J.; Rundell S. R.; Lim H. C.; Wagner J. C.; Morita Y. S.; Swarts B. M.; Siegrist M. S. (2018) Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. eLife 7, e37243 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.; DeMeester K. E.; Hou C. W.; Parent M. A.; Caplan J. L.; Grimes C. L. (2017) Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat. Commun. 8, 15015. 10.1038/ncomms15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt A. J.; O’Grady J. (1996) Drug chirality: a consideration of the significance of the stereochemistry of antimicrobial agents. J. Antimicrob. Chemother. 37, 7–32. 10.1093/jac/37.1.7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.