Flagellum-driven motility has been studied in E. coli and Salmonella for nearly half a century. Over 60 genes control flagellar assembly and function. The expression of these genes is regulated at multiple levels in response to a variety of environmental signals. Cues that elevate c-di-GMP levels, however, inhibit motility by direct binding of the effector YcgR to the flagellar motor. In this study conducted mainly in E. coli, we show that YcgR is the only effector of motor control and tease out the order of YcgR-mediated events. In addition, we find that the σS regulator protein RssB contributes to negative regulation of flagellar gene expression when c-di-GMP levels are elevated.
KEYWORDS: Escherichia coli, RpoS, RssB, Salmonella, YcgR, c-di-GMP, cellulose, flagellar gene regulation, flagellar motor
ABSTRACT
In Escherichia coli and Salmonella, the c-di-GMP effector YcgR inhibits flagellar motility by interacting directly with the motor to alter both its bias and speed. Here, we demonstrate that in both of these bacteria, YcgR acts sequentially, altering motor bias first and then decreasing motor speed. We show that when c-di-GMP levels are high, deletion of ycgR restores wild-type motor behavior in E. coli, indicating that YcgR is the only motor effector in this bacterium. Yet, motility and chemotaxis in soft agar do not return to normal, suggesting that there is a second mechanism that inhibits motility under these conditions. In Salmonella, c-di-GMP-induced synthesis of extracellular cellulose has been reported to entrap flagella and to be responsible for the YcgR-independent motility defect. We found that this is not the case in E. coli. Instead, we found through reversion analysis that deletion of rssB, which codes for a response regulator/adaptor protein that normally directs ClpXP protease to target σS for degradation, restored wild-type motility in the ycgR mutant. Our data suggest that high c-di-GMP levels may promote altered interactions between these proteins to downregulate flagellar gene expression.
IMPORTANCE Flagellum-driven motility has been studied in E. coli and Salmonella for nearly half a century. Over 60 genes control flagellar assembly and function. The expression of these genes is regulated at multiple levels in response to a variety of environmental signals. Cues that elevate c-di-GMP levels, however, inhibit motility by direct binding of the effector YcgR to the flagellar motor. In this study conducted mainly in E. coli, we show that YcgR is the only effector of motor control and tease out the order of YcgR-mediated events. In addition, we find that the σS regulator protein RssB contributes to negative regulation of flagellar gene expression when c-di-GMP levels are elevated.
INTRODUCTION
Escherichia coli and its close relative Salmonella enterica each have multiple enzymes that both synthesize and degrade the signaling molecule c-di-GMP, with the aid of diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively (1). The hallmark of this second messenger in bacteria is its participation in the inverse regulation of biofilms and motility, i.e., when c-di-GMP levels are high, motility is inhibited and exopolysaccharides (EPS) are synthesized, and vice versa (2, 3). c-di-GMP activates biofilm production in E. coli and Salmonella by turning on expression of curli fimbriae, cellulose, or poly-β-1,6-N-acetylglucosamine (PGA) (3, 4). In these bacteria, c-di-GMP is not known to inhibit flagellar motility by inhibiting gene expression but rather by acting directly on the flagellar motor via the c-di-GMP effector YcgR (5–7). Interestingly, genes that regulate the c-di-GMP response (yhjH and ycgR) are included in the flagellar regulon (8–10). Flagellar gene regulation is a complex process in which the master regulator FlhDC, together with the vegetative sigma70, controls the expression of over 50 genes included in 14 operons in a three-tiered regulatory cascade (11). FlhDC, at the top of this cascade, controls sigma28 (FliA) synthesis in the second tier, which in turn controls the third tier. The c-di-GMP regulators YhjH and YcgR are expressed in this bottom tier. YhjH (alternate name, PdeH [12]) is the most active PDE in E. coli and Salmonella (5, 13), and YcgR is a c-di-GMP effector in both (7, 14). Thus, when cells are motile, YhjH degrades c-di-GMP to keep its levels low, while YcgR arrests motor function when environmental conditions activate c-di-GMP synthesis.
The flagellar motors of E. coli and Salmonella consist of a moving rotor and stationary stators (15). The rotor includes the cytoplasmic C ring made of three proteins, attached to a membrane MS ring, the periplasmic rod, and external hook. This basal structure is continuous with the external helical flagellar filament. The stators, which conduct protons to power motor rotation, are positioned at the top of the C ring. As protons travel through, they generate torque at the stator-rotor interface, which drives motor rotation (16). The C ring controls the switching between clockwise (CW) and counterclockwise (CCW) rotor directions in response to chemotaxis signals. E. coli motors spin at ∼125 Hz when attached to the flagellar filament; when the filament is absent (low load), motors rotate at 300 Hz or faster (17–19).
Several studies have shown that YcgR::c-di-GMP affects motor bias and speed in both E. coli and Salmonella (6, 7, 20), primarily by binding to the flagellar rotor (6, 7, 21). Another study in E. coli reported an effect of YcgR on motor speed alone, by binding to the stators (5). In this study, we revisit YcgR action at the motor by the use of both cell tethering and bead assays under regulated expression of YcgR. In both E. coli and Salmonella, we observe that motor bias changes before reduction of speed. We discuss these results in the context of the previously published data. We show that in a yhjH mutant, where c-di-GMP levels are elevated, deletion of ycgR reestablishes normal motor behavior. We investigate why then chemotactic motility assayed in soft agar does not fully recover in the ycgR yhjH double mutant (7, 14). Unlike in Salmonella, where cellulose production induced by c-di-GMP has been shown to be responsible for interfering with flagellar function extracellularly (22, 23), in E. coli we identify the adaptor protein RssB as contributing to the inhibition of flagellar gene expression by an unknown mechanism.
(Part of this research was conducted by V. Nieto in partial fulfillment of the requirements for a Ph.D. from the University of Texas at Austin, Austin, TX, 2013 [24].)
RESULTS AND DISCUSSION
YcgR is the only effector of motor behavior under elevated c-di-GMP levels in E. coli.
Motility assays have been widely used to decipher the action of the c-di-GMP effector YcgR. Cell tethering assays, in which cells are attached to a glass surface by a single flagellum and rotation of the cell body is monitored to investigate motor behavior (25), have shown that in both E. coli and Salmonella, YcgR::c-di-GMP skews the flagellar motor bias in a CCW direction (6, 7, 20) and reduces motor speed in Salmonella (7). Because of the high load on the flagellum in the cell tethering assay, the recorded speeds are low; indeed, this methodology did not detect an effect on motor speed in E. coli (6). YcgR was shown to affect swimming speed in E. coli, as monitored by dark-field microscopy (5). In all of these assays, cellular c-di-GMP levels are elevated by disabling the most active PDE, YhjH (5).
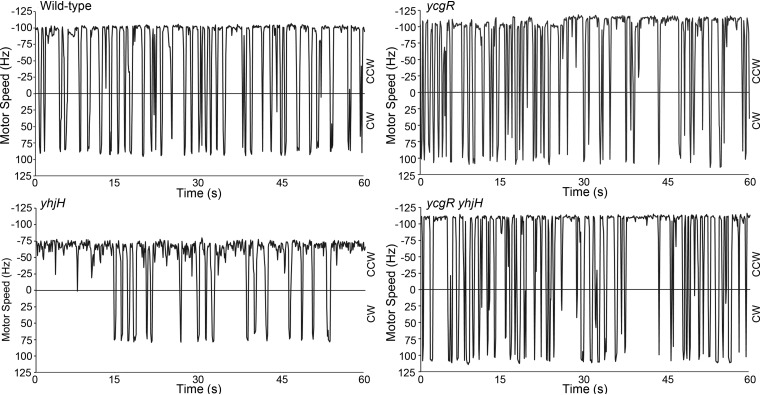
To study the effect of YcgR on motor behavior at a higher resolution, we used a bead assay, in which the bacterial cell body is fixed to the glass surface and a polystyrene bead is attached to a stub of one of its flagellar filaments (26). These experiments focusing on the effects of YcgR in Salmonella and E. coli were first reported by V. Nieto in his Ph.D. dissertation (24). Salmonella motors do not switch as often as those in E. coli, as determined by this assay (27), so we studied the E. coli motor and then committed to this bacterium for a majority of the experiments reported in this study. Wild-type (WT) E. coli motors exhibited speeds and reversals consistent with published studies (26, 28) (Fig. 1). In comparison to the WT, the yhjH mutant had both a CCW bias and lower speeds. Both parameters returned to normal in the ycgR yhjH double mutant, although they averaged slightly higher than those of the WT. Slightly higher motor speeds and reversal frequencies of the ycgR mutant alone indicate that the basal levels of c-di-GMP in WT E. coli (i.e., YcgR+) exert a small inhibitory effect on the motor (Fig. 1). We conclude that under the experimental conditions tested, YcgR is the only effector that controls motor behavior in response to c-di-GMP.
FIG 1.
Behavior of single motors of E. coli strains. Representative motors from E. coli HCB5 (AW405 fliCsticky) and isogenic ycgR, yhjH, and ycgR yhjH mutants were monitored by recording the motion of polystyrene beads attached to filament stubs (see Materials and Methods). Counterclockwise (CCW) and clockwise (CW) speeds are shown. Average CCW speeds (expressed in Hz, unit of frequency that refers to the number of rotations per second) are 76 ± 14, 78.7 ± 14, 56 ± 8, and 83.6 ± 15, and switching frequencies (reversals in motor direction per minute, i.e., when the trace crosses zero) are 40 ± 4.2, 43 ± 6.9, 22.6 ± 4.7, and 46.4 ± 8.3 for WT, ycgR, yhjH, and ycgR yhjH strains, respectively. The profiles are representative of 10 individual motors for each strain.
YcgR acts sequentially to first alter rotational bias and then motor speed in both Salmonella and E. coli.
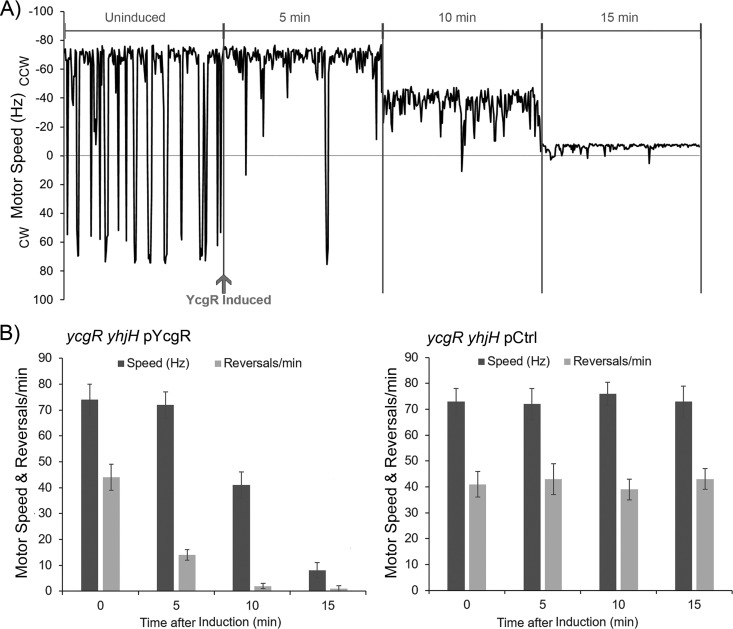
When c-di-GMP levels are high, physiological levels of YcgR are observed to affect both bias and speed (Fig. 1). These effects appeared to be separable in a time course experiment conducted using a cell tethering assay in Salmonella enterica, where controlled induction of YcgR in the yhjH mutant strain appeared to show a change in motor bias before a change in motor speed was evident (24) (see Fig. S1 in the supplemental material). To monitor the temporal separation of these events at a higher resolution, we turned to E. coli. This time we used a ycgR yhjH mutant and recorded the motor behavior in response to controlled expression of YcgR from the lac promoter on a low-copy-number plasmid (Fig. 2). At zero time, without added inducer, the biases and speeds of motors in strains harboring the plasmid with and without YcgR were similar (see the uninduced trace in Fig. 2A as well as zero times in Fig. 2B), showing that there was no leaky expression of YcgR. Within 5 min after the addition of inducer (50 μM isopropyl-β-d-thiogalactopyranoside [IPTG]), the bias of the YcgR+ motors shifted from 44 ± 5 reversals per min to 14 ± 2 reversals per min, while their speeds remained unchanged (72 ± 5 Hz, compared to 74 ± 6 Hz preinduction) (Fig. 2A and B, left panel). The change in motor bias observed at this time point is within range of that seen with native levels of YcgR in the yhjH mutant (Fig. 1) (22.6 ± 4.7 reversals per min); given that there is no effect on motor speed, YcgR levels must be in the near-normal physiological range at this time. By 10 min, the motors were all nearly CCW, and motor speed had reduced approximately 40% (41 ± 5 Hz, compared to 56 ± 8 Hz at native YcgR levels in the yhjH mutant in Fig. 1). After 15 min, the motors turned entirely CCW and their speed had dropped to 8 ± 3 Hz, only 10% of the original speed. The key revelation in this experiment is the behavior of the motor between 0 and 5 min and between 5 and 10 min. The first interval saw a change in bias alone, while the second interval saw an additional change in speed. These two traces clearly show that the YcgR-dependent changes in bias and speed seen in Fig. 1 are separable, revealing a two-step action of YcgR at the motor. In the absence of YcgR, the motor maintained its speed and bias over a similar 15-min period (Fig. 2B, right panel). We conclude that YcgR operates sequentially, first changing motor bias to CCW and then slowing down motor rotation.
FIG 2.
Time course of flagellar motor behavior in a ycgR yhjH mutant. (A) Time course of a single motor trace of strain JP1442 (ycgR yhjH) monitored by the bead assay as described in the legend to Fig. 1. At each of the time points indicated, 60 s of motor activity was recorded, but only 20-s segments are arranged side by side. YcgR was expressed in the cells upon IPTG induction of pJP388, a low-copy-number plasmid. The trace is representative of 15 motors. (B) Average motor speed and reversal frequencies for the 15 motors monitored as described for panel A (left) and for 15 motors of the strain with the vector control plasmid pSEVA224 (right), at four time points during the experiment. In the YcgR+ experiment (left), average motor speeds (expressed in Hz) are 74 ± 6, 72 ± 5, 41 ± 5, and 8 ± 3 and switching frequencies (reversals in motor direction per minute) are 44 ± 5, 14 ± 2, 2 ± 1, and 1 ± 1 at the 0-, 5-, 10-, and 15-min time points, respectively.
A straightforward conclusion from Fig. 2 is that the flagellar rotor is the initial target of YcgR. Several studies to date have demonstrated a change in motor bias as a result of YcgR action (6, 7, 21). A recent study found that YcgR is retained at the motor even after the dissociation of the stators upon de-energizing the cell (21). This large body of evidence is not in keeping with the lone finding that the stators are the only target of YcgR (5). While the data in Fig. 2 are consistent with the proposed two-step model for YcgR action (7), where the initial YcgR binding to the rotor allosterically disrupts the rotor-stator interface to reduce motor speed, the data would also support a model not yet considered: the existence of two independent binding targets of YcgR, i.e., rotor and stators, the latter presenting a lower-affinity target. Another potential model is that the initial YcgR contact with stators reorients the rotor to favor one switch state over another (CCW over CW) but that stator function is not affected until YcgR contacts the rotor. Whatever the detailed mechanism by which bias and speed are sequentially affected, our data clearly show that YcgR impacts both components of motor function.
The YcgR homolog MotI in Bacillus subtilis interacts only with the stators to sever stator-rotor interactions (29). The YcgR homolog of Pseudomonas aeruginosa, FlgZ, is thought to selectively bind and sequester stators that promote swarming motility (30). Neither of these proteins affects motor bias. All three YcgR-like proteins studied to date, however, ultimately generate similar outcomes.
YcgR-independent inhibition of motility in E. coli does not depend on EPS pathways.
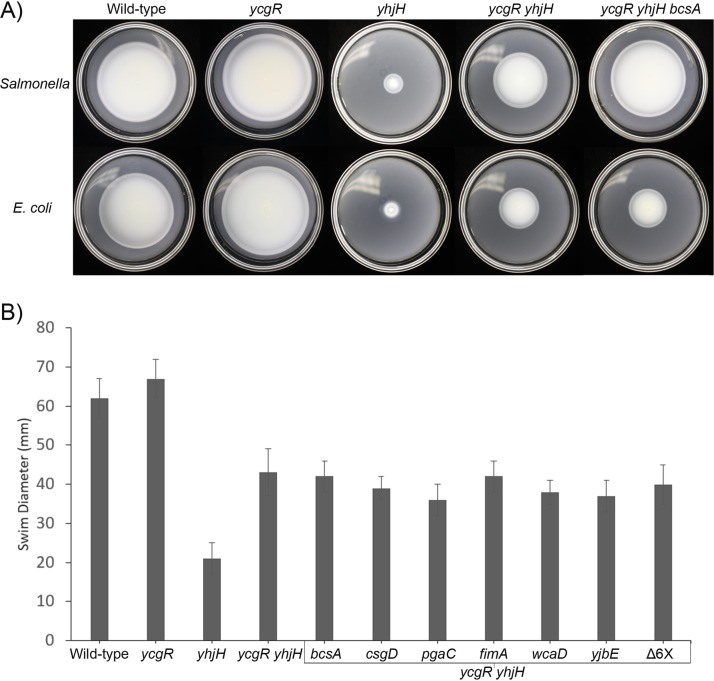
Data presented in Fig. 1 show that in the absence of YcgR in the yhjH mutant, motor behavior returns to normal (compare the yhjH mutant with the ycgR yhjH double mutant and WT). However, in both E. coli and Salmonella, motility assayed in soft agar plates does not (6, 7, 14), suggesting the existence of YcgR-independent pathways that inhibit motility or chemotaxis. These results for the two bacteria are reproduced in Fig. 3A (compare the WT and the yhjH and ycgR yhjH mutants). We note that a lesion in the c-di-GMP-binding site of YcgR is sufficient to abrogate its inhibitory function in the soft agar assay (14), indicating that YcgR does not have a second c-di-GMP-dependent function as measured by this assay. The enhanced soft agar performance of the ycgR mutant in the two bacteria (Fig. 3A, compare the WT and the ycgR mutant) is consistent with the motor data in E. coli, where this mutant shows slightly higher motor speeds and reversal frequencies than the WT (Fig. 1), indicative of an inhibitory effect of basal levels of c-di-GMP in the WT strain.
FIG 3.
Effect of c-di-GMP-induced EPS and curli fimbria production on motility of Salmonella and E. coli, as measured by migration in soft agar. (A) Wild-type Salmonella enterica, E. coli, and their indicated mutant derivatives were inoculated at the center of 0.3% LB swim agar plates and incubated at 30°C for 8 h. (B) Wild-type E. coli and the indicated six EPS/fimbria mutants engineered into the yhjH ycgR strain were assayed as in panel A. Δ6× denotes all six mutations combined into one strain. A paired-sample t test determined that there was no statistically significant difference between the ycgR yhjH mutant and any of the additionally mutated strains.
In two Salmonella enterica serovars (Enteritidis and Typhimurium), motility was fully restored when c-di-GMP-dependent production of cellulose was additionally abolished (23), an observation that we confirmed (Fig. 3A, upper panel, compare the ycgR yhjH mutant to the ycgR yhjH bcsA mutant and WT). The activity of BcsA, a cellulose synthase, is allosterically regulated by c-di-GMP, a pioneering discovery made in Gluconacetobacter xylinus and later confirmed in other bacteria as well (22, 31). Using calcofluor white, a fluorescent blue dye that binds cellulose, we verified that BcsA was functional as a cellulose synthase in E. coli (24). However, we did not observe an inhibitory effect of cellulose on E. coli (Fig. 3A, lower panel, compare the ycgR yhjH and ycgR yhjH bcsA mutants). We confirmed that the residual soft agar motility defect in the ycgR yhjH double mutant was related to c-di-GMP and was not due to a second mutation in this strain by introducing a plasmid expressing either the PDE YhjH from E. coli or the DGC DgcA from Caulobacter crescentus (32) and observing increased or decreased motility, respectively (Fig. S2). Taken together, the data suggest that in the absence of YcgR, c-di-GMP affects E. coli motility in soft agar by some other mechanism, independent of cellulose production.
In P. aeruginosa, c-di-GMP-controlled Pel polysaccharide inhibits motility (30), as does c-di-GMP-controlled EPS production in Vibrio cholerae (33), while in B. subtilis, EPS made by a c-di-GMP-independent pathway has a similar effect (34). In E. coli, production of the adhesive curli fimbriae and the EPS poly-β-1,6-N-acetyl-d-glucosamine (PGA) are also controlled by c-di-GMP (4). To test if these molecules could be involved in the YcgR-independent inhibition of motility in E. coli, we deleted key genes in each of these pathways in the ycgR yhjH mutant, namely, csgD (35) and pgaC (36), respectively. E. coli also makes biofilms by pathways that make type 1 fimbriae, colanic acid, or other EPS under c-di-GMP-independent stresses (37). We inactivated key genes in related pathways as well, namely, fimA (38), wcaD (39), and yjbE (40). Like the bcsA mutation, none of these mutations, alone or in concert with each other, was sufficient to restore wild-type motility to the ycgR yhjH mutant (Fig. 3B). We conclude that EPS pathways of E. coli are not responsible for motility inhibition in the ycgR yhjH double mutant.
YcgR-independent motility inhibition on soft agar is not due to inhibition of chemotaxis.
Migration of bacteria in soft agar plates (Fig. 3) is dependent on both their ability to generate gradients of attractant compounds through consumption of nutrients in the medium and their chemotactic proficiency (41). Thus, a smaller swim diameter of the ycgR yhjH strain in soft agar could be due to a reduced growth rate or to a defect in the chemotaxis signaling pathway, which modulates flagellar rotation bias. The latter possibility seemed plausible, given that c-di-GMP is known to control chemotaxis in other bacteria (42, 43).
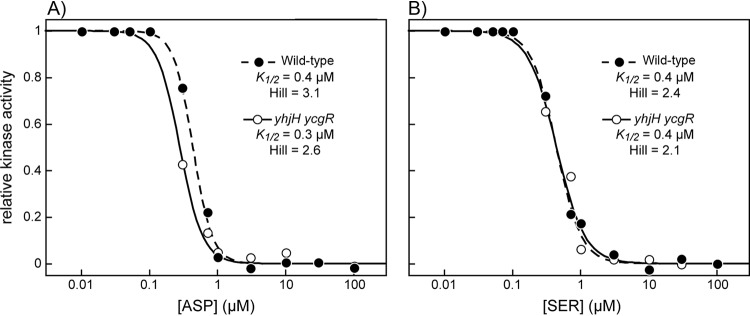
To explore these possibilities, we first ascertained that the growth rate of the ycgR yhjH mutant was not altered compared to the WT (data not shown). Next, we used a Förster resonance energy transfer (FRET)-based in vivo assay to compare attractant responses in the WT and ycgR yhjH strains (44, 45). This assay monitors a FRET interaction between CheY and CheZ, molecules tagged with FRET acceptor and donor fluorophores. In the chemotaxis signaling pathway, CheY obtains phosphoryl groups from the CheA kinase, whose activity is under chemoreceptor control. The phosphorylation state of CheY reflects CheA activity and, in turn, its affinity for the phospho-CheY phosphatase CheZ. The FRET assay thus provides a readout of attractant-induced changes in CheA activity. We tested responses to two attractants, aspartate, which is sensed by the Tar chemoreceptor, and serine, which is sensed by the Tsr chemoreceptor. We found that the WT and ycgR yhjH strains had virtually identical response thresholds and cooperativities to both attractants, which are the major chemoeffectors in Lennox broth (LB) (Fig. 4). We conclude that the soft agar motility defect in the ycgR yhjH strain is not due to a compromised chemotactic response.
FIG 4.
CheA kinase control responses in WT and ycgR yhjH strains. Attractant responses to aspartate (A), mediated by the Tar receptor, and serine (B), mediated by the Tsr receptor, were measured with in vivo FRET kinase assays, as detailed in Materials and Methods.
Suppressor mutations that relieve motility inhibition map to the rssAB operon: suppression does not involve lowering c-di-GMP levels.
Having eliminated differences in motor function, chemotaxis, growth rate, or EPS as causative factors in the slower expansion of ycgR yhjH strains on soft agar, we sought to gain insights into the basis for this effect by isolating and characterizing pseudorevertants of the ycgR yhjH strain with enhanced performance in the soft agar assay. Whole-genome sequencing of six such independent revertant strains identified several potential suppressors (Table 1). The most common mutations (in 5 out of 6 suppressors) were deletions spanning the region including the two-gene operon rssAB (46) and point mutations in nfrA and nfrB (47). The latter comprise overlapping genes that encode the bacteriophage N4 membrane receptor. NfrA is a large outer membrane protein, and NfrB is in the inner membrane. Phage N4 interacts with NfrA directly; an interaction with NfrB is hypothesized to assist in forming a channel for phage entry (48). Whereas identical nfrA and nfrB mutations were found in independent psedorevertants, each rssAB deletion was unique. In E. coli, RssA is annotated as a lipid hydrolase with unknown function, while RssB (formerly called SprE) has been characterized as a response regulator and an adaptor protein for the stationary-phase sigma factor RpoS (σS), targeting it for degradation by the ClpXP protease (49–53). We focused our attention on the rssAB suppressors because of reported inverse coordination between motility and the σS-mediated general stress response (35).
TABLE 1.
Mutational changes in pseudorevertants of a ycgR yhjH strain
| Revertant strain | Mutant gene(s)a | Mutation(s)b | Gene functionc |
|---|---|---|---|
| JP1836d | nfrA | Δ2681–2682 | Bacteriophage N4 receptor |
| rssAB | D893E (GAT→GAA), Δ5–1753 | Regulator of RpoS | |
| JP1837 | nfrA | Δ2681–2682 | Bacteriophage N4 receptor |
| rssAB | D893E (GAT→GAA), Δ1–1807 | Regulator of RpoS | |
| JP1838 | rclB | A74V (GCA→GTA) | Reactive chlorine species stress resistance protein |
| nfrB | E187* (GAG→TAG) | Bacteriophage N4 receptor | |
| JP1839 | rclB | A74V (GCA→GTA) | Reactive chlorine species stress resistance protein |
| nfrB | E187* (GAG→TAG) | Bacteriophage N4 receptor | |
| rssAB | Δ3–1793 | Regulator of RpoS | |
| JP1844 | pitA | L8L (CTG→TTG) | Phosphate transporter |
| rssAB | Δ1–1763 | Regulator of RpoS | |
| JP1852 | nfrA | Δ2681–2682 | Bacteriophage N4 receptor |
| rssAB | D893E (GAT→GAA), Δ1–1784 | Regulator of RpoS |
Mutational changes were identified by whole-genome sequencing of six independent pseudorevertants using breseq (79).
Δ, deletion; deletions indicate the range of nucleotide positions deleted. *, stop codon.
Gene product descriptions are from GenBank annotations. The rssAB deletions refer to nucleotides in the rssAB operon (1,920 nt) rather than the individual genes.
Details of JP strains can be found in Table S1 in the supplemental material.
There are no reports of the rssAB operon influencing c-di-GMP levels. To nonetheless test if recovery of motility in the pseudorevertants was due to reduction in c-di-GMP levels, we used liquid chromatography coupled with tandem mass spectrometry to estimate cellular concentrations of this second messenger in these strains, as well as in an ycgR yhjH strain with an rssAB deletion introduced. As expected, the ycgR yhjH mutant showed an increase in intracellular c-di-GMP (∼40%), with these levels maintained in the revertant strains (Table 2). Thus, suppression does not work by lowering c-di-GMP concentration.
TABLE 2.
Intracellular c-di-GMP levels of pseudorevertant strains
| Strain | Intracellular c-di-GMP concn (nM)a |
|---|---|
| Wild type | 450 ± 30 |
| ycgR yhjH mutant | 850 ± 20 |
| JP1836b | 800 ± 60 |
| JP1837 | 920 ± 30 |
| JP1838 | 940 ± 15 |
| JP1839 | 930 ± 150 |
| JP1844 | 870 ± 0.02 |
| JP1852 | 910 ± 17 |
| ycgR yhjH rssAB mutant | 800 ± 10 |
c-di-GMP concentrations were quantified using liquid chromatography coupled with tandem mass spectrometry as described in Materials and Methods. Samples were prepared and assayed in triplicate; means and standard deviations of the means are shown.
JP strains signify pseudorevertant isolates from the ycgR yhjH double mutant; see Table S1 in the supplemental material. A paired-sample t test was used to calculate P values of <0.05 when wild-type values were compared with any of the other listed strains.
The suppressor phenotype is conferred by RssB and may be related to an altered adaptor function under elevated c-di-GMP.
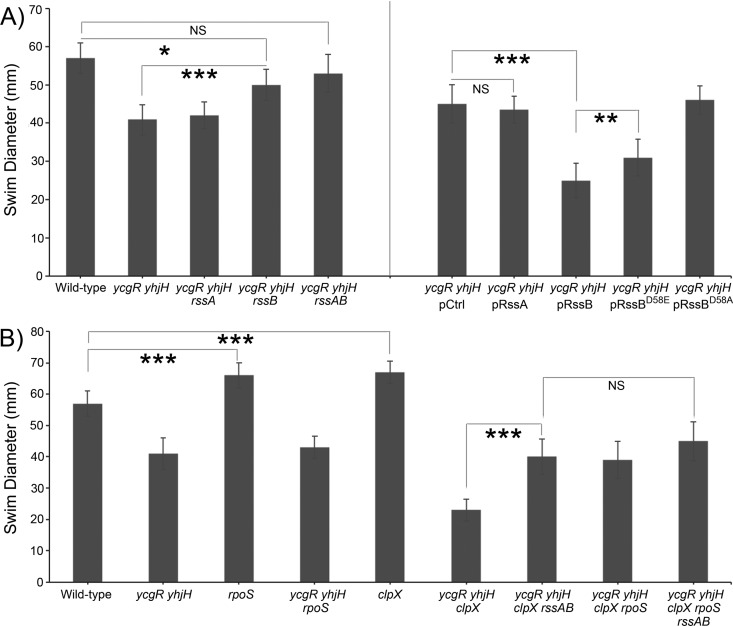
To ask if mutations in both rss genes played a role in motility suppression, we deleted each gene separately (Fig. 5A, left data set). We found that a single deletion of rssB sufficed for suppression; deletion of rssA alone had a negligible effect. When these genes were overexpressed, rssB and not rssA decreased the motility of the ycgR yhjH strain (Fig. 5A, right data set). Thus, loss of RssB function confers the pseudorevertant phenotype.
FIG 5.
Effect of indicated mutations on motility of E. coli ycgR yhjH. (A) Effect of deletions of rssA or rssB (left) and of overproduction of these genes and their mutant derivatives (right) on restoring wild-type levels of motility in the soft agar assay. The two data sets are separated by a vertical line for clarity. (B) Effect in soft agar of rpoS, clpX, and rssAB mutations in WT and ycgR yhjH backgrounds. All strains were inoculated at the center of 0.3% LB swim agar plates and incubated at 30°C for 8 h. For strains carrying expression plasmids, the agar was supplemented with ampicillin (100 μg/ml) and arabinose (0.2% wt/vol). One-way analysis of variance (ANOVA; Tukey’s comparison) using GraphPad Prism 6 is indicated for select comparisons, while those for all pairwise combinations can be found in Table S3 in the supplemental material. Calculated P values are indicated: *, <0.05, **, <0.01, or ***, <0.0001. NS, not statistically significant.
The only known function of E.coli RssB is as an adaptor that targets the general stress sigma factor RpoS (σS) for degradation by the ClpXP protease (50, 51, 53, 54). RssB makes contact with both σS and ClpX, tethering its σS cargo to the ClpXP degradosome (55). The N-terminal domain of RssB is characteristic of the large family of response regulator proteins and, like other response regulators, can be phosphorylated at a conserved aspartic acid residue (D58 in E. coli RssB) (49). Phosphorylation was initially thought to be important for RssB activity (53, 56), but mutants unable to phosphorylate retain a significant amount of activity (57). To test if phosphorylation is important for the motility inhibition phenotype, we overexpressed two different variants of the protein: RssBD58E, which cannot be phosphorylated but is reported to phosphomimic properties that retain partial activity (58, 59), and RssBD58A, a variant that should abrogate response regulator phosphorylation (60). The activity of both alleles is shown in Fig. 5A (right data set). The D58A variant of RssB abolished motility inhibition, while the D58E form showed intermediate levels of inhibition compared to the wild type, suggesting that phosphorylation of RssB is required for this phenotype. We note that the suppressor screen did not identify ArcB, the reported RssB kinase (56).
If the only identified role of RssB is in the degradation of σS, how might it affect motility? Motility gene expression and the general stress response were reported to be inversely coordinated by competition for the core RNA polymerase in E. coli by σS and flagellar sigma factors (σ70, σFlhDC, and σ28) (35). Thus, when σS levels are high, expression from flagellar gene promoters is decreased. Accordingly, deletion of rpoS increased motility (Fig. 5B). In this scheme, given the adaptor role of RssB, its loss should stabilize σS and hence decrease motility by reducing transcription from flagellar gene-specific sigma factors. However, the rssB mutation had an opposite effect, increasing rather than decreasing motility of the ycgR yhjH strain (Fig. 5A, left data set). Given that a variety of stresses regulate the stability of σS, some by producing antiadaptor proteins that interact with RssB to inhibit σS proteolysis (61), it is possible that high c-di-GMP conditions are sensed as a stress signal that directly or indirectly alters interactions between RssB and σS, stabilizing σS against ClpX degradation rather than promoting its degradation. We note that RssB does not have structural motifs likely to participate in c-di-GMP binding (1). If high c-di-GMP levels favor stabilizing interactions between RssB and σS, removal of σS should relieve the motility inhibition, similar to removal of RssB. However, rpoS mutations were not recovered in the reversion analysis. Introduction of an rpoS deletion into the ycgR yhjH strain also did not change its soft agar behavior (Fig. 5B). This result does not necessarily negate the flipped scenario of RssB action proposed above. Given the multiple regulatory pathways controlled by σS, loss of σS may alter cell physiology in a manner that masks its effect on motility under elevated c-di-GMP conditions. Alternatively, RssB might act in a σS-independent manner. To test the latter possibility, a second genetic screen was conducted in JP2173 (Table S2), a ycgR yhjH strain that overproduces RssAB (this experiment was done prior to defining RssB as the motility repressor), looking for suppressors that relieved the motility inhibition shown in Fig. 5A (right data set). The results are tabulated in Table S2. Fifteen of twenty pseudorevertants contained rssAB deletions. Point mutations in pitA were also recovered in 15 of 20 of these revertants, with 12 having both rssAB and pitA mutations. PitA is annotated as a phosphate transporter. It is not obvious to us how PitA function might be related to that of RssB. This larger screen also did not identify ArcB, the reported RssB kinase (56), or σS, the RssB substrate. While this screen did not shed further light on RssB function, it affirms a key role for RssB in reducing soft agar motility under elevated c-di-GMP conditions.
The ClpXP protease targets a large number of substrates in E. coli, regulating many different cellular processes (62). In Salmonella, ClpXP targets the FlhC subunit of the master flagellar regulator for degradation (63, 64). Accordingly, deletion of clpX also improved the motility of the wild-type E. coli strain (Fig. 5B, compare the WT and clpX strains). However, this deletion had an opposite effect on the motility of the ycgR yhjH strain (Fig. 5B, compare the ycgR yhjH and ycgR yhjH clpX strains). One explanation for this result is that loss of ClpX under elevated c-di-GMP conditions might stabilize/enhance σS function via RssB, as proposed above. If this was the case, deletion of rssB should relieve the inhibitory effect of the clpX mutation in the ycgR yhjH background. Surprisingly, it did (Fig. 5B, compare the ycgR yhjH clpX and ycgR yhjH clpX rssAB strains), as did now the deletion of rpoS (Fig. 5B, compare the ycgR yhjH clpX and ycgR yhjH clpX rpoS strains). A clpX rssAB rpoS triple mutation did not improve motility over the clpX rssAB or clpX rpoS double mutation in the ycgR yhjH background, as might be expected if these components acted in the same pathway. While the performance of these strains did not return to wild-type levels, we believe this could be because the deletion of the two important cellular regulators, ClpX and σS, changed the balance of other regulators. It is worth belaboring the complexity of these networks involving sigma factors/RNA polymerase control, motility regulation, and the ever-growing modulon of c-di-GMP. For example, FliA (σ28), implicated by our data as being a part of this system, is a member of an operon that includes FliZ, a regulator that not only contributes to feedback control of several motility components but also functions as an inhibitor of σS (35, 65). Additionally, FlgM acts as a negative regulator of flagellin synthesis by directly binding σ28, serving as a crucial checkpoint in the staggered assembly of the entire flagellin complex (11). Acting as an anti-sigma factor, FlgM is also degraded by the ClpXP complex (66), adding regulatory complexity as these components are removed or accumulate within the system.
If the data presented in Fig. 5B are interpreted in a scenario where RssB stabilizes σS function when c-di-GMP levels are high, then one should observe decreased transcription of flagellar promoters in the ycgR yhjH strain. To test this, we monitored transcription from both the σ70-driven flhDC and σFlhDC-driven σ28 promoters, using β-galactosidase activity as the reporter. With both promoters, transcription decreased in the ycgR yhjH strain and was elevated above that of the WT in the rssAB suppressor (Table 3). Overall, these results point to an altered interaction between RssB, σS, and ClpX under elevated c-di-GMP conditions. A model summarizing these results is diagrammed in Fig. 6. The precise mechanism of RssB involvement in this pathway is a matter for future research.
TABLE 3.
β-Galactosidase reporter assays to test promoter activity of flhDC and fliA genes
| Strain | β-Galactosidase activity (Miller units)a |
|
|---|---|---|
| pPflhD::lacZ | pPfliA::lacZ | |
| Wild type | 2,672 ± 50 | 964 ± 160 |
| ycgR yhjH mutant | 1,684 ± 223 | 507 ± 87 |
| ycgR yhiH rssAB mutant | 3,536 ± 381 | 1,118 ± 198 |
Expression of β-galactosidase from reporter plasmids PflhD::lacZ (pVS182) and PfliA::lacZ (pVS177) was measured in the indicated genetic backgrounds using the Miller assay. Data are averages of experiments carried out in triplicate, with the standard deviation of the mean shown. A paired-sample t test was used to calculate P values of <0.05 when wild-type β-galactosidase activity was compared with the activity of either of the two mutant strains.
FIG 6.
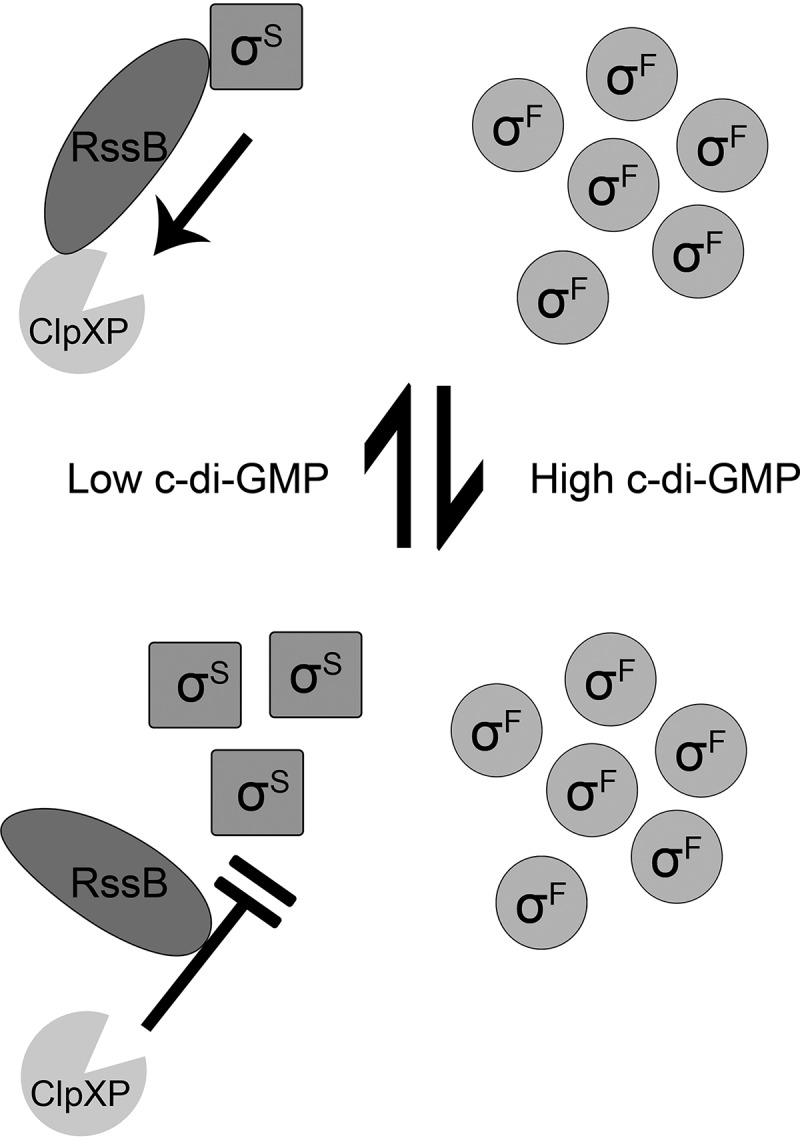
A model for how RssB might inhibit motility under high c-di-GMP conditions. The model is based on the reported inverse coordination between motility and the σS-mediated general stress response (35). In growing cells not exposed to any particular stress, levels of the general stress sigma factor σS are kept low by rapid proteolysis by ClpXP via the adaptor RssB (see the text). Motility is enabled by the activity of multiple flagellar sigmas (collectively indicated by σF) under low-stress conditions. We propose that high c-di-GMP levels are sensed as a stress signal that stabilizes σS by some active process by which RssB prevents σS degradation. The increased competition between σS and σF decreases motility by decreased transcription of the flagellar regulon.
A surprising outcome of this study is the difference between the c-di-GMP-dependent, YcgR-independent mechanisms of motility inhibition in E. coli and Salmonella. Both bacteria encode the cellulose synthase BcsA and make cellulose, and both bacteria encode RssB. Salmonella RssB shares 91% identity with the E. coli protein, and like E. coli RssB, Salmonella RssB promotes σS degradation (59). Yet, cellulose inhibits motility of Salmonella but not E. coli, and conversely, RssB has not been reported to inhibit Salmonella motility. Why this is so is again a matter for future research.
MATERIALS AND METHODS
Strains, growth conditions, mutagenesis, and plasmid constructions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bacterial cultures were grown in Lennox broth (LB) base (20 g/liter) (Invitrogen). For chemotaxis assays, 8 μl of an exponential-phase culture (optical density at 600 nm [OD600], ∼0.6) was inoculated onto swim plates made with 0.3% Bacto agar (Difco) and incubated at 30°C. All plate images shown are representative of three biological replicates, each in triplicate. Where required, the following antibiotics were used: ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), and kanamycin (50 μg/ml). For inducible plasmids, isopropyl-β-d-thiogalactopyranoside (IPTG) and l-arabinose were added at concentrations indicated in the text or figure legends.
Mutant strains of Salmonella and E. coli were constructed by inserting a kanamycin resistance cassette (KAN) into the designated gene as previously described (67) or sourced from the Keio collection (68). Mutations were transferred to fresh backgrounds by phage P22 (HT12/4int103) or phage P1 (P1 Cm) transduction. Excision of the inserted KAN cassettes was achieved by expression of the FLP recombinase encoded by pCP20 (67). The resulting strains were confirmed by DNA sequencing.
Expression plasmids were constructed by amplifying gene sequences from the genomic DNA of wild-type strains by using PCR and appropriate primers (all available upon request), and introduced into their respective vectors (Table S1). All constructs were confirmed by DNA sequencing.
Promoter assays were conducted in the required lacZ backgrounds (Table S1), transformed with either plasmid pVS177 (PfliA::lacZ) or pVS182 (PflhD::lacZ), gifts from Vanessa Sperandio (69) generated using the pRS1551 reporter system (70). The lacZ mutation was generated as described above (67) and moved into the required backgrounds by phage P1 (P1 Cm) transduction. β-Galactosidase activity encoded by lacZ was measured using the Miller assay from cultures grown in LB at 30°C to an OD600 of 0.4 (71).
In vivo FRET CheA kinase assay.
The experimental system closely followed the hardware, software, and methods described by Sourjik et al. (45), with updates and adaptations as described by Lai and Parkinson (72). Briefly, cells containing pVS88, a Förster resonance energy transfer (FRET) reporter plasmid (45), were grown at 30°C to mid-exponential phase in tryptone broth, washed, attached to a round coverslip with polylysine, and mounted in a flow cell (73). The flow cell and all motility buffer test solutions (containing 10 mM Na lactate, 100 μM methionine, and various concentrations of serine) were maintained at 30°C throughout each experiment. Cells were illuminated at the cyan fluorescent protein (CFP) excitation wavelength, and light emission was detected at the CFP (FRET donor) and yellow fluorescent protein (YFP; FRET acceptor) wavelengths with photomultipliers. The ratio of YFP to CFP photon counts accurately reflects CheA kinase activity and changes in response to serine stimuli (44, 74). Fractional changes in kinase activity versus applied serine concentrations were fitted to a multisite Hill equation, yielding two parameter values: K1/2, the attractant concentration that inhibits 50% of the kinase activity, and the Hill coefficient, reflecting the extent of cooperativity of the response (45).
Whole-cell tethering.
Tethering was performed as described in V. Nieto’s thesis (24), based on prior publication of this method (75). Exponentially growing cells were pelleted and resuspended in motility buffer made of 10 mM potassium phosphate (pH 7), 67 mM NaCl, 10 mM sodium lactate, 0.1 mM disodium EDTA, and 0.001 mM l-methionine. They were transferred into a 1-ml sterile syringe connected by its needle (23 gauge) to an identical syringe through 6 to 8 in. of polyethylene tubing (inner diameter, 0.58 mm). Flagella were sheared by gently transferring cells between the two syringes 40 to 50 times with 1-min pauses after every 10 transfers. Forty microliters of the sheared cell suspension was loaded into a chamber created by stacking an 18- by 18-mm coverslip (treated with 0.1% [wt/vol] polylysine solution) over a 24- by 50-mm glass slide, separated by double-sided tape. Cells were incubated at room temperature for 10 min to allow them to attach to the coverslip. The chamber was gently washed 3 times with 40 μl of motility buffer to remove unattached cells. For the time course assay, cells were washed with motility buffer supplemented with 0.2% l-arabinose to induce YcgR expression from pVN5. This buffer supports protein expression from added inducers (76). Cells were observed through phase-contrast microscopy under an Olympus BH-2 microscope at ×40 magnification for a total duration of 45 min, and observations were recorded on an external Sony video recording device. Their rotation patterns were qualitatively categorized by playback of every 5-min interval.
Measurement of single motor rotation by the bead assay.
The bead assay was performed as described previously (26, 28) with modifications (27). E. coli HCB5 (77) expressing FliCsticky from plasmid pFD313 (78) was used for these experiments. Flagella were sheared and the cells were introduced into a chamber as described above. Attached cells were exposed to 40 μl of a 1:50 dilution of polystyrene beads (Polysciences, Warrington, PA; 0.75-μm diameter). The mixture was incubated at room temperature for 10 min to allow the beads to attach to the flagellar filaments. Wash steps were repeated to remove unattached beads, prior to introducing IPTG for induction of YgcR expression. High-speed videos of individual beads were captured and analyzed as described previously (27). Videos were processed using custom analytical programs within LabVIEW 2012 (National Instruments, Austin, TX), provided by Yuichi Inoue (Ishijima Lab, Osaka University, Osaka, Japan).
Isolation and sequencing of pseudoreverants.
The term “pseudorevertant” refers to a situation in which the original mutation remains but a second mutation restores the wild-type phenotype. Such revertants that regained enhanced motility in selected mutant backgrounds were isolated by inoculating the mutant strains in soft agar (swim) plates, allowing migration for an additional 8 h after a WT strain had covered a control plate. “Flares” or regions of enhanced motility could be observed on the periphery of motility haloes. These were purified on LB agar, individual colonies were repurified, their phenotype was reconfirmed, and their genomic DNA was subsequently analyzed by the Genomic Sequencing and Analysis Facility (GSAF) at the University of Texas at Austin. The HiSeq 4000 platform (PE 2 × 150 setup) was used, and data were analyzed using the breseq program (79).
Quantitation of intracellular c-di-GMP concentration.
All c-di-GMP quantifications were analyzed using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Cultures were grown to an OD600 of 0.6, and a 5-ml aliquot of culture was removed and centrifuged for 30 s at 12,000 rpm (15,294 relative centrifugal force [RCF]). The supernatant was immediately removed, and the pellet was resuspended in 100 μl of ice-cold extraction buffer (40% acetonitrile, 40% methanol, 20% water, 0.1 N formic acid) and incubated for 20 min at −20°C. The insoluble fraction was pelleted as described above in a benchtop centrifuge at 4°C for 5 min, and the supernatant was collected and stored at −80°C. Prior to mass spectrometry, the extraction buffer was evaporated using a SpeedVac, and the resulting pellet was resuspended in 100 μl of ultrapure water. Ten microliters of each sample was then analyzed on a Quattro Premier XE mass spectrometer coupled with an Acquity ultraperformance LC system as previously described (80). The intracellular concentration of c-di-GMP was calculated by dividing the intracellular c-di-GMP concentration of the sample by the total volume of the extracted bacteria, which was estimated by multiplying the number of bacterial cells in the extract by the average volume of the bacterial cell (1 × 10−15 liters), with no size differences observed between the strains tested. The average cellular volume for each strain was determined by measuring individual cell dimensions using differential image contrast microscopy and assuming cylindrical cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Blair for helpful comments on the manuscript and David Walker for assistance with whole-genome sequencing analysis.
This work was supported by NIH grant GM118085 and in part by Robert Welch Foundation grant F-1811 to R.M.H. J.S.P. is supported by NIH grant GM19559, and C.W. is supported by NIH grant GM109259.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahlstrom KM, O’Toole GA. 2017. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol 71:179–195. doi: 10.1146/annurev-micro-090816-093325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 4.Povolotsky TL, Hengge R. 2012. ‘Life-style’ control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J Biotechnol 160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 7.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol 188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol 303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Mariconda S, Suzuki A, McClelland M, Harshey RM. 2006. Uncovering a large set of genes that affect surface motility in Salmonella enterica serovar Typhimurium. J Bacteriol 188:7981–7984. doi: 10.1128/JB.00852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hengge R, Galperin MY, Ghigo JM, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 2016. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J Bacteriol 198:7–11. doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HK, Harshey RM. 2016. A diguanylate cyclase acts as a cell division inhibitor in a two-step response to reductive and envelope stresses. mBio 7:e00822-16. doi: 10.1128/mBio.00822-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 15.Berg HC. 2008. Bacterial flagellar motor. Curr Biol 18:R689–691. doi: 10.1016/j.cub.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 17.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 18.Kojima S, Blair DF. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 19.Yuan J, Berg HC. 2008. Resurrection of the flagellar rotary motor near zero load. Proc Natl Acad Sci U S A 105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet 3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Wang F, He R, Zhang R, Yuan J. 2018. The second messenger c-di-GMP adjusts motility and promotes surface aggregation of bacteria. Biophys J 115:2242–2249. doi: 10.1016/j.bpj.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 23.Zorraquino V, Garcia B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, Lasa I, Solano C. 2013. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J Bacteriol 195:417–428. doi: 10.1128/JB.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nieto V. 2013. The c-di-GMP binding protein, YcgR, is the primary inhibitor of motor function in Salmonella and Escherichia coli. PhD thesis. University of Texas, Austin, TX. [Google Scholar]
- 25.Silverman M, Simon M. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 26.Ryu WS, Berry RM, Berg HC. 2000. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403:444–447. doi: 10.1038/35000233. [DOI] [PubMed] [Google Scholar]
- 27.Partridge JD, Nieto V, Harshey RM. 2015. A new player at the flagellar motor: FliL controls both motor output and bias. mBio 6:e02367-14. doi: 10.1128/mBio.02367-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terasawa S, Fukuoka H, Inoue Y, Sagawa T, Takahashi H, Ishijima A. 2011. Coordinated reversal of flagellar motors on a single Escherichia coli cell. Biophys J 100:2193–2200. doi: 10.1016/j.bpj.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian S, Gao X, Dann CE III, Kearns DB. 2017. MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA in Bacillus subtilis. Proc Natl Acad Sci U S A 114:13537–13542. doi: 10.1073/pnas.1716231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker AE, Diepold A, Kuchma SL, Scott JE, Ha DG, Orazi G, Armitage JP, O'Toole GA. 2016. PilZ domain protein FlgZ mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J Bacteriol 198:1837–1846. doi: 10.1128/JB.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross P, Mayer R, Benziman M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol Rev 55:35–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J 32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. 2013. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 35.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22:2434–2446. doi: 10.1101/gad.475808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Preston JF III, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol 186:2724–2734. doi: 10.1128/jb.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr Top Microbiol Immunol 322:249–289. doi: 10.1007/978-3-540-75418-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klemm P. 1984. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Nucleotide sequence and primary structure of the protein. Eur J Biochem 143:395–399. doi: 10.1111/j.1432-1033.1984.tb08386.x. [DOI] [PubMed] [Google Scholar]
- 39.Gottesman S, Trisler P, Torres-Cabassa A. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol 162:1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrieres L, Aslam SN, Cooper RM, Clarke DJ. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153:1070–1080. doi: 10.1099/mic.0.2006/002907-0. [DOI] [PubMed] [Google Scholar]
- 41.Armstrong JB, Adler J, Dahl MM. 1967. Nonchemotactic mutants of Escherichia coli. J Bacteriol 93:390–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, Alexandre G. 2013. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 4:e00001-13. doi: 10.1128/mBio.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xin L, Zeng Y, Sheng S, Chea RA, Liu Q, Li HY, Yang L, Xu L, Chiam KH, Liang ZX. 2019. Regulation of flagellar motor switching by c-di-GMP phosphodiesterases in Pseudomonas aeruginosa. J Biol Chem 294:13789–13799. doi: 10.1074/jbc.RA119.009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sourjik V, Berg HC. 2002. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc Natl Acad Sci U S A 99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sourjik V, Vaknin A, Shimizu TS, Berg HC. 2007. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol 423:365–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 46.Ruiz N, Peterson CN, Silhavy TJ. 2001. RpoS-dependent transcriptional control of sprE: regulatory feedback loop. J Bacteriol 183:5974–5981. doi: 10.1128/JB.183.20.5974-5981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiino DR, Singer MS, Rothman-Denes LB. 1993. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. J Bacteriol 175:7081–7085. doi: 10.1128/jb.175.21.7081-7085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McPartland J, Rothman-Denes LB. 2009. The tail sheath of bacteriophage N4 interacts with the Escherichia coli receptor. J Bacteriol 191:525–532. doi: 10.1128/JB.01423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouche S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. 1998. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol 27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 50.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. 1996. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J 15:1333–1339. doi: 10.1002/j.1460-2075.1996.tb00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratt LA, Silhavy TJ. 1996. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci U S A 93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y, Gottesman S. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol 180:1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. 2001. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev 15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker G, Klauck E, Hengge-Aronis R. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc Natl Acad Sci U S A 96:6439–6444. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hengge R. 2008. The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv Exp Med Biol 631:40–53. doi: 10.1007/978-0-387-78885-2_4. [DOI] [PubMed] [Google Scholar]
- 56.Mika F, Hengge R. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev 19:2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson CN, Ruiz N, Silhavy TJ. 2004. RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J Bacteriol 186:7403–7410. doi: 10.1128/JB.186.21.7403-7410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Battesti A, Majdalani N, Gottesman S. 2015. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci U S A 112:5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreno M, Audia JP, Bearson SM, Webb C, Foster JW. 2000. Regulation of sigma S degradation in Salmonella enterica var typhimurium: in vivo interactions between sigma S, the response regulator MviA(RssB) and ClpX. J Mol Microbiol Biotechnol 2:245–254. [PubMed] [Google Scholar]
- 60.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. 2008. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol 68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 62.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato Y, Takaya A, Mouslim C, Hughes KT, Yamamoto T. 2014. FliT selectively enhances proteolysis of FlhC subunit in FlhD4C2 complex by an ATP-dependent protease, ClpXP. J Biol Chem 289:33001–33011. doi: 10.1074/jbc.M114.593749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomoyasu T, Ohkishi T, Ukyo Y, Tokumitsu A, Takaya A, Suzuki M, Sekiya K, Matsui H, Kutsukake K, Yamamoto T. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar typhimurium. J Bacteriol 184:645–653. doi: 10.1128/jb.184.3.645-653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saini S, Brown JD, Aldridge PD, Rao CV. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol 190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo S, Alshamy I, Hughes KT, Chevance FF. 2014. Analysis of factors that affect FlgM-dependent type III secretion for protein purification with Salmonella enterica serovar Typhimurium. J Bacteriol 196:2333–2347. doi: 10.1128/JB.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sperandio V, Torres AG, Giron JA, Kaper JB. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 183:5187–5197. doi: 10.1128/jb.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 71.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 72.Lai RZ, Parkinson JS. 2014. Functional suppression of HAMP domain signaling defects in the E. coli serine chemoreceptor. J Mol Biol 426:3642–3655. doi: 10.1016/j.jmb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berg HC, Block SM. 1984. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J Gen Microbiol 130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 74.Shimizu TS, Tu Y, Berg HC. 2010. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol 6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolfe AJ, Conley MP, Kramer TJ, Berg HC. 1987. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol 169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scharf BE, Fahrner KA, Turner L, Berg HC. 1998. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci U S A 95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuwajima G. 1988. Construction of a minimum-size functional flagellin of Escherichia coli. J Bacteriol 170:3305–3309. doi: 10.1128/jb.170.7.3305-3309.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol 79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.