Abstract
An oxygen gradient formed along the length of colonic crypts supports stem-cell proliferation at the normoxic crypt base while supporting obligate anaerobe growth in the anoxic colonic lumen. Primary human colonic epithelial cells derived from human gastrointestinal stem cells were cultured within a device possessing materials of tailored oxygen permeability to produce an oxygen-depleted luminal (0.8±0.1% O2) and oxygen-rich basal (11.1±0.5% O2) compartment. This oxygen difference created a stable oxygen gradient across the colonic epithelial cells which remained viable and properly polarized. Facultative and obligate anaerobes Lactobacillus rhamnosus, Bifidobacterium adolescentis, and Clostridium difficile grew readily within the luminal compartment. When formed along the length of an in vitro crypt, the oxygen gradient facilitated cell compartmentalization within the crypt by enhancing confinement of the proliferative cells to the crypt base. This platform provides a simple system to create a physiological oxygen gradient across an intestinal mimic while simultaneously supporting anaerobe co-culture.
Introduction
Humans have co-evolved with their gut microbiota in a symbiotic relationship essential for health, yet how these thousands of species influence human biology remains little understood.1,2 A number of diseases are associated with intestinal dysbiosis, as well as increased susceptibility to bacterial pathogens such as Clostridium difficile.3–6 With this growing awareness, numerous efforts are focused on the gut microbiome for its role in disease and its therapeutic potential.7–10 Despite the importance of the microbiome, the mechanisms underlying the observed disease associations remain obscure, in part because current approaches using fecal sample analysis and animal models have clear limitations. For example, fecal samples represent a limited population of the microbes in the intestine, omitting populations of microbes restricted to various regions and those known to be strictly adhered to the mucosa11 and crypts.12 Animal models are valuable tools for investigating the gut microbiota-host interaction; however, manipulating experimental parameters in animal subjects is difficult and expensive.13 Moreover, animals and humans possess different genetics, anatomy, physiology, metabolism and diet, all of which contribute to shaping the interaction of the host and the gut microbial composition.14 Accordingly, significant interest exists for improved in vitro models of the human gastrointestinal system, in particular models that support human-microbial co-culture.15–17
Co-culture is complicated by the rich luminal microbiota population, >90% of which are obligate anaerobes that die 30–60 min after exposure to room air.18,19 To support the survival of these anaerobes, the colon maintains a very low luminal oxygen tension (PO~0.1–1 mm Hg, <1% O2), yet provides adequate oxygen to support the growth and survival of the epithelial cells lining the intestinal lumen.20–23 In sharp contrast to the luminal microbes, the intestinal epithelial cells have a significant demand for oxygen due to their high turnover and substantial metabolic needs.24 Oxygen delivery to these cells is via an extensive vascular network in the lamina propria.25 Although difficult to accurately assess, the epithelial cells in direct contact with the colonic lumen are believed to be hypoxic (PO<10 mm Hg, 1.4% O2) while the microenvironment at the crypt base where stem and proliferative cells reside is borderline normoxic (PO~80 mm Hg, 5–20% O2).20–23 For these reasons, in vitro co-culture of strict anaerobes with human primary intestinal cells presents a challenge. Thus a simple, robust, and self-sustaining platform for the co-culture of colonic anaerobes over a human primary colonic epithelium is highly sought after to understand host/microbiota interactions.26
To experimentally manipulate and take advantage of the complex relationships responsible for the impact of the microbiome on the intestine as well as throughout the body, the ability to control the interplay between primary human intestinal epithelial cells and commensal gut microbiota must be greatly enhanced. However, current microbe-intestinal assay systems remain severely limited either due to the inability to use primary human intestinal cells or significant technical skills required for use of complex methods and devices.8,27 The anaerobic co-culture of gut microbiota with mammalian cells has typically employed human tumor cell lines as an epithelial mimic (e.g. Caco-2) since these tumor cells are rugged. The ‘Human-oxygen-Bacteria anaerobic’ (HoxBan) system consists of a 50-mL centrifuge tube in which Caco-2 cells on a glass coverslip are cultured in proximity to solid agar seeded with an anaerobic bacterium.26 While a simple system, the approach is not amenable to real-time screening and is short lived (<36 hours). Culture devices that attempt to produce an oxygen gradient across a commercial Transwell insert have also been reported.28–30 These systems are placed within an anaerobic chamber to produce anoxic conditions in the luminal compartment while oxygenated medium is supplied to the basal compartment. The oxygen profile in these devices is not stable over time without continuous perfusion.28,29 A new Transwell compatible system has been reported to solve this issue by perfusing oxygen through the basalcompartment.30 These systems have demonstrated co-culture of human colon cancer cells28,29 and human primary intestinal cells30 with obligate anaerobes, but require continual flow as well as placement of the device into an anaerobic chamber in order to maintain a stable oxygen gradient, thus greatly increasing complexity. Over the past few years, complex, multi-layered microfluidic devices have been used to create an oxygen gradient for anaerobic co-culture, again using Caco-2 cells as the human epithelial mimic.31–33 These devices rely on the perfusion of normally oxygenated and deoxygenated media in segregated compartments to maintain the co-culture environment. A similar strategy uses a Caco-2 tissue construct formed on the inner wall of a miniature cylinder-shaped silk scaffold while medium pre-equilibrated with an anaerobic gas mix is perfused inside the cylinder.34 While these systems generate an oxygen gradient, they require significant technical expertise and equipment to fabricate and maintain the flow systems. Other efforts to create an experimental platform with primary human cells have used explanted intestinal tissue, but rapid cell death and bacterial overgrowth have limited their use.35–37 In 2011, the demonstration of long-term human colonic stem-cell cultures as enclosed organoids embedded in Matrigel™ opened new possibilities for the study of the human gastrointestinal tract using primary human cells.38,39 For culture with bacteria, these systems require injection of microbiota through the surrounding hydrogel and into the enclosed lumen of individual organoids. The required technical skill, rapid bacterial overgrowth, and short-lived co-culture have severely limited this approach.40
Recent advances by our group have produced an epithelial mimic of colonic crypts by culture of primary human colonic stem cells on a shaped scaffold spanning a luminal and basal fluid compartments.41 Implementation of a chemical gradient across the 3D scaffold recapitulates the structure and polarization of the colonic crypts.41 The current work extends this platform to produce a simple in vitro culture system that creates and maintains a physiologic oxygen gradient with no requirement for an external anaerobic environment or flowing gas supply to the culture. Initial studies used a confluent human colonic epithelial monolayer to measure oxygen consumption and depletion from a luminal compartment surrounded by oxygen-impermeable walls. Oxygen passively diffusing into the basal compartment then enabled formation of an oxygen gradient across the epithelial cells. Numerous cellular properties including viability, proliferation, mucus production, and tight junction formation of the human colonic epithelial cells under the oxygen gradient were compared to that of cells under fully aerobic and anaerobic conditions. The ability to form an O2 gradient across a 3D in vitro crypt was then assessed as well as the impact of the formed oxygen gradient on cell compartmentalization within the crypt. Finally, co-culture of facultative and obligate anaerobes was demonstrated within the luminal compartment of the oxygen-gradient device in the presence of the cell monolayer and the in vitro crypts. This co-culture system has the potential to transform intestinal microbiome research by enabling the function of host-microbe interactions to be interrogated in a simple, bench-top system that can be readily adopted by any life science laboratory.
Methods
Fabrication of the threaded chamber and the plug
The cell culture inserts and the plugs were fabricated by milling polycarbonate stock using a CNC mill. The cell chamber was designed to have the same culture area as a Corning 12-well Transwell® insert. The cell chamber and the plug were threaded (M12 × 1.75) for efficient sealing of the luminalcompartment. For cell culture, a polytetrafluoroethylene (PTFE) porous membrane (BGCM00010; Millipore, Burlington, MA) was attached to the bottom of the cell chamber inserts using double-side medical tape (#1504XL, 3M) (Supplemental Fig. S1).
A collagen scaffold was prepared in the device prior to cell seeding as previously reported79 with slight modifications. First, collagen gel (250 μL) was formed on the porous membrane in the cell culture insert placed in a 12-well plate by mixing rat tail collagen I (3.32 mg/mL in 0.02 M acetic acid, Corning) with neutralization buffer (8.6 μM NaOH, 29 μM HEPES, 76 μM NaHCO3 in phosphate buffered saline (PBS)) at final concentration of 1 mg/mL and then incubating at 37°C for 1 h. Then the collagen gel was crosslinked from the bottom side by adding 0.5 mL PBS in the luminal compartment of the device and 1.5 mL of 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC): N-hydroxysuccinimide (NHS): 2-(N-morpholino)ethanesulfonic acid (MES) buffer (0.6 M EDC in water, 0.15 M NHS in water, 0.1 M MES in water, pH 5, 1:1:1 volume ratio) in the basal compartment of the well plate and incubating for 1 h at 25°C. Residual EDC and NHS in the collagen gel were removed by immersion in water for at least 16 h at 25°C. The resulting, partially crosslinked, collagen in the chamber was sterilized with 70% ethanol (5 min) and washed extensively with PBS prior to cell seeding.
To initiate the oxygen gradient, the oxygen impermeable plug was threaded into the cell chamber. When the plug was inserted, the air trapped between the plug and the medium was released through a hole in the plug. Then the hole was sealed with an Ethylene Propylene Diene Monomer (EPDM) rubber cap (0.508 mm). For re-use of the cell culture inserts and the plugs, the porous membrane was detached, and the parts were sonicated and washed with detergent in water, thoroughly rinsed with water, dried and then plasma treated for 5 min.
Power and statistical analyses
Power analysis80 was performed using G*Power applet81 (version 3.1.9.2) to calculate the minimum sample size required for distinguishing the cellular properties in an aerobic vs. anaerobic environment with the assumption that the cells under the oxygen gradient would behave in a similar manner. As preliminary data for this analysis, proliferative cell populations of human colonic epithelial cells grown on neutralized collagen in 24-well plate and exposed to either aerobic or anaerobic conditions were used. The cells were grown aerobically in expansion medium (EM, Table S1) for 4 days with the medium exchanged every 2 days then on day 4 the cells were either grown aerobically or anaerobically for 2 more days in EM. To label proliferative cells, EdU was pulsed on day 5. On day 6, the cells were fixed with 4% paraformaldehyde (PFA) and stained for EdU incorporation and with Hoechst 33342 as described below. The cells were imaged using a Nikon Eclipse TE300 inverted epifluorescence microscope with a Cy5 filter for EdU and DAPI filter for Hoechst 33342 observation. The mean of the EDU positive area normalized by Hoechst 33342 labeled area in aerobic and anaerobic conditions was 0.55 and 0.06 respectively and the standard deviation 0.044 and 0.029, resulting the sample size of 2. An α of 0.05 and power (1-β) = 0.95 was used for these calculations.
In all graphs, the data and error bars indicate the mean values and the standard deviation, respectively. A one-way ANOVA analysis was used for all statistical analyses except for the data summarized in Fig. 3C,D where a paired t-test was used to exclude batch-to-batch variability.
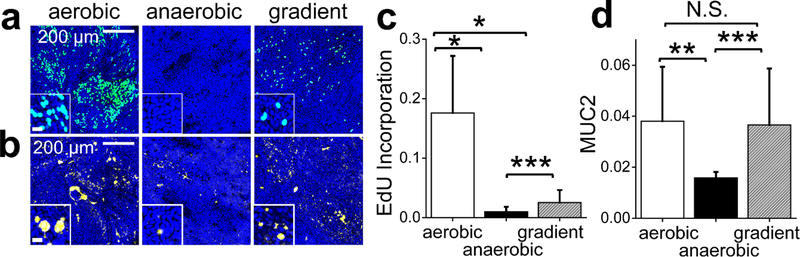
Figure 3.
Measurement of proliferation and goblet cell formation in the monolayer cells exposed to different oxygen environments. a) S-phase cells (aqua or blue-green) as measured by EdU pulse incorporation. Cells were counterstained with Hoechst 33342 (blue). Scale bar in the inset is 20 μm. b) Goblet cell MUC2 (yellow) as measured by immunofluorescence staining of MUC2. Scale bar in the inset is 20 μm. c) Measurement of S-phase cells. The Y axis shows the normalized EdU incorporation calculated by dividing the EdU-positive fluorescent area by the Hoechst 33342-positive area (N=9 independent samples). d) Measurement of MUC2 present within Goblet cells. The Y axis shows the normalized Muc2-positive area calculated by dividing the MUC2 immunofluorescent-positive area by the Hoechst 33342-positive area (N= 3 independent samples). *, **, *** indicate p<0.005, 0.01, or 0.05, respectively.
Human colonic epithelial cell culture
Human primary colon epithelial stem cells from two different sources were used in this study-one was obtained from a colonic biopsy specimen (52 years old male, Meadowmont Endoscopy Center at the University of North Carolina Hospital with consent of the patient [Institutional Review Board approval #14–2013]). The second source was from a cadaveric donor (23 years old male). The colonic crypts were isolated and expanded in EM on collagen gel in a 6-well plate as described previously.82 For routine maintenance, the cells were subcultured every 5–7 days in 6-well plate for up to 16 passages by digesting the underlying collagen with collagenase and then dissociation of the cells from the collagen by incubation with using 0.5 mM Ethylenediaminetetraacetic acid (EDTA) as described previously.82 Karyotyping of the cells at passage 7 and 15 confirmed that the chromosomes of the cells were normal.
For the experiments with the different oxygen environments in the cell culture inserts, the cells were passaged from a 6 well plate to the cell culture insert with collagen prepared as described above (from a 6 well plate to 6 inserts) and cultured for up to 10 days under aerobic conditions as follows. The human colonic stem cells were expanded to form a monolayer in expansion media (EM, Table S1, 0.5 mL placed in the luminal compartment and 2.5 mL placed into the basal compartment) for 6 days with medium exchange every other day until a confluent monolayer was formed. On day 6, the luminal medium was replaced with differentiation medium (DM, Table S1, 0.4 mL) and the basal medium was replaced with stem medium (SM, Table S1, 3 mL). Then the cells were incubated at 37°C for 4 days with medium replacement every other day. On day 10, the luminal plug was installed in the cell culture insert to initiate the oxygen gradient. The cells under the oxygen gradient were further incubated for 2 more days and assayed on day 12 as described below. Aerobic culture samples were prepared in the same manner except that the plug was not used to seal the luminal compartment. For anaerobic culture samples, the culture media was deoxygenated prior to anaerobic culture (<1% O2) and the media were replaced in an anaerobic chamber under 100% N2, or 5% CO2 + 95% N2. The cells were then placed in a glasslock container filled with an anaerobic gas mixture (5% CO2, 10% H2, 85% N2). The glasslock container also contained water to maintain a humidified environment to minimize evaporation of the cell media.
Oxygen sensing and transepithelial electrical resistance (TEER) measurement
The oxygen level was measured using a Microx4 oxygen sensor (PreSens, Germany) with a needle oxygen probe (PreSens, NTH-PSt7) according to the manufacturer’s instructions. To measure the oxygen level in the luminal compartment of the device, the oxygen probe was placed through the rubber cap used to seal the port in the polycarbonate plug so that the tip of the probe was located 2 mm above the surface of the cells. For the oxygen measurements in the basal compartment, a basal reservoir that has the same inner diameter (22.1 mm) and well volume (7 mL) as a 12-well plate was fabricated by milling a polycarbonate slab and then a hole was drilled into the side of the reservoir 7 mm above the well bottom so that the probe was immediately adjacent to the porous membrane (Supplemental Fig. S1). The oxygen probe was inserted through this hole that was then sealed with the rubber cap. For the oxygen level measurement of the basal medium, the cell culture insert was placed in the basal reservoir and the oxygen measurements were acquired every 5 min for 16 h. TEER was measured using a volt-ohmmeter (EVOM2, World Precision Instruments, FL) and a chopstick electrode. TEER was measured immediately after placing cells in an aerobic environment. For TEER measurement on the cells in the oxygen gradient condition, the plug was removed before TEER measurement. The cells in fully anaerobic condition were taken out of the anaerobic environment immediately before the TEER measurement.
Viability assay
Propidium iodide (PI, 2 μM) and Hoechst 33342 (12.5 μM) in PBS were added to the luminal compartmentof the cell cultures and incubated for 30 min to label dead cells (PI positive) and all cells (Hoechst 33342 positive). For detecting PI and Hoechst 33342-stained cells, the labeled monolayers were sampled with a biopsy tool (5 mm in diameter), transferred onto glass slides and the entire sample imaged with an Olympus Fluoview FV3000 confocal microscope (10× objective, 0.4 N.A.) with 561 nm and 405 nm lasers for excitation of PI and Hoechst 33342, respectively. Emissions at 610–710 nm and 430–470 nm were detected for PI and Hoechst 33342, respectively.
Pimonidazole staining
To assay intracellular oxygen depletion, cells were incubated for 2 h with pimonidazole hydrochloride (200 μM, Hypoxyprobe, HP1–100Kit) that was added to the basal compartment. The cells were fixed with 3% glyoxal in PBS (pH 5)83 for 20 min at −20°C. To permeabilize the cells, the sample was incubated for 10 min at 25°C with 0.5% Triton X-100 in PBS for 20 min at 25°C. The sample was then blocked using bovine serum albumen (BSA, 3% in PBS) for 1 h at 25°C. The samples were subsequently incubated with mouse antibody against pimonidazole (Hypoxyprobe, HP1–100Kit) diluted in 3% BSA (1:50) at 4°C for 15 h. The cells were washed with 3% BSA in PBS ×3 and then incubated with anti-mouse antibody conjugated with Alexa Fluor 647 (1:500, 715–605-150, Jackson ImmunoResearch) diluted in 3% BSA (1:500) for 1 h at 25°C. Finally, the cells were washed with 3% BSA in PBS ×2 and then washed with PBS. For imaging, the collagen gels with adherent cells were transferred onto glass slides and imaged using Olympus Fluoview FV3000 confocal microscope (640 nm excitation, emission 650 nm-750 nm). The fluorescence intensity of pimonidazole adduct as indicative of oxygen depletion was measured using Fiji.84 The amount of pimonidazole adduct formed was normalized to the total number of cells by dividing the pimonidazole adduct fluorescence intensity by the Hoeschst 33342 fluorescence intensity.
Immunofluorescence and EdU staining
The cells were fixed with 3% glyoxal solution for ZO-1 immunostaining. All other immunostaining employed fixation in 4% paraformaldehyde for 15–20 min. Cryosection samples were prepared by fixing the cells on collagen with 4% PFA, soaked with 30% sucrose in water for 1 day and then embedded in optimal cutting temperature (OCT) compound (Tissue-Tek O.C.T. Compound, VWR, Radnor, PA). The frozen tissue samples were sectioned into 10 μm thick slices using a cryostat and transferred onto glass slides. The fixed samples and the sectioned samples were permeabilized with 0.5% Triton X-100 in PBS for 20 min, blocked with 3% BSA in PBS for 1 h at 25°C and then incubated with primary antibody at 4°C for at least 15 h. Primary antibodies for MUC2 (Santa Cruz Biotechnology, SC-515032), ZO-1(Proteintech, 21733–1-AP), and ezrin (Thermo, PA5–17518) were diluted in 3% BSA at 1:200 and incubated with the samples at 4°C overnight. The samples were then washed with 3% BSA in PBS ×3 and incubated with fluorophore conjugated secondary antibody. Alexa Fluor 488 donkey anti-mouse (Jackson ImmunoResearch, 715–545-150) and Alexa Fluor 594 donkey anti-rabbit ReadyProbes (ThermoFisher scientific, R37119) were used for MUC2 and ZO-1 respectively at 1:100 dilution in 3% BSA in PBS with incubation for 1 h at 25°C. Hoechst 33342 (2 μM) was added with the secondary antibody to stain DNA. Finally, the cells were washed with 3% BSA ×2 and then with PBS.
5-ethynyl-2´-deoxyuridine (EdU) was used to stain for S phase (proliferative) cells following the manufacturer’s protocol (Click-iT EdU Alexa Fluor 647 Imaging kit, C10340, Thermo Fisher Scientific). The cells were incubated for 24 h in the presence of EdU (10 μM) in the basal medium. The cells were fixed with 4% paraformaldehyde for 15 min at 25°C, permeabilized for 20 min with 0.5% Triton X-100 in PBS. EdU incorporated into the cellular DNA was stained with Cy5 conjugated azide for 1 h at 25°C. For imaging, the samples were either taken with a biopsy punch (5 mm) or picked up with forceps and then transferred on glass slides and imaged using an Olympus Fluoview FV3000 confocal microscope (Exc. 405 nm, 488 nm, 561 nm, 640 nm lasers for Hoechst 33342, Alexa Fluor 488, Alexa Fluor 594, Cy5, respectively). For quantification, the fluorescent area of each stain above an empirically set threshold was calculated using Fiji image processing software.84 To normalize the measurement to the total number of cells present, the fluorescence-positive area was divided by the Hoechst fluorescence area i.e. area above an empirically set threshold.
Bacterial co-culture
L. rhamnosus GG (LGG) (ATCC 53103, obtained from Microbiologics, 01090P) was grown from a single colony in De Man, Rogosa and Sharpe (MRS) broth under aerobic conditions in a tissue culture incubator (5% CO2, 69.3% N2, 18.6% O2) at 37°C. C. difficile 630Δerm was grown from a single colony in brain heart infusion medium (BHIS) supplemented with 5% w/v yeast extract broth in an anaerobic chamber (Coy Laboratories, Grass Lake, MI, 5% CO2, 10% H2, 85% N2). B. adolescentis Reuter ATCC 15703 (obtained from ATCC) was grown in MRS broth supplemented with 5% L-cysteine in an anaerobic environment using the BD GasPak EZ Gas Generating System. All bacterial cultures were initiated one day before co-culture.
For LGG co-culture, the human colonic epithelial cells were grown as described above for 10 days prior to co-culture except that on day 8, the culture medium was replaced with antibiotic free medium. On day 10, the medium was replaced with fresh antibiotic-free medium, and the epithelial cell culture placed into the oxygen gradient device with the oxygen impermeable plug installed. Aerobic control samples were prepared at the same time, but without installation of the plug to leave the luminal fluid compartment open to air. The cells were incubated for 1 day followed by addition of the bacterial sample to the luminal compartment. On day 11, harvested LGG were diluted in MRS broth at 1:1000, and 5 μL of the diluted LGG sample was inoculated in the luminal compartment, resulting in 3×105 colony forming unit (CFU)/mL in the luminal compartment. For the oxygen gradient samples, the rubber cap in the plug was removed, then LGG was inoculated through the port, and the rubber cap was placed again to seal the luminal space. After 24 h, the supernatant was collected and the number of viable LGG in the supernatant was estimated by plating serial dilutions of a known volume onto MRS agar plates. To estimate the number of LGG that adhered to the human cells, the epithelial cells cultured in the presence of the bacteria were washed with PBS to remove loosely attached bacteria. The epithelial monolayer and underlying collagen scaffold were then transferred to a tube and incubated with collagenase for 10–15 min at 37°C. The solution was then vortexed vigorously ×3 and then serial dilutions plated on MRS agar plates to quantify the number of CFU/mL.
For epithelial cell and C. difficile co-culture, the human colonic epithelial cells were grown for 10 days as described above, then transferred into an anaerobic chamber for further manipulation. For samples cultured in the oxygen-gradient device and an anaerobic environment, the medium in the luminal compartments was replaced with antibiotic-free deoxygenated DM (<0.5% O2), and the plug was installed in an anaerobic chamber. The medium within the basal compartment was replaced with normoxic SM in both anaerobic and the oxygen gradient conditions. For the fully anaerobic co-culture samples the basal media was pre-incubated in an anaerobic environment prior to co-culture. Inoculation of C. difficile was performed within an anaerobic chamber. C. difficile were placed into BHIS medium and 4 μL (1×104 CFU/mL) was inoculated into the luminal compartments (0.4 mL), through the port in the plug for the oxygen-gradient gradient device or directly into the luminal compartment for the cells to be cultured under an anaerobic condition. Then the oxygen-gradient device with epithelial and bacterial cells was removed from the anaerobic chamber and incubated in a standard tissue-culture incubator (5% CO2, 69.3% N2, 18.6% O2) at 37°C. The control sample to remain anaerobic was retained within the anaerobic chamber and incubated at 37°C in a glasslock container with a humidified environment. At 6 and 24 h, the contents of the luminal chamber (supernatants, collagen scaffold and epithelial cells) were placed into a tube and incubated with collagenase for 15 min at 37°C. The bacterial suspension was then serially diluted in BHIS medium and plated onto BHIS agar for enumeration of C. difficile.
For B. adolescentis co-culture with the in vitro crypts, the crypts were prepared as described above (7 days in EM then 4 days in DM/SM) except the last medium replacement on day 9 was with antibiotic-free medium. On day 11, the oxygen gradient was generated by placing deoxygenated DM (<0.5% O2) in the luminal compartment followed by installation of the plug while the device was within an anaerobic chamber (5% CO2, 95% N2). Normoxic SM was loaded into the basal compartment. Aerobic crypt-bacteria co-cultures were prepared in the same way except normoxic DM was used and the plug was not installed so that the luminal fluid remained open to the surrounding environment. The luminal compartment of both systems was inoculated with 2.5×104 CFU of B. adolescentis followed by placement of the devices in a standard tissue-culture incubator (5% CO2, 69.3% N2, 18.6% O2) at 37°C. After 24 h of co-culture, the luminal medium containing B. adolescentis was collected and serial dilutions were plated on MRS agar plates in anaerobic chamber BD GasPak EZ Gas Generating System for colony enumeration.
Scanning electron microscopy (SEM)
The epithelial cells without bacteria and the co-culture samples were fixed with 4% paraformaldehyde for 20 min and 40 min, respectively. The samples were washed with PBS, then dehydrated by incubation in solutions with gradually increasing ethanol to water ratios until 100% ethanol was attained. The samples were dried with a critical point dryer (Autosamdri®−931, Toursimis, MD) coated with 15 nm of platinum using a sputter coater (Cressington, UK) and observed by SEM (Hitachi S-4700). The contrast of the image was enhanced using the Enhance Local Contrast (CLAHE) plugin on the Fiji image processing package.84
In vitro crypt formation and culture
The shaped collagen scaffold used for crypt formation was prepared as described previously.41 Briefly, a PTFE membrane was attached to the cell culture device designed for the oxygen gradient using double sided medical tape as described above. Then a COC film with a 3 mm circular opening at the center was attached under the porous membrane. Rat tail collagen (5 mg/mL in 0.1 M MES buffer) was mixed with EDC and NHS at a final concentration of 60 mM and 15 mM, respectively. The collagen mixture (50 μL) was pipetted into the culture insert and the collagen was molded into a crypt shape using a PDMS stamp. The PDMS stamp possessed a 10×10 micropillar array with each micropillar 125 μm diameter (at the largest width), 500 μm length, and a distance between the micropillars of 200 μm. Molding was accomplished in a pressurized chamber filled with N2 gas for 1 h. The stamp was then removed leaving a microwell array with each microwell of similar length and diameter to in vivo human crypts. The shaped collagen scaffold was soaked in water for 16 h to remove excess cross-linking reagents and then sterilized in 70% ethanol and washed with PBS. Human collagen I (10 μg/mL, VitroCol®, Advanced BioMatrix, CA) was coated on the shaped scaffold to enhance cell adhesion. Human primary colon epithelial cells were then plated on the collagen scaffold and cultured in EM for 7 days. The medium was exchanged every other day. On day 7, the growth factor gradient was formed by changing the luminal and basal media to DM (without the growth factors) and SM (with the growth factors), respectively. The cells were grown in the growth factor gradient for 4 days with the medium exchanged every other day. On day 11, the oxygen gradient was generated by placing the plug into the luminal compartment as described above. Corresponding aerobic samples were prepared at the same time by omitting the plug. The cells were then cultured for 2 more days with DM and SM in the luminal and basal compartments, respectively. An EdU incorporation assay was performed in the same way as for the monolayers described above by adding EdU (10 μM) to the basal medium for 24 h. The tissue constructs were then fixed on day 12 with 4% paraformaldehyde for 20 min at 25°C and EdU incorporation measured as described above. For assay of pimonidazole adduct formation, the cultures were continued to day 13 when pimonidazole hydrochloride (200 μM, Hypoxyprobe) was added to the basal medium followed by incubation for 2 h. The cells were fixed on day 13 with 3% glyoxal fixative for 20 min at 25°C and adduct formation assayed as described above.
Results
1. Design and simulation of the oxygen-gradient device
The device incorporated an oxygen-impermeable luminal plug, a culture insert supporting the cells on their scaffold and enclosing the luminal compartment, and a basal compartment into which oxygen diffused passively. The device possessed dimensions similar to that of a commercial Transwell (Fig. 1A). This format was employed due to its familiarity to the research community, its ease of use, and the accessibility of the basal and luminal compartments for sampling and reagent addition. The gas impermeable plug inserted into the luminal compartment blocked oxygen influx from above the cell monolayer as the cells consumed oxygen. The threaded plug was fabricated from polycarbonate due to its low oxygen permeability, transparency, and ease of fabrication by milling. A port in the plug enabled gas release during sealing, sampling of the luminal contents, addition of reagents or bacteria, and measurement of oxygen saturation. During cell culture, the port was sealed with a rubber cap. A thin porous membrane formed the oxygen permeable base of the polycarbonate device insert. A collagen-based hydrogel scaffold (~2 mm thick) was formed on the porous membrane to support the epithelial cell monolayer. The thick collagen scaffold was preferred over a simple porous membrane because when properly shaped the collagen scaffold supports formation of either a cell monolayer or a fully polarized in vitro crypt.41 The wells of a 12-well microtiter plate formed the basal compartment. The basal compartment below the monolayer was supplied with oxygen by leaving the fluid in this compartment in contact with the aerobic atmosphere. This system design was chosen to permit development of an anaerobic luminal compartment by minimizing the influx of oxygen into the luminal compartment and utilizing cell respiration to scavenge oxygen molecules existing within or entering the luminal compartment. It was reasoned that supply of oxygen passively through the basal compartment would then lead to development of a self-sustaining oxygen gradient over time.
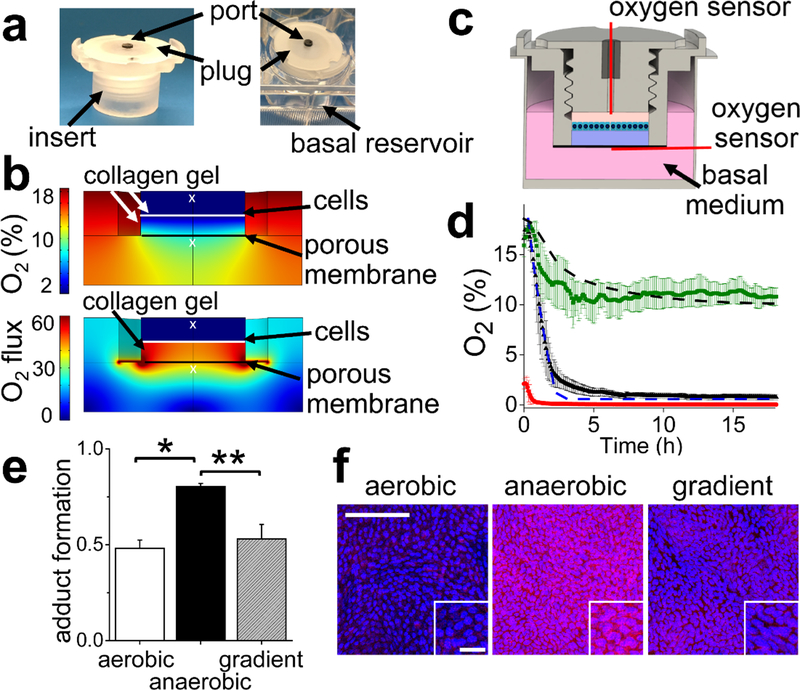
Figure 1.
Design and testing of the oxygen gradient device. a) An image of the device insert (left panel) and the assembled device (insert plus basal compartment, right panel). The insert possessed a gas-impermeable threaded plug to isolate the luminal compartment from the external atmosphere. A port in the plug was used for addition of reagents and measurement of the oxygen saturation. A single well of a 12-well plate was used as the basal reservoir. b) The simulated oxygen saturation (top panel) and oxygen flux (lower panel) in the device at 24 h after the plug was placed. The white “×”s indicate the location of the oxygen probe in the experimental measurements. The Y axis units for O2 flux is (mol/m2∙s). c) Schematic of a coronal section through the device. The red lines indicate the locations of the oxygen probes during the experimental measurements. d) The measured and simulated luminal and basal oxygen profiles over time. The dashed black line and green squares represent the simulated and average measured, basal oxygen saturation, respectively, when the basal and luminal compartments are initially filled with normoxic medium. The dashed blue line and black triangles represent the simulated and average measured, luminal oxygen saturation, respectively, when the luminal and basal compartments are initially filled with normoxic medium. The red circles represent the average oxygen saturation in the luminal compartment when it is initially filled with deoxygenated medium. 3 independent samples were used for all measured values and the error bars depict a single standard deviation of the data points. e) The formation of pimonidazole adducts plotted against the oxygen culture condition. The Y axis displays the normalized adduct formation calculated as the intensity of immunofluorescence staining of the adducts divided by the fluorescence intensity of the Hoechst 33342 dye (N=3 independent samples for all conditions). * and ** indicate p<0.001 or 0.005, respectively. f) Representative images of pimonidazole adduct formation (red) as detected by immunofluorescence staining in aerobic, anaerobic and the oxygen-gradient environments. Hoechst 33342 fluorescence is shown in blue. Scale bar in the large image is 100 μm and in the inset 20 μm.
The oxygen concentration over time within the device in the presence of cells was modeled using a COMSOL simulation. Critical to precision was accurate, experimentally measured values for the oxygen diffusion coefficient within the collagen hydrogel and the oxygen consumption rate (OCR) of primary intestinal epithelial cells. The oxygen diffusion coefficient in collagen was calculated by fitting the time varying oxygen concentration measured within a thick collagen gel to Fick’s second law of diffusion (Supplementary Methods). The collagen slab was initially deoxygenated and then placed into contact with fully oxygenated medium. The oxygen diffusion coefficient in the collagen gel was 1.2±0.1×10−9 m2/s, which is lower than the previously reported oxygen diffusion coefficient in water (3×10−9 m2/s).42 The OCR of human primary colon epithelial cells was calculated by measuring the oxygen concentration over time within the medium in a closed culture vessel containing a cell monolayer on a collagen gel (Supplementary Methods). The experimental measurements were fit to the integrated Michaelis-Menten kinetic equation to obtain a maximum OCR of 14±4 amol/cell∙s at 37°C, which is within an order of magnitude of previously reported values measured for various adherent human cells (1.5–120 amol/cell∙s).43 For Caco-2 cells, the mitochondrial respiration rates of undifferentiated44 (70 amol/cell·s) and semi-differentiated (33 amol/cell·s) 45 cells are within the order of magnitude of the OCR measured for the primary intestinal cells. The difference in cell type (cancer cell line vs. primary cells), differentiation state, and culture medium may contribute to the measured difference in metabolism and OCRs of the various cell types. The measured OCR in this work represents both mitochondrial respiration and glycolysis since the measurement was performed under typical culture conditions, i.e. extracellular matrix and intestinal culture medium. This OCR value is therefore expected to closely resemble that of cells in the described intestinal monolayers and in vitro crypts providing an accurate value for the simulation.
To predict whether an oxygen gradient could be generated in the device using epithelial cell respiration, the oxygen level and flux in the device over time was simulated (Fig. 1B). In the simulation, the walls of the cell culture insert, the plug, and the well plate were modeled as oxygen impermeable since they were comprised of polycarbonate (culture insert, plug) or polystyrene (multiwell plate). Free oxygen diffusion was assumed through porous membrane below the collagen scaffold. The model suggested that luminal hypoxia (<1% O2) could be attained within 3 h after the plug was installed in the device while the oxygen level below the collagen gel would decrease over time reaching a steady state at ~10% O2 (Fig. 1B, supplementary movies 1 and 2). In this simulation, the oxygen gradient across the cell layer and collagen gel (2 mm thick) was sustained over time due to oxygen consumption by the cells suggesting the feasibility of forming an oxygen gradient across the cells and scaffold without the need to flow gas or media into the device.
2. Experimental measurement of the oxygen gradient formed by an epithelial monolayer in the device
The simulated oxygen gradient was experimentally validated by forming a confluent monolayer of primary human epithelial cells across the collagen scaffold (see Methods). The luminal medium was then replaced with oxygenated medium (18.6% O2) or deoxygenated medium (<1% O2) followed by placement of the luminal plug. For both conditions, the basal medium was initially oxygenated (18.6% O2), and the entire device was placed into a standard tissue culture incubator (5% CO2, 69.3% N2, 18.6% O2) at 37°C. The oxygen saturation in both the luminal and basal compartments was measured over time and compared to that of the model (Fig. 1C,D). When the luminal medium was not deoxygenated in advance of device setup, the oxygen saturation in the luminal compartment decreased at a rate of 8.8 ± 0.5% O2/h (from 15 min to 2 h) after installation of the plug. By 2, 3, and 6.5 h after plug insertion, the oxygen saturation was 2.2 ± 0.7%, 1.3 ± 0.4%, and 0.8 ± 0.1%, respectively. When deoxygenated medium was placed at time zero into the luminal compartment, the luminal oxygen level reached a steady state level at 2 h of 0.07 ± 0.02% at 2 h. After placement of the device into the tissue culture incubator, the oxygen saturation of the basal medium also decreased at the rate of 4.1 ±1.1% O2/h, but reached a stable value of 11.1± 0.5% O2 at 4 h. The oxygen saturation decrease in the basal compartment was most likely due to the lack of mixing in the basal compartment with consequent establishment of an oxygen gradient across the liquid medium in the basal compartment itself. When O2 gradients in the basal compartment were eliminated by stirring the medium, the oxygen saturation was stable at 19.1±0.7%. These data matched that predicted by the model and demonstrate that an oxygen gradient was formed across the epithelial cell monolayer. Cellular respiration combined with oxygen-impermeable walls depleted oxygen in the luminal compartment, while the basal compartment remained oxygenated due to exposure of the fluid to the ambient environment.
The presence of pimonidazole adducts, which form on thiols of cellular proteins in the presence of low oxygen saturation (<10 mm Hg or <1.4% O2 under the current conditions), was assessed.46,47 Cells cultured in an anaerobic medium displayed significantly greater adduct formation than those in oxygenated medium (Fig. 1E,F). Cells cultured in the oxygen gradient possessed a level of adduct formation that was not significantly different from that of the cells in the oxygenated medium, but significantly lower than that of the cells under anaerobic conditions. These data suggest that the cells under the oxygen gradient experienced an oxygen influx sufficiently high to minimize the pimonidazole adduct formation in contrast to that in fully anaerobic conditions (Fig. 1B). These data indicate that an oxygen gradient can be created and maintained across the human intestinal epithelial cells by limiting oxygen influx from the luminal face. Because the cells are supplied with oxygen from the basal compartment, the cells were not anoxic under these conditions.
3. Characterization of cell physiology in the oxygen gradient device
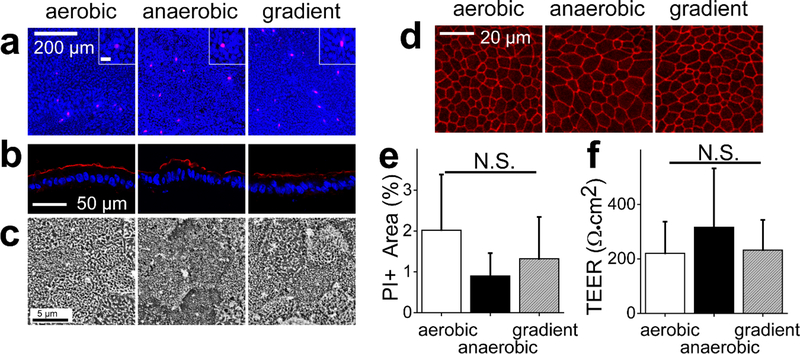
Intestinal epithelial cells, in contrast to epithelial cells derived from other organs, maintain their barrier integrity despite their hypoxic microenvironment.48,49 Indeed, a recent report demonstrated that hypoxia might improve barrier function in intestinal epithelium.50 However, these observations were made using colon cancer cell lines, which can respond to environmental stimuli quite differently than primary cells. To assess the response of human primary intestinal cells to different oxygen levels, cells were cultured aerobically to produce a confluent monolayer on the collagen scaffold and then were placed either under fully aerobic conditions, fully anaerobic conditions, or an oxygen gradient. After 2 days, >95% of the cells were viable without a significant difference between the 3 oxygen conditions (Fig. 2A,E). In vivo, intestinal epithelial cells are polarized, exhibiting distinct luminal and basal surface, and this polarization is critical for proper function such as water, salt, and metabolite transport.51 Ezrin, a brush border protein enriched within the luminal microvilli of absorptive colonocytes, was present at high density on the luminal aspect of the epithelia under all three culture conditions (Fig. 2B). Microvilli on the luminal cell aspect were also clearly visible by electron microscopy suggesting that polarized colonocytes readily formed under the different conditions (Fig. 2C). Colonic epithelial cells form tight intercellular junctions to protect against luminal content leakage into the underlying tissue. The tight junction marker ZO-1 was appropriately localized at intercellular interfaces under all 3 oxygen conditions, suggesting that the cells formed an impermeable monolayer (Fig. 2D). The transepithelial electrical resistance (TEER) for cells under the varying oxygen conditions did not differ significantly (Fig. 2F). The TEER values of the epithelial cell monolayers were consistent with those published for in vivo human colon tissue (100–400 Ω·cm2),52,53 unlike the values for Caco-2 tumor-cell monolayers (>1000 Ω·cm2).32,53,54 These data suggest that the oxygen gradient did not impair the ability of primary human intestinal cells to form a tight barrier when cultured as a monolayer.
Figure 2.
Monolayer cell properties in different oxygen environments. a) Representative fluorescence images of monolayers stained with PI (magenta) and Hoechst 33342 (blue). Scale bar in the inset is 20 μm. b) Representative images of the human colonic epithelial cells cryosectioned and immunostained for ezrin (red: ezrin, blue: Hoechst 33342). d) Representative SEM images of the human colonic epithelial cells in the various oxygen conditions. d) Representative images of ZO-1 immunofluorescence (red) in the human colonic epithelial cells in aerobic, anaerobic, and oxygen-gradient environments. e) Measurement of dead cells under the various oxygen environments. The Y-axis is the PI-positive area divided by the Hoechst 33342 positive area. The data were obtained from 3 independent experiments. f) Monolayer TEER in the various oxygen environments. The aerobic conditions utilized 21 independent samples while the for anaerobic condition and oxygen gradient conditions used 20 independent samples
Oxygen influences growth and fate decisions in various stem-cell niches including embryonic, mesenchymal, hematopoietic, and neural stem cells,55,56 but oxygen’s influence on the proliferation and formation of differentiated intestinal epithelial cells is not known. To evaluate the influence of oxygen depletion on proliferation, cells were pulsed with EdU for 24 h and then assayed for EdU incorporation into DNA. Cells under the oxygen gradient or anaerobic conditions demonstrated significantly lower incorporation of EdU relative to the cells under a fully aerobic environment (Fig. 3A,C). The decreased proliferation in the absence of a high oxygen saturation is consistent with the behavior of other stem cells57 and colon cancer cells,58 but contrary to the increased proliferation of mesenchymal or neural stem cells in hypoxic conditions.55 This result may suggest that the observed decreased proliferation in response to oxygen depletion may be an attribute of the intestinal epithelium. Differentiation to mucus-producing goblet cells was detected by immunofluorescence using antibodies specific to mucin2 (MUC2). The expression of MUC2 was not statistically different for the cells cultured in a fully aerobic environment or under the oxygen gradient, but MUC2 was significantly decreased in cells cultured in the absence of oxygen (Fig. 3B,D). This is consistent with the reduced goblet cell population in ulcerative colitis, where lowered blood oxygen level (hypoxemia) causes an inflammatory hypoxia.59,60 Thus oxygen saturation clearly impacts cell proliferation as well as the ability to form the differentiated cell types.
4. Co-culture with a facultative anaerobe
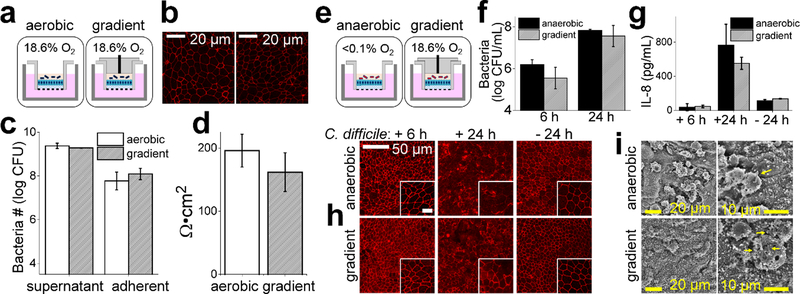
To understand whether the luminal environment of the gradient device was able to support the growth of colonic bacteria, the facultative anaerobe Lactobacillus rhamnosus GG (LGG) was co-cultured with human colonic epithelial cells and compared to co-cultures under fully aerobic conditions. LGG was selected since it is one of the most common constituents of probiotics. LGG (1.5×105 colony forming units, CFU) were inoculated into the luminal chamber above the epithelial cell monolayer. After co-culture for 24 h, the number of viable LGG under the oxygen gradient increased 15,000 fold which was not significantly different from the increase observed for bacteria in the fully aerobic environment (Fig. 4C), consistent with previous reports that LGG exhibits similar growth in aerobic and low oxygen conditions.61 Since LGG binding to human cells is suggested to mediate the probiotic effect of LGG,62 the association of LGG with the epithelial cell surface was examined in the presence and absence of the oxygen gradient. Surface-adherent LGG were assessed after removal of the luminal medium containing the non-adherent LGG. There was no significant difference in the percentage of viable LGG bound to human cells within the fully aerobic (3.6% ± 3.8) and oxygen gradient (6.8% ± 4.0) co-cultures. The proportion of adhered LGG was similar to that observed in prior reports of adhesion to Caco-2 tumor cells.63 Immunostaining of the tight junction protein ZO-1 revealed the expected patterning with intense, but contiguous, staining at the cell-cell borders (Fig. 4B). After 24 h of culture, the TEER of the monolayers under the aerobic and the oxygen gradient conditions was not significantly different suggesting that the barrier function of the monolayer cultured in the presence of LGG was not disrupted (Fig. 4D), and that on these timescales the impact of the nearby bacteria on the epithelial cells was minimal.
Figure 4.
Co-culture of anaerobic bacteria with the epithelial cell monolayers under differing oxygen conditions. a) Schematic diagrams of the co-culture of LGG with the epithelial cell monolayer. The left panel shows the normoxic or oxygenated conditions in which the fluid in both the luminal and basal reservoirs is exposed to the oxygen environment of a standard tissue culture incubator. The right panel illustrates the oxygen gradient environment in which only the fluid in the basal reservoir is exposed to the external environment. b) Immunofluorescence staining of ZO-1 in the cells co-cultured with LGG for 24 h aerobically (left) and under the oxygen gradient (right). c) Shown is the number of viable LGG bacteria in either the supernatant (luminal reservoir) or adhered to the human cells at 24 h (N=3 technical replicates). d) TEER of the epithelial monolayers after 24 h of co-culture with LGG (N=3 technical replicates. e) Schematic diagrams of the co-culture of C. difficile with the epithelial monolayer. The left panel shows the anoxic setup in which the fluid in both the luminal and basal reservoirs is exposed to the de-oxygenated environment of an anaerobic chamber. The right panel illustrates the oxygen gradient environment in which the fluid in the basal reservoir is exposed to the oxygen environment of a standard tissue culture incubator. f) Growth of C. difficile after 6 h and 24 h of co-culture with the epithelial cell monolayer from 3 independent samples for each condition. g) [IL-8] secreted by the epithelial cells into the basal reservoir with (+) and without (−) C. difficile co-culture after 6 and 24 h. from 3 independent samples for each condition except for the control samples without C. difficile in which 2 independent samples were used. h) Immunofluorescence staining of ZO-1 in the human colonic epithelial cells in the absence and presence of C. difficile for 6 or 24 h. Scale bar in the inset is 20 μm. i) SEM images of the human colonic epithelial cell monolayer after 24 h of C. difficile co-culture under the different oxygen environments. The yellow arrows highlight individual rod-shaped C. difficile.
5. Co-culture with the obligate anaerobe, Clostridium difficile (C. difficile)
C. difficile (strain 630Δerm) was chosen as a model obligate anaerobe due to its growing clinical significance in causing intestinal disease when gut microbiota is disrupted by antibiotic treatment or other means. After establishing the epithelial monolayer, the luminal reservoir of the oxygen-gradient device was filled with deoxygenated medium (<1% O2), the device was moved into an anaerobic chamber, and C. difficile at ~5×103 CFU was added to the luminal chamber. The device was then placed in a standard tissue-culture incubator at 37°C. As a positive control for C. difficile growth, co-culture samples were prepared similarly, but the device was placed into an anaerobic chamber. At 6 h and 24 h post-inoculation, bacteria were collected and C. difficile were enumerated. C. difficile expanded 40-and 4000-fold after 6 h and 24 h of co-culture, respectively. There was no significant difference between the number of C. difficile within the anaerobic and oxygen-gradient co-culture at either the 6 h or 24 h time post-inoculation (Fig. 4F). Thus, C. difficile anaerobic bacterial growth was supported in the luminal microenvironment created on the oxygen-gradient device by the colonic epithelial cell monolayer in combination with the gas-impermeable luminal reservoir walls.
The toxicity of C. difficile is largely due to production of the toxins TcdA and TcdB, which act to impair epithelial barrier function, as well as by inciting inflammatory responses.64–66 In particular, C. difficile infection in vivo or addition of the above toxins to cell cultures increases secretion of inflammatory cytokines such as IL-8 from the basal face of the colonic epithelium.67,68 After culture of the epithelium under oxygen-gradient conditions in the presence or absence of C. difficile for 24 h, epithelial cells co-cultured with C. difficile secreted significantly more IL-8 than cultures without the bacteria (Fig. 4G). Epithelial monolayers cultured in the presence of C. difficile under the oxygen gradient or within a fully anaerobic environment demonstrated no significant difference in the amount of IL-8 secreted. Epithelial cells in both culture conditions increased their IL-8 production as the exposure time to C. difficile increased (Fig. 4G).
A major mechanism of action of the C. difficile toxins is cytoskeletal disruption through glycosylation and inactivation of the Rho family of GTPases, which interferes with tight junction assembly.65,66 To investigate whether tight junction disruption occurred in the model epithelium and whether different oxygen conditions might alter this process, human intestinal epithelial cells were co-cultured with C. difficile either under the oxygen gradient or anaerobic conditions, and then assessed for cellular morphology and ZO-1. At 6 h post-inoculation, ZO-1 was properly localized to tight junctions without obvious disruption under either condition (Fig. 4H), similar to the ZO-1 localization in the fully oxygenated cultures (Fig. 2D). However, after 24 h in the presence of C. difficile, the ZO-1 localization in the monolayers cultured in an anaerobic environment was irregular and without clear demarcation of the intercellular borders, suggesting that the tight junctions might be disrupted (Fig. 4H).69–71 Cells under the oxygen gradient with C. difficile showed mixed ZO-1 localization with some ZO-1 staining clearly defined, but other areas with patchy localization (Fig. 4H). When viewed by SEM, the cells in a fully anaerobic environment with the bacterium often adopted a rounded morphology with exposed collagen scaffold in many locations suggesting cell death and detachment from the scaffold (Fig. 4I). Cells cultured under the oxygen-gradient with C. difficile predominantly exhibited a cobblestone morphology with few apparent rounded cells and much less exposed scaffold. When non-adherent bacteria were removed, significantly more C. difficile remained attached to the epithelial cell surface in the oxygen gradient condition (4.2 ± 3.6 cells/100μm2) compared to that in the anaerobic condition (0.14 ± 0.16 cells/100 μm2). These data demonstrate that the oxygen gradient formed in the device support growth of the obligate anaerobic bacteria C. difficile, which mediated secretion of the inflammatory cytokine IL-8, disruption of tight junctions and alteration of cell morphology.
6. Application of an oxygen gradient along the length of an in vitro crypt.
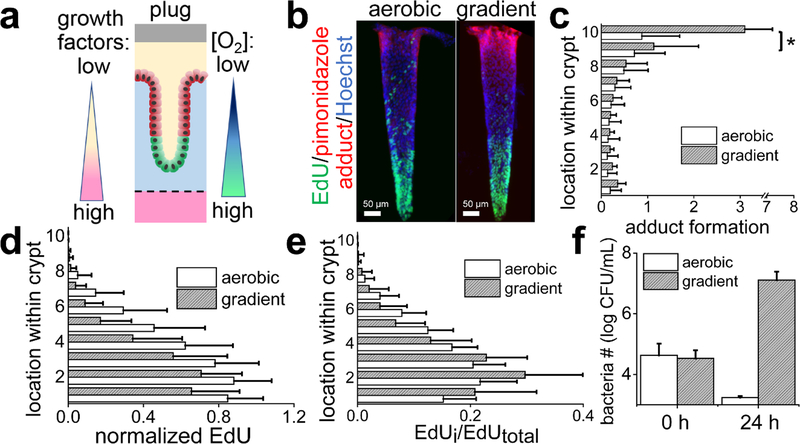
An oxygen gradient was applied across the long axis of 3D in vitro crypts to recreate the oxygen profile thought to exist in vivo, i.e. an oxygenated stem cell niche with a nearby hypoxic differentiated cell zone.41 In this model system, chemical gradients of growth factors along the in vitro crypt axis induce cell compartmentalization with a stem/proliferative zone at the crypt base and differentiated cells localized to the luminal surface of the tissue (Fig. 5A).41 To evaluate the influence of the oxygen gradient on cell compartmentalization, in vitro crypts were formed as previously described41 and then cultured within the oxygen-gradient device or under fully oxygenated conditions for an additional 2 days. Both oxygenation environments demonstrated higher pimonidazole adduct formation at the luminal tissue surface and minimal formation at the crypt base (Fig. 5B); however, adduct formation on the luminal surface of crypts under the oxygen gradient was significantly greater than that for crypts without an imposed oxygen gradient (Fig. 5C). This pattern is consistent with that formed within crypts in vivo.72 The gradient in pimonidazole adduct formation observed in the crypts that were under fully oxygenated conditions suggests that the luminal-facing differentiated cells themselves locally depleted oxygen even when in contact with the oxygenated luminal reservoir (Fig. 5B). Interestingly the stem/proliferative cells at the crypt base did not exhibit pimonidazole adducts despite being a greater distance from an oxygenated medium relative to the differentiated cells. A likely explanation is the metabolic shift that occurs between stem cells and differentiated cells. A shift in metabolism from aerobic glycolysis (less oxygen consumption) to oxidative phosphorylation (greater oxygen demand) occurs in many cell types73–75 as they move from a proliferative, less differentiated state to a non-proliferative, mature phenotype. Intestinal epithelial cells undergo a similar metabolic shift as the stem and transit-amplifying cells become differentiated.76,77 The differentiated cells with their higher oxygen consumption would then create a greater local oxygen depletion along the luminal crypt surface than their counterpart proliferative, undifferentiated cells occupying the crypt base.
Figure 5.
Application of an oxygen gradient across an in vitro crypt. a) Schematic diagram of a single in vitro crypt with application of a chemical and oxygen gradient. Stem/proliferative cells are depicted in green while the differentiated cells in red. A single layer of cells lines the surface of the crypt-shaped scaffold. b) Whole-mount fluorescence images of a side view of two in vitro crypts. In the left panel, the crypt under a chemical gradient, but no applied oxygen gradient, i.e aerobic conditions. In the right panel, the crypt under both a chemical and oxygen gradient. The crypts were stained for pimonidazole adducts (red), EdU incorporation (green), and Hoechst 33342 (blue). c) The normalized pimonidazole adduct formation plotted against the location of the cells along the crypt long axis under different oxygenation conditions. The Y-axis shows the cell location with 0 being the base of the crypt (i.e. basal side) and 10 representing the top of the crypt at the luminal reservoir. Total crypt length was 600 μm. A total 25 individual crypts from 3 different arrays were analyzed for each condition. * represents p<0.01. d) The normalized EdU incorporation plotted against cell location along the crypt axis for different oxygen environments. The normalized EdU incorporation was calculated by dividing the EdU fluorescence intensity by the Hoechst 33342 fluorescence intensity. The aerobic and gradient data were significantly different (p<0.01) for all distances between 0 and 0.8 of the crypt length. A total of 30 (for aerobic) and 32 (for gradient) individual crypts from 3 different arrays were analyzed. e) The relative distribution of the EdU-labeled cells with respect to the location of the cell along the crypt axis. Three different arrays were analyzed for the aerobic (30 crypts total) and the gradient (32 crypts total) samples. Between 0–0.2 and 0.4–0.7 of the crypt length, the p value for the data comparisons p<0.05. f) Growth of B. adolescentis at 0 and 24 h of culture above the in vitro crypts under the fully oxygenated or oxygen gradient environment from 2 technical replicates for the aerobic condition and 3 for the gradient condition.
When grown as a monolayer, the intestinal epithelial cells respond to an anaerobic environment by decreasing cell proliferation (Fig. 3A,C). To assess how this might impact cell compartmentalization within the platform, in vitro crypts (with growth factor gradients) were cultured for 2 days under an oxygen-gradient or in an oxygenated environment and then pulsed with EdU for 24 h. In both conditions, the EdU-labeled (S-phase) cells were located near the base of the crypt (Fig. 5B,D,E). However, significantly fewer proliferative cells were present within the crypts under the oxygen gradient compared to that in the fully oxygenated system (Fig. 5D). Additionally, the proliferative cells under the oxygen gradient were located closer to the crypt base than that under the fully oxygenated condition. When the total area of the crypt positive for EdU incorporation was summed, a significantly greater fraction of the EdU positive crypt resided in the lower 1/3 of the crypt (73 ± 17%) for the oxygen-gradient samples relative to that in the fully oxygenated environment (57 ± 13 %) (Fig. 5E). Imposition of an oxygen gradient across the crypt facilitated compartmentalization of the crypt cells enhancing confinement of the proliferative cells to the crypt base as well as reducing their numbers.
Finally, to determine whether luminal growth of colonic bacteria might be supported within the oxygen-gradient device, the 3D crypts were co-cultured with the obligate anaerobe Bifidobacterium adolescentis. In contrast to C. difficile, B. adolescentis is a nonpathogenic, commonly used probiotic.78 B. adolescentis was co-cultured for 24 h in the luminal reservoir under an oxygen-gradient or fully oxygenated conditions. The number of viable B. adolescentis increased >100-fold after 24 h of co-culture in the oxygen-gradient device, while only 3% survived under the fully oxygenated conditions (Fig.5F). This clearly demonstrates the importance of the oxygen gradient in the co-culture of obligate anaerobic bacteria in the 3D crypt model.
Discussion
In this work we demonstrated a simple method for generating a physiologic oxygen gradient across a primary human colonic epithelium that enabled its co-culture with obligate anaerobic bacteria while the platform was housed under normal atmospheric conditions. By using oxygen impermeable materials to block gas diffusion in the luminal reservoir combined with oxygen consumption of the epithelial cells, the oxygen gradient was formed in less than 2 h. The lack of continuous perfusion of either a liquid or gas simplifies the system making it attractive and accessible to a broad group of users. Oxygen depletion did not impair viability, polarization, or epithelial integrity of the primary human colonic epithelial cells on the times scales measured. In the presence of an oxygen gradient the luminal compartment possessed a sufficiently low oxygen saturation to support obligate anaerobe growth. When formed across 3D in vitro crypts, the applied oxygen gradient assisted cell localization to distinct zones enhancing confinement of the proliferative cells to the crypt base. This suggests that disruption of the oxygen gradient during breach of the colonic epithelium may support barrier repair by enabling proliferative cells to form further up the crypt long axis and more rapidly replace damaged luminal cells. The luminal hypoxia achieved in the oxygen gradient supported the growth of facultative and obligate anaerobic bacteria above both 2D and 3D epithelial formats. C. difficile-induced monolayer damage was decreased in the presence of the oxygen gradient relative to a fully anaerobic epithelial monolayer although it is unclear whether this difference is due to altered microbe-cell interactions or an oxygen-susceptible behavior of C. difficile. The ability to co-culture of pathogenic anaerobes as well as commensal bacteria in proximity to an in vitro crypt will enable novel investigations into the interplay of gut microbes and primary human intestinal epithelial cells. This concept of generating oxygen gradient can be expanded to other tissue models when the same direction of oxygen gradient is desired. The self-forming oxygen gradient cassette offers a simple, bench-top platform with both luminal and basal access to yield a close mimic of the physiological oxygen microenvironment in the colon with support of physiologic and pathogenic bacterial residents of the colonic lumen.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health under R01DK109559 awarded to N.L.A., and R01AI107029 to R.T. We thank Scott Magness’ Laboratory for kindly providing human colon specimens. We thank Sam Hinman and Mallory Maurer for technical assistance. This work was performed in part at the Chapel Hill Analytical and Nanofabrication Laboratory, CHANL, a member of the North Carolina Research Triangle Nanotechnology Network, RTNN, which is supported by the National Science Foundation, Grant ECCS-1542015, as part of the National Nanotechnology Coordinated Infrastructure, NNCI.
Footnotes
Competing interests
Y.W., C.E.S., and N.L.A. have a financial interest in Altis Biosystems. The other authors have no conflicts.
References
- 1.Blaser MJ The microbiome revolution. J Clin Invest 124, 4162–4165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint HJ, Scott KP, Louis P. & Duncan SH The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9, 577–589 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Aw W. & Fukuda S. Understanding the role of the gut ecosystem in diabetes mellitus. Journal of diabetes investigation 9, 5–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaner O. et al. The Gut Microbiome Profile in Obesity: A Systematic Review. International journal of endocrinology 2018, 4095789–4095789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, Covington A. & Pamer EG The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunological reviews 279, 90–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickard JM, Zeng MY, Caruso R. & Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunological reviews 279, 70–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aitoro R. et al. Gut Microbiota as a Target for Preventive and Therapeutic Intervention against Food Allergy. Nutrients 9, E672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum HE The human microbiome. Adv Med Sci 62, 414–420 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Brunkwall L. & Orho-Melander M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia 60, 943–951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane ER, Zisman TL & Suskind DL The microbiota in inflammatory bowel disease: current and therapeutic insights. J Inflamm Res 10, 63–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg PB et al. Diversity of the Human Intestinal Microbial Flora. Science 308, 1635 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SM et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner PV The role of the gut microbiota on animal model reproducibility. Animal Model Exp Med 1, 109–115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen TL, Vieira-Silva S, Liston A. & Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech 8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ et al. The human microbiome project. Nature 449, 804–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirzaei MK & Maurice CF Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15, 397–408 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Kang M. & Martin A. Microbiome and colorectal cancer: Unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Seminars in Immunology 32, 3–13 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Loesche WJ Oxygen sensitivity of various anaerobic bacteria. Appl Microbiol 18, 723–727 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen-Vercoe E. Bringing the gut microbiota into focus through microbial culture: recent progress and future perspective. . Curr Opin Microbiol 16, 625–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espey M. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radical Biol Med 55, 130–140 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Albenberg L. et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063. e1058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeitouni NE, Chotikatum S, von Köckritz-Blickwede M. & Naim HY The impact of hypoxia on intestinal epithelial cell functions: consequences for invasion by bacterial pathogens. Molecular and cellular pediatrics 3, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L, Kelly CJ & Colgan SP Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. American Journal of Physiology-Cell Physiology 309, C350–C360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward JB, Keely SJ & Keely SJ Oxygen in the regulation of intestinal epithelial transport. The Journal of physiology 592, 2473–2489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kachlik D, Baca V. & Stingl J. The spatial arrangement of the human large intestinal wall blood circulation. Journal of Anatomy 216, 335–343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadabad MS et al. A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci Reports 5:17906 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritz JV, Desai MS, Shah P, Schneider JG & Wilmes P. From meta-omics to causality: experimental models for human microbiome research. Microbiome 1, 14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulluwishewa D. et al. Live Faecalibacterium prausnitzii in an apical anaerobic model of the intestinal epithelial barrier. Cellular Microbiology 17, 226–240 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Maier E, Anderson RC & Roy NC Live Faecalibacterium prausnitzii Does Not Enhance Epithelial Barrier Integrity in an Apical Anaerobic Co-Culture Model of the Large Intestine. Nutrients 9, 1349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fofanova TY et al. A novel human enteroid-anaerobe co-culture system to study microbial-host interaction under physiological hypoxia. bioRxiv, 555755 (2019). [Google Scholar]
- 31.Shah P. et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nature Communications 7, 11535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin W. et al. A Robust Longitudinal Co-culture of Obligate Anaerobic Gut Microbiome With Human Intestinal Epithelium in an Anoxic-Oxic Interface-on-a-Chip. Frontiers in Bioengineering and Biotechnology 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jalili-Firoozinezhad S. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature Biomedical Engineering (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W. et al. Multifunctional Bioreactor System for Human Intestine Tissues. ACS Biomaterials Science & Engineering 4, 231–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grivel JC & Margolis L. Use of human tissue explants to study human infectious agents. Nat Protoc 4, 256–269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haller D. et al. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut 47, 79–87 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsilingiri K. et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex-vivo organ culture model. Gut 61, 1007–1015 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Jung P. et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med 17, 1225–1227 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Sato T. & Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Williamson IA et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cellular and Molecular Gastroenterology and Hepatology 6, 301–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y. et al. Formation of Human Colonic Crypt Array by Application of Chemical Gradients Across a Shaped Epithelial Monolayer. Cellular and Molecular Gastroenterology and Hepatology 5, 113–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchwald P. FEM-based oxygen consumption and cell viability models for avascular pancreatic islets. Theoretical Biology and Medical Modelling 6, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner BA, Venkataraman S. & Buettner GR The rate of oxygen utilization by cells. Free radical biology & medicine 51, 700–712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decleer M. et al. Oxygen Consumption Rate Analysis of Mitochondrial Dysfunction Caused by Bacillus cereus Cereulide in Caco-2 and HepG2 Cells. Toxins 10, 266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.JanssenDuijghuijsen LM et al. Mitochondrial ATP Depletion Disrupts Caco-2 Monolayer Integrity and Internalizes Claudin 7. Frontiers in Physiology 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gross MW, Karbach U, Groebe K, Franko AJ & Mueller‐Klieser W. Calibration of misonidazole labeling by simultaneous measurement of oxygen tension and labeling density in multicellular spheroids. International journal of cancer 61, 567–573 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Varia MA et al. Pimonidazole: a novel hypoxia marker for complementary study of tumor hypoxia and cell proliferation in cervical carcinoma. Gynecologic oncology 71, 270–277 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Furuta GT et al. Hypoxia-inducible factor 1–dependent induction of intestinal trefoil factor protects barrier function during hypoxia. Journal of Experimental Medicine 193, 1027–1034 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colgan SP, Dzus AL & Parkos CA Epithelial exposure to hypoxia modulates neutrophil transepithelial migration. Journal of Experimental Medicine 184, 1003–1015 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muenchau S. et al. Hypoxic environment promotes barrier formation in human intestinal epithelial cells through regulation of miRNA-320a expression. Molecular and Cellular Biology, MCB. 00553–00518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneeberger K, Roth S, Nieuwenhuis EE & Middendorp S. Intestinal epithelial cell polarity defects in disease: lessons from microvillus inclusion disease. Disease models & mechanisms 11, dmm031088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carra GE, Ibanez JE & Saravi FD Electrogenic transport, oxygen consumption, and sensitivity to acute hypoxia of human colonic epithelium. International journal of colorectal disease 26, 1205–1210 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Srinivasan B. et al. TEER measurement techniques for in vitro barrier model systems. J Lab Autom 20, 107–126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HJ, Huh D, Hamilton G. & Ingber DE Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a chip 12, 2165–2174 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Mohyeldin A, Garzón-Muvdi T. & Quiñones-Hinojosa A. Oxygen in Stem Cell Biology: A Critical Component of the Stem Cell Niche. Cell Stem Cell 7, 150–161 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Nombela-Arrieta C. & Silberstein LE The science behind the hypoxic niche of hematopoietic stem and progenitors. ASH Education Program Book 2014, 542–547 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Hubbi ME & Semenza GL Regulation of cell proliferation by hypoxia-inducible factors. American Journal of Physiology-Cell Physiology 309, C775–C782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaidi A, Williams AC & Paraskeva C. Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nature cell biology 9, 210–217 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Gersemann M. et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation 77, 84–94 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Tsujij M. et al. Colonic mucosal hemodynamics and tissue oxygenation in patients with ulcerative colitis: Investigation by organ reflectance spectrophotometry. Journal of Gastroenterology 30, 183–188 (1995). [DOI] [PubMed] [Google Scholar]
- 61.Zotta T. et al. Assessment of Aerobic and Respiratory Growth in the Lactobacillus casei Group. PLOS ONE 9, e99189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishiyama K. et al. Adhesion properties of Lactobacillus rhamnosus mucus-binding factor to mucin and extracellular matrix proteins. Bioscience, biotechnology, and biochemistry 79, 271–279 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Lebeer S. et al. Functional Analysis of Lactobacillus rhamnosus GG Pili in Relation to Adhesion and Immunomodulatory Interactions with Intestinal Epithelial Cells. Applied and Environmental Microbiology 78, 185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young VB Old and new models for studying host-microbe interactions in health and disease: C. difficile as an example. American journal of physiology. Gastrointestinal and liver physiology 312, G623–g627 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Nusrat A. et al. Clostridium difficile Toxins Disrupt Epithelial Barrier Function by Altering Membrane Microdomain Localization of Tight Junction Proteins. Infection and Immunity 69, 1329–1336 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aktories K, Schwan C. & Jank T. Clostridium difficile Toxin Biology. Annual Review of Microbiology 71, 281–307 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Yu H. et al. Cytokines Are Markers of the Clostridium difficile-Induced Inflammatory Response and Predict Disease Severity. Clinical and vaccine immunology : CVI 24, e00037–00017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahida YR, Makh S, Hyde S, Gray T. & Borriello SP Effect of Clostridium difficile toxin A on human intestinal epithelial cells: induction of interleukin 8 production and apoptosis after cell detachment. Gut 38, 337–347 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janvilisri T, Scaria J. & Chang Y-F Transcriptional Profiling of Clostridium difficile and Caco-2 Cells during Infection. The Journal of Infectious Diseases 202, 282–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaban L. et al. A 3D intestinal tissue model supports Clostridioides difficile germination, colonization, toxin production and epithelial damage. Anaerobe 50, 85–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leslie JL et al. Persistence and Toxin Production by Clostridium difficile within Human Intestinal Organoids Result in Disruption of Epithelial Paracellular Barrier Function. Infection and Immunity 83, 138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly CJ et al. Fundamental role for HIF-1α in constitutive expression of human β defensin-1. Mucosal Immunology 6, 1110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, Nuebel E, Daley George Q., Koehler Carla M. & Teitell Michael A. Metabolic Regulation in Pluripotent Stem Cells during Reprogramming and Self-Renewal. Cell Stem Cell 11, 589–595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tormos Kathryn V. et al. Mitochondrial Complex III ROS Regulate Adipocyte Differentiation. Cell Metabolism 14, 537–544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heiden MGV, Cantley LC & Thompson CB Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fan YY et al. A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. American journal of physiology. Gastrointestinal and liver physiology 309, G1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Litvak Y, Byndloss MX & Bäumler AJ Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimamura S. et al. Relationship Between Oxygen Sensitivity and Oxygen Metabolism of Bifidobacterium Species. Journal of Dairy Science 75, 3296–3306 (1992). [DOI] [PubMed] [Google Scholar]
- 79.Gunasekara DB et al. A Monolayer of Primary Colonic Epithelium Generated on a Scaffold with a Gradient of Stiffness for Drug Transport Studies. Analytical Chemistry 90, 13331–13340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cohen, J. (Hillsdale, NJ: erlbaum, 1988).
- 81.Faul F, Erdfelder E, Lang A-G & Buchner A. G* Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior research methods 39, 175–191 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Wang Y. et al. Self-renewing Monolayer of Primary Colonic or Rectal Epithelial Cells. Cell Mol Gastroenterol Hepatol 4, 165–182.e167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Richter KN et al. Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy. The EMBO journal 37, 139–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.