Significance
Single-cell transcriptional profiling has become a widespread tool in cell identification, particularly in the nervous system, based on the notion that genomic information determines cell identity. However, many cell-type classification studies are unconstrained by other cellular attributes (e.g., morphology, physiology). Here, we systematically test how accurately transcriptional profiling can assign cell identity to well-studied anatomically and functionally identified neurons in 2 small neuronal networks. While these neurons clearly possess distinct patterns of gene expression across cell types, their expression profiles are not sufficient to unambiguously confirm their identity. We suggest that true cell identity can only be determined by combining gene expression data with other cellular attributes such as innervation pattern, morphology, or physiology.
Keywords: qPCR, RNA-seq, stomatogastric, expression profiling
Abstract
Understanding circuit organization depends on identification of cell types. Recent advances in transcriptional profiling methods have enabled classification of cell types by their gene expression. While exceptionally powerful and high throughput, the ground-truth validation of these methods is difficult: If cell type is unknown, how does one assess whether a given analysis accurately captures neuronal identity? To shed light on the capabilities and limitations of solely using transcriptional profiling for cell-type classification, we performed 2 forms of transcriptional profiling—RNA-seq and quantitative RT-PCR, in single, unambiguously identified neurons from 2 small crustacean neuronal networks: The stomatogastric and cardiac ganglia. We then combined our knowledge of cell type with unbiased clustering analyses and supervised machine learning to determine how accurately functionally defined neuron types can be classified by expression profile alone. The results demonstrate that expression profile is able to capture neuronal identity most accurately when combined with multimodal information that allows for post hoc grouping, so analysis can proceed from a supervised perspective. Solely unsupervised clustering can lead to misidentification and an inability to distinguish between 2 or more cell types. Therefore, this study supports the general utility of cell identification by transcriptional profiling, but adds a caution: It is difficult or impossible to know under what conditions transcriptional profiling alone is capable of assigning cell identity. Only by combining multiple modalities of information such as physiology, morphology, or innervation target can neuronal identity be unambiguously determined.
Unambiguous classification of neuronal cell types is a long-standing goal in neuroscience with the aim to understand the functional components of the nervous system that give rise to circuit dynamics and, ultimately, behavior (1–6). Beyond that, agreement upon neuronal cell types provides the opportunity to greatly increase reproducibility across investigations, allows for evolutionary comparisons across species (7, 8), and facilitates functional access to, and tracking of, neuron types through developmental stages (9). To this end, attempts at defining neuronal identity have been carried out using morphology, electrophysiology, gene expression, spatial patterning, and neurotransmitter phenotypes (10–18). Since the earliest efforts to capture the transcriptomes of single neurons, using linear or PCR amplification of messenger RNA (mRNA) followed by either cDNA library construction (19) or microarray hybridization (10, 20, 21), single-cell RNA sequencing (scRNA-seq) (22) has become the method of choice for many genome-scale investigations into neuron cell type. Advances in microfluidics, library preparation, and sequencing technologies have propelled an explosion of molecular profiling studies seeking to use unique gene expression patterns to discriminate neuronal types from one another, whether for discovery of new types or further classification of existing ones (23–36).
Molecular profiling approaches to tackle the problem of neuronal cell identity have many advantages: First, single-cell transcriptomic data contain thousands of measurements in the form of gene products that can be used both in a qualitative (in the form of marker genes) and quantitative (in the form of absolute transcript counts) manner (6). Second, scRNA-seq allows for very high-throughput processing of samples with hundreds, if not thousands, of single cell transcripts simultaneously using barcoding techniques (37). Third, these techniques can be applied to species that lack well-annotated transcriptomic information, as the cost to generate de novo reference transcriptomes has decreased dramatically in recent years (38). Even the sequencing of heterogeneous tissues from the central nervous system (CNS) can be used in conjunction with predictive modeling to reconstruct markers for major classes of CNS cell types, as has been done with oligodendrocytes, astrocytes, microglia, and neurons, in both humans and mice (39). Classifying neurons into different major categories (such as excitatory vs. inhibitory, parvalbumin+ vs. parvalbumin−, etc.) using qualitative expression measures is an easier task than quantitative approaches that separate neurons into smaller subclasses, but runs into limitations as to how far further classification can proceed. Subclasses of neuron types likely require greater depth of sequencing to resolve, and these neurons are more likely to be defined by the expression of multiple genes rather than unique markers (40). Yet this also is an inherent limitation of scRNA-seq: Low abundance transcripts are often missed or inaccurately classified as differentially expressed (41), and methods to dissociate and isolate cells can alter their transcriptomic profiles before they are even measured (42, 43).
There have now been many studies seeking to determine how many transcriptomically defined cell types might be present in a given part of the brain. For instance, an initial study of the cell-type diversity of the mouse primary visual cortex revealed 42 neuronal and 7 nonneuronal cell types (29). More recent work from the same group identified 133 transcriptomic cell types (44). Work in the retina has led the way as an example of generating a cell-type consensus with an unknown endpoint. Multimodal information of retinal ganglion cell properties, including morphology, physiology, gene expression, and spatial patterning, has converged on over 65 cell types in the macaque fovea and peripheral retina (45). However, not all systems have the same technical advantages as the retinal ganglion cells (such as uniform spatial patterning) that can be indicative of cell type, and multimodal information can be more difficult to obtain than high-throughput transcriptomic profiling methods. Therefore, the reliability of transcriptomic profiling with respect to neuronal identity requires additional evaluation.
In this study, we validate and compare transcriptional profiling via scRNA-seq and quantitative RT-PCR (qRT-PCR) methods, using supervised and unsupervised analyses, in 2 model systems in which neurons are unambiguously identified based on electrophysiological output, synaptic connectivity, axonal projection, and innervation target: The stomatogastric (STG) and cardiac ganglia (CG) of the crab, Cancer borealis. This approach allows us to test directly how much of the known functional and anatomical identity of a neuron is captured in the transcriptomic profile of single neurons within a given network.
Results
Molecular Profiling of Single Identified STG and CG Neurons by RNA-Seq.
Because of their large individual cell body size and our ability to manually collect single identified STG neurons (Fig. 1), we generated transcriptomes for pyloric dilator (PD; n = 11), gastric mill (GM; n = 11), lateral pyloric (LP; n = 8), and ventricular dilator (VD; n = 8) neurons by typical library preparations rather than more automated procedures such as Drop-seq, Split-seq, or 10× Genomics (46). Sequencing data were mapped to the C. borealis nervous system transcriptome (47). After removing transcripts for which there was no expression in any cell type, the dataset contained 28,459 distinct contigs (contiguous sequences) in the complete RNA-seq dataset. These contigs represent more than the full set of genes transcribed in these cells, as multiple contigs may map to a single gene but during transcriptome assembly the intervening sequence could not be resolved to assemble these distinct fragments (see ref. 48). We began our analysis of these data using unbiased hierarchical clustering methods, as is commonly done. Using the complete dataset (referred to as “all expressed contigs”), hierarchical clustering (with data centered and scaled across contigs) resulted in 5 clusters (Fig. 2A) that appeared not to segregate by cell type. One exception was observed among PD cells. All but 2 PD cells fell within 1 distinct cluster, albeit with a GM cell also identified in this cluster (Fig. 2A). While not surprising, the complete cellular transcriptome on its own does not distinguish cell types.
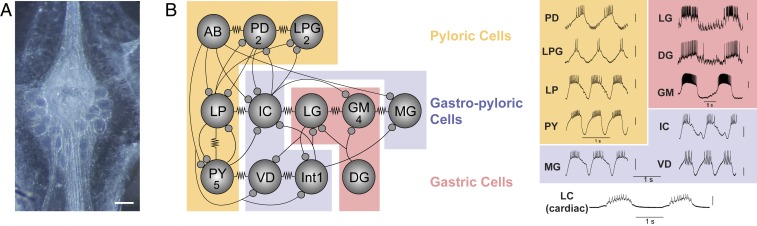
Fig. 1.
(A) Photomicrograph of the stomatogastric ganglion. (Scale bar, 200 μm.) (B) Circuit map of the STG. The STG contains 12 cell types that innervate the pylorus and gastric mill of the crab stomach. These cells are individually identifiable, and their chemical (closed circles) and electrical (resistor symbols) synaptic connections are all known. We used 10 of these 12 STG cell types (not AB or Int1) for this study, as well as motor neurons of the cardiac ganglion as an outgroup for comparison. Example traces were taken from intracellular recordings of each of the 11 identified neuron types used in this study. Neurons are involved in 3 different networks/circuits in the crab, C. borealis: the pyloric network (anterior burster (AB), PD, LPG, LP, and PY; orange box), the gastric network (LG, DG, and GM; red box) and the cardiac ganglion network (Bottom). Note the time scale difference in the long-lasting bursts of the gastric cells (red box) relative to the pyloric cells (orange box). Some neurons (interneuron 1 (INT1), IC, VD, and MG) participate in both gastric and pyloric network activity and are noted in the purple box. LC motor neurons of the cardiac ganglion are used as an “outgroup” to compare expression patterns of motor neurons from a distinct ganglion (cardiac ganglion). Each of the representative recordings is independent as an example of individual cell output, and simultaneous network activity is not plotted here. Thus, none of the phase relationships of these units within their respective rhythms is implied in any of the recordings.
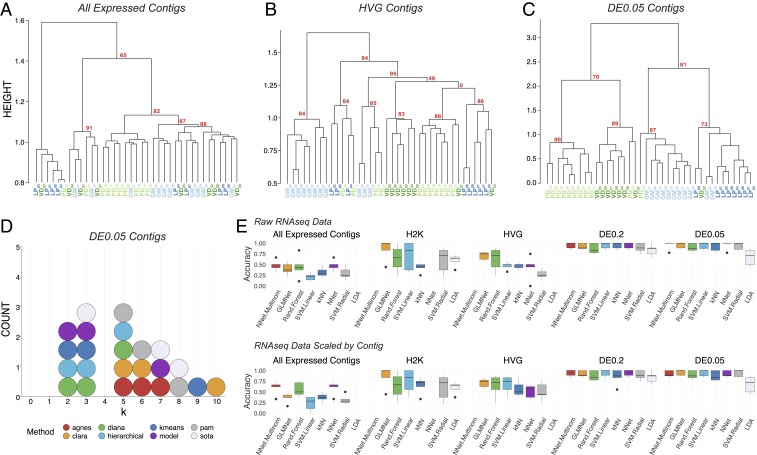
Fig. 2.
Post hoc recapitulation of cell identity via single-cell RNA-seq with hierarchical clustering and sML algorithms. (A) Hierarchical clustering of cell type with correlation as the distance metric, Ward.D2, as the clustering method, and data centered and scaled by contig for all expressed contigs, (B) HVG dataset, and (C) DE contigs at the q < 0.05 level. Each cell type is color coded, and AU P values are noted for each of the major nodes. Cells are identified by type (LP, PD, GM, VD) and a subscript that denotes a unique sample identifier. (D) Dotplot of the top 3 predicted number of clusters (k values) for 8 algorithms. None of these algorithms correctly predicted the expected 4 distinct clusters that would represent the 4 different cell types in this assay. (E) Accuracy (proportion of correctly identified cells) of cell-type prediction using 8 different methods of sML (GLM, kNN, NN, MNN, RF, SVML, SVMR, and LDA) for each of the datasets. Box and whisker plots show the efficacy of these methods to recapitulate cell identity from these 2 sets of contigs as estimated by cross-validation (5 folds). To assess the efficacy of these methods on the full RNA-seq dataset, we used PCA for dimensionality reduction (i.e., >28,000 contigs to 38 PCs) while retaining 99% of the variance. Results are shown for raw data (Top row) and data scaled across contigs (Bottom row).
We identified and extended our unbiased analysis to the most variably expressed genes in the RNA-seq dataset. The first subset represents the top 2,000 most variable contigs (referred to as the “2000 Highest Variability (H2K) contigs” and the second subset includes variable genes identified using a method described by Brennecke et al. (49), assuming a false discovery rate (FDR) of 0.2, which resulted in 922 contigs (referred to as highly variable gene contigs [HVG contigs]). Focusing on variably expressed contigs improved clustering with respect to cell identity, with the HVG dataset outperforming the H2K. In the HVG clustering (Fig. 2B), 8/11 GM cells, 5/8 VD cells, 5/8 PD cells, and 5/8 LP cells formed distinct clusters. However, these nodes are not perfectly segregated by cell type and cells of each kind fail to appropriately cluster. If blind to these cell types, the HVG clustering analysis yields 5 to 6 distinct cell-type clusters, rather than the appropriate 4 (Fig. 2B).
Although differential expression (DE) analysis can only be carried out with a priori knowledge of cell identity or some other post hoc feature by which samples can be grouped, in an attempt to achieve the best performance possible with scRNA-seq clustering analyses we unblinded the analyses to cell type and selected only differentially expressed transcripts. We selected 2 pools of differentially expressed transcripts: Those with a q value <0.2 (referred to as “DE0.2”) or q value <0.05 (“DE0.05”). DE analysis with a q-value cutoff of 0.2 identified 137 transcripts (DE0.2), while a q value of 0.05 identified only 45 transcripts (DE0.05). Hierarchical clustering of the DE0.2 dataset resulted in better clustering but still failed to faithfully recapitulate cell identity. Hierarchical clustering was greatly improved by using the DE0.05 dataset (Fig. 2C) but remained imperfect.
To reveal which preprocessing and clustering methods best recapitulate the predicted number of clusters based on known cell identity, we applied 8 cluster estimation algorithms (optCluster package) (50) on the DE0.05 dataset (centered and scaled by contig, Ward.D2 linkage, and a correlation dissimilarity matrix; Fig. 2D). The highest performing clusterings using the DE0.05 data resulted from using Ward.D with a correlation distance metric, resulting in a Jaccard index of 0.738. The results of cluster estimation differed based on the preprocessing of the datasets. Cluster estimation algorithms were selected from a set of 10 algorithms for use with continuous data as they all yielded usable output. We retained the top 3 predicted k values from each. When data were centered and scaled by contig (Fig. 2D), the mode number of clusters estimated was 3 (5 indices) and 5 (5 indices), and none predicted the correct number of 4 clusters.
Finally, to assess whether unblinded analyses could predict cell type, we tested the ability of 8 supervised machine learning (sML) classification algorithms (generalized linear model [GLM], k-nearest neighbors [kNNs], neural network [NN], multinomial neural network [MNN], random forest [RF], support vector machine with a linear kernel [SVML], support vector machine with a radial kernel [SVMR], and linear discriminant analysis [LDA]) to sort cells based on their transformed or untransformed mRNA abundances. Each model’s accuracy on new data were estimated using 5-fold cross-validation. To capture the variation in the All Expressed Contigs dataset, we transformed the data with principal component analysis (PCA) and used the first 38 principal components, which accounted for over 99% of the variation. The sML mean accuracies on the All Expressed Contigs (PCA transformed) dataset were extremely low, with a maximum mean accuracy of 48.6% (Fig. 2E). sML accuracies improved substantially when classifying the RNA-seq data preprocessed to identify variably expressed contigs (H2K, HVG) and DE contigs (DE0.2, DE0.05), often producing 100% accuracy for several folds during 5-fold cross-validation (Fig. 2E). It should be noted that no method classified all 5 folds with complete accuracy, even with only DE contigs—most methods ranged between 75 and 100% accuracy. While these results are encouraging, even under optimal conditions (transcriptomic data, selection of transcripts by differential expression, ability to use supervised methods) we were unable to consistently classify these neurons with 100% accuracy.
Principal Component Analysis of scRNA-Seq Datasets.
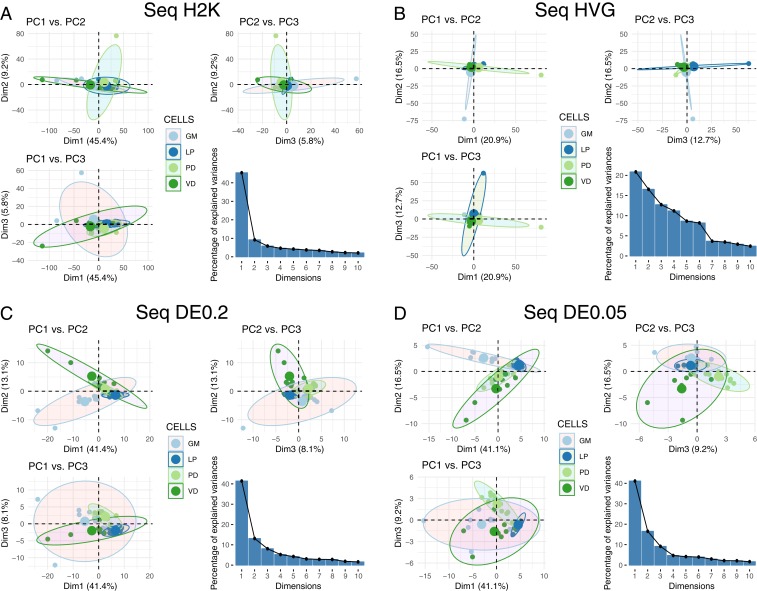
PCA is often used to determine whether the variance seen among transcript abundances can be used to separate cells into discrete types. Thus, we performed PCA on the 4 RNA-seq datasets (H2K, HVG, DE0.2, and DE0.05) to examine the ability of this approach to discriminate among cell types (Fig. 3). For most of these datasets, the first principal component (PC1) accounted for >40% of the explained variance, with the exception of the HVG dataset (Fig. 3). As such, we have listed the top 10 contigs contributing to variation in PC1 for all 4 datasets in SI Appendix, Table S1. We generated pairwise plots of all 3 PCs in attempts to visualize separation of samples into distinct cell types. There is little ability to resolve cell-type differences in the H2K and HVG datasets (Fig. 3 A and B). However, the differentially expressed transcripts allow for some separation of cell type (Fig. 3 C and D), with PD becoming somewhat distinct, for example, in the DE0.05 dataset (Fig. 3D).
Fig. 3.
PCA for 4 different RNA-seq datasets. We performed PCA using (A) the 2,000 contigs with the highest variance in expression (H2K), (B) the HVG and DE contigs at the (C) q < 0.2 (DE0.2), and (D) q < 0.05 (DE0.05) levels. For each panel we have plotted pairwise comparisons of PC1, PC2, and PC3, as well as a scree plot representing the percentage of variance explained by PCs 1 through 10.
Gene Ontology Analyses of RNA-Seq Datasets.
To determine the types of genes represented in the most variable (H2K and HVG) and differentially expressed (DE0.2, DE0.05) datasets among cell populations, we performed gene ontology (GO) enrichment analysis using analysis tools from the PANTHER Classification System (51). Because there is relatively little gene annotation work in the crab, we performed GO analysis by first using BLAST to find the top Drosophila ortholog for a given contig, and then retrieving the GO terms associated with this ortholog for analysis. Thus while this analysis provides interesting insight into cell-type-specific differences in gene expression, there are limitations to the interpretation, particularly with regards to fold enrichment in Drosophila relative to crab. The most robust expression differences (highest fold enrichment) in the H2K molecular function dataset were those of ATP-synthase activity and clathrin binding (SI Appendix, Table S2). Others of note include mRNA 3′-UTR binding, cell adhesion molecule, and calcium ion binding (SI Appendix, Table S2). More resolution is gained by examining the biological process category, where H2K contigs were most overrepresented for “regulation of short-term neuronal synaptic plasticity,” “positive regulation of neuron remodeling,” “substrate adhesion-dependent cell spreading,” and “clathrin-dependent synaptic vesicle endocytosis” categories (SI Appendix, Table S3) among many others. The HVG dataset shows relatively few enriched categories (SI Appendix, Tables S4 and S5) with FDR correction employed, including ATP binding and transferase activity (related to acetylcholine synthesis).
The differentially expressed contigs of the DE0.2 dataset showed no significantly enriched contigs with FDR employed. Without any P value correction, a number of molecular function categories appear as enriched (SI Appendix, Table S6). However, this is less an appropriate enrichment analysis (due to the relatively small number of contigs) and more a description of gene categories present in the DE0.2 contigs. The top several hits are all indicative of transmitter phenotype, particularly acetylcholine synthesis (SI Appendix, Table S6). However, other receptor activity is represented, such as GABA-gated chloride channel and GABA-A receptor activity. Finally, cell–cell adhesion mediator activity appears once again in this list.
Molecular Profiling of Single Identified STG and CG Neurons Using Candidate Genes.
One class of genes that we were surprised to not see represented in DE analyses was the voltage-gated ion channels. A recent study found that 3 classes of neuronal effector genes—ion channels, receptors, and cell adhesion molecules—have the greatest ability to distinguish among morphologically distinct mouse cortical cell populations (52). Our previous work also suggests that differential expression of ion channel mRNAs in STG cells may give rise to their distinct firing properties (53–55). We therefore examined these scRNA-seq data for expression of ion channel mRNAs. Overall, while the sequencing captured most of the known voltage-gated channel subtypes known in C. borealis, raw counts were very low (SI Appendix, Fig. S1). Therefore, we decided to use a qRT-PCR approach to directly test the hypothesis that channels and transmitter receptors are effective genes of interest to differentiate known neuron subtypes.
To examine the molecular profile of individual identified neurons with qRT-PCR, we targeted the following transcripts: ion channels, receptors, gap junction innexins, and neurotransmitter-related transcripts. These cellular components are responsible for giving neurons much of their unique electrophysiological outputs. As such, we predicted that correspondingly unique expression patterns for this gene set would be present in each neuron type. Using multiplex qRT-PCR, we measured the absolute copy number of 65 genes of interest (SI Appendix, Table S7) from 124 individual STG neurons of 11 different types (10 STG neuron types: pyloric dilator [PD], lateral posterior gastric [LPG], ventricular dilator [VD], gastric mill [GM], lateral pyloric [LP], pyloric [PY], inferior cardiac [IC], lateral gastric [LG], median gastric [MG], dorsal gastric [DG], and the large cell [LC] motor neurons from the cardiac ganglion) (n = 10 to 15 per type). We used various methods of unsupervised clustering to generate the “best” clustering of these cells based on a priori known number of cell types. This included substituting any missing values in the qRT-PCR dataset via median interpolation.
We then used k-means, unsupervised hierarchical, and shared nearest neighbor-Cliq (SNN-Cliq) clustering to generate unbiased clustering analyses based on expression of these genes of interest. Initial interrogation focused on data transformations with a fixed hierarchical clustering scheme (Ward.D2, correlation dissimilarity matrix as for the scRNA-seq analysis). Unscaled data, as well as data centered and scaled by gene, resulted in different hierarchical clustering patterns. Using unscaled data, hierarchical clustering performed rather poorly in terms of generating distinct clusters that match known cell identity. Performance—as assessed by Jaccard index—was improved by scaling data across genes, generating 8 distinct nodes with high bootstrap support in hierarchical clustering that capture some of the features of known cell identity (LC, IC, LG, LPG, VD, GM, LP, and PD; Fig. 4A). However, multiple cell types fall into clusters that either do not show any separation by neuron identity (DG, MG, and PY) or show no bootstrap support based on hierarchical clustering (approximately unbiased [AU] P value = 0).
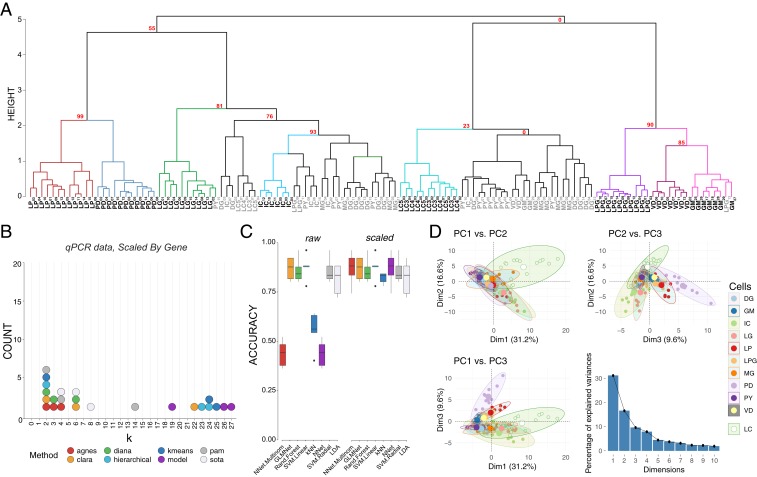
Fig. 4.
Post hoc recapitulation of cell identity via qRT-PCR expression with hierarchical clustering and sML algorithms. (A) Hierarchical clustering of cell type with correlation as the distance metric and Ward.D2 as the clustering method for data centered and scaled across genes. AU P values for a given node are noted in red. Each node that has >80% support by AU P value is color coded, and cell types that form a largely coherent group are noted in bold. Cells that do not appear to cluster by type are noted in gray. Cells are identified by type and a subscript that denotes a unique sample identifier. (B) Dotplot of the top 3 predicted number of clusters based on 8 different prediction algorithms. None of these methods correctly predicted 11 distinct clusters that would represent the 11 different cell types in this assay. (C) Accuracy of cell-type prediction using 8 different methods of sML for each of the datasets. Box and whisker plots show efficacy of each method across 5 cross-validation folds. (D) PCA for qRT-PCR data. Pairwise comparisons of PC1, PC2, and PC3 are shown in each panel as in Fig. 3. PC1 accounted for 31.2% of the variance, PC2 accounted for 16.6%, and PC3 accounted for 9.6% of the total variance across samples. A scree plot shows the amount of variance explained by PCs 1 through 10.
We sought to determine the upper bound for clustering performance with this dataset. If the known anatomical and physiological cell identity is reflected in the ion channel and receptor mRNA profile of STG neurons, then clustering analyses performed on these mRNA data should yield 11 distinct clusters. To determine the feasibility of clustering to sort cell types, we tested 291 clusterings (varying clustering methods, distance metrics, and neighbors considered) for each dataset. Each clustering was compared against the known cell identities with the Jaccard index, which ranges from 0 to 1, where 1 is perfect correspondence between clusterings—in this case, the clustering and cell identity. The best performing combination was data scaled by target and processed using Ward.D2 hierarchical clustering with a correlation distance matrix (Jaccard = 0.636). By contrast, the next best clusterings, Ward.D on correlations and Ward.D on data scaled and PCA transformed using Canberra distance, only achieved Jaccard indices of 0.592 and 0.509, respectively. The 3 least performant methods were single-linkage hierarchical clustering with distance metric of uncentered sample correlation (0.087), maximum distance (0.088), and correlation distance (0.089) metrics. Examining the best performing clustering reveals that LP, PD, LG, IC, DG, LC, PY, GM, LPG, and VD separate fairly well.
Given that an a priori known number of cell types represented in a sample is rare, we tested whether we would have arrived at the correct number of cell types in the sample had we been blind to their identity. We used the best performing transformations from the clustering analysis, i.e., data centered and scaled by gene and a correlation dissimilarity matrix, and 8 cluster determination indices provided by the optCluster package (50). We allowed a minimum of 2 and a maximum of 32 clusters for this and later cluster determination analyses. The mode of the top 3 predicted k values for 8 different methods of cluster estimation was 2 (6 indices), followed by 4 (the expected number of clusters), and 6 (3 indices each) (Fig. 4B). If a researcher were using any 1 of these, or a majority vote of several, the chance they would conclude the correct number of 11 clusters are present would be vanishingly low.
We repeated sML analyses on the qRT-PCR data to examine the “best case scenario” performance for clustering analyses. Performance varied substantially between algorithms (e.g., NN achieved a mean accuracy of 43.5%, whereas SVML produced a mean accuracy of 87.5%) and was affected by whether the data were centered and scaled (e.g., NN improved by 43.5%, SVML did not improve) (Fig. 4C). The highest mean accuracy we achieved was 87.5% (SVML, either with or without scaling). We considered a principal component transformation as well, but it improved the maximum mean accuracy little (NN, 87.9%) and worsened the previously most performant methods (SVML decreased from 87.5 to 66.5%, unscaled and 67.4%, scaled). Although neither produces the highest mean accuracy, RF (87.2 to 83.2%), GLM (86.6 to 79.2%), and LDA (81.9 to 77.7%) performed consistently across transformations, but clearly not equally well. Overall, the top performing accuracy methods involved centering and scaling the data across genes, and yielded similar efficacies across algorithms (Fig. 4C).
Finally, we repeated the PCA to determine if the variance seen among transcript abundances can be used to separate these 11 cell types into discrete clusters. The first 2 principal components (PC1 and PC2) generated from the qRT-PCR data accounted for 31.2% and 16.6% of the variance, respectively (Fig. 4D). PC3 accounted for 9.6% of the variance across samples. The top 10 mRNAs contributing to each of these PCs are listed in SI Appendix, Table S1. We generated pairwise plots of all 3 PCs in attempts to visualize separation of samples into distinct cell types. The most consistent result across all comparisons was that LC neurons from the cardiac ganglion formed a cluster that had less overlap with STG neurons than STG neurons did with each other, particularly in the dimension of PC1 vs. PC2 (Fig. 4D). Visualizations of PC1 vs. PC3 and PC2 vs. PC3 also give some indication that even with these target genes of interest, we are able to resolve some separation of these groups (Fig. 4D). However, without such extensive a priori knowledge about cell type overall, it is difficult to see how PCA would be effective in separating these 11 cell types based on the expression data at hand.
Comparison of qRT-PCR and RNA-Seq Results.
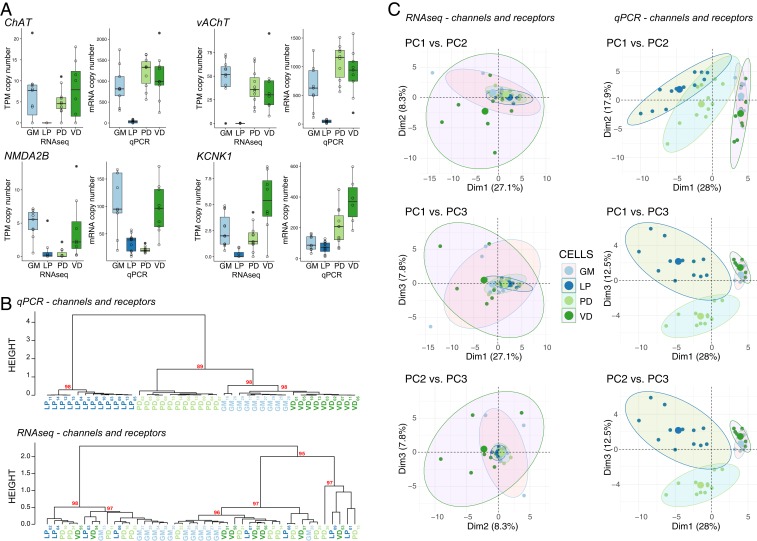
To ensure that the RNA-seq and qRT-PCR data were producing comparable expression results, we identified 4 different transcripts that were represented both in the DE dataset from the RNA-seq and the qRT-PCR dataset for the 4 cell types used in RNA-seq (PD, LP, GM, and VD). Overall, there is very strong agreement in expression patterns for all 4 genes (Fig. 5A), adding confidence to the quality of both datasets with respect to capturing native expression patterns. We then extracted the RNA-seq expression data for all 65 of the transcripts used in the qRT-PCR dataset. When we performed hierarchical clustering analysis and PCA using these 65 channel and receptor transcripts, the qRT-PCR clustered with nearly 100% success (with the exception of 2 GM neurons) into nodes that contain the 4 known distinct cell types, while the RNA-seq dataset using the same transcripts failed to generate coherent cell type clusters (Fig. 5 B and C). As we examined this further, we realized that the 4 transcripts in Fig. 5A (ChAT, vAChT, NMDA2B, and KCNK1) represent somewhat higher abundance transcripts that were differentially expressed and showed consistent patterns between qPCR and RNA-seq methods. Other highly expressed transcript types were not differentially expressed (e.g., NaV and INX1–3), and therefore do not contribute strongly to distinguishing cell identity. Conversely, many of the other transcripts in the qRT-PCR dataset that were distinct across cell types had very low levels of detected expression in the RNA-seq dataset (SI Appendix, Fig. S1).
Fig. 5.
Comparison of expression levels and clustering between qRT-PCR and RNA-seq data. (A) Expression levels of 4 different genes (choline acetyltransferase [ChAT], vesicular acetylcholine transporter [vAChT], NMDA receptor subtype 2B [NMDA2B], and K+ two-pore-domain channel subfamily K member 1 [KCNK1]) between the RNA-seq and qRT-PCR datasets. Data shown are medians, quartiles, and each individual value from a given animal. Each individual data point is also represented as open circles. RNA-seq data are presented as TPM while qRT-PCR data as absolute copy number per cell. (B) Hierarchical clustering comparison between qRT-PCR (Top) and RNA-seq (Bottom) for the same 65 genes represented in the genes of interest pool shown in Fig. 1. Each cell type is color coded, and nodes are labeled with AU values as in previous figures. (C) PCA for scRNA-seq versus qRT-PCR channel and receptor data. Pairwise comparisons of PC1, PC2, and PC3 are shown in each panel as in Fig. 3.
Discussion
Many projects currently attempting to describe neuronal cell types begin with the acquisition of molecular profiles from populations of unidentified neurons (27, 29, 56). Our results demonstrate the strengths and limitations of both unsupervised and supervised methods that rely solely on a molecular profile to recapitulate neuron identity by working “backwards” from an unambiguously known cell identity in a system with a rich history of single-cell neurophysiological characterization, the crustacean stomatogastric ganglion. The analyses clearly demonstrate that even with the most complete a priori knowledge of cell type, there are limitations to determining cell identity through mRNA expression profiles alone. However, these data add to compelling supporting evidence that the molecular profile can partially indicate identity, particularly once supervised methods incorporating known cell identification are employed.
Physiological Insights into STG Network Function and Cell Identity.
It can be problematic to infer physiological properties associated with mature protein function from steady-state mRNA levels. Nevertheless, we did make some observations by comparing gene expression profiles to known STG neuron physiology that could have broader implications. First, despite the fact that PD and LPG cells are strongly electrically coupled and fire in a tightly phase-locked fashion when the gastric mill rhythm is not active (48, 57, 58), PD and LPG were about as different from one another based on the outcome of hierarchical clustering of qRT-PCR data as they could be (Fig. 4A). One might predict that cells with very similar physiological outputs would likely have similar patterns of channel and receptor expression (53), either because their similar physiology reflects common ontogeny (59), or activity-dependent feedback shapes expression in a conserved fashion (see ref. 55); however, neither of these are supported by the data. Futhermore, we would not predict from our results that PD and LPG have a similar developmental trajectory—although their outputs are quite similar—nor are these data consistent with common rules for activity-dependent feedback to the level of steady-state mRNA (54, 60, 61).
Second, among the full set of STG neurons in the qRT-PCR dataset, we did not see clustering that was faithful to neurotransmitter phenotype. For example, 2 of the most closely related neurons in terms of clustering were PD and LP (Fig. 4A). Yet these 2 neurons are cholinergic and glutamatergic, respectively. Therefore, it raises a thought-provoking question regarding cell identity. That is, if 2 neurons were similar in most characteristics, yet release distinct transmitters, then should these be considered more distinct classes of cells than those that release a common transmitter but share far fewer other characteristics? Transmitter phenotype is a common distinguishing feature for assigning cell identity (62); yet even this defining feature is not necessarily fixed for the life of the cell (63).
Finally, the RNA-seq data and subsequent gene ontology analysis yielded a strong indication that some of the most commonly differentially expressed transcripts represented biological processes associated with synaptic plasticity and neuronal and substrate/cell adhesion remodeling (SI Appendix, Table S3). This is in contrast to the lack of differential expression in this dataset among gene families more directly associated with direct membrane voltage and physiological output, such as channels and receptors. This suggests that a key feature of these networks may reside more in the ability to tune and adapt synaptic connectivity to generate and maintain appropriate network output, rather than to tune individual neuronal excitability (64)—although these are certainly not mutually exclusive (65, 66).
General Insights.
There is increasing evidence that discrete classes of genes may distinguish cell types. For example, genes underlying synaptic transmission machinery were crucial for separating mouse cortical GABAergic neurons into different types (67). Sets of genes that are regulated together that can be thought of as a “gene batteries” have also been shown to be indicative of cell type. For example in Caenorhabditis elegans there is expression of neuron-type-specific combinations of transcription factors (62). Recently, 3 classes of neuronal effector genes—ion channels, receptors, and cell adhesion molecules—were determined to have the greatest ability to distinguish among genetically and anatomically defined mouse cortical cell populations (52). Consistent with this work, GO analysis of the 2,000 most variable contigs in the scRNA-seq dataset (H2K) revealed that the top 5 biological process terms that were significantly enriched included “regulation of short-term neuronal synaptic plasticity,” “substrate adhesion-dependent cell spreading,” and “clathrin-dependent synaptic vesicle endocytosis.” Specifically, the differentially expressed contigs dataset (DE0.2) revealed molecular function enrichment for terms related to transmitter identity (“choline:sodium symporter activity” and “acetylcholine transmembrane transporter activity” among others), specifically identified 2 GABA receptor function terms (“GABA-gated chloride ion channel activity” and “GABA-A receptor activity”) and also included “cell–cell adhesion mediator activity.” Finally, our entire qRT-PCR experiment focused on the expression of ion channels, receptors, gap junction innexins, and neurotransmitter-related transcripts. While these 65 genes were not sufficient for classifying cells perfectly into known types, this modest number of transcripts discriminated neuron types fairly well. Thus, categorical families of neuronally expressed genes may yield the most useful data for subdividing neurons into distinct classes or subtypes.
Not every system has the same challenges or advantages in assigning neuronal cell identity. Mouse retinal ganglion cells of the same type are regularly and uniformly spaced throughout the retina, while cells belonging to different types do not exhibit spatial patterning relative to one another and are more randomly distributed (68). Molecular classification of neurons in C. elegans found that anatomically distinct neurons have correspondingly distinct molecular profiles >90% of the time (69). However, 146 distinct molecular profiles were identified from the 118 anatomically distinct neuron classes, indicating the potential for molecular subclassification. This classification relied on hierarchical clustering that was carried out solely on identified reporter genes (most prominently transcription factors) and G protein-coupled receptor ([GPCR]-type sensory receptors) known to be differentially expressed across the 302 neurons of C. elegans from Wormbase.org (70) and not whole transcriptome molecular profiles. It is reassuring that the expression of a wide variety of reporter genes known to be differentially expressed across a population of neurons can recapitulate cell identity. But, this relies on having an established definition of neuron type to constrain hierarchical clustering, as differential expression analysis can only be carried out by assigning samples to different populations. Our results are consistent with these findings, in that clustering is most reliable when differentially expressed targets are used as the transcriptomic dataset. Further, these data also demonstrate that without separating cell types a priori by such additional criteria, molecular cell classification can generate unreliable results, particularly with neurons that belong to the same network.
What are the sources of variability that could mask molecular identification of neuronal identity? Most common high-throughput molecular profiling techniques require destructive sampling to acquire mRNA abundances, which generates only a snapshot of the profile at a single point in time. Gene expression has stochastic characteristics (71, 72); transcription takes place not continually, but in bursts of expression (73) (reviewed in ref. 74); and steady-state mRNA abundances are the result of rates of expression, degradation, and mRNA stability (75). Single-cell transcriptomes can be altered biologically as a consequence of activity (76), injury (77), long-term memory formation (25), differentiation (78), and aging (23, 79), as well as being affected by technical noise (49). Cells also belong to different transcriptional states under certain conditions, with the major distinction between a cell type and cell state being that state is a reversible condition, whereas type is more constant and includes neuronal states (80). Neuron types exist in a continuum, exhibiting variation in expression patterns within defined cell types, increasing difficulty in discreetly drawing the cutoff of one type from another (81). Thus, the assertion that a given neuron has a single transcriptomic profile is an oversimplification and simply represents a moment in time in the life of a given cell.
The present study has limitations. The expression of the focal gene set of ion channels, receptors, gap junction innexins, and neurotransmitter-related transcripts examined here ultimately discriminated neuron types fairly well, using supervised methods taking into account known neuron identity. This same gene set did not perform well in the same cell types using RNA-seq (Fig. 5), where a lack of low-abundance transcripts (such as transcription factors and ion channels) may have prevented us from robustly identifying cell-type-specific expression patterns; thus, depth of sequencing is always an ambiguity in every RNA-seq study (82). Furthermore, while we sampled the mRNA transcriptome of individual neurons, we have not measured other gene products that could drive unique identity, including noncoding RNA species such as microRNA (miRNA) and long noncoding RNA (lncRNA) (83). Epigenetic modifications have also been implicated in neuronal cell identity (84), which were not considered in this study. Further, there are numerous other methods and statistical analyses being applied to molecular profiles to distinguish cell type. We focused on the more commonly employed analyses (PCA, hierarchical clustering, and machine learning algorithms) in the literature. Finally, although we are confident in our ability to identify and harvest the targeted neuron types, we cannot entirely rule out the possibility of an occasional misidentified or wrongly isolated cell, as well as the potential presence of adherent support cells.
This present study reveals the circular nature of using transcriptomics to identify cell types: Molecular profiling is most effective when cells are separated into distinct types a priori, yet this is often not possible in many systems. So then how can we most effectively use molecular profiling on unknown populations of cells? The clear answer is to provide as much multimodal data as possible in the analysis. Here, the additional data were an a priori separation into cell type based on electrophysiological output, synaptic connectivity, axonal projection, and muscle innervation target (85). While it has been more difficult to achieve multimodal data integration in systems such as cortex, the approach is gaining traction and proving effective (86). For example, supervised clustering methods proved superior to unsupervised algorithms in separating pyramidal neurons from interneurons in the mouse neocortex based on morphological phenotypes (87). Genetically and anatomically defined cell populations in the mouse cortex have revealed much finer resolution and confidence in molecular profiling (52), and combined physiological and transcriptomic approaches have yielded valuable insights into spinal interneuron diversity as well (88). Much like a circuit’s connectome alone is insufficient to predict network output and function (89), so too the transcriptome alone is insufficient to generate a definitive cell type. Yet it also is clear that transcriptome profiling provides valuable insight into understanding the functional role of individual neurons and neuron types in a network. Therefore, increasing evidence indicates that transcriptomic approaches will benefit from integration with other modalities of cell-defining characteristics to gain more accurate distinctions among cell types. scRNA-seq data on their own should be viewed with caution with respect to a definitive cell identity assessment until more studies with multimodal integration become available.
Conclusion
Classification and characterization of cell types often has been performed ad hoc within the context of specific studies or species rather than based on a systematic approach. Without a more systematic attempt to define cell type, it will be challenging to use the extensive data being generated in a comparative fashion to its fullest potential (90). Acknowledging that cell types and their diversity are the product of evolution, Arendt et al. (91) defined a cell type as “a set of cells in an organism that change in evolution together, partially independent of other cells, and are evolutionarily more closely related to each other than to other cells.” As a consequence, cells of a given type use certain genomic information—both coding and noncoding—that determines cell identity and is not used by other cells. This suggests that single-cell gene expression profiling is a valuable approach to attain a comprehensive understanding of an organism’s cellular physiology. As such, cell classification schemes are susceptible to similar limitations as phylogenetic studies. For example, the species concept continues to be an area of active discussion among evolutionary biologists (92), and prokaryotic species assignment shares many of the same challenges as single-cell eukaryotic cell-identity approaches (93). Yet there are lessons to be carried across these diverse disciplines. Just as the application of molecular characters in phylogenetic analyses was initially met with skepticism, ultimately this approach became an essential scientific discipline, in part due to the value of combined molecular, morphological, and behavioral data (94). Transcriptomic approaches to cell identity already are broadly embraced. However, to fully leverage these kinds of data, it seems prudent to generate a more systematic definition and approach to classifying neuron identity. This definition should strive to combine multiple modalities of data, both to increase confidence in the transcriptomic identification as well as refine and better standardize the definition of what constitutes distinct cell types or unique cell identity.
Methods
Cell Collection and RNA Preparation.
All animal experiments were approved by the Animal Care and Use Committees at University of Missouri-Columbia and Brandeis University. Adult male Jonah crabs, C. borealis, were purchased from the Fresh Lobster Company (Gloucester, MA) and Commercial Lobster (Boston, MA). Animals were allowed to acclimate to their tanks and kept in filtered artificial seawater tanks chilled at 10 °C to 13 °C on a 12/12 light:dark cycle until use. Prior to dissection, crabs were put on ice for 30 min to induce anesthetization.
The complete stomatogastric nervous system (STNS) was dissected and pinned out in a dish coated in Sylgard (Dow Corning) with chilled (12 °C) physiological saline (composition in mM/l: 440.0 NaCl, 20.0 MgCl2, 13.0 CaCl2, 11.0 KCl, 11.2 Trizma base, and 5.1 maleic acid pH = 7.4 at 23 °C in RNase-free water). Recordings were made of the spontaneously active stomatogastric rhythms, and all were confirmed to be generating healthy and robust output equivalent to the standard in the extensive literature on this preparation (85). This ensured all preparations used in this study were within the realm of normal physiological function. Following desheathing of the STG, neurons were identified by simultaneous intra- and extracellular recordings (48, 57). Ten neuron types identified in the STG of C. borealis were targeted for this study: PD, LPG, LP, IC, LG, MG, GM, PY, VD, and DG. Identified neurons were extracted as previously described (95). More information is provided in SI Appendix, Supplemental Methods. Identified neurons (Fig. 1) were immediately placed in a cryogenic microcentrifuge tube containing 400 µL lysis buffer (Zymo Research) and stored at −80 °C until RNA extraction. Total RNA was extracted using the Quick-RNA MicroPrep kit (Zymo Research) per the manufacturer’s protocol.
Library Preparation and Single-Cell RNA-Seq.
Library construction and RNA-sequencing services were carried out by the University of Texas at Austin Genomic Sequencing and Analysis Facility (Austin, TX). Extracted single-cell RNA from identified neurons from the STG was used to generate cDNA libraries using TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA). Libraries were sequenced in a paired-end 150-bp (2 × 150 bp) configuration on the NextSeq 500 Illumina platform (Illumina). Raw reads were processed and analyzed on the Stampede Cluster at the Texas Advanced Computing Center. Read quality was checked using the program FASTQC. Low-quality reads and adapter sequences were removed using the program Cutadapt (96). The 40 identified neurons used in this study all had at least 4 million uniquely mapped reads per sample, comprising 11 PD, 11 GM, 8 LP, and 8 VD cell types. These sequencing reads are deposited in the National Center for Biotechnology Information (NCBI) BioProject archive (PRJNA524309) with the following identifiers: BioSample: SAMN11022125; sample name: STG neurons; SRA: SRS4411333.
Mapping and Differential Expression.
The software package Kallisto (97) (v0.43.1) was used in the quantification of RNA-seq abundances through the generation of pseudoalignments of paired-end fastq files to the C. borealis annotated nervous system transcriptome (47). While a fully annotated genome represents the best reference for mapping, there is no genome yet available for C. borealis. In general, decapod crustacean genomes are severely lacking. The only published decapod genome likely to be of high enough quality for such mapping is that of the marbled crayfish, Procambaurs fallax f. virginalis (98). However, as this species likely last shared a common ancestor on the order of 350 mya, we did not feel mapping was likely to be successful. Therefore, we elected to use what is a fairly high-quality transcriptome from C. borealis. Bootstrapping of the quantification was performed iteratively for 100 rounds. Resulting counts were normalized through the transcripts per kilobase million (TPM) method. Differential expression analysis was carried out using the software package Sleuth (99) (v0.30.0) using TPM normalized counts for each cell type.
Gene Ontology Enrichment Analysis.
Because C. borealis lacks a well-curated reference genome, GO terms were assigned to the C. borealis transcriptome based on best BLASTX hits through reciprocal queries between crab sequence and the Drosophila melanogaster NCBI RefSeq database (release 93). BLAST annotation was carried out based on Drosophila protein sequence using the BLAST2GO (version 5.1) software suite with the blastx-fast alignment with an E value threshold = 1.0E-3 to generate D. melanogaster NCBI gene IDs associated with each C. borealis contig. This produced 1,348 and 252 annotated gene IDs for the H2K and HVG datasets, respectively. These IDs were used as input for statistical overrepresentation tests using the PANTHER Gene Ontology Classification System (v14.1) with default settings using D. melanogaster as the reference species. Molecular function and biological process GO terms were examined for enrichment in these datasets, and results reported reflect FDR correction except where noted.
Multiplex Primer and Probe Design.
Multiplex primer and probe sequences targeting C. borealis genes were generated using the RealTimeDesign qPCR assay design software from LGC Biosearch Technologies (Petaluma, CA) for custom assays. Multiplex cassettes were designed as a unit to ensure minimal interference in simultaneous qPCR reactions. Probe fluorophore/quencher pairs used in this study are as follows: FAM-BHQ1, CAL Fluor Gold 540-BHQ1, CAL Fluor Red 610-BHQ2, Quasar 670-BHQ2, and Quasar 705-BHQ2. Forward and reverse primer pair, as well as associated probe, sequences can be found in SI Appendix, Table S7.
cDNA Synthesis and Preamplification.
Following RNA extraction, individual neuron RNA samples were reverse transcribed into cDNA using qScript cDNA SuperMix (QuantaBio, Beverly, MA) primed with random hexamers and oligo-dT per the manufacturer’s protocol in 20-µL reactions. Half of each resulting cDNA pool (10 µL) was preamplified using PerfeCTa PreAmp Supermix (QuantaBio) with a 14-cycle RT-PCR primed with a pool of target-specific primers (SI Appendix, Table S7) in a 20-µL reaction per the manufacturer’s protocol to allow for enough product to carry out 15 multiplex qPCR reactions per individual neuron sample. Amplified and unamplified target abundances were compared to ensure minimal amplification bias in the preamplification of samples (SI Appendix, Fig. S2).
Quantitative Single-Cell RT-PCR.
Following preamplification of cDNA, samples were diluted 7.5× in nuclease-free water (150 µL final volume) to allow for the quantification of 73 unique gene products across 15 multiplex assays, each able to measure 4 to 5 different transcripts (SI Appendix, Table S7). Reactions were carried out in triplicate on 96-well plates with 10-µL reactions per well using a CFX96 Touch Real-Time PCR Detection System from Bio-Rad (Hercules, CA). Cycling conditions for qPCR reactions were as follows: 95 °C for 3 min; 40 cycles of 95 °C for 15 s and 58 °C for 1 min. Fluorescent measurements were taken at the end of each cycle. The final concentration of primers in each multiplex qPCR reaction was 2.5 µM and 0.3125 µM for each probe.
To quantify absolute mRNA abundances, standard curves were developed for each qRT-PCR multiplex assay using custom gBlock gene fragments (Integrated DNA Technologies, Coralville, IA). Standard curves were generated using a serial dilution of gBlock gene fragments from 1 × 106 to 1 × 101 copies for each reaction assay and shown to be linear and reproducible. Copy numbers were calculated using the efficiency and slope generated from the standard curves and accounting for the 14-cycle preamplification and subsequent cDNA dilution described above.
Statistical Analysis.
Expanded details on these analyses are provided in the SI Appendix, Supplemental Methods. All statistical analyses were performed using R version 3.5.3 (2019-03-11) “Great Truth” (100).
We used single-cell RNA-seq data to evaluate our methods under expected and near best case scenarios. To this end, we reduced the dimensionality of the data (28,695 contigs) by selecting the 2,000 most variable contigs and by selecting 922 highly variable contigs. We selected those contigs differentially expressed at an alpha of 0.2 or 0.05, centered and scaled these datasets, and used PCA to determine if any of the cell types were visually separable across these subsets of the data.
Next, we performed cluster estimation using the optClust() function of the optCluster package (50). To assess the performance of unsupervised machine learning methods, we tested several clustering algorithms and clustering methods and selected the high-performing clustering methods based on the Jaccard index calculated against cell identity. We selected one of the best performing combinations (Ward’s method with correlation as the distance metric) for visualization.
Finally, we applied several supervised machine-learning methods to evaluate predictive power of expression data in ideal circumstances (i.e., prior knowledge of a given cell type’s molecular identity). For each of the models, we tested a variety of tuning parameters and selected the most effective parameter set before comparison with other methods. Methods were evaluated by using cross-validation (with 5 folds) to produce the expected accuracy on new data. The same approaches were applied to the single-cell qRT-PCR dataset, with a few caveats. Given its relatively smaller size, dimensionality reduction was not necessary to overcome technical or practical hurdles. Thus, we tested both the raw and centered and scaled dataset in addition to PCA transformations of the same. We also increased the maximum k allowed in cluster estimation to 32.
Availability of Data and Materials.
All sequence data can be accessed in the NCBI BioProject archive (PRJNA524309) with the following identifiers: BioSample: SAMN11022125; sample name: STG neurons; SRA: SRS4411333. Accession numbers for crab channel and receptor sequences targeted in qRT-PCR experiments are provided in SI Appendix, Table S7.
Supplementary Material
Acknowledgments
We thank members of the D.J.S., H.A.H., and E.M. laboratories for helpful discussions. We thank the Genomic Sequencing and Analysis Facility (The University of Texas [UT] at Austin) for library preparation and sequencing and the bioinformatics consulting team at the UT Austin Center for Computational Biology and Bioinformatics for helpful advice. This work was supported by National Institutes of Health grant R01MH046742-29 (to E.M. and D.J.S.) and the National Institute of General Medical Sciences T32GM008396 (support for A.J.N.) and National Institute of Mental Health grant 5R25MH059472-18 and the Grass Foundation (support for Neural Systems and Behavior Course at the Marine Biological Laboratory).
Footnotes
The authors declare no competing interest.
Data deposition: All sequence data can be accessed in National Center for Biotechnology Information BioProject archive (PRJNA524309) with the following identifiers: BioSample: SAMN11022125; sample name: STG neurons; SRA: SRS4411333. Accession numbers for crab channel and receptor sequences targeted in qRT-PCR experiments are provided in SI Appendix, Table S7.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911413116/-/DCSupplemental.
References
- 1.Masland R. H., Neuronal cell types. Curr. Biol. 14, R497–R500 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Zeng H., Sanes J. R., Neuronal cell-type classification: Challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Tasic B., Single cell transcriptomics in neuroscience: Cell classification and beyond. Curr. Opin. Neurobiol. 50, 242–249 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Stevens C. F., Neuronal diversity: Too many cell types for comfort? Curr. Biol. 8, R708–R710 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Cuevas-Diaz Duran R., Wei H., Wu J. Q., Single-cell RNA-sequencing of the brain. Clin. Transl. Med. 6, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo L., Callaway E. M., Svoboda K., Genetic dissection of neural circuits: A decade of progress. Neuron 98, 256–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tessmar-Raible K., et al. , Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell 129, 1389–1400 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Tomer R., Denes A. S., Tessmar-Raible K., Arendt D., Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142, 800–809 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Mehta P., et al. , Functional access to neuron subclasses in rodent and primate forebrain. Cell Rep. 26, 2818–2832.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitaker K. W., et al. , Serotonergic modulation of startle-escape plasticity in an African cichlid fish: A single-cell molecular and physiological analysis of a vital neural circuit. J. Neurophysiol. 106, 127–137 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Ho H., et al. , A guide to single-cell transcriptomics in adult rodent brain: The medium spiny neuron transcriptome revisited. Front. Cell. Neurosci. 12, 159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parmhans N., Sajgo S., Niu J., Luo W., Badea T. C., Characterization of retinal ganglion cell, horizontal cell, and amacrine cell types expressing the neurotrophic receptor tyrosine kinase Ret. J. Comp. Neurol. 526, 742–766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha B. R., et al. , Sensory neuron diversity in the inner ear is shaped by activity. Cell 174, 1229–1246.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung S., et al. , Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 545, 477–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Södersten E., et al. , A comprehensive map coupling histone modifications with gene regulation in adult dopaminergic and serotonergic neurons. Nat. Commun. 9, 1226 (2018). Erratum in: Nat. Commun. 9, 4639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadwell C. R., et al. , Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeisel A., et al. , Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Boldog E., et al. , Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 21, 1185–1195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberwine J., et al. , Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. U.S.A. 89, 3010–3014 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tietjen I., et al. , Single-cell transcriptional analysis of neuronal progenitors. Neuron 38, 161–175 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Esumi S., et al. , Method for single-cell microarray analysis and application to gene-expression profiling of GABAergic neuron progenitors. Neurosci. Res. 60, 439–451 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Tang F., et al. , mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Davie K., et al. , A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174, 982–998.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulin J.-F., Tasic B., Hjerling-Leffler J., Trimarchi J. M., Awatramani R., Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Crocker A., Guan X.-J., Murphy C. T., Murthy M., Cell-type-specific transcriptome analysis in the Drosophila mushroom body reveals memory-related changes in gene expression. Cell Rep. 15, 1580–1596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macosko E. Z., et al. , Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Usoskin D., et al. , Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Shin J., Ming G. L., Song H., Decoding neural transcriptomes and epigenomes via high-throughput sequencing. Nat. Neurosci. 17, 1463–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tasic B., et al. , Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Defining cell types and states with single-cell genomics. Genome Res. 25, 1491–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cembrowski M. S., Wang L., Sugino K., Shields B. C., Spruston N., Hipposeq: A comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5, e14997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner A., Regev A., Yosef N., Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gokce O., et al. , Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 16, 1126–1137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle J. P., et al. , Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou T., Matsunami H., Lessons from single-cell transcriptome analysis of oxygen-sensing cells. Cell Tissue Res. 372, 403–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg A. B., et al. , Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reuter J. A., Spacek D. V., Snyder M. P., High-throughput sequencing technologies. Mol. Cell 58, 586–597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley K. W., Nakao-Inoue H., Molofsky A. V., Oldham M. C., Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobert O., Carrera I., Stefanakis N., The molecular and gene regulatory signature of a neuron. Trends Neurosci. 33, 435–445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sha Y., Phan J. H., Wang M. D., Effect of low-expression gene filtering on detection of differentially expressed genes in RNA-seq data. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2015, 6461–6464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Brink S. C., et al. , Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Harris R. M., Kao H.-Y., Alarcon J. M., Hofmann H. A., Fenton A. A., Hippocampal transcriptomic responses to enzyme-mediated cellular dissociation. Hippocampus 29, 876–882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasic B., et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y.-R., et al. , Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziegenhain C., et al. , Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Northcutt A. J., et al. , Deep sequencing of transcriptomes from the nervous systems of two decapod crustaceans to characterize genes important for neural circuit function and modulation. BMC Genomics 17, 868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weimann J. M., Meyrand P., Marder E., Neurons that form multiple pattern generators: Identification and multiple activity patterns of gastric/pyloric neurons in the crab stomatogastric system. J. Neurophysiol. 65, 111–122 (1991). [DOI] [PubMed] [Google Scholar]
- 49.Brennecke P., et al. , Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013). Erratum in: Nat. Methods11, 210 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Sekula M., Datta S., Datta S., optCluster: An R Package for Determining the Optimal Clustering Algorithm. Bioinformation 13, 101–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi H., Muruganujan A., Thomas P. D., PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugino K., et al. , Mapping the transcriptional diversity of genetically and anatomically defined cell populations in the mouse brain. eLife 8, e38619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz D. J., Goaillard J.-M., Marder E. E., Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proc. Natl. Acad. Sci. U.S.A. 104, 13187–13191 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Temporal S., Lett K. M., Schulz D. J., Activity-dependent feedback regulates correlated ion channel mRNA levels in single identified motor neurons. Curr. Biol. 24, 1899–1904 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Santin J. M., Schulz D. J., Membrane voltage is a direct feedback signal that influences correlated ion channel expression in neurons. Curr. Biol. 29, 1683–1688.e2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., et al. , Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell 171, 1206–1220.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hooper S. L., et al. , The innervation of the pyloric region of the crab, Cancer borealis: Homologous muscles in decapod species are differently innervated. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 159, 227–240 (1986). [DOI] [PubMed] [Google Scholar]
- 58.Weimann J. M., Marder E., Switching neurons are integral members of multiple oscillatory networks. Curr. Biol. 4, 896–902 (1994). [DOI] [PubMed] [Google Scholar]
- 59.Farrell J. A., et al. , Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Leary T., Williams A. H., Caplan J. S., Marder E., Correlations in ion channel expression emerge from homeostatic tuning rules. Proc. Natl. Acad. Sci. U.S.A. 110, E2645–E2654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Leary T., Williams A. H., Franci A., Marder E., Cell types, network homeostasis, and pathological compensation from a biologically plausible ion channel expression model. Neuron 82, 809–821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deneris E. S., Hobert O., Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17, 899–907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spitzer N. C., Neurotransmitter switching in the developing and adult brain. Annu. Rev. Neurosci. 40, 1–19 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Prinz A. A., Bucher D., Marder E., Similar network activity from disparate circuit parameters. Nat. Neurosci. 7, 1345–1352 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Turrigiano G., Homeostatic synaptic plasticity: Local and global mechanisms for stabilizing neuronal function. Cold Spring Harb. Perspect. Biol. 4, a005736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schulz D. J., Lane B. J., Homeostatic plasticity of excitability in crustacean central pattern generator networks. Curr. Opin. Neurobiol. 43, 7–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paul A., et al. , Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171, 522–539.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanes J. R., Masland R. H., The types of retinal ganglion cells: Current status and implications for neuronal classification. Annu. Rev. Neurosci. 38, 221–246 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Hobert O., Glenwinkel L., White J., Revisiting neuronal cell type classification in Caenorhabditis elegans. Curr. Biol. 26, R1197–R1203 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Harris T. W., et al. , WormBase 2014: New views of curated biology. Nucleic Acids Res. 42, D789–D793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li G.-W., Xie X. S., Central dogma at the single-molecule level in living cells. Nature 475, 308–315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raj A., van Oudenaarden A., Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 135, 216–226 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y., Ni T., Wang W., Liu F., Gene transcription in bursting: A unified mode for realizing accuracy and stochasticity. Biol. Rev. Camb. Philos. Soc., 10.1111/brv.12452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Symmons O., Raj A., What’s luck got to do with it: Single cells, multiple fates, and biological nondeterminism. Mol. Cell 62, 788–802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pérez-Ortín J. E., Genomics of mRNA turnover. Brief. Funct. Genomics Proteomics 6, 282–291 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Benito E., Barco A., The neuronal activity-driven transcriptome. Mol. Neurobiol. 51, 1071–1088 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Llorens-Bobadilla E., et al. , Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Olivera-Martinez I., et al. , Major transcriptome re-organisation and abrupt changes in signalling, cell cycle and chromatin regulation at neural differentiation in vivo. Development 141, 3266–3276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moroz L. L., Kohn A. B., Single-neuron transcriptome and methylome sequencing for epigenomic analysis of aging Methods Mol. Biol. 1048, 323–352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tasic B., Levi B. P., Menon V., “Single-cell transcriptomic characterization of vertebrate brain composition, development, and function” in Decoding Neural Circuit Structure and Function, Çelik A., Wernet M. F., Eds. (Springer International Publishing, 2017), pp. 437–468. [Google Scholar]
- 81.Cembrowski M. S., Menon V., Continuous variation within cell types of the nervous system. Trends Neurosci. 41, 337–348 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Rizzetto S., et al. , Impact of sequencing depth and read length on single cell RNA sequencing data of T cells. Sci. Rep. 7, 12781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sheng K., Cao W., Niu Y., Deng Q., Zong C., Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat. Methods 14, 267–270 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Mito M., et al. , Cell type-specific survey of epigenetic modifications by tandem chromatin immunoprecipitation sequencing. Sci. Rep. 8, 1143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marder E., Bucher D., Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 69, 291–316 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Kim D.-W., et al. , Multimodal analysis of cell types in a hypothalamic node controlling social behavior. Cell 179, 713–728.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guerra L., et al. , Comparison between supervised and unsupervised classifications of neuronal cell types: A case study. Dev. Neurobiol. 71, 71–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bikoff J. B., et al. , Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell 165, 207–219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gutierrez G. J., O’Leary T., Marder E., Multiple mechanisms switch an electrically coupled, synaptically inhibited neuron between competing rhythmic oscillators. Neuron 77, 845–858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tosches M. A., Laurent G., Evolution of neuronal identity in the cerebral cortex. Curr. Opin. Neurobiol. 56, 199–208 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Arendt D., et al. , The origin and evolution of cell types. Nat. Rev. Genet. 17, 744–757 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Freudenstein J. V., Broe M. B., Folk R. A., Sinn B. T., Biodiversity and the species concept-lineages are not enough. Syst. Biol. 66, 644–656 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Konstantinidis K. T., Ramette A., Tiedje J. M., The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1929–1940 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Avise J. C., Molecular Markers, Natural History and Evolution (Springer, 1994). [Google Scholar]
- 95.Schulz D. J., Goaillard J.-M., Marder E., Variable channel expression in identified single and electrically coupled neurons in different animals. Nat. Neurosci. 9, 356–362 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J 17, 10 (2011). [Google Scholar]
- 97.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Gutekunst J., et al. , Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat. Ecol. Evol. 2, 567–573 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Pimentel H., Bray N. L., Puente S., Melsted P., Pachter L., Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 100.R Core Team R: A language and environment for statistical computing (Version 3.5.3, R Foundation for Statistical Computing, Vienna, Austria, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data can be accessed in the NCBI BioProject archive (PRJNA524309) with the following identifiers: BioSample: SAMN11022125; sample name: STG neurons; SRA: SRS4411333. Accession numbers for crab channel and receptor sequences targeted in qRT-PCR experiments are provided in SI Appendix, Table S7.