Hyperactivation of endoplasmic reticulum stress response decreases lipid (fat) levels to extend life span.
Abstract
Longevity is dictated by a combination of environmental and genetic factors. One of the key mechanisms to regulate life-span extension is the induction of protein chaperones for protein homeostasis. Ectopic activation of the unfolded protein response of the endoplasmic reticulum (UPRER) specifically in neurons is sufficient to enhance organismal stress resistance and extend life span. Here, we find that this activation not only promotes chaperones but also facilitates ER restructuring and ER function. This restructuring is concomitant with lipid depletion through lipophagy. Activation of lipophagy is distinct from chaperone induction and is required for the life-span extension found in this paradigm. Last, we find that overexpression of the lipophagy component, ehbp-1, is sufficient to deplete lipids, remodel ER, and promote life span. Therefore, UPR induction in neurons triggers two distinct programs in the periphery: the proteostasis arm through protein chaperones and metabolic changes through lipid depletion mediated by EH domain binding protein 1 (EHBP-1).
INTRODUCTION
The homeostatic regulation of protein folding (proteostasis), which is monitored in specific subcellular compartments, is an integral player in stress resistance and longevity. The endoplasmic reticulum (ER), in particular, is a central regulator of stress monitoring as it controls nearly a third of the cell’s proteins, provides an internal medium for lipid homeostasis and cell signaling, and communicates directly with all other organelles to maintain cellular secretion. Thus, cells have evolved numerous quality control machineries dedicated to protecting the ER both under basal and stressed conditions.
Notably, the ER has evolved three primary branches of its unfolded protein response (UPRER) to maintain proper secretion, protein folding, and lipid homeostasis. The most studied of the three UPRER branches involves the membrane localized/endoribonuclease inositol-requiring protein-1 (IRE-1), which is activated via oligomerization upon unfolded protein stress in the ER lumen or lipid disequilibrium in the ER membrane. This results in splicing of a specific intron from the mRNA of the transcription factor, X-box binding protein-1 (XBP-1), to create the spliced version of xbp-1 (xbp-1s), which then up-regulates the expression of protein degradation, protein folding, and lipid metabolism gene targets. Another branch of the UPRER is the membrane-localized protein kinase, R (PKR)-like ER kinase (PERK), which, upon ER stress, globally reduces protein translation via phosphorylation of the translation initiation factor, eukaryotic initiation factor 2 (eIF2α). Transcripts that promote protein folding and apoptosis, however, are preferentially translated. The last of the three UPRER branches is the ER membrane–localized activating transcription factor-6 (ATF6), which, upon luminal stress, moves to the Golgi to become a mature transcription factor capable of up-regulating canonical proteostasis genes (1, 2).
While these pathways have been intensively studied for the past two decades, much less is known about the adaptive responses of the ER under long-lived conditions. The small nematode, Caenorhabditis elegans, is an excellent model to simultaneously study the genetic underpinnings of cell-cell communication under adaptive conditions while also enabling the monitoring of its cell biology and organismal health (e.g., life span and stress resistance). Work with C. elegans has shown that its cells become less capable in protein folding and also less able to induce stress responses to proteotoxicity with advanced age (3–9). Overexpression of xbp-1s, specifically in neurons (heretofore referred to as neuronal xbp-1s), extends organismal life span and increases ER stress tolerance in a cell nonautonomous manner (9). While the precise, small ER stress signal was not identified, small clear vesicles are required for this beneficial effect, which could be host to numerous neurotransmitters. The intestine, the putative target tissue in this paradigm, induces chaperones in an XBP-1–dependent manner. However, it is well understood that UPRER induction not only alters protein homeostasis through chaperones but also is an important mechanism for metabolic change and handling of lipid stress (10).
We hypothesized that induction of the UPRER in neurons, which reverses the age-dependent loss of ER proteostasis, also enacts a marked restructuring of ER morphology, which, in turn, imparts a beneficial metabolic change and promotes longevity. Although whole-organismal metabolic restructuring has been a topic of intense study in the aging field, much less is known about the adaptive responses of organelles in long-lived conditions. Here, we find that neuronal xbp-1s animals have notable ER restructuring and lipid depletion. These changes are distinct from chaperone induction and are dependent on the conserved lipophagy complex, RAB-10/EHBP-1/RME-1 (ras-related GTP-binding protein/EH domain binding protein/receptor mediated endocytosis). Thus, we argue that the beneficial effects of nonautonomous UPRER are dependent on two independent, yet equally important, arms of UPRER: the protein homeostasis arm, including chaperone induction and the metabolic arm, which induces ER remodeling and lipophagy.
RESULTS
Ectopic UPRER induction results in ER remodeling
To determine whether preserving ER protein homeostasis would alter age-associated changes in the ER structure, we interrogated ER morphology and organization in the peripheral tissue (intestine) of neuronal xbp-1s animals (Fig. 1A). We visualized the ER during various stages of adulthood in C. elegans using an mCherry::HDEL fusion protein that localizes to the ER lumen through a SEL-1 (Suppressor/Enhancer of Lin-12) signal sequence. At day 1 of adulthood, the ER fills the intestinal lumen and can be visualized as a sponge-like, interconnected, and uniform structure. As early as day 4, intestinal ER is less uniform and begins to collapse, forming disorganized, aggregated structures with increased severity by day 7. Neuronal xbp-1s animals display no difference in intestine ER structures at day 1 compared with wild-type animals. Unexpectedly, at day 4, when wild-type animals begin to exhibit collapse of the ER, neuronal xbp-1s animals have a spherical ER morphology that is not seen at any age in wild-type animals. These structures are visible as early as day 2; by day 7, these structures disappear, and the ER network collapses in a manner indistinguishable from wild-type animals. Therefore, constitutive UPRER in neurons is not sufficient to protect ER quality and structure during aging and only show significant differences in early adulthood.
Fig. 1. Neuronal xbp-1s promotes ER remodeling and increased ER function in peripheral tissue.
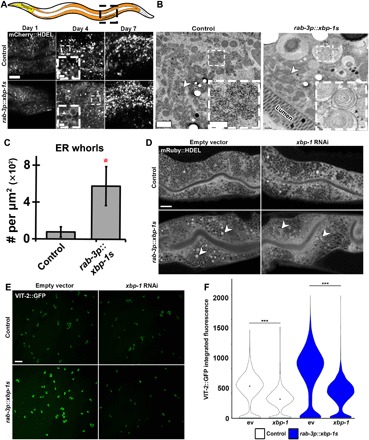
(A) Representative confocal micrographs of ER morphology (via an intestine-specific vha-6p::mCherry::HDEL using a sel-1 signal sequence) at various times in adulthood (“young,” day 1; “middle-aged,” day 4; “old,” day 7) under control and “neuronal xbp-1s” (rab-3p::xbp-1s) conditions. In the cartoon, the intestine is shown in orange, and the pharynx is shown in yellow. Arrowhead indicates representative ER remodeling. Scale bars, 10 μm. (B) Electron micrographs of intestine from control and neuronal xbp-1s–overexpressing animals (rab-3p::xbp-1s) at late day 2 of adulthood. Arrowheads mark rough ER. Scale bars, 1 μm (0.25 μm for inset). (C) Quantification of ER whorls per square micrometer in control versus rab-3p::xbp-1s animals. *P ≤ 0.05, using t test. Plots are representative of three biological replicates and analyzed from 10 to 30 images per sample. (D) Representative confocal micrographs of ER morphology using an alternative labeling method (vha-6p::mRuby::HDEL using an hsp-4 signal sequence). Animals were grown on either empty vector (ev) or xbp-1 RNA interference (RNAi) from hatch and imaged at late day 2. Arrowheads indicate representative ER remodeling. Scale bar, 10 μm. (E) Representative images of eggs expressing VIT-2::GFP. Transgenic control or rab-3p::xbp-1s animals carrying VIT-2::GFP were grown on either empty vector or xbp-1 RNAi from hatch until day 1 adulthood and then bleached, and eggs were imaged. Scale bar, 100 μm. (F) Quantification of data from (E) where control animals are represented as white and neuronal xbp-1s animals are blue. Plots are representative of three biological replicates and n > 2000 eggs per condition. ***P < 0.001, using nonparametic Mann-Whitney test.
To better understand the nature of ER remodeling in neuronal xbp-1s animals, we performed high-magnification transmission electron micrograph imaging and identified denser patches of rough ER compared with controls and an increase in the number of ER whorls (Fig. 1, B and C). ER whorls have been previously observed upon up-regulation of UPRER/ER-phagy (11–13). To ensure that the changes in ER structure and organization were not due to artifacts of the imaging methods, we interrogated ER morphology in neuronal xbp-1s animals using two additional ER makers: mRuby::HDEL localized to the ER through an HSP-4 (heat shock protein) signal sequence (Fig. 1D) and a green fluorescent protein (GFP) fusion to an ER transmembrane protein, SP12 (fig. S1A). Both ER markers showed ER remodeling in neuronal xbp-1s animals similar to mCherry::HDEL and electron microscopy. Moreover, the formation of these structures is partially dependent on xbp-1s in the periphery, as RNA interference (RNAi) knockdown of xbp-1 partially resolved these structures (Fig. 1D and fig. S1A). Strong overexpression of xbp-1s directly in the intestine is sufficient to induce ER remodeling and is dependent on xbp-1 (fig. S1B). However, systemic activation of the UPRER through ER stress by treatment with the chemical agent tunicamycin results in collapsed ER structure distinct from ER restructuring in neuronal and intestinal xbp-1s (fig. S1C). These data suggest that ectopic xbp-1s expression induces a unique phenomenon distinct from direct activation of UPRER under conditions of proteotoxic stress.
As a critical organelle in secretory vesicle formation, we hypothesized that the major ER remodeling found in neuronal xbp-1s animals could result in increased intestinal secretion. To measure secretion, we compared VIT-2::GFP deposition in neuronal xbp-1s animals compared with control animals. VIT-2::GFP is a fluorescently tagged version of vitellogenin, which is a common reporter used to measure protein secretion as a proxy for ER function in the intestine of C. elegans (14, 15). This protein is integral to maternal yolk formation and, under normal conditions, is secreted solely by the intestine and endocytosed by developing eggs. We find that neuronal xbp-1s animals have increased VIT-2::GFP deposition in their eggs. Moreover, this increase is dependent on xbp-1, suggesting that neuronal xbp-1s animals have a significant increase in ER secretory function in the intestine (Fig. 1, E and F).
ER remodeling upon UPRER induction is concomitant with lipid depletion
A massive increase in secretion would be expected to result in lipid depletion due to the expansion of ER membranes. We tested whether intestinal ER remodeling of neuronal xbp-1s animals occurred concurrently with lipid depletion. We measured intestinal lipids using a combination of whole-organism and tissue-specific quantification. Our lab had previously developed a methodology and software, called LAMPro, to exploit underused positional information inherent to BioSorter data, which takes fluorescent traces of many animals at once and defines key regions of interest to various cellular phenomena (16). This enabled us to overcome the practical limits of microscopy (e.g., low n) to reliably measure the distribution of total lipids across animals. Using LAMPro technology, we measured whole-body neutral lipid content (via BODIPY 409/503 staining) of several hundred animals per condition. Briefly, animals were staged when ER remodeling was prominent, fluorescent integrated intensity was measured across the length of BODIPY-stained worms, and data were represented as a heat map of median profiles of relative neutral lipid distribution. Notably, we found that neuronal xbp-1s animals had a marked depletion of lipids compared with wild-type animals (Fig. 2A).
Fig. 2. ER remodeling in neuronal xbp-1s animals is concomitant with lipid depletion.
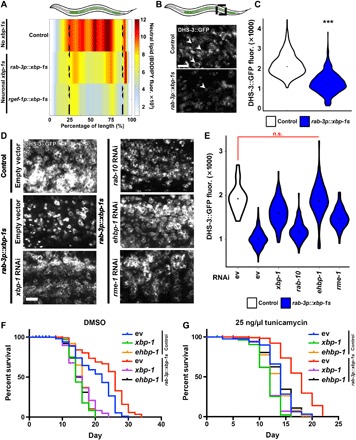
(A) Median profiles of nematode neutral lipid distribution (via BODIPY 409/503 stain) of control animals and neuronal xbp-1s animals using two different neuron-specific promoters (rab-3p and rgef-1p) following alignment. Data are shown as a heat map with red as the highest amount of lipid and white as the lowest. All animals are positioned with the anterior side to the left. (B) Representative confocal micrographs of intestinal lipid droplets (LDs) (via dhs-3p::DHS-3::GFP) in day 2 adult control and neuronal xbp-1s animals. Scale bar, 5 μm. Arrowheads indicate representative LDs. (C) Quantification of LDs (via DHS-3::GFP fluorescence) in day 2 adults in control (white) and neuronal xbp-1s (blue) animals. ***P < 0.001, using nonparametric Mann–Whitney test, against age-matched control. Plots are representative of three biological replicates and n = 460 (control) and 754 (rab-3p::xbp-1s). (D) Representative confocal micrographs of LDs (DHS-3::GFP) from staged, day 2 adults. Animals were grown on RNAi from hatch. Scale bar, 5 μm. (E) Quantification of LDs (via DHS-3::GFP fluorescence) from (D). n.s., P > 0.10 using nonparametric Mann-Whitney test. Plots are representative of three biological replicates and n = 136 to 185 organisms per strain. Direct comparisons to xbp-1 and ehbp-1 knockdown in wild-type animals are available in fig. S4. (F and G) Life spans of nematodes under control [dimethyl sulfoxide (DMSO) (F)] and under ER stress [tunicamycin (25 ng/μl) (G)]. Animals were grown from hatch on ev, xbp-1, or ehbp-1 RNAi and moved onto either 1% DMSO or tunicamycin (25 ng/μl) plates at day 1. Life spans of (F) and (G) were performed simultaneously and separated for ease of visibility. Data are representative of three independent trials. See table S1 for life-span statistics.
Next, we tested whether the decline in lipid content is due to the loss of lipid droplets (LDs), the primary organelle that stores intestinal lipids in C. elegans (17). We visualized LDs with the intestine-specific, fluorescently labeled, short-chain dehydrogenase DHS-3::GFP fusion protein, which is abundant on the surface of C. elegans intestinal LDs (18, 19). To ensure that LAMPro technology could reliably measure changes in lipid content using DHS-3::GFP, we used sbp-1 RNAi as a positive control for decreased lipid content. sbp-1 is the C. elegans ortholog of SREBP1 (sterol regulatory element–binding protein 1) and is essential for fat accumulation (20), and our LAMPro methods can reliably detect a decrease in lipid levels as expected in sbp-1 knockdown animals (fig. S2). Using this marker, we found that LD content was significantly lower in neuronal xbp-1s animals compared with wild-type animals (Fig. 2, B and C). Last, we find that lipid depletion in neuronal xbp-1s animals was partially dependent on xbp-1, as RNAi knockdown of xbp-1 partially suppressed this phenotype, similar to ER remodeling (Fig. 2, D and E).
A lipophagy complex driven by EHBP-1 promotes lipid depletion and is required for life-span extension of neuronal xbp-1s animals
Another study has also presented that neuronal xbp-1s animals have significant lipid depletion, which is dependent on lysosomal expansion and increased activity of lysosomal lipases in the intestine (21), yet independent of the core autophagy machinery. Similarly, we find an expansion of lysosomes in neuronal xbp-1s animals and that these lysosomes colocalize with distinct ER membranes that occur during ER remodeling (fig. S3, A and B). Under conditions of UPRER activation and ER expansion, induction of ER phagy and lipophagy can aid in the redistribution of lipids (12, 22). While Imanikia et al. (21) investigated the role of lysosomal lipases in lipid depletion, here we questioned whether the lipid depletion in neuronal xbp-1s animals was dependent on the elements of the RAB-10/EHBP-1/RME-1 lipophagy complex. We find that RNAi knockdown of ehbp-1 fully suppressed the lipid depletion found in neuronal xbp-1s animals, even more than RNAi knockdown of xbp-1 (Fig. 2, D and E). While xbp-1 and ehbp-1 RNAi alone both increase lipids in wild-type animals, neuronal xbp-1s fails to deplete lipids under xbp-1 or ehbp-1 knockdown, providing evidence that both xbp-1 and ehbp-1 are essential for the lipid depletion phenotype found in this animal (fig. S4). RNAi knockdown of rme-1 only partially suppressed the lipid depletion, and knockdown of rab-10 had no effect. The failure of rme-1 and rab-10 knockdown could be due in part to the fact that they have been associated with induction of autophagy (23). Moreover, knockdown of rab-10 and rme-1 causes phenotypes associated with calorie restriction, including egg laying and growth defects, and an increased life span, which could confound a lipid rescue. Together, these data suggest that EHBP-1, potentially through the RAB-10/RME-1/EHBP-1 complex, is involved in the ER remodeling and lipid depletion found in neuronal xbp-1s animals.
Changes in metabolism, specifically lipid depletion, are directly downstream of UPRER induction by xbp-1s. Therefore, we sought to determine whether RAB-10, RME-1, and EHBP-1 were linked to canonical chaperone induction mediated by xbp-1s. To our surprise, RNAi knockdown of any of these lipophagy components did not affect induction of chaperones, as measured by the reporter hsp-4p::GFP (fig. S5, A to D). However, and most unexpectedly, knockdown of ehbp-1 was sufficient to suppress the life-span extension found in neuronal xbp-1s animals, similar to RNAi knockdown of xbp-1s (Fig. 2F), even though these animals could still induce the canonical chaperone components. Since ehbp-1 knockdown decreased life span in wild-type animals, to control for the possibility that life span is suppressed due to changes in protein homeostasis, we measured life span of ehbp-1 knockdown animals on tunicamycin. Similar to animals grown in the absence of protein misfolding stress, RNAi knockdown of ehbp-1 suppressed life-span extension of neuronal xbp-1s animals. However, ehbp-1 knockdown did not increase sensitivity of wild-type animals to tunicamycin-induced protein misfolding stress (Fig. 2G). These data provide evidence that the life-span extension of neuronal xbp-1s animals is dependent on lipophagy mediated by ehbp-1. Moreover, the activation of lipophagy in these animals is distinct and separable from chaperone induction. However, we also found that life-span extension of neuronal xbp-1s animals is dependent on chaperone induction, as knockdown of hsp-4 suppressed life-span extension similar to knockdown of ehbp-1 (fig. S5E).
Last, to determine whether EHBP-1 was sufficient to drive the phenotypes described in neuronal xbp-1s signaling, we overexpressed ehbp-1 specifically in the target tissue (intestine). ehbp-1 overexpression in the intestine is sufficient to drive depletion of lipids, similar to neuronal xbp-1s overexpression (Fig. 3, A and B). Intestinal ehbp-1 overexpression also resulted in changes in ER structure, reminiscent of xbp-1s overexpression, and displays life-span extension comparably to neuronal xbp-1s overexpression (Fig. 3, C and D). However, unlike xbp-1s overexpression, overexpression of ehbp-1 is not sufficient to induce chaperone induction, although there is a mild increase in stress-induced UPRER (Fig. 3, E and F). Therefore, increased lipophagy via EHBP-1 does not promote chaperone induction. These data suggest that overexpression of ehbp-1 is sufficient to phenocopy the metabolic signaling downstream of xbp-1s, providing further evidence that the lipid depletion and extension of life span found in metabolic remodeling through xbp-1s signaling are driven by EHBP-1–mediated lipid depletion distinct from canonical chaperone signaling.
Fig. 3. Intestinal ehbp-1 overexpression is sufficient to drive lipid depletion, ER remodeling, and life-span extension.
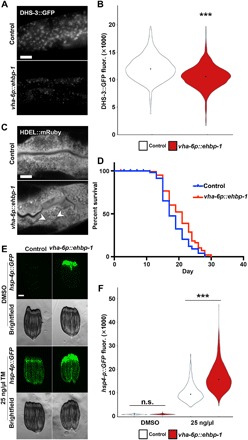
(A) Representative compound micrographs of intestinal LD (via dhs-3p::DHS-3::GFP) in day 2 adult control and vha-6p::ehbp-1 animals. Scale bar, 10 μm. (B) Quantification of LDs (via DHS-3::GFP fluorescence) from (A). ***P < 0.001; using nonparametric Mann-Whitney test against age-matched control on corresponding RNAi. Plots are representative of three biological replicates and n = 2560 for control and n = 324 for vha-6p::ehbp-1. (C) Representative micrographs of ER morphology (mRuby::HDEL) in the intestine of control and vha-6p::ehbp-1 animals. ER remodeling is depicted by arrowheads. (D) Life-span data for wild-type N2 animals or vha-6p::ehbp-1 animals grown on empty vector RNAi from hatch. See table S1 for life-span statistics. (E) Fluorescent light micrographs of hsp-4p::GFP UPRER reporter animals (control and vha-6p::ehbp-1) treated with DMSO or tunicamycin (TM; 25 ng/μl) at L4 for 4 hours and then imaged as day 1 adults. Note that myo-2p::GFP was used as a coinjection marker, and high GFP signal in the pharynx is not an artifact or cause of EHBP-1 overexpression. Scale bar, 100 μm. (F) Quantification of hsp-4p::GFP fluorescence in control (white) and vha-6p::ehbp-1 (red) nematodes on DMSO or tunicamycin (25 ng/μl), using a BioSorter, following various RNAi treatments. The pharynx signal from the myo-2p::GFP coinjection marker was removed from quantification during analysis. ***P < 0.001; n.s., P > 0.10; using Mann-Whitney test. Data are representative of three independent trials with n = 172 to 3205 per strain.
DISCUSSION
Our lab and others have developed cell nonautonomous signaling models (i.e., neuron to intestine) to provide new paradigms in the study of longevity and organismal adaptation to stress. Previously, we developed a model in which neuronal xbp-1s mediates increased life span and ER stress resistance via a neuron-specific, secreted ER stress signal. By characterizing the intestine, the target of this signal, we uncovered that this neuronal signal results in significant ER remodeling and lipid depletion in the intestine. These phenomena are chaperone independent and necessary and sufficient for life-span extension in this paradigm. Together, these data provide evidence that nonautonomous UPRER mediated by xbp-1s activates two distinct pathways in distal tissue: a protein homeostasis arm through chaperone induction and a metabolic arm that involves ER remodeling and lipid depletion (Fig. 4).
Fig. 4. Nonautonomous xbp-1s promotes two independent arms of UPRER: protein homeostasis via chaperones and metabolic changes via EHBP-1.
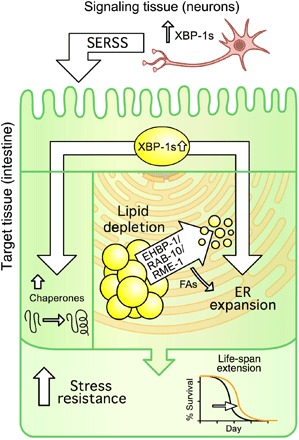
Controlled induction of the xbp-1s in neurons leads to a UPRER-dependent restructuring of the ER morphology in target cells (i.e., intestine). This restructuring coincides with the lipophagic depletion of intestinal LDs and is necessary for the metabolic change and longevity seen in this model. Moreover, loss of lipophagy reverses the lipid depletion, as well as life-span extension in this paradigm. The longevity conferred by lipophagy-mediated lipid depletion is independent of the canonical chaperone induction by XBP-1s, and both are necessary for the life-span extension found in neuronal xbp-1s animals. Last, activation of lipophagy by overexpression of ehbp-1 is sufficient to promote lipid depletion, remodel ER, and extend life span. FAs, fatty acids; SERSS, secreted ER stress signal.
When we characterized the intestinal ER morphology of neuronal xbp-1s adult animals, we found that the changes in ER structure closely resembled endocytic structures, similar to ER whorls identified in previous studies (24). The major structural change in the ER can be due to two potential reasons: increased secretion from the ER or increased autophagic flux of the ER in these animals. ER whorls have been identified during conditions of hypersecretion (25, 26) and during stress for ER turnover (24). Here, we found that neuronal xbp-1s animals exhibited increased secretion as measured by vitellogenin secretion, and thus, it is possible that changes in ER morphology occur during early to mid-adulthood to keep up with the secretory demands of egg formation.
A second possibility in the changes to intestinal ER morphology is through increased autophagy. Autophagy has been observed to be up-regulated in response to ER stress (27–29). Specifically, induction of ER stress, either through phenobarbital treatment, or Tm treatment, or induction of unstable dimers of membrane proteins in the ER, is associated with a concomitant ER expansion and induction of autophagy (11–13, 26, 30). We find that the unique ER structures observed in neuronal xbp-1s animals colocalized with lysosomal markers and that these animals exhibited an expansion of lysosomes. Similarly, another study has identified that lysosomal expansion found in neuronal xbp-1s animals is critical for the life-span extension of this paradigm (21). Therefore, it is possible that there is also an increase in autophagic flux of the ER, which may also contribute to the ER remodeling and lysosomal expansion.
To accommodate for the ER expansion and remodeling, the adaptive UPRER stress response may also massively up-regulate gene expression of membrane lipid enzymes. For example, ectopic expression of xbp-1s increases lysosomal lipases and Δ9 desaturases to drive lipid depletion (21). Here, we find that lipid depletion in neuronal xbp-1s animals is dependent on the conserved lipophagy machinery, RAB-10/EHBP-1/RME-1. Whether this complex works in parallel or in the same molecular pathway as lysosomal lipases has yet to be determined. However, since we find that hyperactivation of lipophagy via overexpression of ehbp-1 is sufficient to drive all the phenotypes described here—lipid depletion, ER remodeling, and increased life span—independent of chaperone induction, it is likely that the phenomenon described here is independent of the previously described mechanism involving lysosomal lipases and Δ9 desaturases. Last, because knockdown of rab-10 and rme-1 causes phenotypes associated with calorie restriction, it is difficult to determine their roles in lipid metabolism. Since overexpression of ehbp-1 is sufficient to drive lipid depletion while its knockdown is sufficient to increase lipids, it is possible that EHBP-1 can regulate lipid levels outside of its canonical role in lipophagy through the RAB-10/EHBP-1/RME-1 complex.
Last, although this phenotype was described in C. elegans, the phenomena of cell nonautonomous UPRER transmission—from the brain to the liver, for instance—is a conserved and well-described process (1, 31–33). It is envisioned, due to the conservation of the RAB-10/EHBP-1/RME-1 complex, that these findings might also apply to understanding the health-promoting effects seen in the liver responding to signals from the nervous system under conditions of activated UPRER.
METHODS
Strains and maintenance
All C. elegans strains used are derivatives of N2 from the Caenorhabditis Genetics Center (CGC) and are listed in table S1. All worms were maintained at 15°C on nematode growth media (NGM) agar plates and fed the OP50 Escherichia coli B strain of bacteria. To avoid genetic drift, all worms were maintained for a maximum of 4 months (16 to 20 generations) before a fresh thaw was performed. For experimentation, worms were bleached to synchronize by degrading carcasses with bleach solution (1.8% sodium hypochlorite and 0.375 M KOH), then washing four times intact eggs with M9 solution (22 mM KH2PO4 monobasic, 42.3 mM Na2HPO4, 85.6 mM NaCl, and 1 mM MgSO4), and plating eggs onto RNAi bacteria on NGM agar plates containing 1 μM isopropyl-β-d-thiogalactopyranoside, carbenicillin (100 μg/ml), and tetracycline (10 μg/ml) until the desired stages of adulthood. RNAi bacteria are derived from the HT115 E. coli K12 strain of bacteria containing pL4440 empty vector control or expressing double-stranded RNA containing the sequence of the target gene. RNAi strains were all isolated from Vidal or Ahringer libraries and sequence verified before use. All RNAi used in this study have been knockdown verified previously (7, 34–36).
rgef-1p::xbp-1s worms were synthesized by injecting N2 worms with pRT5 (for rgef-1p::xbp-1s) at 25 ng/μl, pEK2 (myo-2p::tdtomato) at 2.5 ng/μl, and pD64 (100 ng/μl) vehicle as filler DNA. Worms positive for myo-2p::tdtomato were selected to identify for stable arrays. Integration was performed by gamma irradiation. Briefly, L4 worms were irradiated with 4000 to 4400 rem (roentgen-equivalent-man) of radiation, and integrants were identified by selecting animals that maintained myo-2p::tdtomato at 100% frequency in the F3 generation. Two independent lines were isolated and backcrossed to N2 animals eight times to eliminate mutations, and animals with the most similar phenotypes to the array animals were used for experimentation. vha-6p::ehbp-1 worms were synthesized by injecting N2 worms with pSM1 (for vha-6p::xbp-1s) at 25 ng/μl, pEK1 (myo-2p::GFP) at 2.5 ng/μl, and pD64 vehicle (100 ng/μl) as filler DNA. Worms positive for myo-2p::GFP were selected to identify for stable arrays. All experiments were performed by positively selecting for array animals.
Staging of animals for all ER and lipid imaging and quantification was performed at late day 2/early day 3 of adulthood, as this is the earliest time point when changes to ER morphology were detected. This is the stage of adulthood where egg laying is at its maximum, approximately 90 to 100 hours after hatch at 20°C in standard growth conditions.
Sample preparation and microscopy
Worm fixing protocol and sample imaging for confocal microscopy
Staged worms were rinsed off plates with M9 and added to 15-ml conical tubes. Animals were washed three times with M9 to remove bacteria [do not exceed ~1000 g; 1000 relative centrifugal force (RCF) on an Eppendorf centrifuge 5702] and then transferred to microcentrifuge tubes using a glass Pasteur pipette (note that glass is always recommended for transferring of worms since they will adhere to plastic surfaces). Animals were pelleted, excess M9 was aspirated to 100 μl, and 100 μl of buffered paraformaldehyde (PFA) was added directly to worms [4% PFA, 340 μM CaCl2, 1.2% glucose, 40 mM NaH2PO4, and 60 mM Na2HPO4 (pH 7.4)]. Tubes were rocked at room temperature for 20 min, ~100 μl of liquid was removed, and 1.3 ml of M9 was added. Animals were then left at 4°C overnight. Animals were then spun down, solution was aspirated down to 100 μl, and 200 μl of 70% glycerol buffered in phosphate-buffered saline was mixed in. Animals were stored at −20°C until being imaged. For imaging, samples were mounted directly on glass slides, and a coverslip was placed directly on the specimen and sealed with nail polish. Mounted slides were kept at 4°C for at least 2 days before imaging to allow for settling of specimen and rehydration with glycerol. Fixed animals were viewed on a ZEISS LSM700 inverted confocal microscope, with constant acquisition settings when comparing specimens within a given experiment. Unless otherwise noted, images are maximum intensity projections (using the “Processing” tab in Zen software) of stacks spanning the width of various tissues within a single transgenic animal. Neutral lipid (BODIPY 409/503) staining was performed as previously described in Klapper et al. (37).
Live-cell imaging using wide-field/compound microscopy
When preparing samples for live-cell microscopy, individual worms were picked off plates and mounted directly on microscope slides in M9 containing 100 nM sodium azide. A coverslip was placed directly on the sample, sealed with nail polish, and imaged immediately. Specimens were viewed on a ZEISS AxioObserver.Z1 microscope equipped with a lumencor SOLA light engine and a ZEISS Axiocam 506 camera, driven by ZEISS ZEN Blue software using a 63×/1.4 Plan Apochromat objective and standard GFP filter (ZEISS filter set 46 HE), and a standard dsRed filter was used (ZEISS filter set 43).
BioSorter analysis
Staged worms were washed off plates and resuspended in M9 into 15-ml conical tubes. We used a Union Biometrica COPAS (complex object parameter analysis sorter) BioSorter (product no. 350-5000-000) using both 561- and 488-nm light sources. BioSorter calibration, cleaning, and sample running were performed as described in (16). For vha-6p::ehbp-1 worms, myo-2p::GFP was used as a coinjection marker. Therefore, all BioSorter analysis was performed in the lower 70% of the animal, cutting off signal from the top 30% (i.e., the contribution from the pharynx GFP).
Software specifications and data analysis
Worm profile data were collected using the Biosort 5401.1 software provided for use with the BioSorter machine. Before running an orientation algorithm [“LAMPro” (16)], all profiles were converted from one of multiple .dat file(s) into a single .txt file using the “Export as Text” function in the Profile Reader 16.1 software (also sold by Union Biometrica) and then saved in a folder alongside the .lmd and .dat files that were saved by the BioSorter at the time of data acquisition. All orientation algorithms were written in Perl5 and can be interpreted in Cygwin (a Linux application programming interface that can run on Windows) through additional Perl5 packages. Cygwin can be downloaded at the following address: Cygwin.com/install.html. During Cygwin installation, users will be given the option to install the additional Perl5 packages. Data analysis, significance testing, and data plotting were run using MATLAB scripts (version R2015a), which have been integrated into a single graphical user interface (“LAMPro Suite 2.0”). To quantify the number and/or size of fluorescently labeled cell markers (e.g., MLS::GFP, LMP-1::GFP, and GFP::RAB-10 punctae), stacks were normalized to the same threshold on a dark background using ImageJ. A smooth function eliminated scattered pixels. The area containing the intestine was highlighted, and the “Analyze Particles” function was used to give a count. These counts were normalized to “control” images (e.g., rab-3p::xbp-1s signal divided by the count from animals without rab-3p::xbp-1s). Enrichment relative to controls was then calculated for at least three biological replicates, and a t test was calculated to measure if this was above or below 1. Colocalization was calculated using the “JACoP” (Just another Colocalization Plugin) for experiments that contained at least three biological replicates, and a t test was performed to see if the variance of the set was above or below matched controls. All violin plots were made in R.
Life-span analysis
Life-span measurements were performed on solid NGM agar plates spotted with RNAi bacteria (HT115 E. coli strain K12). Worms were synchronized by bleaching, plated onto RNAi from hatch, and grown to adulthood at 20°C. Adult worms were moved away from progeny by moving worms onto fresh RNAi plates every day until days 7 to 10 when progeny were no longer visible. Animals were then scored every 1 to 2 days for death until all animals were scored. Animals with bagging, vulval explosion, or other age-unrelated deaths were censored and removed from quantification. Prism 5 software was used for statistical analysis. Log-rank (Mantel-Cox) method was used to determine significance.
Epifluorescence microscopy
Staged worms were picked at random (under white light) from a population and immobilized in 100 nM sodium azide. Immobilized worms were aligned on a solid NGM agar plate, and images were captures using a Leica M250FA automated fluorescent stereo microscope equipped with a Hamamatsu ORCA-ER camera.
Induction of ER stress
Protocol was adapted from Tian et al. (36). Briefly, synchronized L4 stage animals were treated with solution (25 ng/μl) of tunicamycin in M9 buffer for 4 hours. Control animals were treated in an equivalent solution of dimethyl sulfoxide (DMSO) (1%) in M9 buffer. Animals were rotated in DMSO or tunicamycin in 15-ml conical tubes. Drugs were washed three times with M9, and animals were plated on OP50 to recover overnight at 20°C and then imaged at day 1.
Electron microscopy
Whole-worm samples were processed for electron microscopy as previously reported (38). Briefly, staged worms were subjected to high-pressure freezing (BAL-TEC HPM 010) and then freeze substituted with an acetone/resin series (25% resin, then 50, 75, and 100%). Worms were cured in pure resin, then sectioned (70-nm sections), and then imaged (FEI Tecnai 12 Transmission Electron Microscope) on formvar-coated mesh grids.
Supplementary Material
Acknowledgments
We are grateful to L. Daniele for the figure illustrations and to R. Bar-Ziv, H. Henderson, A. Murley, and A. Frakes for the careful reading of the manuscript and helpful suggestions. We thank M. E. Sanabria for the essential excel macros for data analysis. Funding: This work was supported by grant 5F32AG053023-02 through the National Institute of Aging (NIA) and the Glenn Foundation for Aging Postdoctoral Fellowship to R.H.-S., the National Science Foundation Graduate Research Fellowship to J.W.P., 1F31AG060660-01 through NIA to M.G.M., and 4R01AG042679-04 through the NIA and the Howard Hughes Medical Institute to A.D. We thank the CGC for availability of strains. Author contributions: J.R.D. developed the story, performed life spans, performed confocal and BioSorter imaging and data analysis, and helped in preparing figures and writing the manuscript. R.H.-S. developed the story, performed life spans, completed wide-field, confocal, and BioSorter imaging and data analysis, prepared the figures, and wrote the manuscript. J.D. and S.M. helped in developing the story and performed life spans, confocal imaging, BioSorter imaging, and strain construction. V.R. helped in the UPRER activation experiments and strain construction. M.S. performed all sample preparation and imaging for electron microscopy. S.U.T., N.K., M.G.M., and P.A.F. helped in life spans and imaging. G.G. performed strain construction for ER imaging. C.B. and M.Z. helped in the data analysis. D.J.E. made essential additions to the LAMPro software, which enabled data analysis in this paper to be performed. L.J. and A.D. helped in developing the story and assisted in manuscript preparation. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from authors. All strains synthesized in this manuscript are derivatives of N2 or other strains from CGC and are either made available on CGC or available upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/1/eaaz1441/DC1
Fig. S1. ER remodeling of the intestine through UPRER activation requires neuronal nonautonomous signaling.
Fig. S2. LAMPro technology can be reliably used to quantify LDs using DHS-3::GFP.
Fig. S3. ER remodeling is coincident with more intestinal lysosomes in neuronal xbp-1s animals.
Fig. S4. Lipid depletion in neuronal xbp-1s is dependent on xbp-1 and ehbp-1.
Fig. S5. Knockdown of lipophagic components does not affect chaperone induction by xbp-1s.
Table S1. Statistics for all life-span data.
Table S2. Organisms and strains used.
Sequences used for cloning
REFERENCES AND NOTES
- 1.Frakes A. E., Dillin A., The UPRER: Sensor and coordinator of organismal homeostasis. Mol. Cell 66, 761–771 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ben-Zvi A., Miller E. A., Morimoto R. I., Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. U.S.A. 106, 14914–14919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M. K., Naidoo N., The endoplasmic reticulum stress response in aging and age-related diseases. Front. Physiol. 3, 263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillin A., Hsu A.-L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A. G., Kamath R. S., Ahringer J., Kenyon C., Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Dillin A., Crawford D. K., Kenyon C., Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830–834 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Durieux J., Wolff S., Dillin A., The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higuchi-Sanabria R., Paul J. W. III, Durieux J., Benitez C., Frankino P. A., Tronnes S. U., Garcia G., Daniele J. R., Monshietehadi S., Dillin A., Spatial regulation of the actin cytoskeleton by HSF-1 during aging. Mol. Biol. Cell 29, 2522–2527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor R. C., Dillin A., XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen X., Ellis R. E., Sakaki K., Kaufman R. J., Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLOS Genet. 1, e37 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingwood D., Schuck S., Ferguson C., Gerl M. J., Simons K., Generation of cubic membranes by controlled homotypic interaction of membrane proteins in the endoplasmic reticulum. J. Biol. Chem. 284, 12041–12048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuck S., Gallagher C. M., Walter P., ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127, 4078–4088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuck S., Prinz W. A., Thorn K. S., Voss C., Walter P., Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187, 525–536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant B., Hirsh D., Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10, 4311–4326 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens J., Spang A., N-glycosylation is required for secretion and mitosis in C. elegans. PLOS ONE 8, e63687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniele J. R., Esping D. J., Garcia G., Parsons L. S., Arriaga E. A., Dillin A., High-throughput characterization of region-specific mitochondrial function and morphology. Sci. Rep. 7, 6749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemieux G. A., Ashrafi K., Neural regulatory pathways of feeding and fat in Caenorhabditis elegans. Annu. Rev. Genet. 49, 413–438 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Na H., Zhang P., Chen Y., Zhu X., Liu Y., Liu Y., Xie K., Xu N., Yang F., Yu Y., Cichello S., Mak H. Y., Wang M. C., Zhang H., Liu P., Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochim. Biophys. Acta 1853, 2481–2491 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Zhang P., Na H., Liu Z., Zhang S., Xue P., Chen Y., Pu J., Peng G., Huang X., Yang F., Xie Z., Xu T., Xu P., Ou G., Zhang S. O., Liu P., Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol. Cell. Proteomics 11, 317–328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura T., Horikawa M., Shimamura S., Hashimoto T., Sakamoto K., Fat accumulation in Caenorhabditis elegans is mediated by SREBP homolog SBP-1. Genes Nutr. 5, 17–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imanikia S., Sheng M., Castro C., Griffin J. L., Taylor R. C., XBP-1 remodels lipid metabolism to extend longevity. Cell Rep. 28, 581–589.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vevea J. D., Garcia E. J., Chan R. B., Zhou B., Schultz M., Di Paolo G., McCaffery J. M., Pon L. A., Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen M., Chandra A., Mitic L. L., Onken B., Driscoll M., Kenyon C., A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLOS Genet. 4, e24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernales S., McDonald K. L., Walter P., Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLOS Biol. 4, e423 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolender R. P., Weibel E. R., A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of treatment. J. Cell Biol. 56, 746–761 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman D., Swarm R. L., Becker J., Ultrastructural study of rat liver and liver neoplasms after long-term treatment with phenobarbital. Cancer Res. 41, 2151–2162 (1981). [PubMed] [Google Scholar]
- 27.Kroemer G., Mariño G., Levine B., Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyriakakis E., Charmpilas N., Tavernarakis N., Differential adiponectin signalling couples ER stress with lipid metabolism to modulate ageing in C. elegans. Sci. Rep. 7, 5115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yorimitsu T., Nair U., Yang Z., Klionsky D. J., Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borgese N., Francolini M., Snapp E., Endoplasmic reticulum architecture: Structures in flux. Curr. Opin. Cell Biol. 18, 358–364 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Y., Wang Z. V., Tao C., Gao N., Holland W. L., Ferdous A., Repa J. J., Liang G., Ye J., Lehrman M. A., Hill J. A., Horton J. D., Scherer P. E., The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J. Clin. Invest. 123, 455–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higuchi-Sanabria R., Frankino P. A., Paul J. W. III, Tronnes S. U., Dillin A., A futile battle? Protein quality control and the stress of aging. Dev. Cell 44, 139–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams K. W., Liu T., Kong X., Fukuda M., Deng Y., Berglund E. D., Deng Z., Gao Y., Liu T., Sohn J.-W., Jia L., Fujikawa T., Kohno D., Scott M. M., Lee S., Lee C. E., Sun K., Chang Y., Scherer P. E., Elmquist J. K., Xbp1s in pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 20, 471–482 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrano A. C., Liu Z., Dillin A., Hunter T., A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature 460, 396–399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkwirth C., Jovaisaite V., Durieux J., Matilainen O., Jordan S. D., Quiros P. M., Steffen K. K., Williams E. G., Mouchiroud L., Tronnes S. U., Murillo V., Wolff S. C., Shaw R. J., Auwerx J., Dillin A., Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165, 1209–1223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Y., Garcia G., Bian Q., Steffen K. K., Joe L., Wolff S., Meyer B. J., Dillin A., Mitochondrial stress induces chromatin reorganization to promote longevity and UPRmt. Cell 165, 1197–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klapper M., Ehmke M., Palgunow D., Böhme M., Matthäus C., Bergner G., Dietzek B., Popp J., Döring F., Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J. Lipid Res. 52, 1281–1293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDONALD K. L., Webb R. I., Freeze substitution in 3 hours or less. J. Microsc. 243, 227–233 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/1/eaaz1441/DC1
Fig. S1. ER remodeling of the intestine through UPRER activation requires neuronal nonautonomous signaling.
Fig. S2. LAMPro technology can be reliably used to quantify LDs using DHS-3::GFP.
Fig. S3. ER remodeling is coincident with more intestinal lysosomes in neuronal xbp-1s animals.
Fig. S4. Lipid depletion in neuronal xbp-1s is dependent on xbp-1 and ehbp-1.
Fig. S5. Knockdown of lipophagic components does not affect chaperone induction by xbp-1s.
Table S1. Statistics for all life-span data.
Table S2. Organisms and strains used.
Sequences used for cloning


