Abstract
Pharmacogenomics (PGx) studies how individual gene variations impact drug response phenotypes, which makes PGx-related knowledge a key component towards precision medicine. A significant part of the state-of-the-art knowledge in PGx is accumulated in scientific publications, where it is hardly reusable by humans or software. Natural language processing techniques have been developed to guide experts who curate this amount of knowledge. But existing works are limited by the absence of a high quality annotated corpus focusing on PGx domain. In particular, this absence restricts the use of supervised machine learning. This article introduces PGxCorpus, a manually annotated corpus, designed to fill this gap and to enable the automatic extraction of PGx relationships from text. It comprises 945 sentences from 911 PubMed abstracts, annotated with PGx entities of interest (mainly gene variations, genes, drugs and phenotypes), and relationships between those. In this article, we present the corpus itself, its construction and a baseline experiment that illustrates how it may be leveraged to synthesize and summarize PGx knowledge.
Subject terms: Health care, Genetics
| Measurement(s) | gene_variant • response to drug • textual entity • chemical entity • haplotype • gene • Pharmacogenomics • Pharmacogenetics • abbreviation textual entity • Pharmacokinetics • Pharmacodynamics • phenotype |
| Technology Type(s) | digital curation |
| Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.11323724
Background & Summary
Pharmacogenomics (or PGx) studies how individual gene variations impact drug response phenotypes1. This is of particular interest for the implementation of precision medicine, i.e. a medicine that tailors treatments (e.g. chosen drugs and dosages) to every patient, in order to reduce the risk of adverse effects and optimize benefits. Indeed, examples of PGx knowledge have already yielded clinical guidelines and practices2,3 that recommend considering individual genotypes when prescribing some particular drugs. For example, patients with the allele *57:01 of the HLA gene are at high risk to present a hypersensitivity reaction if treated with abacavir, an anti-retroviral, and thus should be genotyped for this gene before prescription4.
Many scientific publications report the impact of gene variants on drug responses, and the size of Medline (30 million articles as of Sept. 2019) makes it hard for humans or machines to get a full understanding of the state of the art in this domain. NLP (Natural Language Processing) techniques have been consequently developed and used to structure and synthesize PGx knowledge5,6. Previous works mainly investigated rule-based approaches7–9 and unsupervised learning10,11, because of the absence of an annotated corpus. Supervised learning has also been experimented12–16, but without appropriate corpora, most studies build train and test sets on the basis of PharmGKB, which is the reference database for this domain17. Because it is manually curated, PharmGKB provides a high quality referential for such a task. Annotations provided by PharmGKB (i.e. 2 associated entities and the identifier of the PubMed article in support) result of the consideration by human curators of various knowledge sources: article text; tables and figures; and curator’s own knowledge of the domain. Consequently PharmGKB annotations result from a high level process that can hardly be compared to an NLP-only approach. In particular, most NLP efforts are restricted to open-access texts only, without considering background knowledge. In this sense, systems evaluated in comparison with PharmGKB are tested on how they may guide the curation process, but not on how they can capture what is stated in texts.
In domains close to PGx, corpora have been annotated with biomedical entities of interest for PGx, but never with the three PGx key entities (i.e. drugs, genomic variations and phenotypes) in the same portion of text, and never with relationships specific to PGx. Hahn et al.6 made a panorama of corpora related to PGx. In the next section of this article, we present and compare the most pertinent ones with PGxCorpus. In contrast with existing corpora PGxCorpus encompasses all three key entities of interest in PGx (i.e. drugs, genomic variations and phenotypes) both at the corpus level (the corpus encompasses the three) and at the sentence level (most sentences encompass the three). In addition, both entities and relationships are labeled using detailed hierarchies, allowing the capture of pharmacogenomic relationships with a finest level of granularity. Finally, PGxCorpus is larger than other corpora: it is 2 to 3 times larger in term of annotated relationships than other corpora that include genomic entities. Despite the existence of reference resources, in particular PharmGKB, and of alternatives to classical supervised learning such as weak supervision or active learning, we believe that a high quality training set remains an asset to a domain and that PGxCorpus will serve both the PGx and the Bio-NLP communities.
This manuscript first presents PGxCorpus itself; its construction, in Methods; and a baseline experiment, in Technical Validation.
Position in regards to existing corpora
In domains closely related to PGx, corpora have been annotated with biomedical entities, but only few of them include relationships (see Hahn et al.6 for a panorama, plus18–21). More relevant corpora are related to pharmacovigilance or genetic traits, and then focus on drug–adverse response or SNP–phenotype associations. To our knowledge, no corpus includes annotations of all three PGx key entities, i.e. drugs, genomic variations and drug response phenotypes; and no corpus annotates PGx relationships between these entities. Developed for pharmacovigilance, EU-ADR22 is a corpus composed of three disjoint subcorpora of 100 PubMed abstracts each. Sentences are annotated with pairs of entities either drug-disease, drug-target or target-disease. In the same vein, ADE-EXT23 consists of 2,972 MEDLINE case reports annotated with drugs, conditions (e.g. diseases, signs and symptoms) and their relationships. SNPPhenA24 is a corpus of 360 PubMed abstracts annotated with single nucleotide polymorphisms (SNPs), phenotypes and their relationships. Domains covered by EU-ADR, ADE-EXT or SNPPhena are related to PGx because they encompass entites of interest in PGx, however they only partially fit the purpose of PGx relation extraction because sentences contains only two types of entities and because their annotated relations are only rarely related to PGx. In particular, EU-ADR and ADE-EXT annotate drug reactions without considering genetic factors. SNPPhena does not focus on drug response phenotypes, but on general phenotype and symptoms, and it only considers SNPs whereas other genomic variations are also important in PGx. In addition, the size of EU-ADR sub-corpora and SNPPhena are relatively small (with only a few hundred annotated sentences), which limits the use of supervised learning approaches that require large train sets such as TreeLSTM25. These elements motivated us to construct a new corpus, focused on PGx, and large enough to train deep neural network models.
Table 1 proposes a comparison of PGxCorpus with five related corpora. In contrast with existing corpora and in particular those mentioned above, PGxCorpus encompasses all entities of interest in PGx (i.e. drugs, genomic variations and phenotypes) both at the corpus level and at the sentence level. Indeed, 47% of its annotated relations (1,353 out of 2,871) involve these three types of entities. The ratio of sentences with no relation in PGxCorpus is only 2.7%, which is rather low in comparison with larger corpora such as ADE-EXT where it is up to 71.1%. PGxCorpus contains more types of relationships and entities, allowing a finest level of granularity. Indeed, while PGxCorpus uses 10 types of entities and 7 types of relations (all organized in hierarchies), SNPhenA, EU-ADE and ADE-EXT only use one type of relation (i.e. the relation occurs or not), but specify one of three modalities (positive, hypothetical or negative) in the case of EU-ADR. In the case of SNPPhena five modalities (neutral, weak confidence, moderate confidence, strong confidence and negative) are reported. In PGxCorpus, relationship modality (one out of the four values: positive, hypothetical, negative or both) is also captured and complements the relation type. SemEval DDI uses four types of drug-drug relations; however those are specific to drug-drug interactions and thus are irrelevant to PGx. Unlike the other corpora, PGxCorpus includes nested and discontiguous entities, which allows the capture of complex entities and their relationships. Finally, PGxCorpus is larger than other corpora that include genomic entities (required in PGx): it is 2 to 3 times larger in term of annotated relationships than SNPPhena and EU-ADR.
Table 1.
Main characteristics of PGxCorpus in comparison with related corpora.
| Corpus name | Subcorpus | Corpus size | Sent. w/o rel. | Key entities | #Ent. types | #Rel. types | # Mod. | Nested entities | Discont. entities | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sent. | rel. | % | nb. | Dru. | Gen. | Phe. | |||||||
| SNPPhenA | — | 483 | 1300 | 0 | 0 | ✓ | ✓ | 2 | 1 | 5 | |||
| EU-ADR | drug-disease | 244 | 176 | 0 | 0 | ✓ | ✓ | 2 | 1 | 3 | |||
| drug-target | 247 | 310 | 0 | 0 | ✓ | ✓ | 3 | 1 | 3 | ||||
| target-disease | 355 | 262 | 0 | 0 | ✓ | ✓ | 3 | 1 | 3 | ||||
| SemEval | DrugBank | 5,675 | 3,805 | 65.9 | 3739 | ✓ | 4 | 4 | 1 | ||||
| DDI | MEDLINE | 1,301 | 232 | 87.1 | 1133 | ✓ | 4 | 4 | 1 | ||||
| ADE-EXT | — | 5,939 | 6,701 | 28.9 | 1719 | ✓ | ✓ | 2 | 1 | 1 | |||
| PGxCorpus | — | 945 | 2,871 | 2.7 | 26 | ✓ | ✓ | ✓ | 10 | 7 | 4 | ✓ | ✓ |
Sizes of corpora are reported in term of number of sentences (sent.) and annotated relationships (rel.). The number of sentences without any annotated relation (Sent. w/o rel.) is reported both as a percentage (%) and an absolute number of sentences (nb.). The specific presence of PGx key entities, i.e. drugs (Dru.), genetic factors (Gen.) and phenotypes (Phe.) is reported under the Key entities column. Overall numbers of types of entities and relations used in annotations are reported as #Ent. and #Rel. types respectively. #Mod. refers to the number of modalities for the annotation of relations (e.g. positive, hypothetical, negative).
Despite the existence of reference resources, in particular PharmGKB, and of alternative to classical supervised learning such as weak supervision or active learning, we believe that a high quality training set remains an asset to a domain and thus that the PGx community will benefit from PGxCorpus.
Statistics
PGxCorpus encompasses 945 sentences, from 911 distinct PubMed abstracts, annotated with 6,761 PGx entities and 2,871 relationships between them. Detailed statistics on the type of entities and relationships annotated are provided in Tables 2 and 3, respectively. Note that we distinguish two types of particular entities: nested and discontiguous ones. Nested entities are entities that fully or partially encompass at least one other entity in their offset. In Fig. 1, the phenotype “acenocoumarol sensitivity” is an example of nested entity since it encompasses the “acenocoumarol” drug. Discontiguous entities are entities with a discontiguous offset, such as “VKORC1 genotypes” in Fig. 1. 874 out of the 945 sentences of PGxCorpus (92%) contain the three key entities of pharmacogenomic (i.e., drug, genomic factor and phenotype). Figure 2 presents two additional examples of sentences of PGxCorpus. More examples can be browsed at https://pgxcorpus.loria.fr/. All the corpus abstracts were published between 1952 and 2017.
Table 2.
Numbers of entities annotated in PGxCorpus, by type.
| PGxCorpus entity | Simple | Nested | Discont. | Both N&D | Total |
|---|---|---|---|---|---|
| Chemical | 1,512 | 192 | 2 | 12 | 1,718 |
| Genomic_factor | 21 | 68 | 7 | 3 | 99 |
| ↳Gene_or_protein | 1,685 | 20 | 3 | 0 | 1,708 |
| ↳Genomic_variation | 14 | 37 | 3 | 0 | 54 |
| ↳Limited_variation | 237 | 537 | 98 | 47 | 919 |
| ↳Haplotype | 15 | 112 | 4 | 6 | 137 |
| Phenotype | 282 | 330 | 60 | 27 | 699 |
| ↳Disease | 460 | 143 | 14 | 18 | 635 |
| ↳Pharmacodynamic_phenotype | 157 | 390 | 60 | 25 | 632 |
| ↳Pharmacokinetic_phenotype | 31 | 109 | 14 | 6 | 160 |
| Total | 4,414 | 1,938 | 265 | 144 | 6,761 |
Because nested and discontiguous (Discont.) entities are dealt with differently in our baseline experiments, we report numbers of “simple” annotations, i.e. those that are neither nested nor discontiguous. Nested and Discont. refers to annotations that are either nested or discontiguous. “Both N&D” refers to entities both nested and discontiguous. Every entity is only counted within its most specific type. An entity that appears several times is counted as many times it appears.
Table 3.
Numbers of relations annotated in PGxCorpus, by type.
| isAssociatedWith | 733 |
| ↳influences | 937 |
| ↳causes | 168 |
| ↳decreases | 263 |
| ↳increases | 243 |
| ↳treats | 238 |
| isEquivalentTo | 293 |
| Total | 2,875 |
Every relationship is only counted once within its most specific type.
Fig. 1.
Example of sentence annotated with PGx key and composite entities. The key entities, in red, correspond to entities retrieved by PubTator. Composite entities, in green, were obtained using the PHARE ontology. The syntactic dependency analysis is presented on the bottom of the figure and the entities on top.
Fig. 2.
Two annotated sentences of PGxCorpus. Sentence (a) encompasses a relationship of type influences and of modality hypothetical, denoted by the blue color. Sentence (b) is a title, with two annotated relationships. The first is a relationship of type influences and of modality hypothetical. It is hypothetical because the title states that the paper studies the relation, but not that it is valid. The second relationship is of type causes and annotates a nominal group.
PGxCorpus covers a fair part of the domain of PGx and in particular includes 81.8% of the very important pharmacogenes (or “VIP” genes) as listed by PharmGKB at https://www.pharmgkb.org/vips. The detailed number of occurrences of each VIP gene is in the Supplementary Table S3.
Methods
In this section, we detail the steps of the construction of PGxCorpus, as overviewed by Fig. 3. This construction consists in two main phases: (1) the automatic pre-annotation of named entities and (2) the manual annotation that encompasses the correction of the pre-annotation and the addition of typed relationships between named entities.
Fig. 3.
Overview of the construction of PGxCorpus.
We followed the good practices proposed in26, as well as practical examples provided by EU-ADR, ADE-EXT, SNPPhena and other corpora used in NLP shared tasks such as GENIA27, SemEval DDI28. We considered in particular reports on the MERLOT corpus, which focuses on its annotation guidelines29,30 and inter-annotator agreement31.
Abstract retrieval and sentence splitting
The very first step consists in retrieving, from PubMed32, abstracts of publications related with PGx. This was performed with the tool EDirect33 queried with:
 |
Box 1 |
This query aims at retrieving article abstracts concerned with PGx or containing at least one treatment and one genetic factor. It has been built by browsing manually the hierarchy of the MeSH vocabulary, which annotates PubMed entries. The use of MeSH terms allows PubMed to retrieve articles using synonyms and descendant terms of those used in the query. The query was designed to be general in order to retrieve a large set of abstracts that might mention PGx relationships.
Every retrieved abstract is subsequently split into its constitutive sentences, using GeniaSS34.
Automated pre-annotation
To facilitate the manual annotation of PGx relationships, we automatically pre-annotate sentences with various types of entities of interest for PGx. This pre-annotation is performed in two steps. First, PGx key entities, i.e. Gene, Mutation, Disease and Chemicals, are recognized and annotated with a state-of-the-art Named Entity Recognition (NER) tool. Second, these annotations are extended when they are contained in the description of a PGx composite entity, such as a gene expression or a drug response phenotype.
Recognition of key PGx entities
Pre-annotation is initiated using PubTator35, which recognizes the following biomedical entities from PubMed abstracts: chemicals, diseases, genes, mutations and species. PubTator integrates multiple challenge-winning text mining algorithms, listed in Table 4 along with their performances on various benchmark corpora. Disease recognition is performed with DNorm, which uses BANNER36, a trainable system using Conditional Random Fields (CRF) and a rich feature set for disease recognition. For genes, GeneTUKit uses a combination of machine learning methods (including CRFs) and dictionary-based approaches. For mutations, tmVar also uses a CRF-based model with a set of features including dictionary, linguistic, character, semantic, case pattern and contextual features. PubTator was chosen for three reasons: it offers a wide coverage of PGx key entities; it provides an easy-to-use API to recover PubMed abstracts along with entity types and their boundaries; and it includes high performance NER tools.
Table 4.
Performances reported for PubTator.
| Entity type | Tool | Evaluated on | Performance | ||
|---|---|---|---|---|---|
| P | R | F1 | |||
| Chemicals | Dictionary-based49 | n/a | n/a | n/a | 53.82 |
| Disease | DNorm50 | NCBI Disease | 82.8 | 81.9 | 80.9 |
| Corpus | |||||
| Gene | GeneTUKit51 | n/a | n/a | n/a | 82.97 |
| GNAT-100 | 43.0 | 56.7 | 48.9 | ||
| Mutation | tmVar52 | MutationFinder | 98.80 | 89.62 | 93.98 |
| Corpus | |||||
PubTator is the NER tool used during the pre-annotation step of PGxCorpus. P, R and F1 stand for Precision, Recall and F1-score, respectively. n/a denotes we were not able to find information to fill the cell.
Extension of the annotations with the PHARE ontology
The second phase of the pre-annotation consists in automatically extending key entity annotations, when possible, with the PHARE (PHArmacogenomic RElationships) ontology7. This ontology encompasses frequent terms that, associated in nominal structure with PGx key entities, form PGx composite entities. These terms were obtained by analyzing dependency graphs of nominal structures in which a key entity syntactically modifies another term, and in turn were structured in the PHARE ontology. In the example provided in Fig. 1, the drug name acenocoumarol syntactically modifies the term sensitivity. According to the PHARE ontology, the term sensitivity, when modified by a drug, forms a composite entity that belongs to the DrugSensitivity class. Since this class is a subclass of the Phenotype class, acenocoumarol sensitivity may also be typed as a Phenotype. Following this principle, annotations of PGx key entities made by PubTator are extended, when possible, to PGx composite entities, then typed with classes of the PHARE ontology. To this end, the dependency graph of each sentence is constructed with the Stanford Parser37 and in each graph, the direct vicinity of key entities is explored during the search for terms defined in PHARE.
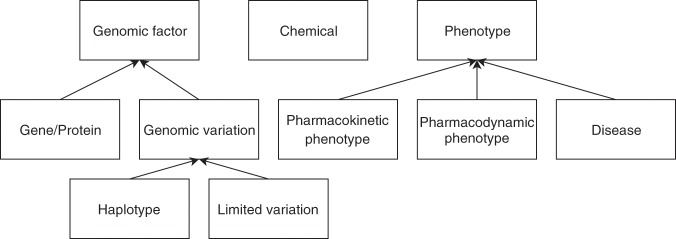
To homogenize the types of entities in PGxCorpus, we first defined a reduced set of entities of interest, listed in Fig. 4 and then defined mappings from PubTator entities and PHARE classes on one side to the types allowed in PGxCorpus on the other side. These mappings are reported in Table 5. Note that we decided to use a type Chemical, instead of Drug: first because we rely on PubTator that recognizes chemicals (without distinguishing them from drugs), second because it allows to broadly include more candidate entities that may be involved in PGx relationships, such as drug metabolites or not yet approved drugs. Furthermore, we decided on a type named Gene_or_protein, broader than Gene, because it is hard to disambiguate between gene and protein names in NLP, and because it is commonly assumed that the task of gene name recognition is indeed a gene-or-protein name recognition38.
Fig. 4.
Types of entities annotated in PGxCorpus and their hierarchy.
Table 5.
Mapping between PubTator entities types, PHARE classes and PGxCorpus entity types.
| Origin | Initial type | Type in PGxCorpus |
|---|---|---|
| PubTator | Chemical | Chemical |
| Disease | Disease | |
| Gene | Gene_or_protein | |
| Mutation | Limited_variation | |
| PHARE | Drug | Chemical |
| DrugMetabolite | Chemical | |
| Gene | Gene_or_protein | |
| GenomicRegion | Genomic_factor | |
| GenomicVariation | Genomic_variation | |
| GeneProduct | Gene_or_protein | |
| Mutation | Limited_variation | |
| Phenotype | Phenotype |
Manual annotations
Before the manual annotation itself, malformed sentences (sentence tokenization errors) and sentences that did not contain at least one drug and one genetic factor, according to PubTator or PHARE are filtered out.
Out of the remaining sentences, we randomly select 1,897 of them to be manually annotated. The annotation process is realized by 11 annotators, out of which 5 are considered senior annotators. Annotators are either pharmacists (3), biologists (3) or bioinformaticians (5). Each sentence is annotated in three phases: first, it is independently annotated by two annotators (senior or not); second, their annotations are, in turn, compared and revised by a third, senior annotator; last, a homogenization phase ends the process.
During the first phase, annotators are provided with sentences and entity pre-annotations. At this stage, they correct pre-annotations, add potential relationships between them, and discard sentences which are ambiguous or not related to PGx domain. Sentences discarded by at least one annotator are not considered for the second phase. During both first and second phases, sentences are randomly assigned to annotators, but we ensure that senior annotators only revise sentences they did not annotate in the first phase.
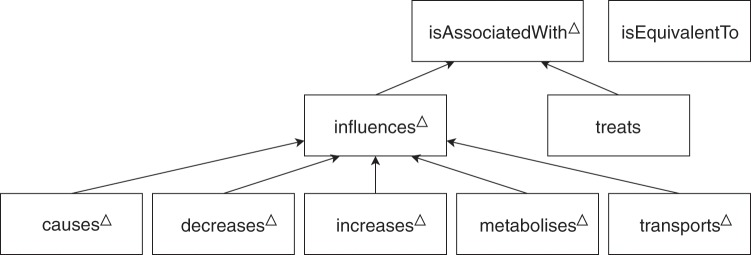
In order to ensure the consistency of the manual annotations, annotators are provided with detailed guidelines39. Those describe the type of entities and relationships to annotate (given in Figs. 4 and 5), relationship modality (affirmed, negated, hypothetical), the main rules to follow, along with examples. Entity and relationship types are organized in simple hierarchies. Some of the relationship types are directly related to PGx (denoted by Δ in Fig. 5), whereas some have a broader scope (i.e. isEquivalentTo and treats). Guidelines also provide an how-to-use guide for the annotation tool and answers frequently-asked questions. The first version of the guidelines was written before the first phase of the annotation. Examples and clarifications were regularly added to the document during the first phase of the annotation. Guidelines were subject to an important revision between the two first annotation phases, to clarify how to annotate ambiguous cases, which had been raised by annotators or by the evaluation of an agreement score between annotators (see Section Inter-annotator agreement).
Fig. 5.
Types of relationships annotated in PGxCorpus and their hierarchy. Types directly related to PGx are marked with Δ, wheras isEquivalentTo and treats have a broader scope.
The final phase of homogenization ends the corpus construction process to reduce the heterogeneity that remains in the annotations after the second phase. Two expert annotators review together sentences in two times: the first time is a complete pass on all annotated sentences to identify sources of heterogeneity. The second time consists in (a) listing sentences associated with each source of heterogeneity using programmatic scripts and keywords, (b) reaching a consensus for their annotation, and (c) modifying the annotations accordingly. Sources of heterogeneity identified at this stage include: the annotation of drug combinations, of dose-related phenotypes, of mutation-related cancer types (e.g. p53-positive breast cancer), of behavior-related phenotypes (e.g. drug abuse, drug dependence), of genomic factors (e.g. exons, promoters, regulatory regions), of treated conditions (e.g. transplantations or post-surgery treatments) and uncommon types of relationships. To address the latter, annotations made with uncommon types (i.e. ‘metabolizes’ and ‘transports’) are turned into their upper-level type of annotations (i.e. ‘influences’). This explains why ‘metabolizes’ and ‘transports’ types are present in Fig. 5, but not in Table 3. We explain the heterogeneity in annotations by the fact that, in some cases, guidelines were specific but disregarded by annotators; in other cases they were caused by unexpected examples that were absent from the guidelines.
Data Records
Statistics on the preparation of PGxCorpus
PubMed has been queried with our initial query (Box 1) in July 2017, to retrieve 86,520 distinct abstracts, split out in 657,538 sentences. Statistics of pre-annotations obtained with PubTator and PHARE on these sentences are provided in Tables 6 and 7, respectively. After filtering malformed sentences and sentences that do not contain at least one genomic factor and one drug, we obtain 176,704 sentences, out of which we randomly pick 1,897 sentences that are subsequently manually annotated. This number of sentences is chosen in regards to constraints of the distribution of the annotation task. These sentences come from 1,813 distinct abstracts.
Table 6.
Type and number of entities recognized by PubTator in the pre-annotation.
| PubTator entity | Number recognized |
|---|---|
| Chemical | 90,816 |
| Disease | 125,487 |
| Gene | 196,460 |
| Mutation | 25,417 |
Table 7.
Number of entities pre-annotated after extending PubTator annotation with the PHARE ontology.
| PHARE entity | Discontiguous | All |
|---|---|---|
| Chemical | 430 | 87,764 |
| Disease | 0 | 29,589 |
| Gene_or_protein | 4,690 | 10,1326 |
| Genomic_variation | 8,698 | 13,601 |
| Phenotype | 10,935 | 16,770 |
Because discontiguous entities are excluded from our baseline experiments (see Section Technical Validation), their number is specified. No disease entity is discontiguously annotated first because PubTator is not generating discontiguous annotation, and second because the extension of annotations with PHARE (which may be discontiguous) is not producing disease annotations, but phenotype annotations.
The first phase of manual annotation, by 11 annotators, took roughly four months. The mean number of sentences annotated by an annotator is 344.73 (standard deviation = 126.33) sentences for this phase. The second phase, by 5 senior annotators, took four other months. Each senior annotator revised 258.6 (sd = 0.54) sentences. Annotations were made on a voluntary basis, which explains the relatively long length of this process.
Sentences that were clearly not about PGx or that presented obvious problems such as incompleteness, typos or ambiguity were asked to be discarded. Accordingly, annotators discarded 952 sentences out of the 1,897 randomly picked, leaving 945 sentences. The main reason for those discards was the scope of sentences. A typical example is the large number of sentences about genetic therapies annotators had to discard, since those also contain both a drug and a gene name and then were selected according to our filtering criteria.
Technical Validation
In this section we present an inter-annotator agreement analysis and results of a baseline experiment of relation extraction where PGxCorpus is used as the training set of a neural network model.
Inter-annotator agreement
Metrics
The annotation task considered for PGxCorpus is particularly rich: it employs 10 entity types, 9 relation types and 3 relation modalities (sometimes named attributes); in addition, entities may be nested or discontiguous. Given this richness, metrics to control the variability of the annotations have been evaluated, in particular at the end of the first phase of the manual annotation, when each sentence has been annotated independently by two annotators. We compute an agreement score that evaluates how much annotators agreed with each others using the F1-score, following40,41. In this case, the agreement or F1-score, is measured using one annotator as a reference and the other as a prediction. Note that inverting the reference and the prediction only inverts the precision and the recall but has no effect on the F1-score itself. We preferred the F1-score instead of other conventional measures such as the kappa coefficient42 because of the complexity of our annotation task. Kappa coefficient is designed to evaluate inter-annotator agreements while taking into account the probability that the agreement might be due to random guess. It is adapted when annotators select a category, out of a set, to annotate already marked-up entities. Then, the larger the set is, the less probable an agreement occurs by chance. In our case, the annotators not only need to select a category, but also to identify the boundaries of these potential entities. In this setting, the probability of a by-chance agreement within the kappa coefficient is low and unadapted. The F1-score is defined as the harmonic mean of the precision and recall , i.e. .
Entity agreement
Agreement on entity annotations is determined in four ways, in regards to two parameters: (a) using exact or partial match; (b) considering the entity hierarchy or not.
An exact match occurs when two annotators agree on both the entity type and their boundaries. A partial match is more flexible since it occurs when two annotators agree on the entity type, but annotation boundaries only overlap. Note that an annotation from the first annotator may overlap with multiple annotations from the second annotator, and vice versa. Considering every overlapping entities as a match would artificially increase the recall and the precision because only one can indeed reflect an agreement between the two annotators. We ensure in this case that an entity from the first annotator is matched with at most one entity from the second annotator using the Hopcroft-Karp algorithm43. In this case, the problem is seen as a maximum matching problem in a bipartite graph, where each set of annotations, one for each annotator, represents a sub-graph. The edges between the two sets represent possible overlaps between one annotation from the first annotator and another from the second.
We also consider a more flexible setting where the agreement takes into account upper hierarchies of entities and relationships, as defined in Figs. 4 and 5. For instance, if a first annotator annotates an entity as Pharmacokinetic phenotype (PK) and a second as Pharmacodynamic phenotype (PD), we consider they agreed to some extent, since both are subtype of Phenotype. In this setting, it can be considered that an entity (or relationship) is indeed annotated with several types: the one specified by an annotator and its parents in the hierarchy. In practice, if we consider the first annotator as the reference and the second as the prediction, we can distinguish three cases: (1) The prediction is more specific than the reference. In this case, common annotations shared by reference and prediction are counted as true positives, while annotations of the prediction that are too specific are false positives. For instance if the reference is Phenotype and the prediction is PD; we count one false positive in the evaluation of PD predictions, but the additional Phenotype annotation, inferred from the hierarchy, enables to count one true positive for Phenotype predictions. (2) The prediction is less specific than the reference. In this case, common annotations shared by reference and prediction are counted as true positives, while classes from the reference that are missed by the prediction are false negative. For instance if the reference is PD and the prediction is Phenotype, we count one true positive for Phenotype prediction, but one false negative in the prediction of PD. (3) The reference and the prediction do not have a direct hierarchy relationships, but a common ancestor (like PD and PK). In this case classes that are shared by the prediction and reference (i.e. the common ancestors) are counted as true positive, but too specific predictions are counted as false positives and missed predictions are counted as false negatives. For instance if the reference is PD and the prediction is PK, we count one true positive for the prediction of Phenotype (i.e. the only common ancestor), one false positive for the prediction of PK and one false negative for the prediction of PD.
Table 8 presents the inter-annotator entity agreement scores, obtained after the first phase of manual annotation, depending on settings (a) and (b). We observe that for relatively simple entities such as chemicals, genes, haplotypes or diseases, F1-score overpasses 70%, even on the strictest constraints (exact match, no hierarchy). We also observe that for more complex entities such as phenotype or genomic variations, annotators tend to agree on the presence of an entity, but not on its offset. This lead to some very low agreement. This motivates us to update the annotation guidelines between the two annotation phases, to clarify on how to decide on entity offsets. For instance it was clarified that adjective qualifying phenotypes should not be annotated, except if they are part of the name of the disease or symptom, but not if they simply qualify its localisation or gravity. Accordingly when encountering “acute pain” annotators should not annotate acute but only pain; whereas encountering “Acute Myeloid Leukemia”, a specific family of leukemia, annotators have to annotate all three words. When considering the hierarchy, the performances for the leaves of the hierarchy should not be affected. However, a slight drop is observed due to the use of the Hopcroft-Karp algorithm. Indeed, when using the hierarchy more potential matches can be observed between prediction and reference annotations which generate more edges in the associated bipartite graph. The Hopcrof-Karp algorithm then removes some of the correct matches between leaves, causing a slight drop in the recall.
Table 8.
Inter-annotator agreement (F1-score) for entity annotations.
| Entity matching: (exact or partial) | exact | exact | partial | partial |
|---|---|---|---|---|
| Considering hierarchy: (yes or no) | no | yes | no | yes |
| Chemical | 76.8 | 76.8 | 82.1 | 82.1 |
| Genomic_factor | 38.6 | 72.6 | 38.8 | 85.7 |
| ↳Gene_or_protein | 85.3 | 85.3 | 90.0 | 89.4 |
| ↳Genomic_variation | 32.9 | 49.3 | 53.0 | 76.8 |
| ↳Limited_variation | 50.8 | 50.8 | 69.0 | 66.2 |
| ↳Haplotype | 76.2 | 76.2 | 77.2 | 76.1 |
| Phenotype | 30.5 | 51.0 | 53.9 | 72.6 |
| ↳Disease | 71.3 | 71.0 | 80.9 | 79.1 |
| ↳Pharmacokinetic_phenotype | 48.2 | 48.2 | 57.0 | 57.0 |
| ↳Pharmacodynamic_phenotype | 31.7 | 31.7 | 47.0 | 47.0 |
| Macro average | 57.4 | 63.8 | 68.7 | 76.1 |
Four different settings enabling more or less flexibility are presented. The agreement score is computed after the first phase of manual annotation.
Relation agreement
Regarding the inter-annotator agreement on relation annotations, we consider the same two settings, plus an additional one: (a) using exact or partial match, which applies in this case to the two entities involved in the relation; (b) the consideration of the hierarchy, which applies in this case to both the hierarchy of entities and relations (see Figs. 4 and 5); (c) whether the direction of the relation is considered or not. Resulting agreements are presented in Table 9.
Table 9.
Inter-annotator agreement (F1-score) for the annotation of relations. Five different settings are presented.
| Entity matching: | exact | exact | partial | partial | partial |
|---|---|---|---|---|---|
| Considering hierarchies: | none | both | none | both | both |
| Considering direction: | yes | yes | yes | yes | no |
| isAssociatedWith | 12.6 | 14.3 | 13.2 | 33.3 | 33.3 |
| ↳influences | 12.8 | 12.8 | 17.7 | 29.3 | 29.8 |
| ↳causes | 35.8 | 35.2 | 37.6 | 37.2 | 39.6 |
| ↳decreases | 25.8 | 26.8 | 33.6 | 36.7 | 36.7 |
| ↳increases | 14.5 | 15.6 | 27.4 | 30.2 | 30.2 |
| ↳metabolizes | 59.0 | 59.0 | 61.5 | 61.5 | 61.5 |
| ↳transports | 83.1 | 83.1 | 83.1 | 83.1 | 83.1 |
| ↳treats | 33.2 | 34.7 | 36.3 | 37.3 | 37.3 |
| isEquivalentTo | 39.6 | 40.2 | 40.7 | 41.3 | 62.5 |
| Macro average | 47.3 | 47.1 | 50.3 | 53.8 | 57.0 |
Although the agreement on the relations is low, note that a relation can be considered correct only if an initial agreement on the two entities in relation has been reached.
Pre-annotation correction
To evaluate how much annotators had to correct the automatically computed pre-annotations, we measured an agreement score between automatic pre-annotations and final annotations. Just like for the inter-annotator agreement, this is computed in the form of an F1-score. In this case, the final annotation is considered as the gold standard, and the F1-score measures how automatic pre-annotation performs in regards to the final annotation ground truth. Table 10 presents F1-score agreements in four distinct settings, which allows to consider how much the boundaries or the type of pre-annotations (or both) needed corrections. The precision of 94.1% when boundaries and type are relaxed means that pre-annotations point at offsets where something is indeed to annotate (94.1% of the time). The relatively low recall in the relaxed setting (64%) illustrates the amount of fully new annotations added by annotators. When considering stricter setting, with an exact match or a same type or both we observe that precision is getting lower, illustrating that entity boundaries and types frequently require corrections. Note that this agreement is only relevant for named entity, but not for relations because the latter are not pre-annotated, but fully done manually.
Table 10.
Agreement (F1-score) between pre- and final annotations.
| Entity matching: | Exact | Exact | Partial | Partial |
|---|---|---|---|---|
| Type matching: | same type | potentially different | same type | potentially different |
| F1-Score (P;R) | 54.2 (66.9;45.5) | 59.0 (72.9;49.6) | 64.4 (79.6;54.1) | 76.2 (94.1;64.0) |
This agreement evaluates the amount of manual corrections that was required after the automatic pre-annotation phase. Precision (P) and recall (R) are given between parenthesis.
Baseline experiments
In this section, we report on baseline experiments with PGxCorpus, which quantitatively evaluate its usefulness for extracting PGx entities and relations from text. The task evaluated here is composed of a first step of named entity recognition (NER) and a second one of relation extraction (RE). The NER is achieved with a variant of a Convolutional Neural Network (CNN) model, whereas the RE is processed with a multichannel CNN (MCCNN). Both models are detailed in the Supplementary Methods section of the Supplementary Material. Source code of the experiments is available at https://github.com/practikpharma/PGxCorpus/.
In a related work44, we used a preliminary, partial and naive set of annotations to test the feasibility of extracting relations and incorporating them in a knowledge network. This included only 307 sentences (out of 945), annotated with a simplified schema of only 4 entity types and 2 relation types. The associated model for RE was simplistic, since it only aimed at empirically demonstrating feasibility. The baseline experiment reported here considers all sentences of PGxCorpus and has been done with more advanced annotation schema and models.
Baseline performances
The objective of these experiments was not to reach the best performances but rather to propose a baseline for future comparisons, as well as to empirically demonstrate the usefulness of PGxCorpus for extracting PGx entities and relations from text.
Named entity recognition
Performances for the named entity recognition experiments, evaluated with a 10-fold cross validation, are reported in Table 11. A main limitation of the NER model is that discontiguous entities were not considered. This may hurt the performance even for contiguous entities since discontiguous entities were considered as negative, even though they might be very similar (from the model point of view) to contiguous entities.
Table 11.
Performances of the task of named entity recognition in terms of F1-score (and its standard deviation in brackets, for the last setting).
| Entity matching: (exact or partial) | exact | exact | partial | partial |
|---|---|---|---|---|
| Considering hierarchy: (yes or no) | no | yes | no | yes |
| Chemical | 76.07 | 76.07 | 82.67 | 82.67 (7.24) |
| Genomic_factor | 22.86 | 71.41 | 27.68 | 83.19 (5.90) |
| ↳Gene_or_protein | 85.72 | 85.72 | 90.58 | 90.05 (3.89) |
| ↳Genomic_variation | 2.67 | 49.13 | 3.83 | 71.18 (9.55) |
| ↳Limited_variation | 47.08 | 47.02 | 72.71 | 71.57 (9.50) |
| ↳Haplotype | 66.97 | 66.97 | 72.47 | 72.47 (19.34) |
| Phenotype | 31.76 | 50.80 | 48.48 | 69.57 (5.40) |
| ↳Disease | 66.90 | 66.88 | 75.68 | 72.59 (7.30) |
| ↳Pharmacokinetic_phenotype | 29.30 | 29.30 | 36.47 | 36.27 (19.40) |
| ↳Pharmacodynamic_phenotype | 38.54 | 38.50 | 58.84 | 58.18 (10.11) |
| Macro average | 49.15 | 59.11 | 59.76 | 71.93 (5.64) |
Balance between precision and recall, as well as details on standard deviations are provided in Supplementary Table S1.
Several observations can be made from the results reported in Table 11. First, the best performances were obtained for Chemical, Gene_or_protein and Disease types, for which (1) the number of training samples is high, (2) PubTator annotations are available and (3) the ratio between normal entities and nested and/or discontiguous entities is low (see Table 2). Note that the definition for the Limited_variation entity used in our corpus is broader than the Mutations recognized by PubTator. PubTator recognizes precises descriptions of variations such as “VKORC1:C > A”, but not general ones such as “a VKORC1 polymorphism”, which we consider. This explains why the performances obtained for Limited_variation were lower than those obtained with PubTator (see Table 4). Even though the number of training samples for Pharmacokinetic_phenotype and Haplotype is low, we obtained reasonable performances. This may be due to a rather homogeneous phrasing and syntax whenever these entities are mentioned. Not considering the hierarchy in cases like Genomic_variation or Genomic_factor types for which few training samples are available and a high heterogeneity is observed led to poor performances. Lastly we note that, as expected, the standard deviation for classes with only few examples annotated was high or very high (above 19 for Haplotype and Pharmacokinetic_phenotype). The random distribution of these “rare” examples between train and test sets, in the 10-fold cross validation, had a strong impact on performances, and explains large standard deviations. Concerning concepts that are leaves of the hierarchy, we observed a slight drop in performances when considering the hierarchy. This is due to the use of the Hopcroft-Karp algorithm as mentioned in the Entity agreement Subsection.
Relation extraction
Performances for the relation extraction (RE) experiments, evaluated with a 10-fold cross validation, are reported in Table 12. The RE model faced several limitations. (1) For a given sentence along with identified entities, the relation predictions were independent. This is obviously too simplistic and the prediction should be made globally. (2) We considered relationships annotated as negated or hypothetical by annotators like regular relationships.
Table 12.
Performances of the task of relation extraction in terms of F1-score (and standard deviation).
| Considering hierarchies: (yes or no) | no | yes |
|---|---|---|
| is Associated WithΔ | 30.89 | 51.71 (4.02) |
| ↳influencesΔ | 36.55 | 46.45 (5.17) |
| ↳causesΔ | 41.91 | 41.91 (13.35) |
| ↳decreasesΔ | 29.47 | 29.47 (9.85) |
| ↳increasesΔ | 17.94 | 17.94 (15.20) |
| ↳treats | 39.97 | 39.97 (12.60) |
| isEquivalentTo | 79.76 | 79.76 (7.69) |
| Macro average | 45.67 | 49.56 (4.51) |
| PGx relations only(Δ), no hierarchy | 54.04 (3.31) | |
The last line provides results of an experiment for which only one category is considered, merging all the types specific to PGx (marked with Δ). For leaves, performances are unchanged when considering the hierarchy. Balance between precision and recall, as well as details on standard deviations are provided in Supplementary Table S2.
Several observations can be made about the RE results reported in Table 12. First, the fact that the model had to deal with multiple, complex and associated classes made the classification problem difficult and the performances relatively low. The experiment in which we considered the hierarchy showed that, even if it was difficult to identify a specific type of relation, is was easier for the model to determine whether there was a relation between two entities or not. In other words, many mis-classifications were in fact predictions for types that belong to the same branch of the hierarchy. Like for the NER, types of relations with less examples tended to be associated with poorer performances and higher standard deviations (except for the isEquivalentTo relationship, which is very homogeneous). To build upon these observations, and particularly to avoid the impact of isEquivalentTo type that is not specific to PGx, we evaluated how PGxCorpus can be used to train a model for any relation specific to PGx (denoted with Δ in Table 12), without considering sub-types nor the hierarchy. Results of this experiment is provided on the last line of Table 12.
Several enhancements could be proposed to improve this baseline model. First, in our implementation, the hierarchy was not considered during the training phase. Accordingly, learning to predict a leaf penalized all the other categories, even those that were considered correct at test time. This explains why the “PGx Relations only” experiment led to better performances than individual classifications with or without hierarchy. On the other hand, considering the hierarchy at training would increase the number of examples for the higher categories of the hierarchy, potentially harming performances for the leaves. A model enabling multiclass labeling and a weighting dependent on the size of classes should balance this bias.
Building upon PGxCorpus
We assembled a manually annotated corpus for pharmacogenomics, named PGxCorpus, and provide an experimental validation of its usefulness for the tasks of NER and RE in this domain.
Unlike existing corpora, PGxCorpus encompasses all three key entities involved in PGx relationships (drugs, genomic factors and phenotypes) and provides a fine-grained hierarchical classification for both PGx entities and relationships. By making this corpus freely available, our objective is to enable the training of supervised PGx relation extraction systems and to facilitate the comparison of their performances. Furthermore, the baseline experiment illustrates that PGxCorpus allows the study of many challenges inherent with biomedical entities and relationships such as discontiguous entities, nested entites, multilabeled relationships, heterogenous distributions, etc. In particular, PGxCorpus offers both a training resource for supervised approaches and a reference to evaluate and compare to future efforts. Out of PGx, such a corpus may serve connected domains by the use of transfer learning approaches, as illustrated by45. For all these reasons, we think that the tasks of PGx NER and RE, supported by the novel existence of PGxCorpus, are well suited for proposing NLP Challenges and shared tasks. By this mean we expect that PGxCorpus will stimulate NLP research as well as facilitate the synthesis of PGx knowledge.
Usage Notes
PGxCorpus is made available under the Creative Commons Attribution 4.0 International Public License (CC-BY). The programmatic code of our baseline experiments is available at https://github.com/practikpharma/PGxCorpus/tree/master/baseline_experiment.
Supplementary information
Acknowledgements
The authors acknowledge their funding institutions: the French National Research Agency (ANR) supports the PractiKPharma project (ANR-15-CE23-0028) and the University of Lorraine program “Lorraine UniversitéExcellence” (15-IDEX-0004); Inria supports the Snowball Inria Associate Team.
Author contributions
J.L. conducted the annotation campaign, designed and conducted baseline experiments, and wrote the manuscript. R.G. conducted the annotation campaign. A.C., C.B., C.J.L., J.L., K.D., M.D.D., M.S.T., N.C.N., N.P., R.G., W.D. annotated the corpus and reviewed the manuscript. P.R. advised on technical aspects of the project and set up the annotation servers. Y.T. advised on the design of the project and in the writing of the manuscript. A.C. designed the study, supervised the annotation campaign and wrote the manuscript. All authors read and approved the final manuscript.
Data availability
PGxCorpus is available in the BioNLP shared task file format46 at three locations:
figshare, an open access data repository47
A BRAT server48, enabling a user friendly online visualization of the annotations: https://pgxcorpus.loria.fr/
A Git repository of the whole project that also includes the annotation guidelines and programmatic code of the baseline experiments presented in Technical Validation https://github.com/practikpharma/PGxCorpus/.
Code availability
A Git repository of the project is accessible at https://github.com/practikpharma/PGxCorpus/. It includes the annotation guidelines, the corpus itself and the programmatic code of the baseline experiments presented in Technical Validation.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-019-0342-9.
References
- 1.Xie H-G, Frueh FW. Pharmacogenomics steps toward personalized medicine. Pers. Med. 2005;2:325–337. doi: 10.2217/17410541.2.4.325. [DOI] [PubMed] [Google Scholar]
- 2.Caudle KE, et al. Incorporation of pharmacogenomics into routine clinical practice: the clinical pharmacogenetics implementation consortium (CPIC) guideline development process. Curr. Drug Metab. 2014;15:209–217. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.2018. U.S. Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labelling. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm
- 4.Martin MA, et al. Clinical pharmacogenetics implementation consortium guidelines for HLAB genotype and abacavir dosing: 2014 update. Clin. Pharmacol. Ther. 2014;95:499–500. doi: 10.1038/clpt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulet A, Cohen KB, Altman RB. The state of the art in text mining and natural language processing for pharmacogenomics. J. Biomed. Inform. 2012;45:825–826. doi: 10.1016/j.jbi.2012.08.001. [DOI] [Google Scholar]
- 6.Hahn U, Cohen KB, Garten Y, Shah NH. Mining the pharmacogenomics literature - a survey of the state of the art. Brief. Bioinform. 2012;13:460–494. doi: 10.1093/bib/bbs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulet A, Shah NH, Garten Y, Musen M, Altman RB. Using text to build semantic networks for pharmacogenomics. J. Biomed. Inform. 2010;43:1009–1019. doi: 10.1016/j.jbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rance B, Doughty E, Demner-Fushman D, Kann MG, Bodenreider O. A mutationcentric approach to identifying pharmacogenomic relations in text. J. Biomed. Inform. 2012;45:835–841. doi: 10.1016/j.jbi.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Friedman C, Finkelstein J. Automated metabolic phenotyping of cytochrome polymorphisms using PubMed abstract mining. AMIA Annu. Symp. Proc. 2017;2017:535. [PMC free article] [PubMed] [Google Scholar]
- 10.Percha B, Altman RB. Learning the structure of biomedical relationships from unstructured text. PLoS Computational Biology. 2015;11:e1004216. doi: 10.1371/journal.pcbi.1004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kissa M, Tsatsaronis G, Schroeder M. Prediction of drug gene associations via ontological profile similarity with application to drug repositioning. Methods. 2015;74:71–82. doi: 10.1016/j.ymeth.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Chang JT, Altman RB. Extracting and characterizing gene–drug relationships from the literature. Pharmacogenet. Genomics. 2004;14:577–586. doi: 10.1097/00008571-200409000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Rinaldi F, Schneider G, Clematide S. Relation mining experiments in the pharmacogenomics domain. J. Biomed. Inform. 2012;45:851–861. doi: 10.1016/j.jbi.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Pakhomov SVS, et al. Using PharmgKB to train text mining approaches for identifying potential gene targets for pharmacogenomic studies. J. Biomed. Inform. 2012;45:862–869. doi: 10.1016/j.jbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Wang Q. A knowledge-driven conditional approach to extract pharmacogenomics specific drug-gene relationships from free text. J. Biomed. Inform. 2012;45:827–834. doi: 10.1016/j.jbi.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, et al. Deep learning of mutation-gene-drug relations from the literature. BMC Bioinform. 2018;19:21. doi: 10.1186/s12859-018-2029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whirl-Carrillo M, et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karimi S, Metke-Jimenez A, Kemp M, Wang C. Cadec: A corpus of adverse drug event annotations. J. Biomed. Inform. 2015;55:73–81. doi: 10.1016/j.jbi.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Lee K, et al. Bronco: Biomedical entity relation oncology corpus for extracting gene-variantdisease- drug relations. Database. 2016;2016:baw043. doi: 10.1093/database/baw043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson P, et al. Annotation and detection of drug effects in text for pharmacovigilance. J. Cheminformatics. 2018;10:37. doi: 10.1186/s13321-018-0290-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zolnoori, M. et al. Development of an adverse drug reaction corpus from consumer health posts for psychiatric medications. In Proceedings of the 2nd Workshop on Social Media Mining for Health Research and Applications @ AMIA 2017 (SMM4H) 19–26 (2017).
- 22.van Mulligen EM, et al. The EU-ADR corpus: Annotated drugs, diseases, targets, and their relationships. J. Biomed. Inform. 2012;45:879–884. doi: 10.1016/j.jbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Gurulingappa H, Mateen-Rajpu A, Toldo L. Extraction of potential adverse drug events from medical case reports. J. Biomed. Semant. 2012;3:15. doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokharaeian B, Esteban AD, Taghizadeh N, Chitsaz H, Chavoshinejad R. SNPPhena: A corpus for extracting ranked associations of single-nucleotide polymorphisms and phenotypes from literature. J. Biomed. Semant. 2017;8:14:1–14:13. doi: 10.1186/s13326-017-0116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai, K. S., Socher, R. & Manning, C. D. Improved semantic representations from treestructured long short-term memory networks. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics, ACL 2015 1556–1566 (2015).
- 26.Leech, G. In Developing Linguistic Corpora: A Guide to Good Practice, Vol. 92 (ed. Wynne, M.) Adding linguistic annotation. (Oxbow Books, 2005).
- 27.Kim J-D, Ohta T, Tsujii J. Corpus annotation for mining biomedical events from literature. BMC Bioinform. 2008;9:10. doi: 10.1186/1471-2105-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrero-Zazo M, Segura-Bedmar I, Martínez P, Declerck T. The DDI corpus: An annotated corpus with pharmacological substances and drug-drug interactions. J. Biomed. Inform. 2013;46:914–920. doi: 10.1016/j.jbi.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Campillos L, et al. A French clinical corpus with comprehensive semantic annotations: development of the medical entity and relation LIMSI annotated text corpus (MERLOT) Lang. Resour. Eval. 2017;52:1–31. [Google Scholar]
- 30.Campillos, L. et al. Annotation scheme for the MERLOT French clinical corpus, https://cabernet.limsi.fr/annotation_guide_for_the_merlot_french_clinical_corpus-Sept2016.pdf (2016).
- 31.Deléger, L., Ligozat, A.-L., Grouin, C., Zweigenbaum, P. & Névéol, A. Annotation of specialized corpora using a comprehensive entity and relation scheme. In Proceedings of the Ninth International Conference on Language Resources and Evaluation, LREC 2014, 1267–1274 (2014).
- 32.Lu Z. PubMed and beyond: A survey of web tools for searching biomedical literature. Database. 2011;2011:baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kans, J. In Entrez Programming Utilities Help Entrez direct: E-utilities on the Unix command line, https://www.ncbi.nlm.nih.gov/books/NBK179288/ (National Center for Biotechnology Information, 2013).
- 34.Sætre R, et al. AKANE system: Protein-protein interaction pairs in BioCreAtIvE 2 challenge, PPI-IPS subtask. Proceedings of the second BioCreAtIvE challenge workshop. 2007;209:212. [Google Scholar]
- 35.Wei C-H, Kao H-Y, Lu Z. PubTator: A web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013;41:W518–W522. doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leaman R, Gonzalez G. BANNER: An executable survey of advances in biomedical named entity recognition. Biocomputing 2008, Proceedings of the Pacific Symposium. 2008;2008:652–663. [PubMed] [Google Scholar]
- 37.de Marneffe, M., MacCartney, B. & Manning, C. D. Generating typed dependency parses from phrase structure parses. In Proceedings of the Fifth International Conference on Language Resources and Evaluation, LREC 2006, 449–454 (2006).
- 38.Yeh A, Morgan A, Colosimo M, Hirschman L. BioCreAtIvE task 1A: Gene mention finding evaluation. BMC Bioinform. 2005;6:S2. doi: 10.1186/1471-2105-6-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legrand, J. et al. PGxCorpus – Annotation guidelines, https://github.com/practikpharma/PGxCorpus/blob/master/annotation_guidelines.pdf (2017).
- 40.Gurulingappa H, et al. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform. 2012;45:885–892. doi: 10.1016/j.jbi.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. J. Am. Med. Inform. Assoc. 2005;12:296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 43.Hopcroft JE, Karp RM. An nˆ5/2 algorithm for maximum matchings in bipartite graphs. SIAM J. Comput. 1973;2:225–231. doi: 10.1137/0202019. [DOI] [Google Scholar]
- 44.Monnin P, et al. PGxO and PGxLOD: a reconciliation of pharmacogenomic knowledge of various provenances, enabling further comparison. BMC Bioinform. 2019;20:139. doi: 10.1186/s12859-019-2693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legrand, J., Toussaint, Y., Raïssi, C. & Coulet, A. Syntax-based transfer learning for the task of biomedical relation extraction. In Proceedings of the Ninth International Workshop on Health Text Mining and Information Analysis, LOUHI 2018, 149–159 (2018).
- 46.Pyysalo S, et al. Overview of the ID, EPI and REL tasks of BioNLP Shared Task 2011. BMC Bioinform. 2012;13:S2. doi: 10.1186/1471-2105-13-S11-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Legrand J, 2019. PGxCorpus: a manually annotated corpus for pharmacogenomics. figshare. [DOI] [PMC free article] [PubMed]
- 48.Stenetorp, P. et al. Brat: a webbased tool for nlp-assisted text annotation. In Proceedings of the Demonstrations at the 13thConference of the European Chapter of the Association for Computational Linguistics, EACL 2012 102–107 (2012).
- 49.Wiegers TC, Davis AP, Mattingly CJ. Collaborative biocuration—text-mining development task for document prioritization for curation. Database. 2012;2012:bas037. doi: 10.1093/database/bas037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leaman R, Islamaj Doğan R, Lu Z. Dnorm: Disease name normalization with pairwise learning to rank. Bioinformatics. 2013;29:2909–2917. doi: 10.1093/bioinformatics/btt474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang M, Liu J, Zhu X. GeneTUKit: A software for document-level gene normalization. Bioinformatics. 2011;27:1032–1033. doi: 10.1093/bioinformatics/btr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei C-H, Harris BR, Kao H-Y, Lu Z. tmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. 2013;29:1433–1439. doi: 10.1093/bioinformatics/btt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2018. U.S. Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labelling. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm
- Legrand J, 2019. PGxCorpus: a manually annotated corpus for pharmacogenomics. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
PGxCorpus is available in the BioNLP shared task file format46 at three locations:
figshare, an open access data repository47
A BRAT server48, enabling a user friendly online visualization of the annotations: https://pgxcorpus.loria.fr/
A Git repository of the whole project that also includes the annotation guidelines and programmatic code of the baseline experiments presented in Technical Validation https://github.com/practikpharma/PGxCorpus/.
A Git repository of the project is accessible at https://github.com/practikpharma/PGxCorpus/. It includes the annotation guidelines, the corpus itself and the programmatic code of the baseline experiments presented in Technical Validation.