Abstract
Purpose
To develop a deep convolutional neural network (CNN) model to categorize multiphase CT and MRI liver observations using the Liver Imaging Reporting and Data System (LI-RADS) (version 2014).
Methods
A pre-existing dataset comprising 314 hepatic observations (163 CT, 151 MRI) with corresponding diameters and LI-RADS categories (LR-1–5) assigned in consensus by two LI-RADS steering committee members was used to develop two CNNs: pre-trained network with an input of triple-phase images (training with transfer learning) and custom-made network with an input of quadruple-phase images (training from scratch). The dataset was randomly split into training, validation, and internal test sets (70:15:15 split). The overall accuracy and area under receiver operating characteristic curve (AUROC) were assessed for categorizing LR-1/2, LR-3, LR-4, and LR-5. External validation was performed for the model with the better performance on the internal test set using two external datasets (EXT-CT and EXT-MR: 68 and 44 observations, respectively).
Results
The transfer learning model outperformed the custom-made model: overall accuracy of 60.4% and AUROCs of 0.85, 0.90, 0.63, 0.82 for LR-1/2, LR-3, LR-4, LR-5, respectively. On EXT-CT, the model had an overall accuracy of 41.2% and AUROCs of 0.70, 0.66, 0.60, 0.76 for LR-1/2, LR-3, LR-4, LR-5, respectively. On EXT-MR, the model had an overall accuracy of 47.7% and AUROCs of 0.88, 0.74, 0.69, 0.79 for LR-1/2, LR-3, LR-4, LR-5, respectively.
Conclusion
Our study shows the feasibility of CNN for assigning LI-RADS categories from a relatively small dataset but highlights the challenges of model development and validation.
Keywords: Carcinoma, Hepatocellular; Deep Learning; Tomography, X-Ray Computed; Magnetic Resonance Imaging
Introduction
The Liver Imaging Reporting and Data System (LI-RADS) was developed to standardize terminology and criteria for interpreting and reporting findings of CT and MRI examinations of the liver in patients at risk for developing hepatocellular carcinoma (HCC) [1]. Reducing variability in the interpretation of imaging findings was one of key motivations for its development, with the goal of improving clarity of communication between radiologists and other specialists caring for these patients. Fowler et al. recently reported that the overall interobserver agreement of LI-RADS categorization was substantial among 113 radiologists (intraclass correlation coefficient of 0.67 for CT and 0.73 for MRI) [2]. On the other hand, Schellhaas et al. [3], Barth et al. [4], and Davenport et al. [5] have reported lower and more variable interobserver agreement values (κ = 0.35–0.61), using different study designs. Even though LI-RADS offers a standardized diagnostic algorithm, there remains variability in categorizing observations.
Deep convolutional neural networks (CNNs) are among several state-of-the-art machine learning techniques that can perform similarly to or sometimes even surpass human experts in medical image classification tasks [6–10]. In theory, CNNs can be developed to mimic expert performance on classification tasks by training them with expert-assigned labels. Consequently, a well-trained CNN algorithm with a dataset labeled by expert radiologists could have the potential to objectively and reproducibly provide LI-RADS categories concordant with the expert classification. Such algorithms may ultimately reduce interpretation variability related to relative expertise of the LI-RADS user and potentially bridge the gap between the novice and expert radiologist.
In this study, we trained CNN models for the classification of hepatic observations on multiphase CT and MRI in patients at risk for HCC according to LI-RADS v2014 with the dataset of images previously reported by Fowler et al. [2], in which consensus categories had been assigned by two members of the LI-RADS Steering Committee. We assessed model performance in a held-out internal test set as well as in two externally collected datasets.
Materials and Methods
Development and Internal Evaluation of the Algorithm
Dataset for Model Development
For model development, we used a pre-existing de-identified HIPAA-compliant dataset [2] comprising axial multiphase contrast-enhanced images in JPEG format and displayed in a PowerPoint file of 381 unique hepatic observations (193 on CT and 188 on MRI). All images were acquired with an extracellular space intravenous contrast agent or gadobenate dimeglumine. PowerPoint slides were annotated with corresponding observation diameters. Two members of the LI-RADS Steering Committee (** and **) assigned LI-RADS categories in consensus using the LI-RADS v2014 algorithm including ancillary features and threshold growth [1] based on only the multiphase images in the PowerPoint file. This yielded 61 LR-1, 35 LR-2, 65 LR-3, 69 LR-4, 102 LR-5, 32 LR-5V, and 17 LR-M observations. While Fowler et al. [2] previously evaluated interreader reliability of LI-RADS using this dataset, in our current study, we used the dataset for training a deep learning model to categorize liver observations according to LI-RADS. Thus, we created a final cohort (hereafter referred to as the “LR-Atlas”) by excluding observations without either pre-contrast images or arterial phase images, observations without available diameter measurements, and observations categorized as either LI-RADS (LR-) 5V or LR-M. The former was excluded because this study focused on parenchymal observation assessment. The latter was excluded because the number of LR-M observations in the given dataset was small (n = 17) with a risk of severe class imbalance.
Image Preprocessing for Model Development
A radiologist (**, with 10 years of experience in liver CT and MRI interpretation), who was blinded to the consensus LI-RADS categories, manually cropped the observations in the PowerPoint file using the following standards: 1) the same square regions of interest (ROIs) were used across multiphase images in a given exam, 2) cropping size was chosen so that the longest observation diameter was greater than 50% of the size of the final cropped image, 3) ROIs included a margin of peri-observation liver parenchyma and excluded non-liver structures as much as possible. Cropped images were saved in bitmap format to retain the pixel resolution of the images (ranging from about 30 × 30 to about 120 × 120 pixels) and then resized to a 224 × 224 matrix.
Model Development
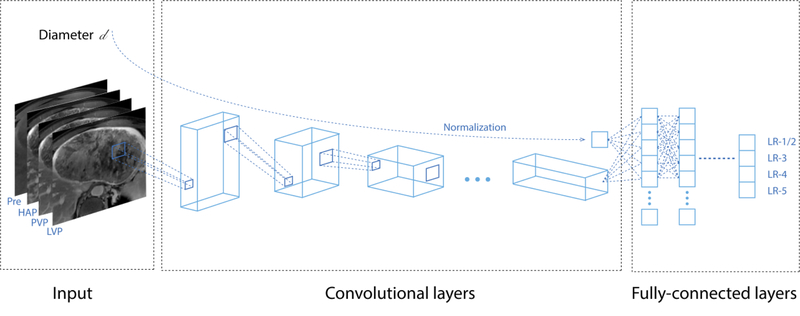
Two distinct models were developed using two CNN architectures: 1) a VGG16 [11] pre-trained network built on ImageNet [12] with triple phase images as the input (pre-contrast, late arterial and delayed phases); and 2) a custom-made network with quadruple phase images as the input (pre-contrast, late arterial, portal venous, and delayed phases). Observation diameters were included in the networks, processed through a fully-connected neural network, and concatenated with the bottleneck layers of the networks (Figure 1). Model output was a vector of probabilities for four classes representing LI-RADS categories (LR-1/2, LR-3, LR-4, LR-5), where the consensus LI-RADS categories were considered as ground truths. LR-1 and LR-2 were pooled into LR-1/2 because they represent observations that are probably or definitely benign and would require no follow-up [1].
Fig. 1.
Model architecture overview. Multiphase CT or MR images as well as observation diameters served as the input, and consensus categories assigned by LI-RADS Steering Committee members served as ground truth labels. Quadruple phase images are shown as an input in this figure for display purpose. Abbreviations: HAP, hepatic arterial phase; LI-RADS, the Liver Imaging Reporting and Data System; LVP, late venous phase; Pre, pre-contrast phase; PVP, portal venous phase
Appendix E1 (see Electronic Supplementary Material 1) details the model architectures and training process. Briefly, for the custom-made network model, we trained the CNN from scratch solely on the LR-Atlas dataset, whereas the pre-trained VGG16 model was trained using a fine-tuning transfer learning method [13]. The LR-Atlas dataset was randomly split into training, validation, and test sets at a ratio of 70:15:15. All training images were augmented using on-the-fly technique. For the custom-made network model, if either a portal venous or delayed phase image was unavailable (n = 4 and 7, respectively), the 4th channel was a copy of the other available venous/delayed phase. Prior examinations and image sets other than the dynamic contrast-enhanced images were not included. Each model was trained on the training set, while hyperparameter tuning and model selection were performed on the validation set. As a sub-analysis, another transfer learning-based network was developed with each of the two distinct subsets of the LR-Atlas dataset: a subset that consisted solely of CT images and a subset that consisted solely of MR images. All the code and analyses were written and performed by one of the authors (**) and reviewed and confirmed externally by another author (**) from a separate institution.
Evaluating the Algorithm with External Datasets
Dataset for External Validation
Model performance was tested independently at two institutions (institution 1: **, and institution 2: **). The model architecture and its weights were provided to these institutions in a Jupyter notebook format [14] and hierarchical data format. At each institution, the Institutional Review Board approved and waived the requirement for informed consent for the external validation study. Two external datasets with at least 40 observations each were created: At institution 1, CT cases between January 2017 and June 2017 were selected consecutively. At institution 2, MRI cases between February 2013 and April 2015 were selected randomly. The inclusion criteria were patients at risk for HCC with quadruple phase dynamic contrast-enhanced CT or MR images available. Following the creation of the datasets, an abdominal radiologist at each institution (** at institution 1, ** at institution 2), each with 9 years of post-fellowship experience, who were blinded to the outputs of the developed CNN, created ground truths by assigning LI-RADS categories according to the LI-RADS v2014 criteria to each observation based on only the multiphase images without referring to prior examinations or other image sets. To determine the final external validation cohort at each institution (hereafter called “EXT-CT” and “EXT-MR”), patients with no observations, observations with histories of prior interventions, examinations with severe artifacts that make images uninterpretable, non-measurable observations, examinations with hepatobiliary (liver-specific) MR contrast agents, and observations categorized LR-5V or LR-M were excluded.
Image Preprocessing for External Validation
Manual image preprocessing was performed on the Picture Archiving and Computer System by a single radiologist at each institution (** for institution 1, and ** for institution 2, with 10 and 3 years of experience in liver CT and MRI interpretation, respectively). For each observation, the radiologist selected the representative axial slice with maximum observation diameter for each phase. The radiologist selected the window width and level at his or her discretion to improve the contrast between the observation and liver parenchyma. Images were magnified and cropped to achieve a pixel resolution similar in scale to that of the training dataset and resized to 224 × 224 as in the training dataset.
Statistical Analysis
To evaluate classification performance, overall accuracy, confusion matrix, and area under receiver operating characteristic curve (AUROC) were calculated for the 4-class classification (LR-1/2, LR-3, LR-4, and LR-5). Overall accuracy was defined as the number of correctly classified observations divided by the total number of observations and was unweighted. Both models (custom-made network and transfer learning-based network) were tested on the held-out internal test set derived from LR-Atlas. Based on performance results across all performance metrics on the internal test set only, the model with the better performance was further evaluated on the external validation sets EXT-CT and EXT-MR. In order to investigate the dependency of the experiments on random state in splitting dataset, the average performance over five experiments with five different random state values for the data splitting was evaluated for the better performing model.
Quadratic weighted κ statistics were also calculated to evaluate radiologist agreement with reference to the consensus LI-RADS categories. We considered a κ value greater than 0.81 as denoting almost perfect agreement and values of 0.61–0.80, 0.41–0.60, 0.21–0.40, 0.00–0.20, and < 0.00 as denoting substantial, moderate, fair, slight, and poor agreement, respectively [15].
Differences in the data distributions between LR-Atlas and EXT-CT/MR were also summarized in terms of frequencies of each LI-RADS category, diagnostic entities in LR-1/2, and observation diameter. LI-RADS category and observation characteristics in LR-1/2 were tested with Fisher’s exact test, whereas observation diameter was compared with Student’s t-test. All P values were assessed at an alpha of 0.05.
Statistical analysis was performed using the Python programming language (version 3.6.4, Python Software Foundation, https://www.python.org/) with the Scipy version 1.0.0 [16] and scikit-learn version 0.19.1 packages [17], and R version 3.5.1 [18].
Results
Datasets
In total, the LR-Atlas dataset included 314 unique observations excluding 12 observations without pre-contrast images, 9 observations without available diameters, 32 observations categorized as LR-5V, and 15 observations categorized as LR-M. The distribution of observation categories was: LR-1/2 (n=89), LR-3 (n=62), LR-4 (n=65), and LR-5 (n=98). In total, the external validation dataset consisted of 112 unique observations (68 on the EXT-CT dataset from 47 scans of 41 unique patients, 44 on the EXT-MR dataset from 15 scans of 11 unique patients). Table 1 shows the distribution of LI-RADS categories for each dataset, with the following category distribution: LR-1/2 (n= 25), LR-3 (n=26), LR-4 (n=29), and LR-5 (n=32).
Table 1.
LI-RADS category distribution in the datasets
| LR-Atlas | EXT-CT | EXT-MR | |||
|---|---|---|---|---|---|
| Total | CT | MRI | CT | MRI | |
| LR-1/2 | 89(28.3) | 36(22.1) | 53(35.1) | 19(27.9) | 6 (13.6) |
| LR-3 | 62(19.7) | 41(25.2) | 21(13.9) | 15(22.1) | 11(25.0) |
| LR-4 | 65(20.7) | 35(21.5) | 30(19.9) | 15(22.1) | 14(31.8) |
| LR-5 | 98(31.2) | 51(31.3) | 47(31.1) | 19(27.9) | 13(29.5) |
| Total | 314 | 163 | 151 | 68 | 44 |
Data represent the number of observations with percentages in parentheses.
Abbreviation: LI-RADS and LR-, the Liver Imaging Reporting and Data System
Model performance
Internal Validation (Model Performance on Held-out Test Set Derived from LR-Atlas)
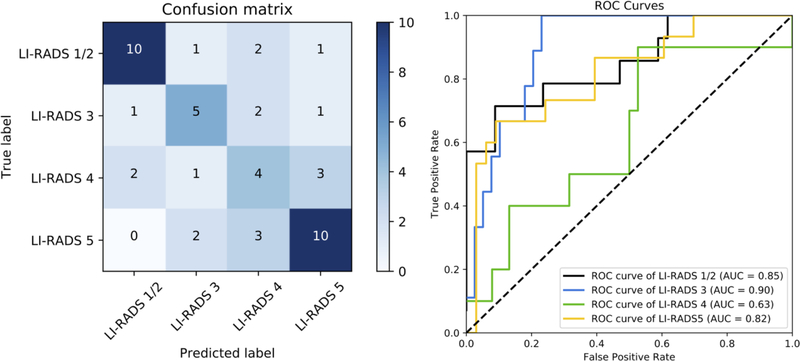
On the LR-Atlas dataset, the transfer learning-based network had an overall accuracy of 60.4%, AUROC of 0.85, 0.90, 0.63, 0.82 for LR-1/2, LR-3, LR-4, LR-5, respectively, and weighted κ of 0.65 (Figure 2). The average performance of the transfer learning-based network over five experiments was as follows: a mean overall accuracy of 59.6%, mean AUROCs of 0.79, 0.84, 0.68, 0.86 for LR-1/2, LR-3, LR-4, LR-5, respectively, and mean weighted κ of 0.58. The custom-made network had an overall accuracy of 37.5%, AUROC of 0.78, 0.65, 0.48, 0.77 for LR-1/2, LR-3, LR-4, LR-5, respectively, and weighted κ of 0.54. For the sub-analysis, the transfer learning-based model trained and tested solely on CT images in the LR-Atlas had an overall accuracy of 60.0%, weighted κ of 0.55, and AUROCs of 0.84, 0.77, 0.82, 0.84 for LR-1/2, LR-3, LR-4, LR-5, respectively; and the transfer learning-based model trained and tested solely on MR images in the LR-Atlas had an overall accuracy of 43.5%, weighted κ of 0.18, and AUROCs of 0.78, 0.47, 0.84, 0.66 for LR-1/2, LR-3, LR-4, LR-5, respectively.
Fig. 2.
Left panel: Confusion matrix for the transfer learning model on the held-out test set from LR-Atlas, which shows the number of observations for each combination of ground truth and predicted LI-RADS category. The numbers of correct categorization are on the diagonal line from the top-left triangle to the bottom-right triangle. Wrong decisions due to overcategorization are in the bottom-left triangle, and wrong decisions due to undercategorization are in the top-right triangle. Right panel: An ROC diagram with corresponding AUCs for the transfer learning model on the held-out test set from LR-Atlas. An ROC curve was created for each LI-RADS category versus all other categories by sweeping a threshold over the predicted probability of the particular LI-RADS category.
Abbreviations: AUC, area under the curve; LI-RADS, the Liver Imaging Reporting and Data System; ROC, receiver operating characteristic curve
External Validation (Model Performance on EXT-CT and EXT-MR)
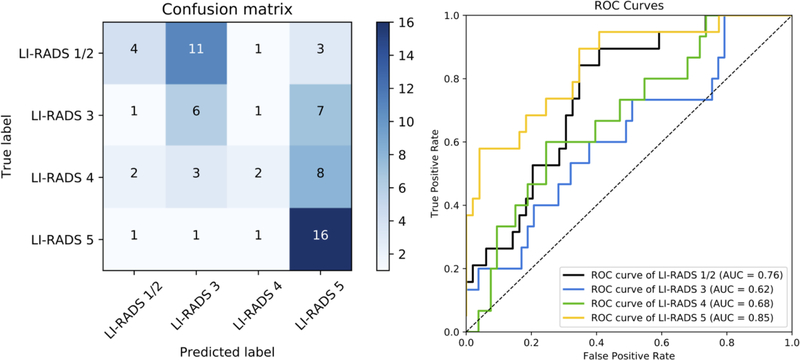
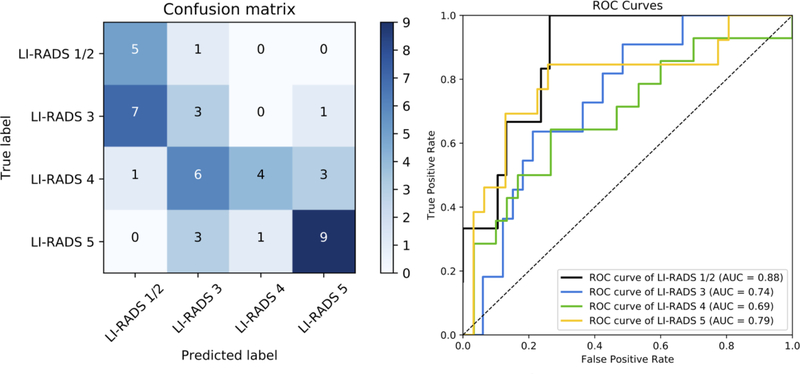
Based on the better performance of the transfer learning-based network with the LR-Atlas dataset, this model was subsequently tested with the externally collected datasets. On the EXT-CT dataset, the model had an overall accuracy of 41.2%, AUROC of 0.76, 0.62, 0.68, 0.85 for LR-1/2, LR-3, LR-4, LR-5, respectively, and weighted κ of 0.46 (Figure 3). On the EXT-MR dataset, the model had an overall accuracy of 47.7%, AUROC of 0.88, 0.74, 0.69, 0.79 for LR-1/2, LR-3, LR-4, LR-5, respectively, and weighted κ of 0.67 (Figure 4). Figure E1 (see Electronic Supplementary Material 2) shows the accuracy of the model plotted as a function of a threshold for the probability of the predicted LI-RADS category. Greater accuracy was achieved at increasing thresholds for both EXT-CT and EXT-MR datasets. Examples of correctly classified and misclassified observations in the external datasets are shown in Figures 5 and 6, respectively.
Fig. 3.
Left panel: Confusion matrix for the transfer learning model on the EXT-CT external test set, which shows the number of observations for each combination of ground truth and predicted LI-RADS category. The numbers of correct categorization are on the diagonal line from the top-left triangle to the bottom-right triangle. Wrong decisions due to overcategorization are in the bottom-left triangle, and wrong decisions due to undercategorization are in the top-right triangle. Right panel: An ROC diagram with corresponding AUCs for the transfer learning model on EXT-CT external test set. An ROC curve was created for each LI-RADS category versus all other categories by sweeping a threshold over the predicted probability of the particular LI-RADS category (right).
Abbreviations: AUC, area under the curve; LI-RADS, the Liver Imaging Reporting and Data System; ROC, receiver operating characteristic curve
Fig. 4.
Left panel: Confusion matrix for the transfer learning model on the EXT-MR external test set, which shows the number of observations for each combination of ground truth and predicted LI-RADS category. The numbers of correct categorization are on the diagonal line from the top-left triangle to the bottom-right triangle. Wrong decisions due to overcategorization are in the bottom-left triangle, and wrong decisions due to undercategorization are in the top-right triangle. Right panel: An ROC diagram with corresponding AUCs for transfer-learned model on the EXT-MR external test set. An ROC curve was created for each LI-RADS category versus all other categories by sweeping a threshold over the predicted probability of the particular LI-RADS category (right).
Abbreviations: AUC, area under the curve; LI-RADS, the Liver Imaging Reporting and Data System; ROC, receiver operating characteristic curve
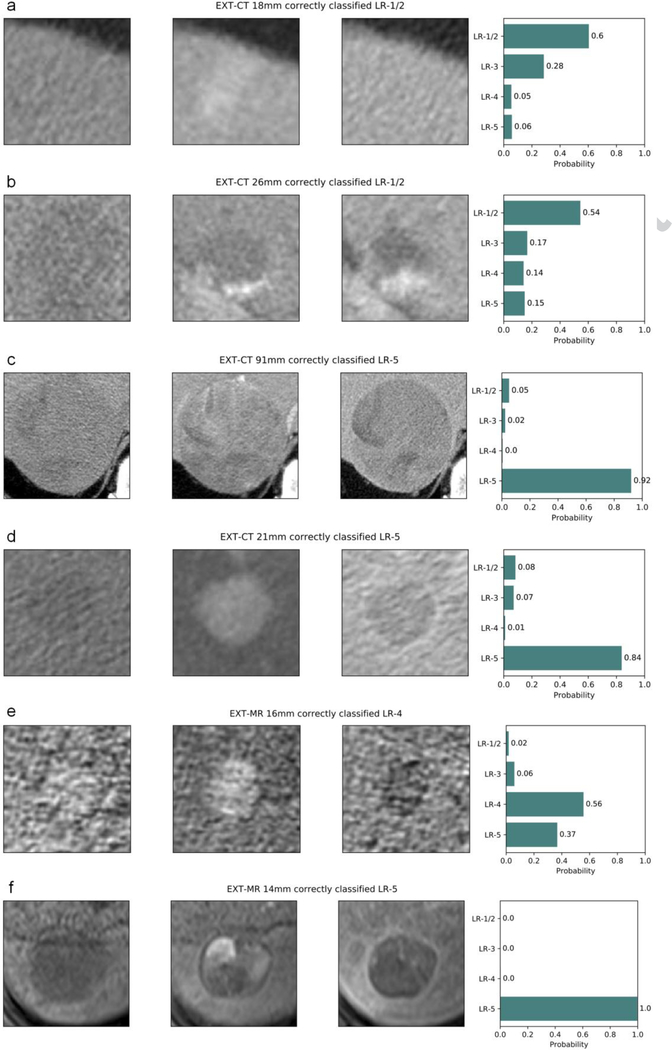
Fig. 5.
Examples of correctly categorized (LI-RADS v2014) observations on external datasets: (a–d) EXT-CT and (e, f) EXT-MR. Each plot title includes the dataset name, observation diameter, ground truth, and predicted LI-RADS category. Pre-contrast, late arterial, and delayed phase cropped images are displayed from left to right, and the rightmost bar plot shows the output probability for each LI-RADS category. (a) transient arterial phase hyperenhancement (LR-1/2), (b) typical hemangioma (LR-1/2), (c, d, f) HCC (LR-5), (e) probable HCC (LR-4).
Abbreviations: HCC, hepatocellular carcinoma, LI-RADS and LR-, the Liver Imaging Reporting and Data System
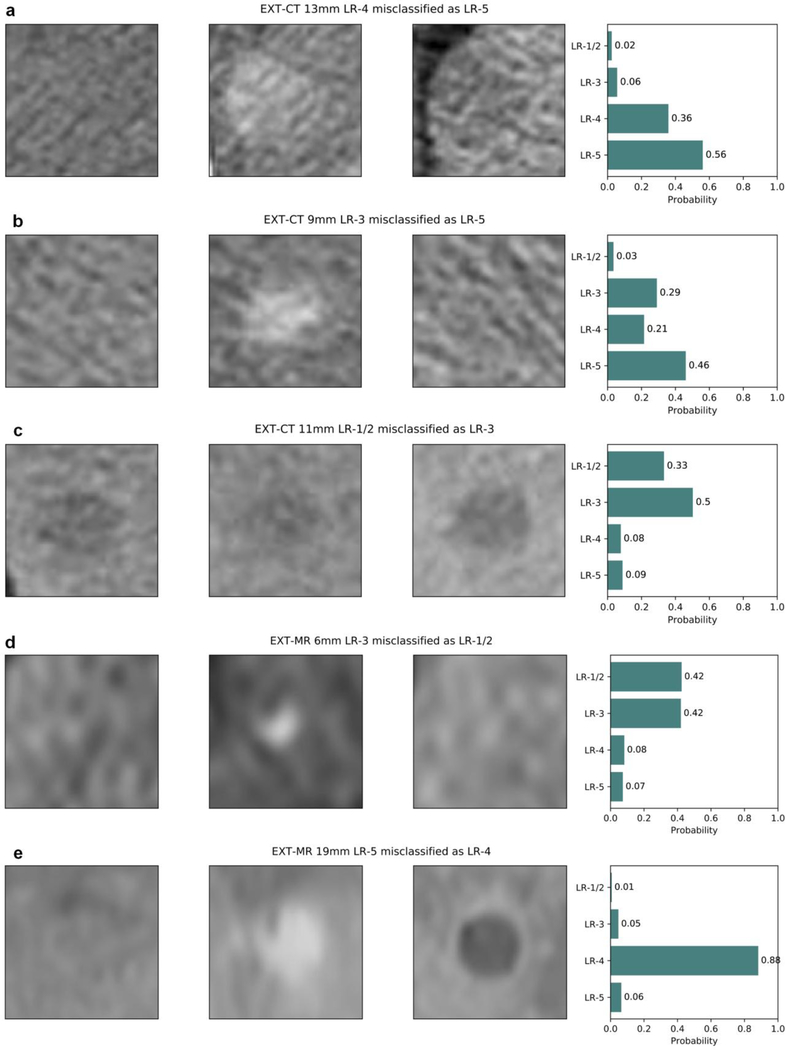
Fig. 6.
Examples of misclassified LI-RADS v2014 observations on external datasets: (a–d) EXT-CT and (e, f) EXT-MR. Each plot title includes the dataset name, observation diameter, ground truth, and predicted LI-RADS category. Pre-contrast, late arterial, and delayed phase cropped images are displayed from left to right, and the rightmost bar plot shows the output probability for each LI-RADS category. (a) An LR-4 observation with a diameter of 13 mm was misclassified as LR-5 by the model, which falls in the cell having two options, LR-4 and LR-5 according to the LI-RADS version 2014 diagnostic table; this, however, should be categorized as LR-5 by the updated LI-RADS version 2018. Probabilities for LR-1/2 and LR-3 were low (2% and 6%, respectively). (b) A case of nodule-like arterial phase hyperenhancement with a diameter of 9 mm had a ground truth label of LR-3 but was misclassified as LR-5 by the model. A major feature of arterial hyperenhancement is evident, whereas washout or enhancing capsule appearance is not apparent. However, the central part of the delayed phase image might appear slightly hypodense compared with the peripheral area. The probability for LR-1/2 was only 3%. (c) A small cyst had a ground truth label of LR-1/2 but was misclassified as LR-3 by the model. Cropped low resolution image due to small observation size may make the observation blurry and hard to be correctly categorized (specifically on late arterial phase image for this particular case). Probabilities for LR-4 and LR-5 were low (8% and 9%, respectively). (d) A 6 mm observation with arterial hyperenhancement had a ground truth label of LR-3 but was misclassified as LR-1/2 by the model, where the probabilities for LR-1/2 and LR-3 was almost the same. Interobserver variability for such observations probably exist even among experts depending on their experience. (e) A 19 mm HCC with a ground truth label of LR-5 was misclassified as LR-4 with a probability of 0.88. HCC exhibited all of the major imaging features: arterial hyperenhancement, washout, and enhancing capsule.
Abbreviations: HCC, hepatocellular carcinoma; LI-RADS and LR-, the Liver Imaging Reporting and Data System
Difference in Data Distribution Between Datasets
The differences in the frequency of each LI-RADS category between LR-Atlas and EXT-CT was not significant (p = 0.67), whereas the distribution among MR entries in LR-Atlas and EXT-MR was significantly different (p = 0.01).
Table 2 shows the distribution of observation characteristics in the LR-1/2 class in LR-Atlas, EXT-CT, and EXT-MR. The distribution of characteristics was significantly different between LR-Atlas and EXT-CT (p = 0.0006). Cirrhosis-related nodules were present (n = 4, 21.1%) in EXT-CT but not present in LR-Atlas. In addition, liver cysts were more frequent in EXT-CT compared with LR-Atlas. Although the EXT-MR dataset only had 6 LR-1/2 entries, the distribution of characteristics was not significantly different compared with LR-Atlas (p = 0.64).
Table 2.
Observation characteristics in LR-1/2 entries
| LR-Atlas | EXT-CT | EXT-MR | |||
|---|---|---|---|---|---|
| LR-1/2 | Total | CT | MRI | CT | MRI |
| cyst | 7(7.9) | 5(13.9) | 2(3.8) | 10(52.6) | 1(16.7) |
| hemangioma | 6(6.7) | 4(11.1) | 2(3.8) | 1(5.3) | 0(0) |
| steatosis | 9(10.1) | 5(13.9) | 4(7.5) | 0(0) | 0(0) |
| perfusion alteration | 44(49.4) | 16(44.4) | 28(52.8) | 4(21.1) | 4(66.7) |
| fibrosis | 9(10.1) | 3(8.3) | 6(11.3) | 0(0) | 1(16.7) |
| pseudomass | 5(5.6) | 3(8.3) | 2(3.8) | 0(0) | 0(0) |
| distinctive nodule without malignant | |||||
| features | 9(10.1) | 0(0) | 9(17.0) | 4(21.1) | 0(0) |
| Total | 89 | 36 | 53 | 19 | 6 |
Data represent the number of observations with percentages in parentheses.
Abbreviation: LR-, the Liver Imaging Reporting and Data System
LR-Atlas, EXT-CT, and EXT-MR had a mean observation diameter of 27.2 mm (range: 4–170 mm), 22.8 mm (range: 5–167 mm), and 19.4 mm (range: 6–61 mm), respectively. Table 3 shows the distribution of the observation diameters in LR-Atlas, EXT-CT, and EXT-MR. Among CT entries, LR-1/2 and LR-4 observations for LR-Atlas had significantly larger diameters compared with that of the EXT-CT dataset. Among MR entries, the diameter for LR-Atlas was significantly different than that of EXT-MR in all LI-RADS classes but LR-1/2.
Table 3.
Statistical results in the differences of observation diameter between the datasets
| EXT-CT | EXT-MR | |
|---|---|---|
| LR-Atlas | 0.2687 (−2.934496, 10.443996) | 0.004522 (2.633242, 14.137109) |
| LR-1/2 | 0.00813 (4.432317, 28.038271) | 0.4449 (−11.34848, 23.77878) |
| LR-3 | 0.5049 (−7.232346, 3.629094) | 0.007659 (1.788302, 10.722520) |
| LR-4 | 0.03953 (−0.1958626, 7.6517564) | 0.01301 (−10.227108, −1.325273) |
| LR-5 | 0.6713 (−22.09857, 14.48867) | 0.009196 (4.44123, 29.80754) |
Data represent p values with 95% confidence intervals in parentheses.
Abbreviation: LR-, the Liver Imaging Reporting and Data System
Discussion
In this study, we developed a CNN model for assigning LI-RADS categories to liver observations on multiphase CT and MRI using a relatively small annotated dataset of JPEG images through transfer learning and data augmentation techniques. The transfer learning model outperformed the CNN model trained from scratch for a LI-RADS categorization task, which is expected given the relatively small sample size of the training dataset. Our results suggest that although medical images are different from natural images on which pre-trained models are trained, features from pre-trained models are still useful for medical imaging classification tasks and a CNN has the potential to categorize hepatic observations without defining explicit hand-crafted imaging features.
As Yasaka et al. showed, data augmentation techniques can be used to improve the performance of a CNN model trained on a relatively small dataset [19]. Unlike natural images, medical images often involve subsets of data covering the same anatomy but with different technical or physiological parameters. In our study, our dataset involved different phases of contrast enhancement. As there is no standard deep learning architecture for image classification with multiphase images yet, we assigned each imaging phase into an input channel for CNN in the same manner as Yasaka et al. [19].
We found that the transfer learning CNN had an inferior performance on external CT and MRI datasets than on the internal held-out test set. Zech et al. [20] recently demonstrated that CNN models trained to detect pneumonia on chest radiographs had lower performance when tested on external datasets with different disease burden and prevalence. Park [21] emphasized the difference between a diagnostic case-control study and a diagnostic cohort study as an external validation methodology for real-world clinical practice. In our study, we found that the LR-Atlas dataset demonstrated significant differences in case selection compared with the external validation sets. The Atlas was not created to reflect typical cases encountered in clinical practice; instead, it had an over-representation of rare liver entities hand-selected as exemplary cases, including geographic fat deposition, confluent fibrosis, hypertrophic pseudomass, nodule like arterial phase hyperenhancement, and distinctive nodule without malignant features for LR-1/2 observations [22]. In contrast, the external datasets consisted of consecutive or random cases at two different centers. As a result, the specific entities and categories differed between the datasets. This study highlights the challenges in developing clinically relevant models with an appropriate training dataset that is representative of a clinical population and methodology. To develop a LI-RADS CNN for clinical use, creating a larger database of observations more typically seen in a LI-RADS population may be beneficial in the long run.
On the external datasets, the transfer learning model correctly categorized a substantial proportion of the LR-5 observations, whereas model performance was consistently worse for LR-3 and LR-4 observations. This may be due in part to the overlap in major imaging features present for LR-3 and LR-4 with LR-5 such as arterial phase hyperenhancement and washout appearance [1]. In addition, interobserver variability among the expert radiologists who assigned ground truth labels for both training and external datasets might have also influenced the lower model performance on these in-between categories. Davenport et al. previously reported lower interobserver agreements for LR-3 and LR-4 compared with LR-1 and LR-5 [5]. In addition, the low performance on LR-4 might also be due to the lack of testing of ancillary features in external datasets, which may have impacted the consensus ground truth in LR- Atlas. Regarding LR-1/2 observations, 57.9% of LR-1/2 observations were miscategorized as LR-3 on the EXT-CT dataset in contrast to the EXT-MR dataset. A majority (54.5%) of those miscategorized observations were nodule-like arterial phase hyperenhancement or distinctive nodule without malignant features [22]. Since radiologists use their experience or prior knowledge to categorize an observation as LR-1/2 rather than the explicitly algorithmic approach used for categorizing LR-3, LR-4, and LR-5, the poor performance for LR-1/2 may be expected.
This study had several limitations. First, the LR-Atlas dataset had some shortcomings for training a CNN model. It was small for deep learning purposes, despite data augmentation; it included a mixture of compressed CT and MR images in JPEG format at a single representative section with nonuniform but fixed window widths and levels and varying magnification; no patient-wise information for the observations was available, such as whether any two of the observations originate from a single scan or patient; and categories were assigned using LI-RADS v2014 and not the latest v2018. The choice of this dataset was based mainly on the immediate availability of an expertly annotated dataset. As for the first point, we developed models with a dataset that had an admixture of CT and MR images, which was based on an assumption that such models had the potential to learn a common imaging characteristic that is applicable to both CT and MR images, such as dynamic contrast kinetics. The sub-analysis showed that the performance was similar between the model trained on the admixture of CT and MR images and the model trained solely on CT images except for AUROC for LR-4; on the other hand, the model trained solely on MR images was inferior compared to the other two in most of the evaluation metrics, which could suggest a hypothesis that an admixture of CT and MR images might help models learn more generic features in hepatic observation classification such as dynamic contrast kinetics, though this should be further investigated in the future. Our model was also trained on single representative axial images, analogous to those used by Yasaka et al [19]; however, a future CNN model that provides prediction based on more images per observation and larger sections of the liver may yield a superior performance. Second, our model does not take into account ancillary features or threshold growth because of the restriction of the shape of the network input; these same features may have been taken into account during the creation of consensus categories for the LR-Atlas. In the LR-Atlas dataset, the number of cases with positive threshold growth were 10 (5 for LR-4 and 5 for LR-5). Among them, only 6 (3 for LR-4 and 3 for LR-5) affected the final LR categories. The proportion of such cases was small (3/65 [4.6%] for LR-4 and 3/98 [3.1%] for LR-5), and thus the influence of threshold growth on the model was likely small. Third, our method incorporated a number of manual image preprocessing procedures at the radiologists’ discretion, such as image magnification, windowing, and cropping as well as visual registration of multiphase images, which could limit the model performance and reproducibility of the present study. Training an additional network to localize observations with bounding boxes may benefit a future classification model using similar inputs. In addition, the datasets used in the present study incorporated heterogeneous data loss and interpolation depending on the observation size, which may not be insignificant in terms of model performance, especially when the observations are small. Additional limitations were that the ground truths were created by different expert radiologists with slightly different methods between datasets and the assignment of LI-RADS categories had unavoidable subjectivity.
In terms of model training for the LI-RADS classification task, having the consensus categories from the LR-Atlas as ground truths was considered the most practical reference standard available. Although pathologically proven HCC could be used as an alternative, a significant proportion of HCCs are currently not resected and most non-malignant lesions are not pathologically proven so selection bias would limit the clinical application of such prediction models. Even though our model performed inferiorly on external datasets, our results provided insights into the importance of training dataset characteristics. Further investigations on a larger multi-institutional database of consecutive observations that reflect their actual clinical prevalence with the latest version of LI-RADS are warranted.
In conclusion, our study shows the feasibility of using CNN to assign LI-RADS categories to liver observations on multiphase CT and MRI from a relatively small sample of images but highlights the challenges of using a non-representative training sample. Long-term creation of a larger LI-RADS database for CNN training would be helpful and may be best achieved with cases from consecutive patients.
Supplementary Material
Acknowledgements:
We thank Joanne Chin for editorial assistance.
Funding: Supported by JSPS Overseas Research Fellowships (R.Y.) (Japan Society for the Promotion of Science (JSPS/OT/290125)) and the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748 (R.Y. and R.K.G.D.).
Abbreviations
- LI-RADS
Liver Imaging Reporting and Data System
- HCC
hepatocellular carcinoma
- CNN
convolutional neural network
- ROI
region of interest
- AUROC
area under receiver operating characteristic curve
Footnotes
Compliance with Ethical Standards
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent: For this type of study formal consent is not required.
Conflict of interest: The authors declare that they have no conflict of interest.
Reference list
- 1.American College of Radiology ACR LI-RADS v2014. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS/LI-RADS-v2014. Accessed December 11, 2018.
- 2.Fowler KJ, Tang A, Santillan C, Bhargavan-Chatfield M, Heiken J, Jha RC, Weinreb J, Hussain H, Mitchell DG, Bashir MR, Costa EAC, Cunha GM, Coombs L, Wolfson T, Gamst AC, Brancatelli G, Yeh B, Sirlin CB (2018) Interreader Reliability of LI-RADS Version 2014 Algorithm and Imaging Features for Diagnosis of Hepatocellular Carcinoma: A Large International Multireader Study. Radiology 286 (1): 173–185. doi: 10.1148/radiol.2017170376 [DOI] [PubMed] [Google Scholar]
- 3.Schellhaas B, Hammon M, Strobel D, Pfeifer L, Kielisch C, Goertz RS, Cavallaro A, Janka R, Neurath MF, Uder M, Seuss H (2018) Interobserver and intermodality agreement of standardized algorithms for non-invasive diagnosis of hepatocellular carcinoma in high-risk patients: CEUS-LI-RADS versus MRI-LI-RADS. European radiology 28 (10):4254–4264. doi: 10.1007/s00330-018-5379-1 [DOI] [PubMed] [Google Scholar]
- 4.Barth BK, Donati OF, Fischer MA, Ulbrich EJ, Karlo CA, Becker A, Seifert B, Reiner CS (2016) Reliability, Validity, and Reader Acceptance of LI-RADS-An In-depth Analysis. Academic radiology 23 (9): 1145–1153. doi: 10.1016/j.acra.2016.03.014 [DOI] [PubMed] [Google Scholar]
- 5.Davenport MS, Khalatbari S, Liu PS, Maturen KE, Kaza RK, Wasnik AP, Al-Hawary MM, Glazer DI, Stein EB, Patel J, Somashekar DK, Viglianti BL, Hussain HK (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272 (1): 132–142. doi: 10.1148/radiol.14131963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR (2016) Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. Jama 316 (22):2402–2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 7.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542 (7639): 115–118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak J, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Hass C, Bruni E, Wong Q, Halici U, Oner MU, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venancio R (2017) Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. Jama 318 (22):2199–2210. doi: 10.1001/jama.2017.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, Mahajan V, Rao P, Warier P (2018) Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet (London, England) 392 (10162):2388–2396. doi: 10.1016/s0140-6736(18)31645-3 [DOI] [PubMed] [Google Scholar]
- 10.De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, Askham H, Glorot X, O’Donoghue B, Visentin D, van den Driessche G, Lakshminarayanan B, Meyer C, Mackinder F, Bouton S, Ayoub K, Chopra R, King D, Karthikesalingam A, Hughes CO, Raine R, Hughes J, Sim DA, Egan C, Tufail A, Montgomery H, Hassabis D, Rees G, Back T, Khaw PT, Suleyman M, Cornebise J, Keane PA, Ronneberger O (2018) Clinically applicable deep learning for diagnosis and referral in retinal disease. Nature medicine 24 (9):1342–1350. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 11.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv. https://arxiv.org/abs/1409.1556. vol 1409.1556. [Google Scholar]
- 12.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L (2015) ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision 115 (3):211–252. doi: 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 13.Yamashita R, Nishio M, Do RKG, Togashi K (2018) Convolutional neural networks: an overview and application in radiology. Insights into imaging 9 (4):611–629. doi: 10.1007/s13244-018-0639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kluyver T, Ragan-Kelley B, Pérez F, Granger BE, Bussonnier M, Frederic J, Kelley K, Hamrick JB, Grout J, Corlay S (2016) Jupyter Notebooks-a publishing format for reproducible computational workflows. In: Loizides F, Scmidt B (eds) Positioning and Power in Academic Publishing: Players, Agents and Agendas. IOS Press, pp 87–90. doi: 10.3233/978-1-61499-649-1-87 [DOI] [Google Scholar]
- 15.Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33 (1): 159–174. doi: 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 16.Jones E, Oliphant T, Peterson P, et al. SciPy: open source scientific tools for Python, 2001-, http://www.scipy.org/. Accessed on December 10, 2018.
- 17.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V (2011) Scikit-learn: Machine learning in Python. Journal of machine learning research 12:2825–2830. doi:Not available [Google Scholar]
- 18.R Development Core Team (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 19.Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 286 (3):887–896. doi: 10.1148/radiol.2017170706 [DOI] [PubMed] [Google Scholar]
- 20.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK (2018) Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS medicine 15 (11):e1002683. doi: 10.1371/journal.pmed.1002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SH (2019) Diagnostic Case-Control versus Diagnostic Cohort Studies for Clinical Validation of Artificial Intelligence Algorithm Performance. Radiology 290 (1):272–273. doi: 10.1148/radiol.2018182294 [DOI] [PubMed] [Google Scholar]
- 22.Jha RC, Mitchell DG, Weinreb JC, Santillan CS, Yeh BM, Francois R, Sirlin CB (2014) LI-RADS categorization of benign and likely benign findings in patients at risk of hepatocellular carcinoma: a pictorial atlas. AJR American journal of roentgenology 203 (1):W48–69. doi: 10.2214/ajr.13.12169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.