Abstract
Aging humans display an increased prevalence and severity of periodontitis, although the mechanisms underlying these findings remain poorly understood. This report examined antigenic diversity of P. gingivalis related to disease presence and patient demographics. Serum IgG antibody to P. gingivalis strains ATCC33277, FDC381, W50 (ATCC53978), W83, A7A1–28 (ATCC53977) and A7436 was measured in 426 participants [periodontally healthy (n=61), gingivitis (N=66) or various levels of periodontitis (N=299)]. We hypothesized that antigenic diversity in P. gingivalis could contribute to a lack of “immunity” in the chronic infections of periodontal disease. Across the strains, the antibody levels in the oldest age group were lower than in the youngest groups, and severe periodontitis patients did not show higher antibody with aging. While 80% of the periodontitis patients in any age group showed an elevated response to at least one of the P. gingivalis strains, the patterns of individual responses in the older group were also substantially different than the other age groups. Significantly greater numbers of older patients showed strain-specific antibody profiles to only 1 strain. The findings support that P. gingivalis may demonstrate antigenic diversity/drift within patients and could be one factor to help explain the inefficiency/ineffectiveness of the adaptive immune response in managing the infection.
Keywords: antibody, aging, periodontitis, antigenic drift, adaptive immunity
1.1. INTRODUCTION
Alterations in both innate and adaptive immunity have been universally observed in aging populations, and have led to the concept of such terms as “immune aging” or “immunosenescence” to reflect the deteriorating nature of the immune system [1]. Adaptive immune responses have been consistently reported to be adversely affected by aging, and present as increased incidence and severity of a wide array of infectious diseases, as well as the prevalence of development of various cancers [2–5]. Recent reports document the parallel loss of capacity and regulation of normal innate and adaptive immune response with aging that effectively alter immunocompetence and promote the pathogenesis of this diversity of diseases [6, 7]. Nevertheless, the actual cause(s) of immunosenescence and “inflammaging” remain to be established [7, 8].
Aging has been documented to display an increased prevalence and severity of periodontitis, although the underlying causes remain poorly understood. Epidemiological data supports demographic differences in rates of periodontitis the disuse prevalence increased in males, older subjects, and in racial/ethnic minority populations [9]. Among biologically plausible mechanisms for these findings, both innate immune and adaptive immune cells isolated from aged individuals exhibit intrinsic defects that could predispose the elderly to dysregulated immune and inflammatory responses underpinning the exacerbated clinical features of disease with aging [10]. With our current level of knowledge, the true significance and function of the adaptive immune response under normal circumstances remains to be elucidated, let alone how aging specifically modulates the effectiveness of these responses.
The humoral adaptive immune response clearly changes from gingival health to periodontal disease and these responses vary with progressing disease and post-therapy [11, 12]. Measurement of serum IgG antibody from patients with chronic adult periodontitis [13–15] versus healthy controls showed the most consistent elevation to P. gingivalis [16–18]. However, the conundrum of existing data is why there appears to be a coincidence of chronic oral infection with accompanying periods of exacerbated disease and an active, often substantial specific local and systemic immune response in periodontitis [11, 19]. Nevertheless, various studies supported the potential for the elevated serum antibody to P. gingivalis and other oral pathogens to lower the burden of the overall microbial biofilm challenge or minimize emergence of pathogens and maintain their colonization levels below a threshold that is necessary to induce destructive processes [20–22].
However, phenotypic changes in bacteria are paralleled by the dramatic ability of certain pathogens to alter the surface characteristics that they present to their environment(s). Variation in antigenic composition of pathogens generally comes in two forms, antigenic “variation” (shift) and “diversity” (drift). Antigenic variation is a specific strategy which requires the ability to abruptly replace one type of surface antigen with another. It generally requires multiple nonallelic genes for surface proteins and usually some form of gene rearrangement [23–25]. The antigenic differences within species are termed antigenic diversity (drift) and are often related to surface structures which are exposed to the defense mechanisms of the host [26–31]. Antigenic diversity occurs by the gradual accumulation of mutations in genes that code for the targets of the immune system which in Gram-negative bacteria are frequently the outer membrane components. These mutations result in considerable population heterogeneity and a drift towards selection of an antigenic type that is stable in the particular host-parasite interaction. As such, it has been suggested that lack of protective immunity to reinfection is due to the host immune response being limited to antigens on existing strain(s). This lack of clear “immunity” in periodontal disease may be related to antigenic drift and diversity in the oral opportunistic pathogens that chronically colonize and are subject to host immune pressure, leading to a selective advantage that accompanies antigenic alterations of the bacteria within the subgingival ecology.
As importantly, description of P. gingivalis antigens and measurement of host responses to P. gingivalis have essentially only used either type strains or human isolates that have been cultivated over extended time intervals [13, 15, 32–36]. Thus, a critical concept addressed in this report is based upon P. gingivalis representing a chronic opportunistic infection of susceptible hosts. Implied in this concept is that characteristics (eg. antigenic diversity/drift) of P. gingivalis and/or the susceptible host must be favorable for the maintenance of this colonization and emergence of the pathogen to a threshold level that contributes to eliciting disease symptoms. Thus, the aim of the study was to document the existence of antigenic diversity in P. gingivalis that could contribute to a lack of “immunity” in the chronic infections of periodontal disease.
2.1. MATERIALS & METHODS
2.1.1. Samples
The demographics of the biorepository serum samples from 426 patients are depicted in Table 1. Healthy subjects were defined by <10% of sites with bleeding on probing (BOP) ≥1 [37], <5% sites with probing pocket depth (PPD) ≥4mm, and no sites with PPD ≥5mm. Gingivitis was defined by >10% sites with BOP ≥1, <10% sites with PPD ≥4mm, and no sites with PPD ≥5mm. Periodontitis was defined by >10% BOP ≥1, ≥10% of sites with PPD ≥4 mm, and ≥3% sites with clinical attachment loss (CAL) >2 mm. Periodontitis was categorized into mild (mean PPD <3 mm), moderate (mean PPD 3–4 mm), and severe (mean PPD >4 mm) [38, 39]. Inclusion/exclusion criteria for the subjects have been reported previously [40–44].
Table 1:
Demographics of population
| Group | Age Mean (range) | Age Group (Y:M:O) % | Gender M:F | Race/Ethnicity (C:B:H:A) % | % Smokers | % Sites BOP Mean (range) | Mean PPD mm (range) | % Sites PPD ≥4 mm (range) | % Sites PPD ≥5 mm (range) |
|---|---|---|---|---|---|---|---|---|---|
| Health (n=61) | 39.5 (21–65) | 43:36:21 | 26:35 | 66:12:23:0 | 8.2 | 1.2 (0–2.9) | 2.1 (1.7–2.7) | 1.8 (0–3.2) | 0 |
| Gingivitis (n=66) | 36.9 (24–60) | 45:41:14 | 30:36 | 65:12:18:5 | 19. ‘ | 27.2 (9.3–71.4) | 2.2 (1.6–2.3) | 4.4 (1.9–8.1) | 0 |
| Periodontitis/Total (299) | 40.4 (21–74) | 30:54:16 | 187:112 | 51:21:20:8 | 28.1 | 41.1 (5.4–100) | 3.5 (2.2–5.9) | 29.2 (1.1–86.8) | 13.6 (0–67.3) |
| Mild (n=85) | 39.1 (22–74) | 48:44:8 | 40:45 | 72:13:9:8 | 18.8 | 21.1 (10.1–78.6) | 2.8 (2.2–3.0) | 15.6 (1.1–86.8) | 3.1 (0–13.7) |
| Moderate (n=152) | 41.0 (21–62) | 18:66:16 | 94:58 | 48:21:22:9 | 27.0 | 43.0 (5.4–96.2) | 3.5 (3.0–4.0) | 29.2 (2.4–84.3) | 12.0 (0–40.5) |
| Severe (n=62) | 40.8 (23–66) | 34:37:29 | 45:17 | 29:36:30:5 | 43.5 | 62.8 (12.9–100) | 4.5 (4.0–5.9) | 47.2 (3.2–81.3) | 31.6 (2.3–67.3) |
Age group identified as Y=≤35; M=36–50; O=>50 years of age
Race/ethnicity identified as Caucasian:Black:Hispanic:Asian
Sites demonstrating BOP ≥1 and pocket depth ≥5 mm were defined as disease sites and sampled as described previously from 124 participants within the larger cohort (Dawson et al. 2009). All plaque specimens were stored at −80°C and assayed within six months of collection. Bacterial plaque samples in sterile PBS were pelleted, DNA was extracted, and quantified as described previously [45]. Real-time PCR was performed using a LightCycler 2.0. and quantification analysis was performed using LightCycler 4.0 software using sample “crossing points” (CP) to determine the presence and the concentration of the target DNA in known and unknown samples after amplification. DNA isolated from a pure culture of P. gingivalis ATCC 33277 was used in generating a standard curve for the universal primers [46, 47]. Quantification of the individual target bacteria and total bacteria from the experimental samples were calculated using the standard curves.
2.1.2. Bacterial cultivation, antigens, and analyses
The strains of P. gingivalis included ATCC33277 [48, 49] originally obtained from the ATCC, FDC381 [48, 50], W50 [ATCC53978; https://www.ncbi.nlm.nih.gov/biosample/SAMN00792205/ [48]], and W83 [48, 51], were originally obtained from Dr. A. Tanner (The Forsyth Institute, Boston, MA). A7A1–28 [ATCC53977; [48, 52]] and A7436 [48, 53] were obtained from Dr. Anne Progulske-Fox (University of Florida), and all have been maintained in our laboratory for over 20 years. The bacteria were cultured in brain heart infusion (Becton Dickinson and Company, Sparks, MD) medium supplemented with 5 μg hemin ml−1 and 1 μg menadione ml−1 under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37°C [54]. For whole bacterial antigens, the bacteria were harvested by centrifugation, washed with phosphate-buffered saline, and formalin-fixed to detect serum IgG antibody levels using an ELISA [35]. The quantification of the antibody was based upon comparison of antibody levels in the experimental samples to standard curves using purified human IgG incorporated into each plate [55]. All samples were tested in triplicate and re-evaluated if the coefficient of variation was >15%. Interplate comparability was provided by analysis of the standard IgG curves based on determining no statistical difference in the slopes of the curves and reactivity of maximum IgG concentration.
2.1.3. Statistical analysis
Analyses of any differences among inflammatory mediators and IgG antibody levels, was conducted via a Kruskal-Wallis ANOVA with post hoc testing of paired groups using a Dunn’s method (SigmaStat, Systat Software, Inc., Richmond, CA). Evaluation of the significance of correlation data was performed using the Spearman Correlation test. Results with an alpha of <0.05 (after being adjusted for the multiple comparisons) were accepted as statistically significant. Significant differences in the slopes of the standard curves was determined using the Real Statistics add in for Excel (http://www.real-statistics.com/regression/hypothesis-testing-significance-regresson-line-slope/).
3.1. RESULTS
3.1.1. Characteristics of serum antibody to P. gingivalis strains with aging
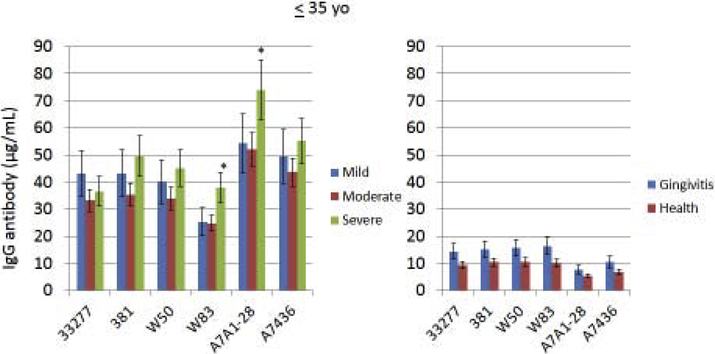
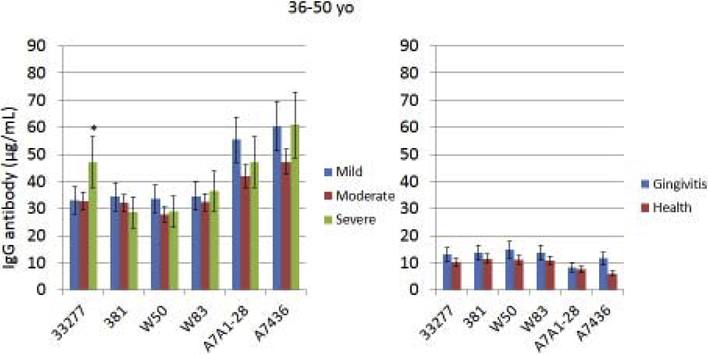
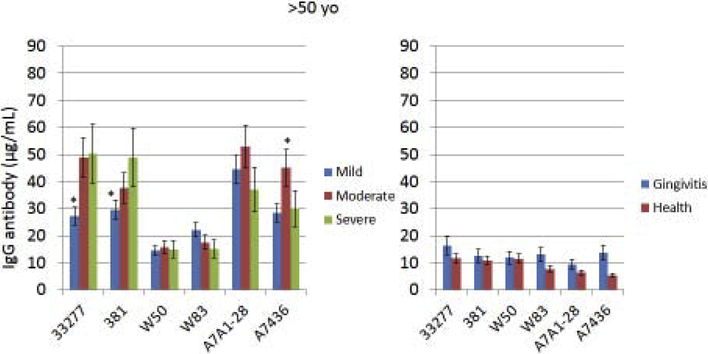
Serum IgG antibody response levels reacting with various strains of P. gingivalis were determined in the population stratified based upon age. This included subjects with periodontitis, as well as in healthy and gingivitis subjects (Fig. 1A–C). First, irrespective of age, antibody levels to all of the strains were low in the health and gingivitis patients with little variation across the strains. Generally, the youngest group of periodontitis patients demonstrated somewhat higher antibody compared to other age groups (increased by 30–260%) to all of the strains, with those individuals exhibiting severe periodontitis having the highest levels. Additionally, the antibody levels to the two more recent clinical isolates (A7A1–28, A7436; [48]) were elevated (increased by 30–200%) in this group compared to any of the more historical laboratory strains. As was noted with the younger group, responses in the 36–50 year old subset were higher to the recent clinical isolates, although the severe periodontitis patients did not show higher antibody. The pattern of responses in the oldest group (>50 yo) was substantially different than the other age groups. First, antibody levels to 2 of the older strains (33277, 381; [48]) were as high as the recent clinical isolates. Also, the levels to these isolates were significantly lower in mild periodontitis patients. Antibody to the older laboratory isolates that were not originally obtained from periodontitis lesions (W50, W83; [48]), were significantly lower than to all other strains. Finally, antibody in the oldest group to the recent clinical isolates was significantly lower in t e severe periodontitis patients. In addition to stratifying the population into specific age groups that provided insights into differences in antibody responses to the various P. gingivalis strains, a correlation analysis was performed to examine the direct relationship of age to antibody response levels (Table 2). In the overall population, generally the antibody levels were negatively correlated with age to each of the strains, consistent with the stratification results; however, within the individual age groups, few statistically significant values were identified.
Figure 1A-C:
Serum antibody levels to individual P. gingivalis strains in patients in different age categories stratified based upon periodontitis severity. The bars denote group means and the vertical brackets enclose 1 SD. The asterisk (*) denotes statistically different from other disease groups at least at p<0.05.
Table 2:
Correlation analysis of antibody responses with age to the various P. gingivalis strains. Correlation coefficients in bold are significantly correlated at least at p<0.05 using a Spearman Rank analysis. SUM denotes correlation of age and antibody as a summation of levels to all strains in each subject.
| Group (yrs.) | 33277 | 381 | W50 | W83 | A7A1–28 | A7436 | SUM |
|---|---|---|---|---|---|---|---|
| Total | 0.0292 | −0.1469 | −0.2730 | −0.0134 | −0.0312 | −0.1186 | −0.1382 |
| ≤35 | 0.0756 | −0.1007 | −0.0194 | 0.0741 | −0.0372 | −0.1728 | −0.0530 |
| 36–50 | 0.0339 | −0.0748 | −0.0439 | 0.0075 | −0.1795 | −0.1194 | −0.1187 |
| >50 | −0.0357 | 0.0343 | 0.2773 | 0.1104 | 0.1421 | −0.0584 | 0.0762 |
We also explored the antigenic conservation among the P. gingivalis strains when cultivated in vitro as antigens. In this study, 3 strains were cultivated on 3 separate occasions approximately 1 month apart under similar conditions. They were used to prepare formalinized bacterial antigens and then sera from 3 patients with varied antibody levels to the strains were examined for the reproducibility of the antibody reactions. As shown in Supplemental Fig. 1, the level of IgG antibody in each serum was reasonably consistent with generally <15% CV suggesting that the antigens displayed on the strains prepared on different occasions were generally stable. Thus, differences in strain reactivities of different samples suggested fundamental antigenic differences across the P. gingivalis strains, and supported substantial differences in the P. gingivalis antigenic repertoire in patients inducing significantly different antibody responses profiles.
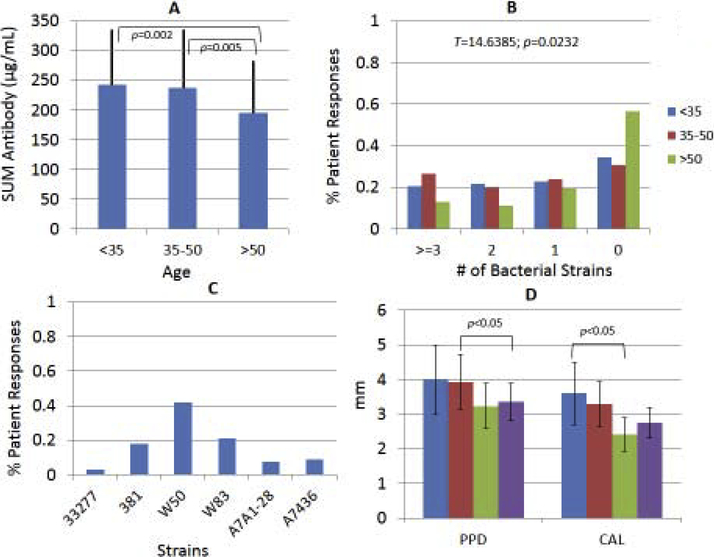
Exploring the characteristics of the population responses to the individual strains showed interesting distributions of elevated antibody to specific strains. Fig. 2A shows that the overall antibody responses to the array of P. gingivalis strains was significantly greater in the younger and middle-age group compared to the oldest patients. Fig. 2B demonstrates the results examining the level of response to each strain in individual patients. Antibody levels were stratified into quartiles for each strain across the population. These quartiles were identified for each patient and summarized as the percentage of patients with antibody in the top quartile to 3 or more strains, 2 strains, only 1 strain, and to none of the strains tested. The results demonstrated a significantly different distribution of strain-specific antibody with the oldest patient group showing significantly fewer patients with high levels of antibody to more than 1 strain and nearly 60% having antibody in the top quartile of the population to none of these strains. Additionally, we addressed the strain characteristics of those patients, in whom antibody was highly elevated to only 1 strain. This included 67/297 patients (22.64%) and identified some patients primarily responsive to one of the 6 strains with over 40% of the patients with an elevated response to strain W50 (Fig. 2C).
Figure 2A-D:
(A) The overall antibody level to all 6 strains within the different age groups of periodontitis patients. The bars denote the group means and the vertical line is 1 SD. (B) Distribution of antibody reactivities in different age groups in which each patient’s sample was determined to be in the top tertile of antibody of the entire population to each strain. The oldest group had significantly fewer patients with antibody responses in the top tertile across all the P. gingivalis strains. (C) Depiction of the % of patients whose antibody levels were in the top tertile of only 1 strain. (D) Relationship of patients with antibody levels in the top tertile to 0, 1, 2, or ≥3 strains. The bars denote the group means of probing pocket depth and clinical attachment level within the response categories. The vertical brackets denote 1 SD.
Finally, the clinical presentation of the patients related to stratification based upon top quartile antibody levels across the P. gingivalis strains is presented. No differences were observed in the clinical parameters of mean probing pocket depth or clinical attachment level in the younger or middle-age group (data not shown). However, we observed that in the oldest group of participants (Fig. 2D) the samples with elevated antibody to 1 or none of the strains tended to have lower clinical disease measures than subjects with elevated antibody to multiple P. gingivalis strains.
Table 2 provides a summary of correlation analyses across the periodontitis population of antibody to each of the P. gingivalis strains. The youngest group of subjects demonstrated significant correlations of the antibody particularly reflected in the recent clinical isolates. In contrast, these relationships were lost in the oldest group, suggesting some alterations in the antibody repertoire in the oldest patients, in which certain antigenic specificities may have been lost.
3.1.2. Race/ethnicity and aging effects on P. gingivalis antibody patterns
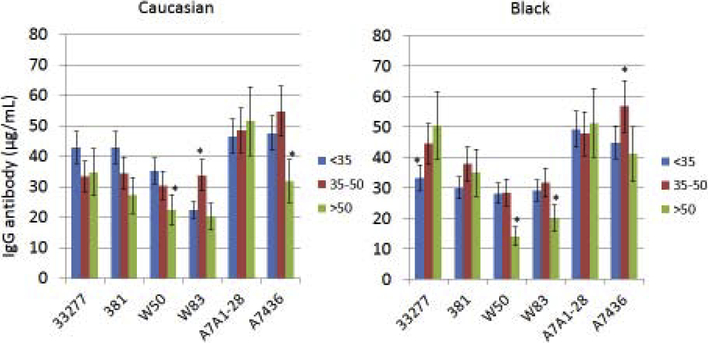
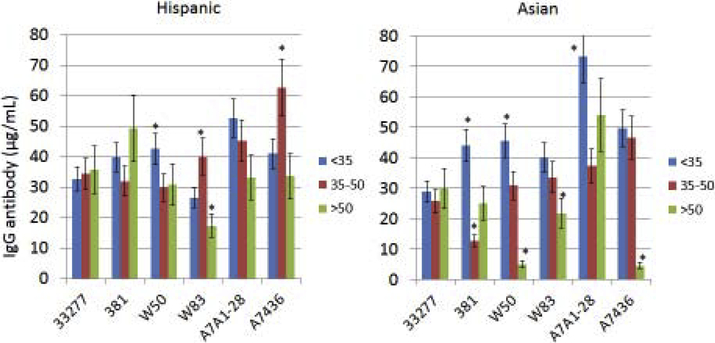
Existing studies have generally not integrated aging into evaluation of the characteristics of antibody responses to oral bacteria related to race/ethnicity. Fig. 3A–B displays the results in categorizing the periodontitis patients into various racial/ethnic groups and stratifying according to age to assess antibody to the various P. gingivalis strains. Older Caucasian patients had decreased levels of antibody to all of the P. gingivalis strains, except the A7A1–28 strain. The Black oldest patient group showed elevated antibody to the 33277 strain, and low antibody to both W50 and W83. Hispanic patients tended to have similar levels of antibody to the various strains across the age groups, albeit the response to W83 was significantly lower in the oldest Hispanic group. Also, a substantial increase in antibody level to the A7436 strain was observed in the middle-age Hispanic group. Finally, the notable pattern in the Asian patients was elevated antibody to multiple strains in the younger group, and significantly decreased antibody to both the W50 and A7436 strains in the oldest group.
Figure 3A-B:
Levels of serum antibody to the P. gingivalis strains (Pg277 – 33277, PgA7 – A7A1–28, Pg7436 – A7436) with patients categorized into racial/ethnic groups and stratified by age. The bars denote group means and the vertical brackets enclose 1 SD. The asterisk (*) denotes statistically different from other age groups at least at p<0.05.
3.1.3. Sex and aging effects on P. gingivalis antibody patterns
While sex differences have been reported with periodontal disease, the biologic underpinnings of this observation remain to be determined. We examined antibody responses to P. gingivalis strains with the periodontitis subjects dichotomized on sex to determine if differences in response characteristics could provide any insight into this clinical observation. No significant differences in antibody to any of the strains based on sex were found (data not shown). However, grouping the patients based on age did reveal some sex differences across the strains. Generally little difference with age or sex was seen in antibody levels to 33277, 381, and A7A1–28 strains. Both female and male oldest patients showed significantly decreased antibody to W50, W83 and A7436 (Supplemental Fig. 2).
3.1.4. Smoking and aging effects on P. gingivalis antibody patterns
We also explored the potential impact of smoking with age in the distribution of antibody responses to determine if the antibody diversity appeared to be adversely affected by smoking. Supplemental Fig. 3 demonstrates a rather minimal impact on the antibody levels across the strains in periodontitis subjects under 50 years of age. However, while the extent and severity of periodontitis is significantly increased with aging [56], antibody to 4 of the 6 strains was significantly decreased in the oldest smoking subset of the population.
3.1.5. Antibody diversity to P. gingivalis strains
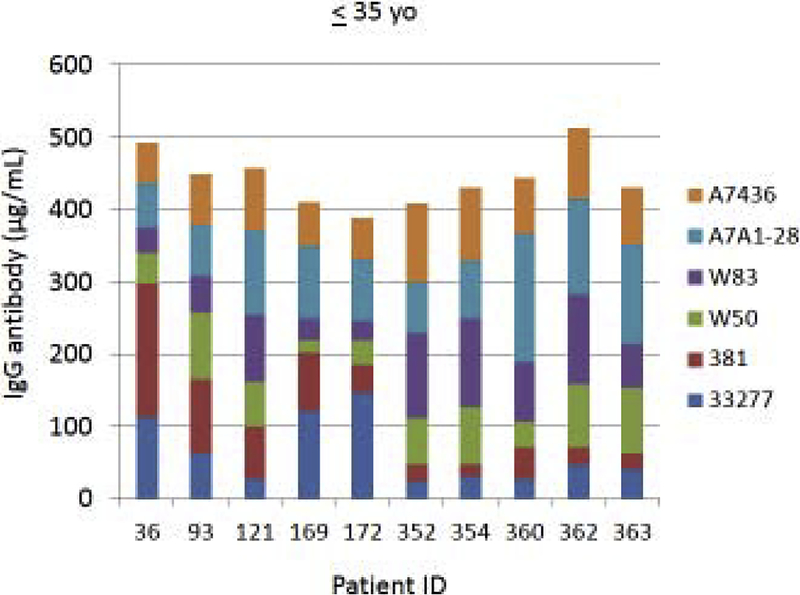
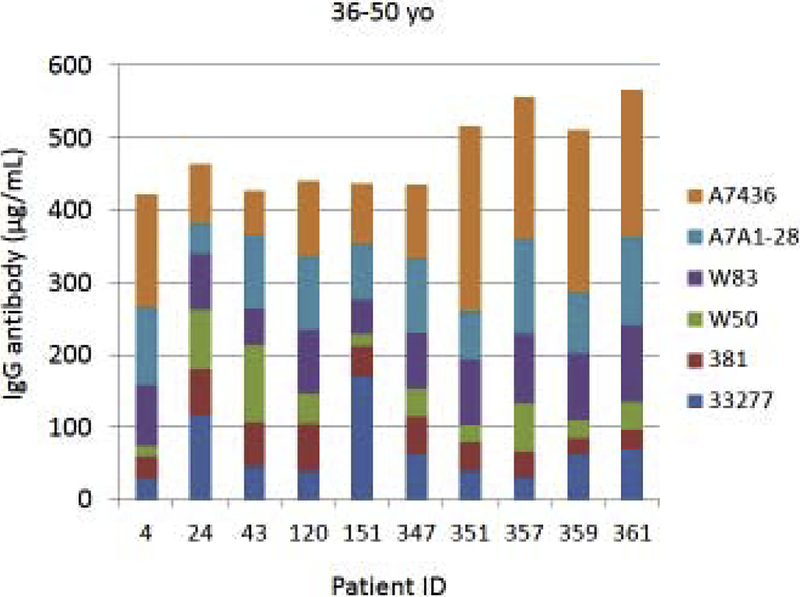
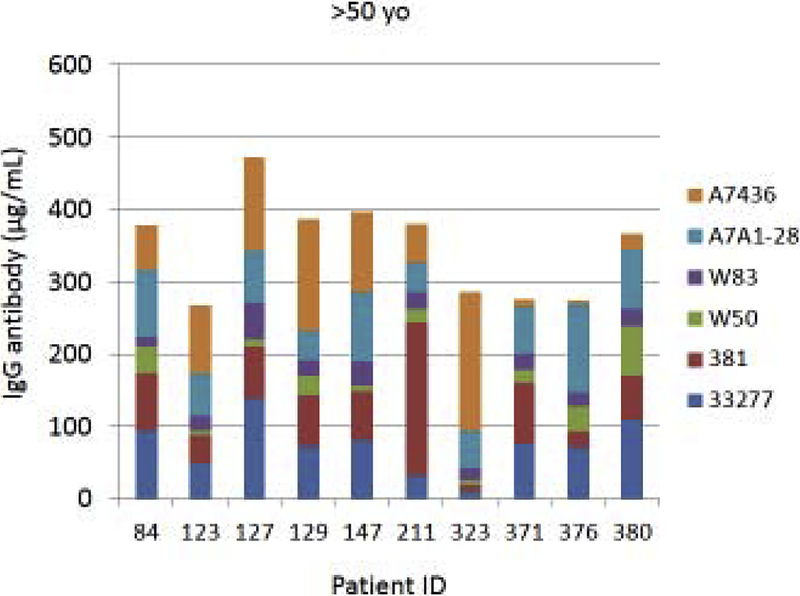
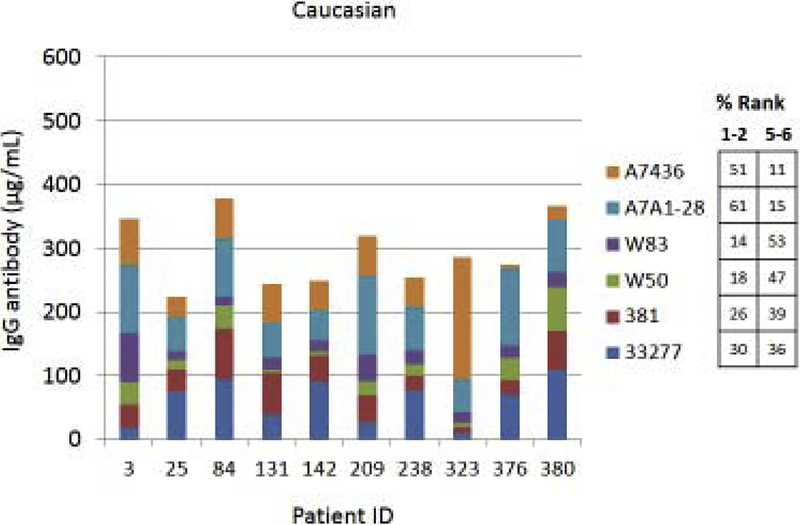
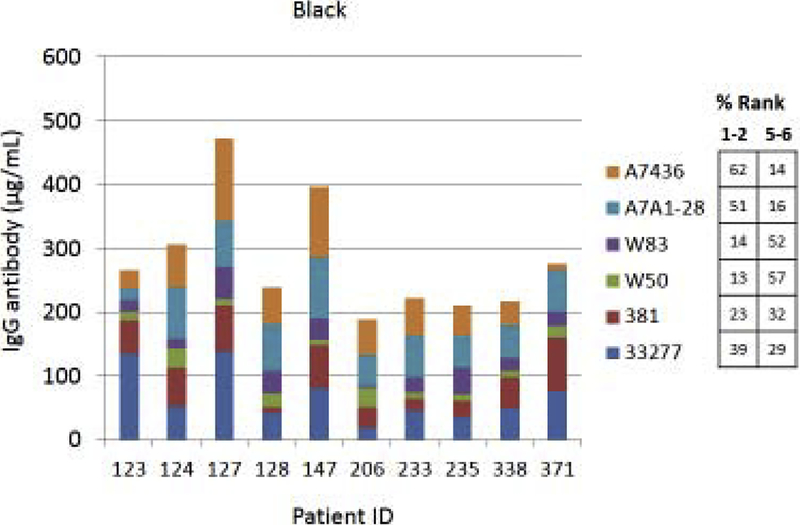
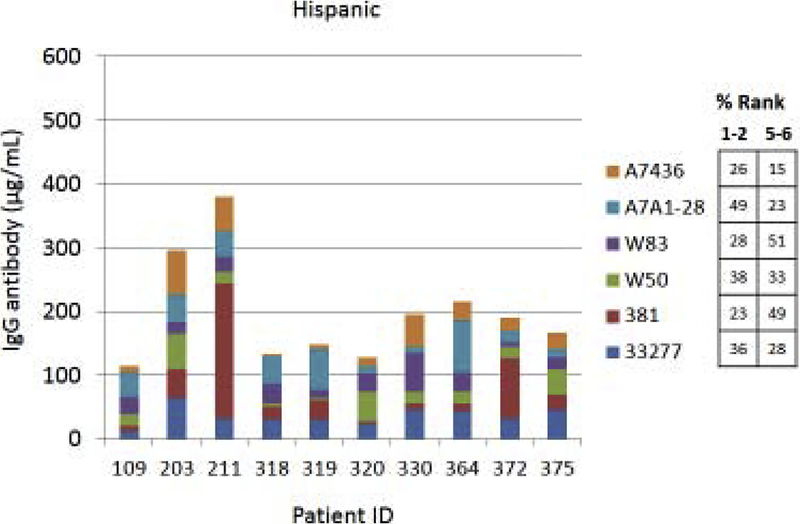
Fig. 4A–C provides examples of the strain specificity of the serum antibody as it related to patient age. The figure includes the antibody distribution to the 6 strains in 10 patients from each age group who demonstrated the highest overall antibody responses as a summation of antibody to all of the strains. While this analysis was carried out for all patients, only the top 10 are depicted, which generally reflects the profile variations within the age grouping. First of note is the difference in the level of antibody to individual strains within the patients. Across all age categories some patients clearly have a predisposition for responding to antigens that are expressed on one strain (eg. ID 36, 360, 351, 359, 211, 323), while others show a very limited response to certain individual strains (eg. ID 169, 354, 4, 151, 84, 123). While the middle age group showed this similar type of variation, a rather striking feature was the dominance of antibody to the more recent clinical isolates in most of these patients (70%). In the oldest group, it is first notable that the summation of antibodies to these strains is generally lower than the other age groups. Also noted was that in many of the patients, antibody levels to the recent clinical isolates (A7A1–28, A7436) were much less than to the older laboratory strains. The exceptions were patients 129 and 323, whose major responses were to antigens presented by the more recent clinical isolates. A similar patient specific analysis is presented in Fig. 5A–C stratifying the subjects based upon race/ethnicity. Generally, the individual subject level responses in the top 10 sum of antibody responses are lower across the P. gingivalis strains in the Hispanic population; however 2 of the individuals (ID 203, 211) showed robust responses to this group of strains. The Caucasian and Black subjects showed the greatest frequency of elevated responses to strain ATCC 33277 and A7A1–28 with Hispanics dominating in responses to strains 381 and A7A1–28. At the individual level, the Caucasian and Black groups showed principal responses to A7A1–28 and A7436 in 50–60% of the patients. In contrast, the dominant responses in 40–50% of Hispanic subjects were to strains A7A1–28 and ATCC 33277. Ranking the level of responses to the various P. gingivalis strains across the racial/ethnic background of individual subjects also provided an interesting comparison. As shown in the figure, the distribution of dominant versus lowest strain responses in the Hispanics was significantly different (Caucasian: X2=45.895, df=5, p<0.0001; Black: X2=83.086, df=5, p<0.0001), while no difference was noted between the Caucasian and Black subgroups.
Figure 4A-C:
Serum antibody levels to the 6 P. gingivalis strains in individual patients based upon age category. The top 10 patients with summed antibody to all the strains are presented in each age grouping.
Figure 5A-C:
Serum antibody levels to the 6 P. gingivalis strains in individual patients based upon race/ethnicity category. The top 10 patients with summed antibody to all the strains are presented in each grouping. The table describes the frequency distribution across all subjects within the racial/ethnic category, with strain responses ranked 1 through 6 for each individual. The numbers denotes percentage of subjects with strain ranked 1 or 2 (highest level) versus the percentage ranked 5 or 6 (lowest level).
3.1.6. Antibody diversity to P. gingivalis strains and P. gingivalis colonization
The study design also allowed us to explore the relationship between the level of P. gingivalis in subgingival plaque from a subse, of the periodontitis patients and the distribution of antibody levels to the various P. gingivalis strains. Supplemental Figure 4 summarizes the relationship and demonstrates decreasing antibody levels to all strains except 381 with decreased levels of P. gingivalis colonization. Similarly, Supplementary Table 1 provides an estimate of the correlation of the antibody levels across the population and within different age categories to P. gingivalis colonization. The results showed a rather limited number of significant correlations of the levels of P. gingivalis and antibody to and individual P. gingivalis strain.
4.1. DISCUSSION
This report developed data from an extensive cross-sectional analysis demonstrating substantial variations in antigenic makeup of various P. gingivalis strains, as reflected by the variation in host antibody reactivity to the strains. Moreover, we demonstrated that antibody to the P. gingivalis strains was decreased with aging, and the results reflected not simply a decrease in antibody levels, but a seemingly more limited repertoire of antibody to the range of P. gingivalis strains. A conundrum of observations with certain chronic infections is why the pathogen/infection can remain associated with the host in the presence of an acquired immune response. While this is a feature of many chronic infections, explanations of the continued infection have often been linked to novel intracellular strategies that protect the pathogen from the immune biomolecules/cells, low antigenicity eliciting an ineffective immune response in quantity or quality, structural/chemical characteristics of the pathogen which minimize the impact of immune components, and alterations of the pathogens’ antigens, presenting the host immune apparatus an ever changing target and limiting the effectiveness of the responses [57–65]. This last facet of pathogen strategy has been described as antigenic variation, antigenic drift, and antigenic diversity depending upon the pathogenic species [23–28, 30, 66–70].
While the literature has recognized for decades a robust immune (antibody) response to periodontal pathogens in subsets of the affected populations, it also describes clear exacerbations and remissions from active, progressing disease, generally related to the presence of the same species of pathogen [19, 20, 71–73]. This report presents data suggesting the P. gingivalis has the ability to alter expression of potentially critical antigens that are recognized by the host and may be important in resulting protection or susceptibility to future disease episodes. Description of diversity of P. gingivalis antigens has been rather limited, generally reflected by reports considering antigenic differences in the capsular polysaccharide across various strains [66, 68, 74–77]. Additionally, these capsular antigenic types have been described to have some limited representation in an individual patient, albeit, no reports exist describing longitudinal changes in the capsule antigen type with disease exacerbations [78–80].
Historically, over nearly 4 decades, P. gingivalis has been identified as a consistent hallmark microorganism in the microbial biofilms of periodontitis lesions [71, 81–83]. It has been shown to increase in proportion, albeit remaining at an overall low percentage, in the biofilms that transition from health to disease [84, 85]. Importantly, published findings support a wide clonal genetic diversity of P. gingivalis strains, and with 19 strains having been sequenced, genome sizes vary by up to 200,000 base pairs, and predicted protein coding genes range from 1,774 to 2,392. Interestingly, Chen et al. [48] just recently published an extensive evaluation of the genomic diversity of P. gingivalis, and provided a phylogenetic tree of the existing strains that have been sequenced. A critical component of this analysis was also that across the 19 strains, nearly ½ of the protein coding genes showed sequence heterogeneity with 2000–3000 predicted protein coding genes showing distribution in only one of the these strains. While this report also identified selected proteins from this list that were unique in the various strains, lacking was: (1) any assessment of the impact on antigenic aspects of these genomic differences, and (2) understanding of how these genomic differences could be expressed within a patient overtime, since these strains were individual isolates obtained a one point in time from 19 different patients.
Our results extend these findings by demonstrating either a unique antigenic portfolio expressed by various P. gingivalis strains (likely, based on Chen et al. [48]) or similar antigenic composition across all the strains with variations in individual responses that are host genetically regulated [20, 86–88]. While the latter of these options is possible, based upon the variation we noted across this population and the relationship to age, race/ethnicity, and sex, it seems less likely. Importantly, the very unique antibody response patterns that were noted within individual patients to the array of P. gingivalis strains support the likelihood of antigenic diversity of P. gingivalis. In a cross-sectional study as this, the data cannot discriminate whether this reflects the strains that originally colonized the individual patient, or antigenic differences arising through genomic alterations that reflect the host-bacterial interactions and immune pressure that would occur with chronic colonization by this opportunistic pathogen. Additionally, within this framework, consideration of genomic modulation within a patient through episodes of disease exacerbations that could explain the novel response patterns remains to be determined.
Beyond this circumstantial evidence antigenic diversity or drift, it was clear that the response repertoire to the P. gingivalis strains was affected by aging, more substantially than race/ethnicity or sex. This is an important observation, since clear epidemiologic evidence supports that racial/ethnic minorities [89–91] and males [9, 92] exhibit an elevated prevalence and severity of periodontitis with aging. Nevertheless, novel differences in population antibody levels to the various P. gingivalis strains were noted across the racial/ethnic distribution of the population. The findings with somewhat distinctive response patterns in Caucasian, Black, Hispanic, and Asian populations to the various P. gingivalis strains may be a reflection of the different interactions of this pathogen with the host immune system within these racial/ethnic groups and could contribute to the variations in expression of periodontitis that has been identified in certain populations [9, 91]. The results suggest that P. gingivalis with different antigenic composition may colonize different groups of individuals, resulting in a more pathogenic relationship due to a more limited antibody response, or features of the genetic control of the antibody response repertoire that is affected to a greater degree by aging related to the race/ethnicity of the individual. Sorting out this relationship and understanding if this feature is being driven by targeted antigenic changes in the pathogen, or altered capacity of the host immune system to control the chronic infection is an important facet of future approaches to improved precision in periodontal diagnosis and therapy [73, 93, 94].
We have reported previously that females with periodontitis showed higher levels of antibody to P. gingivalis than males affected by this disease [95]. However, those data were limited to examination of responses to only P. gingivalis strain 33277. This study expanded the characterization of responses in the disease population related to sex of the individual. Across most strains, older males tended to show lower antibody levels, except to strain A7A1–28. Moreover, comparison of antibody levels between males and females within each strain showed elevated antibody in females to 33277, 381 and W50, with similar antibody levels to strains W83, A7A1–28 and A7436. Generally, the differences were driven by the oldest group of females and males. Epidemiological evidence also clearly documents substantial adverse effects of smoking on many health parameters, including periodontitis [9, 88]. However, a similar analysis of our data related to smoking and age demonstrated a limited impact of strain response differences that only occurred in the oldest periodontitis group to some of the strains. Thus, it is not clear that antigenic diversity of P. gingivalis is particularly pronounced related to sex and smoking, compared to the effects noted with age and race/ethnicity.
Finally, we obtained an initial evaluation of the relationship of P. gingivalis colonization at disease sites and antibody responses to individual P. gingivalis strains. At a population level it did appear that antibody to 5 of the 6 strains decreased with lower proportions of P. gingivalis in the subgingival plaque. This type of information has been reported previously, albeit generally those studies only used one P. gingivalis strain as the antigen detection system [96–98]. A more patient focused approach showed a rather limited number of correlations between the proportion of P. gingivalis and antibody levels of the individual strains. Moreover, we specifically examined this correlation focusing on the individual patient’s predominant response across the strains and found few additional relationships (data not shown). These results reinforced the likelihood of more individualized responses to an antigenic repertoire of homologous P. gingivalis isolates with each patient, and inferred that further studies linking patient specific isolates parallel with the specific responses to these isolates would confirm antigenic heterogeneity across the disease population and confirming diversity overtime as contributing to the disease process.
5.1. CONCLUSIONS
Our findings support that P. gingivalis may, in fact, demonstrate antigenic drift within patients and could be one factor to help explain the inefficiency/ineffectiveness of the adaptive immune response in managing the infection and allowing new antigenic phenotypes to emerge in the biofilms resulting in initiation and progression of disease episodes. This option would also elucidate potential biologic variations of more frequent/rapid occurrences of sufficient antigenic drift that contributes to variations in onset and severity of disease, as suggested by clinical terminology of “rapidly progressive periodontitis” and “refractory periodontitis”. These clinical descriptions could incorporate curtailed development of the antibody repertoire having the capacity to recognize this antigenic drift, as an important basis that enables P. gingivalis to express its pathogenic properties across decades within individual patients. The results supported a loss of antibody breadth with aging in the periodontitis patients. This could be interpreted as a loss of the fundamental aspects of the B cell repertoire in response to P. gingivalis with aging. This type of response alteration has been suggested to occur in other studies of immunity in aging, but data is lacking with responses to chronic antigenic challenge, such as to members of the oral microbiome. Nevertheless a limitation of this study is the lack of a detailed assessment of age effects via isolation of P. gingivalis from individual subjects and critical determination of within subject antibody specificity to their, homologous isolates.
A striking component of periodontitis is the extent of variation across the population in age of onset, rate of progression, and extent/severity of the disease. While there remains an age association with these clinical measures, there is minimal capacity to predict any of these at the individual patient level. Moreover, as a chronic disease that is clinically managed, the explanation for the variation in frequency of episodes of disease overtime, generally related to a similar profile of oral pathogen(s) [99] remains enigmatic. These findings support a novel concept regarding aging impacts on the quality of adaptive immune responses to specific oral pathogens and the interplay between antigenic drift of the pathogen, and basis for episodic disease.
Supplementary Material
Table 3:
Correlation analysis of antibody responses in different age groups to antigens presented by the various P. gingivalis strains. Correlation coefficients in bold are significantly correlated at least at p<0.05 using a Spearman Rank analysis.
| Age Group | Pg Strain | Pg277 | Pg381 | PgW50 | PgW83 | PgA7 | Pg7436 |
|---|---|---|---|---|---|---|---|
| ≤35 years | 33277 | 1 | 0.466 | −0.101 | 0.055 | 0.161 | 0.244 |
| 381 | 1 | 0.090 | 0.052 | 0.080 | 0.197 | ||
| W50 | 1 | 0.420 | 0.298 | 0.412 | |||
| W83 | 1 | 0.472 | 0.748 | ||||
| A7A1–28 | 1 | 0.668 | |||||
| A7436 | 1 | ||||||
| 36–50 years | 33277 | 1 | 0.383 | 0.084 | 0.207 | 0.192 | 0.292 |
| 381 | 1 | 0.131 | 0.320 | 0.160 | 0.256 | ||
| W50 | 1 | 0.209 | −0.002 | 0.098 | |||
| W83 | 1 | 0.292 | 0.566 | ||||
| A7A1–28 | 1 | 0.491 | |||||
| A7436 | 1 | ||||||
| >50 years | 33277 | 1 | 0.307 | 0.324 | 0.411 | 0.431 | 0.234 |
| 381 | 1 | 0.158 | 0.180 | 0.064 | 0.200 | ||
| W50 | 1 | 0.023 | 0.356 | −0.056 | |||
| W83 | 1 | 0.327 | 0.208 | ||||
| A7A1–28 | 1 | 0.169 | |||||
| A7436 | 1 |
Highlights.
Study of antigenic diversity of P. gingivalis in periodontal disease.
Antibody levels in the oldest age group were lower than in the youngest groups.
The older group had fewer with high levels of antibody to more than 1 strain.
P. gingivalis may demonstrate antigenic diversity/drift with these chronic infections.
6.1. ACKNOWLEDGEMENTS
We want to acknowledge the support of U.S.P.H.S. grant RR020145 and GM103538 to the Center for Biomedical Research Excellence, and funding from the Center for Oral Health Research at the University of Kentucky College of Dentistry. Also, expert technical assistance was provided by M.J. Steffen and Dr. R. Peyyala in providing the bacterial antigens and supporting the antibody measurements. The substantial contributions of the clinical support staff in the Delta Dental of Kentucky Clinical Research Center are also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].O’Connor JE, Herrera G, Martinez-Romero A, de Oyanguren FS, Diaz L, Gomes A, Balaguer S, Callaghan RC, Systems Biology and immune aging, Immunol Lett 162(1 Pt B) (2014) 334–45. [DOI] [PubMed] [Google Scholar]
- [2].Krone CL, van de Groep K, Trzcinski K, Sanders EA, Bogaert D, Immunosenescence and pneumococcal disease: an imbalance in host-pathogen interactions, The Lancet. Respiratory medicine 2(2) (2014) 141–53. [DOI] [PubMed] [Google Scholar]
- [3].Fulop T, Larbi A, Kotb R, de Angelis F, Pawelec G, Aging, immunity, and cancer, Discovery medicine 11(61) (2011) 537–50. [PubMed] [Google Scholar]
- [4].Nikolich-Zugich J, The twilight of immunity: emerging concepts in aging of the immune system, Nat Immunol 19(1) (2018) 10–19. [DOI] [PubMed] [Google Scholar]
- [5].El Chakhtoura NG, Bonomo RA, Jump RLP, Influence of Aging and Environment on Presentation of Infection in Older Adults, Infect Dis Clin North Am 31(4) (2017) 593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Castelo-Branco C, Soveral I, The immune system and aging: a review, Gynecol Endocrinol 30(1) (2014) 16–22. [DOI] [PubMed] [Google Scholar]
- [7].Franceschi C, Campisi J, Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases, The journals of gerontology. Series A, Biological sciences and medical sciences 69 Suppl 1 (2014) S4–9. [DOI] [PubMed] [Google Scholar]
- [8].Fulop T, Witkowski JM, Pawelec G, Alan C, Larbi A, On the immunological theory of aging, Interdisciplinary topics in gerontology 39 (2014) 163–76. [DOI] [PubMed] [Google Scholar]
- [9].Eke PI, Wei L, Thornton-Evans GO, Borrell LN, Borgnakke WS, Dye B, Genco RJ, Risk Indicators for Periodontitis in US Adults: NHANES 2009 to 2012, J Periodontol 87(10) (2016) 1174–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hajishengallis G, Aging and its Impact on Innate Immunity and Inflammation: Implications for Periodontitis, Journal of oral biosciences / JAOB, Japanese Association for Oral Biology 56(1) (2014) 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ebersole JL, Humoral immune responses in gingival crevice fluid: local and systemic implications, Periodontol 2000 31 (2003) 135–66. [DOI] [PubMed] [Google Scholar]
- [12].Johnson V, Johnson BD, Sims TJ, Whitney CW, Moncla BJ, Engel LD, Page RC, Effects of treatment on antibody titer to Porphyromonas gingivalis in gingival crevicular fluid of patients with rapidly progressive periodontitis, J Periodontol 64(6) (1993) 559–65. [DOI] [PubMed] [Google Scholar]
- [13].Shet U, Oh HK, Chung HJ, Kim YJ, Kim OS, Lim HJ, Shin MH, Lee SW, Humoral immune responses to periodontal pathogens in the elderly, J Periodontal Implant Sci 45(5) (2015) 178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McArthur WP, Bloom C, Taylor M, Smith J, Wheeler T, Magnusson NI, Antibody responses to suspected periodontal pathogens in elderly subjects with periodontal disease, J Clin Periodontol 22(11) (1995) 842–9. [DOI] [PubMed] [Google Scholar]
- [15].Vlachojannis C, Dye BA, Herrera-Abreu M, Pikdoken L, Lerche-Sehm J, Pretzl B, Celenti R, Papapanou PN, Determinants of serum IgG responses to periodontal bacteria in a nationally representative sample of US adults, J Clin Periodontol 37(8) (2010) 685–96. [DOI] [PubMed] [Google Scholar]
- [16].Baranowska HI, Palmer RM, Wilson RF, A comparison of antibody levels to Bacteroides gingivalis in serum and crevicular fluid from patients with untreated periodontitis, Oral Microbiol Immunol 4(3) (1989) 173–5. [DOI] [PubMed] [Google Scholar]
- [17].Ebersole JL, Taubman MA, Smith DJ, Gingival crevicular fluid antibody to oral microorganisms. II. Distribution and specificity of local antibody responses, J Periodontal Res 20(4) (1985) 349–56. [DOI] [PubMed] [Google Scholar]
- [18].Martin SA, Falkler WA Jr., Suzuki JB, Hawley CE, Mackler BF, Local and systemic immunoglobulins reactive to Bacteroides gingivalis in rapidly progressive and adult periodontitis, J Periodontal Res 21(4) (1986) 351–64. [DOI] [PubMed] [Google Scholar]
- [19].Ebersole JL, Dawson DR 3rd, Morford LA, Peyyala R, Miller CS, Gonzalez OA, Periodontal disease immunology: ‘double indemnity’ in protecting the host, Periodontol 2000 62(1) (2013) 163–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ebersole JL, Dawson D 3rd, Emecen-Huja P, Nagarajan R, Howard K, Grady ME, Thompson K, Peyyala R, Al-Attar A, Lethbridge K, Kirakodu S, Gonzalez OA, The periodontal war: microbes and immunity, Periodontol 2000 75(1) (2017) 52–115. [DOI] [PubMed] [Google Scholar]
- [21].Tatakis DN, Kumar PS, Etiology and pathogenesis of periodontal diseases, Dent Clin North Am 49(3) (2005) 491–516, v. [DOI] [PubMed] [Google Scholar]
- [22].Kinane DF, Bartold PM, Clinical relevance of the host responses of periodontitis, Periodontol 2000 43 (2007) 278–93. [DOI] [PubMed] [Google Scholar]
- [23].Vink C, Rudenko G, Seifert HS, Microbial antigenic variation mediated by homologous DNA recombination, FEMS microbiology reviews 36(5) (2012) 917–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bai X, Borrow R, Genetic shifts of Neisseria meningitidis serogroup B antigens and the quest for a broadly cross-protective vaccine, Expert review of vaccines 9(10) (2010) 1203–17. [DOI] [PubMed] [Google Scholar]
- [25].Koomey M, Bacterial pathogenesis: a variation on variation in Lyme disease, Curr Biol 7(9) (1997) R538–40. [DOI] [PubMed] [Google Scholar]
- [26].Riddle MS, Guerry P, Status of vaccine research and development for Campylobacter jejuni, Vaccine 34(26) (2016) 2903–2906. [DOI] [PubMed] [Google Scholar]
- [27].Roggen EL, De Breucker S, van Dyck E, Piot P, Antigenic diversity in Haemophilus ducreyi as shown by western blot (immunoblot) analysis, Infect Immun 60(2) (1992) 590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Valvano MA, Pathogenicity and molecular genetics of O-specific side-chain lipopolysaccharides of Escherichia coli, Can J Microbiol 38(7) (1992) 711–9. [DOI] [PubMed] [Google Scholar]
- [29].Caugant DA, Levin BR, Orskov I, Orskov F, Svanborg Eden C, Selander RK, Genetic diversity in relation to serotype in Escherichia coli, Infect Immun 49(2) (1985) 407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vinogradov E, King JD, Pathak AK, Harvill ET, Preston A, Antigenic Variation among Bordetella: Bordetella bronchiseptica strain MO149 expresses a novel o chain that is poorly immunogenic, J Biol Chem 285(35) (2010) 26869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lam JS, Taylor VL, Islam ST, Hao Y, Kocincova D, Genetic and Functional Diversity of Pseudomonas aeruginosa Lipopolysaccharide, Frontiers in microbiology 2 (2011) 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mooney J, Kinane DF, Humoral immune responses to Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in adult periodontitis and rapidly progressive periodontitis, Oral Microbiol Immunol 9(6) (1994) 321–6. [DOI] [PubMed] [Google Scholar]
- [33].Whitney C, Ant J, Moncla B, Johnson B, Page RC, Engel D, Serum immunoglobulin G antibody to Porphyromonas gingivalis in rapidly progressive periodontitis: titer, avidity, and subclass distribution, Infect Immun 60(6) (1992) 2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Murray PA, Burstein DA, Winkler JR, Antibodies to Bacteroides gingivalis in patients with treated and untreated periodontal disease, J Periodontol 60(2) (1989) 96–103. [DOI] [PubMed] [Google Scholar]
- [35].Ebersole JL, Taubman MA, Smith DJ, Frey DE, Haffajee AD, Socransky SS, Human serum antibody responses to oral microorganisms. IV. Correlation with homologous infection, Oral Microbiol Immunol 2(2) (1987) 53–9. [DOI] [PubMed] [Google Scholar]
- [36].Mouton C, Hammond PG, Slots J, Genco RJ, Serum antibodies to oral Bacteroides asaccharolyticus (Bacteroides gingivalis): relationship to age and periondontal disease, Infect Immun 31(1) (1981) 182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ebersole JL, Nagarajan R, Akers D, Miller CS, Targeted salivary biomarkers for discrimination of periodontal health and disease(s), Frontiers in cellular and infection microbiology 5 (2015) 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Preshaw PM, Novak MJ, Mellonig J, Magnusson I, Polson A, Giannobile WV, Rowland RW, Thomas J, Walker C, Dawson DR, Sharkey D, Bradshaw MH, Modified-release subantimicrobial dose doxycycline enhances scaling and root planing in subjects with periodontal disease, J Periodontol 79(3) (2008) 440–52. [DOI] [PubMed] [Google Scholar]
- [39].Novak MJ, Dawson DR 3rd, Magnusson I, Karpinia K, Polson A, Polson A, Ryan ME, Ciancio S, Drisko CH, Kinane D, Powala C, Bradshaw M, Combining host modulation and topical antimicrobial therapy in the management of moderate to severe periodontitis: a randomized multicenter trial, J Periodontol 79(1) (2008) 33–41. [DOI] [PubMed] [Google Scholar]
- [40].Syndergaard B, Al-Sabbagh M, Kryscio RJ, Xi J, Ding X, Ebersole JL, Miller CS, Salivary Biomarkers Associated with Gingivitis and Response to Therapy, J Periodontol (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Al-Sabbagh M, Alladah A, Lin Y, Kryscio RJ, Thomas MV, Ebersole JL, Miller CS, Bone remodeling-associated salivary biomarker MIP-1alpha distinguishes periodontal disease from health, J Periodontal Res 47(3) (2012) 389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dawson DR 3rd, Wang C, Danaher RJ, Lin Y, Kryscio RJ, Jacob RJ, Miller CS, Salivary levels of Epstein-Barr virus DNA correlate with subgingival levels, not severity of periodontitis, Oral Dis 15(8) (2009) 554–9. [DOI] [PubMed] [Google Scholar]
- [43].Ebersole JL, Steffen MJ, Thomas MV, Al-Sabbagh M, Smoking-related cotinine levels and host responses in chronic periodontitis, J Periodontal Res 49(5) (2014) 642–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hayman L, Steffen MJ, Stevens J, Badger E, Tempro P, Fuller B, McGuire A, Al-Sabbagh M, Thomas MV, Ebersole JL, Smoking and periodontal disease: discrimination of antibody responses to pathogenic and commensal oral bacteria, Clin Exp Immunol 164(1) (2011) 118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kirakodu SS, Govindaswami M, Novak MJ, Ebersole JL, Novak KF, Optimizing qPCR for the Quantification of Periodontal Pathogens in a Complex Plaque Biofilm, Open Dent J 2 (2008) 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martin FE, Nadkarni MA, Jacques NA, Hunter N, Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis, J Clin Microbiol 40(5) (2002) 1698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Doungudomdacha S, Rawlinson A, Douglas CW, Enumeration of Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in subgingival plaque samples by a quantitative-competitive PCR method, Journal of medical microbiology 49(10) (2000) 861–74. [DOI] [PubMed] [Google Scholar]
- [48].Chen T, Siddiqui H, Olsen I, In silico Comparison of 19 Porphyromonas gingivalis Strains in Genomics, Phylogenetics, Phylogenomics and Functional Genomics, Frontiers in cellular and infection microbiology 7 (2017) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, Duncan MJ, Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis, J Bacteriol 186(16) (2004) 5473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chastain-Gross RP, Xie G, Belanger M, Kumar D, Whitlock JA, Liu L, Raines SM, Farmerie WG, Daligault HE, Han CS, Brettin TS, Progulske-Fox A, Genome Sequence of Porphyromonas gingivalis Strain 381, Genome Announc 5(2) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM, Complete genome sequence of the oral pathogenic Bacterium porphyromonas gingivalis strain W83, J Bacteriol 185(18) (2003) 5591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xie G, Chastain-Gross RP, Belanger M, Kumar D, Whitlock JA, Liu L, Farmerie WG, Zeng CL, Daligult HE, Han CS, Brettin TS, Progulske-Fox A, Genome Sequence of Porphyromonas gingivalis Strain A7A1–28, Genome Announc 5(10) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chastain-Gross RP, Xie G, Belanger M, Kumar D, Whitlock JA, Liu L, Farmerie WG, Daligault HE, Han CS, Brettin TS, Progulske-Fox A, Genome Sequence of Porphyromonas gingivalis Strain A7436, Genome Announc 3(5) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Peyyala R, Kirakodu SS, Ebersole JL, Novak KF, Novel model for multispecies biofilms that uses rigid gas-permeable lenses, Applied and environmental microbiology 77(10) (2011) 3413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ebersole JL, Novak MJ, Michalowicz BS, Hodges JS, Steffen MJ, Ferguson JE, Diangelis A, Buchanan W, Mitchell DA, Papapanou PN, Systemic immune responses in pregnancy and periodontitis: relationship to pregnancy outcomes in the Obstetrics and Periodontal Therapy (OPT) study, J Periodontol 80(6) (2009) 953–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Eke PI, Wei L, Borgnakke WS, Thornton-Evans G, Zhang X, Lu H, McGuire LC, Genco RJ, Periodontitis prevalence in adults >/= 65 years of age, in the USA, Periodontol 2000 72(1) (2016) 76–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hajishengallis G, Immune evasion strategies of Porphyromonas gingivalis, Journal of oral biosciences / JAOB, Japanese Association for Oral Biology 53(3) (2011) 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baetz A, Zimmermann S, Dalpke AH, Microbial immune evasion employing suppressor of cytokine signaling (SOCS) proteins, Inflamm Allergy Drug Targets 6(3) (2007) 160–7. [DOI] [PubMed] [Google Scholar]
- [59].Thompson SA, Campylobacter surface-layers (S-layers) and immune evasion, Annals of periodontology / the American Academy of Periodontology 7(1) (2002) 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Storisteanu DM, Pocock JM, Cowburn AS, Juss JK, Nadesalingam A, Nizet V, Chilvers ER Evasion Neutrophil Extracellular Traps by Respiratory Pathogens, American journal of respiratory cell and molecular biology 56(4) (2017) 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gunn JS, Bakaletz LO, Wozniak DJ, What’s on the Outside Matters: The Role of the Extracellular Polymeric Substance of Gram-negative Biofilms in Evading Host Immunity and as a Target for Therapeutic Intervention, J Biol Chem 291(24) (2016) 12538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shin S, Brodsky IE, The inflammasome: Learning from bacterial evasion strategies, Seminars in immunology 27(2) (2015) 102–10. [DOI] [PubMed] [Google Scholar]
- [63].Fedele G, Bianco M, Ausiello CM, The virulence factors of Bordetella pertussis: talented modulators of host immune response, Arch Immunol Ther Exp (Warsz) 61(6) (2013) 445–57. [DOI] [PubMed] [Google Scholar]
- [64].Cunha LD, Zamboni DS, Subversion of inflammasome activation and pyroptosis by pathogenic bacteria, Frontiers in cellular and infection microbiology 3 (2013) 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Confer AW, Ayalew S, The OmpA family of proteins: roles in bacterial pathogenesis and immunity, Vet Microbiol 163(3–4) (2013) 207–22. [DOI] [PubMed] [Google Scholar]
- [66].Hall LM, Fawell SC, Shi X, Faray-Kele MC, Aduse-Opoku J, Whiley RA, Curtis MA, Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis, Infect Immun 73(7) (2005) 4253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Grogono-Thomas R, Blaser MJ, Ahmadi M, Newell DG, Role of S-layer protein antigenic diversity in the immune responses of sheep experimentally challenged with Campylobacter fetus subsp. fetus, Infect Immun 71(1) (2003) 147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sims TJ, Ali RW, Brockman ES, Skaug N, Page RC, Antigenic variation in Porphyromonas gingivalis ribotypes recognized by serum immunoglobulin G of adult periodontitis patients, Oral Microbiol Immunol 14(2) (1999) 73–85. [DOI] [PubMed] [Google Scholar]
- [69].DiRita VJ, Mekalanos JJ, Genetic regulation of bacterial virulence, Annual review of genetics 23 (1989) 455–82. [DOI] [PubMed] [Google Scholar]
- [70].Ebersole JL, Hall EE, Steffen MJ, Antigenic diversity in the periodontopathogen, Actinobacillus actinomycetemcomitans, Immunological investigations 25(3) (1996) 203–14. [DOI] [PubMed] [Google Scholar]
- [71].Oliveira RR, Fermiano D, Feres M, Figueiredo LC, Teles FR, Soares GM, Faveri M, Levels of Candidate Periodontal Pathogens in Subgingival Biofilm, J Dent Res 95(6) (2016) 711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Nibali L, Henderson B, Sadiq ST, Donos N, Genetic dysbiosis: the role of microbial insults in chronic inflammatory diseases, Journal of oral microbiology 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Slots J, Periodontology: past, present, perspectives, Periodontol 2000 62(1) (2013) 7–19. [DOI] [PubMed] [Google Scholar]
- [74].Zeller I, Hutcherson JA, Lamont RJ, Demuth DR, Gumus P, Nizam N, Buduneli N, Scott DA, Altered antigenic profiling and infectivity of Porphyromonas gingivalis in smokers and non-smokers with periodontitis, J Periodontol 85(6) (2014) 837–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nagano K, Abiko Y, Yoshida Y, Yoshimura F, Genetic nd antigenic analyses of Porphyromonas gingivalis FimA fimbriae, Mol Oral Microbiol 28(5) (2013) 392–403. [DOI] [PubMed] [Google Scholar]
- [76].Kobayashi T, Kaneko S, Tahara T, Hayakawa M, Abiko Y, Yoshie H, Antibody responses to Porphyromonas gingivalis hemagglutinin A and outer membrane protein in chronic periodontitis, J Periodontol 77(3) (2006) 364–9. [DOI] [PubMed] [Google Scholar]
- [77].Ebersole JL, Steffen MJ, Human antibody responses to outer envelope antigens of Porphyromonas gingivalis serotypes, J Periodontal Res 30(1) (1995) 1–14. [DOI] [PubMed] [Google Scholar]
- [78].Bostanci N, Belibasakis GN, Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen, FEMS Microbiol Lett 333(1) (2012) 1–9. [DOI] [PubMed] [Google Scholar]
- [79].Laine ML, Appelmelk BJ, van Winkelhoff AJ, Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients, J Dent Res 76(12) (1997) 1840–4. [DOI] [PubMed] [Google Scholar]
- [80].van Winkelhoff AJ, Appelmelk BJ, Kippuw N, de Graaff J, K-antigens in Porphyromonas gingivalis are associated with virulence, Oral Microbiol Immunol 8(5) (1993) 259–65. [DOI] [PubMed] [Google Scholar]
- [81].Mysak J, Podzimek S, Sommerova P, Lyuya-Mi Y, Bartova J, Janatova T, Prochazkova J, Duskova J, Porphyromonas gingivalis: major periodontopathic pathogen overview, Journal of immunology research 2014 (2014) 476068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME, Porphyromonas gingivalis: keeping the pathos out of the biont, Journal of oral microbiology 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Holt SC, Ebersole JL, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis, Periodontol 2000 38 (2005) 72–122. [DOI] [PubMed] [Google Scholar]
- [84].Teles FR, Teles RP, Uzel NG, Song XQ, Torresyap G, Socransky SS, Haffajee AD, Early microbial succession in redeveloping dental biofilms in periodontal health and disease, J Periodontal Res 47(1) (2012) 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ, Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray, J Periodontol 80(9) (2009) 1421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhan Y, Zhang R, Lv H, Song X, Xu X, Chai L, Lv W, Shang Z, Jiang Y, Zhang R, Prioritization of candidate genes for periodontitis using multiple computational tools, J Periodontol 85(8) (2014) 1059–69. [DOI] [PubMed] [Google Scholar]
- [87].Divaris K, Monda KL, North KE, Olshan AF, Reynolds LM, Hsueh WC, Lange EM, Moss K, Barros SP, Weyant RJ, Liu Y, Newman AB, Beck JD, Offenbacher S, Exploring the genetic basis of chronic periodontitis: a genome-wide association study, Human molecular genetics 22(11) (2013) 2312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Reynolds MA, Modifiable risk factors in periodontitis: at the intersection of aging and disease, Periodontol 2000 64(1) (2014) 7–19. [DOI] [PubMed] [Google Scholar]
- [89].Borrell LN, Crawford ND, Social disparities in periodontitis among US adults: the effect of allostatic load, Journal of epidemiology and community health 65(2) (2011) 144–9. [DOI] [PubMed] [Google Scholar]
- [90].Jenkins WM, Kinane DF, The ‘high risk’ group in periodontitis, British dental journal 167(5) (1989) 168–71. [DOI] [PubMed] [Google Scholar]
- [91].Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ, Cdc GDRP Piriodontal Disease Surveillance workgroup: James Beck, Prevalence of periodontitis in adults in the United States: 2009 and 2010, J Dent Res 91(10) (2012) 914–20. [DOI] [PubMed] [Google Scholar]
- [92].Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, Taylor GW, Page RC, Beck JD, Genco RJ, Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 – 2012, J Periodontol (2015) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lamster IB, Geriatric periodontology: how the need to care for the aging population can influence the future of the dental profession, Periodontol 2000 72(1) (2016) 7–12. [DOI] [PubMed] [Google Scholar]
- [94].Slots J, Periodontitis: facts, fallacies and the future, Periodontol 2000 75(1) (2017) 7–23. [DOI] [PubMed] [Google Scholar]
- [95].Ebersole JL, Al-Sabbagh M, Gonzalez OA, Dawson DR 3rd, Ageing effects on humoral immune responses in chronic periodontitis, J Clin Periodontol 45(6) (2018) 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Takeuchi K, Furuta M, Takeshita T, Shibata Y, Shimazaki Y, Akifusa S, Ninomiya T, Kiyohara Y, Yamashita Y, Serum antibody to Porphyromonas gingivalis and periodontitis progression: the Hisayama Study, J Clin Periodontol (2015). [DOI] [PubMed] [Google Scholar]
- [97].Furuta M, Shimazaki Y, Tanaka S, Takeuchi K, Shibata Y, Takeshita T, Nishimura F, Yamashita Y, Gender-specific associations of serum antibody to Porphyromonas gingivalis and inflammatory markers, BioMed research international 2015 (2015) 897971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ramseier CA, Kinney JS, Herr AE, Braun T, Sugai JV, Shelburne CA, Rayburn LA, Tran HM, Singh AK, Giannobile WV, Identification of pathogen and host-response markers correlated with periodontal disease, J Periodontol 80(3) (2009) 436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ebersole JL, Frey DE, Taubman MA, Haffajee AD, Socransky SS, Dyn; mics of systemic antibody responses in periodontal disease, J Periodontal Res 22(3) (1987) 184–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.