Abstract
The tolerance of fish to fasting offers a model to study the regulatory mechanisms and changes produced when feeding is restored. Gilthead sea bream juveniles were exposed to a 21-days fasting period followed by 2 h to 7-days refeeding. Fasting provoked a decrease in body weight, somatic indexes, and muscle gene expression of members of the Gh/Igf system, signaling molecules (akt, tor and downstream effectors), proliferation marker pcna, myogenic regulatory factors, myostatin, and proteolytic molecules such as cathepsins or calpains, while most ubiquitin-proteasome system members increased or remained stable. In bone, downregulated expression of Gh/Igf members and osteogenic factors was observed, whereas expression of the osteoclastic marker ctsk was increased. Refeeding recovered the expression of Gh/Igf system, myogenic and osteogenic factors in a sequence similar to that of development. Akt and Tor phosphorylation raised at 2 and 5 h post-refeeding, much faster than its gene expression increased, which occurred at day 7. The expression in bone and muscle of the inhibitor myostatin (mstn2) showed an inverse profile suggesting an inter-organ coordination that needs to be further explored in fish. Overall, this study provides new information on the molecules involved in the musculoskeletal system remodeling during the early stages of refeeding in fish.
Subject terms: Molecular biology, Physiology
Introduction
Gilthead sea bream (Sparus aurata Linnaeus 1758) is one of the most important marine fish species in Mediterranean aquaculture, which has expanded over the past two decades1 in parallel with the scientific research and the knowledge of its physiology. Unlike mammals, fish are able to adapt to relatively long periods of starvation and it is possible to use fasting as a practice to improve product quality by reducing body lipid content, and refeeding as a way to induce compensatory growth2–4. Under normal feeding conditions, fish grow and store energy reserves, while in fasting body stores are mobilized to maintain life processes5. During fasting, the metabolism switches to a catabolic status, resulting in low growth rate, and the following refeeding reverts the situation towards a hyper-anabolic phase when organisms attempt to accelerate the growth rate6. Both approaches, fasting and refeeding, can be very informative in fish basic and applied research.
The effects of fasting and refeeding on body growth, metabolism, protein biosynthesis and hormonal responses have been largely studied in fish6,7. The muscle mass of fish species is an important tissue that considerably depends during fasting and refeeding on protein degradation and synthesis. During these stages of nutritional changes, metabolism and growth are adapted to resist the restrictions and rapidly adjust to the arrival of new nutrients. All these determine changes on the endocrine status and in the regulation of substrates mobilization by muscle and bone6,8. In compensatory growth studies, it has been described that refeeding stimulates proliferation of fish myogenic cells9,10.
The growth hormone and insulin-like growth factors (Gh/Igfs) are both, in vertebrates including fish, key factors regulating growth. Muscle and bone are widely regulated by this system and the presence of Gh and Igf1 receptors (Ghrs and Igf1Rs) and Igf isoforms as well as binding proteins (Igfbps) are well described in fish, especially in these tissues11,12. Moreover, in gilthead sea bream, the function of Gh/Igfs and its response to diverse conditions has been characterized and the ratio of the circulating levels of Gh and Igf1 is considered a good marker of growth quality in terms of its relation with body indexes or muscle fibers composition12,13.
Muscle growth is also controlled by myogenic regulatory factors (MRFs) (MyoD, Myf5, Myog and Mrf4), and the negative endogenous regulator myostatin, as well as other factors that control sequentially the process of development and growth6,14,15. The effects of fasting and refeeding on myogenesis have been studied in diverse fish species6,10,16 demonstrating the function of the different MRFs during the metabolic arrest caused with food limitation and the subsequent activation when feeding is restored. Furthermore, during fasting and refeeding, proteolytic molecules play a remarkable role to adapt to the changes in amino acids supply. This is more important in fish that have a specific and enhanced protein metabolism. The main endogenous proteolytic systems, each one performing specific degradative or regulatory functions according to the nutritional conditions are, calpains, cathepsins and ubiquitin-proteasome, all of which are well-known in fish, especially salmonids17–19, but also in gilthead sea bream20–22.
In addition to muscle, bone is also an important tissue for skeletal and locomotion functions, but also, as a reservoir of minerals that is clearly affected when nutrition is restricted. Essential during development, synchronicity between bone and muscle is required for proper musculoskeletal growth23,24. Besides to being induced by the Gh/Igf system, the process of osteogenesis is also regulated by skeleton-derived factors that control specific stages of osteoblasts development and bone building. Although less known in fish than in mammals, most of these molecules have been identified in gilthead sea bream25,26. Thus, while the Runt-related transcription factor 2 (runx2) and the structural molecule fibronectin 1a (fib1a) have a key role in the early development of the tissue, the non-collagenic molecules such as osteopontin (op) and osteocalcin (ocn), play a main role in bone maturation and matrix mineralization. However, little information exists concerning the effects of fasting and refeeding in the regulation of these molecular factors and bone development in fish species.
The objective of this study is to analyze the effects that fasting and refeeding provoke in muscle and bone in S. aurata, characterizing the expression pattern of various genes of interest of the musculoskeletal system to improve our knowledge of the regulatory mechanisms involved in these processes, and to explore their contribution to vertebrate compensatory growth. Specifically, we were interested in the early refeeding (within 24 h) when the mechanisms of tissue remodeling are triggered by the arrival of the first nutrients after 21 days of fasting, which offers an interesting scenario of regulatory molecules that restart their activity gradually after a period of latency.
Materials and Methods
Animals, experimental design and ethics statement
Gilthead sea bream (Sparus aurata L.) juveniles (initial body weight 50 ± 3 g; standard length 15.3 ± 0.68 cm; sexually immature) were obtained from a commercial hatchery (Piscimar, Burriana, Spain) and kept in the facilities of the Faculty of Biology (Universitat de Barcelona, Spain) during four weeks before acclimation period. Forty-two fish were randomly distributed into six two-hundred-liters tanks (7 fish per tank) with a recirculation system. Fish were kept at 23 ± 1 °C and a photoperiod of 12 h of light and 12 h of dark during the whole experiment. During the acclimation period (2 weeks), the fish were fed ad libitum twice a day with a commercial diet (Optibream, Skretting, Burgos, Spain).
After the acclimation period, a 28-day experiment was designed as previously described27. Briefly, it consisted in two parts: a 21-days fasting period and a 7-days refeeding period. Samples were taken at the beginning of fasting (-21 days), and at the end of it at 0, 2, 5, and 24 h and 7 days after refeeding. During the refeeding period, the fish were given the same commercial diet (Optibream, Skretting) once a day until apparent satiety. The -21 days, 24 h and 7 days samplings were performed 24 h after the last feeding. During the experiment, all the environmental conditions (temperature, photoperiod, salinity: 21 °C; 12 L: 12D; 38‰, respectively) were maintained stable.
In each sampling, six individuals (one per tank) were anesthetized and sacrificed per condition. The fish were anesthetized with MS-222 (0.08 g/L) (Sigma-Aldrich, Tres Cantos, Spain), body length (standard) and body weight were measured and blood was drawn from the caudal vein. The plasma levels of Gh and Igf1 were measured by corresponding radioimmunoassays as previously described27. Then, the fish were sacrificed by an overdose of the same anesthesia (0.3 g/L) and, bone from the vertebral column and skeletal white muscle from the dorsal area were dissected and snap frozen in liquid nitrogen immediately. All samples were stored at −80 °C until further analysis. This study was carried out in accordance with the recommendations of the EU, Spanish and Catalan Government-established norms and procedures. The specific protocol was approved by the Ethics and Animal Care Committee of the University of Barcelona (permit numbers CEEA 110/17 and DAAM 9488).
Gene expression analyses
As previously described27, total RNA was extracted from 100 mg of skeletal white muscle and vertebral column samples using TRI Reagent Solution (Applied Biosystems, Alcobendas, Spain) and a Precellys® Evolution and Cryolys system (Bertin Technologies, Montigny-le-Bretonneux, France) for tissue homogenization. Quantity, quality and integrity were determined with a NanoDrop2000 (Thermo Scientific, Alcobendas, Spain), and a 1% agarose gel (w/v), respectively. Then, one µg RNA was treated with DNase I (Life Technologies, Alcobendas, Spain) and retrotranscribed with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Sant Cugat del Vallès, Spain).
Next, the mRNA transcript levels were examined by quantitative real time PCR (qPCR) according to the requirements of the MIQUE guidelines28 in a CFX384TM Real-Time System (Bio-Rad, El Prat de Llobregat, Spain). All reactions were performed in the conditions previously described27. The primers used for each tissue are listed in Supplementary Table 1. Moreover, in both tissues the reference genes elongation factor 1 alpha (ef1α), ribosomal protein S18 (rps18), beta actin (β-actin) and ribosomal protein L27 (rpl27) were analyzed and the combination of the two most stable (ef1α and rps18) was used to calculate the relative expression of the genes of interest following the Pfaffl method29. Both, reference genes stability and relative expression calculation were determined with the implemented Bio-Rad CFX Manager Software (v2.1).
Protein expression analyses
Protein was extracted from 100 mg of either muscle or bone samples. Tissue homogenates were made in 1 mL of RIPA buffer supplemented with phosphatase (PMSF and NA3VO4) and protease inhibitors (P8340, Santa Cruz) using the Precellys® Evolution coupled to a Cryolys cooling system (Bertin Technologies, Montigny-le-Bretonneux, France).
Protein quantification was performed following the Bradford’s method using BSA (Sigma Aldrich, Tres Cantos, Spain) for the standard curve. Next, 15 µg of the soluble protein fraction were prepared in a loading buffer (containing SDS and β-mercaptoethanol), heated at 95 °C for 5 min and run in a 12% polyacrylamide gel. Following, the proteins were transferred overnight to Immobilon® PVDF-FL 0.2 μm Transfer Membranes (Merck Millipore Ltd., Tullagreen, Cork, Ireland), previously activated in methanol. Total transferred protein was determined by 5 min incubation with REVERTTM Total Protein Stain (LI-COR, Lincoln, Nebraska, USA) and reading at 700 nm using the Odyssey Fc Imaging System (LI-COR). Membranes were blocked in Odissey Blocking Buffer (diluted 1:1 in TBS) (LI-COR) for 1 h at room temperature, and then overnight at 4 °C and in agitation in the corresponding diluted primary antibody. The primary antibodies used were as follows: Goat polyclonal anti cathepsin L (anti-Ctsl D-20 antibody; catalog no. sc-6501), goat polyclonal anti-cathepsin D (anti-Ctsd; catalog no. sc-6486) all from Santa Cruz Biotechnolgy (Santa Cruz, California, USA), rabbit polyclonal anti-phospho Akt (cat- no. 9271), anti-total Akt (cat. no. 9272) and anti-phospho Tor (cat. no. 2971) from Cell Signaling Technology (Beverly, MA, USA) and anti-total Tor (cat. no. T2949) from Sigma-Aldrich (Tres Cantos, Spain). All these antibodies have been previously demonstrated to cross-react successfully with the proteins of interest in gilthead sea bream22,23,30. Subsequently after washing with TBS-T, the membranes were incubated with the corresponding secondary antibodies: goat anti-rabbit (Cat. No. 925-32211, Servicios Hospitalarios) or donkey anti-goat (Cat. No. 925-32214, Servicios Hospitalarios) diluted at 1:10000 in the same blocking buffer. After incubation, membranes were washed with TBS-T and fluorescence signal was measured at 800 nm using the Odyssey Fc Imaging System (LI-COR). Stripping was performed using a commercial stripping buffer (NewBlot PVDF 5X Stripping Buffer (LI-COR). Detailed information about Western blot membranes distribution can be found in Supplementary Information 3.
Statistical analyses
The data obtained was analyzed using IBM SPSS Statistics vs. 22 and is presented as a mean ± standard error of the mean (SEM). Normality and homogeneity of the variances were tested with the Shaphiro-Wilk’s test and the Levene’s test respectively. When the data did not show a normal distribution or homoscedasticity, were transformed by logarithm. Differences among groups were tested by one-way analysis of variance (ANOVA) followed by Tukey HSD or LSD, as post-hoc tests. In the case that normality and/or homoscedasticity were not found even after logarithmic data transformation, the non-parametric Kruskal-Wallis test or the Dunnett’s T3 as post-hoc, were used. The confidence interval for all analyses was set at 5%.
Results
Somatic parameters and GH/IGF-I ratio
As shown in Supplementary Table 2, fasting for 21 days provoked an arrest of growth (i.e. body weight and condition factor, CF) that tended to recover after 7 days refeeding. Similarly, hepatosomatic index (HSI) felt down significantly during fasting to reach normal values after 7 days refeeding. Moreover, the viscerosomatic index (VSI) was also significantly reduced by fasting, and refeeding induced an acute increase to recover the basal levels after 1 day. Fasting resulted also in a high Gh/Igf1 ratio that only recovered basal values after 7 days refeeding. In fact, as we previously reported27, plasma Gh levels raised from 0,77 ± 0,03 to 32,77 ± 0,79 ng/ml in response to 21 days of fasting with a progressive decrease to basal levels with refeeding; while circulating Igf1 levels showed an inverse pattern, decreasing from 15,35 ± 2,51 to 5,9 ± 1,32 ng/ml after fasting and recovering only after a long-term refeeding.
Muscle responses to fasting and refeeding
GH and IGF family
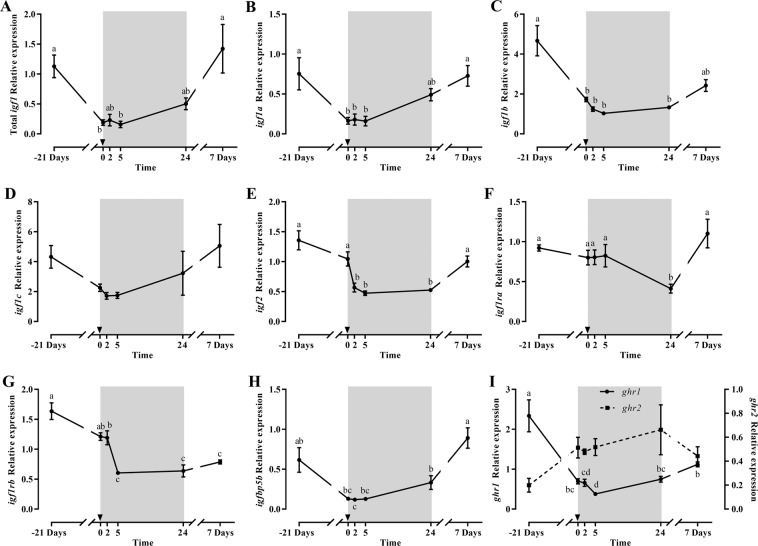
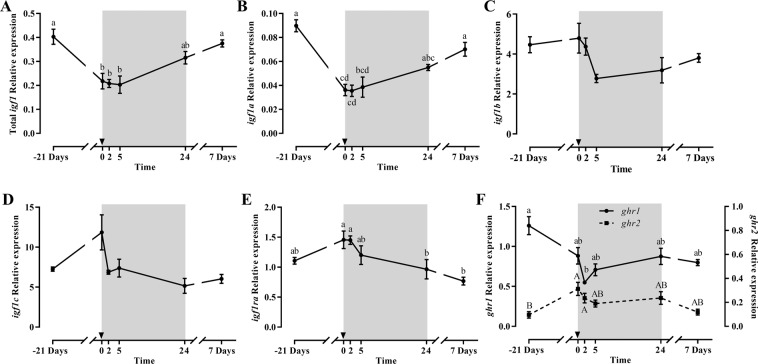
Total igf1 gene expression showed a significant decrease after 21 days of fasting recovering basal levels by day 7 of refeeding (Fig. 1A); the same effect was observed for the igf1 splice variants igf1a and igf1b, but not for igf1c (Fig. 1B–D). The igf2 (Fig. 1E) profile was similar although a significant decrease was observed from 2 h refeeding and basal levels recovered at 7 days refeeding. Between the igf1rs, igf1ra was not affected by fasting but decreased significantly at 24 h refeeding recovering later (Fig. 1F), while igf1rb decreased during fasting, and continued dropping at 2 and 5 h refeeding reaching then significantly the lowest levels that remained low after 7 days refeeding (Fig. 1G). The expression of the igfbps, igfbp1 and igfbp4 was unaltered (data not shown), but igfbp5b decreased significantly at 2 h refeeding, and recovered basal levels 7 days after refeeding (Fig. 1H). Concerning ghrs, they showed an inverse profile, with ghr1 decreasing significantly during fasting and at 2 and 5 h refeeding, to increase at 1 and 7 days refeeding; while ghr2, although not significantly, progressively increased through the experiment up to 24 h of refeeding, decreasing afterwards at day 7 (Fig. 1I).
Figure 1.
Relative gene expression of skeletal white muscle total igf1 (A), igf1a (B), igf1b (C), igf1c (D), igf2 (E), igf1ra (F), igf1rb (G), igfbp5 (H) and ghr1 and ghr2 (I) in gilthead sea bream during the fasting and refeeding experiment. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Signaling molecules
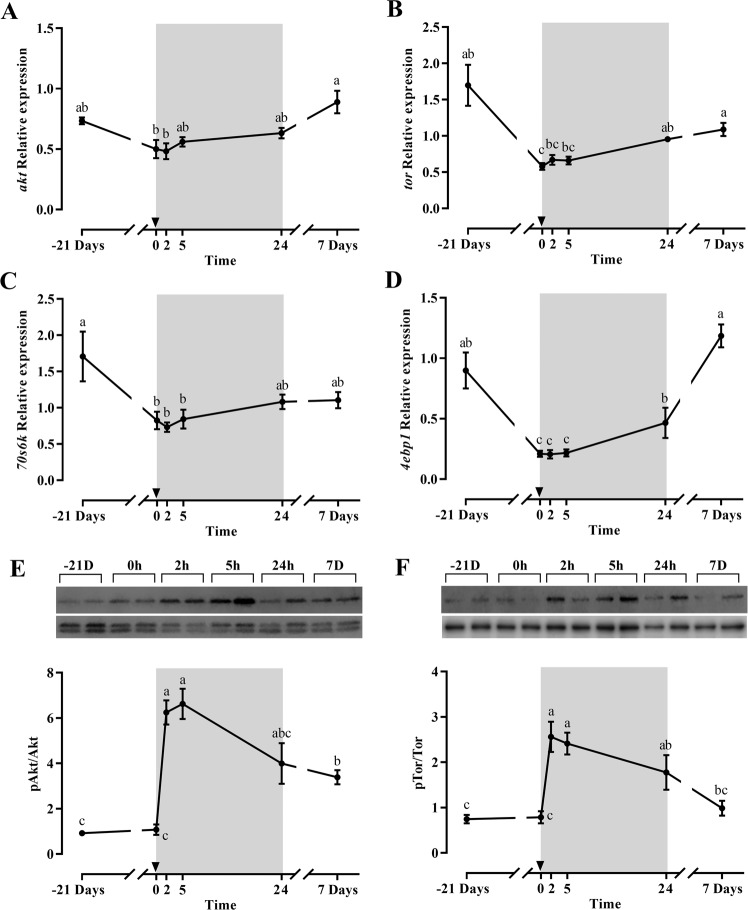
The mRNA levels of akt, tor, and the downstream molecules 70s6k and 4ebp1 (Fig. 2A–D) showed a similar gene expression profile. With the exception of akt, all molecules exhibited a significant decrease with fasting and a progressive increase during the refeeding period, significant at day 7 for akt, tor and 4ebp1. Interestingly the postprandial response for Akt and Tor was much faster when their phosphorylation was studied (Fig. 2E,F), being significantly increased at 2 and 5 h refeeding compared to time zero and decreasing later at 1 and 7 days. foxo3 did not show any significant difference along the experiment (data not shown).
Figure 2.
Relative gene expression of skeletal white muscle akt (A), tor (B), 70s6k (C) and 4ebp1 (D) and representative blot and densiometric analysis of the phosphorylation ratios of Akt (E) and Tor (F) in gilthead sea bream during the fasting and refeeding experiment. For the Western blots, the same membranes cropped in two were used to analyze Tor (top part) and Akt (bottom part). The phosphorylated forms were analyzed first and after stripping, the corresponding total forms were determined in the same membranes. The intensity of the phosphorylated form was normalized by its total form, and the intensity of each specific band was normalized by the total transferred protein for the corresponding well. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Muscle growth-related factors
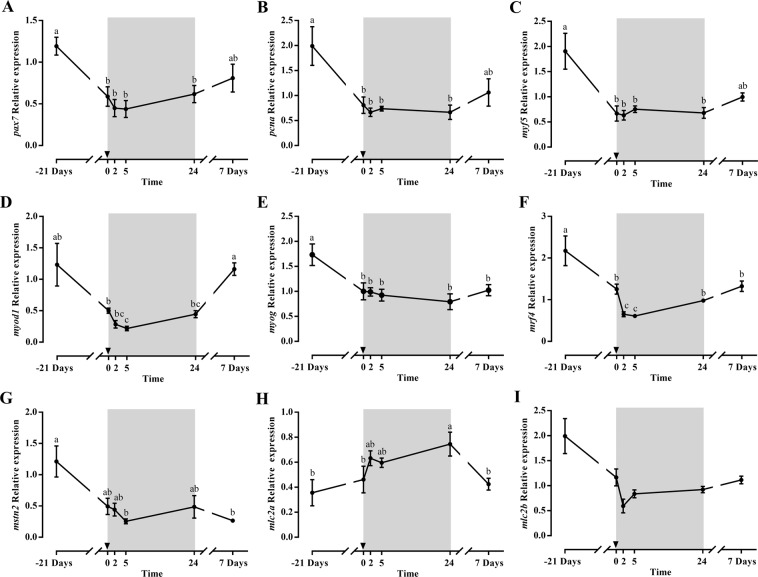
The profiles of pax7 and pcna expression were similar and correlated very well with those of myf5 and myod1, decreasing significantly during fasting and partially recovering after 7 days refeeding (Fig. 3A–D). The expression of other myogenic genes also decreased significantly during fasting or early refeeding (myog, mrf4, mstn2) (Fig. 3E–G), or showed similar tendencies (myod2, mstn1, data not shown). At 7 days of refeeding, mrf4 started to increase again while myog and mstn2 remained at low levels. Moreover, the structural myosin light chains, mlc2a and mlc2b showed an inverse profile with a significant increase for mlc2a at 24 h refeeding, returning to basal levels at day 7, and maintained expression for mlc2b (Fig. 3H–I).
Figure 3.
Relative gene expression of skeletal white muscle pax7 (A), pcna (B), myf5 (C), myod1 (D), myog (E), mrf4 (F), mstn2 (G), mlc2a (H) and mlc2b (I) in gilthead sea bream during the fasting and refeeding experiment. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Proteolytic systems’ genes
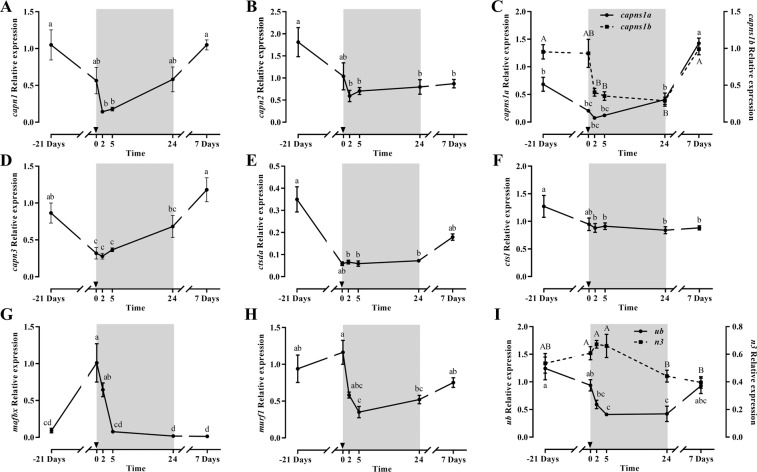
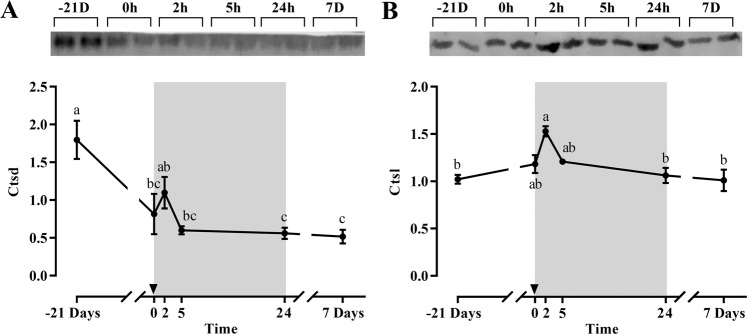
Most of the proteolytic genes expression was significantly downregulated during fasting or early refeeding, recovering basal levels after 7 days refeeding (Fig. 4). This was the case for capn1, capns1b, capn3, ctsda and ub, and a similar tendency was observed for capns1a. Moreover, capn2 and ctsl expression was also significantly downregulated with fasting but after 7-days refeeding were not able to recover basal values. It is remarkable how mafbx and murf1 increased or were maintained after 21 days of fasting to significantly decrease after 5 or 24 h refeeding, reaching basal levels at the end of the experiment, while n3 showed only minor changes on expression along the whole experiment (Fig. 4G–I). Regarding protein expression, only Ctsd decreased significantly with fasting, but both Ctsd and Ctsl presented similar patterns postprandially with a single peak at 2 h (Fig. 5).
Figure 4.
Relative gene expression of skeletal white muscle capn1 (A), capn2 (B), capns1a and capns1b (C), capn3 (D), ctsda (E), ctsl (F), mafbx (G), murf1 (H) and ub and n3 (I) in gilthead sea bream during the fasting and refeeding experiment. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Figure 5.
Representative blot and densiometric protein levels of skeletal white muscle Ctsd (A) and Ctsl (B) in gilthead sea bream during the fasting and refeeding experiment. Each protein was analyzed in cropped membranes of different Western blots along with other proteins (data not shown). The intensity of each specific band was normalized by the total transferred protein. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Bone responses to fasting and refeeding
GH and IGF family
Bone total igf1 and igf1a mRNA levels showed significant lower values after the 21 days of fasting, returning to basal levels at 24 h and 7 days refeeding (Fig. 6A,B). Differently, igf1b and igf1c expression was not significantly affected by fasting or refeeding (Fig. 6C,D); and igf1ra expression was maintained at fasting but significantly decreased at 1 and 7 days refeeding (Fig. 6E). Neither igf1rb nor igfbps were affected by fasting or refeeding (data not shown). ghrs expression presented, as observed in muscle, a reverse profile. Thus, while ghr1 expression decreased significantly during fasting and early refeeding, ghr2 expression increased significantly during the same period. Then, both ghrs returned to initial levels at 24 h or 7 days refeeding (Fig. 6F).
Figure 6.
Relative gene expression of bone total igf1 (A), igf1a (B), igf1b (C), igf1c (D), igf1ra (E) and ghr1 and ghr2 (F) in gilthead sea bream during the fasting and refeeding experiment. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Bone-related genes
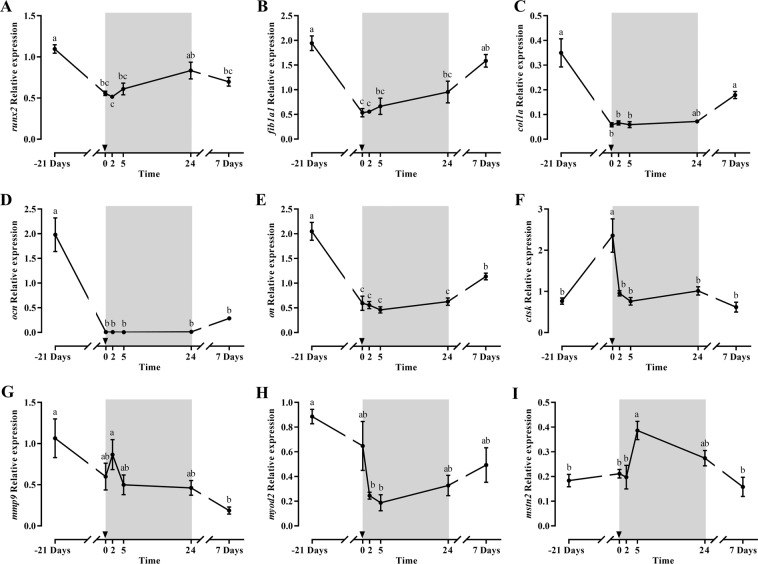
The expression of the osteogenic factors runx2, fib1a, col1a1, ocn and on showed a significant diminution during fasting or early refeeding (Fig. 7A–E). Then, runx2, fib1a and col1a1 recovered the basal mRNA levels after 1 or 7 days of refeeding, while ocn and on presented a similar tendency but without reaching basal levels and still remaining low at day 7.
Figure 7.
Relative gene expression of bone runx2 (A), fib1a (B), col1a1 (C), ocn (D), on (E), ctsk (F), mmp9 (G), myod2 (H) and mstn2 (I) in gilthead sea bream during the fasting and refeeding experiment. The postprandial period is shown in grey and the time in hours. Data are shown as means ± SEM (n = 6). Letters indicates significant differences (p < 0.05) by one-way ANOVA, LSD and Tukey HSD test.
Concerning the osteoclastogenic genes, the expression level of ctsk increased after 21 days of fasting, to recover the basal levels at 2 h refeeding, while mmp9 showed a tendency to decrease at 21 days fasting, peak at 2 h refeeding to then significantly decrease at 7 days refeeding (Fig. 7F,G). trap showed a very similar profile to mmp9 but without significant changes throughout the experiment (data not shown).
Muscle growth-related factors
First, pax7 gene expression was studied in bone samples to confirm purity of the tissue and the results obtained showed very low mRNA levels, close to undetectable showing a basal line without changes throughout the experiment (data not shown). Then, the other myogenic genes studied in bone (i.e. myod1, myod2, mrf4, mstn1 and mstn2) did not show significant responses to either fasting or refeeding (data not shown) with the exception of myod2 and mstn2 (Fig. 7H,I). myod2 expression decreased during fasting showing significant low levels at early refeeding (2 and 5 h), and recovered basal values at the end of the experiment; mstn2 showed an almost contrary profile, since it was stable during fasting but peaked at 5 h postprandial to progressively return to basal values at 24 h and 7 days refeeding.
Discussion
Fish are well adapted to resist fasting periods and this characteristic has been used to study the mechanisms that regulate the balance between anabolism and catabolism in these vertebrates7. The present study continues the previous one already published by Perelló-Amorós and coworkers27 in which the function of ghrelin during fasting and refeeding was characterized in gilthead sea bream. That paper reported also the effects on gh and igf1 pituitary and liver expression, hormone plasma levels and the fish biometric indexes. Briefly, fasting provoked an increase in ghrelin plasma levels and a decrease on liver igf1 expression, while refeeding reversed progressively the condition. In the present study, we focused on the Gh/Igf axis and other regulatory systems in muscle and bone, providing valuable information on the regulation of the musculoskeletal system, especially in bone, a tissue still poorly explored in fish. Refeeding reversed the fasting condition and the sequential sampling permitted to analyze how regulatory mechanisms returned gradually to the growth condition. Although the absence of a pair-fed control group does not allow differentiating among refeeding and postprandial effects, the present data suggest that early refeeding after 21-days fasting amplifies the responses likely occurring postprandially at an attenuated magnitude. Thus, the experimental model would emphasize the main steps of skeletal muscle remodeling that take place in gilthead sea bream when food is restored.
Igfs play an important role on growth regulation in vertebrates12,31–33 and during adverse conditions are downregulated to preserve the survival of the animal over the body growth. Gh secretion is dependent of nutrients and both, Gh and Igf1 regulate metabolism in fish7,34. In gilthead sea bream, 21 days of fasting resulted in a general decrease in the muscular gene expression of total igf1 and its splice variants. This is in agreement with the significant decrease of liver total igf1 expression previously described in the same animals27, indicating altogether that both, systemic and local Igf1 production, might contribute to arrest muscle growth under these catabolic conditions. Similarly, Peterson and coworkers33 in channel catfish (Ictalurus punctatus) reported that fasting for 30 days reduced fish weight by approximately 60% and decreased igf1 mRNA in muscle, and this has been also seen in other fish species11,35. This decrease in Igf1 during a nutrient-starvation period can stop muscle proliferation and development as observed in an in vitro muscular model of gilthead sea bream under specific amino acids limitation (i.e. lysine)36. In the current study, both igfrs were affected differently; thus, igf1ra did not respond to fasting but to refeeding, recovering the expression levels at day 7, while the decreased expression levels of igf1rb in fasting and early refeeding were maintained until the end of the experiment. Such a differential response has been reported in several species using in vivo or in vitro models8,10,32,36,37 and suggests a functional split between both igfrs isoforms. Furthermore, these findings point out that during a fasting or a low food intake period, Igf1 production decreases, but contrarily, some of the igfrs isoforms can be maintained, suggesting a rise in Igf sensitivity8,32,38,39.
Muscle igfbps also responded differently to the treatment and while expression of igfbp1 or igfbp4 was unchanged, igfbp5 significantly decreased in fasting and recovered progressively after 7 days of refeeding. This is in agreement with the anabolic function attributed to igfbp5, which has been identified as a good growth marker in gilthead sea bream40. Similar responses were detected in some species41,42; but not in others35,43,44, although such a decrease is analogous to the Igfbp3 plasma levels determined in fasted coho and chinook salmon45–47, suggesting a similar response of these two igfbps to long term food deprivation in different fish species. Thus, Igfbps in teleost are differently affected by the various metabolic conditions42 and in the case of gilthead sea bream muscle, igfbp5b seems to be the most sensitive form to food availability. Moreover, ghrs expression showed in this study a dual role, with the decrease of ghr1 until 5 h refeeding and a partial increase at 24 h and 7 days refeeding and the increase of ghr2 expression up to 24 h refeeding. These patterns coincide with the differential function attributed to each receptor in gilthead sea bream and other fish species, being Ghr1 anabolic and Ghr2 catabolic23,48. Thus, in gilthead sea bream the growth enhancement caused by sustained exercise was followed by an increase in ghr1 expression and a decrease in ghr2 expression in muscle40. Similarly in this species, Gh treatment induced in muscle an increase of ghr1 expression but not ghr223. In summary, fasting depresses the components involved in growth promotion (ghr1, igf1, igf1rb, igfbp5), while refeeding reverts progressively the situation to activate muscle recovery.
Next, the expression of key elements of the main signaling pathways regulating protein synthesis and muscle growth (akt and tor) was significantly diminished during fasting. Such a scenario has been also observed in different fish species exposed to food deprivation11,22. The present study also demonstrates the sequential activation of those pathway components with the recovery of nutrition; however, it is very interesting to compare such a progressive response in gene expression with the quick phosphorylation of Akt and Tor within 2 h post-feeding. Similar responses were previously observed in fine flounder (Paralichthys adspersus)11, rainbow trout19,49 and even in gilthead sea bream both in vivo50, as well as in vitro30. Thus, the comparison of akt and tor mRNA levels with their corresponding protein phosphorylation status during fasting and refeeding in the present study corroborates the different timely regulation of these pathways at protein and gene levels. Nevertheless, according to our results, it has to be taken into account that while refeeding is likely a major factor in the gene/protein expression and/or phosphorylation responses, they may also occur to some degree (especially those related with protein activation), during the introduction of nutrients in a common postprandial period. Therefore, future studies should consider these overlapping responses to specifically identify the dynamic nature of how nutrients regulate these processes in fish.
The information on MRFs under nutritional restriction in gilthead sea bream is scarce and current results could be useful to understand muscle growth regulation in such catabolic situations. Similarly to that observed in cultured muscle cells with lysine deficiency36, in this study the expression of myod1, myf5, myog, mrf4 and mstn2 decreased during fasting or early refeeding, while late refeeding recovered basal expression of myods and myf5. Thus, making noticeable that the sequence of MRFs up-regulation during refeeding follows the same characteristic order as during myogenesis activation15. The parallelism between the profile of myf5, myod1 and myod2 with pcna or pax7 agrees with the involvement of these MRFs in the first stage of myogenesis. Interestingly, similar patterns of recovery after starvation have been reported for several MRFs, mostly myod and myog, in salmonids16,32,51,52 and in juvenile Nile tilapia (Oreochromis niloticus)6; overall, confirming the beginning of muscle remodeling at that early stage of refeeding. On the other hand, mstn2 expression decreased with fasting and maintained significant low levels after 7 days refeeding, which is in agreement with previous results found in rainbow trout52 and sea bass (Dicentrarchus labrax)53. Thus refeeding provoked a clear down-regulation of mstn gene expression to favor muscle recovery, in parallel to the increase observed of pax7, pcna, and MRFs expression.
Concerning muscle structural components, mlc2a and mlc2b showed an inverse profile, with mlc2a expression not changing with fasting but showing a significant increase at 24 h refeeding to return to basal levels at day 7. Bower and Johnston37 also found in Atlantic salmon an increase of mlc2 after 14 days of refeeding, and previous studies in gilthead sea bream22,23,54 showed that an increase in specifically, mlc2a expression, indicates a condition that favors muscle proliferation. In this sense, in the present study the mlc2a peak at 24 h refeeding paralleled the tendency to enhance the expression of pax7, pcna, tor and the MRFs involved in early myogenesis (myod2 and myf5), facilitating the muscle to grow during this period of compensatory growth.
Several members of the main proteolytic systems have been characterized and their responses upon different experimental challenges investigated in different fish species18,55–60, including gilthead sea bream20–22; however, due to the different experimental conditions (i.e. duration of periods, fish age, etc.) among studies, it is difficult to reach clear explanation of their respective roles. In the present study, two different expression patterns were observed concerning proteolytic genes; those that were downregulated during fasting and recovered with refeeding, like calpains and some cathepsins, in a similar trend to that observed for igf1 or MRFs; and those belonging to the ubiquitin-proteasome system, like murf1 or mafbx, which increased in response to fasting and decreased with refeeding.
Regarding the calpains, in agreement with our findings, channel catfish fasted for 35 days presented a strong down-regulation of capn1 and capn3 genes, which recovered basal expression after a refeeding period58. Contrarily, Salmerón and coworkers20 did not find significant effects on calpains expression in gilthead sea bream and suggested that these molecules could have a secondary role in the adaptation to food deprivation; and in fact, observed later that ctsd and ctsl increased significantly under the same conditions21, similarly as observed in halibut for ctsb and ctsd8. Although we did not observe an increase in the gene expression of ctsda or ctsl in this study, amino acids limitation in in vitro myocytes upregulated both cathepsins expression61. Moreover, in the present study, Ctsl protein expression increased in fasting or early refeeding while Ctsd decreased, suggesting this different response certain distribution of their regulatory functions. Concerning the ubiquitin-proteasome system, in a previous experiment in gilthead sea bream, 30 days of fasting upregulated the expression of the same genes that were increased in the present study21. Different authors have found similar responses of these genes with fasting19,44,50,60,62 and, specific amino acids limitation increased also the expression of mafbx and murf161. Interestingly, results were also supported at a protein level in vivo, since fasted rainbow trout muscle presented an increase in the total amount of polyubiquitinated proteins19.
As an overview, in the present fasting and refeeding model, it seems that there is a clear difference between the expression of genes of the three major proteolytic pathways, being the ubiquitin-proteasome system the one that may have a stronger role in response to food restriction. Refeeding reverts the condition with calpains recovering high basal levels, while cathepsins present a variable role depending on the molecule considered. All this suggests a coordinated distribution of the proteolytic functions during the mobilization of reserves (fasting) and the remodeling associated with the compensatory muscle-growth induced by refeeding.
The skeletal system has multiple physiological functions in vertebrates63, and the high incidence of skeletal deformities is still an important bottleneck for the sustainability of aquaculture64. Information about the effects of fasting and refeeding on bone has been mainly studied in cellular-boned fish, like the rainbow trout65,66, while in fish species with acellular bone such as gilthead sea bream knowledge is limited. In mammals, it is well known that Igfs are important mediators of bone growth67,68, being present at high concentrations in the bone matrix and, any deficiency on the Igf genes affecting skeletal growth69. Igfs stimulate differentiation of osteoblasts regulating the balance between bone accretion and resorption, which occurs throughout life70. So, in a fasting condition, or under severe chronic undernutrition, Igf1 concentrations decrease in association with low bone turnover and significant bone loss71,72. In fish, early studies also demonstrated the growth stimulatory effect of Igf1 in branchial arches of Japanese eel (Anguilla japonica)69 and more recently, in an in vitro model of bone-derived cells from vertebra of gilthead sea bream, Igf1 as well as insulin, were demonstrated to stimulate cell proliferation70.
In the current study, the effects of fasting and refeeding in bone were, in terms of the Gh/Igf system, similar to those found in muscle. Thus, the expression of ghr1 and ghr2 was inverse at fasting and refeeding, but the profiles were similar between the two tissues, supporting the conservation of Ghrs’ role as well as their coordination in muscle and bone to adapt to the changes in alimentary conditions. About peptides, fasting resulted in a decrease of total igf1 and igf1a expression, supporting reduced bone growth during catabolic conditions, while the other splice variants or the igfbps did not respond to the treatments. In agreement to these data, several authors previously demonstrated the proliferative effects of Igf1 in vitro in gilthead sea bream osteoblasts, and in embryonic zebrafish ZF-4 cells73–75. Thus, the bone total igf1 downregulated expression observed in fasting is parallel to the decrease in igf1 and pcna expression in muscle and, the general arrest of growth. On the other hand, during refeeding the upregulated expression of total igf1 and ghr1 in bone as in muscle, pointed out also to the importance of the coordination between the two tissues in this period, for the harmonic growth of the muscle-skeletal system for the fish to properly increase its size.
Several genes regulate the osteogenic process, and runx2 is the key transcription factor determining bone lineage and inducing the expression of genes more involved in matrix production and mineralization (i.e. fib1a, col1a1, ocn and on)26,76. The results of the current study showed that during the fasting period, all osteogenic genes decreased in agreement with results on biometric indexes, MRFs in muscle and, the Gh/Igf system. During starvation, active osteoblasts were not observed in rainbow trout pharyngeal bone, and in Nile tilapia, an imbalance between bone formation and resorption (i.e. osteoblasts versus osteoclasts activity) was reported, resulting in reduced bone mass65,77. Moreover, Van der Velde and coworkers78 suggested also that during fasting, the orexigenic hormone ghrelin increases osteoclastogenesis in mice. In the animals of the present study, ghrelin levels increased during fasting to diminish in an acute way at 5 h post-feeding27. Thus, during the fasting period, the levels of circulating ghrelin can contribute to arrest the osteogenic process increasing bone degradation. The decrease in osteoclastic activity fits well with the significant enhancement of ctsk expression observed after 21 days of fasting and its down-regulation in refeeding, since Ctsk is secreted by osteoclasts and is an important factor of bone resorption79. In this sense, the present study demonstrates such an equal role in gilthead sea bream.
In osteoblast-like cell lines of this species, fish serum is more suitable than bovine serum to induce bone formation causing changes in col1a, ocn and on gene expression80. In the present study, the expression of osteogenic genes was restored with refeeding in a sequential order, being runx2 the first gene upregulated while the other genes followed progressively (fib1a, col1a1, on and ocn) in a sense that resembled the osteogenic process26, indicating that bone growth and matrix mineralization were being reinstated. In agreement, in goldfish (Carassius auratus), it was found that while col1a continually increased after day 7 of scale regeneration, ocn increased only after day 1481. Based on our results, 7 days of refeeding were not enough to recover the latest genes on and ocn, like in muscle for myog or mrf4, suggesting that both tissues recovered in parallel to guarantee an harmonic growth; besides indicating that more than 7 days of refeeding are necessary for complete restoration of the musculoskeletal system anabolism.
The expression of different myogenic genes in mammalian bone is known and their role in coordinating both skeletal tissues has been described82,83 but these studies in fish are scarce. In the present study, pax7, a marker of muscle satellite cells15,84, was evaluated in order to verify the purity of the tissue, which was corroborated since pax7 expression was undetectable in the bone samples. Then, the gene expression of other MRFs was determined. García de la serrana and coworkers85 in a transcriptional study in gilthead sea bream, found for the majority of genes analyzed a faster and more pronounced response to leucine injection in skeletal muscle than in bone. A similar tendency was found in the present study when comparing both tissues, since myod2 expression showed in bone still low levels at 5 and 24 h or 7 days refeeding, while in muscle the levels were already recovered by the end of the period studied. On the other hand, bone mstn2 expression, showed an opposite response compared to muscle with a significant increase at 5 h post-feeding, which could be inhibiting bone growth at this specific stage. In humans, the role of myostatin in bone resorption and rheumatoid arthritis pathology was recently demonstrated82; thus, the inhibitory function of Mstn2 in fish, could serve also in this early refeeding period as a regulator to an overly rapid recovery of bone growth. In fact, the inter-tissue inhibitory role of myostatin has been already described in mammals83,86 serving as a coordinator among skeletal tissues. This function still needs to be demonstrated in fish, but myostatin could contribute not only to control muscle mass, but also to coordinate the growth of both tissues avoiding miss-matches and potentially reducing the appearance of skeletal deformities.
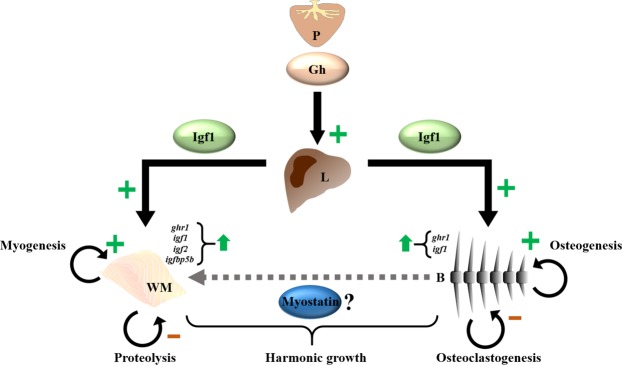
In summary, 21 days of fasting in gilthead sea bream depressed Igf1 as well as most of the synthesis, myogenic and osteogenic mechanisms at the same time that activated several proteolytic molecules from the ubiquitin-proteasome system to mobilize muscle resources. Refeeding triggered rapidly compensatory mechanisms starting with the Igf1 system, and progressively activated the regulatory factors of myogenesis and osteogenesis in a sequence that repeats the processes of development and growth in either tissue, overall restoring proper musculoskeletal growth (Fig. 8). Furthermore, the expression in bone of myostatin suggests an interesting tissue coordinative function that deserves future investigation in this and other fish species. All this can help to better understand the role of these regulatory factors and their crosstalk in the early phase of somatic growth recovery after a period of food limitation.
Figure 8.
Schematic representation of the proposed changes occurring during early refeeding in gilthead sea bream. The Gh/Igf system recovers the synthetizing role with the Gh plasma levels still elevated activating now the hepatic expression/secretion of igf1, in parallel with the progressive up-regulation of the anabolic system components (ghr1, igf1, igf2 and igfbp5b). This condition contributes to the activation of the myogenic (pax7, myf5, myod1 and mrf4) and osteogenic (runx2, fib1a, col1a1 and on) genes, while downregulates the proteolytic (mafbx and murf1) and osteoclastogenic (ctsk) genes in muscle and bone, respectively. This early stage of refeeding may require a fine regulation of the different molecules involved, being myostatin a good candidate for bone and muscle crosstalk to assure harmonic musculoskeletal growth. P: pituitary; L: liver; WM: white muscle; B: bone.
Supplementary information
Acknowledgements
F.L. was supported by a grant from the Department of Scholarship and Students’ Affairs Abroad from the Ministry of Science, Research and Technology of I.R. Iran. M.P-A., E.J.V. and N.R.-H. were supported by predoctoral fellowships (BES-2016–078697, BES-2013-062949 and BES-2015-074654) from the “Ministerio de Economía y Competitividad” (MINECO) from the Spanish Government. The study was funded by projects from MINECO, Spain (AGL2014-57974-R and AGL2017-89436-R to E.C. and I.N. and AGL2015-70679-R to J.G.) and the “Generalitat de Catalunya” (XRAq, 2014SGR-01371 and 2017SGR-1574 to J.G.).
Author contributions
J.G. conceptualized the study; F.L., M.P.-A., E.J.V., A.S.-M., S.B.-P., N.R.-H., J.F.-B. and J.B. performed the sampling; F.L. and M.P.-A. performed the laboratory analyses; F.L., M.P.-A., E.J.V., E.C. and J.G. analyzed and interpreted the data; J.B., I.N., E.C. and J.G. acquired funding; F.L., M.P.-A., E.J.V., E.C. and J.G. drafted and critically reviewed the manuscript. All authors read and approved the final paper.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: F. Lavajoo, M. Perelló-Amorós, E. Capilla and J. Gutiérrez.
Supplementary information
is available for this paper at 10.1038/s41598-019-57013-6.
References
- 1.FAO. The State of Fisheries and Aquaculture in the world 2018. Fao.Org (2018).
- 2.Einen O, Waagan B, Thomassen MS. Starvation prior to slaughter in Atlantic salmon (Salmo salar) Aquaculture. 1998;166:85–104. doi: 10.1016/S0044-8486(98)00279-8. [DOI] [Google Scholar]
- 3.Rasmussen RS, Ostenfeld TH, McLean E. Growth and feed utilisation of rainbow trout subjected to changes in feed lipid concentrations. Aquac. Int. 2000;8:531–542. doi: 10.1023/A:1009291227550. [DOI] [Google Scholar]
- 4.Fauconneau B, Paboeuf G. Effect of fasting and refeeding on in vitro muscle cell proliferation in rainbow trout (Oncorhynchus mykiss) Cell Tissue Res. 2000;301:459–463. doi: 10.1007/s004419900168. [DOI] [PubMed] [Google Scholar]
- 5.Zhang XD, Zhu YF, Cai LS, Wu TX. Effects of fasting on the meat quality and antioxidant defenses of market-size farmed large yellow croaker (Pseudosciaena crocea) Aquaculture. 2008;280:136–139. doi: 10.1016/j.aquaculture.2008.05.010. [DOI] [Google Scholar]
- 6.Nebo C, et al. Short periods of fasting followed by refeeding change the expression of muscle growth-related genes in juvenile Nile tilapia (Oreochromis niloticus) Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2013;164:268–274. doi: 10.1016/j.cbpb.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Navarro, I. & Gutiérrez, J. Chapter 17 Fasting and starvation. in Metabolic biochemistry (eds. Hochachka, P. W. & Mommsen, T. P. B. T.-B. and M. B. of F.) 4, 393–434 (Elsevier, 1995).
- 8.Hagen Ø, Fernandes JMO, Solberg C, Johnston IA. Expression of growth-related genes in muscle during fasting and refeeding of juvenile Atlantic halibut, Hippoglossus hippoglossus L. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2009;152:47–53. doi: 10.1016/j.cbpb.2008.09.083. [DOI] [PubMed] [Google Scholar]
- 9.Rescan PY, et al. Dynamic gene expression in fish muscle during recovery growth induced by a fasting-refeeding schedule. BMC Genomics. 2007;8:1–18. doi: 10.1186/1471-2164-8-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montserrat N, et al. Distinct role of insulin and IGF-I and its receptors in white skeletal muscle during the compensatory growth of gilthead sea bream (Sparus aurata) Aquaculture. 2007;267:188–198. doi: 10.1016/j.aquaculture.2007.04.024. [DOI] [Google Scholar]
- 11.Fuentes EN, Valdés JA, Molina A, Björnsson BT. Regulation of skeletal muscle growth in fish by the growth hormone - Insulin-like growth factor system. Gen. Comp. Endocrinol. 2013;192:136–148. doi: 10.1016/j.ygcen.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Vélez EJ, et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture. 2017;467:28–40. doi: 10.1016/j.aquaculture.2016.07.004. [DOI] [Google Scholar]
- 13.Pérez-Sánchez, J. et al. Somatotropic axis regulation unravels the differential effects of nutritional and environmental factors in growth performance of marine farmed fishes. Front. Endocrinol. (Lausanne). 9 (2018). [DOI] [PMC free article] [PubMed]
- 14.Johnston IA, Bower NI, Macqueen DJ. Growth and the regulation of myotomal muscle mass in teleost fish. J. Exp. Biol. 2011;214:1617–1628. doi: 10.1242/jeb.038620. [DOI] [PubMed] [Google Scholar]
- 15.García de la serrana D, et al. Characterisation and expression of myogenesis regulatory factors during in vitro myoblast development and in vivo fasting in the gilthead sea bream (sparus aurata) Comp. Biochem. Physiol. - A Mol. Integr. Physiol. 2014;167:90–99. doi: 10.1016/j.cbpa.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Bower, N. I. & Johnston, I. A. Transcriptional regulation of the IGF signaling pathway by amino acids and insulin-like growth factors during myogenesis in Atlantic salmon. PLoS One5 (2010). [DOI] [PMC free article] [PubMed]
- 17.Seiliez I, et al. Amino acids downregulate the expression of several autophagy-related genes in rainbow trout myoblasts. Autophagy. 2012;8:364–375. doi: 10.4161/auto.18863. [DOI] [PubMed] [Google Scholar]
- 18.Seiliez I, et al. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss) Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2010;157:258–266. doi: 10.1016/j.cbpb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Seiliez I, et al. Feeding status regulates the polyubiquitination step of the ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss) muscle. J. Nutr. 2008;138:487–491. doi: 10.1093/jn/138.3.487. [DOI] [PubMed] [Google Scholar]
- 20.Salmerón Cristina, García de la serrana Daniel, Jiménez-Amilburu Vanesa, Fontanillas Ramón, Navarro Isabel, Johnston Ian A., Gutiérrez Joaquim, Capilla Encarnación. Characterisation and Expression of Calpain Family Members in Relation to Nutritional Status, Diet Composition and Flesh Texture in Gilthead Sea Bream (Sparus aurata) PLoS ONE. 2013;8(9):e75349. doi: 10.1371/journal.pone.0075349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmerón C, Navarro I, Johnston IA, Gutiérrez J, Capilla E. Characterisation and expression analysis of cathepsins and ubiquitin-proteasome genes in gilthead sea bream (Sparus aurata) skeletal muscle. BMC Res. Notes. 2015;8:149. doi: 10.1186/s13104-015-1121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vélez, E. J. et al. Moderate and sustained exercise modulates muscle proteolytic and myogenic markers in gilthead sea bream (Sparus aurata). Am. J. Physiol. - Regul. Integr. Comp. Physiol. 312, R643–R653 (2017). [DOI] [PubMed]
- 23.Vélez EJ, et al. Recombinant bovine growth hormone (rBGH) enhances somatic growth by regulating the GH-IGF axis in fingerlings of gilthead sea bream (Sparus aurata) Gen. Comp. Endocrinol. 2018;257:192–202. doi: 10.1016/j.ygcen.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Ytteborg Elisabeth, Torgersen Jacob, Baeverfjord Grete, Takle Harald. Health and Environment in Aquaculture. 2012. The Atlantic Salmon (Salmo salar) Vertebra and Cellular Pathways to Vertebral Deformities. [Google Scholar]
- 25.Vieira FA, et al. Comparative analysis of a teleost skeleton transcriptome provides insight into its regulation. Gen. Comp. Endocrinol. 2013;191:45–58. doi: 10.1016/j.ygcen.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 26.Riera-Heredia N, et al. Temperature responsiveness of gilthead sea bream bone; an in vitro and in vivo approach. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-29570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perelló-Amorós M, et al. Ghrelin and Its Receptors in Gilthead Sea Bream: Nutritional. Regulation. Front. Endocrinol. (Lausanne). 2018;9:1–14. doi: 10.3389/fendo.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustin SA, et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45e–45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vélez EJ, et al. IGF-I and amino acids effects through TOR signaling on proliferation and differentiation of gilthead sea bream cultured myocytes. Gen. Comp. Endocrinol. 2014;205:296–304. doi: 10.1016/j.ygcen.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 31.Sjögren K, et al. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc. Natl. Acad. Sci. USA. 1999;96:7088–92. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chauvigné F, Gabillard JC, Weil C, Rescan PY. Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen. Comp. Endocrinol. 2003;132:209–215. doi: 10.1016/S0016-6480(03)00081-9. [DOI] [PubMed] [Google Scholar]
- 33.Peterson BC, Waldbieser GC. Effects of fasting on IGF-I, IGF-II, and IGF-binding protein mRNA concentrations in channel catfish (Ictalurus punctatus) Domest. Anim. Endocrinol. 2009;37:74–83. doi: 10.1016/j.domaniend.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Pierce AL, Fukada H, Dickhoff WW. Metabolic hormones modulate the effect of growth hormone (GH) on insulin-like growth factor-I (IGF-I) mRNA level in primary culture of salmon hepatocytes. J. Endocrinol. 2005;184:341–349. doi: 10.1677/joe.1.05892. [DOI] [PubMed] [Google Scholar]
- 35.Gabillard JC, Kamangar BB, Montserrat N. Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss) J. Endocrinol. 2006;191:15–24. doi: 10.1677/joe.1.06869. [DOI] [PubMed] [Google Scholar]
- 36.Azizi S, et al. Lysine and Leucine Deficiencies Affect Myocytes Development and IGF Signaling in Gilthead Sea Bream (Sparus aurata) PLoS One. 2016;11:e0147618. doi: 10.1371/journal.pone.0147618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bower NI, Taylor RG, Johnston IA. Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Front. Zool. 2009;6:1–13. doi: 10.1186/1742-9994-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernández-Sánchez C, et al. Differential regulation of insulin-like growth factor-I (IGF-I) receptor gene expression by IGF-I and basic fibroblastic growth factor. J. Biol. Chem. 1997;272:4663–4670. doi: 10.1074/jbc.272.8.4663. [DOI] [PubMed] [Google Scholar]
- 39.Imsland AK, Björnsson BT, Gunnarsson S, Foss A, Stefansson SO. Temperature and salinity effects on plasma insulin-like growth factor-I concentrations and growth in juvenile turbot (Scophthalmus maximus) Aquaculture. 2007;271:546–552. doi: 10.1016/j.aquaculture.2007.07.007. [DOI] [Google Scholar]
- 40.Vélez EJ, et al. Effects of sustained exercise on GH-IGFs axis in gilthead sea bream (Sparus aurata) Am. J. Physiol. - Regul. Integr. Comp. Physiol. 2016;310:R313–R322. doi: 10.1152/ajpregu.00230.2015. [DOI] [PubMed] [Google Scholar]
- 41.Zheng GD, et al. Two grass carp (Ctenopharyngodon idella) insulin-like growth factor-binding protein 5 genes exhibit different yet conserved functions in development and growth. Comp. Biochem. Physiol. Part - B Biochem. Mol. Biol. 2016;204:69–76. doi: 10.1016/j.cbpb.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 42.de la Serrana DG, Macqueen DJ. Insulin-like growth factor-binding proteins of teleost fishes. Front. Endocrinol. (Lausanne). 2018;9:1–12. doi: 10.3389/fendo.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bower NI, Li X, Taylor R, Johnston IA. Switching to fast growth: the insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J. Exp. Biol. 2008;211:3859–3870. doi: 10.1242/jeb.024117. [DOI] [PubMed] [Google Scholar]
- 44.Valente Luísa M. P., Bower Neil I., Johnston Ian A. Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. British Journal of Nutrition. 2012;108(12):2148–2157. doi: 10.1017/S0007114512000396. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu M, Swanson P, Hara A, Dickhoff WW. Purification of a 41-kDa insulin-like growth factor binding protein from serum of chinook salmon, Oncorhynchus tshawytscha. Gen. Comp. Endocrinol. 2003;132:103–111. doi: 10.1016/S0016-6480(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu M, Swanson P, Dickhoff WW. Free and protein-bound insulin-like growth factor-I (IGF-I) and IGF- binding proteins in plasma of coho salmon, Oncorhynchus kisutch. Gen. Comp. Endocrinol. 1999;115:398–405. doi: 10.1006/gcen.1999.7328. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu M, Hara A, Dickhoff WW. Development of an RIA for salmon 41 kDa IGF-binding protein. J. Endocrinol. 2003;178:275–283. doi: 10.1677/joe.0.1780275. [DOI] [PubMed] [Google Scholar]
- 48.Saera-Vila A, Calduch-Giner JA, Prunet P, Pérez-Sánchez J. Dynamics of liver GH/IGF axis and selected stress markers in juvenile gilthead sea bream (Sparus aurata) exposed to acute confinement. Differential stress response of growth hormone receptors. Comp. Biochem. Physiol. - A Mol. Integr. Physiol. 2009;154:197–203. doi: 10.1016/j.cbpa.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Lansard M, et al. Hepatic protein kinase B (Akt)target of rapamycin (TOR)-signalling pathways and intermediary metabolism in rainbow trout (Oncorhynchus mykiss) are not significantly affected by feeding plant-based diets. Br. J. Nutr. 2009;102:1564–1573. doi: 10.1017/S000711450999095X. [DOI] [PubMed] [Google Scholar]
- 50.Garcia de la serrana, D. et al. Development Temperature Has Persistent Effects on Muscle Growth Responses in Gilthead Sea Bream. PLoS One7 (2012). [DOI] [PMC free article] [PubMed]
- 51.Johansen, K. A. & Overturf, K. Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. 144, 119–127 (2006). [DOI] [PubMed]
- 52.Montserrat N, Gabillard JC, Capilla E, Navarro MI, Gutiérrez J. Role of insulin, insulin-like growth factors, and muscle regulatory factors in the compensatory growth of the trout (Oncorhynchus mykiss) Gen. Comp. Endocrinol. 2007;150:462–472. doi: 10.1016/j.ygcen.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 53.Terova G, Bernardini G, Binelli G, Gornati R, Saroglia M. cDNA encoding sequences for myostatin and FGF6 in sea bass (Dicentrarchus labrax, L.) and the effect of fasting and refeeding on their abundance levels. Domest. Anim. Endocrinol. 2006;30:304–319. doi: 10.1016/j.domaniend.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Georgiou S, et al. Transient up- and down-regulation of expression of myosin light chain 2 and myostatin mRNA mark the changes from stratified hyperplasia to muscle fiber hypertrophy in larvae of gilthead sea bream (Sparus aurata L.) Cell Tissue Res. 2016;363:541–554. doi: 10.1007/s00441-015-2254-0. [DOI] [PubMed] [Google Scholar]
- 55.Salem M, Nath J, Rexroad CE, Killefer J, Yao J. Identification and molecular characterization of the rainbow trout calpains (Capn1 and Capn2): Their expression in muscle wasting during starvation. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2005;140:63–71. doi: 10.1016/j.cbpc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 56.MacQueen DJ, Delbridge ML, Manthri S, Johnston IA. A newly classified vertebrate calpain protease, directly ancestral to CAPN1 and 2, episodically evolved a restricted physiological function in placental mammals. Mol. Biol. Evol. 2010;27:1886–1902. doi: 10.1093/molbev/msq071. [DOI] [PubMed] [Google Scholar]
- 57.Lepage SE, Bruce AEE. Characterization and comparative expression of zebrafish calpain system genes during early development. Dev. Dyn. 2008;237:819–829. doi: 10.1002/dvdy.21459. [DOI] [PubMed] [Google Scholar]
- 58.Preziosa Elena, Liu Shikai, Terova Genciana, Gao Xiaoyu, Liu Hong, Kucuktas Huseyin, Terhune Jeffery, Liu Zhanjiang. Effect of Nutrient Restriction and Re-Feeding on Calpain Family Genes in Skeletal Muscle of Channel Catfish (Ictalurus punctatus) PLoS ONE. 2013;8(3):e59404. doi: 10.1371/journal.pone.0059404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cleveland, B. M. & Weber, G. M. Effects of insulin-like growth factor-I, insulin, and leucine on protein turnover and ubiquitin ligase expression in rainbow trout primary myocytes. Am. J. Physiol. Integr. Comp. Physiol. 298, R341–R350 (2010). [DOI] [PubMed]
- 60.Bower NI, de la serrana DG, Johnston IA. Characterisation and differential regulation of MAFbx/Atrogin-1 α and β transcripts in skeletal muscle of Atlantic salmon (Salmo salar) Biochem. Biophys. Res. Commun. 2010;396:265–271. doi: 10.1016/j.bbrc.2010.04.076. [DOI] [PubMed] [Google Scholar]
- 61.Vélez EJ, et al. Proteolytic systems’ expression during myogenesis and transcriptional regulation by amino acids in gilthead sea bream cultured muscle cells. PLoS One. 2017;12:1–21. doi: 10.1371/journal.pone.0187339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, et al. Molecular characterization of the MuRF genes in rainbow trout: Potential role in muscle degradation. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2011;158:208–215. doi: 10.1016/j.cbpb.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Lall SP, Lewis-McCrea LM. Role of nutrients in skeletal metabolism and pathology in fish - An overview. Aquaculture. 2007;267:3–19. doi: 10.1016/j.aquaculture.2007.02.053. [DOI] [Google Scholar]
- 64.Boglione C, et al. Skeletal anomalies in reared European fish larvae and juveniles. Part 2: Main typologies, occurrences and causative factors. Rev. Aquac. 2013;5:121–167. doi: 10.1111/raq.12016. [DOI] [Google Scholar]
- 65.Takagi Y, Moriyama S, Hirano T, Yamada J. Effects of growth hormones on bone formation and resorption in rainbow trout (Oncorhynchus mykiss), as examined by histomorphometry of the pharyngeal bone. Gen. Comp. Endocrinol. 1992;86:90–95. doi: 10.1016/0016-6480(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 66.Persson P, Johannsson SH, Takagi Y, Björnsson BT. Estradiol-17β and nutritional status affect calcium balance, scale and bone resorption, and bone formation in rainbow trout, Oncorhynchus mykiss. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 1997;167:468–473. doi: 10.1007/s003600050098. [DOI] [Google Scholar]
- 67.Schoenle E, Zapf J, Humbel RE, Froesch ER. Insulin-like growth factor I stimulates growth in hypophysectomized rats. Nature. 1982;296:252–253. doi: 10.1038/296252a0. [DOI] [PubMed] [Google Scholar]
- 68.He J, Rosen CJ, Adams DJ, Kream BE. Postnatal growth and bone mass in mice with IGF-I haploinsufficiency. Bone. 2006;38:826–835. doi: 10.1016/j.bone.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 69.Beattie J, et al. Insulin- like growth factor-binding protein action in bone tissue: A key role for pregnancy- associated plasma protein-A. Front. Endocrinol. (Lausanne). 2018;9:1–10. doi: 10.3389/fendo.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohlsson C, et al. Growth hormone and bone metabolism. Acta Endocrinol. (Copenh). 1998;128(Suppl):14–8. [PubMed] [Google Scholar]
- 71.Merimee TJ, Zapf J, Froesch ER. Insulin-Like Growth Factors in the Fed and Fasted States*. J. Clin. Endocrinol. Metab. 1982;55:999–1002. doi: 10.1210/jcem-55-5-999. [DOI] [PubMed] [Google Scholar]
- 72.Crane JL, Cao X. Function of matrix IGF-1 in coupling bone resorption and formation. J. Mol. Med. 2014;92:107–115. doi: 10.1007/s00109-013-1084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Capilla E, Teles-García Á, Acerete L, Navarro I, Gutiérrez J. Insulin and IGF-I effects on the proliferation of an osteoblast primary culture from sea bream (Sparus aurata) Gen. Comp. Endocrinol. 2011;172:107–114. doi: 10.1016/j.ygcen.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 74.Pozios KC, Ding J, Degger B, Upton Z, Duan C. IGFs stimulate zebrafish cell proliferation by activating MAP kinase and PI3-kinase-signaling pathways. Am. J. Physiol. Integr. Comp. Physiol. 2001;280:R1230–R1239. doi: 10.1152/ajpregu.2001.280.4.R1230. [DOI] [PubMed] [Google Scholar]
- 75.Tiago DM, Cancela ML, Laizé V. Proliferative and mineralogenic effects of insulin, IGF-1, and vanadate in fish osteoblast-like cells. J. Bone Miner. Metab. 2011;29:377–382. doi: 10.1007/s00774-010-0243-7. [DOI] [PubMed] [Google Scholar]
- 76.Aubin JE, Triffitt JT. Mesenchymal Stem Cells and Osteoblast Differentiation. Princ. Bone Biol. Two-Volume Set. 2002;1:85–107. [Google Scholar]
- 77.Takagi Y. Effects of Starvation and Subsequent Refeeding on Formation and Resorption of Acellular Bone in Tilapia, Oreochromis niloticus. Zoolog. Sci. 2001;18:623–629. doi: 10.2108/zsj.18.623. [DOI] [Google Scholar]
- 78.Van Der Velde M, et al. An age-dependent interaction with leptin unmasks Ghrelin’s bone-protective effects. Endocrinology. 2012;153:3593–3602. doi: 10.1210/en.2012-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuo TR, Chen CH. Bone biomarker for the clinical assessment of osteoporosis: Recent developments and future perspectives. Biomark. Res. 2017;5:5–13. doi: 10.1186/s40364-017-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosa J, Tiago DM, Dias J, Cancela ML, Laizé V. Serum-specific stimulation of proliferation and mineralization of fish bone-derived cells. J. Appl. Ichthyol. 2010;26:251–256. doi: 10.1111/j.1439-0426.2010.01414.x. [DOI] [Google Scholar]
- 81.Thamamongood TA, et al. Expression of osteoblastic and osteoclastic genes during spontaneous regeneration and autotransplantation of goldfish scale: A new tool to study intramembranous bone regeneration. Bone. 2012;50:1240–1249. doi: 10.1016/j.bone.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 82.Dankbar B, et al. Myostatin is a direct regulator of osteoclast differentiation and its inhibition reduces inflammatory joint destruction in mice. Nat. Med. 2015;21:1085–1090. doi: 10.1038/nm.3917. [DOI] [PubMed] [Google Scholar]
- 83.Karsenty G, Mera P. Molecular bases of the crosstalk between bone and muscle. Bone. 2018;115:43–49. doi: 10.1016/j.bone.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seiliez I, Froehlich JM, Marandel L, Gabillard JC, Biga PR. Evolutionary history and epigenetic regulation of the three paralogous pax7 genes in rainbow trout. Cell Tissue Res. 2015;359:715–727. doi: 10.1007/s00441-014-2060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia de la serrana D, Mareco EA, LA Vieira V, Power DM, Johnston IA. Comparison of the transcriptional responses of skeletal muscle and bone to a flooding dose of leucine in the gilthead sea bream (Sparus aurata) Comp. Biochem. Physiol. Part - B Biochem. Mol. Biol. 2016;199:50–57. doi: 10.1016/j.cbpb.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 86.Karsenty G, Olson EN. Bone and Muscle Endocrine Functions: Unexpected Paradigms of Inter-organ Communication. Cell. 2016;164:1248–1256. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).