ABSTRACT
DNA methylation is dynamically modulated during postnatal brain development, and plays a key role in neuronal lineage commitment. This epigenetic mark has also recently been implicated in the development of neural sex differences, many of which are found in the hypothalamus. The level of DNA methylation depends on a balance between the placement of methyl marks by DNA methyltransferases (Dnmts) and their removal, which is catalyzed by ten-eleven translocation (Tet) methylcytosine dioxygenases. Here, we examined developmental changes and sex differences in the expression of Tet and Dnmt enzymes from birth to adulthood in two hypothalamic regions (the preoptic area and ventromedial nucleus) and the hippocampus of mice. We found highest expression of all Tet enzymes (Tet1, Tet2, Tet3) and Dnmts (Dnmt1, Dnmt3a, Dnmt3b) in newborns, despite the fact that global methylation and hydroxymethylation were at their lowest levels at birth. Expression of the Dnmt co-activator, Dnmt3l, followed a pattern opposite to that of the canonical Dnmts (i.e., was very low in newborns and increased with age). Tet enzyme activity was much higher at birth than at weaning in both the hypothalamus and hippocampus, mirroring developmental changes in gene expression. Sex differences in Tet enzyme expression were seen in all brain regions examined during the first week of life, whereas Dnmt expression was more balanced between the sexes. Neonatal testosterone treatment of females only partially masculinized enzyme expression. Thus, Tet expression and activity are elevated during neonatal brain development, and may play important roles in sexual differentiation of the brain.
KEYWORDS: Tet methylcytosine dioxygenases, DNA methyltransferases, DNA hydroxymethylation, DNA methylation, 5-methylcytosine, 5-hydroxymethylcytosine, hypothalamus, hippocampus
Introduction
DNA methylation on the 5-carbon position of cytosine, also referred to as 5-methylcytosine (5mC), is the best studied epigenetic mark in mammalian cells and plays an important role in cell lineage commitments during development [1]. Recently, DNA methylation has also been implicated in the development of sex differences in the rodent brain [2,3]. For example, whole genome sequencing reveals sex differences in DNA methylation in the neonatal and adult hypothalamus and striatum [2,4]. In addition, a neonatal inhibition of DNA methylation disrupts the development of sex differences in dendritic spine density in the preoptic area of the hypothalamus (POA) of rats [2], as well as sexually dimorphic gene expression in the POA and ventromedial nucleus of the hypothalamus (VMH) of mice [3].
DNA methylation is controlled by a family of DNA methyltransferases (Dnmts) (also referred to as DNA methylation ‘writers’). In general, Dnmt3a and Dnmt3b are responsible for placing new methylation marks, and are therefore defined as de novo methyltransferases, whereas Dnmt1 serves as a maintenance methyltransferase [5,6]. In addition, Dnmt3l, which lacks a catalytic methyltransferase domain, facilitates methylation by Dnmt3a and Dnmt3b and plays important roles in maternal genomic imprints during development [7,8].
Although 5mC was originally considered quite stable, this notion has been revolutionized in recent years. For example, the brain exhibits dynamic changes in DNA methylation during development and with learning [9–12], suggesting the presence of an active methylation/demethylation cycle [13]. In 2009, ten-eleven translocation 1 (Tet1), was identified as a 2-oxoglutarate- and Fe(II)-dependent enzyme that catalyzes the conversion of 5mC to 5-hydroxymethylcytosine (5hmC) in the mammalian genome [14]. These findings provided a completely new perspective of an active DNA demethylation process, which may be fundamental for mammalian brain development [15].
Tet enzymes comprise a family of three genes (Tet1, Tet2 and Tet3) that catalyze the iterative oxidation of 5mC to 5hmC and its downstream derivatives, including 5-formylcytosine and, finally, 5-carboxylcytosine [16]. The latter two products can be excised from DNA through base excision repair, resulting in the reversion to unmethylated cytosine [16,17]. 5hmC was originally considered only an intermediate step in the removal of 5mC marks, with global DNA methylation presumably reflecting a balance between DNA methylation and demethylation. More recent evidence, however, suggests that 5hmC also serves as a stable and independent epigenetic mark with unique roles in mammalian brains [10,18,19]. In general, DNA methylation, particularly on CpG islands of gene promoters, is associated with gene repression, whereas hydroxymethylation on gene bodies is usually linked to active transcription [11,20,21].
5hmC is highly enriched in neurons [22], and the global level of 5hmC is approximately ten-fold higher in the central nervous system than in somatic tissues [23,24]. Nonetheless, the role of hydroxymethylation in sexual differentiation of the brain has not been explored. Many sex differences in rodents depend on exposure to testosterone during a critical perinatal period [25]. Here, we examined the mRNA expression of 5mC ‘writers’ (Dnmts) and ‘erasers’ (Tets), Dnmt and Tet enzyme activity, as well as global levels of 5mC and 5hmC in the postnatal mouse brain. We focused on two hypothalamic regions in which sex differences are well established (the POA and VMH), and also examined the hippocampus, where developmental changes in 5mC, 5hmC and Tet enzyme expression have previously been reported [10,11,26].
Results
Tet gene expression is highest in newborns
We first used quantitative RT-PCR to examine the expression of Tet genes from birth to early adulthood (data from males and females combined) in micropunches of the POA, VMH and hippocampus of C57Bl/6 mice. In each brain region, there was a highly significant effect of age for all three Tet enzymes, with the highest expression found in newborns, and a 75–80% drop in expression between P1 and adulthood (Figure 1).
Figure 1.
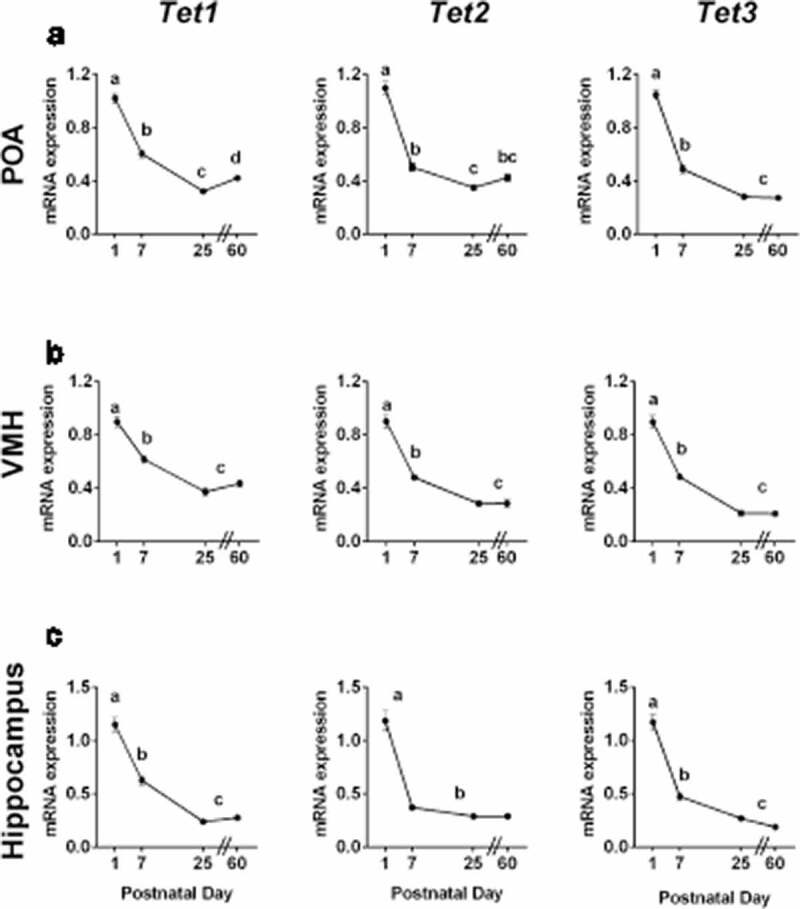
Tet enzyme gene expression is highest at birth. Expression of Tet1, Tet2 and Tet3 mRNA at postnatal days 1, 7, 25, and 60 in a) the preoptic area of the hypothalamus (POA); b) the ventromedial nucleus of the hypothalamus (VMH); and c) the hippocampus. The expression of all three Tet enzymes was highest in newborns, and abruptly decreased by day 7. Different lower-case letters indicate statistically significant differences (P < 0.05). Data are mean ± SEM. N = 19–25 mice per age.
Specifically, in the POA, there was a significant effect of age on expression of Tet1 (F3,78 = 123.6, P < 0.0001), Tet2 (F3,78 = 125.0, P < 0.0001) and Tet3 (F 3,78 = 210.9, P < 0.0001; Figure 1a), with highest expression on P1. Expression of all three Tet enzymes decreased 40–50% between P1 and P7 (P < 0.0001 in each case) and there was a further significant decline between P7 and P25. There was, however, a later increase in Tet1 expression in the POA from P25 to P60 (P < 0.03).
Similarly, expression of Tet enzymes in the VMH was highest in newborns. There was a significant effect of age for Tet1 (F3,79 = 54.85, P < 0.0001), Tet2 (F3,79 = 75.56, P < 0.0001), and Tet3 (F3,79 = 110.0, P < 0.0001; Figure 1b). The expression of all Tet enzymes decreased by ~50% between P1 and P7 (P < 0.0001 in each case) and further decreased between P7 and P25.
In the hippocampus, one-way ANOVA again found significant effects of age for Tet1 (F3,82 = 94.05, P < 0.0001), Tet2 (F3,81 = 68.85, P < 0.0001), and Tet3 (F3,82 = 106.2, P < 0.0001; Figure 1c), with highest levels at P1. Expression of all Tet enzymes decreased 40–60% between P1-P7; Tet2 reached adult levels by P7, whereas Tet1 and Tet3 continued to decrease between P7 and P25 (P < 0.01).
Newborns express high levels of Dnmt1, Dnmt3a and Dnmt3b, but low levels of the coactivator Dnmt3l
The canonical Dnmts were also expressed at high levels in the newborn brain (Figure 2). Dnmt1 and Dnmt3b exhibited the same pattern in all regions examined: there was a main effect of age (F > 100, P < 0.0001 in all cases) with highest levels on P1; expression decreased between P1 and P7 (P < 0.05) and further decreased between P7 and P25 (P < 0.001). Dnmt3a also exhibited a main effect of age in all three regions (F > 76.6 in all cases, P < 0.0001), however, expression remained elevated through P7, before falling to adult levels by P25 in all areas.
Figure 2.
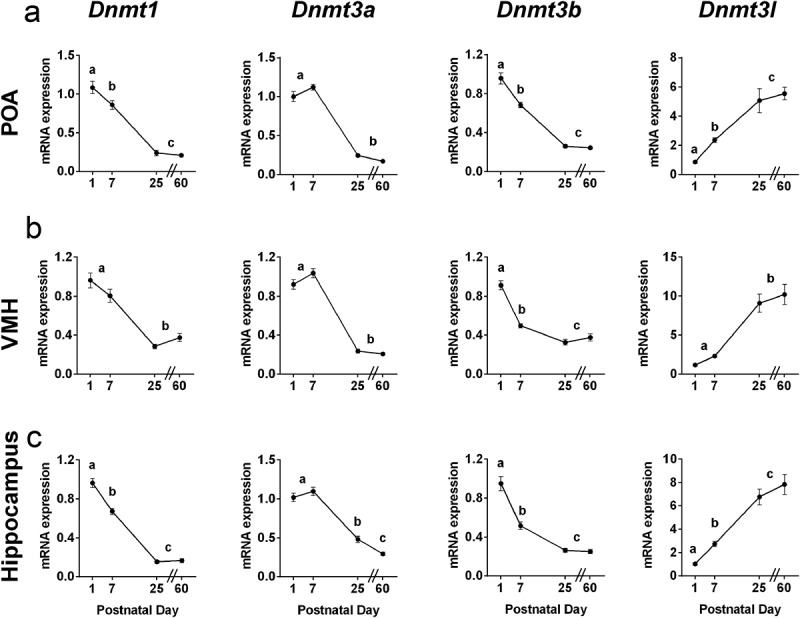
Canonical Dnmt gene expression is highest during neonatal life, whereas expression of the co-activator Dnmt3l increases during postnatal life. Dnmt1, Dnmt3a, Dnmt3b and Dnmt3l mRNA expression at postnatal days 1, 7, 25 and 60 in a) the preoptic area of the hypothalamus (POA); b) the ventromedial nucleus of the hypothalamus (VMH); and c) the hippocampus. The expression of Dnmt1 and Dnmt3b was highest on P1, then dropped to lower levels by P7, whereas expression of Dnmt3a remained elevated throughout the first postnatal week in all brain regions examined. Expression of Dnmt3l increased 6- to 10-fold between postnatal days 1 and 60 in all brain regions. Different letters indicate significant differences. Data are mean ± SEM. N = 19–20 mice per age.
Strikingly, we found the opposite pattern for the methyltransferase co-activator Dnmt3l, which increased 6- to 10-fold in expression between P1 and P60 in all brain regions (Figure 2; POA, F3,68 = 23.00, P < 0.0001; VMH, F3,68 = 26.13, P < 0.0001; and hippocampus: F3,71 = 38.18, P < 0.0001).
Sex differences in Tet expression are seen in all brain regions examined, and are limited to neonatal life
We next compared the expression of Tet enzymes between males, females, and females treated with testosterone for all ages and brain regions. The results of all comparisons are presented in Supplementary Figure 1–3; significant differences were seen only in neonates, and are detailed here.
In the POA, males had higher expression of Tet2 (t9.38 = 8.08; P < 0.0001) and Tet3 (t9.76 = 14.04; P < 0.0001) on P7 than did females (Figure 3). This was especially prominent for Tet3, where the difference exceeded 3-fold. Testosterone treatment of females on P0 and P1 partially masculinized Tet3 expression (t7.17 = 2.35; P = 0.051 for control females vs. testosterone-treated females), but did not significantly affect expression of Tet2.
Figure 3.
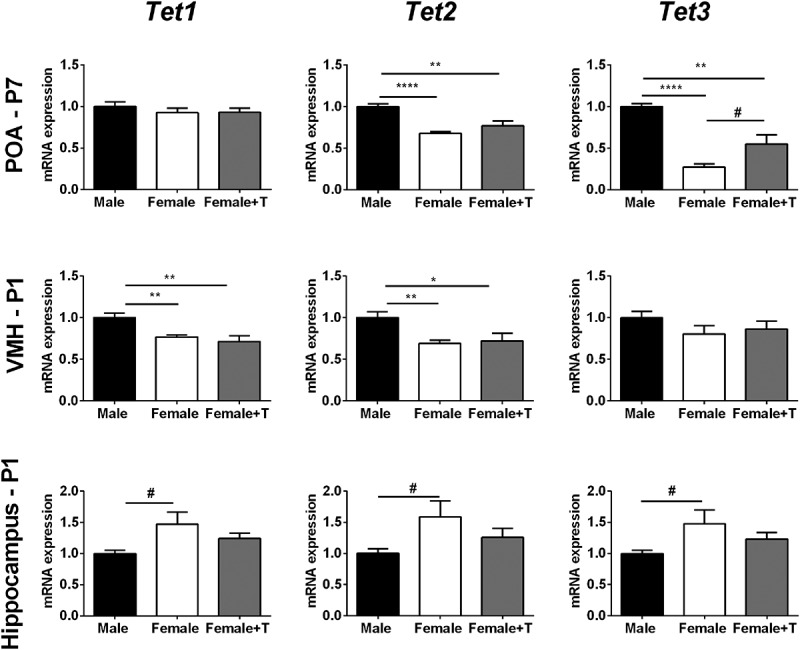
Sex differences in Tet gene expression are observed during neonatal life. In the preoptic area of the hypothalamus (POA), Tet2 and Tet3 expression are higher in males than in females on postnatal day (P) 7. Neonatal testosterone treatment partially masculinized Tet3 expression in females, but did not affect the expression of Tet2. N = 5–7 per group. In the ventromedial nucleus of the hypothalamus (VMH), expression of Tet1 and Tet2 is higher in males on P1, and neonatal testosterone treatment of females did not eliminate the sex difference. N = 6 in each female group, N = 13 males. In the hippocampus, females have marginally higher expression of each Tet enzyme on P1; across all three Tet enzymes, expression was significantly higher in females than in males (P < 0.01). Expression in females treated neonatally with testosterone was intermediate, and not significantly different from that of control males or females. N = 6 in each female group, N = 13 in males. #P < 0.1, *P < 0.05; **P < 0.01; ****P < 0.0001. Data are mean ± SEM.
Males also had higher Tet1 (t16.14 = 3.95; P < 0.01) and Tet2 (t16.81 = 3.92; P < 0.01) expression than females in the VMH, although in this case the sex differences were seen on P1 (Figure 3). Testosterone treatment of females did not affect Tet1 or Tet2 expression, and there were no group differences in Tet3 expression.
In the hippocampus, we found the reverse sex difference, with a trend for higher expression of each Tet gene in females on P1 (Tet1: t5.81 = 2.29; P = 0.06; Tet2: t5.84 = 2.21; P = 0.07; Tet3: t5.62 = 2.1; P = 0.08; Figure 3). Although none of the comparisons alone reached significance, a repeated measures ANOVA indicates a significant effect of group for overall Tet expression in the P1 hippocampus (F2,22 = 5.87; P < 0.01), with higher expression in females than in males (P < 0.01). Expression of Tet enzymes in females treated with neonatal testosterone was intermediate: not significantly different from control females, but also not different from control males (Figure 3).
Few sex differences in DNMT expression
Canonical Dnmt expression was very similar in males and females in all regions and ages (Supplementary Figures 4–6), with one exception: females had higher expression of Dnmt1 than males in the POA on P7 (t9.99 = 3.32; P < 0.01; Figure 4). A similar trend was seen on P1, but did not reach significance (P < 0.1; Supplementary Figure 4). Females also had higher expression of the Dnmt coactivator, Dnmt3l, in the POA on P7 (t8.18 = 2.74; P < 0.03; Figure 4).
Figure 4.
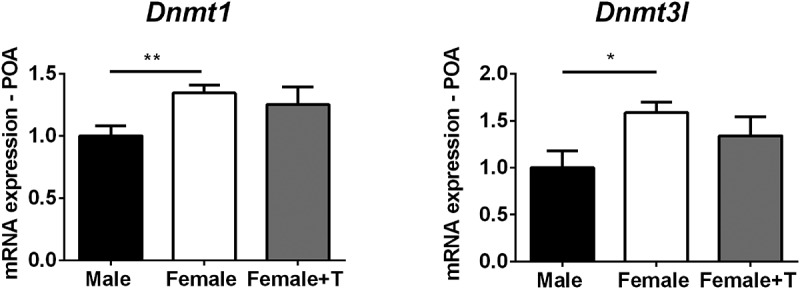
Females have higher expression of Dnmt1 and Dnmt3l in the preoptic area of the hypothalamus (POA) at postnatal day (P) 7. Dnmt1 expression in females treated with testosterone was intermediate and not significantly different from that of control males or females. *P < 0.05. Data are mean ± SEM. N = 5 vehicle females, N = 7 males and females + testosterone.
Tet enzyme activity is elevated in newborns
Higher expression of Tet and Dnmt genes in newborns does not necessarily mean elevated enzyme activity. For example, Nugent and colleagues report a mis-match between canonical Dnmt gene expression and enzyme activity in the POA of newborn rats [2], and we found an inverse relationship between expression of the canonical Dnmts and the co-activator, Dnmt3l, which makes it difficult to predict overall Dnmt activity. We therefore used fluorometric assays to measure 5mC-hydroxylase (Tet) and Dnmt activity at P1 and P25 in a new cohort of male and female mice. Whole hypothalamus and whole hippocampus were dissected from each animal to obtain sufficient protein for enzyme assays. We found two- to three-fold higher Tet enzyme activity at P1 than at P25 in both the hypothalamus (main effect of age: F1,14 = 27.58; P < 0.0001) and hippocampus (F1,14 = 13.44; P < 0.003; Figure 5), consistent with the higher Tet gene expression in newborns. Dnmt activity in the hypothalamus was marginally higher on P1 than on P25 (main effect of age: F1,14 = 3.94; P < 0.07), while in the hippocampus there was an age-by-sex interaction (F1,14 = 5.48; P < 0.05). Dnmt activity in the hippocampus was elevated in females, but not males, on P1 relative to either sex on P25. This result should be interpreted cautiously, however, given the low n per sub-group in this analysis.
Figure 5.
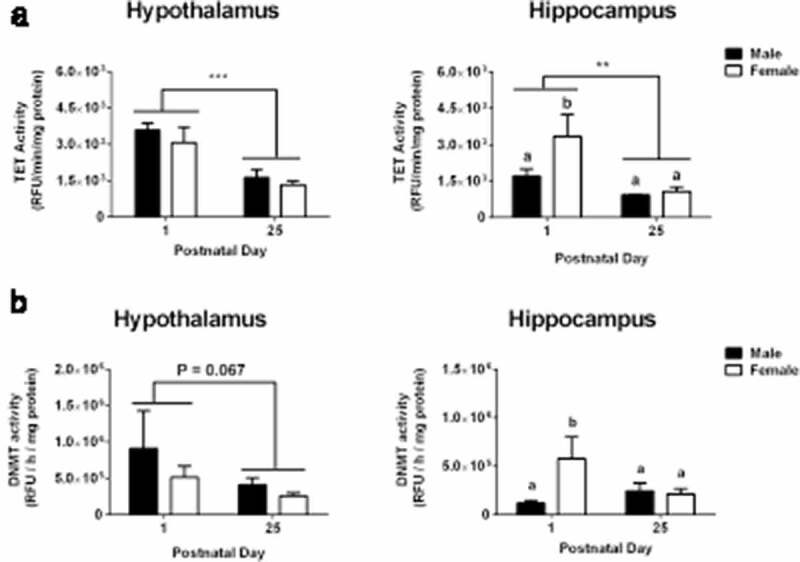
Tet enzyme activity is elevated at birth. a) The activity of demethylating (Tet) enzymes was two-to three-fold higher on postnatal day (P) 1 than on P25 in both the hypothalamus and hippocampus. b) There was a trend for higher methylating (Dnmt) activity in the hypothalamus on P1 compared to P25. In the hippocampus, elevated Dnmt activity on postnatal day 1 was seen in females only. Different letters indicate significant differences. **P < 0.01; ***P < 0.001. Data are mean ± SEM. N = 3 for P1 male and female groups, N = 6 for P25 male and female groups.
Global levels of DNA methylation are higher in the female POA
In the analyses above, females had higher expression of the maintenance methyltransferase Dnmt1 and the Dnmt coactivator Dnmt3l, as well as lower expression of the demethylating enzymes Tet2 and Tet3 in the POA on P7. This suggested that the balance is tipped towards higher DNA methylation in the neonatal female POA. We tested this using dot blots to examine global DNA methylation in micropunches of the POA, and found a main effect of group (F2,45 = 6.182, P < 0.005): females had about two times higher 5mC than did males on P1 and P7, although the difference reached significance only at P7 (P < 0.05; Figure 6). Testosterone treatment of females did not alter global 5mC levels.
Figure 6.
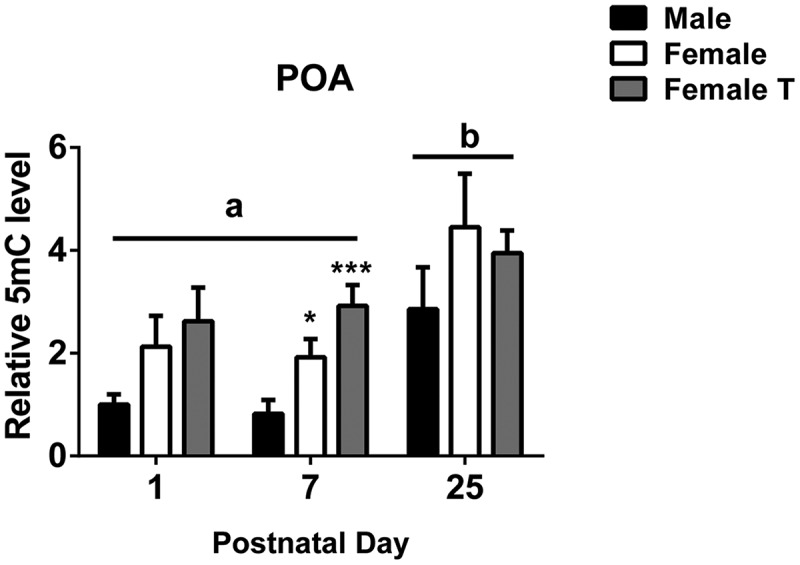
Global levels of 5-methylcytosine (5mC) in the preoptic area of the hypothalamus (POA) increase across development. Global 5mC significantly increased between postnatal day 7 and 25 (P < 0.001). A sex difference was found at postnatal day 7, with two-fold higher global methylation in females than in males. Neonatal testosterone treatment of females did not masculinize global methylation. Different letters indicate significant differences. *P < 0.05 and ***P < 0.001, compared to males on postnatal day 7. Data are mean ± SEM. N = 6 in all groups.
We also found a main effect of age on global 5mC in the POA (F2,45 = 9.842, P < 0.0004), such that levels were higher at P25 than at P1 or P7 (P < 0.005 in both cases; Figure 6).
Global levels of hydroxymethylation increase with age in all brain regions
The very high expression of Tet genes and enzyme activity seen during neonatal life was somewhat surprising, because hydroxymethylation is low at birth and increases with age in the cerebral cortex and cerebellum [10]. Using dot blots, we confirmed this basic observation by comparing 5hmC in the POA on P1, P7 and P25, and in the VMH and hippocampus on P1 and P25 (Figure 7). We found significant main effects of age in all cases, with higher levels of 5hmC at P25 than at the earlier age(s) in the POA (F2,25 = 15.17, P < 0.0001), VMH (F1, 30 = 206.5, P < 0.0001), and hippocampus (F1, 30 = 62.87, P < 0.0001; Figure 7). No significant effects of group or group-by-age interactions were found for global 5hmC, although given the reduced n per group in these analyses, we may not have had sufficient power to detect subtle sex differences.
Figure 7.
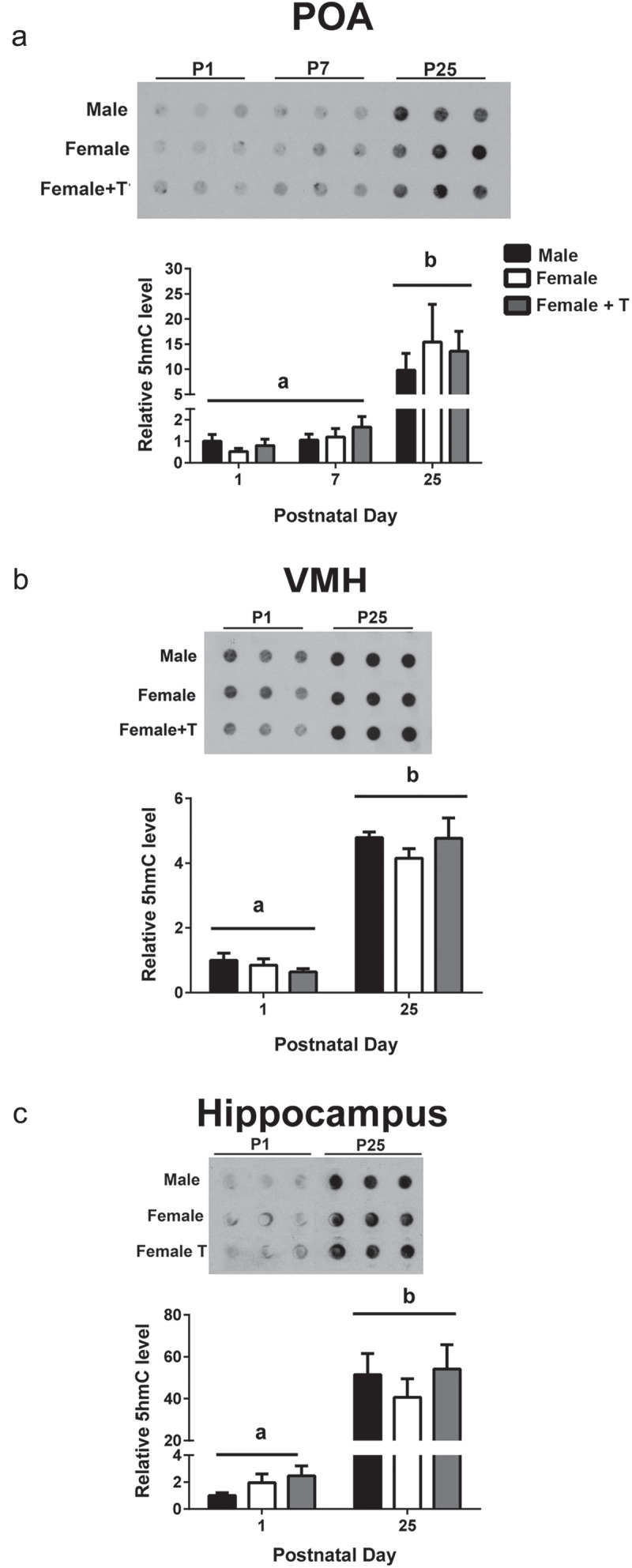
Global 5-hydroxymethylcytosine (5hmC) accumulates across development. Representative dot blots and quantification illustrate that hydroxymethylation significantly increases between postnatal days 1 and 25 in a) the preoptic area of the hypothalamus (POA); b) the ventromedial nucleus of the hypothalamus (VMH); and c) the hippocampus (P < 0.001 in all cases). Different letters indicate significant differences. Data are mean ± SEM. N = 6 in all groups.
Discussion
Key neurodevelopmental processes, such as refinement of the molecular identities of neurons and glial cells, occur during late embryonic through early postnatal life in mammals and depend at least in part on dynamic changes in DNA methylation [11,22,27]. Many sex differences in the brain also depend on events occurring perinatally, and inhibition of DNA methylation on the day of birth disrupts sex differences in morphology and neurochemical phenotype in the hypothalamus of rats and mice [2,3]. The dynamic modulation of 5mC levels during development presumably depends on both the placement of methyl marks by Dnmts, and Tet-mediated conversion of 5mC to 5hmC as the first step in demethylation. Nonetheless, the role of Tet enzymes and 5hmC in brain development are just beginning to be explored. We found highest expression of Tet genes and enzyme activity, as well as sex differences in Tet gene expression, in the hypothalamus and hippocampus during neonatal life. This suggests that Tet enzyme activity is important for neonatal brain development in general and the development of sex differences in particular. There are known sex differences in gene expression in the hypothalamus of rodents [2,3,28,29] that presumably are required for sex-specific functions, and that may rely on sex-specific DNA methylation/demethylation.
In the first analysis of the genome-wide distribution of 5hmC in the brain, Szulwach and colleagues found that 5hmC accumulates in developmentally activated genes between P7 and P60 in the hippocampus and cerebellum of mice [10]. Our results confirm the increase in global 5hmC with age in the hippocampus and extend this pattern to two hypothalamic regions, suggesting that this may be a general feature of brain development. However, despite the low level of 5hmC in newborns, we found higher Tet enzyme activity, as well as higher expression of the three Tet enzyme genes, at birth than in adulthood in all brain regions examined. A similar pattern for Tet gene expression was very recently reported in mouse cerebellum, hippocampus and cortex [26]. Low global 5hmC combined with high expression and activity of Tet enzymes suggests that Tet enzymes may primarily be involved in de-methylation, rather than the placement of stable hydroxymethyl marks in the newborn brain. Alternatively, this pattern might suggest that DNA is less accessible to Tet enzymes perinatally due to a condensed chromatin environment. For example, genetic knockdown or pharmacological inhibition of two histone demethylases, Jmjd2a and Jmjd3, caused the formation of heterochromatin and prevented Tet1/Tet2 binding to key genes involved in osteoblast differentiation [30]. It is also possible that Tet enzymes are less active in the perinatal brain due to the availability of key Tet binding partners [31,32], although our observations on enzyme activity argue against this interpretation.
We also found that global 5mC was low at birth in the hypothalamus and increased by P25, which is consistent with a previous report in the mouse and human cortex [11]. The increase in global 5mC with age in the cortex is due almost exclusively to methylation in a non-CG context [11]. Although most DNA methylation occurs on a cytosine followed by a guanine (CG), non-CG methylation (especially, methylation of a cytosine followed by adenine; CA) also occurs, and this increases with age specifically in neurons [11,33–35] . Our dot blots used a 5mC antibody that detects 5mC equally well in CG and CA contexts (see Methods). Thus, the higher levels of 5mC found in the hypothalamus at P25 might be due to an accumulation of methyl marks in a non-CG context, although this remains to be confirmed.
We found the highest expression of the canonical Dnmt genes in the POA and VMH of newborns, yet Dnmt enzyme activity in whole hypothalamus was only marginally higher on P1 than on P25. The hypothalamus is a highly heterogeneous brain region, and it is possible that enzyme activity in whole hypothalamus does not reflect activity in subregions such as the POA or VMH. Indeed, Nugent et al. [2] found elevated Dnmt activity in the POA of newborn rats, with a large drop in activity at P7, consistent with highest Dnmt activity in newborns (although ages after P7 were not examined). It is also possible that Dnmt3l contributes to the muted effect of age on Dnmt activity. Dnmt3l expression in the hypothalamus and hippocampus increased between birth and P25; as a co-activator of Dnmt3a and Dnmt3b, Dnmt3l may boost enzyme activity to offset decreased Dnmt3a and Dnmt3b gene expression. If such compensation does occur, however, it may be brain region specific because Dnmt3l expression was reported to be low at all ages examined in the mouse cortex [11].
As far as we are aware, sex differences in Tet enzymes in the brain have not previously been reported. We found sex differences in Tet expression in all regions examined that were limited to the neonatal critical period for sexual differentiation. Newborn males had higher expression of Tet enzymes than females in the POA and VMH, whereas females had higher expression of all Tet enzymes in the hippocampus. Although many of the best studied brain sex differences are found in the hypothalamus [25], sex differences in neonatal hippocampus have also recently been reported [36–38]. Males also had lower expression of Dnmt1 and Dnmt3l than females in the POA on P7, and lower global methylation in the newborn male POA. Similarly, more fully methylated CpG sites were found in the POA of P2 female rats than of males [2]. Our present results suggest that greater de-methylation in males could contribute to this finding.
Many sex differences in the mammalian brain are caused by a transient, perinatal exposure to gonadal testosterone in males and can be eliminated by administration of testosterone to females at birth [39]. The dose of testosterone we used here effectively masculinized neuroanatomy in our previous study [40], yet we found only partial or no masculinization of Tet and Dnmt expression in females treated with testosterone in the current study. Some sex differences in the brain are independent of gonadal hormones and due to sex chromosome complement [41–44], or to an interaction of gonadal and sex chromosome factors [45], which may explain the limited effect of testosterone in this study. However, because males are exposed to elevated testosterone from about E18 to P0 [46,47], both pre- and postnatal testosterone may be required to fully masculinize Tet and Dnmt expression in females. In fact, testosterone causes an increase in Tet2 expression and a decrease in global methylation in mouse embryonic neural stem cells in vitro [48], suggesting that effects of testosterone on Tet expression may begin prenatally.
Despite strong sex differences in Tet2 and Tet3 gene expression in the neonatal POA, global 5hmC levels did not differ between the sexes. It is possible that this reflects the limited sensitivity of dot blots, which are not able to detect small differences between groups. Alternatively, the lack of sex differences in neonatal 5hmC may again suggest that higher Tet expression in newborn males is primarily related to a de-methylation event and rapid conversion of 5mC to an unmodified cytosine rather than the placement of stable 5hmC marks. This would result in a sex difference in global 5mC, but not 5hmC, which is what was found. We, and others, have reported sex differences in DNA methylation in a genome-wide survey and examination of steroid receptor genes in the hypothalamus [4,9]. Taken together, our present results indicate that sex differences in DNA methylation in the POA could primarily be a consequence of sex differences in demethylation driven by Tet enzymes.
Materials and methods
Animals
C57BL/6 adult mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Breeding pairs were maintained on a 12-h:12-h light:dark cycle at 22°C with ad libitum access to food and water and were checked daily for the delivery of pups. All procedures were approved by the Institutional Animal Care and Use Committee of Georgia State University and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Neonatal treatment and brain collection
Brains were collected on postnatal days (P) 1, P7, P25 and P60 for analyses of Tet and Dnmt expression, and on P1, P7 and P25 from a separate cohort of mice to evaluate global levels of 5mC and 5hmC. Three groups were included at each age: control males and females that received a subcutaneous injection of peanut oil (25 µl) on P0 and P1, and females that received testosterone propionate (100 µg in 25 µl of peanut oil, Sigma, St Louis, MO, USA) on P0 and P1. For collection on P1, animals were sacrificed 6h after the last injection. Brains were stored at −80°C until processing.
Micro-punch of regions of interest
Brains were maintained at −18°C on a cryostat, and coronally sectioned until reaching the areas of interest. Tissue punches were collected of the POA, the VMH, and the hippocampus. Punch size was 1 mm inner diameter, except for the VMH and hippocampus at P60 (2 mm inner diameter). Tissue was kept at −80°C until processing for RNA or DNA purification.
RNA purification and qRT-PCR
Tissue was homogenized in TRIzol (Invitrogen, Carlsbad, CA, USA) by multiple passes through a 30-gauge needle at 4°C. Total RNA purification proceeded according to the manufacturer’s instructions, and quantity and purity was analyzed by spectrophotometry (NanoDrop 2000, Thermo Fisher Scientific, GA, USA). One µg of total RNA was reverse transcribed to cDNA in a 20 µl reaction with Superscript IV (Invitrogen, Invitrogen, Carlsbad, CA, USA) and random primers (Invitrogen). Quantitative real-time PCR (qPCR) was performed using verified primer sets (Qiagen Inc., Valencia, CA, USA) for Gapdh, Tet1, Tet2, Tet3, Dnmt1, Dnmt3a and Dnmt3b genes. We also examined expression of Dnmt3l using the following primer sequences: forward, CTGCTGACTGAGGATGACCA; reverse, ACCCGCATAGCATTCTGGTA [49]. Each sample was assayed in duplicate using FastStart Essential DNA Green Master (Roche, Basel, Switzerland) in a final volume of 10 µl. Amplification was performed in a Step OneTM Real Time PCR System (Applied Biosystems Inc., Foster City, CA, USA), and amplification efficiency was established by 5-point calibration curves. All primer-pairs were verified to be 90–110% efficient and amplified a single product determined by melting curve analysis. Relative quantifications of mRNA levels were measured by the ΔΔCt method. For the analysis of Tet and Dnmt enzyme expression across development (Figures 1 and 2), the mean of males on P1 was used as the calibrator group. When sex differences within age was analyzed (Figures 3 and 4, and Supplementary Figures 1–6), the mean of the male group for that age was again used as a calibrator.
Enzyme activity assays
The hypothalamus and hippocampus were manually dissected from untreated P1 and P25 male and female mice and frozen at −80°C. Nuclear proteins were purified with the EpiQuik™ Nuclear Extraction Kit I (Epigentek Cat. No OP-0002) and quantified using the Pierce™ BCA Protein Assay Kit (Thermo Scientific Cat. No 23,225) following the instructions provided by the manufacturer.
Total Tet activity was analyzed with the fluorometric Epigenase™ 5mC Hydroxylase TET Activity/Inhibition Assay Kit Assay Kit (Epigentek Cat. No P-3087) following the manufacturer’s instructions. Briefly, the kit provides a methylated substrate stably coated onto microplate wells. Tet enzymes present in the samples convert the methylated substrate to hydroxymethylated products in a 90-minute incubation. The final enzyme product is then recognized using specific antibodies provided with the kit and fluorometrically measured. The enzyme activity was calculated using the following formula: TET Activity (RFU/min/mg protein) = [(Sample RFU – Blank RFU)/(Protein Amount (μg) x 90 min)] x 1000 where RFU are the relative fluorescent units measured.
Total Dnmt Activity was analyzed using the EpiQuik™ DNMT Activity/Inhibition
Assay Ultra Kit (Epigentek Cat. No P-3010). The general procedure and principle mirror the procedure of the Tet Activity kit. The Dnmt activity was calculated using the formula: DNMT Activity (RFU/h/mg protein) = [(Sample RFU – Blank RFU)/(Protein Amount (μg)* x 2 hours)] x 1000.
DNA purification and dot blots
Genomic DNA was purified from brain punches with the EZNA DNA kit (Omega Biotek Inc, Atlanta, GA, USA) and quantified by spectrophotometry. 5hmC- or 5mC-specific dot blots were performed using a Bio-Dot Apparatus (Bio-Rad, Hercules, CA, USA) as described previously [50]. Briefly, DNA was spotted on an Amersham Hybond-N+ membrane (GE Healthcare, Atlanta, GA, USA) and was fixed to the membrane by incubation at 85°C in a hybridizer for 30 min. The membrane was incubated in 5% blotting-grade blocker (Bio Rad), followed by rabbit anti-5hmC antibody (Active Motif, Cat. No. 39,791, 1:2000) or mouse anti-5mC antibody (Active Motif, Cat. No. 39,649, 1:1000) overnight at 4°C. We confirmed that this antibody detects methylated cytosine in both a CG and non-CG context (Supplementary Figure 7). Membranes were then incubated for 1h at room temperature in horseradish peroxidase–conjugated secondary antibodies (anti-rabbit: 1:5000, Cat. No 7074S; anti-mouse: 1:4000, Cat. No 7076P2, Cell Signaling), and signal was visualized by enhanced chemiluminescence. The density of each signal was quantified by ImageJ software.
Statistical analysis
One-way ANOVA was used to evaluate the mRNA expression of Tet and Dnmt genes across development. Two-tailed independent t-tests with Welch’s correction for unequal sample sizes or repeated-measures ANOVA were used to evaluate sex differences within age. Two-way ANOVA (sex-by-age) was used to evaluate global levels of 5hmC and 5mC and enzyme activities during development. ANOVA was followed by Fisher’s least significance difference (LSD) post hoc test when appropriate.
Funding Statement
This work was supported by a Brains & Behavior Seed Grant from Georgia State University, a National Science Foundation Graduate Research Fellowship (to LRC), the National Institutes of Health (R01-MH117122) and an Emory University Alzheimer’s Disease Research Center Pilot grant (P50 AG025688) (to BY). CDC is supported by the Georgia State University Next Generation New Scholars Program.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Juliandi B, Abematsu M, Nakashima K.. Epigenetic regulation in neural stem cell differentiation. Dev Growth Differ. 2010;52:493–504. [DOI] [PubMed] [Google Scholar]
- [2].Nugent BM, Wright CL, Shetty AC, et al. Brain feminization requires active repression of masculinization via DNA methylation. Nat Neurosci. 2015;18:690–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mosley M, Weathington J, Cortes LR, et al. Neonatal inhibition of DNA methylation alters cell phenotype in sexually dimorphic regions of the mouse brain. Endocrinology. 2017;158:1838–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ghahramani NM, Ngun TC, Chen P-Y, et al. The effects of perinatal testosterone exposure on the DNA methylome of the mouse brain are late-emerging. Biol Sex Differ. 2014;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ooi SKT, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. [DOI] [PubMed] [Google Scholar]
- [7].Hata K, Okano M, Lei H, et al. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. [DOI] [PubMed] [Google Scholar]
- [8].Bourc’his D, Xu GL, Lin CS, et al. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294(80):2536–2539. [DOI] [PubMed] [Google Scholar]
- [9].Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology. 2010;151:4871–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Szulwach KE, Li X, Li Y, et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].JJ D, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ma DK, Jang M-H, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(80):930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yao B, Christian KM, He C, et al. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17:537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Niehrs C, Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–227. [DOI] [PubMed] [Google Scholar]
- [18].Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li X, Wei W, Zhao Q-Y, et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci. 2014;111:7120–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mellén M, Ayata P, Dewell S, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mellén M, Ayata P, Heintz N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci U S A. 2017;114:E7812–E7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Penn NW, Suwalski R, O’Riley C, et al. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Globisch D, Münzel M, Müller M, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mccarthy M, De Vries G, Forger N. Sexual differentiation of the brain: A fresh look at mode, mechanisms, and meaning. In: Pfaff DW, Joëls M, editors. Hormones, Brain and Behavior, 3rd ed. UK: Elsevier Inc; 2017. p. 3–32. [Google Scholar]
- [26].Tao H, Xie P, Cao Y, et al. The dynamic DNA demethylation during postnatal neuronal development and neural stem cell differentiation. Stem Cells Int. 2018;2018:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Luo C, Keown CL, Kurihara L, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357(80):600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu X, Coats JK, Yang CF, et al. Modular genetic control of sexually dimorphic behaviors. Cell. 2012;148:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Walker DM, Kirson D, Perez LF, et al. Molecular profiling of postnatal development of the hypothalamus in female and male rats. Biol Reprod. 2012;87:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sepulveda H, Villagra A, Montecino M. Tet-mediated DNA demethylation is required for SWI/SNF-dependent chromatin remodeling and histone-modifying activities that trigger expression of the Sp7 osteoblast master gene during mesenchymal lineage commitment. Mol Cell Biol. 2017;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Costa Y, Ding J, Theunissen TW, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sardina JL, Collombet S, Tian TV, et al. Transcription factors drive Tet2-mediatede enhancer demethylation to reprogram cell fate. Cell Stem Cell. 2018;23:727–741.e9. [DOI] [PubMed] [Google Scholar]
- [33].Xie W, Barr CL, Kim A, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mo A, Mukamel EA, Davis FP, et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ramsahoye BH, Biniszkiewicz D, Lyko F, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci. 2000;97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Isgor C, Sengelaub DR. Effects of neonatal gonadal steroids on adult CA3 pyramidal neuron dendritic morphology and spatial memory in rats. J Neurobiol. 2003;55:179–190. [DOI] [PubMed] [Google Scholar]
- [37].Zhang JM, Konkle ATM, Zup SL, et al. Impact of sex and hormones on new cells in the developing rat hippocampus: A novel source of sex dimorphism? Eur. J Neurosci. 2008;27:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zup SL, Edwards NS, McCarthy MM. Sex- and age-dependent effects of androgens on glutamate-induced cell death and intracellular calcium regulation in the developing hippocampus. Neuroscience. 2014;281:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Forger NG. Epigenetic mechanisms in sexual differentiation of the brain and behaviour. Philos Trans R Soc B Biol Sci. 2016;371:20150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hisasue S, Seney ML, Immerman E, et al. Control of cell number in the bed nucleus of the stria terminalis of mice: role of testosterone metabolites and estrogen receptor subtypes. J Sex Med. 2010;7:1401–1409. [DOI] [PubMed] [Google Scholar]
- [41].De Vries GJ, Rissman EF, Simerly RB, et al. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci Off J Soc Neurosci. 2002;22:9005–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Carruth LL, Reisert I, Arnold AP. Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci. 2002;5:933–934. [DOI] [PubMed] [Google Scholar]
- [43].Cisternas CD, Tome K, Caeiro XE, et al. Sex chromosome complement determines sex differences in aromatase expression and regulation in the stria terminalis and anterior amygdala of the developing mouse brain. Mol Cell Endocrinol. 2015;414:99–110. [DOI] [PubMed] [Google Scholar]
- [44].Cisternas CD, Cabrera Zapata LE, Arevalo MA, et al. Regulation of aromatase expression in the anterior amygdala of the developing mouse brain depends on ERβ and sex chromosome complement. Sci Rep. 2017;7:5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Arnold AP, Chen X, Itoh Y. What a difference an X or Y makes: sex chromosomes, gene dose, and epigenetics in sexual differentiation. Handb Exp Pharmacol. 2012;214:67–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].O’Shaughnessy PJ, Baker P, Sohnius U, et al. Fetal development of Leydig cell activity in the mouse is independent of pituitary gonadotroph function. Endocrinology. 1998;139:1141–1146. [DOI] [PubMed] [Google Scholar]
- [47].Konkle ATM, McCarthy MM. Developmental time course of estradiol, testosterone, and dihydrotestosterone levels in discrete regions of male and female rat brain. Endocrinology. 2011;152:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bramble MS, Roach L, Lipson A, et al. Sex-specific effects of testosterone on the sexually dimorphic transcriptome and epigenome of embryonic neural stem/progenitor cells. Sci Rep. 2016;6:36916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Miozzo F, Arnould H, de Thonel A, et al. Alcohol exposure promotes DNA methyltransferase DNMT3A upregulation through reactive oxygen species-dependent mechanisms. Cell Stress Chaperones. 2018;23:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li X, Yao B, Chen L, et al. Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat Commun. 2017;8:15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


