Abstract
Background
The clinicopathological features and prognosis of breast cancer in Asia are different from those in the Western countries. Tumor‐infiltrating immune cells can influence the outcome of patients with breast cancer, but they have not been systemically evaluated in Asian patients with breast cancer.
Methods
We compared the immune score, composition, and prognostic impact of infiltrating immune cells between Asian and Western patients with breast cancer by analyzing gene expression profiles from eight Gene Expression Omnibus data sets and The Cancer Genome Atlas data set. The Estimation of Stromal and Immune Cells in Malignant Tumours Using Expression Data (ESTIMATE) and Cell Type Identification by Estimating Relative Subsets of Known RNA Transcripts (CIBERSORT) algorithms were used to determine the immune score and composition of tumor‐infiltrating immune cells, respectively.
Findings
This study included 462 Asian patients and 2,186 Western patients. Tumors of Asian patients had significantly higher immune score, particularly in the luminal B and HER2‐enriched subtypes. High immune score was associated with favorable prognosis in both Asian and Western patients, and Asian race with a high ESTIMATE immune score provided additional power to predict longer disease‐free survival. Activated CD4 T cells and M2 macrophages were the most strongly associated with survival in both Asian and Western patients.
Interpretation
Our study highlights the difference in tumor immune microenvironments between Asian and Western patients. The higher ESTIMATE immune score, which represents more abundant tumor‐infiltrating immune cells, in tumors of Asian patients partly explains their favorable prognosis.
Implications for Practice
The tumor microenvironment serves as an interface that affects the human body's reaction to cancer cells. Evidence has revealed that tumor‐infiltrating immune cells were associated with patient prognosis. This study demonstrated the disparity of tumor microenvironments and their prognostic impact between Asian and Western patients with breast cancer. The differences in immune score partially explained the racial survival differences noted in recent studies. Integrated analysis of tumor cells, tumor microenvironment, and racial effect may significantly improve recurrence risk prediction for patients with stage I–III breast cancer. Because the effect of tumor microenvironment varies across different populations, a model of interaction between immune score and race/ethnicity is recommended in accessing the risk of patients with cancer.
Keywords: Immune cell, Composition, Breast cancer, Asia, Prognosis
Short abstract
Statistics indicate that the incidence of breast cancer in Asia has increased in the past three decades. This article evaluates tumor‐infiltrating immune cells of breast cancer, comparing a representative amount and the composition of infiltrating immune cells in breast cancer between Asian and Western patients.
Introduction
The incidence of breast cancer in Asia is typically lower than that in the West. However, breast cancer registry statistics indicated that this incidence has rapidly increased in the past three decades in Asian countries, and breast cancer is presently the most frequently diagnosed cancer and the fourth leading cause of cancer death in Asian women 1, 2. The westernized lifestyle is intuitively considered the major cause of such an increase in Asia 3, 4, and the biology of breast cancer in Asian patients has been thought to be similar to that of their Western counterparts.
However, some studies have reported discrepancies in the pathological features, molecular subtypes, and prognosis of breast cancer between patients in Asian and Western countries 5, 6, 7, 8. Specifically, some studies have shown more vigorous host responses through more intense lymphocytic infiltration in breast cancer in Japanese women, compared with that in white women, and reported a more favorable prognosis 5, 9, 10, 11. Our prior studies showed a high prevalence of the hormone receptor‐positive and human epidermal growth factor receptor 2 (HER2)‐negative breast cancer in young women in Taiwan, and these young patients had relatively favorable prognosis 7, 8. The finding of favorable prognosis in Asian patients was supported by two studies of immigrants in the U.S. These studies, based on U.S. Surveillance, Epidemiology, and End Results (SEER) 9 and SEER18 data, showed that Asian American women with breast cancer survived longer than those of other racial groups, including white non‐Hispanic Americans, and suggested ethnic differences in breast tumor biology 12, 13.
In recent years, numerous studies have reported an association between prognosis and the levels of tumor‐infiltrating immune cells in many solid tumors, as reviewed by Fridman et al. 14. Several studies have reported a positive prognostic and predictive relevance between the abundance of tumor‐infiltrating lymphocytes (TILs) and patients’ outcome or treatment response in breast cancer 15, 16, 17, 18, 19. However, the prognostic and predictive roles among different TIL subsets vary. For example, CD4‐positive, CD25‐positive regulatory T cells express forkhead box P3 (FOXP3) and exhibit suppressor activity, and infiltration by high FOXP3‐positive regulatory T cells has been shown to be associated with shorter survival in several solid tumors 20. In addition, breast cancer commonly involves tumor‐associated macrophages, which have also been shown to be associated with shorter survival 21, 22.
The amount, composition, and prognostic impact of tumor‐infiltrating immune cells of breast cancer have not been systemically evaluated in Asian patients. We used bioinformatics tools to analyze gene expression profiles and systemically compared the representative amount and composition of infiltrating immune cells in breast cancer between Asian and Western patients. Furthermore, we evaluated the prognostic value of infiltrating immune cells in both populations.
Materials and Methods
Data Acquisition
Gene expression microarray data from primary breast tumors were obtained from the Gene Expression Omnibus (GEO) 23, 24 and The Cancer Genome Atlas (TCGA) 25 databases. From the GEO database, we selected breast cancer data sets that analyzed surgical specimens using noncustomized expression arrays with case numbers of more than 100 patients and publication date after 2010. Cancer stage, patient age, and disease‐free survival (DFS) and/or overall survival (OS) were the required clinical variables. Patients with stage IV disease or with unavailable age or staging information and studies from clinical trials were excluded from the analysis. Duplicated cases were analyzed using average expression levels. The basic information of the obtained data sets is summarized in supplemental online Table 1. In total, six data sets from cohorts of white, Western patients (GSE21653, GSE22219, GSE25066, GSE37751, GSE42568, and GSE58644), one data set from a cohort of Asian patients (GSE20685), and two data sets from cohorts of patients of multiple races (GSE45255 and TCGA) were retrieved 26, 27, 28, 29, 30, 31, 32, 33. The microarray platforms used in the data sets included the Affymetrix Human Genome U133A Array, Affymetrix Human Genome U133 Plus 2.0 Array, Affymetrix Human Gene 1.0 ST Array, Illumina humanRef‐8 v1.0 expression beadchip, and Illumina Hiseq. Cancer staging was obtained directly from the data sets or indirectly from tumor size and lymph node status.
Molecular Subtyping, Immune Score, and Compositions of Immune Cells
Probes mapping across microarray platforms was done using Entrez Gene ID. Raw data in. CEL files were downloaded and normalized with the robust multiarray average algorithm in the Affy package of the R program 34. Quantile normalization algorithm was used in the U133 Plus 2.0 Array to obtain the reference baseline data. The remaining data sets were adjusted to the reference data to reduce systematic bias. Gene annotation was based on the Genome Reference Consortium Human Build 38 patch release 7. Molecular subtyping of breast cancer was determined on the basis of gene expression levels using the Prediction Analysis of Microarray 50 (PAM50) algorithm of the genefu package 35. The Estimation of Stromal and Immune Cells in Malignant Tumours Using Expression Data (ESTIMATE) algorithm was used to estimate the immune score, which inferred the abundance of infiltrating immune cells 36. The compositions of infiltrating immune cells were estimated using the Cell Type Identification by Estimating Relative Subsets of Known RNA Transcripts (CIBERSORT) algorithm, which can calculate the proportions of 22 types of immune cells 37 and has been used to determine the prognostic effects of individual immune cell subsets across various cancers 38.
Statistical Analysis
Categorical variables are presented as the frequencies and percentages, and their distributions between Asian and Western patients were compared using the chi‐square test. An unequal variance t test was applied to compare the immune scores. A linear mixed effect model with the study platform variable as random intercept was performed to minimize the biases introduced by different mRNA quantification methods. A proportional z test with Benjamini‐Hochberg adjustment was used to compare the proportion of each immune cell subset between Asian and Western patients. To analyze the prognostic effects, the immune score and each immune subset were individually dichotomized into two groups (high and low) by the median value of all analyzed subjects. The survival outcomes were estimated using the Kaplan‐Meier method. In the univariate analysis, the effects of each potential prognostic factor for the DFS or OS outcomes were examined using the log‐rank test. Multivariate analysis was conducted by fitting Cox proportional hazards models to estimate the adjusted effects of prognostic factors on the DFS and OS outcomes. In exploratory analyses, p < .05 was used to indicate statistical significance, and all tests were two‐tailed. All statistical analyses were performed using the R program version 3.3.2.
Results
Demographics, Molecular Subtypes, and Survival
A total of 462 Asian and 2,186 Western patients with breast cancer were included in this study. Among them, 1,356 and 1,788 cases had information on OS and DFS, respectively (Fig. 1). The demographic characteristics of Asian and Western patients are shown in supplemental online Table 2. Compared with Western patients, Asian patients were younger and had lower frequencies of the luminal A and basal subtypes but higher frequencies of the luminal B and HER2‐enriched subtypes. Univariate analysis showed that Asian patients had significantly longer OS (supplemental online Fig. 1A) and DFS (supplemental online Fig. 2A) when compared with Western patients, and the difference was statistically significant in the luminal A and basal subtypes (supplemental online Fig. 1B, E) in terms of OS and in the luminal B and basal subtypes in terms of DFS (supplemental online Fig. 2C, E). In multivariate analysis, Asian patients presented lower hazards of death (hazard ratio [HR], 0.61; 95% confidence interval [CI], 0.46 − 0.83) and disease relapse (HR, 0.64; 95% CI, 0.51 − 0.80) after adjustment for tumor stage and molecular subtype (supplemental online Table 3).
Figure 1.
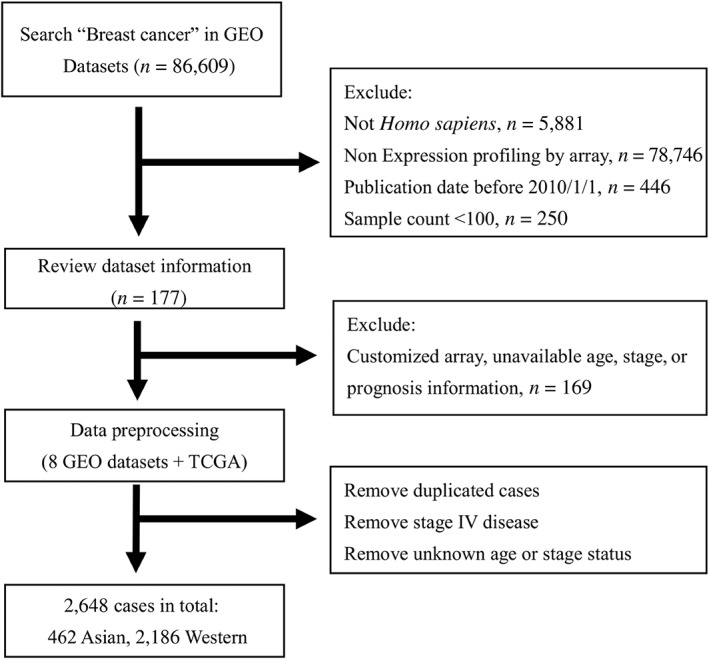
Study flow diagram. By applying searching filter, 177 studies were found. A total of eight data sets met selection criteria in further evaluation.
Abbreviations: GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
ESTIMATE Immune Score and Prognostic Effects
Compared with those of Western patients, breast tumors of Asian patients had a significantly higher immune score (1,187 vs. 1,014; p < .0001), and the difference was statistically significant in the luminal B subtype (1,091 vs. 823, p = .0001) and marginally significant in the HER2‐enriched subtype (1,513 vs. 1,275; p = .0593; Table 1). The linear mixed model showed that race and PAM50 subtype were the determinants for the ESTIMATE immune score. Specifically, tumors of Asian patients and of the HER2‐enriched, basal, and normal‐like subtypes were associated with higher immune score (supplemental online Table 4).
Table 1.
Comparison of Estimation of Stromal and Immune Cells in Malignant Tumours Using Expression Data (ESTIMATE) immune scores of breast tumors between Asian and Western patients
| Tumor subtype | Asian patients | Western patients | p value |
|---|---|---|---|
| All subtypes | 1,187 | 1,014 | <.0001 |
| Luminal A | 958 | 866 | .2009 |
| Luminal B | 1,091 | 823 | .0001 |
| HER2‐enriched | 1,513 | 1,275 | .0593 |
| Basal | 1,477 | 1,381 | .4242 |
| Normal‐like | 1,317 | 1,187 | .5053 |
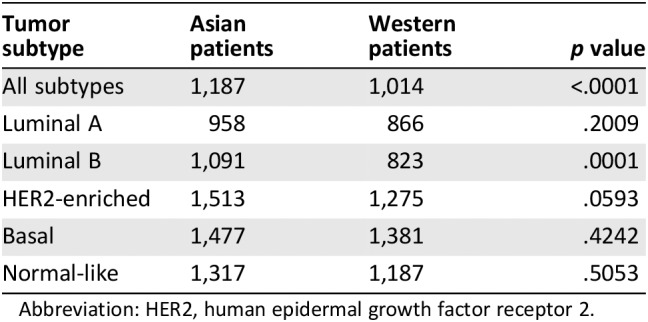
Abbreviation: HER2, human epidermal growth factor receptor 2.
The OS and DFS according to the race and level of the ESTIMATE immune score are shown in Figure 2A and supplemental online Figure 3A, respectively. The OS and DFS curves according to the individual PAM50 subgroups are shown in Figure 2B−F and supplemental online Figure 3B−F, respectively. The prognostic values of the ESTIMATE immune score in all subtypes and among individual PAM50 subtypes after adjustment of age, stage, and/or PAM50 subtype are shown in Table 2. A higher ESTIMATE immune score was significantly associated with longer DFS and OS in both the Asian and Western subgroups (HR ranged from 0.45 to 0.75; Table 2). In the Asian subgroup, there was a statistically significant positive association between the ESTIMATE immune score and survival in the basal subtype in terms of OS and DFS, and there was a trend toward positive association in the luminal B subtype in terms of OS and in the HER2‐enriched subtype in terms of DFS. In the Western subgroup, there was a statistically significant positive association between the ESTIMATE immune score and survival in the HER2‐enriched and basal subtypes in terms of DFS. In contrast, a higher ESTIMATE immune score was associated with shorter DFS in the luminal A subtype of the Western subgroup (HR, 2.09; p = .0206; Table 2).
Figure 2.
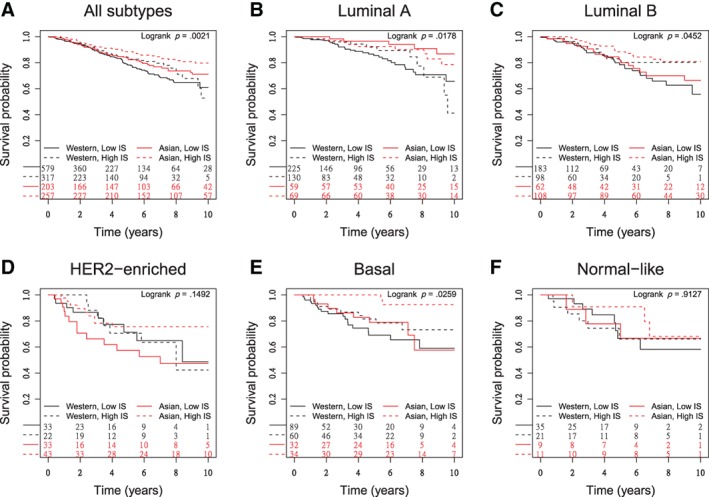
Overall survival by race and Estimation of Stromal and Immune Cells in Malignant Tumours Using Expression Data (ESTIMATE) immune core across all subtypes and individual Prediction Analysis of Microarray 50 (PAM50) subtypes. (A): All subtypes. Asian race and high immune score were associated with longer survival. (B–F): PAM50 subtypes, including luminal A (B), luminal B (C), HER2‐enriched (D), basal (E), and normal‐like (F). The prognostic effects of Asian race and high immune score remained significant in patients with luminal A and basal breast cancer. A similar trend can be seen in luminal B subtype (p = .05). In HER2‐enriched and normal‐like subtypes, the prognostic effects of Asian race and high immune score were insignificant.
Abbreviation: IS, immune score.
Table 2.
Associations of Estimation of Stromal and Immune Cells in Malignant Tumours Using Expression Data (ESTIMATE) immune score (high versus low) with OS and DFS, stratified by race and PAM50 subtype (adjusted for age and stage)
| Race, PAM50 subtype | OS | DFS | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Asian | ||||
| All subtypesa | 0.45 (0.28–0.71) | <.0001 | 0.51 (0.34–0.77) | .0014 |
| Luminal A | 1.34 (0.46–3.91) | .5979 | 0.83 (0.35–1.95) | .6713 |
| Luminal B | 0.51 (0.24–1.09) | .0831 | 0.76 (0.39–1.48) | .4200 |
| HER2‐enriched | 0.53 (0.21–1.39) | .1995 | 0.44 (0.19–1.04) | .0610 |
| Basal | 0.05 (0.01–0.37) | .0037 | 0.06 (0.01–0.44) | .0056 |
| Normal‐like | 0.46 (0.03–7.98) | .5925 | NRb | |
| Western | ||||
| All subtypesa | 0.64 (0.44–0.94) | .0206 | 0.75 (0.60–0.92) | .0061 |
| Luminal A | 0.80 (0.39–1.61) | .5273 | 2.09 (1.11–3.94) | .0226 |
| Luminal B | 0.66 (0.29–1.52) | .3304 | 0.92 (0.65–1.29) | .6245 |
| HER2‐enriched | 0.66 (0.23–1.94) | .4544 | 0.40 (0.24–0.69) | .0010 |
| Basal | 0.58 (0.28–1.22) | .1532 | 0.60 (0.42–0.85) | .0040 |
| Normal‐like | 0.61 (0.20–1.84) | .3781 | 1.33 (0.38–4.65) | .6525 |
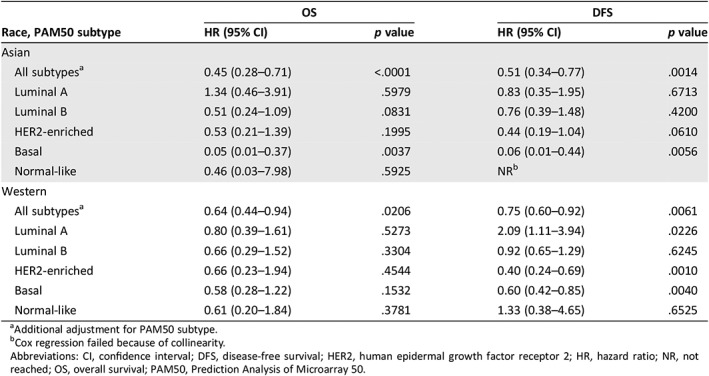
Additional adjustment for PAM50 subtype.
Cox regression failed because of collinearity.
Abbreviations: CI, confidence interval; DFS, disease‐free survival; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; NR, not reached; OS, overall survival; PAM50, Prediction Analysis of Microarray 50.
To explore whether a higher ESTIMATE immune score in tumors contributed to the more favorable prognosis of Asian patients than Western patients, we used the interaction between the ESTIMATE immune score and race as a variable in the Cox survival analysis. Asian race with a high ESTIMATE immune score provided additional power to predict longer DFS but did not reach statistical significance for OS (supplemental online Table 5). The stratified analysis showed that, for all PAM50 subtypes, Asian patients had significantly longer OS in the high and low immune score strata and longer DFS in the low immune score stratum when compared with Western patients. In addition, Asian patients had significantly longer DFS in the strata of luminal B subtype with high immune score and longer OS and DFS in the strata of basal‐like subtype with high immune score (supplemental online Table 6).
Infiltrating Immune Cell Subsets and Prognostic Effects
After adjustment for multiple comparisons, the compositions of infiltrating immune cells were not significantly different between the tumors of Asian and Western patients (Fig. 3). The adjusted analysis of prognostic effects of each immune cell subset on OS and DFS are shown in Figure 4 and supplemental online Figure 4, respectively. In terms of OS, the prognostic impact of each immune cell subset had the same trend in both the Asian and Western subgroups. Among them, high level of activated CD4 T cells and low level of M2 macrophages were significantly associated with longer OS in both Asian and Western patients (Fig. 4A, B). Memory B cells and CD8 T cells were significantly associated with longer OS in Asian patients (Fig. 4A). Resting dendritic cells, resting mast cells, M1 macrophages, CD8 T cells, and gamma‐delta T cells were significantly associated with longer OS, and activated mast cells were associated with shorter OS in the Western subgroup (Fig. 4B). In terms of DFS, activated CD4 T cells were significantly associated with longer DFS in both Asian and Western patients (supplemental online Fig. 4A, B). Memory B cells were associated with longer DFS, and M2 macrophages, neutrophils, and activated dendritic cells were associated with shorter DFS in the Asian subgroup (supplemental online Fig. 4A). Naive CD4 T cells, M0 macrophages, and activated natural killer cells were associated with shorter DFS in the Western subgroup (supplemental online Fig. 4B).
Figure 3.
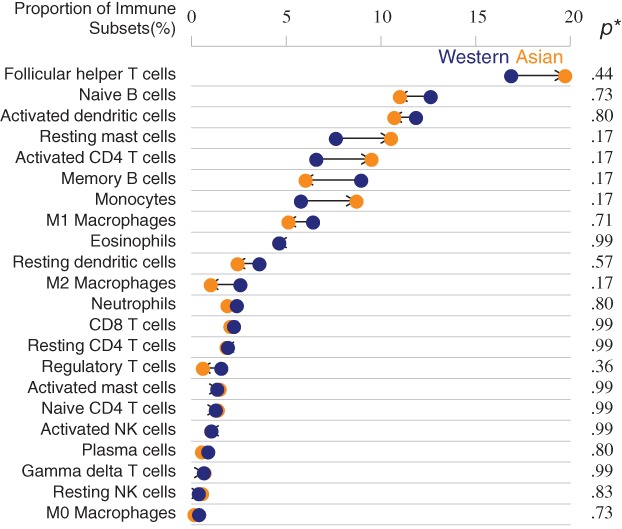
Proportions of immune cell subsets in Asian and Western breast tumors. Monocytes, M1 macrophages, gamma‐delta T cells, naive CD4 T cells, and regulatory T cells exhibited different percentage between races, but statistical analysis was insignificant. * indicates p value calculated by proportional z test with Benjamini‐Hochberg adjustment.
Abbreviation: NK, natural killer.
Figure 4.
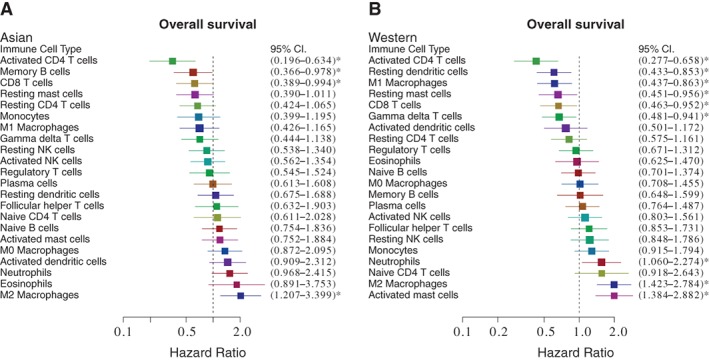
Prognostic effects of individual immune cell subsets on overall survival. (A): Effects of cell subsets on Asian patients. Adjusted for age, stage, and Prediction Analysis of Microarray 50 (PAM50) subtype, activated CD4 T cells, memory B cells, and CD8 T cells were associated with better prognosis, whereas M2 macrophage was associated with worse prognosis. (B): Effects of cell subsets on Western patients. The effects of activated CD4 T cells, CD8 T cells, and M2 macrophages were similar in Western patients with breast cancer. * indicates p < .05.
Abbreviations: CI, confidence interval; NK, natural killer.
Discussion
We demonstrated that Asian patients with breast cancer had significantly longer OS and DFS than Western patients. The breast tumors of Asian patients had significantly higher ESTIMATE immune scores. A high immune score was associated with longer OS and DFS in both Asian and Western patients, and Asian race with a high ESTIMATE immune score had additional impact on longer DFS. The composition of tumor‐infiltrating immune cells was not significantly different between the tumors of Asian and Western patients. The prognostic effects of individual immune cell subsets were generally consistent across different races and survival endpoints. Among them, activated CD4 T cells and M2 macrophages had the most prominent effect on prognosis and were associated with longer and shorter survival, respectively.
The univariate and multivariate analyses showed that Asian patients had significantly longer OS and DFS when compared with Western patients (supplemental online Figs. 1 and 2; supplemental online Table 3). This finding was consistent with that of previous immigration studies based on U.S. SEER9 and SEER18 data, which showed that Asian American women with breast cancer had longer survival than other racial groups, including white non‐Hispanic Americans, and suggested a genetic background effect 12, 13. In this study, the survival differences were more prominent in the basal subtype, with statistical significance in OS and DFS (supplemental online Figs. 1E and 2E) and in the luminal B subtype, with a strong trend in OS (supplemental online Fig. 1C) and statistical significance in DFS (supplemental online Fig. 2C). These novel findings warrant further validation and may be relevant in clinical practice.
This was the first study to report a higher ESTIMATE immune score, which is a surrogate of the amount of tumor‐infiltrating immune cells, in tumors of Asian patients than in those of Western patients. The finding was consistent with that of previous pathological studies, which consistently reported much more intense lymphoid or plasma cell infiltration in breast tumors in Japanese women than in their American counterparts 9, 10, 11, 39, and a multiomic study, which showed that breast cancers from younger Asian patients harbor more immune‐active microenvironments than breast cancers from Western patients 40. In addition, the translational study of the Cleopatra trial, a randomized phase III trial to evaluate the efficacy of pertuzumab in HER2‐positive metastatic breast cancer, showed significantly higher TIL values in Asian participants than in white participants 41. Consistently, our study showed a strong trend of higher immune scores in HER2‐enriched tumors of Asian patients than in their Western counterparts (1,513 vs. 1,275; p = .0593; Table 1).
The reason for the difference in ESTIMATE immune score between Asian and Western patients remains unclear. Genetic factor is likely to be the cause, but nongenetic factors such as diet or microbiome could also contribute to the disparity in tumor immune microenvironment. For example, numerous studies have shown the crosstalk of gut microbiome and the immune system, and the influence of the microbiome on the cancer immunotherapy response 42. An association of the diversity of gastrointestinal microbiome with the expression of TILs in breast cancer was recently reported 43. In addition, the prior studies showed gut microbiota diversity across ethnicities in the U.S. 44 and between adult twins in Korean and the U.S. 45.
In this study, the adjusted analyses showed that a higher ESTIMATE immune score was associated with longer OS and DFS in both the Asian and Western subgroups, and the associations were more prominent in the basal, HER2‐enriched, and luminal B subtypes. In contrast, a higher ESTIMATE immune score was associated with shorter DFS in Western patients with luminal A breast cancer (Table 2). The ESTIMATE immune score represents the amount of tumor‐infiltrating immune cells, and approximately half of these cells were TILs. Our findings were consistent with prior studies that showed the positively prognostic relevance of TILs in triple‐negative and/or HER2‐positive breast cancer 15, 16, 17, 18, 19. For luminal breast cancer, a translational study of neoadjuvant trials by the German Breast Cancer Group reported the unusual finding of shorter OS in patients with high TILs in luminal‐HER2‐negative disease, which were based on immunohistochemistry and in situ hybridization (for HER2 2+) 46. Our study defined the molecular subtypes by gene expressions, and the adjusted univariate analysis showed that high immune score was associated with shorter DFS in the luminal A subtype of the Western subgroup but with a trend toward longer OS in the luminal B subtype of the Asian subgroup. These findings suggest that there are differences in the immune microenvironment of the luminal A and luminal B breast tumors.
Our study revealed no significant difference in the immune cell compositions of breast tumors between the Asian and Western patients. The statistical analysis (proportional z test with Benjamini‐Hochberg adjustment) was relatively conservative and did not account for the absolute number of each immune subset; thus we cannot completely exclude the existence of some minor differences between these two subgroups. For example, the percentages of plasma cells, follicular T cells, and M1 and M2 macrophages were numerically higher in the breast tumors of Asian patients. Our study revealed the complex prognostic effects of immune compositions. This complexity was similar to that reported in recent two studies on breast cancer conducted using the same methodology 47, 48. Ali et al. highlighted that regulatory T cells, M0 macrophages, and M2 macrophages were the most strongly associated with poor outcomes, irrespective of their estrogen receptor (ER) status, and that CD8 T cells and activated memory CD4 T cells were associated with longer survival in ER‐positive tumors 47. Bense et al. reported that higher proportions of regulatory T cells, activated mast cells in HER2‐positive tumors, and M0 macrophages in ER‐positive tumors were associated with shorter survival and that an increased proportion of gamma‐delta T cells in all subtypes was related to a higher pathological complete response rate and prolonged survival 48. In concordance with these studies, our study showed that activated CD4 T cells and M2 macrophages were the most strongly associated with survival in both Asian and Western patients (Fig. 4 and supplemental online Fig. 4).
Finally, we acknowledge that this study has some limitations. First, ethnicity information was not available in some databases. A small percentage of Asian patients included in the Western databases could be misclassified as Western patients. Second, DFS or OS data were not available in some of the databases. Third, the Western patients were relatively older, which may contribute to their shorter OS when compared with Asian patients. To minimize the bias, we adjusted age and stage in the survival analyses. Fourth, tumor grade, a known prognostic factor, was only available for 56% of patients. Therefore, we did not include tumor grade in the survival analysis.
Conclusion
As a first step toward determining the role of tumor‐infiltrating immune cells in Asian patients with breast cancer, we showed that the tumors of Asian patients had higher ESTIMATE immune scores than their Western counterparts. The higher ESTIMATE immune scores in breast tumors of Asian patients partially explain the more favorable prognosis of Asian patients compared with Western patients. Further studies are warranted to validate the difference in immune compositions and the prognostic effects of immune subsets in breast tumors of Asian patients.
Author Contributions
Conception/design: Ching‐Hsuan Chen, Yen‐Shen Lu, Ann‐Lii Cheng, Tzu‐Pin Lu, Ching‐Hung Lin
Provision of study material or patients: Ching‐Hsuan Chen, Tzu‐Pin Lu
Collection and/or assembly of data: Ching‐Hsuan Chen, Chiun‐Sheng Huang, Ming‐Yang Wang, Chun‐Wei Kuo, Ching‐Hung Lin
Data analysis and interpretation: Ching‐Hsuan Chen, Yen‐Shen Lu, Ann‐Lii Cheng, Wen‐Hung Kuo, Ming Chao, Tzu‐Pin Lu, Ching‐Hung Lin
Manuscript writing: Ching‐Hsuan Chen, Yen‐Shen Lu, Ann‐Lii Cheng, I‐Chun Chen, Chun‐Wei Kuo, Tzu‐Pin Lu, Ching‐Hung Lin
Final approval of manuscript: Ching‐Hsuan Chen, Yen‐Shen Lu, Ann‐Lii Cheng, Chiun‐Sheng Huang, Wen‐Hung Kuo, Ming Chao, I‐Chun Chen, Chun‐Wei Kuo, Tzu‐Pin Lu, Ching‐Hung Lin
Disclosures
Yen‐Shen Lu: Novartis, Merck Sharp & Dohme, Roche (RF—clinical study grants), Boehringer Ingelheim, Pfizer, Roche, Novartis (C/A), GlaxoSmithKline, Merck Sharp & Dohme, Pfizer, Roche (RF—contracted research), Novartis, Merck Sharp & Dohme, Pfizer, Roche, Eisai (other—speaker fees); Tzu‐Pin Lu: Amwise Diagnostic (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Figures
Supplemental Tables
Acknowledgments
The results presented here are in part based on data generated by The Cancer Genome Atlas's research network (http://cancergenome.nih.gov/). This study was supported by grants from the Ministry of Health and Welfare, Taiwan (MOHW106‐TDU‐B‐211‐144005), National Taiwan University Hospital (NTUH‐ 003822), and the Ministry of Science and Technology, Taiwan (MOST 106‐2314‐B‐002‐212).
Disclosures of potential conflicts of interest may be found at the end of this article.
Contributor Information
Tzu‐Pin Lu, Email: tplu@ntu.edu.tw.
Ching‐Hung Lin, Email: chinghlin@ntu.edu.tw.
References
- 1. Youlden DR, Cramb SM, Yip CH et al. Incidence and mortality of female breast cancer in the Asia‐Pacific region. Cancer Biol Med 2014;11:101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shin HR, Joubert C, Boniol M et al. Recent trends and patterns in breast cancer incidence among Eastern and Southeastern Asian women. Cancer Causes Control 2010;21:1777–1785. [DOI] [PubMed] [Google Scholar]
- 3. Porter P. “Westernizing” women's risks? Breast cancer in lower‐income countries. N Engl J Med 2008;358:213–216. [DOI] [PubMed] [Google Scholar]
- 4. Chia KS, Reilly M, Tan CS et al. Profound changes in breast cancer incidence may reflect changes into a Westernized lifestyle: A comparative population‐based study in Singapore and Sweden. Int J Cancer 2005;113:302–306. [DOI] [PubMed] [Google Scholar]
- 5. Stemmermann GN. The pathology of breast cancer in Japanese women compared to other ethnic groups: A review. Breast Cancer Res Treat 1991;18(suppl 1):S67–S72. [DOI] [PubMed] [Google Scholar]
- 6. Sakamoto G, Sugano H. Pathology of breast cancer: Present and prospect in Japan. Breast Cancer Res Treat 1991;18(suppl 1):S81–S83. [DOI] [PubMed] [Google Scholar]
- 7. Lin CH, Liau JY, Lu YS et al. Molecular subtypes of breast cancer emerging in young women in Taiwan: Evidence for more than just westernization as a reason for the disease in Asia. Cancer Epidemiol Biomarkers Prev 2009;18:1807–1814. [DOI] [PubMed] [Google Scholar]
- 8. Lin CH, Chuang PY, Chiang CJ et al. Distinct clinicopathological features and prognosis of emerging young‐female breast cancer in an East Asian country: A nationwide cancer registry‐based study. The Oncologist 2014;19:583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chabon AB, Takeuchi S, Sommers SC. Histologic differences in breast carcinoma of Japanese and American women. Cancer 1974;33:1577–1579. [DOI] [PubMed] [Google Scholar]
- 10. MacMahon B, Morrison AS, Ackerman LV et al. Histologic characteristics of breast cancer in Boston and Tokyo. Int J Cancer 1973;11:338–344. [DOI] [PubMed] [Google Scholar]
- 11. Morrison AS, Black MM, Lowe CR et al. Some international differences in histology and survival in breast cancer. Int J Cancer 1973;11:261–267. [DOI] [PubMed] [Google Scholar]
- 12. Clegg LX, Li FP, Hankey BF et al. Cancer survival among US whites and minorities: A SEER (Surveillance, Epidemiology, and End Results) Program population‐based study. Arch Intern Med 2002;162:1985–1993. [DOI] [PubMed] [Google Scholar]
- 13. Iqbal J, Ginsburg O, Rochon PA et al. Differences in breast cancer stage at diagnosis and cancer‐specific survival by race and ethnicity in the United States. JAMA 2015;313:165–173. [DOI] [PubMed] [Google Scholar]
- 14. Fridman WH, Zitvogel L, Sautes‐Fridman C et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–734. [DOI] [PubMed] [Google Scholar]
- 15. Mahmoud SM, Paish EC, Powe DG et al. Tumor‐infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 2011;29:1949–1955. [DOI] [PubMed] [Google Scholar]
- 16. Loi S, Sirtaine N, Piette F et al. Prognostic and predictive value of tumor‐infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node‐positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin‐based chemotherapy: BIG 02‐98. J Clin Oncol 2013;31:860–867. [DOI] [PubMed] [Google Scholar]
- 17. Dieci MV, Mathieu MC, Guarneri V et al. Prognostic and predictive value of tumor‐infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann Oncol 2015;26:1698–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denkert C, Loibl S, Noske A et al. Tumor‐associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 2010;28:105–113. [DOI] [PubMed] [Google Scholar]
- 19. Yu X, Zhang Z, Wang Z et al. Prognostic and predictive value of tumor‐infiltrating lymphocytes in breast cancer: A systematic review and meta‐analysis. Clinical and translational oncology 2016;18:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shang B, Liu Y, Jiang SJ et al. Prognostic value of tumor‐infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta‐analysis. Sci Rep 2015;5:15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leek RD, Lewis CE, Whitehouse R et al. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res 1996;56:4625–4629. [PubMed] [Google Scholar]
- 22. Campbell MJ, Tonlaar NY, Garwood ER et al. Proliferating macrophages associated with high grade, hormone receptor negative breast cancer and poor clinical outcome. Breast Cancer Res Treat 2011;128:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barrett T, Wilhite SE, Ledoux P et al. NCBI GEO: Archive for functional genomics data sets – update. Nucleic Acids Res 2013;41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cancer Genome Atlas N . Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sabatier R, Finetti P, Adelaide J et al. Down‐regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS One 2011;6:e27656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buffa FM, Camps C, Winchester L et al. microRNA‐associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 2011;71:5635–5645. [DOI] [PubMed] [Google Scholar]
- 28. Hatzis C, Pusztai L, Valero V et al. A genomic predictor of response and survival following taxane‐anthracycline chemotherapy for invasive breast cancer. JAMA 2011;305:1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terunuma A, Putluri N, Mishra P et al. MYC‐driven accumulation of 2‐hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 2014;124:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clarke C, Madden SF, Doolan P et al. Correlating transcriptional networks to breast cancer survival: A large‐scale coexpression analysis. Carcinogenesis 2013;34:2300–2308. [DOI] [PubMed] [Google Scholar]
- 31. Tofigh A, Suderman M, Paquet ER et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep 2014;9:129–142. [DOI] [PubMed] [Google Scholar]
- 32. Kao KJ, Chang KM, Hsu HC et al. Correlation of microarray‐based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 2011;11:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagalla S, Chou JW, Willingham MC et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol 2013;14:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gautier L, Cope L, Bolstad BM et al. affy‐‐analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–315. [DOI] [PubMed] [Google Scholar]
- 35. Gendoo DM, Ratanasirigulchai N, Schroder MS et al. Genefu: An R/Bioconductor package for computation of gene expression‐based signatures in breast cancer. Bioinformatics 2016;32:1097–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshihara K, Shahmoradgoli M, Martinez E et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newman AM, Liu CL, Green MR et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gentles AJ, Newman AM, Liu CL et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosen PP, Ashikari R, Thaler H et al. A comparative study of some pathologic features of mammary carcinoma in Tokyo, Japan and New York, USA. Cancer 1977;39:429–434. [DOI] [PubMed] [Google Scholar]
- 40. Kan Z, Ding Y, Kim J et al. Multi‐omics profiling of younger Asian breast cancers reveals distinctive molecular signatures. Nat Commun 2018;9:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luen SJ, Salgado R, Fox S et al. Tumour‐infiltrating lymphocytes in advanced HER2‐positive breast cancer treated with pertuzumab or placebo in addition to trastuzumab and docetaxel: A retrospective analysis of the CLEOPATRA study. Lancet Oncol 2017;18:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gopalakrishnan V, Helmink BA, Spencer CN et al. The Influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi J, Geng C, Sang M et al. Effect of gastrointestinal microbiome and its diversity on the expression of tumor infiltrating lymphocytes in breast cancer. Oncol Lett 2019;17:5050–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brooks AW, Priya S, Blekhman R et al. Gut microbiota diversity across ethnicities in the United States. PLoS Biol 2018;16:e2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee S, Sung J, Lee J et al. Comparison of the gut microbiotas of healthy adult twins living in South Korea and the United States. Appl Environ Microbiol 2011;77:7433–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Denkert C, von Minckwitz G, Darb‐Esfahani S et al. Tumour‐infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19:40–50. [DOI] [PubMed] [Google Scholar]
- 47. Ali HR, Chlon L, Pharoah PD et al. Patterns of immune infiltration in breast cancer and their clinical implications: A gene‐expression‐based retrospective study. PLoS Med 2016;13:e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bense RD, Sotiriou C, Piccart‐Gebhart MJ et al. Relevance of tumor‐infiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst 2017;109:djw192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Figures
Supplemental Tables


