Significance
In RNA metabolism, many proteins act on diverse cellular RNAs, but avoid certain RNA subsets. To understand how this is accomplished, we investigate the exonuclease Rrp6p, which acts on many, but not all RNAs. We measure degradation kinetics at single-nucleotide resolution and find that Rrp6p discriminates against RNAs ending in CCA-3′. This sequence occurs at the 3′ termini of uncharged tRNAs, a class of abundant RNAs that Rrp6p does not degrade in the cell. Our data provide an unexpected perspective on substrate specificity of RNA-interacting proteins by showing that specificity is used not only to enable preferential action on certain RNAs, but also to selectively avoid RNAs.
Keywords: RNA, kinetics, specificity
Abstract
The exoribonuclease Rrp6p is critical for RNA decay in the nucleus. While Rrp6p acts on a large range of diverse substrates, it does not indiscriminately degrade all RNAs. How Rrp6p accomplishes this task is not understood. Here, we measure Rrp6p–RNA binding and degradation kinetics in vitro at single-nucleotide resolution and find an intrinsic substrate selectivity that enables Rrp6p to discriminate against specific RNAs. RNA length and the four 3′-terminal nucleotides contribute most to substrate selectivity and collectively enable Rrp6p to discriminate between different RNAs by several orders of magnitude. The most pronounced discrimination is seen against RNAs ending with CCA-3′. These RNAs correspond to 3′ termini of uncharged tRNAs, which are not targeted by Rrp6p in cells. The data show that in contrast to many other proteins that use substrate selectivity to preferentially interact with specific RNAs, Rrp6p utilizes its selectivity to discriminate against specific RNAs. This ability allows Rrp6p to target diverse substrates while avoiding a subset of RNAs.
The 3′-5′ exoribonuclease Rrp6p is important for quality control of virtually all RNA types in the nucleus (1–3) and for the 3′ processing of snoRNAs and rRNA (4, 5). Rrp6p is conserved in eukaryotes (5). The enzyme is part of the nuclear exosome, where it binds to the 9-subunit exosome core ring on the opposite site from another exoribonuclease, Rrp44p (6–10). Rrp6p is thought to be critical for initial RNA binding and positioning of the RNA substrates for processing by the exosome (7, 9, 11, 12). The enzyme also interacts with components of the TRAMP complex, which appends short adenylate tails to RNAs that are targeted for degradation by the exosome (13, 14). Transcriptome-wide and cellular localization studies further suggest functions of Rrp6p independent of the exosome (15, 16). In addition, it has been proposed that yeast Rrp6p functions outside the exosome during the control of poly(A)-tail length of certain mRNAs through interactions with Nab2p and during snoRNA processing and degradation of prematurely terminated Pol-II transcripts through interactions with Nab3p of the Nrd1-Nab3-Sen1 (NNS) complex (4, 17–19).
Yeast Rrp6p contains 733 amino acids, arranged in at least 4 distinct functional domains (20–22). The N-terminal PMC2NT domain interacts with exosome-associated factors (13, 14). The EXO domain harbors the active site of the DEDD-family exoribonuclease and, together with the adjacent HRDC domain, forms the catalytic module (20, 23). The C-terminal domain binds RNA and interacts with the exosome (24). Several high-resolution structures of Rrp6p with and without exosome components have provided a detailed picture of its architecture, RNA interacting regions, and positioning of the RNA for nucleolytic cleavage (6, 7, 9, 10, 20), during which Rrp6p releases nucleoside 5′-monophosphates (25).
To perform its biological roles, Rrp6p targets a large range of substrates, but it does not indiscriminately degrade all RNAs. How Rrp6p accomplishes this task is not known. Substrate selection by Rrp6p is likely affected by other protein factors, analogous to the Ccr4-Not recruitment by Puf3 (26). However, biochemical data have also shown differing Rrp6p activities toward different RNAs (11, 23, 25). The mechanistic basis for these activity variations is not known. Varying activities toward different substrates indicate inherent substrate specificity and thus preferential action on a subset of substrates (27). This, however, seems to be at odds with the required function of Rrp6p on a broad range of diverse RNAs. How does Rrp6p leverage discrimination between different RNAs for a function that requires action on many diverse substrates, but not on all RNAs? Answering this question is important for understanding the molecular basis for nuclear RNA metabolism, given the central roles of Rrp6p in the nucleus. Answering this question is also important for understanding the function of other enzymes that act on many substrates, but not on all RNAs, including other nucleases, nontemplated polymerases, and helicases (27).
Here, we systematically investigate how Rrp6p discriminates between RNA substrates. To accomplish this goal, we developed a quantitative approach to determine Rrp6p–RNA binding and degradation kinetics in vitro at single-nucleotide resolution. The data obtained with this approach indicate that Rrp6p recognizes, degrades, and distinguishes RNAs in an unexpectedly complex fashion. We detect marked substrate selectivity at the substrate binding and, to a smaller extent, at the degradation steps. In addition, we find that Rrp6p interacts with RNA not only in a productive, but also in a nonproductive binding mode. Both binding modes are impacted by RNA length and sequence. Most notably, the results show that Rrp6p discriminates against RNAs ending in CCA-3′, which occur at the 3′ termini of uncharged tRNAs, a class of abundant RNAs that Rrp6p does not degrade in the cell (28). Our data identify CCA-3′ as an antideterminant for Rrp6p and reveal an unappreciated role of substrate selectivity for a protein that interacts with RNA. In contrast to other proteins, which use substrate selectivity to preferentially interact with certain RNAs, Rrp6p utilizes selectivity to discriminate against specific RNAs. This ability allows Rrp6p to target diverse substrates while avoiding a subset of RNAs.
Results
Rrp6p Activity at Single-Nucleotide Resolution.
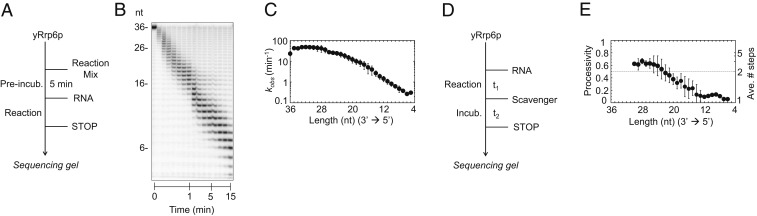
To systematically define how Rrp6p activity differs between RNA substrates, we set out to quantitatively determine Rrp6p–RNA binding and degradation kinetics in vitro at single-nucleotide resolution. We first measured degradation of an RNA with 36 adenylates (A36) under pre–steady-state conditions and determined observed degradation rate constants (kobs) at single-nucleotide resolution (Fig. 1 A–C). The rate constants decreased with the progressive shortening of the substrate (Fig. 1C). This observation indicated scaling of Rrp6p activity with RNA length.
Fig. 1.
Degradation of A36 by Rrp6p at single-nucleotide resolution. (A) Reaction scheme for a pre–steady-state reaction. (B) Representative PAGE for a reaction of Rrp6p (220 nM) with A36 (1 nM). Aliquots were removed at 0.083, 0.16, 0.33, 0.5, 0.66, 0.83, 1, 2, 3, 4, 5, 6, 8, 10, and 15 min. (C) Observed rate constants (kobs) at single-nucleotide resolution for the reaction shown in B. Datapoints represent an average of 3 independent measurements. Error bars show one SD. (D) Reaction scheme for a pulse-chase reaction. (E) Processivity of Rrp6p at single-nucleotide resolution, measured for the A36 substrate. The average number of steps (right axis) was calculated according to P = (N − 1)∙N−1 (P, processivity; N, number of steps) (29). Datapoints represent an average of 3 independent measurements. Error bars show one SD.
We next performed pulse-chase experiments to determine the processivity of Rrp6p on the A36 substrate (Fig. 1D and SI Appendix, Fig. S1). The processivity is determined by the ratio of the rate constant for the degradation step and for dissociation at each nucleotide and correlates with the number of steps the enzyme takes on average per binding event (29, 30). On RNAs with more than 25 nt, Rrp6p proceeded for roughly 3 degradation steps before dissociating from the substrate (Fig. 1E). For RNAs with fewer than 12 nt, Rrp6p is more than 10 times as likely to dissociate than to cleave the terminal nucleotide (Fig. 1E). The data show that Rrp6p is not a strictly distributive enzyme, but able to degrade several nucleotides per binding event for RNAs with more than 25 nt. However, the processivity decreased with progressive shortening of the substrate, providing more evidence that RNA length impacted Rrp6p activity.
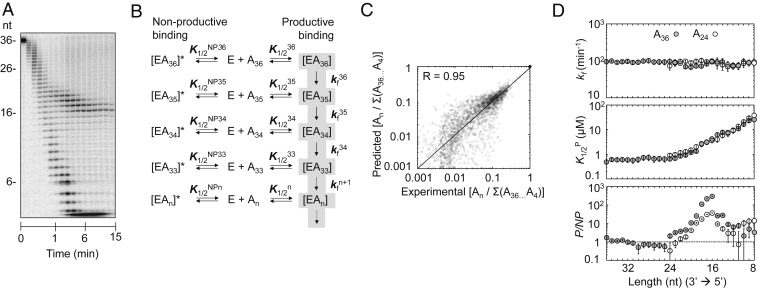
Building on the data from the pre–steady-state and the pulse-chase experiments, we next set out to establish a basic kinetic framework that described the nuclease reaction at single-nucleotide resolution in terms of enzyme–RNA binding and degradation steps. To assemble the framework, we measured reaction timecourses at varying enzyme and substrate concentrations. For reactions with [Rrp6p] > 350 nM, we noted an accumulation of RNA species with 16 to 19 nt (Fig. 2A), consistent with previous observations (12). The accumulation of 16- to 19-nt RNA species increased with rising Rrp6p concentrations and was seen under all tested reaction conditions, including variations in substrate concentrations, reaction temperature, pH, monovalent salt, and Mg2+ concentrations (SI Appendix, Fig. S2). The accumulation of the RNA species was not caused by catalysis of nucleotide addition by Rrp6p (SI Appendix, Fig. S3). Based on this observation and on the increase of the accumulation with the Rrp6p concentration, we incorporated a nonproductive binding step for each nucleotide into the kinetic framework (Fig. 2B). Nonproductive binding of Rrp6p to RNA is consistent with crystal structures of Rrp6p, which have shown 2 RNA-binding modes, one of which is not compatible with cleavage (9).
Fig. 2.
Kinetic framework for RNA degradation by Rrp6p. (A) Representative degradation reaction of A36 (1 nM) at 870 nM Rrp6p. (B) Basic kinetic model for the degradation reaction by Rrp6p. E, Rrp6p; An, substrate or degradation intermediate with n nucleotides; K1/2n, apparent dissociation constant for productive binding for each substrate/intermediate; K1/2NP, apparent dissociation constant for nonproductive binding for each substrate/intermediate; kfn, degradation rate constant for each nucleotide; [EAn], productive enzyme–substrate/intermediate complex; [EAn]*, nonproductive enzyme–substrate/intermediate complex. (C) Correlation between experimental values (fractions of reaction intermediates: An/∑(A36...A4)) for reactions with A36 and values calculated with the kinetic scheme. (D) Kinetic parameters for the degradation of A36 (solid circles) and A24 (open circles) for individual nucleotides (Top, degradation rate constant (kf); Middle, apparent dissociation constant for productive binding (K1/2P); Bottom, ratio between apparent productive and nonproductive dissociation constants (P/NP)). Data show results of the global data fit (SI Appendix, Fig. S4). Error bars mark the SEs of the global fit.
We computed rate and equilibrium constants for the kinetic scheme using a pipeline of explicit and global datafits that combined data from timecourses measured with enzyme and substrate excess and from pulse-chase reactions (SI Appendix, Fig. S4A). For the A36 substrate, we utilized more than 2,800 datapoints to compute for each nucleotide equilibrium constants for productive (K1/2P) and nonproductive binding (K1/2N) of Rrp6p to the RNA and a rate constant for the cleavage step (kf). The kinetic parameters faithfully describe the experimental data over the entire range of reaction conditions, from pre-steady state to quasi-steady state (R = 0.95, Fig. 2C).
We detected no significant impact of the RNA length on the cleavage rate constant (kfn) (Fig. 2 D, Top). In contrast, productive binding affinity of Rrp6p (K1/2P) decreased markedly for RNA species with fewer than 20 nt (Fig. 2 D, Middle). Substrates with 36 and 24 As showed virtually identical affinity profiles (Fig. 2D), indicating that the length dependence of Rrp6p affinity is largely unaffected by the initial length of the substrate. The nonproductive RNA affinity of Rrp6p (K1/2NP(n)) increased sharply between 23 and 12 nt, compared to the productive Rrp6p affinity (Fig. 2 D, Bottom and SI Appendix, Fig. S2B). For RNA species with 16 nt, nonproductive affinity of Rrp6 was roughly 2 orders of magnitude larger than the productive affinity (Fig. 2 D, Bottom). The increase in nonproductive affinity was also largely independent of the length of the initial substrate and explains the accumulation of intermediates with 16 to 19 nt (Fig. 2D). Collectively, analysis of Rrp6p activity on poly(A) substrates with the kinetic framework reveals RNA length-dependent productive and nonproductive Rrp6p binding modes and a cleavage rate constant that is largely unaffected by substrate length.
Sequence Impacts Rrp6p Activity.
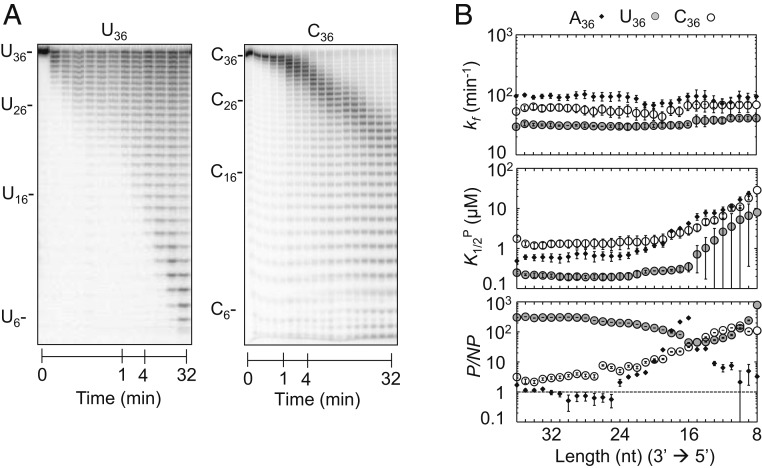
We next examined how substrate sequence impacted the kinetic parameters for homopolymeric U and C substrates (Fig. 3A). Homopolymeric G substrates were not tested because of their propensity to form G-quadruplex structures (31). Under pre–steady-state conditions, Rrp6p activity on U36 and C36 homopolymers scaled with RNA length (Fig. 3A and SI Appendix, Fig. S5 A and B), as seen for A36 (Fig. 1B). At identical conditions, degradation of U36 and C36 was markedly slower than for A36 (Figs. 1B and 3A and SI Appendix, Fig. S5 A and B). Analysis of the degradation reactions of U36 and C36 with our kinetic framework indicated that degradation rate constants (kf) for U36 and C36 did not significantly change with the RNA length (Fig. 3 B, Top), as observed for A36. However, the rate constants for U36 and C36 were lower than for A36 (Fig. 3B). Productive substrate affinities (K1/2P) for U36 and C36 decreased for RNAs with fewer than 20 nt, similarly but not identically to the scaling of K1/2P seen for A36 (Fig. 3 B, Middle). K1/2P values for U36 were lower than for C36 and A36 over the entire range of substrate lengths (Fig. 3B). Nonproductive affinities for U36 were significantly higher than for A36 and C36 (Fig. 3 B, Bottom and SI Appendix, Figs. S6 and S2B). It is possible that the weaker base stacking in poly(U), compared to other sequences, contributes to the high affinities for poly(U) substrates. Collectively, the data indicated that RNA sequence impacts Rrp6p activity and thus provided direct evidence of substrate selectivity of Rrp6p. In addition, the data showed that RNA length impacts Rrp6p binding, regardless of sequence, while cleavage rate constants do not scale with RNA length.
Fig. 3.
Degradation of U36 and C36. (A) Representative degradation reaction of U36 (1 nM; Rrp6p, 110 nM) and C36 (1 nM; Rrp6p, 110 nM). (B) Kinetic parameters for the degradation of U36 (solid circles) and C36 (open circles) for individual nucleotides (Top, degradation rate constant (kf); Middle, apparent dissociation constant for productive binding (K1/2P); Bottom, ratio between apparent productive and nonproductive dissociation constants (P/NP)). Data show results of the global data fit (SI Appendix, Fig. S4). Error bars mark the SEs of the global fit. The corresponding data for A36 (Fig. 2D) are shown for comparison.
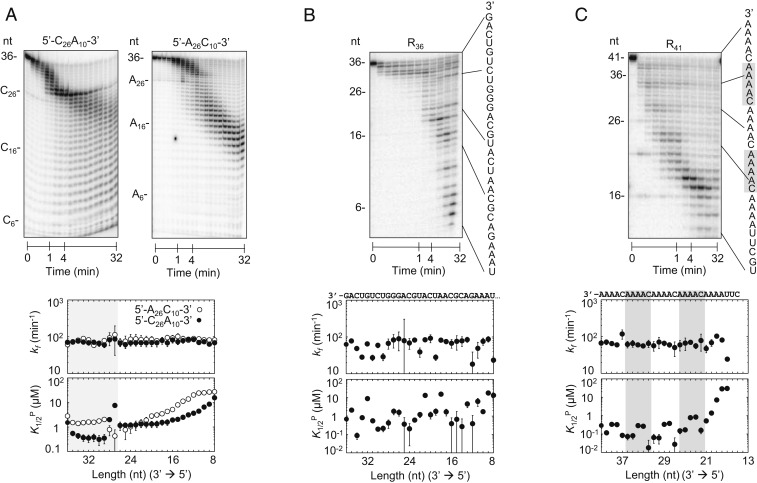
To elucidate RNA features that govern the substrate selectivity of Rrp6p, we determined the kinetic parameters for degradation reactions of a series of substrates with increasing sequence complexity. We started by characterizing 36-nt substrates with fused C and A homopolymers (5′-C26A10-3′, 5′-A26C10-3′; Fig. 4A). We observed a clear difference in the observed degradation rates for the respective A and C regions (Fig. 4A and SI Appendix, Fig. S5 C and D). As seen for the homopolymers, regions composed of A were degraded faster than regions composed of C (Fig. 4A and SI Appendix, Fig. S5 C and D). Kinetic parameters for the respective 3′ regions of the substrates were similar to the parameters for the corresponding homopolymers. RNA-length dependences of functional and nonproductive affinities were also similar to those seen for the corresponding homopolymers (Figs. 3B and 4A and SI Appendix, Fig. S7A). These observations indicated that the RNA sequences in the 3′-terminal region of the progressively shortening substrates affected the degradation activity. This notion was further supported by the kinetic parameters for the regions 3′ of the C/A and A/C border, which differed from those for homopolymeric substrates (Fig. 4A).
Fig. 4.
Degradation of substrates with increasing sequence diversity. (A, Upper) Representative degradation reactions of 5′-C26A10-3′ (1 nM; Rrp6p, 110 nM) and 5′-A26C10-3′ (1 nM; Rrp6p, 110 nM). (A, Lower) kinetic parameters for the degradation of 5′-C26A10-3′ (solid circles) and 5′-A26C10-3′ (open circles) for individual nucleotides (Upper plot, degradation rate constant (kf); Lower plot, apparent dissociation constant for productive binding (K1/2P). Data show results of the global data fits, and error bars mark the SEs of the global fits. Shading indicates the transition between A and C sequence blocks. (B, Upper) Representative degradation reactions of a 36-mer RNA (R36,1 nM; Rrp6p, 110 nM) with mixed sequence (indicated on the right). (B, Lower) Kinetic parameters for the degradation of R36, as in A. (C, Upper) Representative degradation reactions of a 41-mer RNA (R41,1 nM; Rrp6p, 110 nM) with mixed sequence, including four 3′-A4C-5′ repeats (sequence indicated on the right). (C, Lower) Kinetic parameters for the degradation of R41, as in A. Shading indicates the 3′-A4C-5′ repeats.
We next examined a 36-nt substrate with mixed sequence (Fig. 4B). Compared to homopolymeric and dipolymeric substrates (Figs. 2–4) we observed significantly larger fluctuations in cleavage rate constants and processivities and in productive and nonproductive substrate affinities (Fig. 4B and SI Appendix, Figs. S5E and S7B). These data were also consistent with an impact of the 3′-terminal substrate region that Rrp6p encounters as it degrades the substrate. To directly test this notion, we measured degradation of a 41-nt substrate containing four 3′-A4C-5′repeats fused to a mixed-sequence RNA (Fig. 4C and SI Appendix, Fig. S5F). For the repeat region of the RNA, we observed little change in the cleavage rate constants (Fig. 4C), as seen for homo- and dipolymeric substrates (Figs. 2, 3, and 4A). However, we detected 4 similar phases for productive and nonproductive affinities for the repeat region (Fig. 4C and SI Appendix, Fig. S7C). This observation indicated a clear impact of the 3′-terminal substrate region that Rrp6p encounters as it degrades the substrate. In addition, the 4 phases coincide with the four 5-nt repeats in the substrate, suggesting that Rrp6p recognized 5 or fewer nucleotides of the 3′-terminal sequence.
The kinetic data for substrates with varying sequences indicated a multilayered influence of several substrate features on the substrate selectivity of Rrp6p. The sequence of the 3′-terminal region exerts a pronounced impact on productive and nonproductive binding and, to a lesser, but notable extent on the cleavage rate constant. In addition, the length of the RNA and possibly sequence distant from the 3′ end impacted Rrp6p binding.
Rrp6p Recognizes Four 3′-Terminal Nucleotides.
To decipher the rules of sequence selectivity by Rrp6p, it was next critical to dissect the layers of substrate features that impact Rrp6p activity. To accomplish this goal, it was important 1) to determine how many nucleotides in the progressively shortening RNA were critical for the impact on Rrp6p activity, 2) to deconvolute effects of the 3′ termini of the progressively shortening substrates from effects caused by upstream RNA, and 3) to delineate rules according to which 3′-terminal sequences and upstream RNA affected activity.
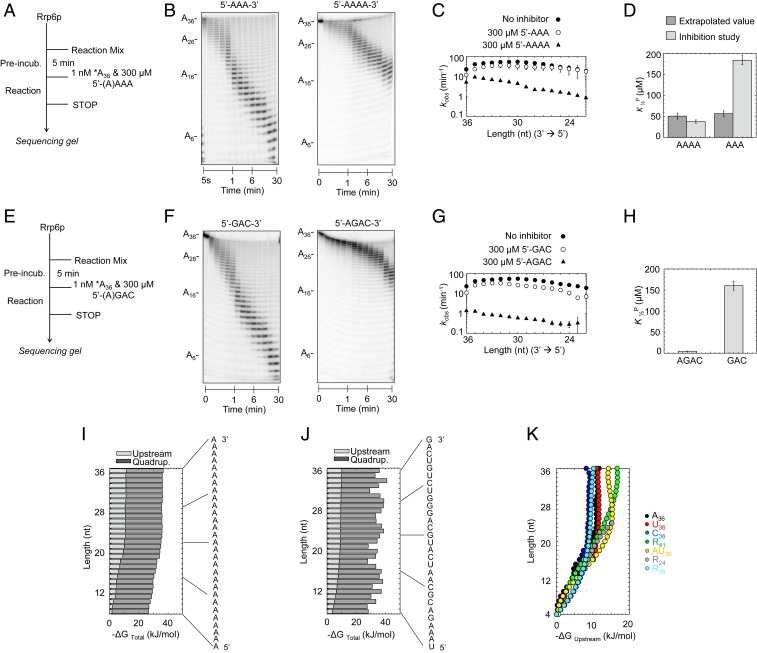
As noted, the phasing of kinetic parameters for the substrate with 5-nt repeats suggested that Rrp6p recognized 5 or fewer 3′-terminal nucleotides (Fig. 4C). To determine the exact number of 3′-terminal nucleotides that critically affected Rrp6p function, we performed competition experiments with 3-nt and 4-nt oligonucleotides (Fig. 5 A and B). At identical concentrations, a 3-nt RNA (A3) impacted the degradation of A36 markedly less than a 4-nt RNA (A4) (Fig. 5 B and C). The affinity for A4 determined from the competition experiments was highly similar to the extrapolated affinity (K1/2P) for the 4-nt fragment in the degradation reaction of A36 (Fig. 5D). In contrast, the affinity for A3 determined from competition experiments was markedly lower than the extrapolated affinity (K1/2P) for the 3-nt fragment in the degradation reaction of A36 (Fig. 5D). We therefore hypothesized that Rrp6p recognized 4 nt in the 3′ terminus.
Fig. 5.
Deconvolution of RNA length and sequence effects on Rrp6p activity. (A) Reaction scheme for competition experiments. (B) Representative competition reactions of A3 (300 μM; A36, 1 nM; Rrp6p, 220 nM) and A4 (300 μM; A36, 1 nM; Rrp6p, 220 nM). (C) Observed rate constants (kobs) for the degradation of A36 at single-nucleotide resolution in the absence (solid circles) and presence of A3 (open circles) and A4 (triangles), under conditions noted in A. Datapoints represent an average of 3 independent measurements. Error bars show one SD. (D) Apparent dissociation constants for productive binding (K1/2P) of A3 and A4 obtained experimentally and calculated values assuming similar scaling of inhibition by A3 and A4 (SI Appendix, Fig. S6). (E) Reaction scheme for competition experiments. (F) Representative competition reactions of 5′-GAC-3′ (300 μM; A36, 1 nM; Rrp6p, 220 nM) and 5′-AGAC-3′ (300 μM; A36, 1 nM; Rrp6p, 220 nM). (G) Observed rate constants (kobs) for the degradation of A36 at single-nucleotide resolution in the absence (solid circles) and presence of 5′-GAC-3′ (open circles) and 5′-AGAC-3′ (triangles), under conditions noted in E. Datapoints represent an average of 3 independent measurements. Error bars show one SD. (H) Apparent dissociation constants for binding of 5′-GAC-3′ and 5′-AGAC-3′ obtained from competitive inhibition experiments. (I) Contributions of upstream RNA (light gray) and 3′-terminal quadruplet (dark gray) to free binding energy for productive binding for each nucleotide for A36. (J) Contributions of upstream RNA (light gray) and 3′-terminal quadruplet (dark gray) to free binding energy for productive binding for each nucleotide for R36 (sequence on the right). (K) Contributions of the upstream RNA component to free binding energy for productive binding for each nucleotide for substrates indicated on the right.
To test this hypothesis, we performed competition experiments with a 3-nt and 4-nt RNA of another sequence (Fig. 5 E and F). The 3-nt RNA (5′GAC) impacted the degradation of A36 markedly less than the corresponding 4-nt RNA (5′AGAC) (Fig. 5 F–H), mirroring the data obtained for A3 and A4 (Fig. 5 B–D). Rrp6p bound 5′-AGAC with higher affinity than A4 (Fig. 5 D and H), further indicating that 4 nt were sufficient to affect sequence selectivity. Collectively, the data indicated a minimal functional binding-site size of Rrp6p with 4 nt. This functional binding-site size is remarkably consistent with crystal structures of Rrp6p, which also show a RNA-binding site with 4 ordered nucleotides (7).
Separation of 3′-Terminal from Upstream RNA Effects on Rrp6p Affinity.
Having determined that 4-nt RNA were sufficient to influence Rrp6p activity, it was next important to separate the impact of the 3′ termini from the upstream effects of RNA. To accomplish this goal, we converted productive RNA affinities (K1/2P) into free energies (ΔG°P). For each substrate length, we considered the overall binding energy as the sum of the binding energies contributed by the 3′ quadruplet and the upstream RNA. For A36, which contained exclusively A quadruplets whose binding energy we had directly measured (Fig. 5D and SI Appendix, Fig. S8), we obtained binding energy terms for upstream RNA that decreased with the number of nucleotides in the RNA (Fig. 5I). For substrates with more than 23 nt, upstream RNA contributed ∼30% of the binding energy. The upstream RNA contribution decreased to near zero for RNAs with fewer than 7 nt (Fig. 5I). Since the substrate contained only A, we conclude that RNA length has a notable impact on the effects of upstream RNA.
We next calculated contribution of upstream RNA to the binding energies for U36 and C36, following the approach described for A36 (SI Appendix, Fig. S9 A and B). Affinities for the minimal U4 and C4 quadruplets differed from A4 (SI Appendix, Fig. S9 A and B). The contribution of upstream RNA to the binding energies differed slightly with sequence, but the overall scaling of this contribution with the number of nucleotides was remarkably similar to A36 (Fig. 5I). This trend was also seen for substrates with fused homopolymeric A and C regions and substrates containing sequence repeats (SI Appendix, Fig. S9D).
To calculate the upstream RNA contribution to the free binding energy for substrates with more complex sequences, we used the free binding energies for quadruplets determined from homopolymeric substrates to deconvolute upstream RNA and quadruplet contributions to overall free binding energies (SI Appendix, Fig. S9 C and D). Again, we observed that contributions of upstream RNA length to the free binding energies differed somewhat with sequence, while the overall scaling of this length contribution with the number of nucleotides was similar to the scaling seen with homo- and dipolymeric sequences (Fig. 5 J and K). The data for these diverse substrates collectively indicate that contribution of upstream RNA to the free binding energy increases from nearly zero for substrates with fewer than 7 nt to 30% to 35% for substrates with more than ∼23 nt. The substrate sequence moderately impacts the effect of upstream RNA on binding energies, accounting for roughly 30% of the variation in free binding energy contributed by upstream RNA (Fig. 5). RNA length contributes the major part of binding energy provided by upstream RNA. For nonproductive binding, upstream RNA affected binding energies for substrates with complex sequences similarly to the patterns seen for productive binding (SI Appendix, Fig. S10 A and B). However, for U and A homopolymers RNA length impacted binding energies differently (SI Appendix, Fig. S10), reflected by the accumulation of RNA species with 16 to 19 nt for poly(A) substrates (Fig. 2). Contacts between upstream RNA and Rrp6p, presumably through one or more domains outside the exonuclease domain, are likely responsible for the impact of upstream RNA on both productive and nonproductive affinities of Rrp6p.
Rrp6p Discriminates Against CCA-3′ Ends.
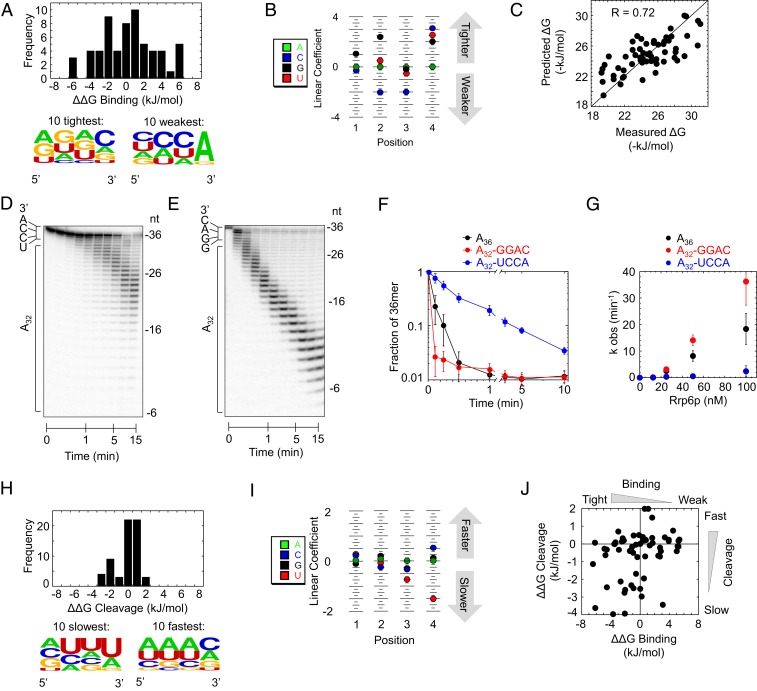
The deconvolution of contributions of upstream RNA and 3′-terminal quadruplets to substrate binding indicated a range of contributions of different quadruplets to binding energy (Fig. 6 A, Top). Our experiments provided free binding-energy contributions for 61 different quadruplets, which we used to calculate an affinity distribution for productive binding with A4 as a reference point with [ΔΔGBinding (A4) = 0 kJ·mol−1]. The range in free binding energy between weakest and tightest binding quadruplets (ΔΔGP = 12 kJ·mol−1, Fig. 6A) corresponds to a range in affinities of more than 2 orders of magnitude.
Fig. 6.
Impact of 3′-terminal quadruplets on sequence selectivity of Rrp6p. (A) Distribution of free energy contributions (ΔΔGBinding) for all experimentally tested 3′-terminal quadruplet sequences to productive binding, compared to the A4 variant [ΔΔGBinding(A4) = 0]. (Bottom) Sequence probability logo for the 10 tightest (Left) and the 10 weakest (Right) bound sequence variants. (B) PWM for the free energy contributions of quadruplet sequence variants. Higher linear coefficients indicate higher energy contribution. (C) Correlation between experimentally measured free energy contributions of 3′-terminal quadruplet sequences to productive binding and corresponding values calculated with the PWM in B. (D) Representative degradation reaction of A32-UCCA-3′ substrate (1 nM RNA, 100 nM Rrp6). (E) Representative degradation reaction of A32-GGAC-3′ substrate (1 nM RNA, 100 nM Rrp6). (F) Representative timecourses of cleavage of the 3′-terminal nucleotide from A36 (black), A32-GGAC-3′ (red), and A32-UCCA-3′ (blue) substrates under conditions used in D and E. (G) Observed rate constants of cleavage of the 3′-terminal nucleotide from A36 (black), A32-GGAC-3′ (red), and A32-UCCA-3′ (blue) substrates as function of Rrp6p concentration. Datapoints represent an average of 3 independent reactions; error bars represent one SD. (H) Distribution of free energies of the cleavage step (ΔΔGCleavage) for all experimentally tested 3′-terminal quadruplet sequences, compared to the A4 variant [ΔΔGCleavage (A4) = 0]. (Bottom) Sequence probability logo for the 10 slowest (Left) and the 10 fastest (Right) reacting sequence variants. (I) PWM for the free energies of quadruplet sequence variants during the cleavage step. Higher linear coefficients indicate higher energy contribution. (J) Correlation between free energies of the cleavage step (ΔΔGCleavage) and free energy contributions to productive binding (ΔΔGBinding) for all experimentally tested quadruplet sequences.
We detected sequence signatures for the weakest- and strongest-bound quadruplets (Fig. 6 A, Bottom). These signatures suggested rules for the sequence selectivity of Rrp6p. To define these rules, we calculated a position weight matrix (PWM) from the free energies for the 61 quadruplets measured with our substrates (Fig. 6B). A PWM considers nucleotide contributions at each position independent of each other (32, 33). More complex binding models were not examined, because experimental data of only 61 of 256 possible quadruplets precluded the use of binding models with more variables than a PWM (34). Nevertheless, the PWM accurately described the experimental data (R = 0.72, Fig. 6C). The PWM indicated that the sequence of the fourth nucleotide from the 3′ end did not significantly impact the affinity of Rrp6p, indicating a nonspecific contribution of this nucleotide position to the binding energy. While the fourth nucleotide is important, its sequence is not critical.
Our analysis revealed that Rrp6p bound weakest to quadruplets ending in CCA-3′. This is the 3′ sequence of uncharged tRNAs (35), an abundant class of nuclear RNAs that Rrp6p does not degrade. Our data suggested that Rrp6p discriminates against binding to the 3′ ends of uncharged tRNAs. This feature appears biologically relevant, because indiscriminate degradation of uncharged tRNAs would likely impact multiple processes of RNA metabolism in an uncontrolled manner (36). Our data indicate that CCA-3′ acts as an antideterminant for Rrp6p. CCA-3′ is also an antideterminant for 3′tRNAse, a tRNA-specific endonuclease (37), suggesting that multiple nucleases avoid CCA-3′.
It was next important to experimentally validate the PWM. To this end, we substituted the 3′ quadruplet of the A36 substrate with a UCCA-3′ and GGAC-3′ segment, one of the weakest and tightest binding quadruplets, respectively. At subsaturating enzyme concentrations, Rrp6p degraded the A32-UCCA-3′ substrate significantly slower than the A32-GGAC substrate, and also markedly slower than the A36 substrate (Fig. 6 D–G). Discrimination occurs primarily at the binding step, consistent with the PWM (SI Appendix, Fig. S11). The data directly show the discrimination against CCA-3′ termini and thus experimentally validate the PWM. We also detected Rrp6p substrate selectivity for quadruplets at the nonproductive binding step (SI Appendix, Fig. S12). These selectivity patterns moderately correlated with those seen for productive binding (R = 0.53; SI Appendix, Fig. S12) and also show discrimination against CCA-3′ termini.
Moderate Substrate Selectivity at the Cleavage Step.
We next quantified substrate selectivity during the cleavage step, which was largely unaffected by upstream RNA and did not scale with substrate length (Figs. 2–4). We could therefore directly calculate PWMs for 3′-terminal quadruplets (Fig. 6I). Energy differences between slowest and fastest cleaved quadruplets (ΔΔGC = 6 kJ·mol−1, Fig. 6H) correspond to a difference in cleavage rate constants of roughly one order of magnitude. While we detected no sequence signature for fast-cleaving quadruplets, we noted an overrepresentation of 3′ U for slow-cleaving quadruplets (Fig. 6I). The PWM revealed that only the 3′-terminal nucleotide impacts substrate selectivity at the cleavage step. The penultimate nucleotide contributed less and the third and fourth nucleotides showed virtually no contribution, consistent with the notion that cleavage occurs between the terminal nucleotides.
A Quantitative Model for Substrate Selectivity of Rrp6p.
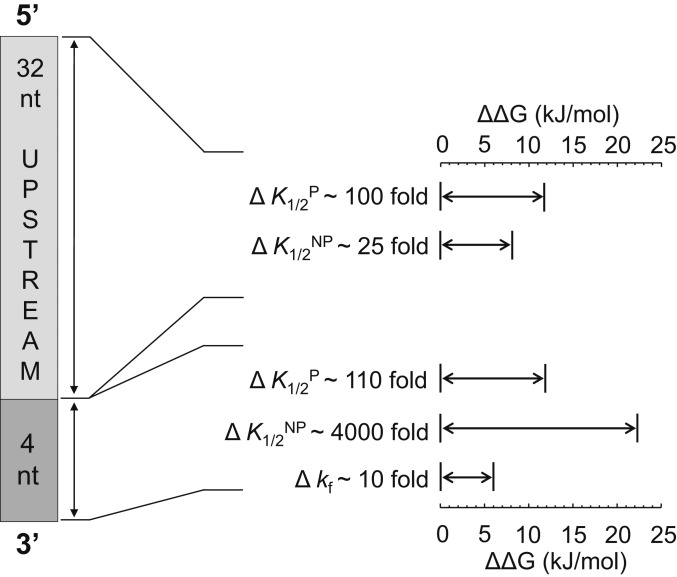
Collectively, our kinetic data revealed a complex mode for substrate selectivity by Rrp6p (Fig. 7). We detected substrate selectivity at the cleavage step and for productive and nonproductive binding. Selectivity at the cleavage step is mainly determined by the 3′-terminal nucleotide, predominantly by discrimination against 3′-U. Productive and nonproductive binding by Rrp6p is determined by upstream RNA and the 3′-terminal quadruplet sequence that Rrp6p encounters at each step, as it degrades the RNA 3′ to 5′. From a free energy point, substrate binding impacts selectivity to a larger degree than the cleavage step (Fig. 6 A and H). Selectivity for productive and nonproductive binding correlates moderately (SI Appendix, Fig. S12C). There is little correlation between the selectivity seen for productive binding and cleavage step (Fig. 6J). The most striking pattern is the discrimination against quadruplets ending in CCA-3′. Given that CCA-3′ ends are present on an abundant class of RNAs that Rrp6p does not degrade, this observation raises the possibility that the substrate selectivity of Rrp6p has evolved to prevent uncontrolled degradation of uncharged tRNAs.
Fig. 7.
Quantitative model for substrate selectivity of Rrp6p: range of free energy contributions (ΔΔG) of upstream RNA and 3′-terminal quadruplets to productive binding (K1/2P), nonproductive binding (K1/2NP), and cleavage step (kf). The variation from weakest- to tightest-bound substrate variant for K1/2 values and slowest to fastest substrate variant for kf values is also given as fold change. Scaling of K1/2P and K1/2NP with RNA length ranges from 4 nt (zero) to 36 nt (maximal value).
Discussion
A High-Resolution Kinetic Approach for Exonuclease Activity.
To systematically analyze substrate selectivity of Rrp6p, we developed and applied an approach to kinetically characterize the exonuclease Rrp6p at single-nucleotide resolution. The approach accesses critical biochemical parameters of nuclease reactions that have remained beyond the scope of current approaches, which monitor only changes in starting substrate and final products. The experiments require only widely available molecular biology equipment and no advanced computational skills are needed for data analysis. Both experimental approach and framework for data analysis can readily be applied to other exonucleases and complexes containing these enzymes.
For Rrp6p, the high-resolution kinetic approach illuminated the activity of the nuclease at unprecedented detail and thereby revealed a use of substrate selectivity that has not been widely appreciated for proteins that interact with RNA. Similar high-resolution, biochemical analyses might reveal unexpected features for other nucleases and thereby help to illuminate aspects of RNA metabolism that are beyond the scope of other approaches.
Processivity and Nonproductive Substrate Binding by Rrp6p.
We detected several additional biochemical features of Rrp6p. First, processivity of Rrp6p is more nuanced than the traditional view of the enzyme as largely distributive (20, 23, 25). Our data show that processivity scales with RNA length and is impacted by 3′-terminal quadruplets (Fig. 1E and SI Appendix, Fig. S5). For RNA (A36) with more than 25 nt, the processivity (P ∼ 0.65) is equivalent to the degradation of roughly 3 nt per binding event. For RNAs with fewer than 12 nt (P < 0.1) Rrp6p requires more than 10 binding events to degrade a single nucleotide (Fig. 1E). The processivity of Rrp6p for longer RNAs is lower than for nucleases considered highly processive, such as Rrp44p, which can degrade more than 10 nt per binding event (25, 38–40). Nevertheless, the ability to degrade multiple nucleotides without dissociating from the RNA might be significant for Rrp6p’s biological function, perhaps for roles outside the exosome (4, 17).
Our biochemical framework further revealed a previously undetected, nonproductive substrate-binding mode of Rrp6p. In this mode, Rrp6p binds RNA without cleaving the terminal nucleotide. Nonproductive RNA binding has a large impact on the biochemical behavior of Rrp6p. For a given RNA substrate, nonproductive affinity is generally higher than productive affinity. Like the productive RNA affinity of Rrp6p, the nonproductive RNA affinity scales with RNA length and sequence. The impact of RNA length and sequence correlates moderately between productive and nonproductive binding modes (Fig. 5K and SI Appendix, Figs. S10B and S12C). For substrates with adenylate tracts of more than 20 nt, the nonproductive binding leads to accumulation of species with 16 to 18 nt (Fig. 2D).
A recent crystal structure of the exosome with Rrp6p and poly(U) shows the RNA bound to only the HRDC domain of Rrp6 and not positioned for substrate cleavage on the nuclease domain (9). This configuration might represent nonproductively bound RNA. Our data also show an impact of the 3′-terminal nucleotides on nonproductive affinity (SI Appendix, Fig. S12 A and B). The HRDC domain in the crystal structure contacts the RNA not bound at the active site (9). Analogously, nonproductive RNA binding might involve sites in and perhaps beyond the HRDC domain. The scaling of the nonproductive affinity with RNA length, up to ∼25 nt, raises the possibility that contacts between RNA and Rrp6p extend beyond the HRDC domain. Consistent with this notion, a segment of the C-terminal domain, the “lasso,” associates with RNA and possibly contributes to nonproductive binding (12).
What might be the biological role of nonproductive RNA binding by Rrp6p? It has been proposed that an important role of Rrp6p in the exosome is to guide the RNA substrate through the central channel toward the exonuclease Rrp44p (7, 9, 10). This function would not be possible if Rrp6p degrades the RNA upon each encounter. A binding mode that does not result in RNA degradation is therefore essential. Our data provide direct, functional evidence for such a binding mode. It remains to be tested whether and how nonproductive substrate binding by Rrp6p is modulated in the exosome.
Substrate Selectivity and Discrimination Against CCA-3′ Termini.
A striking insight from our study is the degree of substrate selectivity by Rrp6p, which results in the discrimination against RNAs with CCA-3′ termini. The data show that substrate selectivity is conferred predominantly at the RNA-binding step, which is impacted by RNA length, upstream sequence, and the 3′-terminal quadruplet (Fig. 7). Affinities between shortest (4 nt) and longest substrates vary by a factor of roughly 100 and affinities between weakest- and tightest-bound 3′-terminal quadruplet by a factor of more than 100 (Fig. 7). In combination, upstream RNA and the 3′-terminal quadruplet can confer discrimination between different substrates by a factor of >104. This degree of selectivity places Rrp6p in the vicinity of RNA-interacting proteins that are considered highly specific (27, 41). Although Rrp6p is unlikely to encounter substrates with fewer than 10 nt in the cell, longer RNAs are presumably bound to other proteins, which can restrict access for Rrp6p. The enzyme most likely senses these RNPs as short RNAs, and our insights for short RNAs might thus be directly relevant for biological settings.
The impact of the 3′-terminal quadruplet on productive substrate binding is consistent with crystal structures of Rrp6p, which show 4 nt bound to the nuclease domain (7). Our data indicate that 4 nt are required for stable functional substrate association (Fig. 5 A–H). However, the sequence of only three 3′-terminal nucleobases impacts substrate binding (Fig. 6B). Although Rrp6p functionally binds to 4 nt, it senses the sequence of only a 3′ triplet.
In this context, it is notable that Rrp6p discriminates against RNAs with terminal CCA-3′. The finding raises the possibility that Rrp6p has evolved to selectively avoid the 3′ ends of uncharged tRNAs, which are abundant in the nucleus (36). While the nuclear exosome with Rrp6p degrades improperly processed tRNA precursors (42), once the CCA is added, even improperly processed tRNAs are protected from nuclear degradation (43). The selective discrimination of Rrp6p against CCA-3′ termini conceivably precludes uncontrolled degradation of uncharged tRNAs by the enzyme and thus prevents undesirable, global alterations of tRNA levels and protein synthesis (44). Moreover, CCA-3′ termini are abundant, given the tRNA levels in the nucleus (36). Selective avoidance of abundant termini prevents sequestration of Rrp6p and thereby ensures availability of the enzyme for its intended functions. CCA-3′ can be seen as an antideterminant for Rrp6p. Interestingly, CCA-3′ has also been identified as antideterminant for 3′-tRNAase (37), suggesting that prevention of degradation of CCA-3′ of uncharged tRNAs is biologically important.
Our findings highlight a previously unappreciated role of substrate selectivity for a protein that interacts with RNA. Substrate selectivity is traditionally viewed as a means for proteins to interact preferentially, or specifically, with a subset of RNA variants (27). To accomplish this, the proteins “utilize” the high-affinity segment of their affinity distribution toward all possible substrate variants (27). In contrast, Rrp6p utilizes the low-affinity range of the affinity distribution to discriminate against CCA-3′ (Fig. 6A). Biological use of the low-affinity end of the affinity distribution is an efficient way to define antideterminants for enzyme activities or protein binding. It is possible that antideterminants reported for RNAseIII (45) also coincide with the low-affinity end of the sequence distribution. The substrate selectivity allows Rrp6p to act on many diverse, but not on all RNAs. Proteins with biological tasks similar to Rrp6p might also employ intrinsic substrate selectivity to discriminate against RNA subsets or even against very specific RNAs.
Materials and Methods
Protein Expression and Purification.
Yeast Rrp6p was expressed in Escherichia coli BL21 (DE3) CodonPlus (CP)-RIL (Stratagene) and purified as described (46). Rrp6p concentration was determined by averaging results from Bradford assay measurements and Coomassie staining using BSA as a standard (47).
RNA Substrates.
RNA substrates were purchased from Sigma and Dharmacon. RNA substrates were 5′ radiolabeled with [γ32P] ATP and T4 polynucleotide kinase (NEB) and purified with denaturing PAGE (48). RNA concentration was quantified by scintillation counting.
Substrate sequences are shown in Table 1.
Table 1.
Substrates used in this study
| Substrate name | Substrate sequence |
| A36 | 5′AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA 3′ |
| U36 | 5′UUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUUU3′ |
| C36 | 5′CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC3′ |
| A26C10 | 5′AAAAAAAAAAAAAAAAAAAAAAAAAACCCCCCCCCC3′ |
| C26A10 | 5′CCCCCCCCCCCCCCCCCCCCCCCCCCAAAAAAAAAA3′ |
| R24 | 5′GUACCCACACUACACAUAGCCACC3′ |
| R36 | 5′AGCACCGUAAAGACGCAAUCAUGCAGGGUCUGUCAG3′ |
| AU36 | 5′UUAUUUAUUAUUUAUUUAUUUAUUAUUUAUUUAUUA3′ |
| R41 | 5′GCGUCUUUACGGUGCUUAAAACAAAACAAAACAAAACAAAA3′ |
| A32-UCCA-3′ | 5′AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAUCCA3′ |
| A32-GGAC-3′ | 5′AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGGAC3′ |
| Triplet oligos | 5′AAA3′ or 5′GAC3′ |
| Quadruplet oligos | 5′AAAA3′ or 5′AGAC3′ |
Pre–Steady-State and Quasi–Steady-State Nuclease Reactions.
Degradation reactions were performed in 30 μL at 30 °C in a temperature-controlled aluminum block in buffer containing 40 mM Mops, pH 6.5, 100 mM NaCl, 0.5 mM MgCl2, 5% glycerol, 0.01% Nonidet P-40, 2 mM DTT. To minimize the impact of condensation on the tube lids, reactions that took less than 30 min were spun down in a minicentrifuge every 3 min. Reactions that took more than 30 min were performed by immersing the tubes in a temperature-controlled water bath. Prior to the reaction, Rrp6p at concentrations indicated was preincubated for 5 min in the reaction buffer.
Pre–steady-state reactions were started by addition of 5′-radiolabeled RNA substrate to a final concentration of [RNA*] = 1 nM. Quasi–steady-state reactions were performed under identical conditions with excess substrate over Rrp6p (combining radiolabeled RNA at [RNA*] = 1 nM with appropriate excess of identical, unlabeled RNA). Aliquots were removed from the reaction at times indicated. The reaction was stopped by addition to an equal volume of stop buffer (80% formamide, 0.1% xylene cyanol, 0.1% bromophenol blue). Samples were stored on ice for same-day PAGE analysis or at −20 °C for analysis at a later date.
Samples were applied to denaturing polyacrylamide sequencing gels (40 cm × 33 cm × 0.4 mm) (20% acrylamide:bis-acrylamide at 29:1, 7 M urea, 1× TBE). The gels were run on a Model S2 Sequencing Gel Apparatus (Life Technologies) with a Bio-Rad PowerPac 3000. Prior to loading, samples were heated to 95 °C for 5 min and the gels were prerun in 1× TBE for 1 h at a constant 40 W until the gel felt warm to the touch. Samples were loaded and gels were run at a constant 40 W for ∼4 h. Glass plates were removed and gels were dried and exposed overnight to a PhosphorImager cassette (43 cm × 35 cm; Amersham Biosciences). Individual bands were visualized on a Typhoon 9400 PhosphorImager (Amersham Biosciences) and quantified using the SAFA software (49). For a typical reaction with a 36-nt substrate and 15 timepoints, we quantified species with more than 3 nt and obtained ∼495 datapoints.
Pulse-Chase Nuclease Reactions.
Pulse-chase reactions were performed under conditions identical to those described for pre–steady-state reactions in a reaction volume of 20 μL. Reactions were initiated as described above and allowed to proceed for a defined period of time (t1). At this time, an excess of cold scavenger RNA (U36, in reaction buffer) was added to a final concentration of 10 μM. Control reactions confirmed complete prevention of Rrp6p rebinding at these scavenger concentrations used (SI Appendix, Fig. S1). After incubation for the indicated time t2, aliquots were removed and added to stop buffer. Samples were applied to and separated on denaturing PAGE as described above. Individual bands were visualized on a Typhoon 9400 PhosphorImager (Amersham Biosciences) and quantified using the SAFA software.
Calculation of Kinetic Parameters.
Kinetic parameters were obtained by global datafit of pre–steady-state, quasi–steady-state, and pulse-chase experiments, according to the strategy outlined in SI Appendix, Fig. S4A. Initial parameters for the global fit were observed rate constants (kobs) for each degradation step at various Rrp6p concentrations, obtained from pre–steady-state reactions, and processivity values (Pi) for each degradation step, obtained from pulse-chase reactions.
Values for kobs were determined with the GFIT software (50), which calculates rate constants for a series of irreversible, pseudo–first-order reactions. Starting values for the first 3 to 4 degradation products were determined by direct fitting of degradation timecourses to
| [1] |
where A0 ... Ai are substrate species (A0 corresponds to A36 for a 36-nt substrate). The derivation of Eq. 1 has been described in detail in a previous paper (51).
Values for kobs for each degradation step were plotted versus Rrp6p concentration and fitted to a binding isotherm to determine kobs at Rrp6p saturation (kobsmax):
| [2] |
For a subset of substrates (e.g., A36), the reaction was too fast for accurate measurements of kobs values at saturating Rrp6p concentrations at 30 °C. To nevertheless obtain these values, we performed reactions at lower temperatures (15, 20, and 25 °C) and extrapolated the kobsmax at 30 °C using the Eyring–Polanyi equation (52, 53).
Processivity values for individual steps were determined as described from the distribution of substrate species before and after pulse chase (51), according to
| [3] |
where A0 to Ai are relative concentrations of substrate species and P1 to Pi+1 are processivities of individual steps (P1 corresponds to the processivity of the degradation of the first nucleotide, e.g., A36 → A35). Eq. 1 was implemented into a computer routine using the Mathematica 6 platform (51). Experimental replicates for all analytical functions (SI Appendix, Table S1) were fitted separately and are reported as mean ± SD.
Global data fitting was performed using Kintek Global Kinetic Explorer, version 6.1 (54, 55). Data were fitted to the kinetic model in Fig. 2C. Values for kobsmax, K′1/2, and Pi, which corresponds to the ratio between cleavage and dissociation rate constant for each step (29), were transformed into initial parameters for association (konP), dissociation (koffP), and forward (kf) rate constants for the global data fit. Initial parameters for nonproductive binding (konNP and koffNP) were found using the Dynamic Simulation feature of the KINTEK software, using experimentally determined parameters for the respective productive reaction steps. The global data fit was performed multiple times for alternating combinations of fixed and floated variables until an overall fit with the lowest possible χ2 value was reached (for examples, see SI Appendix, Fig. S4). The overall quality of the obtained fit was further assessed by plotting the experimental datasets for distributions of substrate species vs. corresponding data calculated with the obtained fit parameters (SI Appendix, Fig. S4 C–E). FitSpace analysis was performed to determine the lower and upper boundaries for each kinetic parameter (55) (SI Appendix, Fig. S4 F and G). For a typical substrate, roughly 2,200 individual datapoints were used to calculate ∼90 kinetic parameters. Given error bars in the plots show the SEM for the respective datapoint for the global fit. Replicates for a given set of experiments are listed (SI Appendix, Table S1). Uncertainty values were calculated for each kinetic parameter by performing a FitSpace Analysis (55). Examples are given for A36 (SI Appendix, Fig. S4F) and R36 (SI Appendix, Fig. S4G).
Competitive Inhibition Experiments.
Inhibition studies (Fig. 5) were performed under conditions identical to those described for pre–steady-state reactions. Competitive inhibitors (3- or 4-nt oligos, final concentrations 0, 10, 25, 50, 75, 150, and 300 μM) were added with the substrate (A36, final concentration 1 nM) to a reaction mix containing 220 nM Rrp6p. Timepoints were collected over 30 min. Apparent Ki values were calculated by performing a global datafit to the kinetic model described above, augmented with a competitive inhibition step for each substrate species with a single Ki value. Only the Ki value was allowed to float in the fit; the other parameters were fixed with the values obtained in the global datafit described above.
Deconvolution of the Impact of Upstream RNA and 3′-Terminal Quadruplet to Free Binding Energy.
To define the impact of upstream RNA and the 3′-terminal quadruplet on the binding of Rrp6p, we calculated contributions from upstream RNA (ΔGUpstream) and from the 3′-terminal quadruplet (ΔGQuadrupl.) to overall free binding energy (ΔGTOTAL), according to
| [4] |
Overall free binding energy is related to the measured dissociation constants (K1/2) according to
| [5] |
For homopolymers, free energy (ΔGTOTAL) for each nucleotide was plotted and values for the respective 3′-terminal quadruplet were subtracted, yielding the remaining contribution of the upstream RNA (Fig. 5I and SI Appendix, Fig. S9 A and B). For substrates with repetitive sequences, free energy (ΔGTOTAL) values were plotted vs. nucleotide. The curves were fitted with polynomial terms (SI Appendix, Fig. S9 C and D). Values of ΔGQuadrupl. that were known for a given quadruplet were subtracted, yielding the ΔGUpstream contributions at the corresponding RNA length. Assuming a smooth function of ΔGUpstream vs. RNA length, values for ΔGUpstream for all RNA lengths of the substrates were obtained. Values for ΔGQuadrupl. for the quadruplets in the respective substrates were calculated according to Eq. 4. For substrates with more complex sequences, ΔGUpstream and ΔGQuadrupl. were obtained according to the strategy used for the substrates with repetitive sequences, using values of ΔGQuadrupl. for quadruplet variants determined from homopolymeric substrates and substrates with repetitive sequences. This analysis with the substrates used provided ΔGQuadrupl. values for 61 different quadruplet variants (Fig. 6A).
PWMs were calculated as previously described (32, 33, 56), as linear regression against the ΔGQuadrupl. values. A at each position was used as a reference (linear coefficient: 0).
Data Availability.
All data are available upon request from the corresponding author.
Supplementary Material
Acknowledgments
We thank Ulf-Peter Guenther and Xuan Ye for expert assistance in calculating the position weight matrices. This work was supported by National Institutes of Health Grants R35GM118088 (to E.J.), T32GM008056-32 (to A.A.), and R35GM118080 (to C.D.L). C.D.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913236117/-/DCSupplemental.
References
- 1.Zinder J. C., Lima C. D., Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31, 88–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid M., Jensen T. H., Controlling nuclear RNA levels. Nat. Rev. Genet. 19, 518–529 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Bresson S., Tollervey D., Surveillance-ready transcription: Nuclear RNA decay as a default fate. Open Biol. 8, 170270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan K. P., Butler J. S., Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 36, 6645–6655 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs M. W., Burkard K. T., Butler J. S., Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273, 13255–13263 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Makino D. L., Baumgärtner M., Conti E., Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 495, 70–75 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wasmuth E. V., Januszyk K., Lima C. D., Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature 511, 435–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonneau F., Basquin J., Ebert J., Lorentzen E., Conti E., The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139, 547–559 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Makino D. L., et al. , RNA degradation paths in a 12-subunit nuclear exosome complex. Nature 524, 54–58 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Zinder J. C., Wasmuth E. V., Lima C. D., Nuclear RNA exosome at 3.1 Å reveals substrate specificities, RNA paths, and allosteric inhibition of Rrp44/Dis3. Mol. Cell 64, 734–745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasmuth E. V., Lima C. D., Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell 48, 133–144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasmuth E. V., Lima C. D., The Rrp6 C-terminal domain binds RNA and activates the nuclear RNA exosome. Nucleic Acids Res. 45, 846–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuch B., et al. , The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 33, 2829–2846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedic E., et al. , Structural analysis of the yeast exosome Rrp6p-Rrp47p complex by small-angle X-ray scattering. Biochem. Biophys. Res. Commun. 450, 634–640 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Schneider C., Kudla G., Wlotzka W., Tuck A., Tollervey D., Transcriptome-wide analysis of exosome targets. Mol. Cell 48, 422–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomecki R., et al. , The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 29, 2342–2357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid M., et al. , Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol. Cell 47, 267–280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasken M. B., Laribee R. N., Corbett A. H., Nab3 facilitates the function of the TRAMP complex in RNA processing via recruitment of Rrp6 independent of Nrd1. PLoS Genet. 11, e1005044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox M. J., Gao H., Smith-Kinnaman W. R., Liu Y., Mosley A. L., The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1-Nab3 pathway. PLoS Genet. 11, e1004999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midtgaard S. F., et al. , Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl. Acad. Sci. U.S.A. 103, 11898–11903 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler J. S., Mitchell P., Rrp6, Rrp47 and cofactors of the nuclear exosome. Adv. Exp. Med. Biol. 702, 91–104 (2010). [PubMed] [Google Scholar]
- 22.Januszyk K., Lima C. D., Structural components and architectures of RNA exosomes. Adv. Exp. Med. Biol. 702, 9–28 (2010). [PMC free article] [PubMed] [Google Scholar]
- 23.Januszyk K., Liu Q., Lima C. D., Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA 17, 1566–1577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasmuth E. V., Lima C. D., The Rrp6 C-terminal domain binds RNA and activates the nuclear RNA exosome. Nucleic Acids Res. 45, 846–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q., Greimann J. C., Lima C. D., Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127, 1223–1237 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Webster M. W., Stowell J. A., Passmore L. A., RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not. eLife 8, e40670 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jankowsky E., Harris M. E., Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 16, 533–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phizicky E. M., Hopper A. K., tRNA processing, modification, and subcellular dynamics: Past, present, and future. RNA 21, 483–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali J. A., Lohman T. M., Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–380 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Jankowsky E., Gross C. H., Shuman S., Pyle A. M., The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature 403, 447–451 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Sengar A., Heddi B., Phan A. T., Formation of G-quadruplexes in poly-G sequences: Structure of a propeller-type parallel-stranded G-quadruplex formed by a G15 stretch. Biochemistry 53, 7718–7723 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Stormo G. D., Modeling the specificity of protein-DNA interactions. Quant. Biol. 1, 115–130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stormo G. D., Zhao Y., Determining the specificity of protein-DNA interactions. Nat. Rev. Genet. 11, 751–760 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y., Stormo G. D., Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol. 29, 480–483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibba M., Soll D., Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee K., Nostramo R. T., Wan Y., Hopper A. K., tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim. Biophys. Acta. Gene Regul. Mech. 1861, 373–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohan A., Whyte S., Wang X., Nashimoto M., Levinger L., The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA 5, 245–256 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dziembowski A., Lorentzen E., Conti E., Séraphin B., A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14, 15–22 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Lorentzen E., Basquin J., Tomecki R., Dziembowski A., Conti E., Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell 29, 717–728 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Lee G., Bratkowski M. A., Ding F., Ke A., Ha T., Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science 336, 1726–1729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominguez D., et al. , Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell 70, 854–867.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadaba S., Wang X., Anderson J. T., Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12, 508–521 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hopper A. K., Huang H. Y., Quality control pathways for nucleus-encoded eukaryotic tRNA biosynthesis and subcellular trafficking. Mol. Cell. Biol. 35, 2052–2058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson G., Coller J., Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K., Nicholson A. W., Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. U.S.A. 94, 13437–13441 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greimann J. C., Lima C. D., Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 448, 185–210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 48.Jankowsky E., Putnam A., Duplex unwinding with DEAD-box proteins. Methods Mol. Biol. 587, 245–264 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Das R., Laederach A., Pearlman S. M., Herschlag D., Altman R. B., SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levin M. K., Hingorani M. M., Holmes R. M., Patel S. S., Carson J. H., Model-based global analysis of heterogeneous experimental data using gfit. Methods Mol. Biol. 500, 335–359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia H., et al. , The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell 145, 890–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eyring H., The activated complex in chemical reactions. J. Chem. Phys. 3, 107–115 (1935). [Google Scholar]
- 53.Evans M. S., Polanyi M., Some applications of the transition state method to the calculation of reaction velocities, especially in solution. Trans. Faraday Soc. 31, 875–894 (1935). [Google Scholar]
- 54.Johnson K. A., Simpson Z. B., Blom T., Global kinetic explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Johnson K. A., Simpson Z. B., Blom T., FitSpace explorer: An algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 387, 30–41 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Guenther U. P., et al. , Hidden specificity in an apparently nonspecific RNA-binding protein. Nature 502, 385–388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the corresponding author.