Significance
There is great interest in selecting for host-associated microbiomes that confer particular functions to their host, and yet it remains unknown whether selection for a robust and stable microbiome is possible. Here, we use a microbiome passaging approach to measure the impact of host-mediated selection on the tomato phyllosphere (above-ground plant surfaces) microbiome. We find robust community responses to selection across replicate lines that are shaped by plant host genotype in early passages, but are genotype-independent in later passages. Work such as ours is crucial to understanding the general principles governing microbiome assembly and adaptation and is widely applicable to both sustainable agriculture and microbiome-related medicine.
Keywords: microbiome assembly, microbiome selection, microbiome engineering, experimental evolution, phyllosphere
Abstract
There is increasing interest in the plant microbiome as it relates to both plant health and agricultural sustainability. One key unanswered question is whether we can select for a plant microbiome that is robust after colonization of target hosts. We used a successive passaging experiment to address this question by selecting upon the tomato phyllosphere microbiome. Beginning with a diverse microbial community generated from field-grown tomato plants, we inoculated replicate plants across 5 plant genotypes for 4 45-d passages, sequencing the microbial community at each passage. We observed consistent shifts in both the bacterial (16S amplicon sequencing) and fungal (internal transcribed spacer region amplicon sequencing) communities across replicate lines over time, as well as a general loss of diversity over the course of the experiment, suggesting that much of the naturally observed microbial community in the phyllosphere is likely transient or poorly adapted within the experimental setting. We found that both host genotype and environment shape microbial composition, but the relative importance of genotype declines through time. Furthermore, using a community coalescence experiment, we found that the bacterial community from the end of the experiment was robust to invasion by the starting bacterial community. These results highlight that selecting for a stable microbiome that is well adapted to a particular host environment is indeed possible, emphasizing the great potential of this approach in agriculture and beyond. In light of the consistent response of the microbiome to selection in the absence of reciprocal host evolution (coevolution) described here, future studies should address how such adaptation influences host health.
The study of microbiomes (diverse microbial communities and their collective genomes) spans both basic and applied research in human health, agriculture, and environmental change. As our understanding of the ability of the microbiome to influence host health and shape host traits deepens, there is increasing interest in selecting and/or designing microbiomes for specific traits or functions. Such trait-based selection of microbiomes has the potential to shape the future of agriculture and medicine (1–3). In agriculture, below-ground microbiota have already proven capable of shifting the flowering time of plant hosts (4), enhancing drought resistance (5, 6), improving plant fitness (7), and even altering above-ground herbivory (8). However, long-term, repeatable success of future efforts will rely on a fundamental understanding of the assembly of, selection within, and coevolution among microbiota within these communities. One of the challenges facing successful, rational microbiome manipulation and assembly is disentangling the forces naturally shaping the community stability, including both host characteristics and microbial immigration. For example, in both humans and plants, there is conflicting evidence as to the relative importance of the environment versus host genotype in shaping the microbiome (9–17), and dispersal has been shown to override host genetics in an experimental zebrafish system (18).
One powerful but underutilized approach to understand and experimentally control for the factors shaping microbiome composition and diversity is experimental evolution. Measuring changes of populations or communities over time under controlled settings in response to a known selection pressure has proved a powerful force in gaining fundamental understanding of both host–pathogen (co)evolution (19) and microbial evolution (20). Here, we harness an experimental evolution approach in order to study how an entire microbial community can be selected upon in a plant host environment that varies across disease resistance-associated genotypes. We test the fundamental, yet relatively untested, assumption that a microbiome can be selected to adapt to its host in a robust fashion. We do so in the absence of selection on a particular plant-associated trait (e.g., flowering time or fecundity) in an attempt to capture how an entire community might naturally change over time to become well adapted to a host environment. To do this, we employ a microbiome passaging approach using the phyllosphere microbiome of tomato (Solanum) as a model system to select for a community that is capable of growth in this relatively oligotrophic environment and is resilient to perturbation via competition with a non-“adapted,” but more diverse community. The phyllosphere, defined as the aerial surfaces of the plant, is a globally important microbial habitat (21) and can shape important plant traits such as protection against foliar disease (22, 23) and growth (24, 25). Successful trait-based selection on the phyllosphere (previously undemonstrated) could therefore allow for enhancement of plant health, but this critically depends on the ability to select for a well-adapted microbial community that is relatively stable against invasion, particularly in open environments in which dispersal from neighboring hosts or the surrounding environment is inevitable.
We collected a diverse phyllosphere microbiome from tomatoes grown in an agricultural setting and transplanted it onto greenhouse-grown plants using a transplantation method shown to be effective for lettuce (26). We serially passaged this diverse microbiome on each of 4 cohorts of tomato plants (6 lines per cohort) of 5 different genotypes (pairs of near-isogenic Solanum lycopersicum genotypes that differed at known disease resistance loci, as well as a wild tomato accession, Solanum pimpinellifolium) for a total of 180 d. On each plant, during each passage, community assembly and dynamics might be driven by neutral processes or reflect positive or negative selection of specific taxa by the plant, dispersal of taxa from the greenhouse environment, and/or the other microbial taxa present. We therefore sought to characterize the relative importance of neutral versus deterministic processes both computationally using a neutral model and empirically using community coalescence experiments (27), in which communities from different passaged lines were combined together and reinoculated onto host plants in a common garden experiment. Overall, we were able to measure and characterize the response of the phyllosphere microbiome to selection in the plant host environment under greenhouse conditions, and our findings suggest selection for a stable and well-adapted plant-associated microbiome.
Results
Serial Passaging Experiment.
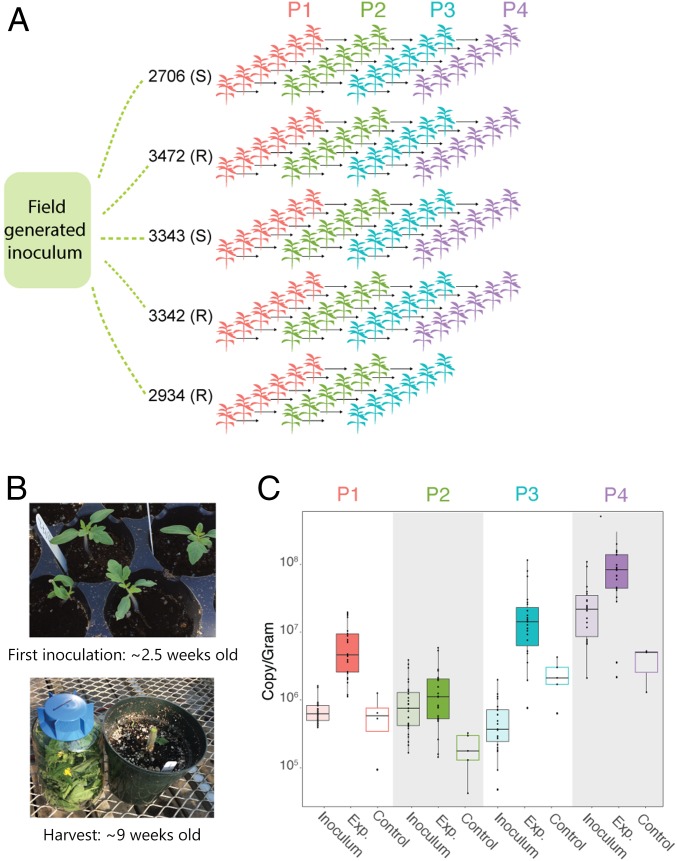
A diverse starting inoculum was collected from field-grown, mature tomato plants. This field microbiome was spray-inoculated onto 30 tomato plants of 5 different genotypes, with 6 replicates each (Fig. 1A). Two-week-old tomato plants were spray-inoculated once per week for 5 wk and then sampled in their entirety 10 d after the final inoculation (Fig. 1B). The phyllosphere microbiome of each plant was then individually passaged on these genetically distinct hosts over the course of 4 45-d passages: passage 1 (P1), P2, P3, and P4 (Fig. 1A; see Materials and Methods for details). Microbiomes were not pooled across plants within a given plant genotype, resulting in 30 independent selection lines. Control plants were inoculated with an equal volume of either heat-killed inoculum (P1) or sterile buffer (subsequent passages) every week. At the end of each passage, bacterial density was measured and normalized to the weight of each plant (Fig. 1C), and communities were sequenced by using 16S ribosomal RNA (rRNA) amplicon sequencing.
Fig. 1.
Serial passaging of the phyllosphere microbiome. (A) Experimental design of serial passaging experiment in which microbial inoculum from an agricultural tomato field was inoculated onto replicates of 5 genotypes and passaged for 4 passages. (B) Plants were first inoculated when they were ∼2.5 wk old, and the entire plant was sampled at ∼9 wk old. (C) Bacterial abundance was measured at the end of each passage from experimental (Exp.) and control plants by using ddPCR and normalized to the weight of each plant. Inoculum density was calculated as well. Note that our measures of bacterial growth likely overestimate the starting densities and do not account for population turnover (as a result of cell death and replacement within a passage) and are therefore highly conservative.
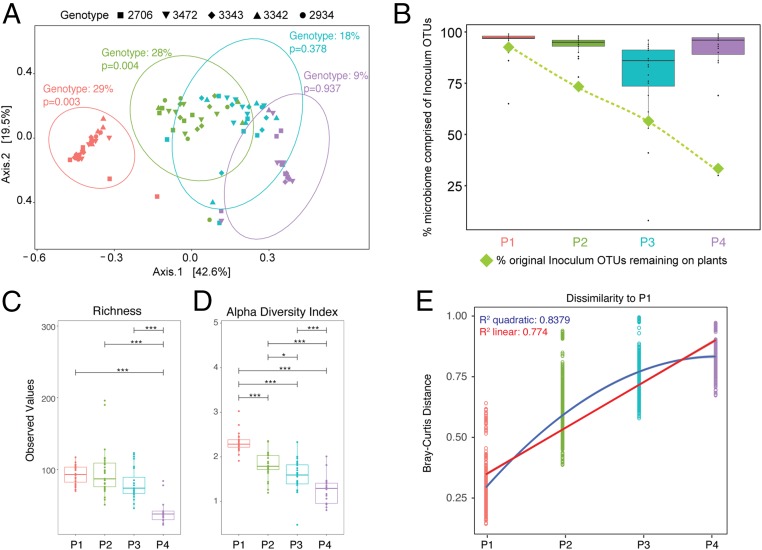
We first measured the impact of host genotype on bacterial community structure (Fig. 2A). Using Bray–Curtis dissimilarity measures, we performed multivariate permutational ANOVA tests (PERMANOVA) at each passage using the Adonis function in the Vegan R package (28, 29). We found that in P1, plant genotype explained 29% of dissimilarity between microbiomes (F4,27 = 2.331, P = 0.003). This result is robust to the removal of an outlying sample (see SI Appendix, Extended Methods for statistical results of that model). In P2, plant genotype similarly explains 28% of the variation in bacterial community dissimilarity (F4,24 = 1.906, P = 0.004). However, genotype becomes an insignificant driver of community composition in both P3 (R2 = 0.18, F4,23 = 1.018, P = 0.378) and P4 (R2 = 0.09, F3,19 = 0.527, P = 0.937). The 5 genotypes can be classified as pathogen “resistant” or “susceptible” based on known loci, and despite the overall effect of genotype at P1 and P2, there was no significant effect of disease resistance on Bray–Curtis dissimilarities, either overall or in any single passage. In some passages, an unequal number of samples across genotypes were analyzed due to exclusion of samples with poor sequencing quality. In order to account for this and ensure that the genotype effect observed in P1 and P2 was not due to heterogeneous dispersion of samples within a group, we tested for homogeneity of multivariate dispersions using the “betadisper” function in Vegan (30, 31). The betadispersion results were insignificant in both P1 (P = 0.234) and P2 (P = 0.231), indicating that the significant effects of genotype observed above were likely not an artifact of dispersion and do indeed reflect biological differences. To further test the robustness of these findings, we removed replicate lines from accession 2934 and reanalyzed the data. We did so because lines from accession 2934 were lost after P3 due to a stem-rot fungal pathogen present in the original inoculum that seemingly only infected this genotype. Significance of genotype in all passages was unchanged by exclusion of these lines from the dataset (see SI Appendix, Extended Methods for statistical details).
Fig. 2.
Bacterial community change over time. (A) Principal coordinates analysis (PCoA) plot of Bray–Curtis dissimilarity among samples shows a significant effect of genotype in P1 and P2 (determined by PERMANOVA tests). Ellipses indicate 95% confidence around the clustering. (B) The percent of original inoculum OTUs present at each passage was calculated (green diamonds), and the reads/sample of inoculum OTUs out of total reads was calculated for each plant at every passage and displayed on a box plot. (C and D) Plots of richness (C) and Shannon’s alpha diversity index (D) at each passage show a significant decrease over time. (E) Bray–Curtis dissimilarities between microbiomes in P1 were compared to those in P1, P2, P3, and P4, and linear and quadratic models were fit to the data. Corrected P values for multiple pairwise comparisons in C and D are indicated on the graph. *P ≤ 0.05; ***P ≤ 0.001.
We next sought to determine if there were more subtle influences of host genotype on the community that were not uncovered through analyzing Bray–Curtis dissimilarity alone. From the original inoculum sample, we identified 10 Operational Taxonomic Units (OTUs) using linear discriminant analysis effect-size (32) that were significantly associated with particular genotypes in P1 and P2. We compared their presence/absence at the end of P4 to those OTUs that were not found to be associated with genotype. Interestingly, those OTUs that were significantly associated with particular genotypes at the start of the experiment were significantly more likely to be present at the end of the experiment than those not associated with genotype (Fisher’s exact test, P = 0.013), suggesting that the loss of genotype effect observed was not driven by loss of particular genotype-associated OTUs.
In addition to genotype effects, we were interested in what other factors were driving the observed change in community composition. Using a multivariate PERMANOVA test on Bray–Curtis dissimilarity, we found that both the number of passages on tomato plants and the sample type (e.g., experimental, control, or inoculum) strongly shaped the microbial community (SI Appendix, Fig. S1: passage: R2 = 0.408, F3, 110 = 27.764, P = 0.001; sample type: R2 = 0.043, F5, 110 = 4.379, P = 0.001). Again, we found that the results of a betadispersion test were insignificant for both passage and sample type, indicating that the observed significant effects were likely not an artifact of unequal dispersion (passage: F3, 112 = 1.501, P = 0.201; sample type: F2, 113 = 1.457, P = 0 0.213). When inoculum and control samples were removed from the analysis, there remained both a significant effect of passage number and an overall effect of plant genotype (passage: R2 = 0.514, F3, 89 = 34.191, P = 0.001; genotype: R2 = 0.040, F4, 89 = 1.999, P = 0.001). In this model, we took into account that individual microbiome lines were passaged and sampled at each passage by performing the multivariate PERMANOVA test with Line ID used as strata. Note: We were unable to conduct a true nested time-series analysis with our multivariate data due to limitations of currently available statistical tests (see Materials and Methods for specific models and further discussion). As above, we performed a betadispersion test and found no significant effect of dispersion regarding genotype or passage (genotype: F4, 92 = 0.725, P = 0.58; passage: F3, 93 = 2.359, P = 0.077). Taken together, the results of these models indicate that the reported findings are robust to differences arising due to both repeated sampling of the same lines at each passage and unequal sample sizes between genotypes and passages.
We next sought to determine the role of dispersal of taxa among tomato plants on the greenhouse bench in shaping the phyllosphere microbiome over time. We did this by directly comparing the communities found on experimental and control plants. We calculated the proportion of OTUs on control plants that were from the inoculum that was sprayed onto experimental plants. At every passage, over 50% of inoculum OTUs were detectable on control plants, suggesting that dispersal in the greenhouse was occurring. Despite this, control and experimental plants were found to host significantly different communities at every passage (univariate PERMANOVAs: all P values < 0.04) with control plants hosting significantly lower bacterial abundance overall (Fig. 1C). Taken together, these data suggest that the effects of low levels of dispersal of taxa among plants in the experiment (as might be expected due to the plants’ proximity to one another and their randomization on the greenhouse bench) were minimal relative to the effects resulting from inoculations.
To better understand how the original, diverse, field inoculum changed over 4 passages on plants in the greenhouse, we calculated the percentage of OTUs in the original inoculum that were detectable over the course of the experiment (Fig. 2B, green diamonds). At the end of P1, 92% of the field inoculum OTUs were still present on the plants, but by P4, this number (when calculated across all plants) was reduced to 29%. We then calculated if the decrease in original community-member diversity was the result of replacement by noninoculum taxa (i.e., those that colonized plants over the course of the experiment). In this case, we observed that the proportion of sequencing reads (divided by total reads) representing the original inoculum OTUs remained above 78% (Fig. 2B, box plots). This indicates poor persistence of the majority of the original taxa from the field-grown plant inoculum, but those that remained seemed to dominate the community. This also suggests that a relatively small percentage of the community was made up of OTUs that colonized plants from the greenhouse environment. Furthermore, there was no visual indication (heat map presented in SI Appendix, Fig. S2) that a large portion of these noninoculum OTUs arrived and persisted on the plants for multiple passages. Of note, some OTUs considered “noninoculum” were likely present in the initial inoculum, but in too low of abundance to detect. To account for the impact of the small percentage of arriving species on community composition, we reanalyzed the dataset using only those OTUs that were observed to be present in the initial inoculum (SI Appendix, Fig. S3A). Using the same multivariate PERMANOVA models as above with permutations limited to within Line IDs, we found that passage number and genotype remained significant drivers of community dissimilarity (passage: R2 = 0.546, F3, 87 = 38.192, P = 0.001; genotype: R2 = 0.039, F4, 87 = 2.062, P = 0.001).
We next measured changes in bacterial density and diversity over the course of passaging and across lines. In P1, we estimated the fold change of bacterial abundance on control plants that were sprayed with heat-killed inoculum and found an average change of 0.76, which is significantly lower than the averaged 11-fold change for experimental plants which received live inoculum (Welch’s 2-sample t test, P < 0.0001). Using a repeated-measures ANOVA, we found an overall significant decrease in both OTU richness and alpha diversity over time across all plant genotypes (P < 0.001 for both). Significant differences between each passage were determined by multiple comparisons of means, and corrected P values (using Bonferroni corrections) are illustrated in Fig. 2 C and D and SI Appendix, Fig. S3B. Neither genotype nor overall disease resistance had a significant effect on richness and diversity at any passage. Importantly, the overall drop in diversity from P1 to P4 does not correspond to a decrease in overall bacterial abundance on plants (Fig. 1C). To test whether this decrease in richness and diversity could be driven by replacement of slower-growing taxa with fast-growing competitors, we analyzed 16S rRNA mean copy number as an indicator of bacterial ecological strategies (33–35). At each passage, we analyzed taxa that made up 95% of total reads. For each taxon, we recorded the mean 16S copy number for that particular family using the Ribosomal RNA Operon Copy Number Database (rrnDB) (36) and calculated the “copy number to relative abundance” ratio for each taxon at each passage (1 through 4). We found that there was no significant effect of passage on the “copy number to relative abundance” ratio (ANOVA: F3, 54 = 0.735, P = 0.536). There was also not a significant effect of passage on “copy number” (where copy number is not normalized to relative abundance of that taxon; F3, 54 = 0.738, P = 0.534). Finally, although passaging was performed in a controlled-temperature greenhouse, outside high and low temperatures and humidity all varied significantly across passages (SI Appendix, Fig. S4; ANOVA, P < 0.001 for all measures), which may have impacted the observed differences in both abundance and growth across passages.
With the knowledge that communities were drastically changing over time, we sought to determine if the rate at which the communities were changing was consistent. To do this, we calculated Bray–Curtis dissimilarity of microbiomes in each passage to P1 microbiomes (Fig. 2E). As we similarly observed through ordination plots in Fig. 1, the communities became more dissimilar to P1 over time. We then fit both a linear and quadratic regression to these data, and we found that both were significant, but there was a better fit of a quadratic model than linear as evidenced by higher R2 and lower Akaike information criterion (AIC) values (linear R2: 0.774, AIC: −3,563.231; quadratic R2: 0.8379, AIC: −4,414.637). When the regression models were compared using an ANOVA, we found that the quadratic model was a significantly better fit for the data (P < 0.0001), suggesting that the rate of community change may be slowing down. However, when we calculated Bray–Curtis dissimilarity across passages for each microbiome line, we observed no significant effect of “passage comparison” on Bray–Curtis dissimilarity (SI Appendix, Fig. S5; ANOVA: F1, 17 = 0.332, P = 0.572), suggesting that the community change may be slowing with respect to comparison to P1, but rate of change from one passage to another seems more constant. From the same model, we also found a moderately significant effect of Line ID on dissimilarity, indicating that some lines may have changed at a different rate than others (F26, 17 = 1.396, P = 0.052). We did not find there to be a significant interaction between Line ID and passage comparison (F20, 17 = 1.396, P = 0.246).
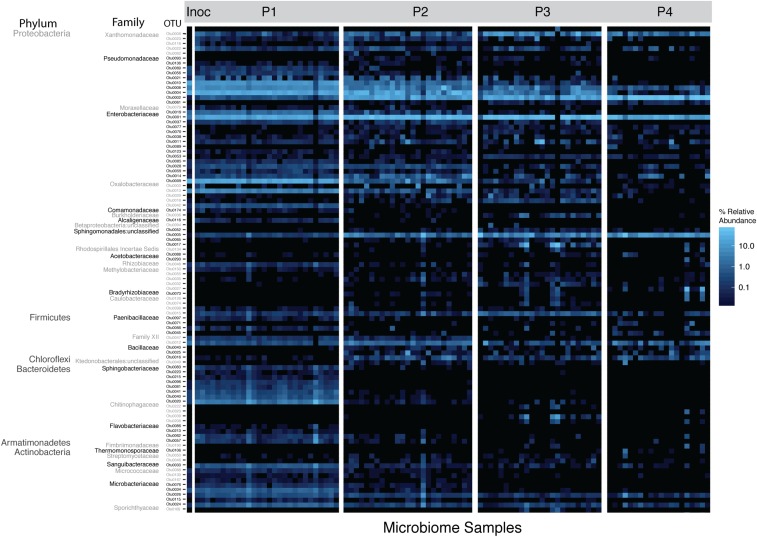
We next observed changes in relative abundance of specific taxa within lines over time (Fig. 3, top 100 OTUs plotted). At each passage, there were numerous taxa that were differentially abundant compared to other passages. In some cases, there was evidence for replacement of OTUs within taxonomic groups. For example, within the family Pseudomonadaceae, there were 3 OTUs that were differentially abundant between P1 and P4.
Fig. 3.
Changing relative abundance of top 100 OTUs. A heat map showing relative abundance of the top 100 OTUs illustrates the changing community composition at multiple taxonomic levels. Full taxonomy of OTUs is found in SI Appendix, Table S1. Inoc, inoculum.
Two Pseudomonads (OTU0010 and 0004) were in significantly higher relative abundance in P1 compared to P4 (paired-samples Wilcoxon test: P < 0.0001). As visualized in Fig. 3, these taxa gradually decreased in relative abundance over the course of passaging. An unclassified Pseudomonadaceae (0002) was significantly more abundant in P4 as compared to P1 (paired-samples Wilcoxon test: P < 0.0001). All 3 OTUs were present in the initial spray inoculum, although OTU0002 represented only 0.03% of rarified spray inoculum reads, whereas Pseudomonas OTU0004 represented 27%, and Pseudomonas OTU0010 represented 21%.
To better understand how bacterial community dynamics were changing over the course of the 4 passages, we utilized a recently developed cohesion metric to quantify connectivity of a microbial community (37). In brief, community cohesion is a computational method used to predict within-microbiome dynamics by quantifying connectivity of microbial communities based on pairwise correlations and relative abundance of taxa. Changes in community cohesion over time are suggestive of biotic interactions, where connectivity can arise from either, or both, positive and negative interactions resulting from cross-feeding (positive) or competition (negative) as well as environmental cofiltering. When applied to our dataset (SI Appendix, Fig. S6A), we found a minor but significant increase in positive cohesion values (among 200 permutations) from P1 to P4 (R2 = 0.19, P < 0.0001). Consistent with positive cohesion values showing increased biotic interactions, there were also increasingly negative cohesion values from P1 to P4, which, again, is minor but significant (R2 = 0.257, P < 0.0001). To further test our hypothesis that community change was due to deterministic processes, a null prediction was generated based on the known community composition of inocula applied at each passage, and we compared our observed communities to the predicted neutral community using a recently developed approach (38) (see Materials and Methods for complete details). We found that Bray–Curtis dissimilarities between predicted (null) and observed communities moderately increased over time (R2 = 0.261, P < 0.0001; SI Appendix, Fig. S6B), as would be expected if community change over the course of the experiment is the result of deterministic rather than stochastic processes.
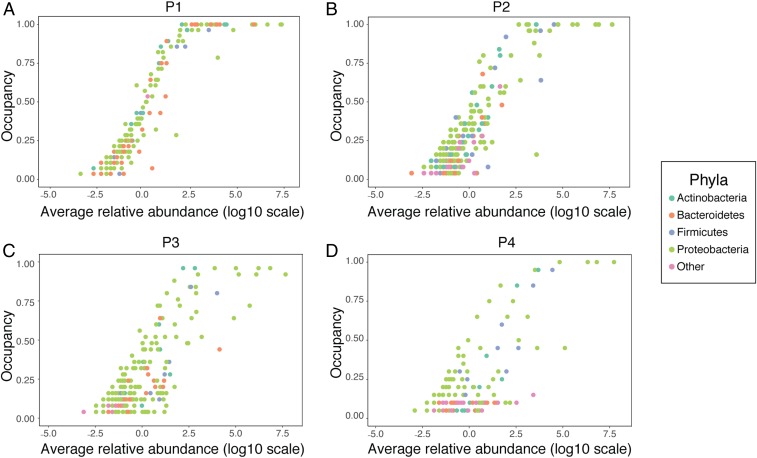
Further evidence for a shift away from neutrality can be observed by using occupancy–abundance curves in which the occupancy, or proportion of individuals in which an OTU is found, is plotted against its relative abundance (Fig. 4). A positive correlation between the 2 is expected to occur by chance, as observed in a neutrally assembled community, but a change in distribution of individuals may indicate a community shaped by deterministic processes (39, 40). When our data were visualized in this manner, we saw that in P1 (Fig. 4A), the most abundant taxa also occupied the highest proportion of plants, as you would expect in a neutral community not undergoing niche selection. However, this trend collapsed by P4 (Fig. 4D), with many abundant taxa occupying far fewer individuals than would be expected under neutrality. When regressions were fit to these distributions, there was an overall decrease in correlation between occupancy and abundance, regardless of whether a linear or polynomial regression was used. Fit to a linear model decreased from an R2 of 0.88 at P1 to 0.60 at P4. There was a significant effect of phyla on the linear model fit across all passages (ANOVA: F4, 12 = 5.318, P = 0.0107). Overall, a polynomial (n = 4) regression was a better fit to the data (P1: R2 = 0.96; P4: R2 = 0.66), but the effect of phyla in this case was insignificant (ANOVA: F4, 12 = 2.566, P = 0.0924).
Fig. 4.
Occupancy–abundance curves. For each OTU, its occupancy (or proportion of plant hosts in which it was found) is plotted against the log(10) of its relative abundance. OTUs belonging to a phylum other than those in the top 4 phyla are classified as “other.” (A) P1. (B) P2. (C) P3. (D) P4.
We next designed an experiment in which we could explicitly test the robustness of the shift away from neutrality by comparing empirical results to model predictions. The experimental design (SI Appendix, Fig. S7A) was to pool together all lines from the end of P4 and reinoculate this single inoculum onto replicate tomato plants across genotypes, mimicking the inoculation procedure from the 1st passage and allowing for a direct comparison to neutral models assuming a shared species pool. Bacterial communities on plants that received the P4-combined inoculum clustered apart from the uncombined P4 communities (SI Appendix, Fig. S7B). A PERMANOVA test on Bray–Curtis dissimilarity demonstrated that P4 bacterial communities were significantly different than P4-combined (R2 = 0.48, F1, 44 = 40.672, P = 0.001). Within just the P4-combined samples, we tested for the effect of genotype on Bray–Curtis dissimilarity. Unlike in P1, we did not observe an effect of genotype on the communities assembled from this combined inoculum (R2 = 0.045, F4, 24 = 1.339, P = 0.241). In the same multivariate PERMANOVA test, we also found that 76% of the variation between samples (R2 = 0.755, F1, 24 = 90.369, P = 0.001) was driven by an exceptional situation of introduction of a greenhouse taxon (OTU0003) to the plants (SI Appendix, Fig. S7C). To test if neutral processes were driving community structure in this experiment, we applied the Sloan neutral community model (41) to our data. This model assumes equal dispersal among hosts (and thus it could not be used for analysis of P2 to P4 data, as microbiomes were passaged without pooling.) In this case, as with P1, the assumption of equal dispersal potential among plants was met. In 200 iterative predictions, the fit of the neutral model was significantly higher in P1 (R2 = 0.87 ± 0.01) than P4-combined (R2 = 0.52 ± 0.05; Student’s t test, P <0.01), suggesting that neutral processes were dictating the community structure after the 1st passage, but not in the P4-combined experiment (SI Appendix, Fig. S7D). We also saw the occupancy–abundance relationship break down in P4-combined when compared to P1 directly (SI Appendix, Fig. S7E).
Mycobiome.
In an effort to understand how the fungal community changed overall from the 1st to the final passage, we used internal transcribed spacer (ITS) region amplicon sequencing to describe the fungal communities across lines in P1 and P4. We observed patterns that were similar in some regards to the bacterial communities. Using multivariate PERMANOVA tests as were performed for the bacterial dataset, we again found both a significant effect of passage number and sample type on fungal communities (SI Appendix, Fig. S8A; passage: R2 = 0.42, F1, 43 = 34.3948, P = 0.001; sample type: R2 = 0.048, F2, 43 = 1.976, P = 0.043). The significant effect of passage number remained after inoculum, control samples, and accession 2934 were removed, and Line ID were used as strata for permutations (SI Appendix, Fig. S8B; R2 = 0.472, F1, 38 = 34.021, P = 0.001). However, unlike in the bacterial community analysis, we found no significant differences in community composition between control and experimental plants when this was tested at each passage using a series of univariate PERMANOVAs (P1: F1, 21 = 2.1057, R2 = 0.09113, P = 0.066; P4: F1, 24 = 0.6479, R2 = 0.02629, P = 0.612). Additionally, we did not find an effect of host genotype at either passage (F4, 16 = 0.87756, R2 = 0.17992, P = 0.595; F3, 19 = 0.92402, R2 = 0.12732, P = 0.53). We also observed a significant decrease in both OTU richness (paired-samples Wilcoxon tests, P = 0.0316) and Shannon’s diversity (P = 0.0067) between P1 and P4 across all genotypes (SI Appendix, Fig. S8 C and D). In all analyses, there were no significant effects of disease resistance. Finally, analysis of the 5 most common taxa overall identified a single OTU, identified as Rhodosporidiobolus nylandii, which was not detectable in the inoculum or P1, but which dominated the fungal community in P4 (SI Appendix, Fig. S8E).
Testing Microbiome Adaptation Using Community Coalescence.
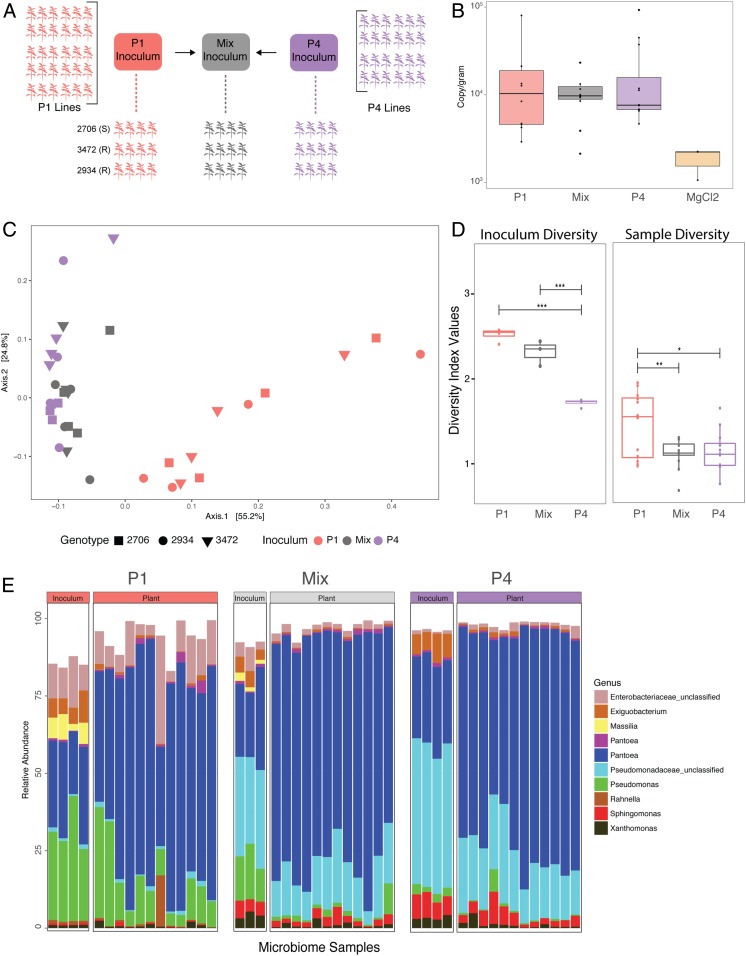
The similarity of changes in community structure both across replicates and genotypes over the course of the passaging experiment (Figs. 1–4) led us to predict that these microbiomes were becoming well adapted to the local plant conditions (by which we mean that the taxa present were positively selected for over time). To further determine if the community changes we observed from P1 to P4 were due to habitat selection rather than neutral processes, we employed a community coalescence competition experiment. In this experiment (Fig. 5A), phyllosphere communities from the end of P1 (pooled across all lines) and the end of P4 (again, pooled across lines) were inoculated onto a new cohort of plants, either on their own or in an ∼50:50 mixture of live cells (as determined by using live/dead propidium monoazide [PMA] treatment followed by droplet digital PCR [ddPCR]; see Materials and Methods for complete details). To ensure that our method for the Mixed inoculum was effective, we sequenced multiple replicates of the P1, P4, and Mixed inocula and began by comparing just these original inoculum samples. We found that inoculum source (e.g., P1, P4, or Mixed) explained 88% of dissimilarity among inocula (PERMANOVA: F2, 8 = 30.196, P = 0.002). A betadispersion test was insignificant, indicating that differences in inoculum samples were not due to heterogeneous variance (F2, 8 = 1.536, P = 0.28). To confirm that the Mixed inoculum was significantly different from both P1 and P4 separately, we compared P1 and Mixed inocula directly and found that 75% of the difference between samples can be explained by this variable (PERMANOVA: F1, 5 = 15.138, P = 0.022). Similarly, when P4 and Mixed were compared directly, 74% of variation in the community was explained (PERMANOVA: F1,5 = 13.999, P = 0.032). This consistent difference among the starting inocula allowed us to compare the communities colonizing plants from each treatment.
Fig. 5.
Testing microbiome adaptation. (A) Plants were inoculated with pooled, passaged microbiomes from the end of P1, P4, or a 50:50 mix of the 2. (B) Bacterial abundance was measured by using ddPCR. (C) A PCoA plot of Bray–Curtis dissimilarity (colored by inoculum source) shows that P1 plants have bacterial communities that are significantly different from P4 and Mixed plants, which are indistinguishable. (D) Shannon’s alpha diversity of the inoculum and experimental plants show significant differences between samples. (E) A bar graph illustrating composition of the top 10 OTUs shows differences in taxa among both the inoculum and experimental plants. Corrected P values for multiple pairwise comparisons in D are indicated on the graph. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Mix, Mixed.
We first measured final bacterial abundance and found that colonization was lower on these plants than in previous experiments, but did not significantly differ among treatments (ANOVA: F3, 32 = 0.971, P = 0.419), apart from control plants, where bacterial colonization was greatly reduced (Fig. 5B). We then compared bacterial communities again using 16S amplicon sequencing. Plants that received P1 inoculum have distinctly different communities than those that received either P4 or the Mixed inoculum (Fig. 5C). Plants that received the Mixed inoculum clustered together with those receiving P4 and were relatively indistinguishable. Using a multivariate PERMANOVA, we determined that inoculum source can explain 45% of Bray–Curtis dissimilarity among samples (F2, 31 = 13.486, P = 0.001), but there was no effect of plant genotype (R2 = 0.034, F2, 31 = 1.017, P = 0.376; although note that only 3 genotypes were used in this experiment). In a pairwise analysis between P1 and Mixed, inoculum source explained 39% of the community dissimilarity (PERMANOVA: F1, 22 = 13.988, P = 0.001). In contrast, inoculum source did not explain any significant variation in dissimilarity among P4 and Mixed inoculum plants (PERMANOVA: F1, 22 = 2.4378, P = 0.103). Together, these results suggest that the plants receiving the 50:50 Mixed inoculum were indistinguishable in community composition from those receiving the pooled, P4-passaged microbiomes, and thus that these selected communities were not invadable by the microbial communities from the start of the experiment. Consistent with our results from the passaging experiment itself, alpha diversity was found to be highest in P1 plants compared to both P4 and Mixed plants (Fig. 5D). Alpha diversity did not differ among communities colonizing plants from the P4 and Mixed inoculums, despite being different between the 2 inocula themselves. We also examined compositional makeup of the communities (Fig. 5E), and consistent with P1 to P4 passaging results, we saw differentially abundant taxa between groups (SI Appendix, Fig. S9). Again, 2 Pseudomonas OTUs were more abundant in P1 plants as compared to P4 and Mixed, in which there was an unclassified Pseudomonaceae that was higher in relative abundance.
Discussion
The impact of a microbiome on host health and fitness depends not only on which microbial organisms are present in the community, but also on how they interact with one another within the microbiome (42). Unlocking the great potential of microbiome manipulation and prebiotic/probiotic treatment in reshaping host health will therefore depend on our ability to understand and predict these interactions. We took a microbiome-passaging approach, inspired by classic experimental evolution, to test how selection for growth in the tomato phyllosphere under greenhouse conditions would impact microbiome diversity and adaptation across genotypes that differ in disease-resistance genes.
Across independently selected lines passaged on 5 tomato genotypes, we observed a dramatic shift in community structure and composition, accompanied by a loss of alpha diversity (Figs. 1 and 2). We cannot differentiate the relative contribution of evolutionary versus ecological change to the communities, but we expect both to have occurred within the time scale of these experiments. We also found that host genotype shapes bacterial community composition early in passaging (P1 and P2), explaining over 24% of variation among samples, but diminishes over time. We had originally predicted that disease resistance would impact the microbiome as a whole as a result of differing interactions with the host immune system. Interestingly, however, we did not observe an overall effect of resistance in shaping community composition. This suggests that there were other genetic differences among hosts that were driving the effect of genotype on microbiome composition in P1 and P2. In general, the relative importance of host genotype and environment in shaping microbiome composition remains highly debated. Our results suggest that the relative importance of genotype versus other factors, such as the growth environment or strength of within-microbiome interactions, changes over the course of passaging on a constant host background. It is possible that genotype-driven differences may become subtler after selection, and thus we are unable to detect them by OTU analysis. Future studies taking a more fine-scale resolution may be able to detect subtler effects when overall taxa richness decreases. We did find that even in the absence of a strong genotype effect, there remains a legacy of genotype effect, in that OTUs found to be significantly associated with particular genotypes early on are more likely to be present at the end of passaging than those that did not exhibit any host preference.
In order to test if the phyllosphere microbiome undergoes habitat filtering, we chose to begin the experiment with a diverse inoculum. This starting community generated from field-grown tomato plants likely contained microbes from other surrounding plant species, dust, soil, and other sources. In particular, neighboring plants have been shown to contribute to both the density and composition of local airborne microbes (43). We found that although the total number of these field inoculum OTUs decreased over the course of the experiment, the taxa that remained consistently made up 78 to 95% of the community. This provides strong evidence that the original spray inoculum underwent niche selection over the course of the experiment. We also saw evidence for niche selection through changing occupancy–abundance distributions. Increased incidents of high-abundance, low-occupancy taxa in P4, or “clumping” (39), is suggestive of niche selection. Gonzalez et al. (40) found a similar breakdown of occupancy–abundance relations in animal communities using miniature moss microcosms. The authors predicted that this was due to dispersal limitation, as their experimental design created habitat fragmentation, and they did not observe this similar decline in correlation in communities that were connected by “habitat corridors.”
In this work, we detect some evidence for dispersal both among plants and from the environment onto the plants. Specifically, we consistently find OTUs on control plants that originated from the spray inoculum that experimental plants received, indicating that some taxa were spread among all plants via, for example, water splash, touching leaves, or insects. We also find a continual influx of taxa from the greenhouse environment onto tomato plants (SI Appendix, Fig. S2), but these taxa do not appear to be establishing themselves on the plants and displacing resident microbes. Taken together, we conclude that dispersal was present in our system, but not sufficient to explain the patterns we observe. Importantly, the key findings that microbiomes vary among genotypes in P1 and P2 and that the communities are well adapted to their environment after 4 passages, are robust to the low levels of dispersal that are likely to have occurred. Future experiments should include filter traps or “fake plants” in order to explicitly test the prevalence and importance of dispersal in the system. Such controls could also be used to measure the role of ecological drift in shaping a community over time, independent of the host.
To directly test the alternative hypothesis that community changes were due to neutral processes such as bottlenecking, ecological drift, or random dispersal as discussed above, we first fit our data to neutral and null models, finding a poorer fit over time. We next experimentally tested for nonneutral microbiome adaption by conducting a community coalescence experiment to measure fitness of passaged microbiomes as compared to those from the start of the experiment. The results of this experiment strongly support the idea that these phyllosphere microbiomes adapted to the plant host environment over the course of 4 passages (Fig. 5). Independent of overall bacterial abundance, P4 microbiomes were able to outcompete the less-adapted P1 microbiomes. One potential explanation for this ability of P4 communities to outcompete P1 is that the taxa that do particularly well in this environment, and are able to reach higher abundances at the end of P4, outcompete the taxa from P1 because they are at higher densities in the Mixed inoculum. However, it is not clear how these possible density effects could be distinguished from the possibility that they are better adapted to the environment. Future work focusing on bacterial functional traits and/or culture-based experiments in which taxa are applied in different relative abundances could help shed insight as to whether the observed competitive interactions were the result of density-dependent effects, competition, or both.
The community coalescence approach (27) allowed us to demonstrate nonneutral selection of a bacterial community that is independent of host genotype and resistant to invasion by a more diverse, nonselected community. This approach was used by others in a study conducted on methanogenic bacterial communities (44). The authors found that when multiple methanogenic communities were combined, a single dominant community emerged from the mix. This emergent dominant community resembled the single community with the highest methane production that went into the combination, suggesting that the most-fit community is capable of reassembly, even in the presence of other community members.
While adaptation to both the local host environment (tomato plants and host genotype) and the larger environment (the greenhouse) were likely driving the increasingly nonneutral selection over time, the strength of within-microbiome biotic interactions likely also increased over the course of the experiment. We saw evidence for this through both increasing positive and negative community cohesion values. We also uncovered a strong effect of a greenhouse-acquired taxon on the community in 1 of the experiments (SI Appendix, Fig. S6). Though we were not able to determine what drove certain plants to be more colonized by this taxon than others, we did observe strong shifts in community composition associated with its relative abundance that may be due to spatial organization of plants in the greenhouse and/or stochastic initial colonization events. In a greenhouse study conducted on Arabidopsis thaliana phyllosphere communities, the authors found that abundance of certain dominant taxa could be tied to spatial organization of the plants that was likely driven by early stochastic events (15).
Although we focused primarily on the bacterial portion of the microbiome, the mycobiome changed over the course of passaging as well (SI Appendix, Fig. S7). Similar to the bacterial community, we observed a significant decrease in diversity and richness from P1 to P4, and we also saw changing community composition. We did not observe any effect of genotype on the fungal community, but the low richness of fungi we recovered from leaf surfaces may have impeded our ability to detect genotype-driven differences. It may be the case that the dominant fungal taxa analyzed (epiphytic yeasts) were not impacted by host genotype. Previous work that demonstrates plant genotype influences the fungal community has primarily included endophytes in addition to epiphytes in their collection and analysis (45–47). The overall low richness of fungi we uncovered may be attributed to our experimental methods, particularly the process of collecting microbes via sonication, which may have biased passaging toward bacterial taxa and fungal epiphytes. Yeasts are thought to be the dominant epiphytical fungal group in the phyllosphere (48), and, indeed, we find yeast to be in the highest relative abundance compared to filamentous fungi. Although it is possible that multikingdom interactions played a role in shaping community composition [as has been demonstrated in A. thaliana (49)], we were unable to perform these analyses due to the relatively few number of fungal taxa that our analyses included. Similarly, our passaging method (e.g., pelleting and removing supernatant at each passage) would have selected against any free viruses—bacteriophages, mycovirsues, or others. Thus, any effect of viruses on the microbiome were eliminated from this study, although we previously found that bacteriophages are capable of altering both abundance and composition in the tomato phyllosphere (50). It is possible that within-microbiome interactions may be contributing to the parallel changes observed over time in the passaged lines. For this reason, and because there is increasing interest in taking a multikingdom approach to studying the microbiome, future work should be designed in a way that enhances the collection and analysis of the complete microbiome, although technical limitations often hinder our ability to do so.
Given the naturally distinct spatial structure, ease of sampling, high culturability, and demonstrated role in plant health (24, 51), the phyllosphere microbiome is an ideal model for testing theories of niche selection and microbiome adaptation, as we have done here. Through spray inoculation, the environment can be evenly saturated with diverse inoculum, and it is possible to sample the successfully colonized community its entirety. Moreover, bacterial abundance and growth can be tracked by using ddPCR, and communities can be described by using next-generation sequencing. We were able to use the phyllosphere model to not only select upon entire host-associated microbial communities, but to then experimentally test our hypotheses regarding microbiome adaption in subsequent experiments. These results also underscore the need for proper no-selection control lines in any study evolving microbiomes that confer a particular host-level trait.
Through this work, we also shed light on a notable challenge in microbiome research. One intriguing interpretation of our data is that when describing the microbiome of an open environment, such as plant surfaces, many of the taxa found there may be transient visitors. In the case of the phyllosphere, there are microbes on leaf surfaces that may have emigrated from air, soil, surrounding plants, or other nonplant habitats and do not necessarily represent an adapted community that is capable of growth and persistence. Passaging of microbiomes in the absence of specific trait-based selection, as we have done here, is a powerful way of differentiating those taxa that are, or can rapidly become, well adapted to the plant host environment. It also raises the question as to if a microbiome should be defined as the community that is found upon sampling and sequencing, or if a true microbiome is one that is adapted to its host or environment.
Overall, we were able to show robust habitat selection of these communities over relatively short plant-host time scales. These results uncover great promise of this approach and system for answering fundamental questions about the forces shaping microbiome assembly over time and also pave the way for selecting stable, uninvadable host-associated microbiomes, which may inform rational microbiome manipulation and probiotic design. Experiments such as these are crucial if we are to understand general principles governing microbiome assembly and adaptation and use this knowledge for transformative applications in both medicine and agriculture.
Materials and Methods
See SI Appendix for extended methods.
Tomato Accessions.
Tomato accessions were obtained from the Tomato Genetics Resource Center (TGRC). Five tomato genotypes were used: S. lycopersicum money maker disease susceptible (TGRC 2706); S. lycopersicum money maker disease resistant (TGRC 3472); S. lycopersicum Rio Grande disease susceptible control for TGRC 3342 (TGRC 3343); S. lycopersicum Rio Grande disease resistant (TGRC 3342); and S. pimpinellifolium wild ancestor (2934).
Tomato Germination and Growth.
Seeds were surface-sterilized by using TGRC recommendations and then were transferred onto 1% water agar plates and placed in the dark at 21 °C until emergence of the hypocotyl and then moved into a growth chamber and allowed to continue germination for 1 wk. After approximately 1 wk, seedlings were planted in sunshine mix #1 soil in seedling trays. After approximately 1 more week of growth, seedlings were transplanted into 8” diameter pots, making the plants ∼2.5 to 3 wk old at the first time of microbial inoculation. Age of inoculation varied slightly from experiment to experiment, but it was kept identical among genotypes within an experiment.
Inoculation Preparation, 1st Passage.
Microbial inoculum for the 1st passage of the experiment was generated from field-grown tomato plants from the University of California, Davis (UC Davis) Student Organic Farm collected in September and October of 2016. Above-ground plant material was collected from various genotypes of tomatoes across 9 different sites spread through 4 fields. Plant material was submerged in sterile phosphate freezing buffer and sonicated for 10 min in a Branson M5800 sonicating water bath. The resulting leaf wash from each site was pooled and divided into 6 aliquots and stored in glycerol freezing buffer. For each inoculation in the 1st passage, an aliquot was thawed, and cells were pelleted and resuspended in 200 mL of 10 mM MgCl2 buffer. Of this, 40 mL was heat-killed in an autoclave for a 30 min at 121 °C. Both live and heat-killed inoculum was plated. There was no growth from heat-killed inoculum, and the live-inoculum concentration was calculated to be 1.1 × 106 colony-forming units/mL. Soil from each site, which had been stored at −20 °C, was combined in a sterile bucket and thoroughly mixed before inoculation.
Inoculation Procedure.
Soil inoculation.
The top layer of every pot was supplemented with 40 g of UC Davis Farm Soil. Soil inoculation was only performed once and only for the 1st passage of plants.
Spray inoculation.
Each plant was sprayed with 4.5 mL of inocula using misting spray tops. Control plants from passage 1 were inoculated with the heat-killed inocula. Control plants from P2 onward were inoculated with sterile 10 mM MgCl2. Immediately after inoculation, plants were placed in a random order in a high-humidity misting chamber for 24 h. After 24 h, the plants were moved to a greenhouse bench. Plants were inoculated once per week in the same manner and were placed in the misting chamber for 24 h after every inoculation.
Plant Sampling and Inoculation Preparation for Passaging Lines.
Ten days after the final spray inoculation, plants were sampled. With the exception of the P4-Combined experiment, all plants were cut off at the base and immediately placed into sterile 1-L bottles individually. By the end of P4-Combined, the plants had grown too large to sample the entire plant, and, instead, roughly 2/3 of the plant material was sampled from each plant, with care taken to sample the same age of branches from every plant. After collection, plant material was weighed, sterile buffer was added, and the entire bottle was sonicated as above. Half of the volume from each plant was pelleted and resuspended in ∼1 mL of 1:1 KB broth glycerol and stored at −80 °C for inoculation of the subsequent passage. The other half of the volume was pelleted and stored as a pellet at −20 °C for DNA extractions. To prepare inoculation of the next passage, microbiome glycerol stocks were thawed, briefly pelleted to remove glycerol, and resuspended in sterile 10 mM MgCl2.
Inoculation Preparation, Combination of P4 Microbiomes (SI Appendix, Fig. S7).
Frozen microbiomes from all plants from the end of passage 4 were thawed, and half the volume was removed from each aliquot. These aliquots were combined into 1 pooled metainoculum. This was divided into 6 aliquots. One was used immediately, and the rest of the aliquots were stored at −20 °C in KB glycerol and thawed by aliquot for each week of inoculation, as above.
P1, P4 Coalescence Experiment (Fig. 5).
Genotypes 2706, 3472, and 2934 were used for this experiment, and 4 plants of each genotype received each treatment (P1, P4, and Mixed). One control plant of each genotype was spray-inoculated with MgCl2. To prepare the inoculum, each passaged microbiome line from the end of P1 was combined. The same was done for each passaged microbiome line from the end of P4. In order to quantify only live cells, we used PMA treatment, using a method adapted from others (52), prior to ddPCR quantification (see below). Bacterial concentration was matched to 7.7 × 106 cells/mL. For the P1/P4 Mixed inoculum, pooled P1 and P4 microbiomes were combined at 50:50 ratio, based on bacterial abundance. Final total concentration was equal to that of P1 and P4. Plants were inoculated for 3 wk and harvested 10 d after the final inoculation as described.
Bacterial Quantification Using ddPCR.
The Bio-Rad QX200 system was used for culture-independent quantification of bacteria. Complete ddPCR methods are described elsewhere (50). Bacterial abundance was measured directly after microbes were sonicated off plant surfaces into sterile buffer. For consistency, the same region of the 16S gene used below for amplicon sequencing was used for bacterial quantification. Peptide nucleic acids (PNAs) were used as well to limit any background amplification of plant mitochondrial or chloroplast DNA. All data were normalized to weight, in grams, and concentrations are reported as 16S copy number/gram.
DNA Extractions.
DNA was extracted from microbial pellets by using the Qiagen PowerSoil DNA-extraction kit. A buffer control extraction was included for every set of extractions in order to identify and exclude taxa present in the dataset due to buffer contamination.
The 16S Libraries.
The 16S rRNA gene was amplified by using dual-indexed primers designed for the V3–V4 region (53) using the following primers: 341F (5-CCTACGGGNBGCASCAG-3) and 785R (5-GACTACNVGGGTATCTAATCC-3) (54). Additionally, we also used PNAs (55) to decrease amplification of plant mitochondrial and chloroplast DNA. Negative buffer controls and PCR controls were sequenced along with experimental samples. Amplicons from each sample were pooled in equimolar concentrations and cleaned by using an AMpure bead clean-up kit. Libraries were prepared for paired 300-paired-end reads in Illumina’s MiSeq V3 platform (Illumina) at The California Institute for Quantitative Biosciences (QB3) at the University of California, Berkeley (UC Berkeley).
ITS Libraries.
By using the same DNA as above, the ITS2 region was amplified using ITS9-F: GAACGCAGCRAAIIGYGA and ITS4-R: TCCTCCGCTTATTGATATGC following a protocol published online by the Joint Genome Institute (56). A 2 PCR was performed (7 cycles) in order to anneal MiSeq illumunia adapters and barcodes onto the amplicons. PCRs were carried out in duplicate and pooled before they were prepared for sequencing by the QB3 sequencing facility as described above.
Sequence Processing and Data Analysis.
MiSeq sequencing files were demultiplexed by the QB3 sequencing facility. Fastq files for all samples are deposited in the NCBI BioProject database (BioProject ID PRJNA57876; ref. 57). Bacterial reads were combined into contigs by using VSearch (58), and the remainder of the analysis was carried out in Mothur (59) following their MiSeq standard operating procedure (60) (see SI Appendix for specifics). We used a 97% similarity cutoff for defining OTUs and the Silva reference database (61) for taxonomic assignment. Bacterial reads were rarified to 8,000 reads per sample. For the fungal community, an OTU table was generated from the fungal community sequencing data by using QIIME2 (Version 2018.8) (see SI Appendix for specifics). Reads were clustered into OTUs at 97% identity and assigned taxonomy by using the UNITE database and the feature-classifier plug-in (62). Fungal reads were rarified to 415 reads per sample. Once bacterial and fungal OTU tables were generated in Mothur and QIIME2, the remainder of the analysis was performed in R by using the following packages: PhylosEq (63), vegan (28), ampvis2 (64), and MicrobiomeSeq (65). Occupancy–abundance curves were generated by using the “Trifolium nodule microbiome analysis script” (66).
Incorporation of Repeated Measures into Statistical Models.
In the serial passaging experiment, each microbiome line was independently passaged across 4 cohorts of tomato plants, and each microbiome line was sampled at the end of each passage. Although the microbiomes were never sampled multiple times from the same tomato plant, the data structure is similar to what one would find in time series experiment. Thus, wherever possible, “Line ID” was incorporated into models to take this into account. The following linear mixed-effects model was utilized for determining significant changes in diversity over time: lmer(Values ∼ Passage + (1|LineID)). In the case of PERMANOVA tests, the strata term was used to limit permutations within Line IDs to test for the main effect of Passage. Statistics are presented by using the following models with the use of strata: adonis(bray.matrix ∼ Passage + Genotype, permutations = 999, strata = LineID). The adonis2 test with the by = ”margin” term was used whenever the strata term was not included in the model. The following model was utilized in these cases: adonis(bray.matrix ∼ Passage + SampleType, by = ”margin”, permutations = 999). See SI Appendix for further discussion.
Community Cohesion Metrics.
The estimations of positive and negative cohesion values follows the cohesion metrics approach proposed by Herren and McMahon (37). We modified their method to estimate cohesion values by using 2 relative abundance profiles of a training set and test set. See SI Appendix for further details.
Neutral Model.
The neutral model we used was proposed by Sloan et al. (41) to describe both microbial diversity and taxa-abundance distribution of a community. Burns et al. (18) have developed a R package based on Sloan’s neutral model to determine the importance of neutral processes to community assembly. In brief, the neutral model creates a potential neutral community by a single free parameter describing the migration rate, m, based on 2 sets of abundance profiles—a local community and metacommunities. The local community describes the observed relative abundance of OTUs, while the metacommunity is estimated by the mean relative abundance across all local communities. The estimated migration rate is the probability of OTU dispersal from the metacommunity to replace a randomly lost individual in the local community. The migration rate can be interpreted as dispersal limitation. In each microbiome passage, half of the samples were randomly selected and the relative abundance profile at the OTU level was used. The neutral model fit and migration rate were estimated in the resolution results of 200 iterations for P1, P2, P3, P4, and P4 Combined.
Null Model Predictions.
We applied a null model approach on the serial passaging data P1 to P4 to characterize the changes of stochastic process driving the assembly of plant microbiome over time. Lines that had high-quality sequencing data at every time point (13 in total) were used for this analysis. The null scenario for each line at each passage was generated by using the data for that same line at the previous passage. The null scenario of P1 was generated by using the original field inoculum sample. The null model approach was based on community pairwise dissimilarity proposed by Chase and Myers (67) and extended by Stegen et al. (68) to incorporate species abundance. We also incorporated species relative abundance into the procedure proposed by Zinger et al. (38) has developed R code for the null model and applied the null model approach on the soil microbiome. This approach does not require a priori knowledge of the local community condition and determines if each plant microbiome at the current passage deviates from a null scenario generated by that same microbiome at the previous passage. See SI Appendix for further details.
Data Availability.
All raw sequences files can be found on NCBI using the BioProject ID PRJNA578761. R code that was used for analysis can be provided upon request.
Supplementary Material
Acknowledgments
We thank the UC Davis Student Farm, who provided access to the fields from which the original inoculum was generated; Christina Winstrom and the Oxford Tract greenhouse staff for their role in plant care throughout the experiments; Shirley Zhang for assistance with plant inoculation; members of the Coleman-Derr laboratory at UC Berkeley for their helpful input on analysis of these data; and Dylan Smith and Shana McDevitt for their continued support with sequencing efforts for the experiment. We thank Academia Sinica, Taiwan and the affiliated faculty Dr. Daryi Wang, which provided the International Research Short-Term Visiting Scholarship Fund for F.C.-H.W. to conduct research at UC Berkeley. The work was supported by NSF Grant DEB 1754494.
Footnotes
The authors declare no competing interest.
Data deposition: Data have been deposited in the NCBI BioProject database, https://www.ncbi.nlm.nih.gov/bioproject (BioProject ID PRJNA578761).
See Commentary on page 808.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908600116/-/DCSupplemental.
References
- 1.Gopal M., Gupta A., Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 7, 1971 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orozco-Mosqueda M. D. C., Rocha-Granados M. D. C., Glick B. R., Santoyo G., Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 208, 25–31 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mimee M., Citorik R. J., Lu T. K., Microbiome therapeutics—Advances and challenges. Adv. Drug Deliv. Rev. 105, 44–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panke-Buisse K., Poole A. C., Goodrich J. K., Ley R. E., Kao-Kniffin J., Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9, 980–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marasco R., et al. , A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7, e48479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolli E., et al. , Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 17, 316–331 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Fitzpatrick C. R., Mustafa Z., Viliunas J., Soil microbes alter plant fitness under competition and drought. J. Evol. Biol. 32, 438–450 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Pineda A., Kaplan I., Bezemer T. M., Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 22, 770–778 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Wagner M. R., et al. , Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 7, 12151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodenhausen N., Bortfeld-Miller M., Ackermann M., Vorholt J. A., A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 10, e1004283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello E. K., et al. , Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson A. K., et al. , Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18933–18938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spor A., Koren O., Ley R., Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 9, 279–290 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Micallef S. A., Channer S., Shiaris M. P., Colón-Carmona A., Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal. Behav. 4, 777–780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maignien L., DeForce E. A., Chafee M. E., Eren A. M., Simmons S. L., Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 5, e00682-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothschild D., et al. , Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Laforest-Lapointe I., Messier C., Kembel S. W., Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome 4, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns A. R., et al. , Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl. Acad. Sci. U.S.A. 114, 11181–11186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebert D., Experimental evolution of parasites. Science 282, 1432–1435 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Buckling A., Craig Maclean R., Brockhurst M. A., Colegrave N., The Beagle in a bottle. Nature 457, 824–829 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Morris C. E., Kinkel L., “Fifty years of phyllosphere microbiology: Significant contributions to research in related fields” in Phyllosphere Microbiology, Lindow S. E., Hecht-Poinar E. L., Elliott V., Eds. (APS Press, St. Paul, MN, 2002), pp. 365–375. [Google Scholar]
- 22.Berg M., Koskella B., Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 28, 2487–2492.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Innerebner G., Knief C., Vorholt J. A., Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3210 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fürnkranz M., et al. , Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J. 2, 561–570 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Stone B. W. G., Weingarten E. A., Jackson C. R., The role of the phyllosphere microbiome in plant health and function. Annu. Plant Rev. Online 2018, 1–24 (2018). [Google Scholar]
- 26.Williams T. R., Marco M. L., Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. MBio 5, e01564-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rillig M. C., et al. , Interchange of entire communities: Microbial community coalescence. Trends Ecol. Evol. 30, 470–476 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Dixon P., Palmer M. W., VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 29.Anderson M. J., A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001). [Google Scholar]
- 30.Anderson M. J., Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Anderson M. J., Ellingsen K. E., McArdle B. H., Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9, 683–693 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Segata N., et al. , Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klappenbach J. A., Dunbar J. M., Schmidt T. M., rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roller B. R. K., Stoddard S. F., Schmidt T. M., Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1, 16160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrestha P. M., Noll M., Liesack W., Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ. Microbiol. 9, 2464–2474 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Stoddard S. F., Smith B. J., Hein R., Roller B. R., Schmidt T. M., rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herren C. M., McMahon K. D., Cohesion: A method for quantifying the connectivity of microbial communities. ISME J. 11, 2426–2438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zinger L., et al. , Body size determines soil community assembly in a tropical forest. Mol. Ecol. 28, 528–543 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Wright D. H., Correlations between incidence and abundance are expected by chance. J. Biogeogr. 18, 463–466 (1991). [Google Scholar]
- 40.Gonzalez A., Lawton J. H., Gilbert F. S., Blackburn T. M., Evans-Freke I., Metapopulation dynamics, abundance, and distribution in a microecosystem. Science 281, 2045–2047 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Sloan W. T., Woodcock S., Lunn M., Head I. M., Curtis T. P., Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb. Ecol. 53, 443–455 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Gould A. L., et al. , Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. U.S.A. 115, E11951–E11960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lymperopoulou D. S., Adams R. I., Lindow S. E., Contribution of vegetation to the microbial composition of nearby outdoor air. Appl. Environ. Microbiol. 82, 3822–3833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sierocinski P., et al. , A single community dominates structure and function of a mixture of multiple methanogenic communities. Curr. Biol. 27, 3390–3395.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Qian X., et al. , Host genotype strongly influences phyllosphere fungal communities associated with Mussaenda pubescens var. alba (Rubiaceae). Fungal Ecol. 36, 141–151 (2018). [Google Scholar]
- 46.Sapkota R., Knorr K., Jørgensen L. N., O’Hanlon K. A., Nicolaisen M., Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol. 207, 1134–1144 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Bálint M., et al. , Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS One 8, e53987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whipps J. M., Hand P., Pink D., Bending G. D., Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 105, 1744–1755 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Agler M. T., et al. , Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14, e1002352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morella N. M., Gomez A. L., Wang G., Leung M. S., Koskella B., The impact of bacteriophages on phyllosphere bacterial abundance and composition. Mol. Ecol. 27, 2025–2038 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Elbeltagy A., et al. , Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67, 5285–5293 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carini P., et al. , Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 16242 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Naylor D., DeGraaf S., Purdom E., Coleman-Derr D., Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 11, 2691–2704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M., Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 9, e105592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lundberg D. S., Yourstone S., Mieczkowski P., Jones C. D., Dangl J. L., Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Daum C., iTag sample amplification QC. http://1ofdmq2n8tc36m6i46scovo2e.wpengine.netdna-cdn.com/wp-content/uploads/2016/04/iTag-Sample-Amplification-QC-v1.3.pdf. Accessed XXX.
- 57.Morella N. M., et al. , Tomato phyllosphere 16S and ITS amplicon sequencing. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA578761. Deposited 21 November 2019.
- 58.Rognes T., et al. , VSEARCH: A versatile open source tool for metagenomics. PeerJ, e2584 (2016). [DOI] [PMC free article] [PubMed]
- 59.Schloss P. D., et al. , Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D., Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quast C., et al. , The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bokulich N. A., et al. , Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMurdie P. J., Holmes S., phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skytte Andersen K. S., Kirkegaard R. H., Karst S. M., Albertsen M., ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. bioRxiv:10.1101/299537 (11 April 2018).
- 65.Ssekagiri A., Sloan W. T., Ijaz U. Z., Data from “An R package for microbial community analysis in an environmental context.” Github. https://github.com/umerijaz/microbiomeSeq. Accessed 1 July 2019.
- 66.Shetty P., Trifolium nodule microbiome analysis script. http://rstudio-pubs-static.s3.amazonaws.com/266780_cac4994322494658904507a7606b1dd8.html. Accessed 1 July 2019.
- 67.Chase J. M., Myers J. A., Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2351–2363 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stegen J. C., et al. , Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequences files can be found on NCBI using the BioProject ID PRJNA578761. R code that was used for analysis can be provided upon request.