Abstract
More than a century ago, the centrosome was discovered and described as “the true division organ of the cell”. Electron microscopy revealed that a centrosome is an amorphous structure or pericentriolar protein matrix that surrounds a pair of well-organized centrioles. Today, the importance of the centrosome as a microtubule-organizing center and coordinator of the mitotic spindle is questioned, because centrioles are absent in up to half of all known eukaryotic species, and various mechanisms for acentrosomal microtubule nucleation have been described. This review recapitulates the known functions of centrosome movements in cellular homeostasis and discusses knowledge gaps in this field.
Keywords: Biological sciences, Cell biology, Cytoskeleton, Developmental biology, Membrane, Molecular biology, Proteins, Centrosome, Nuclear envelope, Cell division, Gamma-tubulin, Microtubule nucleating complex
Biological sciences; Cell biology; Cytoskeleton; Developmental biology; Membrane; Molecular biology; Proteins; Centrosome; Nuclear envelope; Cell division; Gamma-tubulin; Microtubule nucleating complex
1. Introduction
Late in the nineteenth century, separate publications by Edouard Van Beneden, Walther Flemming, and Theodor Boveri described a centrosome as an independent organelle, which, through self-replication, was passed on to new born cells [1, 2]. In short, the centrosome was regarded as part of the cell with clear responsibility for mediating nuclear and cellular division [2], and the discovery of the centrosome was considered to be as important as the discovery of the nucleus [3]. Still, the early enthusiasm among researchers declined over a period of decades, because studies using electron microscopy revealed that a centrosome is an amorphous structure, or pericentriolar protein matrix (PCM), that surrounds a pair of well-structured centrioles and thus has a composition that is difficult to define. Over time, considerable progress has been made in understanding the functions, the structure, and the biology of the centrioles [3].
This review explores the cellular importance of the centrosome in various species and the possible function of this organelle as a component of the γ-tubulin meshwork.
2. Main text
2.1. The presence of centrioles
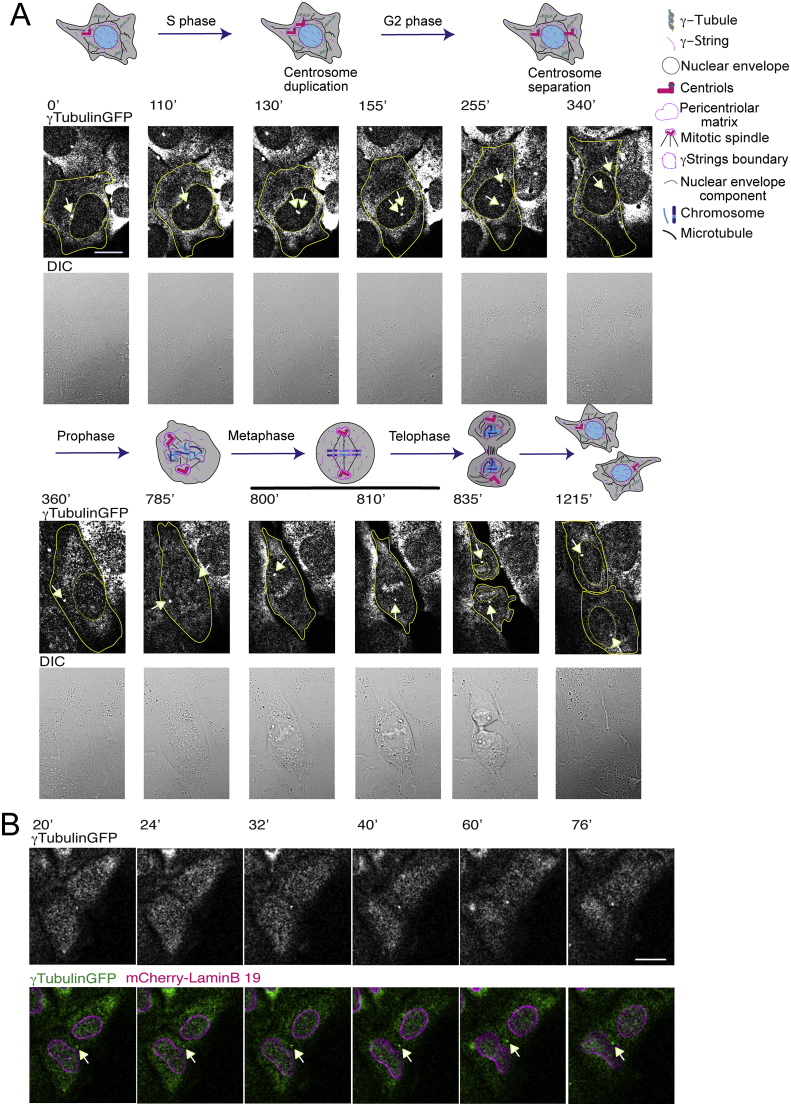
In animal cells, a centrosome has the following components: a two microtubule-based barrel-shaped centrioles and a surrounding amorphous network of proteins or PCM that includes a large number of microtubule-nucleating complexes. In dividing cells, at G1–S transition, a procentriole is formed adjacent to each parental centriole and continues to grow during S phase. At G2–M, the daughter centriole matures, the two centrosomes separate, and both of the centrosomes instruct and determine the formation and positioning of the mitotic spindle during symmetric and asymmetric cell division (Figure 1 and Movie 1; this data is from unpublished works by Alvarado-Kristensson et al.) [4, 5].
Figure 1.
Centrosome positioning during cell division. (A) Time-lapse series of differential interference contrast (DIC)/fluorescence images showing a U2OS cell stably expressing TUBG-shRNA and co-expressing GFP-tagged sh-resistant γ-tubulin1 (γTubulinGFP). The image series presents chosen frames illustrating the changes in the position of the centrosome(s) during cell division. The outer membrane of the cell (yellow lines) and the nucleus (dotted lines) are indicated by solid and dotted lines, respectively. The model [19] depicts the changes known to occur in the centrosome(s) during cell division: the centrosome duplicates in S phase; the two centrosomes separate at the end of G2 phase; the disassembly of the nuclear envelope permits the repositioning of centrosomes to regulate the formation of the mitotic spindle in prophase; the chromosomes are aligned in the middle of the cell and microtubules emanating from the centrosomes are attached to the chromosomes in metaphase; the mitotic spindle pulls apart the chromatin between the newborn cells in telophase. A nuclear membrane is formed around each set of chromosomes, and the division of the cytoplasm between new born cells is ongoing (cytokinesis) for the final generation of two cells. The images shown were collected every 5 min. See also Movie 1. (B) Time-lapse fluorescence images of a U2OS cell stably expressing the following constructs: TUBG-shRNA, GFP-tagged sh-resistant γ-tubulin1 (γTubulinGFP), and mCherry-lamin B1. The images represent selected frames showing the changes in location that the centrosomes undergo during cytokinesis. Here, after furrow ingression, one of the centrosomes transiently moved to the growing furrow to allow the completion of cell division. Images were collected every 4 min. See also Movie 2. (A and B) Stable cell lines were obtained and time-lapse experiments were performed as previously described [20, 25, 68, 69]. In both A and B an arrow indicates a centrosome. Scale bars: 10 μm. This data is from unpublished works by Alvarado-Kristensson et al.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.heliyon.2020.e03238.
The following is the supplementary data related to this article:
Related to Figure 1A. Fluorescence images from a time-lapse confocal microscopy using a laser-scanning confocal microscopy of a U2OS cell stably co-expressing TUBG-shRNA and GFP-tagged sh-resistant γ-tubulin1. This data is from unpublished works by Alvarado-Kristensson et al.
Centrioles in resting cells can bear flagella and cilia, and therefore such centrioles are usually referred to as basal bodies [6]. Also, all species that have cilia-bearing cells at some stage in their life cycle have centrioles [7, 8], which agrees with the role of centrioles in the organization of microtubules in cilia. In Drosophila melanogaster, the absence of centrioles leads to early death after eclosion due to the lack of cilia, whereas spindle formation and chromosome segregation are unaffected [9]. Nonetheless, oocytes in metazoan species lack centrosomes [10], and it has been shown that the first divisions in early mouse embryo development occur in the absence of centrioles [11]. Moreover, the asymmetric inheritance of old and new mother centrioles in mouse cells is necessary for neuron development, and, consequently, for development of the animal itself [5].
Spindle pole bodies in yeast (equivalent to centrosomes in animals) lack centrioles but have a PCM [12]. Indeed, centrioles are absent in up to half of all known eukaryotic species, including most fungi, protists, and vascular plants, as well as many algae [13, 14]. Cells that lack centrioles have microtubule-nucleating complexes that control the formation of the mitotic spindle [15].
2.2. Centrosomes, microtubule-nucleating complexes, and microtubule formation
Centrosomes are microtubule-organizing centers, and both microtubules and centrosomes are highly enriched in a protein family of GTPases called the tubulins [16]. Tubulins are the major components of both microtubules and the γ-tubulin meshwork in eukaryotes [17, 18, 19]. Heterodimers of α- and β-tubulin assemble into microtubules, whereas γ-tubulin assembles into γ-strings [20, 21] and γ-tubules [22]. The γ-tubulin small complex (γ-TuSC) and the γ-tubulin ring complex (γ-TuRC) are two important γ-tubulin-containing complexes that assist in nucleation of microtubules and γ-tubules [17, 18, 22, 23]. In humans, γ-TuSC consists of two γ-tubulin molecules combined with one γ-tubulin complex protein 2 (GCP2) and with one GCP3. γ-TuSCs, together with additional GCPs, form the larger complex γ-TuRC. A microtubule nucleates on the γ-tubulin ring in a γ-TuRC, and several γ-TuRCs together with pericentrin form γ-tubules [22]. Furthermore, α- and β-tubulin are found in the cytoplasm and centrioles, whereas γ-tubulin occurs in the cytoplasm, and centrosomes and is also associated with cellular membranes and chromatin [17, 18, 20, 23, 24, 25].
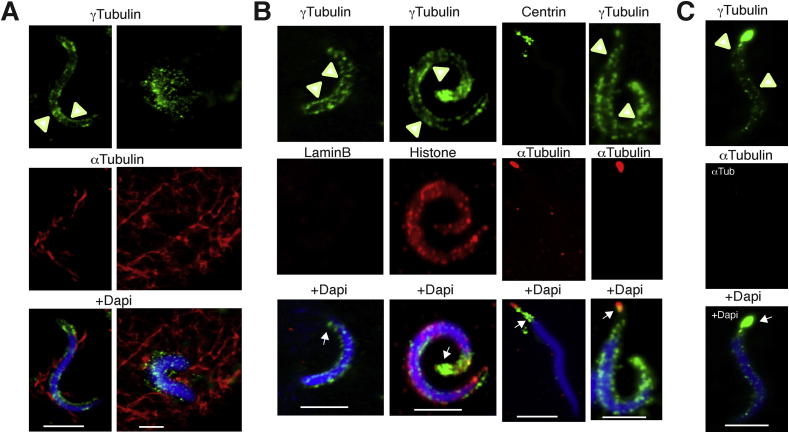
In plants, the acentrosomal nucleation of microtubules occurs at scattered sites in a γTuRCs -dependent manner [26, 27]. Also, in various species, acentrosomal microtubule nucleation takes place on chromatin [28] and is generated by the small GTPase Ran (Ras-related nuclear protein) that locally activates microtubule-associated proteins, including γTuRCs (Figure 2A; this data is from unpublished works by Alvarado-Kristensson et al.) [29, 30, 31]. In addition, the augmin pathway targets γTuRCs to a pre-existing microtubule that assists in nucleation of a new microtubule from the recruited γTuRCs [32, 33, 34]. Thus, a prerequisite for centrosomal and acentrosomal microtubule nucleation is the presence of γ-tubulin.
Figure 2.
A centrosome remains attached to chromatin despite the absence of actin, microtubules, the lamina and the nuclear envelope. A–C. α-Tubulin (centriole and microtubule component), lamin B (nuclear membrane marker), centrin (centrosome marker), histone (chromatin marker), and γ-tubulin (PCM and γ-tubulin nuclear boundary marker) were immunofluorescence stained in demembranated Xenopus laevis sperm that were isolated in the presence of cytochalasin B (depolymerized actin; A and B) or both cytochalasin B and colcemid (depolymerized actin and microtubules; C) and were subsequently pelleted on a cushion containing glycerol to remove debris, and then immunostained as previously described [20]. Chromatin was stained with DAPI. (A) To study the nucleation of microtubules on demembranated sperm, egg extracts were incubated with such sperm before fixation [20]. Immnunostaining with an anti-α-tubulin antibody showed that addition of egg extracts (A) to demembranated sperm (B) triggered the assembly of microtubules on chromatin (A and B). (C) Isolation of demenbranated X. laevis sperm in the presence of colcemid and subsequent fixation and immunostaining with the indicated antibodies showed that colcemid treatment removed centrioles (negative α-tubulin staining) from the PCM, but the PCM still remained attached to the chromatin. (A–C) Arrows and arrowheads indicate the location of PCM and the γ-tubulin nuclear boundary, respectively. Scale bars: 10 μm. This data is from unpublished works by Alvarado-Kristensson et al.
2.3. The centrosome and the nuclear compartment
Spontaneous TUBG1 gene mutation that results in mutation of the amino acid Leu387Pro is associated with brain malformations [35]. In yeast, γ-tubulinLeu387Pro mutation in the DNA-binding domain of γ-tubulin affects the positioning of the nucleus [19, 35, 36]. In eukaryotic cells, a boundary of γ-strings around chromatin coordinates formation of the nucleus [20] and around centrioles assists in the nucleation of microtubules in the PCM [37]. Also, the structures that contain γ-tubulin (i.e. γ-tubules and centrosomes) affect the shape of the nuclear envelope. γ-Tubulin was recently found to be a major component of a novel cytoskeletal element named γ-tubules. In mammalian cells, γ-tubules can emanate from centrosomes and interlace to produce a macro-γ-tubule that changes the shape of the nucleus [22, 38]. In addition, centrosomes can shape the nuclear envelope by residing within an invagination of the nuclear envelope [39].
In time-lapse images of living U2OS cells stably expressing both TUBG-shRNA (which reduces the endogenous γ-tubulin pool by ~ 50 % [25, 40]) and a C-tagged TUBG1-green fluorescence protein (GFP) shRNA-resistant gene (which fluorescence labels the PCM) [20] (Figure 1, Movies 1 and 2; this data is from unpublished works by Alvarado-Kristensson et al.), it is apparent that, during interphase, centrosomes are located on the cytosolic side of the nuclear envelope, and their position on the nuclear envelope is constantly changing (Figure 1A and Movie 1). It can also be noted that the interactions of PCM with the nucleus are sufficiently dynamic to permit independent movements around the nuclear envelope (Figure 1A and Movie 1) and within the cytoplasm (360 min, Figures 1A and 1B) [41]. During G2, the centrosomes separate prior to the onset of mitosis, and this occurs with the mechanical support of myosin II and actin filaments [42]. In higher eukaryotes, entrance into mitosis is characterized by disassembly of the nuclear envelope that permits the repositioning of centrosomes to guide formation of the mitotic spindle (Figure 1A) [43, 44].
Supplementary video related to this article can be found at https://doi.org/10.1016/j.heliyon.2020.e03238.
The following is the supplementary data related to this article:
Related to Figure 1B. Fluorescence images from a time-lapse confocal microscopy using a laser-scanning confocal microscopy of a U2OS cell in late mitosis stably co-expressing TUBG-shRNA, γTubulinGFP (green), and mCherry-lamin B1 (magenta). The white ellipse indicates a centrosome. This data is from unpublished works by Alvarado-Kristensson et al.2
However, in many lower eukaryotes (e.g. fungi, protists, and unicellular algae), the nuclear attachment is necessary for the centrosome to cycle in and out of the nuclear envelope to achieve a closed mitosis. This implies that local openings within the nuclear envelope, close to the microtubule-organizing centers, are sufficient to allow microtubules to gain access to the chromosomes, as outlined by Drechsler and McAinsh [45].
In budding yeast, exit from mitosis must occur after partitioning of chromosomes between the daughter cells. A checkpoint mechanism monitors the coupling between nuclear and cytoplasmic division by causing the GTP-binding protein Tem1, which is a regulator of mitotic exit, to localize to the spindle pole body. Tem1 is activated by the exchange factor Lte1, which is localized to the bud. Consequently, the migration of the spindle pole body to the neck of the bud brings Tem1 in contact with Lte1, which in turn activates Tem1 to trigger mitotic exit [46]. In mammalian cells, immediately before exit from cell division, but after furrow ingression between the two new born cells, one of the centrosomes transiently moves to the growing furrow to allow the completion of cell division (Figure 1B, Movie 2) [41]. Absence of centrosomes leads to defects in the final step of mitosis, cytokinesis [41], which suggests that centrosome movements represent a conserved centrosome-dependent pathway that integrates spatial and temporal cues during cell division.
2.4. Connection of the centrosome to the nucleus
The centrosome is an organelle that is constantly in motion around the nuclear envelope (Figure 1 and Movies 1 and 2), but what is it that keeps the centrosomes close to the nuclear membrane? Previous research has shown that γ-tubulin and the centrosomes interact with KASH (Klarsicht, ANC-1, Syne homology), SUN (Sad1 and UNC-84), and lamin (a component of the nucleoskeleton) [20, 47, 48, 49, 50, 51]. The interactions of centrosomes with SUN and KASH with the centrosome were identified through genetic screens for mispositioned nuclei in model organisms [49, 50, 51]. SUN is an integral component of the inner nuclear membrane, and KASH is a C-tail-anchored membrane protein found in the outer membrane of the nuclear envelope. SUN and KASH domains interact in the perinuclear space to form a bridge that spans the nuclear envelope to connect the cytoskeleton to the nucleoskeleton [48, 49].
A second mechanism of centrosomes interaction with the nucleus can be described as occurs in Dictyostelium and mouse embryonic stem cells, in which microtubule- and lamin-based mechanisms maintain the central position of a centrosome by dynein-mediated forces [52, 53], and the Kif9 kinesin provides a mechanical linkage between the nucleus and centrosome [54].
In demenbranated Xenopus laevis sperm (which lack actin, membranes, and a lamina), a centrosome is part of the chromatin-associated γ-strings boundary (Figure 2B; this data is from unpublished works by Alvarado-Kristensson et al.). In such cells, the centrosome remains attached to chromatin despite the absence of actin, the lamina, and the nuclear envelope. Furthermore, preparation of the sperm in the presence of the tubulin inhibitor colcemid [22, 55, 56], removes the centrioles from the centrosome, while the PCM still remains attached to the chromatin. This implies that attachment of the PCM to chromatin in these cells is not dependent on the presence of microtubules, actin, the nuclear envelope, or the lamina (Figure 2C; this data is from unpublished works by Alvarado-Kristensson et al.).
3. Conclusion and future perspectives
Since the discovery of the centrosome late in the nineteenth century, considerable progress has been made in understanding the function, structure, and replication of centrosomes. Today, these organelles are considered to be microtubule-organizing centers, as well as signal transduction hubs that associate with proteins involved in microtubule, actin, and γ-tubule nucleation, as well as cell cycle progression, checkpoint activation, and DNA repair [19, 22, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67]. Still, our knowledge is limited regarding how positioning of centrosomes integrates spatial and temporal cues during interphase. Thus, the aim of the present review is to summarize the known functions of the centrosome positioning in cellular homeostasis, and also to identify knowledge gaps in the field.
Live imaging of cells in interphase have shown constant changes in the positioning of the centrosomes on the surface of the nuclear envelope. Part of this motion has been described as playing a role in cell differentiation and mitosis [5, 41, 43, 44]. Centrosome movements are necessary for guiding of the mitotic spindle and exit from mitosis. However, mitosis is the final step in cell division, so why do the centrosomes move around the nuclear envelope during interphase, and what is the purpose of those movements? At present, we have no answers to these questions. Therefore, further insights are still needed to elucidate the mechanical signals affecting the spatial positioning of the centrosomes and the loose attachment to the nuclear compartment that controls centrosome movements, and also to explain a possible impact on cellular homeostasis.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This work was supported by the Swedish Cancer Society, the Swedish Childhood Cancer Fund and the Skåne University Hospital in Malmö Cancer Research Fund.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2020.e03238.
Acknowledgements
The author thanks Patricia Ödman for editorial assistance.
References
- 1.Paweletz N. Walther Flemming: pioneer of mitosis research. Nat. Rev. Mol. Cell Biol. 2001;2:72–75. doi: 10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- 2.Scheer U. Historical roots of centrosome research: discovery of Boveri's microscope slides in Würzburg. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schatten H. The mammalian centrosome and its functional significance. Histochem. Cell Biol. 2008;129:667–686. doi: 10.1007/s00418-008-0427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg E.A. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Niu Y., Liu T., Tse G.M. Increased expression of centrosomal alpha, gamma-tubulin in atypical ductal hyperplasia and carcinoma of the breast. Cancer Sci. 2009;100:580–587. doi: 10.1111/j.1349-7006.2008.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gönczy P. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 2012;13:425–435. doi: 10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- 7.Baldauf S.L., Roger A.J., Wenk-Siefert I., Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 8.Marshall W.F. Centriole evolution. Curr. Opin. Cell Biol. 2009;21:14–19. doi: 10.1016/j.ceb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basto R., Lau J., Vinogradova T. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 10.Megraw T.L., Kaufman T.C. The centrosome in Drosophila oocyte development. Curr. Top. Dev. Biol. 2000;49:385–407. doi: 10.1016/s0070-2153(99)49019-2. [DOI] [PubMed] [Google Scholar]
- 11.Courtois A., Schuh M., Ellenberg J., Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 2012;198:357–370. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balczon R. The centrosome in animal cells and its functional homologs in plant and yeast cells. Int. Rev. Cytol. 1996;169:25–82. doi: 10.1016/s0074-7696(08)61984-1. [DOI] [PubMed] [Google Scholar]
- 13.Wasteneys G.O. Microtubule organization in the green kingdom: chaos or self-order? J. Cell Sci. 2002;115:1345–1354. doi: 10.1242/jcs.115.7.1345. [DOI] [PubMed] [Google Scholar]
- 14.Vaughn K.C., Harper J.D. Microtubule-organizing centers and nucleating sites in land plants. Int. Rev. Cytol. 1998;181:75–149. doi: 10.1016/s0074-7696(08)60417-9. [DOI] [PubMed] [Google Scholar]
- 15.Binarová P., Cenklová V., Procházková J. Gamma-tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in Arabidopsis. Plant Cell. 2006;18:1199–1212. doi: 10.1105/tpc.105.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutcher S.K. The tubulin fraternity: alpha to eta. Curr. Opin. Cell Biol. 2001;13:49–54. doi: 10.1016/s0955-0674(00)00173-3. [DOI] [PubMed] [Google Scholar]
- 17.Moritz M., Braunfeld M.B., Sedat J.W., Alberts B., Agard D.A. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y., Wong M.L., Alberts B., Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 19.Alvarado-Kristensson M. γ-tubulin as a signal-transducing molecule and meshwork with therapeutic potential. Signal. Transduct. Target Ther. 2018;3:24. doi: 10.1038/s41392-018-0021-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosselló C.A., Lindström L., Glindre J., Eklund G., Alvarado-Kristensson M. Gamma-tubulin coordinates nuclear envelope assembly around chromatin. Heliyon. 2016;2:e00166. doi: 10.1016/j.heliyon.2016.e00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chumová J., Trögelová L., Kourová H. γ-Tubulin has a conserved intrinsic property of self-polymerization into double stranded filaments and fibrillar networks. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:734–748. doi: 10.1016/j.bbamcr.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Lindström L., Alvarado-Kristensson M. Characterization of gamma-tubulin filaments in mammalian cells. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:158–171. doi: 10.1016/j.bbamcr.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Moritz M., Zheng Y., Alberts B.M., Oegema K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ríos R.M., Sanchís A., Tassin A.M., Fedriani C., Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Lindström L., Li T., Malycheva D. The GTPase domain of gamma-tubulin is required for normal mitochondrial function and spatial organization. Commun. Biol. 2018;1:37. doi: 10.1038/s42003-018-0037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Z., Hotta T., Lee Y.R., Horio T., Liu B. The {gamma}-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell. 2010;22:191–204. doi: 10.1105/tpc.109.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura M., Yagi N., Kato T. Arabidopsis GCP3-interacting protein 1/MOZART 1 is an integral component of the γ-tubulin-containing microtubule nucleating complex. Plant J. 2012;71:216–225. doi: 10.1111/j.1365-313X.2012.04988.x. [DOI] [PubMed] [Google Scholar]
- 28.Hayward D., Wakefield J.G. Chromatin-mediated microtubule nucleation in Drosophila syncytial embryos. Commun. Integr. Biol. 2014;7:e28512. doi: 10.4161/cib.28512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- 30.Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C., Hughes M., Clarke P.R. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J. Cell Sci. 1999;112(Pt 14):2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
- 32.Goshima G., Mayer M., Zhang N., Stuurman N., Vale R.D. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawo S., Bashkurov M., Mullin M. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 34.Hsia K.C., Wilson-Kubalek E.M., Dottore A. Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol. 2014;16:852–863. doi: 10.1038/ncb3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirier K., Lebrun N., Broix L. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Höög G., Zarrizi R., von Stedingk K., Jonsson K., Alvarado-Kristensson M. Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression. FASEB J. 2011;25:3815–3827. doi: 10.1096/fj.11-187484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dictenberg J.B., Zimmerman W., Sparks C.A. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosselló C.A., Lindström L., Eklund G., Corvaisier M., Kristensson M.A. γ-Tubulin⁻γ-Tubulin interactions as the basis for the formation of a meshwork. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19103245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hulspas R., Houtsmuller A.B., Bauman J.G., Nanninga N. The centrosome moves out of a nuclear indentation in human lymphocytes upon activation. Exp. Cell Res. 1994;215:28–32. doi: 10.1006/excr.1994.1310. [DOI] [PubMed] [Google Scholar]
- 40.Ehlén Å., Rosselló C.A., von Stedingk K. Tumors with nonfunctional retinoblastoma protein are killed by reduced γ-tubulin levels. J. Biol. Chem. 2012;287:17241–17247. doi: 10.1074/jbc.M112.357038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piel M., Nordberg J., Euteneuer U., Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 42.Rosenblatt J., Cramer L.P., Baum B., McGee K.M. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 43.Collas P. Sequential PKC- and Cdc2-mediated phosphorylation events elicit zebrafish nuclear envelope disassembly. J. Cell Sci. 1999;112(Pt 6):977–987. doi: 10.1242/jcs.112.6.977. [DOI] [PubMed] [Google Scholar]
- 44.Peter M., Nakagawa J., Dorée M., Labbé J.C., Nigg E.A. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-p. [DOI] [PubMed] [Google Scholar]
- 45.Drechsler H., McAinsh A.D. Exotic mitotic mechanisms. Open Biol. 2012;2:120140. doi: 10.1098/rsob.120140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bardin A.J., Visintin R., Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 47.Larsson V.J., Jafferali M.H., Vijayaraghavan B., Figueroa R.A., Hallberg E. Mitotic spindle assembly and γ-tubulin localisation depend on the integral nuclear membrane protein Samp1. J. Cell Sci. 2018:131. doi: 10.1242/jcs.211664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., Lei K., Yuan X. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starr D.A., Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 50.Puckelwartz M.J., Kessler E., Zhang Y. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyerzon M., Fridolfsson H.N., Ly N., McNally F.J., Starr D.A. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koonce M.P., Köhler J., Neujahr R., Schwartz J.M., Tikhonenko I., Gerisch G. Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 1999;18:6786–6792. doi: 10.1093/emboj/18.23.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y., Zheng Y. Lamins position the nuclear pores and centrosomes by modulating dynein. Mol. Biol. Cell. 2015;26:3379–3389. doi: 10.1091/mbc.E15-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leo M., Santino D., Tikhonenko I., Magidson V., Khodjakov A., Koonce M.P. Rules of engagement: centrosome-nuclear connections in a closed mitotic system. Biol. Open. 2012;1:1111–1117. doi: 10.1242/bio.20122188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chinen T., Liu P., Shioda S. The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nat. Commun. 2015;6:8722. doi: 10.1038/ncomms9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friesen D.E., Barakat K.H., Semenchenko V. Discovery of small molecule inhibitors that interact with γ-tubulin. Chem. Biol. Drug Des. 2012;79:639–652. doi: 10.1111/j.1747-0285.2012.01340.x. [DOI] [PubMed] [Google Scholar]
- 57.Horejsi B., Vinopal S., Sladkova V. Nuclear gamma-tubulin associates with nucleoli and interacts with tumor suppressor protein C53. J. Cell. Physiol. 2012;227:367–382. doi: 10.1002/jcp.22772. [DOI] [PubMed] [Google Scholar]
- 58.Hsu L.C., White R.L. BRCA1 is associated with the centrosome during mitosis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12983–12988. doi: 10.1073/pnas.95.22.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hubert T., Vandekerckhove J., Gettemans J. Cdk1 and BRCA1 target gamma-tubulin to microtubule domains. Biochem. Biophys. Res. Commun. 2011;414:240–245. doi: 10.1016/j.bbrc.2011.09.064. [DOI] [PubMed] [Google Scholar]
- 60.Lesca C., Germanier M., Raynaud-Messina B. DNA damage induce gamma-tubulin-RAD51 nuclear complexes in mammalian cells. Oncogene. 2005;24:5165–5172. doi: 10.1038/sj.onc.1208723. [DOI] [PubMed] [Google Scholar]
- 61.Starita L.M., Machida Y., Sankaran S. BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol. Cell. Biol. 2004;24:8457–8466. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S., Hemmerich P., Grosse F. Centrosomal localization of DNA damage checkpoint proteins. J. Cell. Biochem. 2007;101:451–465. doi: 10.1002/jcb.21195. [DOI] [PubMed] [Google Scholar]
- 63.Morris V.B., Brammall J., Noble J., Reddel R. p53 localizes to the centrosomes and spindles of mitotic cells in the embryonic chick epiblast, human cell lines, and a human primary culture: an immunofluorescence study. Exp. Cell Res. 2000;256:122–130. doi: 10.1006/excr.2000.4800. [DOI] [PubMed] [Google Scholar]
- 64.Kanai M., Tong W.M., Sugihara E., Wang Z.Q., Fukasawa K., Miwa M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chouinard G., Clement I., Lafontaine J., Rodier F., Schmitt E. Cell cycle-dependent localization of CHK2 at centrosomes during mitosis. Cell Div. 2013;8:7. doi: 10.1186/1747-1028-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farina F., Gaillard J., Guerin C. The centrosome is an actin-organizing centre. Nat. Cell Biol. 2016;18:65–75. doi: 10.1038/ncb3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chumová J., Kourová H., Trögelová L., Halada P., Binarová P. Microtubular and nuclear functions of γ-tubulin: are they LINCed? Cells. 2019;8 doi: 10.3390/cells8030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alvarado-Kristensson M. A simple and fast method for fixation of cultured cell lines that preserves cellular structures containing gamma-tubulin. MethodsX. 2018;5:227–233. doi: 10.1016/j.mex.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindström L., Villoutreix B.O., Lehn S. Therapeutic targeting of nuclear γ-tubulin in RB1-negative tumors. Mol. Cancer Res. 2015;13:1073–1082. doi: 10.1158/1541-7786.MCR-15-0063-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1A. Fluorescence images from a time-lapse confocal microscopy using a laser-scanning confocal microscopy of a U2OS cell stably co-expressing TUBG-shRNA and GFP-tagged sh-resistant γ-tubulin1. This data is from unpublished works by Alvarado-Kristensson et al.
Related to Figure 1B. Fluorescence images from a time-lapse confocal microscopy using a laser-scanning confocal microscopy of a U2OS cell in late mitosis stably co-expressing TUBG-shRNA, γTubulinGFP (green), and mCherry-lamin B1 (magenta). The white ellipse indicates a centrosome. This data is from unpublished works by Alvarado-Kristensson et al.2