Abstract
The Apicomplexa phylum comprises diverse parasitic organisms that have evolved from a free-living ancestor. These obligate intracellular parasites exhibit versatile metabolic capabilities reflecting their capacity to survive and grow in different hosts and varying niches. Determined by nutrient availability, they either use their biosynthesis machineries or largely depend on their host for metabolite acquisition. Because vitamins cannot be synthesized by the mammalian host, the enzymes required for their synthesis in apicomplexan parasites represent a large repertoire of potential therapeutic targets. Here, we review recent advances in metabolic reconstruction and functional studies coupled to metabolomics that unravel the interplay between biosynthesis and salvage of vitamins and cofactors in apicomplexans. A particular emphasis is placed on Toxoplasma gondii, during both its acute and latent stages of infection.
Keywords: parasite metabolism, metabolomics, vitamin, Toxoplasma gondii, Plasmodium, parasitology, cofactor, Cryptosporidium, metabolic pathway, nutrient acquisition, Apicomplexa
Introduction
Members of the Apicomplexa encompass a large number of parasites exhibiting a great level of diversity in their life cycles, with morphologically distinct stages in one or more hosts. The phylum includes coccidians, hemosporidians, piroplasms, Cryptosporidia, and gregarines that occupy divergent niches (1). Toxoplasma gondii is the most successful zoonotic parasite of the cyst-forming subclass of coccidians. The proliferative tachyzoites infect and replicate in most cell types and are responsible for an acute infection, whereas the dormant cyst-forming bradyzoites are responsible for chronic infection, predominantly in the brain and striated muscles (2, 3). Plasmodium falciparum is the deadliest form of the human malaria parasites that proliferate in erythrocytes and hepatocytes. T. gondii and malaria parasites replicate intracellularly within a parasitophorous vacuole membrane that is permeable to small metabolites (4–8). In contrast, Theileria and Babesia species that belong to the genera of piroplasms rapidly escape the vacuole and proliferate freely in the cytoplasm of lymphocytes and red blood cells, respectively, with a more direct access to host nutrients (9, 10). Cryptosporidium, an enteric pathogen that relies only on a single host for both its sexual and asexual reproduction, develops in an extracytoplasmic compartment confined to the apical surfaces of epithelial cells and in a vacuole connected to the host cell via an extensively folded membrane structure called the feeder organelle (11). In humans, the causative agents of malaria, toxoplasmosis, and cryptosporidiosis are responsible for over a million deaths each year. From an evolutionary point of view, it is useful to compare the needs and capabilities between the closely related alveolates from the Apicomplexa and Chromerida phylum that group species capable of photosynthesis (12).
Our knowledge of apicomplexan metabolism has greatly benefited from the assembly of parasite genomes and has advanced through functional studies, in particular of T. gondii and Plasmodium spp. A necessary step toward a global understanding of the central carbon metabolism as well as the synthesis and uptake of amino acids, lipids, vitamins, and cofactors involves the use of in silico methods capable of predicting essential reactions, genes, and synthetic lethal pairs (13–16).3 Currently available genome-scale computational models for T. gondii and the malaria parasites (14–17)3 have recently been challenged by an impressive series of genome-wide gene fitness screens (17–19) and stage-specific transcriptomics data (20–22). These global approaches have turned out to be instrumental for the curation and validation of computational networks. Ultimately, incorporating functional analyses of metabolic pathways with molecular biology and metabolomic techniques will improve the accuracy of computational predictions.
In the recent past, several studies have illustrated the power of combining genetic and metabolomics approaches to understand metabolic functions in T. gondii. To summarize, it was shown that glucose and glutamine are the major carbon sources utilized by T. gondii tachyzoites (23, 24) and that glycolysis is essential for bradyzoites (25). The gluconeogenic enzyme fructose bisphosphatase was essential to regulate glycolytic flux in a futile cycle with phosphofructokinase (26). Uniquely, acetyl-CoA in the mitochondrion was shown to be produced via the branched-chain α-ketoacid dehydrogenase complex and not the canonical pyruvate dehydrogenase (PDH)4 complex (27). PDH is required for a functional fatty acid (FA) synthase complex, also known as the FASII, in the apicoplast that produces medium-chain FAs, further elongated at the endoplasmic reticulum to form long monounsaturated FAs (28, 29).
Given the availability of large-scale data sets, systems-wide analysis of parasite metabolism offers a great opportunity to identify essential metabolic functions for targeted drug intervention. In a recent study,3 a well-curated computational genome-scale model, iTgo (in silico T. gondii), was generated. iTgo contains 556 metabolic genes and integrates all available data sets to serve as a valuable platform for model-guided investigations. To harmonize the model with the genome-wide fitness scores for metabolic genes, additional constraints on substrate availabilities from the host as well as reaction utilization based on transcriptomics data were applied (16, 30). The workflow led to a model, 80% consistent with experimentally observed phenotypes,3 allowing for reliable hypothesis generation for experimental validation. The two previous metabolic reconstructions (13, 15) identified several essential metabolic functions and differences within the clonal strains of T. gondii that display distinct virulence profiles. Within the apicomplexans, the most studied and comprehensive metabolic reconstructions were generated for P. falciparum and the rodent malaria parasite, Plasmodium berghei (14, 16, 31). Constant modeling efforts with the incorporation of physiological parameters, such as metabolomics and fluxomics, continue to expand our knowledge of the metabolic versatility of the apicomplexans. Although challenging, future models should consider the kinetic properties of reactions, allowing the simulation of altered enzymatic activities in both the host and parasite (31). Ideally, as complementary constituents of an iterative process, both computational and experimental efforts will ultimately lead to the identification of potential drug targets, mechanisms of drug action and complex host-pathogen interactions.
Among the indispensable pathways for parasite proliferation and persistence, the biosynthesis of vitamins and cofactors offers potential targets for intervention. Vitamins are essential precursors for the production of cofactors and, in humans, can be acquired solely through the diet (32). To date, 13 metabolites are classified as vitamins, required for the functioning of a mammalian cell, facilitating numerous enzymatic reactions. Nine of the 13 vitamins are known to be utilized by the apicomplexans, with three of them (vitamins B5, B6, and B9) being de novo–synthesized by some parasites (33). The vitamins that can be synthesized de novo are probably low in abundance in one or more niches and cannot be sufficiently salvaged. Comparison across the phylum can reveal interesting insights into the origins and subsequent loss of several pathways in certain genera, such as the Cryptosporidia and piroplasms (34–36) (Fig. 1). Both genera possess limited biosynthesis capabilities, reflecting their lifestyle in a nutrient-rich environment and adaptation to mechanisms for metabolite acquisition from the host. Concordantly, the genome of Cryptosporidium hominis was shown to encode more than 80 genes with strong similarity to known transporters and several hundred genes with transporter-like properties (37). Cryptosporidia are also in close contact with the microbiome in the intestinal gut, thus expanding their capacity for nutrient acquisition (38).
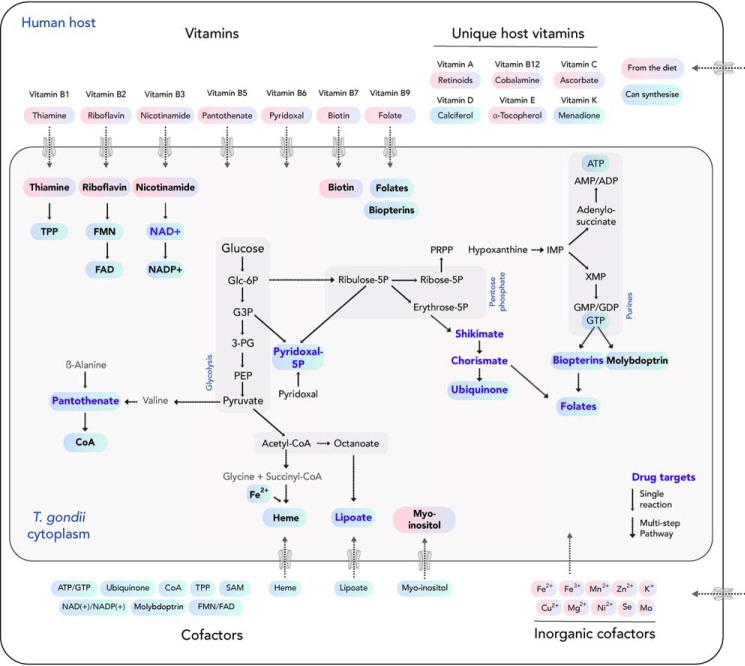
Figure 1.
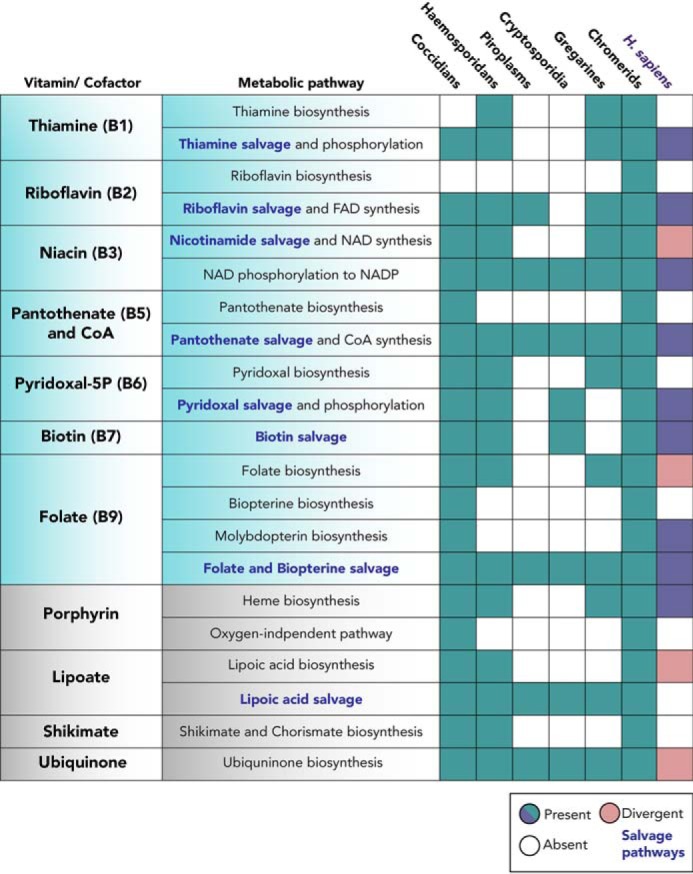
Conservation of vitamin and cofactor biosynthesis or scavenge pathways within the apicomplexans and the human host. The presence or absence of metabolic pathways within the Apicomplexan and Chromerida phylum and the human host, Homo sapiens, is summarized. The gene identifiers and enzyme names in each pathway can be found in Table S1. For each genus, representative organisms were chosen: coccidians (T. gondii), hemosporidians (P. falciparum), piroplasms (Babesia bovis and Theileria annulata), Cryptosporidia (Cryptosporidium muris), gregarines (Gregarina niphandrodes), and chromerida (C. velia and V. brassicaformis).
In the next sections, we review the progress made in T. gondii and apicomplexans in general, to better understand the interrelationship of de novo synthesis and scavenge routes for vitamins and cofactors and their utilization in different life cycle stages. An overview of the pathways in both T. gondii and its mammalian host is presented in Fig. 2. Further, the latest observations are discussed in the context of long-standing questions on the roles of the metabolic pathways for latency and their potential as drug targets.
Figure 2.
Vitamins and cofactors biosynthesis versus scavenge pathways in T. gondii and its mammalian host. Metabolites that can either be de novo produced (blue) or must be salvaged (pink) from an external source are depicted. Enzymes for the production of metabolites (boldface blue type) are potential drug targets, given the unique synthesis capability of the parasite, but not the host.
Vitamin B1
Vitamin B1, or thiamine, is an important precursor for its metabolically active form, thiamine pyrophosphate (TPP). TPP acts as a cofactor for enzymes implicated in carbohydrate and amino acid metabolism, such as the PDH complex, 2-oxoglutarate dehydrogenase, pyruvate decarboxylase, and dihydrolipoamide dehydrogenase. In T. gondii, these enzymes are either residents of the secondary endosymbiotic organelle, called the apicoplast, or the mitochondrion, suggesting a need for the cofactor within these subcellular compartments. Like their mammalian host, the parasites do not possess the pathway for thiamine biosynthesis and must therefore acquire it. Hemosporidians (in particular P. falciparum) are the only apicomplexans that possess the enzymes to synthesize thiamine, like bacteria, plants, and fungi (39–41). The genes implicated in the synthesis of TPP are, however, expressed only in the mosquito vector (salivary gland sporozoites) stage (42). Despite the ability to synthesize thiamine, Plasmodium spp., like other apicomplexans, harbor the key enzyme thiamine diphosphokinase (TPK) to convert the scavenged thiamine into TPP. TPK is expressed in all stages of the Plasmodium life cycle, and several studies have shown that parasite replication is inhibited by thiamine analogues that generate toxic anti-metabolites (43, 44). Deduced from the genome-wide CRISPR-Cas9 screen for T. gondii performed in vitro, TPK is critical for in vitro tachyzoite survival with a high negative fitness score (FS) (−3.28) (Fig. 3). FS are experimentally observed measures (ranging from −7 to +3) and assess the fitness cost of a given gene for parasite survival (17).
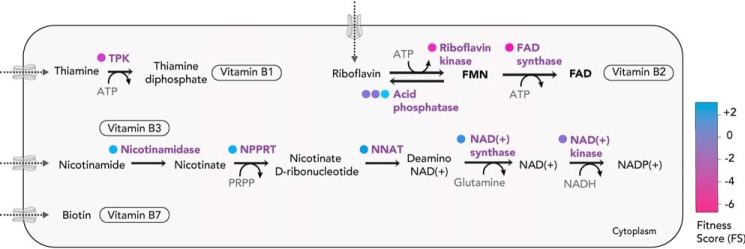
Figure 3.
The scavenge pathways and bioconversion of vitamins (B1, B2, B3, and B7). T. gondii must uptake vitamins B1, B2, B3, and B7 via unknown transport mechanisms and subsequently convert them into the cofactors for utilization within the parasite. FS for the enzymes for the bioconversion are color-coded (in circles). NPPRT, nicotinate phosphoribosyltransferase; NNAT, nicotinate-nucleotide adenylyl transferase.
The mechanism by which thiamine is taken up and translocated across organelles where it is needed is yet to be determined. In humans the thiamine transporters, hThTr1 and hThTr2 have been well-characterized (45, 46), but no obvious orthologs within the parasite's genome could be identified. Interestingly, in certain apicomplexans, such as Cryptosporidia and piroplasms, salvage of the phosphorylated form (TPP) must occur.
Vitamin B2
Vitamin B2, or riboflavin, is crucial for flavin-dependent processes occurring in all subcellular compartments. FMN and FAD participate in redox reactions and play an essential role for the proper functioning of the electron transport chain (ETC), tricarboxylic acid cycle, and fatty acid biosynthesis. Like their mammalian hosts, most apicomplexans are unable to synthesize riboflavin but possess the capacity to convert riboflavin into FMN and FAD. The two genes coding for their synthesis, riboflavin kinase and FAD synthase, are present in T. gondii and are fitness-conferring with an FS of −3.97 and −4.87, respectively (Fig. 3). Exceptionally, Cryptosporidia appear to lack these enzymes and therefore must take up both FMN and FAD, suggesting an exquisite adaptability to scavenge phosphorylated cofactors. Outside the Apicomplexa phylum, the absence of an FMN/FAD synthase can be seen in obligate intracellular α-proteobacteria, Rickettsiae (47).
Vitamin B3
Vitamin B3, or nicotinic acid, also known as niacin, is essential for generation of coenzymes NAD+ and NADP+, which act as key electron carriers in a cell. The apicomplexans are unable to synthesize nicotinate or nicotinamide de novo, indicating that these metabolites are salvaged from the host. All apicomplexans possess the enzymes for the subsequent conversion into NAD+ and NADP+, although the corresponding genes appear dispensable for T. gondii tachyzoites, based on their FS (NAD+ synthase, +0.03; NAD+ kinase, −1.33) (Fig. 3). Of relevance, the CRISPR-Cas9 screen was performed with cultured human foreskin fibroblasts grown in rich media containing an abundance of amino acids, vitamins, salts, and sugars. This may allow certain genes to seem dispensable than they actually would be in a physiological environment more restricted in nutrients. Fitness scores might also vary, depending on the metabolic rates and capabilities of different host cell types (in vitro or in vivo).
In P. falciparum, infected erythrocytes showed a 10-fold increase in NAD+ content compared with uninfected cells, suggesting an efficient and functional biosynthesis pathway in the parasite (48). Due to the substantial release of NAD+ from Plasmodium-infected erythrocytes, NAD+ has been proposed as a potential clinical biomarker for malaria (49). The impact of blocking the parasite nicotinate mononucleotide adenylyl transferase, which synthesizes NAD+ from nicotinate, validates the biosynthesis pathway as an antimalarial target (50).
Vitamin B5
Vitamin B5, or pantothenate (PAN), is the precursor for the biosynthesis of the essential cofactor, CoA. PAN synthesis takes place in most bacteria plants and fungi, but not in animals. The biosynthesis of CoA from PAN, on the other hand, is present in almost all organisms. The de novo synthesis of PAN requires three enzymatic activities: hydroxymethyl transfer to ketoisovalerate (KPHMT), α-ketopantoate reduction (KPR) to pantoate, and pantoate-β-alanine ligation (PBAL). Interestingly, T. gondii encodes the pathway in two sequences conserved within the coccidians, which include Hammondia, Neospora, Besnoitia, Cyclospora, and Eimeria genera. The PAN synthesis pathway has been partially characterized in T. gondii, and its essentiality has been proposed based on the use of chemical inhibitors (51). However, the tested drugs had been developed for Mycobacterium tuberculosis homologue (panC), and off-target effects cannot be excluded.
In T. gondii, the first enzyme in the PAN synthesis pathway is bifunctional, encoding the first two enzymatic steps ketopantoate hydroxymethyl transferase and ketopantoate reductase (KPHMT-KPR). The KPHMT and KPR domains of the protein present conserved key catalytic residues (52, 53) when compared with Escherichia coli panB and panE, respectively. The fusion of the two catalytic domains into one ORF can also be found outside the Apicomplexa phylum in Dinoflagellates (Perkinsus marinus) and free-living photosynthetic Chromerida (Vitrella brassicaformis and Chromera velia), where, interestingly, a single ORF comprises all three enzymes for the synthesis of PAN (Table S1).
The final step of PAN synthesis is catalyzed by PBAL, which ligates pantoate with β-alanine. Sequence comparison with E. coli panC indicates that >30% of the catalytic domain and all catalytic residues (54) are conserved in T. gondii PBAL, pointing to a possible conservation of function. In all members of the phylum, the protein presents an extended N and C terminus (the latter conserved >45% within Neospora, Hammondia, and Besnoitia genera), although no known molecular function has been associated to date. The respective FS of KPHMT-KPR (+0.09) and PBAL (+0.72) indicate in vitro dispensability for PAN synthesis, suggesting that T. gondii, as demonstrated for P. falciparum (55), utilizes host derived PAN for CoA synthesis. Except for the coccidians, the apicomplexans lack PAN synthesis enzymes, and attempts to identify a PAN transporter by orthology have proven difficult (56–58).
CoA, the end product of the pathway, is essential for a broad range of metabolic functions. It provides activated acyl groups for various metabolic pathways, such as the tricarboxylic acid cycle, fatty acid synthesis, and heme synthesis, as well as for gene regulation and post-translational modification of proteins (59). Pantothenate kinase (PanK), which catalyzes the first step in CoA synthesis, has been extensively characterized in P. falciparum (60), allowing pantothenamides (pantothenate mimetic compounds) to be catabolized into CoA antimetabolites (61) with deleterious effects for the parasite (62). Interestingly, of the five enzymes required for CoA synthesis, phosphopantetheine-cysteine ligase and phosphopantothenoylcysteine decarboxylase, which catalyze the second and third step, respectively, are dispensable in both the rodent malaria parasites Plasmodium yoelii and P. berghei (19, 63). This observation could be explained by the promiscuous activity of PanK (64), allowing usage of pantetheine (an intermediate) scavenged from the host cell (65). In T. gondii, the FS of all of the enzymes of the CoA synthesis pathway indicate essentiality (including the two different genes encoding for PanK) (Fig. 4). We have recently identified the gene coding for the final step, dephospho-CoA kinase, previously thought to be missing from the genome, and have shown that the activity is essential for parasite survival by conditional disruption.5
Figure 4.
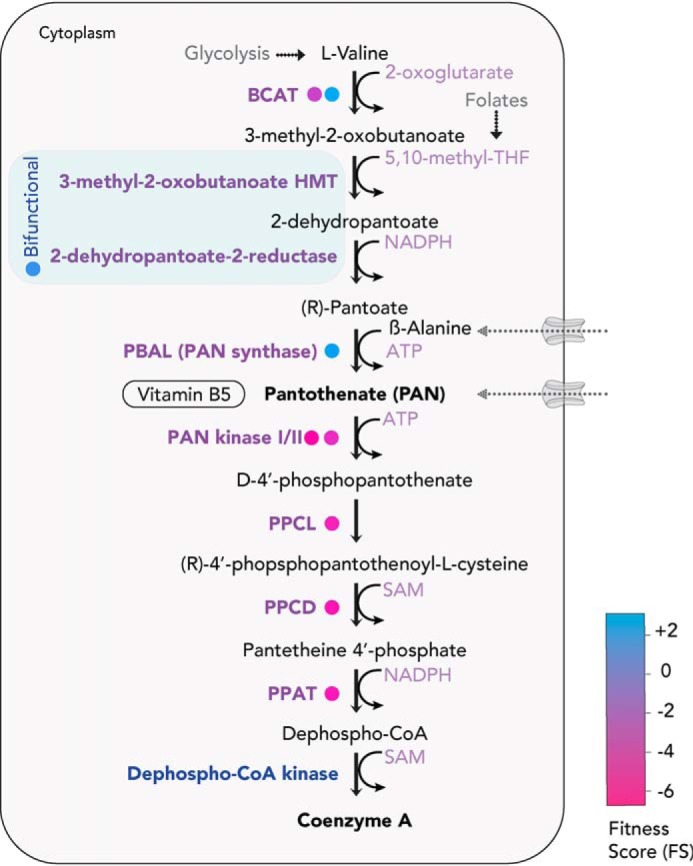
PAN (vitamin B5) and CoA biosynthesis pathway. T. gondii can de novo–synthesize or uptake PAN and subsequently convert it into CoA within the parasite. The bifunctional enzyme for PAN synthesis is shown in blue. FS for the enzymes are color-coded (in circles). BCAT, branched-chain amino acid transaminase; HMT, hydroxymethyltransferase; PPCL, phosphopantetheine-cysteine ligase; PPCD, phosphopantothenoylcysteine decarboxylase; PPAT, pantetheine-phosphate adenylyl transferase.
Taken together, it appears that most apicomplexans share the capability to scavenge PAN from their host. Hence, the retention of the PAN synthesis pathway among the coccidians is intriguing. It is likely that PAN synthesis is required in life cycle stages where exogeneous PAN levels are limiting, such as in sporozoites or in the cyst-enclosed bradyzoites of T. gondii. Importantly, PAN synthesis requires β-alanine, for which no synthesis pathway has been clearly identified in the genome of T. gondii. Thus, the parasite would have to acquire this metabolite from its environment. Further research is necessary to delineate the relevance of PAN synthesis in coccidians.
Vitamin B6
Vitamin B6 is part of the essential vitamin B group of molecules, consisting of pyridoxal, pyridoxamine, and pyridoxine. The metabolically active form is pyridoxal 5′-phosphate (PLP). PLP is a crucial cofactor for the activity of over 140 enzymes, several of them involved in amino acid metabolism (66, 67). Two different routes for the de novo synthesis of PLP exist in organisms: 1-deoxy-d-xylulose 5-phosphate (DOXP)-dependent and DOXP-independent (68). The DOXP-dependent route occurs in proteobacteria and most other bacteria, whereas eukaryotes, including the apicomplexans, utilize the DOXP-independent route. In this route, PLP is synthesized via the activity of two enzymes, PDX1 (PLP synthase subunit) and PDX2 (class I glutamine amidotransferase). Free vitamin B6 forms can also be phosphorylated via the action of pyridoxal kinase (PLK or PDXK). The subsequent conversion of pyridoxamine-5P and pyridoxine-5P to PLP can be performed via a different enzyme, pyridoxal 5′-phosphate synthase (PLP synthase).
Both coccidians and hemosporidians possess all of the enzymes for de novo synthesis as well as scavenge of the vitamin (69, 70). The FS for the genes coding for PDX1 (+0.59), PDX2 (+0.08), PLP synthase (−0.33), and PLK (−0.41) indicate dispensability in vitro (Fig. 5), indicating redundancy between synthesis and salvage for PLP production. In a recent study, disrupting de novo biosynthesis of PLP via conditional knockdown of PDX1 was detrimental in parasites lacking the PLK gene.3 The synthetic lethality showed that blocking both routes for cofactor generation is deleterious, and several PLP-dependent enzymes must become inactive. One such enzyme is glycogen phosphorylase, which breaks down the storage polysaccharide amylopectin (71). In T. gondii, loss of glycogen phosphorylase is associated with amylopectin accumulation and lethal for both tachyzoites and bradyzoites (72). Indeed, amylopectin accumulation was observed in mutants depleted of PLP.3 Although PLP requirement for several enzymes is fulfilled with either the biosynthesis or scavenge pathway in vitro, contrastingly, the deletion of PDX1 alone was sufficient to abolish T. gondii virulence in mice.3 This points to limited or insufficient amounts of pyridoxal in the organs or tissues infected with T. gondii in vivo (73). The sole reliance on the de novo pathway for PLP production in vivo makes PDX1 an attractive drug target or candidate for an attenuated live vaccine.
Figure 5.
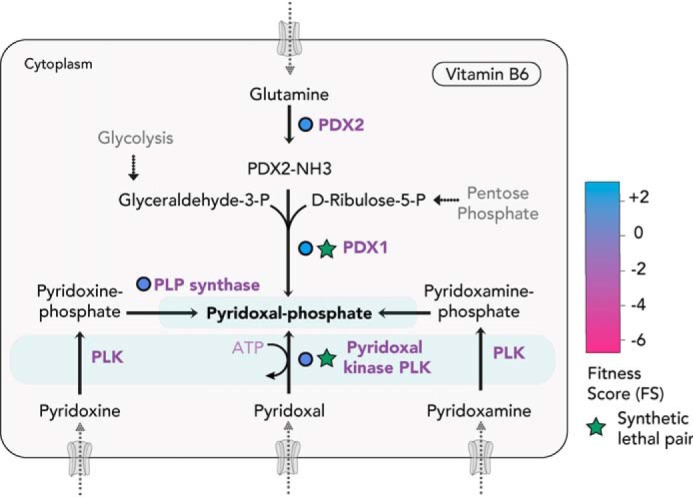
Pyridoxal-5P (vitamin B6) biosynthesis and scavenge pathways. T. gondii can de novo synthesize PLP or uptake the vitamers to subsequently convert them into PLP within the parasite. PLK (in blue) can phosphorylate any of the vitamers—pyridoxal, pyridoxamine, or pyridoxine—and is synthetically lethal with the synthesis enzyme, PDX1. FS for the enzymes for the bioconversion are color-coded (in circles). Experimentally validated enzymes are circled in black.3
If the biosynthesis pathway is the major route for PLP production in T. gondii in vivo, the presence and role of PLK is puzzling. To test its role during latency, mice infected with parasites lacking PLK were examined for cyst formation.3 No reduction in cyst number was observed, compared with the WT, suggesting its dispensability for the chronic stage. It is plausible that the enzyme has a role during the sexual or oocyst stages, recycling any free pyridoxal in the cell and preventing toxic accumulation of the vitamers. How the vitamers enter the parasite remains unknown, and the absence of the biosynthesis of PLP in Cryptosporidia and piroplasms further indicates an unusual salvage mechanism for the phosphorylated cofactor.
In P. falciparum, both PLP biosynthesis and salvage pathways have been shown to be functional. The two genes (encoding for PDX1 and PDX2) are expressed throughout the intraerythrocytic and gametocyte development and have been explored as potential drug targets (74–77). Prodrugs such as pyridoxyl-tryptophan chimeras that interfere with PLP-dependent enzymes and poison the parasite have also been investigated as antimalarials (78, 79). For organisms that lack biosynthesis capabilities, identification of the transporter of pyridoxal and its derivatives would be of significant interest.
Vitamin B7
Vitamin B7 or biotin can be synthesized by bacteria, plants, and some fungi, but not by animals. The apicomplexans also lack the biosynthesis capability for biotin. Biotin is an important cofactor for the enzyme acetyl-CoA carboxylase (ACCase), of which ACCase1 was found in the apicoplast of T. gondii (80). In bacteria, biotin covalently attaches to the ϵ-amino group of specific lysine residues in the carboxylases via the action of a biotin-ligase (81). A putative biotin-ACC-ligase, with similarity to the E. coli biotin operon repressor (BirA) was found in the genome of most apicomplexans. If its role is similar to that of BirA for sensing biotin levels and regulating transcription is unknown (82). How biotin is acquired from the host and transported into the apicoplast, where ACCase1 resides, also remains to be understood. Biotin uptake is mediated by solute transporters in prokaryotes (83) and via a monocarboxylate transporter (MCT1) in mammalian cells (84).
Vitamin B9
Vitamin B9 or folate is crucial for DNA replication, cell division, and synthesis of several amino acids. The folate derivative, 5,10-methylenetetrahydrofolate, is essential for the production of dTMP and dUMP nucleotides. In addition to the de novo folate biosynthesis pathway from shikimate and chorismate, most apicomplexans can also salvage folate from the host via dedicated BT1 or FT transporters (85, 86) (Fig. 6). The high-affinity folate transporters were shown to take up radiolabeled exogenous folic acid in T. gondii (85). If folates are taken up to sustain the acute stage of T. gondii, the existence of the biosynthesis pathway is likely relevant for downstream metabolite production or for a different life cycle stage where the parasite encounters limited access to folates or its precursors. Numerous studies have shown the effects of targeting the folate pathway (87, 88). Several anti-parasitic drugs are currently in use, such as sulfonamides targeting dihydropteroate synthase in combination with inhibitors of the dihydrofolate reductase-thymidylate synthase. Although the anti-folates are thought to be safe, recent studies in P. falciparum have shown emerging resistance to the once potent drug combination. Future studies would have to unravel the molecular mechanisms of resistance and enable future development of alternative strategies targeting the crucial biosynthesis and scavenge pathways (89). In recent in vivo experiments, the contributions of para-amino benzoic acid (pABA), a precursor for folate synthesis, were also re-examined (90, 91). pABA is synthesized with the action of two enzymes, aminodeoxychorismate synthase and aminodeoxychorismate lyase. The two genes were knocked out in the rodent malaria parasite P. berghei, and the deletions were shown to be dispensable for parasite propagation in mice fed with a conventional diet. However, in mice fed with milk (lacking pABA), the mutants displayed a severe growth phenotype, abolished with the supplementation of pABA (90, 91). In the liver stage, the lack of aminodeoxychorismate synthase was dispensable, suggesting an active salvage, given the folate-rich environment of the liver. The results therefore indicate a combination of salvage and synthesis in Plasmodium parasites, to ensure the folate requirements for the fast-growing asexual stages are met.
Figure 6.
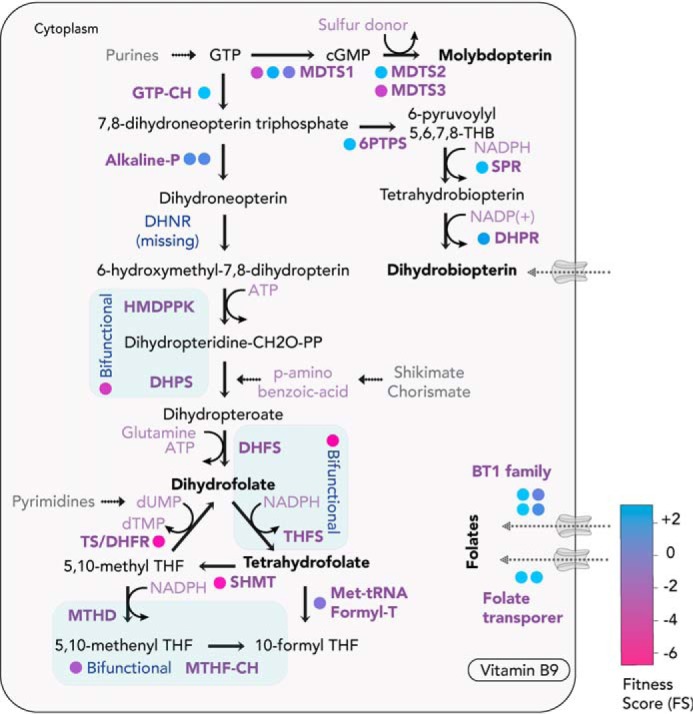
Folate (vitamin B9) and biopterin biosynthesis and scavenge pathways. T. gondii can de novo–synthesize or uptake folates and biopterins. FS for the enzymes for the bioconversion are color-coded (in circles). Enzymes in blue are bifunctional, capable of catalyzing two subsequent reaction steps. TS, thymidylate synthase; DHFR, dihydrofolate reductase; DHPS, dihydropteroate synthase; GTP-CH, GTP cyclohydrolase; MDTS1, molybdopterin cofactor synthesis protein 1 (MOCS1); MDTS2, molybdopterin cofactor synthesis protein 2 (MOCS2/MoaE); MDTS3, molybdopterin cofactor synthesis protein 3 (MOCS3/MoaB); 6PTPS, 6-pyruvoyltetrahydropterin synthase; SPR, sepiapterin reductase; DHPR, 6,7-dihydropteridine reductase; DHPS, dihydropteroate synthase; DHFR, dihydrofolate reductase; TS, thymidylate synthase; MTHD, methylenetetrahydrofolate dehydrogenase; MTHF-CH, methenyl-tetrahydrofolate cyclohydrolase; SHMT, serine hydroxymethyltransferase; DHFS, dihydrofolate synthase; THFS, tetrahydro-folylpolyglutamate synthase; Met-tRNA, methionyl-tRNA formyl-transferase.
Heme
Heme is an essential cofactor required for the function of various enzymes, including cytochromes, catalases, peroxidases, hemoglobin, and others. Heme alternates between an oxidized and reduced state, enabling heme-containing enzymes to catalyze electron transfer reactions in the ETC and other pathways. Heme can be synthesized de novo, via a highly conserved eight-step pathway (92, 93). Alternatively, it can be salvaged via heme-binding proteins and porphyrin transporters, which have been partially identified in protozoan parasites such as trypanosomes but remain elusive in apicomplexans (93–95). Whereas Trypanosoma cruzi and Trypanosoma brucei are unable to synthesize heme, Leishmania spp. have acquired the last three enzymes of the biosynthesis pathway via horizontal gene transfer, possibly acquiring and converting heme precursors from the host (93, 96, 97). Within the Apicomplexa, Cryptosporidia have lost all enzymes required for heme synthesis, relying entirely on an uptake mechanism. Coccidians and hemosporidians encode all enzymes necessary for de novo synthesis of heme (93). They possess a peculiar synthesis pathway, which spans three subcellular compartments, the mitochondrion, apicoplast, and cytosol, and comprises enzymes with distinct ancestral origins (93, 98) (Fig. 7). The parasites utilize the so-called C4 pathway of α-proteobacterial origin, in which the heme precursor δ-aminolevulinic acid (ALA) is synthesized through condensation of succinyl-CoA and glycine in the mitochondrion. δ-ALA is transported to the apicoplast, where the four-step conversion into coproporphyrinogen III occurs, catalyzed by enzymes originating from the algal endosymbiont (98, 99). Coproporphyrinogen III is exported from the apicoplast to the cytosol, where it is converted to protoporphyrinogen IX by a coproporphyrinogen III oxidase (CPO). Protoporphyrinogen IX is subsequently transported to the mitochondrion and converted to heme through the activity of protoporphyrinogen oxidase and ferrochelatase (FC). The contribution of heme uptake versus its de novo synthesis has been investigated in depth in Plasmodium spp. In its blood stages, Plasmodium parasites deal with very high levels of heme, which are released during the digestion of hemoglobin. P. falciparum detoxifies heme by depositing it in a large crystalline pigment termed hemozoin. Hemozoin formation is mediated by a multiprotein complex in the food vacuole, which contains several proteases and a heme detoxification protein (100). Whereas protein-driven hemozoin formation has been postulated before (101), lipid-driven mechanisms (102, 103) and an autocatalytic process have also been proposed (104). Unsurprisingly, heme synthesis is not essential for Plasmodium during the intraerythrocytic development, but the pathway becomes fitness-conferring during liver stages and is essential for development in the mosquito (105–109). Specifically, the loss of FC impairs male gamete formation and ablates oocyst formation in mosquitoes, indicating that Plasmodium can utilize salvaged heme but relies on its synthesis when levels of exogeneous heme become limiting within the insect vector (105, 106).
Figure 7.
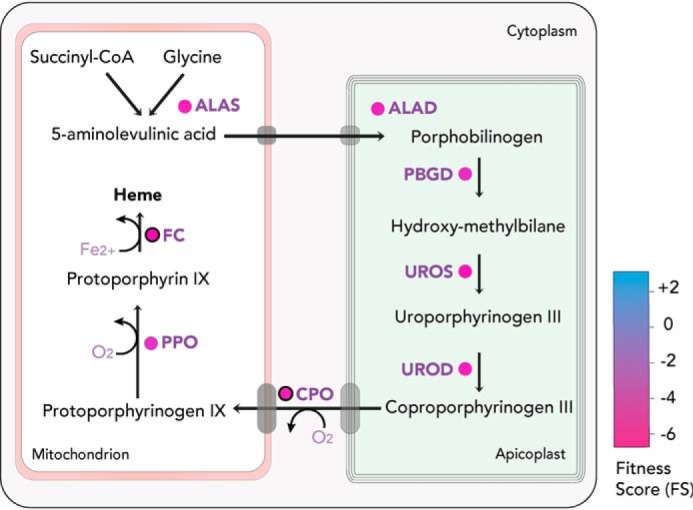
Heme biosynthesis pathway. T. gondii can de novo–synthesize heme in a complex pathway, compartmentalized between the mitochondrion, cytosol, and apicoplast. FS for the enzymes are color-coded (in circles). Experimentally validated enzymes are circled in black.3 ALAS, aminolevulinate synthase; ALAD, aminolevulinate dehydratase; PBGD, porphobilinogen deaminase; UROS, uroporphyrinogen synthase; UROD, uroporphyrinogen decarboxylase; PPO, protoporphyrinogen oxidase.
Heme has also been intensely researched for its role in determining sensitivity of the parasite to the antimalarial drug artemisinin. Heme-bound iron derived from de novo synthesis or hemoglobin digestion reacts with artemisinin, forming active cytotoxic artemisinin radicals (110–112). It has been shown that enhancing heme synthesis, by providing excess heme precursors, increases the sensitivity of Plasmodium to artemisinin. Conversely, the reduction of heme synthesis by genetic means or through pharmacological inhibition decreases sensitivity of both T. gondii and P. falciparum to artemisinin (113, 114).
Whereas T. gondii does not have to deal with copious amounts of heme as in the intra-erythrocytic stage of P. falciparum, it is also expected to encounter varying levels of heme during its complex life cycle. Based on their FS, all enzymes implicated in the heme synthesis pathway appear highly fitness-conferring (17) (Fig. 7), indicating that in vitro tachyzoites are unable to scavenge sufficient amounts of heme from their host. The enzyme catalyzing the second step of the pathway, ALA dehydratase or porphobilinogen synthase, has been characterized biochemically (115). Its crystal structure revealed that the enzyme functions as an octamer in T. gondii and does not contain any metal ions in the active site, although Mg2+ ions are present at the intersections between pro-octamer dimers (116). This metal-independent catalysis is unique to apicomplexans and could render the enzyme an attractive target for intervention.
Interestingly, T. gondii also encodes two putative and distinct types of coproporphyrinogen oxidases, a CPO and a bacterial-type coproporphyrinogen III dehydrogenase (CPDH) (117, 118). Whereas CPO appears to be highly fitness-conferring based on its FS (−4.64), the oxygen-independent CPDH (+2.29) is the only dispensable enzyme associated with the pathway. Its role in the heme synthesis of T. gondii is still unknown, as it may function as the active CPO in a life cycle stage where oxygen levels are limiting. Consistent with this, RNA-Seq data revealed a striking stage specificity, with CPDH being more than 2-fold up-regulated in bradyzoites, oocysts, and sporozoites (20). The role of both enzymes was recently investigated through the generation and characterization of mutant parasites lacking CPO, CPDH, or both. The results confirmed that in the absence of CPO, parasites are severely impaired in their cell division and the overall lytic cycle is compromised. Contrastingly, parasites lacking CPDH grow normally as tachyzoites and are not affected in stage conversion to bradyzoites or in cyst formation in mice.3 Furthermore, no aggravation of the phenotype was observed in parasites lacking both enzymes, CPO and CPDH. Overexpression of CPDH in parasites lacking CPO further confirmed a lack of compensation, possibly due to the differential localization of the two enzymes (CPDH in the mitochondrion and CPO in the cytosol).3 Together, these findings indicate that CPDH is dispensable for both tachyzoites and bradyzoites, highlighting that oxygen levels at these stages are sufficient for the oxygen-dependent CPO to function. Importantly, the activity of CPDH has to date not been formally demonstrated, and misannotations of SAM-dependent enzymes have been reported previously (119). Hence, it remains unclear whether the enzyme truly functions as a CPDH in sporozoites, oocysts, or gametes or whether it functions in a different pathway.
Importantly, although parasites lacking CPO were severely impaired, they remained viable. On the other hand, depletion of the final enzyme, FC, was not tolerated. Mass spectrometry and fluorescence analyses revealed that cells lacking CPO have 10-fold lower heme levels than WT parasites, but 10-fold higher levels of its precursor protoporphyrin IX (ProtoIX).3 These findings indicate that T. gondii likely does not salvage heme itself but rather its precursors ProtoIX or protoporphyrinogen IX from its host. Hence, FC is absolutely essential for the integration of iron into ProtoIX. Conversion of salvaged ProtoIX or protoporphyrinogen IX to heme appears to be inefficient, leading to the described phenotype. This was further supported by the observation that δ-ALA supplementation rescues the growth defect of T. gondii lacking CPO. δ-ALA supplementation leads to a drastic increase in host ProtoIX levels, probably boosting its uptake by T. gondii and allowing it to restore heme levels.
In parasites lacking CPO, the lack of heme and accumulation of its precursor are expected to cause deleterious impacts on T. gondii metabolism and development. Heme is crucial for multiple cellular processes; most notably, it serves as an essential cofactor in several enzymes of the ETC, including cytochrome bc1 of complex III, soluble cytochrome c, and the Cox I subunit of Complex IV (120). It has been proposed that oxidative phosphorylation is the main energy source of tachyzoites and accounts for >90% of the ATP generated in egressed tachyzoites (23). We found that heme depletion in parasites lacking CPO largely disables mitochondrial respiration, although residual low levels of respiration were detected, and parasites devoid of CPO remained sensitive to atovaquone treatment, which inhibits the cytochrome bc1 complex of the ETC.3 Strikingly, these parasites appear to survive through markedly increased rates of glycolysis and are unable to survive in the absence of glucose. These observations highlight the importance of de novo heme synthesis in T. gondii but also demonstrate its astonishing flexibility to adapt and survive solely on an inefficient precursor salvage pathway and rewiring its central carbon metabolism. Given the absence of the heme biosynthesis pathway in Cryptosporidia and piroplasms, future research should focus on the identification of heme or hemoprotein transport mechanisms.
Lipoate
Lipoate, or lipoic acid, is an essential cofactor and, in most eukaryotes, is synthesized in the mitochondrion and transported to other subcellular compartments. In apicomplexans, at least four metabolic complexes use the lipoic acid as a cofactor: PDH, which resides in the apicoplast (121), as well as the α-ketoglutarate dehydrogenase, branched-chain α-ketoacid dehydrogenase, and glycine cleavage complex, which reside in the mitochondrion (27, 122). The coccidians and hemosporidians are able to synthesize and scavenge lipoic acid, whereas the pathways are absent in Cryptosporidia and Piroplasmida. Unlike plants, which have two isoenzymes, LipA and LipB, for lipoylation in the chloroplast and mitochondria, respectively, apicomplexan genomes encode LipA and LipB. Both enzymes are localized to the apicoplast, and a second enzyme, LplA, is found in the mitochondrion (123). Lipoylation of mitochondrial proteins is dramatically reduced when the parasites are grown in lipoic acid–deficient media without affecting the lipoylation of apicoplast proteins (124). Contrastingly, the reduced lipoylation of mitochondrial proteins could be rescued via exogenous supplementation of lipoate in the media, indicating the salvage pathway primarily supplies lipoate for this organelle (124). As seen by the FS of the LplA gene (−2.60), mitochondrial lipoylation seems essential, whereas LipA (−0.97) and LipB (−1.74) (17) in the apicoplast seem dispensable. In the absence of a lipoylated PDH complex, the parasites likely compensate by taking up fatty acids from the host (Fig. 8). Similar observations were reported during the intraerythrocytic stage of P. falciparum (125, 126). The plasma membrane and organellar transporters involved in lipoate salvage have not yet been identified. It is plausible that lipoate is directly scavenged from the host mitochondria, which is in close contact with the parasitophorous vacuole (127).
Figure 8.
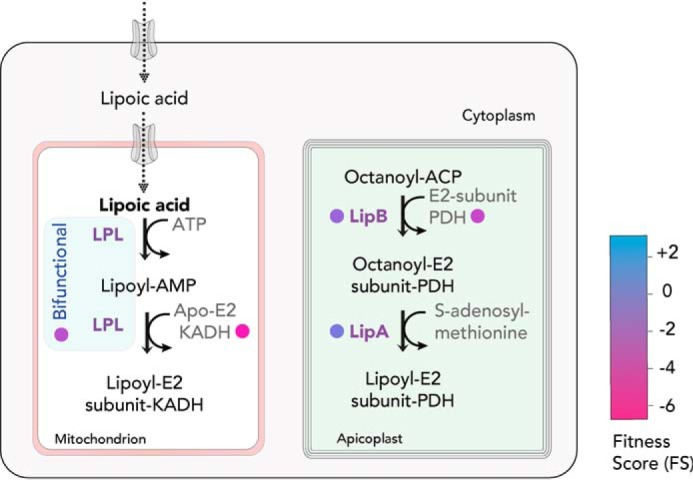
Lipoic acid biosynthesis. T. gondii can de novo–synthesize lipoic acid in the apicoplast but also scavenge the metabolite from its host for its requirement within the mitochondrion. The bifunctional LPL enzyme (in blue) utilizes the scavenged lipoate for the posttranslational modification of branched-chain keto-acid dehydrogenase. LipB and LipA generate lipoate for the modification of the E2 subunit of the apicoplast-resident PDH complex. FS for the enzymes are color-coded (in circles). LipB, lipoyl (octanoyl)-ACP-protein N-lipoyl (octanoyl) transferase; LPL, lipoate-protein ligase; LipA, lipoic acid synthase; KADH, branched-chain keto-acid dehydrogenase.
Shikimate
Shikimate is an important metabolite found in bacteria, plants, and fungi but is absent in animals. It is important for several biosynthetic processes, including the biosynthesis of folate, aromatic amino acids, and ubiquinone. Shikimate is primarily synthesized from erythrose 4-phosphate and phosphoenolpyruvate and subsequently converted to chorismate in a seven-step reaction. Steps 2–6 for chorismate biosynthesis are carried out by a pentafunctional protein (Fig. 9). In most apicomplexans, including the coccidians, hemosporidians, and Cryptosporidia, a single gene of fungal origin exists, called the AROM complex, encoding for all five activities in a single large polypeptide (128, 129). The presence of all functional domains in T. gondii has been verified with bioinformatic analyses (130, 131), although in P. falciparum the sequence similarity to the yeast homolog could not be verified for the first two enzymatic activities. However, evidence for the presence of a shikimate pathway was supported in both T. gondii tachyzoites and the erythrocytic stage of P. falciparum, by treating the parasites with the herbicide glyphosate, inhibitor of the 5-enolpyruvylshikimate-3-phosphate synthase, resulting in a growth defect (132–134). The effect was reversible with the addition of pABA or folate in the media, suggesting an essential role of shikimate in providing precursors for the biosynthesis of folates (133). The role of chorismate in folate biosynthesis has been demonstrated in several studies, but its importance for ubiquinone biosynthesis has not been fully defined. Further, the high negative FS of all enzymes involved in the pathway confirms its essentiality for in vitro T. gondii tachyzoites (AROM complex, −5.22; chorismate synthase, −2.84) and could be targeted for intervention against the coccidians and hemosporidians.
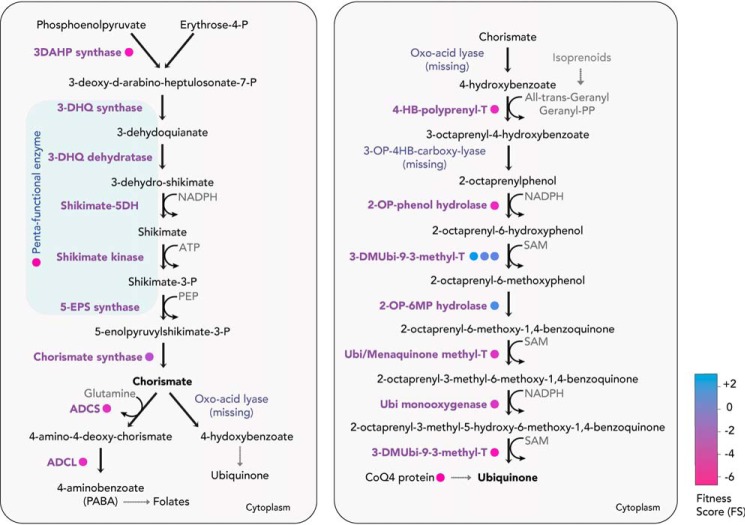
Figure 9.
Shikimate, chorismate, and ubiquinone biosynthesis pathway. T. gondii can de novo–synthesize shikimate and chorismate via a pentafunctional AROM complex, catalyzing the initial five steps (shaded in light blue) and chorismate synthase respectively. Chorismate is a precursor for the biosynthesis of ubiquinone, and the FS for the enzymes are color-coded (in circles). 3DAHP, 3-deoxy-D-arabinoheptulosonate 7-phosphate; 5-EPS, 5-enolpyruvylshikimate-3-phosphate; 3-DHQ, 3-dehydroquinate; ADCS, aminodeoxychorismate synthase; ADCL, aminodeoxychorismate lyase; PEP, phosphoenolpyruvate; 4-HB, 4-hydroxybenzoate.
Ubiquinone
Ubiquinone, also known as coenzyme Q, is an integral component of the electron transport chain for the transfer of electrons from NADH dehydrogenase (complex I) and succinate dehydrogenase (complex II) to cytochrome bc1 complex (complex III). In most organisms, ubiquinone is synthesized from chorismate in nine enzymatic steps. Most of the pathway is conserved among all apicomplexans, with two enzymes, oxo-acid lyase and 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, missing from the genome, based on bioinformatic approaches. The divergence of these enzymes cannot be ruled out, because a functional synthesis pathway in P. falciparum was shown by detecting differences in the ubiquinone side chains when compared with the host (135). The 4-hydroxybenzoate backbone of ubiquinone receives an isoprenoid side chain via the 4-HB-prenyl-transferase, which has been well-characterized in P. falciparum, and localized to the apicoplast (136). The production of long-chain isoprenoids, however, occurs in the mitochondrion via farnesyl pyrophosphate synthase (137), which could subsequently be utilized for the synthesis of ubiquinone and other compounds. It was further shown that fosmidomycin, a drug that inhibits the apicoplast-resident isoprenoid biosynthesis pathway, leads to a decline in ubiquinone synthesis (74). In T. gondii tachyzoites, the last three steps of the pathway (Fig. 9) display highly negative FS (−3.61, −3.62, and −4.49), highlighting their importance for in vitro proliferation.
Conclusion
Apicomplexans possess versatile metabolic capabilities to adapt and adjust to their diverse host environments. Understanding the parasite's requirements for intracellular replication and the contribution of biosynthesis versus uptake of essential metabolites is therefore crucial for the identification of new candidate drug targets (Fig. 10). Whereas the genome sequences of the disease-causing pathogens provide us clues on their metabolic capabilities at a global level, an in-depth understanding of the needs at each life cycle stage is vital. Pathways and enzymes that are essential for proliferation during acute infection may be dispensable upon stage conversion to latency and vice versa. Recent studies encompassing computational, molecular, and metabolomic tools have advanced our understanding of metabolic pathways for the production of key vitamins and cofactors, paving the way for targeted drug development. A few commercially available compounds targeting vitamin and cofactor pathways, such as pyrimethamine and sulfonamides, already exist to treat toxoplasmosis or malaria. With the rise in drug resistance, however, identification of new enzymes absent in the mammalian host may be useful for a target-directed intervention against the apicomplexans.
Figure 10.
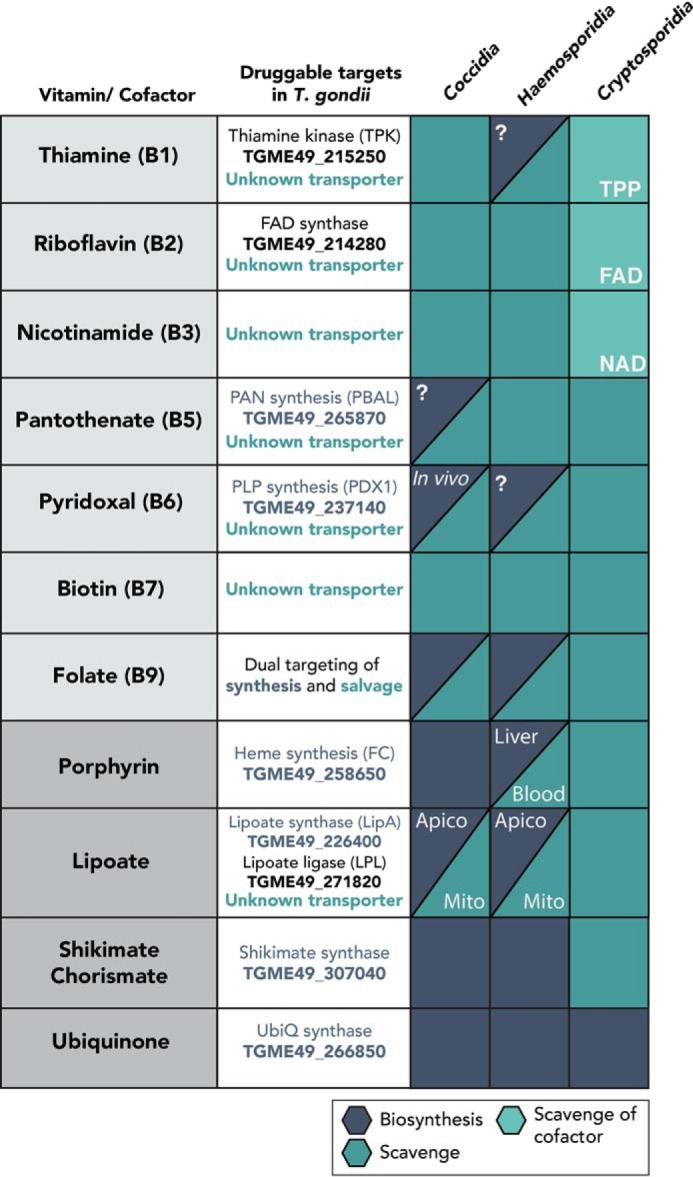
List of potential drug targets in the vitamin and cofactor biosynthesis and salvage pathways within a selected class of apicomplexans. Gene IDs for known genes in T. gondii are listed with unknown transporters. Essentialities of the enzymes for known life cycle stages, in vivo conditions, or intracellular organelles are marked in white. A question mark indicates the presence of a biosynthesis enzyme, although its essentiality for a different life cycle stage of the parasite is unknown. Apico, apicoplast; Mito, mitochondrion.
Supplementary Material
This work was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program under Grant Agreement 695596 (to J. K. and M. L.).
This article contains Table S1.
Krishnan, et al., Functional and computational genomics reveal unprecedented flexibility in stage-specific Toxoplasma metabolism. Cell Host & Microbe., in press.
M. Lunghi, J. Kloehn, and D. Soldati-Favre, unpublished observations.
- PDH
- pyruvate dehydrogenase
- TPK
- thiamine diphosphokinase
- PBAL
- pantoate-β-alanine ligase or PAN synthase
- PAN
- pantothenate
- PLP
- pyridoxal 5′-phosphate
- PLK
- pyridoxal kinase or PdxK
- CPO
- coproporphyrinogen oxidase
- CPDH
- coproporphyrinogen dehydrogenase
- FC
- ferrochelatase
- FA
- fatty acid
- TPP
- thiamine pyrophosphate
- FS
- fitness score(s)
- ETC
- electron transport chain
- KPHMT
- Ketopantoate hydroxymethyltransferase
- KPR
- α-ketopantoate reductase
- PanK
- pantothenate kinase
- DOXP
- 1-deoxy-d-xylulose 5-phosphate
- ACCase
- acetyl-CoA carboxylase
- ALA
- δ-aminolevulinic acid
- ProtoIX
- protoporphyrin IX.
References
- 1. Plattner F., and Soldati-Favre D. (2008) Hijacking of host cellular functions by the Apicomplexa. Annu. Rev. Microbiol. 62, 471–487 10.1146/annurev.micro.62.081307.162802 [DOI] [PubMed] [Google Scholar]
- 2. Dubey J. P., Lindsay D. S., and Speer C. A. (1998) Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin. Microbiol. Rev. 11, 267–299 10.1128/CMR.11.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dubey J. P. (1988) Long-term persistence of Toxoplasma gondii in tissues of pigs inoculated with T. gondii oocysts and effect of freezing on viability of tissue cysts in pork. Am. J. Vet. Res. 49, 910–913 [PubMed] [Google Scholar]
- 4. Polonais V., and Soldati-Favre D. (2010) Versatility in the acquisition of energy and carbon sources by the Apicomplexa. Biol. Cell. 102, 435–445 10.1042/BC20100005 [DOI] [PubMed] [Google Scholar]
- 5. Gold D. A., Kaplan A. D., Lis A., Bett G. C. L., Rosowski E. E., Cirelli K. M., Bougdour A., Sidik S. M., Beck J. R., Lourido S., Egea P. F., Bradley P. J., Hakimi M.-A., Rasmusson R. L., and Saeij J. P. J. (2015) The toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe 17, 642–652 10.1016/j.chom.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garten M., Nasamu A. S., Niles J. C., Zimmerberg J., Goldberg D. E., and Beck J. R. (2018) EXP2 is a nutrient-permeable channel in the vacuolar membrane of Plasmodium and is essential for protein export via PTEX. Nat. Microbiol. 3, 1090–1098 10.1038/s41564-018-0222-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sherling E. S., and van Ooij C. (2016) Host cell remodeling by pathogens: the exomembrane system in Plasmodium-infected erythrocytes. FEMS Microbiol. Rev. 40, 701–721 10.1093/femsre/fuw016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baumeister S., Winterberg M., Duranton C., Huber S. M., Lang F., Kirk K., and Lingelbach K. (2006) Evidence for the involvement of Plasmodium falciparum proteins in the formation of new permeability pathways in the erythrocyte membrane. Mol. Microbiol. 60, 493–504 10.1111/j.1365-2958.2006.05112.x [DOI] [PubMed] [Google Scholar]
- 9. Mehlhorn H., and Shein E. (1984) The piroplasms: life cycle and sexual stages. Adv. Parasitol. 23, 37–103 10.1016/s0065-308x(08)60285-7 [DOI] [PubMed] [Google Scholar]
- 10. Jalovecka M., Hajdusek O., Sojka D., Kopacek P., and Malandrin L. (2018) The complexity of piroplasms life cycles. Front. Cell. Infect. Microbiol. 8, 248 10.3389/fcimb.2018.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Hara S. P., and Chen X.-M. (2011) The cell biology of Cryptosporidium infection. Microbes Infect. 13, 721–730 10.1016/j.micinf.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moore R. B., Oborník M., Janouškovec J., Chrudimský T., Vancová M., Green D. H., Wright S. W., Davies N. W., Bolch C. J. S., Heimann K., Šlapeta, Hoegh-Guldberg J. O., Logsdon J. M., and Carter D. A. (2008) A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451, 959–963 10.1038/nature06635 [DOI] [PubMed] [Google Scholar]
- 13. Song C., Chiasson M. A., Nursimulu N., Hung S. S., Wasmuth J., Grigg M. E., and Parkinson J. (2013) Metabolic reconstruction identifies strain-specific regulation of virulence in Toxoplasma gondii. Mol. Syst. Biol. 9, 708 10.1038/msb.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiappino-Pepe A., Tymoshenko S., Ataman M., Soldati-Favre D., and Hatzimanikatis V. (2017) Bioenergetics-based modeling of Plasmodium falciparum metabolism reveals its essential genes, nutritional requirements, and thermodynamic bottlenecks. PLoS Comput. Biol. 13, e1005397 10.1371/journal.pcbi.1005397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tymoshenko S., Oppenheim R. D., Agren R., Nielsen J., Soldati-Favre D., and Hatzimanikatis V. (2015) Metabolic needs and capabilities of Toxoplasma gondii through combined computational and experimental analysis. PLoS Comput. Biol. 11, e1004261 10.1371/journal.pcbi.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stanway R. R., Bushell E., Chiappino-Pepe A., Roques M., Sanderson T., Franke-Fayard B., Caldelari R., Golomingi M., Nyonda M., Pandey V., Schwach F., Chevalley S., Ramesar J., Metcalf T., Herd C., et al. (2019) Genome-Scale Identification of Essential Metabolic Processes for Targeting the Plasmodium Liver Stage. Cell 179, 1112–1128 10.1016/j.cell.2019.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sidik S. M., Huet D., Ganesan S. M., Huynh M.-H. H., Wang T., Nasamu A. S., Thiru P., Saeij J. P. J., Carruthers V. B., Niles J. C., and Lourido S. (2016) A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell 166, 1423–1435.e12 10.1016/j.cell.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang M., Wang C., Otto T. D., Oberstaller J., Liao X., Adapa S. R., Udenze K., Bronner I. F., Casandra D., Mayho M., Brown J., Li S., Swanson J., Rayner J. C., Jiang R. H. Y., and Adams J. H. (2018) Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360, eaap7847 10.1126/science.aap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R. E., Mather M. W., McFadden G. I., Parts L., Rutledge G. G., Vaidya A. B., et al. (2017) Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272.e8 10.1016/j.cell.2017.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hehl A. B., Basso W. U., Lippuner C., Ramakrishnan C., Okoniewski M., Walker R. A., Grigg M. E., Smith N. C., and Deplazes P. (2015) Asexual expansion of Toxoplasma gondii merozoites is distinct from tachyzoites and entails expression of non-overlapping gene families to attach, invade, and replicate within feline enterocytes. BMC Genomics 16, 66 10.1186/s12864-015-1225-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otto T. D., Böhme U., Jackson A. P., Hunt M., Franke-Fayard B., Hoeijmakers W. A. M., Religa A. A., Robertson L., Sanders M., Ogun S. A., Cunningham D., Erhart A., Billker O., Khan S. M., Stunnenberg H. G., et al. (2014) A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 12, 86 10.1186/s12915-014-0086-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caldelari R., Dogga S., Schmid M. W., Franke-Fayard B., Janse C. J., Soldati-Favre D., and Heussler V. (2019) Transcriptome analysis of Plasmodium berghei during exo-erythrocytic development. Malar. J. 18, 330 10.1186/s12936-019-2968-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. MacRae J. I. I., Sheiner L., Nahid A., Tonkin C., Striepen B., and McConville M. J. J. (2012) Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe 12, 682–692 10.1016/j.chom.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nitzsche R., Zagoriy V., Lucius R., and Gupta N. (2016) Metabolic cooperation of glucose and glutamine is essential for the lytic cycle of obligate intracellular parasite Toxoplasma gondii. J. Biol. Chem. 291, 126–141 10.1074/jbc.M114.624619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shukla A., Olszewski K. L., Llinás M., Rommereim L. M., Fox B. A., Bzik D. J., Xia D., Wastling J., Beiting D., Roos D. S., and Shanmugam D. (2018) Glycolysis is important for optimal asexual growth and formation of mature tissue cysts by Toxoplasma gondii. Int. J. Parasitol. 48, 955–968 10.1016/j.ijpara.2018.05.013 [DOI] [PubMed] [Google Scholar]
- 26. Blume M., Nitzsche R., Sternberg U., Gerlic M., Masters S. L., Gupta N., and McConville M. J. (2015) A Toxoplasma gondii gluconeogenic enzyme contributes to robust central carbon metabolism and is essential for replication and virulence. Cell Host Microbe 18, 210–220 10.1016/j.chom.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 27. Oppenheim R. D., Creek D. J., Macrae J. I., Modrzynska K. K., Pino P., Limenitakis J., Polonais V., Seeber F., Barrett M. P., Billker O., McConville M. J., and Soldati-Favre D. (2014) BCKDH: the missing link in apicomplexan mitochondrial metabolism is required for full virulence of Toxoplasma gondii and Plasmodium berghei. PLoS Pathog. 10, e1004263 10.1371/journal.ppat.1004263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramakrishnan S., Docampo M. D., Macrae J. I., Pujol F. M., Brooks C. F., van Dooren G. G., Hiltunen J. K., Kastaniotis A. J., McConville M. J., Striepen B., Kalervo J. H., Kastaniotis A. J., McConville M. J., and Striepen B. (2012) Apicoplast and endoplasmic reticulum cooperate in fatty acid biosynthesis in apicomplexan parasite Toxoplasma gondii. J. Biol. Chem. 287, 4957–4971 10.1074/jbc.M111.310144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramakrishnan S., Docampo M. D., MacRae J. I., Ralton J. E., Rupasinghe T., McConville M. J., and Striepen B. (2015) The intracellular parasite Toxoplasma gondii depends on the synthesis of long-chain and very long-chain unsaturated fatty acids not supplied by the host cell. Mol. Microbiol. 97, 64–76 10.1111/mmi.13010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pandey V., Hernandez Gardiol D., Chiappino Pepe A., and Hatzimanikatis V. (2019) TEX-FBA: a constraint-based method for integrating gene expression, thermodynamics, and metabolomics data into genome-scale metabolic models. bioRxiv 10.1101/536235 [DOI] [Google Scholar]
- 31. Tymoshenko S., Oppenheim R. D., Soldati-Favre D., and Hatzimanikatis V. (2013) Functional genomics of Plasmodium falciparum using metabolic modelling and analysis. Brief. Funct. Genomics 12, 316–327 10.1093/bfgp/elt017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bender D. A. (2003) Nutritional Biochemistry of the Vitamins, 2nd Ed., pp. 1–8, Cambridge University Press, Cambridge, UK [Google Scholar]
- 33. Müller S., and Kappes B. (2007) Vitamin and cofactor biosynthesis pathways in Plasmodium and other apicomplexan parasites. Trends Parasitol. 23, 112–121 10.1016/j.pt.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seeber F., and Soldati-Favre D. (2010) Metabolic pathways in the apicoplast of Apicomplexa. Int. Rev. Cell Mol. Biol. 281, 161–228 10.1016/S1937-6448(10)81005-6 [DOI] [PubMed] [Google Scholar]
- 35. Hung S. S., and Parkinson J. (2011) Post-genomics resources and tools for studying apicomplexan metabolism. Trends Parasitol. 27, 131–140 10.1016/j.pt.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 36. Fleige T., Limenitakis J., and Soldati-Favre D. (2010) Apicoplast: keep it or leave it. Microbes Infect. 12, 253–262 10.1016/j.micinf.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 37. Xu P., Widmer G., Wang Y., Ozaki L. S., Alves J. M., Serrano M. G., Puiu D., Manque P., Akiyoshi D., Mackey A. J., Pearson W. R., Dear P. H., Bankier A. T., Peterson D. L., Abrahamsen M. S., Kapur V., Tzipori S., and Buck G. A. (2004) The genome of Cryptosporidium hominis. Nature 431, 1107–1112 10.1038/nature02977 [DOI] [PubMed] [Google Scholar]
- 38. Burgess S. L., Gilchrist C. A., Lynn T. C., and Petri W. A. (2017) Parasitic protozoa and interactions with the host intestinal microbiota. Infect. Immun. 85, e00101–17 10.1128/IAI.00101-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hellgren O., Bensch S., and Videvall E. (2018) De novo synthesis of thiamine (vitamin B1) is the ancestral state in Plasmodium parasites—evidence from avian haemosporidians. Parasitology 145, 1084–1089 10.1017/S0031182017002219 [DOI] [PubMed] [Google Scholar]
- 40. Wrenger C., Eschbach M.-L., Müller I. B., Laun N. P., Begley T. P., and Walter R. D. (2006) Vitamin B1 de novo synthesis in the human malaria parasite Plasmodium falciparum depends on external provision of 4-amino-5-hydroxymethyl-2-methylpyrimidine. Biol. Chem. 387, 41–51 10.1515/BC.2006.007 [DOI] [PubMed] [Google Scholar]
- 41. Wrenger C., Knöckel J., Walter R. D., and Müller I. B. (2008) Vitamin B1 and B6 in the malaria parasite: requisite or dispensable? Braz. J. Med. Biol. Res. 41, 82–88 10.1590/S0100-879X2008005000006 [DOI] [PubMed] [Google Scholar]
- 42. Tarun A. S., Baer K., Dumpit R. F., Gray S., Lejarcegui N., Frevert U., and Kappe S. H. I. (2006) Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int. J. Parasitol. 36, 1283–1293 10.1016/j.ijpara.2006.06.009 [DOI] [PubMed] [Google Scholar]
- 43. Chan X. W. A., Wrenger C., Stahl K., Bergmann B., Winterberg M., Müller I. B., Saliba K. J., Muller I. B., and Saliba K. J. (2013) Chemical and genetic validation of thiamine utilization as an antimalarial drug target. Nat. Commun. 4, 2060 10.1038/ncomms3060 [DOI] [PubMed] [Google Scholar]
- 44. Zilles J. L., Croal L. R., and Downs D. M. (2000) Action of the thiamine antagonist bacimethrin on thiamine biosynthesis. J. Bacteriol. 182, 5606–5610 10.1128/JB.182.19.5606-5610.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rajgopal A., Edmondnson A., Goldman I. D., and Zhao R. (2001) SLC19A3 encodes a second thiamine transporter ThTr2. Biochim. Biophys. Acta 1537, 175–178 10.1016/S0925-4439(01)00073-4 [DOI] [PubMed] [Google Scholar]
- 46. Subramanian V. S., Marchant J. S., Parker I., and Said H. M. (2003) Cell biology of the human thiamine transporter-1 (hTHTR1). Intracellular trafficking and membrane targeting mechanisms. J. Biol. Chem. 278, 3976–3984 10.1074/jbc.M210717200 [DOI] [PubMed] [Google Scholar]
- 47. Driscoll T. P., Verhoeve V. I., Guillotte M. L., Lehman S. S., Rennoll S. A., Beier-Sexton M., Rahman M. S., Azad A. F., and Gillespie J. J. (2017) Wholly Rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. MBio 8, e00859–17 10.1128/mBio.00859-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zerez C. R., Roth E. F. Jr., Schulman S., and Tanaka K. R. (1990) Increased nicotinamide adenine dinucleotide content and synthesis in Plasmodium falciparum-infected human erythrocytes. Blood 75, 1705–1710 10.1182/blood.V75.8.1705.bloodjournal7581705 [DOI] [PubMed] [Google Scholar]
- 49. Beri D., Ramdani G., Balan B., Gadara D., Poojary M., Momeux L., Tatu U., and Langsley G. (2019) Insights into physiological roles of unique metabolites released from Plasmodium-infected RBCs and their potential as clinical biomarkers for malaria. Sci. Rep. 9, 2875 10.1038/s41598-018-37816-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Hara J. K., Kerwin L. J., Cobbold S. A., Tai J., Bedell T. A., Reider P. J., and Llinás M. (2014) Targeting NAD+ metabolism in the human malaria parasite Plasmodium falciparum. PLoS One 9, e94061 10.1371/journal.pone.0094061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mageed S. N., Cunningham F., Hung A. W., Silvestre H. L., Wen S., Blundell T. L., Abell C., and McConkey G. A. (2014) Pantothenic acid biosynthesis in the parasite Toxoplasma gondii: a target for chemotherapy. Antimicrob. Agents Chemother. 58, 6345–6353 10.1128/AAC.02640-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ciulli A., Chirgadze D. Y., Smith A. G., Blundell T. L., and Abell C. (2007) Crystal structure of Escherichia coli ketopantoate reductase in a ternary complex with NADP+ and pantoate bound. J. Biol. Chem. 282, 8487–8497 10.1074/jbc.M611171200 [DOI] [PubMed] [Google Scholar]
- 53. von Delft F., Inoue T., Saldanha S. A., Ottenhof H. H., Schmitzberger F., Birch L. M., Dhanaraj V., Witty M., Smith A. G., Blundell T. L., and Abell C. (2003) Structure of E. coli ketopantoate hydroxymethyl transferase complexed with ketopantoate and Mg2+, solved by locating 160 selenomethionine sites. Structure 11, 985–996 10.1016/S0969-2126(03)00158-8 [DOI] [PubMed] [Google Scholar]
- 54. von Delft F., Lewendon A., Dhanaraj V., Blundell T. L., Abell C., and Smith A. G. (2001) The crystal structure of E. coli pantothenate synthetase confirms it as a member of the cytidylyltransferase superfamily. Structure 9, 439–450 10.1016/S0969-2126(01)00604-9 [DOI] [PubMed] [Google Scholar]
- 55. Saliba K. J., Horner H. A., and Kirk K. (1998) Transport and metabolism of the essential vitamin pantothenic acid in human erythrocytes infected with the malaria parasite Plasmodium falciparum. J. Biol. Chem. 273, 10190–10195 10.1074/jbc.273.17.10190 [DOI] [PubMed] [Google Scholar]
- 56. Augagneur Y., Jaubert L., Schiavoni M., Pachikara N., Garg A., Usmani-Brown S., Wesolowski D., Zeller S., Ghosal A., Cornillot E., Said H. M., Kumar P., Altman S., and Ben Mamoun C. (2013) Identification and functional analysis of the primary pantothenate transporter, PfPAT, of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 288, 20558–20567 10.1074/jbc.M113.482992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kehrer J., Singer M., Lemgruber L., Silva P. A. G. C., Frischknecht F., and Mair G. R. (2016) A putative small solute transporter is responsible for the secretion of G377 and TRAP-containing secretory vesicles during Plasmodium gamete egress and sporozoite motility. PLoS Pathog. 12, e1005734 10.1371/journal.ppat.1005734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hammoudi P.-M., Maco B., Dogga S. K., Frénal K., and Soldati-Favre D. (2018) Toxoplasma gondii TFP1 is an essential transporter family protein critical for microneme maturation and exocytosis. Mol. Microbiol. 109, 225–244 10.1111/mmi.13981 [DOI] [PubMed] [Google Scholar]
- 59. Pietrocola F., Galluzzi L., Bravo-San Pedro J. M., Madeo F., and Kroemer G. (2015) Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 10.1016/j.cmet.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 60. Tjhin E. T., Spry C., Sewell A. L., Hoegl A., Barnard L., Sexton A. E., Siddiqui G., Howieson V. M., Maier A. G., Creek D. J., Strauss E., Marquez R., Auclair K., and Saliba K. J. (2018) Mutations in the pantothenate kinase of Plasmodium falciparum confer diverse sensitivity profiles to antiplasmodial pantothenate analogues. PLoS Pathog. 14, e1006918 10.1371/journal.ppat.1006918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Villiers M., Spry C., Macuamule C. J., Barnard L., Wells G., Saliba K. J., and Strauss E. (2017) Antiplasmodial mode of action of pantothenamides: pantothenate kinase serves as a metabolic activator not as a target. ACS Infect. Dis. 3, 527–541 10.1021/acsinfecdis.7b00024 [DOI] [PubMed] [Google Scholar]
- 62. Schalkwijk J., Allman E. L., Jansen P. A., Vries L. E, de Jackowski S., Botman P. N., Beuckens-Schortinghuis C. A., Koolen K. M., Bolscher J. M., Vos M. W., Miller K., Reeves S., Pett H., Trevitt G., Wittlin S., et al. (2018) Antimalarial pantothenamide metabolites target acetyl-CoA synthesis in Plasmodium falciparum. bioRxiv 10.1101/256669 [DOI] [PubMed] [Google Scholar]
- 63. Hart R. J., Abraham A., and Aly A. S. I. (2017) Genetic characterization of coenzyme A biosynthesis reveals essential distinctive functions during malaria parasite development in blood and mosquito. Front. Cell. Infect. Microbiol. 7, 260 10.3389/fcimb.2017.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. de Villiers M., Barnard L., Koekemoer L., Snoep J. L., and Strauss E. (2014) Variation in pantothenate kinase type determines the pantothenamide mode of action and impacts on coenzyme A salvage biosynthesis. FEBS J. 281, 4731–4753 10.1111/febs.13013 [DOI] [PubMed] [Google Scholar]
- 65. Srinivasan B., Baratashvili M., van der Zwaag M., Kanon B., Colombelli C., Lambrechts R. A., Schaap O., Nollen E. A., Podgoršek A., Kosec G., Petković H., Hayflick S., Tiranti V., Reijngoud D.-J., Grzeschik N. A., and Sibon O. C. M. (2015) Extracellular 4′-phosphopantetheine is a source for intracellular coenzyme A synthesis. Nat. Chem. Biol. 11, 784–792 10.1038/nchembio.1906 [DOI] [PubMed] [Google Scholar]
- 66. Percudani R., and Peracchi A. (2003) A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 4, 850–854 10.1038/sj.embor.embor914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hoegl A., Nodwell M. B., Kirsch V. C., Bach N. C., Pfanzelt M., Stahl M., Schneider S., and Sieber S. A. (2018) Mining the cellular inventory of pyridoxal phosphate-dependent enzymes with functionalized cofactor mimics. Nat. Chem. 10, 1234–1245 10.1038/s41557-018-0144-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fitzpatrick T. B., Amrhein N., Kappes B., Macheroux P., Tews I., and Raschle T. (2007) Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem. J. 407, 1–13 10.1042/BJ20070765 [DOI] [PubMed] [Google Scholar]
- 69. Knöckel J., Müller I. B., Bergmann B., Walter R. D., and Wrenger C. (2007) The apicomplexan parasite Toxoplasma gondii generates pyridoxal phosphate de novo. Mol. Biochem. Parasitol. 152, 108–111 10.1016/j.molbiopara.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 70. Gengenbacher M., Fitzpatrick T. B., Raschle T., Flicker K., Sinning I., Müller S., Macheroux P., Tews I., and Kappes B. (2006) Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: biochemical and structural insights. J. Biol. Chem. 281, 3633–3641 10.1074/jbc.M508696200 [DOI] [PubMed] [Google Scholar]
- 71. Palm D., Klein H. W., Schinzel R., Buehner M., and Helmreich E. J. M. (1990) The role of pyridoxal 5′-phosphate in glycogen phosphorylase catalysis. Biochemistry 29, 1099–1107 10.1021/bi00457a001 [DOI] [PubMed] [Google Scholar]
- 72. Sugi T., Tu V., Ma Y., Tomita T., and Weiss L. M. (2017) Toxoplasma gondii requires glycogen phosphorylase for balancing amylopectin storage and for efficient production of brain cysts. MBio 8, e01289–17 10.1128/mBio.01289-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Van de Kamp J. L., Westrick J. A., and Smolen A. (1995) B6 vitamer concentrations in mouse plasma, erythrocytes and tissues. Nutr. Res. 15, 415–422 10.1016/0271-5317(95)00009-7 [DOI] [Google Scholar]
- 74. Cassera M. B., Gozzo F. C., D'Alexandri F. L., Merino E. F., del Portillo H. A., Peres V. J., Almeida I. C., Eberlin M. N., Wunderlich G., Wiesner J., Jomaa H., Kimura E. A., and Katzin A. M. (2004) The methylerythritol phosphate pathway is functionally active in all intraerythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 279, 51749–51759 10.1074/jbc.M408360200 [DOI] [PubMed] [Google Scholar]
- 75. Wrenger C., Eschbach M.-L., Müller I. B., Warnecke D., and Walter R. D. (2005) Analysis of the vitamin B6 biosynthesis pathway in the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 280, 5242–5248 10.1074/jbc.M412475200 [DOI] [PubMed] [Google Scholar]
- 76. Müller I. B., Hyde J. E., and Wrenger C. (2010) Vitamin B metabolism in Plasmodium falciparum as a source of drug targets. Trends Parasitol. 26, 35–43 10.1016/j.pt.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 77. Knöckel J., Müller I. B., Butzloff S., Bergmann B., Walter R. D., and Wrenger C. (2012) The antioxidative effect of de novo generated vitamin B6 in Plasmodium falciparum validated by protein interference. Biochem. J. 443, 397–405 10.1042/BJ20111542 [DOI] [PubMed] [Google Scholar]
- 78. Kronenberger T., Lindner J., Meissner K. A., Zimbres F. M., Coronado M. A., Sauer F. M., Schettert I., and Wrenger C. (2014) Vitamin B6-dependent enzymes in the human malaria parasite Plasmodium falciparum: a druggable target? Biomed. Res. Int. 2014, 108516 10.1155/2014/108516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Müller I. B., Wu F., Bergmann B., Knöckel J., Walter R. D., Gehring H., and Wrenger C. (2009) Poisoning pyridoxal 5-phosphate-dependent enzymes: a new strategy to target the malaria parasite Plasmodium falciparum. PLoS One 4, e4406 10.1371/journal.pone.0004406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jelenska J., Crawford M. J., Harb O. S., Zuther E., Haselkorn R., Roos D. S., and Gornicki P. (2001) Subcellular localization of acetyl-CoA carboxylase in the apicomplexan parasite Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 98, 2723–2728 10.1073/pnas.051629998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chapman-Smith A., and Cronan J. E. Jr. (1999) Molecular biology of biotin attachment to proteins. J. Nutr. 129, 477S–484S 10.1093/jn/129.2.477S [DOI] [PubMed] [Google Scholar]
- 82. Beckett D. (2005) The Escherichia coli biotin regulatory system: a transcriptional switch. J. Nutr. Biochem. 16, 411–415 10.1016/j.jnutbio.2005.03.019 [DOI] [PubMed] [Google Scholar]
- 83. Hebbeln P., Rodionov D. A., Alfandega A., and Eitinger T. (2007) Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. U.S.A. 104, 2909–2914 10.1073/pnas.0609905104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Daberkow R. L., White B. R., Cederberg R. A., Griffin J. B., and Zempleni J. (2003) Monocarboxylate transporter 1 mediates biotin uptake in human peripheral blood mononuclear cells. J. Nutr. 133, 2703–2706 10.1093/jn/133.9.2703 [DOI] [PubMed] [Google Scholar]
- 85. Massimine K. M., Doan L. T., Atreya C. A., Stedman T. T., Anderson K. S., Joiner K. A., and Coppens I. (2005) Toxoplasma gondii is capable of exogenous folate transport: a likely expansion of the BT1 family of transmembrane proteins. Mol. Biochem. Parasitol. 144, 44–54 10.1016/j.molbiopara.2005.07.006 [DOI] [PubMed] [Google Scholar]
- 86. Salcedo-Sora J. E., Ochong E., Beveridge S., Johnson D., Nzila A., Biagini G. A., Stocks P. A., O'Neill P. M., Krishna S., Bray P. G., and Ward S. A. (2011) The molecular basis of folate salvage in Plasmodium falciparum: characterization of two folate transporters. J. Biol. Chem. 286, 44659–44668 10.1074/jbc.M111.286054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hyde J. E. (2005) Exploring the folate pathway in Plasmodium falciparum. Acta Trop. 94, 191–206 10.1016/j.actatropica.2005.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nzila A., Ward S. A., Marsh K., Sims P. F. G., and Hyde J. E. (2005) Comparative folate metabolism in humans and malaria parasites (part I): pointers for malaria treatment from cancer chemotherapy. Trends Parasitol. 21, 292–298 10.1016/j.pt.2005.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Heinberg A., and Kirkman L. (2015) The molecular basis of antifolate resistance in Plasmodium falciparum: looking beyond point mutations. Ann. N.Y. Acad. Sci. 1342, 10–18 10.1111/nyas.12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Matz J. M., Watanabe M., Falade M., Tohge T., Hoefgen R., and Matuschewski K. (2019) Plasmodium para-aminobenzoate synthesis and salvage resolve avoidance of folate competition and adaptation to host diet. Cell Rep. 26, 356–363.e4 10.1016/j.celrep.2018.12.062 [DOI] [PubMed] [Google Scholar]
- 91. Mather M. W., and Ke H. (2019) para-Aminobenzoate synthesis versus salvage in malaria parasites. Trends Parasitol. 35, 176–178 10.1016/j.pt.2019.01.010 [DOI] [PubMed] [Google Scholar]
- 92. Hamza I., and Dailey H. A. (2012) One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta 1823, 1617–1632 10.1016/j.bbamcr.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kořený L., Oborník M., and Lukeš J. (2013) Make it, take it, or leave it: heme metabolism of parasites. PLoS Pathog. 9, e1003088 10.1371/journal.ppat.1003088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Huynh C., Yuan X., Miguel D. C., Renberg R. L., Protchenko O., Philpott C. C., Hamza I., and Andrews N. W. (2012) Heme uptake by Leishmania amazonensis is mediated by the transmembrane protein LHR1. PLoS Pathog. 8, e1002795 10.1371/journal.ppat.1002795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cabello-Donayre M., Orrego L. M., Herráez E., Vargas P., Martínez-García M., Campos-Salinas J., Pérez-Victoria I., Vicente B., Marín J. J. G., and Pérez-Victoria J. M. (2019) Leishmania heme uptake involves LmFLVCRb, a novel porphyrin transporter essential for the parasite. Cell. Mol. Life Sci. 10.1007/s00018-019-03258-3 10.1007/s00018-019-03258-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Koŕený L., Lukeš J., and Oborník M. (2010) Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? Int. J. Parasitol. 40, 149–156 10.1016/j.ijpara.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 97. Tripodi K. E. J., Menendez Bravo S. M., and Cricco J. A. (2011) Role of heme and heme-proteins in trypanosomatid essential metabolic pathways. Enzyme Res. 2011, 873230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Oborník M., and Green B. R. (2005) Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 22, 2343–2353 10.1093/molbev/msi230 [DOI] [PubMed] [Google Scholar]
- 99. Koreny L., Sobotka R., Janouskovec J., Keeling P. J., and Oborník M. (2011) Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant Cell. 23, 3454–3462 10.1105/tpc.111.089102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chugh M., Sundararaman V., Kumar S., Reddy V. S., Siddiqui W. A., Stuart K. D., and Malhotra P. (2013) Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 110, 5392–5397 10.1073/pnas.1218412110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sullivan D. J. Jr., Gluzman I. Y., and Goldberg D. E. (1996) Plasmodium hemozoin formation mediated by histidine-rich proteins. Science 271, 219–222 10.1126/science.271.5246.219 [DOI] [PubMed] [Google Scholar]
- 102. Bendrat K., Berger B. J., and Cerami A. (1995) Haem polymerization in malaria. Nature 378, 138–139 10.1038/378138a0 [DOI] [PubMed] [Google Scholar]
- 103. Fitch C. D., Cai G. Z., Chen Y.-F., and Shoemaker J. D. (1999) Involvement of lipids in ferriprotoporphyrin IX polymerization in malaria. Biochim. Biophys. Acta 1454, 31–37 10.1016/S0925-4439(99)00017-4 [DOI] [PubMed] [Google Scholar]
- 104. Dorn A., Stoffel R., Matile H., Bubendorf A., and Ridley R. G. (1995) Malarial haemozoin/β-haematin supports haem polymerization in the absence of protein. Nature 374, 269–271 10.1038/374269a0 [DOI] [PubMed] [Google Scholar]
- 105. Nagaraj V. A., Sundaram B., Varadarajan N. M., Subramani P. A., Kalappa D. M., Ghosh S. K., and Padmanaban G. (2013) Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PLoS Pathog. 9, e1003522 10.1371/journal.ppat.1003522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ke H., Sigala P. A., Miura K., Morrisey J. M., Mather M. W., Crowley J. R., Henderson J. P., Goldberg D. E., Long C. A., and Vaidya A. B. (2014) The heme biosynthesis pathway is essential for Plasmodium falciparum development in mosquito stage but not in blood stages. J. Biol. Chem. 289, 34827–34837 10.1074/jbc.M114.615831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rizopoulos Z., Matuschewski K., and Haussig J. M. (2016) Distinct prominent roles for enzymes of Plasmodium berghei heme biosynthesis in sporozoite and liver stage maturation. Infect. Immun. 84, 3252–3262 10.1128/IAI.00148-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Goldberg D. E., and Sigala P. A. (2017) Plasmodium heme biosynthesis: to be or not to be essential? PLoS Pathog. 13, e1006511 10.1371/journal.ppat.1006511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sigala P. A., Crowley J. R., Henderson J. P., and Goldberg D. E. (2015) Deconvoluting heme biosynthesis to target blood-stage malaria parasites. Elife 10.7554/eLife.09143 10.7554/eLife.09143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Meshnick S. R., Thomas A., Ranz A., Xu C. M., and Pan H. Z. (1991) Artemisinin (qinghaosu): the role of intracellular hemin in its mechanism of antimalarial action. Mol. Biochem. Parasitol. 49, 181–189 10.1016/0166-6851(91)90062-B [DOI] [PubMed] [Google Scholar]
- 111. Tilley L., Straimer J., Gnädig N. F., Ralph S. A., and Fidock D. A. (2016) Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol. 32, 682–696 10.1016/j.pt.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang J., Zhang C.-J., Chia W. N., Loh C. C. Y., Li Z., Lee Y. M., He Y., Yuan L.-X., Lim T. K., Liu M., Liew C. X., Lee Y. Q., Zhang J., Lu N., Lim C. T., et al. (2015) Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 6, 10111 10.1038/ncomms10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang S., and Gerhard G. S. (2009) Heme mediates cytotoxicity from artemisinin and serves as a general anti-proliferation target. PLoS One 4, e7472 10.1371/journal.pone.0007472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Harding C. R., Sidik S. M., Petrova B., Gnädig N. F., Okombo J., Ward K. E., Markus B. M., Fidock D. A., and Lourido S. (2019) Genetic screens reveal a central role for heme biosynthesis in artemisinin susceptibility. bioRxiv 10.1101/746974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shanmugam D., Wu B., Ramirez U., Jaffe E. K., and Roos D. S. (2010) Plastid-associated porphobilinogen synthase from Toxoplasma gondii: kinetic and structural properties validate therapeutic potential. J. Biol. Chem. 285, 22122–22131 10.1074/jbc.M110.107243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jaffe E. K., Shanmugam D., Gardberg A., Dieterich S., Sankaran B., Stewart L. J., Myler P. J., and Roos D. S. (2011) Crystal structure of Toxoplasma gondii porphobilinogen synthase. J. Biol. Chem. 286, 15298–15307 10.1074/jbc.M111.226225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Layer G., Moser J., Heinz D. W., Jahn D., Schubert W.-D. (2003) Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 22, 6214–6224 10.1093/emboj/cdg598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Choby J. E., and Skaar E. P. (2016) Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 428, 3408–3428 10.1016/j.jmb.2016.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dailey H. A., Gerdes S., Dailey T. A., Burch J. S., and Phillips J. D. (2015) Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc. Natl. Acad. Sci. U.S.A. 112, 2210–2215 10.1073/pnas.1416285112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. van Dooren G. G., Stimmler L. M., and McFadden G. I. (2006) Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 30, 596–630 10.1111/j.1574-6976.2006.00027.x [DOI] [PubMed] [Google Scholar]
- 121. Fleige T., Fischer K., Ferguson D. J. P., Gross U., and Bohne W. (2007) Carbohydrate metabolism in the Toxoplasma gondii apicoplast: localization of three glycolytic isoenzymes, the single pyruvate dehydrogenase complex, and a plastid phosphate translocator. Eukaryot. Cell 6, 984–996 10.1128/EC.00061-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Read M., Müller I. B., Mitchell S. L., Sims P. F., and Hyde J. E. (2010) Dynamic subcellular localization of isoforms of the folate pathway enzyme serine hydroxymethyltransferase (SHMT) through the erythrocytic cycle of Plasmodium falciparum. Malar. J. 9, 351 10.1186/1475-2875-9-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Thomsen-Zieger N., Schachtner J., and Seeber F. (2003) Apicomplexan parasites contain a single lipoic acid synthase located in the plastid. FEBS Lett. 547, 80–86 10.1016/S0014-5793(03)00673-2 [DOI] [PubMed] [Google Scholar]
- 124. Crawford M. J., Thomsen-Zieger N., Ray M., Schachtner J., Roos D. S., and Seeber F. (2006) Toxoplasma gondii scavenges host-derived lipoic acid despite its de novo synthesis in the apicoplast. EMBO J. 25, 3214–3222 10.1038/sj.emboj.7601189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Falkard B., Kumar T. R. S., Hecht L.-S., Matthews K. A., Henrich P. P., Gulati S., Lewis R. E., Manary M. J., Winzeler E. A., Sinnis P., Prigge S. T., Heussler V., Deschermeier C., and Fidock D. (2013) A key role for lipoic acid synthesis during Plasmodium liver stage development. Cell Microbiol. 15, 1585–1604 10.1111/cmi.12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Günther S., Matuschewski K., and Müller S. (2009) Knockout studies reveal an important role of Plasmodium lipoic acid protein ligase A1 for asexual blood stage parasite survival. PLoS One 4, e5510 10.1371/journal.pone.0005510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sinai A. P., Webster P., and Joiner K. A. (1997) Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J. Cell Sci. 110, 2117–2128 [DOI] [PubMed] [Google Scholar]
- 128. Roberts F., Roberts C. W., Johnson J. J., Kyle D. E., Krell T., Coggins J. R., Coombs G. H., Milhous W. K., Tzipori S., Ferguson D. J. P., Chakrabarti D., and McLeod R. (1998) Evidence for the shikimate pathway in apicomplexan parasites. Nature 393, 801–805 10.1038/31723 [DOI] [PubMed] [Google Scholar]
- 129. Peek J., Castiglione G., Shi T., and Christendat D. (2014) Isolation and molecular characterization of the shikimate dehydrogenase domain from the Toxoplasma gondii AROM complex. Mol. Biochem. Parasitol. 194, 16–19 10.1016/j.molbiopara.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 130. Campbell S. A., Richards T. A., Mui E. J., Samuel B. U., Coggins J. R., McLeod R., and Roberts C. W. (2004) A complete shikimate pathway in Toxoplasma gondii: an ancient eukaryotic innovation. Int. J. Parasitol. 34, 5–13 10.1016/j.ijpara.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 131. Richards T. A., Dacks J. B., Campbell S. A., Blanchard J. L., Foster P. G., Mcleod R., and Roberts C. W. (2006) Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5, 1517–1531 10.1128/EC.00106-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schönbrunn E., Eschenburg S., Shuttleworth W. A., Schloss J. V., Amrhein N., Evans J. N., and Kabsch W. (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380 10.1073/pnas.98.4.1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Roberts C. W., Roberts F., Lyons R. E., Kirisits M. J., Mui E. J., Finnerty J., Johnson J. J., Ferguson D. J. P., Coggins J. R., Krell T., Coombs G. H., Milhous W. K., Kyle D. E., Tzipori S., Barnwell J., et al. (2002) The shikimate pathway and its branches in apicomplexan parasites. J. Infect. Dis. 185, S25–S36 10.1086/338004 [DOI] [PubMed] [Google Scholar]
- 134. Keeling P. J., Palmer J. D., Donald R. G. K., Roos D. S., Waller R. F., and McFadden G. I. (1999) Shikimate pathway in apicomplexan parasites. Nature 397, 219–220 10.1038/16618 [DOI] [PubMed] [Google Scholar]
- 135. de Macedo C. S., Uhrig M. L., Kimura E. A., and Katzin A. M. (2002) Characterization of the isoprenoid chain of coenzyme Q in Plasmodium falciparum. FEMS Microbiol. Lett. 207, 13–20 10.1111/j.1574-6968.2002.tb11021.x [DOI] [PubMed] [Google Scholar]
- 136. Tonhosolo R., D'Alexandri F. L., Genta F. A., Wunderlich G., Gozzo F. C., Eberlin M. N., Peres V. J., Kimura E. A., and Katzin A. M. (2005) Identification, molecular cloning and functional characterization of an octaprenyl pyrophosphate synthase in intra-erythrocytic stages of Plasmodium falciparum. Biochem. J. 392, 117–126 10.1042/BJ20050441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Nair S. C., Brooks C. F., Goodman C. D., Sturm A., McFadden G. I., Sundriyal S., Anglin J. L., Song Y., Moreno S. N. J., Striepen B., and Striepen B. (2011) Apicoplast isoprenoid precursor synthesis and the molecular basis of fosmidomycin resistance in Toxoplasma gondii. J. Exp. Med. 208, 1547–1559 10.1084/jem.20110039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.