Abstract
Bacillus cereus is widely distributed in different food products and can cause a variety of symptoms associated with food poisoning. Since ready-to-eat (RTE) foods are not commonly sterilized by heat treatment before consumption, B. cereus contamination may cause severe food safety problems. In this study, we investigated the prevalence of B. cereus in RTE food samples from different regions of China and evaluated the levels of bacterial contamination, antibiotic resistance, virulence gene distribution, and genetic polymorphisms of these isolates. Of the tested retail RTE foods, 35% were positive for B. cereus, with 39 and 83% of the isolated strains harboring the enterotoxin-encoding hblACD and nheABC gene clusters, respectively. The entFM gene was detected in all B. cereus strains. The cytK gene was present in 68% of isolates, but only 7% harbored the emetic toxin-encoding gene cesB. Antimicrobial susceptibility testing revealed that the majority of the isolates were resistant not only to most β-lactam antibiotics, but also to rifamycin. Multilocus sequence typing (MLST) revealed that the 368 isolates belonged to 192 different sequence types (STs) including 93 new STs, the most prevalent of which was ST26. Collectively, our study indicates the prevalence, bacterial contamination levels, and biological characteristics of B. cereus isolated from RTE foods in China and demonstrates the potential hazards of B. cereus in RTE foods.
Keywords: Bacillus cereus, ready-to-eat food, risk assessment, virulence genes, antibiotic resistance, genetic polymorphism
Introduction
Ready-to-eat (RTE) foods, such as cooked meats and poultry, cold vegetable dishes in sauce, cold noodles, and fried rice, are very popular as they are intended for direct consumption. Although they are very convenient for consumers, RTE foods have been shown to be frequently contaminated with pathogenic bacteria such as Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus (Batchoun et al., 2011; Hwang and Park, 2015; Wu et al., 2016; Yang et al., 2016).
Bacillus cereus is a gram-positive bacterium that causes foodborne diseases and is widespread in nature and foods (Marrollo, 2016). B. cereus has been isolated from a variety of foods, particularly RTE foods such as cooked rice and mixed salad (Park et al., 2009; Batchoun et al., 2011; Rahimi et al., 2013; Tewari et al., 2015; Gao et al., 2018; Yu et al., 2019). B. cereus can cause food poisoning even at very low doses, with more than 103 B. cereus g–1 considered unsafe for consumption (Granum and Lund, 1997). Despite safety precautions, numerous food poisoning incidents caused by B. cereus have been reported recently in Spain (Doménech-Sánchez et al., 2011), Belgium (Delbrassinne et al., 2015), Argentina (Lopez et al., 2015), Australia (Sloan-Gardner et al., 2014), England (Nicholls et al., 2016), Austria (Schmid et al., 2016), and France (Glasset et al., 2016).
Bacillus cereus produces a range of virulence factors and can enter the gastrointestinal tract via ingestion, where it causes diarrhea and vomiting (Jensen et al., 2003; Stenfors Arnesen et al., 2008; Song et al., 2019). Diarrhea is associated with four different enterotoxins, the hemolysin BL (HBL, encoded by hblA, hblC, and hblD), non-hemolytic enterotoxin (NHE, encoded by nheA, nheB, nheC), enterotoxin FM (EntFM, encoded by entFM) and the cytotoxin K (CytK, encoded by cytK) (Beecher et al., 1995; Lund and Granum, 1996; Granum et al., 1999; Lund et al., 2000; Bonerba et al., 2010; Tran et al., 2010; Berthold-Pluta et al., 2019). HBL and NHE are both tripartite toxins (Berthold-Pluta et al., 2019). CytK belongs to a member of the family of β-barrel pore forming toxins that can cause serious food poisoning, skin necrosis, hemolysis, and even death (Lund et al., 2000). EntFM is related to cell wall peptidases (CWPs) which can cause diseases such as diarrhea (Tran et al., 2010). Whilst vomiting, or emesis, is induced by a small, heat and acid stable cyclic dodecadepsipeptide ([D-OLeu-D-Ala-L-O-Val-L-Val]3) toxin known as cereulide that is synthesized by non-ribosomal peptide synthetases encoded by ces genes (Ehling-Schulz et al., 2005, 2015). Besides food poisoning, B. cereus is also associated with serious infections such as pneumonia, bacteremia, endophthalmitis, necrotizing fasciitis, osteomyelitis, and endocarditis (Bottone, 2010; Rishi et al., 2013; Ikeda et al., 2015).
Antibiotic treatment is still the main method for treating bacterial infections, including those caused by B. cereus; however, the extensive use of antimicrobials has led to the emergence of antibiotic-resistant strains, including those resistant to multiple antibiotics, which can cause routine treatments to fail (Friedman et al., 2016). Thus, determining the antibiotic resistance profile of B. cereus is important for informing drug selection for treatment regimens.
The contamination of RTE foods by pathogenic bacteria such as B. cereus is a major food safety concern; thus, it is necessary to monitor and characterize B. cereus contamination in RTE foods. This study investigated the potential pathogenicity, contamination levels, molecular characteristics, and antibiotic resistance profiles of B. cereus isolated from RTE foods in China, providing important information about the prevalence of B. cereus in RTE foods.
Materials and Methods
Sample Collection
A total of 860 RTE food samples were collected from retail markets and supermarkets in 39 major Chinese cities (Supplementary Figure S1) between 2011 and 2016 according to the general guidelines of the National Food Safety Standard in Sample Collection (The Hygiene Ministry of China, 2010). The samples included cooked meat (656 samples), cold vegetable dishes in sauce (85 samples), and rice/noodles (119 samples). All samples were placed in separate sterile bags, transferred to the laboratory on ice within 2 days, and kept below 4°C.
Qualitative and Quantitative Detection of B. cereus
Bacillus cereus was qualitatively and quantitatively detected according to the bacteriological analytical manuals of the U.S. Food and Drug Administration and the National Food Safety Standard of China (The Hygiene Ministry of China, 2003; Tallent et al., 2012). In brief, 25 g samples were randomly collected from each RTE food sample and put into sterile blender jar with 225 mL Trypticase Soy Broth (TSB) with polymyxin (Huankai, Guangzhou, China), then blended for 2 min at high speed (10,000 to 12,000 rpm). Homogenates were incubated 48 ± 2 h at 30 ± 2°C. Afterward, a loop of the resulting cultures was streaked onto mannitol egg yolk polymyxin agar plates (MYP) (Huankai), which were incubated 24 h at 30°C. Single colonies were then streaked onto chromogenic B. cereus agar plates (Huankai). Different presumptive colonies from the chromogenic B. cereus agar plates were picked for further biochemical characterization using a B. cereus biochemistry assessor (Huankai) to identify authentic colonies. The most probable number (MPN) method was used for the quantitative detection of B. cereus. A three-tube MPN series was inoculated into TSB with polymyxin, using 1 mL inoculums of 10–1, 10–2, and 10–3 dilutions of each sample, with three tubes at each dilution. The tubes were incubated for 48 ± 2 h at 30 ± 2°C and observed for turbid growth typical of B. cereus. The cultures from turbid, positive tubes were streaked onto MYP agar plates and incubated for 24 h at 30°C. One or more pink, lecithin-positive colonies were selected from each MYP agar plate and further confirmed on chromogenic B. cereus agar plates. The number of tubes confirmed as positive for B. cereus was used to calculate the MPN of B. cereus per g (mL) sample, expressed as MPN/g (mL) using the MPN table.
Virulence Gene Distribution
Genomic DNA was extracted using a genomic DNA extraction kit for gram-positive bacteria (Magen, Guangzhou, China) according to the manufacturer’s instructions. Different virulence genes, including hblA, hblC, hblD, nheA, nheB, nheC, entFM, cytK, and cesB, were detected by PCR using the primers listed in Supplementary Table S1 with previously described thermal profiles (Hansen and Hendriksen, 2001; Ehling-Schulz et al., 2005; Oltuszak-Walczak and Walczak, 2013; Forghani et al., 2014; Yu et al., 2019).
ERIC-PCR
The ERIC-PCR were conducted by using the primers listed in Supplementary Table S1 with previously described thermal profiles (Forghani et al., 2014; Gao et al., 2018). The PCR mixture (25 μL) contained of 50 ng of genomic DNA, 2 μM of each primer, and 12.5 μL of PCR Premix TaqTM (Takara, China). The amplification and agarose gel electrophoresis analysis were performed as described previously (Gao et al., 2018). DNA fingerprints were analyzed by Bionumerics software version 7.6 (Applied Maths, Belgium). The result of ERIC-PCR fingerprinting was used to characterize isolates from the same sample in order to exclude clonal isolates.
Antimicrobial Susceptibility Testing
The sensitivity of B. cereus to 20 antimicrobial agents was tested using the standard Kirby-Bauer disk diffusion method (Park et al., 2009; The Clinical and Laboratory Standards Institute [CLSI], 2010; Kim et al., 2015; Gao et al., 2018; Osman et al., 2018; Yu et al., 2019). The B. cereus isolates were streaked on the nutrition agar plate and grown 16–18 h at 37°C. The colony was then picked and suspended using 0.85% physiological saline to 0.5 McFarland standard and spread on the surface of a Mueller-Hinton agar plate. After the inoculum was dried, the antimicrobial disks were put to the surface of the plates. The Mueller-Hinton agar plates were incubated 16–18 h at 35 ± 2°C, and the inhibition zone was measured. The isolates were classed as susceptible (S), intermediate (I), or resistant (R) according to CLSI guidelines and the inhibition zones were measured and interpreted according to the zone diameter interpretation criteria for S. aureus in Supplementary Table S2.
Housekeeping Gene Amplification, Sequencing, and Sequence Type Determination
The genetic diversity of the B. cereus isolates was characterized using multilocus sequence typing (MLST) with primers specific for seven housekeeping genes (glp, gmk, ilv, pta, pur, pyc, and tpi; Supplementary Table S1). The seven housekeeping genes were sequenced by BGI (Shenzhen, China) and submitted to the PubMLST database for allele number identification. Each isolate was assigned a sequence type (ST) according to their combination of the seven housekeeping gene alleles. New STs were validated by the MLST database curator. Clonal complexes (CCs) were defined as single locus variants (SLV) of two or more independent isolates that shared identical alleles at six or seven loci (Drewnowska and Swiecicka, 2013) and were identified using the geoBURST tool (Francisco et al., 2009) with bootstrap resampling (1,000). Evolutionary relationships between the isolates were evaluated using a minimum spanning tree constructed by PHYLOViZ 2.0 software (Instituto de Microbiologia, Portugal; Ribeiro-Goncalves et al., 2016).
Results
Prevalence of B. cereus in RTE Foods
In this study, 302 of the 860 collected samples (35%) were positive for B. cereus, including 224 of the 656 cooked meat samples (34%), 59 of the 119 rice/noodle samples (50%), and 19 of the 85 cold vegetable dishes in sauce samples (22%). 368 different strains of B. cereus were identified based on the results of biochemical analysis and ERIC-PCR fingerprinting (Supplementary Figure S2). Based on quantitative analysis, 68% (206/302) of the positive samples were contaminated at levels ranging between 3 and 1100 MPN/g; however, 10% (29/302) of the samples exceeded 1100 MPN/g, including 18 cooked meat samples, 10 rice/noodles samples and one cold vegetable dish in sauce sample (Table 1).
TABLE 1.
Prevalence and contamination level of B cereus in different ready-to-eat foods.
| Type | Contamination rate (%)a | MPN value (MPN/g)b | ||
| MPN < 3 (%) | 3 ≤ MPN < 1100 (%) | 1100 ≤ MPN (%) | ||
| Cooked meat | 224/656(34) | 55/224(25) | 151/224(67) | 18/224(8) |
| Rice/noodles | 59/119(50) | 10/59(17) | 39/59(66) | 10/59(17) |
| Cold vegetable dishes in sauce | 19/85(22) | 2/19(11) | 16/19(84) | 1/19(5) |
| Total | 302/860(35) | 67/302(22) | 206/302(68) | 29/302(10) |
aContamination rate = Number of positive samples/Total samples. bMPN value (MPN/g) = Most probable number of B. cereus per gram sample.
Distribution of Virulence Genes Among B. cereus Isolates
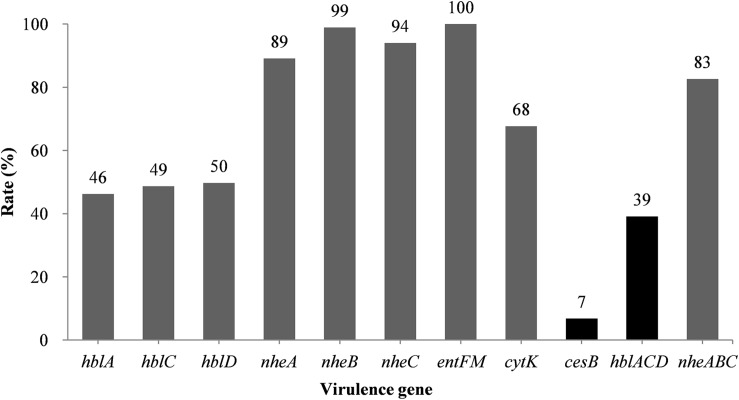
The hblACD gene cluster was found in 39% of the B. cereus isolates (Figure 1 and Supplementary Table S3), with hblA, hblC, and hblD present in 46, 49, and 50% of the isolates, respectively. The NHE genes nheA, nheB, and nheC were found in 89, 99, and 94% of the isolates, respectively. Moreover, 68% of the strains possessed the cytK gene, whereas the cesB gene was the least frequently observed toxin gene, present in just 7% of the strains. However, all of B. cereus isolates possessed the entFM gene, a higher rate than that of other enterotoxin genes.
FIGURE 1.
Distribution of virulence genes in B. cereus isolated from ready-to-eat foods in China. The number at the top of the bars represents the positivity rate for the corresponding toxin genes. hblACD and nheABC indicate strains positive for hblA, hblC, and hblD, or nheA, nheB, and nheC, respectively.
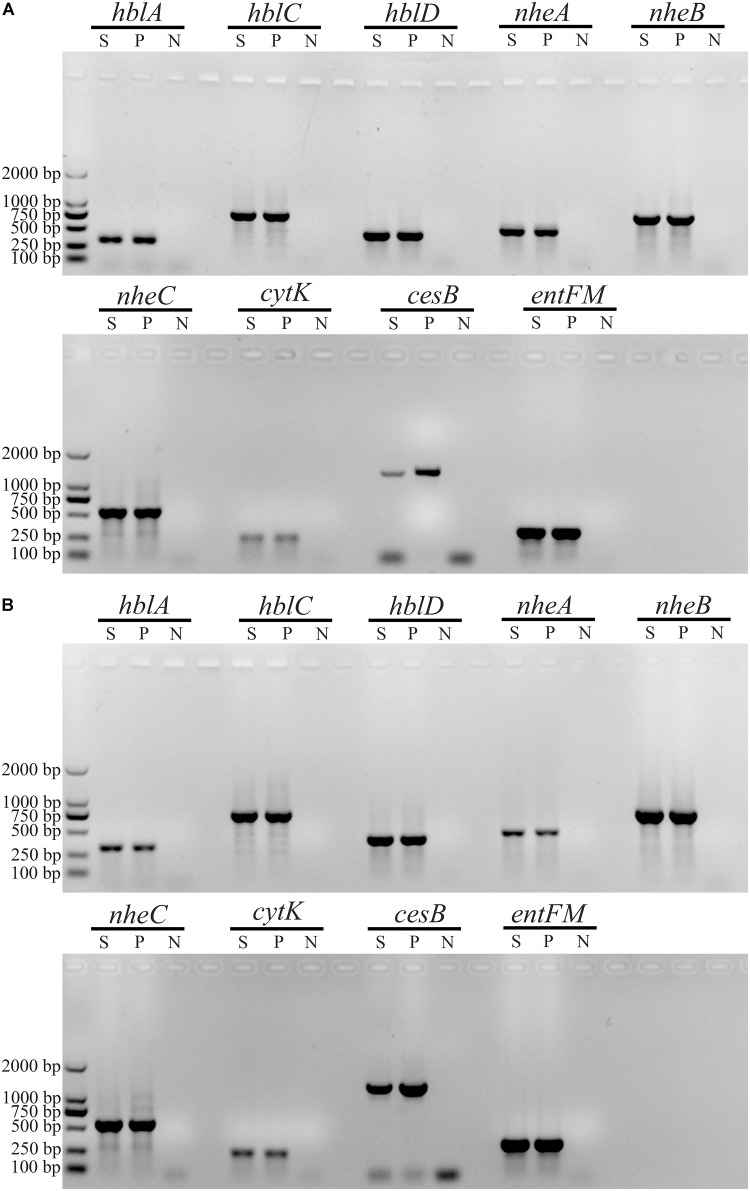
We observed 38 different virulence gene distribution spectra. The most abundant genetic profile, present in 33% of the strains, harbored eight virulence genes (hblA-hblC-hblD-nheA-nheB-nheC-entFM-cytK). Only two isolates (2087-2-Bc and 3709-1A-Bc, Figure 2) contained all nine virulence genes, whereas 27 isolates harbored just three virulence genes each (nheA-nheB-entFM, nheA-entFM-cytK, or nheB-nheC-entFM).
FIGURE 2.
Characterization of virulence genes in B. cereus strains 2087-2-Bc and 3709-1A-Bc. Gel electrophoresis of PCR products of different virulence genes in B. cereus 2087-2-Bc (A) and B. cereus 3709-1A-Bc (B). S, sample; P, positive control; N, negative control.
Antimicrobial Susceptibility of B. cereus Isolates
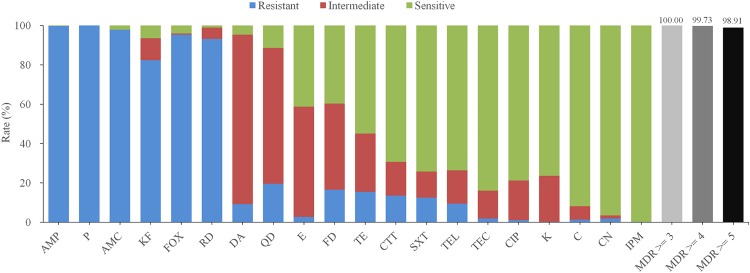
The antimicrobial sensitivity results of the B. cereus isolates are shown in Figure 3 and Table 2. The isolates displayed different degrees of resistance to different antibiotics, particularly β-lactams. The highest rate of resistance was to penicillin (P; 100%), followed by ampicillin (AMP; 99.73%), amoxicillin-clavulanic acid (AMC; 97.83%), cefoxitin (FOX; 95.38%), and cephalothin (KF; 82.34%). Moreover, 13.59% of the strains were resistant to cefotetan (CTT). Most of the isolates were also resistant to rifampin (RD; 93.21%), but far fewer were resistant to quinupristin-dalfopristin (QD; 19.57%), nitrofurantoin (FD; 16.58%), tetracycline (TET; 15.49%) and trimethoprim-sulfamethoxazole (SXT; 12.50%). All isolates were sensitive to other antibiotics, including gentamicin (CN; 96.47%), teicoplanin (TEC; 83.97%), ciprofloxacin (CIP; 78.80%), kanamycin (K; 76.36%), and telithromycin (TEL; 73.64%).
FIGURE 3.
Antibiotic susceptibility of 368 B. cereus strains isolated from ready-to-eat foods in China. Different colors bars (blue, red, and green) represent the proportion of resistant, moderately resistant, and sensitive strains, respectively. The proportion of multidrug resistance (MDR) strains resistant to at least three, four, or five antibiotics is indicated by light gray, gray, or dark bars, respectively. AMP, ampicillin; P, penicillin; AMC, aoxicillin-clavulanic acid; KF, cephalothin; FOX, cefoxitin; RD, rifampin; DA, clindamycin; QD, quinupristin-dalfopristin; E, erythromycin; FD, nitrofurantoin; TE, tetracycline; CTT, cefotetan; SXT, trimethoprim-sulfamethoxazole; TEL, telithromycin; TEC, teicoplanin; CIP, ciprofloxacin; K, kanamycin; C, chloramphenicol; CN, gentamicin; IPM, imipenem.
TABLE 2.
Antibiotic susceptibility of 368 B. cereus strains isolated from ready-to-eat foods in China.
| Category | Antimicrobial class | Antimicrobial agents | Bacillus cereus (n = 368) | |||
| Resistant | Intermediate | Sensitive | ||||
| β-Lactams | Penicillins | I | Ampicillin (10 μg) | 367(99.73%) | 0(0.00%) | 1(0.27%) |
| II | Penicillin (10 units) | 368(100.00%) | 0(0.00%) | 0(0.00%) | ||
| β-Lactam/β-lactamase inhibitor combinations | III | Amoxicillin-clavulanic acid (20 μg/10 μg) | 360(97.83%) | 0(0.00%) | 8(2.17%) | |
| Cephems (parenteral) | IV | Cephalothin (30 μg) | 303(82.34%) | 41(11.14%) | 24(6.52%) | |
| V | Cefoxitin (30 μg) | 351(95.38%) | 2(0.54%) | 15(4.08%) | ||
| VI | Cefotetan (30 μg) | 50(13.59%) | 63(17.12%) | 255(62.29%) | ||
| Penems | VII | Imipenem (10 μg) | 1(0.27%) | 0(0.00%) | 367(99.73%) | |
| Non β-Lactams | Aminoglycosides | VIII | Gentamicin (10 μg) | 7(1.90%) | 6(1.63%) | 355(96.47%) |
| IX | Kanamycin (30 μg) | 1(0.27%) | 86(23.37%) | 281(76.36%) | ||
| Macrolides | X | Erythromycin (15 μg) | 10(2.72%) | 206(55.98%) | 152(41.30%) | |
| Ketolide | XI | Telithromycin (15 μg) | 35(9.51%) | 62(16.85%) | 271(73.64%) | |
| Glycopeptides | XII | Teicoplanin (30 μg) | 7(1.90%) | 52(14.13%) | 309(83.97%) | |
| Quinolones | XIII | Ciprofloxacin (5 μg) | 4(1.09%) | 74(20.11%) | 290(78.80%) | |
| Phenylpropanol | XIV | Chloramphenicol (30 μg) | 5(1.36%) | 25(6.79%) | 338(91.85%) | |
| Tetracyclines | XV | Tetracycline (30 μg) | 57(15.49%) | 109(29.62%) | 202(54.89%) | |
| Folate pathway inhibitors | XVI | Trimethoprim-Sulfamethoxazole (1.25 μg/23.75 μg) | 46(12.50%) | 49(13.32%) | 273(74.18%) | |
| Lincosamides | XVII | Clindamycin (2 μg) | 34(9.24%) | 317(86.14%) | 17(4.62%) | |
| Ansamycins | XVIII | Rifampin (5 μg) | 343(93.21%) | 21(5.71%) | 4(1.09%) | |
| Streptogramins | XIX | Quinupristin-dalfopristin (15 μg) | 72(19.57%) | 254(69.02%) | 42(11.41%) | |
| Nitrofurans | XX | Nitrofurantoin (300 μg) | 61(16.58%) | 161(43.75%) | 146(39.67%) | |
| Pansusceptible | ≥3 Antimicrobia | 100.00% | ||||
| ≥4 Antimicrobia | 99.73% | |||||
| ≥5 Antimicrobia | 98.91% | |||||
Notably, two isolates were resistant to 12 of the antibiotics tested ampicillin (AMP), amoxicillin-clavulanic acid (AMC), penicillin (P), cephalothin (KF), cefoxitin (FOX), cefotetan (CTT), teicoplanin (TEC), trimethoprim-sulfamethoxazole (SXT), clindamycin (DA), rifampin (RD), quinupristin-dalfopristin (QD), nitrofurantoin (FD) and ampicillin (AMP), amoxicillin-clavulanic acid (AMC), penicillin (P), cephalothin (KF), cefoxitin (FOX), erythromycin (E), telithromycin (TEL), trimethoprim-sulfamethoxazole (SXT), clindamycin (DA), rifampin (RD), quinupristin-dalfopristin (QD), nitrofurantoin (FD), respectively. Whilst > 29.35% of the strains were resistant to the six most commonly used antibiotics (AMP-AMC-P-KF-FOX-RD). According to the definition of multidrug resistance (MDR) (Magiorakos et al., 2012), all isolated B. cereus strains were qualified as MDR strains and > 98.91% of the isolates were resistant to five or more antimicrobials.
MLST and Clonal Complex Analysis
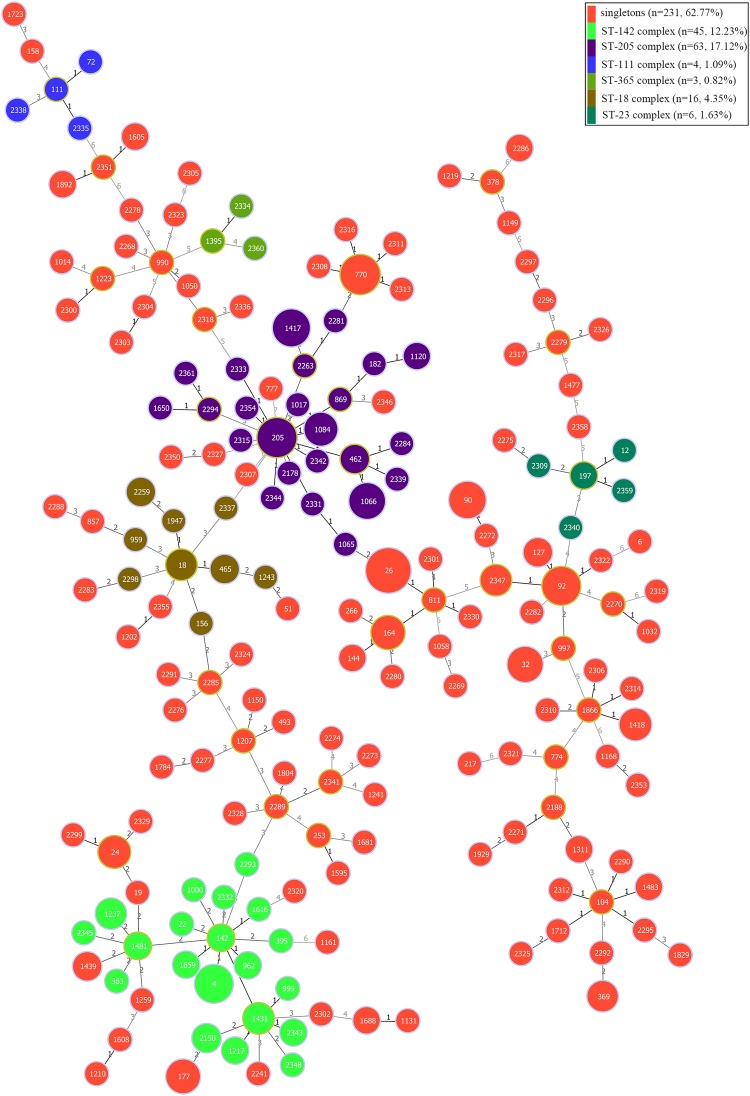
The genetic diversity of the 368 B. cereus isolates was analyzed by MLST with the internal fragment sequences of seven housekeeping genes. In total, 192 different STs were identified, 93 of which were novel and designated ST2150–ST2361. ST26 was the most abundant ST (28 isolates), followed by ST205 (14 isolates). Based on geoBURST analysis, the 192 different STs were grouped into six clonal complexes (CCs; ST-18, ST-23, ST-111, ST-142, ST-205, and ST-365) and 127 singletons. Among these CCs, ST-205, and ST-142 were predominant. Phylogenetic analysis was performed using the concatenated sequences of the seven housekeeping genes; in ST-205 complex, ST205 was the main evolutionary starter and 23 singletons evolved (Figure 4), whereas ST142 was the main evolutionary starter in ST-142 complex.
FIGURE 4.
Minimum spanning tree and genetic diversity of 368 B. cereus strains isolated from ready-to-eat foods in China. Circles filled with red color represent different singletons and with other different colors represent different clonal complexes. The numbers inside the circle indicates the corresponding sequence type (ST). The color grading and corresponding number of the line indicate the change in seven sites between the two strains at both ends of the line. The circle with a larger diameter in each clonal complex is represented the dominant ST.
Discussion
Prevalence and Genetic Diversity of B. cereus Isolates
Bacillus cereus is a foodborne pathogen that causes various symptoms and is found on multiple types of food. In this study, 302 of the 860 RTE samples (35%) were positive for B. cereus, indicating that retailed RTE foods are a potential risk to consumers. The rate of contamination observed in this study was higher than has been reported in some countries, for example Korea (Cho et al., 2011) and Morocco (Merzougui et al., 2014), but lower than in others such as India (Sudershan et al., 2012). Around 90% of the samples had <1100 MPN/g of B. cereus, the generally accepted threshold for B. cereus contamination; however, the remaining samples all exceeded 1100 MPN/g, indicating that some RTE foods may cause food poisoning due to high levels of contamination.
Cooked meats are commonly prepared and sold for direct consumption in different nations. In this study, B. cereus was present in 34% (220/656) of the cooked meat samples, consistent with the frequency reported by Tewari et al. (2015). Research has shown that open-air stalls increases the opportunity for environmental pollution (Ng et al., 2013); for instance, cooked meat sold in open-air stalls can be exposed to dust-containing spores, increasing the chance of B. cereus contamination. This could explain the high prevalence of B. cereus in our cooked meat samples.
Vomiting was first associated with B. cereus in the United Kingdom in 1971 which was caused by fried rice (Mortimer and McCann, 1974). Vomiting due to B. cereus infection is typically associated with starch-containing foods (Fricker et al., 2007; Delbrassinne et al., 2015) which are believed to promote the growth of B. cereus and its production of emetic toxins (Griffiths and Schraft, 2017). In this study, 50% (59/119) of the rice/noodle samples tested positive for B. cereus, which is higher than that reported in other studies of starch-containing foods (Chang et al., 2011; Delbrassinne et al., 2012; Merzougui et al., 2014).
Cold vegetable dishes in sauce, also known as Chinese salad, are a favorite RTE food in China. B. cereus was detected in 22% (19/85) of the cold vegetable dishes in sauce samples, a higher prevalence than observed in other reports (Valero et al., 2007), but lower than that observed in vegetable salads in Korea (Chon et al., 2015). Although cold vegetable dishes in sauce RTEs undergo processing steps, they are usually not subjected to heat processing; thus, microorganisms cannot be completely eliminated on the fresh vegetables. Some studies have indicated that the growth, harvest, processing, and packaging of vegetables are likely to increase the possibility of microbial contamination (Sagoo et al., 2003). Since B. cereus is widespread in nature, particularly in soil, vegetables are easily contaminated by this bacterium; previously, we showed that B. cereus was present in up to 50% of vegetables (Yu et al., 2019). Taken together, these results suggest that the consumption of cold vegetable dishes in sauce made from vegetables contaminated with B. cereus could increase the potential risk to the public.
Some studies have indicated that RTE foods can become contaminated by different bacterial pathogens via food preparation surfaces (Christison et al., 2008; Cho et al., 2011; de Oliveira et al., 2011; Shiningeni et al., 2019). Many pathogenic bacteria such as B. cereus are capable of forming viscous, highly heat- and drought-resistant spores. These properties increase spore retention and make their removal from preparation surfaces on production lines difficult. Following its preparation, the load of B. cereus in an RTE can increase rapidly and reach dangerous levels (above 103 CFU/g or mL); thus, consumers face a higher risk of foodborne illness from RTE foods, which are usually eaten without additional heat treatment. Consequently, regulations have been imposed on the number of B. cereus cells in RTE foods in different countries and regions based on standard guidelines. Hong Kong classifies RTE foods as either satisfactory (<103 CFU/g), acceptable (103–105 CFU/g), or unsatisfactory (>105 CFU/g), and the sale of “unsatisfactory” RTE foods must be halted immediately (Food and Environmental Hygiene Department, 2014). The Food Standards of Australia and New Zealand classes RTEs into four levels: satisfactory, acceptable, unsatisfactory, and potentially harmful (>104 CFU/g of B. cereus) (New South Wales Food Authority, 2009), whereas in England “unsatisfactory” RTE foods are those with >105 CFU/g of B. cereus (Health Protection Agency, 2009). In this study, 29 positive samples (29/302, 10%) had more than 1100 MPN/g of B. cereus, which may exceed some of these guideline limits.
Virulence Gene Profiles of B. cereus Isolates
The most common enterotoxin-encoding genes in the B. cereus strains we identified were the nhe and entFM, consistent with previous reports (Kim et al., 2010; Chon et al., 2015; Hwang and Park, 2015; Glasset et al., 2016; Gao et al., 2018; Yu et al., 2019). CytK is a cytotoxin isolated from a B. cereus strain that caused a severe food poisoning outbreak leading to three deaths in France (Lund et al., 2000). The cytK gene was present in 68% of the B. cereus isolates identified in this study, consistent with earlier studies that found the gene in 40–73% of B. cereus strains isolated from foods (Hwang and Park, 2015; Lee et al., 2017; Gao et al., 2018). Hence, the widespread distribution of diarrhea-causing B. cereus in RTE foods and its potential hazards cannot be ignored.
Approximately 7% (25/368) of the strains isolated in this study were emetic strains, similar to the findings of a study performed in Jordan (Batchoun et al., 2011), lower than the levels reported in Holland and Ghana (Biesta-Peters et al., 2016; Owusu-Kwarteng et al., 2017), but higher than found by studies in Korea (Chon et al., 2015; Hwang and Park, 2015). According to previous studies, starch-containing foods promote toxin production by B. cereus (Griffiths and Schraft, 2017). Interestingly, in this study the 18 B. cereus strains isolated from cooked meat contained the emetic toxin gene cesB. Since vomiting intoxication cases have been known to cause death (Mahler et al., 1997; Dierick et al., 2005; Lopez et al., 2015), even this comparatively small number of cesB positive strains cannot be ignored.
Antimicrobial Susceptibility of B. cereus
Beyond food poisoning, B. cereus is also associated with non-gastrointestinal infections. Antibiotic susceptibility testing can provide a reference for the clinical treatment of food poisoning. In this study, we exhaustively tested the antimicrobial resistance of all B. cereus isolates. More than 82.34% of the isolates were resistant to most of the β-lactam antibiotics, whilst only 13.59% (50/368) and 0.27% (1/368) were resistant to CTT and imipenem (IPM), respectively. This result is unsurprising, since B. cereus can produce a β-lactamase (Chen et al., 2003; Bottone, 2010). According to the results of our antimicrobial sensitivity testing, suspected B. cereus infections should not be treated clinically with broad-spectrum cephalosporins and penicillin, but with the CTT or IPM. We also found that many B. cereus isolates displayed multiple-drug resistance profiles, suggesting that B. cereus infections in RTE foods pose a significant potential risk.
Genetic Diversity of B. cereus in RTE Foods
Epidemiological typing methods, such as MLST, are considered crucial for studying the prevalence of foodborne bacteria. MLST is a nucleotide sequence-based approach for the unambiguous characterization of isolates that has been broadly used for epidemiological typing and risk analyses of many pathogenic bacteria, such as S. aureus (Wu et al., 2018), L. monocytogenes (Wu et al., 2016), Cronobacter spp. (Xu et al., 2015), Salmonella (Matheson et al., 2010), and B. cereus (Cardazzo et al., 2008; Yang et al., 2017; Zhang et al., 2017; Yu et al., 2019). In this study, we used MLST to analyze the genetic polymorphisms of B. cereus strains isolated from RTE foods (Supplementary Table S4). Apart from six CCs, most isolates were assigned as singletons; ST-205 and ST-142 were the predominant CCs, consistent with previous reports (Zhang et al., 2017). Moreover, 28 strains were assigned to ST26, including 18 strains that were found to contain the cesB gene. ST26 is a recognized ST associated with food poisoning that causes vomiting, and includes two clinically isolated strains (NC7401 and F4810/72) (Agata et al., 2002; Priest et al., 2004; Fricker et al., 2007), suggesting that potentially harmful strains may be present in RTE foods in China; however, further studies investigating cereulide production by these strains must be carried out to understand their threat.
Conclusion
In this study, we evaluated the prevalence of B. cereus in RTE foods from different regions in China. Our results indicated that RTE foods are highly contaminated with B. cereus and that the isolated strains contained a variety of pathogenic genes which may increase the potential risk of foodborne diseases. In addition, most of the strains exhibited MDR profiles with important implications for clinical treatment. Together with the diverse genotypic polymorphisms we observed, our findings reveal the potential high risk of B. cereus in RTE foods.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
YD, QW, JW, JZ, and SY conceived the project and designed the experiments. SY, PY, JW, CL, HG, CCL, LK, LY, SW, TL, MC, HZ, RP, YZ, and XW performed the experiments. QW and YD supervised the project. SY and YD analyzed the data and wrote the manuscript. QW, JW, and YD complemented the writing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The authors would like to acknowledge the financial supports of National Key R&D Program of China (Grant Number 2018YFC1602500), the Guangdong Technological Innovation Strategy of Special Funds (Key Areas of Research and Development Program, Grant Number 2018B020205003), the National Natural Science Foundation of China (Grant Numbers 31730070 and 31701195), the Science and Technology Program of Guangzhou, China (Grant Number 201604016068), GDAS’ Special Project of Science and Technology Development (Grant Number 2017GDASCX-0201), the 1000-Youth Elite Program, and the 111 Project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03043/full#supplementary-material
References
- Agata N., Ohta M., Yokoyama K. (2002). Production of Bacillus cereus emetic toxin (cereulide) in various foods. Int. J. Food Microbiol. 73 23–27. 10.1016/S0168-1605(01)00692-4 [DOI] [PubMed] [Google Scholar]
- Batchoun R., Al-Sha’er A. I., Khabour O. F. (2011). Molecular characterization of Bacillus cereus toxigenic strains isolated from different food matrices in Jordan. Foodborne Pathog. Dis. 8 1153–1158. 10.1089/fpd.2011.0853 [DOI] [PubMed] [Google Scholar]
- Beecher D. J., Schoeni J. L., Wong A. C. (1995). Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 63 4423–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold-Pluta A., Pluta A., Garbowska M., Stefanska I. (2019). Prevalence and toxicity characterization of Bacillus cereus in food products from Poland. Foods 8:269. 10.3390/foods8070269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesta-Peters E. G., Dissel S., Reij M. W., Zwietering M. H., in’t Veld P. H. (2016). Characterization and exposure assessment of emetic Bacillus cereus and cereulide production in food products on the Dutch market. J. Food Prot. 79 230–238. 10.4315/0362-028X.JFP-15-217 [DOI] [PubMed] [Google Scholar]
- Bonerba E., Di Pinto A., Novello L., Montemurro F., Terio V., Colao V. (2010). Detection of potentially enterotoxigenic food-related Bacillus cereus by PCR analysis. Int. J. Food Sci. Technol. 45 1310–1315. 10.1111/j.1365-2621.2010.02257.x [DOI] [Google Scholar]
- Bottone E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23 382–398. 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardazzo B., Negrisolo E., Carraro L., Alberghini L., Patarnello T., Giaccone V. (2008). Multiple-locus sequence typing and analysis of toxin genes in Bacillus cereus food-borne isolates. Appl. Environ. Microbiol. 74 850–860. 10.1128/AEM.01495-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. J., Lee J. H., Han B. R., Kwak T. K., Kim J. (2011). Prevalence of the levels of Bacillus cereus in fried rice dishes and its exposure assessment from Chinese-style restaurants. Food Sci. Biotechnol. 20 1351–1359. 10.1007/s10068-011-0186-3 [DOI] [Google Scholar]
- Chen Y., Succi J., Tenover F. C., Koehler T. M. (2003). β-Lactamase genes of the penicillin-susceptible Bacillus anthracis sterne strain. J. Bacteriol. 185 823–830. 10.1128/JB.185.3.823-830.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. I., Lee S. H., Lim J. S., Koh Y. J., Kwak H. S., Hwang I. G. (2011). Detection and distribution of food-borne bacteria in ready-to-eat foods in Korea. Food Sci. Biotechnol. 20 525–529. 10.1007/s10068-011-0073-y [DOI] [Google Scholar]
- Chon J. W., Yim J. H., Kim H. S., Kim D. H., Kim H., Oh D. H., et al. (2015). Quantitative prevalence and toxin gene profile of Bacillus cereus from ready-to-eat vegetables in South Korea. Foodborne Pathog. Dis. 12 795–799. 10.1089/fpd.2015.1977 [DOI] [PubMed] [Google Scholar]
- Christison C. A., Lindsay D., von Holy A. (2008). Microbiological survey of ready-to-eat foods and associated preparation surfaces in retail delicatessens, Johannesburg, South Africa. Food Control 19 727–733. 10.1016/j.foodcont.2007.07.004 [DOI] [Google Scholar]
- de Oliveira M. A., de Souza V. M., Bergamini A. M. M., De Martinis E. C. P. (2011). Microbiological quality of ready-to-eat minimally processed vegetables consumed in Brazil. Food Control 22 1400–1403. 10.1016/j.foodcont.2011.02.020 [DOI] [Google Scholar]
- Delbrassinne L., Andjelkovic M., Dierick K., Denayer S., Mahillon J., Van Loco J. (2012). Prevalence and levels of Bacillus cereus emetic toxin in rice dishes randomly collected from restaurants and comparison with the levels measured in a recent foodborne outbreak. Foodborne Pathog. Dis. 9 809–814. 10.1089/fpd.2012.1168 [DOI] [PubMed] [Google Scholar]
- Delbrassinne L., Botteldoorn N., Andjelkovic M., Dierick K., Denayer S. (2015). An emetic Bacillus cereus outbreak in a kindergarten: detection and quantification of critical levels of cereulide toxin. Foodborne Pathog. Dis. 12 84–87. 10.1089/fpd.2014.1788 [DOI] [PubMed] [Google Scholar]
- Dierick K., Van Coillie E., Swiecicka I., Meyfroidt G., Devlieger H., Meulemans A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43 4277–4279. 10.1128/JCM.43.8.4277-4279.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doménech-Sánchez A., Laso E., Pérez M. J., Berrocal C. I. (2011). Emetic disease caused by Bacillus cereus after consumption of tuna fish in a beach club. Foodborne Pathog. Dis. 8 835–837. 10.1089/fpd.2010.0783 [DOI] [PubMed] [Google Scholar]
- Drewnowska J. M., Swiecicka I. (2013). Eco-genetic structure of Bacillus cereus sensu lato populations from different environments in northeastern Poland. PLoS One 8:e80175. 10.1371/journal.pone.0080175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling-Schulz M., Frenzel E., Gohar M. (2015). Food-bacteria interplay: pathometabolism of emetic Bacillus cereus. Front. Microbiol. 6:704. 10.3389/fmicb.2015.00704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling-Schulz M., Vukov N., Schulz A., Shaheen R., Andersson M., Martlbauer E., et al. (2005). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71 105–113. 10.1128/AEM.71.1.105-113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Environmental Hygiene Department (2014). Microbiological Guidelines for Food. Hong Kong: Food and Environmental Hygiene Department. [Google Scholar]
- Forghani F., Kim J. B., Oh D. H. (2014). Enterotoxigenic profiling of emetic toxin-and enterotoxin-producing Bacillus cereus, isolated from food, environmental, and clinical samples by multiplex PCR. J. Food Sci. 79 M2228–M2293. 10.1111/1750-3841.12666 [DOI] [PubMed] [Google Scholar]
- Francisco A. P., Bugalho M., Ramirez M., Carriço J. A. (2009). Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. 10.1186/1471-2105-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M., Messelhäusser U., Busch U., Scherer S., Ehling-Schulz M. (2007). Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl. Environ. Microbiol. 73 1892–1898. 10.1128/AEM.02219-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. D., Temkin E., Carmeli Y. (2016). The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 22 416–422. 10.1016/j.cmi.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Gao T., Ding Y., Wu Q., Wang J., Zhang J., Yu S., et al. (2018). Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Bacillus cereus isolated from pasteurized milk in China. Front. Microbiol. 9:533. 10.3389/fmicb.2018.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasset B., Herbin S., Guillier L., Cadel-Six S., Vignaud M. L., Grout J., et al. (2016). Bacillus cereus-induced food-borne outbreaks in France, 2007 to 2014: epidemiology and genetic characterisation. Euro Surveill. 21:30413. 10.2807/1560-7917.ES.2016.21.48.30413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granum P. E., Lund T. (1997). Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157 223–228. 10.1016/S0378-1097(97)00438-2 [DOI] [PubMed] [Google Scholar]
- Granum P. E., O’sullivan K., Lund T. (1999). The sequence of the non-haemolytic enterotoxin operon from Bacillus cereus. FEMS Microbiol. Lett. 177 225–229. 10.1111/j.1574-6968.1999.tb13736.x [DOI] [PubMed] [Google Scholar]
- Griffiths M. W., Schraft H. (2017). “‘’Bacillus cereus food poisoning”,” in Foodborne Diseases, eds Dodd C. E. R., Aldsworth T., Stein R. A., Cliver D. O. (London: Academic Press; ), 395–405. 10.1016/b978-0-12-385007-2.00020-6 [DOI] [Google Scholar]
- Hansen B. M., Hendriksen N. B. (2001). Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67 185–189. 10.1128/AEM.67.1.185-189.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Protection Agency (2009). Guidelines for Assessing the Microbiological Safety of Ready-to-Eat Foods Placed on the Market [S/OL]. London: Health Protection Agency. [Google Scholar]
- Hwang J. Y., Park J. H. (2015). Characteristics of enterotoxin distribution, hemolysis, lecithinase, and starch hydrolysis of Bacillus cereus isolated from infant formulas and ready-to-eat foods. J. Dairy Sci. 98 1652–1660. 10.3168/jds.2014-9042 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Yagihara Y., Tatsuno K., Okazaki M., Okugawa S., Moriya K. (2015). Clinical characteristics and antimicrobial susceptibility of Bacillus cereus blood stream infections. Ann. Clin. Microbiol. Antimicrob. 14:43. 10.1186/s12941-015-0104-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen G. B., Hansen B. M., Eilenberg J., Mahillon J. (2003). The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5 631–640. 10.1046/j.1462-2920.2003.00461.x [DOI] [PubMed] [Google Scholar]
- Kim C. W., Cho S. H., Kang S. H., Park Y. B., Yoon M. H., Lee J. B., et al. (2015). Prevalence, genetic diversity, and antibiotic resistance of Bacillus cereus isolated from Korean fermented soybean products. J. Food Sci. 80 M123–M128. 10.1111/1750-3841.12720 [DOI] [PubMed] [Google Scholar]
- Kim J. B., Kim J. M., Kim C. H., Seo K. S., Park Y. B., Choi N. J., et al. (2010). Emetic toxin producing Bacillus cereus Korean isolates contain genes encoding diarrheal-related enterotoxins. Int. J. Food Microbiol. 144 182–186. 10.1016/j.ijfoodmicro.2010.08.021 [DOI] [PubMed] [Google Scholar]
- Lee N., Kim M. D., Chang H. J., Choi S. W., Chun H. S. (2017). Genetic diversity, antimicrobial resistance, toxin gene profiles, and toxin production ability of Bacillus cereus isolates from doenjang, a Korean fermented soybean paste. J. Food Saf. 37 e12363 10.1111/jfs.12363 [DOI] [Google Scholar]
- Lopez A. C., Minnaard J., Perez P. F., Alippi A. M. (2015). A case of intoxication due to a highly cytotoxic Bacillus cereus strain isolated from cooked chicken. Food Microbiol. 46 195–199. 10.1016/j.fm.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Lund T., De Buyser M. L., Granum P. E. (2000). A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38 254–261. 10.1046/j.1365-2958.2000.02147.x [DOI] [PubMed] [Google Scholar]
- Lund T., Granum P. E. (1996). Characterisation of a non-haemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol. Lett. 141 151–156. 10.1111/j.1574-6968.1996.tb08377.x [DOI] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mahler H., Pasi A., Kramer J. M., Schulte P., Scoging A. C., Bar W., et al. (1997). Fulminant liver failure in association with the emetic toxin of Bacillus cereus. N. Engl. J. Med. 336 1142–1148. 10.1056/NEJM199704173361604 [DOI] [PubMed] [Google Scholar]
- Marrollo R. (2016). “Bacillus cereus food-borne disease,” in The Diverse Faces of Bacillus cereus, ed. Savini V. (London: Academic Press; ), 61–72. 10.1016/b978-0-12-801474-5.00005-0 [DOI] [Google Scholar]
- Matheson N., Kingsley R. A., Sturgess K., Aliyu S. H., Wain J., Dougan G., et al. (2010). Ten years experience of Salmonella infections in Cambridge. UK. J. Infect. 60 21–25. 10.1016/j.jinf.2009.09.016 [DOI] [PubMed] [Google Scholar]
- Merzougui S., Lkhider M., Grosset N., Gautier M., Cohen N. (2014). Prevalence, PFGE typing, and antibiotic resistance of Bacillus cereus group isolated from food in Morocco. Foodborne Pathog. Dis. 11 145–149. 10.1089/fpd.2013.1615 [DOI] [PubMed] [Google Scholar]
- Mortimer P. R., McCann G. (1974). Food-poisoning episodes associated with Bacillus cereus in fried rice. Lancet 1 1043–1045. 10.1016/s0140-6736(74)90434-6 [DOI] [PubMed] [Google Scholar]
- New South Wales Food Authority (2009). Microbiological Quality Guide for Ready-to-Eat Foods. A Guide to Interpreting Microbiological Results [S/OL]. [2017-01-20]. NSW/FA/CP028/0906. Newington, CT: New South Wales Food Authority. [Google Scholar]
- Ng Y. F., Wong S. L., Cheng H. L., Yu H. F., Chan S. W. (2013). The microbiological quality of ready-to-eat food in Siu Mei and Lo Mei shops in Hong Kong. Food Control 34 547–553. 10.1016/j.foodcont.2013.05.018 [DOI] [Google Scholar]
- Nicholls M., Purcell B., Willis C., Amar C. F., Kanagarajah S., Chamberlain D., et al. (2016). Investigation of an outbreak of vomiting in nurseries in south east England. May 2012. Epidemiol. Infect. 144 582–590. 10.1017/S0950268815001491 [DOI] [PubMed] [Google Scholar]
- Oltuszak-Walczak E., Walczak P. (2013). PCR detection of cytK gene in Bacillus cereus group strains isolated from food samples. J. Microbiol. Methods 95 295–301. 10.1016/j.mimet.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Osman K. M., Kappell A. D., Orabi A., Al-Maary K. S., Mubarak A. S., Dawoud T. M., et al. (2018). Poultry and beef meat as potential seedbeds for antimicrobial resistant enterotoxigenic Bacillus species: a materializing epidemiological and potential severe health hazard. Sci. Rep. 8:11600. 10.1038/s41598-018-29932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Kwarteng J., Wuni A., Akabanda F., Tano-Debrah K., Jespersen L. (2017). Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 17:65. 10.1186/s12866-017-0975-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. B., Kim J. B., Shin S. W., Kim J. C., Cho S. H., Lee B. K., et al. (2009). Prevalence, genetic diversity, and antibiotic susceptibility of Bacillus cereus strains isolated from rice and cereals collected in Korea. J. Food Prot. 72 612–617. 10.4315/0362-028x-72.3.612 [DOI] [PubMed] [Google Scholar]
- Priest F. G., Barker M., Baillie L. W., Holmes E. C., Maiden M. C. (2004). Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 186 7959–7970. 10.1128/JB.186.23.7959-7970.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi E., Abdos F., Momtaz H., Baghbadorani Z. T., Jalali M. (2013). Bacillus cereus in infant foods: prevalence study and distribution of enterotoxigenic virulence factors in Isfahan province. Iran. Sci. World J. 2013:292571. 10.1155/2013/292571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Goncalves B., Francisco A. P., Vaz C., Ramirez M., Carriço J. A. (2016). PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 44 W246–W251. 10.1093/nar/gkw359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi E., Rishi P., Sengupta S., Jambulingam M., Madhavan H. N., Gopal L., et al. (2013). Acute postoperative Bacillus cereus endophthalmitis mimicking toxic anterior segment syndrome. Ophthalmology 120 181–185. 10.1016/j.ophtha.2012.07.009 [DOI] [PubMed] [Google Scholar]
- Sagoo S. K., Little C. L., Ward L., Gillespie I. A., Mitchell R. T. (2003). Microbiological study of ready-to-eat salad vegetables from retail establishments uncovers a national outbreak of salmonellosis. J. Food Prot. 66 403–409. 10.4315/0362-028x-66.3.403 [DOI] [PubMed] [Google Scholar]
- Schmid D., Rademacher C., Kanitz E. E., Frenzel E., Simons E., Allerberger F., et al. (2016). Elucidation of enterotoxigenic Bacillus cereus outbreaks in Austria by complementary epidemiological and microbiological investigations, 2013. Int. J. Food Microbiol. 232 80–86. 10.1016/j.ijfoodmicro.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Shiningeni D., Chimwamurombe P., Shilangale R., Misihairabgwi J. (2019). Prevalence of pathogenic bacteria in street vended ready-to-eat meats in Windhoek. Namibia. Meat Sci. 148 223–228. 10.1016/j.meatsci.2018.05.014 [DOI] [PubMed] [Google Scholar]
- Sloan-Gardner T. S., Glynn-Robinson A. J., Roberts-Witteveen A., Krsteski R., Rogers K., Kaye A., et al. (2014). An outbreak of gastroenteritis linked to a buffet lunch served at a Canberra restaurant. Commun. Dis. Intell. Q. Rep. 38 E273–E278. [DOI] [PubMed] [Google Scholar]
- Song Z., Zhao Q., Zhu L., Zhang Z., Jiang L., Huang H. (2019). Draft genome sequence of multidrug-resistant β-lactamase-producing Bacillus cereus S66 isolated from China. J. Glob. Antimicrob. Resist. 17 23–24. 10.1016/j.jgar.2019.02.019 [DOI] [PubMed] [Google Scholar]
- Stenfors Arnesen L. P., Fagerlund A., Granum P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32 579–606. 10.1111/j.1574-6976.2008.00112.x [DOI] [PubMed] [Google Scholar]
- Sudershan R. V., Naveen Kumar R., Kashinath L., Bhaskar V., Polasa K. (2012). Microbiological hazard identification and exposure assessment of poultry products sold in various localities of Hyderabad. India. Sci World J. 2012:736040. 10.1100/2012/736040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallent S. M., Rhodehamel E. J., Harmon S. M., Bennett R. W. (2012). Data from: Bacteriological analytical manual; methods for specific pathogens. U. S. Food and Drug Administration. Chapter 14 Bacillus cereus. Available at: https://www.fda.gov/food/laboratory-methods-food/bam-bacillus-cereus (accessed November 7, 2019). [Google Scholar]
- Tewari A., Singh S. P., Singh R. (2015). Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand. India. J. Food Sci. Technol. 52 1796–1801. 10.1007/s13197-013-1162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Clinical and Laboratory Standards Institute [CLSI] (2010). Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. Approved Standard-M100-S20. Wayne, PA: The Clinical and Laboratory Standards Institute. [Google Scholar]
- The Hygiene Ministry of China (2003). National Food Safety Standard. Food Microbiological Examination: Bacillus cereus Test. Beijing: The Hygiene Ministry of China. [Google Scholar]
- The Hygiene Ministry of China (2010). National Food Safety Standard. Food Microbiological Examination: General Guidelines. Beijing: The Hygiene Ministry of China. [Google Scholar]
- Tran S. L., Guillemet E., Gohar M., Lereclus D., Ramarao N. (2010). CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J. Bacteriol. 192 2638–2642. 10.1128/JB.01315-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero M., Hernández-Herrero L. A., Giner M. J. (2007). Survival, isolation and characterization of a psychrotrophic Bacillus cereus strain from a mayonnaise-based ready-to-eat vegetable salad. Food Microbiol. 24 671–677. 10.1016/j.fm.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Wu S., Huang J., Wu Q., Zhang J., Zhang F., Yang X., et al. (2018). Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 9:2767. 10.3389/fmicb.2018.02767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wu Q., Zhang J., Chen M., Guo W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from Chinese retail ready-to-eat food. Front. Microbiol. 7:168. 10.3389/fmicb.2016.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Li C., Wu Q., Zhang J., Huang J., Yang G. (2015). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int. J. Food Microbiol. 204 17–23. 10.1016/j.ijfoodmicro.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang J., Yu S., Wu Q., Guo W., Huang J., et al. (2016). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready-to-eat foods in China. Front. Microbiol. 7:816 10.3389/fmicb.2016.00816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yu X., Zhan L., Chen J., Zhang Y., Zhang J., et al. (2017). Multilocus sequence type profiles of Bacillus cereus isolates from infant formula in China. Food Microbiol. 62 46–50. 10.1016/j.fm.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Yu P., Yu S., Wang J., Guo H., Zhang Y., Liao X., et al. (2019). Bacillus cereus isolated from vegetables in China: incidence, genetic diversity, virulence genes, and antimicrobial resistance. Front. Microbiol. 10:948. 10.3389/fmicb.2019.00948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen J., Feng C., Zhan L., Zhang J., Li Y., et al. (2017). Quantitative prevalence, phenotypic and genotypic characteristics of Bacillus cereus isolated from retail infant foods in China. Foodborne Pathog. Dis. 14 564–572. 10.1089/fpd.2017.2287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.