Significance
Expansions of GAA repeats cause a severe hereditary neurodegenerative disease, Friedreich’s ataxia. In this study, we characterized the mechanisms of GAA repeat contractions in a yeast experimental system. These mechanisms might, in the long run, aid development of a therapy for this currently incurable disease. We show that GAA repeats contract during DNA replication, which can explain the high level of somatic instability of this repeat in patient tissues. We also provided evidence that a triple-stranded DNA structure is at the heart of GAA repeat instability. This discovery highlights the role of triplex DNA in genome instability and human disease.
Keywords: repeat expansion diseases, repeat contractions, Friedreich’s ataxia, DNA triplex, DNA replication
Abstract
Friedreich’s ataxia (FRDA) is a human hereditary disease caused by the presence of expanded (GAA)n repeats in the first intron of the FXN gene [V. Campuzano et al., Science 271, 1423–1427 (1996)]. In somatic tissues of FRDA patients, (GAA)n repeat tracts are highly unstable, with contractions more common than expansions [R. Sharma et al., Hum. Mol. Genet. 11, 2175–2187 (2002)]. Here we describe an experimental system to characterize GAA repeat contractions in yeast and to conduct a genetic analysis of this process. We found that large-scale contraction is a one-step process, resulting in a median loss of ∼60 triplet repeats. Our genetic analysis revealed that contractions occur during DNA replication, rather than by various DNA repair pathways. Repeats contract in the course of lagging-strand synthesis: The processivity subunit of DNA polymerase δ, Pol32, and the catalytic domain of Rev1, a translesion polymerase, act together in the same pathway to counteract contractions. Accumulation of single-stranded DNA (ssDNA) in the lagging-strand template greatly increases the probability that (GAA)n repeats contract, which in turn promotes repeat instability in rfa1, rad27, and dna2 mutants. Finally, by comparing contraction rates for homopurine-homopyrimidine repeats differing in their mirror symmetry, we found that contractions depend on a repeat’s triplex-forming ability. We propose that accumulation of ssDNA in the lagging-strand template fosters the formation of a triplex between the nascent and fold-back template strands of the repeat. Occasional jumps of DNA polymerase through this triplex hurdle, result in repeat contractions in the nascent lagging strand.
Friedreich’s ataxia (FRDA) is the most common hereditary ataxia in humans. This autosomal recessive genetic disease is caused by the presence of an expanded GAA repeat tract in the first intron of the FXN gene, which encodes for frataxin, a protein required for proper mitochondrial function (1). Healthy people usually have 8 to 34 GAA repeats, carriers have 35 to 70 repeats, and affected individuals have more than 70 repeats, commonly hundreds of repeats (2–4).
The number of expanded GAA repeats negatively correlates with disease onset and positively correlates with disease severity (4–6). This is because expanded GAA repeats impede expression of the FXN gene at the transcription level (1, 7–9), which ultimately leads to mitochondrial dysfunction (10, 11). Expanded GAA repeats are also known to stall replication fork progression (7, 12–14) and induce mutagenesis in the surrounding DNA both in patient cells (15) and model organisms (16–18). In a yeast experimental system, they also promote chromosomal fragility (19), mitotic cross-overs (20), and complex genomic rearrangements (21).
Not surprisingly, therefore, long (GAA)n runs are very unstable in length and are prone to both expansions and contractions. Long tracts of GAA repeats are also particularly unstable in somatic tissues (22), with contractions more common than expansions (23, 24). It was hypothesized that the instability of GAA repeats is due to their ability to form an intramolecular DNA triplex, also known as H-DNA (25, 26). This triplex may contain either one purine and two pyrimidine strands (YR*Y triplex) or one pyrimidine and two purine strands (YR*R triplex) (27). GAA repeats form both types of triplexes in supercoiled DNA in vitro, but there are conflicting reports on which of them is more stable under physiological conditions (26, 28–32).
Understanding the mechanism of GAA repeat contractions is very important, as facilitating contractions might help reverse the progression of this currently incurable disease. However, the mechanisms of GAA repeat contractions are not yet understood. Earlier studies in bacteria suggested that contractions of GAA repeats were linked to RecA-mediated replication fork restart (33) or double-strand break (DSB) repair via the single-strand annealing (SSA) pathway (34). In mammalian systems, contractions were counteracted by mismatch repair (MMR) (35) and promoted by base excision repair (BER) (36).
To distinguish between various possible mechanisms that could lead to large-scale GAA repeat contractions, we developed an experimental system to conduct quantitative genetic analysis of this process in yeast, Saccharomyces cerevisiae. We found that repeat contractions predominantly occur during DNA replication, likely in the course of lagging-strand synthesis. Contraction rates of various homopurine-homopyrimidine repeats appear to relate to their ability to form triplexes. We conclude that contractions result from a triplex bypass during lagging-strand synthesis.
Results
An Experimental System to Study Large-Scale GAA Repeat Contractions in Yeast.
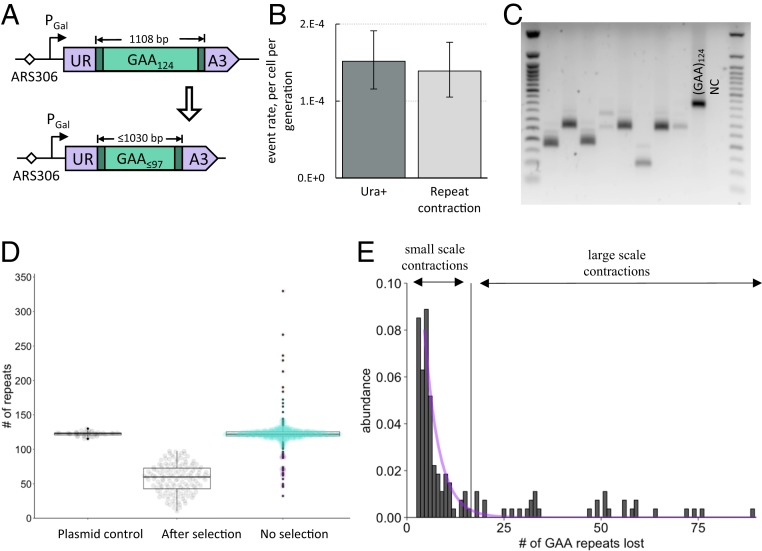
To measure the rate of large-scale contractions of GAA repeats in yeast, we used a genetic cassette containing the (GAA)124 repeat within an artificial intron of the URA3 gene located under the control of the inducible Gal promoter (37). In this setting, the (GAA)124 repeat tract constitutes a template for lagging-strand synthesis and is in the sense strand for URA3 transcription, mimicking the position of the (GAA)n run in the human FXN gene (1). Due to yeast’s inability to splice out long introns, cells containing this cassette are Ura–. Contractions of more than 20 repeats enable splicing, thus making cells Ura+ when galactose is present in the media (Fig. 1A).
Fig. 1.
An experimental system to study large-scale GAA repeat contractions in yeast. (A) Reporter cassette designed to select for large-scale (GAA)n repeat contractions. The (GAA)124 repeat located within an intron of the artificially split URA3 gene precludes splicing, making cells Ura−. A loss of 27 or more repeats restores splicing, allowing for detection of contraction events on media lacking uracil. (B) Fluctuation test results. Fluctuation test was performed as described in Materials and Methods. FluCalc (63) software was used to calculate rates of mutational events. For the Ura+ rate, the total number of Ura+ colonies from each selective plate was used. For GAA repeat contraction rate, the fraction of Ura+ colonies from each selective plate that had repeat contractions, as confirmed by PCR, was used. (C) Typical PCR amplification data showing the lengths of (GAA)n repeat tracts from colonies grown on selective plates. (GAA)124 is a positive control obtained by amplifying the repeat from the plasmid with the (GAA)124 run. NC is a negative control with no template DNA for PCR. (D) Distribution of repeat tract sizes. Plasmid control: Repeat tract amplified from a plasmid bearing the (GAA)124 run (n = 42). After selection: Repeat tract amplified from colonies grown on plates lacking uracil (n = 144). No selection: Repeat tract amplified from colonies grown on YPD plates additionally supplemented with uracil and adenine (n = 607). Different colors represent different types of events as categorized by k-means clustering analysis: Large-scale contractions are in brown, small-scale contractions and expansions are in turquoise, large-scale contractions are in purple. (E) Histogram of repeat lengths in colonies with contractions. The vertical bar denotes the border between small- and large-scale contractions as identified by k-means clustering analysis. The purple line represents exponential distribution fit for the small-scale contractions (Kolmogorov–Smirnov test, P = 0.067).
To confirm that we selected for repeat contractions, we amplified the repeat tract from randomly selected colonies that had grown on the selective Ura– media for 4 d (Fig. 1C). Of 183 analyzed colonies, 91% revealed a repeat tract that was significantly shorter than the original. Thus, the Ura+ rate was practically indistinguishable from the bona fide repeat contraction rate (Fig. 1B), which also was true for the various mutants tested (SI Appendix, Fig. S2 and Table S1). Therefore, we used the Ura+ rate as a proxy for the repeat contraction rate throughout this study.
The distribution of contracted repeat lengths revealed that the median size of the repeat tract in a colony grown on the selective media was 60 GAA repeats (Fig. 1D); that is, roughly half of the repeat tract was lost. This loss could have resulted from a single large-scale contraction event or from consecutive small-scale repeat contractions. To distinguish between these possibilities, we measured the distribution of repeat sizes in yeast colonies that were grown without any selective pressure. If large-scale contractions result from consecutive small-scale events, one should observe a single exponential distribution centered at the original repeat tract length. Alternatively, if large-scale repeat contractions occur independently from small-scale repeat contractions, one would expect to see a disproportionally large number of colonies with short repeats that did not originate from the exponential distribution.
Experimentally, we observed a large number of small-scale repeat contractions (one to three repeats) clustering around the initial repeat tract size in colonies grown without selection. However, there was a distinct additional cluster of repeat lengths around 70 GAA repeats, as well as 7 colonies carrying large repeat expansions (Fig. 1 D and E). The lower half of the distribution corresponding to repeat contractions failed to fit an exponential model (Kolmogorov–Smirnov test, P = 2.3⋅10−13), unless we only included small-scale contractions (Kolmogorov–Smirnov test, P = 0.067) categorized by k-means clustering analysis (Fig. 1 D and E). We conclude that large-scale repeat contractions occur independently from small-scale contractions.
We then compared the size distribution of large-scale repeat contractions from colonies grown with and without selection. The median size of contracted repeats was only slightly smaller under selective conditions than without selection (60 vs. 70 repeats) (Fig. 1B). Thus, selective pressure did not significantly affect the scale of repeat contractions in our system. Overall, these data show that long GAA repeats are prone to large-scale contractions in yeast that seem to occur in one step.
Impaired Lagging-Strand Replication Facilitates GAA Repeat Contractions.
Impaired Okazaki fragment processing is known to destabilize repetitive DNA sequences (38–51). We looked at whether this was true for our system by testing major yeast endonucleases implicated in the processing of single-stranded flaps during Okazaki fragment maturation: Rad27, Exo1, and Dna2.
Rad27 is a 5′ to 3′ exonuclease and 5′ flap endonuclease, which cuts short flaps that are not bound by replication protein A (RPA) (52, 53). We found that knockout of RAD27 gene leads to a drastic increase in the contraction rate of GAA repeats (Fig. 2A). To distinguish which enzymatic function of Rad27 counteracts contractions, we chose to test three rad27 missense mutants with different phenotypes. A phosphate steering mutant, rad27-4A, is defective in both endo- and exonuclease activity of the protein (47). Mutations rad27-G67S and rad27-G240D both lead to a severe exonuclease defect but differ in the extent of endonuclease deficiency, with rad27-G240D being more profoundly impaired (54). We found that all of these mutants exhibited a markedly increased GAA repeat contraction rate (Fig. 2A). Taken together, these data show that there is no single activity of Rad27 that ensures the stable maintenance of GAA repeats, which is consistent with previously reported data (55). Interestingly, the deletion of EXO1, encoding a 5′ to 3′ exonuclease and 5′-flap endonuclease (56) that can compensate for Rad27 (57), did not alter the repeat contraction rate (Fig. 2A).
Fig. 2.
The effect of mutations in DNA replication genes on the GAA repeat contraction rate. Contraction rates were assessed via fluctuation test. Note that the scale differs between separate graphs. Bars represent 95% confidence intervals. Numbers in bars show fold-change relative to WT (wild type), if significant. (A) Mutations in Okazaki fragment maturation factors. (B) Mutations in the RPA subunit Rfa1. *Estimated minimum value is plotted. (C) Effect of RPA overexpression in rad27 mutants. (D) Pol α mutations. (E) Pol δ mutations. (F) TLS polymerases.
To test whether the role of the flap-endonuclease in GAA repeat stability is because of Okazaki fragment maturation or general replication integrity, we tested the interaction of Rad27 with a component of the replication-pausing complex, Tof1. By itself, the tof1∆ mutation leads to a modest (2.5-fold) increase in the GAA repeat contraction rate and it is not synergistic with rad27-4A or rad27-G240D mutations (SI Appendix, Fig. S4), which means that Rad27 and Tof1 function in the same or overlapping genetic pathways to prevent GAA repeat contractions.
Another Okazaki fragment-processing nuclease, Dna2, has single-stranded DNA (ssDNA) endonuclease, DNA-helicase, and ATPase activities. The nuclease domain is essential, and Dna2 is the only known nuclease that can cut long flaps during Okazaki fragment maturation (52, 58). We measured the GAA repeat contraction rate in the dna2-H547A (dna2-24) mutant, which has severely reduced endonuclease activity but unaltered ATPase or helicase activities (59). This mutant demonstrated a sevenfold increase in the repeat contraction rate, comparable with that in rad27 mutants (Fig. 2A).
We therefore hypothesized that the elevated level of repeat contractions in rad27 and dna2 mutants could be explained by the accumulation of unprotected ssDNA during DNA replication. To test this possibility, we measured the GAA repeat contraction rate in the mutants of RPA, the main eukaryotic ssDNA binding protein. We tested the effect of two well-characterized mutations in the RFA1 gene, which encodes for the main RPA subunit: rfa1-S351P (rfa1-t6) and rfa1-K45E (rfa1-t11). A temperature-sensitive mutation rfa1-S351P is in the protein’s DNA binding subdomain, so mutant yeast fail to complete replication at a restrictive temperature. Mutation rfa1-K45E (rfa1-t11) is in the N-terminal subdomain involved in RPA interactions with other proteins (60, 61); it has little, if any, growth defect, but is UV- and methyl methane sulfonate-sensitive (62).
The repeat contraction rate increased in both rfa1 mutants. However, the effect of the rfa1-S351P mutation was much more striking than the effect of the rfa1-K45E mutation. Whereas the latter showed a fourfold increase in the contraction rate, the repeat completely contracted in every colony that contained the rfa1-S351P mutation even at a permissive (23 to 26 °C) temperature. In other words, independent rfa1-S351P clones were unable to retain a full-length repeat for more than a few cell divisions. Our conjectural estimate using FluCalc (63) suggested that, to explain these results, the rfa1-S351P mutation should increase the contraction rate at least 100-fold (Fig. 2B).
To further confirm that accumulation of unprotected ssDNA is the cause of repeat instability in Okazaki fragment maturation mutants, we introduced a multicopy plasmid containing all three RFA genes into rad27∆ and rad27-G240D mutants and verified RPA overexpression in the resultant strains by qRT-PCR (SI Appendix, Fig. S6). In accord with our hypothesis, RPA overexpression strongly decreased the repeat contraction rate in both rad27 mutants (Fig. 2C).
What remained uncertain, however, was whether GAA repeat contractions happen during the leading- or lagging-strand synthesis. We therefore decided to test the role of lagging-strand polymerases, Pol α and Pol δ, in GAA repeat contractions.
Pol α is an essential subunit of the polymerase α-primase complex, which synthesizes RNA–DNA primers for each Okazaki fragment (64). We integrated our contraction cassette into two DNA polymerase α-mutants that are characterized by low efficiency and fidelity and are prone to bypass DNA lesions (16, 65–67). The contraction rate was elevated in both of these mutants as compared to the WT strain, particularly strongly (12×) in the pol1-Y869A mutant (Fig. 2D).
For Pol δ, we analyzed an exonuclease dead mutant pol3-D520V, which reduces its proofreading activity (68–70), and pol32∆, a knockout of the Pol δ processivity subunit (71). The pol3-D520V mutation did not affect the repeat contraction rate. In contrast, deletion of the POL32 gene led to a 3.3-fold increase in the contraction rate (Fig. 2E). We therefore concluded that processivity, rather than fidelity of Pol δ, ensures proper replication and stability of GAA repeats.
Taken together, our data strongly argue that ssDNA intermediates, formed during lagging-strand synthesis, are at the heart of GAA repeat contractions.
Catalytic Activity of Rev1 Prevents GAA Repeat Contractions.
Does translesion DNA synthesis (TLS) contribute to GAA repeat replication and stability? TLS may promote GAA repeat contractions by hurdling lagging-strand synthesis through a DNA secondary structure, in which case one would expect a decrease in repeat contractions in TLS-deficient mutants. Alternatively, translesion polymerases might support lagging-strand synthesis across the repeat, which would lead to an increased contraction rate in TLS-deficient mutants.
There are three TLS polymerases in S. cerevisiae: Pol ζ (REV3), Pol η (RAD30), and Rev1 (72). We found that neither Pol ζ nor Pol η, either alone or in combination, play a substantial role in GAA repeat contractions (Fig. 2F). In contrast, a REV1 knockout showed an increased contraction rate, which was epistatic and similar in scale to the pol32Δ mutant (Fig. 2F). These data indicate that Rev1 somehow prevents contractions during lagging-strand synthesis. Rev1 is a deoxycytidyl transferase capable of inserting a C opposite to a G or a lesion on the template (73). We hypothesized that this activity of Rev1 promotes lagging-strand synthesis through the (GAA)n run, thus preventing repeat contractions. Supporting this idea, we demonstrated that the catalytic dead mutant rev1-CD (74) elevated the rate of repeat contractions similarly to that in rev1Δ (Fig. 2F).
DNA Repair Pathways Do Not Play a Major Role in GAA Repeat Contractions.
DNA repair has previously been implicated in GAA repeat contractions (34–36). We therefore wondered if any of the DNA repair or DNA damage tolerance (DDT) pathways can act downstream or in parallel with DNA replication in the process of repeat contraction. Since expanded GAA repeats are fragile and prone to DSB formation (19, 75, 76), it is foreseeable that repair of a DSB within the repeat by homologous recombination (HR) or nonhomologous end-joining (NHEJ) might promote repeat contractions. We, thus, measured the contraction rate in the knockouts of a set of HR genes—namely MUS81, MRE11, SRS2, SGS2, RAD51, RAD52, and RAD59—as well as a NHEJ gene, YKU70. The contraction rate was not significantly different from the WT in the majority of these mutants. Only knockouts of RAD52 and RAD59 genes, responsible for the SSA pathway in yeast (77), as well as YKU70, showed mildly increased rates over the WT (Fig. 3 and SI Appendix, Fig. S5). This means that DSB repair does not contribute to GAA repeat contractions. If anything, the SSA and NHEJ pathways might mildly protect against them.
Fig. 3.
The effect of mutations in various DNA repair genes on GAA repeat contraction rate. Contraction rates were assessed via fluctuation test. Bars represent 95% confidence intervals. Dotted black lines represent significant difference threshold. WT stands for the wild type.
DDT pathways were previously implicated in the instability of hard-to-replicate DNA repeats, such as GAA (46), ATTCT (78), and CAG (79). Therefore, we tested several DDT master regulator genes in our system. Knockout of RAD18 had no effect on the contraction rate, which was surprising given the effect of Rev1. At the same time, the rad6∆ and rad5∆ strains exhibited a slightly increased contraction rate (Fig. 3 and SI Appendix, Fig. S5), suggesting that template switching might counteract GAA repeat contractions, albeit mildly.
We also tested the role of BER, which might induce GAA repeat contractions in human cells (36), as well as nucleotide excision repair (NER), which affects the instability of CAG repeats in a variety of model systems (80–83). These pathways do not seem to promote contractions in our system, as the contraction rate was not changed upon treating yeast with H2O2 (SI Appendix, Fig. S3). Similarly, knocking out major BER and NER players did not affect the contraction rate, with a single exception of the rad2∆ strain, which showed a very modest increase (Fig. 3).
Finally, substantial literature implicates MMR in GAA repeat instability. Various MMR proteins were shown to accumulate in the FXN locus (35, 84), promote GAA repeat expansions (84–87) and fragility (19), as well as protect against GAA repeat contractions (35). Of all MMR genes studied in our system, only three genes showed a significant change in the contraction rate. MSH2 and MSH3 knockouts exhibited a small (twofold) decrease, while MLH3 knockout showed a slight increase (Fig. 3). Thus, MutSβ promotes GAA repeat contractions in our system, and Mlh3 counteracts them, albeit role of either is mild. Taken together, our data argue that the bulk of GAA repeat contractions happen during DNA replication, while the contribution from DNA repair or DNA damage avoidance pathways is at best modest.
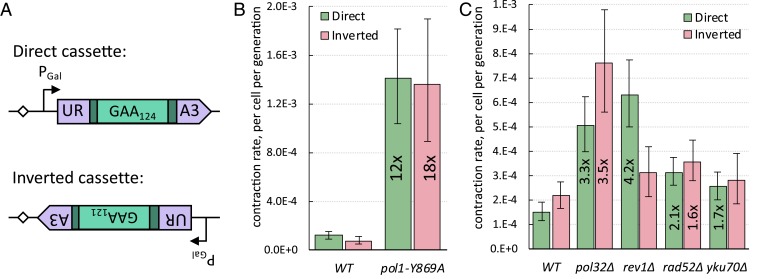
Lagging-Strand Replication Facilitates GAA Repeat Contractions in the Inverted Repeat Orientation.
Could the repeat become more stable when the cassette is flipped, placing the (TTC)n run onto the lagging-strand template, given that there is no fork stalling in this orientation (46)? We tested this idea by measuring the repeat contraction rates when our cassette was flipped relative to the ARS306 origin of replication (Fig. 4A). The transcription level of the URA3 reporter appeared to be indistinguishable between the two cassette orientations (SI Appendix, Fig. S9). Contrary to our expectations, the repeat contraction rate in the inverted cassette turned out to be nearly identical to that in the direct cassette (Fig. 4 B and C and SI Appendix, Fig. S7); furthermore, the mean number of deleted repeats was similar between the two orientations (SI Appendix, Fig. S8).
Fig. 4.
Comparison of genetic controls of GAA repeat contractions between the inverted and direct cassettes. (A) Similarities and differences between direct and inverted cassettes. In both cassettes, the (GAA)n tract is in the sense strand of transcription. In the direct cassette, the (GAA)124 tract serves as the lagging-strand template during DNA replication. In the inverted orientation, the (GAA)121 tract serves as the leading-strand template during DNA replication and there is a possibility for head-on transcription-replication collisions. (B) The effect of pol1-Y869A mutation on GAA repeat contraction rate in strains with direct versus inverted cassettes. Bars represent 95% confidence intervals. Numbers in bars show fold-change relative to corresponding WT (wild type). (C) Genetic control of GAA repeat contraction rate in strains with direct versus inverted cassettes. Bars represent 95% confidence intervals. Numbers in bars show fold-change relative to corresponding WT, if significant.
Since the inverted cassette faces replication going from the ARS306 head-on, we were wondering whether transcription–replication collisions contribute to repeat contractions in this orientation. However, galactose induction of transcription did not change the repeat contraction rate in the inverted orientation (SI Appendix, Fig. S7) any more than it did in the direct orientation (37). Therefore, the high rate of contractions in the inverted cassette orientation is not caused by transcription–replication collisions. Nor is it caused by a DSB formation, as the rate of repeat contractions in the rad52Δ and yku70Δ mutants did not decrease compared to the WT rate for the inverted cassette, just like in the direct cassette (Fig. 4C).
We then looked at the effect of genes involved in lagging-strand DNA synthesis that were major players in repeat stability for the direct cassette. Both the pol1-Y869A and pol32Δ mutations led to a similar increase in the contraction rate in the inverted cassette as they did in the direct cassette (Fig. 4 B and C). We believe, therefore, that in the inverted cassette, repeats likely contract during lagging-strand synthesis along the (TTC)n template. One notable difference between the two cassettes, however, is the role of Rev1: Its knockout has no effect on repeat contraction in the inverted cassette (Fig. 4C). This further suggests that contractions happen during lagging-strand synthesis, since the presence of the (TTC)n run in the lagging-strand template makes a templated insertion of C by Rev1 impossible. We concluded that impaired lagging-strand replication boosts GAA repeat contractions independent of the repeat orientation relative to the replication direction.
Triplex Formation Facilitates GAA Repeat Contractions.
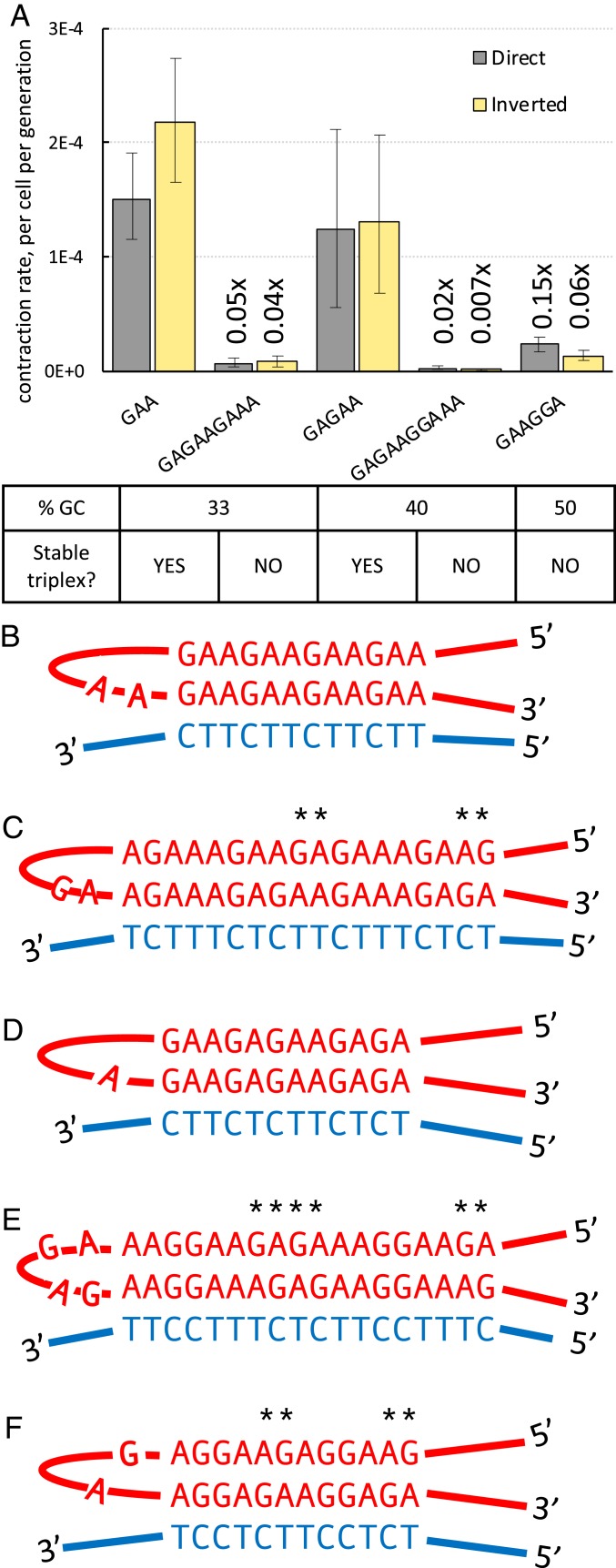
Because GAA repeats can form triplex H-DNA in vitro (26, 30–32), we hypothesized that this structure could be involved in large-scale repeat contractions in yeast, as was previously proposed for bacterial plasmids (25). To test this idea, we took advantage of the fact that H-DNA can only be formed by homopurine-homopyrimidine sequences that are mirror repeats. We, thus, constructed a new cassette bearing a (GAGAAGAAA)41GAG repeat tract. Similar to the (GAA)124 run, it is a 372-bp-long homopurine-homopyrimidine repeat with a 33% GC content, but it lacks mirror symmetry required for H-DNA formation (Fig. 5 B and C). We reasoned that if H-DNA promotes repeat contractions, we should see fewer contractions of the GAGAAGAAA repeat compared to the GAA repeat. Indeed, the rates of GAGAAGAAA repeat contractions were more than an order-of-magnitude less than that of the GAA repeat in either orientation of the cassette (Fig. 5A).
Fig. 5.
Effect of a repeat mirror symmetry on its contraction rate. (A) Contraction rates of various homopurine-homopyrimidine repeats in both orientations relative to the replication direction. Bars represent 95% confidence intervals. Numbers on bars show fold-change relative to the (GAA)124 in corresponding orientation. (B) A triplex formed by (GAA)n repeat. (C) (GAGAAGAAA)n repeat can only form a triplex that contains multiple mismatches, indicated by asterisks. (D) A triplex formed by (GAGAA)n repeat. (E) (GAGAAGGAAA)n repeat can only form a triplex that contains multiple mismatches, indicated by asterisks. (F) (GAAGGA)n repeat can only form a triplex that contains multiple mismatches, indicated by asterisks.
Furthermore, we tested another pair of homopurine-homopyrimidine repeats that had identical lengths and CG contents (40%), but differed in their mirror symmetry: (GAGAA)74GA and (GAGAAGGAAA)37GA (Fig. 5 D and E). The mirror GAGAA repeat appeared to have practically the same contraction rate as the GAA repeat in both cassette orientations. In contrast, the GAGAAGGAAA repeat, which lacks mirror symmetry, showed a dramatically decreased rate of contraction, similarly to what we observed for the GAGAAGAAA repeat (Fig. 5A).
Finally, we measured the contraction rate of the (GAAGGA)64 repeat, which was previously found in a patient with mild and late-onset FRDA. It is not a mirror repeat (Fig. 5F), and its inability to form a triplex was previously confirmed in vitro (28). Consistent with our hypothesis, it was markedly more stable than the GAA repeat (Fig. 5A). We conclude, therefore, that triplex-forming ability is responsible for GAA repeat contractions.
We were so far unable to identify proteins that unravel triplexes during DNA replication and as such help to maintain the integrity of GAA repeats. Deletion of either STM1 or CHL1 genes, whose products were reported to bind or unwind DNA triplexes in vitro (88, 89), did not affect the rate of GAA repeat contractions in our system (SI Appendix, Fig. S10). Further studies are needed to find proteins promoting triplex bypass during DNA replication.
Discussion
FRDA is a severe degenerative disease caused by massive expansions of GAA repeats in the first intron of the FXN gene. Importantly, biopsies of FRDA patients show high heterogeneity of repeat sizes: In some tissues, up to 70% of an expanded (GAA)n repeat tract can be lost during somatic cell division, while in other tissues there is a bias toward expansions (22–24). Since repeat size is a primary factor in disease progression, understanding the mechanisms of repeat contractions is of principle importance and, in the long run, could potentially benefit FRDA patients. To identify these mechanisms, we created a new experimental system to quantitatively measure the rate and scale of expanded GAA repeat contractions in yeast, S. cerevisiae.
Remarkably, our system allowed us to detect large-scale contractions of the starting (GAA)124 repeat, where the cells lose roughly half of the repeat tract (Fig. 1D). How can a gene lose 60 trinucleotide repeats? One possibility could be a sequential accumulation of small-scale contractions. Such a pattern was observed for GAA repeat expansions in human induced pluripotent stem cells (84). Alternatively, a large-scale contraction could occur in one step. As we wanted to distinguish between these two possibilities, we analyzed how GAA repeats contract in our system when no selection was applied. Although the vast majority of events were small-scale contractions, in the direct cassette there was a statistically distinct cluster of large-scale contractions similar in scale to large-scale contractions observed under selective pressure (Fig. 1 D and E). We believe that large-scale contractions occur in one-step events during yeast growth in nonselective media in this orientation. The interpretation of the contraction distribution data for the inverted cassette is less obvious (SI Appendix, Fig. S1). While we saw an accumulation of large-scale contractions without any selection, similar to the direct cassette, statistically speaking the distribution of contractions in the inverted cassette can be modeled by a single exponential distribution. Thus, in this orientation we cannot formally reject the null hypothesis that large-scale contractions could arise from consequent small-scale events. We are skeptical that this is the case, however, given that: 1) The subset containing only small scale contractions is better modeled by an exponential distribution than all contractions (SI Appendix, Fig. S1), 2) the scale of large-scale contractions is virtually identical between the two cassette orientations, and 3) the genetic controls of repeat contractions are the same for both cassettes (see below).
What molecular mechanisms could account for large-scale repeat contractions? Although the precise mechanisms of GAA repeat contraction in eukaryotes were not established before, previous studies suggested a role for DNA repair (34–36, 87). We have tested numerous DNA repair pathways, such as NER, BER, and MMR, as well as DSB repair pathways, namely SSA, HR, and NHEJ. Our data show that these DNA repair pathways do not play a major role in GAA repeat contractions in yeast.
It was also previously suggested that GAA repeat contractions might occur while a replication fork transverses the repeat. One study in bacteria suggested that slippage of the two repetitive strands during DNA replication could result in GAA repeat contractions (90). Another bacterial study suggested that formation of the YR*Y triplex in the course of leading or lagging-strand synthesis results, respectively, in repeat contractions or expansions (25). Our data demonstrate that large-scale contractions of the GAA repeat occur during DNA replication in eukaryotic cells.
The following arguments indicate that repeat contractions happen during lagging-strand synthesis regardless of repeat orientation. First, knocking out Pol32, a processivity subunit of the lagging-strand DNA polymerase δ, leads to an increase in repeat contractions (Fig. 2E). Second, the repeat contraction rate is elevated in mutants of DNA polymerase α, the main role of which is in lagging-strand synthesis. Third, the repeat contraction rate skyrockets in the temperature-sensitive rfa1-S351P mutant, which is defective for ssDNA-binding (Fig. 2B). Finally, the second strongest repeat contraction phenotype was observed when we mutated Rad27, a key enzyme involved in Okazaki fragment maturation (Fig. 2A). While we cannot rule out that contractions can also occur during leading-strand synthesis, overall our data argue that the bulk of contractions happen during lagging-strand synthesis.
It was long known that the yeast Rad27 flap-endonuclease protein protects various DNA repetitive sequences from instability (38–51). The original model postulated that a long flap that forms in the absence of Rad27 cleavage gets incorporated into the newly synthesized Okazaki fragment, resulting in a repeat expansion (91). This model cannot explain, however, how the persistence of long flaps in rad27 mutants could lead to repeat contractions, as observed by us and others (38, 40, 41, 43–45, 48–51). Our current data show no direct correlation between flap-endonuclease activity and the repeat contraction rate in various rad27 mutants. For example, the rad27-G240D mutant, severely defective in flap cleavage activity in vitro, has a smaller GAA repeat contraction rate than the more moderate rad27-G67S mutant (Fig. 2A). Along the same line, Dna2 is involved in processing very long Okazaki flaps by a mechanism that is distinct from the Rad27 pathway. Yet the dna2-H547A mutation elevates contraction rate similarly to rad27 mutations.
To account for these discrepancies, we wondered whether the failure of Rad27 or Dna2 to cleave Okazaki fragment flaps does not promote repeat contractions directly. Instead, we hypothesized that elevated contraction rates in rad27 and dna2 mutants could be due to the depletion of RPA. RPA could be titrated out by the ssDNA gaps (45, 92) or long flaps (93) observed in rad27∆ mutants. Indeed, overexpression of the bacterial ssDNA-binding protein was shown to rescue the viability of cells depleted of both Rad27 and Dna2 (94). To directly test our hypothesis, we overexpressed all three subunits of yeast RPA in various rad27 mutants. In accord with our expectations, RPA overexpression reversed the high contraction phenotype observed in those mutants (Fig. 2C). We suggest, therefore, that in the absence of fully functional Rad27 or Dna2, RPA is titrated out from the (GAA)n repeat, impeding lagging-strand DNA synthesis and resulting in repeat contractions.
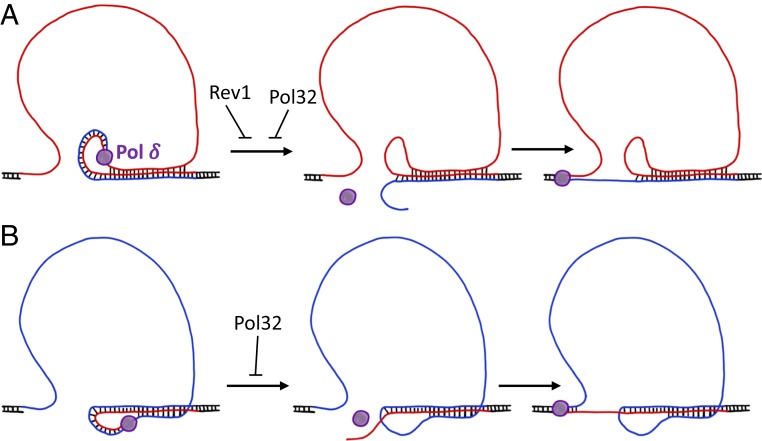
How could the accumulation of ssDNA lead to GAA repeat contractions? Since this repeat can form a triplex, we assumed that triplex formation between the lagging-strand template and the nascent lagging strand could be involved (95, 96). The strength of such a triplex would depend on the size of the single-stranded portion of the lagging-strand template. Since triplex formation is known to impede DNA polymerization (95–97), we hypothesized that Pol δ could occasionally jump through the triplex hurdle, which would result in repeat contraction after the next round of replication (Fig. 6).
Fig. 6.
Model of GAA repeat contraction in direct (A) and inverted (B) cassettes. Homopurine (GAA)n strand shown in red; polypyrimidine (TTC)n strand shown in blue. Flanking DNA sequences shown in black. When Pol δ progresses along the single-stranded (GAA)n or (TTC)n run, the template DNA strand may transiently fold back to form a dynamic triplex structure with the nascent strand. This can lead to Pol δ’s dissociation, particularly in the absence of its processivity subunit, Pol32. Lagging-strand synthesis may resume after the nascent strand partially unwinds and reanneals to the downstream part of the repeat, ultimately resulting in contraction. In the direct cassette, Rev1 may counteract contractions by periodically switching with Pol δ, allowing it to transverse through this dynamic triplex.
To check this hypothesis, we compared the contraction rate within two pairs of homopurine-homopyrimidine repeats with identical lengths and CG contents. In each pair, however, only one repeat entertains mirror symmetry (i.e., can form a stable intramolecular triplex) (98). Since another repeat lacks mirror symmetry, it cannot form a stable intramolecular triplex due to the presence of multiple mismatched triads (Fig. 5 C–F). In accord with our hypothesis, repeats with mirror symmetry were more than an order of magnitude more prone for contractions than those without mirror symmetry (Fig. 5A). We also measured the contraction rate in the GAAGGA repeat, which was discovered in a patient with mild, late-onset FRDA. This repeat does not have mirror symmetry, inhibit reporter gene expression, form triplex in vitro, or exhibit instability during intergenerational transmissions (28, 99) or in Escherichia coli (100). Similar to other asymmetrical repeats, the rate of GAAGGA repeat contractions was much lower than that of the GAA repeat in our system (Fig. 5A). We conclude that the triplex-forming ability of the GAA repeat is at the heart of its instability.
Our working model for GAA repeat contraction is shown in Fig. 6. We propose that an ssDNA stretch in the lagging-strand template in front of the DNA polymerase δ forms a triplex composed of a nascent and a fold-back template strand corresponding to the repeat. Importantly, the GAA repeat was previously shown to form both YR*Y and YR*R triplexes in superhelical DNA, albeit their relative stability remained in dispute (25, 26, 31, 32). These observations are consistent with our data on the lack of orientation-dependency of the GAA repeat contraction rate. Either a YR*Y or a YR*R triplex could be formed depending on which repetitive run (GAA)n or (TTC)n is in the lagging-strand template. This finding is different from what was previously proposed for the GAA repeat instability in E. coli, in which the type of triplex formed within the repetitive run defined whether the repeat would contract or expand (25). Nonetheless, since either triplex can impede DNA polymerization (95–97), we suggest that triplex formation promotes Pol δ dissociation from its template. Occasional unwinding of the nascent strand (either spontaneously or driven by a yet-to-be-identified DNA helicase) and its reannealing to the downstream repetitive sequence can resume the lagging-strand synthesis resulting in a much shorter repeat. In agreement with this model, we observed that Pol32, a processivity subunit of Pol δ, prevents Pol δ dissociation from the template, counteracting repeat contraction (Fig. 2E).
Our data also provide some unforeseen insight into the role of Rev1 in the replication of structure-forming DNA repeats. It has long been debated whether the catalytic activity of Rev1 plays a significant biological role. On the one hand, Rev1 plays a structural role in recruiting Pol ζ to conduct TLS (101–103). On the other hand, the catalytic subunit of Rev1 is evolutionarily conserved (73). Furthermore, it is Rev1, but not Pol ζ or, that ensures efficient replication across G-quadruplexes (104) and counteracts CTG repeat instability (105). Our data demonstrate that catalytically active Rev1 stabilizes the GAA repeat when the (GAA)n run is in the lagging-strand template, but not in the opposite orientation (Figs. 2F and 4C). We hypothesize that it could insert a C opposite a G when Pol δ gets jammed on the repetitive template, thus allowing it to resume lagging-strand synthesis. Interestingly, it seems like the catalytic activity of Rev1 in this scenario is not regulated by the ubiquitination of proliferating cell nuclear antigen as evidenced by unchanged contraction rates in the rad18∆ mutant (Fig. 3). Obviously, this cannot happen when the (TTC)n run is in the lagging-strand template, which would explain the null effect of knocking out REV1 in the inverted cassette (Fig. 4C).
Materials and Methods
Fluctuation Tests.
Individual colonies of several independent clones of each genotype grown on YPDUA were dissolved in 200 μL of sterile water, serially diluted, and plated on YPD and Gal+Ura− media. The cells from the first dilution were pelleted, and then their DNA was extracted and subjected to PCR to amplify the repeat tract. Colonies in which the repeat tract size was different from the starting number of repeats were excluded from the analysis. After the colonies grew and were counted, the contraction rate was estimated using FluCalc software (63). We considered the rates of (GAA)124 repeat contraction to be significantly different if the 95% confidence intervals for the rate values did not overlap between two samples.
Additional information regarding cassette cloning, yeast strain construction, spot tests, measuring of repeat distribution sizes, and qRT-PCR can be found in the SI Appendix.
Data Availability.
The authors declare that all data supporting the findings of this study are available within the paper and SI Appendix. All plasmids and yeast strains constructed in this study will be available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Catherine Freudenreich, Duncan Smith, Mitch McVey, Elina Radchenko, and Alexander Neil for many fruitful discussions; Matthew Cook for helping with mutant strain construction; and Ethan Hoffman for proofreading the manuscript. This study was supported by NIH Grant R35GM130322 and by a generous contribution from the White family (to S.M.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913416117/-/DCSupplemental.
References
- 1.Campuzano V., et al. , Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Cossée M., et al. , Evolution of the Friedreich’s ataxia trinucleotide repeat expansion: Founder effect and premutations. Proc. Natl. Acad. Sci. U.S.A. 94, 7452–7457 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dürr A., et al. , Clinical and genetic abnormalities in patients with Friedreich’s ataxia. N. Engl. J. Med. 335, 1169–1175 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Filla A., et al. , The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am. J. Hum. Genet. 59, 554–560 (1996). [PMC free article] [PubMed] [Google Scholar]
- 5.Montermini L., et al. , Phenotypic variability in Friedreich ataxia: Role of the associated GAA triplet repeat expansion. Ann. Neurol. 41, 675–682 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Delatycki M. B., et al. , Clinical and genetic study of Friedreich ataxia in an Australian population. Am. J. Med. Genet. 87, 168–174 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Ohshima K., Montermini L., Wells R. D., Pandolfo M., Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 273, 14588–14595 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Bidichandani S. I., Ashizawa T., Patel P. I., The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 62, 111–121 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krasilnikova M. M., et al. , Effects of Friedreich’s ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 35, 1075–1084 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koutnikova H., et al. , Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet. 16, 345–351 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Anzovino A., Lane D. J., Huang M. L., Richardson D. R., Fixing frataxin: ‘Ironing out’ the metabolic defect in Friedreich’s ataxia. Br. J. Pharmacol. 171, 2174–2190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasilnikova M. M., Mirkin S. M., Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 24, 2286–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandok G. S., Patel M. P., Mirkin S. M., Krasilnikova M. M., Effects of Friedreich’s ataxia GAA repeats on DNA replication in mammalian cells. Nucleic Acids Res. 40, 3964–3974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Follonier C., Oehler J., Herrador R., Lopes M., Friedreich’s ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat. Struct. Mol. Biol. 20, 486–494 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Bidichandani S. I., et al. , Somatic sequence variation at the Friedreich ataxia locus includes complete contraction of the expanded GAA triplet repeat, significant length variation in serially passaged lymphoblasts and enhanced mutagenesis in the flanking sequence. Hum. Mol. Genet. 8, 2425–2436 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Shah K. A., et al. , Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2, 1088–1095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang W., Dominska M., Gawel M., Greenwell P. W., Petes T. D., Genomic deletions and point mutations induced in Saccharomyces cerevisiae by the trinucleotide repeats (GAA·TTC) associated with Friedreich’s ataxia. DNA Repair (Amst.) 12, 10–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saini N., et al. , Fragile DNA motifs trigger mutagenesis at distant chromosomal loci in Saccharomyces cerevisiae. PLoS Genet. 9, e1003551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H. M., et al. , Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 27, 2896–2906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W., et al. , Friedreich’s ataxia (GAA)n•(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisae. PLoS Genet. 7, e1001270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinty R. J., et al. , Nanopore sequencing of complex genomic rearrangements in yeast reveals mechanisms of repeat-mediated double-strand break repair. Genome Res. 27, 2072–2082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellenbroich Y., Schwinger E., Zühlke C., Limited somatic mosaicism for Friedreich’s ataxia GAA triplet repeat expansions identified by small pool PCR in blood leukocytes. Acta Neurol. Scand. 103, 188–192 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Sharma R., et al. , The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum. Mol. Genet. 11, 2175–2187 (2002). [DOI] [PubMed] [Google Scholar]
- 24.De Biase I., et al. , Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics 90, 1–5 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Gacy A. M., et al. , GAA instability in Friedreich’s ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell 1, 583–593 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto N., et al. , Sticky DNA: Self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol. Cell 3, 465–475 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Frank-Kamenetskii M. D., Mirkin S. M., Triplex DNA structures. Annu. Rev. Biochem. 64, 65–95 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Ohshima K., et al. , A nonpathogenic GAAGGA repeat in the Friedreich gene: Implications for pathogenesis. Neurology 53, 1854–1857 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Grabczyk E., Usdin K., The GAA*TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 28, 2815–2822 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariappan S. V., Catasti P., Silks L. A. 3rd, Bradbury E. M., Gupta G., The high-resolution structure of the triplex formed by the GAA/TTC triplet repeat associated with Friedreich’s ataxia. J. Mol. Biol. 285, 2035–2052 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Potaman V. N., et al. , Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 32, 1224–1231 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergquist H., et al. , Structure-specific recognition of Friedreich’s ataxia (GAA)n repeats by benzoquinoquinoxaline derivatives. ChemBioChem 10, 2629–2637 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Pollard L. M., Chutake Y. K., Rindler P. M., Bidichandani S. I., Deficiency of RecA-dependent RecFOR and RecBCD pathways causes increased instability of the (GAA*TTC)n sequence when GAA is the lagging strand template. Nucleic Acids Res. 35, 6884–6894 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard L. M., Bourn R. L., Bidichandani S. I., Repair of DNA double-strand breaks within the (GAA*TTC)n sequence results in frequent deletion of the triplet-repeat sequence. Nucleic Acids Res. 36, 489–500 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezzatizadeh V., et al. , The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol. Dis. 46, 165–171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai Y., et al. , Base excision repair of chemotherapeutically-induced alkylated DNA damage predominantly causes contractions of expanded GAA repeats associated with Friedreich’s ataxia. PLoS One 9, e93464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neil A. J., Liang M. U., Khristich A. N., Shah K. A., Mirkin S. M., RNA-DNA hybrids promote the expansion of Friedreich’s ataxia (GAA)n repeats via break-induced replication. Nucleic Acids Res. 46, 3487–3497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Y., Counter C., Alani E., Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics 151, 499–509 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., et al. , Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell 48, 254–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Refsland E. W., Livingston D. M., Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast. Genetics 171, 923–934 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopes J., Debrauwère H., Buard J., Nicolas A., Instability of the human minisatellite CEB1 in rad27Delta and dna2-1 replication-deficient yeast cells. EMBO J. 21, 3201–3211 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweitzer J. K., Livingston D. M., Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7, 69–74 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Omer S., Lavi B., Mieczkowski P. A., Covo S., Hazkani-Covo E., Whole genome sequence analysis of mutations accumulated in rad27Δ yeast strains with defects in the processing of Okazaki fragments indicates template-switching events. G3 (Bethesda) 7, 3775–3787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maleki S., Cederberg H., Rannug U., The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41, 333–341 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Parenteau J., Wellinger R. J., Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19, 4143–4152 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shishkin A. A., et al. , Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol. Cell 35, 82–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsutakawa S. E., et al. , Phosphate steering by flap endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat. Commun. 8, 15855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White P. J., Borts R. H., Hirst M. C., Stability of the human fragile X (CGG)(n) triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol. 19, 5675–5684 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson R. E., Kovvali G. K., Prakash L., Prakash S., Role of yeast Rth1 nuclease and its homologs in mutation avoidance, DNA repair, and DNA replication. Curr. Genet. 34, 21–29 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Freudenreich C. H., Kantrow S. M., Zakian V. A., Expansion and length-dependent fragility of CTG repeats in yeast. Science 279, 853–856 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Kokoska R. J., et al. , Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol. 18, 2779–2788 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bae S. H., Bae K. H., Kim J. A., Seo Y. S., RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Gloor J. W., Balakrishnan L., Campbell J. L., Bambara R. A., Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1. Nucleic Acids Res. 40, 6774–6786 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y., Zhang H., Veeraraghavan J., Bambara R. A., Freudenreich C. H., Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 24, 4049–4064 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh P., Zheng L., Chavez V., Qiu J., Shen B., Concerted action of exonuclease and gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 282, 3465–3477 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Tran P. T., Erdeniz N., Dudley S., Liskay R. M., Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst.) 1, 895–912 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., Burgers P. M., Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 283, 34129–34140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levikova M., Cejka P., The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 43, 7888–7897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K. H., et al. , The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 28, 2873–2881 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Binz S. K., Wold M. S., Regulatory functions of the N-terminal domain of the 70-kDa subunit of replication protein A (RPA). J. Biol. Chem. 283, 21559–21570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y., Vaithiyalingam S., Shi Q., Chazin W. J., Zinkel S. S., BID binds to replication protein A and stimulates ATR function following replicative stress. Mol. Cell. Biol. 31, 4298–4309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umezu K., Sugawara N., Chen C., Haber J. E., Kolodner R. D., Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics 148, 989–1005 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radchenko E. A., McGinty R. J., Aksenova A. Y., Neil A. J., Mirkin S. M., Quantitative analysis of the rates for repeat-mediated genome instability in a yeast experimental system. Methods Mol. Biol. 1672, 421–438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plevani P., et al. , Polypeptide structure of DNA primase from a yeast DNA polymerase-primase complex. J. Biol. Chem. 260, 7102–7107 (1985). [PubMed] [Google Scholar]
- 65.Niimi A., et al. , Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol. Cell. Biol. 24, 2734–2746 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki M., et al. , PCNA mono-ubiquitination and activation of translesion DNA polymerases by DNA polymerase alpha. J. Biochem. 146, 13–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pavlov Y. I., Shcherbakova P. V., Kunkel T. A., In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics 159, 47–64 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nick McElhinny S. A., Stith C. M., Burgers P. M., Kunkel T. A., Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 282, 2324–2332 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.St Charles J. A., Liberti S. E., Williams J. S., Lujan S. A., Kunkel T. A., Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair (Amst.) 31, 41–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin Y. H., et al. , The 3′→5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. U.S.A. 98, 5122–5127 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson E., Garg P., Burgers P. M., The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 279, 1907–1915 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Goodman M. F., Woodgate R., Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol. 5, a010363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Masuda Y., et al. , Deoxycytidyl transferase activity of the human REV1 protein is closely associated with the conserved polymerase domain. J. Biol. Chem. 276, 15051–15058 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Zhou Y., Wang J., Zhang Y., Wang Z., The catalytic function of the Rev1 dCMP transferase is required in a lesion-specific manner for translesion synthesis and base damage-induced mutagenesis. Nucleic Acids Res. 38, 5036–5046 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao J., et al. , Distinct mechanisms of nuclease-directed DNA-structure-induced genetic instability in cancer genomes. Cell Rep. 22, 1200–1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGinty R. J., et al. , A defective mRNA cleavage and polyadenylation complex facilitates expansions of transcribed (GAA)n repeats associated with Friedreich’s ataxia. Cell Rep. 20, 2490–2500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ivanov E. L., Sugawara N., Fishman-Lobell J., Haber J. E., Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142, 693–704 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cherng N., et al. , Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 2843–2848 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Daee D. L., Mertz T., Lahue R. S., Postreplication repair inhibits CAG.CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 27, 102–110 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szwarocka S. T., Staczek P., Parniewski P., Chromosomal model for analysis of a long CTG/CAG tract stability in wild-type Escherichia coli and its nucleotide excision repair mutants. Can. J. Microbiol. 53, 860–868 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Lin Y., Wilson J. H., Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 27, 6209–6217 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hubert L. Jr, Lin Y., Dion V., Wilson J. H., Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum. Mol. Genet. 20, 4822–4830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Concannon C., Lahue R. S., Nucleotide excision repair and the 26S proteasome function together to promote trinucleotide repeat expansions. DNA Repair (Amst.) 13, 42–49 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Du J., et al. , Role of mismatch repair enzymes in GAA·TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J. Biol. Chem. 287, 29861–29872 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halabi A., Fuselier K. T. B., Grabczyk E., GAA·TTC repeat expansion in human cells is mediated by mismatch repair complex MutLγ and depends upon the endonuclease domain in MLH3 isoform one. Nucleic Acids Res. 46, 4022–4032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ezzatizadeh V., et al. , MutLα heterodimers modify the molecular phenotype of Friedreich ataxia. PLoS One 9, e100523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Halabi A., Ditch S., Wang J., Grabczyk E., DNA mismatch repair complex MutSβ promotes GAA·TTC repeat expansion in human cells. J. Biol. Chem. 287, 29958–29967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nelson L. D., Musso M., Van Dyke M. W., The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J. Biol. Chem. 275, 5573–5581 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Guo M., et al. , A distinct triplex DNA unwinding activity of ChlR1 helicase. J. Biol. Chem. 290, 5174–5189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wells R. D., et al. , Small slipped register genetic instabilities in Escherichia coli in triplet repeat sequences associated with hereditary neurological diseases. J. Biol. Chem. 273, 19532–19541 (1998). [DOI] [PubMed] [Google Scholar]
- 91.Gordenin D. A., Kunkel T. A., Resnick M. A., Repeat expansion—All in a flap? Nat. Genet. 16, 116–118 (1997). [DOI] [PubMed] [Google Scholar]
- 92.Parenteau J., Wellinger R. J., Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics 162, 1583–1594 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu B., Hu J., Wang J., Kong D., Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J. Biol. Chem. 292, 4777–4788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kahli M., Osmundson J. S., Yeung R., Smith D. J., Processing of eukaryotic Okazaki fragments by redundant nucleases can be uncoupled from ongoing DNA replication in vivo. Nucleic Acids Res. 47, 1814–1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baran N., Lapidot A., Manor H., Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc. Natl. Acad. Sci. U.S.A. 88, 507–511 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dayn A., Samadashwily G. M., Mirkin S. M., Intramolecular DNA triplexes: Unusual sequence requirements and influence on DNA polymerization. Proc. Natl. Acad. Sci. U.S.A. 89, 11406–11410 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krasilnikov A. S., et al. , Mechanisms of triplex-caused polymerization arrest. Nucleic Acids Res. 25, 1339–1346 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mirkin S. M., et al. , DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature 330, 495–497 (1987). [DOI] [PubMed] [Google Scholar]
- 99.Vetcher A. A., Napierala M., Wells R. D., Sticky DNA: Effect of the polypurine.polypyrimidine sequence. J. Biol. Chem. 277, 39228–39234 (2002). [DOI] [PubMed] [Google Scholar]
- 100.Sakamoto N., et al. , GGA*TCC-interrupted triplets in long GAA*TTC repeats inhibit the formation of triplex and sticky DNA structures, alleviate transcription inhibition, and reduce genetic instabilities. J. Biol. Chem. 276, 27178–27187 (2001). [DOI] [PubMed] [Google Scholar]
- 101.Acharya N., Johnson R. E., Prakash S., Prakash L., Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol. Cell. Biol. 26, 9555–9563 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Acharya N., et al. , Complex formation of yeast Rev1 and Rev7 proteins: A novel role for the polymerase-associated domain. Mol. Cell. Biol. 25, 9734–9740 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.D’Souza S., Walker G. C., Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol. Cell. Biol. 26, 8173–8182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sarkies P., Reams C., Simpson L. J., Sale J. E., Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40, 703–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collins N. S., Bhattacharyya S., Lahue R. S., Rev1 enhances CAG.CTG repeat stability in Saccharomyces cerevisiae. DNA Repair (Amst.) 6, 38–44 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and SI Appendix. All plasmids and yeast strains constructed in this study will be available from the corresponding author upon request.