Significance
The polymerization of the protein actin into helical filaments generates forces that power various cellular movements and overall cell motility. Cofilin is an essential actin cofactor that regulates this process by severing actin filaments. However, the mechanism of cofilin-mediated severing has remained unclear. A particular challenge to visualizing molecular details of this process is that cofilin binding breaks the helical symmetry of an actin filament, impeding the use of existing cryo-EM structure determination methods. Here, we use cryo-electron microscopy methods to determine high-resolution 3D structures of cofilin-actin severing intermediates. Our data reveal how cofilin-induced defects in the actin filament give rise to severing and show how multiple cofilins can work together to recruit additional cofilins to the filament.
Keywords: cofilin, actin, cytoskeleton, phosphorylation
Abstract
Members of the cofilin/ADF family of proteins sever actin filaments, increasing the number of filament ends available for polymerization or depolymerization. Cofilin binds actin filaments with positive cooperativity, forming clusters of contiguously bound cofilin along the filament lattice. Filament severing occurs preferentially at boundaries between bare and cofilin-decorated (cofilactin) segments and is biased at 1 side of a cluster. A molecular understanding of cooperative binding and filament severing has been impeded by a lack of structural data describing boundaries. Here, we apply methods for analyzing filament cryo-electron microscopy (cryo-EM) data at the single subunit level to directly investigate the structure of boundaries within partially decorated cofilactin filaments. Subnanometer resolution maps of isolated, bound cofilin molecules and an actin-cofilactin boundary indicate that cofilin-induced actin conformational changes are local and limited to subunits directly contacting bound cofilin. An isolated, bound cofilin compromises longitudinal filament contacts of 1 protofilament, consistent with a single cofilin having filament-severing activity. An individual, bound phosphomimetic (S3D) cofilin with weak severing activity adopts a unique binding mode that does not perturb actin structure. Cofilin clusters disrupt both protofilaments, consistent with a higher severing activity at boundaries compared to single cofilin. Comparison of these structures indicates that this disruption is substantially greater at pointed end sides of cofilactin clusters than at the barbed end. These structures, with the distribution of bound cofilin clusters, suggest that maximum binding cooperativity is achieved when 2 cofilins occupy adjacent sites. These results reveal the structural origins of cooperative cofilin binding and actin filament severing.
Members of the ADF/cofilin family of essential actin filament regulatory proteins, herein referred to as cofilin, modulate actin cytoskeletal dynamics by severing actin filaments, which increases the number of filament ends to which subunits add or dissociate (1, 2). Cofilin binds actin filaments cooperatively (3–6), forming clusters of contiguously bound cofilin (3, 5, 7, 8) (hereafter referred to as a “cluster”). Cofilin occupancy alters the filament twist (9) and subunit tilt (10, 11) and also renders filaments more compliant in bending (12–14) and twisting (15, 16).
Severing occurs preferentially at boundaries between cofilin-decorated (cofilactin) and bare actin segments (2, 3, 13, 17, 18), where structural and mechanical discontinuities exist (2, 18, 19). Cluster growth (20) and filament severing (7, 8, 21) is biased and occurs more readily at the pointed end side of clusters compared to the barbed end side. These activities of cofilin are regulated by phosphorylation; phospho-cofilin and phosphomimetic mutants bind and sever filaments weakly (22).
Previous cryo-electron microscopy (cryo-EM) studies of stable, saturated cofilactin filaments revealed that cofilin binds between longitudinal neighboring actin subunits and rotates the outer domain of actin with respect to the inner domain (10, 11, 23). This subunit tilting displaces the actin D-loop, compromising longitudinal subunit interactions (10, 11, 23, 24). Bound cofilins bridge neighboring actin filament subunits, compensating for the loss of stabilizing D-loop interactions.
The molecular mechanism of cooperative cofilin binding and filament severing has been limited by lack of structural data and atomic level models of individual, bound cofilin and boundaries where severing occurs. These structures have remained elusive due to their scarcity as well as ongoing challenging in three-dimensional (3D) reconstruction of helical assemblies with introduced asymmetry (e.g., boundaries). Here, we use a recently developed method to analyze cryo-EM samples of actin filaments partially decorated with cofilin. We developed models of single, bound cofilin and cofilactin boundaries at subnanometer resolution, which has allowed us to directly investigate conformational states that promote severing and cooperative binding.
Results
High-Resolution Reconstructions of Bare Actin and Cofilactin Filament Segments Derived from a Heterogeneous Sample.
We equilibrated cofilin and actin filaments at concentrations yielding a binding density (v) of 0.5, corresponding to 1 bound cofilin per 2 actin subunits, at acidic pH (6.6) to minimize filament depolymerization (25), and prepared vitrified cryo-EM specimens. The sparse population of boundaries (∼1 boundary per 5 micrographs) necessitated collection of a large dataset (∼3,000 micrographs) to obtain a sufficient number of particles to perform 3D structure analysis. To facilitate filament identification, we used a recently devised template-matching procedure (26) to automatically detect filament paths and extract contiguous, overlapping box segments centered on consecutive 27.5-Å repeats along the filament (SI Appendix, Fig. S1).
Filaments were heterogeneous and featured contiguous stretches of bare actin or cofilactin (SI Appendix, Fig. S1), consistent with positive cooperative binding. We performed IHRSR single particle filament structure refinement (27) using a single, common reference volume to refine all filament segments (bare and cofilactin), which preserved the geometric relationship between all subunits in a given filament, and thus allowed the relative positions of all subunits to be calculated (28). The resulting 3D reconstruction refined to a nominal resolution of 3.6 Å, but the map quality was compromised due to averaging of cofilactin and bare actin segments, and featured weak density for bound cofilin, as expected due to substoichiometric cofilin occupancy.
We applied a recently developed method to separate actin and cofilactin classes on a single subunit basis (28, 29). Cofilin-occupied sites in filaments were distinguished from bare actin by applying maximum-likelihood 3D classification (with fixed particle orientations and translations) using a combination of signal subtraction and 3D masking to focus on the central subunit of each filament box segment (30). This procedure yielded cofilactin (nominal resolution 4.1 Å) and bare actin (nominal resolution 4.4 Å) structural classes.
We improved the resolution of cofilactin to 3.4 Å (SI Appendix, Figs. S2 and S4) and bare actin to 3.5 Å (SI Appendix, Figs. S3 and S4) with independent refinement of these 2 classes. Despite originating from a heterogeneous mixture, these maps are largely indistinguishable from comparable or slightly lower resolution structural models derived from homogeneous samples of bare actin (31) or cofilactin (10, 23). Cofilin bridges longitudinally adjacent filament subunits, changes the filament twist, and tilts the actin outer domain, which displaces and disorders the actin D-loop (SI Appendix, Fig. S2). Bare actin filaments retain the untilted conformation and maintain extensive intersubunit D-loop contacts (SI Appendix, Fig. S3).
A Single Cofilin Tilts and Disorders the D-Loop of 1 Actin Subunit.
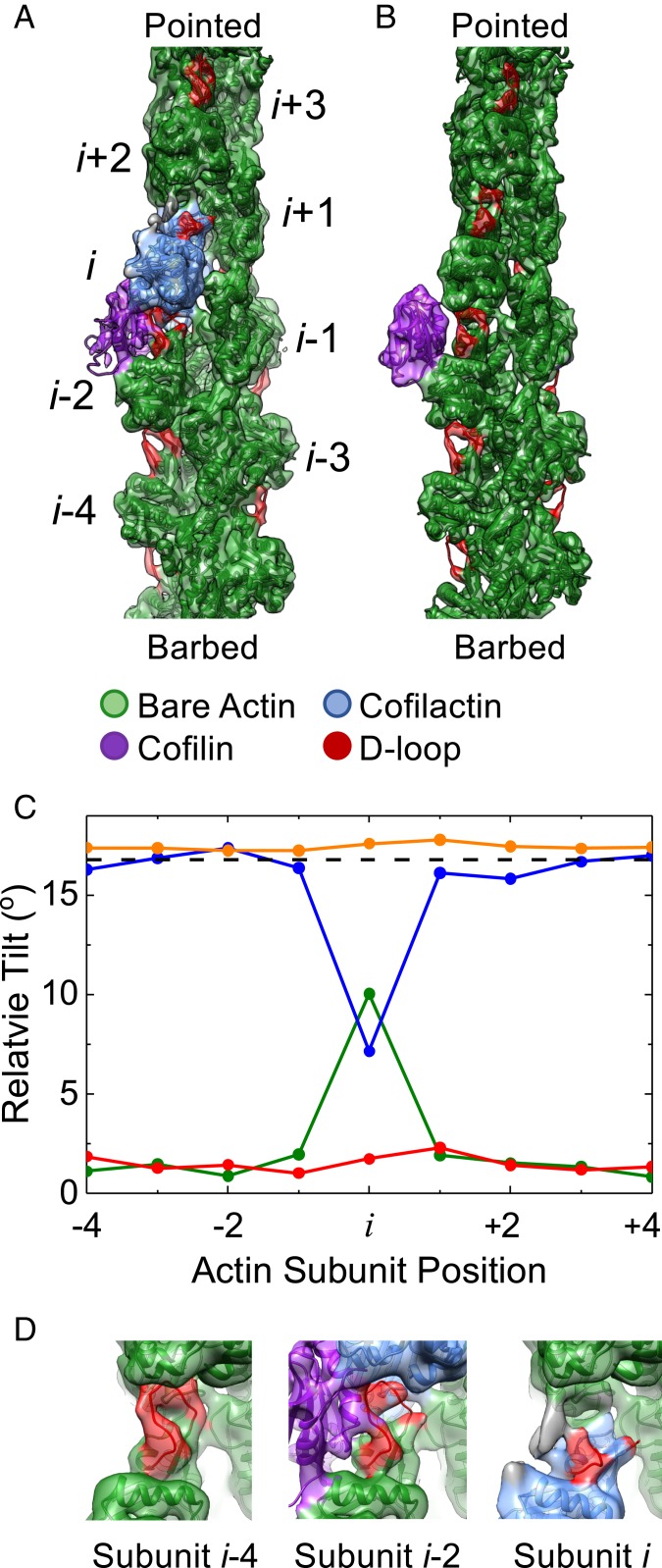
This single subunit classification procedure allowed us to determine the structure of an actin filament with single bound cofilin. We reconstructed a 7.8-Å resolution cryo-EM density map from 8,917 particles (Fig. 1A and SI Appendix, Fig. S4), revealing a unique configuration of actin in which the single, bound cofilin induces highly local perturbations in the actin filament (Fig. 1C).
Fig. 1.
A single, bound WT cofilin disrupts intersubunit contacts between the actin subunit to which it is bound (subunit i) and its nearest longitudinal neighbor (subunit i − 2). (A) Cryo-EM density map reconstructed from filament segments (n = 8,917) containing ≥5 bare sites, followed by 1 cofilin-occupied site, followed by ≥4 bare sites progressing from the barbed to the pointed end of the filament. PDB models of actin and cofilactin were fit into the cryo-EM density. The cofilactin models are colored blue (PDB ID code 5YU8), bare actin is colored green (PDB ID code 6DJO), and cofilin is colored purple (PDB ID code 5YU8). The actin D-loop is colored red. (B) Cryo-EM reconstruction of 24,000 isolated, bound S3D-cofilin molecules identified by 3D classification on 5 actin subunit-long segments, colored as in A. (C) Fitted outer domains into the WT and S3D-cofilin maps were compared with reference cofilactin (WT, blue; S3D, orange) or bare actin (WT, green; S3D, red) models that were aligned to the actin inner domain region of the map. The rotation angle between the outer domains of the reference models is also shown (dashed black line). Only the actin subunit bound to WT cofilin toward the pointed end of the filament (subunit i) adopts a cofilactin-like conformation; all other subunits resemble bare actin. (D) Close-up view of the D-loops from actin subunits i, i − 2, and i − 4 from the single, bound WT cofilin fragment from identical orientations. Cofilin disrupts the D-loop cryo-EM density of the 2 actin subunits adjacent to it (subunits i and i − 2) while the D-loop cryo-EM density of all other D-loops remains properly positioned and relatively strong. Some unaccounted additional density (gray) appears near the D-loop of subunit i, which may originate from conformational mixing with an alternative structural state where cofilin is incompletely bound and/or fails to tilt subunit i.
Cofilin compromises the integrity of the filament in this structure by perturbing a longitudinal actin–actin interface without forming a compensating bridging interaction across the same interface (Fig. 1). As with canonical cofilactin, the individual, bound cofilin bridges 2 neighboring actin subunits (i, i − 2) along a single protofilament. However, independent rigid body fitting of the actin inner and outer domains into this map reveals that only the pointed end binding partner (subunit i) tilts (Fig. 1C)—its outer domain tilts in the same direction and by nearly as much (∼10° vs. 18°) as in cofilactin (SI Appendix, Fig. S2). The second subunit in contact with cofilin (subunit i − 2) retains an untilted, bare actin conformation, as do all other actin subunits in the reconstruction (Fig. 1C). Minor discrepancies (∼2° to 3°) between the tilt magnitude of untilted and canonical bare actin subunits (Fig. 1C) likely reflect uncertainties in the rigid body fits of the atomic models into the density map, due to limited resolution.
Actin subunit (i) tilting by cofilin disrupts canonical interactions between filament subunits i and i + 2. (Fig. 1 A and D). Repositioning and disordering of the subunit i D-loop upon tilting detaches it from the adjacent actin in the pointed end direction (subunit i + 2) (Fig. 1 C and D). Loss of this longitudinal contact introduces a structural “defect” in the protofilament that could potentially compromise filament integrity.
A single bound cofilin also disrupts canonical interactions between subunits i and i − 2, but this perturbation is compensated by cofilin “bridging” of these 2 subunits (Fig. 1). These cofilin bridging interactions are distinct from those in large cofilin clusters (i.e., saturated cofilactin) (SI Appendix, Fig. S2) due to the untilted conformation of subunit i − 2. The untilted subunit i − 2 forms a previously unidentified contact with the cofilin with its D-loop (Fig. 1D). Subunit bridging by a single cofilin, while distinct from that observed in large cofilin clusters, should stabilize the subunit i and i − 2 interface.
A Single S3D-Cofilin Adopts a Unique Binding Mode That Does Not Alter the Actin Conformation.
The phosphomimetic S3D-cofilin mutant binds and severs actin filaments more weakly than wild-type (WT) cofilin (22), but the fully decorated S3D-cofilin:actin complex is indistinguishable from WT cofilactin at ∼8 Å resolution (i.e., actin is in the tilted conformation) (22), leaving the mechanism behind weak binding and severing unclear. To investigate further, we reanalyzed a published dataset (22) of actin filaments partially decorated with S3D-cofilin (referred to as S3D-cofilactin) using a modified classification scheme (Materials and Methods) and identified 23,918 particles of isolated S3D-cofilactin subunits. This was a sufficient number of particles to reconstruct a map of single, bound S3D-cofilin (Fig. 1B).
The resulting structure reveals that, in contrast to WT cofilin, a single, isolated S3D-cofilin fails to detectably perturb the filament; all subunits maintain the bare actin conformation (Fig. 1C). Moreover, S3D-cofilin primarily contacts subunit i − 2 and forms no detectable contacts with subunit i (Fig. 1B). The S3D-cofilin contacts with actin subunit i − 2 resemble those observed with fully decorated cofilactin. However, since subunit i − 2 remains in the untilted bare actin conformation, bound S3D-cofilin tilts away from subunit i and the filament long axis (Fig. 1B). In this arrangement, the S3D-cofilin N terminus resides >10 Å from its binding site on subunit i (Fig. 1B), consistent with the cofilin N terminus undocking from actin subunit i (22). Thus, a single S3D-cofilin forms fewer contacts with actin and does not visibly affect the filament structure, consistent with its reported weak binding and negligible severing activity (22).
Classification and 3D Reconstruction of Boundaries Between Bare and Cofilactin Filament Segments.
Due to actin filament polarity, a given cluster has 2 distinct boundaries between bare and cofilactin segments—one on the barbed end side of the cluster, and the other on the pointed end side. We previously reported that these boundaries could be identified and distinguished from each other in cryo-EM images of partially decorated filaments (28), but limited sample size precluded 3D reconstruction. Here, we performed 3D structural analysis of the boundaries after having collected a considerably larger dataset.
We identified barbed and pointed end boundaries by searching the single-subunit occupancy data for cofilin clusters (n ≥ 5) adjacent to a bare actin segment (n ≥ 5). Here, we define boundaries according to contacts made with bound cofilin: actin subunits in direct contact with cofilin are considered part of a cofilactin segment and all other actin subunits are considered part of a bare actin segment. This procedure identified 671 WT cofilin barbed end boundaries (bare actin at the cluster barbed end) and 146 WT cofilin pointed end boundaries (bare actin at the cluster pointed end) within the 1,117,338 particles defining the dataset. The resolution achieved by these reconstructions (9.5 Å and 24 Å for the barbed and pointed end boundaries, respectively) (SI Appendix, Fig. S4) permitted analysis of conformational changes at the barbed end boundary.
Attempts to identify S3D-cofilin barbed and pointed end boundaries were unsuccessful. The binary classification scheme utilized for WT cofilactin did not yield a decorated class, which we attribute to higher background noise in the S3D images due to a large excess of unbound cofilin. In addition, the higher cooperativity of S3D-cofilin compared to WT cofilin introduces fewer boundaries (22), which presumably reduced the observed boundary population. We devised an alternate classification scheme to detect isolated, bound S3D-cofilin molecules (Materials and Methods) by including multiple adjacent cofilin sites within the focused mask region. This scheme also failed to detect boundaries involving multiple bound cofilins but does not rule out their presence in the S3D sample.
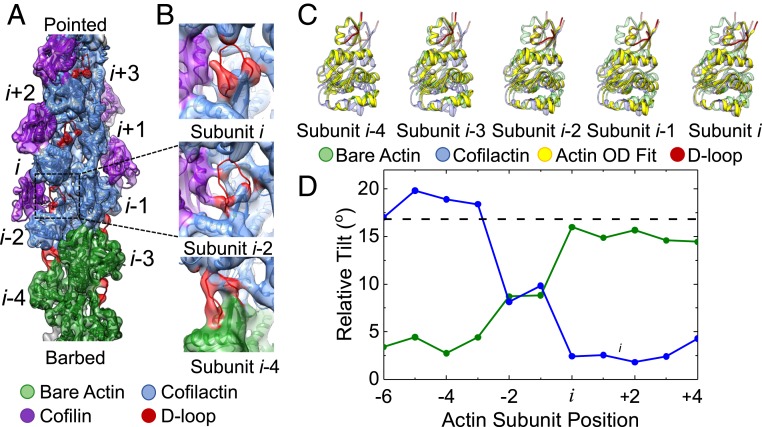
Longitudinal D-Loop Contacts Persist at the Barbed End Side of Cofilin Clusters, Despite an Abrupt Transition in Actin Conformation.
Analysis of a 3D reconstruction of the barbed end boundary indicates that the inherent twist of cofilactin persists to the actin subunits cofilin directly contacts and not further (SI Appendix, Fig. S5). That is, neither does the cofilactin twist propagate from the boundary into bare actin segments, nor do these bare actin segments alter the intrinsic twist of the adjoining cofilactin segment. This result confirms and refines our previous conclusion, derived from alignment parameters from two-dimensional (2D) image processing, that the filament twist changes abruptly (within 1 to 2 subunits) at barbed end boundaries (28). The current results make it clear that a cofilin cluster includes all actin subunits in direct contact with cofilin (including subunits i − 1 and i − 2) (SI Appendix, Fig. S5) and that there is no twist propagation beyond this boundary.
Measurements of subunit tilt at the barbed end boundary revealed an abrupt transition in actin conformation that coincides with the terminal cofilin-bound actin subunits (Fig. 2C). However, in contrast to single, bound WT cofilin, which only converts its pointed end binding partner to the tilted conformation (Fig. 1C), both actin subunits in contact with the terminal cofilin molecule of a cluster adopt the tilted conformation at barbed end boundaries (Fig. 2C). As a consequence, the abrupt tilt transition in the actin filament at the barbed end boundary is displaced from the cofilin-bound region where it is not bridged by bound cofilin (Fig. 2). This lack of a bridge, combined with an abrupt tilt transition, introduces a potential weakness in the filament lattice at the (i − 1, i − 2)–(i − 3, i − 4) interface.
Fig. 2.
Actin conformation changes abruptly at the barbed end boundary of large cofilin clusters. (A) Cryo-EM density map reconstructed from filament segments (n = 671) containing ≥5 bare sites followed by ≥5 cofilin-occupied sites progressing from the barbed to the pointed end of the filament. PDB models of actin and cofilactin were fit into the cryo-EM density. The cofilactin models are colored blue (PDB ID code 5YU8), bare actin is colored green (PDB ID code 6DJO), and cofilin is colored purple (PDB ID code 5YU8). The actin D-loop is colored red. (B) Close-up view of the D-loops from actin subunits i, i − 2, and i − 4 along the segment shown from identical orientations. The D-loop cryo-EM density indicates a loss of order for all subunits directly in contact with cofilin. (C) The orientation of the outer domain of an actin subunit (solid yellow) changes abruptly from the cofilactin (transparent blue) to the bare actin conformation (transparent green) at the boundary. Fitted outer domains were compared with reference cofilactin (transparent blue) or bare actin (transparent green) models that were fitted into the actin inner domain region of the map. The visible portions of the D-loop are colored red. (D) The rotation angle between the fitted and reference bare actin (green) and cofilactin (blue) outer domains shown in C. The rotation angle between the outer domains of the reference models is also shown (dashed black line).
Cluster barbed end boundaries retain an ordered D-loop that forms substantial longitudinal intersubunit contacts (Fig. 2B), but these appear strained when compared to those of canonical (bare) actin due to the abrupt tilt transition (Fig. 2). This contrasts with the cluster pointed end observed in isolated, bound WT cofilin where D-loop contacts are completely broken (Fig. 1). This observed reduction in longitudinal contacts at pointed end boundaries compared with barbed end boundaries in our structures is consistent with pointed end boundaries severing more readily than those at the barbed end side of cofilin clusters (7, 8, 21). The greater disruption at the pointed end side of a cluster may also facilitate cofilin binding, thereby contributing to preferential growth on this side of a cluster (20).
A similarly detailed analysis was not feasible for the pointed end boundary due to the lower resolution observed for this reconstruction. However, features of this boundary can be inferred from the single bound cofilin reconstruction (Discussion).
The Probability Distribution of Bound Cofilin Suggests 2 Contiguously Bound Cofilins “Nucleate” Cooperative Cluster Growth.
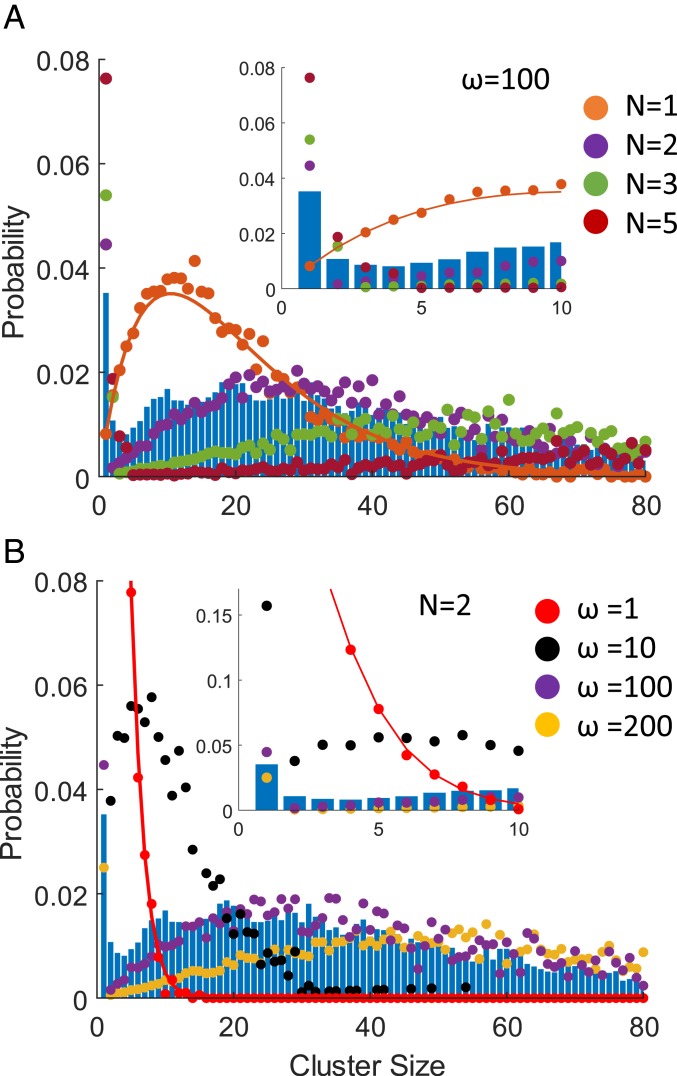
The single-subunit cofilin occupancy data developed here permits analysis of the cofilin cluster size (C) distribution (Fig. 3). A significant population of single bound cofilin is readily observed at a cofilin binding density (v) of 0.5 (Fig. 3), from which we determined the structure of an isolated, bound cofilin (Fig. 1A). In contrast, substantially fewer cofilin clusters of C > 1 were identified in these samples (Fig. 3).
Fig. 3.
Probability of a bound (WT) cofilin being in a cluster of given size (C). (A) The size distribution of clusters formed from contiguously bound cofilin molecules observed in cryo-EM specimens (blue histogram) compared with distributions predicted by Monte Carlo simulations for nucleus sizes (N) of 1 (orange), 2 (purple), 3 (green), or 5 (red) with a cooperativity (ω) of 100 and a binding density (v) of 0.5. The orange line through the simulated data for n = 1 represents the analytical solution for a nucleus size (N) of 1. Analytical solutions for higher nucleus sizes are not available. (Inset) A magnified view of the same plot for cluster sizes (C) 0 to 10. (B) Cluster size distribution observed in cryo-EM specimens (blue histogram) compared with distributions predicted by Monte Carlo simulations for cooperativity (ω) of 1 (red), 10 (black), 100 (purple), or 200 (yellow) with a critical cluster size (N) of 2. (Inset) A magnified view of the same plot for cluster sizes (C) 0 to 10.
The observed distribution of bound cofilin places strong constraints on the value of the critical cluster size needed for full cooperative binding interactions (i.e., “nucleus” size required to activate cooperative interactions [N]). At a given cofilin binding density, 2 binding parameters significantly influence the distribution profile at equilibrium (or steady-state): the degree of cooperativity (given by the cooperativity parameter ω) and the nucleus size (N) required for cooperative interactions. The observation of small clusters (C < N) places constraints on the possible nucleus size (N), and the distribution profile of large clusters (C > N) places constraints on the degree of cooperativity (ω). Because we are analyzing an equilibrium distribution of bound cofilin, the cluster size distribution is independent of the binding kinetics.
At a cofilin binding density of 0.5, values of ω = 100 and nucleus size n = 2 predict bound cofilin distributions that qualitatively match our experimentally observed distribution (Fig. 3), displaying a prominent population at C = 1, followed by a sharp reduction in frequency of C = 2, 3, 4…, before gradually increasing to a maximum at C ≈ 20, followed by a subsequent reduction (Fig. 3). A nucleus size (N) of 1 predicts relatively few singly bound cofilin (Fig. 3A), independent of the cooperativity, and a nucleus size of n = 3 predicts a distribution that does not coincide with that observed in our dataset. Thus, a nucleus size (N) of 2 best accounts for the experimentally observed cofilin cluster size distribution (Fig. 3), suggesting that 2 contiguously bound cofilin molecules serve as the nucleus for cooperative cofilin binding. Furthermore, lower or higher cooperativity values (ω) also predict distributions that do not coincide at all with the observed distribution (data for a nucleus size [N] of 2 shown in Fig. 3B). Thus, n = 2 and ω = 100 are consistent with the probability distribution of a bound cofilin being in a cluster of a given size.
Discussion
Conformational Changes in Actin Are Limited to Subunits in Direct Contact with Cofilin.
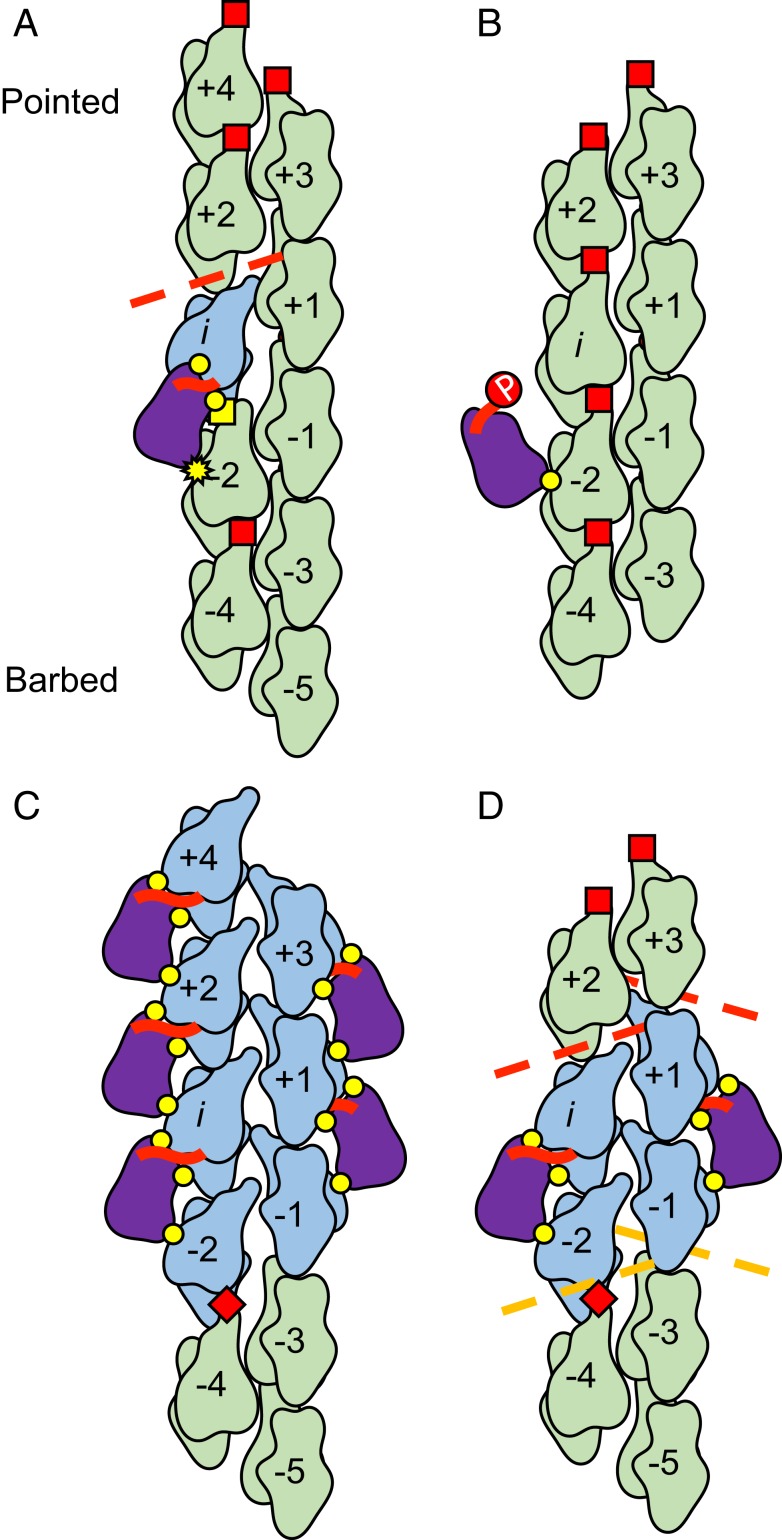
The subnanometer boundary reconstructions presented here demonstrate that only actin subunits in direct contact with cofilin undergo substantial conformational changes; actin subunits not directly contacting cofilin retain the bare actin conformation (Fig. 4). Thus, the geometry of cofilin cluster boundaries is determined by local, nearest neighbor interactions, rather than long-range, nonnearest neighbor effects extending from the boundary. This presumably explains why nearest neighbor models of cooperative cofilin binding can account for much of the equilibrium and kinetic data reported in the literature, including the correlation of boundary density with severing activity (ref. 32 and references therein).
Fig. 4.
Cooperative cofilin binding model on actin filaments. (A) Rotation of the outer domain of actin (subunit i; blue) breaks the D-loop contacts (dashed red line) with the longitudinal neighbor (subunit i + 2) and allows the N terminus of isolated, bound wild-type cofilin (thick red line) to make significant contacts with the filament (i.e., “docking”). The barbed end of the neighboring actin (subunit i − 2) does not tilt, but its D-loop is blocked from forming native intersubunit contacts by bound cofilin, with which it forms a novel interaction (yellow square). Intact D-loop interactions with adjacent neighbors are represented by red squares, and canonical cofilin-actin contacts are represented by yellow circles. The yellow star represents a cofilin:actin contact perturbed by a lack of tilting in subunit i − 2. (B) Phosphorylation (red circle labeled “P”) of the cofilin N terminus prevents its interaction with the actin outer domain, significantly weakening the binding interaction and disfavoring rotation of the outer domain. (C) At a cluster barbed end, outer domain rotation by the terminal cofilins deforms (dashed orange lines) but fails to break native D-loop contacts with adjacent neighbors in the bare region (red diamonds). (D) In a cluster, illustrated as the minimal cluster (i.e., nucleus) that occupies both protofilaments, all actin subunits directly contacting cofilin adopt the tilted conformation. (A–D) We emphasize for the general reader that the filament is illustrated as an untwisted helix to better visualize all of the intersubunit interfaces, some of which would be out of the plane of view if the filament were illustrated with a realistic helical twist.
The lower resolution of the pointed end boundary reconstruction leaves open the possibility that conformational changes could propagate further into the bare actin segment than in the case of isolated, bound cofilin. However, it appears unlikely that conformational changes would propagate multiple (n > 3) subunits away from the boundary in the absence of direct cofilin-binding interactions, given the abrupt twist transition observed at pointed end cluster boundaries (28) and recent MD results (33) that indicate the tilted conformation of actin is inherently unstable. Therefore, we conclude that the pointed end boundary of cofilin clusters resembles the pointed end protofilament boundary of an isolated, bound cofilin, in that stable cooperative conformational changes do not persist beyond subunits in direct contact with cofilin.
Maximal Cooperative Cofilin Binding Requires a Nucleus of 2 Adjacent, Contiguously Bound Cofilins.
Analysis of the cluster size (C) distribution in our dataset strongly favors a model in which the critical cluster size, or nucleus (N), for cooperative cofilin binding interactions is 2 contiguously bound cofilins (Fig. 3). While in principle these cofilins could be laterally or longitudinally adjacent, our simulations are based on a linear, 1-dimensional filament lattice model and do not consider 2 longitudinally adjacent cofilins (i − i ± 2) to be contiguously bound. We note that the reduced population of both laterally and longitudinally adjacent bound cofilin pairs in our observed cluster size distribution raises the possibility that both clusters could serve as nuclei for cooperative cluster growth.
A cluster of C = 2 could achieve full cooperativity if its boundaries closely resembled those of large clusters, where the 2 cofilins at the barbed end side of the cluster tilt the adjacent, unoccupied actin subunits on each protofilament (Figs. 2 and 4C). This scenario could be readily achieved with a laterally adjacent dimer (Fig. 4D). However, similar cooperativity may be achieved via other configurations of C = 2 clusters if, for example, tilting of multiple subunits in one protofilament leads to compensatory tilting in the other protofilament (23). A common feature of these nuclei would be conversion (e.g., tilting) of (at least) 4 contiguous actin subunits in the filament.
A single, bound WT cofilin tilts 1 of the 2 actin subunits it directly contacts (Figs. 1 and 4A). This tilting displaces SD2 from the neighboring longitudinal subunit (i + 2) at its pointed end side, which is expected to facilitate cofilin binding at this neighboring site (5, 6, 31). Thus, a single, bound cofilin molecule could potentially exert cooperative binding effects with its longitudinal neighbors. We anticipate that the degree of cooperativity is less than that of 2 adjacent cofilins, given that the observed cofilin cluster size (C) distribution (Fig. 3) features a substantial population of isolated, bound cofilin (this population would be diminished if isolated, bound cofilins functioned as nuclei).
Our results do not explain the different cooperativity of the S3D mutant compared to wild-type cofilin (22, 34). Although isolated, singly bound S3D-cofilin has no detectable effect on the filament structure (Fig. 1B), clusters of contiguously bound S3D-cofilin together do tilt filament subunits (22), which presumably arises from the additional binding energy associated with binding of multiple S3D-cofilins.
Implications for Filament Severing.
Asymmetry between the pointed and barbed end boundary structures presented here offer a structural explanation to how severing activity can be biased toward the pointed end side of bound cofilin clusters. Tilted cofilactin subunits have compromised longitudinal interactions with neighboring, untilted actin subunits (Fig. 4). These compromised interactions can be viewed as structural defects in the filament lattice. The more dramatic loss of these interactions on the pointed end side of bound cofilin clusters, compared to the barbed end side (Fig. 4), would render these filament subunit interfaces more susceptible to fragmentation by external loads (35–38) or thermally driven (7, 13, 17) fluctuations in filament shape.
Our results indicate that isolated WT cofilin disrupts the filament lattice, suggesting that it has a capacity to promote severing. The substantial loss of subunit i D-loop density and contacts compromises longitudinal intersubunit contacts with subunit i + 2 (Figs. 1 A and D and 4A). However, as this disruption only occurs on 1 protofilament, the severing activity is likely significantly weaker than at cluster boundaries, where both protofilaments are compromised. This observation is consistent with a single, bound WT cofilin having severing activity (3, 4, 22) but is at variance with studies concluding that clusters of bound cofilin must reach a critical size to sever filaments (7, 8). The discrepancy between our conclusions, derived from an equilibrium analysis, and these other studies may result from severing being in a kinetic competition with cluster growth in the latter cases, such that small cofilin clusters grew more rapidly than filaments severed. It is also possible that larger clusters (C > 3) sever filaments more readily than smaller clusters although we consider this unlikely, for reasons stated above.
The Cofilin N Terminus Plays a Critical Role in Actin Binding and Severing.
The structures presented here suggest that direct contact with the N terminus of cofilin is strongly linked to actin subunit tilting and thus presumably to the changes in filament mechanical properties (12–16, 39) associated with conversion to the cofilactin-like conformation. The N terminus of single, bound WT cofilin is positioned to interact with the pointed end binding partner of cofilin (subunit i), which adopts the tilted conformation (Figs. 1 and 4A); in contrast, the barbed end binding partner (subunit i − 2), which does not interact with the cofilin N terminus, retains an untilted conformation. These observations are accounted for by steric occlusion of cofilin by actin SD1 in the untilted conformation (10); tilting allows cofilin to bind and its N terminus to insert.
Chemically modifying the cofilin N terminus to mimic phospho-cofilin (S3D-cofilin) displaces the N terminus of an isolated, bound cofilin from the actin filament, allowing the actin subunits to which it is bound to retain their native actin conformation (Figs. 1 and 4B). This binding mode accounts for the weak binding and severing activity of single, isolated S3D-cofilin compared to WT cofilin (22). However, this unique binding mode is not observed in clusters of S3D-cofilin, which appear indistinguishable from clusters of WT cofilin at ∼8 Å resolution, despite distinct mechanical properties and severing activities (22). The structures presented here do not address the origins of this behavior.
Partial tilting is observed in the barbed end binding partners of the terminal cofilin molecules in a barbed end boundary (subunits i − 1, i − 2) (Figs. 2 and 4C), which are positioned distal from the cofilin N terminus where they cannot interact. Thus, actin subunit tilting is necessary for cofilin N terminus insertion, but subunits can partially tilt without cofilin N-terminal insertion. This observation is consistent with cofilin trapping a partially tilted thermal conformer of actin (5, 6), with N-terminal insertion leading to the fully tilted conformation. Collectively, these observations suggest that cooperative cofilin incorporation into a growing cluster is mediated by partial tilting of subunits immediately adjacent to the boundary (i.e., nearest neighbors).
Phosphate Binding to Actin Is Tightly Coupled to Subunit Tilting and Cofilin Occupancy.
A recent structural study (31) offered an explanation to how Pi dissociates from an ADP-Pi-actin subunit and how bound Pi weakens cofilin binding to filaments. Our cofilactin (SI Appendix, Fig. S2) and bare actin (SI Appendix, Fig. S3) maps are consistent with their interpretations and further suggest that subunit tilting also contributes.
Actin subunit tilting, which is tightly linked to cofilin occupancy, repositions the β-carbon of actin S14 to overlap with the Pi binding site of ADP-Pi-actin. Thus, tilting and Pi binding are mutually exclusive (i.e., the tilted conformation and bound Pi cannot coexist) (SI Appendix, Fig. S6), predicting that bound Pi obstructs subunit tilting, and tilting obstructs (i.e., weakens) Pi binding. The observed kinetic acceleration of Pi release from actin by cofilin (17, 40) therefore implies that cofilin accelerates and/or induces subunit tilting, rather than passively selecting for subunits in the tilted conformation (SI Appendix, Fig. S6).
Materials and Methods
Protein expression and purification, sample preparation, particle alignment, and single subunit classification of partially decorated actin filaments were performed as described (28). Boundaries were selected from the set of classified filament subunits using a custom script coded in the awk computer language. Subunit tilt and intersubunit twist measurements were calculated by rigid body fitting cofilin and actin atomic models into cryo-EM density maps with UCSF Chimera (41). Simulated equilibrium cofilin cluster size distributions were obtained through Monte Carlo simulations of a nearest neighbor cooperative binding model (3). A detailed description of the above procedures can be found in SI Appendix, Materials and Methods.
Data Availability.
The cryo-EM maps have been deposited with the EMDataBank, www.emdataresource.org (accession codes EMD-20711 [cofilactin], EMD- 20719 [bare actin], EMD- 20721 [single, bound WT cofilin], EMD- 20724 [single, bound S3D-cofilin], and EMD- 20726 [barbed end boundary]). The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6VAO [cofilactin], 6VAU [bare actin], 6UBY [single, bound WT cofilin], 6UC0 [single, bound S3D-cofilin], and 6UC4 [barbed end boundary]). Python scripts for interpolation and smoothing of filament coordinates are available on request.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health R01 Grants GM097348 (to E.M.D.L.C.) and GM110533001 (to C.V.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Dr. Shenping Wu (Yale Cryo-EM Resource) for assistance with microscopy and data collection. We thank the staff at the Yale School of Medicine Center for Cellular and Molecular Imaging and at the High-Performance Computing facility for expert support and maintenance of these facilities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM maps have been deposited with the EMDataBank (accession codes EMD-20711, EMD-20719, EMD-20721, EMD-20724, and EMD-20726). The atomic coordinates and structure factors have been deposited in the Protein Data Bank, (PDB ID codes 6VAO, 6VAU, 6UBY, 6UC0, and 6UC4).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915987117/-/DCSupplemental.
References
- 1.Kanellos G., Frame M. C., Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 129, 3211–3218 (2016). [DOI] [PubMed] [Google Scholar]
- 2.De La Cruz E. M., How cofilin severs an actin filament. Biophys. Rev. 1, 51–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De La Cruz E. M., Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J. Mol. Biol. 346, 557–564 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Andrianantoandro E., Pollard T. D., Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Cao W., Goodarzi J. P., De La Cruz E. M., Energetics and kinetics of cooperative cofilin-actin filament interactions. J. Mol. Biol. 361, 257–267 (2006). [DOI] [PubMed] [Google Scholar]
- 6.De La Cruz E. M., Sept D., The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys. J. 98, 1893–1901 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gressin L., Guillotin A., Guérin C., Blanchoin L., Michelot A., Architecture dependence of actin filament network disassembly. Curr. Biol. 25, 1437–1447 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Wioland H., et al. , ADF/Cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 27, 1956–1967.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGough A., Pope B., Chiu W., Weeds A., Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 138, 771–781 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galkin V. E., et al. , Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 20568–20572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galkin V. E., Orlova A., Lukoyanova N., Wriggers W., Egelman E. H., Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 153, 75–86 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullough B. R., Blanchoin L., Martiel J. L., De la Cruz E. M., Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J. Mol. Biol. 381, 550–558 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCullough B. R., et al. , Cofilin-linked changes in actin filament flexibility promote severing. Biophys. J. 101, 151–159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaendtner J., De La Cruz E. M., Voth G. A., Actin filament remodeling by actin depolymerization factor/cofilin. Proc. Natl. Acad. Sci. U.S.A. 107, 7299–7304 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prochniewicz E., Janson N., Thomas D. D., De la Cruz E. M., Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 353, 990–1000 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Fan J., et al. , Molecular origins of cofilin-linked changes in actin filament mechanics. J. Mol. Biol. 425, 1225–1240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suarez C., et al. , Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 21, 862–868 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang H., et al. , Site-specific cation release drives actin filament severing by vertebrate cofilin. Proc. Natl. Acad. Sci. U.S.A. 111, 17821–17826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De La Cruz E. M., Gardel M. L., Actin mechanics and fragmentation. J. Biol. Chem. 290, 17137–17144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngo K. X., Kodera N., Katayama E., Ando T., Uyeda T. Q. P., Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 4, e04806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin S. M., Jansen S., Goode B. L., TIRF microscopy analysis of human Cof1, Cof2, and ADF effects on actin filament severing and turnover. J. Mol. Biol. 428, 1604–1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elam W. A., et al. , Phosphomimetic S3D cofilin binds but only weakly severs actin filaments. J. Biol. Chem. 292, 19565–19579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka K., et al. , Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 9, 1860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galkin V. E., et al. , ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 163, 1057–1066 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeoh S., Pope B., Mannherz H. G., Weeds A., Determining the differences in actin binding by human ADF and cofilin. J. Mol. Biol. 315, 911–925 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Huber S. T., Kuhm T., Sachse C., Automated tracing of helical assemblies from electron cryo-micrographs. J. Struct. Biol. 202, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egelman E. H., A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Huehn A., et al. , The actin filament twist changes abruptly at boundaries between bare and cofilin-decorated segments. J. Biol. Chem. 293, 5377–5383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mentes A., et al. , High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc. Natl. Acad. Sci. U.S.A. 115, 1292–1297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheres S. H., RELION: Implementation of a bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elam W. A., Kang H., De la Cruz E. M., Biophysics of actin filament severing by cofilin. FEBS Lett. 587, 1215–1219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oda T., Takeda S., Narita A., Maéda Y., Structural polymorphism of actin. J. Mol. Biol. 431, 3217–3228 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Pope B. J., Gonsior S. M., Yeoh S., McGough A., Weeds A. G., Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J. Mol. Biol. 298, 649–661 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Schramm A. C., et al. , Actin filament strain promotes severing and cofilin dissociation. Biophys. J. 112, 2624–2633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramm A. C., Hocky G. M., Voth G. A., Martiel J. L., De La Cruz E. M., Plastic deformation and fragmentation of strained actin filaments. Biophys. J. 117, 453–463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De La Cruz E. M., Martiel J. L., Blanchoin L., Mechanical heterogeneity favors fragmentation of strained actin filaments. Biophys. J. 108, 2270–2281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X. F., et al. , Regulation of axon growth by myosin II-dependent mechanocatalysis of cofilin activity. J. Cell Biol. 218, 2329–2349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De La Cruz E. M., Roland J., McCullough B. R., Blanchoin L., Martiel J. L., Origin of twist-bend coupling in actin filaments. Biophys. J. 99, 1852–1860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanchoin L., Pollard T. D., Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps have been deposited with the EMDataBank, www.emdataresource.org (accession codes EMD-20711 [cofilactin], EMD- 20719 [bare actin], EMD- 20721 [single, bound WT cofilin], EMD- 20724 [single, bound S3D-cofilin], and EMD- 20726 [barbed end boundary]). The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6VAO [cofilactin], 6VAU [bare actin], 6UBY [single, bound WT cofilin], 6UC0 [single, bound S3D-cofilin], and 6UC4 [barbed end boundary]). Python scripts for interpolation and smoothing of filament coordinates are available on request.