Non-catalytic subunits of the heteromeric acetyl-CoA carboxylase facilitate the assembly of an activate enzyme and hence accelerate the ability to generate the key precursor of fatty acid biosynthesis, malonyl-CoA.
Abstract
Arabidopsis (Arabidopsis thaliana), like most dicotyledonous plants, expresses a multicomponent, heteromeric acetyl-CoA carboxylase (htACCase), which catalyzes the generation of the malonyl-CoA precursor of de novo fatty acid biosynthesis. This enzyme consists of four catalytic subunits: biotin carboxylase (BC), carboxyltransferase (CT)-α, CT-β, and biotin carboxyl carrier protein (BCCP1 or BCCP2). By coexpressing combinations of components in a bacterial expression system, we demonstrate noncatalytic BADCs facilitate the assembly and activation of BCCP–BADC–BC subcomplexes catalyzing the bicarbonate-dependent hydrolysis of ATP, which is the first half-reaction catalyzed by the htACCase enzyme. Although BADC proteins do not directly impact formation of the CT–αβ subcomplex, the BADC-facilitated BCCP–BADC–BC subcomplex can more readily interact with the CT–αβ subcomplex to facilitate the generation of malonyl-CoA. The Arabidopsis genome encodes three BADC isoforms (BADC1, BADC2, and BADC3), and BADC2 and BADC3 (rather than BADC1), in combination with BCCP1, best support this quaternary-structural organization and catalytic activation of the htACCase enzyme. Physiological genetic studies validate these attributes as Arabidopsis double mutants singularly expressing BADC2, BADC3, or BADC1 present increasingly greater deleterious impacts on morphological and biochemical phenotypes. Specifically, plants expressing only BADC2 develop normally, plants only expressing BADC3 suffer a stunted root-growth phenotype, and plants expressing only BADC1 are embryo-lethal. The latter phenotype may also be associated with the distinct suborganelle localization of BADC1 in plastids as compared to the localization of the other two BADC homologs. These finding can inspire novel strategies to improve the biological sources of fats and oils for dietary and industrial applications.
Biotin-containing enzymes that catalyze carboxylation, decarboxylation, or transcarboxylation reactions have been the focus of many biochemical and phylogenetic studies, which have revealed a complex evolutionary relationship (Cronan and Waldrop, 2002; Nikolau et al., 2003; Tong, 2013). The overall reactions catalyzed by these enzymes can be divided into two half-reactions, which are catalyzed at two physically distinct subsites, the biotin carboxylase (BC) and carboxyltransferase (CT) subsites. These catalytic subsites are biochemically linked by the biotin prosthetic group, which is carried by the biotin carboxyl-carrier protein (BCCP). These enzymes have served as models to understand the evolutionary assembly of these catalytic subsites as multifunctional enzymes via gene duplication, gene fusion, and adaptation mechanisms (Knowles, 1989; Faris et al., 2001; Huang et al., 2002; Jordan et al., 2003; Chalupska et al., 2008; Fan et al., 2009; Li et al., 2010, 2011b; Lombard and Moreira, 2011; Huerlimann et al., 2015).
There are a range of quaternary structural organizations for these catalytic subsites, ranging from homomeric, heteromeric, and intermediate organizations. Specifically, in homomeric enzymes the three functionalities (BC, CT, and BCCP) are domains on a single large polypeptide, whereas in heteromeric structures these three functionalities are associated with three distinct proteins (Tong, 2013). Enzymes that are organized as intermediate-type quaternary structures have two of these functionalities as domains on one protein, and the third functionality is on another protein. Exemplary of this structural diversity is the enzyme acetyl-CoA carboxylase (ACCase), which catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA, a reaction that is of significance in regulating fatty acid and polyketide biosynthesis (Cronan and Waldrop, 2002; Nikolau et al., 2003; Tong, 2013). The heteromeric ACCase (htACCase) occurs uniquely in bacteria, the homomeric ACCase (hmACCase) occurs in fungi and animals (Nikolau et al., 2003; Sasaki and Nagano, 2004; Wakil and Abu-Elheiga, 2009), and the intermediate form occurs in discrete biological clades, particularly in bacteria and archaea (Hügler et al., 2003; Gago et al., 2011). The quaternary structure of these latter forms of ACCase are often stabilized by the occurrence of a third noncatalytic subunit, often termed the “ε-subunit” (Gago et al., 2011). Uniquely, most plants (with the exception of the Gramineae) express both htACCase and hmACCase (Nikolau et al., 2003), with the former being localized in plastids, and the latter occurring in the cytosol.
A common primary sequence-motif that characterizes these enzymes is the universal biotin-attachment motif, which defines the Lys residue that is biotinylated in the BCCP-functionality. This cofactor is covalently bound, via an amide bond, to the ε-amino group of the specific Lys residue located within the conserved motif (Ala/Val)-Met-Lys-(Met/Leu). This motif also shares similarity with lipoylation sites, and thus the motif has been annotated as the biotin/lipoyl attachment domain (PF00364; http://pfam.xfam.org/family/PF00364; Finn et al., 2016). In plants, exemplified by Arabidopsis (Arabidopsis thaliana), this motif occurs in 12 genes encoding subunits of methylcrotonyl-CoA carboxylase (AT1G03090), two hmACCase isoforms (AT1G36160 and AT1G36180), two subunit isoforms of a htACCase (AT5G16390 and AT5G15530), three mitochondrial pyruvate dehydrogenase isoforms (AT3G52200, AT3G13930, and AT1G54220), a plastidial pyruvate dehydrogenase subunit (AT3G25860), an α-ketoglutarate dehydrogenase subunit (AT5G55070), a branched-chain α-keto acid dehydrogenase subunit (AT3G06850) and the H subunit of the Gly decarboxylase complex (GDC; AT2G35370; Roesler et al., 1994; Choi et al., 1995; Yanai et al., 1995; Weaver et al., 1995; Ke et al., 1997; Taylor et al., 2004; Ewald et al., 2007; Song and Liu, 2015).
In addition, the Arabidopsis genome contains three genes (AT1G52670 [BADC2], AT3G15690 [BADC3], and AT3G56130 [BADC1]), which encode proteins that share homology with these biotinylated or lipoylated proteins, but they lack the recognizable biotinylation/lipoylation defining motif; these proteins have been called Biotin/lipoyl Attachment Domain Containing (BADC) proteins (Salie et al., 2016), and previously they had been called BCCP-Like Proteins (BLPs; Ding, 2008; Mentzen et al., 2008; Chen et al., 2009; Feria Bourrellier et al., 2010; Joyard et al., 2010; Olinares et al., 2010). The in vitro characterization of extracts containing BADC–BCCP complexes (Salie et al., 2016) and in vivo analysis of plants carrying mutations in these genes (Keereetaweep et al., 2018) have been interpreted to indicate BADCs may be inhibitory of ACCase activity, providing a mechanism for regulating catalytic activity. Further analyses have indicated the expression of these genes is under the transcriptional regulation of WRINKLED1, facilitating coordinate expression with other catalytic components of central metabolism and fatty acid biosynthesis (Liu et al., 2019).
In this study, we have purified eight distinct BCCP–BADC–BC subcomplexes and determined their enzymological properties. In addition, we have developed an htACCase reconstitution system, which catalyzes the overall ATP-dependent, carboxylation of acetyl-CoA. These characterizations reveal BADCs activate ACCase catalytic activity by facilitating the assembly of BCCP–BADC–BC subcomplexes, which one could rationalize as enabling the channeling of substrates between the catalytic sites, particularly in the first half-reaction catalyzed by the BC subunit. Complementary genetic and biochemical strategies are consistent with this model and they dissect the physiological function of these three apparently redundant proteins.
RESULTS
Prior characterizations of the htACCase purified from its native sources (e.g. Escherichia coli or plants) has indicated this enzyme can be fractionated into two catalytically active subcomplexes (i.e. a subcomplex of BC and BCCP, and the CT–αβ subcomplex), which can be remixed to produce a catalytically competent enzyme (Alberts and Vagelos, 1968; Dimroth et al., 1970; Sasaki et al., 1993; Reverdatto et al., 1999). We therefore evaluated the ability to reconstitute such subcomplexes by coexpressing the Arabidopsis htACCase subunits in E. coli. In all of these coexpression experiments, only one subunit carried a small affinity tag (i.e. Hexa-His-tag), facilitating purification of potential complexes formed with the other coexpressed subunits.
BADC3 Facilitates the Formation of BC–BCCP Complexes
Coexpressing BC (AT5G35360) with BCCP1 (AT5G16390) or with BCCP2 (AT5G15530), with the latter proteins being His-tagged, led to the recovery of BC–BCCP1 or BC–BCCP2 complexes, which were purifiable by affinity chromatography with a Ni-NTA column. These recovered complexes were primarily composed of either BCCP1 or BCCP2, with BC being a very minor component of the preparations (<5%; Fig. 1A, first two lanes). However, the triple coexpression of BC, BADC3, and BCCP1 led to the formation of a complex that could also be purified via the His-tag, which contained all three subunits at near equal molar levels (Fig. 1A). Gel filtration chromatography indicated the resulting complex has a molecular mass of 930 ± 110 kD.
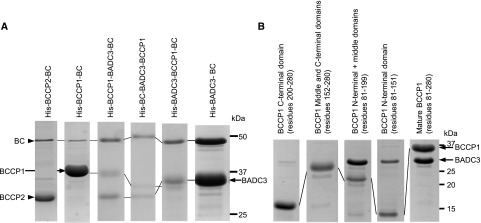
Figure 1.
BADC3 facilitates the assembly of a BCCP–BADC3–BC subcomplex. A, SDS-PAGE analysis of BCCP–BADC3–BC subcomplexes, which were coexpressed and copurified by Ni-NTA affinity chromatography, facilitated by the genetic fusion of a His-tag to each of the indicated subunits. The fusion of the His-tag to these proteins is the cause of the variation in migration of each protein band. B, Interactions between BADC3 and BCCP1 subdomains. SDS-PAGE analysis of BADC3–BCCP1 complexes, which were copurified by Ni-NTA affinity chromatography, facilitated by the genetic fusion of a His-tag to the mature BCCP1 (residues 81–280 of full-length protein), the C-terminal domain of BCCP1 (residues 200–280 of full-length protein), the middle and C-terminal domain of BCCP1 (residues 152–280 of full-length protein), N-terminal and middle domain of BCCP1 (residues 81–199 of full-length protein), or the N-terminal domain of BCCP1 (residues 81–151 of full-length protein). The figures are representative results of triplicate experiments that were sequentially conducted.
Using the BADC3 isoform as the model, we further explored the specific physical interactions that occur among the components of the BCCP1–BADC3–BC complex, utilizing the same coexpression and copurification strategy, but moving the His-tag among the three components of the complex. Specifically, independent of whether the His-tag was associated with BC or BADC3, affinity chromatography always purified a complex that contained the three components (BC, BADC3, and BCCP1; Fig. 1A). Collectively therefore, these experiments indicate BADC3 acts as the “glue” to assemble the BCCP1–BADC3–BC complex, and irrespective of which subunit carried the His-tag for purification, an ∼1 MDa BCCP1–BADC3–BC complex was recovered (Supplemental Fig. S1).
The interactions between BCCP1 and BADC3 that facilitate this complex formation were further dissected by genetically fragmenting the mature BCCP1 protein into three domains: (1) the N-terminal domain (residues 81–151); (2) the Ala/Pro-rich middle domain (residues 152–199); and (3) the C-terminal domain (residues 200–280). Of these three domains, the C-terminal domain shares the highest sequence similarity with other BCCP proteins and contains the characteristic biotinylation site sequence, Ala-Met-Lys-Met (Sasaki et al., 1993; Chapman-Smith and Cronan, 1999; Sasaki and Nagano, 2004). The N-terminal and Ala/Pro-rich middle domains are considerably more diverse in sequence among characterized BCCP proteins. However, although diverse in sequence, the middle domain is rich in Ala and Pro residues, which is a common feature of BCCPs and of intrinsically disordered proteins (Uversky, 2016; Ruan et al., 2019).
Physical interactions between BADC3 and these three BCCP1 domains were investigated by coexpressing His-tagged BCCP1-derivatives that lacked one or two of these domains and evaluating if the coexpressed BADC3 protein could be copurified. Figure 1B shows the BCCP1 derivatives that lack the N-terminal domain, or those that lack the N-terminal and middle domains, cannot associate with and copurify BADC3. However, the BCCP1 derivatives that lack the C-terminal domain or lack the C-terminal and middle domains can successfully associate with and copurify BADC3. These data therefore establish BADC3 physically interacts with the N-terminal domain of the BCCP1.
Interactions between CT-α and CT-β Subunits Require a CT-β Subunit Mutation and Are Independent of BADC3
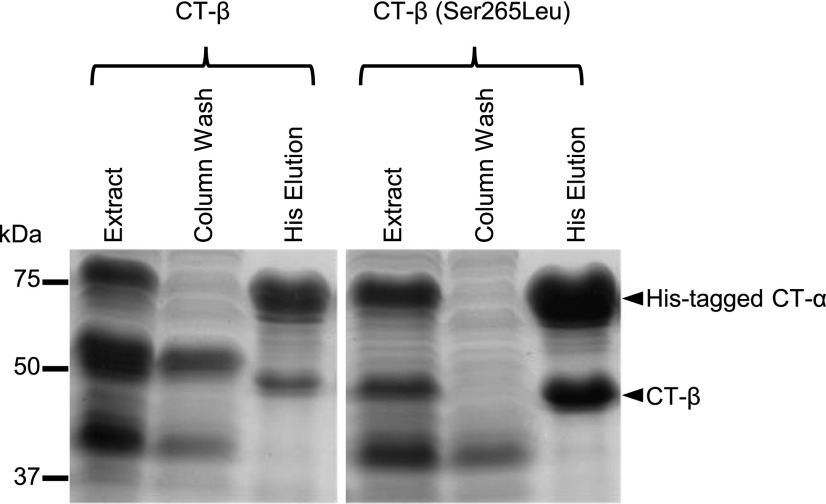
Having established the role of BADC3 in facilitating the formation of the BCCP1–BADC3–BC subcomplex, we explored whether this facilitates interactions between CT-α (AT2G38040) and/or CT-β (ATCG00500) subunits. The constitution of a CT–αβ subcomplex was highly favored when the CT-β subunit was mutated to replace residue Ser-265 with Leu. Thus, when the His-tagged CT-α subunit was coexpressed with CT-β(Ser-265Leu), the former was successful in copurifying the CT–αβ subcomplex (Fig. 2), and this occurred even in the absence of BADC3. Prior studies have identified the importance of altering the Ser-265 residue for htACCase catalytic function and that this modification occurs via a posttranscriptional RNA-editing mechanism (Sasaki et al., 2001; Robbins et al., 2009; Yu et al., 2009). The recovered CT–αβ subcomplex appears to have an equimolar ratio of the two subunits with a molecular mass of ∼1,040 ± 120 kD, suggesting this is an α8/β8 heteromeric complex.
Figure 2.
The Ser-265Leu mutation in CT-β facilitates the formation of the CT–αβ subcomplex. SDS-PAGE analysis of extracts from E. coli strains coexpressing His-tagged CT-α with either wild-type CT-β or the CT-β(Ser-265Leu) mutant. The extracts were passed through a Ni-NTA affinity chromatography column, and the non-bound proteins were collected in the “Column Wash” fraction, while proteins that bound to the column were eluted with a His-containing buffer. Proteins were visualized by staining gels with Coomassie Brilliant Blue. The figure is a representative result of triplicate experiments that were sequentially conducted.
In Vitro Reconstitution of a Catalytically Competent htACCase
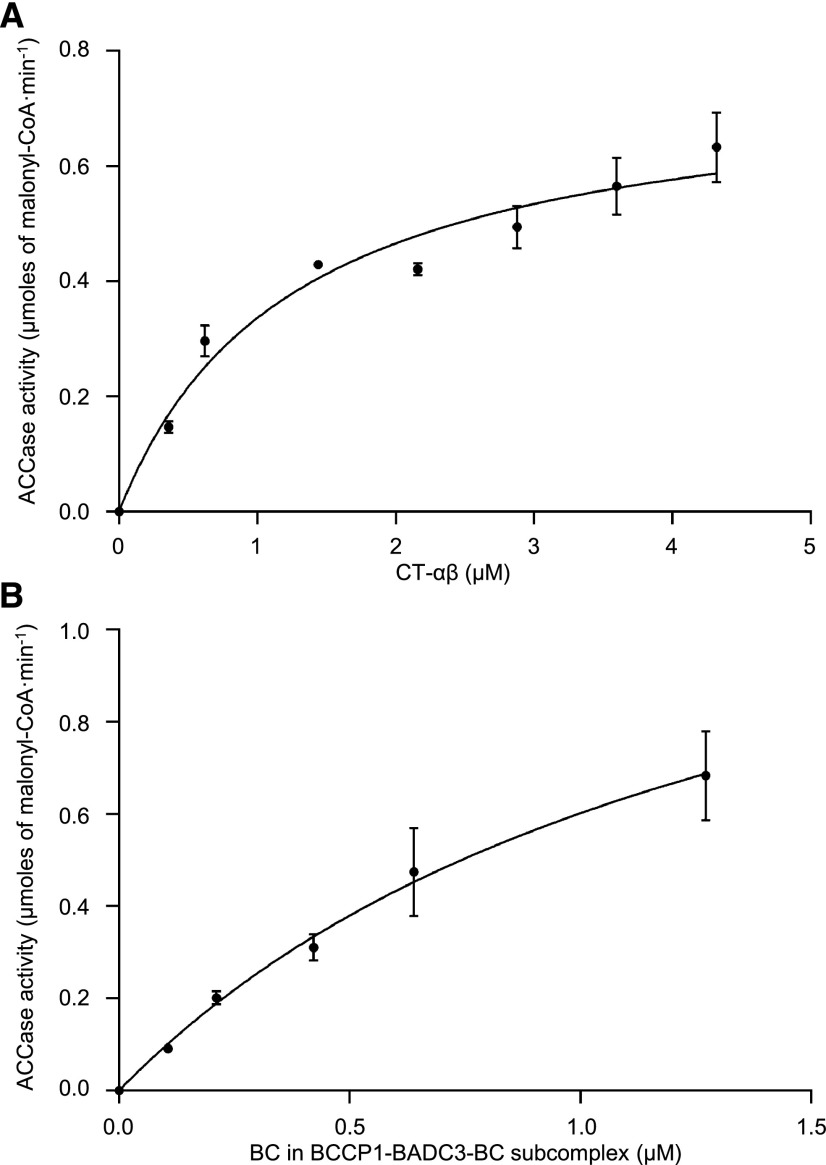
We explored the interrelationship between the CT–αβ subcomplex and the BCCP1–BADC3–BC subcomplex and their combined competence to catalyze the acetyl-CoA carboxylation reaction. Specifically, a constant concentration of either the purified BCCP1–BADC3–BC subcomplex was titrated in vitro with an increasing amount of the purified CT-αβ subcomplex (Fig. 3A), or vice versa (Fig. 3B), and the resulting mixtures were assayed for the ability to catalyze the formation of malonyl-CoA from acetyl-CoA. In both titration experiments, the reaction rate of acetyl-CoA carboxylation increased with increasing concentration of the titrating subcomplex. More specifically, maximal activity appeared to be reached when the CT–αβ subcomplex was titrated to be more than 2-fold higher molar excess than the BC subunit contained in the BCCP1–BADC3–BC subcomplex (Fig. 3A).
Figure 3.
Interdependence between BCCP1–BADC3–BC and CT–αβ subcomplexes to support the carboxylation of acetyl-CoA. A, Increasing concentration of purified CT–αβ subcomplex was titrated against a constant concentration of BC (0.62 μm) in the purified BCCP1–BADC3–BC subcomplex. B, Increasing concentration of purified BCCP1–BADC3–BC subcomplex was titrated against a constant concentration of purified CT–αβ subcomplex (2.18 μm). htACCase activity was measured as the rate of malonyl-CoA appearance using the MCR-coupled assay. Each data point represents the mean ± se (n = 3), and the experiment was duplicated with analogous results.
Role of BADC Isoforms in Catalytic Competence of htACCase
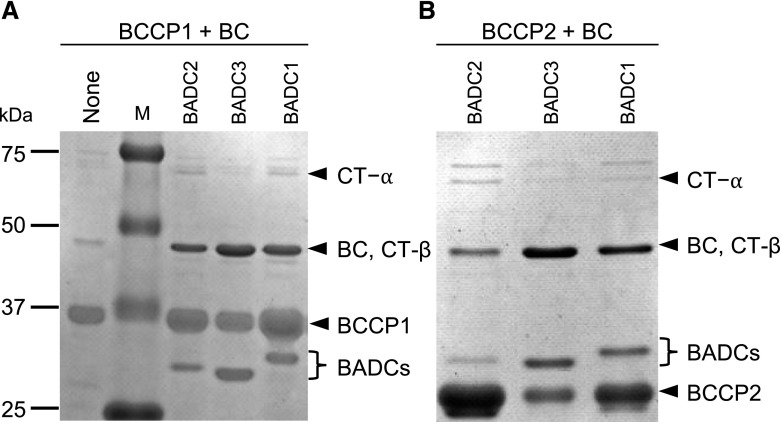
We further explored whether all three BADC isoforms could facilitate this physical interaction among the BC and BCCP1 or BCCP2 subunits and the relative catalytic capabilities of the resulting reconstituted enzymes. In these experiments, His-tagged BCCP1 or His-tagged BCCP2 were coexpressed and copurified with the other three catalytic subunits (i.e. BC, CT-α, and CT-β) in the presence or absence of each of the three BADC isoforms. Purification via the His-tag indicated any one of the three BADC isoforms facilitated the assembly of complexes that contain BCCP and BC (Fig. 4), but not the CT-α and CT-β subunits.
Figure 4.
BADC-facilitated assembly of htACCase subcomplexes. SDS-PAGE analysis and Coomassie Brilliant Blue staining of proteins copurified with His-tagged BCCP1 (A) or His-tagged BCCP2 (B) by Ni-NTA affinity chromatography from E. coli protein extracts coexpressing BC, CT-α, and CT-β in the absence or presence of each BADC isoform. The identity of each htACCase subunit was determined by immunoblot analysis, including the protein band at ∼50 kD, which is the comigrating BC and CT-β subunits. The figure is a representative result of triplicate experiments that were sequentially conducted. M, molecular weight markers.
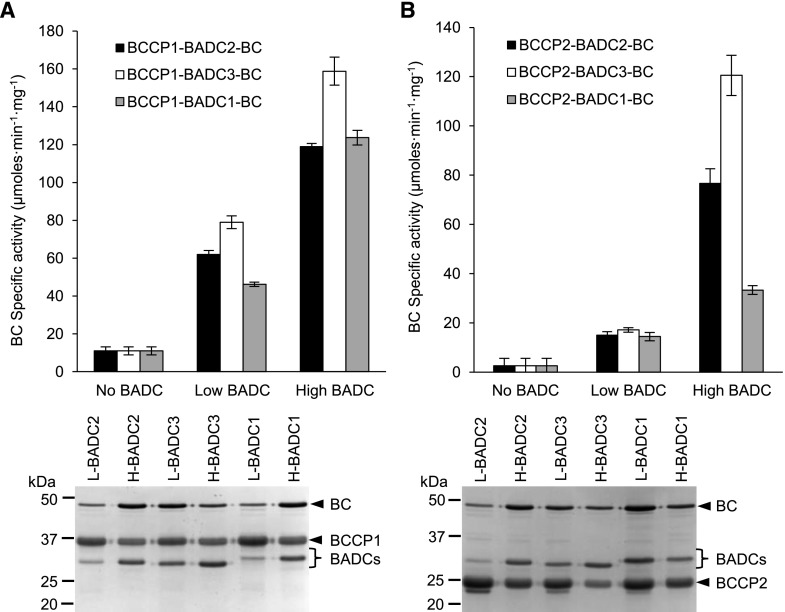
Subsequent experiments characterized the eight potential complexes that could be assembled between BCCP1 or BCCP2 with BC and in the presence or absence of either BADC2, BADC3, or BADC1. Using the described strategy, the affinity purified complexes were analyzed by SDS-PAGE and in parallel assayed for BC activity (i.e. the bicarbonate-dependent ATPase activity that is characteristic of the ACCase first half-reaction).
In the absence of any BADC isoforms, the molar ratios of BC:BCCP1 and BC:BCCP2 in the recovered complexes were 0.03:1 and 0.02:1, respectively, and these preparations supported very low BC catalytic activity (in the range of 2–14 µmoles·min−1·mg−1 of BC; Fig. 5). However, when the complexes were reconstituted in the presence of any individual BADC isoform, the recovery of BC in the complex increased so that the BC:BCCP ratios ranged between 0.1:1 and 0.4:1, and this was accompanied by a 4- to 7-fold increase in BC specific activity catalyzed by the recovered preparations.
Figure 5.
BADC-facilitated assembly of BCCP–BADC–BC subcomplexes are activated in the ability to catalyze the bicarbonate-dependent hydrolysis of ATP in the first half-reaction of htACCase. Heterologous coexpression and copurification of either low- or high-level expression of individual BADCs with BC and BCCP subunits. Subcomplexes copurified by Ni-NTA affinity chromatography from E. coli protein extracts expressing His-tagged BCCP1 (A) or His-tagged BCCP2 (B) coexpressed with BC in the absence or presence of each BADC isoform expressed from a low-copy or high-copy number expression plasmid. Replicate assays formed at five different concentrations of BC protein, in the range of 0.5–10 µg per assay, determined the rate of bicarbonate-dependent appearance of ADP via the PK/LDH coupling reactions. L, proteins expressed from low copy number vectors; H, proteins expressed from high copy number expression vectors. Each data-point represents the mean ± se (n = 3), and the experiments were duplicated with analogous results.
Additionally, by increasing the expression of BADCs in the coexpressing strains, the recovery of BC in the BCCP–BADC–BC subcomplex was further increased (Fig. 5). In these latter experiments the expression levels of BADCs were controlled by using either a low-copy vector (i.e. pACYCDuet-1, which is maintained at 10–12 copies per cell) or high-copy vector (i.e. pETDuet-1, which is maintained at ∼40 copies per cell; Novy and Morris, 2001; Sektas and Szybalski, 2002; Held et al., 2003). The expression of BADCs from the high-copy vector resulted in a 2- to 5-fold increase in the expression of the three BADC isoforms as compared to the expression of these proteins from the low-copy expression vector (Fig. 5). In the presence of larger quantities of expressed BADCs, the recovery of BC that is associated with BCCP and BADC was 2- to 6-fold higher, hence the molar ratio of BC:BCCP in the recovered complexes ranged between 0.2:1 and 0.8:1 (Table 1).
Table 1. Molar ratio of subunits in recovered BCCP–BADC–BC subcomplexes.
Molar ratios were estimated using the tool Image Lab (Bio-Rad) in reference to the amount of either BCCP1 or BCCP2 via SDS-PAGE gels loaded with copurified proteins and stained with Coomassie Brilliant Blue. Data are the average ± se (n = 3).
| Subcomplex | Low-Level BADC Expressiona | High-Level BADC Expressionb | ||||
|---|---|---|---|---|---|---|
| BCCP | BADCs | BC | BCCP | BADCs | BC | |
| BCCP1–BC | 1.00 ± 0.03 | — | 0.030 ± 0.001 | 1.00 ± 0.02 | — | 0.030 ± 0.001 |
| BCCP1–BADC2–BC | 1.00 ± 0.05 | 0.16 ± 0.01 | 0.11 ± 0.01 | 1.00 ± 0.03 | 0.94 ± 0.03 | 0.81 ± 0.02 |
| BCCP1–BADC3–BC | 1.00 ± 0.07 | 0.46 ± 0.03 | 0.38 ± 0.03 | 1.00 ± 0.04 | 1.28 ± 0.05 | 0.41 ± 0.02 |
| BCCP1–BADC1–BC | 1.00 ± 0.08 | 0.11 ± 0.01 | 0.100 ± 0.001 | 1.00 ± 0.03 | 0.64 ± 0.02 | 0.64 ± 0.02 |
| BCCP2–BC | 1.00 ± 0.05 | — | 0.020 ± 0.001 | 1.00 ± 0.05 | — | 0.020 ± 0.001 |
| BCCP2–BADC2–BC | 1.00 ± 0.06 | 0.14 ± 0.01 | 0.14 ± 0.01 | 1.00 ± 0.07 | 0.34 ± 0.02 | 0.34 ± 0.02 |
| BCCP2–BADC3–BC | 1.00 ± 0.08 | 0.14 ± 0.01 | 0.19 ± 0.02 | 1.00 ± 0.01 | 1.00 ± 0.01 | 0.500 ± 0.003 |
| BCCP2–BADC1–BC | 1.0 ± 0.2 | 0.30 ± 0.06 | 0.25 ± 0.05 | 1.00 ± 0.03 | 0.22 ± 0.01 | 0.22 ± 0.01 |
Low-level expression of BADC was from the low-copy number expression vector, pACYCDuet-1.
High-level expression of BADC was from the high-copy number expression vector, pETDuet-1.
Concomitant with the enhanced association of BC in the BCCP–BADC–BC complexes, the presence of any of the three BADC isoforms substantially enhanced the ability of the resulting complex to catalyze the BC-catalyzed reaction (Fig. 5). The stimulation of BC-specific activity varies depending upon the combination of BCCP and BADC isozymes that constitute the BCCP–BADC–BC complexes. The most active complexes are those that are assembled with BADC3, irrespective of whether BCCP1 or BCCP2 provides the biotin prosthetic group required in the BC-catalyzed reaction. The specific activity expressed by the BCCP1–BADC3–BC and BCCP2–BADC3–BC complexes were 16- and 40-fold greater than the complex that formed in the absence of any BADC.
The BCCP1-containing complex was more responsive to BADC-activation, enhancing BC activity by 4- to 7-fold with low-level expression of BADCs, and this activity was further enhanced by 2-fold with the higher level of BADC expression (Fig. 5A). In contrast, the activation of the BCCP2-containing complex by any of the three BADCs required the higher-expressing BADC system (Fig. 4B). Furthermore, the efficacy of this activation differed among the three BADC isoforms, with BADC3 being most effective and BADC1 being the least effective in this activation, and this is particularly apparent with the BCCP2-containing complex.
Enzymological Properties of Reconstituted htACCase
Measures of substrate specificity differences among the eight reconstituted htACCase isozymes was obtained by determining the kinetic Michaelis–Menten constants for bicarbonate, ATP, and acetyl-CoA with each of the isozymes (Supplemental Figs. S2–S4). The inclusion of any BADC isoform greatly increased catalytic activity, enabling more accurate determination of Km and Vmax values for these substrates. The inclusion of BADC2 or BADC3 was most efficient in increasing this catalytic activity, while the inclusion of BADC1 was less impactful in supporting catalysis. In contrast, in the absence of any BADCs, the isolated BCCP1–BC and BCCP2–BC subcomplexes supported very low catalytic activity, and therefore the Michaelis–Menten constants determined with these subcomplexes had larger errors (Table 2).
Table 2. Michaelis–Menten kinetic constants for ATP and potassium bicarbonate.
Because of substandard enzymatic activity in the absence of BADC, enzyme activity of htACCase of non-BADC containing subcomplexes was not determinable in some cases. Rate of formation of malonyl-CoA by htACCase was measured using the MCR-coupled assay. Data are the average ± se (n = 3). ND, not determined.
| Subcomplex | ATP kinetic constants | KHCO3 kinetic constants | ||||
|---|---|---|---|---|---|---|
| Vmax (μmoles·min−1·mg−1) | Km (mm) | Vmax/Km | Vmax (μmoles·min−1·mg−1) | Km (mm) | Vmax/Km | |
| BCCP1–BC | 1.6 ± 0.6 | 0.6 ± 0.5 | 2.8 ± 2.7 | 0.30 ± 0.09 | 0.4 ± 0.9 | 0.70 ± 1.22 |
| BCCP1–BADC2–BC | 27.9 ± 1.5 | 0.4 ± 0.1 | 70.0 ± 11.6 | 25.4 ± 1.3 | 4.6 ± 0.5 | 5.5 ± 0.7 |
| BCCP1–BADC3–BC | 27.3 ± 2.0 | 0.4 ± 0.1 | 69.5 ± 15.4 | 18.5 ± 0.9 | 3.7 ± 0.5 | 5.0 ± 0.7 |
| BCCP1–BADC1–BC | 2.3 ± 1.9 | 1.5 ± 2.3 | 1.5 ± 2.7 | 1.7 ± 0.3 | 1.5 ± 0.7 | 1.2 ± 0.5 |
| BCCP2–BC | 0.7 ± 0.1 | 0.6 ± 0.1 | 1.2 ± 0.3 | ND | ND | ND |
| BCCP2–BADC2–BC | 26.4 ± 1.1 | 0.30 ± 0.04 | 94.4 ± 12.8 | 31.7 ± 2.6 | 10.7 ± 1.7 | 3.0 ± 0.5 |
| BCCP2–BADC3–BC | 11.4 ± 0.9 | 0.4 ± 0.1 | 28.8 ± 6.6 | 9.6 ± 0.9 | 4.3 ± 1.1 | 2.3 ± 0.6 |
| BCCP2–BADC1–BC | 4.3 ± 0.5 | 0.3 ± 0.3 | 13.5 ± 13.6 | 4.3 ± 2.3 | 5.4 ± 5.9 | 0.8 ± 1.0 |
The Michaelis–Menten constants for bicarbonate and ATP were determined using the malonyl-CoA reductase (MCR)-coupled assay. These analyses were individually conducted with different combinations of either BCCP1 or BCCP2, in association with each of the three BADC isoforms. In all combinations evaluated, the BCCP1-containing subcomplex showed ∼2-fold higher Vmax/Km(bicarbonate) values than the BCCP2-containing subcomplexes (Table 2). In every case this activation was primarily due to an average 15-fold increase in Vmax values as compared to the BADC-free subcomplexes. In addition, the Km(bicarbonate) values increased 4–10 times in the presence of the BADCs, possibly indicating the affinity for this substrate may decrease in the presence of the noncatalytic subunits (Table 2).
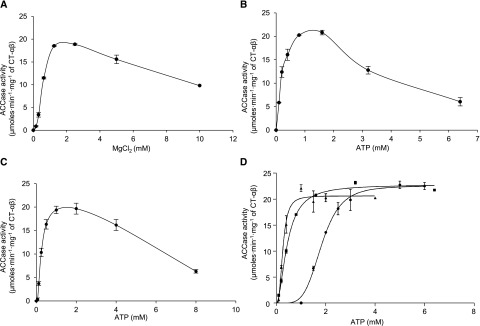
As with many ATP-dependent enzymes, including ACCase, there is an interdependence between ATP and Mg2+ concentrations in the reaction catalyzed by the htACCase (Sauer and Heise, 1983, 1984; Nikolau and Hawke, 1984; Harwood, 1988; Sasaki et al., 1997). These experiments were conducted with reconstituted mixtures of the two htACCase subcomplexes, the BCCP1–BADC3–BC subcomplex and the CT–αβ subcomplex at the optimal ratio as determined in Figure 3. Three separate sets of assays were used to explore these interrelationships.
In the first set of assays, the concentration of ATP was kept constant at 1 mm, and a range of increasing MgCl2 concentrations was used in the assays (Fig. 6A). In response to increasing MgCl2 concentration, ACCase activity increased sigmoidally, peaking at 1.25 mm of MgCl2, and activity was inhibited at concentrations >2.5 mm MgCl2. In the second set of assays, the concentration of MgCl2 was kept constant at 1.25 mm and ATP concentration was increased. In these titration assays, ACCase activity increased hyperbolically and peaked at 0.8 mm of ATP with higher concentrations of ATP leading to inhibition (Fig. 6B). In combination, these two sets of assays establish optimal ACCase activity occurs when the MgCl2:ATP ratio is between 1.2 and 1.5, and an excess of either MgCl2 or ATP is inhibitory to catalysis.
Figure 6.
The interdependence of htACCase catalytic activity with ATP and MgCl2. A, The effect of increasing MgCl2 concentration in the presence of 1 mm ATP. B, The effect of increasing ATP concentration in the presence of 1.25 mm of MgCl2. C, The effect of increasing ATP and MgCl2 concentrations while maintaining a constant ratio of [ATP]:[MgCl2] = 1:2.5. D, The effect of increasing ATP and MgCl2 concentrations while maintaining the following relationships: [MgCl2] = [ATP] − 1 mm (●); [MgCl2] = [ATP] (▪), and [MgCl2] = [ATP] + 2.5 mm (▲). htACCase activity was measured as the rate of malonyl-CoA appearance using the MCR-coupled assay. Each data point represents the mean ± se (n = 3), and the experiment was duplicated with analogous results.
In the third series of assays, ACCase activity was determined with increasing ATP concentration while the molar ratio MgCl2:ATP was kept at 2.5. In these assays maximal activity occurred at 0.5 mm ATP, and concentrations higher than 4 mm were inhibitory (Fig. 6C). In a final set of assays, ACCase activity was determined in the presence of increasing ATP concentration but in the presence of MgCl2 concentrations that were either 1 mm less than that of ATP, equal to ATP, or 2.5 mm greater than ATP (London and Steck, 1969; Storer and Cornish-Bowden, 1976; Nikolau and Hawke, 1984). The activity response curve in the first case was sigmoidal and became increasingly hyperbolic as the concentration of MgCl2 increased relative to ATP. This characteristic was quantified by the calculation of the Khalf for ATP, which decreased from 1.8 to 0.26 mm, and the Hill coefficient, which decreased from >4 to ∼2 (Fig. 6D; Table 3).
Table 3. Hill analysis of the cooperativity between ATP and MgCl2 in the carboxylation of acetyl-CoA catalyzed by htACCase.
ACCase activity was measured as the rate of malonyl-CoA appearance using the MCR-coupled assay. Data are the average ± se (n = 3).
| Terms | [MgCl2] = [ATP] | [MgCl2] = [ATP] − 1.0 mm | [MgCl2] = [ATP] + 2.5 mm |
|---|---|---|---|
| Vmax | 22.8 ± 0.3 | 22.5 ± 0.8 | 20.6 ± 0.5 |
| n | 1.9 ± 0.1 | 4.7 ± 0.7 | 2.8 ± 0.4 |
| Khalf | 0.45 ± 0.01 | 1.80 ± 0.07 | 0.26 ± 0.01 |
Collectively, these assays indicated the Mg∙ATP complex is the true substrate of the htACCase enzyme, and that excess free Mg2+ or ATP inhibited catalytic activity (Fig. 6C). Ultimately, the Mg∙ATP complex also became inhibitory (above 4 mm), but that occurred at concentrations well above the inhibitory effects of free Mg2+ or ATP. In addition, the changes in the value of the Hill coefficient in response to the altered ATP/MgCl2 levels is indicative of an allosteric cooperativity between ATP and Mg2+ in affecting the activity of the htACCase.
Table 2 presents the values of Vmax, Km(ATP), and Vmax/Km(ATP) for the eight htACCase isoforms reconstituted with either BCCP1 or BCCP2, individually integrating one of the three BADCs. The activation of htACCase measured as the Vmax/Km(ATP) values was maximal with BADC2 and BADC3, which is primarily due to a 20- to 30-fold increase in Vmax with little effect on Km(ATP). The ability of BADC1 to activate ACCase activity was lower, although its presence was still able to increase Vmax by 2- and 5-fold without affecting Km(ATP).
Finally, the Michaelis–Menten constants for acetyl-CoA were determined using the most active BADC-containing subcomplex, namely BCCP1–BADC3–BC. The Km for acetyl-CoA was 1.2 ± 0.1 mm with a Vmax of 39.8 ± 1.7 µmoles·min−1·mg−1 of CT-αβ subcomplex.
Physiological Functions of the BADCs
In the light of the in vitro biochemical characterization of the BADC proteins, which established the function of these proteins as facilitators of htACCase complex formation and thus as activators of catalytic activity, we examined the in planta physiological functions of these proteins by reverse genetic strategies. In these experiments, we characterized mutant plants that either cannot express one of the three BADC isoforms or double mutant plants that singularly express only one of the three BADC isoforms (Supplemental Fig. S5).
Mutant plants that lack any single BADC isoform grow indistinguishably from wild-type siblings. This conclusion is based on observations that compared such physiological traits as the germination rate, the physical appearance of the growing seedlings and mature plants, leaf and bolt growth rates, and seed size and color. In parallel, the biochemical consequence of these singular mutations on fatty acid accumulation is also minimal (Keereetaweep et al., 2018). We surmise therefore that the three BADC genes share considerable functional redundancy, and plants missing any single isoform grow normally, which implies the expression of the remaining two isoforms is sufficient for growth and viability.
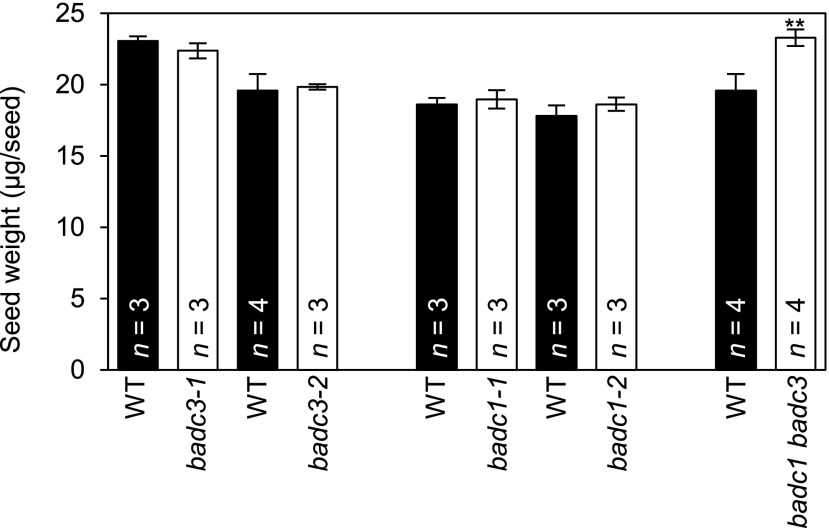
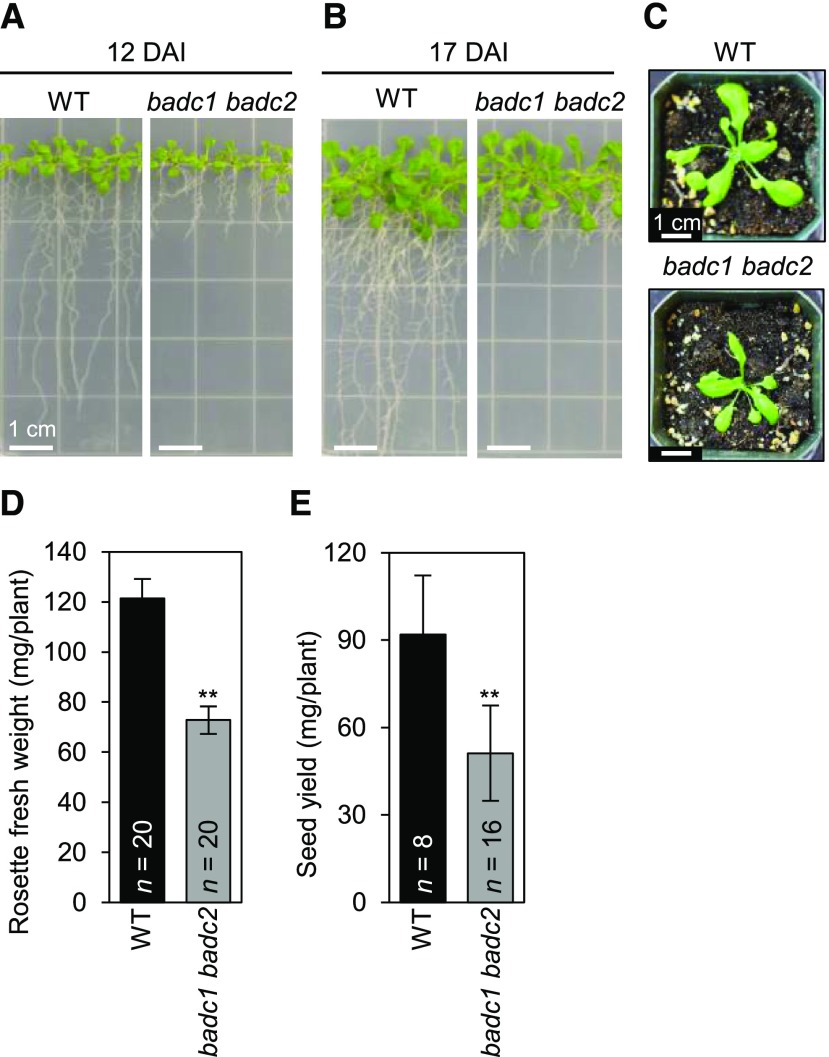
This functional redundancy was eliminated in the double mutant plants expressing only one of the three BADC isoforms. Three different phenotypes were presented by such double mutant plant lines: (1) those that express only BADC1 are not recoverable and are embryo-lethal (Table 4; Supplemental Fig. S6), (2) those that express only BADC2 are near normal, with the exception that these plants produce seeds that are ∼20% heavier than the wild type (Fig. 7; Supplemental Data S1), and (3) those that express only BADC3 manifest a complex reduced growth phenotype (Fig. 8; Supplmental Data S2 and S3) these latter multiple effects may be a secondary consequence of reduced root growth, which was the first detectable phenotypic effect that developed in plants expressing only BADC3. Indeed, such complex phenotypes occur in a number of other fatty acid metabolism mutants that primarily first affect root growth (Shelley et al., 2013; Ding et al., 2015).
Table 4. Segregation analysis on badc mutant alleles.
| Allele | Number of Heterozygous Parent Plants Tested | Genotype of Progeny | Number of Progeny in Each Category |  |
P Value |
|---|---|---|---|---|---|
| badc2-1 | 1 | badc2-1/badc2-1 | 11 | 1.00 | 0.61 |
| badc2-1/BADC2 | 22 | ||||
| BADC2/BADC2 | 15 | ||||
| badc2-2 | 3 | badc2-2/badc2-2 | 16 | 1.10 | 0.58 |
| badc2-2/BADC2 | 34 | ||||
| BADC2/BADC2 | 12 | ||||
| badc3-1 | 2 | badc3-1/badc3-1 | 28 | 3.66 | 0.16 |
| badc3-1/BADC3 | 36 | ||||
| BADC3/BADC3 | 18 | ||||
| badc3-2 | 2 | badc3-2/badc3-2 | 22 | 4.75 | 0.09 |
| badc3-2/BADC3 | 39 | ||||
| BADC3/BADC3 | 10 | ||||
| badc1-1 | 2 | badc1-1/badc1-1 | 24 | 0.40 | 0.82 |
| badc1-1/BADC1 | 41 | ||||
| BADC1/BADC1 | 21 | ||||
| badc1-2 | 2 | badc1-2/badc1-2 | 11 | 3.12 | 0.21 |
| badc1-2/BADC1 | 35 | ||||
| BADC1/BADC1 | 21 | ||||
| badc2-1/BADC2 badc3-1/badc3-1 | 3 | badc2-1/badc2-1 badc3-1/badc3-1 | 0 | — | — |
| badc2-1/BADC2 badc3-1/badc3-1 | 200 | 178.38 | <0.01 | ||
| BADC2/BADC2 badc3-1/badc3-1 | 185 | — | — |
Figure 7.
Effect of badc mutations on seed biomass. The weight of individual seeds was calculated by weighing aliquots of a known number of seeds (∼100 seeds per aliquot). The number of plants from which the seeds were collected (n) is indicated, and each plant was tested three times. The data represents the mean, and error bars indicate the mean ± se. Student’s t test was used to determine the significance in the difference between the wild type (WT) and each indicated mutant (**P < 0.01).
Figure 8.
Phenotypes of the badc1 badc2 double mutant. A, Seedlings at 12 d after imbibition (DAI). B, Seedlings at 17 DAI. C, Rosettes at 28 DAI. D, Rosette fresh weight at 28 DAI. E, Seed biomass yield. In (D) and (E), the data represent the mean for the indicated number of replicates (n), and error bars indicate the mean ± se. Student’s t test was used to determine the significance in the difference between the wild type (WT) and each indicated mutant (**P < 0.01).
Collectively these characterizations led to the conclusion that BADC2 and BADC3 share the highest functional redundancy, but that redundancy is not symmetrical, with BADC2 being able to replace almost all BADC3 functions, but not vice versa. BADC1 however, appears to have a distinct function, or is not sufficient to replace BADC2 or BADC3, and thus plants only expressing BADC1 were not viable through seed development.
The segregation of this defective seed development phenotype expressed by the badc2 badc3 double mutant could be due to the inability to produce viable gametes and/or the inability to develop viable seeds after fertilization. The former would result in ovules that are not fertilized, and the latter would result in aborted seeds. Distinguishing between these possibilities was tested by reciprocal genetic crosses using pollen from either a wild-type plant or one with the genotype badc2-1/BADC2 badc3-1/badc3-1 (Table 5). The wild-type plants generate haploid pollen gametes that all carry wild-type BADC2 and BADC3 alleles, but the latter plants generate haploid pollen gametes that carry either BADC2 or badc2-1 alleles in coupling with the mutant badc3-1 allele. In both crosses, normal Mendelian segregation predicts the progeny should be equally divided into two genotypic categories: (1) badc2-1/BADC2 badc3-1/BADC2 and (2) BADC2/BADC2 badc3-1/BADC3. As expected, individual F2 progeny from each of these reciprocal crosses were heterozygous at the BADC3 locus. Furthermore, 50% of the progeny were heterozygous at the BADC2 locus when pollen was obtained from wild-type plants. However, this genotype was recovered at only half of the expected rate when pollen was obtained from badc2-1/BADC2 badc3-1/badc3-1 plants (Table 5). Therefore, these data establish the transmission of male gametes is reduced by 50% when only the BADC1 isoform is expressed.
Table 5. Transmission of badc2-1 badc3-1 gametes.
| Cross | Total Number of Progeny Tested (from n Siliques) | Number of Progeny in Each Genotype Category |  |
P Value | ||
|---|---|---|---|---|---|---|
| badc2-1/BADC2 badc3-1/BADC3 | BADC2/BADC2 badc3-1/BADC3 | |||||
| badc2-1/BADC2 badc3-1/badc3-1♀ | BADC2/BADC2 BADC3/BADC3♂ | 160 (n = 9) | 78 | 82 | 0.1 | 0.8 |
| BADC2/BADC2 BADC3/BADC3♀ | badc2-1/BADC2 badc3-1/badc3-1♂ | 232 (n = 10) | 81 | 151 | 21.1 | <0.01 |
Collectively therefore, these genetic studies establish plants singularly expressing only BADC1 cannot support seed development and can only partially support male gamete transmission. In contrast, plants singularly expressing only BADC2 or BADC3 were viable throughout the plant’s life cycle, but the latter plants expressing only BADC3 displayed discrete morphological growth phenotypes, particularly associated with root development.
Biochemical Characterization of badc Mutants
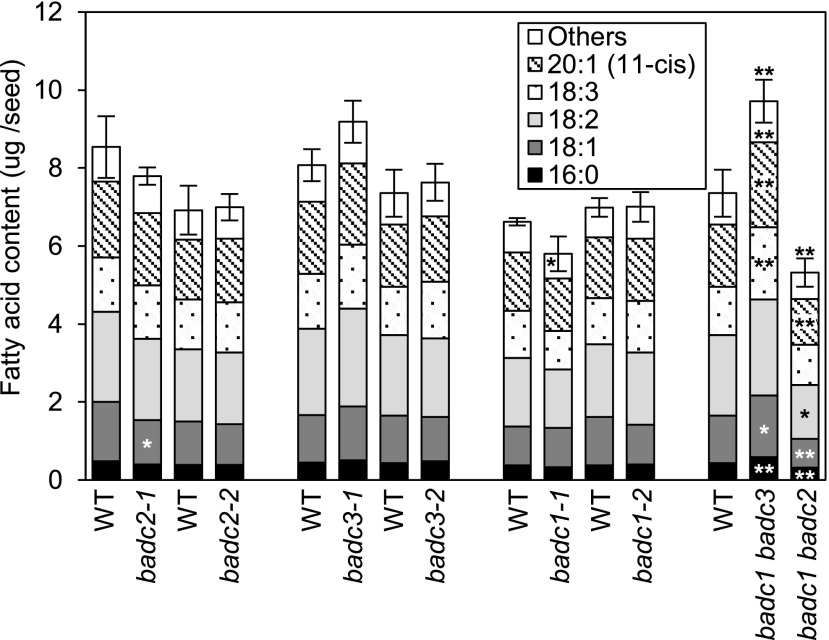
Because the htACCase enzyme is crucial to de novo fatty acid biosynthesis, we evaluated the biochemical phenotype of the badc mutants by assaying mature seeds for fatty acid composition and content. In contrast to the single badc mutants that were viable and did not show any detectable changes in fatty acid profiles, the viable double mutant strains displayed clear changes in fatty acid titers. In the badc1 badc2 double mutant (which showed a root growth phenotype), the fatty acid content per seed weight was reduced by ∼25% from the wild type (Fig. 9; Supplemental Data S4), but as noted earlier, this effect maybe a secondary consequence of the reduced root growth (Shelley et al., 2013; Ding et al., 2015; Liu et al., 2019). The effect of the other viable double mutant (i.e. badc1 badc3) was the opposite, with a 30% increase in total fatty acid content per seed (Fig. 9). In both mutants, the seed fatty acid composition profiles were minimally different from the wild-type.
Figure 9.
Effect of single and double badc mutants on fatty acid accumulation in mature seeds. Fatty acids were extracted from samples of 100 seeds. Each value is the mean of three to six biological replicates, and error bars indicate the mean ± se of the total fatty acid content. The sum of the minor fatty acid components (18:0, 20:0, 20:1[13-cis], 20:2, and 22:1) is indicated as “Others.” Student’s t test was used to determine the significance in the difference between the wild type (WT) and each indicated mutant (*P < 0.05; **P < 0.01). Asterisks labeled within the data bars represent the comparisons for individual fatty acid components, and asterisks labeled on top of the data bars represent the comparison for total fatty acid content.
We also determined the fatty acid profiles and titers of the organs that were phenotypically affected by the badc1 badc2 double mutants (i.e. roots and rosette leaves). Even though the rosette leaves of this double mutant were smaller than the wild-type leaves (Fig. 8, C and D), the fatty acid profile and titer was unaffected (Supplemental Fig. S7A; Supplemental Data S5 and S6). Similarly, there was no detectable alteration in the faty acid profile and fatty acid titer of the roots of badc1 badc2 double mutants (Supplemental Fig. S7B). Therefore, because these organs were smaller in the double mutant lines, one can reason that biomass productivity by these plants may be limited by the availability of fatty acids to support membrane lipid deposition, and thus maintaining a constant fatty acid titer per unit biomass. A similar conclusion was deduced by the analysis of mutant plants that showed reduced expression of BCCP1 and BCCP2 (Li et al., 2011a).
Spatial and Temporal Expression and Expression Crosstalk among BADC and htACCase Genes
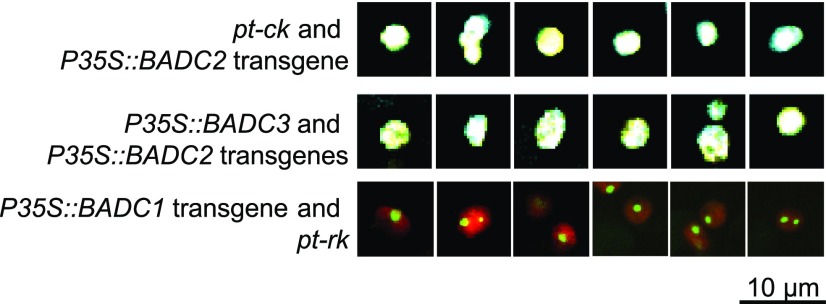
As components of the htACCase enzyme, the BADC proteins are targeted to plastids. This is computationally predicted by ChloroP and has been experimentally validated by subcellular fractionation studies (Feria Bourrellier et al., 2010; Olinares et al., 2010). Moreover, these fractionation studies have confirmed BADCs are in a complex that also contains the catalytic htACCase subunits and the PII protein (Feria Bourrellier et al., 2010; Olinares et al., 2010). We expanded on these findings by transgenically expressing fluorescently tagged BADCs. These experiments revealed the individual BADC isozymes segregate to distinct locations within plastids. Specifically, as shown in Figure 10 and Supplemental Figure S8, BADC2 and BADC3 occupied the same volume within the imaged plastids, which was also visualized by the plastid-marker carried by pt-ck (cyan fluorescent protein [CFP]; Nelson et al., 2007), and all three proteins appeared to fill the entire organelle.
Figure 10.
Subcellular localization of BADCs. Merged confocal micrographs of mesophyll cells of rosette leaves from transgenic plants carrying the indicated transgenes. The notations pt-ck or pt-rk are vectors that carry plastid markers fused with CFP or mCherry.
In contrast, BADC1 shows a very distinct pattern of distribution within the plastids, being focused in discrete structures within the organelles that were ∼1 µm in diameter, and these occurred either singly or sometimes in pairs in each plastid. This distinct pattern of localization mirrors the distribution of the PII protein in plastids, which is also associated with the megadalton htACCase complex (Baud et al., 2010). Similar discrete subplastidic localization of proteins has previously been reported for other enzymes associated with lipid metabolism, including enzymes involved in fatty acid biosynthesis (e.g. 3-ketoacyl-ACP reductase; Mueller et al., 2014), and the metabolism of fatty acids that generate oxylipids (e.g. two allene oxide synthase isozymes, the lipoxygenase H3 isozyme, and hydroperoxide lyase; Farmaki et al., 2007).
At the level of individual organs, Figure 11 shows data addressing the question of whether changes in the expression of individual BADCs and htACCase genes are altered in response to the missing BADC proteins in each of the mutants that were characterized. These experiments evaluated mRNA expression patterns by reverse transcriptase-quatitative PCR (RT-qPCR) with isolated RNA templates and expression of individual subunit-proteins by immunological immunoblot analysis of megadalton htACCase-containing complexes (Olinares et al., 2010) isolated in parallel from the same tissue samples.
Figure 11.
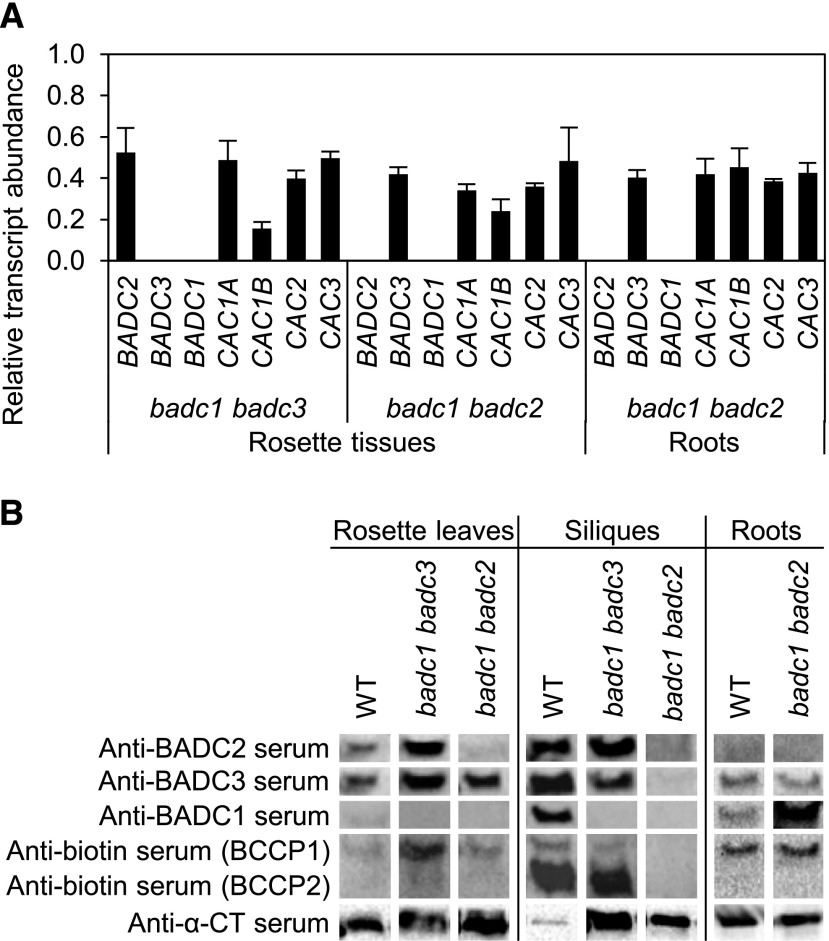
Effect of badc double mutants on the expression of BADCs and htACCase subunit genes. A, Relative mRNA levels were determined by RT-qPCR. RNA templates for these assays were isolated from 30 d-after-imbibition (DAI) rosettes and 15 DAI roots. Each value is the mean of three biological replicates and represents the relative level of the indicated mRNA in the double mutant in relation to the wild-type (WT) tissues. Error bars indicate the mean ± se. B, Accumulation of BADC proteins and htACCase subunits. The megadalton fraction was isolated from rosettes 30 DAI, siliques 9 d after flowering, and roots 15 DAI from wild type, badc1 badc2 double mutant, and badc1 badc3 double mutant plants. The megadalton fractions were subjected to SDS-PAGE immunoblot analysis. Each membrane was sequentially probed with each of the indicated antisera after the prior visualization reagents were removed; antibiotin serum was used as the terminal visualization reagent. The figure is a representative result of triplicate experiments that were sequentially conducted.
Consistent with the known physiology of seed fatty acid accumulation patterns in wild-type plants, the highest expression of the htACCase catalytic subunits (Choi et al., 1995; Ke et al., 1997, 2000; Sun et al., 1997; Thelen et al., 2001) and of the noncatalytic BADC subunits occurred in developing seeds within siliques (Fig. 11B). In the two viable mutants (i.e. badc1 badc2 and badc1 badc3) expressing only a single BADC paralog, the mRNA abundance of the remaining paralog was reduced by ∼50% from the wild-type state in the organs whose growth was reduced by these mutations. These reductions were associated with similar reductions in the level of the mRNAs encoding the htACCase catalytic subunits (Fig. 11A), indicative of the crosstalk among these genes to coordinate expression.
The parallel immunoblot analysis of the megadalton htACCase-containing complex (Olinares et al., 2010) isolated from the same tissues indicated a similar crosstalk mechanism in the accumulation of the individual subunit proteins (Fig. 11B) but discoordinate from the changes observed at the level of mRNA accumulation. The interpretation of these immunoblot data are more complex because of the cross-reactivity among the three BADC antisera that we generated. However, by standardizing the reactivity of each antiserum with recombinantly expressed, pure individual BADC proteins, and evaluating the signal generated by each antiserum with megadalton htACCase preparations isolated from mutant plants only expressing a single BADC isoform, we were able to deduce the effect on the accumulation of each isoform.
First, the analyses with recombinantly expressed, purified individual BADC proteins established that anti-BADC1 serum was near monospecific in reacting with BADC1, whereas anti-BADC3 serum cross-reacted with both BADC2 and BADC3, and anti-BADC2 serum cross-reacted with all three BADC isoforms (Supplemental Fig. S9). Because of the near-monospecific reactivity of anti-BADC1 serum, the immunoblot analyses of the megadalton htACCase preparations with this antiserum were the simplest to interpret. Specifically, in wild-type plants, BADC1 was undetectable in rosette leaves, whereas it was highly expressed in developing siliques and occurred at intermediate levels in roots.
Because of the cross-reactivity that is displayed by the anti-BADC2 and anti-BADC3 sera, we compared the immunoblot signals obtained with the megadalton htACCase preparations isolated from the double mutant plants. Thus, the megadalton htACCase preparations isolated from the badc1 badc3 double mutant revealed the accumulation of only the BADC2 isoform. Hence, using either the anti-BADC2 or the anti-BADC3 sera indicated accumulation of BADC2 increased from wild-type levels in both rosette leaves and siliques of the badc1 badc3 double mutant (Fig. 11B). Similar deductions with megadalton htACCase preparations isolated from badc1 badc2 double mutants indicated the accumulation of the BADC3 isoform also increased in rosette leaves and roots of this mutant (Fig. 11B).
In parallel with these alterations in the expression of the BADC isoforms, the accumulation of the catalytic subunits of htACCase were also altered in these mutants, but these latter effects were tissue-specific and only affected a subset of the subunits evaluated. Specifically, in rosette leaves, BCCP1 accumulation was enhanced only when BADC2 was present, while in developing siliques BCCP1 and BCCP2 accumulation was nearly abolished when only BADC3 was present. The level of CT-α subunit increased in mutant rosette and silique tissues that individually expressed either BADC2 or BADC3 (Fig. 11B).
DISCUSSION
Biotin-containing enzymes, such as ACCase, have been considered exemplary of evolutionary-generated modular arrangement of catalytic subsites, which coordinate to facilitate the overall reaction of the enzyme (Toh et al., 1993; Jitrapakdee and Wallace, 2003; Lombard and Moreira, 2011). The structural organization of fatty acid synthases and polyketide synthases also support this supposition (Smith and Sherman, 2008; Chan and Vogel, 2010; Bukhari et al., 2014). Structural characterization of these enzymes indicate catalytic coordination is facilitated by the quaternary organization of the catalytic subsites, which have to be iteratively visited by intermediates of the overall reaction (Broussard et al., 2013; Tran et al., 2015; Wei and Tong, 2015; Hunkeler et al., 2016).
Central to this coordination among catalytic subsites of ACCase is the biotin prosthetic group, carried by the BCCP subunit or domain, which alternatively visits two catalytic subsites, BC and CT, to complete the formation of malonyl-CoA per visitation cycle. One can envision therefore, that the organization of catalytic subsites in near adjacent positions, as facilitated by domains on the single polypeptide of the hmACCase, enable more efficient overall catalysis than occurs with enzymes that maintain these subsites on separate and distinct proteins, as occurs with the htACCase.
The Role of BADC Paralogs in BC-BCCP Subcomplex Formation and Effect on ACCase Catalysis
Similar to the bacterial ACCase, typified by the well-characterized E. coli enzyme (Tong, 2013), the plant htACCase is composed of four catalytic subunits, three of which are nuclear-encoded (BC, BCCP, and CT-α subunits) and one encoded by the plastome (CT-β; Nikolau et al., 2003). Distinct from the bacterial htACCase, however, the plant enzyme also incorporates noncatalytic subunits, BADCs (Olinares et al., 2010). Prior characterizations that evaluated the effect of expressing BADCs in E. coli, or adding these proteins in vitro to plant extracts that contain the preformed htACCase enzyme complex have been interpreted to indicate BADCs are inhibitors of ACCase catalysis (Salie et al., 2016). However, our coexpression, copurification, and reconstitution experiments conducted with all plant-derived purified subunits establish BADCs facilitate the formation of a complex between BC and BCCP, and the resulting BCCP–BC–BADC subcomplexes show enhanced competence in catalyzing the first half-reaction of the htACCase, the bicarbonate-dependent hydrolysis of ATP.
The BADC-mediated protein-protein interactions that facilitate the formation of the BCCP–BADC–BC subcomplexes occur between the N-terminal and middle domains of the BCCP subunit and the BADC protein. This latter middle domain is rich in Pro and Ala residues, and structural studies of homologous BCCP proteins indicate this domain is unstructured in solution. These are features common to intrinsically disordered proteins, which act as flexible linkers, facilitating the stabilization of transient complexes (Uversky, 2016; Ruan et al., 2019). In contrast, the C-terminal domain of BCCP, which carries the biotin prosthetic group, takes a distinct stable globular fold whose structure can be determined by NMR and x-ray crystallographic methods (Athappilly and Hendrickson, 1995; Yao et al., 1997; Roberts et al., 1999; Reddy et al., 2000). Although the BADC-mediated interactions do not affect the assembly of a CT–αβ subcomplex, the formation of two subcomplexes appears to be a prerequisite to the ability of the holoenzyme to catalyze the overall acetyl-CoA carboxylation reaction. Rather, the formation of the CT–αβ subcomplex is facilitated by a Ser-265Leu mutation in the CT-β subunit, which has previously been identified to be an in planta generated posttranscriptional RNA-editing modification (Robbins et al., 2009). We thereby conclude in the absence of BADC, BCCPs are incapable of forming the subcomplex with BC, and the presence of a BADC isozyme is essential for the catalytic competence of the htACCase. Therefore, the data presented herein indicate the role of the BADCs is to enable the assembly of a catalytically competent complex between the BCCP and BC subunits. Hence BADCs are positive regulators of the htACCase, and they achieve this by facilitating the formation of a catalytically more efficient complex between the BCCP–BC–BADC and CT–αβ subcomplex.
It is interesting to note that phylogenetic analyses indicate BADC-type proteins do not occur in all organisms expressing a htACCase type enzyme (Salie et al., 2016); no BADC homologs are detectable in prokaryotic organisms, which also express a htACCase enzyme. One can speculate the evolutionary appearance of BADCs conferred a selective advantage facilitated by the assembly of a more active ACCase enzyme, which may be rationalized as conferring the evolutionary ability leading to oleaginous organisms that use a htACCase to commit carbon to fatty acid biosynthesis. For example, except for bacteria belonging to the actinomycetes group (e.g. Mycobacterium, Rhodococcus, Nocardia, and Streptomyces), most prokaryotes do not accumulate large quantities of fatty acid-based lipids (Wältermann et al., 2005). Rather the non-actinomycetes bacteria accumulate polyhydroxyalkanoates to serve as storage reservoirs for energy and carbon (Możejko-Ciesielska and Kiewisz, 2016), and polyhydroxyalkanoate biosynthesis does not utilize malonyl–CoA as an intermediate, and is thus independent of ACCase catalysis. However, in the actinomycetes group that utilizes fatty acid-based lipids to store carbon and energy, a different structural organization of the ACCase has evolved, which facilitates assembly and catalytic efficiency by fusing the BC, BCCP, and CT-α and CT-β subunits on to two polypeptide subunits (α and β, respectively), whose assembly into a supramolecular structure is facilitated by a noncatalytic, ε-subunit (Gago et al., 2011).
Catalytic Properties of BADC-Assembled htACCase
The catalytic capabilities of the BADC-containing, reconstituted htACCase enzyme were explored by comparing the rates of the first half-reaction (i.e. the BC-catalyzed ATP-hydrolysis) with the overall reaction of the holoenzyme complex (i.e. the rate of malonyl-CoA formation). These comparisons established the inclusion of BADC3 facilitates the formation of the BCCP1-BADC3–BC subcomplex, enhancing the BC-catalyzed ATP-hydrolysis reaction by 15-fold. Furthermore, mixing the BADC3-containing BCCP1–BADC3–BC subcomplex with the CT-αβ subcomplex further enhanced the reaction rate by more than 2-fold (from 160 ± 7 μmol·min−1·mg−1 of BC to 353 ± 37 μmol·min−1·mg−1 of BC) the rate of acetyl-CoA carboxylation, measured as the rate of malonyl-CoA formation. Thus, the presence of BADC appears to facilitate the assembly of the holoenzyme complex, avoiding the potential wasteful expenditure of energy (i.e. ATP hydrolysis) without the coupled formation of malonyl-CoA.
The reconstituted htACCase complex exhibits classical Michaelis–Menten kinetic behavior, and the BADC-mediated activation of the enzyme is primarily due to enhancing the Vmax of the enzyme and minimally affecting the Km for each substrate. Moreover, these characterizations establish the Mg × ATP complex is the true substrate of the enzyme and that free Mg2+ is an activator of htACCase activity. This latter enzymological behavior is consistent with this enzyme’s role in regulating fatty acid biosynthesis in chloroplasts and ensuring it is a light-mediated process (Sauer and Heise, 1983, 1984; Nikolau and Hawke, 1984; Harwood, 1988; Sasaki et al., 1997).
In Vivo Physiological Functions of BADCs
The ability to adjudicate physiological functions of genes based on the characterization of mutant phenotypes is confounded when individual components of a multisubunit complex are encoded by paralogous gene families; for example, three BADC and two BCCP isoforms constitute the plastid-localized htACCase. Dissecting this complexity is now enhanced with the integrated understanding for the first time: (1) the functional ability of each BADC paralog to positively support ACCase catalysis; (2) the spatial and temporal expression patterns of each paralog, particularly in tissues whose growth and development are affected by mutations; and (3) the stoichiometry of the subunits in the htACCase complex, and the potential in vivo instability of the complex.
Earlier studies dissected the role of the two BCCP paralogs and established that, whereas BCCP2 is highly expressed during seed development (Thelen et al., 2001), the BCCP1 isoform provides the essential functionality crucial for successfully completing seed development (Li et al., 2011a). Consistent with the expected high demand for malonyl-CoA generation to enable the accumulation of large quantities of fatty acids in seeds, in this study we demonstrate the expression of the three BADC paralogs parallel the expression of the htACCase catalytic subunits (with the exception of the plastome-encoded β–CT subunit; Ke et al., 1997, 2000), with maximal expression occurring in developing siliques and seeds. Additionally, several observations extracted from the combined dataset that compare expression of the BADC genes at the mRNA and protein levels indicate complex posttranscriptional and posttranslational controlling mechanisms that discoordinate the accumulation of the individual BADC subunits and the levels of each BADC mRNA. Moreover, such mechanisms have to be invoked to explain complex regulatory crosstalk among the three BADC paralogs and the genes encoding the catalytic subunits of the htACCase.
Interpreting the phenotypes of the badc mutants is confounded by the apparent genetic and biochemical redundancy in the BADC-encoding genes. For example, there is the possibility that in the single or double badc mutant lines the expression of the still functional BADC gene(s) may be enhanced to offset for the missing BADC function(s). In fact, our expression studies in the badc mutant lines reveal there is no such compensatory mechanism to replace the missing BADC functions in these mutants. Thus, we can interpret the effect of the mutation(s) as the removal of the expression of that specific gene function. However, the observations reported herein indicate failure to express any single BADC isoform results in the depression in the expression of the functional BADC gene(s) and the depression in the expression of the genes coding for the catalytic htACCase subunits. These observations are consistent with the role that BADCs have in assembling a functional htACCase megadalton complex. Namely, in the absence of a BADC, the ability to assemble the megadalton htACCase complex is reduced, leading to reduced accumulation of the catalytic subunits, possibly associated with turnover of the proteins not associated with the complex.
A second confounding issue is the possibility of generating many different isoforms of the htACCase, which contain diverse mixtures of catalytic (one of two BCCPs) and noncatalytic (one or more of three BADCs) subunits. Addressing this question requires prior knowledge of the subunit stoichiometry in the megadalton htACCase complex, which was obtained in our in vitro reconstitution titration experiments. These experiments established a 2-fold molar excess of the CT–αβ subcomplex over that of BC in the BCCP–BADC–BC subcomplex is adequate for optimal activity of the holoenzyme. We estimated therefore, that optimal activity occurs with each subunit being at a molar ratio of 1:2:3:2:2 (BC:BCCP1:BADC3:CT-α:CT-β). This parallels the molar ratio of the bacterial htACCase, which assembles in the absence of BADCs to a ratio of 1:2 for the BC:BCCP subcomplex (Broussard et al., 2013) and 1:1 for the CT-α:CT-β subcomplex (Tong, 2013).
Genetic Dissection of BADC Gene Redundancy
The data obtained from the biologically fabricated htACCase subcomplexes, containing either BCCP1 or BCCP2 and BC, with or without one of the three BADC paralogs, were used to guide interpretation of the phenotypes presented by mutants that lack specific combinations of BADC isoforms. Specifically, plants expressing only the BADC2 isoform were near normal, but plants expressing only the BADC3 isoform suffered a root growth penalty, and plants expressing only the BADC1 isoform were inviable, showing an embryo lethal phenotype. This latter finding parallels the nonsymmetrical redundancy that occurs between BCCP1 and BCCP2; namely plants that only express BCCP1 are near-normal and viable, whereas plants only expressing BCCP2 are inviable and show an embryo lethal phenotype (Li et al., 2011a).
The experiments that fabricated BCCP–BADC–BC subcomplexes clarified why the bccp2 mutant is viable through the process of embryogenesis, whereas the bccp1 mutant aborts embryogenesis at the 16-cell stage (Li et al., 2011a). Specifically, the BC specific activity displayed by the BCCP1–BADC–BC subcomplexes is 3- to 5-fold higher than those of the BCCP2–BADC–BC subcomplexes. Thus, despite the fact that BCCP2 is expressed uniquely during embryogenesis, the resulting subcomplex is not sufficiently active to compensate for the activity provided by the BCCP1-containing BCCP–BC–BADC subcomplex, ultimately leading to failed embryogenesis, probably due to a constraint in the supply of fatty acids for complex lipid assembly. Similarly, the embryo lethality of the badc2 badc3 double mutant plants, which only express BADC1, is associated with the significantly lower BC specific activity displayed by the BADC1-containing BCCP–BC–BADC subcomplexes, which would again constrain the supply of malonyl-CoA for fatty acid biosynthesis and thus not enable embryogenesis. An additional feature of BADC1 expression that may also contribute to the embryo-lethality of the badc2 badc3 double mutant is the distinct subcellular localization of this protein in the chloroplasts. Namely, in contrast to BADC2 and BADC3, BADC1 appears to discretely colocalize in plastids to distinct structures with another htACCase-associated protein (i.e. PII; Baud et al., 2010), and these associations may become essential during embryogenesis.
The root-growth phenotype associated with plants only expressing BADC3 may be ascribed to the fact that this paralog is expressed at low levels in roots. Thus, despite the fact that the BADC3 isoform is the most effective paralog in supporting the in vitro formation and activation of the BC–BADC–BCCP complex, the level of BADC3 that remains active in the badc1 badc2 double mutant appears to be insufficient to support ample formation of active htACCase complex, thus reducing the supply of malonyl-CoA for fatty acid biosynthesis needed to support the normal growth of this organ. Finally, the singular expression of only BADC2 (i.e. the badc1 badc3 double mutant) is sufficient to support growth throughout the plant’s life cycle without any significant phenotypic defects. Therefore, BADC1 and BADC3 appear to be superfluous in relation to BADC2 throughout the life cycle of the organism.
Revised Physiological BADC Functions
Prior characterizations of mutations in the three BADC genes have been interpreted to inaccurately propose BADC proteins are inhibitors of htACCase activity (Salie et al., 2016). This latter inhibitory model for BADC-protein action was based on the in vitro mixing of BADC1 with E. coli and plant preparations of htACCase and evaluating the effect on catalytic activity. As we indicate herein, the htACCase complex is stabilized in the presence of BADC proteins, and the addition of excess BADC1 could be perceived as inhibitory if there is an exchange with the complex-bound BADC components replacing BADC2 or BADC3, which are more competent in stabilizing and activating the htACCase complex than BADC1. It is more difficult to rationalize the result obtained in mixing BADC1 with the E. coli htACCase, as in this heterologous system these components have not “seen” each other since the evolutionary separation of bacteria from plants.
Apart from the badc3 mutants, the physiological effect of the badc mutations reported herein are similar with those reported by prior characterizations (Salie et al., 2016; Keereetaweep et al., 2018; Liu et al., 2019). However, based on the activation model for BADC functions, these physiological effects have been reinterpreted. In the case of the badc3 mutants, there are inconsistencies in the accumulation of fatty acids reported by the different studies. For example, whereas we found badc3 mutations did not affect fatty acid levels, Keereetaweep et al., (2018) reported increased fatty acid level in these mutants. Furthermore, there is a similar small discrepancy between our findings and those of Keereetaweep et al., (2018) in the badc1 badc2 double mutants. We suggest two potential explanations in these differences, (1) an environmental effect associated with the lighting regime used to grow the plants in the two studies; and (2) the genotypes of the mutant stocks used in these characterizations. Specifically, we grew the plants under continuous illumination, which avoids complexities associated with a diurnal light regime, whereas Keereetaweep et al. (2018) grew plants using a 16-h illuminated/8-h darkness lighting regime. Such illumination regimes are known environmental modulators of fatty acid accumulation traits (Nakamura et al., 2014; Kim et al., 2017) and may be the source of the different findings in the two studies. In addition, in our study and in the study by Keereetaweep et al. (2018), the identical badc3-1 allele was used in both studies; the GABI_170E12 allele obtained from Gabi-Kat and CS2103834, respectively, and these are one and the same (see https://abrc.osu.edu/stocks?search%5Btaxon%5D=Arabidopsis+thaliana&search%5Bsearch_text%5D=at3g15690&search%5Bmaterial_type%5D%5B%5D=seed&search%5Bsearch_fields%5D=AGI+locus+identifier). However, the stocks carrying this allele also carry a second mutation in the AT1G78920 gene, (http://signal.salk.edu/cgi-bin/tdnaexpress?JOB=TEXT&TYPE=DATA&QUERY=GABI_170E12). As we indicated in the “Materials and Methods,” by back-crossing the original GABI_170E12 stock with a wild-type Col-0 stock, we generated a line which only carried the badc3-1 allele and not the mutant AT1G78920 allele, and this was the line that was used in the subsequent analyses. This was not done in the Keereetaweep et al. (2018) study, and hence, it’s also possible this genetic difference may contribute to the difference in fatty acid content reported by the two studies.
In conclusion, this study illustrates the importance of integrating an accurate model of the in vitro capabilities of the components of multisubunit complexes to fully understand the functionality of gene products as dissected by in vivo genetic manipulations.
MATERIALS AND METHODS
Biological Materials
Recombinant proteins were expressed in Escherichia coli strains BL21 (DE3), and DH5α. The MCR gene from Chloroflexus aurantiacus was donated by Dr. Georg Fuchs (University Freiburg, Germany). E. coli-expression (with a pTrc99A vector) and purification of MCR was conducted as described in Kroeger et al. (2011), and the purified enzyme was flash-frozen and stored at −20°C.
The procedures and conditions used for planting and growing Arabidopsis (Arabidopsis thaliana) plants were described in Li et al. (2011a). Seed stocks of wild-type Arabidopsis (ecotype Col-0) and transfer-DNA (T-DNA) tagged mutant lines carrying badc1-1 (SALK_000817), badc1-2 (SALK_150657), badc2-1 (SALK_021108), and badc2-2 (SALK_121810) alleles were obtained from the Arabidopsis Biological Resource Center (Alonso et al., 2003). The seed stocks carrying the badc3-1 allele (GABI_170E12, ecotype Columbia) were obtained from GABI-Kat (https://www.gabi-kat.de/; Rosso et al., 2003). This original stock also carried a T-DNA insertion at a second gene locus (AT1G78920). This latter T-DNA tagged allele was removed by back-crossing the GABI_170E12 stock with the wild-type Col-0 stock and selecting progeny only carrying the badc3-1 allele and not the AT1G78920-associated T-DNA allele. All lines were confirmed to be homozygous for the mutant alleles by scoring the genotypes of a segregating selfed population. The genotypes of each allele in individual plants were determined by PCR-based assays as described in Sussman et al. (2002).
Plasmid Expression Vectors
Individual BADC proteins were expressed in E. coli strains using either pET30a (BADC1) or pET30b (BADC2 and BADC3) vectors. The open reading frame (ORF) for each BADC isoform was PCR amplified from the following complementary DNA clones: (1) BADC1 from RAFL04-09-B10, GenBank accession number AV821358, corresponding to GenBank reference sequence NM_115471.2); (2) BADC2 from M53C9STM (GenBank accession number BE524711, corresponding to GenBank reference sequence NM_104145.3); and (3) BADC3 from 224O9T7 (GenBank accession number N65015, corresponding to GenBank reference sequence NM_112439.3). The sequences of primers used in these subcloning experiments are listed in Supplemental Table S1.
Individual BADC isoforms were coexpressed in E. coli with different combination of htACCase subunits using the pETDuet-1, pCDFDuet-1, pACYCDuet-1, and pRSFDuet-1 expression vectors (MilliporeSigma). Because each of these vectors carry compatible origins of replications and different antibiotic resistance genes, different combinations of vectors could be maintained in E. coli BL21 (DE3) strains. The sequences coding for the chloroplast-targeting peptide sequence of each BADC and each htACCase subunit were identified using ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 2000), and these were removed when the ORF coding each mature protein was PCR-amplified (see Supplemental Table S1 for primer sequences). In the case of the CT-β subunit, the S265L mutation, which is a product of the in planta posttranscriptional modification of the CT-β transcript (Upton, 2014), was introduced using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies; see Supplemental Table S1 for the primer sequences).
Four deletion-derivatives of the mature BCCP1 protein (residues 81–280) were coexpressed with htACCase catalytic subunits using the pET30F vector (Jing, 2013). These derivative proteins were: (1) the N-terminal domain, residues Pro-81 to Gln-151; (2) the N-terminal and Ala/Prorich middle domains, residues Pro-81 to Ala-199; (3) the Ala/Pro-rich and C-terminal domains, residues Ala-152 to Pro-280; and (4) the C-terminal domain, residues Lys-200 to Pro-280.
Fluorescently tagged BADC proteins were expressed in planta using pEarleyGate recombinant expression constructs (Earley et al., 2006). Each BADC ORF was cloned into pEarleyGate 101, 102, and 103 vectors, using Gateway technology (Thermo Fisher Scientific). Primers used for PCR in these constructions are listed in Supplemental Table S1, and the resultant pEarley-BADC2-YFP, pEarley-BADC3-CFP, and pEarley-BADC1-GFP constructs were sequence-confirmed. Plant transformations were conducted by an Agrobacterium tumefaciens (strain C58C1)-mediated floral-dip method (Li et al., 2011b).
BADC Antisera
Individual recombinant BADC proteins were used to generate antiserum by immunizing New Zealand White female rabbits (Oryctolagus cuniculus) as described in Ke et al. (2000). Each of the resulting BADC antisera were evaluated by immunoblot analysis using SDS-PAGE gels loaded with equal amounts of purified BADC1, BADC2, and BADC3 proteins.
Isolation of the Arabidopsis Megadalton Holo–htACCase Complex
The holo-htACCase complex was isolated by a procedure modified from that of Olinares et al. (2010). One to 10 g of seedling tissue, flash-frozen in liquid nitrogen, was homogenized on ice in a 15-mL glass Dounce tissue grinder (MilliporeSigma) with 10 mL of ice cold extraction buffer composed of 25 mm of HEPES at pH 8.0, 10 mm of NaCl, 10 mm of MgCl2, 20% (v/v) glycerol, 1% (v/v) Triton X-100, and Plant Protease Inhibitor Cocktail (MilliporeSigma). The homogenate was centrifuged at 53,000g for 30 min, and the supernatant was filtered using a 0.45-μm syringe filter. This protein extract was concentrated to ∼20 mg/mL using a chilled 10-kD molecular cut-off Amicon Ultra-15 Centrifugal Filter (MilliporeSigma) by centrifugation at 3,900g in a swinging bucket rotor. The protein concentration was determined by the Bradford method (Bradford, 1976). An aliquot of this extract containing 2 mg of protein (in a volume of 100 μL) was loaded onto a Superose 6, 10/300 GL gel filtration column (GE Healthcare Bio-Sciences) using an ÄKTA FPLC system (GE Healthcare Bio-Sciences). The mobile phase was composed of 25 mm of HEPES at pH 8.0, 10 mm of NaCl, and 10 mm of MgCl2, and chromatography was performed at a flow rate of 0.25 mL/min. The A280 of the eluent was continuously monitored. Eight 2.1-mL fractions were collected between the elution volumes of 7 mL and 23.8 mL, corresponding to the molecular mass range of between 2,000 and 1 kD. These fractions were flash-frozen in liquid nitrogen and lyophilized to dryness. Fraction 2, corresponding to the molecular mass range of 700–2,000 kD, was used for immunoblot analyses to detect the htACCase subunits and the BADCs. These samples were redissolved in 133 μL 1× SDS-PAGE sample buffer and subjected to electrophoresis using 20% (w/v) polyacrylamide gels.
Copurification of E. coli-Reconstituted htACCase Complexes
Using the DUET vectors described above, different combinations of catalytic subunits of the htACCase were coexpressed in E. coli with either His-tagged BCCP1 or His-tagged BCCP2 and without and in the presence of individual BADC isoforms. The resulting cell pellets were lysed by sonication in buffer A containing 20 mm Tris-HCl at pH 8.0, 500 mm of NaCl with 5 mm of imidazole, and after centrifugation at 18,000g for 30 min, the supernatant was saved and loaded on to a Ni-NTA resin-based affinity column (G-Biosciences) pre-equilibrated with buffer A. The non-bound proteins were washed with 5–10 column volumes of buffer A, and subsequently, bound proteins were eluted by washing the column with buffer A containing increasing concentrations of imidazole up to 120 mm of imidazole. The fractions containing the eluted proteins were pooled and dialyzed overnight against 25 mm of HEPES buffer at pH 8.0, 150 mm of NaCl, and 10% (v/v) glycerol. The purified protein fractions were concentrated with an Amicon Ultra-15 Centrifugal Filter (MilliporeSigma), analyzed by SDS-PAGE, and assayed for catalytic capabilities.
RNA Analyses
RNA extraction from plant tissues and RT-qPCR procedures were performed as described in Ding et al. (2012). RT-qPCR primers were designed to detect the three BADC sequences that span the position of the T-DNA insertions (Supplemental Table S1), and PCR products were analyzed by agarose gel electrophoresis and confirmed by sequencing. RT-qPCR was performed as described in Jin et al. (2012), using three biological replicates for each sample. Each biological replicate represents the average of four technical replicates. UBQ10 (AT4G05320) was used as the reference gene for quantification with primer sequences adapted from Czechowski et al. (2005). All primer sequences are listed in Supplemental Table S1.
Protein Analysis
Unless otherwise noted, protein extracts were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue R-250 or subjected to immunoblot analysis as described in Ding et al. (2012). HtACCase preparations, purified from E. coli extracts, were analyzed by SDS-PAGE, and after staining with Coomassie Brilliant Blue R-250, the intensity of the protein bands was quantified with ChemiDoc XRS+ using the software Image Lab 6.0 (Bio-Rad). The intensity of the recovered BC-band was compared to standards of known quantities of purified BC, which was analyzed on the same gel, in parallel with the analysis of purified htACCase preparations. Similar analyses were used to quantify the recovery of BCCP1 and BCCP2.
For immunoblot analysis, after electrophoresis proteins were transferred to a nitrocellulose membrane, specific antisera were used to identify BADC1 (1:3,000 dilution), BADC2 (1:2,000 dilution), and BADC3 (1:5,000 dilution).
Microscopic Analyses
Confocal microscopy was conducted with a model no. TCS SP5 system (Leica Microsystems; http://www.leica-microsystems.com/home/), and an HCX PL APO CS 100.0×1.40 OIL UV immersion objective was used to collect images (Leica Microsystems). The laser wavelengths of excitation and emission for each fluorescent tag were as follows, respectively: for YFP, 514 nm and 525–580 nm; for CFP, 405 nm and 450–485 nm; for GFP, 489 nm and 500–533 nm; and for mCherry, 552 nm and 563–599 nm. All the images were analyzed using the Leica Application Suite (Leica Microsystems).
Stereo-microscopic images of siliques were obtained using a model no. SZH-10 stereo-microscope (Olympus; http://www.olympus-global.com/) with an AxioCam HRC digital camera (Zeiss; http://www.zeiss.com/). The detailed procedures were described in Ding et al. (2012).
Fatty Acid Analysis
Arabidopsis seeds (∼100 seeds), lyophilized rosette tissue (∼10 mg dry weight) or lyophilized root tissue (∼5 mg dry weight) were each spiked with 10 μL of 4-mg/mL glyceryl triheptadecanoate (MilliporeSigma) as an internal standard and were homogenized using a 1-mL glass homogenizer with 200 μL of HPLC grade hexane (Thermo Fisher Scientific). After incubation at room temperature for 30 min, the hexane extract was transferred to a gas chromatography (GC) vial, and 500 μL of 1-m sodium methoxide in methanol (MilliporeSigma) was added. After incubation at room temperature for 10 min with occasional mixing by vortexing, 150 μL of water was added to stop the methylation reaction. After extraction with 1 mL of hexane, the upper hexane layer was directly analyzed with a model no. 7890A GC system (Agilent Technologies) equipped with an SP-2330 capillary column (MilliporeSigma) measuring 15 m in length, 0.25 mm i.d. with a 0.2-μm film thickness. Helium was used as the carrier gas at a flow rate of 0.8 mL/min. The initial temperature of the GC oven was maintained at 190°C for 3.6 min, which was then increased at a rate of 50°C/min to 240°C, and maintained there for 1.5 min. Elution of the fatty acid methyl esters (FAMEs) was monitored with a flame ionization detector, and fatty acids were identified by comparing the retention times with standard C8-C24 FAME mixture (MilliporeSigma). Further verification of the FAMEs was performed using a GC–mass spectrometry system as described in Guan et al. (2015).
Enzymatic Assays
Two assays were used to assess the catalytic abilities of isolated htACCase complexes: (1) the BC first half-reaction of the enzyme; and (2) overall acetyl-CoA carboxylation reaction to form malonyl-CoA. The specific activity of BC was determined by coupling the appearance of ADP to the oxidation of NADH using pyruvate kinase and lactate dehydrogenase. This results in a change in A340, which was monitored with a model no. EL808 microplate reader (BioTek Instruments). Assays were conducted at 37°C, and the 50-µL reaction mixture contained 100 mm of HEPES buffer at pH 8.0, 10 mm of MgCl2, 0.5 mm of PEP, 0.5 mm of DTT, 0.5 mm of NADH, 1 mm of ATP, 10 mm of KHCO3, and ∼5 units of pyruvate kinase (MilliporeSigma) and 7 units of lactate dehydrogenase (MilliporeSigma). Negative control did not contain KHCO3. To ensure this latter condition, reagents were prepared free of dissolved CO2. This was achieved by boiling water for several minutes and subsequently cooling under a nitrogen gas atmosphere. This water preparation was used to prepare 100 mm of HEPES buffer at pH 8.0 in an anaerobic chamber (with 95% argon and 5% hydrogen atmosphere). The buffer was stored in vials containing a septum and was used in assays for determining kinetic constants associated with the bicarbonate-dependence of the ACCase reaction. All other assay components, except the enzymes, were bubbled with nitrogen gas to remove traces of dissolved CO2 from the reaction mix.
ACCase activity was determined by coupling the appearance of the product, malonyl-CoA, to the biochemical reduction of NADP+ using MCR (Kroeger et al., 2011), resulting in a change in A340. The assays were conducted in microtiter dishes with 50 μL of reaction mixtures consisting of 3.0 μg of purified MCR, 0.5 mm of DTT, 5 mm of MgCl2, 2mM of ATP, 0.5 mm of NADPH, 15 mM of KHCO3, and different amounts of ACCase enzyme preparations. The mixtures were initially incubated at 37°C for 10 min, and the reaction was started by the addition of 2 mm of acetyl-CoA. The rate of the reaction was monitored as the rate of change of A340 using a model no. EL808 microplate reader (BioTek).
Accession Numbers
Sequence data for the genes examined in this study can be found in the GenBank database under accession numebrs: BCCP1 (AT5G16390), BCCP2 (AT5G15530), BC (AT5G35360), CT-a (AT2G38040), CT-b (ATCG00500), BADC1 (AT3G56130), BADC2 (AT1G52670), and BADC3 (AT3G15690).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gel filtration chromatography of purified htACCase subcomplexes.
Supplemental Figure S2. The ATP dependence of the acetyl-CoA carboxylation reaction catalyzed by different subcomplex combinations.
Supplemental Figure S3. The bicarbonate dependence of the acetyl-CoA carboxylation reaction catalyzed by different subcomplex combinations.
Supplemental Figure S4. The acetyl-CoA dependence of the acetyl-CoA carboxylation reaction catalyzed by the combination of the BCCP1–BADC3–BC and CT–αβ subcomplexes.
Supplemental Figure S5. Molecular characterization of badc mutant alleles.
Supplemental Figure S6. Effect of the badc2 badc3 double mutant on seed development.
Supplemental Figure S7. Fatty acid content in rosette and root tissues.
Supplemental Figure S8. Subcellular localization of BADCs.
Supplemental Figure S9. Cross reactivity of antisera raised against individual BADC1, BADC2, and BADC3 recombinant proteins.
Supplemental Table S1. DNA primer sequences used in this study.
Supplemental Data S1. Data for Figure 7.
Supplemental Data S2. Data for Figure 8D.
Supplemental Data S3. Data for Figure 8E.
Supplemental Data S4. Data for Figure 9.
Supplemental Data S5. Data for Supplemental Figure 7A.
Supplemental Data S6. Data for Supplemental Figure 7A.
Acknowledgments
Any opinions, findings, and conclusions or recommendations expressed here are those of the authors, and do not necessarily reflect the views of the U.S. National Science Foundation or the Department of Energy. We thank Drs. M. Ann Perera and Lucas Showman (W.M. Keck Metabolomics Research Facility, Iowa State University) for guidance on fatty acid analysis, Dr. Gustavo C. MacIntosh (Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University) for the use of the FPLC system, and Margie Carter and Randall Den Adel (Roy J. Carver High Resolution Microscopy Facility, Iowa State University) for help in subcellular localization analyses and stereomicroscopy.
Footnotes
This work was supported in part by the United States National Science Foundation (EEC-0813570) and the Office of Science, Bioimaging Technology Program of the Department of Energy (DE-SC0014038).
Articles can be viewed without a subscription.
References
- Alberts AW, Vagelos PR (1968) Acetyl CoA carboxylase. I. Requirement for two protein fractions. Proc Natl Acad Sci USA 59: 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Athappilly FK, Hendrickson WA (1995) Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure 3: 1407–1419 [DOI] [PubMed] [Google Scholar]
- Baud S, Feria Bourrellier AB, Azzopardi M, Berger A, Dechorgnat J, Daniel-Vedele F, Lepiniec L, Miquel M, Rochat C, Hodges M, Ferrario-Méry S (2010) PII is induced by WRINKLED1 and fine-tunes fatty acid composition in seeds of Arabidopsis thaliana. Plant J 64: 291–303 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Broussard TC, Kobe MJ, Pakhomova S, Neau DB, Price AE, Champion TS, Waldrop GL (2013) The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase. Structure 21: 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari HST, Jakob RP, Maier T (2014) Evolutionary origins of the multienzyme architecture of giant fungal fatty acid synthase. Structure 22: 1775–1785 [DOI] [PubMed] [Google Scholar]
- Chalupska D, Lee HY, Faris JD, Evrard A, Chalhoub B, Haselkorn R, Gornicki P (2008) Acc homoeoloci and the evolution of wheat genomes. Proc Natl Acad Sci USA 105: 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DI, Vogel HJ (2010) Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J 430: 1–19 [DOI] [PubMed] [Google Scholar]
- Chapman-Smith A, Cronan JE Jr. (1999) In vivo enzymatic protein biotinylation. Biomol Eng 16: 119–125 [DOI] [PubMed] [Google Scholar]
- Chen M, Mooney BP, Hajduch M, Joshi T, Zhou M, Xu D, Thelen JJ (2009) System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol 150: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Yu F, Wurtele ES, Nikolau BJ (1995) Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol 109: 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE Jr., Waldrop GL (2002) Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res 41: 407–435 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimroth P, Guchhait RB, Stoll E, Lane MD (1970) Enzymatic carboxylation of biotin: molecular and catalytic properties of a component enzyme of acetyl CoA carboxylase. Proc Natl Acad Sci USA 67: 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G. (2008) Molecular, genetic, physiological and biochemical studies of 3methylcrotonyl-CoA carboxylase and biotin carboxyl carrier protein-like proteins in Arabidopsis thaliana. Doctoral dissertation. Iowa State University, Ames, IA [Google Scholar]
- Ding G, Che P, Ilarslan H, Wurtele ES, Nikolau BJ (2012) Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J 70: 562–577 [DOI] [PubMed] [Google Scholar]
- Ding W, Lin L, Zhang B, Xiang X, Wu J, Pan Z, Zhu S (2015) OsKASI, a β-ketoacyl-[acyl carrier protein] synthase I, is involved in root development in rice (Oryza sativa L.). Planta 242: 203–213 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Ewald R, Kolukisaoglu U, Bauwe U, Mikkat S, Bauwe H (2007) Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol 145: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Sha LN, Yang RW, Zhang HQ, Kang HY, Ding CB, Zhang L, Zheng YL, Zhou YH (2009) Phylogeny and evolutionary history of Leymus (Triticeae; Poaceae) based on a single-copy nuclear gene encoding plastid acetyl-CoA carboxylase. BMC Evol Biol 9: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J, Sirikhachornkit A, Haselkorn R, Gill B, Gornicki P (2001) Chromosome mapping and phylogenetic analysis of the cytosolic acetyl-CoA carboxylase loci in wheat. Mol Biol Evol 18: 1720–1733 [DOI] [PubMed] [Google Scholar]
- Farmaki T, Sanmartín M, Jiménez P, Paneque M, Sanz C, Vancanneyt G, León J, Sánchez-Serrano JJ (2007) Differential distribution of the lipoxygenase pathway enzymes within potato chloroplasts. J Exp Bot 58: 555–568 [DOI] [PubMed] [Google Scholar]
- Feria Bourrellier AB, Valot B, Guillot A, Ambard-Bretteville F, Vidal J, Hodges M (2010) Chloroplast acetyl-CoA carboxylase activity is 2-oxoglutarate-regulated by interaction of PII with the biotin carboxyl carrier subunit. Proc Natl Acad Sci USA 107: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. (2016) The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res 44(D1): D279–D285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago G, Diacovich L, Arabolaza A, Tsai S-C, Gramajo H (2011) Fatty acid biosynthesis in actinomycetes. FEMS Microbiol Rev 35: 475–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Chen H, Abramson A, Man H, Wu J, Yu O, Nikolau BJ (2015) A phosphopantetheinyl transferase that is essential for mitochondrial fatty acid biosynthesis. Plant J 84: 718–732 [DOI] [PubMed] [Google Scholar]
- Harwood JL. (1988) Fatty acid metabolism. Annu Rev Plant Physiol Plant Mol Biol 39: 101–138 [Google Scholar]
- Held D, Yaeger K, Novy R (2003) New coexpression vectors for expanded compatibilities in E. coli. Innovations 18: 4–6 [Google Scholar]
- Huang S, Sirikhachornkit A, Faris JD, Su X, Gill BS, Haselkorn R, Gornicki P (2002) Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol Biol 48: 805–820 [DOI] [PubMed] [Google Scholar]
- Huerlimann R, Zenger KR, Jerry DR, Heimann K (2015) Phylogenetic analysis of nucleus-encoded acetyl-CoA carboxylases targeted at the cytosol and plastid of algae. PLoS One 10: e0131099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M, Krieger RS, Jahn M, Fuchs G (2003) Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur J Biochem 270: 736–744 [DOI] [PubMed] [Google Scholar]
- Hunkeler M, Stuttfeld E, Hagmann A, Imseng S, Maier T (2016) The dynamic organization of fungal acetyl-CoA carboxylase. Nat Commun 7: 11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Song Z, Nikolau BJ (2012) Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. Plant J 70: 1015–1032 [DOI] [PubMed] [Google Scholar]
- Jing F. (2013) Characterization of acyl-ACP thioesterases for the purpose of diversifying fatty acid synthesis pathway. Doctoral dissertation. Iowa State University, Ames, IA [Google Scholar]
- Jitrapakdee S, Wallace JC (2003) The biotin enzyme family: Conserved structural motifs and domain rearrangements. Curr Protein Pept Sci 4: 217–229 [DOI] [PubMed] [Google Scholar]
- Jordan IK, Henze K, Fedorova ND, Koonin EV, Galperin MY (2003) Phylogenomic analysis of the Giardia intestinalis transcarboxylase reveals multiple instances of domain fusion and fission in the evolution of biotin-dependent enzymes. J Mol Microbiol Biotechnol 5: 172–189 [DOI] [PubMed] [Google Scholar]
- Joyard J, Ferro M, Masselon C, Seigneurin-Berny D, Salvi D, Garin J, Rolland N (2010) Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog Lipid Res 49: 128–158 [DOI] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ (2000) The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol 123: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Choi JK, Smith M, Horner HT, Nikolau BJ, Wurtele ES (1997) Structure of the CAC1 gene and in situ characterization of its expression. The Arabidopsis thaliana gene coding for the biotin-containing subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol 113: 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keereetaweep J, Liu H, Zhai Z, Shanklin J (2018) Biotin attachment domain-containing proteins irreversibly inhibit acetyl CoA carboxylase. Plant Physiol 177: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Kim HS, Choi SH, Jang JY, Jeong MJ, Lee SI (2017) The importance of the circadian clock in regulating plant metabolism. Int J Mol Sci 18: E2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles JR. (1989) The mechanism of biotin-dependent enzymes. Annu Rev Biochem 58: 195–221 [DOI] [PubMed] [Google Scholar]
- Kroeger JK, Zarzycki J, Fuchs G (2011) A spectrophotometric assay for measuring acetyl-coenzyme A carboxylase. Anal Biochem 411: 100–105 [DOI] [PubMed] [Google Scholar]
- Li X, Ilarslan H, Brachova L, Qian H-R, Li L, Che P, Wurtele ES, Nikolau BJ (2011a) Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. Plant Physiol 155: 293–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-G, Yin W-B, Guo H, Song L-Y, Chen Y-H, Guan R-Z, Wang J-Q, Wang RR-C, Hu Z-M (2010) Genes encoding the α-carboxyltransferase subunit of acetyl-CoA carboxylase from Brassica napus and parental species: Cloning, expression patterns, and evolution. Genome 53: 360–370 [DOI] [PubMed] [Google Scholar]
- Li Z-G, Yin W-B, Song L-Y, Chen Y-H, Guan R-Z, Wang J-Q, Wang RR-C, Hu Z-M (2011b) Genes encoding the biotin carboxylase subunit of acetyl-CoA carboxylase from Brassica napus and parental species: Cloning, expression patterns, and evolution. Genome 54: 202–211 [DOI] [PubMed] [Google Scholar]
- Liu H, Zhai Z, Kuczynski K, Keereetaweep J, Schwender J, Shanklin J (2019) WRINKLED1 regulates BIOTIN ATTACHMENT DOMAIN-CONTAINING proteins that inhibit fatty acid synthesis. Plant Physiol 181: 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard J, Moreira D (2011) Early evolution of the biotin-dependent carboxylase family. BMC Evol Biol 11: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London WP, Steck TL (1969) Kinetics of enzyme reactions with interaction between a substrate and a (metal) modifier. Biochemistry 8: 1767–1779 [DOI] [PubMed] [Google Scholar]
- Mentzen WI, Peng J, Ransom N, Nikolau BJ, Wurtele ES (2008) Articulation of three core metabolic processes in Arabidopsis: Fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biol 8: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Możejko-Ciesielska J, Kiewisz R (2016) Bacterial polyhydroxyalkanoates: Still fabulous? Microbiol Res 192: 271–282 [DOI] [PubMed] [Google Scholar]
- Mueller SJ, Lang D, Hoernstein SNW, Lang EGE, Schuessele C, Schmidt A, Fluck M, Leisibach D, Niegl C, Zimmer AD, Schlosser A, Reski R (2014) Quantitative analysis of the mitochondrial and plastid proteomes of the moss Physcomitrella patens reveals protein macrocompartmentation and microcompartmentation. Plant Physiol 164: 2081–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Andrés F, Kanehara K, Liu YC, Coupland G, Dörmann P (2014) Diurnal and circadian expression profiles of glycerolipid biosynthetic genes in Arabidopsis. Plant Signal Behav 9: e29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Hawke JC (1984) Purification and characterization of maize leaf acetyl-coenzyme A carboxylase. Arch Biochem Biophys 228: 86–96 [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Ohlrogge JB, Wurtele ES (2003) Plant biotin-containing carboxylases. Arch Biochem Biophys 414: 211–222 [DOI] [PubMed] [Google Scholar]
- Novy R, Morris B (2001) Use of glucose to control basal expression in the pET system. Innovations 13: 8–10 [Google Scholar]
- Olinares PDB, Ponnala L, van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy DV, Shenoy BC, Carey PR, Sönnichsen FD (2000) High resolution solution structure of the 1.3S subunit of transcarboxylase from Propionibacterium shermanii. Biochemistry 39: 2509–2516 [DOI] [PubMed] [Google Scholar]
- Reverdatto S, Beilinson V, Nielsen NC (1999) A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol 119: 961–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15: 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EL, Shu N, Howard MJ, Broadhurst RW, Chapman-Smith A, Wallace JC, Morris T, Cronan JE Jr., Perham RN (1999) Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry 38: 5045–5053 [DOI] [PubMed] [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB (1994) Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene. Plant Physiol 105: 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Ruan H, Sun Q, Zhang W, Liu Y, Lai L (2019) Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov Today 24: 217–227 [DOI] [PubMed] [Google Scholar]
- Salie MJ, Zhang N, Lancikova V, Xu D, Thelen JJ (2016) A family of negative regulators targets the committed step of de novo fatty acid biosynthesis. Plant Cell 28: 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R (1993) Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem 268: 25118–25123 [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M (1997) Link between light and fatty acid synthesis: Thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA 94: 11096–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Ohmori A, Iguchi H, Nagano Y (2001) Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J Biol Chem 276: 3937–3940 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y (2004) Plant acetyl-CoA carboxylase: Structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Sauer A, Heise K-P (1983) On the light dependence of fatty acid synthesis in spinach chloroplasts. Plant Physiol 73: 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer A, Heise K-P (1984) Regulation of acetyl-coenzyme a-carboxylase and acetyl-coenzyme a-synthetase in spinach chloroplasts. Zeitschrift Naturforsch C J Biosci 39: 268–275 [Google Scholar]
- Sektas M, Szybalski W (2002) Novel single-copy pETcoco vector with dual controls for amplification and expression. Innovations 14: 10–12 [Google Scholar]
- Shelley IJ, Nishiuchi S, Shibata K, Inukai Y (2013) SLL1, which encodes a member of the stearoyl-acyl carrier protein fatty acid desaturase family, is involved in cell elongation in lateral roots via regulation of fatty acid content in rice. Plant Sci 207: 12–17 [DOI] [PubMed] [Google Scholar]
- Smith JL, Sherman DH (2008) An enzyme assembly line. Science 321: 1304–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Liu D (2015) Mutations in the three Arabidopsis genes that encode the E2 subunit of the mitochondrial pyruvate dehydrogenase complex differentially affect enzymatic activity and plant growth. Plant Cell Rep 34: 1919–1926 [DOI] [PubMed] [Google Scholar]
- Storer AC, Cornish-Bowden A (1976) Concentration of MgATP2- and other ions in solution. Calculation of the true concentrations of species present in mixtures of associating ions. Biochem J 159: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ke J, Johnson JL, Nikolau BJ, Wurtele ES (1997) Biochemical and molecular biological characterization of CAC2, the Arabidopsis thaliana gene coding for the biotin carboxylase subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol 115: 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2002) The Arabidopsis knockout facility at the University of Wisconsin–Madison: Fig. 1. Plant Physiol 24: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Mekhedov S, Ohlrogge JB (2001) Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol 125: 2016–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Kondo H, Tanabe T (1993) Molecular evolution of biotin-dependent carboxylases. Eur J Biochem 215: 687–696 [DOI] [PubMed] [Google Scholar]
- Tong L. (2013) Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70: 863–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Hsiao YS, Jo J, Chou CY, Dietrich LEP, Walz T, Tong L (2015) Structure and function of a single-chain, multi-domain long-chain acyl-CoA carboxylase. Nature 518: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton B. (2014) Biochemical characterization of the biotindependent carboxylases, acetyl-CoA carboxylase and 3-methylcrotonyl-CoA carboxylase. Doctoral dissertation. Iowa State University, Ames, IA [Google Scholar]
- Uversky VN. (2016) Dancing protein clouds: The strange biology and chaotic physics of intrinsically disordered proteins. J Biol Chem 291: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil SJ, Abu-Elheiga LA (2009) Fatty acid metabolism: Target for metabolic syndrome. J Lipid Res 50(Suppl): S138–S143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stöveken T, von Landenberg P, Steinbüchel A (2005) Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol 55: 750–763 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Lebrun L, Franklin A, Huang L, Hoffman N, Wurtele ES, Nikolau BJ (1995) Molecular cloning of the biotinylated subunit of 3-methylcrotonyl-coenzyme A carboxylase of Arabidopsis thaliana. Plant Physiol 107: 1013–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Tong L (2015) Crystal structure of the 500-kDa yeast acetyl-CoA carboxylase holoenzyme dimer. Nature 526: 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N (1995) Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: Tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol 36: 779–787 [DOI] [PubMed] [Google Scholar]
- Yao X, Wei D, Soden C Jr., Summers MF, Beckett D (1997) Structure of the carboxy-terminal fragment of the apo-biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase. Biochemistry 36: 15089–15100 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023 [DOI] [PubMed] [Google Scholar]