SUMMARY
Many proteins in their unbound structures have cryptic sites that are not appropriately sized for drug binding. We consider here 32 proteins from the recently published CryptoSite set with validated cryptic sites, and study whether the sites remain cryptic in all available X-ray structures of the proteins solved without any ligand bound near the sites. It was shown that only few of these proteins have binding pockets that never form without ligand binding. Sites that are cryptic in some structures but spontaneously form in others are also rare. In most proteins the forming of pockets is impacted by mutations or ligand binding at locations far from the cryptic site. To further explore these mechanisms, we applied adiabatic biased molecular dynamics (ABMD) simulations to guide the proteins from their ligand-free structures to ligand-bound conformations, and studied the distribution of druggability scores of the pockets located at the cryptic sites.
Keywords: induced fit, conformational selection, binding pocket, druggability, ligand binding, allostery
eTOC Blurb
Sun et al. provided structure-based analysis of cryptic site opening. Understanding the mechanisms of such site formation is important for the identification of novel druggable targets. Through analyses of X-ray structures and biased molecular dynamics, three types of cryptic site opening were discussed: “genuine”, spontaneous, and allosterically-impacted.
INTRODUCTION
The binding of proteins to small molecules is central to various biological functions, including enzyme catalysis, receptor activation, and drug action, and thus detection, comparison and analyses of binding pockets are pivotal to structure-based drug design (Perot et al., 2010). In many proteins significant differences in protein conformation exist between the unbound and bound states, and in some cases the binding site is not even detectable in ligand-free structures. These so-called cryptic sites can be important for drug discovery because they can provide previously undescribed pockets and thus enable targeting of proteins that would otherwise be considered undruggable. For example, it was predicted that considering cryptic sites of the structurally characterized proteins increases the size of the potentially “druggable” disease-associated human proteome from ~40% to ~78% (Cimermancic et al., 2016). Thus, targeting of cryptic binding sites represents an attractive but somewhat underexplored approach to modulating protein function with small molecules (Acker et al., 2017; Cimermancic et al., 2016). An important related question is whether the pockets are already present in some of the unliganded structures, since this information affects the choice of methods used for the identification of such sites.
The search for cryptic sites has been intensified with the improving performance of molecular dynamics (MD) simulation methods that have a history of successful applications (Durrant et al., 2010; Durrant and McCammon, 2011; Grant et al., 2011; Wagner et al., 2016; Wassman et al., 2013). More recently, the development of Markov state models (MSMs) provided an even more powerful tool and stronger motivation for the discovery of cryptic sites (Bowman et al., 2015; Bowman and Geissler, 2012; Hart et al., 2017; Knoverek et al., 2019; Porter et al., 2019). MSMs are built from extensive MD simulations to describe a protein’s intrinsic dynamics, and provide a reduced view of the ensemble of spontaneous fluctuations the molecule undergoes at equilibrium, thereby identifying transient pockets and their probabilities (Bowman and Geissler, 2012). Recent MSM simulations revealed that the forming of ligand binding pockets at cryptic sites requires large cooperative changes to the surface of the protein, and that this property helps to identify such sites (Porter et al., 2019).
The goal of this paper is to consider a set of proteins with validated cryptic sites, and to study whether the sites remain always cryptic without ligand binding, or pockets already form in some of the structures. In order to answer this question with some generality we want to study a substantial number of proteins rather than only a few. In spite of advances in methodology and computer speed, MD or MSM simulations are computationally still too demanding for a large-scale study, and hence we primarily investigate X-ray structures from the Protein Data Bank (PDB). However, for three proteins the results of the empirical analysis are supported by performing adiabatic biased molecular dynamics (ABMD) simulations (Harvey and Gabb, 1993; Marchi and Ballone, 1999; Paci and Karplus, 1999).
The starting point of our analysis is the CryptoSite set of protein pairs developed for benchmarking cryptic site detection algorithms (Cimermancic et al., 2016). Each of the of 93 bound-unbound pairs in this set included an unbound structure without a well-formed pocket and another structure co-crystallized with a biologically relevant ligand bound at the same location. A limitation of the CryptoSite set is that each pair contained only a single unbound structure, although to determine whether a site can be considered genuinely cryptic it would be important to consider the full range of ligand-free conformations available to the protein. Therefore in our previous work we extended the set by adding all structures in the Protein Data Bank having at least 95% sequence identity and no ligand bound within the 5 Å neighborhood of the cryptic site (Beglov et al., 2018). All structures in this extended set were mapped using the FTMap program (Kozakov et al., 2015a), and it was shown that the vicinity of the cryptic site included a strong binding hot spot in some of the unbound structures for over 90% of the 93 proteins (Beglov et al., 2018). Since binding hot spots disproportionately contribute to the binding free energy of any ligand (DeLano, 2002; Hall et al., 2015), and some attractive forces are clearly required for ligand binding, this result was not unexpected. However, binding hot spots can be located both in relatively flat surface regions and in crevices that are too tight to accommodate drug-sized ligands, and we did not investigate whether appropriate pockets were actually formed in any of the unbound structures. In fact, FTMap is not even suitable for such analysis, since its results are relatively invariant to conformational changes (Kozakov et al., 2015a; Kozakov et al., 2011).
To examine the statistics of pockets before any ligand binds, we considered the proteins in the CryptoSite set that had at least 10 apo structures in the Protein Data Bank (PDB). To characterize the pockets in the structures we calculated a druggability score (DS) at the cryptic site using the Fpocket program (Le Guilloux et al., 2009; Schmidtke et al., 2010). Fpocket is more sensitive to conformational changes than FTMap. The Fpocket DS values depend on shape, size, and polarity of the pocket (see Star Methods) and vary between zero (no pocket) and 1.0 (pocket ideal for binding druglike small molecules). The number of structures for each protein is generally much higher than 10, and having multiple ligand-free X-ray structures enabled us to generate histograms of Fpocket druggability scores (Schmidtke and Barril, 2010). We considered DS = 0.5 as the lower threshold for a well-formed pocket, and disregarded any protein if the cryptic pocket had DS < 0.5 in the ligand-bound structure. We also omitted any protein if none of its unliganded structures satisfied the FTMap druggability conditions (Kozakov et al., 2015b). These selection criteria reduced the number of proteins considered in this study to 32.
As will be shown, the 32 protein can be grouped into the three different types. The first group includes eight proteins with cryptic sites that, based on the available X-ray structures, can be considered “genuine” since the pocket at the site does not form without ligand binding. In contrast, the apo structures of six proteins in the second group exhibit binding pockets that seem to spontaneously form in a substantial fraction of structures. Finally, in the largest group of 18 proteins forming of a pocket is impacted by off-site mutations or ligand binding, thus emphasizing the role of allosteric communication in the opening of the cryptic site. We assume that the X-ray structure of a protein correspond to the free energy minimum of the crystal under the condition of crystallization. However, the protein has an ensemble of slightly higher energy conformations (Hilser et al., 2012; Motlagh et al., 2014; Wrabl et al., 2011), and changes in the conditions of crystallization, introducing site directed mutations, or mutating some residues all perturb the free energy landscape and thereby can alter the X-ray structure. While analyzing the unliganded structures in the PDB provides some chance for capturing alternative structures, some possibly with better formed pockets, we readily admit that this approach is far from systematic. However, we will demonstrate that the results show the substantial information available in the PDB on the opening of cryptic sites.
To further explore how cryptic sites are formed, we selected one typical protein from each of the three groups and applied adiabatic biased molecular dynamics (ABMD) simulations (Harvey and Gabb, 1993; Marchi and Ballone, 1999; Paci and Karplus, 1999). The simulations use a biasing force to guide the proteins from their ligand-free structures to ligand-bound conformations. ABMD is similar to targeted molecular dynamics (Schlitter et al., 1994), but it is more gentle because the biasing force is only applied when the system is diverging from its path towards the target structure. Guiding the structures toward well-formed pockets enables rigorous sampling the transitions between the two states, generating a distribution of druggability scores of the pocket located at the cryptic site. By varying the value of a force constant we can assess the extent of how energetically demanding such conformational transitions are. As mentioned, the three proteins studied by ABMD represent different pocket opening mechanisms. From the first group we consider the higher affinity phosphotyrosine (pTyr) binding pocket of protein tyrosine phosphatase 1B (PTP1B), which does not seem to form without binding a charged ligand. Accordingly, considerable force is needed in the simulation to guide the structure toward the ligand-bound conformation. In contrast, the active site of beta-secretase 1 (BACE1) is defined by a loop that can open and close essentially on its own, and hence not much force is needed to move it between the two states. The third protein we study is TEM-1 β-lactamase, in which the cryptic allosteric site is formed by moving apart two helices. As will be shown, the results of these simulations confirm the trends of pocket formation observed in the X-ray structures.
RESULTS
Proteins in the CryptoSite set
The increasing number of X-ray structures determined under different conditions for the same proteins enabled us to study conformational variations, including the potential opening of cryptic pockets, in a large set of proteins, and thus arrive at conclusions that may have some level of generality. As already mentioned, the starting point of our study is the CryptoSite set of X-ray structures of proteins with validated cryptic binding sites (Cimermancic et al., 2016). For each bound structure in this set we added all unbound structures with at least 95% sequence identity in the Protein Data Bank that had nothing bound within the 5 Å neighborhood of the cryptic site (Beglov et al., 2018). The extended CryptoSite set was filtered to consider only good quality X-ray structures for the analysis of druggability score histograms. Structures with a resolution lower than 3.5 Å and all structures that were determined by cryo-EM or NMR were discarded (see Methods). In addition, we restricted the analysis to 32 proteins that had at least 10 unbound structures satisfying the above criteria (Table 1). The number of retained unbound structures per protein varied from 10 to 249.
Table 1.
Proteins with cryptic sides studied
| Apoa | Holoa | Ligb | Name | Nc | Figure | Site |
|---|---|---|---|---|---|---|
| 2CM2_A | 2H4K_A | 509 | PTP1B | 19 | 1 | High affinity pTyr binding site |
| 1 PKL_B | 3HQP_P | ATP | Pyruvate kinase enzyme | 10 | 3A | ATP+Oxalate binding site |
| 1RTC_A | 1BR6_A | PT1 | Ricin | 23 | 3B | Pteroic acid binding at the active site |
| 1RHB_A | 2W5K_B | NDP | Ribonuclease A. | 83 | 3C | NADPH binding at the active site |
| 3CJ0_A | 2BRL_A | POO | HCV polymerase NS5B | 249 | 3D | Between fingers and thumb domains |
| 2F6V_A | 1T49_A | 892 | PTP1B | 108 | 3E | Allosteric site under the C-terminal helix |
| 1ZAH_B | 2OT1_D | N3P | Fructose aldolase | 36 | 3F | Competitive inhibitor binding site |
| 1 G24_D | 1GZF_C | NIR | Rho ADP-Ribosyl. Enz. | 15 | S1 | Structure also contain NAD and ADP |
| 1 W50_A | 3IXJ_C | 586 | BACE-1 protease | 19 | 4 | Active site, too open in apo structure |
| 1BSQ_A | 1 GX8_A | RTL | Bovine Beta-lactoglobulin | 34 | 5A | Retinol binding in the central cavity |
| 1HAG_E | 1GHY_H | 121 | Thrombin | 52 | 5B | Pocket is too open with flexible loops |
| 3F74_C | 3BQM_C | BQM | Alpha-L (Integrin) domain | 25 | 5C | Active site with disordered C terminus |
| 1MY0_B | 1N0T_D | AT1 | Glutamate receptor 2 | 26 | 5D | Stabilizes the open form of the receptor |
| 1XCG_B | 10W3_B | GDP | Transforming protein | 12 | S2 | GDP interacts only with RhoA |
| 1 JWP_A | 1 PZ0_A | CBT | TEM β-lactamase | 21 | 6 | Allosteric site between two helixes |
| 2BLS_B | 3GQZ_A | GF7 | AMPc beta-lactamase | 35 | 8A | Weak peripheral allosteric site |
| 2BU8_A | 2BU2_A | TF1 | Pyruvate dehyd. kinase | 11 | 8B | Allosteric inhibitor site |
| 3CJ0_A | 3FQK_B | 79Z | HCV polymerase NS5B | 143 | 8C | Binding near the active site. |
| 2BRK_A | 2GIR_B | NN3 | HCV polymerase NS5B | 186 | S3A | Non-nucleotide inhibitor (thumb) site |
| 1FXX_A | 3HL8_A | BBP | Exodeoxyribonuclease I | 14 | 8D | BBP prevents Exol/SSB interactions |
| 1OK8_A | 10KE_B | BOG | Dengue 2 virus envelope | 15 | 8E | Site is between two domains |
| 2AKA_A | 1 YV3_A | BIT | Myosin II | 30 | 8F | Narrow planar ligand binding site |
| 3MN9_A | 3EKS_A | CY9 | Monomer. actin with toxin | 36 | S3B | Binding to the barbed end of filaments |
| 1 NUW_A | 1 EYJ_B | AMP | Fruct. 1,6- bisphosphatase | 25 | S3C | AMP binding site |
| 3PUW_E | 1FQC_A | GLO | Maltodextrin/maltose BP | 19 | S3D | Interdomain binding |
| 3KQA_B | 3LTH_A | UD1 | MurA dead-end complex | 13 | S3E | Interdomain binding |
| 3GXD_B | 2WCG_A | MT5 | Acid-beta-glucosidase | 26 | S3F | Active site |
| 1BNC_B | 2V5A_A | LZL | Biotin carboxylase | 18 | S4A | ATP competitive inhibitor site |
| 1MY1_C | 1FTL_A | DNQ | Glutamate receptor 2. | 26 | S4B | Interdomain binding |
| 2AX9_A | 2PIQ_A | RB1 | Androgen receptor | 23 | S4C | Allosteric inhibitor binds on surface |
| 2ZB1_A | 2NPQ_A | BOG | P38 MAP kinase | 144 | S4D | Helix 253–261 moves outward |
| 2AIR_H | 1ZA1_D | CTP | Aspartate transcarbamylase |
60 | S5 | Binds CTP at the flexible N-terminal |
PDB ID of the apo and holo structures in the CryptoSite database.
Name of the ligand binding at the cryptic site.
Number of structures considered.
Table 1 shows the Protein Data Bank (PDB) IDs of the unbound and bound structures, the three letter PDB code of the ligand bound at the cryptic site and considered in the CryptoSite set (Cimermancic et al., 2016), the name of the protein, the number of unliganded structures in the extended set that satisfy our selection criteria, the figure that shows the DS histogram, and a short comment. Since we studied 32 proteins, detailed discussion had to be limited. However, we provide extended comments (Table S1), DS histograms that are not shown in the main text (Figures S1, S2, and S3), and the complete list of selected ligand free structures and their calculated DS values for all 32 proteins (Data S1).
Proteins that require ligand binding for forming a pocket at the cryptic site
A binding site can be considered genuinely cryptic if the binding pocket never forms without a bound ligand, and thus the DS distribution is strongly skewed toward small values, i.e., DS < 0.5 in all structures. Based on the X-ray structures of the 32 proteins considered, it appears that such proteins are relatively rare. In addition, even for these proteins there generally exist a limited number of exceptions. As will be discussed, proteins that have no detectable pockets in almost all unbound structures still may have such pockets due to either a mutation or ligand binding at a distant site that lead to opening the cryptic site without a bound ligand. Therefore we indicate if the protein is a mutant or if it is a complex with a ligand or protein binding at a distant (non-cryptic) site. It is generally helpful that the structures in the PDB are supplemented by publications that provide information on the origins of cryptic site properties and help to explain why the exception may occur.
To demonstrate that some proteins are unlikely to form a pocket at the cryptic site without ligand binding we selected protein tyrosine phosphatase 1B (PTP1B), an extremely well-studied protein, in which the most important subsite of the active site is cryptic. This pocket is known as the site of the high affinity phosphotyrosine binding (Puius et al., 1997). In the CryptoSite set this site is represented by the unbound structure 2CM2 and by the structure 2H4K, co-crystallized with a small inhibitor. In Figure 1A we copied the inhibitor from the bound structure into the unbound one to show that in the latter the binding site is broad and open rather than a drug-sized cavity. Such cavity forms in the inhibitor-bound structure (Figure 1B). Without ligand the binding site is open because loop 179 – 188 turns away from the site (Figure 2A). The loop moves closer to the site and forms a tight pocket in all bound structures, with the side chain of F182 acting as the lid (Figures 1B and 2A). The monocyclic thiophene inhibitor in 2H4K has low affinity (Ki = 1300– 3200 nM), but the same pocket binds an inhibitor with Ki = 4 nM in the PDB structure 2QBP. Although the pocket is very important for binding active site inhibitors and the phosphotyrosine moiety of substrates, it has a druggability score DS > 0.1 only in two unbound structures. The first is the C215D mutant (PDB ID 1PA1, DS = 0.468) and the second is the low resolution structure 2HNP (DS = 0.338). We note that some of the 19 “unliganded” PTP1B structures in the PDB are mutants or have inhibitors binding far from the active site, but the pocket remains too open in all such structures.
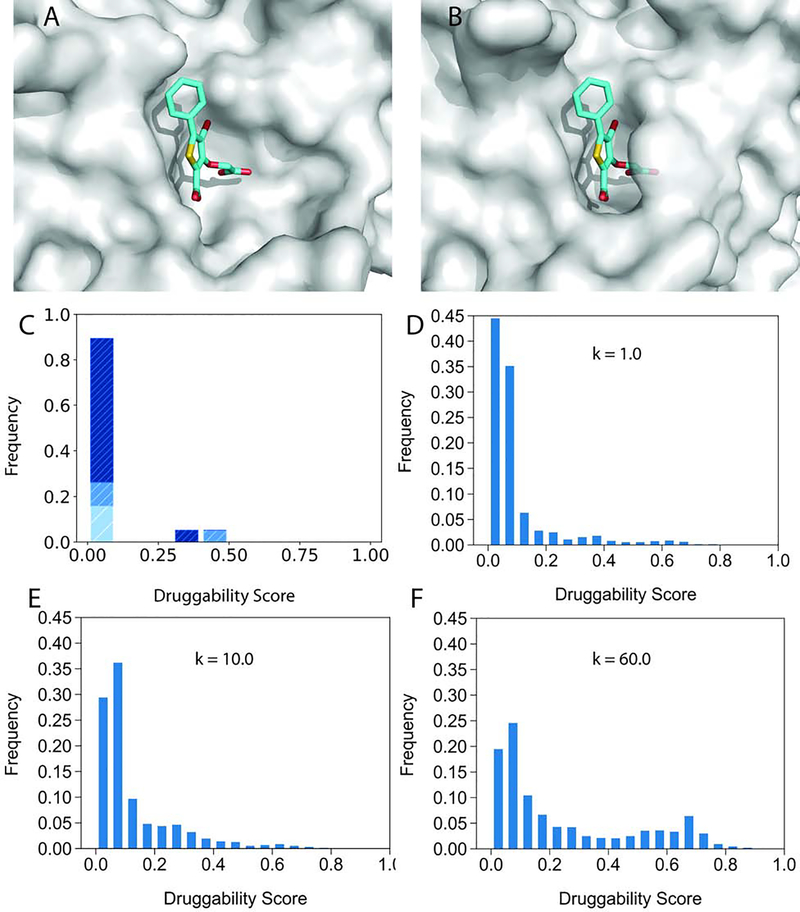
Figure 1.
Forming the pocket at the site of high affinity phosphotyrosine binding in PTP1B. A. Unbound PTP1B structure 2CM2 shown as grey surface. The inhibitor 509 from the ligand-bound structure 2H4K (cyan sticks) is shown for refence, demonstrating that the site is too open. B. In the ligand-bound structure 2H4K the pocket binding the inhibitor 509 is well formed. The protein is shown as partially transparent surface for improved visibility. C. Druggability scores (DSs) of unliganded PTP1B structures in the PDB. The distribution of DS values is shown in dark, light, and medium blue, respectively, for unbound structures, complexes, and mutants. Here “complex” means a protein or ligand binding at a distant site. D. Distribution of druggability score (DS) values obtained by adiabatic biased molecular dynamics (ABMD) simulations of unliganded PTP1B at k=1.0 (kcal/mol)/Å2. E. Distribution of DS values obtained by ABMD simulations of PTP1B at k=10.0 (kcal/mol)/Å2. F. Distribution of DS values obtained by ABMD simulations of PTP1B at k=60.0 (kcal/mol)/Å2.
Figure 2.
Conformational change and a snapshot from the ABMD simulation of protein tyrosine phosphatase 1B (PTP1B). All structures are shown in cartoon representation. A. Loop 179 – 188 in the unbound structure 2CM2 (grey) and in the inhibitor-bound structure 2H4K (orange). B. Loop 179 – 188 from a snapshot at t = 12 ns of the ABMD simulations with k = 60.0 kcal/mol/Å2 (blue).
Since the move of loop 179–188 to form the pocket without ligand binding was observed only in one of the 19 structures, the conformational transition may require overcoming some energy barriers. To test this hypothesis we used adiabatic biased molecular dynamics (ABMD) simulations to guide the protein from its unbound to its ligand-bound state (See Star Methods). The biasing force was proportional to the distance from the target structure, and was only applied when the system was diverging from its path towards this target structure. The “distance” was measured by the mean squared distance (MSD) from the bound conformation. With each biasing force we ran three independent 20 ns ABMD simulations seeded with different initial random velocities. Values were recorded at 40 ps intervals, resulting in 502 frames for each trajectory. Frames from all 3 trajectories were combined for analysis. Since small transitional pockets may be formed in this process, druggabilty scores (DSs) were calculated for all pockets within 5 Å of the ligand superimposed from the bound structure, and the maximum DS value was reported. Figures 1D, 1E, and 1F show the distributions of DS values from the simulations with the biasing force constants k=1.0 kcal/mol/Å2, k=10.0 kcal/mol/Å2, and k=60.0 kcal/mol/Å2, respectively (see Star Methods). At both k=1.0 kcal/mol/Å2 and k=10.0 kcal/mol/Å2 the distributions are heavily skewed toward low DS values, and the pocket is getting formed in a small fraction of snapshots only when the much larger force, k=60.0 kcal/mol/Å2, is applied. Figure 2B shows a snapshot at 12 ns from the latter simulation, attesting that loop 179–188 moves toward its position in the ligand-bound structure. We show DS distributions for six more proteins with cryptic sites that almost never form without bound ligands (Figure 3). Pyruvate kinase from Leishmania Mexicana functions as a homotetramer, each subunit with substantial hinge motion between two domains. The active site of the enzyme has DS < 0.5 in all known ligand-free structures (Figure 3A). Similarly to PTP1B, the site is too open in these structures, and becomes well defined only upon binding to ATP and a substrate that cause the closing of a lid-like domain onto the site. We note that pyruvate kinase also has an allosteric site, which binds FDP (fructose 2,6 bisphosphate), almost 40 Å away from the active site, and some of the structures considered in Figure 3A have FDP bound at the allosteric site. Although FDP is known to act as an allosteric effector that increases the rate of the phosphorylation sevenfold (Morgan et al., 2010), Figure 3A reveals that binding at the allosteric site does not affect the DS at the active site. In fact, the increase in the reaction rate is due to stabilization of the tetramer by FDP binding. In agreement with this result, pyruvate kinase is a known example of allostery without conformational change (Morgan et al., 2010).
Figure 3.
Druggability scores (DSs) of unliganded structures of proteins with DS distributions skewed toward the unbound state. The distributions of DS values are shown in dark, light, and medium blue, respectively, for unbound structures, complexes, and mutants. The label shows the 3-letter code of the ligand bound at the cryptic site, and the name of the ligand is shown in parenthesis here. A. Pyruvate kinase (ATP plus oxalate). B. Ricin (pteroic acid). C. Ribonuclease A (NADPH). D. Hepatitis C virus RNA polymerase NS5B (indole-based allosteric inhibitor binding at the thumb domain). E. Protein tyrosine phosphatase 1B (allosteric inhibitor binding at the C-end). F. Fructose-1,6-bisphosphate aldolase enzyme from rabbit muscle (naphthol AS-E phosphate, a competitive inhibitor.).
The active site of ricin (Figure 3B) is closed in most unbound structures because the side chain of Y80 protrudes into the site, stabilized with H-bond to the backbone O of G121. However, the pocket can be affected by antibody binding at a distant site (PDB ID 4KUC), leading to DS > 0.5 in a few structures (Figure 3B). In ribonuclease A the cryptic site binds NADPH (PDB ID 2W5K), but in most apo structures the side chain of H119 protrudes into the site. The only structures with DS > 0.5 are 3EV3, crystallized in 70% t-butanol (DS = 0.547) and 3EIC (DS = 0.697), which is the F120A mutant. The CryptoSite set includes three cryptic allosteric sites of the hepatitis C virus polymerase NS5B. The first site is occupied by a small alpha-helix in the unbound structure 3CJ0 at the tip of the N-terminal loop that connects the fingers and thumb domains. Inhibitors binding at the site displace the helix and prevent intramolecular contacts between the two domains, thereby precluding their coordinated movements during RNA synthesis. Such conformational change does not occur in unliganded structures or, with the exception of a single complex (PDB ID 3BSC), in structures with inhibitors bound at the other two allosteric sites (Figure 3D). In contrast, it appears that inhibitor binding at this first site affects the pockets at the other two allosteric sites, and hence those will be discussed in the third group of proteins.
We have already discussed the cryptic pocket in the active site of PTP1B. The protein also has a cryptic allosteric site located under its C-terminal helix, which is partially unstructured in the inhibitor bound structure (PDB ID 1T49). The inhibitor binds in a very narrow hydrophobic pocket formed by L192, F196 and F280, more than 20 Å away from the active site (Wiesmann et al., 2004). Binding at the active site does not affect the allosteric site that is closed in the unliganded structures with the C-terminal helix intact. The pocket is partially accessible only in two ligand-free structures, both with a bound Mg2+ ion (Figure 3E). We place two more proteins into the group with genuine cryptic sites, fructose-1,6-bisphosphate aldolase (Figure 3F) and the Rho ADP-ribosylating Clostridium botulinum C3 exoenzyme (Figure S1), with details given in Table S1.
Proteins with spontaneously forming pockets at cryptic sites
As the other extreme we were looking for proteins with sites that were considered cryptic in CryptoSite, but have pockets that seem to spontaneously form in some of the ligand-free structures. Such behavior is seen in beta-secretase 1 (BACE1), represented by unbound and bound structures 1W50 and 3IXJ in the CryptoSite set. In the unbound structures the loop comprising residues 71–74 is turned away from the site, making the pocket too open to score as druggable (Figures 4A and 4E). The loop is closing down on the inhibitor in the bound structure 3IXJ (Figures 4B and 4E), resulting in a well-formed pocket that binds the isophthalamide ligand with high affinity (Bjorklund et al., 2010). The analysis of unbound BACE1 structures shows a broad distribution of druggability scores between conformations resembling the unbound and bound forms (Figure 4C), with 39% of structures with DS > 0.5. Apart from a single complex with an antibody bound far from the active site, all BACE1 structures in the PDB are of the wildtype human protein, and the various X-ray structures differ only in the crystal form and the conditions of crystallization. The overall root mean square deviation (RMSD) of many unbound structures with DS > 0.5 is less than 0.5 Å from the bound structure 3IXJ. Thus, the variation in DS values seems to be the consequence of the variation in loop conformation, indicating significant conformational selection as part of the pocket opening. This hypothesis is supported by the results of biased molecular dynamics simulations. Indeed, simulation at k=1.0 kcal/mol/Å2 and started from the apo state shows that the distribution of DS values is already somewhat skewed to the right, i.e., toward a well-formed binding pocket (Figure 4D), and loop 71–74 is getting close to its position in the ligand-bound state as shown by a snapshot at t = 12 ns (Figure 4F).
Figure 4.
Forming the cryptic ligand binding site in beta-secretase 1 (BACE-1). A. Unbound structure 1W50 (partially transparent grey surface). The inhibitor 586 from the ligand-bound structure 3IXJ of BACE-1 is shown for reference (cyan sticks). The flexible loop 71–74 is shown as blue cartoon. B. Structure 3IXJ of BACE-1 (grey surface), co-crystallized with the inhibitor (cyan sticks). The flexible loop 71–74 is shown as blue cartoon. Based on the surface representation, the loop provides the lid of the inhibitor-binding pocket. C. Druggability scores (DSs) of unliganded BACE-1 structures in the PDB. The distributions of DS values are shown in dark and light blue, respectively, for unbound structures and complexes. All structures are of the wildtype protein, and apart from a single structure with an exosite-binding antibody have no ligand bound. D. Distribution of druggability score (DS) values obtained by adiabatic biased molecular dynamics (ABMD) simulations of BACE-1 at k=1.0 (kcal/mol)/Å2. E. Conformational change of BACE-1 upon ligand binding. Loop 71–74 is shown in the unbound structure 1W50 (grey) and in the inhibitor-bound structure 3IXJ (orange). F. Also shown is loop 71–74 from a snapshot at t = 12 ns of the ABMD simulations with k = 1.0 kcal/mol/Å2 (blue).
We have found only five other proteins with similar properties among the 32 studied. The first is bovine beta-lactoglobulin, which binds retinol in the middle of a β-barrel (PDB ID 1GX8). In a few unbound structures loop 84 to 90 acts as a lid that prevents access to the large and well-formed binding site. However, in most structures the flexible loop is open enough to provide access to the site (Figure 5A). Notice that many of the beta-lactoglobulins in the PDB are from other species rather than bovine, but the mutations do not affect the conclusion that the pocket is almost always well formed. Similarly, in a number of apo structures of human thrombin such as chain E of 1 HAG (which is actually a prothrombin), the active site is too open, but becomes well formed in many apo structures. Although there are mutant thrombins within the 95% sequence identity as well as complexes with ligands binding at distant sites, their impacts do not change the conclusion that the active site of thrombin can form spontaneously before any ligands bind (Figure 5B). The fourth protein in the CryptoSite set that does not seem to have a genuine cryptic site is the ligand binding domain of the alpha-L integrin lymphocyte function-associated antigen-1 (LFA-1). The cryptic site of LFA-1 binds an allosteric inhibitor (PDB ID 3BQM). In some structures without this inhibitor the disordered carboxyl end protrudes into the site, but in others the binding pocket is well formed (Figure 5C). The fifth and sixth proteins in this group are the glutamate receptor 2 protein (Figure 5D) and the complex formed by the transforming protein RhoA and RhoGAP (Figure S2), with details given in Table S1.
Figure 5.
Druggability scores (DSs) of unliganded structures of proteins with a cryptic site that is frequently well formed. The ligand bound at the cryptic site is shown in the label and is listed in parenthesis here. The distributions of DS values are shown in dark, light, and medium blue, respectively, for unbound structures, complexes, and mutants. A. Bovine beta-lactoglobulin (retinol). B. Thrombin (active site inhibitor 121). C. Integrin lymphocyte function-associated antigen-1 (LFA-1) ligand binding (I) domain (inhibitor BQM). D. Glutamate receptor 2 (competitive antagonist ATPO binding to the core of the receptor). \
Impact of mutations on cryptic site opening: TEM-1 β-lactamase
In the remaining 18 proteins the druggability score at the cryptic site substantially depends on mutations and/or on the binding of ligands or proteins at distant sites. Before discussing the other proteins, we focus on the impact of mutations on the opening of the cryptic site in TEM-1 β-lactamase, which is a textbook case of cryptic allosteric sites (Horn and Shoichet, 2004). The active site of TEM-1, with the catalytic residues S70, K73, and K234, is located between the two domains of the protein. In the unbound structures such as 1JWP helices H11 (residues 218–230) and H12 (residues 271–289), located above the active site on different domains, are close to each other (Figure 6A). The X-ray structure 1PZO showed two small inhibitors bound to this region by forcing apart the two helixes (Figures 6B and 7A). Although the center of this cryptic site is 16 Å from the center of the active site of the enzyme, one of the inhibitors has a second binding mode that partly occludes the active site near residues S235, G245, and G236 (Horn and Shoichet, 2004). However, it appears that binding to this second site would only be possible to a structure formed by inhibitor binding to the first, core site. This “opening” of the secondary structure results in major backbone and side-chain rearrangement that exposes mainly hydrophobic surface to the compound.
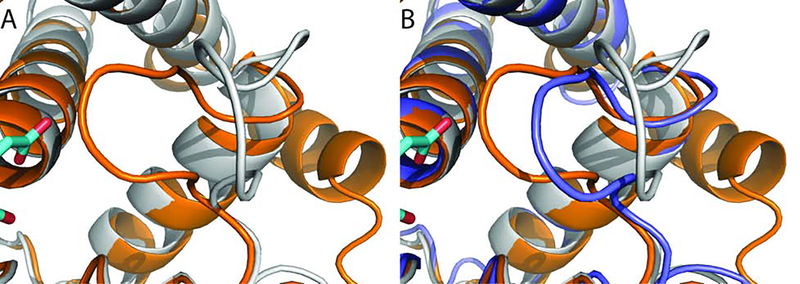
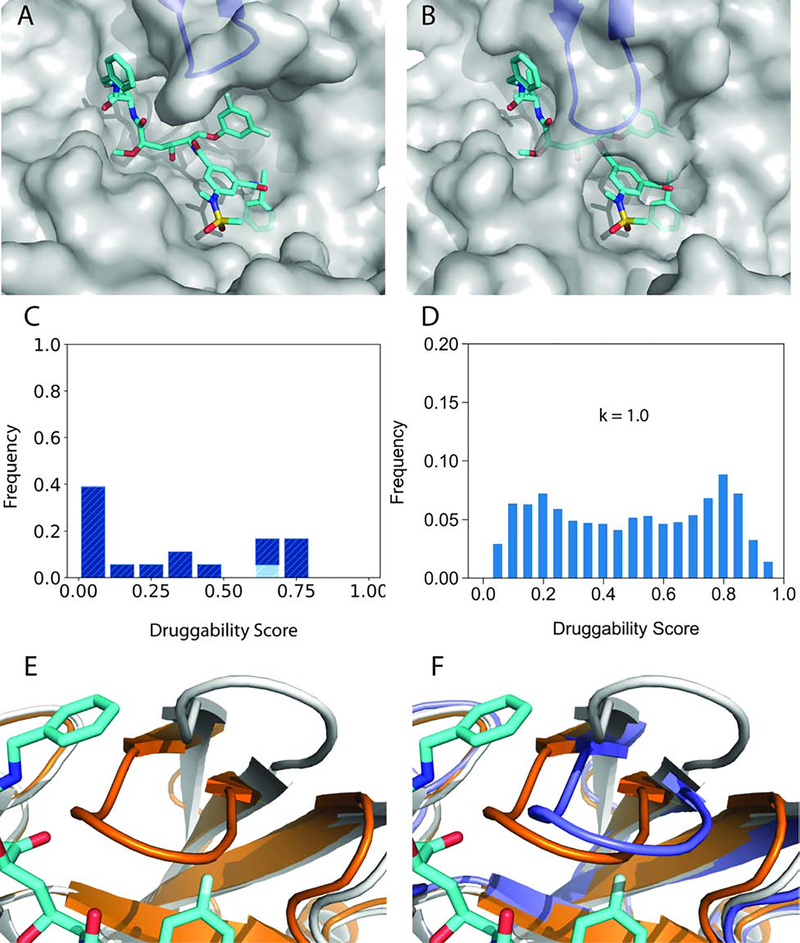
Figure 6.
Opening the cryptic allosteric site in TEM-1 β-lactamase. A. Unbound structure 1JWP of TEM-1 β-lactamase (grey cartoon). Two small allosteric inhibitors from the structure 1PZO are shown for reference (cyan sticks). B. Inhibitor-bound structure 1PZO with two allosteric inhibitors, demonstrating that the two helices lining the allosteric site move apart. C. Druggability scores (DSs) of unliganded TEM-1 β-lactamase structures in the PDB. The distributions of DS values are shown in dark, light, and medium blue, respectively, for unbound structures, complexes, and mutants. Here “complex” means a protein or ligand binding at a distant site. D. Distribution of druggability score (DS) values obtained by adiabatic biased molecular dynamics (ABMD) simulations of TEM-1 β-lactamase at k=1.0 (kcal/mol)/Å2. E. Distribution of DS values obtained by ABMD simulations of TEM-1 β-lactamase at k=10.0 (kcal/mol)/Å2. F. Distribution of DS values obtained by ABMD simulations of TEM-1 β-lactamase at k=30.0 (kcal/mol)/Å2.
Figure 7.
Conformational change and a snapshot from the ABMD simulation of TEM-1 β-lactamase. All structures are shown in cartoon representation. A. Unbound structure 1JWP (grey), superimposed with the bound structure 1PZO (orange). The two allosteric inhibitors bound to 1PZO are shown in cyan. B. Helix H11 from a snapshot at t = 20 ns of the ABMD simulations of TEM-1 β-lactamase with k = 30.0 kcal/mol/Å2 (blue). H11 partially unfolds to open a small but druggable pocket for the binding of ligands.
As shown in Figure 6C, the pocket at the cryptic site is deemed druggable (DS > 0.5) by Fpocket in over 50% of the unliganded lactamase structures. However, 19 of the 21 apo structures have some mutated amino acid residues. Introducing mutations represents the main mechanism by which opportunistic and pathogenic bacteria become resistant to β-lactam antibiotics, and hence many mutants have been generated. A substantial number of studies examined how these mutations affect antibiotic resistance and stability (Abriata et al., 2012; Brown et al., 2010; Dellus-Gur et al., 2013; Kather et al., 2008; Marciano et al., 2008; Modi and Ozkan, 2018; Orencia et al., 2001; Speck et al., 2012; Stec et al., 2005; Thomas et al., 2005; Wang et al., 2002a, b). Here we consider a different question and study how the mutations affect the druggability of the allosteric site. In Table 2 we list the mutations, the DS value, and the melting temperature Tm if available in the literature. These results reveal that the allosteric site is essentially closed, resulting in small DS values, in the TEM β-lactamase variants with the most stabilizing mutations. These variants include the so-called stabilized v.13 version with mutations A42G, N52A, I84V, R120G, M182T, L201A, and T265M (PDB ID 4IBX) (Dellus-Gur et al., 2013), and a second stabilized variant with the mutations P62S, V80I, E147G, M182T, L201P, A224V, I247V, and R275R (PDB ID 3DTM) (Kather et al., 2008), both resulting in a melting temperature Tm around 69°C. We did not find data for the variant with the mutations M182T and V184A, but M182T alone yields Tm = 63.2°C. For comparison, the melting temperature of the wildtype TEM-1 (PDB ID 1ZG4) is Tm = 58.5°C. We did not find Tm value for S70G, but removal of catalytic residues is known to increase stability (Knies et al., 2017). All these stabilized mutants have druggability scores DS < 0.2. The other mutants in Table 2 have both destabilizing and stabilizing mutations that keep Tm in the 52°C to 59°C range (Knies et al., 2017). It is known that mutations improving antibiotic resistance activity are generally destabilizing, but these proteins also acquire additional mutations that restore stability (Latallo et al., 2017; Wang et al., 2002a; Zimmerman et al., 2017). Such mutants include TEM-76, TEM-84, and TEM-52. The pocket in these proteins tends to be more open, with DS > 0.2, increasing to DS > 0.8 in the very unstable mutant L201P. Note that the two wildtype TEM-1 structures in Table 2, 1ZG4 and chain E of 4OQG, have very different druggability values. For 1ZG4 the value DS = 0.390 is in good agreement with the melting temperature Tm = 58.5°C, but DS = 0.629 calculated for 4OQG is too high. In fact, the unit cell for 4OQG includes six chains, and five of the chains have an inhibitor bound at the active sites. Although no bound inhibitor is seen in chain E considered in Table 2, it is very likely that the pocket is still affected, and hence the high DS value is an error. Thus, it appears that the mutations that reduce stability generally also yield a more open allosteric pocket. Since the allosteric site is located between the two domains of the protein, and the interactions between the domains affect both the stability of the protein and the volume of the cryptic site, this observation is not difficult to explain.
Table 2.
TEM β-lactamase structures, druggability scores, mutations, and melting temperatures
| PDB ID | DSa | Tb | Mutation (E. Coli) |
|---|---|---|---|
| 4MEZ_B | 0.032 | M | (M68L, M69T) |
| 4MEZ_A | 0.037 | M | (M68L, M69T) |
| 4IBX_E | 0.057 | M | TEM v.13 (A42G, N52A, I84V, R120G, M182T, L201A, T265M), Tm = 69.0°C |
| 1 ZG6_A | 0.076 | M | (S70G) Catalytic residue mutation expected to improve stability |
| 3DTM_A | 0.129 | M | (P62S, V80I, E147G, M182T, L201P, A224V, I247V, R275R), Tm = 69.2 °C |
| 1JWP_A | 0.186 | M | (M182T, V184A) Strong stabilization, M182T alone yields Tm = 63.2°C |
| 1YT4_A | 0.237 | M | TEM-76 (S130G), Tm = 52.3 °C |
| 1CK3_A | 0.325 | M | TEM-84 (N276D), Tm = 58.0 °C |
| 1ZG4_A | 0.390 | U | None, WT TEM1 beta lactamase, Tm = 58.5 °C |
| 4GKU_A | 0.418 | M | (I84V, V184A), V184A on its own yields Tm = 58.1 °C |
| 3TOI_B | 0.541 | M | First 15 residues removed & (I56V, R120G, M182T, T195S, I208M, A224V, R241 H,T265M), Tm = 59.0 °C |
| 1HTZ_E | 0.571 | M | TEM52 (E104K, M182T, G238S), Tm = 55.6 °C |
| 1HTZ_C | 0.599 | M | TEM52 (E104K, M182T, G238S), Tm = 55.6 °C |
| 1HTZ_B | 0.612 | M | TEM52 (E104K, M182T, G238S), Tm = 55.6 °C |
| 4OQG_E | 0.629 | U | None, WT TEM-1 beta-lactamase: no ligand in chain E, Tm = 58.5 °C |
| 1HTZ_A | 0.640 | M | TEM52 (E104K, M182T, G238S), Tm = 55.6 °C |
| 1HTZ_D | 0.640 | M | TEM52 (E104K, M182T, G238S), Tm = 55.6 °C |
| 3TOI_A | 0.669 | M | First 15 residues removed & (I56V, R120G, M182T, T195S, I208M, A224V, R241 H,T265M), Tm = 59.0 °C |
| 1 LI9_A | 0.698 | M | TEM-34 (M69V), Tm almost identical to or greater than that of TEM-1 |
| 1LHY_A | 0.718 | M | TEM-30 (R244S), Destabilizing |
| 3CMZ_A | 0.849 | M | (L201P), Tm = 53.4 °C |
Druggability score.
Type: M – mutant, U – unbound wild type protein.
Since only two structures are available for the unliganded wild type TEM-1 β-lactamase, simple inspection does not provide information on the forces needed to open the site, and performing MD simulations is particularly important. Markov state models (MSMs) built from hundreds of microseconds of MD simulations have shown that the allosteric pocket was at least partially open for 53% of the simulation time (Bowman and Geissler, 2012). The cryptic pocket identified by the MSM simulations was also used for the design of allosteric modulators (Hart et al., 2017). In contrast, MD simulations of the same protein by Gervasio and co-workers (Oleinikovas et al., 2016) using parallel tempering failed to show appreciable opening of the site when starting from the apo crystal structure. In order to reliably capture the conformational transition from the closed to open allosteric site we have applied the ABMD method to the M182T variant of the β-lactamase. Simulations at k=1.0 kcal/mol/Å2 show that the pocket is already formed in some fraction of conformations, in good agreement with the MSM results (Bowman and Geissler, 2012). However, the pockets are only partially open, with the peak DS value around 0.6 (Figure 6D). It is interesting that the site has a “binary” behavior having either closed or partially open states with limited intermediate conformations. This is in contrast to the site in BACE1, which has an almost flat distribution of DS values (Figure 4D). Increasing to biasing force to k=10.0 kcal/mol/Å2 and then to k=30.0 kcal/mol/Å2 increases the fraction of partially open sites (Figures 6E and 6F). However, even a high DS value does not necessarily mean that the allosteric site is fully open. For example, Figure 7B shows a snapshot at t = 20 ns from the simulation with k=30.0 kcal/mol/Å2. Although this structure has a pocket with DS = 0.8 close to the site that binds the allosteric inhibitors, the pocket is created by some unfolding of the amino end of helix H11, and it can only partially accommodate one of the inhibitors.
Impact of mutations or off-site binding in other proteins
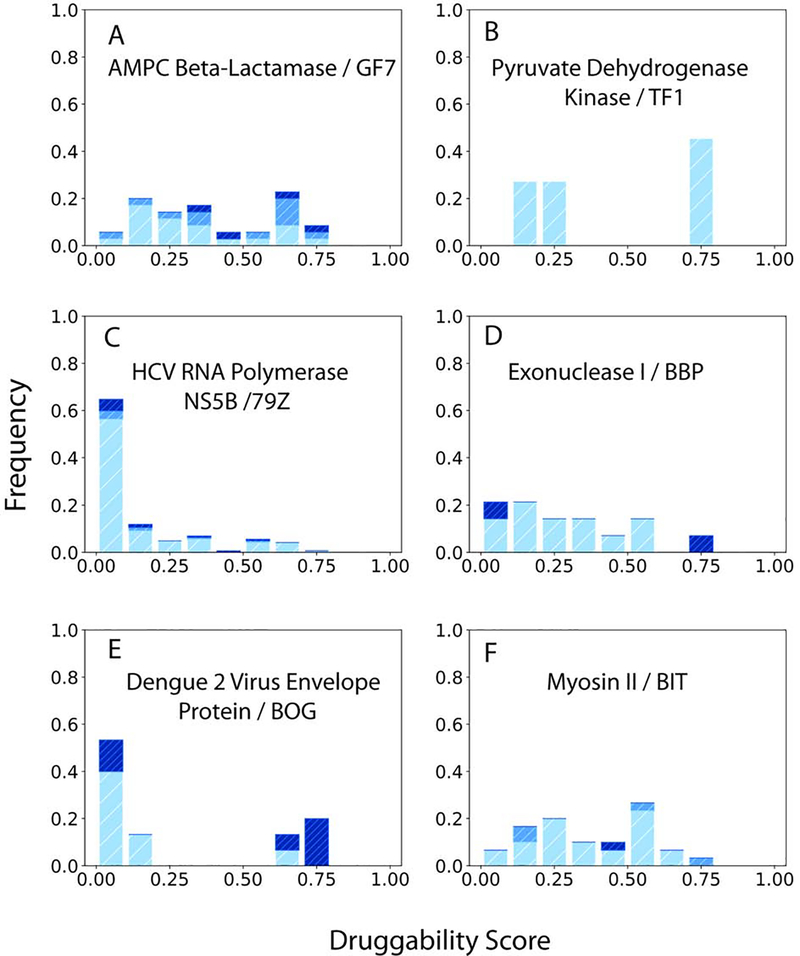
Table 1 shows 17 more proteins with cryptic sites that seem to form in some structures due to mutation, binding of ligands or proteins at locations distant from the cryptic site, or simply due to changes in the conditions of crystallization. Here we describe for six of these proteins why forming the cryptic site depends on such additional factors.
The first example is AMPc beta-lactamase with a mechanism of cryptic site opening that is similar to that of TEM-1 beta-lactamase, although the two proteins exhibit limited sequence or structure similarity. In many unbound structures of the AMPc beta-lactamase residues 289–293 form a small helix protruding into the site. In the presence of fragment-sized inhibitors the same residues form a loop allowing for ligand binding (PDB ID 3GQZ). Although the active site is more than 8 Å from the allosteric site, the two sites are in the same crevice, and binding of active site inhibitors seems to affect the opening of the allosteric site, which can also be impacted by mutations (Figure 8A).
The second protein in this group is human pyruvate dehydrogenase kinase, which has a non-competitive (allosteric) inhibitor site, 33 Å from the ADP binding site (PDB ID 2BU2). Upon binding by the inhibitor TF1, the helix alpha-2 shifts by a hinge motion. The loop of residues 34–37 is found to be very flexible in all structures determined to date. This may be necessary to facilitate the hinge movement of the helix. In spite of the large distance, the opening of the cryptic site is clearly affected by binding at the ADP site, since DS<0.3 in all ADP-bound structures but DS>0.7 in all structures with bound ADP-competitive inhibitors, and thus the binding of the inhibitors helps to open the allosteric site (Figure 8B, see also Data S1).
We have already discussed one allosteric site of the hepatitis C virus polymerase NS5B located between the fingers and thumb domains (PDB ID 2BRL). A second site is near the polymerase active site in an elongated, predominantly hydrophobic pocket, between the primer grip motif (residues 364 –369) and the central sheet (strands 214–219, 319–325, and 310–316), in the core of the palm domain (PDB ID 3FQK). Inhibitor binding at the third site (PDB ID 2GIR) causes a slight shift of residue L419 and a significant rotamer change for M423 relative to the apo-enzyme conformation (Le Pogam et al., 2006). Although the DS distributions for both sites are skewed toward low values (Figures 7C and S1C), it seems that opening is somewhat affected by inhibitor binding at the first site, and the pockets are already formed in a number of structures.
At its cryptic site exodeoxyribonuclease I (ExoI) binds BCBP (PDB ID 3HL8), which inhibits its interaction with bacterial single-stranded DNA-binding proteins. In many unbound structures W245 protrudes into the weak surface site. The pocket is generally not well formed, but there are a few exceptions. Almost all structures are co-crystallized with various oligonucleotides, and such interactions affect the cryptic site, but the highest DS value occurs in a ligand-free structure (Figure 8D).
The cryptic site in the Dengue 2 virus envelope protein is located between two domains and binds the detergent n-octyl--D-glucoside (PDB ID 1OKE). Spontaneous variations may occur between open and closed states. The key change is the local rearrangement of the hairpin formed by residues 268–280, and the concomitant opening up of a hydrophobic pocket. The most open pockets occur in unbound structures, whereas DS is reduced by the binding of antibodies at distant sites (Figure 8E), motivating the placement of the protein in this category.
In Myosin II the cryptic site binds the inhibitor blebbistatin in a very narrow cavity. In the unbound structures the side chains of L262 and Y634 protrude into the pocket. Changes in backbone are small. Many structures bind nucleotides at a location far from the cryptic site. In addition, the protein has a different (allosteric) inhibitor binding site closer to the surface. The binding of these ligands is likely to affect the blebbistatin binding pocket deep in the protein (Figure 8F).
Figure 8.
Druggability scores (DSs) of unliganded structures of proteins with cryptic sites impacted by mutations or binding at distant sites. The ligand bound at the cryptic site shown in parenthesis. The distributions of DS values are shown in dark, light, and medium blue, respectively, for unbound structures, complexes, and mutants. A. AMPc beta-lactamase (Inhibitor GF7). B. Human pyruvate dehydrogenase kinase (Allosteric inhibitor TF1). C. Hepatitis C virus RNA polymerase NS5B (Inhibitor 79Z binding near the active site). D. Exodeoxyribonuclease I (Inhibitor BCBP). E. Dengue 2 virus envelope protein (Detergent n-octyl--D-glucoside). F. Myosin II (inhibitor blebbistatin).
The other 11 proteins in this group are monomeric actin, fructose 1,6-bisphosphatase, maltodextrin/maltose binding protein, the MurA dead-end complex, acid-beta-glucosidase, biotin carboxylase, glutamate receptor 2, androgen receptor, p38 map kinase, and aspartate transcarbamylase. Details for these proteins are given in Table S1, and the DS histograms are shown in Figures S3, S4, and S5, all related to Table 1 and Figure 8.
DISCUSSION
The binding of a ligand molecule is often accompanied by conformational changes of the protein. This is definitely the case if the binding site is cryptic, thus it is not detectable in the unliganded protein. A central question is whether the ligand induces the conformational change via induced-fit, or rather selects and stabilizes a complementary conformation from a pre-existing equilibrium of ground and excited states of the protein via conformational selection (Weikl and von Deuster, 2009). Since the binding proceeds from the free energy minimum of the separate target protein to the free energy minimum of the receptor-ligand complex, the distinction is kinetic rather than thermodynamic. However, the free energy landscape of the protein determines the pathway of the association. In fact, the unbounds state is always an ensemble of conformations (Hilser et al., 2006). If conformations without the pocket formed are at deep free energy minima, then the probability of pocket formation without ligand binding is small. On the other extreme, if the landscape includes minima leading to conformations with pockets formed, then the binding site is most likely cryptic only in a certain fraction of the conformational ensemble.
Molecular dynamics (MD) is increasingly considered as a valuable tool to characterize conformational ensembles of macromolecules. One of the major strengths of this approach is that it provides both thermodynamic and kinetic information (Knoverek et al., 2019). However, as discussed for TEM-1 β-lactamase, the results of simulations depend on a multiplicity of factors (Bowman and Geissler, 2012; Oleinikovas et al., 2016), including the force field parameters (Childers and Daggett, 2018) and the strategy of sampling (Zimmerman et al., 2018). In addition, each timestep is on the order of a femtosecond, while many of the biological processes of interest take a millisecond or longer. Performing over 1012 iterations is computationally expensive, and limits the applicability of the method. The use of Markov state models (MSMs) enables ultra-long MD simulations (Lane et al., 2011), and helps to elucidate functional conformational changes (Chodera and Noe, 2014; Huang et al., 2018). In spite of recent development, MSMs still require substantial computational resources and have been applied only to a few proteins for the analysis of cryptic site opening (Bowman and Geissler, 2012; Knoverek et al., 2019; Maurer et al., 2012; Porter et al., 2019).
The main goal of this paper was to consider unliganded X-ray structures of proteins with validated cryptic sites and to study whether the sites remain always cryptic without ligand binding, or pockets already form in some of the structures. The simple approach of documenting the druggability of pockets at cryptic sites in 32 proteins enabled us to arrive at some fairly general conclusions. First, we have shown that few proteins have even approximately “genuine” cryptic pockets that are unlikely to form without ligand binding. Second, proteins on the other extreme, with spontaneously opening and closing cryptic sites, are also rare. The largest group includes proteins that, under some conditions, have a cryptic pocket with very low druggability, but easily form a more druggable pocket if the conditions change. This behavior is in good agreement with the assumptions that the native state of the protein is defined by an ensemble of conformational states at free energy minima with similar energy levels (Hilser et al., 2006). Even moderate perturbations can change the free energy landscape and thereby impact the distribution of residence probabilities at the various states, also affecting the druggability of pocket at the cryptic site. The practical implication of this finding is that in order to discover cryptic allosteric site it is always advisable to investigate all homologous proteins, As shown for TEM-1 β-lactamase, it is particularly useful to study slightly destabilized versions of a protein. The conclusions from the analysis of X-ray structures were confirmed by adiabatic biased molecular dynamics (ABMD) simulations (Harvey and Gabb, 1993; Marchi and Ballone, 1999; Paci and Karplus, 1999), applied to one protein from each of the three groups.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Sandor Vajda (vaida@bu.edu).
This study did not generate new unique reagents.
METHOD DETAILS
Adiabatic Biased Molecular Dynamics
We applied adiabatic biased molecular dynamics (ABMD) simulations to three proteins, PTP1B, beta-secretase 1, and TEM-1 β-lactamase, all with well-validated cryptic sites. The simulations were performed using the GPU version of Desmond (Bowers et al., 2006) running on Nvidia GTX 1080 graphics cards on a 4-GPU desktop computer. We used the OPLSAA_2005 force field and SPC water in our simulations. Every simulation started with an equilibration protocol including the following steps: (1) Brownian dynamics NVT, T = 10 K, Δt = 1 fs, restraints on solute heavy atoms, t = 100 ps, (2) NVT, T = 10 K, Δt = 1 fs, restraints on solute heavy atoms, t = 12 ps, (3) NPT, T = 10 K, restraints on solute heavy atoms, t = 12ps, (4) NPT, T = 310 K, Δt = 2.5 fs, restraints on solute heavy atoms, t = 12ps, and (5) NPT, T = 310 K, Δt = 2.5 fs, no restraints, t = 24 ps. The production runs were configured NPT using Nose-Hoover chain with a 1 ps relaxation time for thermostat (single temperature group), and Martyna-Tobias-Klein barostat with 2 ps relaxation time and isotropic coupling. We utilized a RESPA integrator with Δt = 2.5 fs for bonded and near nonbonded interactions and Δt = 7.5 fs for far nonbonded interactions. The particle-mesh Ewald algorithm was used with periodic boundary conditions to compute long-range electrostatic interactions with the real space cutoff set to 9 Å for both electrostatic and van der Waals interactions. Water molecules were constrained with SHAKE.
The ABMD simulations were used to guide a protein molecule from apo to holo structure (Harvey and Gabb, 1993; Marchi and Ballone, 1999; Paci and Karplus, 1999). ABMD is similar to targeted molecular dynamics (TMD) (Schlitter et al., 1994), but it is more gentle because the biasing force is only applied when the system is diverging from its path towards the target structure. The “distance” from the target ligand-bound conformation is measured by RMSD and when the system moves toward the target autonomously, no force is applied. The time-dependent ABMD/RMSD biasing potential, U is a function of the conformation of the protein, R, and at a time, t, is given by:
where H is a Heaviside function (H(X) = 1 if X > 0 and H(X) = 0 otherwise), k is a force constant and X(R, t) is:
d(R1, R2 ) denotes the RMSD between conformations R1 and R2, RT is the target structure. By varying the value of the force constant k we were able to assess, qualitatively, the extent of how energetically challenged different conformational transitions were. For each system we ran three independent, short ABMD simulations (20 ns each, seeded with different initial random velocities). Values were recorded at 40 ps intervals, resulting in 502 frames for each trajectory. Frames from all 3 trajectories were combined for analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data Set
The starting point of this paper is a representative set of X-ray structures of proteins with validated cryptic binding sites. This set was originally selected for training and testing the CryptoSite cryptic site prediction protocol (Cimermancic et al., 2016), and hence is referred here as the CryptoSite set. Considering 504,647 candidate pairs of ligand-bound structures with their unbound counterparts, Cimermancic et al. used pocket detection algorithms to retain only pairs with a small pocket score in the unbound form and a substantially larger score in the bound form. Manual inspection of the structures resulted in a dataset of 93 bound-unbound pairs in which each unbound structure had a site considered cryptic due to its low pocket score, and each bound structure had a biologically relevant ligand bound at the site. While the original CryptoSite set included only one unbound structure in each pair, in order to study the information provided by different unbound structures of a given protein, for each bound structure in the set we added all unbound structures with at least 95% sequence identity that were available in the Protein Data Bank (Beglov et al., 2018). Structures determined by NMR or cryo-EM, as well as X-ray structures with lower than 3.5 Å resolution were excluded. The structures were superimposed on the ligand-bound structure and structures with any ligand within 5 Å of the cryptic site ligand were also excluded. Finally we removed all proteins that had less than 10 structures satisfying the above criteria. The number of such unbound structures varied from 10 to 249 per protein (Table 1).
Identification of binding pockets using the Fpocket program
Fpocket is an open source pocket detection package (Le Guilloux et al., 2009; Schmidtke et al., 2010). The method is based on the concept of alpha spheres. An alpha sphere is a sphere that contacts four atoms on its boundary and contains no internal atom. For a protein, very small spheres are located within the protein, large spheres at the exterior, and clefts and cavities correspond to spheres of intermediate radii. Thus, it is possible to filter the ensemble of alpha spheres defined from the atoms of a protein according to some minimal and maximal radii values in order to address pocket detection. Accordingly, the Fpocket algorithm includes three steps. During the first step the whole ensemble of alpha spheres is determined from the protein structure. Fpocket returns a pre-filtered collection of spheres. The second step consists in identifying clusters of spheres close together, to identify pockets, and to remove clusters of poor interest. The third step calculates properties from the atoms of the pocket, in order to score each pocket (Le Guilloux et al., 2009; Schmidtke et al., 2010). The Fpocket program was used with default parameter to identify the ligand binding pockets of the ligand-bound and all unbound X-ray structures of the proteins in the data set. Fpocket was also used to determine the pockets of the structures generated along the trajectories of the ABMD simulations of PTP1B, beta-secretase 1, and TEM-1 β-lactamase. For each value of the force constants k, the program was applied to each of the 1506 frames collected for each of the three proteins. Fpocket generally identifies multiple ligand binding pockets. All pockets within 5 Å of the ligand superimposed from the bound structure were retained for further analysis.
Calculation of the Fpocket druggability scores
The Fpocket druggability score (DS) is a numerical value between 0 and 1 associated to each pocket (Schmidtke and Barril, 2010). This score intends to assess the likeliness of the pocket to bind a small drug like molecule. A low score indicates that drug like molecules are likely to not bind to this pocket. A druggability score of DS = 0.5 (the threshold) indicates that binding of prodrugs or druglike molecules can be possible. DS = 1 indicates that binding of druglike molecules is very likely. The descriptors for calculating the DS value for a pocket are the normalized mean local hydrophobic density, a hydrophobicity score based on a residue based hydrophobicity scale, and a normalized polarity score (Schmidtke and Barril, 2010). To calculate a druggability score, first logistic models were derived for each pocket descriptor. In the next step, predictions coming from these “one descriptor based” logistic models were associated in one common logistic model where statistically nonsignificant descriptors or unstable models were filtered out. To train and validate the druggability score, a set consisting of proteins with druggable cavities, other proteins with nondruggable cavities, and a large number of decoys was used. This set was split in two. The first half of the data was used to train the model by determining the weighting coefficients of the separate contributions to the overall score. Training and internal validation was performed using a 10-fold bootstrap with a one-third/two-third training/validation ratio. The second half of the data was reserved for external validation (selected by random before the bootstrap run). Receiver operator characteristics (ROC) and derivative figures were produced. Druggability scores were calculated for the pockets identified by Fpocket in the X-ray structures and structures generated by the ABMD simulations as described in the previous section. The calculations were restricted to pockets found within 5 A of the ligand superimposed from the bound structure. For each structure, the pocket with the maximum DS value was selected as the predicted ligand binding site, and this maximal DS value was used to create the histogram throughout the paper. All DS values are reported in the file Data S1.
DATA AND CODE AVAILABILITY
Data Sl.xlsx, druggability scores of all proteins studied, related to Table 1. All duggability score (DS) values obtained the by Fpocket program are listed in the file. Each line of the data set provides names of apo and holo structures from the CryptoSite dataset, the PDB ID of the structure of the selected unbound homolog, the distance of the pocket from the ligand at the cryptic site in the holo structure, the Fpocket druggability score (DS), indication whether the structure is unbound (U), mutant (M), or a complex (C), and the name of the protein from the PDB file.
The Fpocket program used here is available at http://fpocket.sourceforge.net/.
Supplementary Material
Highlights.
X-ray structures of proteins provide ample information on cryptic site opening.
Biased MD and druggability scores confirm results from X-ray structures.
“Genuine” and spontaneously formed cryptic sites are both rare.
In most proteins the pocket formation is impacted by mutations or off-site binding.
ACKNOWLEDGMENTS
This investigation was supported by grant R35-GM118078 from the National Institute of General Medical Sciences.
Footnotes
DECLARATION OF INTEREST
Current affiliation of Istvan Kolossvary is Silicon Therapeutics, Boston, MA, 02215. Dmitri Beglov is an employee and shareholder of Acpharis Inc., Holliston, MA 01746, in addition to his affiliation with Boston University. Sandor Vajda is a shareholder of Acpharis Inc.
Supplementary Information
Table S1, related to Table 1 in the main text, provides additioinal information on the mechanism of cryptic site opening for each of the 32 proteins considered in Table 1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abriata LA, Salverda ML, and Tomatis PE (2012). Sequence-function-stability relationships in proteins from datasets of functionally annotated variants: the case of TEM beta-lactamases. FEBS Lett. 586, 3330–3335. [DOI] [PubMed] [Google Scholar]
- Acker TM, Gable JE, Bohn MF, Jaishankar P, Thompson MC, Fraser JS, Renslo AR, and Craik CS (2017). Allosteric inhibitors, crystallography, and comparative analysis reveal network of coordinated movement across human herpesvirus proteases. J. Am. Chem. Soc 139, 11650–11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglov D, Hall DR, Wakefield AE, Luo L, Allen KN, Kozakov D, Whitty A, and Vajda S (2018). Exploring the structural origins of cryptic sites on proteins. Proc. Natl. Acad. Sci. U S A 115, E3416–E3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund C, Oscarson S, Benkestock K, Borkakoti N, Jansson K, Lindberg J, Vrang L, Hallberg A, Rosenquist A, and Samuelsson B (2010). Design and synthesis of potent and selective BACE-1 inhibitors. J. Med. Chem 53, 1458–1464. [DOI] [PubMed] [Google Scholar]
- Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, and Sacerdoti FD (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters. In SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (IEEE), pp. 43–43. [Google Scholar]
- Bowman GR, Bolin ER, Hart KM, Maguire BC, and Marqusee S (2015). Discovery of multiple hidden allosteric sites by combining Markov state models and experiments. Proc. Natl. Acad. Sci. U S A 112, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, and Geissler PL (2012). Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc. Natl. Acad. Sci. U S A 109, 11681–11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, Pennington JM, Huang W, Ayvaz T, and Palzkill T (2010). Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM beta-lactamases. J. Mol. Biol 404, 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers MC, and Daggett V (2018). Validating molecular dynamics simulations against experimental observables in light of underlying conformational ensembles. J. Phys. Chem. B 122, 6673–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodera JD, and Noé F (2014). Markov state models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol 25, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P, Weinkam P, Rettenmaier TJ, Bichmann L, Keedy DA, Woldeyes RA, Schneidman-Duhovny D, Demerdash ON, Mitchell JC, Wells JA, et al. (2016). CryptoSite: Expanding the druggable proteome by characterization and prediction of cryptic binding sites. J. Mol. Biol 428, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002). Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol 12, 14–20. [DOI] [PubMed] [Google Scholar]
- Dellus-Gur E, Toth-Petroczy A, Elias M, and Tawfik DS (2013). What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol 425, 2609–2621. [DOI] [PubMed] [Google Scholar]
- Durrant JD, Keranen H, Wilson BA, and McCammon JA (2010). Computational identification of uncharacterized cruzain binding sites. PLoS Negl. Trop. Dis 4, e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant JD, and McCammon JA (2011). Molecular dynamics simulations and drug discovery. BMC Biol 9, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Lukman S, Hocker HJ, Sayyah J, Brown JH, McCammon JA, and Gorfe AA (2011). Novel allosteric sites on Ras for lead generation. PLoS One 6, e25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DR, Kozakov D, Whitty A, and Vajda S (2015). Lessons from hot spot analysis for fragment-based drug discovery. Trends Pharmacol. Sci 36, 724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart KM, Moeder KE, Ho CMW, Zimmerman MI, Frederick TE, and Bowman GR (2017). Designing small molecules to target cryptic pockets yields both positive and negative allosteric modulators. PLoS One 12, e0178678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SC, and Gabb HA (1993). Conformational Transitions using molecular-dynamics with minimum biasing. Biopolymers 33, 1167–1172. [DOI] [PubMed] [Google Scholar]
- Hilser VJ, Garcia-Moreno EB, Oas TG, Kapp G, and Whitten ST (2006). A statistical thermodynamic model of the protein ensemble. Chem. Rev 106, 1545–1558. [DOI] [PubMed] [Google Scholar]
- Hilser VJ, Wrabl JO, and Motlagh HN (2012). Structural and energetic basis of allostery. Annu. Rev. Biophys. 41, 585–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn JR, and Shoichet BK (2004). Allosteric inhibition through core disruption. J. Mol. Biol 336, 1283–1291. [DOI] [PubMed] [Google Scholar]
- Huang M, Song K, Liu X, Lu S, Shen Q, Wang R, Gao J, Hong Y, Li Q, Ni D, et al. (2018). AlloFinder: a strategy for allosteric modulator discovery and allosterome analyses. Nucleic Acids Res. 46, W451–W458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather I, Jakob RP, Dobbek H, and Schmid FX (2008). Increased folding stability of TEM-1 beta-lactamase by in vitro selection. J. Mol. Biol 383, 238–251. [DOI] [PubMed] [Google Scholar]
- Knies JL, Cai F, and Weinreich DM (2017). Enzyme efficiency but not thermostability drives cefotaxime resistance evolution in TEM-1 beta-lactamase. Mol. Biol. Evol. 34, 1040–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoverek CR, Amarasinghe GK, and Bowman GR (2019). Advanced methods for accessing protein shape-shifting present new therapeutic opportunities. Trends. Biochem. Sci. 44, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, and Vajda S (2015a). The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc 10, 733–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, and Vajda S (2011). Structural conservation of druggable hot spots in protein-protein interfaces. Proc. Natl. Acad. Sci. U S A 108, 13528–13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Napoleon RL, Yueh C, Whitty A, and Vajda S (2015b). New frontiers in druggability. J. Med. Chem. 58, 9063–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TJ, Bowman GR, Beauchamp K, Voelz VA, and Pande VS (2011). Markov state model reveals folding and functional dynamics in ultra-long MD trajectories. J. Am. Chem. Soc 133, 18413–18419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latallo MJ, Cortina GA, Faham S, Nakamoto RK, and Kasson PM (2017). Predicting allosteric mutants that increase activity of a major antibiotic resistance enzyme. Chem. Sci 8, 6484–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guilloux V, Schmidtke P, and Tuffery P (2009). Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pogam S, Kang H, Harris SF, Leveque V, Giannetti AM, Ali S, Jiang WR, Rajyaguru S, Tavares G, Oshiro C, et al. (2006). Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol 80, 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi M, and Ballone P (1999). Adiabatic bias molecular dynamics: A method to navigate the conformational space of complex molecular systems. J. Chem. Phys 110, 3697–3702. [Google Scholar]
- Marciano DC, Pennington JM, Wang X, Wang J, Chen Y, Thomas VL, Shoichet BK, and Palzkill T (2008). Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase. J. Mol. Biol 384, 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. (2012). Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U S A 109, 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi T, and Ozkan SB (2018). Mutations Utilize Dynamic Allostery to Confer Resistance in TEM-1 beta-lactamase. Int. J. Mol. Sci 19, 3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HP, McNae IW, Nowicki MW, Hannaert V, Michels PA, Fothergill-Gilmore LA, and Walkinshaw MD (2010). Allosteric mechanism of pyruvate kinase from Leishmania mexicana uses a rock and lock model. J. Biol. Chem 285, 12892–12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, and Hilser VJ (2014). The ensemble nature of allostery. Nature 508, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinikovas V, Saladino G, Cossins BP, and Gervasio FL (2016). Understanding cryptic pocket formation in protein targets by enhanced sampling simulations. J. Am. Chem. Soc 138, 14257–14263. [DOI] [PubMed] [Google Scholar]
- Orencia MC, Yoon JS, Ness JE, Stemmer WP, and Stevens RC (2001). Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol 8, 238–242. [DOI] [PubMed] [Google Scholar]
- Paci E, and Karplus M (1999). Forced unfolding of fibronectin type 3 modules: An analysis by biased molecular dynamics simulations. J. Mol. Biol 288, 441–459. [DOI] [PubMed] [Google Scholar]
- Perot S, Sperandio O, Miteva MA, Camproux AC, and Villoutreix BO (2010). Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Disc. Today 15, 656–667. [DOI] [PubMed] [Google Scholar]
- Porter JR, Moeder KE, Sibbald CA, Zimmerman MI, Hart KM, Greenberg MJ, and Bowman GR (2019). Cooperative changes in solvent exposure identify cryptic pockets, switches, and allosteric coupling. Biophys. J 116, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puius YA, Zhao Y, Sullivan M, Lawrence DS, Almo SC, and Zhang ZY (1997). Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design. Proc. Natl. Acad. Sci. U S A 94, 13420–13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitter J, Engels M, and Kruger P (1994). Targeted molecular-dynamics - a new approach for searching pathways of conformational transitions. J. Mol. Graph 12, 84–89. [DOI] [PubMed] [Google Scholar]
- Schmidtke P, and Barril X (2010). Understanding and predicting druggability. a high-throughput method for detection of drug binding sites. J. Med. Chem 53, 5858–5867. [DOI] [PubMed] [Google Scholar]
- Schmidtke P, Le Guilloux V, Maupetit J, and Tuffery P (2010). Fpocket: online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 38, W582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck J, Hecky J, Tam HK, Arndt KM, Einsle O, and Muller KM (2012). Exploring the molecular linkage of protein stability traits for enzyme optimization by iterative truncation and evolution. Biochemistry 51, 4850–4867. [DOI] [PubMed] [Google Scholar]
- Stec B, Holtz KM, Wojciechowski CL, and Kantrowitz ER (2005). Structure of the wildtype TEM-1 beta-lactamase at 1.55 angstrom and the mutant enzyme Ser70Ala at 2.1 angstrom suggest the mode of noncovalent catalysis for the mutant enzyme. Acta Cryst. D-Biol. Cryst 61, 1072–1079. [DOI] [PubMed] [Google Scholar]
- Thomas VL, Golemi-Kotra D, Kim C, Vakulenko SB, Mobashery S, and Shoichet BK (2005). Structural consequences of the inhibitor-resistant Ser130Gly substitution in TEM beta-lactamase. Biochemistry 44, 9330–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Lee CT, Durrant JD, Malmstrom RD, Feher VA, and Amaro RE (2016). Emerging computational methods for the rational discovery of allosteric drugs. Chem. Rev. 116, 6370–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Minasov G, and Shoichet BK (2002a). Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol 320, 85–95. [DOI] [PubMed] [Google Scholar]
- Wang X, Minasov G, and Shoichet BK (2002b). The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34. J. Biol. Chem 277, 32149–32156. [DOI] [PubMed] [Google Scholar]
- Wassman CD, Baronio R, Demir O, Wallentine BD, Chen CK, Hall LV, Salehi F, Lin DW, Chung BP, Hatfield GW, et al. (2013). Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat. Commun 4, 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikl TR, and von Deuster C (2009). Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins 75, 104–110. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, et al. (2004). Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct. Mol. Biol 11, 730–737. [DOI] [PubMed] [Google Scholar]
- Wrabl JO, Gu J, Liu T, Schrank TP, Whitten ST, and Hilser VJ (2011). The role of protein conformational fluctuations in allostery, function, and evolution. Biophys. Chem 159, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman MI, Hart KM, Sibbald CA, Frederick TE, Jimah JR, Knoverek CR, Tolia NH, and Bowman GR (2017). Prediction of new stabilizing mutations based on mechanistic insights from markov state models. ACS Cent. Sci 3, 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman MI, Porter JR, Sun X, Silva RR, and Bowman GR (2018). Choice of adaptive sampling strategy impacts state discovery, transition probabilities, and the apparent mechanism of conformational changes. J. Chem. Theory. Comput 14, 5459–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Sl.xlsx, druggability scores of all proteins studied, related to Table 1. All duggability score (DS) values obtained the by Fpocket program are listed in the file. Each line of the data set provides names of apo and holo structures from the CryptoSite dataset, the PDB ID of the structure of the selected unbound homolog, the distance of the pocket from the ligand at the cryptic site in the holo structure, the Fpocket druggability score (DS), indication whether the structure is unbound (U), mutant (M), or a complex (C), and the name of the protein from the PDB file.
The Fpocket program used here is available at http://fpocket.sourceforge.net/.