Abstract
Background
Neurodegenerative diseases are incurable disorders caused by progressive neuronal cell death. Retinitis pigmentosa (RP) is a blinding neurodegenerative disease that results in photoreceptor death and progresses to the loss of the entire retinal network. We previously found that proteomic analysis of the adjacent vitreous served as way to indirectly biopsy the retina and identify changes in the retinal proteome.
Methods
We analyzed protein expression in liquid vitreous biopsies from autosomal recessive (ar)RP patients with PDE6A mutations and arRP mice with Pde6ɑ mutations. Proteomic analysis of retina and vitreous samples identified molecular pathways affected at the onset of photoreceptor death. Based on affected molecular pathways, arRP mice were treated with a ketogenic diet or metabolites involved in fatty-acid synthesis, oxidative phosphorylation, and the tricarboxylic acid (TCA) cycle.
Findings
Dietary supplementation of a single metabolite, ɑ-ketoglutarate, increased docosahexaeonic acid levels, provided neuroprotection, and enhanced visual function in arRP mice. A ketogenic diet delayed photoreceptor cell loss, while vitamin B supplementation had a limited effect. Finally, desorption electrospray ionization mass spectrometry imaging (DESI-MSI) on ɑ-ketoglutarate-treated mice revealed restoration of metabolites that correlated with our proteomic findings: uridine, dihydrouridine, and thymidine (pyrimidine and purine metabolism), glutamine and glutamate (glutamine/glutamate conversion), and succinic and aconitic acid (TCA cycle).
Interpretation
This study demonstrates that replenishing TCA cycle metabolites via oral supplementation prolongs retinal function and provides a neuroprotective effect on the photoreceptor cells and inner retinal network.
Funding
NIH grants [R01EY026682, R01EY024665, R01EY025225, R01EY024698, R21AG050437, P30EY026877, 5P30EY019007, R01EY018213, F30EYE027986, T32GM007337, 5P30CA013696], NSF grant CHE-1734082.
Keywords: Metabolomics, Metabolite supplementation, TCA cycle, Ketogenic diet, ɑ-ketoglutarate, Desorption electrospray ionization mass spectrometry
Research in context.
Evidence before this study
Retinitis pigmentosa (RP) consists of a group of inherited retinal degenerative diseases with high genetic heterogeneity, which can be caused by mutations in more than 60 genes. Despite the precise genetic characterization of RP patients, the underlying mechanisms of photoreceptor cell death is not well understood. Our previous studies have shown that changes in the retinal proteome during disease can be detected through proteomic analysis of vitreous biopsies, as proteins from molecular pathways affected during disease pathogenesis are elevated in the vitreous fluid.
Added value of this study
We applied a liquid biopsy proteomics approach to identify the proteins lost from the retina and elevated in the vitreous during RP progression. We then used these results to determine the molecular pathways most affected at the onset of disease. We found that metabolic and cellular energy pathways were highly disrupted at disease onset. Next, we replenished metabolites found within these disrupted cellular pathways and determined that some metabolites, specifically those involved in the tricarboxylic acid (TCA) cycle, can provide neuroprotection to the photoreceptor cells in our RP mouse model.
Implications of all the available evidence
This study provided key insight into the cellular pathways involved in retinal degeneration. We found that cellular metabolism and energy pathways are highly disrupted at the onset of RP, and that the delivery of metabolites within these critical pathways in the arRP preclinical mouse model showed efficacious neuroprotective effects on the inner retina. The results from this study lay the groundwork for future experiments to address how the TCA cycle and aerobic metabolism influence the photoreceptors and their signaling to the inner retinal network, and thus improve upon neuroprotective approaches targeting photoreceptor cell metabolism. This work also suggests that directly targeting the TCA cycle may be a beneficial approach for future RP therapy.
Alt-text: Unlabelled box
1. Introduction
Neurodegenerative diseases are debilitating, chronic disorders associated with neuronal cell loss in sensory, motor, or cognitive systems [1]. They are typically characterized by abnormal proteostasis, where the cell is unable to maintain regulation of protein folding, translation, and degradation pathways. These defects in proteostasis lead to processes of cell death such as apoptosis and disrupted signaling mechanisms within the cell from oxidative stress, glutamate-mediated cytotoxicity, neuroinflammation, mitochondrial dysfunction, and increased endoplasmic reticulum (ER) stress [2], [3], [4], [5], [6], [7]. However, the early factors involved in neuronal cell death are still not understood. Retinitis pigmentosa (RP) consists of a group of inherited retinal neurodegenerative diseases with high genetic heterogeneity, which can be caused by mutations in more than 60 genes. RP affects approximately 1.5 million people worldwide, and approximately 10% of Americans carry a recessive mutant RP allele. Despite precise genetic characterization of RP patients, identification of causative mutations has had limited impact on therapy. To date, the only treatment for RP is supplementation with vitamin A (15,000 IU/day), but its clinical value is controversial, it does not cure disease, and vitamin A can have adverse effects for other retinal neurodegenerations such as Stargardt macular dystrophy [8]. There have been recent advances to treat RP patients using prosthetics or gene therapy approaches [9,10], but only end-stage RP or patients with the RPE65 gene mutation, respectively, are currently eligible for such therapies. Thus, there is a critical need to understand the underlying mechanisms of photoreceptor death and provide a general therapy independent of specific gene mutations. Delaying photoreceptor cell degeneration, even without a complete cure, can significantly increase the quality of life for RP patients and prolong their ability to live independently.
There is accumulating evidence linking neurodegeneration to energy metabolism, particularly in age-related neurological disorders like Huntington's, Alzheimer's, and Parkinson's disease [1]. Neuronal aging is often accompanied by metabolic and neurophysiologic changes that are associated with impaired function. In the eye, retinal photoreceptors are metabolically robust and require high rates of aerobic ATP synthesis. Defects in retinal cellular metabolism are associated with age-related degenerative retinal diseases, such as rod dystrophy and RP. We have previously shown, using proteomics, that the representation of metabolic pathways and enzymes in the human retina varies by anatomic location (i.e. the peripheral, juxta-macular, and foveomacular regions) [11]. In addition to anatomical differences in metabolic pathway representation, energy consumption in the retinal cell layers is highly compartmentalized. It has been demonstrated that the mammalian inner retina metabolizes a majority (∼70%) of its glucose via the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS). In contrast, the outer retina metabolizes ∼60% of its glucose through glycolysis [12,13]. Thus, defects in aerobic metabolism could preferentially affect inner retinal function. In support of this, photoreceptor cells of the retina are highly susceptible to glycolytic inhibition (via sodium iodoacetate injection) and mature ‘RP’ rats lacking a majority of their photoreceptors display an ∼50% reduction in glycolytic activity compared to normal rat retinas [14,15]. Glucose metabolism plays other non-energetic roles in the retina in addition to meeting the high energy demands of photoreceptors [16,17]. For example, glucose metabolism by the pentose phosphate pathway leads to the production of intracellular glutathione, which prevents neuronal cell death through redox inactivation of cytochrome c-mediated apoptosis pathways [18]. Glucose metabolism also feeds into fatty acid synthesis and lipid metabolism pathways, which are responsible for maintaining proper membrane lipid composition in neuronal cells [19]. Disruption of lipogenesis (by deleting fatty acid synthase) in the neural retina leads to abnormal synaptic structure and apoptosis, resulting in a phenotype resembling RP [20]. Thus, loss of these key metabolic pathways and enzymes would result in abnormal nutrient and metabolite availability, and impaired visual function.
It has been shown that the risk of neuronal cell death is constant, regardless of the underlying cause [21]. Therefore, pathways must exist that can be targeted to generally reduce or protect against neuronal cell death. A potential therapeutic approach for RP is to reprogram metabolic pathways that are altered during rod dystrophy and RP progression. In one such approach, we found that knocking down sirtuin-6 (SIRT6) to reprogram retinal cells toward anabolism delayed photoreceptor cell degeneration in a mouse model of RP [22]. For this study, we applied a liquid biopsy proteomics approach to identify the molecular pathway targets affected during RP progression. Using proteomics, we have previously shown that molecular changes in the neural retina can be detected through proteomic analysis of human and mouse vitreous biopsies, since the retina deposits proteins into the adjacent vitreous extracellular matrix in retinal disease [23, 24]. In this study, we collected vitreous biopsy samples from patients with autosomal recessive retinitis pigmentosa (arRP) carrying mutations in PDE6A, a gene encoding the catalytic ɑ-subunit of the rod-specific cGMP-dependent phosphodiesterase (PDE6), and from mice carrying a Pde6ɑ mutation. Based on proteomic analyses, we targeted diseased metabolic pathways to investigate treatments for RP patients and found key metabolites that prolong photoreceptor cell survival and visual function. We further compared the distribution of these metabolites using desorption electrospray ionization mass spectrometry imaging (DESI-MSI) between untreated Pde6ɑ mice and metabolite-treated mice.
2. Materials and methods
2.1. Study approval
The study protocol was approved by the Institutional Review Board for Human Subjects Research (IRB) at Columbia University and Stanford University, was HIPAA compliant, and adhered to the tenets of the Declaration of Helsinki. All subjects underwent informed consent for study participation. All experiments were performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research and were all approved by the Animal Care and Use Committee at Columbia University and Stanford University.
2.2. Human patient imaging
Clinical examination and testing were performed as previously described [25]. Autofluorescence (AF) images were obtained using a Topcon TRC 50DX camera (Topcon, Pyramus, NJ, USA). Optical coherence tomography imaging was obtained from the spectral-domain Heidelberg HRA2 Spectralis, version 1.6.1 (Heidelberg Engineering, Inc, Vista, CA, USA). Genetic testing was performed as previously described [26].
2.3. Human vitreous sample collection
Pars plana vitrectomy for vitreous biopsy was performed as previously described [27]. We used a single-step transconjunctival 23-gauge trocar cannula system (Alcon Laboratories Inc, Fort Worth, TX). A light pipe and vitreous cutter were inserted into the mid vitreous and the cutter was activated for 30 s without infusion. An undiluted 1.0-cc sample of vitreous was then manually aspirated into a 3-cc syringe. Vitreous samples were immediately centrifuged in the operating room at 15,000 × g for 5 min at room temperature to remove particulate matter, and samples were then stored at −80 °C.
2.4. Mouse lines and husbandry
C57BL/6J-Pde6αnmf363/nmf363 (RRID: MGI:3828994), with a D670G mutation, herein referred to as Pde6αD670G mice, were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Pde6αD670G are co-isogenic in the C57BL/6J (B6) background; therefore, age matched B6 mice (RRID: IMSR_JAX:000664) were used as experimental controls. Mice were bred and maintained at the facilities of Columbia University and Stanford University. Animals were kept on a light–dark cycle (12 h–12 h). Food and water were available ad libitum throughout the experiment, whether treated or untreated. The ketogenic diet was provided by BioServ (Ketogenic Diet, AIN-76A-Modified, High-Fat, Paste; Flemington, NJ) in place of standard chow. The ketogenic diet was stored refrigerated and changed every other day for the mice during the duration of the experiment. Vitamins B2 (riboflavin, Sigma Aldrich) and B3 (nicotinamide, Sigma Aldrich) were provided at a concentration of 2.5 g/L in the mouse drinking water. ɑ-KG (alpha-ketoglutaric acid, Sigma Aldrich) was provided at a concentration of 10 g/L in the mouse drinking water. Melatonin (Sigma Aldrich) was provided at a concentration of 80 mg/L in the mouse drinking water. Resveratrol (Sigma Aldrich) was provided at a concentration of 120 mg/L in the mouse drinking water. All water bottles were covered in foil to reduce light exposure and changed weekly during the duration of the experiment following approved institutional guidelines.
2.5. Mouse autofluorescence/infrared imaging
Autofluorescence/infrared (AF/IR) fundus imaging was obtained with the Spectralis scanning laser confocal ophthalmoscope (OCT-SLO Spectralis 2; Heidelberg Engineering, Heidelberg, Germany) as previously described [28,29]. Pupils were dilated using topical 2.5% phenylephrine hydrochloride and 1% tropicamide (Akorn, Inc., Lakeforest, IL, USA). Mice were anesthetized by intraperitoneal injection of 0.1 mL /10 g body weight of anesthesia [1 mL ketamine–100 mg/ mL (Ketaset III, Fort Dodge, IA, USA) and 0.1 mL xylazine—20 mg/ mL (Lloyd Laboratories, Shenandoah, IA, USA) in 8.9 mL PBS]. AF imaging was obtained at 488-nm absorption and 495-nm emission using a 55° lens. IR imaging was obtained at 790 nm absorption and 830 nm emission using a 55° lens. Images were taken of the central retina, with the optic nerve located in the center of the image.
2.6. Electroretinography (ERG)
Mice were dark-adapted overnight, manipulations were conducted under dim red-light illumination, and recordings were made using Espion or Celeris ERG equipment (Diagnosys LLC, Littleton, MA, USA) as previously described [28,29]. Briefly, adult B6 control mice were tested at the beginning of each session to ensure equal readouts from the electrodes for both eyes before testing the experimental mice. Pupils were dilated using topical 2.5% phenylephrine hydrochloride and 1% tropicamide (Akorn Inc., Lakeforest, IL, USA). Mice were anesthetized by intraperitoneal injection of 0.1 mL/10 g body weight of anesthesia [1 mL ketamine 100 mg/mL (Ketaset III, Fort Dodge, IA, USA) and 0.1 mL xylazine 20 mg/mL (Lloyd Laboratories, Shenandoah, IA, USA) in 8.9 mL PBS]. Body temperature was maintained at 37 °C during the procedure. Both eyes were recorded simultaneously, and responses were averaged for each trial. Responses were taken from the Espion/Celeris readout in microvolts (μV). Statistical significance was set at p < 0.05, and 1-way ANOVA followed by Tukey's multiple comparison's test was used to compare all treated, untreated, and wild-type groups.
2.7. Outer nuclear layer quantification
Mice were sacrificed, and the eyes enucleated as previously described [30,31]. Eyes were embedded in paraffin, sectioned, and stained with hematoxylin and eosin by Excalibur Pathology, Inc. (Norman, OK), before being visualized by light microscopy (Leica DM 5000B, Leica Microsystems, Germany). Quantification of photoreceptor nuclei was conducted on several sections that contained the optic nerve, as follows: the distance between the optic nerve and the ciliary body was divided into three, approximately equal, quadrants on each side of the retina. The number of nuclei in four columns were counted within each single quadrant. These counts were then used to determine the average thickness of the ONL for each individual animal and within each quadrant of the retina, spanning from the ciliary body to the optic nerve head (ONH) and out to the ciliary body. Sectioning proceeded along the long axis of the segment, so that each section contained upper and lower retina as well as the posterior pole. Statistical analysis was performed on the average ONL thickness of each individual animal, taking into account all cell counts for all quadrants, and Student's t-test was performed with statistical significance set at p < 0.05.
2.8. Immunocytochemistry
Unfixed enucleated eyes were embedded in optimal cutting temperature (OCT) medium and frozen using dry ice. Sagittal sections (10 μm thick) of the ipsilateral retina were collected onto superfrost plus glass slides. OCT-embedded sections were placed in precooled 4% paraformaldehyde and brought to room temperature for 20 min to fix the tissue. Sections were blocked for 1 hour with 3% goat serum/0.1% Triton X-100 (cat. no. T8532; Sigma-Aldrich, St. Louis, MO) in PBS. Primary antibody against glial fibrillary acidic protein (GFAP; 1:400; cat. no. Mab360; Millipore, Burlington, MA; RRID: AB_11212597) was diluted in 3% goat serum and PBS overnight at 4 °C. The slides were washed with PBS (3 × 5 min) and incubated with the secondary antibody GAM647 (1:300; cat. no. A-32728; Thermo Fisher Scientific; RRID: AB_2633277) in 3% goat serum in PBS for 1 hour at room temperature. Samples were washed with PBS (3 × 5 min), incubated with DAPI (4 μg/mL; cat. no. D9564; Sigma-Aldrich) for 10 min at room temperature and washed with PBS (3 × 5 min). Slides were mounted with ProLong Gold Antifade Mountant (cat. no. P36934; Thermo Fisher Scientific; RRID: SCR_015961) followed by imaging with confocal microscopy at 25× (SP8 confocal, Leica Microsystems, Wetzlar, Germany).
2.9. Mouse vitreous and retina sample collection
The vitreous and retina from 12 mouse eyes were eviscerated as described previously [32]. Briefly, scleral tissue posterior to the limbus was grasped with 0.22 forceps and a microsurgical blade was used to make a linear incision in the cornea from limbus to limbus. A fine curved needle holder was inserted behind the lens toward the posterior aspect of the globe. The needle holder was partially closed and pulled forward pushing the lens through the corneal incision while leaving the eye wall intact. The vitreous was partially adherent to the lens. The lens-vitreous tissue was then placed into a filtered centrifugation tube containing 20 mL of protease inhibitor cocktail (Roche) dissolved in PBS. The fine curved needle holder was placed as far posterior to the globe as possible, near the optic nerve. The needle holder was partially closed, and pulled forward, pushing the retina forward through the corneal incision. The vitreous appeared as a translucent gel adherent to the retina. The retina-vitreous tissue was placed into the filtered centrifuge tube containing the lens-vitreous tissue. The filtered centrifuge tube was spun at 14,000 × g for 12 min and the eluent (vitreous) was collected. Retina was separated from the lens tissue remaining in the filtered centrifuge tube and collected in PBS containing protease inhibitor cocktail.
2.10. Protein extraction and digestion
The received vitreous and retina samples were diluted in 2% SDS, 100 mM Tris-HCl (pH 7.6), 100 mM DTT to approximately 0.5 mL volume and heated at 95 °C for 10 min. Each sample was then briefly vortexed and sonicated for 10 s using a probe-tip sonicator (Omni International). The samples were then returned to incubate at 95 °C for an additional 10 min. Samples were then transferred to a 30 K Amicon MWCO device (Millipore) and centrifuged at 16,100 × g for 30 min. Then 400 µL of 8 M urea, 100 mM Tris-HCl (pH 7.6) was added to each device and centrifuged as before and the filtrate discarded. This step was repeated once. Then 400 µL of 8 M urea, 100 mM Tris-HCl (pH 7.6), 15 mM iodoacetamide was added to each device and incubated in the dark for 30 min. The samples were then centrifuged as before, and the filtrate discarded. Then 400 µL of 8 M urea, 100 mM Tris–HCl (pH 7.6) was added to each device and centrifuged as before and the filtrate discarded. This step was repeated once more. Then 400 µL of 2 M urea, 100 mM Tris-HCl (pH 7.6) was added to each device along with 2.5 µg trypsin. The devices incubated overnight on a heat block at 37 °C. The devices were then centrifuged, and the filtrate collected. Then 400 µL 0.5 M NaCl was added to each device and centrifuged as before. The filtrate was added to the previously collected filtrate.
2.11. Peptide desalting and fractionation
Digested peptides were desalted using C18 stop-and-go extraction (STAGE) tips. Briefly, for each sample a C18 STAGE tip was activated with methanol, then conditioned with 75% acetonitrile, 0.5% acetic acid followed by 0.5% acetic acid. Samples were loaded onto the tips and desalted with 0.5% acetic acid. Peptides were eluted with 75% acetonitrile, 0.5% acetic acid and lyophilized in a SpeedVac (Thermo Savant) to dryness, approximately 2 hours. Peptides were fractionated using strong anion exchange (SAX) STAGE tips. Briefly, for each sample a SAX STAGE tip was activated with methanol, then conditioned with Britton–Robinson buffer (BRB), pH 3.0 followed by BRB (pH 11.5). Peptides were loaded onto the tips and the flow-through collected followed by and five additional fractions by subsequent application of BRB at pH 8.0, 6.0, 5.0, 4.0 and 3.0. Each fraction was desalted using a C18 STAGE tip and lyophilized as described above.
2.12. Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Each SAX fraction was analyzed by LC-MS/MS. LC was performed on an Agilent 1100 Nano-flow system. Mobile phase A was 94.5% MilliQ water, 5% acetonitrile, 0.5% acetic acid. Mobile phase B was 80% acetonitrile, 19.5% MilliQ water, 0.5% acetic acid. The 150 min LC gradient ran from 5% A to 35% B over 105 min, with the remaining time used for sample loading and column regeneration. Samples were loaded to a 2 cm × 100 µm I.D. trap-column positioned on an actuated valve (Rheodyne). The column was 13 cm × 100 µm I.D. fused silica with a pulled tip emitter. Both trap and analytical columns were packed with 3.5 µm C18 (Zorbax SB, Agilent). The LC was interfaced to a dual pressure linear ion trap mass spectrometer (LTQ Velos, Thermo Fisher) via nano-electrospray ionization. An electrospray voltage of 1.5 kV was applied to a pre-column tee. The mass spectrometer was programmed to acquire, by data-dependent acquisition, tandem mass spectra from the top 15 ions in the full scan from 400–1,400 m/z. Dynamic exclusion was set to 30 seconds.
2.13. LC-MS/MS data processing and library searching
Mass spectrometer MS data files were converted to mzXML format and then to mgf format using msconvert in ProteoWizard and mzXML_to_mgf v4.4, rev 1, respectively. Peak list data were searched using two algorithms: OMSSA v2.1.9 and X!Tandem TPP v4.4, rev 1. The UniProt mouse or human protein sequence library was used in a target-decoy format (derived from UniProt as of 10/22/2012). The mgf files were searched using OMSSA with precursor settings of +/− 2.0 Da and fragment tolerance of +/− 0.8 Da. In X!Tandem, precursor settings were set to −1 Da and +3 Da and the fragment tolerance to 0.8 Da. The number of missed cleavages was set to 1; the fixed modifications were Carbamidomethyl (C); the variable modifications were Oxidation (M). XML output files were parsed using TPP v4.4, rev 1 in the programs PeptideProphet, iProphet, and ProteinProphet. Proteins were required to have 2 or more unique peptides with E-value scores of 0.01 or less. Relative quantitation was performed by spectral counting. Data were normalized based on total spectral counts (hits) per sample.
2.14. LC-MS/MS data analysis and visualization
Results (i.e. protein spectral counts) were saved in Excel as .txt format and were uploaded into the Partek Genomics Suite 6.5 software package as described previously [23,24]. The data was normalized to log base 2 and compared using 1-way ANOVA analysis. All proteins with non-significant (p > 0.05) changes were eliminated from the table. The significant values were mapped using the ‘cluster based on significant genes’ visualization function with the standardization option chosen. Results of the 1-way ANOVA analysis were displayed as a heatmap with relative protein expression between the experimental groups represented on a log scale. Each column of the heatmap represents an individual sample (n = 3 for each group) and each row represents an individual protein. Orange indicates high expression, while green/black indicates low or no expression. PANTHER Pathway Analysis was utilized to determine the most significant molecular pathways affected by the proteins present in each group [33]. Gene ontology (GO) analysis was also performed in PANTHER. Pie charts were created for the visualization of GO distributions within the list of proteins for each GO term category including biological process, molecular function, and cellular component.
2.15. Desorption electrospray ionization mass spectrometry imaging (DESI-MSI)
Mouse retinas were dissected and frozen in optimal cutting temperature (OCT) media. Cryosections (10-µm) were collected (Cryostat Microsystem, Leica Biosystems) with at least three retinal sections per SuperFrost Plus Slide (Fisher Scientific), with at least three slides collected per mouse (at least 9 retinal sections per mouse). Briefly, DESI-MSI was performed in the negative ion mode (−5 kV) from m/z 50–1,000, using LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) coupled to a home-built DESI-source and a two-dimensional (2D) motorized stage. The retinal tissues were raster scanned under impinging charged droplets using a 2D moving stage in horizontal rows separated by a 200-µm (spatial resolution) vertical step. The impinging primary charged droplets were generated from the electrospray nebulization of a histologically compatible solvent system, 1:1 (vol/vol) dimethylformamide/acetonitrile (DMF/ACN, flow rate 1 µL/min). The electrospray nebulization was assisted by sheath gas nitrogen (N2, 170 psi) and a high electric field of −5 kV. The spatial resolution of DESI-MSI is defined by the spray spot size of ∼200 µm. DESI-MSI of all tissue samples were carried out under identical experimental conditions, such as spray tip-to-surface distance ∼2 mm, spray incident angle of 55°, and spray-to-inlet distance ∼5 mm. MSI data acquisition was performed using XCalibur 2.2 software (Thermo Fisher Scientific Inc.). XCalibur 2.2 mass spectra files (.raw files) were converted to .mzML format files using msconvert; then imzMLConverter (version 1.3.0) was used to combine .mzML files into .imzML format files, readable by another software MSiReader (version v1.00). The 2D chemical maps of molecular ions were plotted using Biomap. To identify significantly differentially expressed metabolites, XCalibur raw data files were converted to csv files for statistical analysis. Raw csv data was imported to the R language for further processing. To account for random effects influencing mass spectrometry data, the intensity of each metabolite was normalized by the total ion current for the corresponding pixel. Next, a nearest neighbor clustering method was used to collect the pixel intensities corresponding to the nearest molecular ion peak. To determine which metabolites could distinguish Pde6αD670G mice from ɑ-KG treated mice, we applied the SAM (Significance Analysis of Microarrays) technique using the samr package in R [34]. The metabolites were filtered according to a false discovery rate (FDR) cutoff of 5%. SAM identified 913 potentially significant metabolites (FDR < 0.05), of which 377 metabolites had an absolute fold change greater than 1. Although SAM returned 377 metabolites with notable fold change, to help interpret the biological role of metabolites, we restricted our analysis to peaks for which tandem-MS and high mass resolution analyses were performed by using the LTQ-Orbitrap XL (Thermo Scientific). The relative intensity is equal to the intensity of the metabolite normalized by the total ion current (sum of all intensity values from the detected ions).
2.16. Metabolite/lipid identification
For lipid and metabolite identification, tandem-MS and high-mass resolution analyses were performed using the LTQ-Orbitrap XL (Thermo Scientific). The retinal tissue sections were dissolved in 500 μL of methanol: water (80:20) for metabolite extraction. The undissolved tissue was centrifuged down by 4000 × g for 5 min. The resultant clear solution was sprayed into mass spectrometer for tandem-MS and high-mass resolution analyses, at a flow rate of 2 μL/min, sheath gas pressure (N2, 120 psi), and the spray voltage −5 kV. For the MS/MS studies, a normalized collision energy of 20–40% was applied. The metabolites and lipids were assigned based on high-mass resolution analysis, isotope distribution, and tandem-MS fragmentation patterns. In the case when several detectable fragmentation patterns were generated by isomeric parent ions, the most intense fragmentation peaks were used to assign the molecule identity listed in SI Appendix. The LipidMaps (www.lipidmaps.org/), MassBank (www.massbank.jp), Metlin (https://metlin.scripps.edu/), and the human metabolome database (HMDB) were used for the metabolite identification. The detected species were mostly deprotonated small metabolites related to the tricarboxylic acid (TCA) cycle, and deprotonated lipids including free fatty acids (FAs), fatty acid dimers, phosphatidic acids, and glycerophospholipids.
2.17. Statistical analysis
Differences in experimental groups were determined by the Student's t-test as appropriate, or by one-way ANOVA followed by Tukey's post-hoc multiple comparison's test (see individual methods section). A p value < 0.05 was considered significant. Measurements were done blinded to experimental group. All error bars represent standard deviation unless otherwise listed.
3. Results
3.1. Proteomics of the human vitreous highlight changes occurring in the neural retina during RP disease progression
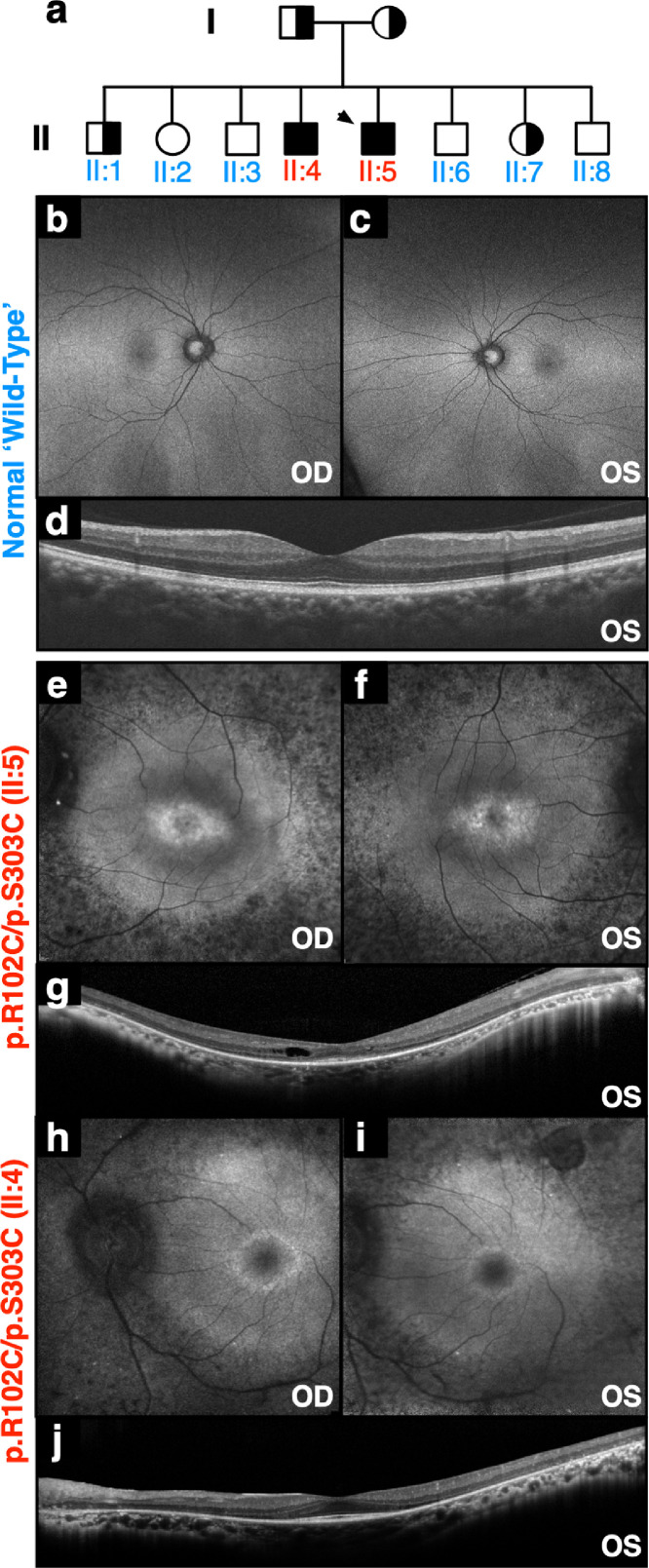
The proband presented to us at age 38 with end-stage arRP. Genetic testing revealed p.R102C/p.S303C mutations in the PDE6A gene (II:5; Fig. 1(a)). Mutations in PDE6 contribute to a significant fraction of RP cases (7–9%; OMIM: 180071) [35]. In comparison to a normal control (Fig. 1(b)–(d)), fundus autofluorescence (AF) revealed high-density central AF rings (Fig. 1(e)–(f)) and optical coherence tomography (OCT) revealed thinning of the neuronal cell layers (Fig. 1(g)). Additionally, the proband's affected brother (II:4; Fig. 1(a)) displayed a similar disease stage with hyper-autofluorescent rings on AF imaging (Fig. 1(h)–(i)). OCT imaging similarly revealed thinning of retinal cell layers and cystoid macular edema (Fig. 1(j)). The affected brothers both developed significant epiretinal membranes (ERMs) and underwent vitrectomy surgery. Liquid vitreous biopsies were collected at the time of surgery (Fig. 2(a) -(b), Table S1). ERM vitreous samples from two patients without RP were used as comparative controls since the two arRP patients underwent vitrectomy surgery for ERM removal (Fig. 2(a)-(b)). Vitreous samples were analyzed using shotgun liquid chromatography-tandem mass spectrometry (LC-MS/MS) to determine proteomic content (Table S2).
Fig. 1.
Two patients with autosomal recessive retinitis pigmentosa carrying mutations in PDE6A: (a) Pedigree of a family with autosomal recessive retinitis pigmentosa (arRP) caused by mutations in the PDE6A gene. The proband is denoted by the arrow. (b–c) Fundus autofluorescence (488 nm) of a control human eye. (d) Spatially corresponding SD-OCT scans through the fovea of a control human eye. (e–f) Fundus autofluorescence (488 nm) of the proband (II:5) revealed confluent areas of hypo-autofluorescence, suggesting RPE loss throughout the periphery, and a central ring of hyper-autofluorescence overlying the parafovea. (g) Spatially corresponding SD-OCT scans through the fovea confirmed marked peripheral outer retinal thinning and RPE loss. Despite macular edema, the fovea was relatively preserved, with a distinguishable ellipsoid zone. (h–i) Fundus autofluorescence of the affected brother (II:4) revealed similar confluent areas of hypo-autofluorescence and a central ring of hyper-autofluorescence overlying the parafovea. (j) Spatially corresponding SD-OCT scans showed similar marked peripheral outer retinal thinning and RPE loss.
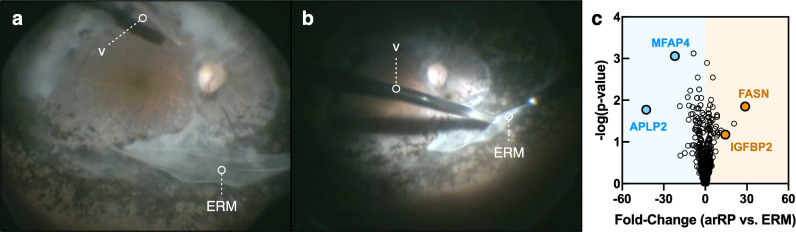
Fig. 2.
Proteomic analysis of PDE6A human vitreous. (a–b) Pars plana vitrectomy and epiretinal membrane (ERM) removal procedure for an PDE6A (arRP) patient (II:5 in Fig. 1). A vitreous biopsy was collected from this patient and the affected brother. Protein content was analyzed by LC-MS/MS. The vitreous cutter is denoted by the letter ‘v.’ (c) Protein spectral counts were analyzed by 1-way ANOVA. Results are represented as a volcano plot. The horizontal axis (x-axis) displays the log2 fold-change value (arRP vs. ERM) and the vertical axis (y-axis) displays the noise-adjusted signal as the -log10 (p-value) from the 1-way ANOVA analysis. Increased levels of FASN and IGFBP2 are denoted by the orange circles. Blue circles indicate decreased levels of MFAP4 and APLP2 in arRP vitreous compared to controls.
It is rare to obtain arRP vitreous samples as the indication for vitrectomy surgery in these patients is exceptional. Despite the limitation of small sample size, descriptive semi-quantitative analysis can provide insight into underlying human disease mechanisms in a rare patient population, and there are no human arRP proteomic reports to date. We therefore performed label-free relative quantification of proteins identified in arRP vitreous relative to ERM, which was defined as the reference group. We detected 612 proteins in arRP vitreous by LC-MS/MS (Table S3). We compared protein expression between arRP and ERM vitreous: there were 110 elevated proteins (with a fold-change ≥2) in arRP vitreous compared to ERM controls (Fig. 2(c)). We observed a significant fraction (36%) of elevated, intracellular proteins in arRP vitreous despite the extracellular location and acellular composition of the vitreous. We previously reported that a significant fraction of proteins in the normal human vitreous are intracellular, particularly in the vitreous core, suggesting that surrounding tissues release proteins into the adjacent vitreous [36]. Among these elevated intracellular proteins were fatty acid synthase (FASN) and insulin-like growth factor-binding protein 2 (IGFBP2). FASN is involved in the synthesis of long-chain fatty acids from acetyl-CoA and NADPH. IGFBP2 has been shown to promote fibroblast differentiation and angiogenesis (Fig. 2(c)) [37,38]. There were 135 depleted proteins (with a fold-change ≤ −2) in arRP vitreous compared to ERM controls. Among these proteins were microfibrillar-associated protein 4 (MFAP4) and amyloid beta precursor like 2 (APLP2; Fig. 2(c)). MFAP4 plays a role in maintaining integrin activation, which is important for maintaining RPE-cell adhesion to Bruch's membrane [39]. Interestingly, APLP2 loss or dysfunction has been shown to result in abnormal amacrine and horizontal cell differentiation [39]. These results suggested that the observed proteomic changes in human arRP vitreous could reflect molecular changes in the neural retina. However, the human patient proteomes reflected samples collected at a very late stage of disease, long after significant photoreceptor death and may not reflect the critical cellular pathways altered at the onset of neurodegeneration. We therefore chose to gain further insight into the earlier disease stages using a preclinical model of arRP.
3.2. Proteomic analysis of Pde6ɑD670G retina and vitreous identifies alterations in global protein expression that may precede cell loss and the RP clinical phenotype
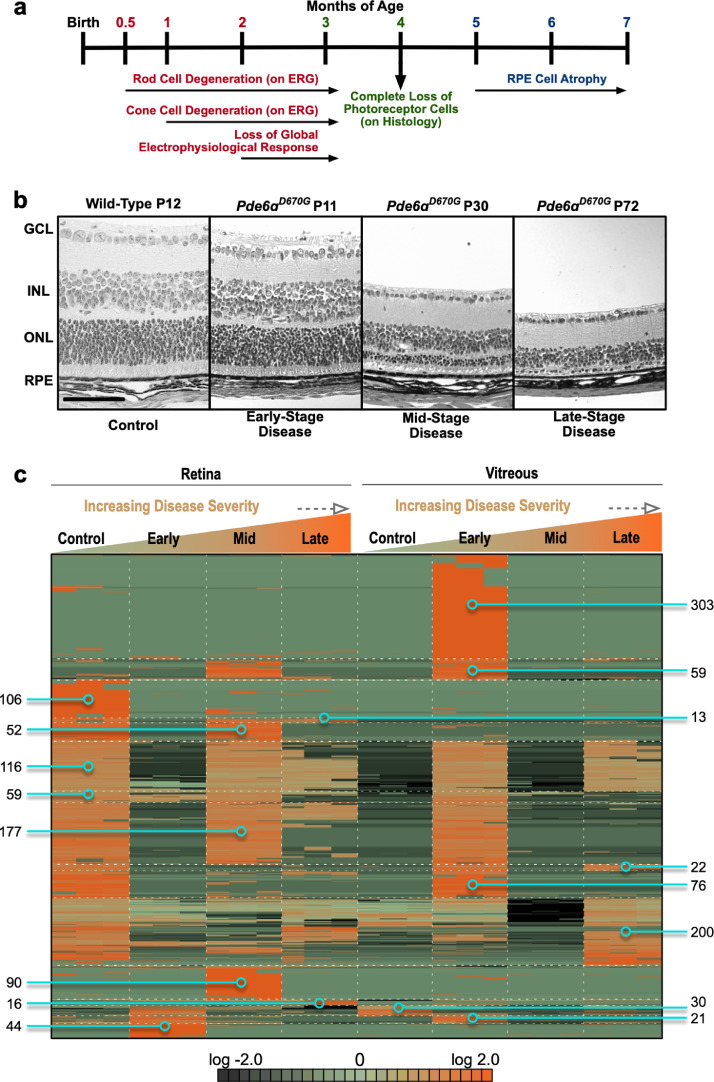
We utilized the Pde6ɑD670G mutant mouse to determine whether any of the cellular pathways found in our human patient proteomes were disrupted at the onset of rod dystrophy, before the clinical RP phenotype is detectable. The Pde6ɑD670G phenotype closely resembles that of patients with arRP. These mice develop early onset severe retinal degeneration, characterized by photoreceptor death, retinal vessel attenuation, pigmented patches, ERG abnormalities, and white retinal spotting [29]. The mice display a loss of rod photoreceptor visual function by ERG analysis as early as one month of age and full loss of global visual function on ERG analysis by two months of age (Fig. 3(a)). In particular, Pde6ɑD670G mice do not display histologically detectable photoreceptor degeneration until after postnatal day 14 (P14; Fig. 3(b)), so retina and vitreous samples were collected from these mice at disease onset (P15), mid-stage (P30) and late-stage disease (P90; Table S4). Retina and vitreous samples for wild-type and Pde6ɑD670G mice were trypsinized and underwent LC-MS/MS analysis (Table S5-6). Notably there were fewer unique proteins in the Pde6ɑD670G mouse retina at 90 days (484 ± 63 individual proteins) than 15 days (1,135 ± 55 individual proteins), suggesting a decrease in protein expression as neurodegeneration progresses and/or increased degradation of these proteins (Table S7). Conversely, we noticed there were more unique proteins in the Pde6ɑD670G mouse vitreous at 15 days (364 ± 30 individual proteins) compared to wild-type vitreous (202 ± 9 individual proteins), suggesting an increase in vitreous protein expression at early stages of neuronal cell death (Table S8).
Fig. 3.
Proteomic analysis of Pde6ɑD670G retina and vitreous at early-, mid-, and late-stage retinitis pigmentosa (RP). (a) Timeline of the clinical disease presentation in Pde6ɑD670G mice. (b) Histological analysis of Pde6ɑD670G mouse retinas at early-(P11), mid-(P30) and late-(P72) stage disease compared to a wild-type mouse at (P12) shows loss of the ONL photoreceptor cells over time. RPE, retinal pigment epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 100 µm. (c) Protein spectral counts were analyzed by 1-way ANOVA followed by hierarchical clustering. A total of 1,384 proteins were differentially expressed in the Pde6ɑD670G vitreous and retina samples compared to controls (p < 0.05 [1-way ANOVA]). Results are represented as a heatmap and display protein expression levels on a logarithmic scale. Orange indicates high expression while dark green/black indicates low or no expression.
Retina and vitreous protein levels (with two or more spectra) from Pde6ɑD670G and wild-type mice were compared using 1-way ANOVA and heatmap clustering (Table S9). A total of 1,384 proteins were differentially expressed in the Pde6ɑD670G retina and vitreous samples compared to controls (p < 0.05 [1-way ANOVA]; Fig. 3(c)). There were 303 proteins highly-expressed in Pde6ɑD670G vitreous at day 15 that were not present in the control retina or vitreous (Fig. 3(c)). A majority of these proteins were intracellular and represented protein synthesis, gene expression, DNA replication, recombination, and repair pathways. Of these 303 proteins, there were 16 downstream effectors of the CD3 receptor: Coronin-1A (CORO1A), DEAD box protein 19A (DDX19A), eukaryotic translation initiation factor 4E (EIF4E), GABA receptor-associated protein-like 2 (GABARAP12), hemopexin (HPX), E3 ubiquitin-protein ligase Huwe1 (HUWE1), Lon protease homolog (LONP1), acyl-protein thioesterase 1 (LYPLA1), metastasis-associated protein (MTA2), alpha-N-acetylgalactosaminidase (NAGA), purine nucleoside phosphorylase (PNP), RNA polymerase II subunit B1 (POLR2A), ubiquitin receptor (RAD23B), thiosulfate sulfurtransferase (TSTA2), thioredoxin reductase 1 (TXNRD1), and alanine-tRNA ligase (AARS). The CD3 co-receptor is expressed on T-cells and activates cytotoxic and helper T-cells [40]. The presence of downstream CD3 effectors in the Pde6ɑD670G vitreous suggests that lymphocytes may be active at early stages of the disease. Further investigation of the RP immune response is required to validate this observation. There were 360 proteins in the control retina that were absent in the degenerating (Pde6ɑD670G) retina at P15. These 360 proteins were absent in the control vitreous but, surprisingly, were detected in the diseased (Pde6ɑD670G) vitreous (Fig. 3(c)). Together, this indicates that these proteins have changed their location from the retina to the vitreous in the arRP mice as compared to wild-type controls.
To obtain a global view of the Pde6ɑD670G retina and vitreous proteomes at different stages of degeneration, a gene ontology (GO) analysis of highly represented proteins in each group was performed (statistically-significantly elevated proteins in each group; p < 0.05 [1-way ANOVA]). The highest represented categories in the retina were cellular process, catalytic activity, and intracellular regions (Fig. S1). Interestingly, there was a decrease in the number of proteins with transporter activity in the Pde6ɑD670G retina at early stage neuronal cell death, consistent with a loss of protein expression at this stage (Fig. S1). When comparing the four total retinal protein profiles, the GO summaries for the control retina were similar to the Pde6ɑD670G retina at P30 and P90. This result suggests that the comparison of the overall ontological distribution of proteins (with respect to cellular component, biological process, or molecular function) is insufficient to capture differences between the groups. GO summaries, for example, do not take into account relative quantitative amounts. Thus, different bioinformatic platforms and more detailed analysis (e.g. pathway and network analysis) might detect critical proteomic differences between the control and Pde6ɑD670G retina at different stages. The GO distributions of the vitreous proteomes were distinct, illustrating that the vitreous and retina contained different functional categories of proteins with respect to their condition. For example, arRP vitreous contained a higher proportion of membrane-bound proteins compared to wild-type (Fig. S2). In addition, the arRP retina contained a higher proportion of the GO category “antioxidant activity” compared to wild-type. Together, this indicated that the arRP retina and vitreous express distinct functional categories of proteins compared to wild-type mice at each stage of disease progression, and that these protein pathways may reflect early diagnostic markers or targets for therapeutics preventing further neuronal cell death and loss of the neural retinal network.
3.3. Oxidative metabolic pathways are downregulated in the Pde6ɑD670G retina at the onset of neuronal cell death
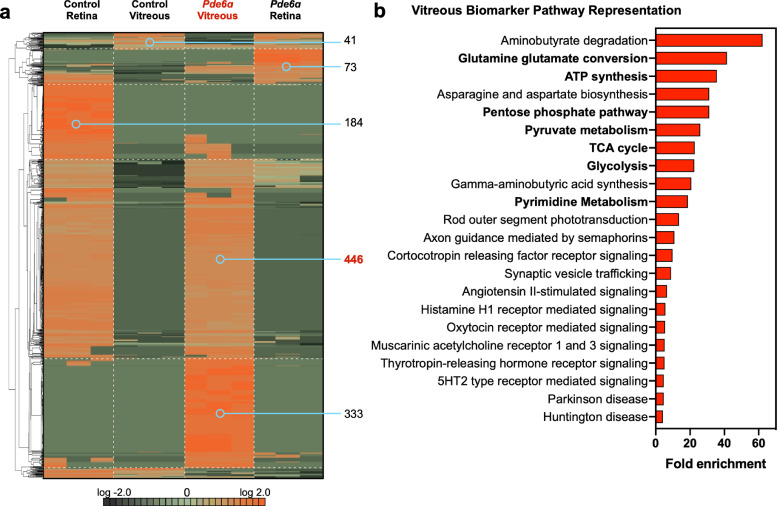
We hypothesized that cellular pathways disrupted at the onset of rod dystrophy might reflect key targets for therapeutic approaches. Therefore, we focused our further investigation on the Pde6ɑD670G mouse proteomes collected at early-stage RP disease (P15). To examine differential protein expression at the onset of neuronal cell death, vitreous and retina protein levels (with two or more spectra) from Pde6ɑD670G (P15) and wild-type mice were compared using 1-way ANOVA and hierarchical heatmap clustering. A total of 1,067 proteins were differentially expressed in the Pde6ɑD670G samples compared to controls (p < 0.05 [1-way ANOVA]; Fig. 4(a); Table S10). Similar to the previous analysis comparing all Pde6ɑD670G stages (Fig. 3(c)), there was a significant decrease in protein expression in Pde6ɑD670G retina at early-stage neurodegeneration (P15). There was also a significant increase in vitreous protein expression at the onset of neuronal cell death in the Pde6ɑD670G mice (779 upregulated proteins; p < 0.05 [1-way ANOVA]). The decrease in retinal protein expression and corresponding increase in vitreous protein expression suggested that the degenerating neural retina may leak proteins into the vitreous, similar to that seen in human patient samples [23,41]. There were 446 proteins in the control retina that were absent in the degenerating (Pde6ɑD670G) retina at P15. These 446 proteins were absent in the control vitreous but, surprisingly, were detected in the diseased (Pde6ɑD670G) vitreous. These results suggest that 446 retinal proteins were depleted from the healthy retina and released into the vitreous at the onset of RP (Fig. 4(a); Table S11). In contrast to the previous analysis comparing all Pde6ɑD670G stages (Fig. 3(c)), we observed a greater number of migrating proteins in the early stage analysis (446 in Fig. 4(a) vs. 360 in Fig. 3(c)). This difference in observed proteins is due to the fact that fewer experimental groups are being compared in Fig. 4(a) (early stage) compared to Fig. 3(c) (all groups). GO analysis categorized a significant fraction of these proteins to be intracellular, further suggesting that they have migrated from the degenerating neural retina into the extracellular vitreous (Fig. S3a). The proteins in this list with the most significant fold-change (FC) were ADP/ATP translocase 1 (SLC25A4; p-value = 1.68e−15 [1-way ANOVA]), malate dehydrogenase 2 (MDH2; p-value = 1.00e−19 [1-way ANOVA]), and ATP synthase (ATP5A1; p-value = 2.06e−13 [1-way ANOVA]). Categorization of these 446 proteins by their biological process revealed that a significant fraction was involved in energetic processes (Fig. S3b).
Fig. 4.
Proteomic analysis of Pde6ɑD670G retina and vitreous identifies global protein expression changes. (a) Protein spectral counts were analyzed by 1-way ANOVA followed by hierarchical clustering. A total of 1,067 proteins were differentially expressed in the Pde6ɑD670G vitreous and retina samples (when comparing protein expression profiles at P15 only) compared to controls (p < 0.05 [1-way ANOVA]). Results are represented as a heatmap and display protein expression levels on a logarithmic scale. Orange indicates high expression while dark green/black indicates low or no expression. Pde6ɑD670G vitreous samples are highlighted by the red font. There were 446 proteins that were depleted in the Pde6ɑD670G retina that were upregulated in the Pde6ɑD670G vitreous. There were 41 proteins that were depleted in the Pde6ɑD670G vitreous compared to the control vitreous. (b) Pathway representation of the 446 candidate biomarkers in the Pde6ɑD670G vitreous. Pathways are ranked by their fold-enrichment obtained from the Mann–Whitney U Test. Metabolic and energetic pathways are denoted by bold-faced text.
We next analyzed the pathway representation of these vitreous biomarkers. Pathway analysis revealed that these proteins represent metabolic (e.g. OXPHOS, TCA cycle), synaptic signaling, and visual transduction pathways (Fig. 4(b); Tables S12–15). As anticipated, we found key proteins involved in the rod photoreceptor transduction pathway to be increased in the arRP vitreous at the onset of rod cell degeneration (P15): rhodopsin (RHO), guanine nucleotide-binding protein alpha and beta subunits (GNAT1 and GNB1), phosducin (PDC), cyclic nucleotide-gated channel beta 1 (CNGB1), rhodopsin kinase (GRK1), recoverin (RCVRN), and S-arrestin (SAG). Taken together, these results suggest that these 446 proteins are candidate biomarkers for early arRP detection and possible targets for neuroprotective therapeutics. We identified proteins involved in glycolysis, TCA cycle, and OXPHOS: hexokinase (HK2), aldolase A, (ALDOA), phosphofructokinase (PFK1), enolase (ENO1), pyruvate kinase (PKM), pyruvate dehydrogenase E1 component subunit (PDHA1), citrate synthase (CS), aconitate hydratase (ACO2), isocitrate dehydrogenase (IDH1 and −3), ɑ-ketoglutarate dehydrogenase (OGDH), succinate-CoA ligase beta subunit (SUCLA2), succinate dehydrogenase flavoprotein subunit (SDHA), and malate dehydrogenase (MDH1 and −2). We also identified proteins involved in fatty acid and amino acid synthesis pathways: fatty acid synthase (FASN) and glutamic-oxaloacetic transaminase (GOT1 and GOT2). This observation suggested that retinal oxidative metabolism was affected in early retinal degeneration. Defects in oxidative metabolism have been previously shown to contribute to photoreceptor degeneration in RP patients [42]. For example, loss-of-function mutations in the IDH3B gene encoding the beta subunit of NAD-specific isocitrate dehydrogenase cause non-syndromic RP [43]. We similarly noted increased levels of FASN in the vitreous of Pde6ɑD670G mice at the onset of photoreceptor degeneration. Loss of FASN in the neural retina has been shown to result in progressive neurodegeneration resembling RP in mouse models [20]. Additionally, we have previously shown that metabolic and antioxidant stress proteins were regionally expressed within the human retina, in particular the OXPHOS and TCA cycle pathways, indicating that the retinal neural network is likely sensitive to changes within the metabolic pathways that occur during disease [11]. Although our human case represented a late-stage of disease, after loss of the rod neuronal cells, the validation and identification of a candidate biomarker with our early-stage Pde6ɑD670G mouse proteome suggested potential target pathways for therapeutic approaches (Fig. 5).
Fig. 5.
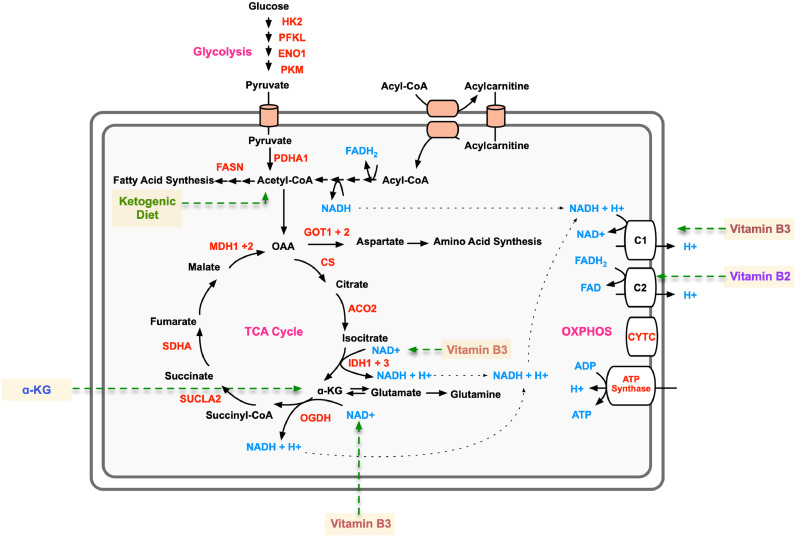
Proteomics-guided metabolite supplementation strategy for arRP. Metabolic pathway diagram highlighting identified significantly elevated metabolic proteins in the Pde6ɑD670G vitreous (red text). Metabolites are represented by blue text. Affected pathways include the TCA cycle, glycolysis, and oxidative phosphorylation (OXPHOS; magenta text). The metabolite/dietary therapies are highlighted in yellow.
3.4. The ketogenic diet delays photoreceptor cell loss in arRP mice
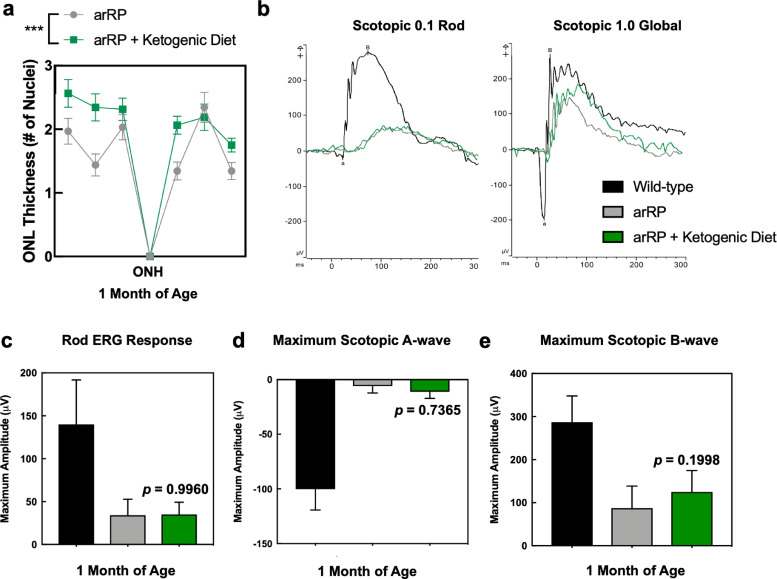
We tested the arRP candidate biomarker, FASN, by targeting this critical protein with a dietary approach. The ketogenic diet has gained public popularity for health and weight loss benefits, as well as being an effective treatment option for young patients with epileptic seizures that do not respond to standard drug treatments [44]. The ketogenic diet provides acetyl-CoA that can feed into both the TCA cycle and the fatty acid synthesis pathways (Fig. 5). We placed arRP mice on a ketogenic diet (6:1:1 fat:protein:carbohydrate) beginning at postnatal day 21 (P21), mid-stage of photoreceptor degeneration. Histological analysis of the outer nuclear layer (ONL) thickness showed mild, but significant, photoreceptor cell survival in mice treated with the ketogenic diet compared to untreated control littermates at one month of age (Fig. 6(a)). We next examined visual function at one month of age. Representative ERG traces of arRP mice treated with the ketogenic diet showed no difference in the rod photoreceptor cell response, but there was a mild rescue of the global visual response compared to littermates on standard chow diet (Fig. 6(b)). Quantification of ERG recordings from arRP mice treated with the ketogenic diet showed no significant difference in ERG responses compared to littermates on a standard chow diet, although there was a trend toward a- and b-wave visual rescue at the maximal scotopic 1.0 ERG setting (Fig. 6(c)–(e)). One limitation of the ketogenic diet was that the diet could not support nursing pups during early development, so it was not initiated until P21, at mid-stage of retinal disease when a rescue effect may be diminished. Therefore, we considered oral supplementation of metabolites that could be initiated earlier.
Fig. 6.
Treatment with the ketogenic diet rescues photoreceptor cell survival in the arRP preclinical mouse. (a) Histological analysis of the retinas of Pde6ɑD670G mice on normal chow diet (gray) compared to Pde6ɑD670G mice provided the ketogenic diet (green) shows a significant increase in the thickness of the outer nuclear layer (ONL) by number of photoreceptor cell nuclei in mice treated with the ketogenic diet at one month of age, one-week post-dietary treatment. Results are displayed as a morphometric quantification (spider graph) of ONL layer thickness superior (left) and inferior (right) to the optic nerve head (ONH). Wild-type mice have an ONL thickness of approximately 10 nuclei. ***p < 0.001 [Student's t-test]. N = 8 eyes each group, with multiple ONL thickness counts per eye as described in Methods. Error bars = SEM. (b) Representative traces of the scotopic 0.1 (flash strength in cd·s·m−2) dim-light rod-specific electroretinography (ERG; left) shows no significant difference in the rod photoreceptor cell response in Pde6ɑD670G mice treated with the ketogenic diet (green) in comparison to untreated Pde6ɑD670G mice (gray) at one month of age. Representative traces of the scotopic 1.0 (flash strength in cd·s·m−2) global electroretinography (ERG; left) shows some rescue of the global visual response in Pde6ɑD670G mice treated with the ketogenic diet (green) in comparison to wild-type controls (black) and untreated Pde6ɑD670G mice (gray) at one month of age. Quantification of ERGs from a cohort of mice shows no significant visual rescue of (c) the rod photoreceptor cell response, (d) the maximum scotopic a-wave response, or (e) the maximum scotopic b-wave response in mice treated with the ketogenic diet (green) in comparison with wild-type controls (black) or untreated Pde6ɑD670G mice (gray) at one month of age, although there was a trend toward an increased a- and b-wave maximal response. N = 5 WT, 6 arRP, 6 arRP + ketogenic diet mice.
3.5. Oral supplementation with single metabolites prolongs neuronal cell survival and vision
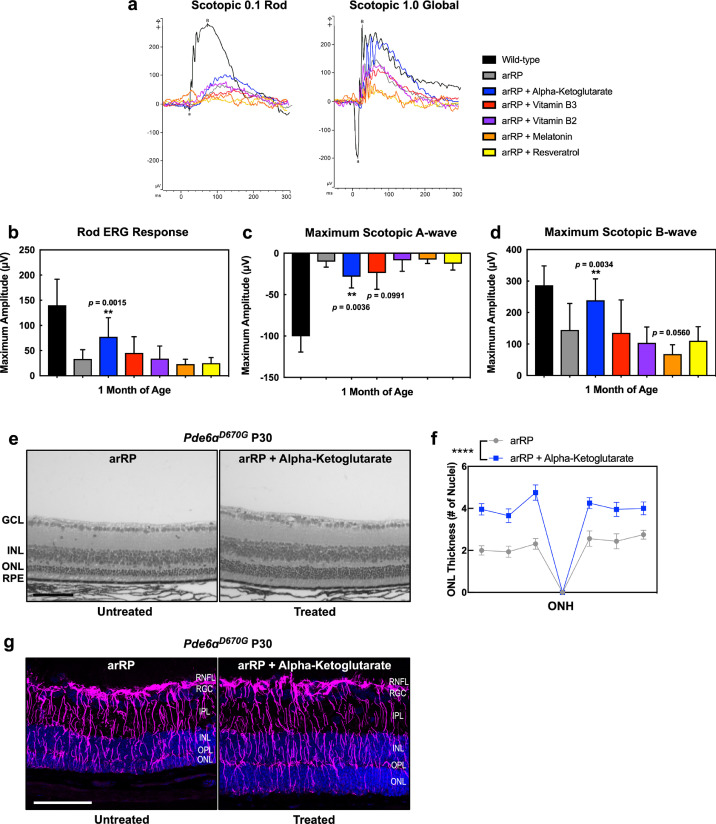
The onset of retinal cell degeneration occurs rapidly in the arRP preclinical model, before weaning age; therefore, metabolites were chosen that were known to transfer into the mother's milk and cross tissue barriers to be fed to the nursing pups [45,46]. Thus, Pde6ɑD670G mice were provided with ɑ-ketoglutarate (ɑ-KG), vitamin B3, vitamin B2, melatonin, or resveratrol in the drinking water beginning at postnatal day 0 (P0) and examined for functional visual rescue by ERG at one-month post-treatment (Fig. 5). Representative ERG traces showed visual rescue both in the rod photoreceptors, rod-cone photoreceptor a-wave response, and global inner retina scotopic ERG response in Pde6ɑD670G mice treated with ɑ-KG (Fig. 7). For vitamin B3 and vitamin B2, some mice showed enhanced ERG visual responses compared to controls, but analysis of a larger cohort of mice did not show statistical significance for photoreceptor cell and inner retina function on ERG (Fig. 7(b)–(d)). Both melatonin and resveratrol did not show an effect on visual function after supplementation in the Pde6ɑD670G arRP mice (Fig. 7(b)–(d)). Since ɑ-KG oral supplementation prolonged visual function in the Pde6ɑD670G mice, we examined the retina by histology (Fig. 7(e)–(f)). Histological analysis confirmed a statistically significant rescue of photoreceptor cells (ONL) and their inner/outer segments after treatment with ɑ-KG (Fig. 7(e)–(f)). It has previously been shown that glial fibrillary acidic protein (GFAP) expression is increased in Müller glial processes that extend throughout the retina in response to retinal stress [47]. The distribution of GFAP immunoreactivity was assessed to evaluate retinal response to injury in untreated arRP mice and arRP mice treated with ɑ-KG. Both groups showed consistently increased GFAP immunoreactivity spanning the length of the retina, suggesting that ɑ-KG supplementation does not inhibit Müller glial activation in this disease model after one month of treatment (Fig. 7(g)). Overall, oral supplementation with a single metabolite, ɑ-KG, provided a significant neuroprotective effect on the rod photoreceptors and neural retinal network for at least one month of age, when treated before disease onset. However, we cannot formally exclude the possibility that the systemic effects from oral supplementation led to a neuroprotective outcome. The rescue effects on photoreceptor cell survival and visual function after treatment with the ketogenic diet or ɑ-KG supported our human and mouse proteomic data, suggesting that the neural retina is sensitive to changes in metabolism, and that targeting the TCA cycle may be a potential therapeutic approach for RP patients.
Fig. 7.
Oral supplementation of single metabolites rescues photoreceptor cell survival and visual function in the arRP preclinical mouse. (a) Representative traces of the scotopic 0.1 (flash strength in cd·s·m−2) dim-light rod-specific electroretinography (ERG; left) and scotopic 1.0 (flash strength in cd·s·m−2) global ERG (right) in Pde6ɑD670G mice treated with ɑ-KG (blue), vitamin B2 (purple), vitamin B3 (red), melatonin (orange) or resveratrol (yellow) supplementation in comparison to untreated Pde6ɑD670G mice (gray) and wild-type controls (black) at one month of age. Quantification of ERGs from a cohort of mice treated with ɑ-KG (blue), vitamin B2 (purple), vitamin B3 (red), melatonin (orange) or resveratrol (yellow) supplementation shows significant visual rescue of (b) the rod photoreceptor cell response, (c) the maximum scotopic a-wave response, and (d) the maximum scotopic b-wave response in mice treated with ɑ-KG supplementation (blue) in comparison with wild-type controls (black) or untreated Pde6ɑD670G mice (gray) at one month of age. **p < 0.01; ***p < 0.001 [1-way ANOVA followed by Tukey's multiple comparison's test]. N = 5 WT, 6 arRP, 11 arRP + ɑ-KG mice, 8 arRP + vitamin B3, 7 arRP + vitamin B2, 8 arRP + melatonin and 7 arRP + resveratrol. (e-f) Histological analysis of the retinas of untreated Pde6ɑD670G mice (left) compared to treated Pde6ɑD670G mice (right) shows a significant increase in the thickness of the outer nuclear layer (ONL) by number of photoreceptor cell nuclei in mice treated with ɑ-KG supplementation (blue) at one month of age. GCL, ganglion cell layer; INL, inner nuclear layer; RPE, retinal pigment epithelium. Scale bar = 100 µm. (f) Results are displayed as a morphometric quantification (spider graph) of ONL layer thickness superior (left) and inferior (right) to the optic nerve head (ONH). Wild-type mice have an ONL thickness of approximately 10 nuclei. ****p < 0.0001 [Student's t-test]. N = 3 wild-type and 4 arRP + ɑ-KG eyes, with multiple ONL thickness counts per eye as described in Methods. Error bars = SEM. (g) Extensive Müller glial cell activation seen in arRP mice with and without ɑ-KG treatment. Retinas were dissected at one month of age from untreated arRP mice and arRP mice treated with ɑ-KG beginning at postnatal day 0. Retinas were stained with glial fibrillary acidic protein (GFAP; purple) and DAPI (blue) to identify retinal nuclei. In both the untreated arRP (left) and ɑ-KG treated (right) arRP mice there is increased expression of GFAP in Müller glia spanning the retina, suggesting that the treatment has no effect on Muller glial activation. N = 3 per group. Images taken at 25× magnification. Scale bar = 100 μm.
3.6. Metabolite/lipid imaging of retinal tissue sections
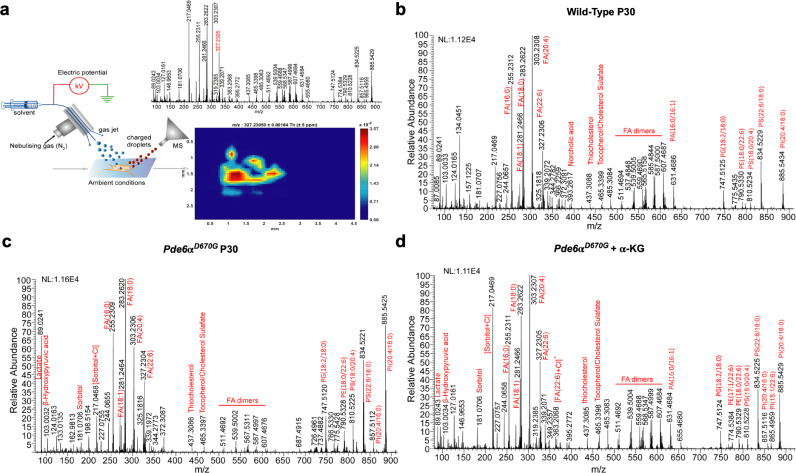
To investigate the lipid and metabolite profiles of Pde6ɑD670G mice with and without ɑ-KG treatment, we selected 35 fresh frozen tissue samples and performed negative ion mode desorption electrospray ionization mass spectrometry imaging (DESI-MSI; Fig. 8(a)). Retina tissue samples were harvested from wild-type (n = 8), untreated Pde6ɑD670G (n = 8), and ɑ-KG treated Pde6ɑD670G mice (n = 14) at one-month of age. Additionally, whole eye (n = 5 for each group) samples were also harvested. For each case, 10 μm frozen sections were used for DESI-MSI. Each pixel (200 µm) in the tissue DESI-MS image is represented with the mass spectrum in the 50–1,000 m/z range. The molecular ions identified were small metabolites related to glycolysis, TCA cycle, fatty acids, and phospholipids (Fig. 8(b)–(d)). The relative intensity distribution of each individual metabolite in the wild type, arRP, and treated tissue of both retina and the whole eye were shown in 2D chemical heatmaps (Fig. S4-S5). Across 35 total tissue samples, 26,341 unique peaks were found in the 50–1,000 m/z range. Significance analysis of microarrays (SAM) identified 377 differentially expressed metabolites between untreated Pde6ɑD670G and ɑ-KG treated mice. Although SAM returned 377 metabolites with notable fold-change (absolute fold change >1), to help interpret the biological role of metabolites, we restricted our analysis to peaks for which tandem-MS and high mass resolution analyses were performed by using the LTQ-Orbitrap XL (Thermo Scientific; Figs. S6–S7; Table S16). To check whether orally supplemented metabolites had reached the retina, we first analyzed the changes in relative intensity for ɑ-KG (m/z 145.014) in our DESI-MSI data. Although DESI-MSI did detect ɑ-KG (Fig. S8) in some pixels, there was limited change between Pde6ɑD670G and ɑ-KG treated groups. This is possible if the orally supplemented ɑ-KG is being consumed for the production of other metabolites in TCA and in glutamate-glutamine conversion pathways, which were ascertained next.
Fig. 8.
DESI-MSI of retinal tissue sections: (a) schematic representation of the DESI-MSI in the range m/z 50–1,000. The impinging primary charged droplets desorb the surface metabolites into the secondary charged droplets, followed by the ion transfer to the mass spectrometer. The 2D-chemical map of docosahexaenoic acid (DHA; peak listed in red font) is shown with pixel size 200 µm. Raw MS spectrum across the pixels in a row is shown for (b) wild type retina tissue (c) arRP retina tissue and (d) ɑ-KG treated retina tissue. Red labels denote peaks assigned to identified metabolites and lipids. NL, normalization level; FA, fatty acid; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine.
Identification of differentially distributed metabolites from SAM revealed several altered pathways between untreated Pde6ɑD670G and ɑ-KG treated groups that were consistent with our proteomic findings: uridine, dihydrouridine, and thymidine (pyrimidine and purine metabolism), docosahexaenoic and arachidonic acid (lipid metabolism), glutamine and glutamyl-alanine (glutamine/glutamate conversion and aminobutryate degradation), and ascorbic acid (oxidative metabolism). We found intermediates in the TCA cycle that were highly elevated in ɑ-KG treated mice compared to the untreated Pde6ɑD670G mice. For example, α-KG is broken down and reduced to succinyl-CoA, which is then converted to succinic acid. From DESI-MSI imaging and SAM, we noted that succinic acid (m/z 117.01) levels were elevated in the treated state (fold change 1.3, FDR < 0.05) compared to the untreated group (Fig. 9(a)). This indicated that the consumption of the supplemental α-KG in the treatment group is reflected in higher levels of succinic acid. Additionally, aconitic acid (m/z 173.0) is an intermediate in the conversion of citrate to isocitrate in the TCA cycle, which can also be converted from ɑ-KG. Aconitic acid levels were elevated in the treated state (fold change 2.5, FDR < 0.05), compared to wild-type and untreated Pde6ɑD670G levels (Fig. 9(b)). Since it is known that ɑ-KG can act as a substrate for amino acid synthesis, particularly glutamine, we investigated whether the consumption of ɑ-KG was reflected in glutamine synthesis pathways. Glutamine levels were elevated (fold change 1.87, FDR < 0.05) in treated tissues compared to untreated Pde6ɑD670G mice (Fig. 9(c)). Glutamate levels were slightly increased in treated tissues compared to untreated Pde6ɑD670G mice (Fig. 9(d)). Therefore, DESI-MSI provided some indication that supplemental ɑ-KG was being consumed to increase levels of TCA metabolites such as aconitic acid and glutamine levels, which likely play a role in the cell's antioxidant response [48].
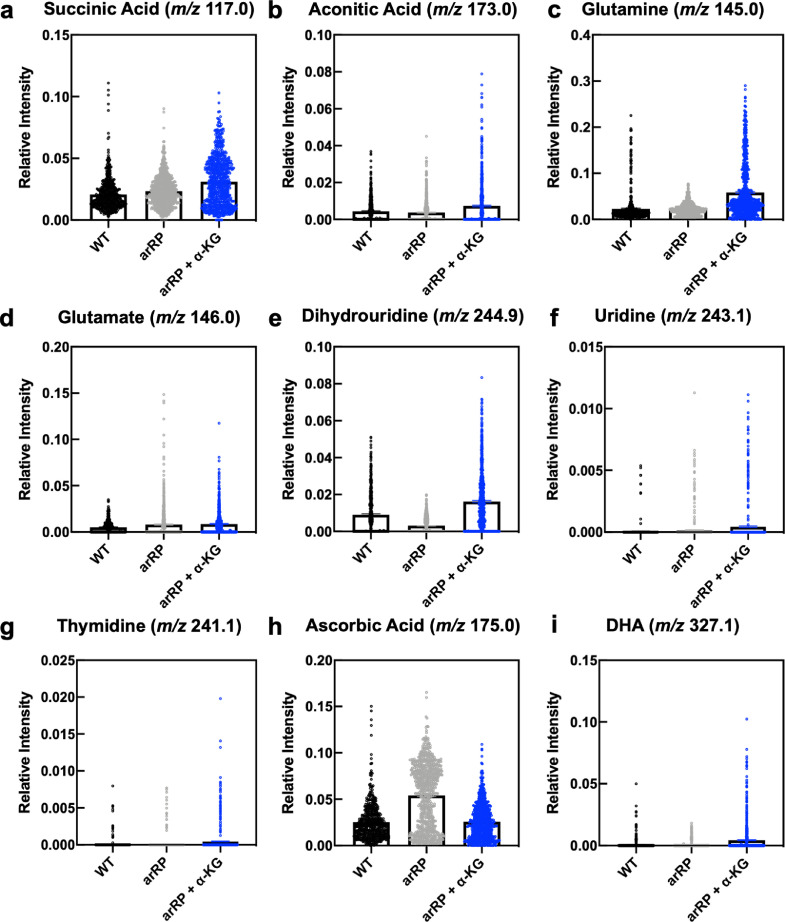
Fig. 9.
Distributions of retinal lipids and metabolite relative intensities across wild-type, arRP, and ɑ-KG treated mice from DESI-MSI data: Distribution of (a) succinic acid (m/z 117.01), (b) aconitic acid (m/z 173.0), (c) glutamine (m/z 145.0), (d) glutamate (m/z 146.0), (e) dihydrouridine (m/z 244.9), (f) uridine (m/z 243.1), (g) thymidine (m/z 241.1), (h) ascorbic acid, and (i) docosahexaenoic acid (DHA; m/z 327.1) across wild-type, arRP, and ɑ-KG treated mice. Results are displayed as scatter plots of the relative intensity of the corresponding metabolite normalized by the total ion current (sum of all intensity values from the detected ions). Boxes represent the mean ± SEM.
Consistent with the pathway representation of our vitreous proteomic biomarkers, DESI-MSI independently detected altered levels of metabolites related to pyrimidine metabolism. Notably, our results from SAM revealed that levels of dihydrouridine (m/z 244.98) were lower in Pde6ɑD670G mice and restored following ɑ-KG treatment (fold change 18.1, FDR < 0.05; Fig. 9(e)). Similarly, levels of thymidine (m/z 241.17) and uridine (m/z 243.16) were elevated in ɑ-KG treated mice compared to Pde6ɑD670G and wild-type mice, further suggesting that ɑ-KG treatment influenced pyrimidine and purine metabolism (Fig. 9(f)–(g)). This trend of altered nucleotide metabolism in arRP has been found in other murine metabolomics analyses [49]. Weiss et al. had previously used broad-spectrum metabolomics to report links between altered levels of thymidine and other pyrimidine nucleotide metabolic products and RP in a different RP murine model [49]. Overall, our work, in conjunction with other previously published efforts, suggests that pyrimidine metabolism is altered in RP and may be related to mitochondrial DNA replication pathways [48].
The proteomics data indicated the downregulation of oxidative pathways in Pde6ɑD670G retina compared to wild-type retina. We therefore looked to determine if any of the identified metabolites from SAM reflected a change in oxidative pathways between the disease and treated groups. Ascorbic acid (m/z 175.06) was found at significantly higher intensities in untreated disease samples compared to the treated group (fold change 1.9, FDR < 0.05; Fig. 9(h)). It is well-known that RP is characterized by rod photoreceptor cell death followed by slower cone death, where cone death is associated with oxidative damage [50]. Our DESI-MSI results revealed that the untreated disease samples had a higher distribution of ascorbic acid, which is a known antioxidant. Additionally, there were metabolites that did not change significantly in the disease state but were elevated by ɑ-KG treatment. For example, docosahexaeonic acid (DHA; m/z 327.12) levels were elevated (fold change 1.9, FDR < 0.05) in the treated mice compared to untreated mice (Fig. 9(i)). DHA accounts for a significant fraction of omega-3 fatty acids in the retina, and studies have found links between reduction in DHA with altered retinal function, including in RP [50,51]. This result is consistent with the numerous suggested protective roles of DHA in the retina, including anti-inflammatory, anti-angiogenesis, and anti-neurotoxic mechanisms [52].
4. Discussion
Liquid biopsies and the analysis of fluid compartments near diseased tissues represents an innovative tool in precision health to overcome the current limitations of tissue biopsies. This advancement is exemplified in the personalized treatment of cancer, where circulating tumor-derived material (e.g. circulating tumor DNA/RNA, extracellular vesicles, and cells) are detected in the blood [53]. These cellular biomarkers of the tumor microenvironment aid clinicians to better determine the appropriate therapeutic approach and predict prognosis. Neurodegenerative diseases, however, currently lack early detection methods for diagnosis and screening. In the case of retinal degenerative diseases, we have previously shown that liquid biopsies of the adjacent vitreous fluid can capture intracellular events in the neural retinal network that can guide diagnosis and treatment [23,41]. Advancements in this approach could allow for serial sampling and longitudinal monitoring of disease progress without the need to biopsy the neurosensory retina [24,36]. Our proteomic analysis of end-stage human arRP vitreous highlighted molecular pathways that might be affected during the progression of neuronal cell death and retinal remodeling. In order to gain further insight into the earlier disease stages, we utilized a preclinical murine model of arRP.
We examined the retina and vitreous proteomes from Pde6ɑD670G mice at early, mid, and late stages of neurodegeneration. Since there was a large number of differentially expressed proteins between Pde6ɑD670G mice and wild-type controls, we chose to focus on the proteins that were highly represented in the pathway analysis (Fig. 4(b)). The identification of related proteins within known pathways (e.g. TCA cycle, pyrimidine metabolism, phototransduction, etc.) allowed for focused interpretation of the data. In the early disease stage, we observed rod phototransduction proteins were depleted in the retina and elevated within the vitreous before significant loss of the photoreceptor cell nuclei is detectable by histological analysis. This suggests that molecular biomarkers are likely to appear in the human vitreous much earlier than the clinical biomarkers currently used for diagnosis. In human patients, liquid biopsies could similarly be used as an early diagnostic to confirm the onset of photoreceptor degeneration, even without diagnosis of a specific genetic mutation by genetic testing. Additionally, we found that the vitreous proteomes differed in the arRP mouse model at early-, mid-, and late-stage of disease. It may be possible to use this liquid biopsy approach to estimate the molecular stage of disease based on the proteomic content of the vitreous. Comparison of proteomes before and after therapeutic approaches can also be studied using liquid biopsy and may have important implications in gene therapy studies. Further, our DESI-MSI data complemented our understanding of key altered pathways detected by our proteomics and pathway analysis approach. Specifically, DESI-MSI provided a precise approach to investigate how changes at the protein level are reflected in the retinal metabolome of arRP mice following ɑ-KG treatment. Using DESI-MSI and SAM, we identified several significantly altered metabolites between disease and treated groups, across multiple pathways, including the TCA cycle, glutamine synthesis, pyrimidine metabolism, oxidative pathways, and fatty acid metabolism. It is likely that DESI-MSI in tandem with studying the vitreous proteome can help identify potential biomarkers that help elucidate effects of metabolite therapy.
At the early timepoint, the Pde6ɑD670G mice lose proteins critical for oxidative phosphorylation and aerobic metabolism, likely depressing protein turnover and leading to the cell death of the rod photoreceptors. Although the number of patient biopsies was limited and the disease stage was late, there were overlapping proteins in the vitreous proteome of the Pde6ɑD670G mice and humans. The loss of proteins involved in critical metabolic pathways from the retina was supported by our previous proteomics of the human retina that highlighted the sensitivity of the neural retinal network to changes within the metabolic pathways [11]. Interestingly, previous research studies have shown that metabolic rewiring is likely to be both a cause and a consequence of photoreceptor degeneration [54], and that the disruption of normal energy metabolism plays a key role in photoreceptor degeneration in the rd10 RP mouse model [49]. We also found that there were significant alterations in purine and pyrimidine nucleotide metabolism pathways at disease onset in our arRP mouse model, similar to the results in the rd10 RP mouse study (Table S12). Taken together, the proteomic profiles suggest targeting the affected metabolic pathways by replenishing them though dietary supplementation.
The most effective therapy tested was ɑ-KG, a key component of the TCA cycle that acts on various downstream metabolic pathways within the cell and determines the overall rate of the energetic process [49] as well as enhancing reductive carboxylation and supporting redox homeostasis in photoreceptors [55,56]. In addition to its metabolic roles, ɑ-KG can be converted to glutamine, and subsequently glutathione GSH, thereby facilitating the synthesis of potent antioxidants in the cell [48]. These antioxidant properties may have beneficial effects in the context of RP therapy. Additional studies have shown that ɑ-KG supplementation can extend the lifespan of C. elegans through the inhibition of ATP synthase and TOR [57] as well as increase circulating plasma levels of insulin and growth hormones in humans [58]. In the mouse, ɑ-KG derivatives have been delivered as a treatment for hypoxic conditions without any notations of adverse side effects [59,60]. We found that oral supplementation with ɑ-KG alone provided significant visual rescue of the rods, cones, and inner retina visual responses through at least one month of age. We chose the dosage of ɑ-KG based on published literature for supplementation in the mouse drinking water [61]. Additional studies are needed to determine the lowest dosage of ɑ-KG and its durability. Previous studies in mice have demonstrated that high systemic doses of ɑ-KG affected body weight and intestinal innate immunity by influencing the intestinal microbiome [61]. In this study, mice treated with ɑ-KG appeared to display a smaller body stature than controls, and higher doses of ɑ-KG would likely be detrimental to the survival of the litters after birth. It is also possible that the rescue effect of ɑ-KG is indirect and a result of changes to the systemic metabolism. One way to distinguish between these effects in future studies would be through intraocular administration of ɑ-KG.
The ketogenic diet has been shown to have neuroprotective effects in the context of several neurodegenerative diseases, such as Alzheimer's and Parkinson's disease [62]. The neuroprotective effect of the ketogenic diet has been hypothesized to be a result of the biochemical changes resulting from glycolytic inhibition, increased acetyl-CoA production, and formation of ketone bodies in the cell [63]. These biochemical changes have proven effective in targeting cancer cells, which primarily use glycolysis as their main source of ATP [64]. Photoreceptors are similarly sensitive to glycolytic inhibition but can attenuate the impact of glycolytic inhibition by using alternative sources of ATP (e.g. fatty acid beta-oxidation) [15,65]. The high rate of aerobic ATP synthesis in photoreceptors can lead to the production of harmful reactive oxygen species (ROS), which are implicated in the progression of retinal degenerative diseases [42]. We have previously shown that different anatomical regions of the human retina are susceptible to oxidative stress, highlighting the importance of antioxidant availability in the retina [11]. Ketone bodies increase mitochondrial respiration via increased ATP production and reduce ROS formation by increasing NADH oxidation, thereby improving the efficiency of the mitochondrial respiratory chain complex [62]. Targeting the TCA cycle and fatty acid synthase pathway by acetyl-CoA production via the ketogenic diet led to increased global retinal visual function compared to controls by one month of age. As the ketogenic diet could not be provided until weaning age, the effect was limited based on beginning the dietary treatment after approximately half of the neuronal photoreceptor cells have already been lost. Nevertheless, significant photoreceptor cell survival was detectable by histological analysis after treatment with the ketogenic diet. This suggests that earlier delivery of metabolites targeting the TCA cycle may lead to prolonged visual rescue in the Pde6ɑD670G mouse model and neuroprotection of the photoreceptor cells and inner retinal network.
Other metabolites tested in our arRP preclinical mouse model were less effective. Nicotinamide adenine dinucleotide (NAD), available from vitamin B3 supplementation, is a critical co-enzyme for aerobic metabolism and OXPHOS. In particular, retinal levels of NAD decrease with age and may leave neuronal cells susceptible to insults and disease [66]. Furthermore, vitamin B3 supplementation has shown positive effects toward enhancing energy metabolism and proper functioning of the central nervous system [67] as well as acting as a protective agent against glaucoma and age-related neurodegeneration [66]. Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), which can be delivered via vitamin B2 supplementation, are critical for enzymatic reactions within the electron transport chain and the TCA cycle. Vitamin B2 acts as an antioxidant by promoting regeneration of glutathione and is indispensable for cellular growth and supplementation with vitamin B2 has shown possible beneficial effects for prevention of hyperhomocysteinemia, cataracts and migraine headaches [68]. We found that targeting OXPHOS by oral supplementation with vitamins B2 or B3 provided an electrophysiological rescue effect in some, but not all, neuronal cells of treated mice compared to their respective controls. Further studies are needed to test combination therapy with our effective treatment groups (ɑ-KG, the ketogenic diet, and/or vitamins B2 and B3) that may have a synergistic effect in rescuing the neuronal cell loss and more closely reproduce a strategy to use in human arRP patients. Longer-acting structural analogs of these metabolites may also provide a longer lasting effect.
We also tested metabolites that act on the sirtuin pathway and may indirectly affect cellular metabolism. Melatonin is produced within the retina and is known to stimulate sirtuin activity in mitochondria, which deacetylates many of the enzymes involved within the TCA cycle. Previous studies have shown that supplementation with melatonin has no known toxicity affects when taken during pregnancy in both humans and mice [69,70]. Additionally, resveratrol is a plant-derived polyphenol that has been widely studied for its potential benefits as a protective agent against cancers, mutagenesis, and a variety of other biological functions [70]. It also acts to stimulate the sirtuin pathway, and this stimulation as well as its antioxidative actions are thought to provide a neuroprotective effect in both rat and mouse studies [70]. Supplementation with resveratrol has no known toxicity in humans even at high dosages, and the only affect noted when taken during pregnancy in mice was a slight increase in the body weight of the pups after birth [71]. However, in our study, oral supplementation with either melatonin or resveratrol did not provide a neuroprotective effect on the photoreceptor cells in our preclinical model of arRP. This suggests that directly targeting the TCA cycle may be the most beneficial approach for future RP therapy.
The proteomic and DESI-MSI approach in this study provided key pathways for targeting and testing therapeutics, and delivery of metabolites within these critical pathways in the arRP preclinical mouse model showed efficacious neuroprotective effects on the inner retina. The results from these studies lay the groundwork for future experiments to address how the TCA cycle and aerobic metabolism influence the photoreceptors and their signaling to the inner retinal network, and thus improve upon neuroprotective approaches targeting photoreceptor cell metabolism. Overall, the key metabolic pathways (OXPHOS and TCA cycle) detected in our proteomics screen are critical targets for therapeutics for PDE6-related RP. Further investigation is required to determine if this approach could be applicable to other forms of RP or other human neurodegenerative disorders.
Funding sources
VBM and AGB are supported by NIH grants [R01EY026682, R01EY024665, R01EY025225, R01EY024698, R21AG050437 and P30EY026877], and Research to Prevent Blindness (RPB), New York, NY. GV is supported by NIH grants [F30EYE027986 and T32GM007337]. Jonas Children's Vision Care and Bernard & Shirlee Brown Glaucoma Laboratory are supported by the National Institute of Health [5P30EY019007, R01EY018213], National Cancer Institute Core [5P30CA013696], the Research to Prevent Blindness (RPB) Physician-Scientist Award, unrestricted funds from RPB, New York, NY, USA. Foundation Fighting Blindness [TA-NMT-0116–0692-COLU], the Research to Prevent Blindness (RPB) Physician-Scientist Award, and unrestricted funds from RPB, New York, NY, USA. S.H.T. is a member of the RD-CURE Consortium and is supported by Kobi and Nancy Karp, the Crowley Family Fund, the Rosenbaum Family Foundation, the Tistou and Charlotte Kerstan Foundation, the Schneeweiss Stem Cell Fund, New York State [C029572], and the Gebroe Family Foundation. RNZ is supported by NSF grant CHE-1734082.
Data and materials availability
All data associated with this study are present in the paper or the Supplementary Materials. The mass spectrometry proteomics data have been deposited to Mendeley Data with the dataset identifiers (DOI): 10.17632/x9mkhv743y.2 and 10.17632/vn5c8zh5wd.2.
Declaration of Competing Interest
Dr. Wert has nothing to disclose. Mr. Velez has nothing to disclose. Dr. Kanchustambham has nothing to disclose. Mr. Shankar has nothing to disclose. Ms. Evans has nothing to disclose. Dr. Sengillo has nothing to disclose. Dr. Zare has nothing to disclose. Dr. Bassuk has nothing to disclose. Dr. Tsang has nothing to disclose. Dr. Mahajan has nothing to disclose.
Acknowledgments
We thank Jing Yang, Daniel A. Machlab, and Teja Chemudupati for technical assistance. We thank Bioproximity for assistance with mass spectrometry data collection. We thank Patsy Nishina for mouse model resources. We thank Jianhai Du and his laboratory members from the University of Washington for advice and discussions.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102636.
Appendix. Supplementary materials
Table S3. Relative quantification of proteins identified in human vitreous samples.
Table S5. Distinct peptides identified in Pde6ɑD670G retina and vitreous samples.
Table S6. Protein groups identified in Pde6ɑD670G retina and vitreous samples.
Table S7. Relative quantification and comparison of proteins identified in Pde6ɑD670G retina samples.
Table S8. Relative quantification and comparison of proteins identified in Pde6ɑD670G vitreous samples.
Table S9. Differentially-expressed Pde6ɑD670G retina and vitreous proteins in all disease stages.
Table S10. Differentially-expressed Pde6ɑD670G retina and vitreous proteins in early disease stages.
References
- 1.Procaccini C., Santopaolo M., Faicchia D., Colamatteo A., Formisano L., de Candia P. Role of metabolism in neurodegenerative disorders. Metabolism. 2016;65(9):1376–1390. doi: 10.1016/j.metabol.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Lehtonen S., Sonninen T.M., Wojciechowski S., Goldsteins G., Koistinaho J. Dysfunction of cellular proteostasis in Parkinson's disease. Front Neurosci. 2019;13:457. doi: 10.3389/fnins.2019.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strohm L., Behrends C. Glia-specific autophagy dysfunction in als. Semin Cell Dev Biol. 2019 doi: 10.1016/j.semcdb.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 4.Olivares-Banuelos T.N., Chi-Castaneda D., Ortega A. Glutamate transporters: gene expression regulation and signaling properties. Neuropharmacology. 2019 doi: 10.1016/j.neuropharm.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 5.Shukla M., Chinchalongporn V., Govitrapong P., Reiter R.J. The role of melatonin in targeting cell signaling pathways in neurodegeneration. Ann NY Acad Sci. 2019;1443(1):75–96. doi: 10.1111/nyas.14005. [DOI] [PubMed] [Google Scholar]
- 6.Ambrogini P., Torquato P., Bartolini D., Albertini M.C., Lattanzi D., Di Palma M. Excitotoxicity, neuroinflammation and oxidant stress as molecular bases of epileptogenesis and epilepsy-derived neurodegeneration: the role of vitamin E. Biochim Biophys Acta Mol Basis Dis. 2019;1865(6):1098–1112. doi: 10.1016/j.bbadis.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Chi H., Chang H.Y., Sang T.K. Neuronal cell death mechanisms in major neurodegenerative diseases. Int J Mol Sci. 2018;19(10) doi: 10.3390/ijms19103082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsang S.H., Sharma T. Stargardt disease. Adv Exp Med Biol. 2018;1085:139–151. doi: 10.1007/978-3-319-95046-4_27. [DOI] [PubMed] [Google Scholar]
- 9.Luo Y.H., da Cruz L. The Argus((R)) II retinal prosthesis system. Prog Retin Eye Res. 2016;50:89–107. doi: 10.1016/j.preteyeres.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velez G., Machlab D.A., Tang P.H., Sun Y., Tsang S.H., Bassuk A.G. Proteomic analysis of the human retina reveals region-specific susceptibilities to metabolic- and oxidative stress-related diseases. PLoS ONE. 2018;13(2) doi: 10.1371/journal.pone.0193250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Tornquist P., Bill A. Glucose metabolism in pig outer retina in light and darkness. Acta Physiol Scand. 1997;160(1):75–81. doi: 10.1046/j.1365-201X.1997.00030.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang L., Tornquist P., Bill A. Glucose metabolism of the inner retina in pigs in darkness and light. Acta Physiol Scand. 1997;160(1):71–74. doi: 10.1046/j.1365-201X.1997.00131.x. [DOI] [PubMed] [Google Scholar]
- 14.Graymore C. Possible significance of the isoenzymes of lactic dehydrogenase in the retina of the rat. Nature. 1964;201:615–616. doi: 10.1038/201615b0. [DOI] [PubMed] [Google Scholar]
- 15.Noell W.K. The effect of iodoacetate on the vertebrate retina. J Cell Comp Physiol. 1951;37(2):283–307. doi: 10.1002/jcp.1030370209. [DOI] [PubMed] [Google Scholar]
- 16.Chertov A.O., Holzhausen L., Kuok I.T., Couron D., Parker E., Linton J.D. Roles of glucose in photoreceptor survival. J Biol Chem. 2011;286(40):34700–34711. doi: 10.1074/jbc.M111.279752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayan D.S., Chidlow G., Wood J.P., Casson R.J. Glucose metabolism in mammalian photoreceptor inner and outer segments. Clin Exp Ophthalmol. 2017;45(7):730–741. doi: 10.1111/ceo.12952. [DOI] [PubMed] [Google Scholar]
- 18.Vaughn A.E., Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome C. Nat Cell Biol. 2008;10(12):1477–1483. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson J.P., Faulkner A., Vernon R.G. Regulation of glycolysis and fatty acid synthesis from glucose in sheep adipose tissue. Biochem J. 1982;206(3):577–586. doi: 10.1042/bj2060577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajagopal R., Zhang S., Wei X., Doggett T., Adak S., Enright J. Retinal de novo lipogenesis coordinates neurotrophic signaling to maintain vision. JCI Insight. 2018;3(1) doi: 10.1172/jci.insight.97076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyal J.S., Gantner M.L., Smith L.E.H. Retinal energy demands control vascular supply of the retina in development and disease: the role of neuronal lipid and glucose metabolism. Prog Retin Eye Res. 2018;64:131–156. doi: 10.1016/j.preteyeres.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., Du J., Justus S., Hsu C.W., Bonet-Ponce L., Wu W.H. Reprogramming metabolism by targeting sirtuin 6 attenuates retinal degeneration. J Clin Invest. 2016;126(12):4659–4673. doi: 10.1172/JCI86905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velez G., Roybal C.N., Colgan D., Tsang S.H., Bassuk A.G., Mahajan V.B. Precision medicine: personalized proteomics for the diagnosis and treatment of idiopathic inflammatory disease. JAMA Ophthalmol. 2016;134(4):444–448. doi: 10.1001/jamaophthalmol.2015.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahajan V.B., Skeie J.M. Translational vitreous proteomics. Proteomics Clin Appl. 2014;8(3–4):204–208. doi: 10.1002/prca.201300062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowell H.A., Bassuk A.G., Mahajan V.B. Monozygotic twins with CAPN5 autosomal dominant neovascular inflammatory vitreoretinopathy. Clin Ophthalmol. 2012;6:2037–2044. doi: 10.2147/OPTH.S40086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahajan V.B., Skeie J.M., Bassuk A.G., Fingert J.H., Braun T.A., Daggett H.T. Calpain-5 mutations cause autoimmune uveitis, retinal neovascularization, and photoreceptor degeneration. PLoS Genet. 2012;8(10) doi: 10.1371/journal.pgen.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skeie J.M., Brown E.N., Martinez H.D., Russell S.R., Birkholz E.S., Folk J.C. Proteomic analysis of vitreous biopsy techniques. Retina. 2012;32(10):2141–2149. doi: 10.1097/IAE.0b013e3182562017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wert K.J., Davis R.J., Sancho-Pelluz J., Nishina P.M., Tsang S.H. Gene therapy provides long-term visual function in a pre-clinical model of retinitis pigmentosa. Hum Mol Genet. 2013;22(3):558–567. doi: 10.1093/hmg/dds466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wert K.J., Sancho-Pelluz J., Tsang S.H. Mid-stage intervention achieves similar efficacy as conventional early-stage treatment using gene therapy in a pre-clinical model of retinitis pigmentosa. Hum Mol Genet. 2014;23(2):514–523. doi: 10.1093/hmg/ddt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wert K.J., Bassuk A.G., Wu W.H., Gakhar L., Coglan D., Mahajan M. CAPN5 mutation in hereditary uveitis: the R243L mutation increases calpain catalytic activity and triggers intraocular inflammation in a mouse model. Hum Mol Genet. 2015;24(16):4584–4598. doi: 10.1093/hmg/ddv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wert K.J., Skeie J.M., Bassuk A.G., Olivier A.K., Tsang S.H., Mahajan V.B. Functional validation of a human CAPN5 exome variant by lentiviral transduction into mouse retina. Hum Mol Genet. 2014;23(10):2665–2677. doi: 10.1093/hmg/ddt661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skeie J.M., Tsang S.H., Mahajan V.B. Evisceration of mouse vitreous and retina for proteomic analyses. J Vis Exp. 2011;(50) doi: 10.3791/2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new panther GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–DD26. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsang S.H., Tsui I., Chou C.L., Zernant J., Haamer E., Iranmanesh R. A novel mutation and phenotypes in phosphodiesterase 6 deficiency. Am J Ophthalmol. 2008;146(5):780–788. doi: 10.1016/j.ajo.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skeie J.M., Roybal C.N., Mahajan V.B. Proteomic insight into the molecular function of the vitreous. PLoS ONE. 2015;10(5) doi: 10.1371/journal.pone.0127567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azar W.J., Azar S.H., Higgins S., Hu J.F., Hoffman A.R., Newgreen D.F. IGFBP-2 enhances VEGF gene promoter activity and consequent promotion of angiogenesis by neuroblastoma cells. Endocrinology. 2011;152(9):3332–3342. doi: 10.1210/en.2011-1121. [DOI] [PubMed] [Google Scholar]
- 38.Park S.H., Kim K.W., Kim J.C. The role of insulin-like growth factor binding protein 2 (IGFBP2) in the regulation of corneal fibroblast differentiation. Invest Ophthalmol Vis Sci. 2015;56(12):7293–7302. doi: 10.1167/iovs.15-16616. [DOI] [PubMed] [Google Scholar]
- 39.Afshari F.T., Kwok J.C., Andrews M.R., Blits B., Martin K.R., Faissner A. Integrin activation or alpha 9 expression allows retinal pigmented epithelial cell adhesion on Bruch's membrane in wet age-related macular degeneration. Brain. 2010;133(Pt 2):448–464. doi: 10.1093/brain/awp319. [DOI] [PubMed] [Google Scholar]
- 40.Malissen B., Schmitt-Verhulst A.M. Transmembrane signalling through the T-cell-receptor-CD3 complex. Curr Opin Immunol. 1993;5(3):324–333. doi: 10.1016/0952-7915(93)90049-x. [DOI] [PubMed] [Google Scholar]
- 41.Velez G., Tang P.H., Cabral T., Cho G.Y., Machlab D.A., Tsang S.H. Personalized proteomics for precision health: identifying biomarkers of vitreoretinal disease. Transl Vis Sci Technol. 2018;7(5):12. doi: 10.1167/tvst.7.5.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punzo C., Xiong W., Cepko C.L. Loss of daylight vision in retinal degeneration: are oxidative stress and metabolic dysregulation to blame? J Biol Chem. 2012;287(3):1642–1648. doi: 10.1074/jbc.R111.304428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartong D.T., Dange M., McGee T.L., Berson E.L., Dryja T.P., Colman R.F. Insights from retinitis pigmentosa into the roles of isocitrate dehydrogenases in the Krebs cycle. Nat Genet. 2008;40(10):1230–1234. doi: 10.1038/ng.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Andrea Meira I., Romao T.T., Pires do Prado H.J., Kruger L.T., Pires M.E.P., da Conceicao P.O. Ketogenic diet and epilepsy: what we know so far. Front Neurosci. 2019;13:5. doi: 10.3389/fnins.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smilowitz J.T., O'Sullivan A., Barile D., German J.B., Lonnerdal B., Slupsky C.M. The human milk metabolome reveals diverse oligosaccharide profiles. J Nutr. 2013;143(11):1709–1718. doi: 10.3945/jn.113.178772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vazquez-Fresno R. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–DD17. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis G.P., Fisher S.K. Up-regulation of glial fibrillary acidic protein in response to retinal injury: its potential role in glial remodeling and a comparison to vimentin expression. Int Rev Cytol. 2003;230:263–290. doi: 10.1016/s0074-7696(03)30005-1. [DOI] [PubMed] [Google Scholar]
- 48.Liu S., He L., Yao K. The antioxidative function of alpha-ketoglutarate and its applications. Biomed Res Int. 2018;2018 doi: 10.1155/2018/3408467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss E.R., Osawa S., Xiong Y., Dhungana S., Carlson J., McRitchie S. Broad spectrum metabolomics for detection of abnormal metabolic pathways in a mouse model for retinitis pigmentosa. Exp Eye Res. 2019;184:135–145. doi: 10.1016/j.exer.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komeima K., Rogers B.S., Lu L., Campochiaro P.A. Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc Natl Acad Sci U S A. 2006;103(30):11300–11305. doi: 10.1073/pnas.0604056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman D.R., DeMar J.C., Heird W.C., Birch D.G., Anderson R.E. Impaired synthesis of DHA in patients with X-linked retinitis pigmentosa. J Lipid Res. 2001;42(9):1395–1401. [PubMed] [Google Scholar]
- 52.Querques G., Forte R., Souied E.H. Retina and omega-3. J Nutr Metab. 2011;2011 doi: 10.1155/2011/748361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Rubis G., Rajeev Krishnan S., Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40(3):172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Du J., An J., Linton J.D., Wang Y., Hurley J.B. How excessive cGMP impacts metabolic proteins in retinas at the onset of degeneration. Adv Exp Med Biol. 2018;1074:289–295. doi: 10.1007/978-3-319-75402-4_35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang L., Shestov A.A., Swain P., Yang C., Parker S.J., Wang Q.A. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin R.M., Fu X., Pai M.Y., Vergnes L., Hwang H., Deng G. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510(7505):397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu N., Yang M., Gaur U., Xu H., Yao Y., Li D. Alpha-Ketoglutarate: physiological functions and applications. Biomol Ther (Seoul) 2016;24(1):1–8. doi: 10.4062/biomolther.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou P., Kuo C.Y., Cheng C.T., Liou J.P., Ann D.K., Chen Q. Intermediary metabolite precursor dimethyl-2-ketoglutarate stabilizes hypoxia-inducible factor-1alpha by inhibiting prolyl-4-hydroxylase PHD2. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0113865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacKenzie E.D., Selak M.A., Tennant D.A., Payne L.J., Crosby S., Frederiksen C.M. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27(9):3282–3289. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen S., Bin P., Ren W., Gao W., Liu G., Yin J. Alpha-ketoglutarate (AKG) lowers body weight and affects intestinal innate immunity through influencing intestinal microbiota. Oncotarget. 2017;8(24):38184–38192. doi: 10.18632/oncotarget.17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wlodarek D. Role of ketogenic diets in neurodegenerative diseases (Alzheimer's disease and Parkinson's disease) Nutrients. 2019;11(1) doi: 10.3390/nu11010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puchalska P., Crawford P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 2017;25(2):262–284. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber D.D., Aminazdeh-Gohari S., Kofler B. Ketogenic diet in cancer therapy. Aging (Albany NY) 2018;10(2):164–165. doi: 10.18632/aging.101382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joyal J.S., Sun Y., Gantner M.L., Shao Z., Evans L.P., Saba N. Retinal lipid and glucose metabolism dictates angiogenesis through the lipid sensor Ffar1. Nat Med. 2016;22(4):439–445. doi: 10.1038/nm.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams P.A., Harder J.M., Foxworth N.E., Cochran K.E., Philip V.M., Porciatti V. Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science. 2017;355(6326):756–760. doi: 10.1126/science.aal0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mills K.F., Yoshida S., Stein L.R., Grozio A., Kubota S., Sasaki Y. Long-Term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 2016;24(6):795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peechakara BV, Gupta M. Vitamin B2 (Riboflavin). StatPearls. Treasure Island (FL)2019. [PubMed]
- 69.Guan S., Xie L., Ma T., Lv D., Jing W., Tian X. Effects of melatonin on early pregnancy in mouse: involving the regulation of StAR, Cyp11a1, and Ihh expression. Int J Mol Sci. 2017;18(8) doi: 10.3390/ijms18081637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramis M.R., Esteban S., Miralles A., Tan D.X., Reiter R.J. Caloric restriction, resveratrol and melatonin: role of SIRT1 and implications for aging and related-diseases. Mech Ageing Dev. 2015;146–148:28–41. doi: 10.1016/j.mad.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 71.Zheng S., Feng Q., Cheng J., Zheng J. Maternal resveratrol consumption and its programming effects on metabolic health in offspring mechanisms and potential implications. Biosci Rep. 2018;38(2) doi: 10.1042/BSR20171741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Relative quantification of proteins identified in human vitreous samples.
Table S5. Distinct peptides identified in Pde6ɑD670G retina and vitreous samples.
Table S6. Protein groups identified in Pde6ɑD670G retina and vitreous samples.
Table S7. Relative quantification and comparison of proteins identified in Pde6ɑD670G retina samples.
Table S8. Relative quantification and comparison of proteins identified in Pde6ɑD670G vitreous samples.
Table S9. Differentially-expressed Pde6ɑD670G retina and vitreous proteins in all disease stages.
Table S10. Differentially-expressed Pde6ɑD670G retina and vitreous proteins in early disease stages.