Summary
Autism spectrum disorders (ASD) are a group of neurodevelopmental disorders with symptoms including social deficits, anxiety, and communication difficulties. However, ASD pathogenic mechanisms are poorly understood. Mutations of CUL3, which encodes Cullin 3 (CUL3), a component of an E3 ligase complex, are thought of as risk factors for ASD and schizophrenia (SCZ). CUL3 is abundant in the brain, yet little is known of its function. Here we showed that CUL3 is critical for neurodevelopment. CUL3 deficient mice exhibited social deficits and anxiety-like behaviors with enhanced glutamatergic transmission and neuronal excitability. Proteomic analysis revealed eIF4G1, a protein for Cap-dependent translation, as a potential target of CUL3. ASD-associated cellular and behavioral deficits could be rescued by pharmacological inhibition of the eIF4G1 function and chemogenetic inhibition of neuronal activity. Thus, CUL3 is critical to neural development, neurotransmission, and excitation-inhibition (E-I) balance. Our study reveals novel insight into pathophysiological mechanisms of ASD and SCZ.
ETOC paragraph:
Mutations of CUL3, a component of an E3 ligase complex, are thought of as risk factors for Autism spectrum disorders (ASD) and schizophrenia. Here Dong and colleagues showed CUL3 deficiency in mice caused social deficits and anxiety-like behaviors and enhanced glutamatergic transmission and neuronal excitability. Proteomic analysis revealed eIF4G1, a protein for Cap-dependent translation, as a potential target of CUL3 deficiency. Pharmacological inhibition of eIF4G1 and chemogenetic inhibition of neuronal activity attenuated ASD-associated cellular and behavioral deficits.
Introduction
Efficient synaptic transmission requires proper synaptic structure and functional machineries that control neurotransmitter release and activation of postsynaptic receptors. Dysregulated synapse formation and synaptic transmission could cause E-I imbalance, a mechanism of various neuropsychiatric disorders (Penzes et al., 2011; Rubenstein and Merzenich, 2003). This includes ASD, which are often associated with hyper-glutamatergic and/or hypo-GABAergic functions (Coghlan et al., 2012; Fatemi, 2008; Fatemi et al., 2009).
ASD are neurodevelopmental disorders that affect 1 in 59 children in the US (Baio et al., 2018). Clinical signs, symptoms that are frequently comorbid with ASD include social deficits, anxiety, communication difficulties and repetitive behaviors (Bryson et al., 2003; Gillott et al., 2001). Also associated are anatomical abnormalities including reduced brain volume (Carper et al., 2002), decreased cortical thickness (Wegiel et al., 2014), alterations in dendritic branching and spine densities (Raymond et al., 1995), and agenesis of corpus callosum (Lau et al., 2013). However, pathological mechanisms of ASD are poorly understood although several genes have been identified including MECP2 for Rett’s Syndrome (Liyanage and Rastegar, 2014), FMR1 for Fragile X Syndrome (Crawford et al., 2001), UBE3A for Angelman syndrome and Prader-Willi Syndrome (Buiting, 2010) and SHANK3 for autism (Moessner et al., 2007). Mice lacking these genes are deficient in social interaction or recognition or display repetitive self-grooming, anxiety and/or seizure that are closely associated with ASD (Auerbach et al., 2011; Banerjee et al., 2014; Bourgeron, 2009; Crawley, 2012; Homberg et al., 2016; Jacquemont et al., 2007; Lee et al., 2017; Rubenstein and Merzenich, 2003; Shepherd and Katz, 2011; Silverman et al., 2010; Takahashi et al., 2012). They are often plagued with neurodevelopmental deficits, abnormal neuronal connectivity, and disrupted E-I balance. However, causal genetic mutations have not been identified in more than 70% of cases (Schaaf and Zoghbi, 2011).
Cullin 3 (CUL3) is a component of the CUL3-RING E3 ubiquitin ligase (CRL) complex, consisting of CUL3, RING-box protein 1 (RBX1), and a Bric-a-brac/Tramtrack/Broad (BTB) protein (Deshaies, 1999; Pickart, 2001; Pintard et al., 2004). It regulates a plethora of cell functions such as anti-oxidation, cell cycle, protein trafficking and signal transduction (Anderica-Romero et al., 2013; Cullinan et al., 2004). Recently, de novo mutations have been identified in CUL3 in exome analyses of parent-child trios exhibiting sporadic ASD by two independent studies using overlapping samples (Kong et al., 2012; O’Roak et al., 2012). In addition, CUL3 is one of the 107 risk genes identified in a genome-wide association study (GWAS) of a large population of autism cases (De Rubeis et al., 2014). Third, CUL3 has been associated with schizophrenia (SCZ) in a multi-stage SCZ GWAS spanning 108 conservatively defined loci; and its mutations overlap with those in ASD (Schizophrenia Working Group Of The Psychiatric Genomics Consortium et al., 2014). Although CUL3 is abundantly expressed in the brain, little is known of its role in the nervous system or pathogenic mechanisms of CUL3 mutations.
In this study, we investigated the function of CUL3 in neural development and transmission. Because ASD-associated mutations of CUL3 are paternal origin nonsense mutations (De Rubeis et al., 2014; Kong et al., 2012; O’Roak et al., 2012) that likely produce truncated, dysfunctional proteins, we focused on Cul3 heterozygous mice. CUL3 deficient mice displayed impairment in social ability, anxiety level, E-I balance and spine density. Proteomic analysis of brain samples from mutant mice identified eIF4G1, a key component of Cap-dependent translation, as a potential target of CUL3. Finally, we explored if phenotypes of mutant mice could be rescued by pharmacological inhibition of eIF4G1 and chemogenetic inhibition of neuronal activity. Results indicated a critical role of CUL3 in neural development, neurotransmission, and E-I balance and reveal insight into pathophysiological mechanisms of ASD and SCZ.
Results
Social deficits and anxiety-like behaviors of CUL3 deficient mice
We crossed Cul3f/f mice with GFAP::Cre mice that express Cre under the promoter of GFAP, a gene that is expressed in neural progenitor cells of both neurons and astrocytes (Kriegstein and Alvarez-Buylla, 2009; Noctor et al., 2001; Zhuo et al., 2001). Cre in GFAP-Cre mice is expressed in the majority of projection neurons in the hippocampus (99%) and cortex (88%)(Madisen et al., 2010; Malatesta et al., 2003; Zhuo et al., 2001). Cul3 level was reduced in gene-dosage-dependent manner in Cul3f/+, GFAP-Cul3f/+ or GFAP-Cul3f/f mice (Figure S1A). The body size and brain weight of GFAP-Cul3f/f mice were decreased, compared with Cul3f/+ (hereafter referred as control) mice (Figure S1B). GFAP-Cul3f/f mice showed premature death before P17 (Figures S1C), with decreased brain size and cortical thickness, disrupted cortical layers, agenesis of corpus callosum and hippocampal deformation (Figures S1D–G). These results indicate that Cul3 is critical for brain development. However, these deficits were not observed in heterozygous (GFAP-Cul3f/+) mice (Figures S1D–G) although Cul3 level was reduced by more than 40% (Figure S1A). GFAP-Cul3f/+ mice were viable and fertile and survived as long as control mice (Figure S1C). Because Cul3 mutations in patients are mostly heterozygous, GFAP-Cul3f/+ (also referred as mt hereafter) mice would serve as a model to study the pathogenic mechanisms of Cul3 loss-of-function.
First, we sought to determine whether Cul3 mutation alters social ability, a major target of ASD and SCZ. Mice were subjected to three-chamber social interaction tests (Figure 1A) for social preference and social memory (Moy et al., 2004). In social preference tests, control mice spent more time with social targets (a stranger mouse, S1), compared with an inanimate object (O) (Figure 1B). However, there was no difference in time spent between social targets and inanimate objects by GFAP-Cul3f/+ mice. The sniffing time of mt mice towards S1 was reduced, compared with control mice. In accord, the social preference index was reduced in mt mice (Fig. 1C), suggesting impaired social preference (Figures 1B, 1C). Next, we compared social memory between control and mt mice. Control mice spent more time with a new stranger mouse (S2), compared with that for a familiar mouse (S1) (Figure 1D). However, GFAP-Cul3f/+ mice spent less time with S2 mice, compared with littermate controls. The sniffing time of mt mice towards S2 was reduced, compared with control mice (Fig. 1D). And, the social novelty index was reduced in mt mice (Fig. 1E), suggesting an impairment in social memory. Notice that control and mt mice showed similar latency to localize buried food pellets in an olfactory sensing test (Figures S2A, S2B), suggesting normal olfactory sensing of mt mice. Together, these results indicate that GAFP-Cul3f/+ mice are impaired in social preference and social memory.
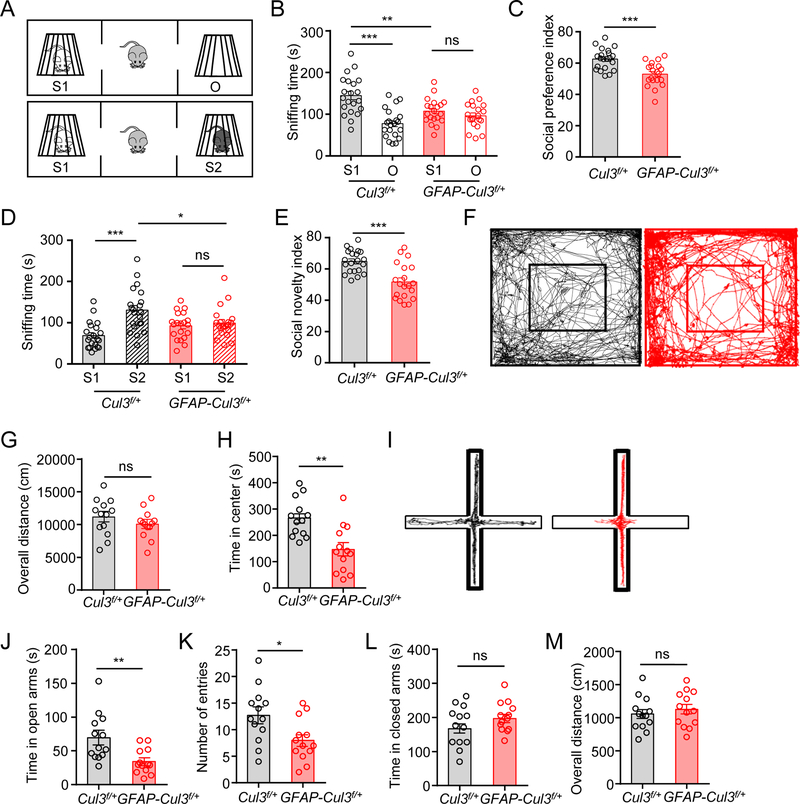
Figure 1. Social deficits and anxiety-like behaviors in CUL3 deficient mice.
(A) Schematic diagram of three-chamber social interaction tests.
(B) Reduced sniffing time with a novel mouse (S1) in GFAP-Cul3f/+ mice, compared with control mice. n = 21 mice for control; n = 20 mice for GFAP-Cul3f/+; control S1 (143 ± 9.4 s) vs control O (77.7 ± 7.5 s), p < 0.001; control S1 vs GFAP-Cul3f/+ S1 (108 ± 6.7 s), p = 0.009; GFAP-Cul3f/+ S1 vs GFAP-Cul3f/+ O (96.2 ± 6.9 s), p > 0.999; Two-way ANOVA followed by Bonferroni’s post hoc test.
(C) Reduced social preference index in GFAP-Cul3f/+ mice. n = 21 mice for control; n = 20 mice for GFAP-Cul3f/+; control (62.5 ± 1.5) vs GFAP-Cul3f/+ (52.9 ± 1.7), p = 0.0002; U = 71; Mann-Whitney test.
(D) Reduced sniffing time with a stranger mouse (S2) in GFAP-Cul3f/+ mice. n = 21 mice for control; n = 20 mice for GFAP-Cul3f/+; control S1 (68.9 ± 7.0 s) vs control S2 (131 ± 11.2 s), p < 0.001; control S2 vs GFAP-Cul3f/+ S2 (98.2 ± 8.4 s), p = 0.015; GFAP-Cul3f/+ S1 (92.5 ± 7.2 s) vs GFAP-Cul3f/+ S2, p > 0.999; Two-way ANOVA followed by Bonferroni’s post hoc test.
(E) Reduced social novelty index in GFAP-Cul3f/+ mice. n = 21 mice for control; n = 20 mice for GFAP-Cul3f/+; control (64.8 ± 1.6) vs GFAP-Cul3f/+ (51.7 ± 2.5), p = 0.0002; U = 73; Mann-Whitney test.
(F) Representative traces of 30 min in open field tests.
(G) No difference in total distance traveled. n = 13 mice for each genotype; control (11200 ± 1010 cm) vs GFAP-Cul3f/+ (10100 ± 789 cm), p = 0.3167; U = 64.5; Mann-Whitney test.
(H) Reduced time spent in the center. n = 13 mice for each genotype; control (266 ± 28.8 s) vs GFAP-Cul3f/+ (146 ± 36.2 s), p = 0.0012; U = 24; Mann-Whitney test.
(I) Representative traces of 10 min in the elevated plus maze test.
(J) Reduced time in open arms. n = 13 mice for each genotype; control (69.3 ± 12.6s) vs GFAP-Cul3f/+ (34.2 ± 7.1 s), p = 0.0037; U = 29.5; Mann-Whitney test.
(K) Reduced entries into open arms. n = 13 mice for each genotype; control (14.1 ± 1.7) vs GFAP-Cul3f/+ (9.5 ± 1.8), p = 0.0211; U = 40; Mann-Whitney test.
(L) No difference in time spent in closed arms. n = 13 mice for each genotype; control (167 ± 23.0 s) vs GFAP-Cul3f/+ (197 ± 17.8 s), p = 0.2593; U = 52; Mann-Whitney test.
(M) No difference in total distance traveled. n = 13 mice for each genotype; control (1100 ± 265 cm) vs GFAP-Cul3f/+ (1130 ± 99.2 cm), = 0.4717; U = 70; Mann-Whitney test.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
In addition, in open field arena, GFAP-Cul3f/+ mice spent less time in the center, compared with Cul3f/+ littermate controls (without Cre), indicating increased anxiety (Figures 1F, 1H). Social deficits and anxiety were not due to a change in locomotion because the total distance traveled in open field tests and latency to fall in accelerated rotarod tests were comparable between the two genotypes (Figures 1G, S2C, S2D). To further test anxiety, mt mice were subjected to elevated plus mazes (EPM) (Pellow et al., 1985). The entries into open arms by mt mice were fewer and the total time they spent in open arms were reduced, compared with control littermates (Figures 1I–K). The time spent in closed arms, as well as overall distance traveled in EPM were similar between the two groups (Figures 1L, 1M). These results indicate that CUL3 deficiency increased anxiety.
To determine whether CUL3 deficiency alters working memory, mice were subjected to Y-Maze tests (Aggleton et al., 1986). GFAP-Cul3f/+ displayed similar numbers of arm entries and spontaneous alterations, compared to control mice (Figures S2E–G), suggesting no obvious impairment of spatial working memory by CUL3 deficiency. Shank3 and Sapap3 mutant mice displayed repeated grooming phenotypes (Peca et al., 2011; Welch et al., 2007). However, GFAP-Cul3f/+ and control mice displayed similar time and numbers of grooming episodes (Figures S2H, S2I), consistent with no change in spontaneous alterations in Y-maze tests, which is also considered as a measure of repetitive behavior (Yadin et al., 1991). These results indicated that CUL3 deficiency specifically increased anxiety and impaired social interaction without altering grooming behavior and working memory.
Increased spines, glutamatergic transmission and E-I imbalance
NeuN-labeled neurons in CA1 and CA3 and at different layers of the neocortex at age of P60 were similar between GFAP-Cul3f/+ and control littlermates (Figures S3A–D), indicating that CUL3 deficiency did not alter the number of neurons in these regions. Golgi staining revealed similar total length and complexity of apical and basal dendrites of CA1 neurons between the two groups (Figures S3E–G). However, the numbers of spines on apical dendrites of CA1 neurons were increased in GFAP-Cul3f/+ mice, compared with control mice at age of P60 (Figures 2A, 2B). Thus, Cul3 joins a group of genes including Nsmf (Kallmann syndrome), Fmr1 (fragile X mental retardation 1) and NRCAM, whose mutation leads to increased number of spines (De Rubeis and Bagni, 2011; Hutcheson et al., 2004; Jiang et al., 2013; Martinez-Cerdeno, 2017).
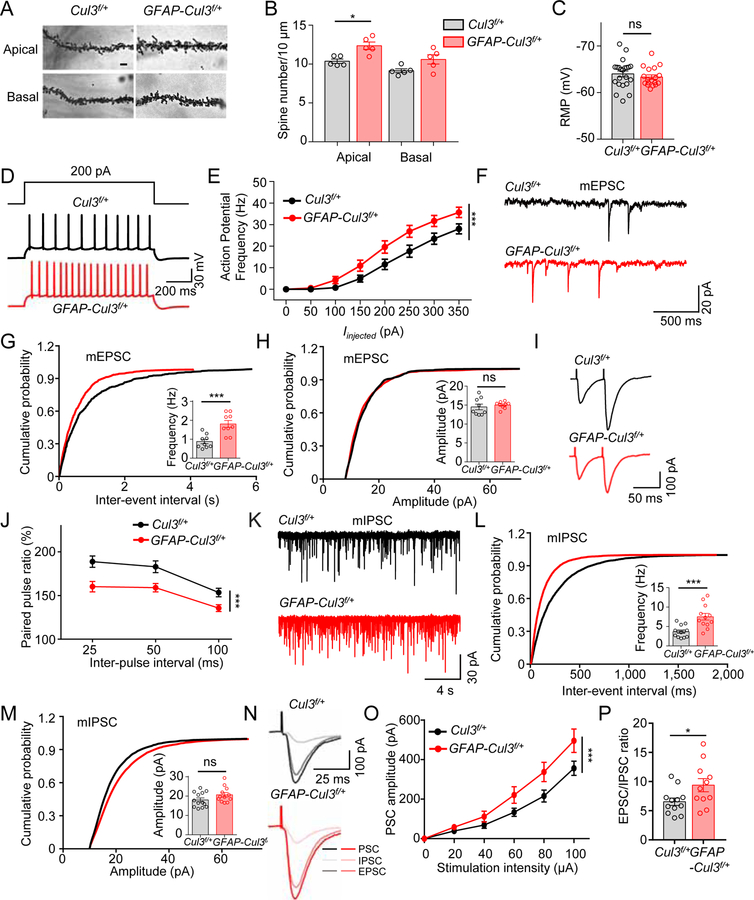
Figure 2. Increased spine density, neuronal excitability, synaptic transmission and disrupted E-I balance in CA1 hippocampal pyramidal neurons of adult GFAP-Cul3f/+ mice.
(A) Representative Golgi staining images of apical and basal dendrites of CA1 pyramidal neurons of P60 control and GFAP-Cul3f/+ mice. Scale bar, 5 µm
(B) Increased spine density in apical dendrites of CA1 GFAP-Cul3f/+ pyramidal neurons. n = 5 mice per genotype; spine density in apical dendrites, control (10.3 ± 0.3) vs GFAP-Cul3f/+ (12.4 ± 0.6), p = 0.0317; U = 2; Mann-Whitney test.
(C) Comparable resting membrane potentials of CA1 pyramidal neurons. n = 22 neurons, 5 mice for Cul3f/+ group; n = 18 neurons, 4 mice for GFAP-Cul3f/+ group; control (−63.9 ± 0.7 mV) vs GFAP-Cul3f/+ (−63.3 ± 0.4 mV), p = 0.3701; U = 164.5; Mann-Whitney test.
(D) Representative traces of spikes evoked by injecting depolarizing currents.
(E) Firing rate plotted against increasing injected currents. n = 13 neurons, 3 mice for Cul3f/+ group; n = 18 neurons, 4 mice for GFAP-Cul3f/+ group; p < 0.0001; F(1, 232) = 25.27; Two-way ANOVA followed by Bonferroni’s post hoc test.
(F) Representative mEPSC traces.
(G) Increased mEPSC frequency. n = 9 neurons, 3 mice for both genotypes; control (0.88 ± 0.16 Hz) vs GFAP-Cul3f/+ (1.8 ± 0.2 Hz), p = 0.0005; U = 4; Mann-Whitney test.
(H) No difference in mEPSC amplitude. n = 9 neurons, 3 mice for both genotypes; control (14.3 ± 1.0 pA) vs GFAP-Cul3f/+ (15.0 ± 0.4 pA), p = 0.4225; U = 31; Mann-Whitney test.
(I) Representative traces of pair-pulse stimulation.
(J) PPRs plotted against inter-stimulus intervals. n = 11 neurons, 3 mice for Cul3f/+ group; n = 13 neurons, 3 mice for GFAP-Cul3f/+ group; p < 0.001; F(1, 66) = 27.26; Two-way ANOVA.
(K) Representative mIPSC traces in CA1 pyramidal neurons.
(L) Increased mIPSC frequency. n = 13 neurons, 3 mice for Cul3f/+ group; n = 12 neurons, 3 mice for GFAP-Cul3f/+ group; control (3.7 ± 0.5 Hz) vs GFAP-Cul3f/+ (7.5 ± 1.0 Hz), p = 0.0003; U = 16; Mann-Whitney test.
(M) No difference in mIPSC amplitude. n = 13 neurons, 3 mice for Cul3f/+ group; n = 12 neurons, 3 mice for GFAP-Cul3f/+ group; control (18.0 ± 1.5 pA) vs GFAP-Cul3f/+ (20.7 ± 1.6 pA), p = 0.1095; U = 48; Mann-Whitney test.
(N) Representative of postsynaptic currents (PSC), EPSCs and IPSCs by sequentially evoked synaptic responses. Neurons were recorded initially for PSCs without inhibitors and then for IPSCs in the presence of CNQX and APV. IPSCs were inhibited by bicuculline (data not shown). EPSCs were derived by subtracting IPSCs from PSCs.
(O) Increased PSC amplitudes. n = 9 neurons, 3 mice for Cul3f/+ group; n = 9 neurons, 3 mice for GFAP-Cul3f/+ group; p = 0.0001; F(1, 96) = 14.45; Two-way ANOVA.
(P) Increased EPSC/IPSC ratio in GFAP-Cul3f/+ mice (n = 12 neurons, 3 mice for Cul3f/+ group; n = 11 neurons, 3 mice for GFAP-Cul3f/+ group; control (6.5 ± 1.0) vs GFAP-Cul3f/+ (9.4 ± 1.6), p = 0.0268; U = 30; Mann-Whitney test).
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
Next, we determined whether CUL3 deficiency alters neuronal properties, including resting membrane potential (RMP), excitabilities and miniature excitatory postsynaptic currents (mEPSCs) and miniature inhibitory postsynaptic currents (mIPSCs). CUL3 deficiency had little effect on RMP of CA1 pyramidal neurons of adult (P60) mice (Figure 2C). However, the frequency of action potentials (APs) in response to injected currents was increased in GFAP-Cul3f/+ mice, compared with controls (Figures 2D, 2E), indicating elevated excitability. As shown in Figures 2F–H, mEPSC frequency, but not amplitude, of GFAP-Cul3f/+ CA1 neurons was increased, in agreement of increased spines. The elevated mEPSC frequency may also be contributed by increased glutamate release probability because paired-pulse ratios were reduced in GFAP-Cul3f/+ slices (Figures 2I, 2J), the decay of NMDAR currents in the presence of MK-801 was faster (Figures S3H–J), and the rate of successful responses and synaptic efficacy (but not potency) to minimal stimulation was higher, compared with control slices (Figures S3K, S3L).
To determine whether CUL3 deficiency alters GABAergic transmission, we measured mIPSCs in CA1 neurons. Compared with control littermates, mIPSC frequency, but not amplitude, was increased in GFAP-Cul3f/+ hippocampus (Figures 2K–M). Together with data described above, these results demonstrate that both glutamatergic and GABAergic transmissions are altered in mt mice. Moreover, E-I ratios were increased in GFAP-Cul3f/+ slices, compared with control slices (Figures 2N–P), revealing a pathological mechanism. These results suggest that Cul3 is necessary for proper neurotransmission.
Similar deficits from CUL3 deficiency in pyramidal neurons
Because the frequencies of both mEPSC and mIPSC were increased in adult CUL3 deficient mice, we next determined which deficit occurred earlier. As shown in Figures S4A–F, the frequency of mEPSC, but not mIPSC, was increased in hippocampal slices of young (~P15) GFAP-Cul3f/+, compared with controls. In agreement with what was observed in adult mice, no difference was detected in mEPSC or mIPSC amplitudes. The excitability of CA1 neurons was also increased in P15 mt mice (Figures S4G, S4H), with comparable RMP (Figure S4I). These results suggest that increased GABAergic transmission in adult mice may be a secondary or compensatory response to elevated glutamatergic activity. This notion was supported by increased spine numbers of CA1 neurons of mt mice at age of P15 (Figures S4J, S4K), but similar inhibitory synapses onto CA1 neurons (Figures S4L–N).
To further test this hypothesis, we specifically deleted Cul3 from pyramidal neurons by crossing Cul3f/f mice with NEX::Cre mice where Cre expression begins at embryonic day 11.5 in pyramidal neurons (Goebbels et al., 2006). CUL3 was reduced in the brain of NEX::Cre;Cul3f/f (NEX-Cul3f/f) mice (Figure S5A), which died prematurely prior to P21 with similar morphologic phenotypes observed in GFAP-Cul3f/f mice (Figures S5B–G). As with heterozygous mice mediated by GFAP-Cre, NEX-Cul3f/+ mice were viable and fertile, with ~30% reduction in CUL3 protein (Figures S5M S5N). NEX-Cul3f/+ mice showed no difference in dendrites of CA1 neurons, but spine numbers were increased (Figures S5H–L). NEX-Cul3f/+ CA1 neurons showed an increase in excitability, mEPSC frequency, mIPSC frequency, but no change in RMP (Figures 3A–I). At the behavioral level, NEX-Cul3f/+ mice exhibited similar deficits as GFAP-Cul3f/+ mice, including anxiety-like behaviors and abnormal social interaction (Figures 3J–Q, S5O, S5P). These results demonstrate a cell-autonomous role of CUL3 in developing pyramidal neurons for synaptic function, E-I balance and behaviors.
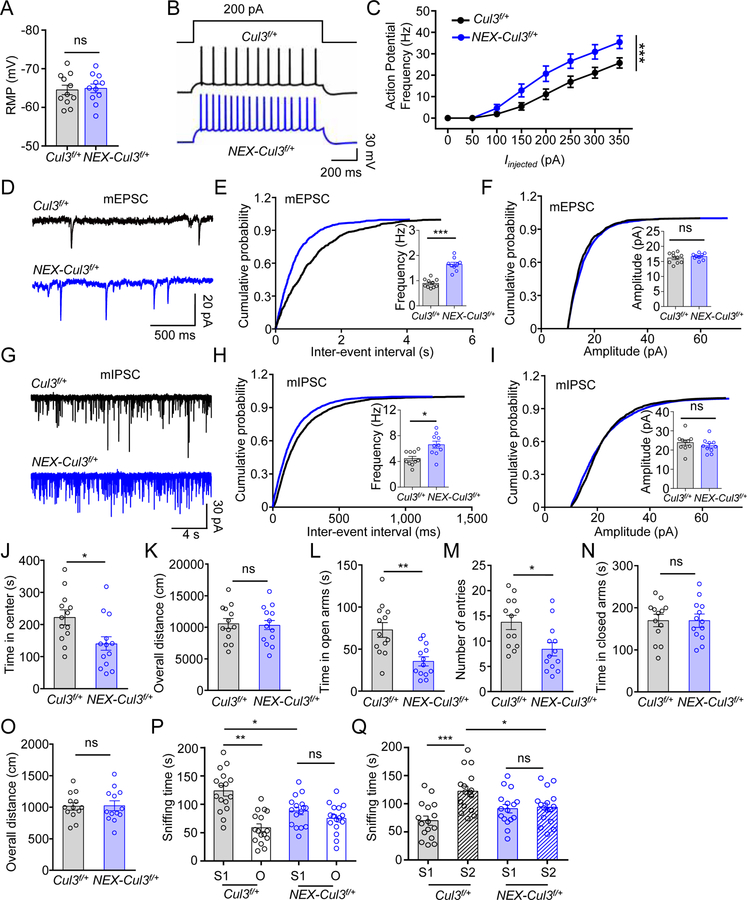
Figure 3. Similar neurotransmission and behavior deficits in adult NEX-Cul3f/+ mice.
(A) Comparable resting membrane potentials of CA1 pyramidal neurons. n = 11 neurons, 3 mice for both genotypes; control (−64.5 ± 1.8 mV) vs NEX-Cul3f/+ (−64.9 ± 1.6 mV), p = 0.7969; U = 56; Mann-Whitney test.
(B) Representative traces of spikes evoked by injecting depolarizing currents.
(C) Firing rates plotted against increasing injected currents. n = 10 neurons, 3 mice for Cul3f/+ group; n = 9 neurons, 3 mice for NEX-Cul3f/+ group; p < 0.0001; F(1, 136) = 28.76; Two-way ANOVA.
(D) Representative mEPSC traces.
(E) Increased mEPSC frequency. n = 10 neurons, 3 mice for Cul3f/+ group; n = 9 neurons, 3 mice for NEX-Cul3f/+ group; control (0.9 ± 0.06 Hz) vs NEX-Cul3f/+ (1.6 ± 0.1 Hz), p < 0.0001; U = 0; Mann-Whitney test.
(F) No difference in mEPSC amplitude. n = 10 neurons, 3 mice for Cul3f/+ group; n = 9 neurons, 3 mice for NEX-Cul3f/+ group; control (16.1 ± 0.6 pA) vs NEX-Cul3f/+ (16.6 ± 0.4 pA), p = 0.5490; U = 37; Mann-Whitney test.
(G) Representative mIPSC traces.
(H) Increased mIPSC frequency. n = 10 neurons, 3 mice for both genotypes; control (4.4 ± 0.4 Hz) vs NEX-Cul3f/+ (6.6 ± 0.8 Hz), p = 0.0107; U = 17; Mann-Whitney test.
(I) No difference in mIPSC amplitude. n = 10 neurons, 3 mice for both genotypes; control (23.7 ± 1.8 pA) vs NEX-Cul3f/+ (22.3 ± 1.6 pA), p = 0.2710; U = 35; Mann-Whitney test.
(J) Reduced time spent in the center. n = 13 mice for each genotype; control (215 ± 22.4 s) vs NEX-Cul3f/+ (149 ± 30.2 s), p = 0.0140; U = 37; Mann-Whitney test.
(K) No difference in total distance traveled. n = 13 mice for each genotype; control (10700 ± 804 cm) vs NEX-Cul3f/+ (10300 ± 718 cm), p = 0.8403; U = 80; Mann-Whitney test.
(L) Reduced time in open arms. n = 13 mice for each genotype; control (74.2 ± 10.7 s) s) vs NEX-Cul3f/+ (34.8 ± 7.0 s), p = 0.0015; U = 25; Mann-Whitney test.
(M) Reduced entries into open arms. n = 13 mice for each genotype; control (14.2 ± 1.8) vs NEX-Cul3f/+ (8.8 ± 1.8), p = 0.0081; U = 34; Mann-Whitney test.
(N) No difference in time spent in closed arms. n = 13 mice for each genotype; control (167 ± 19.6 s) vs NEX-Cul3f/+ (182 ± 17 s), p = 0.8910; U = 81.5; Mann-Whitney test.
(O) No difference in total distance traveled. n = 13 mice for each genotype; control (1030 ± 86.5 cm) vs NEX-Cul3f/+ (1010 ± 241 cm), p = 0.6958; U = 76.5; Mann-Whitney test.
(P) Reduced sniffing time with a novel mouse (S1) in NEX-Cul3f/+ mice than control mice. n = 16 mice per each group; control S1 (122 ± 9.2 s) vs control O (58.5 ± 6.9 s), p < 0.001; control S1 vs NEX-Cul3f/+ S1 (88.3 ± 6.6 s), p = 0.012; NEX-Cul3f/+ S1 vs NEX-Cul3f/+ O (75.2 ± 6.4 s), p > 0.999; F(1, 60) = 27.11; Two-way ANOVA followed by Bonferroni’s post hoc test.
(Q) Reduced sniffing time with a stranger mouse (S2) in NEX-Cul3f/+ mice than control mice. n = 16 mice per each group; control S1 (69.9 ± 8.2 s) vs control S2 (122 ± 9.0 s), p < 0.001; control S2 vs NEX-Cul3f/+ S2 (93.7 ± 7.5 s), p = 0.032; NEX-Cul3f/+ S1 (91.2 ± 7.5 s) vs NEX-Cul3f/+ S2, p > 0.999; F(1, 60) = 11.56; Two-way ANOVA followed by Bonferroni’s post hoc test.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
Increased Cap-dependent translation in CUL3 deficient brains
To investigate mechanisms of CUL3 deficiency, we identified proteins whose levels are altered in CUL3 mt mice by tandem mass tagging (TMT)-based quantitative proteomics analysis (Figure 4A). Of total 5720 proteins identified, 335 proteins were differentially expressed (DE) between GFAP-Cul3f/+ and control samples (p<0.05) and 1466 proteins were differentially expressed between GFAP-Cul3f/f and control samples (p<0.05) (Figures 4B–D, Table S1 and S2). Gene ontology analysis of increased proteins by Cul3 heterozygous mutation implicated neuronal excitability, SNARE complex disassembly, synaptic vesicle localization, synaptic vesicle cycle and presynaptic assembly including SNAP-α, SNAP-β, NSF, AP-3 complex subunit mu-2, CAPS1, Rabphilin-3A, eIF4G1, NLGN1, and PTEN (Figures 4E, S6A, S6B, Tables S3, and S4). However, DE proteins failed to survive FDR correction. Therefore, we performed western blot analysis to validate selected DE proteins. Remarkably, levels of VGLUT1, α/β-SNAP, NSF, VAMP1 were found to be increased in GFAP-Cul3f/+ samples (Figures S7A, S7B), revealing potential mechanisms of synaptic alteration in Cul3 heterozygous mutant mice. Cellular functions altered by Cul3 homozygous mutation included neurogenesis, neuron differentiation, protein localization, providing mechanisms for premature death and neural developmental deficits (Figures S6C, S6D, Tables S5 and S6). Of the genes in SFARI categories 1 and 2, three were differentially expressed in Cul3 mt mice – upregulated PTEN and downregulated BCKDK and CUL3 (Figure S6E). Western blot analysis confirmed higher protein levels of PTEN and SHANK1 (Figures S7A, S7B).
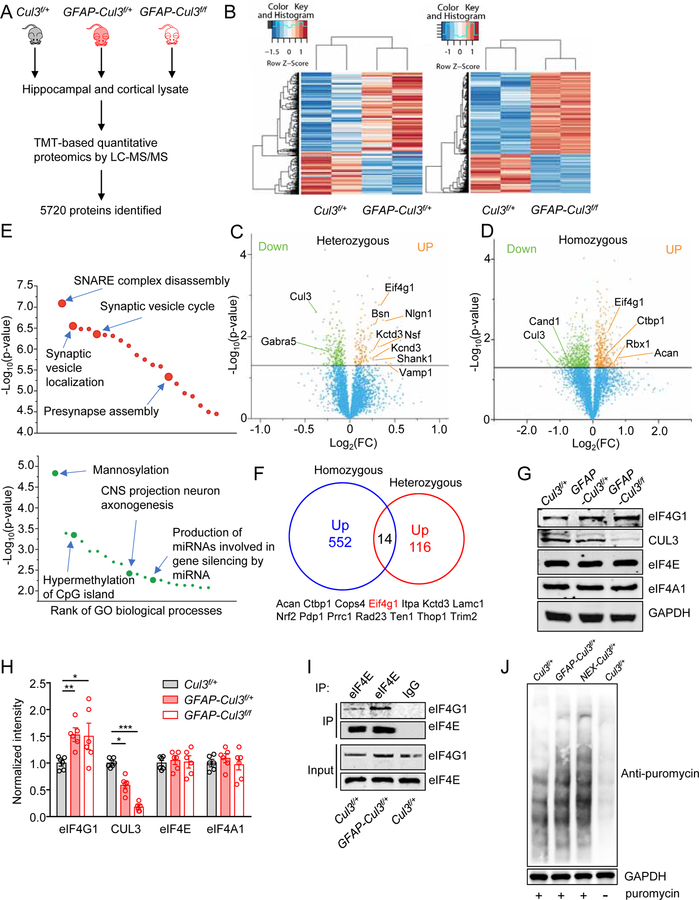
Figure 4. Abnormal expression of synaptic and autism-related proteins in CUL3 deficient brains.
(A) Schematic workflow of quantitative proteomic analysis. Proteomic data were obtained from pooled hippocampus and cortex. n = 3–4 mice for each group (P14, male).
(B) Heat maps of DE proteins in GFAP-Cul3f/+ and GFAP-Cul3f/f brains samples, compared with Cul3f/+ brains.
(C) Volcano plot for DE proteins (116 upregulated, 219 downregulated) in GFAP-Cul3f/+ (heterozygous) brain samples, compared with Cul3f/+ brains. Green and orange dots indicate statistically DE proteins.
(D) Volcano plot for DE proteins (552 upregulated, 914 downregulated) in GFAP-Cul3f/f (homozygous) brain samples, compared with Cul3f/+ brains. Green and orange dots indicate statistically DE proteins.
(E) GO enrichment analysis of DE proteins. DE proteins were enriched in biological processes including ‘SNARE complex disassembly’, ‘Synaptic vesicle localization’, ‘Mannosylation’ and ‘CNS projection neuron axonogenesis’. Full results of the analysis are presented in Supplementary Figures. 8A and 8B.
(F) Proteins upregulated in both GFAP-Cul3f/+ and GFAP-Cul3f/f mice brains.
(G) Representative blots for cortical tissues collected from P14 Cul3f/+ and GFAP-Cul3f/+ and GFAP-Cul3f/f mice.
(H) Quantification analysis of data in (H). Band densities of interested proteins were normalized by the loading control GAPDH; values of control mice were taken as 1; n = 6 mice per each genotype; eIF4G1, Cul3f/+ (1.01 ± 0.15) vs GFAP-Cul3f/+ (1.59 ± 0.14), p = 0.0458; Cul3f/+ vs GFAP-Cul3f/f (1.71 ± 0.3), p = 0.0319; CUL3, Cul3f/+ (1.02 ± 0.11) vs GFAP-Cul3f/+ (0.58 ± 0.07), p = 0.0012, Cul3f/+ vs GFAP-Cul3f/f (0.18 ± 0.03), p < 0.0001; One-way ANOVA followed by Tukey’s post hoc test.
(I) Increased association of eIF4G1 with eIF4E proteins in GFAP-Cul3f/f mice, compared with Cul3f/+ mice. Hippocampus lysates were incubated with mouse eIF4E antibody and immunoprecipitates were probed for eIF4G1.
(J) Increased Cap-dependent translation in DIV14 GFAP-Cul3f/+ and NEX-Cul3f/+ neurons as measured with SUnSET.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
We reasoned that primary targets of CUL3 deficiency should be elevated by both heterozygous and homozygous mutations. Of 552 and 116 proteins that were increased in GFAP-Cul3f/f and GFAP-Cul3f/+ samples, respectively, 14 were identified in both (Figure 4F). Among them was eIF4G1, a key factor that controls Cap-dependent translation initiation and is critical for synaptic protein synthesis (Marcotrigiano et al., 1999). eIF4G1 forms the eIF4F complex which interacts with eIF4A to initiate translation (Gingras et al., 1999; Nielsen and Trachsel, 1988). Therefore, we determined whether Cap-dependent translation initiation components are altered in Cul3 mt mice and found an increase in eIF4G1 (Figures 4G, 4H). This effect appeared to be specific because the mutation had little effect on the levels of eIF4E or eIF4A1. Interestingly, HA-eIF4G1 and GFP-CUL3 co-precipitated in HEK293 cells (Figure S7C), suggesting a possible interaction. Moreover, eIF4G1 level was increased in neurons treated with DI-591, a CUL3 neddylation inhibitor which specifically inhibits CUL3 dependent ubiquitination (Zhou et al., 2017), indicating the involvement of CUL3 in eIF4G1 stability (Figures S7D, S7E). eIF4G1 was also increased by MG132, a proteasome inhibitor of Ub-dependent degradation (Figures S7D, S7E). Finally, ubiquitinated eIF4G1 was reduced in Cul3 mt cortex (Figure S7F). Together, these results support the notion that eIF4G1 is a target of CUL3-dependent ubiquitination.
Co-precipitation of endogenous eIF4E and eIF4G1 was increased in CUL3 deficient brain, compared with controls, indicating increased eIF4E-eIF4G1 interaction (Figure 4I). To test if the increased eIF4E-eIF4G1 complex leads to enhanced protein translation, we incubated hippocampal neurons with puromycin, a tRNA analog which terminates translation and releases prematurely-truncated proteins that can be blotted with anti-puromycin antibody (David et al., 2012; Schmidt et al., 2009). Puromycylation was increased in CUL3 deficient neurons, compared with control neurons (Figure 4J), indicating increased protein synthesis. These data support the notion that CUL3 deficiency increased eIF4G1 and thus Cap-dependent protein synthesis.
Diminishing synaptic deficits by inhibiting eIF4E-eIF4G1 complex
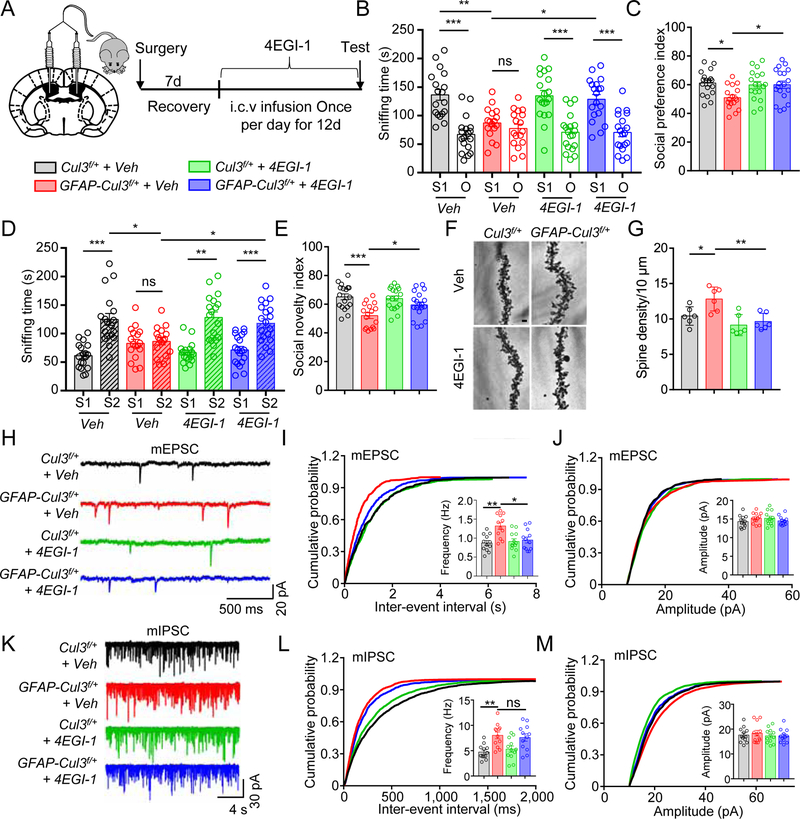
Next, we inhibited Cap-dependent translation by 4EGI-1, an inhibitor of the eIF4E-eIF4G1 complex (Fan et al., 2010). Intra-cerebral ventricular (i.c.v) injection (50 µM, bilateral, 0.5 µl daily, for 12 days) (Santini et al., 2017) diminished social deficits in GFAP-Cul3f/+ mice (Figures 5A–E). However, 4EGI-1 had little effect on anxiety-like phenotypes (Figures S8A–D). Spine densities were similar between Veh- and 4EGI-1-treated control mice, in agreement with a previous report (Santini et al., 2017). However, 4EGI-1 reduced the spines and mEPSC frequency in GFAP-Cul3f/+ mice, compared with Veh-treated GFAP-Cul3f/+ mice (Figures 5F–J). This effect seemed to be selective because 4EGI-1 had little effect on mIPSC frequency (Figures 5K–M). However, increased excitability of CA1 neurons of Cul3 mt mice was not attenuated by 4EGI-1 (Figures S8E–G). These results demonstrated that disrupting the eIF4E-eIF4G1 complex could rescue the abnormality of spines and mEPSC frequency, but not excitability in Cul3 mt mice.
Figure 5. 4EGI-1 ameliorated synaptic and social deficits in CUL3 deficient mice.
(A) Schematic diagram 4EGI-1 i.c.v., bilateral infusion.
(B) 4EGI-1 attenuated social preference deficits in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+; n = 17 mice for Veh-treated GFAP-Cul3f/+; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+; Veh-treated Cul3f/+ S1 (136 ± 9.5 s) vs Veh-treated Cul3f/+ O (66.8 ± 7.3 s), p < 0.001; Veh-treated GFAP-Cul3f/+ S1 (87.2 ± 6.6 s) vs Veh-treated GFAP-Cul3f/+ O (77.4 ± 7.6 s), p > 0.999; 4EGI-1-treated Cul3f/+ S1 (135 ± 9.8 s) vs 4EGI-1-treated Cul3f/+ O (67.6 ± 7.0 s), p < 0.001; 4EGI-1-treated GFAP-Cul3f/+ S1 (129 ± 8.5 s) vs 4EGI-1-treated GFAP-Cul3f/+ O (70.6 ± 7.6 s), p < 0.001; Veh-treated Cul3f/+ S1 vs Veh-treated GFAP-Cul3f/+ S1, p = 0.0012; Veh-treated GFAP-Cul3f/+ S1 vs 4EGI-1-treated GFAP-Cul3f/+ S1, p = 0.012; F(1, 136) = 80.24; Two-way ANOVA followed by Bonferroni’s post hoc test.
(C) 4EGI-1 restored social preference index in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+; n = 17 mice for Veh-treated GFAP-Cul3f/+; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+; Veh-treated Cul3f/+ (61.2 ± 1.9 s) vs Veh-treated GFAP-Cul3f/+ (51.0 ± 1.8 s), p = 0.0125; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (60.1 ± 2.5 s), p = 0.031; One-way ANOVA followed by Tukey’s post hoc test.
(D) 4EGI-1 attenuated social novelty deficits in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+; n = 17 mice for Veh-treated GFAP-Cul3f/+; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+; Veh-treated Cul3f/+ S1 (61.2 ± 5.2 s) vs Veh-treated Cul3f/+ S2 (125 ± 10.3 s), p < 0.001; Veh-treated GFAP-Cul3f/+ S1 (82.9 ± 6.8 s) vs Veh-treated GFAP-Cul3f/+ S2 (86.3 ± 6.1 s), p > 0.999; 4EGI-1-treated Cul3f/+ S1 (66.5 ± 4.3 s) vs 4EGI-1-treated Cul3f/+ S2 (128 ± 9.6 s), p < 0.001; 4EGI-1-treated GFAP-Cul3f/+ S1 (71.4 ± 6.0 s) vs 4EGI-1-treated GFAP-Cul3f/+ S2 (118 ± 7.9 s), p < 0.001; Veh-treated Cul3f/+ S2 vs Veh-treated GFAP-Cul3f/+ S2, p = 0.0135; Veh-treated GFAP-Cul3f/+ S2 vs 4EGI-1-treated GFAP-Cul3f/+ S2, p = 0.0421; F(1, 136) = 59.13; Two-way ANOVA followed by Bonferroni’s post hoc test.
(E) 4EGI-1 restored social novelty index in GFAP-Cul3f/+ mice. n = 18 mice for Veh or 4EGI-1 treated Cul3f/+ group; n = 17 mice for Veh-treated GFAP-Cul3f/+ group; n = 19 mice for 4EGI-1-treated GFAP-Cul3f/+ group; Veh-treated Cul3f/+ (65.1 ± 1.9 s) vs Veh-treated GFAP-Cul3f/+ (52.0 ± 1.8 s), p = 0.0001; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (59.5 ± 2.0 s), p = 0.046; One-way ANOVA followed by Tukey’s post hoc test.
(F) Representative Golgi staining images of apical dendrites of CA1 pyramidal neurons. Scale bar, 5 µm.
(G) Dendritic spine density of GFAP-Cul3f/+ neurons was reduced in 4EGI-1-treated GFAP-Cul3f/+ neurons and similar to that of Veh-treated Cul3f/+ neurons. n = 5 mice for all groups; Veh-treated Cul3f/+ (10.4 ± 0.8) vs Veh-treated GFAP-Cul3f/+ (12.8 ± 1.1), p = 0.0394; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (9.6 ± 0.8), p = 0.0049; One-way ANOVA followed by Tukey’s post hoc test.
(H) Representative mEPSC traces.
(I) mEPSC frequency of GFAP-Cul3f/+ neurons was reduced in 4EGI-1-treated GFAP-Cul3f/+ neurons and similar to that of Veh-treated Cul3f/+ neurons. n = 12 neurons, 3 mice per each group; Veh-treated Cul3f/+ (0.87 ± 0.09 Hz) vs Veh-treated GFAP-Cul3f/+ (1.32 ± 0.15 Hz), p = 0.0071; 4EGI-1-treated GFAP-Cul3f/+ (0.95 ± 0.13 Hz) vs Veh-treated GFAP-Cul3f/+, p = 0.0351; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(J) No effect of 4EGI-1 on mEPSC amplitudes. n = 12 neurons, 3 mice per each group; p > 0.9000 for all comparisons; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(K) Representative mIPSC traces.
(L) mIPSC frequency of GFAP-Cul3f/+ neurons remained increased in 4EGI-1-treated GFAP-Cul3f/+ neurons. n = 12 neurons, 3 mice per each group; Veh-treated Cul3f/+ (4.7 ± 0.58 Hz) vs Veh-treated GFAP-Cul3f/+ (8.0 ± 1.0 Hz), p = 0.0090; 4EGI-1-treated GFAP-Cul3f/+ (7.6 ± 1.02 Hz) vs Veh-treated GFAP-Cul3f/+, p > 0.9999; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(M) No effect of 4EGI-1 on mIPSC amplitudes. n = 12 neurons, 3 mice per each group; p > 0.9999 for all comparisons; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
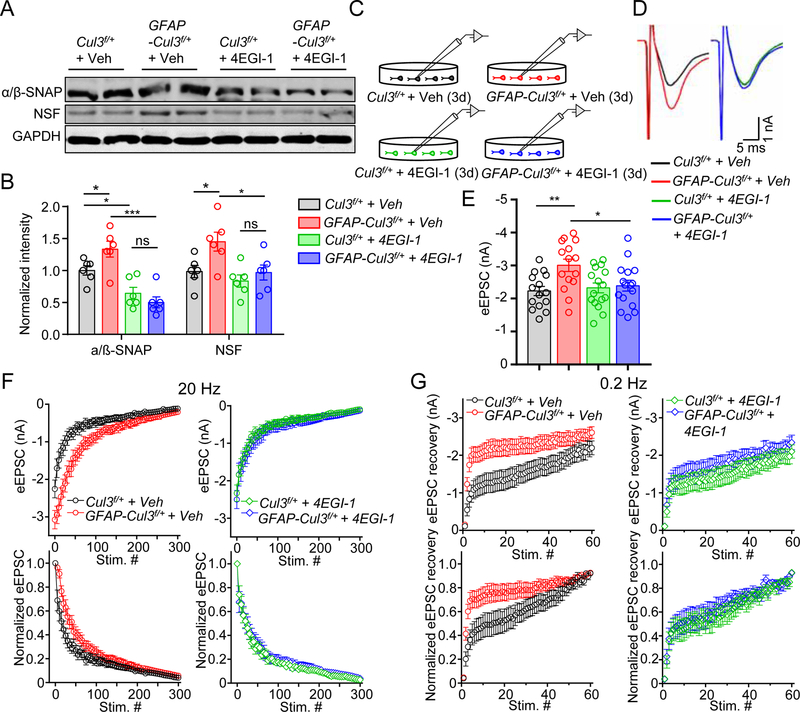
Beside Cap-dependent protein translation, GO analysis also suggested alternations in SNARE disassembly pathways (Figures 4E, S6A). Indeed, SNAP-α, SNAP-β (α/β-SNAP) and NSF were increased in Cul3 mt neurons; and the increase was mitigated by 4EGI-1 (Figures 6A, 6B). Notice that 4EGI-1 was able to reduce α/β-SNAP levels in control neurons (Figures 6A, 6B). Functionally, 4EGI-1 increased PPRs, slowed the decay of NMDA current decay, and reduced currents to minimal stimulation of Cul3 mt slices (Figures S8H–N), suggesting a role of increased protein translation in excessive glutamate release. Moreover, we determined whether synaptic vesicle depletion and recovery is altered in Cul3 mt neurons (Figure 6C). eEPSCs elicited by individual stimuli were increased by CUL3 deficiency (Figures 6D, 6E); and the increase was diminished by 4EGI-1 (Figure 6E). When neurons were subjected to a train of stimuli at 20 Hz (Lou et al., 2012), eEPSC amplitudes were gradually reduced in both control and mt neurons. However, rates of eEPSC reduction in Cul3 mt neurons were slower than controls (Figure 6F, left panels). Furthermore, we elicited eEPSCs by a train of stimuli at 0.2 Hz immediately after the 20-Hz train. eEPSCs of Cul3 mt neurons increased at a rate faster than that in controls (Figure 6G, left panels). These results could suggest slower depletion and/or increased recovery of synaptic vesicles in Cul3 mt mice. Remarkably, both phenotypes were mitigated by 4EGI-1 (Figures 6F, 6G, right panels). Together, our results support a working model that presynaptic deficits may involve increased eIF4E-eIF4G1 complex.
Figure 6. 4EGI-1 diminished hyper-synaptic vesicle turnovers and reduced the increased level of NSF and α/β-SNAP in CUL3 deficiency brain.
(A) Representative blots for the level of NSF and α/β-SNAP in cultured CUL3 deficient neurons.
(B) Quantification analysis of data in (F). Band densities of interested proteins were normalized by the loading control GAPDH; values of control mice were taken as 1; n = 6 mice per each genotype; α/β-SNAP, Cul3f/+ + Veh (1.0 ± 0.18) vs GFAP-Cul3f/+ + Veh (1.3 ± 0.15), p = 0.0464, for Cul3f/+ + Veh vs Cul3f/+ + 4EGI-1 (0.6 ± 0.2), p = 0.0404; Cul3f/+ + 4EGI-1 vs GFAP-Cul3f/+ + 4EGI-1 (0.5 ± 0.11), p = 0.7834; GFAP-Cul3f/+ + Veh vs GFAP-Cul3f/+ + 4EGI-1, p < 0.0001; NSF, Cul3f/+ + Veh (1.0 ± 0.11) vs GFAP-Cul3f/+ + Veh (1.5 ± 0.19), p = 0.0451; Cul3f/+ + 4EGI-1 (0.8 ± 0.12) vs GFAP-Cul3f/+ + 4EGI-1 (1.0 ± 0.14), p = 0.8472; GFAP-Cul3f/+ + Veh vs GFAP-Cul3f/+ + 4EGI-1, p = 0.0365; One-way ANOVA followed by Tukey’s post hoc test.
(C) Schematic diagram for whole cell recording of cultured cortical and hippocampal neuron dissociated from Cul3f/+ and GFAP-Cul3f/+ mice.
(D) Representative eEPSC in cultured cortical and hippocampal neuron.
(E) 4EGI-1 diminished eEPSC amplitude in GFAP-Cul3f/+ hippocampal neuron. n = 15 neurons for Veh-treated Cul3f/+ and GFAP-Cul3f/+ groups; n = 16 neurons for 4EGI-1-treated Cul3f/+ and GFAP-Cul3f/+ groups; Veh-treated Cul3f/+ (−2.2 ± 0.14 nA) vs Veh-treated GFAP-Cul3f/+ (−3.0 ± 0.19 nA), p = 0.0075; Veh-treated GFAP-Cul3f/+ vs 4EGI-1-treated GFAP-Cul3f/+ (−2.3 ± 0.17 nA), p = 0.042; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(F) EPSCs elicited by 300 stimuli at 20 Hz. n = 15 neuron for Veh-treated Cul3f/+; n = 17 neurons for Veh-treated GFAP-Cul3f/+; n = 18 neurons for 4EGI-1-treated Cul3f/+; n = 16 neurons for 4EGI-1-treated GFAP-Cul3f/+. EPSCs were normalized to the peak amplitude of the first EPSC in each AP train.
(G) Time course of EPSC recovery after vesicle depletion. n = 15 neuron for Veh-treated Cul3f/+; n = 17 neurons for Veh-treated GFAP-Cul3f/+; n = 18 neurons for 4EGI-1-treated Cul3f/+; n = 16 neurons for 4EGI-1-treated GFAP-Cul3f/+. EPSCs were normalized to the EPSC peak amplitude of the train.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
Mitigating anxiety phenotypes by chemogenetic inhibition
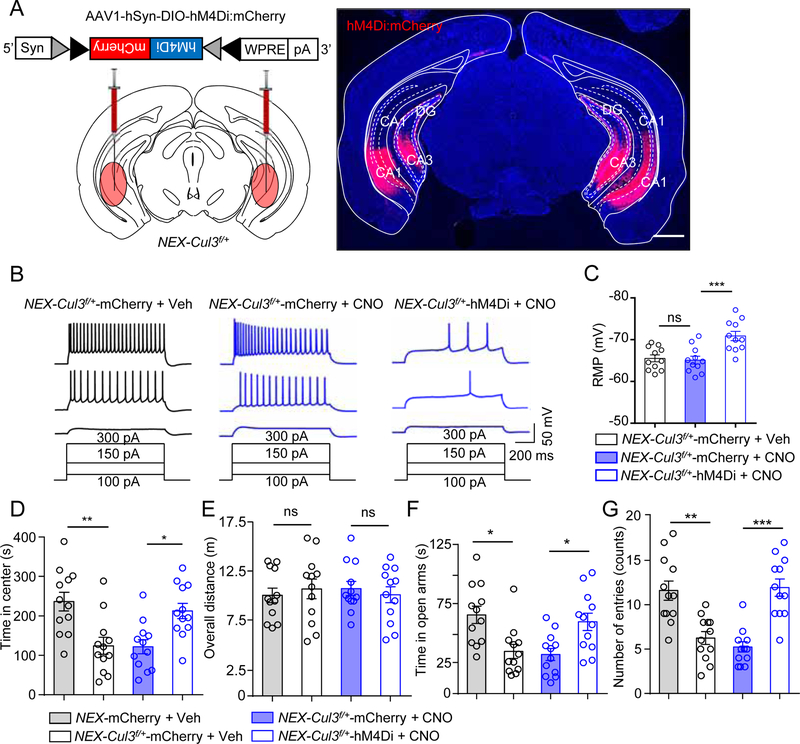
Anxiety level and the excitability of CA1 neurons remained abnormally high after 4EGI-1 treatment, suggesting a mechanism independent of Cap-dependent translation. We sought to reduce the activity of ventral hippocampal (vHPC) pyramidal cells of adult mice by a DREADD (Designer Receptors Exclusively Activated by Designer Drugs) chemical genetic approach. Because Cre in GFAP-Cre adult mice is mostly expressed in astrocytes (Garcia et al., 2004), NEX-Cre mice were used to exclusively target pyramidal neurons. NEX-Cul3f/+ mice were bilaterally injected with AAV1-DIO-hM4Di:mCherry, which expressed hM4Di in Cre-dependent manner (Figure 7A). hM4Di is an engineered M4 muscarinic acetylcholine receptor that, when bound to CNO (Clozapine N-Oxide), can inhibit neurons by Gi-dependent activation of inward potassium channel (Urban and Roth, 2015). Postmortem analysis confirmed the expression of injected virus largely in vHPC (Figures 7A, S9). Hippocampal slices of virus-injected mice were recorded for RMP and excitability. CNO treatment of slices from mice injected with control virus (AAV1-DIO-mCherry) had little effect on RMP or cellular excitability (Figures 7B, 7C). However, CNO reduced the number of APs elicited by injected currents in NEX-Cul3f/+ neurons expressing hM4Di, compared with CNO-treated NEX-Cul3f/+ neurons expressing control virus (Figure 7B). These results indicated that CNO was able to reduce the excitability of CA1 pyramidal neurons in NEX-Cul3f/+ mice.
Figure 7. Chemogenetic inhibition of vHPC pyramidal neurons attenuated anxiety-like behaviors in NEX-Cul3f/+ mice.
(A) Diagram of Cre-DIO-dependent expression (left) and validation of hM4Di-mCherry in vHPC in NEX-Cul3f/+ mice. Scale bar, 1 mm.
(B) Representative spike traces of vHPC CA1 pyramidal neurons. hM4Di or mCherry expressing brain slices were incubated with 10 µM CNO and neurons were stimulated by injected depolarizing currents at 50 pA, 150 pA and 300 pA.
(C) CNO reduced RMP of hM4Di-expressing neurons. n = 11 neurons, 3 mice for all groups; for NEX-Cul3f/+-mCherry+Veh (−65.5 ± 1.2 mV) vs NEX-Cul3f/+-mCherry+CNO (−65.1 ± 1.4 mV); p > 0.9999; for NEX-Cul3f/+-mCherry+CNO vs NEX-Cul3f/+-hM4Di+CNO (−70.9 ± 1.9 mV); p = 0.0033; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(D) CNO (2 mg/kg, i.p.) increased time spent in center of hM4Di-injected NEX-Cul3f/+ mice during open field tests. n = 12 mice for all groups; for NEX-mCherry + Veh (231 ± 25.7 s) vs NEX-Cul3f/+-mCherry + Veh (122 ± 24.4 s), p = 0.0078; NEX-Cul3f/+-mCherry + CNO (118 ± 18.3 s) vs NEX-Cul3f/+-hM4Di + CNO (224 ± 19.7 s), p = 0.0329; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(E) No effect by CNO on overall distance traveled of indicated groups. n = 12 mice for all groups; p > 0.9999 for all comparisons; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(F) CNO increased time spent in open arms of hM4Di-injected NEX-Cul3f/+ mice in EPM tests. n = 12 mice for all groups; for NEX-mCherry + Veh (65.2 ± 8.4 s) vs NEX-Cul3f/+-mCherry + Veh (38.1 ± 6.9 s), p = 0.0268; NEX-Cul3f/+-mCherry + CNO (35.4 ± 3.7 s) vs NEX-Cul3f/+-hM4Di + CNO (61.6 ± 7.4 s), p = 0.0419; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
(G) CNO increased number of entries of hM4Di-injected NEX-Cul3f/+ mice in EPM tests. n = 12 mice for all groups; for NEX-mCherry + Veh (11.4 ± 0.8) vs NEX-Cul3f/+-mCherry + Veh (6.2 ± 0.9), p = 0.0092; NEX-Cul3f/+-mCherry + CNO (5.1 ± 0.4) vs NEX-Cul3f/+-hM4Di + CNO (11.6 ± 0.8), p = 0.0002; Kruskal-Wallis ANOVA followed by Dunn’s post hoc test.
Data were shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significant difference.
Next, we determined if anxiety phenotypes of NEX-Cul3f/+ mice could be altered by suppressing neuronal activity. Mice were treated with CNO (2 mg/kg, i.p.) or Veh 30 min prior to behavioral tests (Zhu et al., 2014). CNO treatment extended the time that NEX-Cul3f/+ mice spent in the center arena during open field tests without altering the overall distance (Figures 7D, 7E). In addition, it increased time NEX-Cul3f/+ mice spent in and entries into the open arm in EPM tests (Figures 7F, 7G). These results suggest that anxiety-like phenotypes in NEX-Cul3f/+ mice could be attenuated by reducing the activity of vHPC pyramidal neurons.
Discussion
The major findings of this paper are as follows. First, homozygous mutation of Cul3 by GFAP-driven Cre (GFAP-Cul3f/f) reduced cortical thickness, hippocampus deformation, and loss of corpus callosum. Heterozygous mutants (GFAP-Cul3f/+) showed increased spine densities of pyramidal neurons. These results suggest a critical role of CUL3 in neural development and reveal a potential mechanism of related brain disorders as increased spine density has been implicated in autism (Penzes et al., 2011). Second, GFAP-Cul3f/+ mice exhibited social interaction deficits and anxiety-like behaviors, major symptoms of ASD. These behavioral deficits were associated with increased glutamatergic transmission, elevated pyramidal neuronal excitability and disrupted E-I balance. These results suggest that CUL3 deficiency impairs social behaviors and anxiety, likely via enhanced glutamatergic activity. Most of the phenotypes were recapitulated in NEX-Cul3f/+ mice, demonstrating a cell-autonomous function of Cul3 in pyramidal neurons. Third, proteomic and biochemical experiments revealed higher levels of eIF4G1 and increased formation of the eIF4E-eIF4G1 complex, which is critical to Cap-dependent translation, in GFAP-Cul3f/+ brains. Inhibiting the eIF4E-eIF4G1 complex by 4EGI-1 ameliorated deficits in social interaction, spine densities, E-I balance, and glutamate release. These results suggest increased Cap-dependent translation as a pathophysiological mechanism. Finally, anxiety-like behaviors in NEX-Cul3f/+ mice were attenuated by chemogenetic inhibition of the activity of vHPC pyramidal neurons. Together, these results reveal a critical role of CUL3 in neural development and synaptic transmission, likely via regulating eIF4E-dependent translation in pyramidal neurons. Given genetic associations between Cul3 and ASD, our data contributes to the understanding of the pathogenesis of a subset of ASD patients carrying CUL3 mutations.
Individuals with ASD often experience seizures and sensory hyper-reactivity (Boyd et al., 2010; Yasuhara, 2010). Glutamate levels were increased in different regions of the brain including hippocampus, amygdala and the blood in individuals with ASD (Aldred et al., 2003). These observations suggest that cortical hyperfunction of glutamatergic pathway as a pathological mechanism of ASD (Fatemi, 2008; Rubenstein and Merzenich, 2003). Underlying cellular mechanisms are likely to be complex. Post-mortem studies of brains of individuals with ASD showed spine abnormality of projection neurons (Raymond et al., 1995). We found increased spine densities in hippocampal neurons in adult as well as immature CUL3 deficient mice (Figures 2A, S7D). In accord, mEPSC frequency was increased (Figures 2G, 3E). In addition, CUL3 deficient pyramidal neurons displayed reduced PPRs, faster decay of NMDA currents in the presence of MK801, and larger currents in response to minimal stimulations (Figures 2J, S4B, S4E), suggesting enhanced probability of glutamate release. Moreover, the intrinsic excitability of pyramidal neurons, which was elicited by injected currents in the presence of blockers of glutamate and GABAA receptors, was increased in CUL3 deficient mice. In addition, eEPSC/eIPSC ratios were elevated, suggesting an E-I imbalance with higher glutamatergic drive in mutant mice. These results indicate a critical role of CUL3 in the development of glutamatergic synapses and regulation of their functions. It is worthy pointing out that increased spine density in CUL3 deficient mice may result from impaired formation of synapses, or a problem in pruning excess synapses. This question warrants future investigation. Together with findings of increased intrinsic excitability and glutamate release of pyramidal neurons in mice lacking Fmr1 or Mecp2, (Contractor et al., 2015; Gibson et al., 2008; Luque et al., 2017; Zhang et al., 2014; Zhang et al., 2010), our results support the notion that hyperfunction of glutamatergic pathways might be a pathological mechanism for ASD.
Synaptic transmission is regulated by SNARE proteins including STX1A, VAMP (also known as synaptobrevin) and SNAP-25 and regulators of SNARE assembly and disassembly including the calcium sensor synaptotagmin, and NSF and α/β-SNAP that facilitate disassembly to control glutamate release. GWAS analysis suggested an association of STX1A with ASD (Nakamura et al., 2008). Protein and/or mRNA levels of STX1A, VAMP2, SNAP-25 or NSF were found abnormal in postmortem brain tissues from SCZ or ASD patients (Durdiakova et al., 2014; Egbujo et al., 2016; Fatemi et al., 2001). We show that VAMP1, NSF, and α/β-SNAP were increased in Cul3 heterozygous mice (Figures S6A, S6B). Interestingly, proteomic analysis of Cul3 heterozygous and homozygous mt brains led to the identification of eIF4G1, a key factor that controls Cap-dependent translation initiation (Figure 4G). eIF4G1 interacts with eIF4E and eIF4A to form a complex (eIF4F) at the mRNA 5’ cap to initiate Cap-dependent protein translation (Gingras et al., 1999; Pelletier and Sonenberg, 1985; Sonenberg et al., 1979). This type of translation is critical for synaptic transmission and plasticity (Marcotrigiano et al., 1999). For example, overexpressing eIF4E or deleting 4EB-P1, an endogenous inhibitor of the eIF4G1-eIF4E complex increases spine density, disrupts E-I balance and impairs social ability in mice (Gkogkas et al., 2013; Huynh et al., 2015; Lee et al., 2017; Martinez-Cerdeno, 2017; Santini et al., 2017; Santini et al., 2013). eIF4G1 could regulate presynaptic assembly in glutamatergic synapses in Drosophila (Menon et al., 2015). De novo missense mutations of eIF4G1 have been identified in ASD patients (O’Roak et al., 2012); and its interacting protein eIF4E is also a risk gene of ASD (Neves-Pereira et al., 2009). Increased translation in ASD is thought to be Cap-dependent (Huynh et al., 2015; Oberer et al., 2005). 4EGI-1, an inhibitor of the eIF4E-eIF4G1 complex, diminishes behavior deficits of mice overexpressing eIF4E, mice lacking eIF4EBP2 (an endogenous inhibitor of the eIF4E-eIF4G1 complex) and Fmr1 mt mice (Gkogkas et al., 2013; Santini et al., 2017; Santini et al., 2013).
The following evidence suggests that Cap-dependent translation was abnormal in CUL3 deficient mice. First, western blot analysis showed increased eIF4G1 levels in CUL3 deficient brains (Figures 4G, 4H). In addition, the eIF4E-eIF4G1 complex was increased in CUL3 deficient brains (Figure 4I). Second, 4EGI-1 diminished abnormalities in spine density, synaptic transmission and release probability (Figures 5E, 5G, 5M). Third, protein levels of SNARE complex disassembly molecules NSF and α/β-SNAP were reduced in 4EGI-1-treated mutant neurons (Figure 6A). Fourth, 4EGI-1 reduced synaptic vesicle turnover that was increased in Cul3 mt neurons (Figures 6F, 6G). Finally, deficits of social recognition and memory in Cul3 mt mice were rescued by 4EGI-1 (Figures 5B–E). Our findings supported a working model that CUL3 deficiency alters neurotransmission via enhancing Cap-dependent protein translation.
Elevated cortical excitability has been implicated in ASD pathology (Rubenstein and Merzenich, 2003). Elevated activity in the anterior hippocampus of humans and rhesus monkeys (equivalent to vHPC in rodents) is associated with sustained anxiety (Hasler et al., 2007; Oler et al., 2010). Anxiety-provoking stimuli increased the expression of c-fos in rodent vHPC (Duncan et al., 1996; Pezzone et al., 1992; Senba et al., 1993; Silveira et al., 1993). Activation of vHPC pyramidal neurons promotes anxiety-related behaviors (Felix-Ortiz et al., 2013; Jimenez et al., 2018; Parfitt et al., 2017) whereas lesion and inactivation of the vHPC attenuate anxiety-related behaviors (Adhikari et al., 2011; Bannerman et al., 2003; Kjelstrup et al., 2002; Padilla-Coreano et al, 2016; Parfitt et al., 2017). Mutation of Fmr1 and Mecp2, genes implicated in Fragile X syndrome (FXS) and Rett’s syndrome, respectively, increased the intrinsic excitability of projection neurons (Gibson et al., 2008; Zhang et al., 2014). Remarkably, anxiety-like behaviors in Cul3 mt mice could be diminished by reducing the activity of vHPC pyramidal neurons with DREADD (Figures 7D–G), in support of the notion that increased neuronal activity serves as a mechanism for anxiety-like behavior in Cul3 mt mice. How CUL3 deficiency increases the neuronal activity remains unclear and warrants future studies. Abnormal excitability remained after treatment with 4EGI-1, which could rescue deficits in neurotransmission and social interaction, suggesting a mechanism independent of Cap-dependent translation. Although by using cell-specific Cre, this study reveals functions of CUL3 and underlying cellular and molecular mechanisms in the nervous system. Because CUL3 is a ubiquitously expressed protein, a germline heterozygous Cul3 mutant mouse line (i.e., with genetic loss in the brain and the body) may also be a valuable model to study relevant ASD.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All unique/stable reagents generated in this study are available from the Lead Contact, Lin Mei (lin.mei@case.edu), with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mice were housed in a room at 24 °C in a 12 h light /dark cycle with access to food and water ad libitum. Male mice, P13-P90 of age, were used in the study. PCR-based genotyping was performed on genomic DNA isolated from tails or cortexes. Cul3f/f mice (#028349) were purchased from The Jackson Laboratory and genotyped with primer 1 (CAG GGC TGT AAT TCT GTC TGG), primer 2 (ATG CTC CCT ACC ATG CAA AC) and primer 3 (AGA CTG CCT TGG GAA AAG CG). Cul3f/f (C57BL) mice were crossed with GFAP::Cre transgenic mice (#004600, Jackson Laboratory) (Zhuo et al., 2001) to produce GFAP-Cul3f/f and GFAP-Cul3f/+ mice. GFAP-Cre mice were genotyped with forward prime (ACT CCT TCA TAA AGC CCT) and reverse primer (ATC ACT CGT TGC ATC GAC CG). Cul3f/f mice were crossed with NEX-Cre mice (kindly provided by Dr. Klaus-Armin Nave) (Goebbels et al., 2006) to produce NEX-Cul3f/f and NEX-Cul3f/+ mice. NEX-Cre mice were genotyped with primer 1 (GAG TCC TGG AAT CAG TCT TTT TC), primer 2 (ATC ACT CGT TGC ATC GAC CG) and primer 3 (CCG CAT AAC CAG TGA AAC AG). Behavior analysis was preformed using P60-P80 male mice. Experimental procedures were approved by the Institutional Animal Care and Use Committee of Augusta University and Case Western Reserve University.
GFAP-Cre homozygous mice are not viable, perhaps due to transgene insertion (see info from https://www.jax.org/strain/004600). However, heterozygous GFAP-Cre mice are viable, fertile, and normal in brain/body size and do not display any gross physical or behavioral abnormalities (Kim et al., 2014; Lim et al., 2009; Muller et al., 2008; Sanchez-Ortiz et al., 2014). Therefore, all studies used heterozygous GFAP-Cre mice. As shown in Figure S13, we did not find difference between Cul3f/+, GFAP-Cre and NEX-Cre mice in social preference (A and B) and social memory (C and D). No anxiety or locomotor deficits were found in GFAP-Cre or NEX-Cre mice (E and F). Moreover, intrinsic excitability and resting membrane potentials of CA1 pyramidal neurons were comparable in all three groups (Figures S10A–C). Furthermore, PPRs were similar in all three groups (Figures S10D, S10E). In addition, no difference was found in the frequency and amplitude in either mEPSC or mIPSC among three groups (Figures S10F–K).
Neuron culture
Neurons were cultured as previously described (Wang et al., 2018). Briefly, hippocampi and cortex were isolated from P0 mice and kept in ice-cold Hank’s balanced salt solution (ThermoFisher, 14025092) and incubated with 20 units/ml papain at 37 °C for 15 min. Dissociated ce lls were suspended in plating media (DMEM + 10% FBS) and plated with a density of 50000–70000/cm2 onto poly-L-lysine–coated 8 mm-cover slips in 12-well plates. Medium was replaced 4 h after initial incubation with the maintenance medium containing neurobasal medium (ThermoFisher, 21103049) supplemented with 2% B-27 supplement (ThermoFisher, 17504044), 1% GlutaMax (ThermoFisher, 35050061), and 1% penicillin/streptomycin. Neurons were placed in incubators with 37 °C in 5% CO 2. Medium was changed by half in every 3 days. Neurons were treated with Vehicle or 4EGI-1 (1.5 nM) for 3 days before electrophysiological or biochemical analysis.
METHOD DETAILS
Immunostaining
Immunostaining was performed as described previously (Wang et al., 2018). In brief, mice were anesthetized with isoflurane and perfused with room temperature (RT) PBS followed by 50 ml, 4 °C, and 4 % paraformaldehyde (PFA). Brains were post-fixed in 4% PFA at 4 °C ove rnight and dehydrated by 30% sucrose at 4 °C for two days. Brains were then embedded in OCT (ThermoFisher, 23–730-571), rapidly frozen and sectioned into 40 µm slices. Sections were blocked and permeabilized in PBS containing 0.3% Triton X-100 and 5% goat serum for 2 h at RT. They were incubated at 4 °C overnight with primary antibodies in PBS containing 5% goat serum and 2% BSA. Sections were washed with PBS and incubated at RT for 1 h with donkey anti-mouse IgG secondary antibody.
Western blotting and co-immunoprecipitation
Western blotting was performed as described previously (Zhao et al., 2017). Cortical tissues and cultured neurons were homogenized in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA,) containing 0.5 % sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 1 mM DTT and protease inhibitor cocktail (ThermoFisher, 11697498001). Lysates were centrifuged at 10,000 × g for 10 min at 4 °C to remove debris, to obtain homogenates. Samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were immunoblotted with antibodies and immunoreactive bands were visualized using the LI-COR Odyssey infrared imaging system. Intensity of immunoreactive bands were quantitated by using ImageJ (NIH). Band density of interested proteins was normalized to loading control (GAPDH).
Homogenate of 100 µg were pre-cleared by 1 h incubating at 4 °C with protein A/G PLUS-Agarose beads (Santa Cruz, SC-2003). Pre-cleared samples were incubated with respective antibodies overnight at 4 °C in lysate buffer and subsequently with protein A/G PLUS-Agarose beads (Santa Cruz) at 4 °C for 4 h. Beads were then washed three times with lysis buffer. Equal volume of 2×SDS sample buffer was used to elute proteins.
SUnSET
Protein translation assay was performed using a previously described SUnSET method (Schmidt et al., 2009). Puromycin-modified proteins were revealed by blotting using the mouse monoclonal antibody 12D10.
Ubiquitination assay
Cortical tissues were isolated from mice and homogenized in a buffer (1 ml per 100 mg of tissue) containing 2% SDS, 150 mM NaCl, 10 mM Tris-HCl (pH 8.0), and protease inhibitors including 1 mM phenylmethylsulfonyl fluoride, 1 µg/µl pepstatin, 1 µg/µl leupeptin, and 2 µg/ml aprotinin. Samples were diluted by adding 9 × volumes of the dilution buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, and 1% Triton. Supernatants were incubated with eIF4G1 antibody overnight at 4°C and then with prot ein G agarose beads. Precipitated proteins were subjected to western blotting and probed with anti-Ub antibody.
Golgi staining
Golgi staining was performed as previously described using a kit from FD NeuroTechnologies (PK401) (Wang et al., 2018). In brief, brains were isolated from anesthetized mice and incubated in 1:1 mixture of FD Solution A:B for 24 h. Brains were transferred into fresh FD Solution A:B for 12 days at RT in dark. Coronal sections (200 µm) were cut in PBS containing 50 % sucrose using a VT-1000S vibratome (Leica Microsystems) and mounted on 3% gelatin-coated slides. Staining procedures were carried out based on the manufacturer’s protocol. Slices were dehydrated in series dilutions of ethanol and mounted with mounting medium. Dendrites were traced, and lengths were measured by the Neurite Tracer implanted in ImageJ Fiji. Spines on the secondary branches of apical and basal dendrites in the CA1 region were counted.
Electrophysiology
Mice were anesthetized with isoflurane; brains were quickly removed to ice-cold oxygenated (95% O2/5% CO2) cutting solution containing (in mM): 120 choline chloride, 2.5 KCl, 7 MgCl2, 0.5 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, and 25 glucose. 300-µm slices of hippocampus using VT1200S Vibratome (Leica Microsystems) as described elsewhere (Bischofberger et al., 2006). Slices were recovered in oxygenated artificial cerebrospinal fluid (ACSF) containing (in mM): 124 NaCl, 2.5 KCl, 2 MgSO4, 2.5 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 glucose; for 30 min at 32 °C and then mainta ined at 25 ± 1 °C for an additional 1 h before recording.
Slices were placed to a recording chamber superfused (2 ml/min) with ACSF at 32–34 °C. Pyramidal neurons in CA1 were vis ualized with infrared optics using an upright fixed microscope equipped with a 40X water-immersion lens (BX51WI, Olympus) and CCD monochrome video camera (C2400–75, Hamamatsu). Patch pipettes were prepared by a horizontal pipette puller (P-1000; Sutter Instruments) with a resistance of 3–5 MK. For mEPSC recording, pyramidal neurons were held at −70 mV in the presence of bicuculline (20 μM) and tetrodotoxin (TTX, 1 μM), with the pipette solution containing (in mM): 125 K-gluconate, 5 KCl, 10 HEPES, 0.2 EGTA, 1 MgCl2, 4 Mg-ATP, 0.3 Na-GTP and 10 phosphocreatine (pH 7.35, 290 mOsm). For mIPSC recording, pyramidal neurons were held at −70 mV in the presence of 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX, 20 μM), DL-2-amino-5-phosphonopentanoic acid (DL-AP5, 50 μM) and TTX (1 μM), with the pipette solution containing (in mM): 130 CsCl, 10 HEPES, 0.2 EGTA, 1 MgCl2, 4 Mg-ATP, 0.3 Na-GTP, 10 phosphocreatine and 5 QX314 (pH 7.35, 290 mOsm).
To evaluate intrinsic excitability, CA1 pyramidal neurons stimulated by injecting a series of depolarizing pulses in the presence of 20 μM CNQX, 50 μM DL-APV and 20 μM bicuculline. Action potential firing was measured by current clamp recording. The pipette solution contained (in mM): 125 K-gluconate, 5 KCl, 10 HEPES, 0.2 EGTA, 1 MgCl2, 4 Mg-ATP, 0.3 Na-GTP and 10 phosphocreatine (pH 7.35, 290 mOsm).
Procedures to measure paired-pulse ratios, EPSC response to minimal stimulations, and MK-801 inhibition of NMDA currents were described before (Sun et al., 2016). In these experiments, EPSCs were evoked by stimulating the SC-CA1 pathway. For PPR recording, pyramidal neurons were clamp at −70 mV in the presence of 20 µM bicuculline. The ratios were calculated as (2nd EPSC/1st EPSC) × 100. For the minimal stimulation recording, pyramidal neurons were held at −70 mV and the stimulus intensity was adjusted to fulfill the following criterias,1) all or none synaptic events were generated, 2) little or no variation in EPSC latency, 3) no change in mean size or shape of EPSCs by a small change in stimulus intensity, 4) complete failure to evoke EPSCs by 10–20% reduction in stimulus intensity. Responses that failed to meet these criteria were rejected. For MK-801 treatment assay, pyramidal neurons were clamp at +40 mV in the presence of 20 µM bicuculline and 20 µM CNQX. Pre-exposure to MK-801 for 5 min is required before recording. EPSC amplitudes were normalized to the first EPSC and fitted with single-exponential functions to calculate decay constants (t, in number of stimuli).
Procedures for E-I ratio measurement were described before (Cheng et al., 2018). In brief, a two-concentric bipolar stimulating electrode (FHC) providing 0.1-Hz stimulation was placed close to the stratum pyramidal layer of the CA1 to obtain evoked synaptic responses. Brain slices were first superfused with ACSF to record total postsynaptic currents (PSC), and then IPSCs in the presence of 20 µM CNQX and 100 µM DL-APV. 20 µM bicuculline was added to confirm the remaining currents are inhibitory. The E-I ratio was calculated as (PSC – IPSC)/IPSC. All data were collected from 3 or more neurons per mouse and 3 or more mice per group.
Procedures for culture cell recording were described before (Milosevic et al., 2011) with the internal solution containing (in mM): 137 K-gluconate, 10 NaCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.3 Na-GTP and 5 Na2-phosphocreatine (pH 7.35, 290 mOsm). The extracellular solution contained (in mM): 122 NaCl, 2.5 KCl, 1 MgCl2, 2 CaCl2, 20 HEPES, 10 glucose, 2 μM bicuculline and 2 μM strychnine (pH 7.35). Neurons were clamped at −70 mV and the electrode was placed at ~200 μm away from the recorded soma to obtain evoked responses. Recordings were performed with MultiClamp 700B amplifier and 1550B digitizer (Molecular Devices). Series resistance was below 20 MK and monitored throughout the experiments. Data were sampled at 10 kHz and filtered at 2 kHz. To ensure a similar number of presynaptic cells were stimulated, equal density of neurons was plated on coverslips. Second, stimulating electrodes were placed in the same distance from the soma to be recorded. Third, same stimulating currents were applied in all experiments. Fourth, to further minimize the effect of potential presynaptic variation, we calculated the ratio by normalizing each evoked EPSC over the first evoked EPSC of the same neuron.
Behavioral tests
Behavioral tests were done blind to genotypes or treatments. Locomotor activity was measured as described previously (Wang et al., 2018). Mice were placed in a chamber (50 × 50 × 10 cm) and monitored for movement for 30 min using an overhead camera and tracking software (EthoVision, Noldus). For working memory, mice were placed at the center of a Y-shaped maze with three arms (35 cm). They were allowed to move freely through the maze for 8 min. The total number and direction of arm entries were recorded. Nonoverlapping entrance sequences (e.g., ABC, BCA) were defined as spontaneous alternations. Social preference and social memory were measured in a black Plexiglas rectangular box that consists of 3 interconnected chambers. In habituation, a mouse was placed in the central chamber and allowed to explore the 3 chambers for 10 min. If it showed preference to a side chamber, the mouse was excluded from the test. To test social preference, the test mouse was placed in the center chamber with both gates to the side chambers closed. A stranger wild-type mouse (S1) was placed in a mesh container in a side chamber while the other chamber had a mesh container with a novel object (O). After opening both gates, the test mouse was monitored for the distance to the S1 and O cages for 10 min. To test social memory, the test mouse was placed in the center chamber with both gates closed; the S1 mouse was placed in the O cage and a second stranger wild-type mouse (S2) was placed in the previous S1 cage. The test mouse was allowed for free exploration and monitored for the distance to either cages for 10 min. The time spent in sniffing of each container was recorded and analyzed as previously described (Nieuwenhuis et al., 2011; Nygaard et al., 2019), to directly compare between different groups as well as within groups. Social preference index and social novelty index were calculated as described (Nygaard et al., 2019).
Tests with elevated plus maze were performed as described previously (Walf and Frye, 2007). Mice were placed in the center of the maze with two open arms (50 × 10 cm) and two perpendicular enclosed arms (50 × 10 × 40 cm). Movement in the maze was monitored for 10 min using an overhead camera and tracking software (EthoVision, Noldus). Motor learning skill was tested on accelerated rotarod by placing mice on a 9 cm diameter rod. Before the training, the mice were habituated to stay on the immobile rod for 5 min. Habituation was repeated every day for 1 min before test. The speed of the rod was accelerated from 4 to 40 rpm. The test was preformed one session per day for three consecutive days and the latency to falling was analyzed.
Buried food-seeking test was performed as described previously (Machado et al., 2018). Mice were food deprived for 24 h before the test. A 2-g pellet of regular chow was buried 8 cm beneath the surface of the fresh bedding in a corner of the test cage. Mice were transferred into the test cage in the opposite corner to the buried pellet. The latency to find food pellet was monitored. Grooming behavior was monitored for 20 min after placing a mouse into a new cage with new bedding. Total time spent in grooming and the number of grooming were scored individually.
Stereotaxic cannulations or injections
For cannulation, mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) and secured in a stereotaxic apparatus (Kopf Instruments, Tujunga, CA). Holes were drilled into the skull and guide cannula (26-gauge) with dummy cannula were bilaterally implanted into the lateral ventricle at coordinates (AP −0.22 mm, ML ±1.00 mm mediolateral, and DV −2.50 mm). Seven days after, mice were injected with 4EGI-1 by inner cannula (33-gauge) (0.5 µl, 50 mM, each side). For virus injection, anesthetized mice were secured in a stereotaxic apparatus. Holes were drilled into the skull and viruses (titer > 1012 particles/ml, 200 nl, each side) were microinjected into vHPC using a glass pipette with fine tip at a rate of 30 nl/min (coordinates: AP −2.90 mm, ML ± 3.00 mm and DV −4.00 mm). After injection, pipettes were left in place for 5 min to allow for diffusion of injected virus before being slowly withdrawn. Mice were tested 3 weeks after injection. Injection locations were validated in each mouse after experiments.
Proteomic analysis
Frozen cortical tissues were homogenized in the buffer containing 8 M urea, 100 mM Tris-HCl, pH 8.0, in a matrix D tube on a FastPrep 24 homogenizer (MP Biomedicals, Solon, OH). 40 µg protein aliquots from each sample were reduced by 8 mM DTT and alkylated by 32 mM iodoacetamide. Samples were precipitated by adding 6 volumes of cold acetone (−20°C) and incubated at −20°C overnight. Precipitated samples were centrifu ged at 10,000 g for 5 min. After removing the supernatant, the pellets were air dried in a fume hood to evaporate acetone and reconstituted in 50 mM triethyl ammonium bicarbonate (TEAB). Samples were digested in 1 µg trypsin (Promega, Madison, WI) in 50 mM TEAB overnight at room temperature. Peptide concentrations after digestion were measured by Pierce™ Quantitative Colorimetric Peptide Assay (Thermo Fisher Scientific, Waltham, MA). 25 µg of peptides from each sample were labeled with TMT10plex Mass Tagging reagents (Thermo Fisher Scientific, Waltham, MA) and quenched with 5% hydroxylamine.
TMT-labeled peptide samples were equally combined and desalted using a C18 column (3.5 µ m particle size, 2.1 mm × 150 mm, Waters) on HPLC at a flow rate of 0.2 ml/min. Peptides were eluted by a stepwise gradient with mobile phase A (10 mM ammonium formate, pH) and mobile phase B (90% acetonitrile, 10 mM ammonium formate, pH 10) over 40 min and collected into 40 collections at 1 collection per minute. Samples were re-combined into multiple fractions and dried in a SpeedVac centrifugal concentrator and reconstituted in 1% acetic acid with final peptide concentration ~ 0.2 µg/ µl. Samples were analyzed on a Dionex Ultimate 3000 UHPLC system interfaced with a ThermoScientific Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Waltham, MA).
MS data were analyzed using Proteome Discoverer V 2.1 (Thermo Fisher Scientific, Waltham, MA), and searched against mouse UniProtKB protein sequence database (25,035 entries) with an automatically generated decoy database (reversed sequences). A false discovery rate (FDR) was set to 1% for both peptide and protein identifications. Oxidation of methionine and acetylation of protein N-terminus were set as dynamic modifications and carbamidomethylation of cysteine and TMT10plex on lysine and N-terminus were set as static modifications in the searching workflow. For peptide quantification, most confident centroid was set as integration method of report ions with 20 ppm integration tolerance using HCD-MS3 scans. Reporter abundance was based on intensity with average reporter S/N threshold set to 10. Total peptide amounts on channels average were used for normalization and scaling. A total of 5,025 proteins were identified in the fractions at 1% FDR. Relative protein abundance ratios between groups were calculated using the average of normalized reporter ion intensities of the group and subjected to a two-tailed Student T-test with the threshold of significance at p < 0.05.
Bioinformatics analyses
Differentially expressed proteins were subjected to Gene Ontology (GO) analysis by using ClueGO, a Cytoscape 3.7.1 plug-in. The murine GO (Biological Processes, version from 4 July 2018) was used with the following settings: type of analysis: single; GO terms level: 3 – 20; GO term restriction: 2 genes and 5%; evidence code: all. A significance threshold level of 0.05 was applied. GO term groups reflecting similar properties or functions were generated and custom terms were created generalizing such groups. The statistical test used for the enrichment was based on two-sided hypergeometric test and adjusted by using the step-down Bonferroni method. Volcano plots and bar graphs were generated by Origin Pro 8.0 software. Predicted ASD-associated gene were obtained from SFARI gene database (https://gene.sfari.org/).
STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism 7.0. Two-tailed Mann-Whitney U test or Student’s t test was used to compare data from two groups in Figures 1C, 1E, 1G, 1H, 1J, 1K, 1L, 1M, 2B, 2C, 2G, 2H, 2L 2M, 2P, 3A, 3E, 3F, 3H, 3I, 3J, 3K, 3L, 3M, 3N, 3O, S2B, S2G, S2H, S2I, S3B, S3D, S3F, S3J, S3L, S4B, S4C, S4E, S4F, S4K, S4M, S4N, S5I, S5J, S5L, S5O, S5N, S5P and S7B. Kruskal-Wallis ANOVA was followed by Dunn’s post hoc test in Figures 5I, 5L, 5M, 6E, 7C, 7D, 7E, 7F, 7G, S5A, S5B, S8A, S8B, S8C, S8D, S8L, S8N, S10B, S10D, S10E and S10F. One-way ANOVA was followed by Tukey’s post hoc test in Figures 4H, 5C, 5E, 5G, 6B, S1A, S1B, S7E, S10M, S10N, S10P and S10Q. Two-way ANOVA was used with more than two parameters and followed by Bonferroni’s post hoc test in Figures 1B, 1D, 2E, 2J, 2O, 3C, 3P, 3Q, 5B, 5D, S4H, S8G, S8I, S10A, S10C, S10H and S10K. n represents the number of animals or cells tested. Data were presented as mean ± SEM. The accepted level of significance was p < 0.05.
Supplementary Material
Table S1. Proteomics analysis of cortex and hippocampus between GFAP-Cul3f/f and control mice. Related to Figure 4.
Table S2. Proteomics analysis of cortex and hippocampus between GFAP-Cul3f/+ and control mice. Related to Figure 4.
Table S3. GO analysis of up-regulated proteins in GFAP-Cul3f/+ samples. Related to Figure 4.
Table S4. GO analysis of down-regulated proteins in GFAP-Cul3f/+ samples. Related to Figure 4.
Table S5. GO analysis of up-regulated proteins in GFAP-Cul3f/f samples. Related to Figure 4.
Table S6. GO analysis of down-regulated proteins in GFAP-Cul3f/f samples. Related to Figure 4.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-eIF4G1 | Bethyl Laboratories | Cat# A300-502A RRID:AB_143249 |
| anti-CUL3 | Bethyl Laboratories | Cat# A301-109A RRID:AB_873023 |
| anti-eIF4E | Novus Biologicals | Cat# NB100-58833 |
| anti-GAPDH | Santa Cruz Biotechnology | Cat# SC-32233 RRID:AB_877748 |
| α/β-SNAP | Santa Cruz Biotechnology | Cat# SC-48349 RRID:AB_628263 |
| anti-NSF | Cell Signaling Technology | Cat# 2145S RRID:AB_2155696 |
| anti-VGLUT1 | Synaptic Systems | Cat# 135 011 RRID:AB_2617088 |
| anti-PKA | Cell Signaling Technology | Cat# #4782 RRID:AB_2170170 |
| anti-SNAP25 | Abcam | Cat# ab5666 RRID:AB_305033 |
| anti-VAMP1 | R&D Systems | Cat# AF4828 RRID:AB_2212446 |
| anti-synaptotagmin1 | Developmental Studies Hybridoma Bank | Cat# mAB30 RRID:AB_2295002 |
| anti-synaptophysin | Cell Signaling Technology | Cat# 4329 RRID:AB_1904154 |
| anti-puromycin | Millipore | Cat# MABE343 RRID:AB_2566826 |
| anti-PTEN | Cell Signaling Technology | Cat# 9559 RRID:AB_390810 |
| anti-SHANK1 | Novus Biologicals | Cat# NB300-167 RRID:AB_2187584 |
| anti-NeuN | Millipore | Cat# MAB377 RRID:AB_2298772 |
| anti-RFP | Rockland | Cat# 600-401-379 RRID:AB_2209751 |
| Bacterial and Virus Strains | ||
| AAV1-hsyn-DIO-mCherry | Gene Therapy Center Vector Core at the University of North Carolina at Chapel Hill | N/A |
| AAV1-hsyn-DIO-hM4Di:mCherry | Gene Therapy Center Vector Core at the University of North Carolina at Chapel Hill | N/A |
| Biological Samples | ||
| mouse brain tissue | Mouse strains listed in the table | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DL-APV | Tocris Bioscience | Cat# 0105 |
| CNQX | Tocris Bioscience | Cat# 0190 |
| Bicuculline | Tocris Bioscience | Cat# 0130 |
| DI-591 | Dr. Shaomeng Wang at the University of Michigan | Zhou et al., 2017 |
| 4EGI-1 | Tocris Bioscience | Cat# 4800 |
| Critical Commercial Assays | ||
| Golgi staining kit | FD NeuroTechnologies | Cat# PK401 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat# CRL-3216 RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| NEX-Cre mice | Dr. Klaus-Armin Nave at the Max Planck Institute | Goebbels et al., 2006 |
| FVB-Tg(GFAP-cre)25Mes/J | The Jackson Laboratory | Cat# 004600 RRID:IMSR_JAX:00 4600 |
| Cul3tm1Jdsr/J | The Jackson Laboratory | Cat# 028349 RRID:IMSR_JAX:02 8349 |
| Tg(Thy1-EGFP)MJrs/J | The Jackson Laboratory | Cat# 007788 RRID:IMSR_JAX:00 7788 |
| Recombinant DNA | ||
| pcDNA3-HA-eIF4G1 | Addgene | Cat #45640 RRID:Addgene_456 40 |
| pcDNA3-GFP-CUL3 | Mei Laboratory | |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.net/WelcomeRRID:SCR_003070 |
| Prism 7 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/RRID:SCR_002798 |
| Zen software | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.htmlRRID:SCR_013672 |
| EthoVision XT | Noldus | https://www.noldus.com/ethovisionRRID:SCR_000441 |
| Origin Pro 8.0 | OriginLab | https://www.originlab.com/ |
| Adobe Illustrator | Adobe | http://www.adobe.com/products/illustrator.htmlRRID:SCR_010279 |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.htmlRRID:SCR_014199 |
| ClueGO | ClueGO | http://www.ici.upmc.fr/cluego/RRID:SCR_005748 |
Highlights:
Cul3 mutant mice exhibits social behavioral deficits and anxiety-like behaviors
CUL3 deficiency impairs neurotransmission, excitability and E-I balance
Protein translation and synaptic vesicle turnover are increased in Cul3 mutant mice
Inhibiting protein translation rescues social behavior and neurotransmission deficits
ACKNOWLEDGMENTS
The authors thank Yi Sun and Shaomeng Wang for providing DI-591. We would like to thank Huabo Su, Yanan Wang, Yao Tian, and Lei Li for kind suggestions; lab members for helpful discussions; and Belinda Willard and Ling Li for assistance in proteomics. This study was supported by a grant from the National Institutes of Health (MH083317 to L.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
Data availability
The data that support the current study are available from the Lead Contact upon reasonable request.
Code availability
Custom codes generated during this study are available at Github: https://github.com/pdpmb/CUL3-deficiency-_code
REFERENCES
- Adhikari A, Topiwala MA, and Gordon JA. (2011). Single units in the medial prefrontal cortex with anxiety-related firing patterns are preferentially influenced by ventral hippocampal activity. Neuron 71, 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggleton JP, Hunt PR, and Rawlins JN. (1986). The effects of hippocampal lesions upon spatial and non-spatial tests of working memory. Behav. Brain Res 19, 133–146. [DOI] [PubMed] [Google Scholar]
- Aldred S, Moore KM, Fitzgerald M, and Waring RH. (2003). Plasma amino acid levels in children with autism and their families. J. Autism Dev. Disord 33, 93–97. [DOI] [PubMed] [Google Scholar]
- Anderica-Romero AC, Gonzalez-Herrera IG, Santamaria A, and Pedraza-Chaverri J. (2013). Cullin 3 as a novel target in diverse pathologies. Redox Biol. 1, 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, and Bear MF. (2011). Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, Kurzius-Spencer M, Zahorodny W, Robinson RC, and White T, et al. (2018). Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill Summ 67, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Riordan M, and Bhat MA. (2014). Genetic aspects of autism spectrum disorders: insights from animal models. Front Cell Neurosci. 8, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannerman DM, Grubb M, Deacon RMJ, Yee BK, Feldon J, and Rawlins JNP. (2003). Ventral hippocampal lesions affect anxiety but not spatial learning. Behav. Brain Res 139, 197–213. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. (2009). A synaptic trek to autism. Curr. Opin. Neurobiol 19, 231–234. [DOI] [PubMed] [Google Scholar]
- Boyd BA, Baranek GT, Sideris J, Poe MD, Watson LR, Patten E, and Miller H. (2010). Sensory features and repetitive behaviors in children with autism and developmental delays. Autism Res. 3, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson SE, Rogers SJ, and Fombonne E. (2003). Autism spectrum disorders: early detection, intervention, education, and psychopharmacological management. Can J. Psychiatry 48, 506–516. [DOI] [PubMed] [Google Scholar]
- Buiting K. (2010). Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet 154C, 365–376. [DOI] [PubMed] [Google Scholar]
- Carper RA, Moses P, Tigue ZD, and Courchesne E. (2002). Cerebral lobes in autism: early hyperplasia and abnormal age effects. Neuroimage 16, 1038–1051. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wang ZM, Tan W, Wang X, Li Y, Bai B, Li Y, Zhang SF, Yan HL, and Chen ZL, et al. (2018). Partial loss of psychiatric risk gene Mir137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat. Neurosci 21, 1689–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan S, Horder J, Inkster B, Mendez MA, Murphy DG, and Nutt DJ. (2012). GABA system dysfunction in autism and related disorders: from synapse to symptoms. Neurosci. Biobehav. Rev 36, 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Klyachko VA, and Portera-Cailliau C. (2015). Altered Neuronal and Circuit Excitability in Fragile X Syndrome. Neuron 87, 699–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Acuna JM, and Sherman SL. (2001). FMR1 and the fragile X syndrome: human genome epidemiology review. Genet Med 3, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN. (2012). Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin. Neurosci 14, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, and Diehl JA. (2004). The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol 24, 8477–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, and Yewdell JW. (2012). Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol 197, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, and Walker S, et al. (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, and Bagni C. (2011). Regulation of molecular pathways in the Fragile X Syndrome: insights into Autism Spectrum Disorders. J. Neurodev. Disord 3, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Duncan GE, Knapp DJ, and Breese GR. (1996). Neuroanatomical characterization of Fos induction in rat behavioral models of anxiety. Brain Res 713, 79–91. [DOI] [PubMed] [Google Scholar]
- Durdiakova J, Warrier V, Banerjee-Basu S, Baron-Cohen S, and Chakrabarti B. (2014). STX1A and Asperger syndrome: a replication study. Mol. Autism 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egbujo CN, Sinclair D, and Hahn CG. (2016). Dysregulations of Synaptic Vesicle Trafficking in Schizophrenia. Curr. Psychiatry Rep 18, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Li Y, Yue P, Khuri FR, and Sun SY. (2010). The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments TRAIL-mediated apoptosis through c-FLIP Down-regulation and DR5 induction independent of inhibition of cap-dependent protein translation. Neoplasia 12, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH. (2008). The hyperglutamatergic hypothesis of autism. Prog. Neuropsychopharmacol. Biol. Psychiatry 32, 911, 912–913. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Earle JA, Stary JM, Lee S, and Sedgewick J. (2001). Altered levels of the synaptosomal associated protein SNAP-25 in hippocampus of subjects with mood disorders and schizophrenia. Neuroreport 12, 3257–3262. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, and Thuras PD. (2009). GABAA Receptor Downregulation in Brains of Subjects with Autism. J. Autism Dev. Disord 39, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix-Ortiz AC, Beyeler A, Seo C, Leppla CA, Wildes CP, and Tye KM. (2013). BLA to vHPC inputs modulate anxiety-related behaviors. Neuron 79, 658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Doan NB, Imura T, Bush TG, and Sofroniew MV. (2004). GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 7, 1233–1241. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Bartley AF, Hays SA, and Huber KM. (2008). Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J. Neurophysiol 100, 2615–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillott A, Furniss F, and Walter A. (2001). Anxiety in High-Functioning Children with Autism. Autism 5, 277–286. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, and Sonenberg N. (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, and Dallaire P, et al. (2013). Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, and Nave KA. (2006). Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44, 611–621. [DOI] [PubMed] [Google Scholar]
- Hasler G, Fromm S, Alvarez RP, Luckenbaugh DA, Drevets WC, and Grillon C. (2007). Cerebral blood flow in immediate and sustained anxiety. J. Neurosci 27, 6313–6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessler NA, Shirke AM, and Malinow R. (1993). The probability of transmitter release at a mammalian central synapse. Nature 366, 569–572. [DOI] [PubMed] [Google Scholar]
- Homberg JR, Kyzar EJ, Scattoni ML, Norton WH, Pittman J, Gaikwad S, Nguyen M, Poudel MK, Ullmann JF, and Diamond DM, et al. (2016). Genetic and environmental modulation of neurodevelopmental disorders: Translational insights from labs to beds. Brain Res. Bull 125, 79–91. [DOI] [PubMed] [Google Scholar]
- Hutcheson HB, Olson LM, Bradford Y, Folstein SE, Santangelo SL, Sutcliffe JS, and Haines JL. (2004). Examination of NRCAM, LRRN3, KIAA0716, and LAMB1 as autism candidate genes. BMC Med. Genet. 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Shah M, Koo SY, Faraud KS, Santini E, and Klann E. (2015). eIF4E/Fmr1 double mutant mice display cognitive impairment in addition to ASD-like behaviors. Neurobiol. Dis 83, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Hagerman PJ, and Leehey MA. (2007). Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 6, 45–55. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, and He M, et al. (2013). Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet 93, 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JC, Su K, Goldberg AR, Luna VM, Biane JS, Ordek G, Zhou P, Ong SK, Wright MA, and Zweifel L, et al. (2018). Anxiety Cells in a Hippocampal-Hypothalamic Circuit. Neuron 97, 670–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Wang Y, Kim SJ, Bornhorst M, Jecrois ES, Anthony TE, Wang C, Li YE, Guan JL, Murphy GG, and Zhu Y. (2014). Transient inhibition of the ERK pathway prevents cerebellar developmental defects and improves long-term motor functions in murine models of neurofibromatosis type 1. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelstrup KG, Tuvnes FA, Steffenach HA, Murison R, Moser EI, and Moser MB. (2002). Reduced fear expression after lesions of the ventral hippocampus. Proc. Natl. Acad. Sci. USA 99, 10825–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, and Jonasdottir A, et al. (2012). Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, and Alvarez-Buylla A. (2009). The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 32, 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YC, Hinkley LB, Bukshpun P, Strominger ZA, Wakahiro ML, Baron-Cohen S, Allison C, Auyeung B, Jeremy RJ, and Nagarajan SS, et al. (2013). Autism traits in individuals with agenesis of the corpus callosum. J. Autism Dev. Disord 43, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Lee J, and Kim E. (2017). Excitation/Inhibition Imbalance in Animal Models of Autism Spectrum Disorders. Biol. Psychiatry 81, 838–847. [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, and Alvarez-Buylla A. (2009). Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VR, and Rastegar M. (2014). Rett syndrome and MeCP2. Neuromolecular Med 16, 231–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Fan F, Messa M, Raimondi A, Wu Y, Looger LL, Ferguson SM, and De Camilli P. (2012). Reduced release probability prevents vesicle depletion and transmission failure at dynamin mutant synapses. Proc. Natl. Acad. Sci. USA 109, E515–E523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque MA, Beltran-Matas P, Marin MC, Torres B, and Herrero L. (2017). Excitability is increased in hippocampal CA1 pyramidal cells of Fmr1 knockout mice. Plos One 12, e185067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C, Reis-Silva T, Lyra C, Felicio L, and Malnic B. (2018). Buried Food-seeking Test for the Assessment of Olfactory Detection in Mice, Vol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, and Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, and Gotz M. (2003). Neuronal or glial progeny: regional differences in radial glia fate. Neuron 37, 751–764. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, and Burley SK. (1999). Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716. [DOI] [PubMed] [Google Scholar]
- Martinez-Cerdeno V. (2017). Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev. Neurobiol 77, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Carrillo RA, and Zinn K. (2015). The translational regulator Cup controls NMJ presynaptic terminal morphology. Mol. Cell Neurosci 67, 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic I, Giovedi S, Lou X, Raimondi A, Collesi C, Shen H, Paradise S, O’Toole E, Ferguson S, Cremona O, and De Camilli P. (2011). Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron 72, 587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, and Scherer SW. (2007). Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet 81, 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, and Crawley JN. (2004). Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes. Brain Behav 3, 287–302. [DOI] [PubMed] [Google Scholar]
- Muller SK, Fagel DM, Stevens HE, Rabenstein RL, Maragnoli ME, Ohkubo Y, Picciotto MR, Schwartz ML, and Vaccarino FM. (2008). Deficiency in inhibitory cortical interneurons associates with hyperactivity in fibroblast growth factor receptor 1 mutant mice. Biol Psychiatry 63, 953–962. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Anitha A, Yamada K, Tsujii M, Iwayama Y, Hattori E, Toyota T, Suda S, Takei N, and Iwata Y, et al. (2008). Genetic and expression analyses reveal elevated expression of syntaxin 1A (STX1A) in high functioning autism. Int. J. Neuropsychopharmacol 11, 1073–1084. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Muller B, Massie D, Williams JH, O’Brien PC, Hughes A, Shen SB, Clair DS, and Miedzybrodzka Z. (2009). Deregulation of EIF4E: a novel mechanism for autism. J. Med. Genet 46, 759–765. [DOI] [PubMed] [Google Scholar]
- Nielsen PJ, and Trachsel H. (1988). The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. Embo. J 7, 2097–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis S, Forstmann BU, and Wagenmakers EJ. (2011). Erroneous analyses of interactions in neuroscience: a problem of significance. Nat Neurosci. 14, 1105–1107. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, and Kriegstein AR. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720. [DOI] [PubMed] [Google Scholar]
- Nygaard KR, Maloney SE, and Dougherty JD. (2019). Erroneous inference based on a lack of preference within one group: Autism, mice, and the social approach task. Autism Res. [DOI] [PMC free article] [PubMed]
- Oberer M, Marintchev A, and Wagner G. (2005). Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes. Dev 19, 2212–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler JA, Fox AS, Shelton SE, Rogers J, Dyer TD, Davidson RJ, Shelledy W, Oakes TR, Blangero J, and Kalin NH. (2010). Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature 466, 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, and Smith JD, et al. (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Coreano N, Bolkan SS, Pierce GM, Blackman DR, Hardin WD, Garcia-Garcia AL, Spellman TJ, and Gordon JA. (2016). Direct Ventral Hippocampal-Prefrontal Input Is Required for Anxiety-Related Neural Activity and Behavior. Neuron 89, 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt GM, Nguyen R, Bang JY, Aqrabawi AJ, Tran MM, Seo DK, Richards BA, and Kim JC. (2017). Bidirectional Control of Anxiety-Related Behaviors in Mice: Role of Inputs Arising from the Ventral Hippocampus to the Lateral Septum and Medial Prefrontal Cortex. Neuropsychopharmacol. 42, 1715–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, and Feng G. (2011). Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, and Sonenberg N. (1985). Insertion mutagenesis to increase secondary structure within the 5’ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526. [DOI] [PubMed] [Google Scholar]
- Pellow S, Chopin P, File SE, and Briley M. (1985). Validation of open:closed arm entries in an elevated plus-maze as a measure of anxiety in the rat. J. Neurosci. Methods 14, 149–167. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, and Woolfrey KM. (2011). Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 14, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzone MA, Lee WS, Hoffman GE, and Rabin BS. (1992). Induction of c-Fos immunoreactivity in the rat forebrain by conditioned and unconditioned aversive stimuli. Brain Res. 597, 41–50. [DOI] [PubMed] [Google Scholar]
- Pickart CM. (2001). Mechanisms Underlying Ubiquitination. Annu. Rev Biochem 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willems A, and Peter M. (2004). Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. Embo. J 23, 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, and Kemper TL. (1995). Hippocampus in autism: a Golgi analysis. Acta. Neuropathol 91, 117–119. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, and Merzenich MM. (2003). Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes. Brain Behav 2, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ortiz E, Cho W, Nazarenko I, Mo W, Chen J, and Parada LF. (2014). NF1 regulation of RAS/ERK signaling is required for appropriate granule neuron progenitor expansion and migration in cerebellar development. Genes Dev 28, 2407–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, Longo F, Koo SY, Mojica E, D’Andrea L, Bagni C, and Klann E. (2017). Reducing eIF4E-eIF4G interactions restores the balance between protein synthesis and actin dynamics in fragile X syndrome model mice. Sci. Signal 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, and Klann E. (2013). Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, and Zoghbi HY. (2011). Solving the autism puzzle a few pieces at a time. Neuron 70, 806–808. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, Holmans PA, Lee P, Bulik-Sullivan B, and Collier DA, et al. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P. (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Senba E, Matsunaga K, Tohyama M, and Noguchi K. (1993). Stress-induced c-fos expression in the rat brain: activation mechanism of sympathetic pathway. Brain Res Bull 31, 329–344. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, and Katz DM. (2011). Synaptic microcircuit dysfunction in genetic models of neurodevelopmental disorders: focus on Mecp2 and Met. Curr. Opin. Neurobiol 21, 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira MC, Sandner G, and Graeff FG. (1993). Induction of Fos immunoreactivity in the brain by exposure to the elevated plus-maze. Behav. Brain Res 56, 115–118. [DOI] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, and Crawley JN. (2010). Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci 11, 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Rupprecht KM, Hecht SM, and Shatkin AJ. (1979). Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. USA 76, 4345–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yoshino A, Yamanaka A, Asanuma C, Satou T, Hayashi S, Masuo Y, Sadamoto K, and Koike K. (2012). Effects of inhaled lavender essential oil on stress-loaded animals: changes in anxiety-related behavior and expression levels of selected mRNAs and proteins. Nat. Prod. Commun 7, 1539–1544. [PubMed] [Google Scholar]
- Urban DJ, and Roth BL. (2015). DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu. Rev. Pharmacol. Toxicol 55, 399–417. [DOI] [PubMed] [Google Scholar]
- Walf AA, and Frye CA. (2007). The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat. Protoc 2, 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu F, Chen W, Sun X, Cui W, Dong Z, Zhao K, Zhang H, Li H, and Xing G, et al. (2018). Genetic recovery of ErbB4 in adulthood partially restores brain functions in null mice. Proc. Natl. Acad. Sci. USA 115, 13105–13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Figueiredo D, Sun XD, Dong ZQ, Chen WB, Cui WP, Liu F, Wang HS, Li HW, and Robinson H, et al. (2018). Controlling of glutamate release by neuregulin3 via inhibiting the assembly of the SNARE complex. Proc. Natl. Acad. Sci. USA 115, 2508–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, Flory M, Kuchna I, Nowicki K, Ma SY, Imaki H, Wegiel J, Cohen IL, London E, Wisniewski T, and Brown WT. (2014). Stereological study of the neuronal number and volume of 38 brain subdivisions of subjects diagnosed with autism reveals significant alterations restricted to the striatum, amygdala and cerebellum. Acta Neuropathol. Commun 2, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, Feliciano C, Chen M, Adams JP, and Luo J, et al. (2007). Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 448, 894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadin E, Friedman E, and Bridger WH. (1991). Spontaneous alternation behavior: an animal model for obsessive-compulsive disorder? Pharmacol Biochem Behav 40, 311–315. [DOI] [PubMed] [Google Scholar]
- Yasuhara A. (2010). Correlation between EEG abnormalities and symptoms of autism spectrum disorder (ASD). Brain Dev. 32, 791–798. [DOI] [PubMed] [Google Scholar]
- Zhang W, Peterson M, Beyer B, Frankel WN, and Zhang ZW. (2014). Loss of MeCP2 from forebrain excitatory neurons leads to cortical hyperexcitation and seizures. J. Neurosci 34, 2754–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cui N, Wu Z, Su J, Tadepalli JS, Sekizar S, and Jiang C. (2010). Intrinsic membrane properties of locus coeruleus neurons in Mecp2-null mice. Am. J. Physiol. Cell Physiol 298, C635–C646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Shen C, Lu Y, Huang Z, Li L, Rand CD, Pan J, Sun X, Tan Z, and Wang H, et al. (2017). Muscle Yap Is a Regulator of Neuromuscular Junction Formation and Regeneration. J. Neurosci 37, 3465–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lu J, Liu L, Bernard D, Yang CY, Fernandez-Salas E, Chinnaswamy K, Layton S, Stuckey J, and Yu Q, et al. (2017). A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat Commun. 8, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Pleil KE, Urban DJ, Moy SS, Kash TL, and Roth BL. (2014). Chemogenetic inactivation of ventral hippocampal glutamatergic neurons disrupts consolidation of contextual fear memory. Neuropsychopharmacol. 39, 1880–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, and Messing A. (2001). hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proteomics analysis of cortex and hippocampus between GFAP-Cul3f/f and control mice. Related to Figure 4.
Table S2. Proteomics analysis of cortex and hippocampus between GFAP-Cul3f/+ and control mice. Related to Figure 4.
Table S3. GO analysis of up-regulated proteins in GFAP-Cul3f/+ samples. Related to Figure 4.
Table S4. GO analysis of down-regulated proteins in GFAP-Cul3f/+ samples. Related to Figure 4.
Table S5. GO analysis of up-regulated proteins in GFAP-Cul3f/f samples. Related to Figure 4.
Table S6. GO analysis of down-regulated proteins in GFAP-Cul3f/f samples. Related to Figure 4.