Two cryo-EM structures of IGF2R at different pH values imply the mechanism for its ligand release in endocytic compartments.
Abstract
Many cell surface receptors internalize their ligands and deliver them to endosomes, where the acidic pH causes the ligand to dissociate. The liberated receptor returns to the cell surface in a process called receptor cycling. The structural basis for pH-dependent ligand dissociation is not well understood. In some receptors, the ligand binding domain is composed of multiple repeated sequences. The insulin-like growth factor 2 receptor (IGF2R) contains 15 β strand–rich repeat domains. The overall structure and the mechanism by which IGF2R binds IGF2 and releases it are unknown. We used cryo-EM to determine the structures of the IGF2R at pH 7.4 with IGF2 bound and at pH 4.5 in the ligand-dissociated state. The results reveal different arrangements of the receptor in different pH environments mediated by changes in the interactions between the repeated sequences. These results have implications for our understanding of ligand release from receptors in endocytic compartments.
INTRODUCTION
The sorting and recycling of cell surface receptors with their endocytosed ligands are essential events for maintaining the homeostasis of specific ligands in the extracellular space and transducing the signal into the cytosol. Receptors capture their ligands on the cell surface, and ligand-receptor dissociation occurs in a low pH endosomal compartment. In some cell surface receptors, the ligand binding domain is composed of multiple repeated sequences. For example, members of the low-density lipoprotein receptor (LDL-R) family contain multiple repeats of 40 amino acids, and each repeat folds into a discrete structure stabilized by multiple internal disulfide bonds (1). Another example is the insulin-like growth factor 2 receptor (IGF2R), which contains 15 β strand–rich repeat domains. IGF2 promotes cell proliferation and differentiation in many somatic tissues during embryonic and fetal development through binding IGF1R and insulin receptor (2–6). It also stimulates tumor growth by promoting cell division (7). To prevent excess IGF2 signaling, IGF2R, acting as a signal antagonist, delivers IGF2 from the extracellular space to the endosome, facilitating IGF2 degradation (8–10). This feature suggests that IGF2R can serve as a tumor suppressor by down-regulating IGF2 levels, potentially preventing the growth of tumors (11–13).
IGF2R also serves as the cation-independent mannose 6-phosphate receptor (CI-MPR), which mediates the delivery of ~60 different newly synthesized mannose 6-phosphate (Man6P)–tagged proteins from the trans-Golgi to prelysosomes via the early endosome (3–5). IGF2R, which can form a complex with various lysosomal enzymes, is transported from the trans-Golgi to the late endosome. The low pH of the endosome triggers the dissociation of these complexes (5). Upon dissociation, the lysosomal enzymes are released into the lysosomes, and IGF2R either returns to the Golgi to repeat this process or distributes on the cell surface to internalize IGF2. Furthermore, there are a number of additional ligands that interact with IGF2R, including transforming growth factor–β1, urokinase-type plasminogen activator receptor, granzyme B, retinoic acid, and others (4). Man6P analogs can be used to deliver specific compounds to lysosomes by targeting IGF2R for future therapies of lysosomal diseases (14). Therefore, structural knowledge of IGF2R is important for understanding the molecular mechanism of lysosomal protein trafficking and IGF2 regulation and facilitating drug development for the treatment of cancers and lysosomal storage diseases.
RESULTS
Structure determination
IGF2R is a type I transmembrane glycoprotein that contains 15 β strand–rich repeat domains, which can bind various cargoes, followed by a transmembrane helix and a small cytosolic domain (Fig. 1A). The structures of parts of IGF2R have been determined by x-ray crystallographic studies. These studies and in vitro mutagenesis have shown that IGF2R uses domain 11 for binding IGF2 (15–18), and it engages a newly synthesized Man6P-tagged proteins using domains 3, 5, and 9 (5, 19). These domains are structural homologs, each consisting of 11 major β strands (Fig. 1, B and C, and fig. S1). Notably, domain 13 also contains a fibronectin type II fold (FNII) that is inserted between β8 and β9 and is required for stabilizing domain 11, allowing it to bind the IGF2 ligand (Fig. 1C) (17). However, the functions of the other domains and how the 15 domains collaborate in ligand recognition have remained a mystery, since the entire structure of IGF2R is not available.
Fig. 1. Overall structure of the apo state of the IGF2R at pH 4.5.
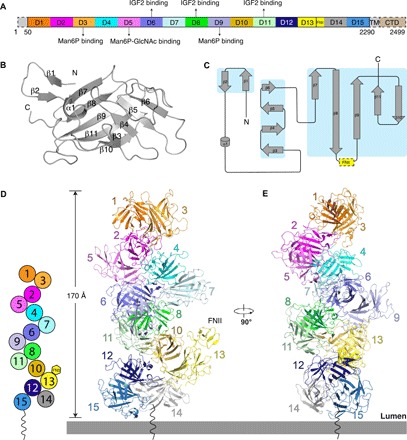
(A) Schematic diagram of full-length IGF2R. Each domain is shown in different colors. The N terminus, transmembrane (TM) helix, and cytosolic domain (CTD) are not seen in the structure and are indicated with dashed boxes. (B) Secondary structure of a single domain. The secondary-structure elements are labeled. (C) Topology diagram for a single domain. All the secondary-structure elements are colored gray, and the β sheets are highlighted in the cyan boxes. The FNII domain of domain 13 is indicated by a yellow dashed box. (D) Diagram of the domain contacts in the apo state and overall structure of apo state IGF2R. The figure illustrates the interactions between the even-numbered domains and the odd-numbered domains at position X + 3. These interactions generate a helix-like shape (left). Domains in the same subgroup are indicated with dark and light colors. (E) Overall structure of apo state IGF2R rotated by 90°.
We purified native full-length IGF2R from bovine liver using previously reported protocols (20, 21). The purified protein is well behaved, suggesting that it is suitable for structural investigation. The protein was first prepared at the physiologic lysosomal pH of 4.5. At this pH, the endogenous cargoes are released from the receptor, revealing the apo state of the IGF2R in the endosome (fig. S2A). Then, we changed the buffer to pH 7.4 and incubated the receptor with IGF2 to capture the structure of the ligand-bound receptor on the cell surface (fig. S2B).
The “helix-like” assembly of apo IGF2R at pH 4.5
The apo-structure was determined at a resolution of 3.46 Å (figs. S3 and S4 and table S1). We were able to determine the structure using our cryo–electron microscopy (cryo-EM) density maps along with the previously resolved crystal structures of the individual domains. In the apo state at pH 4.5, all 15 domains are tightly packed together presenting a helix-like conformation (Fig. 1, D and E); however, the N terminus, transmembrane helix, and cytosolic domain are invisible in the cryo-EM map. The dimensions of the extracellular domain of IGF2R are 170 Å × 80 Å × 80 Å. The 15 domains, except domain 14, can be divided into seven subgroups: domains 1 and 3, domains 2 and 5, domains 4 and 7, domains 6 and 9, domains 8 and 11, domains 10 and 13, and domains 12 and 15. (Fig. 1D).
Further structural comparison shows that these seven subgroups share a similar conformation (fig. S5). Each subgroup is stabilized by interactions between β sheets formed by β strands β7 to β11 of each domain (Fig. 2, A to G), while β strands β1 and β2 mediate packing between neighboring subgroups. The β3 of domains 1, 2, 4, 6, 8, 10, and 12 (the first domain of every subgroup) are also involved in subgroup packing, while β4 to β6 always face the solvent. In addition to the β strands, loops connecting the subgroups and the internal loops of each domain (mostly the loops connecting β9 and β10) provide major interactions between the subgroups (Fig. 2, H to M). Domain 2 interacts with domains 4 and 5 and connects with domains 1 and 3 (Fig. 2, B, H, and I). Domain 8 serves as a hub to bind five domains including domains 6, 7, 9, 10, and 11 (Fig. 2, E, K, and L). Although domain 14 does not form a subgroup with other domains, it still interacts with domains 12 and 15 (Figs. 1D and 2M). Notably, because the FNII domain of domain 13 is on the edge of the complex, it does not bind any structural elements of the other domains (Fig. 1D).
Fig. 2. Assembly pattern for apo state of the IGF2R at pH 4.5.
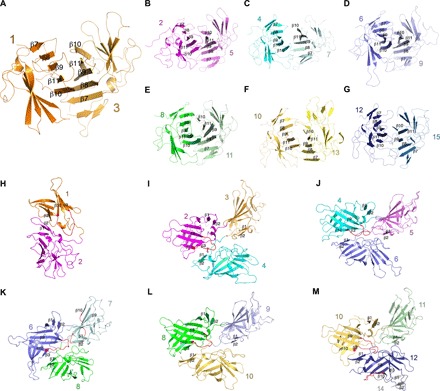
(A to G) The interaction interfaces of the seven subgroups. The β sheets formed by β strands β7 to β11 of each domain are indicated. (H to M) The interaction details of neighboring domains in the apo state. The structural elements involved in the interaction are indicated. The β9-β10 loop is shown in red.
The “pistol-like” assembly of IGF2-bound IGF2R at pH 7.4
When the IGF2R was purified at pH 7.4, negative staining EM revealed a “beads on a string” shape (fig. S6A). Unfortunately, three-dimensional (3D) reconstruction with cryo-EM failed (fig. S6). When purified in the presence of IGF2 at pH 7.4, the IGF2R was stabilized. The IGF2-bound structure was determined at a resolution of 4.32 Å (figs. S7 and S8 and table S1) using cryo-EM. Compared with the apo structure at pH 4.5, the ligand-bound IGF2R showed a marked structural change. The 15 domains are rearranged, and the dimensions changed to 230 Å × 170 Å × 70 Å (Fig. 3, A and B). The conformation resembles a pistol with IGF2 bound at the intersection of the grip and barrel (Fig. 3B). The transmembrane helix and cytosolic domain are undetermined in the map. Domains 1 to 3 and 15 show a lower local resolution than they do in the apo state, suggesting local flexibility in the IGF2-bound state (fig. S7). Because of the lower local resolution, these domains are not included in our structure model.
Fig. 3. IGF2R conformational changes after binding IGF2 in pH 7.4.
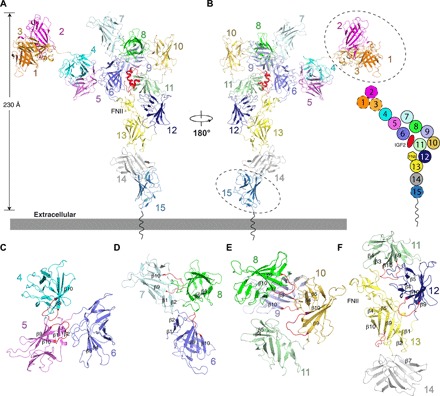
(A) Overall structure of the IGF2R-IGF2 complex. The color scheme of IGF2R is the same as in Fig. 1A. IGF2 is highlighted in red. (B) Overall structure of the IGF2R-IGF2 complex rotated by 180° with respect to (A). The schematic diagram of this state is shown on the right. Because of low resolution, domains 1 to 3 and domain 15 were not built into the experimental map but modeled (indicated by dashed ovals) in (A) and (B) based on the cryo-EM map. (C) The interaction interfaces between domains 4, 5, and 6. (D) The interaction interfaces between domains 6, 7, and 8. (E) The interaction interfaces between domains 8, 9, 10, and 11. (F) The interaction interfaces between domains 11, 12, 13, and 14. All the structural elements involved in interactions are indicated. The β9-β10 loop is shown in red.
In the IGF2-bound state, the seven subgroups have been severed, causing a more extended morphology than in the apo state with consecutive subgroups also adjacent. Similar to the subgroup-subgroup interaction in the apo state, the interactions among the individual domains in the IGF2-bound state are primarily provided by loops, including the domain-connecting loops and the internal loops of each domain. The β9-β10 loop retains its role in mediating the interaction between neighboring domains (Fig. 3, C to F). Domain 5 interacts only with domains 4 and 6 (Fig. 3C), while domain 8 interacts with domains 6, 7, 9, and 10 (Fig. 3, D and E). In the apo state, domain 10 is in the center and connects with domains 8, 9, 11, 12, and 13 (Fig. 1D). However, in the IGF2-bound state, domain 10 has moved to the edge of the complex where it contacts domains 9 and 11 (Fig. 3B). Domain 11 interacts with domains 6, 9, 10, 12, and FNII of domain 13 and accommodates the IGF2 ligand together with domains 6 and 8 (Fig. 3, A, B, E and F).
IGF2R engages IGF2 by domains 6, 8, and 11
The IGF2-bound structure reveals the interaction details between IGF2R and IGF2. The interaction interfaces bury a total solvent-accessible surface area of 4300 Å2 by PISA (Proteins, Interfaces, Structures, and Assemblies) calculation (22). Besides the well-known involvement of domain 11, we found domains 6 and 8 to also participate in the specific recognition of IGF2. These domains embrace IGF2 from three different angles. Domain 6 interacts with the α helices α1 and α2 and loops of IGF2 through β strands, domain 8 interacts mainly with α2 and a loop of IGF2 by its α1-β3 and β4-β5 loops, while domain 11 makes extensive contacts with α1, α3, and loops of IGF2; this interface is structurally almost identical with the IGF2–domain 11 structure, which has been previously reported with a root mean square deviation of 0.775 Å for 123 Cα atoms (Fig. 4, A to C) (17).
Fig. 4. Interaction details of IGF2 with IGF2R.
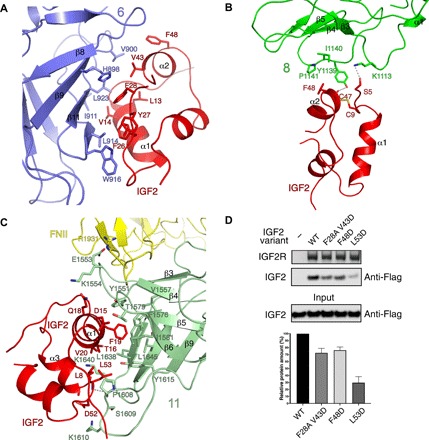
(A) The interface of domain 6 to IGF2. (B) The interface of domain 8 to IGF2. (C) The interface of domain 11 to IGF2. The residues and structural elements involved in the interaction are indicated. The hydrogen bond is represented by a dashed line. The proteins are colored as in Fig. 3B. (D) Pull-down assay between the wild-type (WT) and mutant Flag-IGF2 and the wild-type IGF2R. IGF2R is conjugated onto PMP resin. Top: The IGF2R is shown on SDS–polyacrylamide gel electrophoresis (PAGE) with Coomassie blue staining, while the WT or mutant IGF2 was detected by immunoblotting. Bottom: Quantification of the bands at the top by normalization against IGF2R. The black column represents the amount of WT protein, which is designated as 100%. Gray columns represent the relative amount of mutant proteins. Error bars represent the means ± SD (n = 3).
In the apo state, domains 6 and 9 form one subgroup via their β7 to β11 β sheets (Fig. 2D), while in the IGF2-bound state, IGF2 replaces domain 9 to bind β7 to β11 of domain 6 (Fig. 4A). The subgroup of domains 8 and 11 connects by hydrophobic interactions through their major β strands at pH 4.5 (Fig. 2E), and there is no interaction between domains 11 and 13 in the apo state (Fig. 1D). In the IGF2-bound state, the interaction between domains 8 and 11 has been abolished, allowing them to bind IGF2 (Fig. 4, B and C, respectively). Domain 13 has relocated and its FNII domain interacts with domain 11 to support IGF2 binding by a salt bridge between E1553 and R1931 (Fig. 4C).
To further validate our structure, we generated IGF2 variants following a previously established protocol (23). The double mutations F28A and V43D on the interface between domain 6 and IGF2 reduce binding between IGF2R and IGF2 by 20 to 30% based on a pull-down assay (Fig. 4D). The F48D mutant, which abolishes the interface between domain 8 and IGF2, also reduces binding between IGF2R and IGF2 by 20 to 30% (Fig. 4D). Notably, the L53D mutation causes an interference in the interface between domain 11 and IGF2, abolishing over 60% of the binding between IGF2R and IGF2 (Fig. 4D). These findings directly support our observation that domains 6, 8, and 11 are involved in the ligand recognition of IGF2R. Moreover, previous reports showed that IGF2 mutations such as Y27E, which is involved in interaction with domain 6, L8A, F19A, and D52A, which are involved in interactions with domain 11, affect the binding affinity with IGF2R (17, 24, 25), further supporting our structural observations.
The putative mechanism of IGF2R-mediated Man6P cargo release
In addition to binding IGF2, IGF2R also transports newly synthesized Man6P-containing proteins from the trans-Golgi to prelysosomes via the early endosome. There are two kinds of mannose 6-phosphate receptor: CI-MPR (also called IGF2R) and the cation-dependent mannose 6-phosphate receptor (CD-MPR), which contains only one β strand–rich repeat domain for binding Man6P. The structures of the CD-MPR dimer in different pH values have been determined (26). The lumenal domain of CD-MPR, which shares a similar fold with the IGF2R domains, uses a cavity created by multiple β strands and connected loops to bind Man6P at pH 6.5. The β10-β11 loops in the binding region change their conformations to narrow the cavity to facilitate release of Man6P (fig. S9A). We speculate that IGF2R may similarly rearrange its domains to adopt a conformation poised for capturing Man6P-tagged proteins. Previous work also showed that Man6P-tagged proteins can mediate the dimerization of IGF2R for transport (21).
Previous studies have shown that domains 3, 5, and 9 are required for IGF2R to bind Man6P (5). Comparison of the conformations of domains 5 and 9 at different pH values suggests that a low pH induces a constriction of the cavity lined by the domains, possibly facilitating Man6P release (fig. S9B). Our structural analysis shows that the IGF2 binding site is different from the Man6P binding site. We purified pentamannosyl 6-phosphate (PMP)–labeled bovine serum albumin (BSA), which mimics Man6P-labeled substrates, according to a previous study (27). The pull-down assay shows that at pH 6.5, the wild-type (WT) IGF2 can interact with IGF2R and PMP-labeled BSA simultaneously; however, IGF2 with the L53D mutation cannot bind IGF2R and the PMP-labeled BSA (Fig. 5A). This result is consistent with the result of a previous study that shows that the binding sites of IGF2R to Man6P and IGF2 are independent (28). Notably, lysosomal enzymes that contain Man6P residues, such as β-galactosidase, show competition with IGF2 for receptor binding (29, 30), while bivalent Man6P-based β-glucuronidase shows an increased internalization of IGF2 bound to the IGF2R (31), implying that the binding between IGF2 and IGF2R may be regulated by distinct Man6P-labeled substrates. Further investigations on this mechanism are required.
Fig. 5. The binding sites of IGF2R to Man6P and IGF2 are independent.
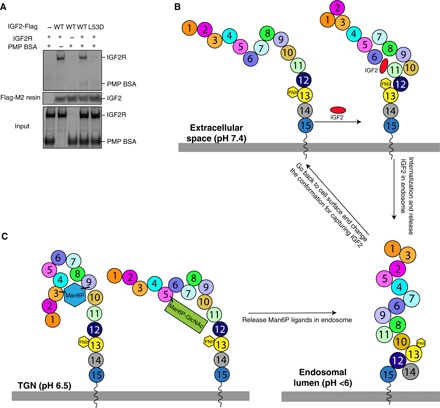
(A) Pull-down assay between IGF2, PMP-labeled BSA, and IGF2R. IGF2 is conjugated onto Flag-M2 resin. The proteins are shown on SDS-PAGE with Coomassie blue staining. (B) Working model of IGF2R for IGF2 recognition, postulating a molecular mechanism of how IGF2R changes its conformation to recognize IGF2 by using its domains at different pH values. (C) A speculation of how Man6P substrates are recognized by IGF2R. TGN, trans-Golgi network.
DISCUSSION
Our structures reveal that the multiple domains of IGF2R have different arrangements in different pH environments, which can allow binding of the IGF2 ligand (Fig. 5B). The structural analysis, along with previous functional assays, reveals that all 15 domains are necessary for the physiological functions of IGF2R. Domains 3, 5, and 9 are essential for binding the Man6P ligand (5, 19). Our structure shows that domains 6, 8, 11, and 13 are required for IGF2 engagement (Fig. 4), and the other domains serve as a scaffold for the conformational change, allowing IGF2 to access its binding sites at pH 7.4. Notably, this conformational change does not require an energy source in the form of adenosine triphosphate or guanosine triphosphate, instead most likely being driven by pH. Histidine residues undergo the charge transitions between pH 4.5 and pH 7.4 to facilitate a pH-dependent conformational change. In addition, the loops between the domains always consist of less than 20 residues. These features facilitate hydrophobic interactions between the domains, allowing them to snugly pack together rather than being flexible as in the endosomal lumen, possibly competing with each other. This tight conformation also forces IGF2 or Man6P-tagged protein to be released from the receptor. Negative stain EM shows that the ligand-free IGF2R in pH 7.4 reveals a beads-on-a-string shape (fig. S6A). At neutral pH, the hydrophilic interactions between the loops of the domains stretch the receptor to create space for ligand binding.
Previous studies have shown that the low pH could trigger ligand dissociation from the receptor and then the receptor can recycle to the cell surface to receive a new ligand (5, 32). In comparison with the IGF2-bound state, the low pH induces a substantial conformational change in IGF2R that releases the ligand, reinforced by changes in the interactions between the repeated sequences. This considerable rearrangement of multiple domains can diminish the interface between IGF2R and IGF2 rather than the subtle conformational change of its loop of CD-MPR to narrow the cavity for dissociating the Man6P ligand (fig. S9). The IGF2R may capture the Man6P substrates by its domains 3, 5, and 9 in the Golgi and deliver them into prelysosomes (Fig. 5C). This perhaps explains why the multiple repeats are required for some cell surface receptors: The repeats interact with the ligand through distinct interfaces that stabilize ligand binding on the cell surface; furthermore, the low pH triggers the repeats to pack together, forcing the ligand to be released from the binding site.
These observations suggest a general molecular mechanism of substrate recognition by multiple-repeat domain containing receptors like LDL-R and vitellogenin receptor. LDL-R uses epidermal growth factor (EGF) repeats for binding the LDL particles on the cell surface. As the internalized LDL-R with the LDL particle is delivered to the lysosome, the low pH may induce a conformational change of the EGF repeats to release the LDL particle into the lysosome (33, 34).
MATERIALS AND METHODS
Preparation of PMP resin
Phosphomannan was prepared from Pichia (Hansenula) holstii NRRL Y-2448 yeast as previously described (35). After obtaining the phosphomannan, PMP fragments of yeast phosphomannan were prepared as previously described (36). The PMP oligosaccharides were derivatized with the bifunctional reagent, p-(aminophenyl)ethylamine (PAPEA), and coupled to CNBr-activated Sepharose. Briefly, 1 g of CNBr-activated resin was incubated with 25 mg of PMP-PAPEA in 10 ml of coupling buffer [0.5 M NaCl and 0.1 M NaHCO3 (pH 8.3)] for 2 hours at room temperature. The resin was then added to a 1.5 cm by 12 cm column and was washed with 20 ml of coupling buffer. It was then quenched with 10 ml of 1 M ethanolamine (pH 8.0) for 2 hours. After quenching, the resin was washed sequentially with 25 ml each of 0.1 M NaHCO3, 0.1 M sodium acetate (pH 5.0), and phosphate-buffered saline (PBS). It was then equilibrated with PBS (pH 7.4) plus 0.1% Triton X-100 (column wash buffer).
Protein purification
The full-length IGF2R was purified from bovine liver as described previously (21). Briefly, 200 g of bovine liver was blended with extraction buffer containing 50 mM imidazole (pH 7), 150 mM NaCl, 5 mM β-glycerol phosphate, 2% Triton X-100, 0.25% sodium deoxycholate, 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), aprotinin (20 μg/ml), and leupeptin (10 μg/ml). After centrifugation at 18,000 rpm for 50 min, the supernatant was filtered by cheesecloth and incubated with affinity PMP-Sepharose resin for 1.5 hours. Then, the resin was washed with wash buffer [50 mM imidazole (pH 7), 150 mM NaCl, 5 mM sodium β-glycerophosphate, and 0.05% Triton X-100]. The protein was eluted with wash buffer plus 10 mM Man6P and further purified by gel filtration using a Superose 6 10/300 column (GE Healthcare) pre-equilibrated with buffer A [150 mM ammonium acetate, 250 mM acetic acid (pH 4.5), and 0.005% GDN (glyco-diosgenin) (Anatrace)] to release the endogenous cargoes. Mass spectrometry confirmed the identity of the protein. The peak fractions were collected and concentrated to 4 to 5 mg/ml for cryo-EM grid preparation. To obtain the ligand-free IGF2R, the protein in pH 4.5 was further purified with gel filtration using a Superose 6 10/300 column pre-equilibrated with buffer B [20 mM Hepes (pH 7.4), 150 mM NaCl, and 0.0042% GDN (Anatrace)]. The peak fractions were collected and diluted to 0.01 mg/ml for negative staining grid preparation. To assemble the IGF2R-IGF2 complex, the IGF2R in the apo state was diluted into buffer B directly and mixed with fivefold excess (molar ratio) of human IGF2 (purchased from R&D Systems, catalog number 292-G2-250) for 1 hour. The complex was applied to a Superose 6 10/300 column pre-equilibrated with buffer B. The peak fractions were collected and concentrated to 4 to 5 mg/ml for cryo-EM grid preparation.
IGF2 was made in Escherichia coli using the previously reported methods (23). Human mature IGF2 was cloned into pET His6 NusA vector (Addgene) with an N-terminal linker containing the amino acids “GAMA” and a C-terminal Flag tag. The plasmid was expressed in E. coli BL21 (DE3) cells, and the transformed cells were grown at 37°C in LB medium until OD600 (optical density at 600 nm) reached 0.8 and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 8 hours at 25°C. The cells were harvested by centrifugation, resuspended in a lysis buffer [50 mM Tris-HCl (pH 8.5), 500 mM NaCl, 0.05% Triton X-100, 10% glycerol, and 1 mM PMSF], and then lysed with sonication. After the cell lysates were centrifuged at 18,000 rpm for 40 min, the supernatant was purified by a Ni–nitrilotriacetic acid column (QIAGEN) with the lysis buffer containing 50 mM Tris-HCl (pH 8.5) and 150 mM NaCl supplemented with 20 mM imidazole and 500 mM imidazole serving as the washing buffer and elution buffer, respectively. After the NusA was cleaved off by TEV (Tobacco Etch Virus), the protein was further purified by gel filtration using a Superdex 200 10/300 column (GE Healthcare) pre-equilibrated with buffer [50 mM Tris-HCl (pH 8.5) and 150 mM NaCl]. The purified protein was of sufficient purity (>95%) and analyzed by SDS–polyacrylamide gel electrophoresis (PAGE). The mutants were constructed using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). All constructs were confirmed by DNA sequencing.
The PMP-labeled BSA was prepared following a previously published procedure (27), and PMP (0.2 M) was coupled to BSA (15 mg/ml) in 50 mM N,N-bis(2-ydroxyethyl) glycine (pH 9.0) with 160 mM NaCNBH3 at 37°C for 5 days. Then, the product was purified on a G-50 Sephadex column and Superdex 200 10/300 column sequentially with PBS. The collected fractions show an obvious larger molecular weight than the original BSA by SDS-PAGE.
Pull-down assay
To verify the functional relevance of the IGF2R-IGF2 interaction, we performed in vitro PMP pull-down assay. The WT and mutant IGF2 were expressed and purified from E. coli as described above. Purified IGF2R protein was immobilized to 20 μl of PMP resin and then incubated with WT or mutant IGF2 for 1 hour at 4°C in buffer B. Then, the resin was spun down and washed twice with the same buffer. The IGF2R was shown with SDS-PAGE with Coomassie blue staining, while the WT or mutant IGF2 was detected by anti-Flag antibody (M185, MBL Life Science).
To see whether PMP-labeled BSA can interact with IGF2R when binding IGF2, purified IGF2 protein was immobilized to anti-Flag M2 resin and then incubated with IGF2R and/or PMP-labeled BSA for 1 hour at 4°C in a buffer containing 20 mM MES (pH 6.5), 150 mM NaCl, and 0.0042% GDN. The beads were washed twice and eluted with 28-μl buffer supplemented with Flag peptide (0.3 mg/ml). Twenty-three microliters of the elution was loaded on SDS-PAGE for detection. Each assay was reproduced at least three times, images were analyzed, and band intensities were quantified using ImageJ. Bar graphs were generated by Prism 8 (GraphPad).
Negative staining EM of ligand-free IGF2R
Aliquots (3.5 μl) of ligand-free IGF2R (0.01 mg/ml) in buffer B were adsorbed to a glow-discharged, 400-mesh, carbon-coated copper grid and stained with 2% (w/v) phosphotungstic acid (pH 7.4). Images were recorded on a Tecnai G2 Spirit transmission electron microscope (FEI) equipped with a LaB6 source at 120 kV using a Gatan Ultrascan charge-coupled device camera.
Cryo-EM sample preparation and imaging
Freshly purified apo state IGF2R (4 to 5 mg/ml) in buffer A and the IGF2R-IGF2 complex (4 to 5 mg/ml) in buffer B were added to Quantifoil R1.2/1.3 400 mesh Au holey carbon grids (Quantifoil), blotted using a Vitrobot Mark IV (FEI), and frozen in liquid ethane. The grid of the apo state IGF2R protein was imaged using a 300-keV Titan Krios (FEI) with a Gatan K2 Summit direct electron detector (Gatan). The data were collected in superresolution mode at a pixel size of 0.5 Å with a dose rate of ~2 electrons per pixel per second. Images were recorded for 10-s exposures in 50 subframes to give a total dose of ~80 electrons/Å2. The grid of the IGF2-IGF2R complex was imaged using a 300-keV Titan Krios (FEI) with a Gatan K3 Summit direct electron detector (Gatan). The data were collected in superresolution mode at a pixel size of 0.43 Å with a dose rate of ~6 electrons per pixel per second. Images were recorded for 3-s exposures in 75 subframes to give a total dose of ~100 electrons/Å2. The grid of ligand-free IGF2R was imaged using a 200-keV Talos Arctica (FEI) with a Gatan K3 Summit direct electron detector (Gatan). The data were collected in superresolution mode at a pixel size of 0.445 Å with a dose rate of ~6 electrons per pixel per second. Images were recorded for 2-s exposures in 40 subframes to give a total dose of ~60 electrons/Å2.
Image processing and 3D reconstruction
For the three datasets, dark subtracted images were normalized by gain reference and binned twofold, which resulted in the original pixel size of 1, 0.86, and 0.89 Å for apo state IGF2R, the IGF2R-IGF2 complex, and ligand-free IGF2R, respectively. The beam-induced motion correction was performed using the program MotionCor2 (37), and the contrast transfer function was estimated using CTFFIND (38). To generate apo state IGF2R templates for auto-picking, ~2000 particles were manually picked and classified by 2D classification in RELION. After auto-picking in RELION (39), the low-quality images and false-positive particles were removed manually. About 271,448 particles were extracted for subsequent 2D and 3D classification. An initial model generated by RELION was used for 3D classification low-pass–filtered to 50 Å. The best class of 3D classification, containing around 128,789 particles, provided a 4.59-Å map after 3D auto-refinement in RELION. Bayesian polishing of particles was then performed on the 128,789 particles using RELION-3. The 3D refinement using a soft mask and solvent-flattened Fourier shell correlations (FSCs) yielded a reconstruction at 3.46 Å before postprocessing. Applying a soft mask in RELION-3 postprocessing yielded a final cryo-EM map of 3.46 Å. Resolution was estimated using the FSC 0.143 criterion.
For the IGF2R-IGF2 complex, ~2000 particles were manually picked and classified by 2D classification in RELION to generate the templates for auto-picking. After auto-picking in RELION, low-quality images and false-positive particles were removed manually. About 349,357 particles were extracted for the first round of 2D and 3D classification using an initial model generated by RELION. Particles from the best 3D class were chosen for the second round of 2D and 3D classification. The best class of the secondary 3D classification, containing 75,821 particles, provided an 8.18-Å map after 3D auto-refinement in RELION. Bayesian polishing of particles was then performed on the 75,821 particles using RELION-3. The 3D refinement using a soft mask and solvent-flattened FSCs yielded a reconstruction at 4.39 Å before postprocessing. Applying a soft mask in RELION-3 postprocessing yielded a final cryo-EM map of 4.32 Å. Resolution was estimated using the FSC 0.143 criterion.
For ligand-free IGF2R, ~2000 particles were manually picked and classified by 2D classification in RELION to generate the templates for auto-picking. The low-quality images and false-positive particles were removed manually after auto-picking. About 79,993 particles were extracted for 2D classification. The particles could not be aligned well, resulting in unclear features of the 2D classes. Particles from the 2D classes with relatively clear backgrounds were chosen for 3D classification using an initial model generated by RELION. None of the 3D classes were qualified for 3D auto-refinement.
Model construction
To obtain better side-chain densities for model building, we sharpened the map of apo state IGF2R using postprocessing in RELION-3. On the basis of previously reported structures including domains 1 to 3 [Protein Data Bank (PDB) code 1Q25], domains 11 to 14 (PDB code 2V5O), domain 5 (PDB code 2KVA), and the predicted structures including domains 4, 6 to 10, and 15 by SWISS-MODEL (https://swissmodel.expasy.org/), each domain was docked into the cryo-EM map using Chimera (40) to generate the initial model. Domain 13, the only domain that contains a featured FNII domain, was first revealed and docked into the map. Then, the other domains were docked into the map based on the connecting loops and structural features. The final model was built in Coot (41). The glycosylation sites of the IGF2R extracellular domains and the three to four pairs of disulfide bonds of each domain helped to confirm the register of the residues. Residues 1 to 50 (N terminus), 999 to 1010 (loop in domain 7), 1814 to 1826 (the N terminus of domain 13), and 2135 to 2142 (the linker between domains 14 and 15) were not resolved or built.
For the IGF2-bound IGF2R, we sharpened the map of the complex using postprocessing in RELION-3. The structures of domains 11 to 14 (PDB code 2V5O) and domains 11 to 13 of the IGF2 complex (PDB code 2V5P) were first docked into the map using Chimera to generate the initial model. Domains 4 to 10 were docked on the basis of the structure of the apo state IGF2R. Model building was performed manually using Coot. Domains 1 to 3 and domain 15 were not built because of poor densities. Residues 1000 to 1011 (loop in domain 7) and 1693 to 1696 (loop in domain 12) of IGF2R and residues 1 to 4 (N terminus), 30 to 40 (α1-α2 linker), and 64 to 67 (C terminus) of IGF2 were not built.
Model refinement and validation
The model was refined in real space using PHENIX (42) and also in reciprocal space using Refmac with secondary-structure restraints and stereochemical restraints (43, 44). For cross-validations, the final model was refined against one of the half-maps generated by 3D auto-refine, and the model versus map FSC curves were generated in the Comprehensive validation module in PHENIX. Local resolutions were estimated using RELION-3. PHENIX and MolProbity (45) were used to validate the final model. Structural figures were generated using PyMOL (www.pymol.org) and Chimera (40).
Supplementary Material
Acknowledgments
We thank R. MacDonald for sharing the protocol for protein preparation. We thank D. Stoddard at the UT Southwestern Cryo-EM Facility (funded, in part, by the CPRIT Core Facility Support Award RP170644), M. Ebrahim and J. Sotiris at The Rockefeller University Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center for data collection, A. Lemoff at the UT Southwestern Proteomics Core for mass spectrometry identification, and M. Brown, E. Debler, and J. Goldstein for discussion during manuscript preparation. Funding: This work was supported by the Endowed Scholars Program in Medical Science of UT Southwestern Medical Center, O’Donnell Junior Faculty Funds, NIH grant P01 HL020948, NIH grant R01GM134700 (to X.L.), and The Rockefeller University (to E.C.). X.Q. is the recipient of a DDBrown Fellow of the Life Sciences Research Foundation. X.L. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-53-19) and a Rita C. and William P. Clements, Jr. Scholar in Biomedical Research at UT Southwestern Medical Center. Author contributions: X.L. conceived the project. R.W. and P.S. prepared the protein. R.W., X.Q., and E.C. carried out cryo-EM work. R.W. and X.Q. built and refined the structures. R.W. carried out the functional studies. All the authors analyzed the data and contributed to manuscript preparation. X.L. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The 3D cryo-EM density maps of apo state IGF2R and the IGF2-IGF2R complex have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-20815 and EMD-20816. Atomic coordinates for the atomic model of apo state IGF2R and IGF2-IGF2R have been deposited in the PDB under accession numbers 6UM1 and 6UM2. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/7/eaaz1466/DC1
Fig. S1. Sequence alignment of different domains in IGF2R.
Fig. S2. Biochemical characterization of apo state IGF2R and IGF2-bound IGF2R.
Fig. S3. Data processing and model quality assessment of apo state IGF2R.
Fig. S4. Cryo-EM map of apo state IGF2R.
Fig. S5. Structural comparison for seven subgroups of apo state IGF2R in two different views.
Fig. S6. Data processing for ligand-free IGF2R.
Fig. S7. Data processing and model quality assessment of IGF2-bound IGF2R.
Fig. S8. Cryo-EM map of IGF2-bound IGF2R.
Fig. S9. Structural comparison of CD-MPR and domains of IGF2R for Man6P.
Table S1. Cryo-EM data collection, refinement, and validation statistics.
REFERENCES AND NOTES
- 1.Brown M. S., Anderson R. G., Basu S. K., Goldstein J. L., Recycling of cell-surface receptors: Observations from the LDL receptor system. Cold Spring Harb. Symp. Quant. Biol. 46, 713–721 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Natowicz M. R., Chi M. M., Lowry O. H., Sly W. S., Enzymatic identification of mannose 6-phosphate on the recognition marker for receptor-mediated pinocytosis of beta-glucuronidase by human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 76, 4322–4326 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald R. G., Pfeffer S. R., Coussens L., Tepper M. A., Brocklebank C. M., Mole J. E., Anderson J. K., Chen E., Czech M. P., Ullrich A., A single receptor binds both insulin-like growth factor II and mannose-6-phosphate. Science 239, 1134–1137 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Ghosh P., Dahms N. M., Kornfeld S., Mannose 6-phosphate receptors: New twists in the tale. Nat. Rev. Mol. Cell Biol. 4, 202–212 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Kim J.-J., Olson L. J., Dahms N. M., Carbohydrate recognition by the mannose-6-phosphate receptors. Curr. Opin. Struct. Biol. 19, 534–542 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergman D., Halje M., Nordin M., Engström W., Insulin-like growth factor 2 in development and disease: A mini-review. Gerontology 59, 240–249 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Fürstenberger G., Senn H. J., Insulin-like growth factors and cancer. Lancet Oncol. 3, 298–302 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Jadot M., Canfield W. M., Gregory W., Kornfeld S., Characterization of the signal for rapid internalization of the bovine mannose 6-phosphate/insulin-like growth factor-II receptor. J. Biol. Chem. 267, 11069–11077 (1992). [PubMed] [Google Scholar]
- 9.Johnson K. F., Chan W., Kornfeld S., Cation-dependent mannose 6-phosphate receptor contains two internalization signals in its cytoplasmic domain. Proc. Natl. Acad. Sci. U.S.A. 87, 10010–10014 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goda Y., Pfeffer S. R., Selective recycling of the mannose 6-phosphate/IGF-II receptor to the trans Golgi network in vitro. Cell 55, 309–320 (1988). [DOI] [PubMed] [Google Scholar]
- 11.Zavorka M. E., Connelly C. M., Grosely R., MacDonald R. G., Inhibition of insulin-like growth factor II (IGF-II)-dependent cell growth by multidentate pentamannosyl 6-phosphate-based ligands targeting the mannose 6-phosphate/IGF-II receptor. Oncotarget 7, 62386–62410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeflich A., Fettscher O., Lahm H., Blum W. F., Kolb H. J., Engelhardt D., Wolf E., Weber M. M., Overexpression of insulin-like growth factor-binding protein-2 results in increased tumorigenic potential in Y-1 adrenocortical tumor cells. Cancer Res. 60, 834–838 (2000). [PubMed] [Google Scholar]
- 13.Byrd J. C., Devi G. R., De Souza A. T., Jirtle R. L., MacDonald R. G., Disruption of ligand binding to the insulin-like growth factor II/mannose 6-phosphate receptor by cancer-associated missense mutations. J. Biol. Chem. 274, 24408–24416 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Gary-Bobo M., Nirde P., Jeanjean A., Morere A., Garcia M., Mannose 6-phosphate receptor targeting and its applications in human diseases. Curr. Med. Chem. 14, 2945–2953 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garmroudi F., MacDonald R. G., Localization of the insulin-like growth factor II (IGF-II) binding/cross-linking site of the IGF-II/mannose 6-phosphate receptor to extracellular repeats 10–11. J. Biol. Chem. 269, 26944–26952 (1994). [PubMed] [Google Scholar]
- 16.Schmidt B., Kiecke-Siemsen C., Waheed A., Braulke T., von Figura K., Localization of the insulin-like growth factor II binding site to amino acids 1508–1566 in repeat 11 of the mannose 6-phosphate/insulin-like growth factor II receptor. J. Biol. Chem. 270, 14975–14982 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Brown J., Delaine C., Zaccheo O. J., Siebold C., Gilbert R. J., van Boxel G., Denley A., Wallace J. C., Hassan A. B., Forbes B. E., Jones E. Y., Structure and functional analysis of the IGF-II/IGF2R interaction. EMBO J. 27, 265–276 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams C., Hoppe H.-J., Rezgui D., Strickland M., Forbes B. E., Grutzner F., Frago S., Ellis R. Z., Wattana-Amorn P., Prince S. N., Zaccheo O. J., Nolan C. M., Mungall A. J., Jones E. Y., Crump M. P., Hassan A. B., An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution. Science 338, 1209–1213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson L. J., Dahms N. M., Kim J.-J., The N-terminal carbohydrate recognition site of the cation-independent mannose 6-phosphate receptor. J. Biol. Chem. 279, 34000–34009 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Brown W. J., Farquhar M. G., The mannose-6-phosphate receptor for lysosomal enzymes is concentrated in cis Golgi cisternae. Cell 36, 295–307 (1984). [DOI] [PubMed] [Google Scholar]
- 21.Byrd J. C., Park J. H., Schaffer B. S., Garmroudi F., MacDonald R. G., Dimerization of the insulin-like growth factor II/mannose 6-phosphate receptor. J. Biol. Chem. 275, 18647–18656 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Williams C., Rezgui D., Prince S. N., Zaccheo O. J., Foulstone E. J., Forbes B. E., Norton R. S., Crosby J., Hassan A. B., Crump M. P., Rezgui D., Prince S. N., Zaccheo O. J., Foulstone E. J., Forbes B. E., Norton R. S., Crosby J., Hassan A. B., Crump M. P., Structural insights into the interaction of insulin-like growth factor 2 with IGF2R domain 11. Structure 15, 1065–1078 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Sakano K., Enjoh T., Numata F., Fujiwara H., Marumoto Y., Higashihashi N., Sato Y., Perdue J. F., Fujita-Yamaguchi Y., The design, expression, and characterization of human insulin-like growth factor II (IGF-II) mutants specific for either the IGF-II/cation-independent mannose 6-phosphate receptor or IGF-I receptor. J. Biol. Chem. 266, 20626–20635 (1991). [PubMed] [Google Scholar]
- 25.Roth B. V., Bürgisser D. M., Lüthi C., Humbel R. E., Mutants of human insulin-like growth factor II: Expression and characterization of analogs with a substitution of TYR27 and/or a deletion of residues 62–67. Biochem. Biophys. Res. Commun. 181, 907–914 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Olson L. J., Hindsgaul O., Dahms N. M., Kim J.-J., Structural insights into the mechanism of pH-dependent ligand binding and release by the cation-dependent mannose 6-phosphate receptor. J. Biol. Chem. 283, 10124–10134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braulke T., Gartung C., Hasilik A., von Figura K., Is movement of mannose 6-phosphate-specific receptor triggered by binding of lysosomal enzymes? J. Cell Biol. 104, 1735–1742 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiess W., Thomas C. L., Sklar M. M., Nissley S. P., β-Galactosidase decreases the binding affinity of the insulin-like-growth-factor-II/mannose-6-phosphate receptor for insulin-like-growth-factor II. Eur. J. Biochem. 190, 71–77 (1990). [DOI] [PubMed] [Google Scholar]
- 29.Kiess W., Thomas C. L., Greenstein L. A., Lee L., Sklar M. M., Rechler M. M., Sahagian G. G., Nissley S. P., Insulin-like growth factor-II (IGF-II) inhibits both the cellular uptake of β-galactosidase and the binding of β-galactosidase to purified IGF-II/mannose 6-phosphate receptor. J. Biol. Chem. 264, 4710–4714 (1989). [PubMed] [Google Scholar]
- 30.Kiess W., Blickenstaff G. D., Sklar M. M., Thomas C. L., Nissley S. P., Sahagian G. G., Biochemical evidence that the type II insulin-like growth factor receptor is identical to the cation-independent mannose 6-phosphate receptor. J. Biol. Chem. 263, 9339–9344 (1988). [PubMed] [Google Scholar]
- 31.York S. J., Arneson L. S., Gregory W. T., Dahms N. M., Kornfeld S., The rate of internalization of the mannose 6-phosphate/insulin-like growth factor II receptor is enhanced by multivalent ligand binding. J. Biol. Chem. 274, 1164–1171 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Grant B. D., Donaldson J. G., Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10, 597–608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J., Structure of the LDL receptor extracellular domain at endosomal pH. Science 298, 2353–2358 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Goldstein J. L., Brown M. S., The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 29, 431–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeanes A., Pittsley J. E., Watson P. R., Dimler R. J., Characterization and properties of the phosphomannan from Hansenula hostii NRRL Y-2448. Arch. Biochem. Biophys. 92, 343–350 (1961). [DOI] [PubMed] [Google Scholar]
- 36.Murray G. J., Neville D. M. Jr., Mannose 6-phosphate receptor-mediated uptake of modified low density lipoprotein results in down regulation of hydroxymethylglutaryl-CoA reductase in normal and familial hypercholesterolemic fibroblasts. J. Biol. Chem. 255, 11942–11948 (1980). [PubMed] [Google Scholar]
- 37.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown A., Long F., Nicholls R. A., Toots J., Emsley P., Murshudov G., Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/7/eaaz1466/DC1
Fig. S1. Sequence alignment of different domains in IGF2R.
Fig. S2. Biochemical characterization of apo state IGF2R and IGF2-bound IGF2R.
Fig. S3. Data processing and model quality assessment of apo state IGF2R.
Fig. S4. Cryo-EM map of apo state IGF2R.
Fig. S5. Structural comparison for seven subgroups of apo state IGF2R in two different views.
Fig. S6. Data processing for ligand-free IGF2R.
Fig. S7. Data processing and model quality assessment of IGF2-bound IGF2R.
Fig. S8. Cryo-EM map of IGF2-bound IGF2R.
Fig. S9. Structural comparison of CD-MPR and domains of IGF2R for Man6P.
Table S1. Cryo-EM data collection, refinement, and validation statistics.


