In the Cairns-Foster adaptive mutation system, lac mutant cells are plated on lactose medium where 50 revertant colonies accumulate over 5 days above a non-growing lawn. A new model attributes this behavior to selective...
Keywords: adaptive mutation, selection, mutagenesis, DinB, copy number variation, selective gene amplification, DNA repair, break-induced replication, recombination-dependent replication, rolling-circle replication, bacterial mating, plasmid transfer
Abstract
The Escherichia coli system of Cairns and Foster employs a lac frameshift mutation that reverts rarely (10−9/cell/division) during unrestricted growth. However, when 108 cells are plated on lactose medium, the nongrowing lawn produces ∼50 Lac+ revertant colonies that accumulate linearly with time over 5 days. Revertants carry very few associated mutations. This behavior has been attributed to an evolved mechanism (“adaptive mutation” or “stress-induced mutagenesis”) that responds to starvation by preferentially creating mutations that improve growth. We describe an alternative model, “selective inbreeding,” in which natural selection acts during intercellular transfer of the plasmid that carries the mutant lac allele and the dinB gene for an error-prone polymerase. Revertant genome sequences show that the plasmid is more intensely mutagenized than the chromosome. Revertants vary widely in their number of plasmid and chromosomal mutations. Plasmid mutations are distributed evenly, but chromosomal mutations are focused near the replication origin. Rare, heavily mutagenized, revertants have acquired a plasmid tra mutation that eliminates conjugation ability. These findings support the new model, in which revertants are initiated by rare pre-existing cells (105) with many copies of the F’lac plasmid. These cells divide under selection, producing daughters that mate. Recombination between donor and recipient plasmids initiates rolling-circle plasmid over-replication, causing a mutagenic elevation of DinB level. A lac+ reversion event starts chromosome replication and mutagenesis by accumulated DinB. After reversion, plasmid transfer moves the revertant lac+ allele into an unmutagenized cell, and away from associated mutations. Thus, natural selection explains why mutagenesis appears stress-induced and directed.
IN the Cairns-Foster system, a population of mutant cells incapable of utilizing lactose is plated on minimal lactose medium. Over the course of 5 days, ∼50 Lac+ revertant colonies accumulate linearly above the nongrowing lawn population. Most revertants show few associated unselected mutations. This phenomenon has raised two questions. First, “How do new mutations form in a nongrowing population?” Second, “How are mutations directed preferentially to lac and away from the rest of the genome?” A controversy has surrounded attempts to explain this system. Supporters of a stress-induced mutagenesis model propose a mechanism that has evolved to facilitate adaptation (Foster 2007; Galhardo et al. 2007). This mechanism, called adaptive mutation, is said to sense the problem and create mutations that restore growth. Mutations are attributed to an error-prone polymerase (polIV, also called DinB), which is regulated by both the global DNA repair system (SOS) and the global system for inducing genes in stationary phase (RpoS) (Wagner et al. 1999; Layton and Foster 2003).
We have proposed that natural selection acts on pre-existing cells with a lac duplication, driving further amplification. Ribosomal frameshifting provides the mutant allele with a low level of function that can support division if the gene is amplified sufficiently. Mutagenesis results when the lac amplification includes dinB, which encodes an error-prone polymerase, and happens to be located near lac on the F’lac plasmid (Roth et al. 2006; Maisnier-Patin and Roth 2015, 2016). The normal role of DinB is to bypass bulky lesions in the template strand by translesion DNA synthesis (Fuchs and Fujii 2013). When overproduced, DinB makes frequent mistakes, including many frameshift mutations, during replication of undamaged DNA (Wagner et al. 1999; Wagner and Nohmi 2000; Kim et al. 2001). Our initial model proposed that growth improves under selection due to amplification of a region including the lac allele and the nearby dinB gene. This amplification mutagenizes the whole genome. This model predicted that revertant colonies should accumulate exponentially, and that reversion should require dinB near lac. It did not explain the apparent direction of mutations to sites that improve growth. This model did not explain the phenomenology.
Here, we describe a new model that explains the observed linear accumulation of revertants and the fact that dinB can be anywhere on the plasmid (Yamayoshi et al. 2018). In the new model, conjugative plasmid transfer underlies the reversion process. The lac function improves due to over-replication and mutagenesis of the plasmid during conjugative transfer between nondividing cells. Selection also favors transfer of the revertant lac+ allele away from deleterious associated mutations, giving the appearance of directed mutation. This selective inbreeding model is described here and tested by analysis of full genome sequences of 59 revertant strains.
Background Details of the Cairns-Foster Parent Strain and the Kinetics of Reversion
The parent cell genotype
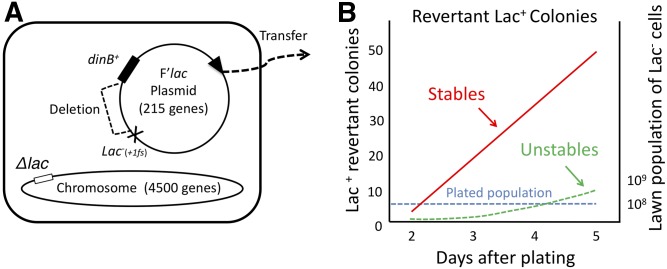
Parent cells have a deletion of the chromosomal lac region and carry a conjugative F’lac plasmid with a hybrid lacI-lacZ fusion gene expressed from a constitutive promoter (iQ) (see Figure 1A). Growth on lactose is blocked by a +1 frameshift in the lacI portion of the hybrid gene. This mutation prevents growth on lactose by reducing the β-galactosidase to ∼2% of the normal parental value. Mutations that correct the reading frame of the fusion gene permit full expression of the LacZ portion of the gene, and allow a single plasmid copy to support growth on lactose. The parent plasmid used here carries a deletion that places the lac operon close (within 200 bp) to the dinB+ gene (Yamayoshi et al. 2018), so that any tandem lac duplication is likely to include the dinB+ gene (see Figure 1A). The residual function of a single copy of the mutant lac region is insufficient to support growth, but growth is allowed if that region is sufficiently amplified by either tandem repeats or increased plasmid copy number.
Figure 1.
Cairns-Foster tester strain and its reversion behavior. (A) Genotype of the E. coli tester strain (TT27001). The lac region is deleted from the chromosome and is carried by a conjugative F’lac plasmid, which is able to transfer from one cell to another. Most tester strains have 1–2 plasmid copies, but 105 of the plated 108 cells have >10 plasmid copies. These cells arise before plating, and initiate all of the revertant colonies that develop under selection (Sano et al. 2014). (B) Course of a reversion experiment, during which the plated 108 lac mutant cells do not grow, but give rise to ∼50 stable Lac+ colonies per plate, which accumulate linearly over the course of 5 days, and are derived from the 105 initiator cells. Of the revertant colonies appearing on day 5, 90% are stably Lac+, and carry a point mutation that corrects the mutant lac allele, while 10% are unstably Lac+, and owe their growth to a tandem amplification of the mutant lac allele, which has low residual function. The dashed blue line indicates the lawn population of 108 plated cells that do not divide under selection.
Time course of the experiment
The tester strain is pregrown on glycerol, and 108 washed cells are plated on lactose medium with 109 scavengers—isogenic cells whose F’lac plasmid carries a deletion of the entire lac operon. Scavenger cells prevent residual growth of testers by consuming any usable carbon sources that might contaminate the medium, or be released by tester cells or revertant colonies. Most importantly, scavenger cells compete with testers during selection. The residual lac activity of testers splits lactose to glucose and galactose. Testers use glucose preferentially and excrete galactose, which is either reassimilated or consumed by the scavenger cells. By competing for galactose, scavenger cells reduce tester growth and the number of revertants that appear under selection (Foster 1994; Andersson et al. 1998; Maisnier-Patin and Roth 2018).
While tester cells cannot divide under selection, the residual lactose metabolism supplied by ribosomal frameshifting of the mutant allele can support conjugation, plasmid over-replication, and formation of lac+ revertant alleles. Lac+ revertant colonies appear and accumulate over time. These revertant colonies fall into stable and unstable Lac+ classes. Stable Lac+ revertants accumulate linearly with time, and account for 90% of total revertants at day 5. Their phenotype is due to a frameshift mutation that restores full activity to the mutant lac allele. The remaining 10% of revertants are unstably Lac+ and owe their growth to a tandem amplification of the mutant lac region within the plasmid (Kugelberg et al. 2006). These unstable revertant colonies accumulate at an accelerating rate and have 10–100 copies of the lac region within a single copy of the F’lac plasmid (see Figure 1B).
The behavior of this system makes it appear as if starvation induces cells to mutagenize their own nonreplicating genome. Based on this behavior, it was suggested that cells might possess an evolved mechanism that senses stress and generally mutagenizes a nonreplicating genome (Cairns et al. 1988; Cairns and Foster 1991; Foster 2007; Galhardo et al. 2007). However, revertants acquire very few associated nonselected mutations, so it appears that mutagenesis is somehow directed preferentially to sites that restore ability to grow on lactose (Foster and Cairns 1992). We have previously favored an alternative idea that revertants are initiated by pre-existing cells with multiple copies of the mutant lac allele. Our initial model suggested that these cells amplify their mutant lac allele and grow slowly under selection until reversion occurs (Hendrickson et al. 2002). Here, we describe and provide evidence to support a replacement for both original models.
The controversy
A huge body of data has accumulated describing this system. The two sides of the controversy agree substantially about the dependencies of reversion, but have interpreted results as support for either stress-induced mutagenesis or response to selection. These results have been reviewed repeatedly (Roth et al. 2006; Foster 2007; Galhardo et al. 2007) but have not resolved the conflict or eliminated either model.
Points of agreement:
Reversion requires low-level residual function of the mutant lac operon, but does not involve growth of the plated mutant lawn under selection (Foster 1994).
Both stable and unstable Lac+ revertant colonies arise only if the mutant lac allele is located on a conjugative F’lac plasmid capable of transfer (Foster and Trimarchi 1995a; Radicella et al. 1995).
Accumulation of both stable and unstable revertants requires RecA-dependent homologous recombination, by the RecABCD pathway (Cairns and Foster 1991; Harris et al. 1994).
Formation of stable revertants requires the mutagenic repair polymerase DinB, which is encoded on the plasmid and expressed under control of the global SOS and RpoS regulatory systems (McKenzie et al. 2000; Lombardo et al. 2004; Yamayoshi et al. 2018).
Formation of unstable revertants does not depend on DinB or its regulatory systems (McKenzie et al. 2001; Yamayoshi et al. 2018).
Points of disagreement:
Recent findings addressed these issues and led to the new model.
Initially it was claimed that reversion was induced by starvation of cells on the selection plate (Cairns et al. 1988; Cairns and Foster 1991). More recent evidence showed that all revertants are initiated by rare cells that arise prior to selection and carry an abnormally high number of plasmid copies (Sano et al. 2014).
It was originally proposed that the dinB+ gene, required for stable reversion, could be located either on the F’lac plasmid or in the chromosome (McKenzie et al. 2001). More recent evidence demonstrates that reversion under selection requires a dinB+ allele that can be located anywhere on the same plasmid as the mutant lac allele (Yamayoshi et al. 2018).
Early in the controversy, it was suggested that reversion required plasmid transfer (Galitski and Roth 1995; Radicella et al. 1995; Peters et al. 1996; Godoy and Fox 2000). Later it was claimed that the F conjugation (tra) machinery was needed only to make single-strand DNA nicks at the plasmid transfer replication origin (OriT). These nicks become double-strand breaks when encountered by the replication fork, and were proposed to be repaired by a mutagenic recombination-dependent repair pathway (Foster and Trimarchi 1995a,b). The most recent evidence suggests that complete plasmid transfer is essential for reversion, but occurs only between identical daughters of rare single plated cells with multiple plasmid copies (Maisnier-Patin and Roth 2018).
Selective inbreeding—A model for reversion under selection in the Cairns-Foster system
The three newest findings, described above, suggested a new model in which selection drives a multi-step reversion process. (1) Initiator cells with high plasmid copy number arise before selection. (2) These cells can divide once under selection and produce identical daughter cells that mate incestuously. (3) Plasmid-plasmid recombination initiates rolling circle plasmid replication. This amplifies the lac and dinB genes during plasmid transfer and mutagenizes the plasmid. (4) Reversion to Lac+ restores a fully Lac+ phenotype, including exponential cell growth and mutagenesis of the chromosome by accumulated DinB protein. (5) Variably mutagenized revertant plasmids and chromosomes segregate at random. (6) Conjugative backcrosses move revertant plasmids into less-mutagenized hosts and separate the selected revertant lac+ allele from deleterious associated mutations. These stages are detailed below.
Generation of variability before selection:
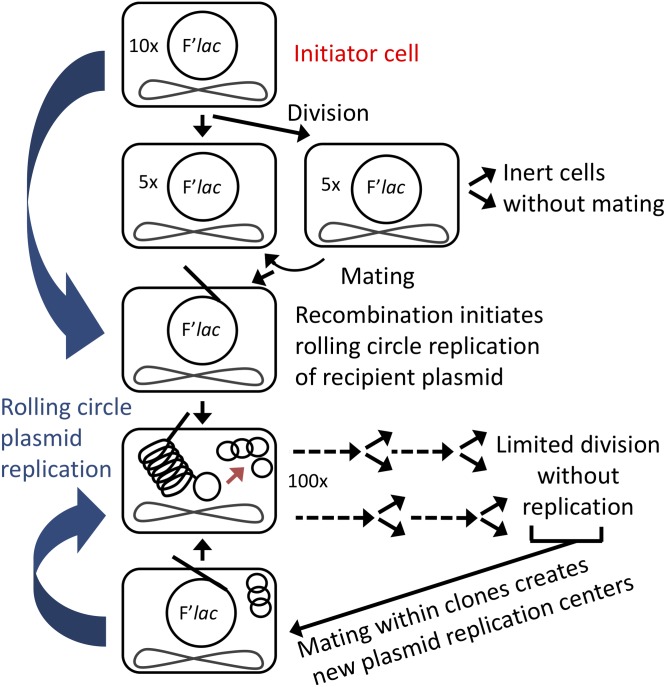
Each revertant is derived from an initiator cell that arises during nonselective pregrowth and thus cannot be stress-induced. These initiator cells are estimated to have ∼10 copies of the F’lac plasmid, which contains both the mutant lac allele and a functional dinB+ gene (Sano et al. 2014). An estimated 105 such initiator cells are included with the 108 cells plated on lactose medium. Because of their elevated plasmid copy number, initiator cells have a sufficient level of lac activity to divide once under selection, producing identical daughter cells (each with multiple copies of F’lac). Mating between them is possible because they both possess a somewhat elevated plasmid copy number and are located close together on the selection plate.
Stage one—amplification and mutagenesis of the plasmid:
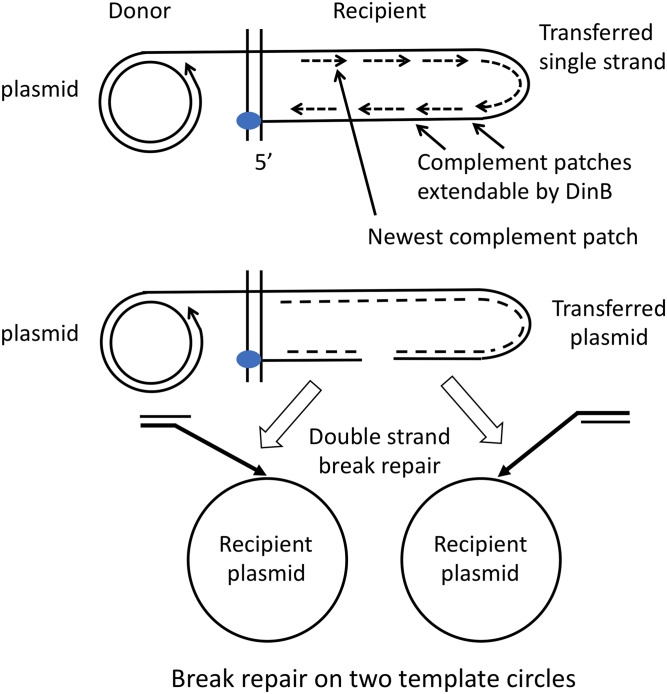
One initiator daughter transfers a single plasmid strand into an identical daughter cell, where a complementary strand is synthesized using single-strand initiation sites located at intervals around the plasmid (Masai and Arai 1997). Any break in the duplex provides ends that are subject to RecA-dependent DNA recombinational repair (Figure 2). The presence of multiple recipient plasmid copies enhances the likelihood that the two broken ends will initiate unopposed rolling circle replication forks on different plasmids rather than being repaired on a single template. Replication forks initiated from recombination intermediates are mutagenic (Kuzminov 1995; Malkova and Haber 2012). High levels of DinB can displace the replicative Pol III polymerase and cause intense mutagenesis during slow replication of the strand displaced from the recipient plasmid.
Figure 2.
Plasmid replication during transfer under selection. The transferred single strand enters 5′ end first, and a complementary strand is synthesized from multiple primer sequences that form gapped duplex intermediates. A double-strand break creates ends that can recombine with recipient plasmids by homologous recombination and initiate recombination-dependent replication (Kuzminov and Stahl 1999; Malkova and Ira 2013).
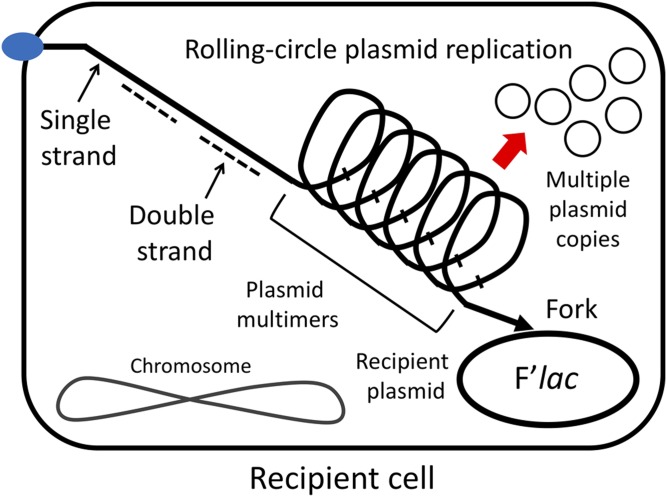
Rolling-circle replication increases plasmid copy number in cells that divide very little. Plasmid circles are produced from the concatemeric replication product by recombination, but their subsequent independent vegetative replication is blocked by plasmid copy control mechanisms (Par) (Figure 3). Increased plasmid copy number provides energy (lac copy number) and amplifies the level of DinB protein to stimulate mutagenesis. Rare cell divisions allowed by lac amplification produce daughter cells with reduced plasmid number that are unable to grow or revert. Plasmid replication continues only in mating pairs and lost pairs are replaced by occasional new matings within the clone (Figure 4). Repeated mating intensifies mutagenesis.
Figure 3.
Concatemers produce independent plasmid circles. The concatemeric product of rolling circle replication breaks down to plasmid circles whose independent replication is prevented by the plasmid copy number control system (Par).
Figure 4.
Plasmid amplification without chromosome replication before reversion. Initiator cells can divide once under selection and daughters can mate. Rolling circle plasmid over-replication amplifies both lac and dinB. Free plasmids generated from the concatemeric replication product cannot replicate due to plasmid copy number control. Divisions reduce plasmid number, and produce cells that grow very little. Only mating pairs replicate and mutagenize the plasmid. Lost mating pairs can be replaced by new mating events.
In stage one, mutagenesis by DinB is focused on the replicating F’lac plasmid, since chromosomes replicate very little. In addition, chromosome replication may be inhibited by accumulated DinB protein (Uchida et al. 2008; Indiani et al. 2009; Mori et al. 2012). The number of replicating plasmids per clone, and the basis for reversion, remains constant over time, explaining why revertant colony number accumulates linearly.
There are previous reports that conjugation is inherently mutagenic (Kunz and Glickman 1983; Christensen et al. 1985) and that SOS induction leads to mutagenic break repair following transfer (Lloyd and Buckman 1995; Matic et al. 1995). The model suggests that conjugation becomes intensely mutagenic when high levels of DinB contribute both to synthesis of a complement for the transferred strand and to rolling circle plasmid replication.
Reversion stage two—reversion to lac+ initiates chromosome replication and mutagenesis:
A reversion event produces a lac+ allele that supplies energy and reinstates exponential cell division. Initial chromosome replication is made highly mutagenic by the DinB protein accumulated during stage one. DinB also slows replication forks, which increases chromosome replication complexity (CRC) and stimulates replication initiation at the origin (Kuzminov 2016).
Reversion stage three—plasmid segregation re-assorts mutagenized chromosomes and plasmids:
As cells divide, the plasmid copy number drops. Chromosomal mutagenesis drops as division dilutes DinB protein and lowers plasmid copy number. Once the copy number reaches one to two, plasmid replication resumes, and cell division generates random combinations of plasmid and chromosome that will persist as the clone expands. Many revertant cells will show impaired growth due to deleterious chromosomal mutations. Selection favors cells with the least deleterious combinations of mutations.
Reversion stage four—backcross of revertant plasmid with nonmutagenized cells:
Selection against deleterious mutations favors secondary backcrosses that move a revertant plasmid into cells in the same clone that have experienced less DinB mutagenesis. Recombination can separate the selected lac+ allele from associated plasmid mutations, yielding cells in which mutagenesis appears to have been directed to adaptive targets. Rare revertants with a lac+ allele may acquire a plasmid tra mutation and lose ability to transfer. These revertant plasmids cannot benefit from this backcross, and retain a lac+ allele associated with a heavily mutagenized chromosome and plasmid.
Development of unstable Lac+ revertants:
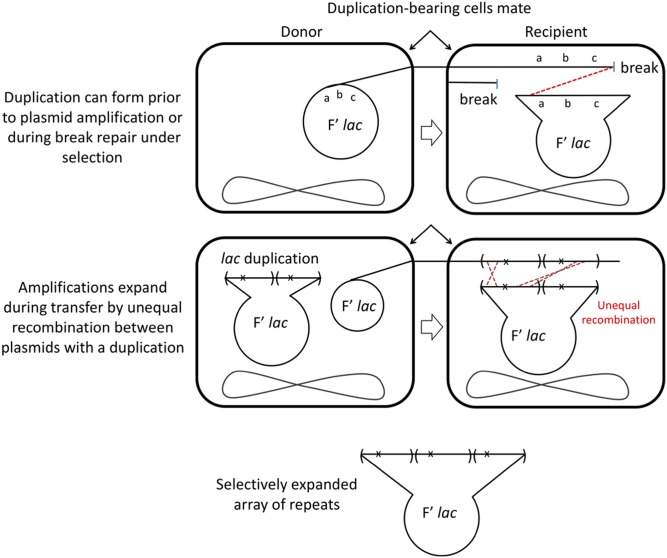
Most unstable revertants are initiated by pre-existing cells with an elevated number of plasmids, each with an internal duplication of the lac operon (Figure 5). Duplications can also form during break repair following mating between daughters of a standard initiator cell. The shift to break-induced replication (BIR) favors template switching, which can result in complex rearrangements (McVey et al. 2016; Sakofsky and Malkova 2017). Evidence for such rearrangements was seen previously in the Cairns-Foster system in that some amplifications formed under selection are derived from tandem inversion duplications (TIDs) (Hastings et al. 2009; Kugelberg et al. 2010). Mating between duplication-bearing cells can stimulate unequal recombination events that amplify repeat copy number. Repeated recombination increases the lac copy number by expanding the lac array under selection. Array expansion requires both mating and recombination, but does not require DinB or mutagenesis, since growth depends entirely on the elevated number of mutant lac alleles.
Figure 5.
Formation of amplifications in unstable revertants Most unstable revertants are initiated by pre-existing cells that carry multiple copies of a plasmid with a tandem duplication of the mutant lac region. Other duplications may form under selection during transfer between standard initiator daughter cells. Transfer of a duplication-bearing plasmid provides conditions for unequal recombination that can expand the tandem array of lac copies.
The upward inflection in accumulation of unstable colonies (Figure 1B) occurs because the expanding tandem array is heritable and allows exponential cell division. Chromosomes in an unstable revertant clone are mutagenized very little because there is less mating and less SOS induction. Each of the cell divisions that accompany expansion of the dinB array causes a twofold dilution of DinB protein and plasmid copy number. Growth reduces the basal expression of the dinB allele, which is induced by RpoS during stationary phase. Each cell division reduces the DinB level caused by dinB gene amplification. When lac amplification is sufficient (perhaps 50 copies) to support full growth, plasmid copy number drops to one to two, and plasmid replication resumes. Tandem amplifications can provide a heritable Lac+ phenotype that is subject to occasional loss of copy number, which explains their instability.
The selective inbreeding model predicts properties of revertant genome sequences
The model described above makes predictions regarding the nature and distribution of mutations in revertant genomes.
Since the model attributes mutagenesis to dinB amplification and accumulation of DinB polymerase, a trivial prediction is that both the selected lac+ mutations and associated unselected mutations should be of types typical for the error-prone DinB polymerase.
Mutagenesis is focused on the plasmid, which over-replicates in nondividing cells during stage one.
Revertants should vary in the distribution of associated mutations between plasmid and chromosome because mutagenized plasmids and chromosomes segregate at random during stage three.
Chromosomal mutations cluster near the origin because inhibition of replication fork by DinB stimulates repeated initiation.
Most stable revertants will have very few associated mutations because backcrosses frequently separate the revertant lac+ allele from accompanying lesions in both the chromosome and the plasmid (stage four).
Rare revertants that acquire a plasmid tra mutation after reversion will show more intense mutagenesis of plasmid and chromosome, because they cannot take part in backcrosses and therefore must maintain their revertant lac+ allele in a heavily mutagenized background.
Unstable revertants are less mutagenized than stable revertants, since heritable lac tandem amplifications support slow but steady cell division. This growth minimizes basal dinB expression, dilutes DinB and reduces plasmid copy number, thereby minimizing mutagenesis of both chromosomes and plasmids.
These predictions are tested here by analysis of whole genome sequences of 59 Lac+ revertants.
Materials and Methods
Bacterial strains
All strains used for reversion are derivatives of Escherichia coli K-12 strain P9OC (Coulondre and Miller 1977). The tester strain, TT27001, carries a rifampicin resistance mutation and an F’lac derivative with a 16 kb deletion, which places the dinB+ promoter ∼180 bp from the divergent lac promoter (Yamayoshi et al. 2018). The scavenger strain FC29 (TR7177) is rifampicin sensitive, and contains an F’lac plasmid with a deletion of the lac region (Cairns and Foster 1991). The F−, pro−, streptomycin-resistant E. coli strain (TT27036) (Maisnier-Patin and Roth 2018) was used as a recipient in mating assays.
Media
Bacterial strains were routinely grown in lysogeny broth (LB). Antibiotics were added at the following concentrations: streptomycin, 200 µg/ml; nalidixic acid 50 µg/ml; rifampicin, 100 µg/ml. Minimal media plates were no-citrate Vogel-Bonner E medium (NCE) with appropriate additions (Berkowitz et al. 1968). The rich medium used to diagnose stable and unstable revertants was agar containing Difco nutrient broth (NB) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; 40 μg/ml).
Lac reversion assay and colony isolation
The tester (TT27001) and scavenger (TR7177) strains were grown overnight in minimum NCE medium supplemented with MgSO4 (2 mM), thiamine (50 µM), and glycerol (0.1%). Before use, each culture was diluted 100-fold into the same minimal NCE medium and grown again to saturation. Cells were then sedimented and resuspended in NCE salts. Selection plates contained NCE, lactose (0.1%), MgSO4 (2 mM), thiamine (50 µM), and X-gal (25 µg/ml) solidified by 1.5% agar. Each plate was prepared by first spreading 109 scavenger cells and incubating 24 hr at 37° to allow consumption of any contaminating carbon sources. Next the prescavenged selection plates were plated with ∼108 washed tester cells. Selection plates were incubated at 37° for 5 days. The number of revertant colonies was scored daily. Colonies appearing on day 5 were plugged, and cells were suspended in 1 ml NCE and frozen at −80° with 10% DMSO.
Determination of revertant phenotype stability
For each revertant, cells from the frozen plugs were diluted in NCE and plated for single colonies on NB plates containing rifampicin and X-gal. Revertants that are stably Lac+ grew as solid blue colonies. Unstable revertants formed sectored blue and white “star” colonies. Once the stability of each plugged colony was determined, 31 stable colonies and 19 unstable colonies were chosen randomly for whole genome sequencing. One stable colony (S8) later proved to be unstable and was renamed U0. Thus 30 random stable and 20 unstable revertants were sequenced. During this process three stable revertants were found to form tiny colonies on rich medium, and were added to the sequenced list.
Genomic DNA preparation and sequencing
Revertants from the frozen plugs were resuspended in NCE and plated onto minimal lactose plates containing rifampicin to prevent growth of scavenger cells. A single colony from each plug was diluted and grown to saturation in liquid minimal lactose medium. DNA was prepared from these cells using Wizard Genomic DNA kits (Promega) per manufacturer’s protocol for gram-negative bacteria. Bar-coded libraries were prepared using Illumina kits (standard PCR-dependent protocol), and paired-read sequencing done on Illumina MiSeq machines at the iBEST DNA sequencing facility (University of Idaho, Moscow, ID).
Processing of sequence data
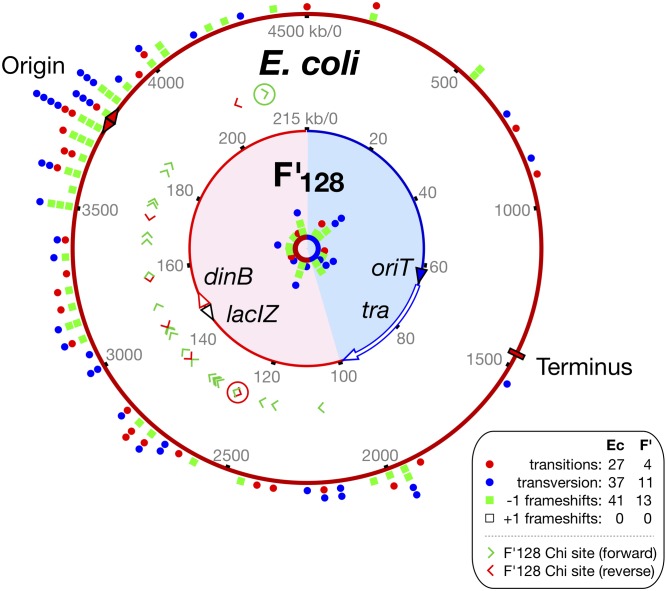
Raw data were processed by us using BASH, AWK, and Python scripts (E.C.K.) and the following bioinformatics tools: bwa (Li and Durbin 2009), samtools (Li 2011), varscan (Koboldt et al. 2012), and makeblastdb/BLASTplus (Camacho et al. 2009). Circular maps were made using BRIG (Alikhan et al. 2011), with subsequent enhancement by Intaglio (https://www.purgatorydesign.com/Intaglio/). The chromosomal reference sequence was based on GenBank UFC4000096, and the plasmid on GenBank AP001918 with insert derived from UFC4000096. Other modifications were made to reflect engineered variations and mutations known to be present in the originating laboratory wild type. Variants were called when 80% or more aligned reads presented the same modification at an identical reference address.
Mating assays
To test F’lac plasmid transfer ability, revertant cells were patched on LB plates and grown overnight at 37°. These patches were then printed to LB plates spread with recipient strain TT27036, a F−, pro−, streptomycin-resistant E. coli strain to allow mating. The mating plates were incubated overnight to allow transfer, and then printed to selective minimal NCE plates supplemented with glucose (0.2%), thiamine (50 µM), MgSO4 (2 mM), and streptomycin (200 µg/ml), selecting for Pro+ StrR cells that had received a plasmid transfer. Mating was scored the next day.
Scoring mutator phenotypes
To test their mutator phenotype, single revertant colonies were used to inoculate 4 ml LB cultures, and incubated at 37° for 24 hr. Cultures were spread on nalidixic acid to score frequency of NalR mutants. Mutant frequencies were determined as the ratio of the number of nalidixic acid-resistant colony-forming units (cfu) per milliliter to the total number viable cells plated.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains used in this study are available upon request. DNA sequencing data are available at NCBI under BioProject accession number PRJNA590198. Supplemental Material, Table S1 available at figshare contains all mutations of the Lac+ revertants. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10848680.
Results
The reversion phenomenon
In the Cairns-Foster system, ∼50 revertant colonies appear above a lawn of 108 nongrowing cells over the course of 5 days. The parental lac mutation reverts at 10−9/cell/division during nonselective growth in liquid medium (Foster and Trimarchi 1994). Consequently, very few reversion events are expected to occur in the plated noninitiator population. If one uses the 105 plated initiator cells as the basis of reversion, and assumes one division, selection appears to increase lac reversion rate from 1/109 cells/division to ∼50/105 cells/division, a five order-of-magnitude increase. It has been hard to explain this remarkable increase, which occurs with very few cell divisions, and very little obvious general mutagenesis of the plated lawn population. The new model attributes the enhanced reversion rate to selection-driven, highly mutagenic localized over-replication of lac within each of 105 initiator clones with very few cell divisions.
The nature of stable lac revertants is consistent with formation by DinB
The parent lac mutation is a +1 frameshift mutation in a run of G/C base pairs within the lacI portion of the chimeric lacI-lacZ gene. Mutations that restore full LacZ function in a stable Lac+ revertant must provide a −1 frameshift that compensates for the parent mutation, and allows transcription and translation to extend into the distal lacZ portion of the fusion gene. Several lac revertants have been characterized previously (Foster and Trimarchi 1994; Rosenberg et al. 1994). Table 1 describes the lac mutations in 39 stable revertants. Most revertants affect the run of G/C pairs at the site of the parental +1 mutation, which is shown at the top of Table 1.
Table 1. Stable Lac+ revertants formed under selection (39 mutations).
| Number of mutations | Mutation that corrects lacIZ reading frame | Site of mutation in lacIZ gene | Sequence changes in the lacI region and revertant numbers |
|---|---|---|---|
| 25 | Single base –1 | In original mutant run | gagacggggca - > gagacgggca S1,S4,S6,S7,S9,S10,S11,S14,S16,S19,S20,S21,S23,S25,S26,S27,S28,S29,S30,S32,S33,A58,A60,A61,A62 |
| 7 | Single base –1 | Sites near original mutant run | TG->T (A57a, A59a), GC->G(S2, S31), AG->A(S24a), CA->(S13), GT->G(S22a) |
| 3 | Multi-base insertions | Sites near original mutant run | +2 bp C->CGT (S34), +5 bp T->TTCACC (S5), +8 bp T->TTTCACCAG (S15) |
| 4 | Multi-base deletions | Sites near original mutant run | −7 bp(S12), -10 bp(S3), –112 bp (S17, S18) removes original site; between repeated GGTTTGC |
Revertants that arose by losing a single base from a monotonous run of three or more bases.
The nature of these lac+ mutations is consistent with known effects of error-prone DinB polymerase, as described above. This polymerase is known to make preferentially −1 frameshift mutations, most frequently in monotonous runs, and often affecting bases following a G/C base pair (Wagner and Nohmi 2000). The −1 frameshift mutations at the original mutant site all remove one base from a run of four G/C pairs, and many of the seven nearby −1 frameshifts (marked by an asterisk) arose in runs of three or more bases. Although the nature of these mutations is consistent with DinB activity, the Lac+ selection constrains the types of lactose mutations recovered. Better evidence for DinB action under selection will be discussed later for the associated unselected mutation whose nature is unconstrained.
Stable revertants fall into multiple classes based on their number of unselected mutations
Random revertant Lac+ colonies were picked on day 5 and single-colony isolated on minimal lactose medium and on NB plates with X-gal to allow easy discrimination of stable (solid blue) and unstable (blue-white sectored) revertant colonies. A total of 30 random stable revertant colonies were picked from lactose plates, grown in liquid lactose medium and genome sequenced. Three additional stable revertants grew very slowly on rich NB plates and were also sequenced. Stable revertants were designated S1–S34. (Revertant S8 proved later to be unstable and was redesignated U0.) The 20 unstable revertants were designated U0–U19 and will be described later. The 33 stable revertants are classified based on their number of unselected mutations, as seen in Table 2, which does not include their selected lac mutations (described above). All sequenced genomes characterized here are described in Table S1.
Table 2. Unselected mutations found in stable revertants (Total of 33).
| Revertant class | Revertant numbera | Chromosomal mutations in each strain | F’lac Mutations in each strain | Total mutations/genome | ||||
|---|---|---|---|---|---|---|---|---|
| SNPs | Indels | Total | SNPs | Indels | Total | |||
| Light mutagenesis (23) | S11, S17 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| S21 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | |
| S2, S3, S5, S27, S31, S32 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | |
| S4, S7, S10, S12, S13, S14, S15, S16, S18, S19, S20, S24. S25, S34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Moderate mutagenesis (6) | S1 | 0 | 0 | 0 | 2 | 2 | 4 | 4 |
| S29 | 3 | 0 | 3 | 1 | 0 | 1 | 4 | |
| S33 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | |
| S28 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | |
| S9 | 1 | 0 | 1 | 2 | 1 | 3 | 4 | |
| S22 | 0 | 0 | 0 | 4 | 1 | 5 | 5 | |
| Intense mutagenesisb (1) | S6 | 15 | 9 | 24 | 6 | 6 | 12 | 36 |
| Slow-growing revertants (3) | S23 | 17 | 3 | 20 | 0 | 1 | 1 | 21 |
| S26 | 1 | 0 | 1 | 9 | 0 | 9 | 10 | |
| S30 | 64 | 41 | 105 | 15 | 13 | 28 | 133 | |
One stable revertant S8 was later found to be unstable and was redesignated U0, which is described below. (The S8 name was dropped.)
One of 30 random revertants (S6) was found to be heavily mutagenized and to have acquired a plasmid tra indel mutation after reverting to lac+. Three other revertants showed slow growth and also were heavily mutagenized. Two of these revertants (S23 and S30) acquired a plasmid tra mutation and lost conjugation ability. Revertant S26 has no tra+ mutation but carries a mutagenized revertant plasmid integrated into a chromosome with only one mutation (see text).
The majority of randomly chosen stable revertants (23/30) fall into one class (designated light mutagenesis), with a total of 0–2 unselected mutations per genome (Table 2.) Of these 30 random revertants, 6 were classified as moderately mutagenized with two to five mutations—an average of one mutation per chromosome and two in their F’lac plasmid. Only 1 of the initial 30 stable revertants (S6) showed more intense general mutagenesis.
Also in Table 2 are three additional stable revertants (of ∼200 screened) that were sequenced because they grew poorly on rich medium. One slow-growing revertant (S23) showed heavy chromosome mutagenesis, and another (S30) showed incredibly intense mutagenesis of both chromosome and plasmid. Both of these strains have plasmid tra mutations that eliminate conjugation ability. The third slow-growing revertant (S26) showed slightly elevated mutagenesis of its plasmid, but had no tra mutation. Its mutagenized revertant plasmid was inserted into a chromosome with only one mutation by a transposition event mediated by the plasmid insertion sequence IS3B.
The uneven distribution of associated mutations among revertant strains was first noted by Rosche and Foster (1999), and is analyzed in more detail here using complete genome sequences. The variable distribution of mutations between chromosome and plasmid in different revertants is predicted by the model described here. The correlation between heavy mutagenesis and acquisition of a transfer defect will be pursued later.
Unstable revertant classes
Table 3 describes the distribution of associated mutations in unstable revertants. All were rather lightly mutagenized. Of the 20 unstable revertants, 18 had only one mutation in either their chromosome or plasmid. Since the dinB+ gene is near lac on the plasmid, we initially expected it to be coamplified with lac under selection and cause some mutagenesis. We think mutagenesis was minimal for several reasons. Since tandem amplifications are heritable, increased lac copy number is expected to permit slow exponential growth early in the history of the clone. Each division reduces plasmid copy number and dilutes DinB protein level. This growth minimizes the RpoS-dependent basal expression of the dinB gene, and, therefore, reduces the mutagenesis caused by its amplification. By minimizing mutagenesis, growth also appears to minimize the tendency to focus mutagenesis on the F’lac plasmid. Mutagenesis is more focused in stable revertants, whose precursor cells divide rarely and experience more extensive plasmid overamplification. Sequencing shows that unstable revertants have ∼50 plasmid copies of lac following overnight growth in liquid lactose medium. We presume that the initial slow growth of unstable revertant precursors under selection occurs with a considerably lower lac copy number.
Table 3. Mutations in Unstable Lac+ revertants.
| Revertant strains (20 total) | Chromosomal mutations in each strain | F’lac plasmid mutations in each strain | |||||
|---|---|---|---|---|---|---|---|
| Class | Strain ID | SNPs | Indels | Total | SNPs | Indels | Total |
| Lightly mutagenized (18) | U0, U6, U9, U11, U12, U16, U19 | 0 | 0 | 0 | 0 | 0 | 0 |
| U1, U8, U14 | 0 | 0 | 0 | 1 | 0 | 1 | |
| U3, U5, U7, U10, U15, U18 | 1 | 0 | 1 | 0 | 0 | 0 | |
| U17 | 0 | 0 | 0 | 0 | 1 | 1 | |
| U2 | 1 | 0 | 1 | 1 | 0 | 1 | |
| Moderately mutagenized (2) | U4 | 2 | 0 | 2 | 0 | 0 | 0 |
| U13 | 6 | 0 | 6 | 1 | 0 | 1 | |
Of the 20 unstable revertants tested, 2 showed slightly more intense mutagenesis. We suggest that these arose from initiator cells with multiple copies of the standard plasmid. In the course of transfer, one plasmid acquired a tandem lac-dinB duplication that was subject to heritable amplification. The initial starvation would allow higher basal dinB gene expression, and more plasmid over-replication during mating before tandem amplification of lac and dinB initiated cell division and chromosome mutagenesis. Neither of these two revertants acquired a tra mutation, suggesting that the mutagenesis experienced by unstable revertants is insufficient to generate heavily mutagenized genomes such as those found in some stable revertants.
Variation in mutagenesis intensity in different Lac+ revertant classes
When compared to unselected populations of E. coli, even revertants with the fewest unselected mutations have experienced potent mutagenesis under selection. The intensity of mutagenesis was estimated by combining the mutations carried by all the revertants in a single class, and calculating the rate required to generate them in one replication of their combined genomes. During unrestricted growth, one expects a single cell division to produce mutations at a rate of ∼0.001mutations/genome/division (Lee et al. 2012). This rate is set as a baseline for estimating the effect of selection on mutation accumulation. This comparison is appropriate because revertant colonies arise above a lawn that shows no apparent growth. Table 4 describes the estimated intensity of mutagenesis in different parts of the genomes of the several classes of revertants described in Table 2 and Table 3.
Table 4. Revertants with various mutation intensities.
| Revertants | Total mutations/aggregate genomes (kb) | Mutation Intensity (MUT) (mutations/gigabase)a | Ratio of MUT on plasmid/chromosomeb | Fold increase caused by selection |
|---|---|---|---|---|
| Stable revertant types (33) | ||||
| Lightly mutagenized (23 pooled revertants): | ||||
| In chromosomes | 6/(4500 × 23) | 58 | 276× | |
| In plasmid | 3/(215 × 23) | 607 | (10) | 2,759× |
| Moderately mutagenized (6 pooled revertants): | ||||
| In chromosomes | 6/(4500 × 6) | 222 | 1,009× | |
| In plasmid | 15/(215 × 6) | 11,628 | (52) | 52,854× |
| Intensely mutagenizedc (4 pooled revertants): | ||||
| In chromosomes | 150/(4500 × 4) | 8,333 | 37,877× | |
| In plasmid | 50/(215 × 4) | 58,139 | (7) | 264,268× |
| Unstable revertant types (20) | ||||
| Lightly mutagenized (18 pooled revertants): | ||||
| In chromosomes | 7/(4500 × 18) | 86 | 390× | |
| In plasmid | 5/(215 × 18) | 1,291 | (15.0) | 5,868× |
| Moderately mutagenized (2 pooled revertants): | ||||
| In chromosomes | 8/(4500 × 2) | 889 | 4,040× | |
| In plasmid | 1/(215 × 2) | 2,325 | (2.6) | 10,568× |
Mutation intensity during nonselective growth = 0.22 MUT (mutations/gigabase/division)
All classes show more intense mutagenesis of their F’lac plasmid than their chromosome.
Three of the intensely mutagenized revertants acquired a tra mutation in their F’lac plasmid following reversion to lac+.
Under selection, the 23 most lightly mutagenized stable revertants together accumulated six chromosomal mutations in aggregate genomes of 23 × 4500 kb, indicating a mutation intensity of 58 MUT (mutations per gigabase). They accumulated three plasmid mutations, indicating a mutation intensity in the plasmid of 3/(23 × 215) or 607 MUT. In these 23 revertants, selection resulted in a 276-fold increase in chromosomal mutation number and about a 3000-fold increase in plasmid mutations over that expected during unrestricted growth. Plasmids were mutagenized ∼10-fold more intensely than chromosomes as previously observed (Foster 1997; Bull et al. 2001). In contrast, moderately mutagenized revertants acquired six chromosomal mutations in a total of 6 × 4500 kb, and thus suffered 222 MUT. The plasmids in these strains acquired 15 mutations in 6 × 215 kb of plasmid DNA or 11,628 MUT. Thus, in this revertant class, selection enabled a 1000-fold increase in chromosomal mutations, and a 50,000-fold increase in plasmid mutations over that expected for one replication of an unselected genome. Thus the moderately mutagenized class shows more intense mutagenesis and a higher ratio of plasmid to chromosome mutagenesis. The more intense plasmid mutagenesis is taken as evidence that the reversion event in moderately mutagenized revertants occurred later during stage one, after more plasmid replication and greater DinB accumulation. Since the chromosome replicates very little during stage one, later reversion results in more intense focusing of mutagenesis on the plasmid.
Only one of the 30 randomly chosen stable revertants (S6) showed intense mutagenesis. This revertant is assumed to have arisen very late in stage one, after more plasmid mutagenesis and more DinB accumulation. Unlike less mutagenized revertants, this plasmid is assumed to have segregated with a heavily mutagenized chromosome rather than being transferred to a host cell that experienced little or no mutagenesis. Reasons for these segregation differences are suggested later.
Taken together, the 18 unstable revertants with lightest mutagenesis experienced seven chromosomal mutations in genomes of 18 × 4500 kb (Table 4). This is an intensity of 86 MUT, roughly the same as that experienced by the majority of stable revertants (60 MUT) (see Table 4). The plasmids of these unstable revertants acquired five mutations and thus suffered an intensity of 5/(215 × 18) or 1291 MUT. The two more intensely mutagenized unstable revertants (U4, U13) acquired eight chromosomal mutations in two genomes of 4500, and, thus, experienced an intensity of 889 MUT, ∼100-fold more mutagenesis than the chromosomes of other unstable revertants. The plasmids of those two revertants acquired a single mutation, and, thus, experienced 1 mutation/2 × 215 kb or 2325 MUT—only ∼threefold more intense mutagenesis than the chromosome. Note that unstable revertants experienced ∼10-fold less intense mutagenesis than the four intensely mutagenized stable revertants. We suggest that this is true because over-replication of the dinB-lac region due to tandem amplification allows cells to grow exponentially (but slowly), and dilute their DinB enzyme level twofold at each cell division. Growth reduces the basal dinB expression, and, thus, reduces the effect of amplification on the mutation rate compared to that seen in the nondividing precursors of stable revertants. This precursor cell division explains the accelerating rate of unstable colony accumulation seen in Figure 1.
Numerology of stable revertant formation
In the Cairns-Foster system, the number of stable revertant colonies increases linearly with time over the course of 5–6 days. If cells capable of experiencing full reversion showed slow exponential growth prior to full reversion, one would expect the number of visible revertant colonies to increase exponentially. The linear increase actually seen is interpreted as evidence that cells divide very little before reversion, but the basis of reversion rate—the number of replicating lac copies and mutation rate—remains constant. The few cell divisions due to increased lac number produce cells that ultimately stop growing due to reduced plasmid copy. Since copy number control prevents free plasmid replication in these cells, they do not produce revertants. The number of cells engaged in mating and rolling circle replication remains approximately constant. Mating pairs that break down are replaced by later mating events in the clone. The constant probability per unit time of forming a revertant explains why revertant colony number accumulates linearly with time.
Reversion events per plate are proportional to the number of plated initiator cells (105) times the number of plasmid replications per clone, times the increase in mutation rate under selection. We predict that in each clone ∼100 plasmid replications occur per day and the mutation rate increases an average of 1000-fold during conjugation and plasmid replication. This combination can produce the observed 10 revertants added to the reversion plate per day (10−9 ×105 × 100 × 1000), and predicts a revertant colony number that increases linearly to ∼50 over 5 days. The mutation rate increase may include effects of conjugation itself (Kunz and Glickman 1983; Christensen et al. 1985), and recombination-dependent over-replication (Kuzminov and Stahl 1999), as well as the effects of amplified DinB protein.
During the reversion process, the commonest and most lightly mutagenized revertants experience a 3000-fold increase in number of plasmid mutations, and a nearly 300-fold increase in chromosomal mutations (Table 4). The added mutations reflect the number of genome replications times the increased mutation rate experienced as the 105 plated initiator cells go through the reversion process. Prior to selection, a parental cell with the ancestral genome divides ∼50 times, with initiators having 10 of these divisions in cells with 10 plasmid copies—providing initiators with about a 140-fold increase in opportunities for reversion (40 at a normal mutation rate of 1, and 10 at a 10-fold elevated mutation rate). Under selection, mutation is further enhanced by multiple lac replications, with an elevated number of dinB gene copies (stage one). The 3000-fold increase in plasmid mutagenesis suggests that the bulk of the stable revertants (23/30) form after 50 replications with an average 60-fold increase in mutation rate. The 300-fold increase in chromosomal mutations suggests that these cells have replicated less than five times with this mutation rate to explain their less intense mutagenesis. The moderately mutagenized revertants may arise from later reversion events. Their 80,000-fold increase in plasmid mutations could have occurred after 400 plasmid replications, with an average 200-fold increase in mutation rate. Their 1000-fold increase in chromosomal mutations suggests that the chromosome replicated around five times at this mutation rate.
The number of mutations seen in revertants that appear to have experienced light or moderate mutagenesis may understate the intensity of mutagenesis actually experienced by their precursors if their revertant plasmid has been transferred into a less-intensely mutagenized cell, or has recombined with an unmutagenized plasmid, as suggested below.
Heavily mutagenized revertants
Of the original 30 random stable revertants, only 1 (S6) showed many associated mutations (see Table 2). Of 200 total revertants screened, 3 were found to grow slowly on rich medium (S23, S26, S30), all of which showed an elevated number of associated mutations. Together, the four heavily mutagenized revertants acquired a total of 150 chromosomal mutations and 50 mutations in their much smaller plasmid. This suggests that their chromosomes experienced a mutational intensity of 150/(4 × 4500 kb) or 8300 MUT. This is ∼40,000 times more intense than expected for one chromosome replication without selection. Their combined plasmids experienced an intensity of 58,139 MUT (50/(4 × 215 kb), which is 264,268 times more than expected for one replication without selection. Thus, in these rare strains, selection increased chromosome mutagenesis ∼40,000-fold and plasmid mutagenesis ∼300,000-fold. While these rare strains appear to have suffered the most intense mutagenesis, we suggest below that this high level of mutagenesis may actually be common, and these strains are unusual in the way their revertant plasmids segregated during clone development.
All four heavily mutagenized strains acquired mutations that affected their ability to transfer the revertant plasmid. Three had plasmid tra mutations and lost all plasmid transfer ability, while in the fourth (S26), the plasmid is integrated into the chromosome, making it more difficult to transfer the entire plasmid sequence to a new recipient. The correlation between tra mutations and the appearance of more intense mutagenesis was surprising, since plasmid transfer is essential for reversion under selection (Maisnier-Patin and Roth 2018). We think that revertants that appear to be heavily mutagenized are ones that fail to benefit from a backcross after reversion, and they are rare because they happened, by chance, to have escaped lethal mutations. If the intense mutagenesis seen in hyper-mutagenized revertants is typical of all revertants prior to the purifying backcross, most strains may have experienced on the order of 1000 plasmid replications with a 1000-fold increase in mutation rate.
Revertants selected for tra defects also show heavier mutagenesis
To test the correlation between tra mutations and apparent heavy mutagenesis, a set of 200 new stable revertants was isolated and tested for conjugation ability. Five of these revertants were found to be conjugation-defective, and three showed increased mutagenesis. (See Table 5). Two had hyper-mutagenized plasmids (A58, A62), and one showed hyper-mutagenesis of both plasmid and chromosome (A60). Two tra mutant revertants showed no increase in associated mutation (A57, A61). Of the extra 200 stable revertants that retained transfer-proficiency (A59), 1 was sequenced as a tra+ control. It resembled the majority lightly mutagenized class of stable revertants described in Table 2.
Table 5. Mutagenesis of conjugation-defective stable Lac+ revertants.
| Revertant numbers (5 of 200)b | Mating ability | Chromosome | F’lac plasmid | Transfer (tra) mutations | ||||
|---|---|---|---|---|---|---|---|---|
| SNPs | Indels | Total | SNPs | Indels | Totala | |||
| A59 (control) | + | 1 | 0 | 1 | 0 | 0 | 0 | None (control) |
| Rev A57 | − | 2 | 0 | 2 | 0 | 1 | 1 | traC indel |
| Rev A58 | − | 1 | 0 | 1 | 21 | 13 | 34 | 2 traG SNPs 1 traD indel 1 traW indel |
| Rev A60 | − | 17 | 5 | 22 | 3 | 4 | 7 | traM indel |
| Rev A61 | − | 0 | 0 | 0 | 0 | 1 | 1 | traC indel |
| Rev A62 | − | 1 | 0 | 1 | 19 | 0 | 19 | traC transversion |
These totals do not include the lac+ reversion mutation.
Revertants in bold type are those with intensive mutagenesis.
Two of the original 23 lightly mutagenized stable revertants shown in Table 2 (S11, S17) also had tra mutations, both of which were SNPs that caused no transfer defect. Taken together, these results show a clear correlation between transfer defects formed under selection and the presence of more associated mutations, but do not imply that the tra defect caused that mutagenesis.
Heavily mutagenized revertants are not mutators
The results above suggested that roughly 3% of revertants appear heavily mutagenized and have tra mutations that block transfer. Several possible explanations for the frequent tra mutations were considered. None of the heavily mutagenized strains was a mutator. Early in the controversy regarding the Cairns-Foster system, it was suggested that reversion under selection might result from mutators with a stable increase in mutation rate. Torkelson et al. (1997) found that roughly 10% of revertants with associated mutations are heritable mutators and show a higher than normal rate of mutation to rifampicin resistance. In light of this possibility, we screened revertants for the presence of mutators by measuring their frequency of acquired nalidixic acid resistance. These tests included the four heavily mutagenized strains in Table 2 (S6, S23, S30, S26), and those isolated later as being defective for conjugation (A60, A62, A58; Table 5). None showed an increased probability of becoming nalidixic acid resistant. We conclude that genetic mutators are not involved in lac reversion under selection, even when associated with hyper-mutagenesis. Apparently, selection favors only transitory increases in mutation rate.
Frequent tra mutations are not due to heavy mutagenesis
A trivial explanation for the tra mutations carried by eight of the sequenced revertants is that the tra gene cluster (32 kb) represents 15% of the 215 kb plasmid genome, and is thus a large target likely to be hit in any strain that experiences intense mutagenesis. This does not explain the correlation between mutation number and transfer defects. If frequent tra mutations were a secondary consequence of high mutagenesis, one would expect tra mutants to be common in strains with a high frequency of plasmid mutations. This is not the case.
The heavily mutagenized revertant S23 acquired 20 chromosomal mutations, and only one on the F’lac plasmid (Table 2). That single mutation is a tra indel mutation. This tra mutation is not associated with heavy plasmid mutagenesis. This is also true of the set of revertants chosen for their failure to transfer (Table 5). Neither revertant A57 nor A61 is heavily mutagenized, but both carry a single plasmid null mutation—in a tra gene. Similarly, revertants with highly mutagenized plasmids (A58, A62) have 34 and 19 plasmid mutations, respectively. Yet, revertant A58 has multiple tra mutations, while A62 has only one. Thus the appearance of tra mutations is not correlated with the intensity of plasmid mutagenesis.
The tra mutations acquired during reversion are not positively selected, since three of the most heavily mutagenized revertants grow very poorly, apparently due to their load of associated mutations. These revertants seem to have survived despite tra defects, not because of them. We suggest below that heavy mutagenesis is experienced by many revertant cells, most of which die. The majority of surviving revertants have a revertant lac allele that has been separated from its mutational load by backcrosses that require late plasmid transfer. These backcrosses are prevented in strains that acquire a tra mutation after reversion, forcing the revertant allele to remain in the heavily mutagenized strain where it originated. This suggests that transfer ability may be a selective advantage and tra mutant strains are rare revertants that lack that advantage but happened to survive despite their defect.
Late backcrosses may explain the correlation of conjugation defects with heavy mutagenesis and the apparent direction of mutations to adaptive targets
Two aspects of mutation distribution are remarkable. First, some revertants appear to be more intensely mutagenized than others. This was attributed above to late occurrence of the reversion event during the period of focused plasmid mutagenesis (stage one). Second, revertants show a variable distribution of mutations between plasmid and chromosome. These observations suggest an explanation for the correlation between high mutagenesis and tra mutations.
Most stable revertants show more intense mutagenesis of plasmid than chromosome (Table 4). This has been attributed to the mutagenic over-replication of plasmids during stage one—a period during which chromosomes replicate very little. Chromosomal replication resumes following a reversion and is made mutagenic by the DinB protein accumulated during stage one. The erratic distribution of mutations between plasmid and chromosome reflects segregation of variably mutagenized plasmids and chromosomes during stage three.
We propose that mutagenesis often produces deleterious, or even lethal, mutations. Cells that can grow best are those in which the least mutagenized revertant plasmids have segregated into host cells with relatively unmutagenized chromosomes. In addition, plasmid transfer can move the revertant plasmid away from its chromosomal mutations and allow recombination to separate the lac+ allele from associated plasmid mutations. Selection favors unmutagenized hosts that receive a revertant lac+ allele without an undo burden of associated plasmid mutations. The first clue that backcrosses may be frequent was the finding that three of the four most intensely mutagenized revertants grow poorly, and have acquired a tra null mutation that eliminates plasmid transfer. We propose that these tra mutations arise after reversion and prevent the backcross, forcing a revertant to form a colony despite its load of deleterious mutations. Such hyper-mutagenized revertants are rare because most succumb to the effects of mutagenesis. This predicts that, while transfer is essential at early stages of lac reversion under selection, it often occurs after reversion, and further improves revertant growth by moving the lac+ allele away from deleterious associated mutations. Most of these backcrosses must be between descendants of the original initiator cell, since revertants rarely show exchanges between different plated cells (Foster and Trimarchi 1995b).
This secondary selected transfer may explain why the Cairns-Foster experiment gives the impression of directing mutations to adaptive sites. That is, lac reversion is extremely rare during nonselective growth (∼10−9/cell/division) but appears to be very common under selection (50/108 plated cells). Mating and staged mutagenesis of plasmid and chromosome explain the enormous increase in lac reversion frequency and also the frequent occurrence of revertant lac+ alleles in relatively clean genetic backgrounds.
Supporters of stress-induced mutation have proposed that the plasmid transfer functions cooperate to nick DNA at the transfer origin OriT, and, thereby, lead to plasmid mutagenesis by damaging DNA near the plasmid tra region. While later evidence has demonstrated the essential role of actual transfer (Maisnier-Patin and Roth 2018), the frequent detection of tra mutations could suggest localized mutagenesis of the OriT-tra region. As described above, this idea is not consistent with distribution of tra mutations vis-à-vis other plasmid mutations. It is also not consistent with the positions of associated mutations on the chromosome and plasmid.
Positional distribution of unselected mutations
The genomic position of all unselected mutations is shown in Figure 6. The main histogram shows 339 mutations, of which 204 are in the chromosome and 135 in the plasmid. The region below the histogram describes the distribution in individual strains or revertant pools. While the histogram shows a rough balance with two-thirds of mutations in the chromosome and one-third in the much smaller plasmid, the mutations in some individual strains are strongly biased toward either chromosome or plasmid locations. An interpretation of this variation is below.
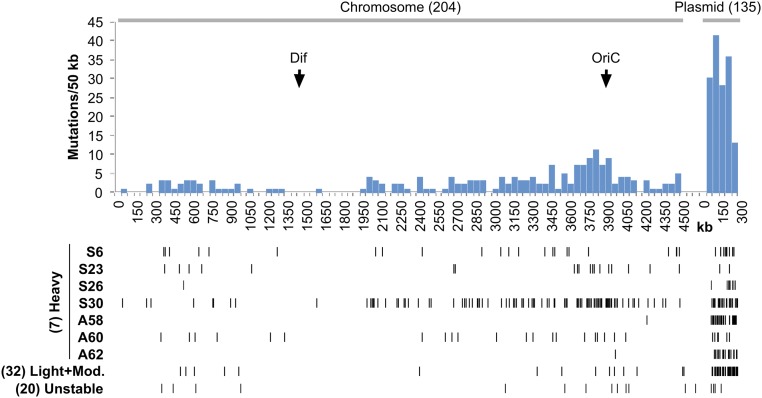
Figure 6.
Position of unselected mutations in the chromosome. The histogram describes unselected mutations carried by all 59 revertants sequenced. Below the histogram are mutations from seven single heavily mutagenized strains, from a pool of 32 stable revertants with light-to-moderate mutagenesis, and from a pool of 20 unstable revertants.
In the histogram, chromosomal mutations are seen to accumulate near the origin of replication (OriC). This pattern is dominated by the single revertant S30, which had 105 chromosomal and 28 plasmid mutations, and is shown below the histogram. Other strains with heavily mutagenized chromosomes (S6, S23, A60) show a similar mutation distribution. Strains with heavy mutagenesis of only the plasmid (S26, A58, A62) have virtually no chromosomal mutations near the origin, suggesting that their chromosomes never experienced DinB mutagenesis.
The model proposes that mutations accumulate near the origin because accumulated DinB protein inhibits replication fork movement. This inhibition causes an increase in chromosome replication complexity (CRC) after reversion occurs. The CRC is the number of initiations that occur per termination (Kuzminov 2016). Normally this number is between two and eight depending on cell growth rate. However, this number can increase to as much as 64 when the fork movement is inhibited. We suggest that accumulated DinB protein strongly inhibits fork movement and thereby stimulates overinitiation. This replication inhibition by DinB has been reported previously (Uchida et al. 2008; Mori et al. 2012). Since DinB replication is poorly processive (Fuchs and Fujii 2013), many of its forks collapse, leaving mutagenized tracks centered at the origin, which can then recombine back into the chromosome. Below the histogram, a pool of 32 light and moderately mutagenized revertants and 20 unstable revertants is shown. These strains show less striking focus of mutations on the origin, perhaps because their reversion occurred at a lower concentration of DinB protein, or their revertant allele was moved by conjugation into strains that had experienced no mutagenesis.
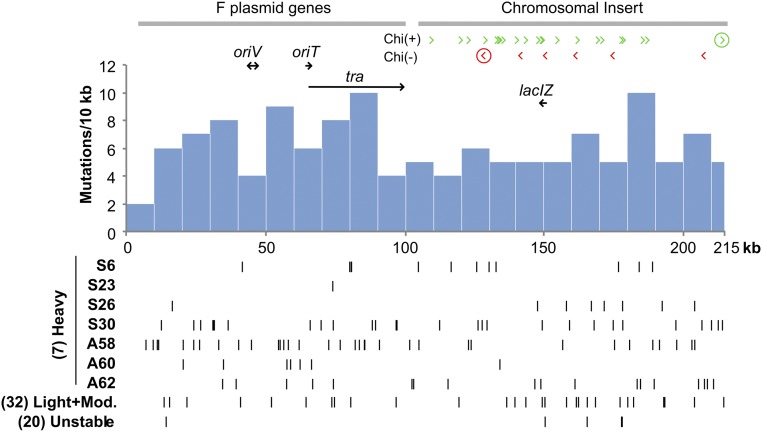
As seen in Figure 7, the distribution of plasmid mutations is fairly even. This is consistent with repeated rolling circle replication of the plasmid and does not support the idea that mutagenesis occurs during repair of a double strand break at OriT. Opponents of plasmid transfer suggested that reversion requires the F’lac element to contribute only by making a single strand nick at OriT, not by actual plasmid transfer (Foster and Trimarchi 1995a). They proposed that this nick became a double-strand break when encountered by a vegetative replication fork, and mutagenesis occurs during repair of this break in the presence of DinB. This would require ends to be resected by RecBCD until activated at a properly oriented chi site, creating two 3′ ends that initiate converging forks on an unbroken template. This might predict intense mutagenesis of the plasmid region between the properly oriented chi sites that flank OriT (Foster and Trimarchi 1995a; Rosenberg et al. 1995).
Figure 7.
Position of associated mutations on F’lac plasmid. The histogram includes all unselected plasmid mutations found in all revertants analyzed. Individual strains and pools are described below the histogram. Lac+ mutations are not included. The position and orientation of all chi sites are indicated above the histogram, where arrow points indicate the 3′ end of the chi site that would initiate replication at a new fork. The two circled chi sites are the closest properly oriented sites flanking the OriT site in the circular plasmid.
However, as seen in Figure 7, there are no chi sites in the F portion of the F’lac plasmid, so the closest chi sites flanking OriT (in the circular plasmid) are those circled in the figure, which are each ∼50 kb away from OriT. These sites are too far away to permit recognition by RecBCD. The region affected by mutagenic repair of a break at OriT would not include lac. The OriT proposal is not supported by the mutation distribution shown here, nor does it explain chromosome mutagenesis.
The plasmid mutation distribution, seen in Figure 7, is consistent with the idea that spontaneous breaks arise near some chi site during transfer and initiate repeated mutagenic rolling circle replication of the whole plasmid of different templates see Figure 2.) The observed mutation distribution is also consistent with the idea that revertant plasmids are frequently transferred into less mutagenized hosts, allowing recombination between plasmids. Such recombination events would be expected to remove associated mutations located farthest from lac+, whose retention is selected. In Figure 7, the pool of lightly and moderately mutagenized revertants show a clear clustering of associated mutations near lac, suggesting that recombination between plasmids moved a lac region and nearby mutations into an unmutagenized plasmid. This may also explain the distribution of mutations in S26, where we suggest a mutagenized plasmid was transferred and recombined in an unmutagenized strain carrying an integrated F’lac; this backcross moved lac+ with a few nearby associated mutations. The clustering of mutations near lac is not seen in the tra defective strains with a hyper-mutagenized plasmid such as S30, A58, and A62, which are unlikely to have gone through a backcross.
Most mutations made under lactose selection are caused by DinB
The first evidence for involvement of DinB in reversion under selection was the finding that a dinB mutation in the tester strain reduces the number of stable revertants ∼10-fold, but does not alter the frequency of unstable revertants (McKenzie et al. 2001; Yamayoshi et al. 2018). In the model described here, the frequency of both lac reversion and associated mutations is greatly enhanced by overproduction of the error-prone DinB polymerase during plasmid over-replication. If the observed mutagenesis is caused by DinB overproduction, one expects both the revertant lesions (lac+) and unselected associated mutations to resemble those known to form during DinB-mediated DNA replication. The set of mutations made by DinB differs in several ways from those formed spontaneously (Table 6).
Table 6. Mutation Spectra With and Without DinB.
| Mutation type | Lee et al. 2012 E. coli mutation accumulation unselected | Wagner and Nohmi, 2000 (phage lambda C gene) | Unselected mutations associated with: | |||
|---|---|---|---|---|---|---|
| Spontaneous | Overproduced DinB | 32a Light-moderate stables | 7b Heavy stables | 20c Light unstables | ||
| Substitutions (total) | 233 (91%) | 55 (59%) | 30 (33%) | 46 (85%) | 189 (67%) | 13 (93%) |
| Transitions | 131 (51%) | 6 (6%) | 12 (13%) | 12 (22%) | 63 (22%) | 4 (29%) |
| A/T->G/C | 49 | 2 | 9 | 5 | 35 | 1 |
| G/C->A/T | 82 | 4 | 3 | 7 | 28 | 4 |
| Transversions | 102 (40%) | 49 (53%) | 18 (20%) | 34 (63%) | 126 (44%) | 9 (64%) |
| A/T->T/A | 17 | 2 | 0 | 5 | 2 | 2 |
| 38 | 0 | 7 | 14 | 38 | 4 | |
| G/C->T/A | 30 | 47 | 6 | 8 | 54 | 4 |
| G/C->C/G | 17 | 0 | 5 | 7 | 32 | 5 |
| Indels (total) | 21 (8%) | 37 (40%) | 59 (66%) | 8 (15%) | 95 (33%) | 1(7%) |
| −1 one base | 13 | 7 | 54 | 6 | 93 | 0 |
| +1 one base | 6 | 28 | 3 | 1 | 0 | 0 |
| indel > 1 base | 2 | 2 | 2 | 1 | 2 | 1 |
| Overall total | 254 | 92 | 89 | 54 | 284 | 14 |
Stable revertants (32) with light or moderate mutagenesis (<5 mutations): S1, S2, S3, S4, S5, S7, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, S19, S20, S21, S22, S24, S25, S27, S28, S29, S31, S32, S33, S34, A57, A59, A61.
Stable revertants (7) with heavy mutagenesis (≥10 mutations): S6, S23, S26, S30, A58, A60, A62.
Unstable revertants (20) with light or moderate mutagenesis (<10 mutations): U0, U1, U2, U3, U4, U5, U6, U7, U8, U9, U10, U11, U12, U13, U14, U15, U16, U17, U18, U19 (There were no heavily mutagenized unstables).
The spectrum of spontaneous mutations arising in unselected growing populations shows two distinctive features. First, ∼90% of total mutations are base substitutions and relatively few (10%) are frameshifts or indels (Lee et al. 2012). Second, base substitution mutations include a slight excess of transitions over transversions (Vogel 1972; Collins and Jukes 1994; Lee et al. 2012; Foster et al. 2015; Sprouffske et al. 2018). These features are seen in the first column of Table 6, which shows a set of 254 spontaneous mutations accumulated during ∼6000 generations of unselected growth (Lee et al. 2012). Notice that indel mutations are only 8% of the total, and include both +1 and −1 types. The bulk of the accumulated mutations (90%) are base substitutions, with transitions (51%) exceeding transversions (40%).
Column two presents results of Wagner and Nohmi (2000), who examined the effect of DinB overproduction on spontaneous mutations in the repressor gene of phage lambda. Loss-of-function mutations were selected following growth in strains with or without a plasmid overexpressing DinB polymerase. The spontaneous spectrum is not identical to that of the E. coli chromosome, due, in part, to the nature of the repressor gene sequence, but primarily to the fact that loss-of-function mutations were selected. Frameshift mutations were common, but their frequency increased substantially (from 40 to 66% of total) in the presence of excess DinB, where the vast majority of DinB-induced frameshift mutations are the −1 type.
These results are compared to those for the unselected mutations accompanying lac+ revertants selected in the Cairns-Foster system (last three columns of Table 6). We examined 54 mutations (not including lac) from 32 strains, including the 29 lightly and moderately mutagenized random revertants described in Table 2, plus the three lightly mutagenized revertants described in Table 5. We also examined the 284 mutations from the seven intensely mutagenized stable revertants. These two mutation sets resembled each other, and were similar to mutations made by DinB. That is, in both sets arising during the lac selection, frequency of frameshift mutations increased over that without selection from 8 to 15% or 30% (light and heavy mutagenesis) of the total, and virtually all were the −1 type. In addition, under lactose selection the frequency of transversion mutations (63% and 44%) clearly exceeded that of transitions (22% and 22%). We conclude that the unselected mutations associated with selected lac reversion are likely to have been made by DinB, and this is true regardless of the intensity of mutagenesis.
Unstable revertants had very few mutations, despite the fact that they develop with multiple tandem copies of the plasmid dinB gene, and show a 400-fold increase in plasmid mutagenesis during selection. To test this, we characterized mutations that formed in the 20 unstable revertants and describe them in the rightmost column in Table 6. Unlike changes in stable revertants that appeared to be DinB-induced, unstable revertants had very few indels and no −1 frameshift mutations. However, as expected for DinB mutations, the 13 SNP mutations included an excess of transversions—nine vs. only four transitions. The unstable revertant with the most highly mutagenized chromosome (U13) had seven unselected mutations—six transversions and one transition. The higher level of transversion mutations suggests that the unselected mutations in unstable revertants are caused by DinB. The lack of frameshift mutations may indicate that frameshift mutations are favored by higher concentrations of DinB protein.
The nature of stable lac revertants
The mutations that confer a stable Lac+ phenotype are listed in Table 1. Mutations that restore full LacZ function must provide a −1 frameshift to compensate for the parent +1 mutation. Of the 39 stable revertants sequenced, most are single base deletions (32/39) in or near the parent +1 mutation, and most of these (29/32) arose in monotonous base runs. The rest were multibase insertions or deletions. These mutations are consistent with those seen for DinB, but since they were demanded by the lac selection, they do not shed light on whether DinB was responsible for their formation.
Discussion
The persistence of the adaptive mutation controversy
The Cairns selection system was first described in 1988 (Cairns et al. 1988) and the final version of the system in 1991 (Cairns and Foster 1991). Despite a vast number of publications and reviews, no model has convincingly explained how this system works. The explanation seemed so obvious—a nongrowing cell population gives rise preferentially to rare adaptive mutations that allow growth on lactose. Many researchers attribute this behavior to an evolved mechanism that senses growth limitation and directs new mutations to sites that improve growth (Foster 1999). This interpretation was widely accepted at face value because it seemed both obvious and revolutionary—it appeared to violate standard genetic theory (Fitzgerald and Rosenberg 2019).
We remained convinced that this phenomenology would reflect natural selection. Our original model proposed that pre-existing cells with a lac duplication grew slowly on lactose and responded to selection by expanding a tandem amplification of the growth-limiting mutant lac allele and the nearby dinB gene on the plasmid (Andersson et al. 1998; Hendrickson et al. 2002; Roth et al. 2003; Slechta et al. 2003). This model predicted that the number of visible revertant colonies should accumulate exponentially over time as clones grow. In fact, the number of stable revertant colonies increases linearly (see Figure 1). Our early model predicted that reversion would require a location of dinB near the lac so both could amplify under selection, which later proved incorrect—dinB can be anywhere on the plasmid (Yamayoshi et al. 2018). This model needed improvement.
The new model supersedes our initial suggestion, and explains the linear increase in colony number by proposing that selection acts on a constant number of replicating F’lac plasmid copies. Occasional cell divisions produce daughter cells with a reduced plasmid number that are unable to grow or revert. Mutagenesis is focused on the plasmid due to selected over-replication of the entire plasmid, regardless of the position of dinB. Rare pre-existing initiator cells with multiple plasmid copies divide a few times under selection to produce daughters that mate and initiate plasmid over-replication. Previous conjugation tests failed to detect mating because they scored mating between different plated cells, which is very rare (Foster and Trimarchi 1995b). Mating between identical daughters of initiator cells are hard to detect. Thus in the Cairns-Foster system, selection acts on a process that is far from obvious. Selection acts on multiple genetic operations, including two acts of genetic transfer, and plasmid over-replication in a nongrowing cell population. While these events rely on peculiarities of the parent strain, the general underlying principles are likely to be broadly applicable to other genetic systems.
The essence of selection in the Cairns-Foster system
Natural selection requires genetic variability that affects the ability of individuals to reproduce. In the Cairns-Foster system, the initial variability is stochastic differences in plasmid copy number that arise during growth prior to plating on selective medium. This variation reflects insufficiency of the plasmid copy control system, which maintains plasmid number at ∼1 per cell, but actually produces a distribution of copy number with roughly 1 in 1000 cells possessing ∼10 or more copies (Sano et al. 2014). After plating, cells with extra copies of the plasmid have energy to support division and incestuous conjugation between identical daughter cells. Conjugation simultaneously increases the copy number of lac and dinB genes and provides energy for repeated recombination, replication, and mutagenesis. Thus, selection for more energy from lactose drives over-replication and localized mutagenesis and leads to a full reversion event.
Very little chromosome replication occurs prior to reversion because cells are starved for lactose, and every division causes a twofold drop in plasmid copy number. The lac reversion event initiates stage two by providing enough energy to support chromosome replication and cell division. Chromosome replication is impaired and made mutagenic by accumulated DinB protein, while free plasmid replication is prevented by the plasmid copy number control mechanism until copy number drops to one. In stage three (plasmid segregation), cell division reduces plasmid copy-number and leads to random segregation of plasmids and chromosomes. Many—if not most—revertants initially have a heavy load of deleterious mutations that limit their growth, but transfer of the revertant lac+ allele into cells with fewer mutations allows revertants to escape noxious mutations, and gives the appearance that mutagenesis was directed to adaptive targets.
The key features of the Cairns-Foster system that underlie its “shocking” behavior
An important peculiarity of this system is that a single copy of the mutant allele has too little residual function to support division under selection, but amplification of that allele allows growth. The hybrid gene under selection, lacIZ, must be located on a conjugative plasmid that also encodes a mutagenic DNA repair polymerase, DinB. Revertants are all initiated by pre-existing initiator cells with an excessive plasmid copy number (Sano et al. 2014). While cells with an F plasmid generally mate better with F− cells than with each other, early work on this system demonstrated efficient transfer between F’lac-bearing cells during starvation (Peters et al. 1996). This work anticipates many aspects of the model described here. In the ultimate Cairns-Foster system (Cairns and Foster 1991), residual tester cell growth is prevented by scavenger cells so mating is limited to daughters of rare pre-existing cells with excessive plasmid copy number (Sano et al. 2014). Repeated plasmid replication following transfer becomes mutagenic due to overproduction of the error prone DinB polymerase, which happens to be encoded by the same plasmid.
Mutations appear directed to adaptive sites for three main reasons. First, during stage one, mutagenesis is focussed on the small F’lac plasmid, whose replication and consequent mutagenesis occurs in cells that undergo very few divisions or chromosome replications. Second, after lac reversion enables cell division, chromosomes are mutagenized to various degrees depending on their accumulated level of DinB (stage two). Third, during stage four, conjugation can move a revertant lac+ allele away from its toxic genetic background into cells of the same clone that have not suffered heavy mutagenesis. Selection favors the plasmid-chromosome combinations with fewest deleterious mutations. This gives the appearance that mutations arise preferentially at adaptive sites.
Broader applicability of the principles underlying the Cairns system
While the critical features of the Cairns-Foster system appear unique, the underlying principles may be broadly applicable to other biological systems, both bacterial and eukaryotic. The main principle is that selection favors cells that over-replicate a particular subset of their genome. This provides more opportunities for mutation, and can be associated with a localized increase in mutation rate per act of replication. In the Cairns system, local amplification occurs during transfer of the limiting genes from one cell to another. Having the gene for an error-prone polymerase on the same plasmid as the growth-limiting lac allele makes this selected coamplification mutagenic.
In viruses, bacteria, and eukaryotic cell populations, selection can favor amplification of growth-limiting genes (Andersson and Hughes 2009; Elliott et al. 2013; Bayer et al. 2018; Gaines et al. 2019). Recently it has been found that many pathogenic bacterial strains appear sensitive to antibiotics based on tests of their low minimal inhibitory concentrations (MIC), but are found to rapidly acquire resistance under selection, which can lead to a failure of therapy. This has been termed “heteroresistance,” because the apparently sensitive populations include rare cells with a gene amplification that provides some antibiotic resistance (Nicoloff et al. 2019). Selection drives expansion of a tandem array leading to highly resistant organisms that overtake the population. It is not clear whether this local amplification is associated with an increased rate of mutation per replication. Thus, heteroresistant strains resemble the Cairns and Foster strain in that the parent population shows very little growth under selection, but includes cells that rapidly develop growth ability under selection. While the error-prone DinB polymerase encoded by F’lac may make the Cairns-Foster system seem very specialized, it is interesting to note that new resistance genes often appear on conjugative plasmids, and that the pSLT plasmid shared by many Salmonella isolates is both conjugative and includes the gene for an error prone translesion polymerase SamAB (Andersson et al. 2010). Supporters of stress-induced mutagenesis have proposed that DinB is part of an evolved system that is both mutagenic and carcinogenic (Xia et al. 2019). We propose that in the Cairns-Foster system, DinB is mutagenic because it is coamplified under selection with the growth-limiting lac allele.
Eukaryotic cells can also respond to selection by over-replication of local genome regions. Initially this was thought to occur by repeated firing of an internal replication origin, leading to “onion skin” structures with repeated copies of a small region that can collapse to create tandem repeats in direct and inverse order (Stark et al. 1979; Schimke et al. 1986). This idea fell out of favor, but has recently gained more support (Green et al. 2010). Furthermore, amplification by rereplication has recently been found to be mutagenic (Bui and Li 2019). The focus of chromosomal mutagenesis near the origin in the Cairns-Foster system may reflect origin stimulation (increases in CRC) when fork movement is inhibited by accumulated DinB (Kuzminov 2016). These parallels may suggest that selective gene amplification is often a means of accelerating formation of adaptive mutations.
Revertants differ in the level of mutagenesis they have experienced
Early in the history of the Cairns-Foster controversy, Rosche and Foster (1999) presented excellent evidence that revertants differ in their levels of associated general mutagenesis. No mechanistic basis for this variation was proposed, but this observation is understandable in terms of the model described here. The intensity of plasmid mutagenesis depends on when reversion occurs during the course of plasmid replication and DinB amplification (stage one). The degree of chromosome mutagenesis depends on the amount of DinB protein that has accumulated before reversion leads to resumption of chromosome replication. Thus, the DinB level dictates how intensely the chromosome is mutagenized. A revertant’s number of chromosomal mutations is determined by whether the revertant plasmid segregates with a highly mutagenized chromosome, or if it is transferred to a lightly mutagenized host before segregation is complete.
Unstable Lac+ cells within stable revertant colonies
Within stable revertant colonies, ∼1 cell in 1000 makes a sectored colony on rich medium, resembling those typical of unstable revertants (Hendrickson et al. 2002; Hastings et al. 2004). Our initial selection model proposed that these cells carried a tandem lac amplification and were precursors of the stable revertants formed during growth under selection. These cells were later found not to contain tandem lac arrays, but to give rise to a mixture of solid blue and solid white colonies when restreaked on rich medium with X-gal (S. Maisnier-Patin and J.R. Roth, unpublished data; Hastings et al. 2004). This unstable Lac+ phenotype was first attributed to chance juxtaposition of revertant and parental cells on the selection plate (Hastings et al. 2004)—a phenomenon that we could not duplicate experimentally. The rare colonies are better explained by the model presented here. We suggest that unstable Lac+ cells found within a stable revertant colony contain a mixture of revertant and mutant plasmids that have not completely segregated. In the first streak, the segregation process goes to completion within each colony, generating some sectors with only a stable revertant plasmid and some with only a mutant plasmid. Restreaking these colonies gives a mixture of the two colony types described above.
The importance of plasmid transfer
Early in the history of the adaptive mutation controversy, evidence was presented that plasmid transfer is essential for reversion under selection (Galitski and Roth 1995; Radicella et al. 1995; Peters et al. 1996; Godoy and Fox 2000). Later, four lines of evidence were used to argue against this conclusion. We think all of these counter-arguments are flawed.
Trimarchi and Foster claimed that all of the conjugation (tra) functions were needed to allow the TraI protein to cut one strand of plasmid DNA at the transfer origin. Reversion required this nicking, but not actual transfer of plasmid DNA from one cell to another (Foster and Trimarchi 1995a,b). This conclusion was based on reversion experiments done with mixtures of genetically marked tester strains that allowed detection of plasmid transfer from one plated tester type to another during the reversion process. Very few revertants carried a revertant F’lac+ plasmid that had moved to a different plated cell type. To be detected, transfer required distinctly marked testers to contact each other on the selection plate and transfer their plasmid before or after reversion. We argue here that this occurs rarely because very few of the plated cells have enough energy to support mating. In our model, mating only occurs between the identical daughters of a rare initiator cell, which can divide because of its high copy of the F’lac plasmid (Maisnier-Patin and Roth 2018). Thus, each revertant clone is initiated by a single plated initiator cell. The experiments of Trimarchi and Foster could not detect this essential mating, which occurs between genetically identical daughters.
-
Opponents of plasmid transfer proposed that the plasmid transfer (tra) functions contributed to reversion only by making single-strand nicks at the transfer origin (OriT). These nicks were said to become double-strand breaks when encountered by a vegetative replication fork (Foster 1999), and mutagenesis occurred during repair of these breaks in the presence of DinB. This idea requires processing of double-strand ends by RecBCD, and activation of these ends at chi sites so that RecA could initiate converging replication forks. Mutagenesis would occur between opposing chi sites that flank a region that included OriT and lac. However, the F portion of the plasmid has no chi sites, and the closest chi sites that flank OriT are ∼50 kb away (see Figure 7). These sites are too far away to support RecBCD-dependent repair of a break at OriT, and, if this occurred, the mutagenized region would not include lac (see Figure 7). The even distribution of mutations seen in Figure 7 is better explained by the new model, which proposes repeated mutagenic rolling circle replication of the whole plasmid during transfer.
A few revertants show intense mutagenesis of the chromosome, but not the plasmid. This is not expected to result from simple nicking of plasmid DNA at OriT, but is explained by the plasmid transfer model, if a revertant lac+ allele loses its associated mutations by recombination with an unmutagenized plasmid.
3. To demonstrate that F plasmid transfer functions contribute by making DNA nicks at OriT (made by TraI), supporters of stress-induced mutagenesis introduced a cutting site on the F’lac plasmid and expressed the nuclease from another plasmid (Rodriguez et al. 2002; Ponder et al. 2005). Overexpression of this nuclease stimulated reversion under selection even in strains that lacked ability to transfer the F’lac plasmid. Induction of this nuclease did not stimulate reversion of a lac frameshift mutation located near a cutting site in the chromosome, because rolling circle over-replication of the circular chromosome does not happen (Shee et al. 2011b). Another genetic system used a tetracycline resistance gene (tet) with a frameshift mutation placed near a cutting site in the chromosome, and studied revertants that accumulated in cells held long term in stationary phase with an expressed nuclease (not under selection for growth improvement) (Shee et al. 2011a). Although a slight stimulation of tet reversion was seen following a period of nuclease induction, cell turnover was not determined, and cell death can lead to an overestimation of mutation rates (Frenoy and Bonhoeffer 2018). We submit that these nuclease experiments shed no light on the Cairns-Foster system. The nuclease is produced at a high level for such a long time that it causes recurring DNA damage and stimulates recombinational damage repair synthesis that can promote reversion without transfer (like UV mutagenesis). We suggest that recurrent DNA damage stimulates reversion by a route that is unrelated to the transfer-dependent events that occur under selection in the Cairns-Foster system.
-
4. McKenzie and Rosenberg claimed that the dinB+ gene, which is required for reversion and induced during growth limitation by the global RpoS or SOS control system, can be located either at its normal chromosomal site or in the F’lac plasmid (McKenzie et al. 2001). We could not replicate their results, and found that reversion under selection occurred only if a functional dinB+ allele is located somewhere on the same F’lac plasmid that carries the mutant lac allele (Yamayoshi et al. 2018). Only at this position can dinB+ be selectively coamplified with the mutant lac allele when the plasmid is over-replicated. The claim that a chromosomal dinB+ allele could support reversion was based on use of a dinB+ allele that had been added to chromosome as part of a lambda specialized transducing prophage inserted near the chromosomal galactose operon (McKenzie et al. 2001). We suspect that the dinB+ gene in these experiments is over-expressed from the gal promoter, which is induced during selection for growth on lactose. Alternatively, the dinB gene may have been selectively coamplified with the whole galactose region. The requirement for dinB+ on the F’lac plasmid was also demonstrated by Kim et al. (2001), who showed clearly that selection did not stimulate lac reversion in strains whose only dinB+ allele is at its normal chromosomal location.
To support the idea of regulation of dinB transcription by global control systems, supporters of stress-induced mutagenesis, showed that reversion under selection was prevented by mutations that reduce basal expression of dinB (either a lexIND or rpoS null mutation) (McKenzie et al. 2000; Lombardo et al. 2004; Yamayoshi et al. 2018). In the new model, mutagenesis is caused by coamplification of dinB with lac under selection, explaining the need for these genes to be colocated on the F’lac plasmid. We suggest that global control mutations reduce the basal level of dinB transcription, and, thereby, render selective amplification of dinB ineffective at mutagenesis.
Revertant S30 illustrates the power of mutagenesis under selection in the Cairns-Foster system
The intensity of mutagenesis that is possible during selection in the Cairns system is graphically demonstrated by revertant S30. Growth of a laboratory strain with a potent chemical mutagen (e.g., nitrosoguanidine) can increase the mutation rate perhaps 1000-fold, and generate survivors with a handful of chromosomal mutations (Adelberg et al. 1965; Miller 1992). It was therefore shocking to find that during lac reversion under selection, revertant S30 acquired 133 mutations—105 chromosomal and 28 in the plasmid. This demonstrates the intense mutagenesis that can be achieved during repeated plasmid transfer with elevated levels of DinB protein. The genome of this revertant is diagrammed in Figure 8.
Figure 8.
Exceptional hypermutagenized revertant. One stable lac+ revertant (S30) accumulated 133 mutations—105 chromosomal and 28 plasmid. The small central circle represents the F’lac plasmid drawn to scale with the chromosome (outer circle). Colored sectors indicate portions of the plasmid derived from the parent F plasmid (blue) and the added bacterial segment (red). The white arrow designates the tra operon. Chi sites that could activate RecBCD are indicated outside the plasmid map. Note that the F-derived segment includes no chi sites. This strain acquired transfer deficiencies (tra) after reversion, and, therefore, escaped the backcrosses that usually separate the revertant lac+ allele from deleterious or lethal associated mutations in the Cairns-Foster system.
The unusual revertant S30 seen in Figure 8 illustrates the several conclusions drawn here. Chromosomal mutations center around the replication origin while mutations are distributed more evenly on the plasmid (center). As expected for mutations caused by DinB, frameshift mutations are common (68% of total), and are all of the −1 type. Base substitution mutations are mostly transversions (61% of SNPs). This strain acquired multiple tra mutations—two traI indels, one traK indel, transversions in traC, S, and T genes—and became transfer defective after reversion. We suggest that this intense mutagenesis is common in the Cairns-Foster system, but is usually lethal. Following reversion, selection acts secondarily to move the revertant lac+ allele into a host that experienced little or no mutagenesis. Thus, selection acts before reversion to generate mutations, and after reversion to minimize the number of deleterious mutations associated with the selected lac+ allele.
Summary
The secret of the Cairns-Foster system is selection, which primarily drives over-replication of lac, and secondarily leads to mutagenesis by over-replicating dinB. Later backcrosses occur under selection to separate the revertant lac+ allele from deleterious associated mutations. Without selective backcrosses, we submit that most revertants in the Cairns system would suffer disastrous and frequently lethal consequences of over-mutagenesis by DinB. The intensity of this mutagenesis probably reflects the ability of DinB polymerase to load on the structures generated during plasmid transfer and break-induced rolling circle replication.
Acknowledgments
We thank Fritz Roth and two anonymous reviewers for helpful comments on the manuscript. Our work was supported by National Institutes of Health grant 5R01GM027068 (to J.R.R.).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.10848680.
Communicating editor: J. Nickoloff
Literature Cited
- Adelberg E. A., Mandel M., and Ching Chen G. C., 1965. Optimal conditions for mutagenesis by N-methyl-N’-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem. Biophys. Res. Commun. 18: 788–795. 10.1016/0006-291X(65)90855-7 [DOI] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L., and Beatson S. A., 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12: 402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., and Hughes D., 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43: 167–195. 10.1146/annurev-genet-102108-134805 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Slechta E. S., and Roth J. R., 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282: 1133–1135. 10.1126/science.282.5391.1133 [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Koskiniemi S., and Hughes D., 2010. Biological roles of translesion synthesis DNA polymerases in eubacteria. Mol. Microbiol. 77: 540–548. 10.1111/j.1365-2958.2010.07260.x [DOI] [PubMed] [Google Scholar]
- Bayer A., Brennan G., and Geballe A. P., 2018. Adaptation by copy number variation in monopartite viruses. Curr. Opin. Virol. 33: 7–12. 10.1016/j.coviro.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz D., Hushon J. M., Whitfield H. J., Roth J. R., and Ames B. N., 1968. Procedures for identifying nonsense mutations. J. Bacteriol. 96: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui D. T., and Li J. J., 2019. DNA rereplication is susceptible to nucleotide-level mutagenesis. Genetics 212: 445–460. 10.1534/genetics.119.302194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull H. J., Lombardo M.-J., and Rosenberg S. M., 2001. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc. Natl. Acad. Sci. USA 98: 8334–8341. 10.1073/pnas.151009798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., and Foster P. L., 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., and Miller S., 1988. The origin of mutants. Nature 335: 142–145. 10.1038/335142a0 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen R. B., Christensen J. R., and Lawrence C. W., 1985. Conjugation-dependent enhancement of induced and spontaneous mutation in the lacI gene of E. coli. Mol. Gen. Genet. 201: 35–37. 10.1007/BF00397983 [DOI] [PubMed] [Google Scholar]
- Collins D. W., and Jukes T. H., 1994. Rates of transition and transversion in coding sequences since the human-rodent divergence. Genomics 20: 386–396. 10.1006/geno.1994.1192 [DOI] [PubMed] [Google Scholar]
- Coulondre C., and Miller J. H., 1977. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J. Mol. Biol. 117: 577–606. 10.1016/0022-2836(77)90059-6 [DOI] [PubMed] [Google Scholar]
- Elliott K. T., Cuff L. E., and Neidle E. L., 2013. Copy number change: evolving views on gene amplification. Future Microbiol. 8: 887–899. 10.2217/fmb.13.53 [DOI] [PubMed] [Google Scholar]
- Fitzgerald D. M., and Rosenberg S. M., 2019. What is mutation? A chapter in the series: how microbes “jeopardize” the modern synthesis. PLoS Genet. 15: e1007995 10.1371/journal.pgen.1007995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 1994. Population dynamics of a Lac- strain of Escherichia coli during selection for lactose utilization. Genetics 138: 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 1997. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 179: 1550–1554. 10.1128/jb.179.5.1550-1554.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 1999. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet. 33: 57–88. 10.1146/annurev.genet.33.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42: 373–397. 10.1080/10409230701648494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., and Cairns J., 1992. Mechanisms of directed mutation. Genetics 131: 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., and Trimarchi J. M., 1994. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science 265: 407–409. 10.1126/science.8023164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., and Trimarchi J. M., 1995a Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. USA 92: 5487–5490. 10.1073/pnas.92.12.5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., and Trimarchi J. M., 1995b Conjugation is not required for adaptive reversion of an episomal frameshift mutation in Escherichia coli. J. Bacteriol. 177: 6670–6671. 10.1128/jb.177.22.6670-6671.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Lee H., Popodi E., Townes J. P., and Tang H., 2015. Determinants of spontaneous mutation in the bacterium Escherichia coli as revealed by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 112: E5990–E5999. 10.1073/pnas.1512136112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenoy A., and Bonhoeffer S., 2018. Death and population dynamics affect mutation rate estimates and evolvability under stress in bacteria. PLoS Biol. 16: e2005056 10.1371/journal.pbio.2005056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R. P., and Fujii S., 2013. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 5: a012682 10.1101/cshperspect.a012682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines T. A., Patterson E. L., and Neve P., 2019. Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance. New Phytol. 223: 1770–1775. 10.1111/nph.15858 [DOI] [PubMed] [Google Scholar]
- Galhardo R. S., Hastings P. J., and Rosenberg S. M., 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42: 399–435. 10.1080/10409230701648502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T., and Roth J. R., 1995. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science 268: 421–423. 10.1126/science.7716546 [DOI] [PubMed] [Google Scholar]
- Godoy V. G., and Fox M. S., 2000. Transposon stability and a role for conjugational transfer in adaptive mutability. Proc. Natl. Acad. Sci. USA 97: 7393–7398. 10.1073/pnas.130186597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. M., Finn K. J., and Li J. J., 2010. Loss of DNA replication control is a potent inducer of gene amplification. Science 329: 943–946 (erratum: Science 330: 448). 10.1126/science.1190966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Longerich S., and Rosenberg S. M., 1994. Recombination in adaptive mutation. Science 264: 258–260. 10.1126/science.8146657 [DOI] [PubMed] [Google Scholar]
- Hastings P. J., Slack A., Petrosino J. F., and Rosenberg S. M., 2004. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2: e399 10.1371/journal.pbio.0020399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J., Ira G., and Lupski J. R., 2009. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5: e1000327 10.1371/journal.pgen.1000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H., Slechta E. S., Bergthorsson U., Andersson D. I., and Roth J. R., 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 99: 2164–2169. 10.1073/pnas.032680899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C., Langston L. D., Yurieva O., Goodman M. F., and O’Donnell M., 2009. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. USA 106: 6031–6038. 10.1073/pnas.0901403106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Matsui K., Yamada M., Gruz P., and Nohmi T., 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266: 207–215. 10.1007/s004380100541 [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D. et al. , 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Kofoid E., Reams A. B., Andersson D. I., and Roth J. R., 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. USA 103: 17319–17324. 10.1073/pnas.0608309103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E., Kofoid E., Andersson D. I., Lu Y., Mellor J. et al. , 2010. The tandem inversion duplication in Salmonella enterica: selection drives unstable precursors to final mutation types. Genetics 185: 65–80. 10.1534/genetics.110.114074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., and Glickman B. W., 1983. The infidelity of conjugal DNA transfer in Escherichia coli. Genetics 105: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A., 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16: 373–384. 10.1111/j.1365-2958.1995.tb02403.x [DOI] [PubMed] [Google Scholar]
- Kuzminov A., 2016. Chromosomal replication complexity: a novel DNA metrics and genome instability factor. PLoS Genet. 12: e1006229 10.1371/journal.pgen.1006229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A., and Stahl F. W., 1999. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 13: 345–356. 10.1101/gad.13.3.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton J. C., and Foster P. L., 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50: 549–561. 10.1046/j.1365-2958.2003.03704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Popodi E., Tang H., and Foster P. L., 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 109: E2774–E2783. 10.1073/pnas.1210309109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., and Buckman C., 1995. Conjugational recombination in Escherichia coli: genetic analysis of recombinant formation in Hfr x F- crosses. Genetics 139: 1123–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M. J., Aponyi I., and Rosenberg S. M., 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166: 669–680. 10.1534/genetics.166.2.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S., and Roth J. R., 2015. The origin of mutants under selection: how natural selection mimics mutagenesis (adaptive mutation). Cold Spring Harb. Perspect. Biol. 7: a018176 10.1101/cshperspect.a018176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S., and Roth J. R., 2016. The adaptive mutation controversy, pp. 26–36 in Encyclopedia of evolutionary biology, edited by Kliman R. M., Academic Press, Oxford: 10.1016/B978-0-12-800049-6.00229-8 [DOI] [Google Scholar]
- Maisnier-Patin S., and Roth J. R., 2018. Selection and plasmid transfer underlie adaptive mutation in Escherichia coli. Genetics 210: 821–841. 10.1534/genetics.118.301347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., and Haber J. E., 2012. Mutations arising during repair of chromosome breaks. Annu. Rev. Genet. 46: 455–473. 10.1146/annurev-genet-110711-155547 [DOI] [PubMed] [Google Scholar]
- Malkova A., and Ira G., 2013. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 23: 271–279. 10.1016/j.gde.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., and Arai K., 1997. Frpo: a novel single-stranded DNA promoter for transcription and for primer RNA synthesis of DNA replication. Cell 89: 897–907. 10.1016/S0092-8674(00)80275-5 [DOI] [PubMed] [Google Scholar]
- Matic I., Rayssiguier C., and Radman M., 1995. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80: 507–515. 10.1016/0092-8674(95)90501-4 [DOI] [PubMed] [Google Scholar]
- McKenzie G. J., Harris R. S., Lee P. L., and Rosenberg S. M., 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97: 6646–6651. 10.1073/pnas.120161797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie G., Lee P., Lombardo M.-J., Hastings P., and Rosenberg S., 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7: 571–579 (erratum: Mol. Cell 7: 1119). 10.1016/S1097-2765(01)00204-0 [DOI] [PubMed] [Google Scholar]
- McVey M., Khodaverdian V. Y., Meyer D., Cerqueira P. G., and Heyer W. D., 2016. Eukaryotic DNA polymerases in homologous recombination. Annu. Rev. Genet. 50: 393–421. 10.1146/annurev-genet-120215-035243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., 1992. A Short Course in Bacterial Genetics, Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Mori T., Nakamura T., Okazaki N., Furukohri A., Maki H. et al. , 2012. Escherichia coli DinB inhibits replication fork progression without significantly inducing the SOS response. Genes Genet. Syst. 87: 75–87. 10.1266/ggs.87.75 [DOI] [PubMed] [Google Scholar]
- Nicoloff H., Hjort K., Levin B. R., and Andersson D. I., 2019. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4: 504–514. 10.1038/s41564-018-0342-0 [DOI] [PubMed] [Google Scholar]
- Peters J. E., Bartoszyk I. M., Dheer S., and Benson S. A., 1996. Redundant homosexual F transfer facilitates selection-induced reversion of plasmid mutations. J. Bacteriol. 178: 3037–3043. 10.1128/jb.178.11.3037-3043.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder R. G., Fonville N. C., and Rosenberg S. M., 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19: 791–804. 10.1016/j.molcel.2005.07.025 [DOI] [PubMed] [Google Scholar]
- Radicella J. P., Park P. U., and Fox M. S., 1995. Adaptive mutation in Escherichia coli: a role for conjugation. Science 268: 418–420. 10.1126/science.7716545 [DOI] [PubMed] [Google Scholar]
- Rodriguez C., Tompkin J., Hazel J., and Foster P. L., 2002. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J. Bacteriol. 184: 5599–5608. 10.1128/JB.184.20.5599-5608.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., and Foster P. L., 1999. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 6862–6867. 10.1073/pnas.96.12.6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S. M., Longerich S., Gee P., and Harris R. S., 1994. Adaptive mutation by deletions in small mononucleotide repeats. Science 265: 405–407. 10.1126/science.8023163 [DOI] [PubMed] [Google Scholar]
- Rosenberg S. M., Harris R. S., and Torkelson J., 1995. Molecular handles on adaptive mutation. Mol. Microbiol. 18: 185–189. 10.1111/j.1365-2958.1995.mmi_18020185.x [DOI] [PubMed] [Google Scholar]
- Roth J. R., Kofoid E., Roth F. P., Berg O. G., Seger J. et al. , 2003. Regulating general mutation rates: examination of the hypermutable state model for Cairnisian adaptive mutation. Genetics 163: 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Kugelberg E., Reams A. B., Kofoid E., and Andersson D. I., 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Microbiol. 60: 477–501. 10.1146/annurev.micro.60.080805.142045 [DOI] [PubMed] [Google Scholar]
- Sakofsky C. J., and Malkova A., 2017. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 52: 395–413. 10.1080/10409238.2017.1314444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano E., Maisnier-Patin S., Aboubechara J. P., Quinones-Soto S., and Roth J. R., 2014. Plasmid copy number underlies adaptive mutability in bacteria. Genetics 198: 919–933. 10.1534/genetics.114.170068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T., Sherwood S. W., Hill A. B., and Johnston R. N., 1986. Overreplication and recombination of DNA in higher eukaryotes: potential consequences and biological implications. Proc. Natl. Acad. Sci. USA 83: 2157–2161. 10.1073/pnas.83.7.2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Gibson J. L., Darrow M. C., Gonzalez C., and Rosenberg S. M., 2011a Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 108: 13659–13664. 10.1073/pnas.1104681108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Ponder R., Gibson J. L., and Rosenberg S. M., 2011b What limits the efficiency of double-strand break-dependent stress-induced mutation in Escherichia coli? J. Mol. Microbiol. Biotechnol. 21: 8–19. 10.1159/000335354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta E. S., Bunny K. L., Kugelberg E., Kofoid E., Andersson D. I. et al. , 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100: 12847–12852. 10.1073/pnas.1735464100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouffske K., Aguilar-Rodriguez J., Sniegowski P., and Wagner A., 2018. High mutation rates limit evolutionary adaptation in Escherichia coli. PLoS Genet. 14: e1007324 10.1371/journal.pgen.1007324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G. R., Dower W. J., Schimke R. T., Brown R. E., and Kerr I. M., 1979. 2–5A synthetase: assay, distribution and variation with growth or hormone status. Nature 278: 471–473. 10.1038/278471a0 [DOI] [PubMed] [Google Scholar]
- Torkelson J., Harris R. S., Lombardo M. J., Nagendran J., Thulin C. et al. , 1997. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 16: 3303–3311. 10.1093/emboj/16.11.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K., Furukohri A., Shinozaki Y., Mori T., Ogawara D. et al. , 2008. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol. Microbiol. 70: 608–622. 10.1111/j.1365-2958.2008.06423.x [DOI] [PubMed] [Google Scholar]
- Vogel F., 1972. Non-randomness of base replacement in point mutation. J. Mol. Evol. 1: 334–367. 10.1007/BF01653962 [DOI] [PubMed] [Google Scholar]
- Wagner J., and Nohmi T., 2000. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182: 4587–4595. 10.1128/JB.182.16.4587-4595.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J., Gruz P., Kim S. R., Yamada M., Matsui K. et al. , 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4: 281–286. 10.1016/S1097-2765(00)80376-7 [DOI] [PubMed] [Google Scholar]
- Xia J., Chiu L. Y., Nehring R. B., Bravo Nunez M. A., Mei Q. et al. , 2019. Bacteria-to-Human protein networks reveal origins of endogenous DNA damage. Cell 176: 127–143.e24. 10.1016/j.cell.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi I., Maisnier-Patin S., and Roth J. R., 2018. Selection-enhanced mutagenesis of lac genes is due to their coamplification with dinB encoding an error-prone DNA polymerase. Genetics 208: 1009–1021. 10.1534/genetics.117.300409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains used in this study are available upon request. DNA sequencing data are available at NCBI under BioProject accession number PRJNA590198. Supplemental Material, Table S1 available at figshare contains all mutations of the Lac+ revertants. Supplemental material available at figshare: https://doi.org/10.25386/genetics.10848680.