Abstract
Bone resorption is a severe consequence of inflammatory diseases associated with osteolysis, such as rheumatoid arthritis (RA), often leading to disability in patients. In physiological conditions, the differentiation of bone-resorbing osteoclasts is delicately regulated by the balance between osteoclastogenic and anti-osteoclastogenic mechanisms. Inflammation has complex impact on osteoclastogenesis and bone destruction, and the underlying mechanisms of which, especially feedback inhibition, are underexplored. Here we identify a novel regulatory network mediated by RBP-J/NFATc1-miR182 in TNF-induced osteoclastogenesis and inflammatory bone resorption. This network includes negative regulator RBP-J and positive regulators, NFATc1 and miR182, of osteoclast differentiation. In this network, miR182 is a direct target of both RBP-J and NFATc1. RBP-J represses while NFATc1 activates miR182 expression through binding to specific open chromatin regions in the miR182 promoter. Inhibition of miR182 by RBP-J servers as a critical mechanism that limits TNF-induced osteoclast differentiation and inflammatory bone resorption. Inflammation, such as that which occurs in RA, shifts the expression levels of the components in this network mediated by RBP-J/NFATc1-miR182-FoxO3/PKR (previously identified miR182 targets) towards more osteoclastogenic, rather than healthy, conditions. Treatment with TNF inhibitors in RA patients reverses the expression changes of the network components and osteoclastogenic potential. Thus, this network controls the balance between activating and repressive signals that determine the extent of osteoclastogenesis. These findings collectively highlight the biological significance and translational implication of this newly identified intrinsic regulatory network in inflammatory osteoclastogenesis and osteolysis.
Keywords: Inflammatory bone resorption, osteoclast, Rheumatoid arthritis, osteoimmunology
INTRODUCTION
The inflammatory bone destruction associated with multiple diseases, such as rheumatoid arthritis (RA), psoriatic arthritis and periodontitis, is a major cause of morbidity and disability in patients. Osteoclasts function as crucial pathogenic cells leading to excessive bone resorption in these inflammatory settings (1–5). The extent of osteoclastogenesis is delicately modulated and determined by the balance between osteoclastogenic and anti-osteoclastogenic mechanisms in physiological conditions (6, 7). In response to the master osteoclastogenic cytokine RANKL, a broad range of signaling cascades mediated by canonical and non-canonical NF-kB pathways, mitogen-activated kinase (MAPK) pathways and calcium signaling, induce positive regulators, such as nuclear factor of activated T cells c1 (NFATc1), c-Fos and B lymphocyte-induced maturation protein-1 (Blimp1), to drive osteoclast differentiation. Meanwhile, intrinsic anti-osteoclastogenic mechanisms mediated by negative regulators, such as interferon regulatory factor (IRF8), v-maf musculoaponeurotic fibrosarcoma oncogene family protein B (MafB) and differentially expressed in FDCP 6 homolog (Def6), provide “a braking system” to prevent excessive osteoclastogenesis and bone resorption (6, 8–16). These mechanisms can interact with each other and form regulatory networks to coordinately control osteoclastogenesis. Identification of these networks will broaden and deepen our understanding of the mechanisms that control osteoclastogenesis and bone metabolism, and help establish optimal and potential novel therapeutic strategies. A great amount of work has focused on individual factors. However, little attention was paid to the osteoclastic regulatory networks, especially in inflammatory conditions.
Tumor necrosis factor- α (TNFα) is an inflammatory cytokine important for immunity and inflammation. The resounding success of TNF blockade therapy has demonstrated a key role for TNFα in the pathogenesis of autoimmune/inflammatory diseases such as RA. TNFα plays a major role, mostly in synergy with RANKL, in promoting pathologic osteoclastogenesis and bone resorption in inflammatory diseases (1, 2, 13, 17–20). TNFα also executes its indirect osteoclastogenic effect through augmentation of RANK expression on osteoclast precursors, induction of M-CSF and RANKL expression, and suppression of OPG (17, 21–24). Interestingly, although TNFα induces a similar signal transduction cascade to that of RANKL, the direct osteoclastogenic capacity of TNFα alone on osteoclast precursors is dramatically weaker than that of RANKL, which has been demonstrated by both genetic evidence and osteoclastogenesis in human CD14 positive cells (25–27). The mechanisms that restrain TNF-induced osteoclastogenesis are much less understood than those that promote osteoclastogenesis in response to RANKL (13, 28).
Recombination signal binding protein for immunoglobulin kappa J region (RBP-J), a nuclear DNA-binding protein, can function as either a transcriptional repressor or activator depending on the partner proteins with which it interacts (29). RBP-J is originally identified and best known as a master transcription factor in the canonical Notch signaling pathway (29). Accumulating evidence now shows that RBP-J acts as a central transcription factor that receives inputs not only from canonical Notch signaling but also from other pathways dependent on cell types, such as the Wnt/β-catenin (30), NF-κB (31, 32), TAK1 (33), TLR (34, 35), TNFα (11), and ITAM signaling pathways (10), and is also targeted by viral proteins (36) and other cellular proteins (37, 38). RBP-J regulates diverse cellular functions, such as differentiation, proliferation, survival and development (29). In myeloid lineage cells, RBP-J has been implicated in inflammatory macrophage activation and function (34, 35, 39), dendritic cell (DC) differentiation, and maintenance of CD8− DC populations (40, 41). Upon interaction with a variety of partners and signaling pathways in different scenarios, the regulatory networks mediated by RBP-J are diverse and context-dependent.
Human genetic evidence revealed the association of RBPJ allelic polymorphisms with RA (42–44). Notably, we found that RBP-J expression level was suppressed in the synovial fluid macrophages/osteoclast precursors isolated from RA patients (10), supporting a pathological relevance of RBP-J to RA. Furthermore, our recent studies have identified RBP-J as a key transcriptional repressor for osteoclastogenesis, especially in response to TNF-induced osteoclast formation and inflammatory bone resorption (11). However, the molecular mechanisms by which RBP-J suppresses inflammatory osteoclastogenesis and bone resorption are far from understood.
Our genome-wide profiling of miRNAs shows that RBP-J significantly suppresses miR182, which is a TNF-inducible miRNA and a critical osteoclastogenic driver in bone remodeling (45, 46). In the present study, we identify a novel and unique regulatory network including the positive (NFATc1) and negative (RBP-J) upstream players that orchestrate miR182 transcription and function to coordinately control TNF-induced osteoclastogenesis. Inflammatory conditions, such as that of RA, disrupt the balance between the positive and inhibitory mechanisms in this network, leading towards excessive bone erosion. This study highlights the importance of a newly identified network in TNF-mediated osteoclastogenesis and shed insights into the translational implications of treating the unbalanced network in osteolytic diseases.
MATERIALS AND METHODS
Animal study
RbpjΔM/ΔM (Rbpjflox/floxLysMcre(+)) and Mir182ΔM/ΔM (Mir182flox/floxLysMcre(+)) Mice have been described previously (11, 46). Mir182ΔM/ΔMRbpjΔM/ΔM double knockout (dKO) mice were generated by crossing Mir182flox/+LysMcre(+) with Rbpjflox/+LysMcre(+) mice. Gender- and age-matched mice with LysMcre(+) genotype were used as wild type controls (referred to as Ctrl) for experiments. Myeloid-specific miR-182 overexpression mice (referred to as Mir182mTg) were described previously (46). These mice were generated by crossing LSL (LoxP-Stop-LoxP)-Mir182 mice (47), in which the mouse Mir182 cDNA was knocked into the ubiquitously expressed Rosa26 locus preceded by a STOP fragment flanked by loxP sites, with LysMcre(+) mice (The Jackson Laboratory) on the C57BL/6 background. Gender- and age-matched Mir182mTg mice and their littermates LysMcre(+) mice as wild type controls (referred to as Ctrl) were used. Nfatc1 knockout mice (Nfatc1flox/floxMx1cre mice, referred to as Nfatc1 KO) were described previously (48). The bone marrow isolated from gender- and age-matched Nfatc1 KO and wild type Nfatc1flox/flox mice used in the experiments were kindly gifted by Dr. Julia Charles (Brigham and Women’s Hospital, Boston). All mice used in this study have been backcrossed to the C57BL/6 background for at least 10 generations. All animal procedures were approved by the Hospital for Special Surgery Institutional Animal Care and Use Committee (IACUC), and Weill Cornell Medical College IACUC.
TNF-induced supracalvarial osteolysis model
TNF-induced supracalvarial osteolysis was performed as previously described (10). TNFα was administrated daily at the dose of 75 μg/kg to the calvarial periosteum of age and gender-matched mice for five consecutive days before the mice were sacrificed. The calvarial bones were subjected to μCT analysis, sectioning, TRAP staining, and histological analysis.
K/BxN Serum Transfer-Induced Arthritis
K/BxN Serum Transfer-Induced Arthritis was performed as previously described (46). K/BxN serum pools were prepared, and arthritis was induced by intraperitoneal injection of 100 μl of K/BxN serum to the female mice on days 0 and 2. The development of arthritis was monitored by measuring the thickness of wrist and ankle joints with digital slide caliper (Bel-Art Products). For each animal, joint thickness was calculated as the sum of the measurements of both wrists and both ankles. Joint thickness was represented as the average for each group. Mice were sacrificed on day 10 and serum and paws were collected. Hind paws were subjected to sectioning, TRAP staining and histological analysis.
Micro-computed tomography (μCT) analysis
Calvarial bones were fixed in 10% buffered formalin and scanned by using a Scanco μCT-35 scanner (SCANCOMedical). 3D images of calvarial bones were reconstructed by using Scanco software according to the manufacturer’s instructions. Calvarial bones were subjected to sectioning, TRAP staining and histological analysis. The Osteomeasure software was used for bone histomorphometry using standard procedures according to the program’s instruction.
Reagents
Murine or human M-CSF, murine or human TNFα were purchased from PeproTech. FK506 was purchased from invitrogen and Cyclosporin A (CsA) was purchased from Millipore.
Cell culture
To obtain bone marrow macrophages (BMMs), mouse bone marrow cells were harvested from tibiae and femora of age and gender-matched mutant and control mice and cultured for 3 days in α-MEM medium (Thermo Fisher Scientific) with 10% FBS (Atlanta Biologicals), glutamine (2.4 mM, Thermo Fisher Scientific), Penicillin-Streptomycin (Thermo Fisher Scientific) and L929 supernatant (condition medium, CM), which contained the equivalent of 20 ng/ml of rM-CSF and was used as a source of M-CSF (10). The attached BMMs were scraped, seeded at a density of 4.5 × 104/cm2, and cultured in α-MEM medium with 10% FBS, 1% glutamine and CM for overnight. Except where stated, the cells were then treated without or with optimized concentrations of TNFα (40 ng/ml) in the presence of CM for times indicated in the figure legends. Culture media were exchanged every three days. Human osteoclast cultures were performed as described previously (46). Briefly, peripheral blood mononuclear cells (PBMCs) from whole blood of healthy volunteers or RA patients were isolated by density gradient centrifugation using Ficoll (Invitrogen Life Technologies, Carlsbad, CA). CD14(+) cells were purified from fresh PBMCs using anti-CD14 magnetic beads (Miltenyi Biotec, Auburn, CA) as recommended by the manufacturer. Human CD14(+) monocytes were cultured in α-MEM medium with 10% FBS in the presence of M-CSF (20 ng/ml; PeproTech, Rocky Hill, NJ) for 2 days to obtain monocyte-derived macrophages, which were further cultured with RANKL for osteoclast differentiation. The RA samples in Fig. 7 were described previously (46). Briefly, the RA CD14(+) PBMCs were from RA patients (age ≥ 18 and <70 years) who fulfilled American College of Rheumatology (ACR) 2010 RA classification criteria with disease duration < 5 years and were under TNFi therapy for the first time (Enbrel, 25mg weekly). Experiments with human cells were approved by the Hospital for Special Surgery Institutional Review Board. Informed consent (PBMC collection) was obtained from all RA patients. TRAP staining was performed with an acid phosphatase leukocyte diagnostic kit (Sigma-Aldrich) in accordance with the manufacturer’s instructions. TRAP-positive multinucleated cells containing 3 or more nuclei were counted as osteoclasts. The cell counts were normalized for size in the quantification. Murine macrophage cell line, RAW264.7 cells were purchased from the American Type Culture Collection. RAW264.7 cells were cultured in α-MEM medium (Thermo Fisher Scientific) with 10% FBS (Atlanta Biologicals), glutamine (2.4 mM, Thermo Fisher Scientific) and Penicillin-Streptomycin (Thermo Fisher Scientific). The cell line was routinely tested for mycoplasma contamination.
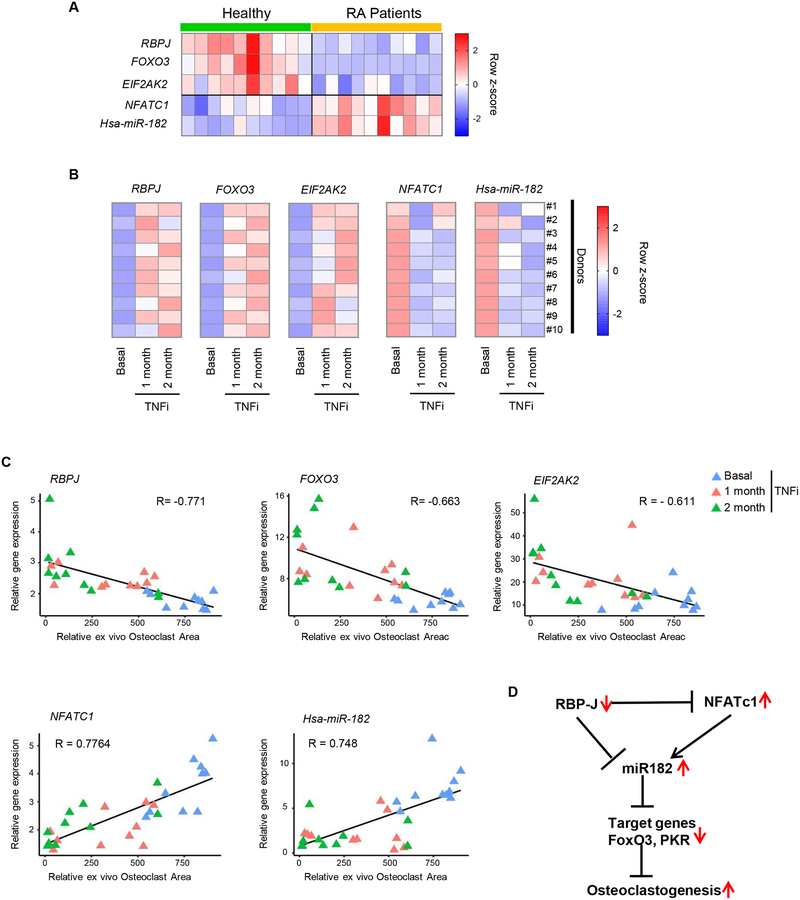
Figure 7. The RBP-J/NFATc1-miR182-PKR/FoxO3 network is significantly correlated with RA.
A. Heat map showing gene expression of RBPJ, FOXO3, EIF2AK2, NFATC1, Hsa-miR-182 in human CD14(+) PBMCs from healthy donors and RA patients. n = 10/group. B. Heat maps showing gene expressions of RBPJ, FOXO3, EIF2AK2, NFATC1, Hsa-miR-182 in human CD14(+) PBMCs from RA patients before (basal) and after TNFi (Enbrel) for 1 and 2 months. n = 10/group. C. Scatter plots showing that the relative TRAP-positive osteoclast area obtained from RA CD14(+) PBMC cell cultures has a significant negative correlation with RBP-J (upper left), FOXO3 (upper middle) or EIF2AK2 (upper right) expression, and a significant positive correlation with NFATC1 (lower left) or Hsa-mir-182 (lower right) expression. Each triangle represents an RA patient in the indicated conditions. Pearson’s R = −0.771 (RBPJ), −0.663 (FOXO3), - 0.611 (EIF2AK2), 0.7764 (NFATC1) and 0.748 (Hsa-miR-182). p value = 0.0000108 (RBPJ), 0.0000658 (FOXO3), 0.000332 (EIF2AK2), 0.000000456 (NFATC1) or 0.00000388 (Hsa-miR-182). D. A model showing the expression changes of the key components of the RBP-J/NFATc1-miR182 network under RA inflammatory conditions, in which the negative regulators RBP-J, FOXO3 and PKR are downregulated while positive osteoclastogenic factors NFATC1 and miR-182 are upregulated, leading to an overall enhanced osteoclastogenesis in RA.
Reverse transcription and real-time PCR
For quantification of mRNA, reverse transcription and real-time PCR were performed as previously described (45). DNA-free RNA was obtained with the RNeasy MiniKit (Qiagen, Valencia, CA) with DNase treatment, and 1 ug of total RNA was reverse-transcribed with random hexamers and MMLV-Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. Real-time PCR was done in triplicate with the QuantStudio 5 Real-time PCR system and Fast SYBR® Green Master Mix (Thermo Fisher Scientific) with 500 nM primers. mRNA amounts were normalized relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. When reverse transcription was omitted, threshold cycle number increased by at least ten, signifying lack of genomic DNA contamination or nonspecific amplification; the generation of only the correct size amplification products was confirmed with agarose gel electrophoresis. The primers for real-time PCR were as follows: For mouse primers, Nfatc1: 5’-CCCGTCACATTCTGGTCCAT-3’ and 5’-CAAGTAACCGTGTAGCTCCACAA-3’; Prdm1: 5’-TTCTTGTGTGGTATTGTCGGGACTT-3’ and 5’-TTGGGGACACTCTTTGGGTAGAGTT-3’; Acp5: 5’-ACGGCTACTTGCGGTTTC-3’ and 5’-TCCTTGGGAGGCTGGTC-3’; Ctsk: 5’-AAGATATTGGTGGCTTTGG-3’ and 5’-ATCGCTGCGTCCCTCT-3’; Acp5: 5’-ACGGCTACTTGCGGTTTC-3’ and 5’-TCCTTGGGAGGCTGGTC-3’; Calcr: 5’-ACATGATCCAGTTCACCAGGCAGA-3’and 5’-AGGTTCTTGGTGACCTCCCAACTT-3’; Atp6v0d2: 5’- GAAGCTGTCAACATTGCAGA-3’and 5’- TCACCGTGATCCTTGCAGAAT-3’and Gapdh: 5’-ATCAAGAAGGTGGTGAAGCA-3’and 5’-AGACAACCTGGTCCTCAGTGT-3’. For human primers, RBPJ: 5’-TTCAAAAACCCCGTTGTCTC-3’ and 5’-CAAAACCAACCAACCAACC-3’; NFATC1: 5’-AAAGACGCAGAAACGACG-3’ and 5’-TCTCACTAACGGGACATCAC-3’; FOXO3: 5’- ACTTCAAGGATAAGGGCGACAG-3’ and 5’- TATGCAGTGACAGGTTGTGC-3’; EIF2AK2: 5’-AATGCCGCAGCCAAATTAGC-3’ and 5’-AGGCCTATGTAATTCCCCATGG-3’; GAPDH: 5’-ATCAAGAAGGTGGTGAAGCA-3’ and 5’- GTCGCTGTTGAAGTCAGAGGA-3’.
For quantification of microRNA, total RNA was isolated, and the small RNA fraction was enriched with the mirVana miRNA Isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For quantitative RT-PCR analysis of miRNA, cDNA was prepared from small RNAs with the TaqMan microRNA Reverse Transcription Kit (Thermo Fisher Scientific). TaqMan MicroRNA assays were used according to the manufacturer’s recommendations (Thermo Fisher Scientific) for real-time PCR. The TaqMan U6 snRNA assay (Thermo Fisher Scientific) was used for normalization of microRNA expression values.
Immunoblot analysis
Total cell extracts were obtained using lysis buffer containing 150 mM Tris-HCl (pH 6.8), 6% SDS, 30% glycerol, and 0.03% Bromophenol Blue; 10% 2-ME was added immediately before harvesting cells. Cell lysates were fractionated on 7.5% SDS-PAGE, transferred to Immobilon-P membranes (Millipore), and incubated with specific antibodies. Western Lightning plus-ECL (PerkinElmer) was used for detection. NFATc1 antibody (556602, 1:1000) was from BD Biosciences; Blimp1 (sc-47732, 1:1000), IRF8 (sc-6058, 1:1000) and GAPDH (sc-25778, 1:3000) antibodies were from Santa Cruz Biotechnology.
Luciferase reporter assay
A 2448 bp DNA fragment of the promoter of mmu-miR182 was cloned into the pGL3 basic Luciferase Reporter Vector (E1751, Promega). The reporter plasmid containing miR182 promoter were co-transfected into RAW264.7cells with CMV-renilla luciferase reporter (as an internal transfection control), together with RBP-J expression vector or a corresponding empty vector as a control using TransIT-TKO transfection reagent (Mirus), in accordance with the manufacturer’s instructions. Forty-eight hours after transfection and TNFα stimulation, the cells were lysed with passive lysis buffer (Promega), and firefly and renilla luciferase activities were measured using the Dual-luciferase reporter assay system (Promega).
Formaldehyde-assisted isolation of regulatory elements (FAIRE) assay
To identify and quantify chromatin compaction/accessibility, FAIRE assay was performed as previously described (49). Cells (10×106 cells per condition) were fixed with 1% formaldehyde for 10 min at room temperature. The reaction was quenched by the addition of 0.125 M glycine for 5 min. Then the cells were washed with ice-cold PBS twice. Fixed cells were lysed in buffer LB1 (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and protease inhibitors) for 10 min on ice. The nuclei were pelleted, resuspended in buffer LB2 (10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitors) and incubated for 10 min on ice. After centrifuge, the nuclei were lysed in buffer LB3 (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, and protease inhibitors). Then the fixed chromatin was sonicated by using a Bioruptor Pico device (Diagenode) for 6 cycles of 30 sec on/30 sec off. Ten percent of sonicated nuclear lysates was taken as input. DNA was extracted by adding an equal volume of phenol-chloroform solution and purified by MinElute PCR Purification Kit (Qiagen). Chromatin accessibility was determined by qPCR and normalized relative to total input. The qPCR primers used in the FAIRE assay were as follows: miR-182 promoter locus 1: 5’-GTGTTGGTATGGCCCAGTTC-3’ and 5’-AGGAGAACCAGAAAGCTATGGC-3’; miR-182 promoter locus 2: 5’-TGCCACTCTCTTCCTTGGTTAC-3’ and 5’-TGCTCTCAAAGGCACTGTACC-3’; miR-182 promoter locus 3: 5’-AGGGCTTGAGGAGGTTTTACAC-3’ and 5’- AGCCAGACCAGTAAGCCTATG-3’; miR-182 promoter (TSS): 5’-TGACATTCCCCAGAGCCTAAAG-3’ and 5’-TGTGGCTTGACAAGGAAGTG-3’; CtsK promoter (TSS): 5’- ACGTTGGAAATGGTGCAGAG-3’ and 5’- ACAGCCCTAGTTGTCTCCATTC-3’;β-Globin locus: 5’-ACATGTGTGTGGGAGGAGTG-3’ and 5’-GGACAATCCCTGAAAAAGCA-3’; Nfatc1#1: 5’-TTGGAATCCTGTAGCAGAAGGC-3’ and 5’- AACAGATGGAGATGCTTGCG-3’; Nfatc1#2: 5’-TAGAACTGGGCCATACCAACAC-3’ and 5’- TAACCAAAGCAGTCCTCAGACC-3’; Nfatc1 promoter (−800bp of TSS): 5’- CCGGGACGCCCATGCAATCTGTTAGTAATT-3’ and 5’- GCGGGTGCCCTGAGAAAGCTACTCTCCCTT-3’.
ChIP assay
Cells (10×106 cells per condition) were crosslinked for 10 min at room temperature with 0.8% formaldehyde solution followed by 5 min quenching with 125 mM glycine. Cells were pelleted at 4°C and washed with ice-cold PBS twice. The crosslinked cells were lysed with buffer LB1 with protease inhibitors on ice for 10 min. The nuclei were pelleted, resuspended in buffer LB2 and incubated for 10 min on ice. The lysis samples were resuspended and sonicated in buffer LB3 using a Bioruptor Pico device (Diagenode) for 6 cycles of 30 sec on/30 sec off. After sonication, samples were centrifuged at 12,000 rpm for 10 minutes at 4°C and 10% of sonicated cell lysates was taken as input. The chromatin lysates were incubated with Protein A/G magnetic beads (Themofisher) with 5 μg of the appropriate antibody overnight at 4°C. RBP-J antibody (#5313) was from Cell Signaling Technology. NFATc1 antibody (#556602) was from BD Biosciences. After overnight incubation, antibody-bound magnetic beads were washed twice with Low salt buffer, twice with High salt buffer, once with LiCl wash buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 1% NP-40), and once with TE with 50 mM NaCl. Cross-links were reversed by overnight incubation at 65°C. Input and ChIP DNA was treated with RNase A and Proteinase K to remove RNAs and proteins. DNA was purified with MinElute PCR Purification Kit (Qiagen). DNA was analyzed by qPCR and normalized relative to total input. The qPCR primers used in the ChIP assay: miR-182 promoter locus 1: 5’-GTGTTGGTATGGCCCAGTTC-3’ and 5’-AGGAGAACCAGAAAGCTATGGC-3’; miR-182 promoter locus 2: 5’-TGCCACTCTCTTCCTTGGTTAC-3’ and 5’-TGCTCTCAAAGGCACTGTACC-3’; miR-182 promoter locus 3: 5’-AGGGCTTGAGGAGGTTTTACAC-3’ and 5’- AGCCAGACCAGTAAGCCTATG-3’; β-Globin locus: 5’-ACATGTGTGTGGGAGGAGTG-3’ and 5’- GGACAATCCCTGAAAAAGCA-3’; Hes1 promoter: 5’-TCTTCCTCCCATTGGCTGAAAG-3’ and 5’-CCCTGGCGGCCTCTATATATATC-3’; Nfatc1#1: 5’-TTGGAATCCTGTAGCAGAAGGC-3’ and 5’- AACAGATGGAGATGCTTGCG-3’; Nfatc1#2: 5’-TAGAACTGGGCCATACCAACAC-3’ and 5’- TAACCAAAGCAGTCCTCAGACC-3’.
Statistical analysis
Statistical analysis was performed using Graphpad Prism® software. Two-tailed Student’s t test was applied when there were only two groups of samples. In the case of more than two groups of samples, one-way ANOVA was used with one condition. ANOVA analysis was followed by post hoc Turkey’s multiple comparisons. p < 0.05 was taken as statistically significant; *p value < 0.05 and **p value < 0.01. The data displayed normal distribution. The estimated variance was similar between experimental groups. Data are presented as the mean ± SD or ±SEM as indicated in the figure legends.
RESULTS
Identification of miR182 as a direct target of RBP-J
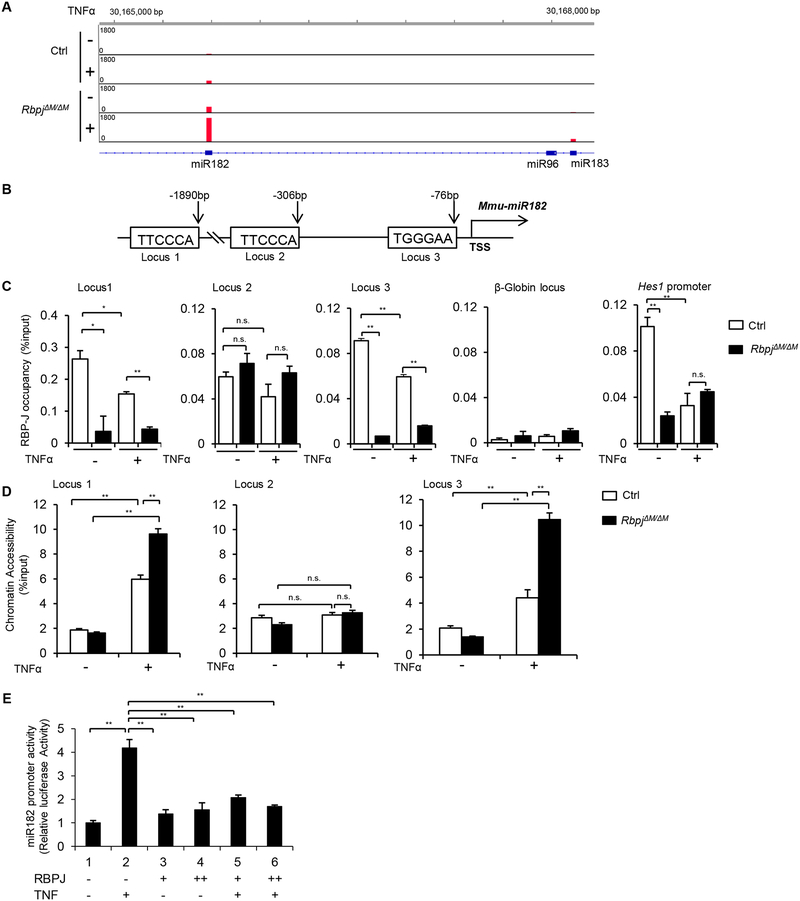
RBP-J is a key repressor of TNF-induced osteoclast differentiation and inflammatory bone resorption (10, 11, 28). To identify downstream targets mediated by RBP-J, we applied a genome-wide, high throughput sequencing of microRNAs (miRNA-seq) to perform global profiling of miRNA expressions in response to TNFα stimulation during osteoclast differentiation in wild-type control and RBP-J-deficient bone marrow-derived macrophages (45). We spotted miR182 that is significantly induced by TNF stimulation but suppressed by RBP-J ((45) and miRNA-seq aligned reads at murine miR182/96/183 locus shown in Fig. 1A). With consideration of the important role for miR182 in promoting osteoclastogenesis in bone remodeling (46), we first asked whether RBP-J directly regulates miR182 expression. Following this line, in silico analysis of the Mir182 promoter region revealed three highly matched RBP-J binding sites located 1.9kb (TTCCCA, locus 1), 306bp (TTCCCA, locus 2) and 76bp (TGGGAA, locus 3) upstream of its transcription start site (TSS) (Fig. 1B). We then tested RBP-J binding states at these loci in the bone marrow derived macrophages (BMMs) from the control or myeloid specific RBP-J deficient (RbpjΔM/ΔM (Rbpjf/f;LysMcre), (11)) mice during TNF induced osteoclastogenesis. Chromatin immunoprecipitation (ChIP) assays showed RBP-J binding signals at locus 1 and 3 of miR182 promoter region, as well as at the promoter of canonical RBP-J target Hes1 (as a positive control), but not at locus 2 of miR182 promoter or the transcriptionally silent β-globin gene locus (as a negative control) (Fig. 1C). These RBP-J binding signals diminished in the RBP-J deficient cells (Fig. 1C). Consistent with the finding that TNFα induces miR182 expression, the RBP-J occupancy level at locus 1 and 3 of miR182 promoter region were reduced by TNF stimulation (Fig. 1C), supporting that RBP-J is a transcriptional inhibitor of miR182 expression. To determine whether the RBP-J binding regions possess open chromatin feature, we performed formaldehyde-assisted isolation of regulatory elements (FAIRE) assay, which identifies nucleosome-depleted regions and active regulatory sequences that are often associated with regulatory factor binding. Indeed, FAIRE signals were induced by TNFα, and were observed at locus 1 and 3, but not 2, of the miR182 promoter region, which correspond to RBP-J binding sites. RBP-J deficiency further enhanced the extent of the open chromatin states at these regions (Fig. 1D), indicating that RBP-J binding directs local chromatin towards a closed and inactive structure, which supports RBP-J as a transcriptional repressor of miR182 expression. Lack of RBP-J increases the expression of miR182 and osteoclastogenic marker genes, such as Cathepsin K (CtsK) (11, 45). In parallel to the gene expression, the enhanced open chromatin states indicated by FAIRE signals were also observed at the TSS of miR182 or CtsK promoter in the absence of RBP-J, but not at the transcriptionally silent β-globin gene locus (Suppl. Fig.1). To determine the functional significance of RBP-J binding on miR182 transcription, we performed miR182 promoter reporter assay. As shown in Fig. 1E, TNFα-induced miR182 promoter activity, which was strikingly suppressed by RBP-J expression. These results collectively demonstrate that miR182 is a new target of RBP-J, which acts as a transcriptional repressor to inhibit miR182 expression.
Figure 1. RBP-J directly targets miR182 and represses its expression.
A. miRNA-seq aligned reads at murine miR182/96/183 locus displayed by Integrative Genomics Viewer (IGV). The Ctrl or RbpjΔM/ΔM BMMs were stimulated or not with TNFα (40 ng/ml) for 48 h, and miRNAs were extracted and subjected to miRNA-seq. A representative of read signals at the miR182/96/183 locus from two independent miRNA-seq datasets (GSE72966) is shown. B. A diagram depicting three putative RBP-J-binding motifs in the mouse miR-182 promoter region. C. ChIP analysis of RBP-J occupancy at the indicated loci in the Ctrl or RbpjΔM/ΔM BMMs stimulated or not with TNFα (40 ng/ml) for 48 h. D. FAIRE analysis of chromatin accessibility at the miR-182 promoter in the Ctrl or RbpjΔM/ΔM BMMs stimulated or not with TNFα (40 ng/ml) for 48 h. E. miR-182 promoter activities measured from the RAW264.7 cells transfected with the miR182 promoter reporter plasmid and/or RBP-J expression plasmid in the absence or presence of TNFα (40 ng/ml) for 48 h (n=3). Data are mean ± SEM. *p < 0.05; **p < 0.01; n.s., not statistically significant.
miR182 is a positive regulator in TNF-induced osteoclastogenesis
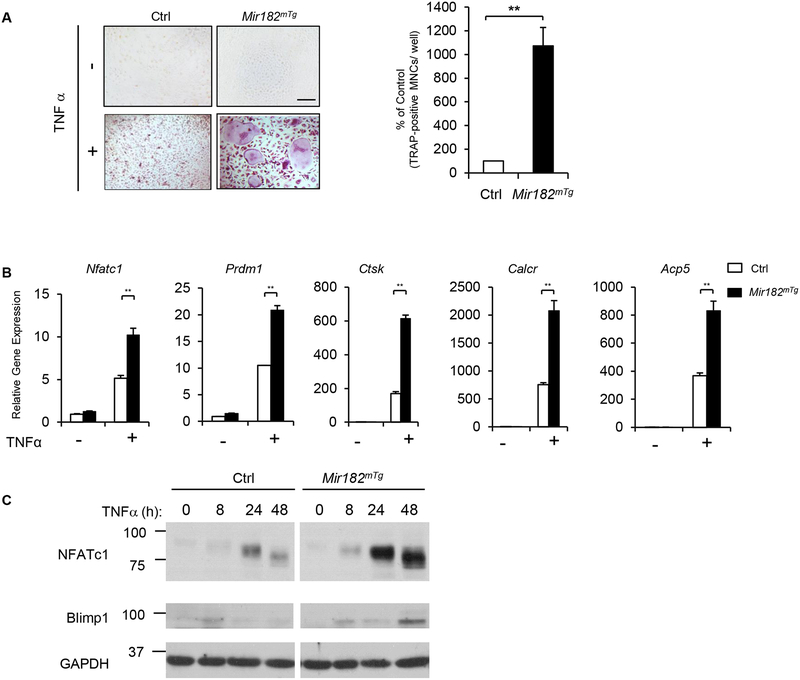
As a downstream target of RBP-J, miR182 expression is constantly suppressed (Fig. 1). To unveil the biological function of miR182, we took a genetic approach by crossing LysMcre mice with LoxP-STOP-LoxP (LSL)–miR-182 mice (47) to generate myeloid lineage conditional Mir182 transgenic (Tg) mice (Mir182mTg), in which Cre expression conditionally induces miR182 overexpression in myeloid lineage osteoclast precursors. The littermate LysMcre mice were used as the controls. We first examined osteoclast differentiation using bone marrow derived macrophages (BMMs) as osteoclast precursors. As expected from our previous data and literature (10, 11, 13, 19, 28), TNFα only induced a low number of small tartrate-resistant acid phosphatase (TRAP)+ multinucleated cells (MNCs) in the control cultures (Fig. 2A). Strikingly, a dramatically greater number of giant osteoclasts were induced to form by TNFα in Mir182mTg cells (Fig. 2A). The enhanced osteoclastogenesis by overexpression of miR182 was further corroborated by the markedly increased expression of osteoclast marker genes in the Mir182mTg cell cultures, such as Ctsk (encoding cathepsin K), Calcr (encoding calcitonin receptor) and Acp5 (encoding TRAP) (Fig. 2B). Moreover, the induction of the positive osteoclastogenic transcription factors, NFATc1 and Blimp1, was significantly enhanced in the Mir182mTg cell cultures (Fig. 2B, C). These results clearly show that miR182 is a positive regulator of TNF-induced osteoclast differentiation. However, in the presence of its upstream repressor RBP-J, both the expression and the osteoclastogenic capacity of miR182 are suppressed.
Figure 2. Overexpression of miR182 promotes TNF-induced osteoclastogenesis.
A. Osteoclast differentiation using BMMs derived from Ctrl and Mir182mTg mice stimulated with or without TNFα (40 ng/mL) for three days. TRAP staining (left panel) was performed and the area of TRAP-positive MNCs (≥ 3 nuclei/cell) per well relative to the control was calculated (right panel). TRAP-positive cells appear red in the photographs. Scale bar: 200 μm. B. Quantitative real-time PCR (qPCR) analysis of mRNA expression of Nfatc1 (encoding NFATc1), Prdm1 (encoding Blimp1), Ctsk (encoding cathepsin K), Calcr (encoding calcitonin receptor) and Acp5 (encoding TRAP) during osteoclastogenesis using BMMs from the Ctrl and Mir182mTg mice treated with or without TNFα for two days. C. Immunoblot analysis of the expression of NFATc1, Blimp1 and IRF8 induced by TNFα at the indicated times. GAPDH was used as a loading control. Data are mean ± SEM. **p < 0.01.
Suppression of miR182 by RBP-J serves as a crucial mechanism restraining TNF-induced osteoclastogenesis and bone resorption
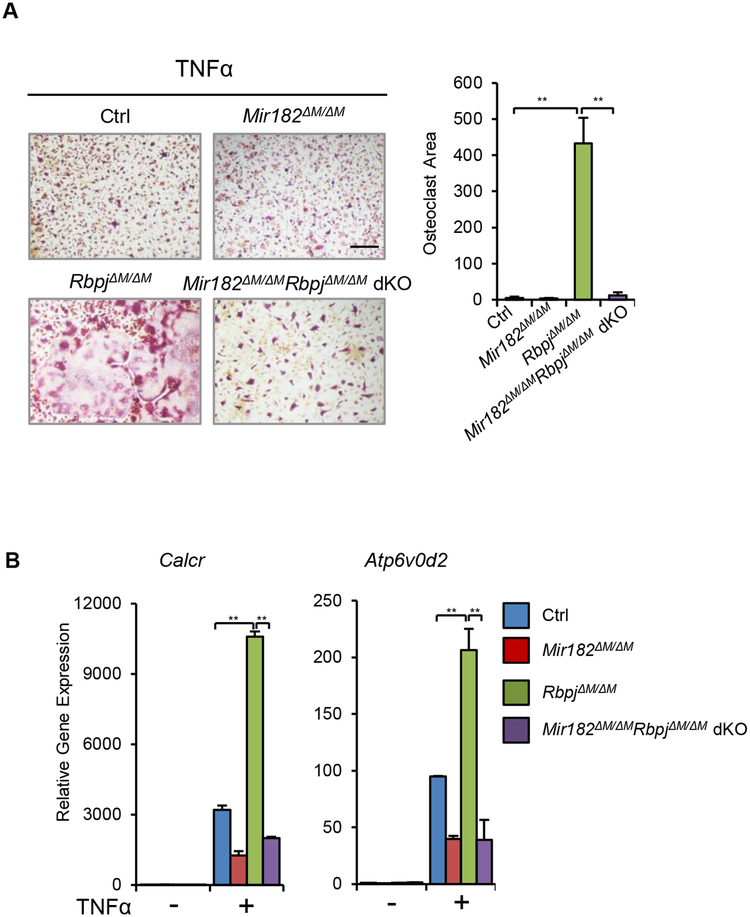
The drastically enhanced TNF-induced osteoclastogenesis phenotype in RbpjΔM/ΔM cells is very similar to that in Mir182mTg cells, suggesting that RBP-J and miR182 may function in an axis in the regulation of osteoclastogenesis. We then sought to provide genetic evidence for the biological importance of the RBP-J-miR-182 axis in vivo in suppressing TNF-induced osteoclastogenesis and bone resorption. We first tested whether miR182 deletion could abolish TNF-enhanced osteoclastogenesis in RBP-J-deficient cells. To this end, we crossed RbpjΔM/ΔM and Mir182ΔM/ΔM mice to generate RbpjΔM/ΔMMir182ΔM/ΔM double knockout mice (dKO), in which both RBP-J and miR-182 were deleted in the myeloid macrophage lineage. As shown in Fig 3A, TNFα induced only a small number of osteoclasts in the control cells, and even less in Mir182ΔM/ΔM cells. RBP-J deficiency strikingly enhanced TNF-induced osteoclast differentiation (Fig. 3A), which is consistent with our previous findings (11). In contrast, there were only a few small TRAP+ osteoclasts formed in the RbpjΔM/ΔMMir182ΔM/ΔM dKO cells (Fig. 3A), indicating that miR182 deletion completely abrogated the enhanced osteoclastogenesis induced by RBP-J deficiency. The expression changes of osteoclast marker genes, such as Calcr and Atp6v0d2 (encoding ATPase H+ transporting v0 subunit d2), in the control, Mir182ΔM/ΔM, RbpjΔM/ΔM and dKO BMM cells in response to TNF are in parallel to and reflect their osteoclast differentiation phenotypes (Fig. 3B). These results demonstrate that miR182 is a key downstream target of RBP-J, responsible for the biological function of RBP-J in osteoclastic inhibition. Suppression of miR182 by RBP-J is required for RBP-J to inhibit TNF-induced osteoclastogenesis.
Figure 3. miR182 deficiency abolishes RBP-J inhibited osteoclastogenesis.
A. Osteoclast differentiation using BMMs derived from Ctrl, Mir182ΔM/ΔM, RbpjΔM/ΔM, and Mir182ΔM/ΔMRbpjΔM/ΔM dKO mice stimulated with TNFα (40 ng/mL) for three days. TRAP staining (left panel) was performed and the area of TRAP-positive MNCs (≥ 3 nuclei/ cell) per well was quantified (right panel). TRAP-positive cells appear red in the photographs. Scale bar: 200 μm. B. qPCR analysis of mRNA expression of Calcr and Atp6v0d2 (encoding V-type proton ATPase subunit d 2) during osteoclastogenesis using BMMs from Ctrl, Mir182ΔM/ΔM, RbpjΔM/ΔM, and Mir182ΔM/ΔMRbpjΔM/ΔM dKO mice stimulated with TNFα (40 ng/mL) for three days. Data are mean ± SEM. **p < 0.01; n.s., not statistically significant.
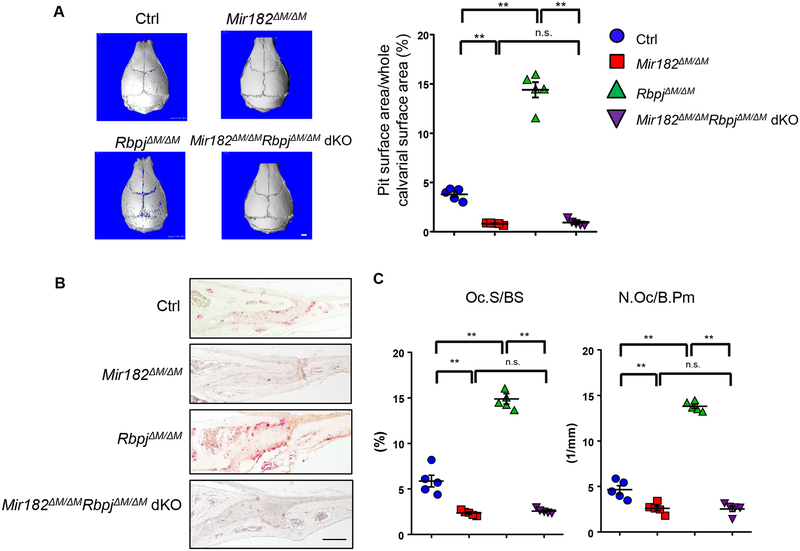
Next, we used a well-established inflammatory calvarial osteolysis model to test the function of RBP-J-miR182 axis in vivo. PBS injection as a negative control did not induce resorptive pit formation on the calvarial bone surfaces (data not shown). Administration of TNFα to the calvarial periosteum resulted in resorptive pit formation on the calvarial bone surface, identifiable by μCT analysis in the control mice, and in the Mir182ΔM/ΔM mice to a lower extent (Fig. 4A). In contrast, a lot of more resorptive pits and osteoclast formation were detected in the RbpjΔM/ΔM mice by μCT analysis (Fig. 4A), and the TRAP staining of the calvarial bone surfaces and histological slices (Fig. 4B, C). Deletion of miR182 in the RbpjΔM/ΔMMir182ΔM/ΔM dKO mice, however, completely abolished these enhanced osteoclast formation and bone erosion resulting from RBP-J deficiency (Fig. 4A, B, C).
Figure 4. miR182 deficiency rescues the TNF-induced bone resorption in RbpjΔM/ΔM mice.
A. μCT images (left panel) and the quantification of pit area (right panel) of the surface of whole calvaria, B. TRAP staining of calvarial histological sections and C. histomorphometric analysis of calvarial slices obtained from Ctrl, Mir182ΔM/ΔM, RbpjΔM/ΔM, and Mir182ΔM/ΔMRbpjΔM/ΔM dKO mice after the application of TNFα daily for five days to the calvarial periosteum. n=5 per group. Oc.S/BS, osteoclast surface per bone surface; N.Oc/B.Pm, number of osteoclasts per bone perimeter. Data are mean ± SEM. **p < 0.01; n.s., not statistically significant. Scale bars: A, 1.0 mm ; B, 200 μm.
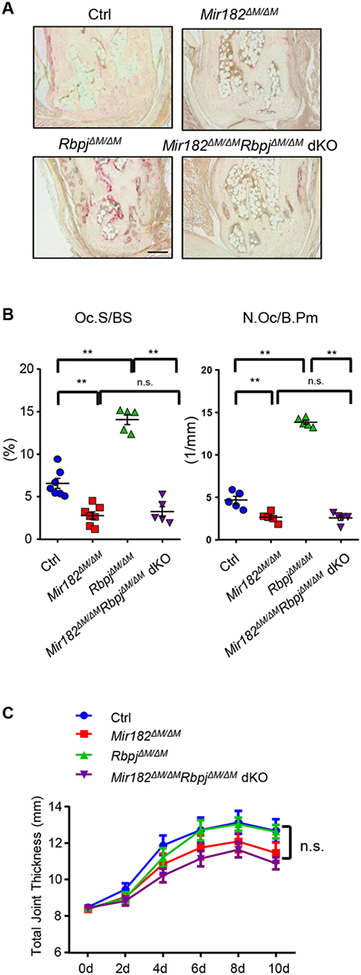
We then used a more pathologically relevant model, K/BxN serum-induced arthritis model (46), which mimics inflammatory peri-articular bone erosion as that which occurs in human RA disease. This mouse model does not need autoimmunity induction and thus allows investigation of inflammatory bone resorption during the inflammatory effector phase of arthritis. Similarly as observed in the calvarial model, miR182 absence completely reversed RBP-J deficiency-enhanced osteoclast formation and joint erosion (Fig. 5A, B). The articular bone in the RbpjΔM/ΔMMir182ΔM/ΔM dKO mice was protected from the inflammation-induced arthritis (Fig. 5A, B). As a note, the clinical course of inflammation, indicated by the joint swelling between the mice with different genotypes, was similar (Fig. 5C), indicating that the RBP-J-miR182 axis does not significantly affect inflammation in this model, but prominently regulates inflammatory osteoclast formation and bone resorption.
Figure 5. miR182 deficiency protects Mir182ΔM/ΔMRbpjΔM/ΔM dKO mice from bone erosion in inflammatory arthritis.
A. TRAP staining of histological sections of tarsal joints (Scale bar: 200 μm) and B. histomorphometric analysis of the tarsal joint sections obtained from the indicated mice that developed K/BxN serum-induced arthritis. ES/BS, erosion surface per bone surface. Oc.S/BS, osteoclast surface per bone surface; N.Oc/B.Pm, number of osteoclasts per bone perimeter. n = 5 per group. Data are mean ± SEM. **p < 0.01; n.s., not statistically significant. C. Time course of joint swelling of inflammatory arthritis developed in Ctrl, Mir182ΔM/ΔM, RbpjΔM/ΔM, and Mir182ΔM/ΔMRbpjΔM/ΔM dKO mice. For each mouse, joint swelling was calculated as the sum of measurements of joint thickness of two wrists and two ankles. n = 5 per group. Joint swelling is represented as the mean ± SD for each group. n.s., not statistically significant.
Collectively, these in vitro and in vivo findings reveal a newly identified RBP-J-miR182 axis that plays a crucial role in the regulation of inflammatory osteoclastogenesis and bone destruction. Suppression of miR-182 by RBP-J is a key intrinsic mechanism that limits TNF-induced osteoclastogenesis and bone resorption.
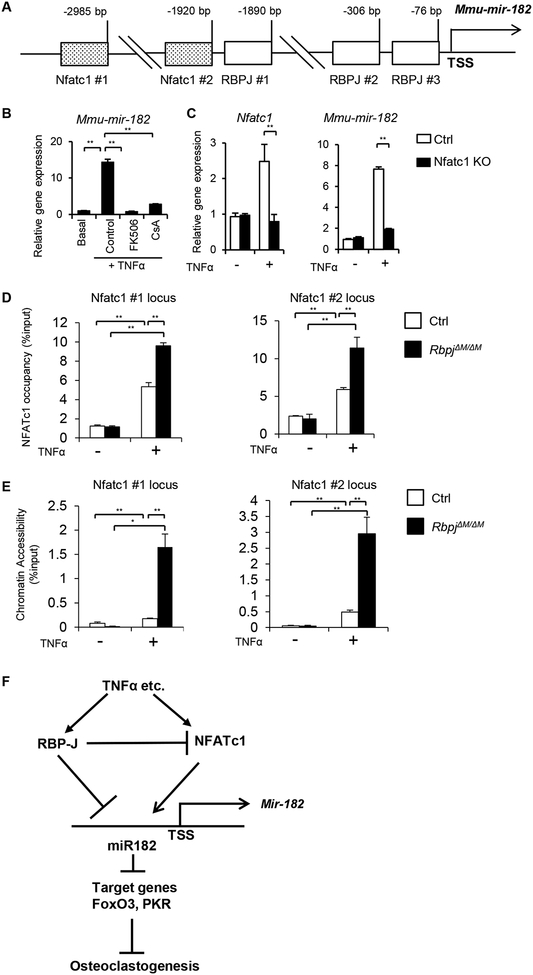
NFATc1 is a key upstream regulator for miR182 induction
It is unclear how miR182 is induced during osteoclast differentiation. In silico analysis of miR182 promoter predicted two NFATc1 binding sites, 2985 bp (site 1) and 1920 bp (site 2) upstream of the TSS of the miR182 gene locus (Fig. 6A). Since NFATc1 is a master osteoclastogenic transcription factor, we asked whether NFATc1 is responsible for miR182 induction. Inhibition of NFATc1 activation using calcineurin inhibitors, FK506 or CsA, strongly suppressed miR182 expression induced by TNFα (Fig. 6B). We further used BMMs isolated from the control or myeloid lineage osteoclast precursor conditional Nfatc1 KO mice (Nfatc1f/f;Mx1cre). As expected, there was no Nfatc1 induction by TNFα in the Nfatc1 KO BMMs (Fig. 6C). miR182 was induced by TNFα in the control cells, but not in the Nfatc1 KO cells (Fig. 6C), indicating that miR182 induction by TNFα is dependent on NFATc1. FAIRE analysis of the miR182 promoter region showed that TNFα relaxed chromatin states at the two NFATc1 binding sites, which became more open in the absence of RBP-J (Fig. 6D). The open chromatin at the NFATc1 binding sites pointed these loci to be active regulatory elements that are often associated with regulator binding. Indeed, ChIP assay demonstrated that TNFα recruited NFATc1 binding to these two sites at miR182 promoter and RBP-J deficiency furthermore drastically enhanced NFATc1 occupancy (Fig. 6E). Similar binding pattern of NFATc1 was observed at its binding site in NFATc1 promoter region (Suppl. Fig. 2), which is used as a positive control and consistent with literature (50). These data provide evidence for the notion that miR182 is also a direct target of NFATc1, which is a key positive regulator for miR182 induction. TNF-induced miR182 expression requires the presence of NFATc1.
Figure 6. NFATc1 directly targets miR182 and activates its expression.
A. A diagram depicting two putative NFATc1 binding sites and three RBPJ binding motifs in the mouse miR-182 promoter region. B. qPCR analysis of mature mouse miR-182 (mmu-mir-182) expression using BMMs from the WT mice treated with FK506 (10 ug/ml), CsA (10 ug/ml) or the control DMSO vehicle for two days in the presence or absence of TNFα. C. qPCR analysis of mature mouse miR182 (mmu-mir-182) expression using BMMs from the control and Nfatc1 KO mice treated with or without TNFα for two days. D. ChIP analysis of NFATc1 occupancy at the indicated loci in the miR182 promoter in the Ctrl or RbpjΔM/ΔM BMMs stimulated or not with TNFα (40 ng/ml) for 48 h. E. FAIRE analysis of chromatin accessibility at the NFATc1 binding sites in the miR-182 promoter in the Ctrl or RbpjΔM/ΔM BMMs stimulated or not with TNFα (40 ng/ml) for 48 h. F. A model showing a regulatory network, in which RBPJ suppresses the expression of NFATc1 and miR182, miR182 as a direct target receives negative regulatory signals from RBP-J but positive signals from NFATc1, and miR182 further regulates osteoclastogenesis via its targets, PKR and FoxO3. Data are mean ± SEM. **p < 0.01.
Take together of our results and previous findings, we identified a novel and unique regulatory network mediated by the RBP-J/NFATc1-miR182 axis in TNF-induced osteoclastogenesis (Fig. 6F). RBP-J, as a key repressor of inflammatory bone resorption, inhibits the expression and function of NFATc1 (11) and miR182, both of which are positive regulators of osteoclast differentiation. miR182, as a direct target, receives positive signals from NFATc1 but negative inputs from RBP-J. miR182 further functions through its downstream targets, FoxO3 and PKR (45, 46), to regulate osteoclast differentiation. Overall, the key osteoclastogenic and anti-osteoclastogenic regulators in this network coordinately control osteoclastogenesis and bone resorption.
The RBP-J/NFATc1-miR182 regulatory network is significantly correlated with RA
We next asked whether the RBP-J/NFATc1-miR182 regulatory network is involved in human inflammatory diseases associated with bone destruction, such as RA. We compared the expression levels of the network components in the human osteoclast precursor CD14 (+) PBMCs isolated from healthy donors and RA patients, and found that the expression of RBPJ, FOXO3 and EIF2AK2 (encoding PKR) was significantly decreased, while the expression of NFATc1 and miR182 was elevated in RA cells relative to the healthy controls (Fig. 7A). These results indicate that the anti-osteoclastic regulators (RBP-J, FOXO3 and PKR) are repressed, whereas osteoclastic regulators (NFATC1 and miR182) are enhanced in the RA setting, which leads towards an overall enhanced osteoclastogenic condition in RA. With TNF blockade therapy of these RA patients with a soluble receptor of human TNF (Enbrel) that specifically blocks TNF activity and inflammation in RA, we observed strikingly upregulated expression levels of RBPJ, FOXO3 and EIF2AK2, but significantly decreased levels of NFATC1 and miR182 in CD14 (+) PBMCs isolated from each patient after one or two months of treatment (Fig. 7B). These data indicate that treatment of RA reverses the altered and unbalanced osteoclastogenic network towards healthy conditions. This was corroborated by the correlation study between the expression changes of the network components and the ex vivo osteoclastogenesis levels (Fig. 7C). We isolated CD14 (+) PBMCs from each patient before and after one or two months of treatment using TNFi and performed ex vivo osteoclast differentiation. Along treatment, osteoclastogenesis was decreased by TNFi. Furthermore, statistical analysis showed that osteoclast differentiation extent (indicated by osteoclast areas) had strong negative correlations with RBPJ, FOXO3 and EIF2AK2 expressions, but positive with NFATC1 and miR182 levels (Fig. 7C). These human RA data provide evidence for the presence of the regulatory network mediated by RBP-J/NFATc1-miR182, which is significantly correlated with the osteoclastogenic levels. Disease conditions, such as RA, can alter this regulatory network towards more osteoclastogenic than healthy conditions. Therefore, these data highlight the clinical relevance of the RBP-J/NFATc1-miR182 regulatory network in the therapeutic strategies for the treatment of diseases involving bone erosion.
DISCUSSION
Inflammatory bone resorption is a major clinical problem and cause of morbidity in diseases such as RA and periodontitis. Inflammatory conditions have complex impacts on osteoclastogenesis and bone remodeling (1, 5, 28). The underlying molecular mechanisms remain largely unknown. This study identified a novel and unique regulatory network mediated by RBP-J/NFATc1-miR182 that plays an important role in TNF-induced osteoclastogenesis and inflammatory bone resorption. miR182, for the first time, was identified as a direct target of both RBP-J and NFATc1. RBP-J suppresses while NFATc1 activates miR182 expression. This new, highly integrated network includes both positive and feedback inhibitory regulators. Crosstalk and balance between regulators in this network, in response to different scenarios, can serve to establish biological switches that control cell differentiation extent. For example, TNFα maintains RBP-J expression level and activates RBP-J activity (11). As a consequence, TNFα is not able to effectively induce the expression of osteoclastogenic regulators NFATc1 and miR182 in the presence of RBP-J. Hence, inhibition of miR182 and NFATc1 by RBP-J is a crucial mechanism limiting TNF-induced osteoclastogenesis and bone resorption. However, in RA disease condition, there are multiple cytokines in addition to TNFα that together lead to a complex inflammatory state. RBP-J activity at sites of inflammation can potentially be attenuated by cytokines that activate Jak-STAT signaling, and are pathogenic in diseases such as RA (10, 51, 52). It is presumably the RA inflammation that overrides the TNF effects on RBP-J expression and leads to lower RBP-J levels in RA than in healthy condition.
Inflammation also induces oxidative stress, and bone resorption leads to calcium release, which in turn regulates inflammation (53, 54). These complex inflammatory states and associated pathological changes could also affect the components of the identified regulatory network, and thereby contributing to the mechanisms underlying pathological osteoclastogenesis and bone resorption. In response to environmental cues, such as inflammation in RA, which decrease RBP-J expression level/activity (10, 51, 52), the regulatory network can shift towards osteoclastogenic. RANKL stimulation downregulates RBP-J expression, which in turn allows RANKL to effectively induce NFATc1 and miR182 (11, 46) that drive osteoclastogenesis. Thus, the distinct regulation of the network by TNFα and RANKL contributes to their different osteoclastogenic capacity. Our previous work and this study introduce a concept that cytokines such as TNFα are subject to ‘brakes’ or feedback inhibitory mechanisms that restrain their osteoclastogenic potential. Augmentation of these intrinsic inhibitory mechanisms could help develop novel therapeutic strategies to treat osteolysis.
In addition to the biochemical evidence that miR182 is a direct target of RBP-J, we investigated and established the biological function of the RBP-J-miR182 axis in regulation of inflammatory bone resorption in this study. Both in vitro osteoclast differentiation and in vivo experiments, using two inflammatory bone resorption models, demonstrate that the RBP-J-miR182 axis is a key pathway whereby TNFα restrains its osteoclastogenic potential. The expression and targets of miRNAs are context-dependent and highly specific to cell and tissue types (55). The targets of miR182 are variable in different cells according to a variety of settings and stimulations. Our prior studies identified FoxO3 and PKR as key miR182 targets that act as osteoclastogenic inhibitors (46). PKR represses osteoclastogenesis through activating autocrine IFNβ-mediated feedback inhibition (46). FoxO1 and FoxO3 are well-defined miR182 targets in several biological settings (56, 57). FoxO family members, FoxO1, 3, and 4 proteins, are involved in osteoclast differentiation (45, 58, 59), but reported with different functions and mechanisms. Some studies show that FoxO1, 3 and 4 proteins are inhibitors of osteoclastogenesis (58), whereas others found FoxO1 as a positive regulator (59). These data indicate that the FoxO family plays an important but complex role in osteoclastogenesis. We found that FoxO3 is an important miR182 target in TNF-mediated osteoclast differentiation (45). Previous study shows that FoxO3 targets catalase and Cyclin D1 to arrest the cell cycle and promote apoptosis in RANKL-induced osteoclastogenesis (58). We did not observe these changes in response to TNFα, suggesting distinct mechanisms by which FoxO3 suppresses TNF-induced osteoclastogenesis. Future work will be needed to elucidate the mechanisms and to test the miR182-FoxO3 axis in vivo.
Current treatments of excessive bone resorption, such as in osteoporosis, using RANK receptor blockers or neutralizing RANKL antibodies are able to effectively inhibit osteoclast formation. However, blocking RANKL signaling could result in potential long-term side effects, such as bone remodeling defects, due to strong inhibition of osteoclast formation. TNF inhibitors treat inflammation and associated joint erosion such as that occurring in RA, but long-term usage has immunorepressive side effects, such as opportunistic infections. Alternative or complementary approaches to control abnormal osteoclastogenesis in disease conditions are therefore needed in order to ameliorate side effects and benefit patients. Our findings in this study revealed the presence of the RBP-J/NFATc1-miR182-PKR/FoxO3 network in human RA and its significant correlation with ex vivo osteoclastogenesis. The RA data clearly show that inflammation shifts the network towards osteoclastogenic, reflected by decreased RBP-J, PKR and FoxO3 expression, but enhanced NFATc1 and miR182 levels. Treatment of inflammation reverses the expression levels of the components in this network close to healthy condition. These findings indicate that this network is responsive and sensitive to environmental cues, and shed insights into the implications of treating the unbalanced network in osteolytic conditions. In addition, genetic evidence show that RBPJ and FOXO3 are closely associated with human inflammatory diseases (42–44, 60, 61), such as RA. The genetic linkage with RA of the two components of the RBP-J/NFATc1-miR182-PKR/FoxO3 network we discovered further supports a role for this network in the pathogenesis of inflammatory diseases. Taken together, our findings discovered a novel regulatory network mediated by RBP-J/NFATc1-miR182 in TNF-induced osteoclastogenesis, demonstrated the biological function of this regulatory network in inflammatory bone resorption, and unveiled the correlation between the network and RA. The results suggest the translational implications of RBP-J/NFATc1-miR182 network in treating inflammatory osteoclastogenesis and bone destruction.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Julia Charles for providing bone marrows of Nfatc1 KO (Nfatc1f/f;Mx1Cre) mice and their control mice. We are grateful to Christine Miller, Mahmoud Elguindy, Shin-ichi Nakano, Zhonghao Deng and Courtney Ng from Dr. Baohong Zhao’s laboratory as well as Drs Yufan Chen and Liang Zhao for their helpful discussions and assistance. This work was supported by grants from the National Institutes of Health (AR062047, AR068970 and AR071463 to B.Z.). The content of this manuscript is solely the responsibilities of the authors and does not necessarily represent the official views of the NIH.
Nonstandard Abbreviations:
- Blimp1
B lymphocyte–induced maturation protein-1
- BMM
bone marrow–derived macrophage
- ChIP
chromatin immunoprecipitation
- FAIRE
Formaldehyde-Assisted Isolation of Regulatory Elements
- FOXO3
Forkhead box class O 3
- μCT
microcomputed tomography
- MNC
multinucleated cell
- M-CSF
macrophage colony-stimulating factor
- NFATc1
nuclear factor of activated T cells c1
- PBMCs
peripheral blood monocytes
- PKR
protein kinase double-stranded RNA-dependent
- RA
rheumatoid arthritis
- RANKL
receptor activator for NF-kB ligand
- RBP-J
Recombination signal binding protein for immunoglobulin kappa J region
- TNFα
Tumor necrosis factor- α
- TNFi
TNF inhibitor
- TRAP
tartrate-resistant acid phosphatase
- WT
wild-type
Footnotes
Disclosures: The authors have no conflict of interest.
REFERENCES
- 1.Schett G, and Gravallese E (2012) Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol 8, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novack DV, and Teitelbaum SL (2008) The osteoclast: friend or foe? Annu Rev Pathol 3, 457–484 [DOI] [PubMed] [Google Scholar]
- 3.Schett G, and Teitelbaum SL (2009) Osteoclasts and arthritis. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 24, 1142–1146 [DOI] [PubMed] [Google Scholar]
- 4.Boyce BF, Yao Z, Zhang Q, Guo R, Lu Y, Schwarz EM, and Xing L (2007) New roles for osteoclasts in bone. Ann N Y Acad Sci 1116, 245–254 [DOI] [PubMed] [Google Scholar]
- 5.Sato K, and Takayanagi H (2006) Osteoclasts, rheumatoid arthritis, and osteoimmunology. Current opinion in rheumatology 18, 419–426 [DOI] [PubMed] [Google Scholar]
- 6.Zhao B, and Ivashkiv LB (2011) Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res Ther 13, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao Q, Chen S, Qin H, Feng J, Liu H, Liu D, Li A, Shen Y, Zhao Y, Li J, and Zong Z (2017) An appropriate Wnt/beta-catenin expression level during the remodeling phase is required for improved bone fracture healing in mice. Scientific reports 7, 2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binder N, Miller C, Yoshida M, Inoue K, Nakano S, Hu X, Ivashkiv LB, Schett G, Pernis A, Goldring SR, Ross FP, and Zhao B (2017) Def6 Restrains Osteoclastogenesis and Inflammatory Bone Resorption. Journal of immunology 198, 3436–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asagiri M, and Takayanagi H (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 10.Li S, Miller CH, Giannopoulou E, Hu X, Ivashkiv LB, and Zhao B (2014) RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. The Journal of clinical investigation 124, 5057–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao B, Grimes SN, Li S, Hu X, and Ivashkiv LB (2012) TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med 209, 319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, Tamura T, Ozato K, Choi Y, Ivashkiv LB, Takayanagi H, and Kamijo R (2009) Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med 15, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyce BF, Xiu Y, Li J, Xing L, and Yao Z (2015) NF-kappaB-Mediated Regulation of Osteoclastogenesis. Endocrinology and metabolism 30, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F, and Teitelbaum SL (2013) Osteoclasts: New Insights. Bone research 1, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakashima T, and Takayanagi H (2011) New regulation mechanisms of osteoclast differentiation. Ann N Y Acad Sci 1240, E13–18 [DOI] [PubMed] [Google Scholar]
- 16.Humphrey MB, Lanier LL, and Nakamura MC (2005) Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev 208, 50–65 [DOI] [PubMed] [Google Scholar]
- 17.Teitelbaum SL (2006) Osteoclasts; culprits in inflammatory osteolysis. Arthritis Res Ther 8, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YH, Heulsmann A, Tondravi MM, Mukherjee A, and Abu-Amer Y (2001) Tumor necrosis factor-alpha (TNF) stimulates RANKL-induced osteoclastogenesis via coupling of TNF type 1 receptor and RANK signaling pathways. The Journal of biological chemistry 276, 563–568 [DOI] [PubMed] [Google Scholar]
- 19.Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, and Teitelbaum SL (2000) TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. The Journal of clinical investigation 106, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller K, Murphy C, Kirstein B, Fox SW, and Chambers TJ (2002) TNFalpha potently activates osteoclasts, through a direct action independent of and strongly synergistic with RANKL. Endocrinology 143, 1108–1118 [DOI] [PubMed] [Google Scholar]
- 21.Cenci S, Weitzmann MN, Roggia C, Namba N, Novack D, Woodring J, and Pacifici R (2000) Estrogen deficiency induces bone loss by enhancing T-cell production of TNF-alpha. The Journal of clinical investigation 106, 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weitzmann MN (2013) The Role of Inflammatory Cytokines, the RANKL/OPG Axis, and the Immunoskeletal Interface in Physiological Bone Turnover and Osteoporosis. Scientifica 2013, 125705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitaura H, Kimura K, Ishida M, Kohara H, Yoshimatsu M, and Takano-Yamamoto T (2013) Immunological reaction in TNF-alpha-mediated osteoclast formation and bone resorption in vitro and in vivo. Clinical & developmental immunology 2013, 181849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh MC, and Choi Y (2014) Biology of the RANKL-RANK-OPG System in Immunity, Bone, and Beyond. Frontiers in immunology 5, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, De Smedt T, Daro E, Smith J, Tometsko ME, Maliszewski CR, Armstrong A, Shen V, Bain S, Cosman D, Anderson D, Morrissey PJ, Peschon JJ, and Schuh J (1999) RANK is essential for osteoclast and lymph node development. Genes & development 13, 2412–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettit AR, Ji H, von Stechow D, Muller R, Goldring SR, Choi Y, Benoist C, and Gravallese EM (2001) TRANCE/RANKL knockout mice are protected from bone erosion in a serum transfer model of arthritis. The American journal of pathology 159, 1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarilina A, Xu K, Chen J, and Ivashkiv LB (2011) TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proceedings of the National Academy of Sciences of the United States of America 108, 1573–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao B (2017) TNF and Bone Remodeling. Current osteoporosis reports 15, 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopan R, and Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu T, Kagawa T, Inoue T, Nonaka A, Takada S, Aburatani H, and Taga T (2008) Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol 28, 7427–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plaisance S, Vanden Berghe W, Boone E, Fiers W, and Haegeman G (1997) Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol Cell Biol 17, 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izumiya Y, Izumiya C, Hsia D, Ellison TJ, Luciw PA, and Kung HJ (2009) NF-kappaB serves as a cellular sensor of Kaposi’s sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J Virol 83, 4435–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swarnkar G, Karuppaiah K, Mbalaviele G, Chen TH, and Abu-Amer Y (2015) Osteopetrosis in TAK1-deficient mice owing to defective NF-kappaB and NOTCH signaling. Proceedings of the National Academy of Sciences of the United States of America 112, 154–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, Outtz H, Kitajewski J, Shi C, Weber S, Saftig P, Li Y, Ozato K, Blobel CP, Ivashkiv LB, and Hu X (2012) Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nature immunology 13, 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, Tateya T, Kang YJ, Han J, Gessler M, Kageyama R, and Ivashkiv LB (2008) Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. In Immunity Vol. 29 pp. 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayward SD (2004) Viral interactions with the Notch pathway. Seminars in cancer biology 14, 387–396 [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi Y, Furukawa T, Tun T, Han H, and Honjo T (1998) LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol 18, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beres TM, Masui T, Swift GH, Shi L, Henke RM, and MacDonald RJ (2006) PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol 26, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foldi J, Shang Y, Zhao B, Ivashkiv LB, and Hu X (2016) RBP-J is required for M2 macrophage polarization in response to chitin and mediates expression of a subset of M2 genes. Protein & cell 7, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang Y, Smith S, and Hu X (2016) Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein & cell 7, 159–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caton ML, Smith-Raska MR, and Reizis B (2007) Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med 204, 1653–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP, Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, and Plenge RM (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, Zhernakova A, Stahl E, Viatte S, McAllister K, Amos CI, Padyukov L, Toes RE, Huizinga TW, Wijmenga C, Trynka G, Franke L, Westra HJ, Alfredsson L, Hu X, Sandor C, de Bakker PI, Davila S, Khor CC, Heng KK, Andrews R, Edkins S, Hunt SE, Langford C, Symmons D, Concannon P, Onengut-Gumuscu S, Rich SS, Deloukas P, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Arlsetig L, Martin J, Rantapaa-Dahlqvist S, Plenge RM, Raychaudhuri S, Klareskog L, Gregersen PK, and Worthington J (2012) High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet 44, 1336–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orozco G, Viatte S, Bowes J, Martin P, Wilson AG, Morgan AW, Steer S, Wordsworth P, Hocking LJ, Barton A, Worthington J, and Eyre S (2013) Novel RA susceptibility locus at 22q12 identified in an extended UK genome wide association study. Arthritis Rheum [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller CH, Smith SM, Elguindy M, Zhang T, Xiang JZ, Hu X, Ivashkiv LB, and Zhao B (2016) RBP-J-Regulated miR-182 Promotes TNF-alpha-Induced Osteoclastogenesis. Journal of immunology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inoue K, Deng Z, Chen Y, Giannopoulou E, Xu R, Gong S, Greenblatt MB, Mangala LS, Lopez-Berestein G, Kirsch DG, Sood AK, Zhao L, and Zhao B (2018) Bone protection by inhibition of microRNA-182. Nature communications 9, 4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD, Cason D, Luo L, Ma Y, Van Mater D, Gladdy R, Lev DC, Cardona DM, and Kirsch DG (2014) MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. The Journal of clinical investigation 124, 4305–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, Ostrowski MC, Olsen BR, and Glimcher LH (2008) NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. The Journal of clinical investigation 118, 3775–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sokhi UK, Liber MP, Frye L, Park S, Kang K, Pannellini T, Zhao B, Norinsky R, Ivashkiv LB, and Gong S (2018) Dissection and function of autoimmunity-associated TNFAIP3 (A20) gene enhancers in humanized mouse models. Nature communications 9, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, Morita I, Wagner EF, Mak TW, Serfling E, and Takayanagi H (2005) Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivashkiv LB, and Hu X (2003) The JAK/STAT pathway in rheumatoid arthritis: pathogenic or protective? Arthritis Rheum 48, 2092–2096 [DOI] [PubMed] [Google Scholar]
- 52.Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, Tateya T, Kang YJ, Han J, Gessler M, Kageyama R, and Ivashkiv LB (2008) Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity 29, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendy GN, and Canaff L (2016) Calcium-sensing receptor, proinflammatory cytokines and calcium homeostasis. Seminars in cell & developmental biology 49, 37–43 [DOI] [PubMed] [Google Scholar]
- 54.Klein GL, Castro SM, and Garofalo RP (2016) The calcium-sensing receptor as a mediator of inflammation. Seminars in cell & developmental biology 49, 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olive V, Minella AC, and He L (2015) Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Science signaling 8, re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, and Hernando E (2009) Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America 106, 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, Heinz GA, Fang Z, Li N, Bissels U, Hatam F, Jahn A, Hammoud B, Matz M, Schulze FM, Baumgrass R, Bosio A, Mollenkopf HJ, Grun J, Thiel A, Chen W, Hofer T, Loddenkemper C, Lohning M, Chang HD, Rajewsky N, Radbruch A, and Mashreghi MF (2010) The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nature immunology 11, 1057–1062 [DOI] [PubMed] [Google Scholar]
- 58.Bartell SM, Kim HN, Ambrogini E, Han L, Iyer S, Serra Ucer S, Rabinovitch P, Jilka RL, Weinstein RS, Zhao H, O’Brien CA, Manolagas SC, and Almeida M (2014) FoxO proteins restrain osteoclastogenesis and bone resorption by attenuating H2O2 accumulation. Nature communications 5, 3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Dong G, Jeon HH, Elazizi M, La LB, Hameedaldeen A, Xiao E, Tian C, Alsadun S, Choi Y, and Graves DT (2015) FOXO1 mediates RANKL-induced osteoclast formation and activity. Journal of immunology 194, 2878–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, Roberts R, Viatte S, Fu B, Peshu N, Hien TT, Phu NH, Wesley E, Edwards C, Ahmad T, Mansfield JC, Gearry R, Dunstan S, Williams TN, Barton A, Vinuesa CG, Consortium UIG, Parkes M, Lyons PA, and Smith KG (2013) Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 155, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gregersen PK, and Manjarrez-Orduno N (2013) FOXO in the hole: leveraging GWAS for outcome and function. Cell 155, 11–12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.