Abstract
Cancer arises from the accumulation of genetic alterations, which can lead to the production of mutant proteins not expressed by normal cells. These mutant proteins can be processed and presented on the cell surface by major histocompatibility complex molecules as neoepitopes, allowing CD8+ T cells to mount responses against them. Using predictive algorithms to identify putative endogenous antitumor T cell responses in solid tumors has resulted in an average 2% of predicted neoepitopes being targeted. This suggests that low mutation burden tumors, which include many pediatric tumors, are poorly immunogenic. Here, we report that pediatric patients with acute lymphoblastic leukemia (ALL) possess tumor-associated neoepitope-specific CD8+ T cells, responding to 86% of tested neoantigens and recognizing 68% of the tested neoepitopes. These responses include a public neoantigen from the ETV6-RUNX1 fusion that is targeted in 7 of 9 tested patients. We characterized phenotypic and transcriptional profiles of CD8+ TILs at the single cell level and found a heterogeneous population that included highly functional effectors. Moreover, we observed immunodominance hierarchies among the CD8+ TILs restricted to one or two putative neoepitopes. Our results indicate that robust antitumor immune responses are induced in pediatric ALL despite their low mutation burdens and emphasize the importance of immunodominance in shaping cellular immune responses. Furthermore, these data suggest that pediatric cancers may be amenable to immunotherapies aimed at enhancing immune recognition of tumor-specific neoantigens.
One Sentence Summary:
Pediatric acute lymphoblastic leukemia elicits a broad, functional, antitumor T cell response, targeting multiple mutations.
INTRODUCTION
Recent insights from animal studies, translational research, and correlative clinical data have highlighted the importance of the immune system as a therapeutic target for cancer treatment (1–4). Among the most promising are immunotherapies aimed at exploiting and co-opting the host’s adaptive immune system, particularly cytotoxic CD8+ T cells (2, 5–8). Currently, several approaches for targeting tumors with specific immune effectors are beginning to bear fruit, including the use of: (1) immunomodulatory monoclonal antibodies blocking inhibitory receptor signaling in endogenous antitumor CD8+ T cells, (2) expanded tumor infiltrating lymphocytes (TILs), (3) T cell receptor (TCR) engineered T cells (TCR-T), and (4) chimeric antigen receptorexpressing (CAR) T cells (9–19). Although immunomodulatory therapies have shown clinical utility in some adult solid tumors, especially those with higher mutation burdens (20–23), attempts to identify tumor-reactive T cell responses against identified mutations have had a relatively low success rate for any given mutation, with only about 2% eliciting a measurable response in patients either functionally or by tetramer staining. This has led to the hypothesis that qualitative features of the mutations, including whether they contain substantial non-self sequences or homology to a pathogen-associated epitope, influence the endogenous T cell response and efficacy of checkpoint blockade immunotherapies (24, 25).
In parallel to the development of immune checkpoint blockade (ICB), CAR and TCR-T approaches that engineer the patient’s own T cells with a single specificity to target the tumor have shown efficacy. These approaches target tumor associated antigens, such as CD19 in B cell malignancies, or tumor specific antigens, such as the H3-K27M mutation in gliomas (26–28). Identifying tumor specific mutations provides an advantage over targeting tumor associated antigens by limiting collateral losses, such as the B cell aplasia observed after CD19 CAR therapy. To date, these approaches have targeted single antigens, demonstrating that a monoclonal immune response is competent for tumor control under appropriate conditions. The identification of additional high-quality targets is a major focus for cell-based therapy research.
It is important to note that many of the aforementioned studies were carried out in adult solid tumors with high mutation rates (29, 30), leaving open the question of whether low mutation burden tumors contain correspondingly poor endogenous T cell responses, a view loosely supported by the observation that checkpoint blockade has generally not been as successful in these tumors (21). This question has particular relevance for pediatric tumors, which generally exhibit markedly fewer somatic mutations. Although recent studies using immunomodulatory therapies in pediatric patients with neuroblastoma appear promising (31, 32), these therapies are hypothesized to enlist endogenous natural killer cells, thus further research is needed to establish whether antitumor CD8+ T cell responses are also present in childhood cancers. In this study, we aimed to establish the endogenous CD8+ T cell response from pediatric patients with ALL, which on average has far fewer nonsynonymous mutations than other tumor types (33), and to identify candidate tumor specific mutations that may be targetable by TCR-T cells. We hypothesized that although leukemias contain lower mutational burdens, the neoantigens that arise in this tumor type might serve as functional targets capable of inducing antitumor T cell responses (34).
RESULTS
Identification of putative neoepitopes in pediatric acute lymphoblastic leukemia
To identify putative patient-specific cancer neoantigens (fig. S1A–C), somatically acquired genetic alterations in the tumor were detected via high-throughput genomic sequencing of diagnostic biopsies and matched germline tissue obtained from a cohort of pediatric patients (table S1) with ALL. Tumor samples were also subjected to mRNA sequencing, which was used to determine human leukocyte antigen (HLA) haplotype and confirm the expression of HLA class I genes within the tumor milieu (consisting of tumor, monocytic, and T cells; table S2). Samples from patients with HLA-A genes (HLA-A30 and HLA-A2) exhibiting lower fragment per kilobase of transcript per million mapped reads (FPKM) values were also subjected to flow cytometric analysis to confirm that proteins were also expressed on the surface of the tumor cells used for sequencing (fig. S1D).
Using patient-specific HLA class I types inferred from mRNA sequencing, we screened tumor-specific somatic missense mutations and gene fusions for their potential to generate neoepitopes (i.e., neoantigens predicted to bind patient-specific HLA proteins). We classified the binding of peptides to HLA alleles as strong putative neoepitopes if their predicted binding affinity (IC50) was below 150 nM, as intermediate to weak putative neoepitopes if their IC50 was between 150 and 500 nM, and as putative nonbinders if their IC50 was greater than 500 nM. Although the number of mutations in our cohort was low in comparison to other cancer types (35), we detected between five and twenty-eight predicted nonamer neoepitopes per patient (fig. S1B), with an average of 8 strong nonamer neoepitopes (range: 2–14) and 6 intermediate nonamer neoepitopes (range: 3–14; fig. S1C; table S3) per patient. Furthermore, we predicted epitopes spanning the fusion junction in 3 of the 4 patients with detected ETV6-RUNX1 genomic fusions in their tumor biopsies. These mutations were of particular interest because similar fusions occur across multiple patients with distinct HLA haplotypes (36).
Functional assays identify a pool of neoepitope-reactive CD8+ T cells
Due to the paucity of studies addressing the role of cytotoxic CD8+ T cells in pediatric malignancies, we first aimed to determine the presence of CD8+ T cells in ALL bone marrow samples and subsequently assessed the differentiation and activation status of CD8+ TILs via flow cytometry (fig. S2). Although the frequency of CD8+ T cells was slightly lower in bone marrow samples from our patient cohort in comparison to a healthy donor (likely due to the high frequency of leukemic CD19+ B cells typical of ALL patients), we detected CD8+ TILs in all samples (fig. S2A,B). These TILs expressed several markers indicative of antigen-specific stimulation (fig. S2A–E), including the upregulation of PD-1, TIM-3, and 4–1BB.
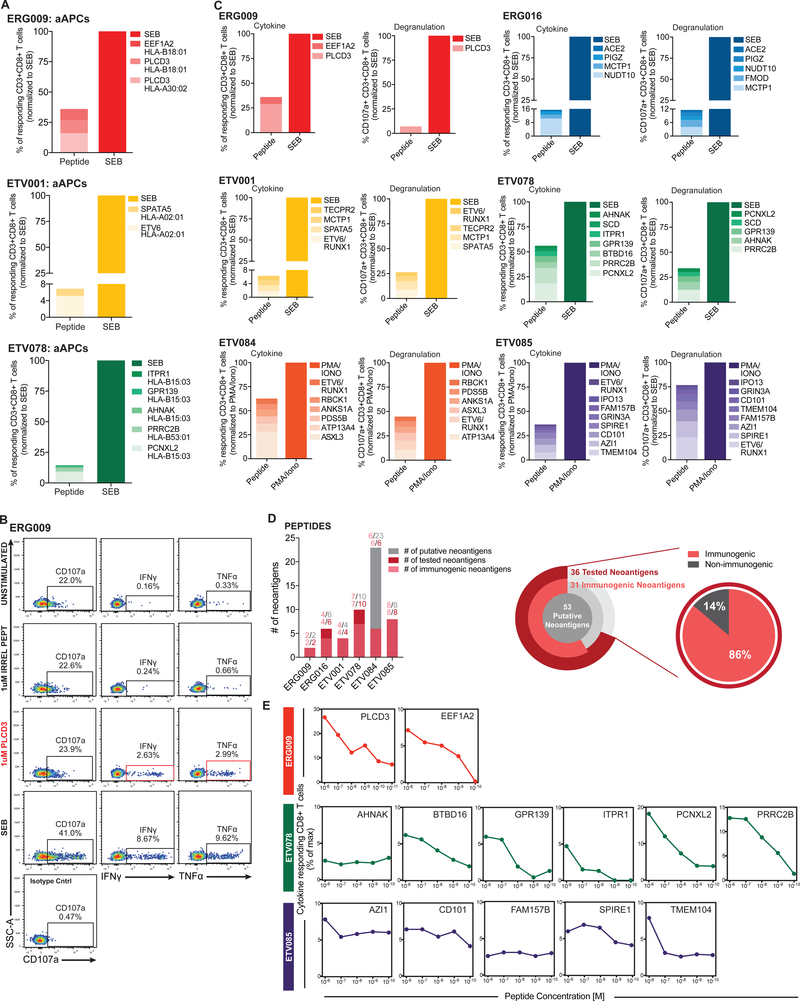
Previous studies on cancer patients with melanoma (4, 6, 37) and non-small cell lung carcinoma (21) have identified T cell responses by tetramer or functional assays, but only a small fraction (0.05–2%) of the putative neoepitopes tested were found to have elicited a detectable response. Although there is a lower mutational burden in ALL compared to these solid tumors, we hypothesized that this might result in a greater frequency of responsive CD8+ T cells due to the limited number of tumor-specific epitopes available for the immune system to target. To test whether CD8+ T cells within the TILs were responsive to mutated neoepitopes directly ex vivo, we engineered 5 artificial antigen-presenting cells (aAPCs), each of which expressed a single patient-specific HLA (fig. S3). Focusing on the three patients with the highest frequency of CD8+ T cells, we pulsed the HLA-specific aAPCs with synthetic 15 amino acid (aa) peptides that corresponded to the predicted neoepitopes, co-cultured them with isolated CD8+ TILs, and monitored for production of interferon gamma (IFNγ) and tumor necrosis factor alpha (TNFα). To control for potential background noise and non-specific stimulation, patients’ CD8+ T cell cytokine production in the presence of putative patient-specific tumor neoantigens were calculated relative to responses to an irrelevant peptide. In addition, we stimulated isolated CD8+ T cells with SEB (Staphylococcal enterotoxin B) as a positive control to determine the maximal response rate of these cells, and subsequent analyses were normalized to this maximum. All three tested peptides in patient ERG009 (two restricted by B*18:01 and one by A*30:02) generated robust responses to peptide-aAPC stimulation, totaling over 30% of the SEB response (Fig. 1A). Similarly, the two A*02:01-binding peptides tested with A*02:01 aAPCs for patient ETV001 generated responses (Fig. 1A). Finally, the five peptides for ETV078 all generated responses above background, but the magnitude of the response was relatively low. In sum, all three patients exhibited CD8+ T cell cytokine production in response to specific peptide stimulation, indicating that the predicted neoepitopes restricted by the patients’ HLAs were indeed generating endogenous responses.
Fig. 1. Putative cancer neoantigens elicit robust T cell responses.
(A) Enriched CD8+ tumor infiltrating lymphocytes (TILs) from 3 patients were independently co-cultured with artificial antigen presenting cells (aAPCs) expressing a single patient-specific human leukocyte antigen (HLA) molecule and pulsed with 1 μg/mL of anti-human CD28/CD49d and either 1 μM of the indicated 15mer peptide or 100 μg /mL staphylococcal enterotoxin B (SEB). Cytokine production by CD8+ TILs was subsequently measured by intracellular cytokine staining (ICS). Normalized frequency (neoantigen peptide compared to SEB) of responding single, live CD8+ lymphocytes that produced cytokines (IFNγ or TNFα) following stimulation is plotted for each acute lymphoblastic leukemia (ALL) patient. (B) Representative flow cytometry plots from one patient depicting single, live CD8+ lymphocytes that are CD107a+ (left panel), IFNγ+ (middle panel), or TNFα+ (right panel) following irrelevant peptide, neoantigen peptide, or polyclonal (SEB; positive control) stimulation. Flow plots for negative controls (unstimulated cells lacking peptide stimulation and isotype controls) are also shown and were used to set gates. (C) For six patients, 1–2 × 106 bone marrow mononuclear cells (BMMCs) were stimulated with 1 μg /mL of anti-human CD28/CD49d and either 1μM of the indicated 15mer peptide, 100 μg /mL SEB, or 1X phorbol 12-myristate 13-acetate (PMA)/Ionomycin cell stimulation cocktail (as a positive control; see Methods) and then subjected to ICS. Normalized frequency (see Methods) of responding single, live CD8+ T cells that produced cytokines (IFNγ or TNFα; left panel) or degranulated (right panel) following stimulation is plotted for each ALL patient. (D) T cell response statistics for all putative neoantigens per patient (bar chart) and at the cohort level (pie charts). (E) For three patients, 2 × 106 BMMCs were pulsed with serial dilutions (1μM to 10 pM) of the indicated 15mer peptide, 100 μg /mL SEB, or 1X PMA/Ionomycin cell stimulation cocktail and subjected to ICS. Normalized frequency of responding CD8+ T cells that produced cytokines (IFNγ or TNFα) following stimulation is plotted.
Given the surprisingly high rate of neoantigen-specific responses, we expanded our analyses to include direct stimulation of the bone marrow without the use of aAPCs or purification of the CD8+ T cells, which allowed us to test more neoantigens across a larger number of patients. Freshly processed autologous bone marrow cells (consisting of tumor, APCs, CD8+ T cells, and other cell types and products in the local tumor milieu) were pulsed with synthetic 15aa peptides to monitor the endogenous response of the CD8+ T cells. Fig. 1B shows the representative gating strategy from one pediatric ALL patient sample that was used to identify cytokine-producing and degranulating (CD107a/b+) neoantigen-specific CD8+ TILs. All six patients responded to at least one neoantigenic peptide in this assay, with relatively high rates of reactivity (ranging from ~6% to ~59%) among the total CD8+ TIL population (Fig. 1C). These analyses identified a spectrum of reactivities by CD8+ TILs ranging from moderate cumulative cytokine responses observed for ERG016 and ETV001, to intermediate responses detected for ERG009 and ETV085, to strong responses measured for ETV078 and ETV084 (Fig. 1C). For ERG016, ~15% of the responding CD8+ TILs displayed reactivity towards four out of six putative mutant neoantigens and for ETV001, ~15% and ~6% of the responding CD8+ TILs displayed reactivity towards four out of six and four out of four putative mutant neoantigens, respectively (Fig. 1D). Among the intermediate responders, CD8+ TIL reactivity for ERG009 and ETV085 was mounted against two out of two and eight out of eight putative neoantigens, which accounted for ~36% and ~37% of the maximal CD8+ TIL cytokine response, respectively. Additionally, for ETV078 and ETV084, ~56% and ~59% of the responding CD8+ TILs were reactive against seven out of ten and six of the twenty-three (only six neoantigens were tested in the assay) putative mutant neoantigens, respectively (Fig. 1D). Strikingly, out of the 36 putative neoantigens tested in our studies, 31 were found to be immunogenic and capable of inducing robust cytokine responses (Fig. 1C,D). To determine whether similar responses would be observed at lower concentrations of neoantigen, where recognition of fewer peptide-MHC complexes may limit reactivity, serial dilutions of mutated peptides were used to monitor the response of CD8+ T cells to peptide-pulsed autologous bone marrow cells. Several neoantigens elicited reactive CD8+ T cell responses at nanomolar (e.g., PLCD3, EEF1A2, BTBD16, PCNXL2, and PRRC2B) and 100 picomolar (e.g., AHNAK, AZI1, CD101, FAM157B, SPIRE1, and TMEM104) concentrations (Fig. 1E).
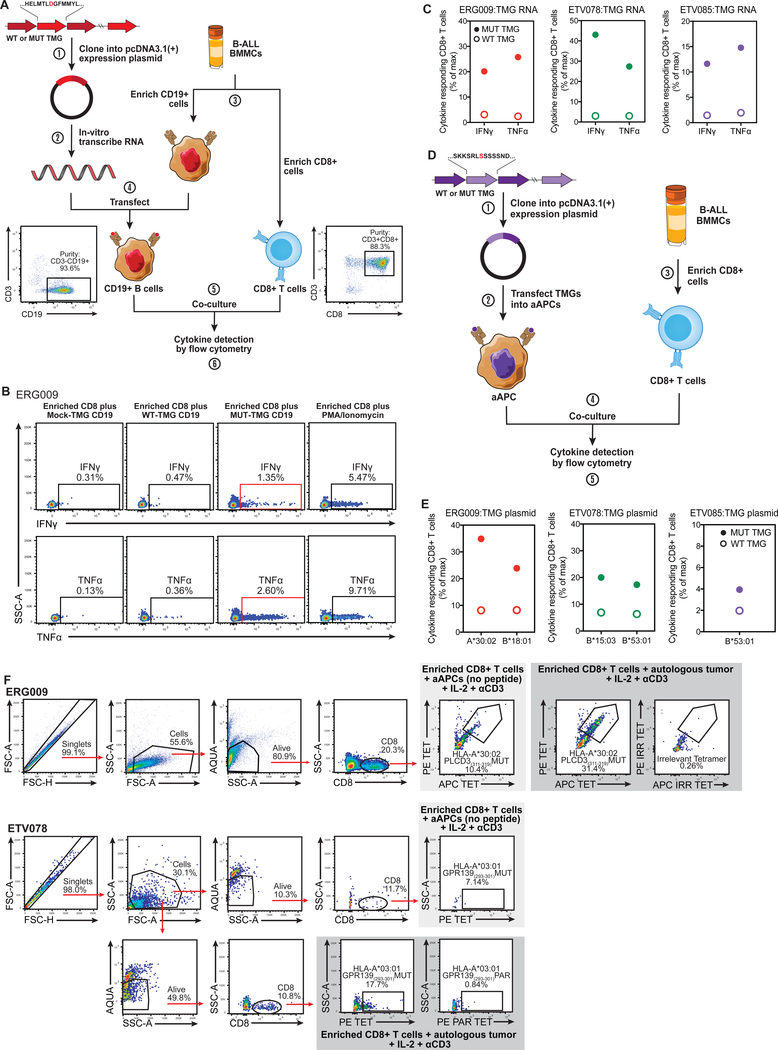
To confirm that the results against exogenously presented peptides were not an artifact of exogenous peptide stimulation and mirrored the responses that would be seen against cells processing and presenting endogenous neoantigens, we used three additional approaches (shown in schematic for tandem minigenes in fig. 2A and 2D, and see Materials and Methods for TIL expansion). First, autologous tumor cells (CD19+ B cells) were transfected with mRNA encoding either wild-type or mutant neoepitopes in a tandem minigene (TMG) configuration (Fig. 2A). Cytokine production by enriched CD8+ T cells co-cultured with autologous tumor cells was measured by intracellular cytokine staining (ICS; Fig. 2A–C). In the second approach, we tested the response of enriched CD8+ T cells against aAPCs transfected with plasmid TMGs encoding wild-type or mutant peptides (Fig. 2D) and expressing patient-specific HLAs. In both assays, CD8+ T cell reactivity was greater against mutant versus wild-type peptide-encoding TMGs (Fig. 2B, C, and E). To test whether healthy individuals also possess comparable frequencies of neoantigen-reactive T cells, CD8+ T cells were enriched from healthy adult donor PBMCs and co-cultured with the mRNA-transfected tumor cells from three patients, and cytokine production was measured by ICS. In all cases, the CD8+ T cell response from healthy donors was negligible against both wild-type and mutant mRNA-transfected tumor cells (fig. S4).
Fig. 2. Endogenous neoantigens are processed, presented, and induce CD8+ T cells responses.
(A) Schematic depicting the generation of tandem minigene (TMG) constructs (step 1) and in-vitro-transcribed (IVT) RNA (step 2) used to screen for recognition of putative somatic mutations. BMMCs were enriched for CD19+ cancer cells and CD8+ T cells (step 3). Cancer cells were then transfected with IVT RNA (step 4) and co-cultured with enriched CD8+ T cells (step 5). Co-cultured CD8+ T cells were subsequently subjected to ICS, and the frequency of cytokine producing cells was determined (step 6). Flow cytometry plots depict the representative purity following selection of CD19+ (left panel) and CD8+ T cells (right panel). (B) Representative flow cytometry plots from one patient depicting single, live CD8+ lymphocytes that are IFNγ+ or TNFα+ following co-culture with autologous CD19+ cancer cells transfected with mock-TMG RNA, wild-type TMG RNA, mutant TMG RNA, or enriched CD8+ T cells stimulated with PMA/Ionomycin. (C) Normalized frequency of IFNγ- and TNFα-producing CD8+ T cells following co-culture with autologous tumor cells transfected with wild-type TMG RNA (open circles) or mutant TMG RNA (filled circles) from three patients. (D) Schematic depicting the generation of TMG constructs (step 1) used to transfect aAPCs (step 2) to screen for recognition of putative somatic mutations. BMMCs were enriched for CD8+ T cells (step 3). aAPCs transfected with TMG plasmid DNA were co-cultured with enriched CD8+ T cells (step 4). Co-cultured CD8+ T cells were interrogated by ICS, and the frequency of cytokine producing cells was determined (step 5). (E) Normalized frequency of IFNγ- and TNFα-producing CD8+ T cells following co-culture with aAPCs transfected with wild-type TMG plasmid DNA (open circles) or mutant TMG plasmid DNA (filled circles) from three patients. (F) 2–3 × 104 sorted CD8+ TILs from 2 patients were independently co-cultured with either 8 × 104 autologous CD19+ tumor cells (dark gray shaded) or 8 × 104 aAPCs expressing patient-specific HLAs (negative control; light gray shaded). CD8+ TIL expansion and tumor reactivity were determined by flow cytometry after 21 days of co-culturing by comparing the frequency of mutant tetramer-positive CD8+ TILs (ERG009: PE and APC conjugated HLA-A*30:02 PLCD3(311–319)MUT; ETV078: PE conjugated HLA-A*03:01 GPR139(293–301)MUT) from autologous tumor and aAPC co-cultures. Tetramer-positive gates were set based on the binding of CD8+ TILs to irrelevant (ERG009: PE and APC conjugated HLA-A*24:02 CD101(884–892)IRR) or parent tetramers (ETV078 PE conjugated HLA-A*03:01 GPR139(293–301)PAR.
To further validate that the CD8+ TILs specifically recognized putative neoepitopes expressed by unmanipulated autologous tumor cells, we sorted purified CD19+ tumor cells and co-cultured them with sorted CD8+ T cells from the diagnostic ALL specimens of two patients (ERG009 and ETV078; see methods) with high frequencies of CD8+ T cells and a strong putative neoepitope that could be targeted (fig. S1; table S3). Following well-established protocols (38–41) for expanding TILs and generating T cell lines driven by endogenously presented antigens, we cultured the sorted endogenous CD8+ T cells with either aAPCs expressing patient-specific HLAs (negative control; non-neoantigen expressing targets) or autologous tumor cells. We then constructed peptide:MHC tetramers corresponding to neoantigens identified as strong binders (table S3) and likely to be immunogenic for these patients and analyzed for the presence of neoepitope-specific CD8+ T cells after 21 days of co-culturing. A control tetramer for patient ERG009 was used containing an irrelevant (but HLA-stabilizing) peptide, whereas the “parent” tetramer was used as a control in patient ETV078, containing the endogenous, non-mutated peptide (see Materials and Methods and table S4). For both patients, co-cultures containing autologous tumor cells resulted in a greater expansion of tetramer positive neoepitope-specific CD8+ T cells when compared to the co-cultures containing aAPCs (Fig. 2F). Collectively, these findings demonstrate that tumor neoantigens are processed and presented by the tumors present in pediatric patients with ALL.
Antitumor T cell responses are neoepitope-specific and form immunodominance hierarchies
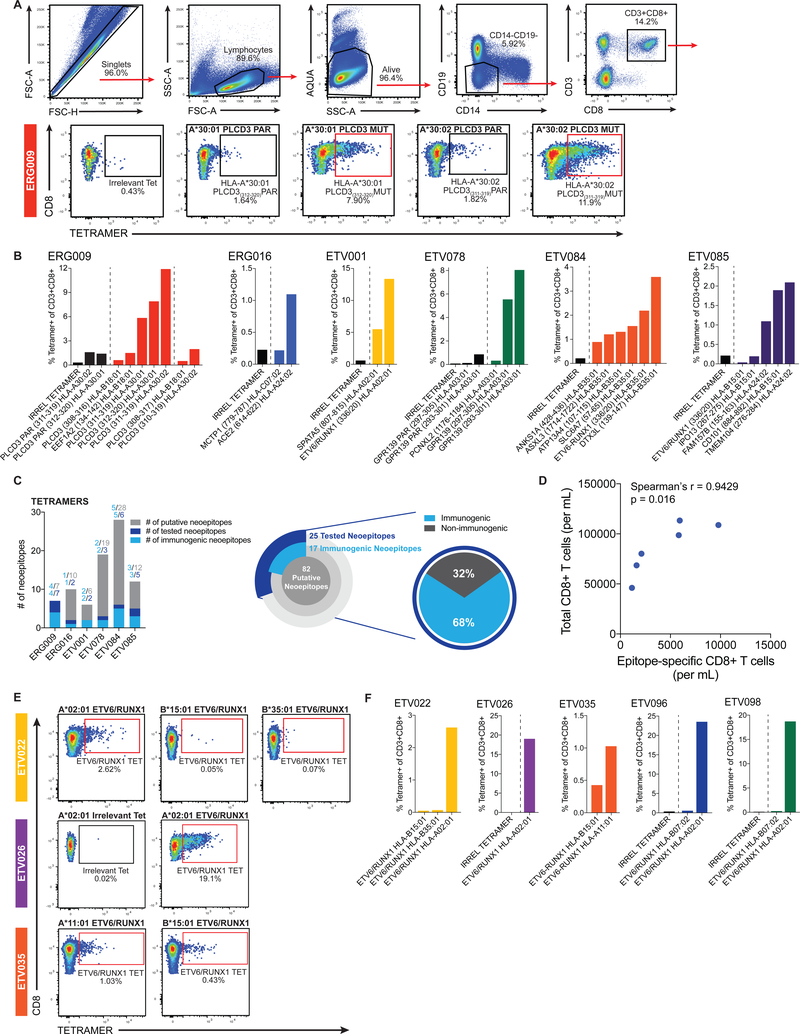
In our phenotypic characterization of ALL-specific CD8+ TILs, we observed some patients (e.g., ETV078, ETV084) with PD-1/Tim-3 positive CD8+ T cells, the characteristic phenotype of terminal exhaustion. Although our aAPC and functional assays provided clear evidence of tumor-reactive T cells, functional assays poorly detect exhausted cells, which can lead to underestimating the overall response magnitude. Thus, to more precisely quantify the extent to which pediatric ALLs contain a pool of endogenous tumor-reactive CD8+ T cells, we mapped TIL responses to specific epitopes by generating twenty-five patient-specific tetramers that correspond to the predicted neoepitopes identified previously (table S3). Importantly, we utilized nine unique HLAA, -B, and -C alleles in order to accurately assess the global neoantigen-specific CD8+ T cell response in pediatric ALL. Fig. 3A shows a representative tetramer gating strategy that was used to identify freshly isolated neoepitope-specific CD8+ TILs. In all six patients, we identified one or more neoepitope-specific CD8+ TIL populations, each of which constituted from 1.1% to 13.4% of the total CD8+ TIL pool (Fig. 3B). Of the 25 tested patient-specific tetramers, 68% bound to TILs above background set by irrelevant HLA-matched tetramers, indicating that a large proportion of the predicted neoepitopes elicited an endogenous antitumor response from CD8+ T cells (Fig. 3C and fig. S5). Within those responses, we observed immunodominance hierarchies among the distinct TIL populations, with a majority of tetramer-bound CD8+ T cells restricted to one or two putative neoepitopes. Importantly, the tetramer-bound CD8+ T cells recognizing these neoepitopes exhibited both recently activated and chronically activated phenotypes (as defined by PD1 and Tim-3 expression), further supporting the specificity of these cells for their corresponding neoantigens and suggesting that variable states of differentiation and functionality may exist within neoantigen-specific CD8+ T cells (fig. S6).
Fig. 3. Antitumor CD8+ T cell responses are neoepitope-specific and form immunodominance hierarchies.
(A) Representative gating strategy from one patient used to identify and quantify neoepitope-specific CD8+ T cells in the bone marrow of patients with ALL. HLA tetramer staining of HLA-restricted CD8+ TILs is depicted for ERG009. (B) Frequency of CD3+CD8+ TILs from six patients binding irrelevant and/or parent tetramers (black bars), and mutant tetramers (colored bars) within the bone marrow of patient samples. Dashed lines distinguish tetramers complexed with irrelevant and/or parent peptides from tetramers complexed with nonamer and decamer mutant peptides. (C) T cell response statistics for all putative neoepitopes per patient (bar chart) and at the cohort level (pie charts). (D) Scatterplot depicting the relationship between the total CD8+ T cell and neoepitope-specific CD8+ T cell response from six patients in our cohort. Correlation coefficient (r) and p-value (p) were calculated using the Spearman rank-order correlation test. (E) Representative tetramer gating from three patients used to identify and quantify neoepitope-specific CD8+ T cells in an additional cohort of patients with the ETV6-RUNX1 gene fusion. (F) Frequency of CD3+CD8+ TILs binding irrelevant (black bars), and ETV6-RUNX1 fusion tetramers (colored bars) within the bone marrow of five additional patients containing the ETV6-RUNX1 gene fusion. Dashed lines distinguish tetramers complexed with irrelevant peptides from tetramers complexed with fusion peptides derived from ETV6-RUNX1. Representative flow cytometry plots depict the frequency of CD8+ TILs binding irrelevant, mutant tetramers (red outline; cancer neoantigen bound to patient-specific HLA), and/or wild-type (parent self-peptide bound to patient-specific HLA) tetramers.
Altogether, these results demonstrate that robust CD8+ T cell responses are observable in pediatric leukemias and that a large proportion of potential neoepitopes are targeted by the immune system. Combining the results from the functional and tetramer binding assays, we find that 86% of the tested neoantigenic peptides and 68% of the generated tetramers elicited a response above background. Using these data to calculate the total number of tumor-specific cells in each patient, we found that the overall magnitude of the CD8+ TIL response in the bone marrow was correlated with the magnitude of the tumor-specific response, suggesting that the antitumor response represents a major component of the TIL population (Spearman’s rank correlation ρ = 0.9429, P = 0.016, Fig. 3D).
In addition to characterizing the neoantigen-specific CD8+ T cell response against patient-specific somatic missense mutations, we also aimed to establish whether common gene fusions found in pediatric ALL could also be targeted by CD8+ T cells. To this end, we focused on a cohort of patients with a characteristic fusion between ETV6 and RUNX1, which is found in ~20–25% of cases of pediatric ALL(42) and is associated with a more favorable prognosis. In our original patient set we observed ETV6-RUNX1-targeted responses in two out of four patients, though the responses were restricted by different HLAs (A02:01 and B35:01). We thus selected five more patients for testing, including four with A02:01 alleles along with several other alleles predicted to bind a fusion-derived peptide. In all five patients, at least one measurable response above background was detected for CD8+ T cells recognizing neoepitopes derived from the ETV6-RUNX1 gene fusion (Fig. 3E,F).
Given the degree of antitumor reactivity observed, we designed additional tetramers in order to test whether the antitumor responses cross-reacted with the endogenous, unmutated self-(“parent”) epitopes. Of the 8 parent tetramers that were designed, 4 failed to fold, likely due to a reduction in peptide:MHC affinity for the parent peptide in comparison to the mutated peptide. However, all four of the folded parent tetramers exhibited reactivity similar to irrelevant background tetramer staining that was lower than the corresponding neoepitope tetramers’ reactivity (fig. S5), verifying that the responses detected by our tetramer assays are highly specific for the mutated neoepitopes. One concern with tetramer-based analyses is the potential for non-specific binding. Although staining with tetramers containing the parent epitope serves as a strong control for this, we also stained each patient with irrelevant tetramers (designed to match the HLA of the patient) in order to rule out any inherent staining that results from binding between the patient’s TCRs and the tetramer’s HLA. The background in each patient to a matched HLA tetramer were consistent with unstained samples, indicating that the high reactivity observed in the tetramer-binding assays did not result from non-specific binding caused by a patient-intrinsic phenomenon. To rule out potential promiscuous binding from the tetramers that bound high frequencies of cells, we used those tetramers to stain cells from patients with matching HLAs but lacking the mutations associated with the tetramer-specific neoepitopes (table S4). Again, none of the tetramers showed substantial off-target binding, further demonstrating the specificity of each patient’s endogenous CD8+ T cell response towards their corresponding neoepitopes.
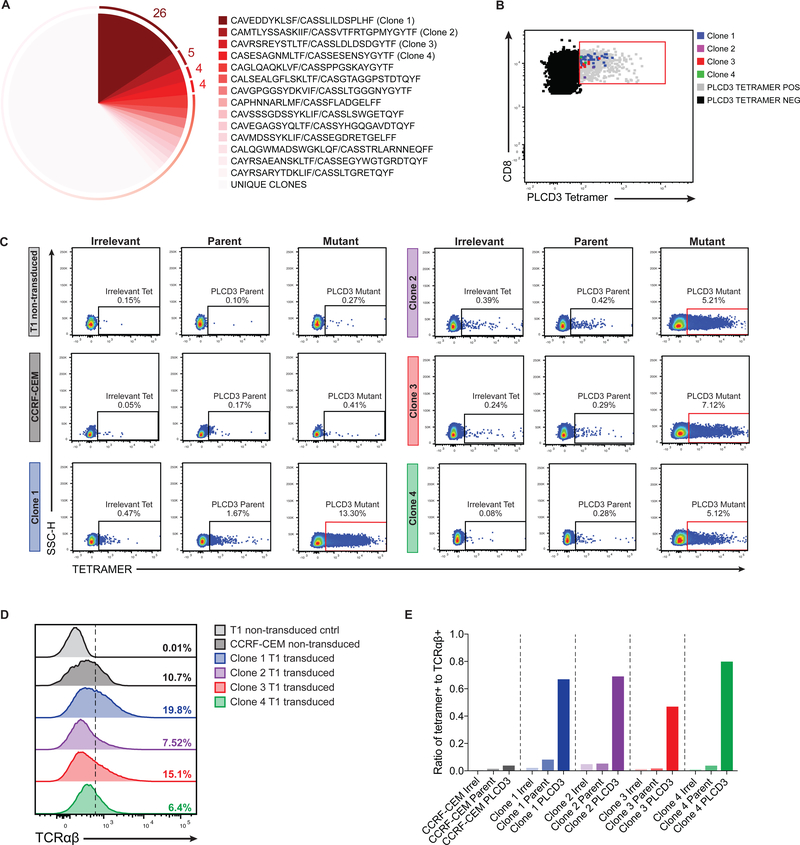
Given the broad spread of mean fluorescence intensities observed in tetramers that bound above background, we set out to confirm that cells from the full range of positive staining were indeed specific. We sequenced the paired αβ T cell receptor (TCR) from 166 PLCD3 tetramer-binding cells from patient ERG009. We observed extensive evidence of clonality, with the top 4 clonal expansions representing over 23% of this epitope-specific CD8+ T cell population (Fig. 4A). Focusing on these 4 clonal expansions, which represented a range of tetramer-staining intensities (Fig. 4B), we cloned their αβ TCRs. We then stably transduced these receptors by lentiviral transduction into the SUP-T1 cell line and confirmed TCR expression. Each epitope-specific CD8+ TCR clone demonstrated considerable tetramer reactivity (fig. 4C–E). Importantly, these receptors bound neither the matched HLA parent tetramer containing the unmutated peptide nor the irrelevant tetramer. Additionally, the specificity of the mutant tetramer for the cloned TCRs was further supported by the lack of tetramer binding observed within the CCRF-CEM cell line that expressed an irrelevant TCR (fig. 4C,E). Thus, all four cloned receptors, representing 39 cells from the original sort and 23% of the cells binding to this tetramer, were confirmed to be neoepitope-specific.
Fig. 4. Characterization and confirmation of the PLCD3-neoepitope TCRαβ repertoire in patient ERG009.
Ex vivo stained CD8+ TILs from ERG009 were single-cell sorted based on PLCD3 tetramer binding (HLA-A*30:02 PLCD3(311–319) mutant tetramer) for TCR analysis. (A) Clonotypic analysis of paired T cell receptor CDR3 regions (CDR3α and CDR3β) using single-cell multiplexed PCR on sorted PLCD3+ CD8+ TILs from patient ERG009. Numbers adjacent to the pie chart slices represent the number of PLCD3 tetramer-binding cells within clonally expanded populations. (B) Flow cytometry analysis of PLCD3 tetramer-binding CD8+ T cells. Dot plot depicts populations of PLCD3 tetramer negative (shaded black) and sorted PLCD3 tetramer positive (gated, shaded gray) CD8+ TILs from ERG009. Events corresponding to the top 4 clonotypes (shaded blue, purple, red, and green) from the sorted cells were overlaid onto the tetramer positive population. (C) Flow cytometry plots depicting the frequency of SUP-T1 non-transduced cells (light gray), CCRF-CEM cells expressing an irrelevant TCR (dark gray), and SUP-T1 TCR transduced cells containing CDR3α and CDR3β corresponding to clones marked in 4A binding either an irrelevant (left panels), parent PLCD3 (middle panels), or mutant PLCD3 tetramer (right panels). (D) Flow cytometry histograms showing the frequency of TCR-expressing cells (as in 4C). (E) Ratio of PLCD3 tetramer-positive cells (from 4C) to TCR-expressing cells (from 4D).
The transcriptional profiles of ALL-specific CD8+ TILs exhibit inter- and intra-patient heterogeneity
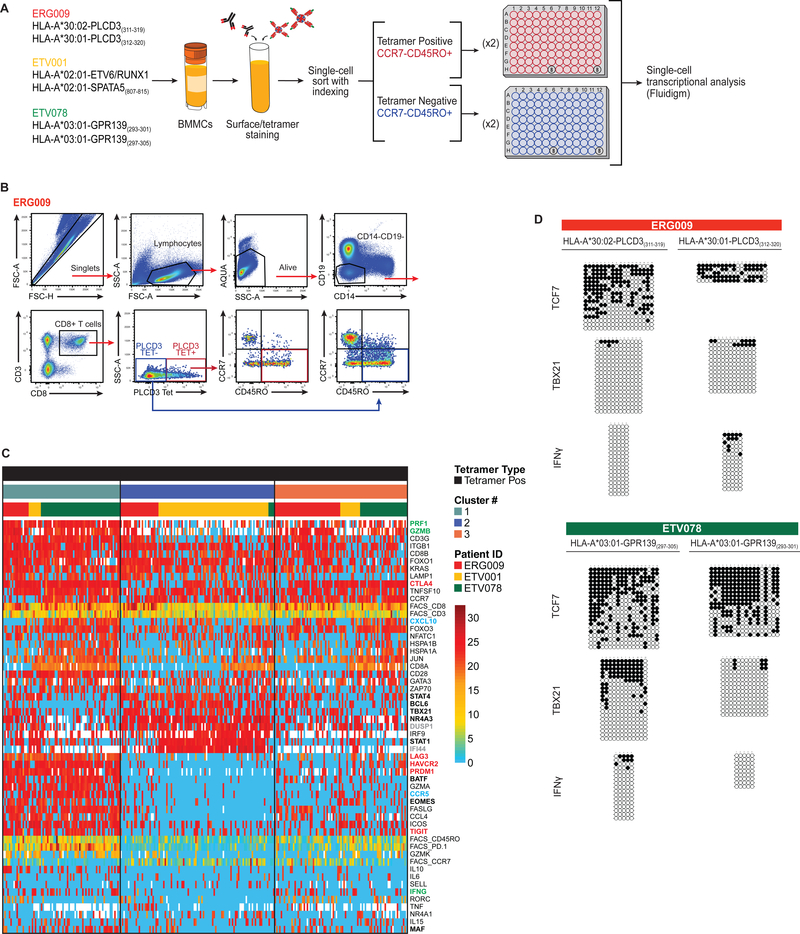
The functional and specificity assays indicated that the tumor-specific CD8+ T cells in our samples had a range of intrinsic activities. For example, adding together all the PLCD3 tetramers tested from patient ERG009 resulted in a predicted frequency of approximately 28% PLCD3-specific cells, and the functional cytokine assay correspondingly indicated a 29% response rate. In contrast, in ETV001 and ETV078, certain epitopes had much higher frequencies associated with tetramer staining than found in the functional assay; for example, the GPR139 tetramer in ETV078 bound to approximately 14% of the CD8+ T cells, while the functional assays only detected about 6% of CD8+ T cells as responsive to the mutated peptide. Hypothesizing that this discordance may be indicative of dysfunction in subsets of these cell populations, we further interrogated the tumor-specific CD8+ T cells via single-cell transcriptional profiling.
To characterize the transcriptional environment of neoepitope-specific TILs, we performed multiplexed quantitative PCR analysis on tetramer-positive and -negative CD8+ populations; by using an index sorting approach, we were able to simultaneously assay transcription and cell-surface phenotype at single-cell resolution (fig. 5A,B; table S5). Due to the scarcity of material available from the tumor biopsies, this analysis was restricted to the three patients with the greatest number of samples and the highest frequency of tetramer-positive CD8+ T cells. The 88 genes we selected for analysis included: transcription factors shown to influence CD8+ T cell differentiation (43), chemokines and chemokine receptors, cytokines, activating and inhibitory co-receptors, and molecules associated with degranulation and cell survival (see Materials and Methods for exclusion criteria and expression-level determination).
Fig. 5. The transcriptional profiles of neoepitope-specific CD8+ TILs exhibit inter- and intra-patient heterogeneity.
(A) Workflow of the experimental strategy for single-cell transcriptional studies. Bone marrow cells from three patients (ERG009, ETV001, and ETV078) were stained using an antibody cocktail and neoepitope-specific tetramers. CCR7−CD45RO+ tetramer-binding CD8+ TILs were single-cell index sorted for transcriptome studies. (B) Representative gating strategy from one patient depicting single-cell sorted CD8+ TIL subsets: tetramer-positive CCR7−CD45RO+ (red outline), and tetramer-negative CCR7−CD45RO+ (blue outline); 94 single cells from these two gates were sorted into 96-well plates. (C) Heatmap visualizing unscaled expression of genes (transcript expression threshold values; Et) and scaled surface protein data (MFI) for single-cell sorted neoepitope-specific CD8+ TILs (white indicates missing data) from three patients. Genes are ordered according to Ward’s method, and two clusters of relatively invariant genes were removed for ease of visualization. Top margin color bars represent, from top to bottom, groupings based on tetramer type (Tetramer positive), hierarchical cluster number (clusters 1–3), and patient IDs (ERG009, ETV001, and ETV078). Bolded gene name colors represent: transcription factors (black), inhibitory receptors (red), functional molecules (green), chemokine/chemokine receptors (blue), and transcriptional regulators (gray). (D) Representative bisulfite sequencing DNA methylation analysis of TCF7, TBX21, and IFNγ loci among bulk CCR7−CD45RO+ neoepitope-specific CD8+ TILs from two patients.
To assess the degree of heterogeneity in tumor-specific CD8+ TILs both within and between patients, we focused on the neoepitope-specific effector-phenotype (CCR7−CD45RO+) TILs from ERG009, ETV001, and ETV078. The expression of activation markers PD1 and Tim3 varied considerably, with ETV078 exhibiting a large proportion of cells with an exhausted phenotype (fig. S7). In line with recent molecular profiling studies (44–46), we observed a spectrum of dynamic transcriptional changes that take place during antigen-specific CD8+ T cell responses. We identified three clusters (hierarchical clusters 1–3) with distinct transcriptional signatures across the sorted TIL populations, which differed in their functional status and in the expression of transcription factors, transcriptional regulators, signaling inhibitors, cytokines, cytotoxic granules, and trafficking molecules (Fig. 5C). Specifically, these clusters were distinguished by transcription factors and regulators associated with functional effector (43, 47) (TBX21 and EOMES), dysfunctional (48, 49) (STAT1, STAT3, STAT4, NR4A2, NR4A3 and BCL6) or exhausted (45, 46) (EOMES, MAF, PRDM1, and BATF) CD8+ T cells. Consistent with published findings, (47, 50, 51) exhausted cells expressed several inhibitory receptors (HAVCR2, LAG3, TIGIT, and CTLA4; Fig. 5C; fig. S7). We observed an exhaustion phenotype (cluster 1) as the majority of tetramer-positive effectors in only a single patient (ETV078), but notably this patient also exhibited cells that lacked the exhaustion signature (cluster 3). In ETV001, the dominant subset of antigen-specific CD8+ T cells exhibited expression profiles concordant with what has been described elsewhere (48) as a dysfunctional phenotype (cluster 2), characterized by a type I IFN signature. In contrast, most cells from ERG009 exhibited an expression profile corresponding with a classical effector phenotype (43, 52–54) (GzmB, IFNG, CXCL10) and lacked exhaustion markers.
It has recently been reported that CD8+ T cell differentiation and exhaustion have been coupled to stable changes in DNA methylation programming at loci associated with developmental plasticity and effector function (e.g., TBX21, TCF7, and IFNγ)(55, 56). Because our data indicated that many of the neoepitope-specific CD8+ TILs were functional, we tested the methylation profiles for ERG009 and ETV078. Consistent with the effector-like molecular signature we observed for ERG009, we found that the IFNγ and TBX21 loci for CD45RO+ sorted cells from this patient were predominantly unmethylated (Fig. 5D). In contrast, the CD45RO+ GPR139(297–305) tetramer-sorted cells from ETV078 displayed a more heterogeneous methylation program for the TBX21 locus, which may reflect a greater progression towards an exhaustion phenotype as might be expected given the greater proportion of exhausted cells we observed in this patient compared to ERG009.
Additionally, the partial demethylation of the TCF7 locus, which has been associated with retention of T cell developmental plasticity (57), supports the notion that this pool of neoepitope-specific CD8+ TILs may contain cells at various stages of differentiation, including functional effectors. Taken together, our results demonstrate that pediatric ALL elicits a potent neoepitope-specific CD8+ T cell response that exhibits molecular hallmarks of functional effector differentiation.
Discussion
There are at least three non-mutually exclusive possibilities to explain the much higher response rate in these ALL tumors compared to reports from adult solid tumors. First, it is hypothetically possible that pediatric patients may mount fundamentally different responses compared to adults. Second, hematologic malignancies, and in particular B cell tumors, may be inherently more immunogenic, as they interact with responding T cells as professional immune cells and may therefore provide greater accessibility to infiltrating T cells by not forming a dense epithelial matrix. Last, lower mutation burdens may force the immune response to target the majority of potential neoantigens; in the context of a highly diverse epitope landscape, immunodominance effects might drive the response to target a small subset of antigens. Although common mechanisms of immune evasion, such as the downregulation of specific HLA molecules, might help explain the skewing of the immune response towards a limited number of neoepitopes, we did not find any evidence of selective downregulation of specific HLA molecules transcriptionally or when staining for the corresponding proteins.
There is some evidence that certain B cell malignancies are highly immunogenic and controllable by T cell responses. Hodgkin’s lymphoma, and in particular those with a viral etiology from EBV or HIV, have been shown to be highly responsive to PD1/PDL1 blockade (58–61). In contrast, ICB has shown limited efficacy in other forms of blood cancers, most notably in multiple myeloma (62, 63). Although responsiveness to checkpoint blockade is not equivalent to immunogenicity in general, the consistent correlation between mutation burden and checkpoint responsiveness has lent support to the idea that mutation burden and immunogenicity are correlated. In the case of ALL, the positive prognosis for standard patients has limited the scope of trials focused on checkpoint blockade. Nonetheless, our data would suggest that the failure of standard checkpoint blockade therapies in ALL is not a consequence of a complete lack of immunogenicity, as we found activated T cells responding to tumor epitopes across every patient. Instead, we suspect this may be a problem of efficiency, with a relatively low effector:target ratio between the antitumor response and the tumor itself, which has an intrinsically high rate of replication. Furthermore, only a minor portion of the neoepitope-specific T cells we examined exhibited an exhausted phenotype and expression of PD1, thus there may not be a clear target for PD1-checkpoint blockade in this disease context. In order to fully validate our findings and further address these questions, studies with larger patient cohorts and across multiple institutions would be needed. Recent efforts (64, 65) to identify alternative checkpoints for hematologic malignancies, such as CD200-CD200R, might also explain the lack of antitumor efficacy of these cells.
In addition to identifying antitumor responses for every patient examined, the other striking feature of these experiments was that several mutations within the same patient elicited a T cell response. In some patients, we were able to account for over 50% of the responsive T cell population in the tumor, though in others the response rate was much lower. A possible explanation for the reduced response rate is that the rules for peptide binding to certain HLA molecules are incompletely defined owing to the smaller datasets available to train the predictive algorithms used to identify the putative neoepitopes used in our screening assays. Mass spectrometry-based screening approaches could be carried out in tandem to gain a more complete picture of the cancer neoantigens that are processed and presented on tumor cells; however, the large cellular input required for this approach was a limiting factor in our studies. Additionally, our focus on fusions and SNV mutations, without consideration of frameshift or indel mutations, may have limited our ability to detect some responses. One possible explanation for the high rate of responsiveness may be immunodominant focusing on the relatively small number of mutations present in the tumor, particularly when compared to adult solid tumors. Immunodominance effects are well-defined in viral models of CD8+ T cell responses and have also been increasingly described in human tumors (8). Immunodominance can create extreme biases, with single, dominant, epitope-targeted T cell populations representing 70–80% of the entire CD8+ T cell response to a complex antigen (66). Strikingly, this remains true even in responses to viruses, where hundreds of potential epitopes are theoretically possible and large numbers of peptides are verifiably processed and presented in infected cells (67, 68). The parameters that determine immunodominant selection have not been definitively determined, but it is clear that peptide-MHC concentration and T cell precursor frequency are implicated. Tumors with large numbers of mutations and a high potential number of neoepitopes might be akin to a large virus, where only a selected number of epitopes are actually targeted due to this immunodominance effect; relatedly, the immune response may target a greater proportion of potential neoepitopes in tumors with low mutation burden, which is consistent with our observations.
Finally, determining the heterogeneity of antigen presentation and the extent of inhibitory receptor expression across the tumor will be necessary to assess the extent to which any particular tumor cell is targetable by these effector responses. In support of this idea, we found interesting parallels between the extent of inhibitory receptor ligand expression by tumor cells (fig. S8) and the frequency of exhausted CD8+ T cells (fig. 5 and fig. S2) in our patient samples. Tumors (e.g., ETV078 and ERG016) with increased frequencies of exhausted CD8+ T cells (characterized by high PD-1 and TIM-3 expression) also expressed greater amounts of the corresponding inhibitory ligands (PDL1 and galectin-9, respectively); however, a more in-depth analysis of these factors would be necessary to fully understand their contribution to limiting the immune response. Co-culture of purified T cells and tumor cells from the diagnostic sample in two patients over three weeks expanded T cells specific for mapped tumor-specific antigens. These data indicate that at least these antigens were actively presented on the ex vivo tumor cells, but the heterogeneity of presentation and the role of cross-presentation in inducing endogenous T cell responses cannot be determined from these experiments. In order to estimate the heterogeneity in antigen presentation, we used mutant allele frequency value cutoffs established previously (69) to determine the clonality of targeted mutations within our tumor samples. Although our analysis indicated that the mutations were present in the majority of ALL blasts and were not sub-clonal (table S7), additional single-cell and longitudinal studies would be needed to address questions of clonality and immunoediting definitively. Additionally, in vitro or in vivo killing assays with patient-derived xenograft models would help determine the potential potency of these antitumor responses.
Our data provide an opportunity to consider adoptive T cell, monoclonal antibody, and targeted TCR therapy for ALL in pediatric patients. Although ALL currently has a high response rate to traditional chemotherapy approaches, the treatment protocols are extended and induce substantial sequelae. The potential utility of immunotherapies in treating ALL relapse is perhaps even more relevant, as relapse in ALL currently leads to particularly poor prognosis. It is notable that in our study, seven out of nine patients with the ETV6-RUNX1 gene fusion generated T cells responses targeting this neoantigen. This raises the possibility of generating off-the-shelf therapeutics that target the primary driver mutation in patients with this and other common fusions. Whereas the public response to these common neoantigens offers more immediate clinical applications, developing a pipeline to rapidly adapt TCRs targeting private neoantigens into therapeutics would be a more generalizable approach for all patients. Although the antitumor T cell responses in these patients did not control their tumors, augmenting antitumor T cell frequencies and providing enhanced activation signals in the context of transgenic TCR therapy may lead to significant improvements in immune control and should be explored in clinical treatments.
Materials and Methods
Study Design
The objective of this study was to investigate the response of endogenous CD8+ T cells against patient-specific cancer neoantigens from diagnostic tumor samples collected from pediatric patients with ALL. We hypothesized that although pediatric leukemias contain lower mutational burdens, the neoantigens that arise in this tumor type likely serve as functional targets due to their increased immunogenicity resulting from the tumor’s microenvironment, cellular origin, and high neoepitope expression (34). To test whether endogenous CD8+ TILs were able to effectively recognize and target these mutated neoepitopes directly ex vivo, we monitored TIL degranulation and production of IFNγ and TNFα after stimulation with both aAPCs and autologous tumor cells. We assessed the global neoantigen-specific CD8+ T cell response by mapping TIL responses to specific epitopes by utilizing patient-specific tetramers that corresponded to predicted neoepitopes across HLA-A, -B, and -C alleles. We further characterized the nature of the endogenous CD8+ T cell response by determining, at the single cell level, the transcriptional profile of tumor-reactive CD8+ TILs and comparing their inter- and intra-patient heterogeneity. Researchers could not be blinded to experimental procedures due to the nature of the neoantigen screening assays requiring patient-specific neoantigens be used to assess endogenous CD8+ T cell functional responses and because of the limited quantity of material. Pediatric ALL specimens were selected based on sample availability and on the cancer type (e.g., B-ALL). The use of human tissues was approved by the institutional review board (IRB #00000029 – Expedited Protocol XPD-13–098) of St. Jude Children’s Research Hospital. Primary data are reported in data file S1.
Statistical analysis
For single-cell expression data, Ct values were recovered from the BioMark HD, and data were analyzed using the Fludigm Real-Time PCR analysis software. The quality threshold was set to 0.65, and a linear derivative was used for baseline correction. Ct values of filtered single-cell expression data were converted to expression thresholds (Et) and analyzed using the MAST statistical framework (70) in R, which utilizes hurdle models designed specifically for single-cell expression data that take into account both the proportion of cells expressing a gene and the expression of the gene. Cells with a cellular detection rate in the 1% and 99% quantiles were filtered from further analysis, and missing data points were considered as zero-values unless otherwise indicated. Additional statistical methods are described in the figure legends and in the relevant methods descriptions; in general, presented data were largely characterized descriptively without the use of explicit statistical analyses. Hierarchical clustering was performed as described in the “Analysis and visualization of single-cell expression data” section of the Materials and Methods in the Supplementary Material.
Supplementary Material
Fig. S1. Experimental pipeline used to identify cancer neoepitopes.
Fig. S2. Phenotypic characterization of CD8+ tumor infiltrating lymphocytes from human bone marrow samples.
Fig. S3. Generation of aAPCs expressing single patient-specific HLA class I molecules.
Fig. S4. Healthy donors exhibit negligible responses against endogenous neoantigens.
Fig. S5. Neoepitope tetramers are patient-specific and exhibit negligible nonspecific binding.
Fig. S6. Phenotypic characterization of neoepitope-specific CD8+ TILs.
Fig. S7. Patient-specific transcriptional profiles.
Fig. S8. Phenotypic characterization of patient-specific CD19+ tumor cells.
Table S1. Patient ALL subtypes.
Table S2. FPKM values for HLA genes.
Table S3. Patient-specific neoepitopes.
Table S4. Tetramers used for specificity assays.
Table S5. Fluidigm primer list.
Table S6. Peptides for functional assays.
Table S7. Mutant allele frequencies (MAF) for sequenced mutations.
Table S8. Single-cell indexed FACS median fluorescence intensity (MFI).
Data file S1. Primary data.
Acknowledgments:
We would like to thank Didier Trono (Global Health Institute at École Polytechnique Fédérale de Lausanne) for the psPAX2 and pMD2.G constructs.
Funding: This work was supported by the National Institutes of Health (grants R01 AI107625 (Thomas), R01AI136514 (Thomas), R35 GM118041 (Chen), R35-CA197695–01A1 (Mullighan), NIGMS 5 P50GM115279–02 Projects 1 and 2 (Mullighan), and CORE C (Zhang), NCI 5 P01CA096832–12 (Zhang), R01 AI11442 (Youngblood)), Key for a Cure Foundation (Thomas and Youngblood), and ALSAC. This work was supported by the National Institutes of Health (grants R01 AI107625 (P.G.T.), R01AI136514 (P.G.T.), R35 GM118041 (Chen), NCI Outstanding Investigator Award R35-CA197695–01A1 (Mullighan), NIGMS 5 P50GM115279–02 Projects 1 and 2 (Mullighan), and CORE C (Zhang), NCI 5 P01CA096832–12 (Zhang), R01 AI11442 (B.A.Y.)), Key for a Cure Foundation (P.G.T and B.A.Y.), and ALSAC.
P.T. and X.Z.G are inventors on patent application WO2017096239A1 submitted by St. Jude Children’s Research Hospital that covers approaches for cloning and expression of T cell receptors. B.A.Y. is an inventor on patent application WO2017079642A1, submitted by Emory University and St. Jude Children’s Research Hospital, related to the manipulation of DNMT3a activity to enhance T cell based immunotherapies, and, with H.A., patent application WO2018104909A2, submitted by St. Jude Children’s Research Hospital that covers bioinformatic tools to assess epigenetic programs associated with T cell differentiation. B.A.Y. is listed on patents related to DNA methylation profile and biomarkers WO2018104909 and DNMT3A Gene Modifications US15/773,783. P.G.T. has received speaking fees from PACT Pharma and Illumina. C.G.M. has declared financial relationships with Pfizer, AbbVie, and Loxo Oncology (research funding); Amgen and Pfizer (speakers bureau); Amgen and Pfizer (travel fees, honoraria). C.G.M. has received advisory/consulting fees from Pfizer and Amgen.
Footnotes
Data and materials availability: All data associated with this study are present in the paper or Supplementary Materials. Single-cell gene expression data have been submitted to the Gene Expression Omnibus (GSE130670) and single-cell protein data is provided in Table S8. All other sequencing data can be accessed via European Genome-Phenome Archive (www.ebi.ac.uk/ega/search/site/PCGP) and St Jude Pediatric Cancer (PeCan) Data Portal (https://pecan.stjude.org/home). The EGA numbers corresponding to the data in this manuscript are EGAD00001002654 and EGAS00001000514.
Code and Data availability
Single-cell gene expression data have been submitted to the Gene Expression Omnibus (GSE130670) and single-cell protein data is provided in Table S8. All other sequencing data can be accessed via European Genome-Phenome Archive (www.ebi.ac.uk/ega/search/site/PCGP) and St Jude Pediatric Cancer (PeCan) Data Portal (https://pecan.stjude.org/home). The EGA numbers corresponding to the data in this manuscript are EGAD00001002654 and EGAS00001000514.
Competing interests: All other authors declare no competing interests.
References and Notes:
- 1.Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM, Targeting neoantigens to augment antitumour immunity, Nat. Rev. Cancer 17, 209–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schumacher TN, Schreiber RD, Neoantigens in cancer immunotherapy, Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Robbins PF, Kassim SH, Tran TLN, Crystal JS, Morgan RA, Feldman SA, Yang JC, Dudley ME, Wunderlich JR, Sherry RM, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee C-CR, Li YF, El-Gamil M, Rosenberg SA, A Pilot Trial Using Lymphocytes Genetically Engineered with an NY-ESO-1–Reactive T-cell Receptor: Long-term Follow-up and Correlates with Response, Clin. Cancer Res. 21, 1019–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strønen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, Donia M, Böschen M-L, Lund-Johansen F, Olweus J, Schumacher TN, Targeting of cancer neoantigens with donor-derived T cell receptor repertoires, Science 352, 1337–1341 (2016). [DOI] [PubMed] [Google Scholar]
- 5.van Buuren MM, Calis JJ, Schumacher TN, High sensitivity of cancer exome-based CD8 T cell neo-antigen identification, Oncoimmunology 3, e28836 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS, Li YF, El-Gamil M, Rosenberg SA, Robbins PF, Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes, J. Clin. Invest. 125, 3981–3991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wölfel C, Huber C, Wölfel T, The response of autologous T cells to a human melanoma is dominated by mutated neoantigens, Proc. Natl. Acad. Sci. U. S. A. 102, 16013–16018 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamora AE, Crawford JC, Thomas PG, Hitting the Target: How T Cells Detect and Eliminate Tumors, J. Immunol. 200, 392–399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Restifo NP, Dudley ME, Rosenberg SA, Adoptive immunotherapy for cancer: harnessing the T cell response, Nat. Rev. Immunol. 12, 269–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadelain M, Rivière I, Riddell S, Therapeutic T cell engineering, Nature 545, 423–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J, Mulder GE, Toebes M, Vesely MD, Lam SSK, Korman AJ, Allison JP, Freeman GJ, Sharpe AH, Pearce EL, Schumacher TN, Aebersold R, Rammensee H-G, Melief CJM, Mardis ER, Gillanders WE, Artyomov MN, Schreiber RD, Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens, Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Lu Y-C, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, Rosenberg SA, Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells, Nat. Med. 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJA, Behjati S, Hilkmann H, El Atmioui D, Nieuwland M, Stratton MR, Kerkhoven RM, Kesmir C, Haanen JB, Kvistborg P, Schumacher TN, Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma, J. Clin. Oncol. 31, e439–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu T-E, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR Jr, Villaruz LC, Havel L, Krejci J, Corral Jaime J, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA, CheckMate 026 Investigators, First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer, N. Engl. J. Med. 376, 2415–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn M-J, Felip E, Lee J-S, Hellmann MD, Hamid O, Goldman JW, Soria J-C, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L, KEYNOTE-001 Investigators, Pembrolizumab for the treatment of non-small-cell lung cancer, N. Engl. J. Med. 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM, Safety and activity of anti-PD-L1 antibody in patients with advanced cancer, N. Engl. J. Med. 366, 2455–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M, Safety, activity, and immune correlates of anti-PD1 antibody in cancer, N. Engl. J. Med. 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maude SL, Teachey DT, Porter DL, Grupp SA, CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia, Blood 125, 4017–4023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stromnes IM, Schmitt TM, Hulbert A, Brockenbrough JS, Nguyen HN, Cuevas C, Dotson AM, Tan X, Hotes JL, Greenberg PD, Hingorani SR, T Cells Engineered against a Native Antigen Can Surmount Immunologic and Physical Barriers to Treat Pancreatic Ductal Adenocarcinoma, Cancer Cell 28, 638–652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA, Genetic basis for clinical response to CTLA-4 blockade in melanoma, N. Engl. J. Med. 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA, Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer, Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA, Genomic correlates of response to CTLA-4 blockade in metastatic melanoma, Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R, Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers, Mol. Cancer Ther. 16, 2598–2608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, Wells DK, Cary CIO, Grbovic-Huezo O, Attiyeh M, Medina B, Zhang J, Loo J, Saglimbeni J, Abu-Akeel M, Zappasodi R, Riaz N, Smoragiewicz M, Kelley ZL, Basturk O, Australian Pancreatic Cancer Genome Initiative, Garvan Institute of Medical Research, Prince of Wales Hospital, Royal North Shore Hospital, University of Glasgow, St Vincent’s Hospital, QIMR Berghofer Medical Research Institute, University of Melbourne, Centre for Cancer Research, University of Queensland, Institute for Molecular Bioscience, Bankstown Hospital, Liverpool Hospital, Royal Prince Alfred Hospital, Chris O’Brien Lifehouse, Westmead Hospital, Fremantle Hospital, St John of God Healthcare, Royal Adelaide Hospital, Flinders Medical Centre, Envoi Pathology, Princess Alexandria Hospital, Austin Hospital, Johns Hopkins Medical Institutes, ARC-Net Centre for Applied Research on Cancer, Gönen M, Levine AJ, Allen PJ, Fearon DT, Merad M, Gnjatic S, Iacobuzio-Donahue CA, Wolchok JD, DeMatteo RP, Chan TA, Greenbaum BD, Merghoub T, Leach SD, Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer, Nature 551, 512–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, Rizvi NA, Merghoub T, Levine AJ, Chan TA, Wolchok JD, Greenbaum BD, A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy, Nature 551, 517–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava S, Riddell SR, Engineering CAR-T cells: Design concepts, Trends Immunol. 36, 494–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davila ML, Brentjens R, Wang X, Rivière I, Sadelain M, How do CARs work?: Early insights from recent clinical studies targeting CD19, Oncoimmunology 1, 1577–1583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chheda ZS, Kohanbash G, Okada K, Jahan N, Sidney J, Pecoraro M, Yang X, Carrera DA, Downey KM, Shrivastav S, Liu S, Lin Y, Lagisetti C, Chuntova P, Watchmaker PB, Mueller S, Pollack IF, Rajalingam R, Carcaboso AM, Mann M, Sette A, Garcia KC, Hou Y, Okada H, Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy, J. Exp. Med. 215, 141–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu Y-C, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, Gartner J, Jia L, Trebska-McGowan K, Somerville RP, Robbins PF, Rosenberg SA, Goff SL, Feldman SA, Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer, Nat. Med. 24, 724– 730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, Zhang W, Sougnez C, Cibulskis K, Sidney J, Stevenson K, Ritz J, Neuberg D, Brusic V, Gabriel S, Lander ES, Getz G, Hacohen N, Wu CJ, Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia, Blood 124, 453–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, Smith M, Anderson B, Villablanca JG, Matthay KK, Shimada H, Grupp SA, Seeger R, Reynolds CP, Buxton A, Reisfeld RA, Gillies SD, Cohn SL, Maris JM, Sondel PM, Children’s Oncology Group, Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma, N. Engl. J. Med. 363, 1324–1334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Federico SM, McCarville MB, Shulkin BL, Sondel PM, Hank JA, Hutson P, Meagher M, Shafer A, Ng CY, Leung W, Janssen WE, Wu J, Mao S, Brennan RC, Santana VM, Pappo AS, Furman WL, A Pilot Trial of Humanized Anti-GD2 Monoclonal Antibody (hu14.18K322A) with Chemotherapy and Natural Killer Cells in Children with Recurrent/Refractory Neuroblastoma, Clin. Cancer Res. 23, 6441–6449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM, Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden, Genome Med. 9, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachireddy P, Burkhardt UE, Rajasagi M, Wu CJ, Haematological malignancies: at the forefront of immunotherapeutic innovation, Nat. Rev. Cancer 15, 201–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW, Cancer genome landscapes, Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang T-C, Carter RA, Li Y, Li Y, Wang H, Edmonson MN, Chen X, Arnold P, Geiger TL, Wu G, Peng J, Dyer M, Downing JR, Green DR, Thomas PG, Zhang J, The Neoepitope Landscape in Pediatric Cancers, Genome Med.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, Hanada K-I, Almeida JR, Darko S, Douek DC, Yang JC, Rosenberg SA, PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors, J. Clin. Invest. 124, 2246–2259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, Platten M, Tabatabai G, Dutoit V, van der Burg SH, Thor Straten P, Martínez-Ricarte F, Ponsati B, Okada H, Lassen U, Admon A, Ottensmeier CH, Ulges A, Kreiter S, von Deimling A, Skardelly M, Migliorini D, Kroep JR, Idorn M, Rodon J, Piró J, Poulsen HS, Shraibman B, McCann K, Mendrzyk R, Löwer M, Stieglbauer M, Britten CM, Capper D, Welters MJP, Sahuquillo J, Kiesel K, Derhovanessian E, Rusch E, Bunse L, Song C, Heesch S, Wagner C, Kemmer-Brück A, Ludwig J, Castle JC, Schoor O, Tadmor AD, Green E, Fritsche J, Meyer M, Pawlowski N, Dorner S, Hoffgaard F, Rössler B, Maurer D, Weinschenk T, Reinhardt C, Huber C, Rammensee H-G, Singh-Jasuja H, Sahin U, Dietrich P-Y, Wick W, Actively personalized vaccination trial for newly diagnosed glioblastoma, Nature 565, 240–245 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Scheper W, Kelderman S, Fanchi LF, Linnemann C, Bendle G, de Rooij MAJ, Hirt C, Mezzadra R, Slagter M, Dijkstra K, Kluin RJC, Snaebjornsson P, Milne K, Nelson BH, Zijlmans H, Kenter G, Voest EE, Haanen JBAG, Schumacher TN, Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers, Nat. Med. 25, 89– 94 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Martin SD, Wick DA, Nielsen JS, Little N, Holt RA, Nelson BH, A library-based screening method identifies neoantigen-reactive T cells in peripheral blood prior to relapse of ovarian cancer, Oncoimmunology 7, e1371895 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran E, Ahmadzadeh M, Lu Y-C, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA, Immunogenicity of somatic mutations in human gastrointestinal cancers, Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shurtleff SA, Buijs A, Behm FG, Rubnitz JE, Raimondi SC, Hancock ML, Chan GC, Pui CH, Grosveld G, Downing JR, TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis, Leukemia 9, 1985–1989 (1995). [PubMed] [Google Scholar]
- 43.Kaech SM, Cui W, Transcriptional control of effector and memory CD8+ T cell differentiation, Nat. Rev. Immunol. 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, Zuniga EI, Goldrath AW, Chang JT, Yeo GW, Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing, Nat. Immunol. 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R, Molecular signature of CD8+ T cell exhaustion during chronic viral infection, Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Doering TA, Crawford A, Angelosanto JM, Paley MA, Ziegler CG, Wherry EJ, Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory, Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arsenio J, Kakaradov B, Metz PJ, Kim SH, Yeo GW, Chang JT, Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses, Nat. Immunol. 15, 365–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speiser DE, Ho P-C, Verdeil G, Regulatory circuits of T cell function in cancer, Nat. Rev. Immunol. 16, 599–611 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, Xia J, Kwon JYH, Nevin J, Herbst RH, Yanai I, Rozenblatt-Rosen O, Kuchroo VK, Regev A, Anderson AC, A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells, Cell 166, 1500–1511.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue H-H, Ahmed R, Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy, Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djenidi F, Adam J, Goubar A, Durgeau A, Meurice G, de Montpréville V, Validire P, Besse B, Mami-Chouaib F, CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients, J. Immunol. 194, 3475–3486 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Li G, Yang Q, Zhu Y, Wang H-R, Chen X, Zhang X, Lu B, T-Bet and Eomes Regulate the Balance between the Effector/Central Memory T Cells versus Memory Stem Like T Cells, PLoS One 8, e67401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peperzak V, Veraar EAM, Xiao Y, Babala N, Thiadens K, Brugmans M, Borst J, CD8+ T cells produce the chemokine CXCL10 in response to CD27/CD70 costimulation to promote generation of the CD8+ effector T cell pool, J. Immunol. 191, 3025–3036 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Reiser J, Banerjee A, Effector, Memory, and Dysfunctional CD8(+) T Cell Fates in the Antitumor Immune Response, J Immunol Res 2016, 8941260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, Youngblood B, De Novo Epigenetic Programs Inhibit PD-1 Blockade-Mediated T Cell Rejuvenation, Cell 170, 142–157.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdelsamed HA, Moustaki A, Fan Y, Dogra P, Ghoneim HE, Zebley CC, Triplett BM, Sekaly R-P, Youngblood B, Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis, J. Exp. Med. 214, 1593–1606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, Pradervand S, Thimme R, Zehn D, Held W, T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections, Immunity 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Merryman RW, Armand P, Wright KT, Rodig SJ, Checkpoint blockade in Hodgkin and non-Hodgkin lymphoma, Blood Adv 1, 2643–2654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carbone A, Gloghini A, Epstein Barr Virus-Associated Hodgkin Lymphoma, Cancers 10 (2018), doi: 10.3390/cancers10060163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ansell SM, Nivolumab in the Treatment of Hodgkin Lymphoma, Clin. Cancer Res. 23, 1623–1626 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P, PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma, N. Engl. J. Med. 372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suen H, Brown R, Yang S, Ho PJ, Gibson J, Joshua D, The failure of immune checkpoint blockade in multiple myeloma with PD-1 inhibitors in a phase 1 study, Leukemia 29, 1621–1622 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Goodman A, Patel SP, Kurzrock R, PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas, Nat. Rev. Clin. Oncol. 14, 203–220 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Wong KK, Khatri I, Shaha S, Spaner DE, Gorczynski RM, The role of CD200 in immunity to B cell lymphoma, J. Leukoc. Biol. 88, 361–372 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Gorczynski RM, Zhu F, Checkpoint blockade in solid tumors and B-cell malignancies, with special consideration of the role of CD200 Cancer Management and Research 9, 601–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hosie L, Pachnio A, Zuo J, Pearce H, Riddell S, Moss P, Cytomegalovirus-specific T cells restricted by HLA-cw* 0702 increase markedly with age and dominate the CD8+ T-cell repertoire in older people, Front. Immunol. 8, 1776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace ME, Keating R, Heath WR, Carbone FR, The cytotoxic T-cell response to herpes simplex virus type 1 infection of C57BL/6 mice is almost entirely directed against a single immunodominant determinant, J. Virol. 73, 7619–7626 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Croft NP, Smith SA, Wong YC, Tan CT, Dudek NL, Flesch IEA, Lin LCW, Tscharke DC, Purcell AW, Kinetics of antigen expression and epitope presentation during virus infection, PLoS Pathog. 9, e1003129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, Zhou X, Ma X, Sioson E, Li Y, Rusch M, Gupta P, Pei D, Cheng C, Smith MA, Auvil JG, Gerhard DS, Relling MV, Winick NJ, Carroll AJ, Heerema NA, Raetz E, Devidas M, Willman CL, Harvey RC, Carroll WL, Dunsmore KP, Winter SS, Wood BL, Sorrentino BP, Downing JR, Loh ML, Hunger SP, Zhang J, Mullighan CG, The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia, Nat. Genet. 49, 1211–1218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data, Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen S-C, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui C-H, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG, The genetic basis of early T-cell precursor acute lymphoblastic leukaemia, Nature 481, 157–163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang Y-L, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen S-C, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen I-M, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrózek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui C-H, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG, Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia, N. Engl. J. Med. 371, 1005–1015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O, OptiType: precision HLA typing from next-generation sequencing data, Bioinformatics 30, 3310–3316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao Y, Smyth GK, Shi W, The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote, Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karosiene E, Lundegaard C, Lund O, Nielsen M, NetMHCcons: a consensus method for the major histocompatibility complex class I predictions, Immunogenetics 64, 177–186 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Zhang GL, Ansari HR, Bradley P, Cawley GC, Hertz T, Hu X, Jojic N, Kim Y, Kohlbacher O, Lund O, Lundegaard C, Magaret CA, Nielsen M, Papadopoulos H, Raghava GPS, Tal V-S, Xue LC, Yanover C, Zhu S, Rock MT, Crowe JE, Panayiotou C, Polycarpou MM, Duch W, Brusic V, Machine learning competition in immunology - Prediction of HLA class I binding peptides, J. Immunol. Methods 374, 1–4 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA, Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival, Genome Res. 24, 743–750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG, T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection, Sci. Transl. Med. 4, 128 ra 42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaujoux R, Seoighe C, A flexible R package for nonnegative matrix factorization, BMC Bioinformatics 11, 367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumaki Y, Oda M, Okano M, QUMA: quantification tool for methylation analysis, Nucleic Acids Res. 36, W170–5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Experimental pipeline used to identify cancer neoepitopes.
Fig. S2. Phenotypic characterization of CD8+ tumor infiltrating lymphocytes from human bone marrow samples.
Fig. S3. Generation of aAPCs expressing single patient-specific HLA class I molecules.
Fig. S4. Healthy donors exhibit negligible responses against endogenous neoantigens.
Fig. S5. Neoepitope tetramers are patient-specific and exhibit negligible nonspecific binding.
Fig. S6. Phenotypic characterization of neoepitope-specific CD8+ TILs.
Fig. S7. Patient-specific transcriptional profiles.
Fig. S8. Phenotypic characterization of patient-specific CD19+ tumor cells.
Table S1. Patient ALL subtypes.
Table S2. FPKM values for HLA genes.
Table S3. Patient-specific neoepitopes.
Table S4. Tetramers used for specificity assays.
Table S5. Fluidigm primer list.
Table S6. Peptides for functional assays.
Table S7. Mutant allele frequencies (MAF) for sequenced mutations.
Table S8. Single-cell indexed FACS median fluorescence intensity (MFI).
Data file S1. Primary data.