Abstract
Background
We report a large family with four successive generations, presenting with a complex phenotype of severe congenital neutropenia (SCN), partially penetrant monocytosis, and hearing loss of varying severity.
Methods
We performed whole exome sequencing to identify the causative variants. Sanger sequencing was used to perform segregation analyses on remaining family members.
Results
We identified and classified a pathogenic GFI1 variant and a likely pathogenic variant in MYO6 which together explain the complex phenotypes seen in this family.
Conclusions
We present a case illustrating the benefits of a broad screening approach that allows identification of oligogenic determinants of complex human phenotypes which may have been missed if the screening was limited to a targeted gene panel with the assumption of a syndromic disorder. This is important for correct genetic diagnosis of families and disentangling the range and severity of phenotypes associated with high impact variants.
Keywords: Neutropenia, Congenital neutropenia, Leukemia predisposition, Polygenic inheritance, Hearing loss
Background
Severe congenital neutropenia (SCN) was first described by Kostmann in 1956 in 14 individuals from 9 consanguineous families [1]. It is usually diagnosed in early childhood and is characterized by chronic neutropenia, susceptibility to bacterial infections and associated with a predisposition to myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML). SCN is genetically heterogeneous with autosomal recessive, autosomal dominant (AD) and X-linked forms described as well as de novo cases [2]. It may also occur as a part of a syndrome with other developmental defects (e.g. Shwachman-Diamond Syndrome) [3]. We describe a family with a complex phenotype of SCN and hearing loss of varying severity.
The genetic basis of SCN is well described and involves mutations in a number of different genes (Supplemental Table S2). Pathogenic variants in ELANE, the gene encoding neutrophil elastase, are the most common cause of AD SCN. Growth factor independent 1 transcriptional repressor (GFI1) germline variants have been reported in four patients with neutropenia; inheritance patterns were consistent with an AD mode of inheritance [4]. HAX1 variants underlie some autosomal recessive forms and were found to be the underlying cause of SCN in the families originally described by Kostmann [5]. It has been suggested that these variants impair neutrophil maturation via defective CSF3R signaling as the number of G-CSF receptors on myeloid precursors of SCN patients is elevated and majority of SCN patients benefit from the administration of pharmacological doses of granulocyte colony stimulating factor (G-CSF) [6].
Depending on the causative genetic lesion, neutropenia can sometimes present with extra-haematopoietic abnormalities such as pancreatic exocrine insufficiency (SBDS, ELF1) and deafness (GATA2). We previously reported heritable variants in GATA2 as predisposing to familial MDS and AML [7]. Since then, a high incidence of GATA2 variants has been found in patients with mild neutropenia who evolve to develop MDS and AML [8]. Interestingly, sensorineural hearing loss and monocytopenia are other manifestations associated with haploinsufficiency of GATA2. Here, we report a large family with multiple generations affected by neutropenia and hearing loss.
Methods
Patient samples
Blood and hair samples were obtained with consent from members of the family as part of the Australia Familial Haematological Cancer Study (AFHCS). gDNA was isolated using the QIAamp DNA Mini Kit (Qiagen).
Whole exome sequencing (WES)
We performed WES (SeqCap EZ MedExome, Roche NimbleGen) on two individuals (III-1 proband and IV-1). Variant annotation was performed through our ACRF Cancer Genomics Facility custom pipeline, which takes into consideration pathogenicity/oncogenicity predictions (CADD> 10, Polyphen 2, SIFT, Mutation Taster, GERP > 2, COSMIC parameters including specific-mutation and gene frequency), population minor allele frequencies (1000 GP, ESP, gnomAD), OMIM, and Gene Ontology. All candidate variants were manually curated to remove errors. The variants were interrogated for known SCN genes (Table S2) and non-syndromic hearing loss (Table S3).
Sanger sequencing
Sanger sequencing was performed on available samples for confirmation of GFI1 and MYO6 variants in family members to perform segregation analysis.
Results
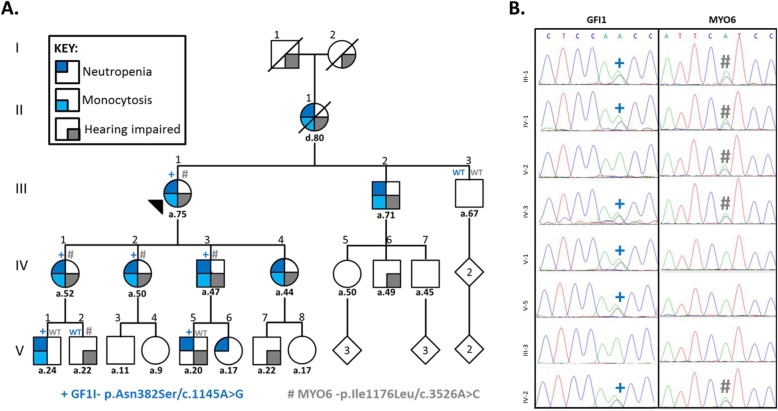
We identified a five-generation kindred with four successive generations affected by congenital neutropenia (10 individuals; eight also had monocytosis – available blood counts in Table S1) and five generations affected by hearing loss of varying severity (13 individuals) (Fig. 1a, Table 1). Sanger sequencing of GATA2 was performed due to partial overlap in the phenotypes observed within the family, but no pathogenic coding variants were found. We therefore performed whole exome sequencing on III-1 and IV-1(see Methods). The variants were interrogated for known SCN genes (Supplemental Table S2). We identified a previously reported variant underlying congenital neutropenia in GFI1 (NM_005263.5, c.1145A > G/p.Asn382Ser, N382S) (Fig. 1b, left panel) [4]. The N382S variant segregated with neutropenia in 6 consenting family members who were tested (Table 1) including V-5 who reported having had low white cells counts (blood reports unavailable). With the addition of these 6 cases to the 3 previously reported individuals, this variant can now be classified as a pathogenic variant, as per ACMG variant classification guidelines (Supplemental Table S4) [9].
Fig. 1.
Family with inherited neutropaenia, monocytosis and hearing impairment associated with mutations in GFI1 and MYO6. Pedigree, phenotypes and mutation status are indicated as per the key provided (a). Causative heterozygous mutations in GFI1 (p.N382S/c.1145A > G) and MYO6 (p.I1176L/c.3526A > C) were identified by whole exome sequencing performed on III-1 and IV-1. Sanger sequencing on available samples from consenting individuals was used for segregation analysis and confirmation of variants in individuals denoted by ‘+’ and ‘#’, respectively (b)
Table 1.
Genotypes and phenotypes of various members in the family
| Individual | GFI1 | Blood | MYO6 | Hearing |
|---|---|---|---|---|
| III-1 | N382S (het) | Neutropenia, monocytosis | I1176L (het) | Impaired |
| III-3 | WT | Normal | WT | Normal |
| IV-1 | N382S (het) | Neutropenia, monocytosis | I1176L (het) | Impaired |
| IV-2 | N382S (het) | Neutropenia, monocytosis | I1176L (het) | Impaired |
| IV-3 | N382S (het) | Neutropenia | I1176L (het) | Impaired |
| V-1 | N382S (het) | Neutropenia, monocytosis | WT | Normal |
| V-2 | WT | Normal | I1176L (het) | Very slight hearing loss |
| V-5 | N382S (het) | Has had low white cell counta | WT | Slight hearing loss |
| I-1 | Unknown | Unknown | Unknown | Impaired |
| I-2 | Unknown | Unknown | Unknown | Impaired in later years |
| II-1 | Unknown | Neutropenia, monocytosis | Unknown | Impaired |
| III-2 | Unknown | Neutropenia, monocytosis | Unknown | Impaired |
| IV-4 | Unknown | Neutropenia | Unknown | Impaired |
| IV-5 | Unknown | Normal | Unknown | Normal |
| IV-6 | Unknown | Unknown | Unknown | Impaired |
| IV-7 | Unknown | Normal | Unknown | Normal |
| V-3 | Unknown | Normal | Unknown | Normal |
| V-4 | Unknown | Normal | Unknown | Normal |
| V-6 | Unknown | Has had low white cell counta | Unknown | Normal |
| V-7 | Unknown | Unknown | Unknown | Slight hearing loss |
| V-8 | Unknown | Unknown | Unknown | Normal |
Abbreviations: WT Wildtype, het Heterozygous
areported in patient questionnaires/interview – no accompanying blood reports available
As GFI1 variants have not previously been associated with hearing loss, we explored additional genetic causes for the hearing loss phenotype observed in this family. Over 100 genes have been reported to underlie hereditary hearing loss. Analysis of the data for variants in genes associated with non-syndromic hearing loss (Supplemental Table S3), identified a novel single nucleotide variant in MYO6 (NM_004999.4, c.3526A > C/p.Ile1176Leu, I1176L) (Fig. 1b, right panel), a gene that has previously been associated with AD hearing loss [10]. The novel MYO6 I1176L variant segregated with hearing loss in all but one individual (V-5) who displayed clinically unconfirmed mild hearing loss (Fig. 1a). Supplemental Table S4 summarizes variant annotation and classification.
Discussion
Congenital neutropenia and monocytosis due to GFI1 variants was first reported in 2003, following the observation that GFI1-deficient mice were unexpectedly neutropenic [4]. GFI1 encodes a zinc finger transcriptional repressor oncoprotein. The N382S variant occurs in a highly evolutionarily conserved region of the GFI1 protein (Supplemental Figure S1). In vitro studies demonstrated that the variant acted in a dominant negative manner, abolishing DNA binding and hence repressor activity of the protein [4]. Interestingly, GFI1 recruits to chromatin the enzyme lysine-specific demethylase-1 (LSD1); pharmacologic inhibition of LSD1 or genetic knock-down of LSD1 skews granulocyte-monocyte progenitor differentiation resulting in neutropenia and monocytosis as seen in this family [10]. LSD1 is currently a target for the treatment of AML (NCT02842827).
SCN can be a pre-leukemic syndrome with evolution to leukemia recognized in patients with ELANE and HAX1 variants, as well as X-linked neutropenia (WAS). Patients with SCN reportedly have a 21% cumulative incidence of myeloid malignancy after 10 years with the risk of leukemia being higher in patients requiring high doses of G-CSF, and is associated with acquired mutations in CSF3R and RUNX1 [11].
To date, there have been no reports of MDS or AML in individuals with GFI1 variants alone (3 with N382S, 1 with K403R, 1 with R412*) [4] with the exception of three individuals who carried germline variants in both GFI1 and ELANE [12]. Our family adds another 6 cases of confirmed GFI1 variant carriers (aged 20–75 years) and 2 likely carriers (over 71 years) without progression to myeloid malignancy, suggesting that the GFI1 variant alone does not confer a high risk of leukemia development.
Variants in myosin genes are known to be involved in several types of syndromic and non-syndromic hearing loss. Variants in myosin VI (MYO6) have been identified in AD and recessive hearing loss [10, 13]. The primary evidence for the association of myosin VI with the hearing process is based on the Snell’s waltzer mice, that exhibit deafness [14]. The myosin VI protein is highly expressed at the base of stereocilia rootlets and in the pericuticular necklace of the inner and outer hair cells of the organ of Corti. MYO6 is required for the proper maturation of inner hair cell ribbon synapses and it has been shown to interact with DFNB9 (responsible for a recessive form of deafness) via the globular domain [15]. The I1176L variant is surrounded by a block of conserved amino acids and resides within a very highly conserved region (Figure S1) showing 91% amino acid sequence identity from p.Asn1165- Lys1285, that encodes the globular domain in the protein [15]. Miyagawa et al. have reported various other variants within this globular domain where the degree of hearing loss ranged from mild to profound.
Intriguingly, GFI1 has also been reported to be essential for inner ear hair cell differentiation [15, 16]. Hence individual V-5, who has reported slight hearing loss and has tested wildtype for MYO6, but carries the GFI1 variant, is an interesting case in this respect. Another curious observation is that family members with severe hearing impairment carry both variants while those with milder hearing problems only carry either of the variants. Though the MYO6 variant is most likely responsible for hearing loss in the family, it remains to be established if the GFI1 variant contributes to the phenotype.
Conclusions
We have presented a family with a complex phenotype of SCN and hearing loss that can be attributed to AD variants in two genes, GFI1 causing SCN, and MYO6 leading to hearing loss. Broader screening may be warranted in cases with complex disease presentations as polygenic inheritance could be missed if testing is restricted to specific gene panels. Although progression to leukemia has not been described to date in the limited number of individuals carrying germline pathogenic GFI1 variants, it remains to be established whether molecular monitoring for acquired variants should be considered as part of a risk management scheme.
Supplementary information
Additional file 1: Table S1. Neutrophil and monocyte counts of individuals where data was available. Table S2. Genes known to be mutated in congenital neutropenia. Table S3. Genes known to be mutated in non-syndromic hearing loss. Table S4. Computational predictions, variant annotation and classification.
Additional file 2: Figure S1. Sequence conservation of mutated residues in GFI1 and MYO, across species.
Acknowledgements
We wish to acknowledge the family for willingness to participate in the Australian Familial Haematological Cancer Study.
Abbreviations
- AD
Autosomal dominant
- AML
Acute myeloid leukemia
- G-CSF
Granulocyte colony stimulating factor
- LSD-1
Lysine-specific demethylase-1
- MDS
Myelodysplastic syndrome
- SCN
Severe congenital neutropenia
Authors’ contributions
LG, PV, CNH, AB and HSS wrote the manuscript. CNH, AB and HSS planned experiments. PV, CF, JF, KP, MB, NP, HYR, AWS, CNH and AB carried out different aspects of data analysis. All authors critically reviewed and approved the manuscript.
Funding
HSS is supported by the Beat Cancer Principal Research Fellowship (PRF0517) and manuscript has been produced with the financial and other support of Cancer Council SA’s Beat Cancer Project on behalf of its donors, and the State Government of South Australia through the Department of Health. This work was supported by grant funding from the National Health and Medical Research Council, Australia (APP1024215, APP1023059) which paid staff salaries and consumables. The funders had no role in the design of the study or in the collection, analysis and interpretation of data or in writing the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study is available in the EGA repository (Study ID: EGAS00001004176). These are accessible via the following links: https://www.ebi.ac.uk/ega/studies/EGAS00001004176; https://www.ebi.ac.uk/ega/datasets/EGAD00001005937.
Ethics approval and consent to participate
The study was carried out under the Australian Familial Haematological Cancer Study (REC1542/02/2022) approved by the Women’s and Children’s Hospital Human Research Ethics Committee in accordance with the Declaration of Helsinki. Individuals included in the study have read and signed the informed consent form.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Parvathy Venugopal, Lucia Gagliardi and Cecily Forsyth are equal first authors
Christopher N Hahn, Anna L Brown and Hamish S Scott are equal senior authors
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-0971-z.
References
- 1.Kostmann R. Infantile genetic agranulocytosis; agranulocytosis infantilis hereditaria. Acta Paediatr Suppl. 1956;45(Suppl 105):1–78. [PubMed] [Google Scholar]
- 2.Skokowa J, Dale DC, Touw IP, Zeidler C, Welte K. Severe congenital neutropenias. Nat Rev Dis Primers. 2017;3:17032. doi: 10.1038/nrdp.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Touw IP. Game of clones: the genomic evolution of severe congenital neutropenia. Hematology Am Soc Hematol Educ Program. 2015;2015:1–7. doi: 10.1182/asheducation-2015.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34(3):308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schaffer AA, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39(1):86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 6.Klimiankou M, Uenalan M, Kandabarau S, Nustede R, Steiert I, Mellor-Heineke S, et al. Ultra-sensitive CSF3R deep sequencing in patients with severe congenital neutropenia. Front Immunol. 2019;10:116. doi: 10.3389/fimmu.2019.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasquet M, Bellanne-Chantelot C, Tavitian S, Prade N, Beaupain B, Larochelle O, et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 2013;121(5):822–829. doi: 10.1182/blood-2012-08-447367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyagawa M, Nishio SY, Kumakawa K, Usami S. Massively parallel DNA sequencing successfully identified seven families with deafness-associated MYO6 mutations: the mutational spectrum and clinical characteristics. Ann Otol Rhinol Laryngol. 2015;124(Suppl 1):148S–157S. doi: 10.1177/0003489415575055. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg PS, Zeidler C, Bolyard AA, Alter BP, Bonilla MA, Boxer LA, et al. Stable long-term risk of leukaemia in patients with severe congenital neutropenia maintained on G-CSF therapy. Br J Haematol. 2010;150(2):196–199. doi: 10.1111/j.1365-2141.2010.08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skokowa J, Steinemann D, Katsman-Kuipers JE, Zeidler C, Klimenkova O, Klimiankou M, et al. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014;123(14):2229–2237. doi: 10.1182/blood-2013-11-538025. [DOI] [PubMed] [Google Scholar]
- 13.Ahmed ZM, Morell RJ, Riazuddin S, Gropman A, Shaukat S, Ahmad MM, et al. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet. 2003;72(5):1315–1322. doi: 10.1086/375122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avraham KB, Hasson T, Sobe T, Balsara B, Testa JR, Skvorak AB, et al. Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice. Hum Mol Genet. 1997;6(8):1225–1231. doi: 10.1093/hmg/6.8.1225. [DOI] [PubMed] [Google Scholar]
- 15.Roux I, Hosie S, Johnson SL, Bahloul A, Cayet N, Nouaille S, et al. Myosin VI is required for the proper maturation and function of inner hair cell ribbon synapses. Hum Mol Genet. 2009;18(23):4615–4628. doi: 10.1093/hmg/ddp429. [DOI] [PubMed] [Google Scholar]
- 16.Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130(1):221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Neutrophil and monocyte counts of individuals where data was available. Table S2. Genes known to be mutated in congenital neutropenia. Table S3. Genes known to be mutated in non-syndromic hearing loss. Table S4. Computational predictions, variant annotation and classification.
Additional file 2: Figure S1. Sequence conservation of mutated residues in GFI1 and MYO, across species.
Data Availability Statement
The datasets generated and/or analysed during the current study is available in the EGA repository (Study ID: EGAS00001004176). These are accessible via the following links: https://www.ebi.ac.uk/ega/studies/EGAS00001004176; https://www.ebi.ac.uk/ega/datasets/EGAD00001005937.