Abstract
Background
It is hypothesised that being a blood-feeding ectoparasite, Argulus foliaceus (Linnaeus, 1758), uses similar mechanisms for digestion and host immune evasion to those used by other haematophagous ecdysozoa, including caligid copepods (e.g. sea louse). We recently described and characterised glands associated with the feeding appendages of A. foliaceus using histological techniques. The work described in the present study is the first undertaken with the objective of identifying and partially characterising the components secreted from these glands using a proteomic approach.
Methods
Argulus foliaceus parasites were sampled from the skin of rainbow trout (Oncorhynchus mykiss), from Loch Fad on the Isle of Bute, Scotland, UK. The proteins from A. foliaceus secretory/excretory products (SEPs) were collected from the supernatant of artificial freshwater conditioned with active adult parasites (n = 5–9 per ml; n = 560 total). Proteins within the SEPs were identified and characterised using LC-ESI-MS/MS analysis. Data are available via ProteomeXchange with identifier PXD016226.
Results
Data mining of a protein database translated from an A. foliaceus dataset using ProteinScape allowed identification of 27 predicted protein sequences from the A. foliaceus SEPs, each protein matching the criteria of 2 peptides with at least 4 contiguous amino acids. Nine proteins had no matching sequence through OmicsBox (Blast2GO) analysis searches suggesting that Argulus spp. may additionally have unique proteins present in their SEPs. SignalP 5.0 software, identified 13 proteins with a signal sequence suggestive of signal peptides and supportive of secreted proteins being identified. Notably, the functional characteristics of identified A. foliaceus proteins/domains have also been described from the salivary glands and saliva of other blood-feeding arthropods such as ticks. Identified proteins included: transporters, peroxidases, metalloproteases, proteases and serine protease inhibitors which are known to play roles in parasite immune evasion/induction (e.g. astacin), immunomodulation (e.g. serpin) and digestion (e.g. trypsin).
Conclusions
To our knowledge, the present study represents the first proteomic analysis undertaken for SEPs from any branchiuran fish louse. Here we reveal possible functional roles of A. foliaceus SEPs in digestion and immunomodulation, with a number of protein families shared with other haematophagous ectoparasites. A number of apparently unique secreted proteins were identified compared to other haematophagous ecdysozoa.
Keywords: Secretions, Immunomodulation, Fish lice, Argulus, Branchiura
Background
Argulus foliaceus (Linnaeus, 1758) is a member of the branchiuran family Argulidae and has a worldwide distribution [1–3]. In the UK, this parasitic arthropod causes the condition argulosis, which has economic impacts for both aquaculture and sports fishing industries [4] and affects wild and cultured fish populations [5, 6].
Argulus spp. attach to their hosts for extended periods using the first maxillae, which form hooks as larvae but are modified into suction discs as adults. It has been hypothesised that to enable long term host contact and feeding, Argulus spp., like other ectoparasitic arthropods such as ticks and salmon lice (Lepeophtheirus salmonis (Krøyer, 1837)), must secrete a range of pharmacologically active components. These components have been suggested to be produced in the parasite’s spinal, proboscis and labial glands and to be delivered through the pre-oral spine and the mouth tube to facilitate blood-feeding [7, 8]. Argulus spp. have been reported to feed on blood [9–11], mucus [12] and host skin [13] and thus have a presumed requirement to modulate host immunity. Pharmacologically active substances delivered in parasite secretions aid in modulating such defence mechanisms including those associated with pain, haemostasis, inflammation, complement activation and tissue repair [14–19]. However, to date there are limited genomic resources available for detailed investigation of this parasite. Furthermore, little formal data have been collected on either the composition or function of Argulus spp. secreted proteins or other active components [20].
During blood-feeding, other haematophagous arthropods inject the host with salivary products rich in digestive enzymes and other pharmacologically active proteins that have anti-haemostatic, anti-inflammatory and immunomodulatory properties. These salivary products help in counteracting the host immune response and wound recovery and hence facilitate feeding [21–28]. Recently, proteomics-based studies have contributed considerably to the identification of proteins from the salivary glands of a number of haematophagous insects including different species of mosquitoes i.e. Anopheles stephensi Liston, 1901, A. campestris-like, Aedes aegypti (Linnaeus in Hasselquist, 1762) and Culex pipiens quinquefasciatus Say, 1823. These studies have provided data relating to the functional roles of these proteins, and have facilitated their molecular and biological characterisation [21]. Recent transcriptomic and proteomic studies of salivary glands from different species of ticks have similarly identified a diverse range of active molecules/proteins that modify the hosts’ physiology [29–31]. Given the success of these studies in identifying active components of the saliva of other haematophagous arthropods, a proteomics approach may prove similarly productive to decipher the composition and role of A. foliaceus SEPs.
In two earlier studies, western blots of whole body extracts of A. siamensis (Wilson, 1926) [32] and A. foliaceus [33] revealed the presence of a number of immunodominant polypeptides recognised by immune rainbow trout (Oncorhynchus mykiss (Walbaum, 1792)) and rabbit serum. Notably, and despite phylogenetic distance, antigenic cross-reactivity between A. foliaceus and parasitic caligid copepod (L. salmonis and Caligus elongatus (von Nordmann, 1832)) antigens was demonstrated using serum antibodies raised against A. foliaceus in rainbow trout [20]. Both caligid sea lice and branchiuran freshwater lice feed on the fish epithelium, mucus and blood to some degree. To allow this, sea lice SEPs comprise a number of immunomodulatory proteins [34, 35] including metallopeptidases, serine proteinases and cysteine proteinases; previously identified in tick saliva [36]. Sea lice SEPs also contain anti-oxidant proteins identified in helminths [37], and serine endopeptidases, e.g. trypsins, identified in flies [38]. Whether the mechanisms involved in parasitism are similar between sea lice and other arthropods parasitising fish has yet to be established.
From the foregoing, the aim of the present study was to identify and characterise protein components of the SEPs produced by A. foliaceus. Characterisation of these proteins is key to establishing the biological function of branchiuran/A. foliaceus SEPs and can assist in identifying potential vaccine candidates or drug targets for the future development of more sustainable argulosis control strategies. To this end, proteomic data generated by LC-ESI-MS/MS were searched against a bespoke protein database assembled using proteins translated from an A. foliaceus transcriptomic dataset (submitted to the EBI with a project number PRJEB34947) and likely biological roles for discovered proteins ascribed.
Methods
Sample collection and incubation in artificial freshwater
Adult argulids were collected directly from fish hosts, rainbow trout, from Loch Fad on the Isle of Bute, Scotland, UK. A total of 560 adult male and female parasites were used. Between five and nine parasites (depending on the parasite sizes) were placed in 1.5 ml sterile Eppendorf tubes containing 1 ml of artificial freshwater, prepared as described by Klüttgen et al. [39]. Artificial freshwater without parasites was used as a negative control (50 × 1.5 ml sterile tubes of 1 ml of artificial freshwater). All the tubes were incubated at 10 °C overnight (18 h). The following day all parasites were alive and active and water samples expected to contain SEPs from A. foliaceus were pooled into 2 × 50 ml sterile Falcon tubes and the negative controls pooled into an additional 50 ml sterile tube. These samples and the control were rapidly chilled to − 70 °C and maintained frozen until used for sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS PAGE
Secretion/excretion samples and the negative control were thawed on ice and centrifuged at 4000×g for 30 min to concentrate proteins of > 3 kDa through 3 kDa cut-off centrifugal filters (Amicon® Ultra, Millipore, Cork, Ireland) prior to sample denaturation. The protein content of concentrated SEPs was measured using a Pierce Bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
A dilution series of A. foliaceus SEP samples for SDS-PAGE analysis [40] was set up for different stains, one for silver stain and one for Coomassie stain. The concentrated sample (1:1) was combined with 2× SDS sample buffer (SDS reducing buffer; 2.5 ml 0.5 M Tris-HCL pH 6.8, 2 ml glycerol, 4 ml 10% SDS, 0.31 g dithiothreitol (DTT), 2 mg bromophenol blue and DW added to give 10 ml). Sequential 2-fold dilutions of 1:4 and 1:8 dilutions were then made with RNAse-free water. The samples were kept on ice then denatured at 100 °C in boiling water for 5 min then centrifuged (Micsolite, Thermo IEC) for 2 min at 16000×g. Five µl of 2–250 kDa mixed range pre-stained molecular weight markers (Precision Plus Protein™ Standards- Bio-Rad, Hemel Hempstead, UK) were used to estimate the size of proteins and loaded into two Precast 12-well Polyacrylamide gels (12% Mini-PROTEAN® TGX™ Precast Protein Gels, Bio-Rad) followed by 15 µl of the samples in each well in descending order of sample concentration. The gels were run at 130 V for 75 min followed by washing with DDW for 5 min. After electrophoresis, fixed proteins were visualised with QC colloidal Coomassie stain (Bio-Rad) and silver stain using a silver staining kit (ProteoSilver ™-PROTSIL1-1KT, St. Louis, USA). The gel was scanned using a benchtop scanner (EPSON expression 1680 Pro) and kept at 4 °C in a sealed polyethylene bag with DDW until sent for gel and liquid chromatography electrospray ionisation tandem mass spectrometry (GeLC-ESI-MS/MS). The lane of the 1:2 sample dilution was selected for MS/MS analysis conducted at the Moredun Research Institute, Midlothian, UK.
LC-ESI-MS/MS
The protein identifications were performed at The Moredun Proteomics Facility, Moredun Research Institute (Scotland, UK). The gel lane was excised and sliced horizontally from top to bottom to yield a series of 24 equal gel slices of 2.5 mm depth. Each of the resulting gel slices was then subjected to standard in-gel destaining, reduction, alkylation and trypsinolysis procedures [41]. Digests were transferred to low-protein-binding HPLC sample vials immediately prior to LC-ESI-MS/MS analysis. Liquid chromatography was performed using an Ultimate 3000 Nano-HPLC system (Dionex, Leeds, England) comprising a WPS-3000 well-plate micro auto sampler, an FLM-3000 flow manager and column compartment, a UVD-3000 UV detector, an LPG-3600 dual-gradient micropump and an SRD-3600 solvent rack controlled by Chromeleon™ chromatography software (Dionex). A micro-pump flow rate of 246 µl min−1 was used in combination with a cap-flow splitter cartridge, affording a 1/82 flow split and a final flow rate of 3 µl min−1 through a 5 cm × 200 µm ID monolithic reversed phase column (Dionex) maintained at 50 °C. Samples of 4 µl were applied to the column by direct injection. Peptides were eluted by the application of 15 min linear gradient from 8–45% solvent B (80% acetonitrile, 0.1% (v/v) formic acid) and directed through a 3 nl UV detector flow cell. LC was interfaced directly with a 3-D high capacity ion trap mass spectrometer (amaZon-ETD, Bruker Daltonics, Bremen, Germany) via a low-volume (50 µl min−1 maximum) stainless steel nebuliser (cat. no. G1946-20260; Agilent, Santa Clara, CA, USA) and ESI. Parameters for tandem MS analysis were based on those described previously [42].
Database mining
The MS/MS data, formatted as Mascot Generic Format (mgf), was imported into ProteinScape™ V3.1 (Bruker Daltonics) proteomics data analysis software for downstream mining of a custom Argulus database. This custom database was constructed using translated proteins from the transcriptome dataset of Argulus foliaceus (in the absence of a full Argulus genome sequence) and comprised 60,257 protein sequences in total (the sequence data for the transcriptome were submitted to the EBI with a project number PRJEB34947 under the title “De novo transcriptome sequencing of branchiuran fish lice, Argulus foliaceus (Linnaeus, 1758) and Argulus coregoni (Thorell, 1865)”). De novo assembly was performed on the cleaned RNA-seq raw data using Trinity v2.1.1 [43]. The selection of the Trinity tool for final assembly was decided upon following a trial with other assembler software such as Velvet and ABySS whereby the use of Trinity was found to give higher numbers of more consistent reads. All the sample reads were merged into a single dataset for each species and the assembly was run. Then Transdecoder v2.0.1 [44] was used to find the coding region within the transcripts. Identification of the coding region transcripts gave open reading frames (ORFs) and amino acid sequences, to prepare the assembled dataset for annotation. Annotation was achieved by (i) BLAST v2.2.31 software [45, 46] using the uniprot/trembl-invertebrates database as query [47]; and (ii) Annotation HMMER v3.1b2 [48] using the Pfam A v29.0 as query [49].
Database searches were conducted utilising the Mascot™ V2.5.1 (Matrix Science) search engine. Mascot search parameters were set in accordance with published guidelines [50] and to this end, fixed (carbamidomethyl “C”) and variable (oxidation “M” and deamidation “N, Q”) modifications were selected along with peptide (MS) and secondary fragmentation (MS/MS) mass tolerance values of 0.5 Da whilst allowing for a single 13C isotope. Protein identifications obtained from each of the 24 individual gel slices were compiled using the “protein list compilation” feature within ProteinScape, which parses the data to remove redundancies. From the compiled protein lists individual identifications deemed significant by MASCOT (score > 35 which indicates P-value < 0.05) (Additional file 1: Table S1) were inspected manually and considered significant only if (i) two peptides were matched for each protein; (ii) peptides were represented by a sequence coverage of > 5%; and (iii) each matched peptide contained an unbroken “b” or “y” ion series represented by a minimum of four contiguous amino acid residues. The compilation of all gel slices and manual validation left a list of 27 proteins significantly identified by LC-ESI-MS/MS analysis from the custom transcriptome derived Argulus database.
Functional analysis
To assign identity and function to the sequences of the proteins passing the criteria for significance following LC-ESI-MS/MS, the sequences were then searched using OmicsBox/ (Blast2Go) analysis. Searches were performed using the whole NCBI Blast and InterProscan for Protein function assignment, and then sequences of the proteins were finally blasted using BLAST P against NCBI ‘Metazoan’ database to get accession numbers with the most reliable hits. Proteins were then assigned into functional groups by searching the InterProScan databases and Gene ontology databases. Annotations from both searches were then merged. The Gene Ontology (GO) terms assigned to each protein were then used to construct pie charts based on biological process, cellular component and molecular function. The number of proteins and percentage were included with each GO term. Separate Pfam searches were conducted and SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/) was used to predict the presence and location of signal peptide cleavage sites in amino acid sequences, which could inform of any associated secretory property of the protein. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016226.
Results
Protein profiles in secretory/excretory products (SEPs) by SDS PAGE
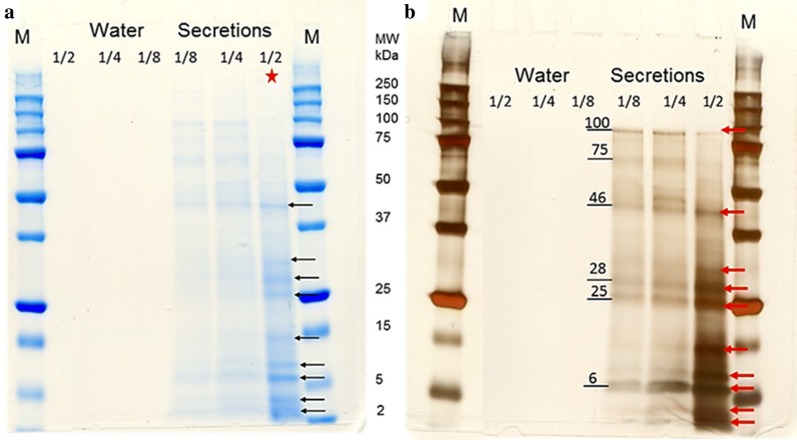
The protein content of harvested A. foliaceus SEPs was approximately 410 μg ml−1. SDS-PAGE analysis showed 10 intense bands of proteins with molecular masses in the range of 3–45 kDa. The use of three different dilutions of the secretions indicated a dilution effect in the intensity and number of bands obtained. The lack of bands observed from the sterilised artificial water (negative control) confirmed that the protein bands resulted from A. foliaceus parasites, either as secretions or excretory products, i.e. minimal environmental contamination (Fig. 1). Notably, there were 4 intense bands even in the most diluted sample; 1:8, with approximate molecular weights of 5, 25, 28 and 46 kDa, and an intense band was seen in the more dilute samples at around 100 kDa compared to the 1:2 diluted sample (Fig. 1b).
Fig. 1.
SDS-PAGE of A. foliaceus secretions. Secretory/excretory products (SEPs) were collected and proteins separated on 12% SDS-PAGE gels and stained with QC colloidal Coomassie stain (a) and silver stain (b) to visualise the protein bands. Numbers on the left indicate the approximate molecular mass (MW; kDa) of the proteins within the most diluted sample. Ten distinct bands (arrows) were observed after staining the gels. Molecular mass (2–250 kDa) marker is shown in the middle between the two gels. Asterisk indicates the lane from the Coomassie stained gel that was selected for GeLC-MS/MS analysis. No bands were seen in the water control
LC-ESI-MS/MS analysis
SEPs were collected from 560 live and active A. foliaceus parasites of mixed sex adult life stages for GeLC-MS/MS analysis and protein identifications were confirmed by positive matches (Additional file 2: Table S2) with transcript sequences (unpublished data). From the compiled identified protein lists obtained from pooled A. foliaceus SEPs, 27 passed the specified “stringent” quality criteria and were associated with 27 annotated proteins (Tables 1, 2). To assign functional identity to these proteins, the amino acid sequences were searched against OmicsBox (Blast2GO) database and in order to find homologous metazoan proteins, the NCBI BLASTP program was used (Tables 1, 2) and the e-values of the proteins shown in the Table 1 were from the OmicsBox output. The InterProScan search assigned functional identity to 18 proteins, listed in Table 1, which shows Pfam functional descriptions, suggested functions from other arthropod studies and signal peptide predictions. The number of peptides, Signal P, molecular weight of proteins and percentage of sequence coverage are also listed (Table 1).
Table 1.
List of proteins identified by GeLC-MS/MS analysis of A. foliaceus secretory/excretory products showing hits with OmicsBOx (Blast2Go) database
| Family/Domain | SeqName | Description | MW (kDa) | No. Peptides | SC (%) | Pro. length | E-value | SignalP | Hits Organism | Function | NCBI ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitellogenin_N | afol_3429.1p | Von Willebrand factor type D domain protein | 172.5 | 55 | 39.4 | 1499 | 8.39E−11 | Yes | Ixodes scapularis | Transporters, osmolality, immunity and clotting [25, 51–53] | XP_029822532.1 |
| afol_51180.1p | Vitellogenin-like isoform X1 | 183.8 | 44 | 30.1 | 1582 | 1.26E−11 | No | Ixodes scapularis | XP_029826448.1 | ||
| Hemocyanin | afol_43548.1p | Hemocyanin subunit type 1 precursor | 81.2 | 18 | 26.3 | 697 | 0 | Yes | Argulus foliaceus | Respiratory, protein storage [54] | CUH82791.1 |
| afol_52001.1p | Hemocyanin A chain | 36.2 | 8 | 30.6 | 314 | 0 | No | Argulus foliaceus | CUH82792.1 | ||
| Astacin | afol_16671.1p | Protein SpAN-like | 72.6 | 11 | 21.2 | 660 | 1.49E−55 | Yes | Saccoglossus kowalevskii | Immunity; antifungal activity [55], food digestion, host penetration, immune evasion or activation [56–58] | XP_002739691.1 |
| afol_52344.2p | Protein SpAN-like | 76.4 | 12 | 17.6 | 687 | 2.85E−50 | No | Penaeus vannamei | XP_027221531.1 | ||
| afol_18725.8p | Blastula protease 10-like | 50.7 | 8 | 18.4 | 462 | 1.44E−17 | No | Penaeus vannamei | XP_027219989.1 | ||
| Serpin | afol_27409.2p | Serpin B6-like | 51.4 | 7 | 20.0 | 461 | 6.15E−66 | No | Rhipicephalus microplus | Anticoagulation activity, modulate host immune response, regulation of host inflammation, antihaemostatic effects, and platelet aggregation [59–64] | AHC98669.1 |
| afol_25414.2p | Leukocyte elastase inhibitor-like | 11.2 | 4 | 35.1 | 94 | 1.05E−16 | No | Ixodes scapularis | EEC05896.1 | ||
| Fasciclin | afol_20127.1p | Beta-ig-h3 fasciclin | 37.1 | 5 | 23.1 | 333 | 1.64E−22 | Yes | Culex quinquefasciatus | Mediate cell adhesion [65, 66] | XP_001847648.1 |
| Trypsin | afol_3246.7p | Transmembrane protease serine 9 | 28.1 | 5 | 21.4 | 262 | 2.81E−12 | No | Penaeus vannamei | Digestion, maintain host-parasite relationship [67] | XP_027224040.1 |
| afol_16878.20p | Trypsin-1 | 32.0 | 9 | 34.5 | 293 | 6.81E−61 | Yes | Ceratitis capitata | XP_004520346.1 | ||
| afol_19181.1p | Serine protease 29 | 47.0 | 7 | 24.5 | 466 | 8.72E−87 | Yes | Bombus terrestris | XP_020722765.1 | ||
| afol_18345.1p | Transmembrane protease serine 9-like | 18.3 | 4 | 25.6 | 164 | 1.38E−17 | No | Drosophila navojoa | XP_030246706.1 | ||
| afol_51127.2p | Trypsin | 25.1 | 3 | 15.7 | 236 | 2.13E−57 | No | Penaeus vannamei | ROT79324.1 | ||
| afol_56688.3p | Serine protease1/2 | 29.1 | 2 | 6.1 | 262 | 3.42E−39 | Yes | Ixodes scapularis | XP_029851465.1 | ||
| Peptidase_M14 | afol_12392.3p | Mast cell carboxypeptidase A | 48.3 | 5 | 16.7 | 424 | 4.21E−88 | No | Penaeus vannamei | Proteolytic-enzyme [68]) | XP_027226524.1 |
| VIT | afol_32337.1p | Inter-alpha-trypsin inhibitor heavy chain H4-like isoform X2 | 98.2 | 4 | 7.4 | 883 | 2.31E−151 | Yes | Lingula anatina | Proteinase inhibitor [36] | XP_013407760.1 |
Note: Description of suggested functions reported in different species of arthropods, and were blasted against Metazoa in NCBI BLASTP to obtain an Accession No. Signal P was used to predict the secretory property of the protein
Abbreviations: MW, molecular weight of protein; SC, sequence coverage; E-value, from the OmicsBox output
Table 2.
Proteins identified from A. foliaceus secretory/excretory products by mass spectrometric analysis that showed no hits in OmicsBox/Blast2GO and were blasted against Metazoa in NCBI BLASTP, signal P
| Protein ID | Protein length | MW (kDa) | SC (%) | E-value (BlastP) | NCBI ID | Signal P |
|---|---|---|---|---|---|---|
| afol_3444.2p | 192 | 20.7 | 40.1 | No | ||
| afol_55421.1p | 140 | 14.9 | 42.1 | 0.57 | XP_008192422.1 | Yes |
| afol_9654.1p | 408 | 44.7 | 16.2 | 5.4 | XP_027221531.1 | Yes |
| afol_25364.1p | 438 | 48.9 | 14.4 | 0.24 | XP_029189514.1 | No |
| afol_15401.2p | 230 | 25.7 | 13 | No | ||
| afol_2470.1p | 111 | 11.7 | 17.1 | 0.006 | XP_017135474.1 | No |
| afol_50565.1p | 388 | 43.8 | 10.6 | Yes | ||
| afol_52850.1p | 399 | 43.7 | 6 | 0.083 | XP_023212714.1 | Yes |
| afol_45298.1p | 189 | 21.5 | 25.4 | 0.49 | EEC08850.1 | Yes |
Abbreviations: MW, molecular weight of protein; SC, sequence coverage; E-value, from the NCBI BlastP
From the initial list 9 proteins did not return any matches from OmicsBox (Blast2GO) and only 6 out of these 9 showed significant hits when blasted against Metazoa in NCBI BLASTP (Table 2).
Signal P analysis showed that out of these 27 proteins 13 were found to have a signal peptide sequence (Tables 1, 2).
Assigned function of SE products
Proteins identified from the SEPs were assigned GO terms within the biological process, cellular component and molecular function domains (Fig. 2). Proteins associated with molecular function accounted for 8% catalytic activity and 50% hydrolase activity. The most represented proteins in the biological process category (Fig. 2b) were assigned to oxidation-reduction processes. Cellular component represented only one GO term (with 2 entries), assigned to cellular anatomical entity (Fig. 2c) indicating the difficulty in characterising parasite SEPs in the absence of an annotated genome. OmicsBOx data (Additional file 3: Table S3) summarised the GO, InterProScan domain, families and IDs distributions data that resulted from OmicsBox hits of SE products, which showed functional data of the 27 identified proteins. This table represents the range of nominal roles assigned to the group of proteins found in SEPs of A. foliaceus.
Fig. 2.
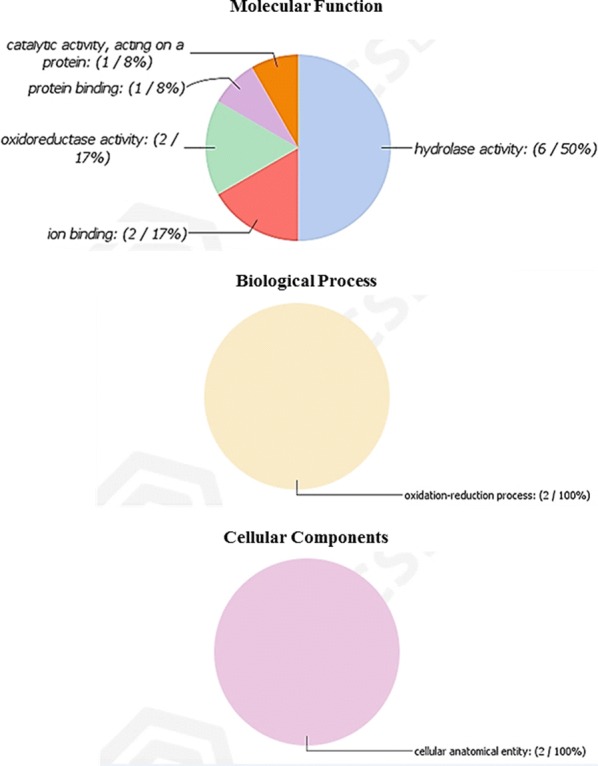
GO distribution of the proteins identified from A. foliaceus secretory/excretory products
Discussion
Identifying the SEPs of A. foliaceus is important for determining how the parasite establishes host attachment and facilitates blood-feeding, as the components of ectoparasite secretions are known to play functional roles in such interactions [51–53]. In this regard, the secretions of Argulus spp. resemble those in the saliva of ticks and other haematophagous arthropods, containing bioactive molecules released to maintain a successful feeding site. Using LC-ESI-MS/MS, proteins such as serpin, trypsin and fascilin have been previously associated with this role in ticks (e.g. Ornithodoros moubata [25]), mosquitoes (e.g. Anopheles culicifacies [54]) and sea lice (e.g. L. salmonis [55]) saliva/salivary glands and SEPs. Sea lice trypsins, vitellogenin-like proteins and proteins with immunomodulatory functions or host adhesion properties have similarly been studied with suggestions of their potential as vaccine antigen candidates [56–58]. In order to feed, Argulus spp. doubtless needs to modulate host immune defence mechanisms (haemostasis and immunity) and inhibit host tissue repair responses in similar ways to other haematophagous arthropods such as ticks [59–62].
SDS-PAGE of the SEPs of A. foliaceus revealed a range of different protein bands between 3–100 kDa. Ruane et al. [33] showed similar, but more numerous, protein profile ranges from whole A. foliaceus homogenates, with proteins of molecular weights between 15–100 kDa; however, the authors suggested that the absence of higher molecular weight proteins may be due to the inability of the Coomassie stain to detect these potentially low abundance proteins in their study. From A. siamensis homogenates [32], protein bands between 16.22–130.55 kDa were detected by SDS-PAGE with intensely stained bands of > 66 kDa. In contrast to these previous studies, analysis in the present study was limited to A. foliaceus SEPs run under denaturing conditions, confirming the presence of secreted proteins, which were detectable in the most concentrated sample preparation (1:2 dilution) by Coomassie stain, mostly in the lower molecular weight range of 5–46 kDa. No proteomic studies on the secretions of any branchiuran including Argulus spp., had previously been conducted prior to this study, especially since genomic resources are limited. Therefore, in order to identify the major proteins of A. foliaceus SEPs, an integrated transcriptomic (manuscript in preparation) and proteomic approach was used. The (LC-ESI-MS/MS) data combined with the A. foliaceus transcriptome, used as a reference database, resulted in the identification of a number of different proteins nominally secreted by the parasite.
Overall, the analysis of the SEPs sample identified 27 proteins, of which only 18 were recognised by OmicsBox (Blast2GO) analysis and the other 9 protein sequences could not be recognised, and therefore could not be assigned a role at the present time. This is far fewer than the number of proteins identified from other parasite secretory products (e.g. 135 in the saliva of the hard tick Haemaphysalis longicornis [36] or 187 in the SEPs of L. salmonis [55]. Parasite SEP protein yields can be enhanced by inducing salivation or stimulation of the salivary glands using dopamine and pilocarpine, but with varying success [34, 36, 51, 55]. The overall number of identified proteins would also be anticipated to increase when a fully annotated genome becomes available. Some of the proteins that were identified are highly abundant in arthropods, particularly ecdysozoans, for example vitellogenins (vitellogenin-N) (e.g. in L. salmonis [63]) and haemocyanins (e.g. in crabs (Cancer magister) [64]) thus may simply be residual. Nonetheless, many of these proteins have previously been suggested to play a role in host infection in other haematophagous arthropods. These include metallopeptidases such as Peptidase_M14 and Astacin (Peptidase family M12A), proteases such as trypsin; and serpin; and other protein domains such as fasciclin and VIT (Vault protein inter-alpha-trypsin domain). Of the 27 identified proteins, 13 proteins carried a predicted signal sequence using Signal P suggesting them to be extracellular proteins and discharged within the secretions of the parasites. Identification of a number of signal peptides, short peptides (~ 16–30 aa) that direct newly synthesized proteins towards the secretory pathway [65], suggests that some of the proteins identified in the present study are secreted products, which notably included some vitellogenins and haemocyanins (Table 1). Such signal peptides target a protein for translocation across the endoplasmic reticulum (ER) membrane in eukaryotes [66]. The lack of a predicted signal peptide for other proteins discovered in this study may not indicate that they were not secreted but may simply result from the partial nature of the protein sequences, which were insufficient for prediction by Signal P; however, further studies need to confirm this.
This study identified a number of putative novel proteins (i.e. with no similarity in metazoan databases) from A. foliaceus secretions. Although the biological functions of these secreted proteins are unknown, they may have properties controlling physiological functions during Argulus attachment. As homologues for some of these proteins could not be found in the OmicsBox (Blast2GO) and NCBI databases, this could suggest that Argulus may have other unique proteins compared to other well characterised haematophagous ecdysozoa, such as insects, ticks and nematodes, for potentially modulating or evading their host’s immune system. However, considering the low SC% values and high e-values of a number of these proteins, their functional role is questionable at the current time. Other proteins, however, were analysed where functional identification was successful in this study and these have also previously been described in other haematophagous arthropods such as in tick salivary proteomes [36, 67, 68]. The role of this latter group of proteins from A. foliaceus secretions may therefore be similar to that played by their homologues in other ectoparasites during host-parasite interactions and supports their importance for A. foliaceus in feeding, digestion and evading host immune defences.
Vitellogenin is a lipoprotein generally related to reproduction in arthropods; however, it has been shown that the production of this protein can be positively associated with the size of blood meals, as is the case in ticks, where vitellogenin binds to the derivative haem from the host to initiate the reproduction cycle [69–71]. This was supported by the findings of Galay et al. [69] who showed that silencing of the secretory ferritin gene of the hard tick H. longicornis affected two vitellogenin genes. Moreover, Rosell-Davis & Coons [71] showed that onset of feeding initiates vitellogenin production.
Recent proteomic analysis of L. salmonis SEPs found a vitellogenin-like protein unique to adult females [55]. Dalvin et al. [63] examining L. salmonis did not observe any transcription of this protein in the ovary but they did localise the presence of these proteins in the haemolymph [63] indicating the protein is circulated through the louse and thus may have a role in reproductive processes following feeding similar to ticks. Further studies to localise this protein in Argulus spp. need to be conducted to give an indication of its reproductive or additional functional roles in this parasite.
Haemocyanin proteins in arthropods have been characterised as the main oxygen transporters in the haemolymph of many species [72]. In addition to their respiratory role, haemocyanin proteins are also involved in a range of other physiological processes including osmoregulation, protein storage and enzymatic activities [73, 74]. Destoumieux-Garzón et al. [75] revealed the importance of this protein to crustacean immunity in terms of the production of antifungal (poly) peptides. Recently, Pinnow et al. [72] identified two haemocyanin subunits from A. foliaceus, which were confirmed in this study, including two haemocyanin protein domains, hemocyanin subunit type 1 precursor and hemocyanin A chain. Although the production of haemocyanins is normal for respiration, Pinnow et al. [72] described haemocyanin 2 as a storage protein. Although apparently secreted proteins, i.e. possessing signal peptides, it should be noted, however, that both vitellogenin and haemocyanin could also be present as a result of damage to individual specimens and subsequent leakage of haemolymph. Therefore, the potential secretory roles of vitellogenin and haemocyanin proteins in Argulus, should they indeed be secreted, needs further investigation.
Metalloendopeptidase astacin and carboxypeptidase M14 identified in A. foliaceus SEPs have also been identified in the saliva of the haematophagous Mediterranean colubrariid (vampire) snail Colubraria reticulata which feeds on the blood of fishes [76] and also in L. salmonis [77]. Members of the astacin family have been considered to maintain blood flow to the lesion site through hydrolysis of fibrinogen and fibronectin [78, 79] leading to local haemorrhage [80]. It has been hypothesised that astacins have digestive and anticoagulation roles, also inactivating prey/host vasoactive peptides [76, 81] to maintain host-parasite relationships [82–85]. However, sea lice astacin is expressed in tegument glands associated with probable roles in lubrication of integument as opposed to feeding [86]. Therefore, the presence of these proteases in Argulus SEPs may be associated with functional roles in haematophagy, but as the feeding activity is similar to L. salmonis, they may also be involved in preventing drag when infecting moving fish.
Trypsins are proteases that have been found overexpressed in Atlantic salmon-fed L. salmonis lice (compared to lice feeding on less susceptible hosts) [77] and were also identified in A. foliaceus SEPs. Trypsins are secretory endopeptidases within the serine protease superfamily, known to facilitate food digestion, host penetration and to help in maintaining the host-parasite relationship [84]. They can also act as anticoagulating proteins [35, 76, 87, 88]. Although proteases have diverse biological functions within different tissues of blood-feeding arthropods [87], the detection of putative secreted trypsins in SEPs, verified by signal peptides, suggests that these trypsins have a vital role in A. foliaceus feeding processes and might play a role in parasite-host interactions. Trypsin-like serine proteases secreted in the salmon louse gut act as a general digestive protease [88, 89]. Trypsin-like proteases have been detected in the sea lice species C. rogercresseyi and L. salmonis and in their SEPs [35, 55, 77, 90, 91]. In addition, trypsin-like proteases have also been detected in the skin mucus of Atlantic salmon infected with L. salmonis, and have been suggested to play a role in facilitating feeding and evasion of the host immune response [92]. Whether the trypsins detected here are derived from salivary glands of A. foliaceus or simply gut contents requires further work.
Serpins, serine proteinase inhibitors, have previously been found to be secreted in arthropod saliva at the feeding site in order to facilitate blood meal acquisition through counteracting host defence mechanisms [36]. Two protein domains of serpin were identified in A. foliaceus SEPs. Serpin-mediated modulation of host immune response is achieved in ticks by impairing the hosts homeostatic and inflammatory responses, platelet aggregation and anticoagulation activity [24, 26, 54, 60, 93–98]. Salivary serpin 6 in the lone star tick, Amblyomma americanum, has been shown to have an inhibitory role in blood clotting and complement activation [60]. Moreover, Ixodes scapularis salivary serpin was found to inhibit the action of thrombin, platelet aggregation and trypsin at the tick-host interface [54]. Such serpins are likely to be inhibitors of pro-inflammatory and pro-coagulant proteases [99], such as Iris2, whereas serpins in Ixodes ricinus have been shown to inhibit inflammation by inhibiting cathepsin G and chymase [26]. Kim et al. [100] concluded that serpin was involved in host defence mechanisms during feeding by the inhibition of host trypsin and trypsin-like proteases. Weakened inflammatory responses have been reported recently in more susceptible carp species infected by A. siamensis [101] and suppression of pro-inflammatory responses by L. salmonis on infected Atlantic salmon is considered to contribute significantly to greater salmonid host susceptibility to salmon lice [102–104].
The presence of serpin in A. foliaceus secretions therefore implies a similar role in facilitating parasitism and modulating host immune responses in argulids. The observed presence of serpins by MS with a predicted MW of 51.4 kDa (afol_27409.2p; Table 1) is supported by SDS-PAGE of the SEPs sample, where an intense band was observed at 46.6 kDa. Similar to other blood-sucking arthropods these findings suggest that serpin in Argulus spp. may be one of the major components involved in evasion of the host defence mechanisms for ingestion of a successful blood meal. We recently used lectin-binding assays to characterise the A. foliaceus glands and have localised proteins with binding affinity to glycoaminoglycans (GAGs) to the spinal gland [7]. Serpins utilise GAGs for protease inhibition activity including modulation of coagulation [105] so it is likely that the spinal glands secretions are involved in A. foliaceus serpin activity.
Fasciclin protein domain from the A. foliaceus SEPs, also known as transforming growth factor-beta-induced protein [84], possessed a signal peptide. This protein has also been identified in the saliva of the argasid tick Ornithodoros moubata with the presence of signal peptides supporting a secretory nature [25], and salivary glands in the mosquito Anopheles culicifacies [106] and L. salmonis [77]. The fasciclin protein function was predicted to be associated with mediation of cell adhesion and signalling [106, 107], although its role in haematophagous arthropods remains unclear.
VIT (Vault protein inter-alpha-trypsin protein) domain was detected in the SEPs of A. foliaceus with an associated signal peptide, albeit with a relatively low SC% of 7.4 (Table 1). Interestingly, this protein domain has been identified recently, associated with von Willebrand factor type A protein domain, in the salivary subset of vampire snail C. reticulata (Mollusca: Gastropoda), feeding on fish blood. VIT has also been reported as the most overexpressed salivary transcript of the feeding-related proteins in the salivary glands of the parasitic snail [76]. This proteinase inhibitor was found, among several proteinase inhibitors, in the saliva of the hard tick H. longicornis, to be secreted into the feeding site to maintaining homeostasis, thus facilitate blood meal acquisition [36]. Due to the presence of VIT with a signal peptide in A. foliaceus SEPs, a similar mode of action may exist to that of H. longicornis VIT, but further validation is required.
In terms of molecular function, GO analysis was very restricted but revealed that the majority of the identified A. foliaceus proteins are nominally involved in catalytic activity acting on a protein (8%) and hydrolase activity (50%), and 17% in ion binding and oxireducatase activity, which were found also from L. salmonis adult SEPs [55]. Notably, in terms of biological function, oxidation-reduction processes accounted for 100% of the A. foliaceus SEPs identified, which may have roles in protection against host-derived reactive oxygen species [55, 108]. Proteins that were detected amongst the A. foliaceus SEPs but for which no biological function could be assigned on account of the absence of homologues in the Pfam and NCBI databases, remain of potential interest as the basis for further study.
The most common SE proteins assigned to molecular function were catalase, peptidases, hydrolases, endopeptidases, serine type endopeptidases, metallopeptidases and oxidoreductase. These proteinases were observed in both adult stages of L. salmonis in the study conducted by Hamilton et al. [55] and were suggested to potentially facilitate host-parasite interactions. For instance, serine peptidases and serine type endopeptidases may be of key importance to the success of the pre-adult- salmon louse in evading the host immune system before going into the final stage in the parasite life-cycle [109]. Catalase and other proteins detected in the saliva from the tick H. longicornis are suggested to play a role in detoxifying generated oxidants during blood meal acquisition and/or host oxidants associated with inflammation [36].
Conclusions
To our knowledge, this study represents the first proteomic analysis undertaken for SEPs from any branchiuran fish louse. Here we reveal possible functional roles of A. foliaceus SEPs in digestion and immunomodulation, with a number of protein families shared with other haematophagous ectoparasites. A number of apparently unique secreted proteins were identified compared to other haematophagous ecdysozoa.
Supplementary information
Additional file 1: Table S1. All the hits initially returned by the MS with highlights to those that fell below the MASCOT assigned score of 35 which indicates a P-value < 0.05, hits above 35 that did not meet validation criteria and proteins identified as positive as they pass all criteria.
Additional file 2: Table S2. Argulus foliaceus positive hits.
Additional file 3: Table S3. OmicsBOx (Blast2Go) analysis, using whole NCBI as Blast search and InterProscan for protein function assignment.
Acknowledgments
The authors would like to acknowledge the contributions of Mr Jimmy Poole on Isle of Bute for his enormous assistance in providing Argulus samples throughout the duration of this study. The authors acknowledge Dr John Taggart and Mrs Jacquie Ireland (Institute of Aquaculture (IoA), University of Stirling (UoS)) for providing support and guidance for all aspects of molecular research. A big thank you to Scott Hamilton (Moredun Research Institute, Midlothian, UK) for his advice on SEP collection and functional protein analysis.
Abbreviations
- LC-ESI-MS/MS
liquid chromatography-electrospray ionisation-tandem mass spectrometry
- NaCl
sodium chloride
- HPLC
high-performance liquid chromatography
- BCA
bicinchoninic acid
- kDa
kilodalton
- EST
expressed sequence tag
- SDSPAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- PGE2
prostaglandin synthase E2
- CaCl2
calcium chloride dihydrate
- DW
distilled water
- NaHCO3
sodium bicarbonate
- VIT/VWA
Vault protein inter-alpha-trypsin domain and von Willebrand factor type A domain
- PPIB
peptidyl-prolyl cis-trans isomerase B
Authors’ contributions
AA, SJM and JB designed and planned the experiments. AA was involved with sample collection. AA and SJM carried out the laboratory experiments and analysed the data. AA wrote the manuscript. KM and NI were involved in proteomics preparation and analysis. MB and SW contributed to the transcriptomic database and bioinformatics. All authors revised the final manuscript. All authors read and approved the final manuscript.
Funding
This work was generously supported by Sultan Qaboos University, Oman and by the Schlumberger Foundation Faculty for the Future programme.
Availability of data and materials
The datasets which support the conclusions are included within the article. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [110] partner repository with the dataset identifier PXD016226 [111, 112].
Ethics approval and consent to participate
The project was ethically approved by the University of Stirling Ethics Committee (Animal Welfare and Ethics Review Board; AWERB).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aisha AmbuAli, Email: hala@squ.edu.om.
Sean J. Monaghan, Email: s.j.monaghan@stir.ac.uk
Kevin McLean, Email: Kevin.McLean@moredun.ac.uk.
Neil F. Inglis, Email: Neil.Inglis@moredun.ac.uk
Michaël Bekaert, Email: michael.bekaert@stir.ac.uk.
Stefanie Wehner, Email: wehner.ste@gmail.com.
James E. Bron, Email: j.e.bron@stir.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-3964-z.
References
- 1.Fryer G. The parasitic Crustacea of African freshwater fishes: their biology and distribution. J Zool London. 1968;156:45–95. doi: 10.1111/j.1469-7998.1968.tb08578.x. [DOI] [Google Scholar]
- 2.Kabata Z. Diseases of fish, book 1: crustacea as enemies of fishes. Snieszko SF, Axelrod H, editor. New Jersey: T.F.H Publications; 1970.
- 3.Byrnes T. Two new Argulus Species (Branchiura: Argulidae) found on Astralian bream (Acanthopagurs spp.) Aust Zool. 1985;21:579–586. [Google Scholar]
- 4.Taylor NG, Sommerville C, Wootten R. A review of Argulus spp. ocuring in UK freshwater. Bristol: Environment Agency; 2005. [Google Scholar]
- 5.Rahman MM. Some aspects of the biology of a freshwater fish parasite Argulus foliaceus (L.) (Argulidae, Branchiura, Crustacea) J Bangladesh J Zool. 1995;23:77–86. [Google Scholar]
- 6.Sahoo P, Mohanty J, Garnayak SK, Mohanty BR, Kar B, Prasanth H, et al. Estimation of loss due to argulosis in carp culture ponds in India. Indian J Fish. 2013;60:99–102. [Google Scholar]
- 7.AmbuAli A, Monaghan SJ, Al-Adawi K, Al-Kindi M, Bron JE. Histological and histochemical characterisation of glands associated with the feeding appendages of Argulus foliaceus (Linnaeus, 1758) Parasitol Int. 2019;69:82–92. doi: 10.1016/j.parint.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Øvergård A-C, Hamre LA, Harasimczuk E, Dalvin S, Nilsen F, Grotmol S. Exocrine glands of Lepeophtheirus salmonis (Copepoda: Caligidae): Distribution, developmental appearance, and site of secretion. J Morphol. 2016;277:1616–1630. doi: 10.1002/jmor.20611. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman GL. Argulus, a branchiuran parasite of freshwater fishes. Washington: US Department of the Interior, Fish and Wildlife Service, Division of Fishery Research; 1977. [Google Scholar]
- 10.Mikheev VN, Mikheev AV, Pasternak AF, Valtonen ET. Light-mediated host searching strategies in a fish ectoparasite, Argulus foliaceus L. (Crustacea: Branchiura) Parasitology. 2000;120:409–416. doi: 10.1017/S0031182099005569. [DOI] [PubMed] [Google Scholar]
- 11.Walker P, Flik G, Bonga S. The biology of parasites from the genus Argulus and a review of the interactions with its host. In: Gert F, Geert W, editors. Host-parasite interactions. London: Taylor & Francis; 2004. pp. 107–129. [PubMed] [Google Scholar]
- 12.LaMarre E, Cochran PA. Lack of host species selection by the exotic parasitic crustacean, Argulus japonicus. J Freshw Ecol. 1992;7:77–80. doi: 10.1080/02705060.1992.9664672. [DOI] [Google Scholar]
- 13.Van der Salm AL, Nolan DT, Spanings FAT, Wendelaar Bonga SE. Effects of infection with the ectoparasite Argulus japonicus (Thiele) and administration of cortisol on cellular proliferation and apoptosis in the epidermis of common carp, Cyprinus carpio L., skin. J Fish Dis. 2000;23:173–184. doi: 10.1046/j.1365-2761.2000.00230.x. [DOI] [Google Scholar]
- 14.Shimura S, Inoue K. toxic effects of extract from the mouthparts of the Argulus coregoni Thorell. (Crustacea: Branchiura) Bull Jap Soc Sci Fish. 1984;50:729. doi: 10.2331/suisan.50.729. [DOI] [Google Scholar]
- 15.Swanepoel JH, Avenant-Oldewage A. Comments on the morphology of the pre-oral spine in Argulus (Crustacea: Branchiura) J Morphol. 1992;212:155–162. doi: 10.1002/jmor.1052120206. [DOI] [PubMed] [Google Scholar]
- 16.Gresty KA, Boxshall GA, Nagasawa K. The fine structure and function of the cephalic appendages of the branchiuran parasite, Argulus japonicus Thiele. Philos Trans R Soc B Biol Sci. 1993;339:119–135. doi: 10.1098/rstb.1993.0009. [DOI] [Google Scholar]
- 17.Møller OS, Olesen J. The little-known Dipteropeltis hirundo Calman, 1912 (Crustacea, Branchiura): SEM investigations of paratype material in light of recent phylogenetic analyses. Exp Parasitol. 2010;125:30–41. doi: 10.1016/j.exppara.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Saha SK, Guha A, Banerjee A. Feeding apparatus and associated glands in the freshwater fish ectoparasite Argulus Siamensis Wilson, 1926 (Branchiura) Crustaceana. 2011;84:1153–1168. doi: 10.1163/156854011X587469. [DOI] [Google Scholar]
- 19.Al-Darwesh AA, Al-Shabbani MAA, Faris BH. Diagnostic and pathological study of Argulus japonicas in goldfish (Carassius Auratus) Glob J Bio-Sci Biotechnol. 2014;3:384–387. [Google Scholar]
- 20.Von Reumont BM, Campbell LI, Jenner RA. Quo vadis venomics? A roadmap to neglected venomous invertebrates. Toxins. 2014;6:3488–3551. doi: 10.3390/toxins6123488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawal R, Vijay S, Kadian K, Singh J, Pande V, Sharma A. Towards a proteomic catalogue and differential annotation of salivary gland proteins in blood fed malaria vector Anopheles culicifacies by mass spectrometry. PLoS ONE. 2016;11:e0161870. doi: 10.1371/journal.pone.0161870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho-costa TM, Mendes MT, Vinicius M, Alvares T, Gomes M, Tiburcio S, et al. Immunosuppressive effects of Amblyomma cajennense tick saliva on murine bone marrow- derived dendritic cells. Parasit Vectors. 2015;8:22. doi: 10.1186/s13071-015-0634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anstead CA, Korhonen PK, Young ND, Hall RS, Jex AR, Murali SC, et al. Lucilia cuprina genome unlocks parasitic fly biology to underpin future interventions. Nat Commun. 2015;6:7344. doi: 10.1038/ncomms8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tirloni L, Seixas A, Mulenga A, Da Silva Vaz I, Termignoni C. A family of serine protease inhibitors (serpins) in the cattle tick Rhipicephalus (Boophilus) microplus. Exp Parasitol. 2014;137:25–34. doi: 10.1016/j.exppara.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Díaz-Martín V, Manzano-Román R, Valero L, Oleaga A, Encinas-Grandes A, Pérez-Sánchez R. An insight into the proteome of the saliva of the argasid tick Ornithodoros moubata reveals important differences in saliva protein composition between the sexes. J Proteomics. 2013;80:216–235. doi: 10.1016/j.jprot.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Chmelar J, Oliveira CJ, Rezacova P, Francischetti IMB, Kovarova Z, Pejler G, et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weston-Davies W, Nuttall P. Parasite saliva as a source of antiallergic agents. Lancet. 2002;359:1067. doi: 10.1016/S0140-6736(02)08075-3. [DOI] [PubMed] [Google Scholar]
- 28.Horn F, dos Santos PC, Termignoni C. Boophilus microplus anticoagulant protein: an antithrombin inhibitor isolated from the cattle tick saliva. Arch Biochem Biophys. 2000;384:68–73. doi: 10.1006/abbi.2000.2076. [DOI] [PubMed] [Google Scholar]
- 29.Ribeiro J, Anderson J, Manoukis N, Meng Z, Francischetti I. A further insight into the sialome of the tropical bont tick, Amblyomma variegatum. BMC Genomics. 2011;12:136. doi: 10.1186/1471-2164-12-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karim S, Singh P, Ribeiro JMC. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS ONE. 2011;6(12):e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francischetti I, Sa-Nunes A, Mans B, Santos I, Ribeiro J. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saurabh S, Mohanty J, Garnayak SK, Sahoo PK. Identification of immunodominant polypeptides of the freshwater fish lice Argulus siamensis (Wilson)—preliminary findings. Indian J Fish. 2012;59:103–106. [Google Scholar]
- 33.Ruane N, McCarthy T, Reilly P. Antibody response to crustacean ectoparasites in rainbow trout, Oncorhynchus mykiss (Walbaum), immunized with Argulus foliaceus L. antigen extract. J Fish Dis. 1995;18:529–537. doi: 10.1111/j.1365-2761.1995.tb00357.x. [DOI] [Google Scholar]
- 34.Fast MD, Ross NW, Craft CA, Locke SJ, MacKinnon SL, Johnson SC. Lepeophtheirus salmonis: characterization of prostaglandin E(2) in secretory products of the salmon louse by RP-HPLC and mass spectrometry. Exp Parasitol. 2004;107:5–13. doi: 10.1016/j.exppara.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Fast MD, Johnson SC, Eddy TD, Pinto D, Ross NW. Lepeophtheirus salmonis secretory/excretory products and their effects on Atlantic salmon immune gene regulation. Parasite Immunol. 2007;29:179–189. doi: 10.1111/j.1365-3024.2007.00932.x. [DOI] [PubMed] [Google Scholar]
- 36.Tirloni L, Islam MS, Kim TK, Diedrich JK, Yates JR, Pinto AFM, et al. Saliva from nymph and adult females of Haemaphysalis longicornis: a proteomic study. Parasit Vectors. 2015;8:338. doi: 10.1186/s13071-015-0918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dzik JM. Molecules released by helminth parasites involved in host colonization. Acta Biochim Pol. 2006;53:33–64. doi: 10.18388/abp.2006_3361. [DOI] [PubMed] [Google Scholar]
- 38.Boulard Chantal. Degradation of bovine C3 by serine proteases from parasites Hypoderma lineatum (Diptera, Oestridae) Vet Immunol Immunopathol. 1989;20:387–398. doi: 10.1016/0165-2427(89)90083-4. [DOI] [PubMed] [Google Scholar]
- 39.Klüttgen B, Dülmer U, Engels M, Ratte H. ADaM, an artificial freshwater for the culture of zooplankton. Water Res. 1994;28:743–746. doi: 10.1016/0043-1354(94)90157-0. [DOI] [Google Scholar]
- 40.Laemmli UK. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batycka M, Inglis NF, Cook K, Adam A, Fraser-Pitt D, Smith DG, et al. Ultra-fast tandem mass spectrometry scanning combined with monolithic column liquid chromatography increases throughput in proteomic analysis. Rapid Commun Mass Spectrom. 2006;20:2074–2080. doi: 10.1002/rcm.2563. [DOI] [PubMed] [Google Scholar]
- 43.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2015;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Philip D, Bowden J, et al. Reference generation and analysis with Trinity. Nat Protoc. 2013;8:1–43. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;9:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The UniProt Consortium UniProt The UniProt Consortium UniProt: a hub protein information. Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eddy SR. Profile Hidden Markov Models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 49.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor GK, Goodlett DR. Rules governing protein identification by mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3420. doi: 10.1002/rcm.2225. [DOI] [PubMed] [Google Scholar]
- 51.Salát J, Paesen GC, Řezáčová P, Kotsyfakis M, Kovářová Z, Šanda M, et al. Crystal structure and functional characterization of an immunomodulatory salivary cystatin from the soft tick Ornithodoros moubata. Biochem J. 2010;429:103–112. doi: 10.1042/BJ20100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Champagne DE, Wasserman HA, Kumar S, Singh S. Pharmacological and immunological properties of saliva of the blood-feeding insects Rhodnius prolixus and Aedes aegypti. Physiol Entomol. 2004;29:269–277. doi: 10.1111/j.0307-6962.2004.00406.x. [DOI] [Google Scholar]
- 53.Gillespie RD, Mbow ML, Titus RG. The immunomodulatory factors of bloodfeeding arthropod saliva. Parasite Immunol. 2000;22:319–331. doi: 10.1046/j.1365-3024.2000.00309.x. [DOI] [PubMed] [Google Scholar]
- 54.Ibelli AMG, Kim TK, Hill CC, Lewis LA, Bakshi M, Miller S, et al. A blood meal-induced Ixodes scapularis tick saliva serpin inhibits trypsin and thrombin, and interferes with platelet aggregation and blood clotting. Int J Parasitol. 2014;44:369–379. doi: 10.1016/j.ijpara.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamilton S, McLean K, Monaghan SJ, McNair C, Inglis NF, McDonald H, et al. Characterisation of proteins in excretory/secretory products collected from salmon lice, Lepeophtheirus salmonis. Parasit Vectors. 2018;11:294. doi: 10.1186/s13071-018-2885-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson SC, Fast MD. Interactions between sea lice and their hosts. In: Gert F, Geert W, editors. Host-parasite interactions. London: Taylor & Francis; 2004. pp. 131–159. [PubMed] [Google Scholar]
- 57.Boxaspen K. A review of the biology and genetics of sea lice. ICES J Mar Sci. 2006;63:1304–1316. doi: 10.1016/j.icesjms.2006.04.017. [DOI] [Google Scholar]
- 58.Boxshall GA, Bravo S. On the identity of the common Caligus (Copepoda: Siphonostomatoida: Caligidae) from salmonid netpen systems in southern Chile. Contrib Zool. 2000;69:137–146. doi: 10.1163/18759866-0690102015. [DOI] [Google Scholar]
- 59.Radulović ŽM, Kim TK, Porter LM, Sze S-H, Lewis L, Mulenga A. A 24-48 h fed Amblyomma americanum tick saliva immuno-proteome. BMC Genomics. 2014;15:518. doi: 10.1186/1471-2164-15-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulenga A, Kim T, Ibelli AMG. Amblyomma americanum tick saliva serine protease inhibitor 6 is a cross-class inhibitor of serine proteases and papain-like cysteine proteases that delays plasma clotting and inhibits platelet aggregation. Insect Mol Biol. 2013;22:306–319. doi: 10.1111/imb.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chmelar J, Calvo E, Pedra JHF, Francischetti IMB, Kotsyfakis M. Tick salivary secretion as a source of antihemostatics. J Proteomics. 2012;75:3842–3854. doi: 10.1016/j.jprot.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro JMC. Role of saliva in blood-feeding by arthropods. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 63.Dalvin S, Frost P, Loeffen P, Skern-Mauritzen R, Baban J, Rønnestad I, et al. Characterisation of two vitellogenins in the salmon louse Lepeophtheirus salmonis: molecular, functional and evolutional analysis. Dis Aquat Organ. 2011;94:211–224. doi: 10.3354/dao02331. [DOI] [PubMed] [Google Scholar]
- 64.Terwilliger NB, Ryan MC. Functional and phylogenetic analyses of phenoloxidases from brachyuran (Cancer magister) and branchiopod (Artemia franciscana, Triops longicaudatus) Crustaceana. Biol Bull. 2006;210:38–50. doi: 10.2307/4134535. [DOI] [PubMed] [Google Scholar]
- 65.Kapp K, Schrempf S, Lemberg MK, Dobberstein B. Post-targeting functions of signal peptides. In: Zimmermann R, editor. Protein Transp into endoplasmic reticulum. Austin, Texas: Landes Bioscience; 2009. pp. 1–6. [Google Scholar]
- 66.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tirloni L, Reck J, Terra RMS, Martins JR, Mulenga A, Sherman NE, et al. Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: a comparison between partially and fully engorged females. PLoS ONE. 2014;9:4. doi: 10.1371/journal.pone.0094831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maritz-olivier C, Stutzer C, Jongejan F, Neitz AWH, Gaspar ARM. Tick anti-hemostatics: targets for future vaccines and therapeutics. Trends Parasitol. 2007;23:397–407. doi: 10.1016/j.pt.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 69.Galay RL, Aung KM, Umemiya-Shirafuji R, Maeda H, Matsuo T, Kawaguchi H, et al. Multiple ferritins are vital to successful blood feeding and reproduction of the hard tick Haemaphysalis longicornis. J Exp Biol. 2013;216:1905–1915. doi: 10.1242/jeb.081240. [DOI] [PubMed] [Google Scholar]
- 70.Donohue KV, Khalil SMS, Sonenshine DE, Roe RM. Heme-binding storage proteins in the Chelicerata. J Insect Physiol. 2009;55:287–296. doi: 10.1016/j.jinsphys.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 71.Rosell-Davis R, Coons LB. Relationship between feeding, mating, vitellogenin production and vitellogenesis in the tick Dermacentor variabilis. Exp Appl Acarol. 1989;7:95–105. doi: 10.1007/BF01200456. [DOI] [PubMed] [Google Scholar]
- 72.Pinnow P, Fabrizius A, Pick C, Burmester T. Identification and characterisation of hemocyanin of the fish louse Argulus (Crustacea: Branchiura) J Comp Physiol B. 2016;186:161–168. doi: 10.1007/s00360-015-0943-3. [DOI] [PubMed] [Google Scholar]
- 73.Paul RJ, Pirow R. The physiological significance of respiratory proteins in invertebrates. Zool J. 1997;100:298–306. [Google Scholar]
- 74.Depledge MH, Bjerregaard P. Haemolymph protein composition and copper levels in decapod crustaceans. Helgoländer Meeresuntersuchungen. 1989;43:207–223. doi: 10.1007/BF02367900. [DOI] [Google Scholar]
- 75.Destoumieux-Garzón D, Saulnier D, Garnier J, Jouffrey C, Bulet P, Bachère E. Crustacean immunity: antifungal peptides are generated from the C terminus of shrimp hemocyanin in response to microbial challenge. J Biol Chem. 2001;276:47070–47077. doi: 10.1074/jbc.M103817200. [DOI] [PubMed] [Google Scholar]
- 76.Modica MV, Lombardo F, Franchini P, Oliverio M. The venomous cocktail of the vampire snail Colubraria reticulata (Mollusca, Gastropoda) BMC Genomics. 2015;16:441. doi: 10.1186/s12864-015-1648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Braden LM, Sutherland BJG, Koop BF, Jones SRM. Enhanced transcriptomic responses in the Pacific salmon louse Lepeophtheirus salmonis oncorhynchi to the non-native Atlantic Salmon Salmo salar suggests increased parasite fitness. BMC Genomics. 2017;18:110. doi: 10.1186/s12864-017-3520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Da Silveira RB, Wille ACM, Chaim OM, Appel MH, Silva DT, Franco CRC, et al. Identification, cloning, expression and functional characterization of an astacin-like metalloprotease toxin from Loxosceles intermedia (brown spider) venom. Biochem J. 2007;406:355–363. doi: 10.1042/BJ20070363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Da Silveira RB, Dos Santos Filho JF, Mangili OC, Veiga SS, Gremski W, Nader HB, et al. Identification of proteases in the extract of venom glands from brown spiders. Toxicon. 2002;40:815–822. doi: 10.1016/S0041-0101(02)00078-8. [DOI] [PubMed] [Google Scholar]
- 80.Trevisan-Silva D, Gremski LH, Chaim OM, da Silveira RB, Meissner GO, Mangili OC, et al. Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles) Biochimie. 2010;92:21–32. doi: 10.1016/j.biochi.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 81.Lun HM, Mak CH, Ko RC. Characterization and cloning of metallo-proteinase in the excretory/secretory products of the infective-stage larva of Trichinella spiralis. Parasitol Res. 2003;90:27–37. doi: 10.1007/s00436-002-0815-0. [DOI] [PubMed] [Google Scholar]
- 82.Bąska P, Wiśniewski M, Krzyżowska M, Długosz E, Zygner W, Górski P, et al. Molecular cloning and characterisation of in vitro immune response against astacin-like metalloprotease Ace-MTP-2 from Ancylostoma ceylanicum. Exp Parasitol. 2013;133:472–482. doi: 10.1016/j.exppara.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 83.Park J-O, Pan J, Möhrlen F, Schupp M-O, Johnsen R, Baillie DL, et al. Characterization of the astacin family of metalloproteases in C. elegans. BMC Dev Biol. 2010;10:14. doi: 10.1186/1471-213X-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim HJ, Kim IS. Transforming growth factor-β-induced gene product, as a novel ligand of integrin αMβ2, promotes monocytes adhesion, migration and chemotaxis. Int J Biochem Cell Biol. 2008;40:991–1004. doi: 10.1016/j.biocel.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 85.Gallego SG, Loukas A, Slade RW, Neva FA, Varatharajalu R, Nutman TB, et al. Identification of an astacin-like metallo-proteinase transcript from the infective larvae of Strongyloides stercoralis. Parasitol Int. 2005;54:123–133. doi: 10.1016/j.parint.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 86.Øvergård AC, Hamre LA, Harasimczuk E, Dalvin S, Nilsen F, Grotmol S. Exocrine glands of Lepeophtheirus salmonis (Copepoda: Caligidae): distribution, developmental appearance, and site of secretion. J Morphol. 2016;227:1616–1630. doi: 10.1002/jmor.20611. [DOI] [PubMed] [Google Scholar]
- 87.Santiago PB, de Araújo CN, Motta FN, Praça YR, Charneau S, Bastos IMD, et al. Proteases of haematophagous arthropod vectors are involved in blood-feeding, yolk formation and immunity—a review. Parasit Vectors. 2017;10:79. doi: 10.1186/s13071-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kvamme BO, Frost P, Nilsen F. The cloning and characterisation of full-length trypsins from the salmon louse Lepeophtheirus salmonis. Mol Biochem Parasitol. 2004;136:303–307. doi: 10.1016/j.molbiopara.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 89.Johnson SC, Ewart KV, Ross N, Murray HM. Molecular cloning of trypsin cDNAs and trypsin gene expression in the salmon louse Lepeophtheirus salmonis (Copepoda: Caligidae) Parasitol Res. 2002;88:789–796. doi: 10.1007/s00436-002-0656-x. [DOI] [PubMed] [Google Scholar]
- 90.Valenzuela-Miranda D, Gallardo-Escárate C. Caligus rogercresseyi serine proteases: Transcriptomic analysis in response to delousing drugs treatments. Aquaculture. 2016;465:65–77. doi: 10.1016/j.aquaculture.2016.08.027. [DOI] [Google Scholar]
- 91.Fast MD, Burka JF, Johnson SC, Ross NW. Enzymes released from Lepeophtheirus salmonis in response to mucus from different salmonids. J Parasitol. 2003;89:7–13. doi: 10.1645/0022-3395(2003)089[0007:ERFLSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 92.Ross NW, Firth KJ, Wang A, Burka JF, Johnson SC. Changes in hydrolytic enzyme activities of naive Atlantic salmon Salmo salar skin mucus due to infection with the salmon louse Lepeophtheirus salmonis and cortisol implantation. Dis Aquat Organ. 2000;41:43–51. doi: 10.3354/dao041043. [DOI] [PubMed] [Google Scholar]
- 93.Mudenda L, Pierlé SA, Turse JE, Scoles GA, Purvine SO, Nicora CD, et al. Proteomics informed by transcriptomics identifies novel secreted proteins in Dermacentor andersoni saliva. Int J Parasitol. 2014;44:1029–1037. doi: 10.1016/j.ijpara.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 94.Yu Y, Cao J, Zhou Y, Zhang H, Zhou J. Isolation and characterization of two novel serpins from the tick Rhipicephalus haemaphysaloides. Ticks Tick Borne Dis. 2013;4:297–303. doi: 10.1016/j.ttbdis.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 95.Prevot PP, Beschin A, Lins L, Beaufays J, Grosjean A, Bruys L, et al. Exosites mediate the anti-inflammatory effects of a multifunctional serpin from the saliva of the tick Ixodes ricinus. FEBS J. 2009;276:3235–3246. doi: 10.1111/j.1742-4658.2009.07038.x. [DOI] [PubMed] [Google Scholar]
- 96.Prevot PP, Adam B, Boudjeltia KZ, Brossard M, Lins L, Cauchie P, et al. Anti-hemostatic effects of a serpin from the saliva of the tick Ixodes ricinus. J Biol Chem. 2006;281:26361–26369. doi: 10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- 97.Steen NA, Barker SC, Alewood PF. Proteins in the saliva of the Ixodida (ticks): pharmacological features and biological significance. Toxicon. 2006;47:1–20. doi: 10.1016/j.toxicon.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 98.Leboulle G, Crippa M, Decrem Y, Mejri N, Brossard M, Bollen A, et al. Characterization of a novel salivary immunosuppressive protein from Ixodes ricinus ticks. J Biol Chem. 2002;277:10083–10089. doi: 10.1074/jbc.M111391200. [DOI] [PubMed] [Google Scholar]
- 99.Tirloni L, Kim TK, Coutinho ML, Ali A, Seixas A, Termignoni C, et al. The putative role of Rhipicephalus microplus salivary serpins in the tick-host relationship. Insect Biochem Mol Biol. 2016;71:12–28. doi: 10.1016/j.ibmb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim TK, Tirloni L, Radulovic Z, Lewis L, Bakshi M, Hill C, et al. Conserved Amblyomma americanum tick Serpin19, an inhibitor of blood clotting factors Xa and XIa, trypsin and plasmin, has anti-haemostatic functions. Int J Parasitol. 2015;45:613–627. doi: 10.1016/j.ijpara.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kar B, Moussa C, Mohapatra A, Mohanty J, Jayasankar P, Sahoo PK. Variation in susceptibility pattern of fish to: do immune responses of host play a role? Vet Parasitol. 2016;221:76–83. doi: 10.1016/j.vetpar.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 102.Johnson SC, Albright LJ. Comparative susceptibility and histopathology of the response of naive Atlantic, chinook and coho salmon to experimental infection with Lepeophtheirus salmonis (Copepoda: Caligidae) Dis Aquat Organ. 1992;14:179–193. doi: 10.3354/dao014179. [DOI] [Google Scholar]
- 103.Fast MD. Fish immune responses to parasitic copepod (namely sea lice) infection. Dev Comp Immunol. 2014;43:300–312. doi: 10.1016/j.dci.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 104.Braden LM, Barker DE, Koop BF, Jones SRM. Differential modulation of resistance biomarkers in skin of juvenile and mature pink salmon, Oncorhynchus gorbuscha by the salmon louse, Lepeophtheirus salmonis. Fish Shellfish Immunol. 2015;47:7–14. doi: 10.1016/j.fsi.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 105.Rein CM, Desai UR, Church FC. Serpin-glycosaminoglycan interactions. Methods Enzymol. 2011;501:105–137. doi: 10.1016/B978-0-12-385950-1.00007-9. [DOI] [PubMed] [Google Scholar]
- 106.Sharma P, Sharma S, Mishra AK, Thomas T, De Das T, Rohilla SL, et al. Unraveling dual feeding associated molecular complexity of salivary glands in the mosquito Anopheles culicifacies. Biol Open. 2015;4:1002–1015. doi: 10.1242/bio.012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gobert GN, Tran MH, Moertel L, Mulvenna J, Jones MK, McManus DP, et al. Transcriptional changes in Schistosoma mansoni during early schistosomula development and in the presence of erythrocytes. PLoS Negl Trop Dis. 2010 doi: 10.1371/journal.pntd.0000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robinson MW, Hutchinson AT, Dalton JP, Donnelly S. Peroxiredoxin: a central player in immune modulation. Parasite Immunol. 2010;32:305–313. doi: 10.1111/j.1365-3024.2010.01201.x. [DOI] [PubMed] [Google Scholar]
- 109.Firth KJ, Johnson SC, Ross NW. Characterization of proteases in the skin mucus of Atlantic salmon (Salmo salar) infected with the salmon louse (Lepeophtheirus salmonis) and in whole-body louse homogenate. J Parasitol. 2000;86:1199–1205. doi: 10.1645/0022-3395(2000)086[1199:COPITS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 110.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Perez-Riverol Y, Xu QW, Wang R, Uszkoreit J, Griss J, Sanchez A, et al. PRIDE inspector toolsuite: Moving toward a universal visualization tool for proteomics data standard formats and quality assessment of proteomexchange datasets. Mol Cell Proteomics. 2016;15:305–317. doi: 10.1074/mcp.O115.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. All the hits initially returned by the MS with highlights to those that fell below the MASCOT assigned score of 35 which indicates a P-value < 0.05, hits above 35 that did not meet validation criteria and proteins identified as positive as they pass all criteria.
Additional file 2: Table S2. Argulus foliaceus positive hits.
Additional file 3: Table S3. OmicsBOx (Blast2Go) analysis, using whole NCBI as Blast search and InterProscan for protein function assignment.
Data Availability Statement
The datasets which support the conclusions are included within the article. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [110] partner repository with the dataset identifier PXD016226 [111, 112].