Abstract
A key goal of geroscience research is to identify effective interventions to extend human healthspan, the years of healthy life. Currently, majority of the geroprotectors are found by screening compounds in model organisms; whether they will be effective in humans is largely unknown. Here we present a new strategy called ANDRU (aging network based drug discovery) to help the discovery of human geroprotectors. It first identifies human aging subnetworks that putatively function at the interface between aging and age-related diseases; it then screens for pharmacological interventions that may “reverse” the age-associated transcriptional changes occurred in these subnetworks. We applied ANDRU to human adipose gene expression data from the Genotype Tissue Expression (GTEx) project. For the top 31 identified compounds, 19 of them showed at least some evidence supporting their function in improving metabolic traits or lifespan, which include type 2 diabetes drugs such as pioglitazone. As the query aging genes were refined to the ones with more intimate links to diseases, ANDRU identified more meaningful drug hits than the general approach without considering the underlying network structures. In summary, ANDRU represents a promising human data-driven strategy that may speed up the discovery of interventions to extend human healthspan.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00106-x) contains supplementary material, which is available to authorized users.
Keywords: Aging, Age-related diseases, Drug repurposing, Network pharmacology, Pharmacogenomics, Geroscience
Introduction
Aging is a major risk factor for age-related diseases (ARDs) and the ultimate cause for most human mortalities (Kennedy et al. 2014). It has been demonstrated in model organisms, that genetic, environmental, and pharmacological interventions capable of extending lifespan are associated with delayed onset and progression of multiple age-related diseases (Everitt et al. 2006). Such observations have laid the foundations for the hypothesis of geroscience (Kennedy et al. 2014), which states that aging is the major modifiable risk factor for most chronic diseases, and it could be possible to simultaneously prevent multiple age-related diseases by targeting the basic biology of aging.
Through drug screening and testing in model organisms, more than four hundred “anti-aging” drugs have been identified (Moskalev et al. 2015; Barardo et al. 2017). Despite the progress, the development of human geroprotectors is facing significant challenges (Kumar and Lombard 2016). Since majority of the geroprotectors were identified in model organisms with little human data support, their effectiveness in promoting human healthspan remains largely unknown. Such uncertainty poses high risk for the pharmaceutical industry to engage in the very costly clinical trials (Kumar and Lombard 2016).
Human aging is a complex process with a large number of genes and pathways being involved. For example, our and other groups have shown that the expression of hundreds to thousands of genes changes with age in various tissues (Glass et al. 2013; Yang et al. 2015). To achieve a holistic understanding of the aging process and to better elucidate the complex interconnections between aging and ARDs, systems and network approaches have been explored (Zhang et al. 2016; Fernandes et al. 2016). For example, Wang et al. showed that age-related disease genes are closer to aging genes in a protein-protein interaction (PPI) network (Wang et al. 2009). We found that age-related disease categories shared functional terms including canonical aging pathways, suggesting that conserved pathways of aging might simultaneously influence multiple ARDs in humans (Johnson et al. 2015). We also developed a network algorithm called GeroNet to prioritize biological processes mediating the connections between aging and ARDs (Yang et al. 2016). Despite these progresses, most existing models neither considered tissue specificity nor incorporated the network dynamics associated with aging. Therefore, they could only capture human aging systems with limited accuracy. Although more recent work has started to address the tissue specificity in human aging (Johnson et al. 2016), quantitative tissue specific aging network models are yet to be developed to allow more accurate characterization of the interplay between aging and ARDs.
In addition to providing biological insights into aging and disease mechanisms, network biology can also be used to facilitate drug discovery (Csermely et al. 2013). A promising area under active development is based on the concept of Connectivity Map (CMap) (Lamb 2007), which uses compound perturbation-induced gene expression changes to identify drug candidates for treating human diseases (Iorio et al. 2010). An increasing number of successful studies have been reported which provide a strong proof-of-concept, which include a study in C. elegans that identified compounds mimicking the effect of caloric restriction on extending lifespan (Dudley et al. 2011; Wagner et al. 2015; Calvert et al. 2016).
Previously, we worked on a large human genomic dataset generated by the Gene-Tissue Expression (GTEx) project (The GTEx Consortium 2015). This dataset allows a comprehensive survey of tissue-specific aging mechanisms and we showed that it could recapitulate multiple well-established aging hallmarks (Yang et al. 2015). In this study, we further demonstrate a novel framework called ANDRU (aging network based drug discovery) to integrate GTEx with other pharmacogenomic datasets to identify possible interventions to extend human healthspan.
Materials and methods
Data collection and processing
Gene expression data
Human tissue gene expression data were obtained from the GTEx portal (The GTEx Consortium 2015). We performed a few data processing steps to consider genes having at least 0.1 RPKM (Reads Per Kilobase Million) in 2 or more individuals followed by a quantile normalization across genes. Similar to published GTEx study (The GTEx Consortium 2015), we adjusted several confounding factors, including: (1) gender, (2) collection center, (3) RIN (RNA Integrity Number), (4) ischemic time, and (5) top 3 genotype principal components (PCs).
Genotype data
To correct for population stratification, the top 3 genotype PCs used as covariates in gene expression analysis were constructed using GCTA (Yang et al. 2011a) on non-autosomal SNPs (Yang et al. 2011b). Several quality control steps were performed using plink to exclude those variants with MAF < 1%, HWE p < 1e-6, and variant missing rate > 5%.
Age-group division and construction of young and old gene co-expression networks
We divided samples into four groups based on donor’s chronological age, namely, Young (age < =35), mid.Young (35 < age < = 50), mid.Old (50 < age < = 65), and Old (age > 65). We used Young and Old groups to define age-associated differentially expressed genes (DEGs) and construct co-expression networks for the ANDRU pipeline. For each gene, its expression values were quantile normalized into a standard normal distribution. We removed outlier samples by a hierarchical clustering provided by the weighted gene correlation network analysis (WGCNA) (Zhang and Horvath 2005). We set the soft power in WGCNA to 5 for adipose and 4 for artery aorta, respectively to construct the co-expression networks.
Differential gene expression and differential connectivity analysis
Read counts were used to call differential expression genes by HTSeq/DESeq (Anders and Huber 2010) and edgeR/limma (Robinson et al. 2010; Law et al. 2014) between young and old samples (adjusted p value less than 0.05 was set as threshold). We used TMM (the weighted trimmed mean of M-values) normalization in edgeR, the batch effect due to collection center was corrected using ComBat in the sva Bioconductor package (Leek et al. 2012). We applied DGCA (Differential Gene Correlation Analysis) for a differential connectivity analysis (McKenzie et al. 2016). DGCA computes differentially correlated gene pairs between two groups of samples. We performed a permutation analysis to evaluate the enrichment of differentially connected gene pairs in each module.
Rank candidate compounds based on drug perturbation-induced gene expression signatures
We used 5170 drug perturbation-induced gene expression signatures collected in Crowd Extracted Expression of Differential Signatures (CREEDS) for our analysis (Wang et al. 2016). We also considered 6100 expression profiles covering 1309 compounds from CMap (Lamb 2007).
Since the two perturbation gene expression signature databases have different data contents, i.e., the CREEDS only provides DEG gene names while CMap contains the whole array expression data, we used different approaches to query and rank candidate compounds. For CMap, we used its web-based query tool to rank candidate drugs. This web-tool used a modified Kolmogorov-Smirnov test statistics to calculate the similarity between drug-perturbation induced gene expression changes and aging DEGs. For signatures from CREEDS, we considered three methods to rank them. These methods included the Signed Jaccard Index as implemented by the CREEDs web query tool; the Gene Set Enrichment Analysis (GSEA), and Fisher’s Exact Test based calculation. For GSEA, we first sorted every gene in a tissue based on their differential expression between old and young samples, so highly up-regulated genes in old individuals were ranked at top and down-regulated in the old individuals were ranked at the bottom. We used the fgsea R package to calculate GSEA enrichment scores and adjusted p-values. A positive enrichment score indicates that drug perturbation signature is enriched in the top ranked genes while a negative enrichment score indicates the opposite case. For Fisher’s Exact Test, we calculated the significance of overlap between up- and down-regulated aging genes with drug perturbation-induced up- and down-regulated genes. A drug was ranked to the top if drug induced genes significantly overlapped with aging DEGs (in each direction).
To jointly consider the output from all methods and further narrow down the candidate list, we prioritized drugs that were ranked at the top by at least two methods. To compare ANDRU with general approach without considering the underlying network structures, we performed queries using either DEGs from turquoise module or all DEGs as input genes and compared the results.
Results
An overview of the ANDRU pipeline
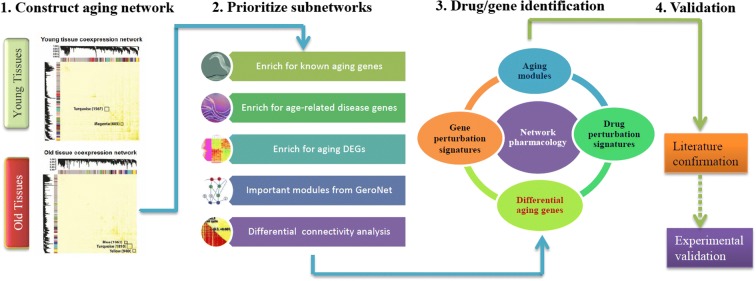
ANDRU takes tissue specific transcriptomic data from young- and old-age human samples as input. It outputs a list of candidate compounds that may help to slow aging and provide geroprotection in the corresponding tissue. A network model is generated to reveal the underlying subnetworks that may function at the interface between aging and ARDs, which will be used to prioritize compounds to achieve geroprotection. As shown in Fig. 1., for a tissue under consideration, we first construct gene co-expression networks (Zhang and Horvath 2005) in young and old tissue samples, respectively. The subnetworks (also called modules) are constructed based on correlations among gene expressions and their topological overlaps. This is a data-driven approach to unbiasedly divide genome/transcriptome into smaller modules that are often enriched for specific biological functions. Second, using aging DEGs, known disease gene sets and a few other criteria, we prioritize subnetworks that are influenced by both aging and diseases (we consider them as the putative interface between aging and diseases). Third, the aging DEGs in the prioritized subnetworks are used to query pharmacogenomic datasets to identify interventions that may “reverse” the age-associated expression changes in the corresponding subnetworks. The top ranked compounds are further evaluated based on independent data and/or literatures.
Fig. 1.
An overview of the ANDRU pipeline. Four major steps are 1). Tissue and age specific aging network construction; 2) Subnetwork prioritization based on multiple criteria to highlight the converging point of aging and age-related diseases; 3) Application of a pharmacogenomic approach to rank candidate drugs or genes. As an example, the down-regulated genes in drug 3’s perturbation signature is highly enriched for the up-regulated aging DEGs in the orange subnetwork, and therefore drug 3 is considered as a hit; and 4) Either literature or experimental based confirmation of the top candidate compounds or genes
We applied ANDRU to subcutaneous adipose tissue samples from GTEx. Adipose is a dynamic tissue showing profound changes with aging and can play important roles in the development of multiple ARDs such as diabetes (Palmer and Kirkland 2016).
Samples in the old-age group showed strongest age-associated gene expression changes compared to samples in other age groups
To gain insight into the overall age-associated gene expression changes, we first divided adipose samples into four groups based on donor’s chronological ages: Young (age < = 35, n = 40), mid.Young (35 < age < = 50, n = 93), mid.Old (50 < age < = 65, n = 171), and Old (age > 65, n = 46) (n is the number of samples). We compared the three older groups (mid.Young, mid.Old and Old) with the Young group to define DEGs in each group. It is of note that the sample size of each group is different which may lead to different statistical power in DEG calculation. To adjust for this, we randomly selected 40 samples in each of the three older groups to ensure same number of samples across all the groups. We then performed DEG calling based on the down-sampled data. The process was repeated 10 times and results are summarized in Tables S1 and S2. Using Old group as an example (which had 46 samples), the number of DEGs based on down-sampling ranged from 673 to 2198 with a mean of 1555.6 and standard deviation(SD) of 578.8. This is slightly smaller than 1619, the number of DEGs called using all Old samples. Although the change in the number of DEGs due to down-sampling is not particularly large for the Old group, the change is more significant in Mid.Old and Mid.Young (Table S2). For example, the number of DEGs using all Mid.Old samples was 3471, but it reduced to 1194.4 ± 830.5 in down-sampling, indicating that sample size indeed has a strong effect on the number of DEGs. Therefore, we used DEGs called based on down-sampling for the cross-group comparison.
Based on DEG results from down-sampling runs, Old group showed the largest number of DEGs (mean ± SD: 1555.6 ± 578.8) (Table S2). The mid.Young group had the least number of DEGs, with only 2.3 DEGs being found on average.
In order to ensure that our grouping of GTEx samples based on age and DEG calling provide biological meaningful results with very few false positives, we performed a randomization test. For each group (Young, mid-Young, mid-Old, and Old group), we randomly split samples into two subgroups of equal size for 100 times, we then performed the DEG analysis by comparing the gene expression between these two subgroups. Except for the mid-Young group in which we detected 1 significant DEG in 2 runs, there were no significant DEGs detected in all the other 3 groups. This indicates that samples within each age group are similar for their age-related gene expression and the between-group DEGs should contain very few false positives.
In addition to DEG analysis, we also compared the three older-age groups with Young using a differential gene correlation analysis (DGCA). DGCA identifies significant change of correlations among gene-pairs under two conditions, which provides information on the transcriptional changes from a different angle compared with the DEG (McKenzie et al. 2016). The DGCA results also indicated a similar pattern as observed in the DEG analysis (Fig. S1), i.e., Old group showed the largest number of significant differential correlations compared with the two middle-age groups (mid.Young and mid.Old). Given that the Old group manifested the greatest aging effect at the transcriptome level, and older individuals are likely to benefit the most from pro-longevity interventions, we decided to focus on comparing the Old and Young groups for the downstream analyses.
For the Young and Old groups, we identified 4 and 2 outlier samples, respectively and they were removed from further analysis. 36 samples in the Young group (age < = 35) and 52 samples in the Old group (age > =65) were used for the following analyses. The sample information of age and sex distribution are shown in Fig. S2.
Deriving and comparing young and old adipose tissue gene co-expression networks
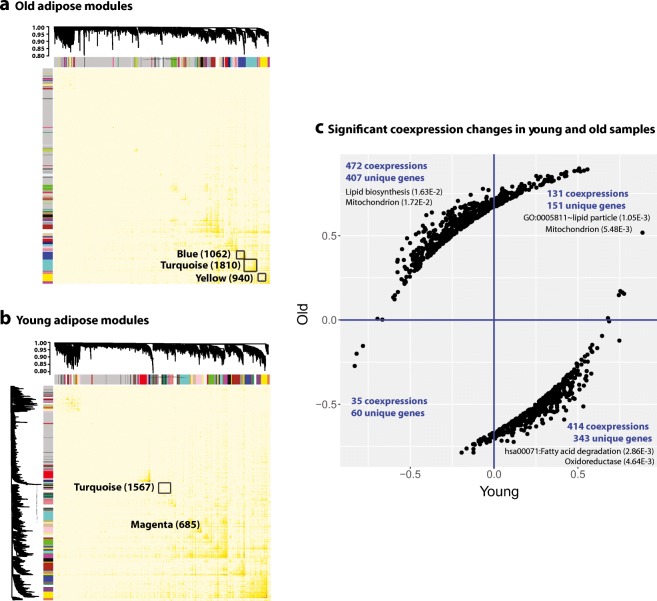
We constructed gene co-expression network using WGCNA and obtained 103 and 85 subnetworks in young and old adipose tissues respectively (Zhang and Horvath 2005). The largest module from the old group is “turquoise” (denoted hereafter as turquoiseold), which consists of 1299 protein-coding genes (see Fig. 2A). This module is significantly enriched for “mitochondrial matrix” (FDR =1.90E-13) and “oxidoreductase” (FDR = 8.34E-13) (Table S3). Other large modules from the old group are enriched for “cell junction”, “Ubl conjugation pathway”, and “signal peptide” (see details in Table S4). The top modules in the young adipose tissue (see Fig. 2B) include “turquoise” module (1010 protein-coding genes), which is enriched for “DNA repair” (FDR = 1.58E-04); “yellow” module (939 protein-coding genes), which is enriched for “WD repeat” (FDR = 1.46E-09), and “magenta” module (604 protein-coding genes, denoted hereafter as magentayoung), which is enriched for “mitochondrion” (FDR = 7.99E-32) (Table S4).
Fig. 2.
WGCNA networks in (A) young and (B) old adipose tissues, a few large size modules are highlighted with their number of genes annotated (both protein coding and non-protein coding genes are counted), and (C) the significant co-expression correlation changes between young and old samples for the turquoiseold module
Since we constructed networks from young and old samples separately, the subnetwork IDs (in color names) were assigned independently and did not match between the young and old networks. To map and compare the global network structures in young and old tissues, we considered gene overlap between the two networks. We found that turquoiseold most significantly overlapped with magentayoung with Jaccard index of 0.12 (see Table S5). Both modules are significantly enriched for “mitochondria” related (“GO:0005759~mitochondrial matrix” for turquioseold and “GO:0005739~mitochondrion” for magentayoung) and “transition peptide” terms (see Table S4). Since the magentayoung and turquoiseold show quite different sizes, this could imply that the co-expression wiring has changed substantially due to aging for genes in this subnetwork. For a further investigation, we performed a differential connectivity analysis (McKenzie et al. 2016). Specifically, we aimed to identify the significant differentially co-expressed gene pairs between young and old networks. From this analysis, 1051 significant differential co-expressions (p value ≤5.0E-4) involving 643 unique genes were found (Fig. 2C. and Dataset 1). The 643 genes are significantly enriched for “oxidoreductase” (FDR of 1.66E-6), “GO:0005759~mitochondrial matrix” (FDR of 5.26E-5), and “hsa00350:tyrosine metabolism” (FDR of 6.85E-3) (Table S6). For these significantly changed co-expression pairs, the majority of them show gained co-expression in the old network. For example, 708 gene-gene pairs showed significant correlations only in old samples but not in young samples. In contrast, only 38 gene-gene pairs are significantly correlated in young samples and no longer significantly correlated in old tissues (loss of correlation). For the 505 genes in the 708 gene pairs with gained correlation in old adipose (regardless of the sign of the correlation), they are enriched for “mitochondrion” (FDR = 5.2E-5), “oxidoreductase” (FDR = 3.6E-5), and “fatty acid degradation” (FDR = 4.5E-2). For the 1051 significant differential co-expressions, majority of them show changes in their co-expression directions. For example, there are 472 negative correlations in the young samples but they become positively correlated in the old samples (in the 2nd quadrant in Fig. 2C). Similarly, 414 positive co-expressions in the young tissues became negatively correlated in the old tissues (in the 4th quadrant in Fig. 2C). This result indicates that substantial network re-wiring occurred in the adipose tissue during aging. The enriched functions of the re-wired genes suggest potential link with age-dependent adipose function declines in insulin sensitivity, lipolytic and fatty acid responsiveness (Tchkonia et al. 2010). Since we had imbalanced number of samples for young vs. old adipose tissues, to ensure the gain of co-expression was not due to larger sample size for old adipose, we performed a down-sampling calculation. Specifically, we randomly selected 36 samples from the old adipose tissue for 20 times, and calculated the co-expression values for the same 1051 gene pairs. We found that the results were consistent with the ones obtained from using all the 52 old adipose samples (Dataset 1). Therefore, the gain of co-expression in the old adipose was not caused by the larger sample size.
Prioritizing subnetworks that may function as the interface between aging and age-related diseases
As co-expression network divides transcriptome into a number of non-overlapping subnetworks, it is unclear if all these subnetworks contribute equally to aging and ARDs. Based on our previous work (Yang et al. 2016), we hypothesize that subnetworks are differentially involved in aging and make distinct contribution to the development of age-related diseases. To evaluate the relative importance of each subnetwork, we considered multiple criteria and applied them to the old tissue sample derived co-expression network. We focused on old network since ARDs mainly occur in older individuals and therefore, modules derived from the old tissue samples are more likely to reveal the interconnection between aging and ARDs. We describe the details of each criterion below.
Criterion 1. Enrichment for GenAge genes: GenAge database (Build 18) contains 305 literature-based manually curated putative human aging genes (Tacutu et al. 2013), and has been used as a reference aging gene list by several studies (Zhang et al. 2016; Wang et al. 2009). We calculated the enrichment of GenAge genes for each subnetwork (see Table S7). The top three overlap were seen in the hotpinkold (FDR = 5.36E-01), turquoiseold (FDR = 5.36E-01), and yellowold (FDR = 5.71E-01). These subnetworks are strongly enriched for cell cycle genes, mitochondrial matrix, and signal peptide/glycoprotein, respectively (Table S7). Although the enrichment for GenAge genes was not particularly strong as FDRs were greater than 0.01, it is still suggestive that some modules are more enriched for human “aging genes” compared to others. It is of note that turquoiseold contains APOE (apolipoprotein E), which is one of the very few genes that are reproducible in genome-wide association studies for human longevity (Table S8) (Schachter et al. 1994).
Criterion 2. Enrichment for differential expressed aging genes: We applied DESeq (Anders and Huber 2010) to identify differentially expressed genes (DEGs) between young and old adipose tissue samples. At FDR 0.05, there were 382 DEGs, among which 265 were protein-coding genes with 148 up-regulated and 117 down-regulated (Table S9). We also performed sample down-sampling to ensure that the DEGs were robust and did not dependent on the difference in sample size (see Supplementary Table S1 and S2). The up-regulated protein coding DEGs are significant enriched for functions like “extracellular matrix” (FDR 6.74E-6), “glycoprotein” (FDR 1.05E-5), and “signal peptide” (FDR 2.43E-3), and the down-regulated genes are significant enriched for “transmembrane” (FDR 2.56E-3) (Table S10). It is of note that the top ranked DEG was CDKN2A, whose expression levels increased more than threefold from young to old adipose tissues (FDR 1.75E-6). CDKN2A encodes p16INK4A, which is a commonly used biomarker for cellular senescence (Sharpless and Sherr 2015). Studies have shown that by clearing p16INK4A-positive cells, mice could have a longer lifespan with delayed onset of age-related disorders (Baker et al. 2016).
We mapped the aging DEGs to the old adipose tissue co-expression network and summarized the results in Table 1. There are 3 modules significantly enriched for the aging DEGs, i.e., turquoiseold (97common genes with FDR 3.02E-42), magentaold (31 genes with FDR 1.49E-13), and yellowold (28 genes with FDR 1.18E-04). Turquoiseold contained about two fifth of all the significant aging DEGs (97out of 265), indicating that aging had a very strong influence on gene expression in this subnetwork among all the subnetworks.
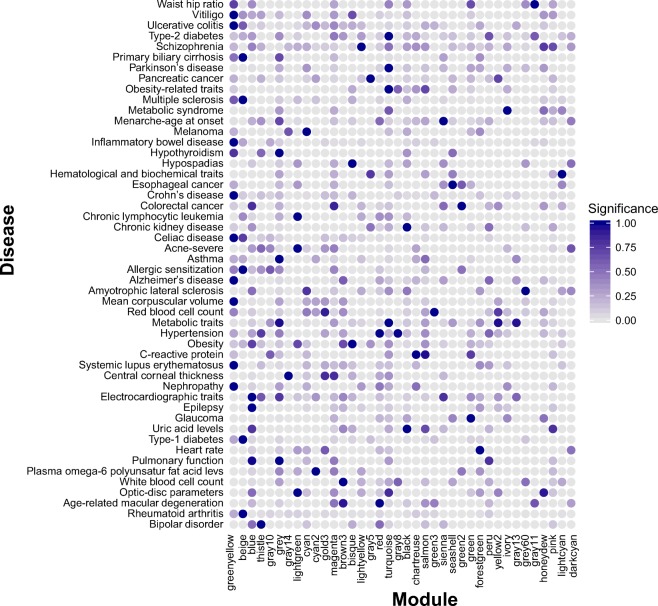
Criterion 3. Enrichment for disease genes: We previously manually curated genes associated with 277 diseases and traits, among which 84 diseases and traits had at least 20 associated genes (Yang et al. 2016). We evaluated the enrichment of disease genes in each subnetwork for all the 84 diseases/traits (Table S11). As shown in Fig. 3, there exists significant enrichment of disease genes in multiple subnetworks. For example, the greenyellowold subnetwork overlapped with multiple immune related diseases (e.g., vitiligo, ulcerative colitis, inflammatory bowel disease, and Alzheimer’s disease) while it is highly enriched for “immunity” (adjusted p value = 4.3E-34). The turquoiseold subnetwork significantly overlapped with genes associated with obesity-related traits, Parkinson’s disease, metabolite traits, and was the only subnetwork that significantly overlapped with type 2 diabetes genes, indicating it may play an important function in mediating the development of these metabolic related diseases.
Criterion 4. Strong connection with age-related diseases by GeroNet calculation: GeroNet is a method that allows the consideration of connections between aging and disease genes through network connections. Different from criteria 3, it considers indirect interactions (i.e., interactions mediated by other genes) between aging and disease genes (Yang et al. 2016). We performed GeroNet analysis using GenAge genes as aging genes and disease genes from 277 disease and trait categories. We used the HPRD (human protein reference database) as a reference PPI network (Yang et al. 2016). The results from GeroNet showed that turquoiseold was the most important subnetwork in mediating the connections between aging and multiple diseases in all the 85 subnetworks. This result further suggests that turquoiseold subnetwork plays a critical role in mediating the connections between aging and age-related disease including metabolic diseases such as type 2 diabetes.
Criterion 5. Enrichment for differential co-expressions: As we have shown in the previous section, we observed profound co-expression re-wiring between magentayoung and turquoiseold modules. To assess the significance level of re-wiring, we performed a permutation analysis. Briefly, for a subnetwork of size n, we randomly selected n genes from all the genes in the transcriptome to form a random subnetwork. Using the same set of young and old tissue samples, we calculated the number of significant differential co-expressions for this “random” subnetwork between young and old ages. By running this permutation test for 1000 times, we estimated a p value for observing the actual number of differentially co-expressed gene-gene pairs in our real data. A gene pair was called differentially connected if p value was less than or equal to 5E-4. Using this permutation estimation, the estimated p value for observing 1051 differential co-expressions was close to 0 (p < 0.001), which indicates that co-expression rewiring in the turquoiseold module was highly significant.
Table 1.
A summary of the top ten largest modules in old adipose tissue and their enrichment for GenAge genes, aging DEGs, Disease genes, Differential co-expression genes and the significance for connecting to ARDs as indicated by GeroNet
| Module | #Gene | Function | GenAge | Differential gene | Disease gene | Differential co-expression gene | GeroNet | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Term | FDR | #Overlap | FDR (Rank*) | #Overlap | FDR (Rank*) | #Significant Disease | Geometric mean P (Rank*) | #mean of permutation significant pairs / #significant pairs | FDR (Rank*) | #Significant diseases | Geometric mean FDR (Rank*) | ||
| turquoise | 1299 | mitochondrial matrix | 1.90E-13 | 34 | 5.36E-01 (2) | 97 | 2.84E-40 (1) | 13 | 4.60E-01 (2) | 259.85/1051 | ~ 0 (1) | 125 | 3.62E-49 (1) |
| magenta | 345 | signal peptide | 2.4E-24 | 3 | 1.00E+00 (48) | 31 | 1.49E-13 (2) | 10 | 5.67E-01 (5) | 12.53/306 | ~ 0 (1) | 80 | 1.53E-29 (17) |
| yellow | 691 | signal peptide | 7.36E-06 | 20 | 5.71E-01 (3) | 28 | 1.18E-04 (3) | 10 | 5.45E-01 (4) | 68.68/486 | ~ 0 (1) | 97 | 5.28E-41 (2) |
| maroon | 85 | Oxidoreductase | 3.9E-4 | 2 | 1.00E+00 (30) | 3 | 1.00E-00 (4) | 5 | 8.10E-01 (29) | 0.83/12 | ~ 0 (1) | 18 | 2.30E-17 (34) |
| salmon | 244 | Disulfide bond | 4.5E-3 | 4 | 1.00E+00 (34) | 6 | 1.00E-00 (5) | 7 | 6.14E-01 (7) | 8.89/21 | 2.56E-03 (50) | 62 | 6.41E-25 (23) |
| green | 360 | 8 | 1.00E+00 (15) | 8 | 1.00E-00 (6) | 6 | 6.89#-01 (11) | 27.70/807 | ~ 0 (1) | 67 | 8.97E-34 (10) | ||
| blue | 763 | Cell junction | 7.86E-18 | 10 | 1.00E+00 (46) | 13 | 1.00E-00 (7) | 16 | 5.27E-01 (3) | 89.94/492 | ~ 0 (1) | 98 | 2.37E-32 (11) |
| lightyellow | 175 | 5 | 1.00E+00 (12) | 3 | 1.00E-00 (8) | 5 | 6.99E-01 (13) | 3.73/102 | ~ 0 (1) | 44 | 4.73E-31 (14) | ||
| red3 | 52 | 0 | 1.00E+00 (80) | 1 | 1.00E-00 (9) | 0 | 8.93E-01 (52) | 0.24/0 | 7.48E-01 (75) | 13 | 2.73E-16 (37) | ||
| bisque | 123 | Proto-oncogene | 4.9E-2 | 5 | 9.33E-01 (6) | 2 | 1.00E-00 (10) | 8 | 7.02E-01 (14) | 1.85/6 | 2.45E-02 (54) | 33 | 3.45E-26 (20) |
*Rank was based on the significance of the subnetwork for the corresponding criterion
Fig. 3.
Enrichment of disease genes for each subnetwork derived from old adipose tissues. Only subnetworks/diseases having at least one significant enrichment (p value <0.02) are shown
To summarize, we listed the results for the top 10 modules with more than 50 protein coding genes in Table 1 (sorted based on the enrichment for aging DEGs). By a joint consideration of all the criteria, particularly on the criteria that reported significant modules (FDR < 0.05), the turquoiseold stood out as the top subnetwork (Table 1) in which aging and a group of metabolic diseases like type 2 diabetes were connected, suggesting it is a key subnetwork function as the interface between aging and ARDs in adipose tissue.
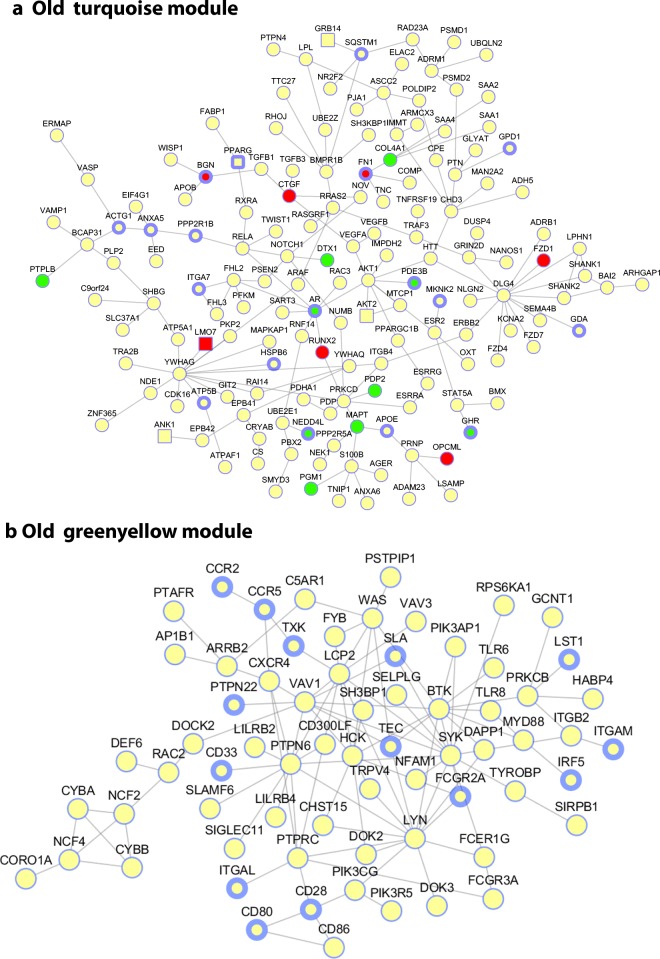
To visualize the turquoiseold subnetwork, we mapped its genes onto the HPRD protein-protein interaction (PPI) network (Fig. S3) and showed the largest connected component in Fig. 4A. We annotated the diabetes genes (square shaped nodes), significantly DEGs between young and old samples (nodes in red color for up-regulated and green color for down-regulated gene expressions in old adipose tissue samples) and APOE-perturbed genes (nodes with thick border, the APOE knockout DEGs were derived from GSE44653) in Fig. 4A. It is of note that signaling pathways important in growth regulation and metabolic function such as the PPARG and GHR/AKT pathway are contained in this PPI subnetwork. Similarly, we show the greenyellowold PPI network in Fig. 4B and genes associated with inflammatory diseases are highlighted with thicker borders.
Fig. 4.
Protein-protein interaction network view of two subnetworks derived from old adipose tissue.a turquoiseold module and its association to aging, diabetes, and APOE perturbed genes. Each node denotes a gene and each edge denotes a protein-protein interaction. The nodes in square shape (e.g., AKT2) are diabetes associated genes obtained from GWAS catalog and OMIM; the nodes with filled red-(up) or green-(down) (e.g., MAPT) are significantly differentially expressed genes (at FDR < = 0.05 by DESeq) between young and old samples; the nodes with blue thick border (e.g., APOE and GDA) are APOE perturbed genes as defined in CREEDS. b greenyellowold module mapped to PPI network and genes associated with inflammatory diseases (nodes with thicker border), including vitiligo, ulcerative colitis, multiple sclerosis, inflammatory bowel disease, Crohn’s disease, celiac disease, Alzheimer’s disease, and systemic lupus erythematosus
Prioritize compounds using a network pharmacogenomic approach
An important goal of human aging and geroscience research is to identify effective interventions to slow aging and delay the onset of age-related diseases. To achieve this goal, we adopt a network pharmacogenomic approach based on the concept of Connectivity Map (CMap) (Lamb 2007). CMap contains more than 6000 drug-perturbation induced gene expression profiles generated from multiple human cell lines, with a coverage of 1309 compounds. The CMap has been used to query various disease-associated gene expression signatures to identify drugs that may “reverse” the gene expression changes observed in the disease conditions. Such identified drugs are considered candidates to treat the corresponding disease. The CMap concept has been successful applied to an increasing number of disease areas (Dudley et al. 2011; Mirza et al. 2017). These success further stimulated efforts to construct even larger scale perturbation-induced expression databases, e.g., The Library of Network-Based Cellular Signatures (LINCS) Program (Duan et al. 2016), and a crowd extracted expression of differential signatures (CREEDS) (Wang et al. 2016). Here, we hypothesize that if a drug or gene perturbation could modulate aging DEGs in our prioritized aging subnetworks toward their youthful states, then this drug/gene is likely to be geroprotective for the tissue under investigation. To test this hypothesis, we considered age-related DEGs in the turquoiseold module (21 up-regulated and 76 down-regulated genes) to query various pharmacogenomic datasets. We also considered using all the DEGs as query input (265 differential genes with 148 up-regulated genes and 117 down-regulated genes) (Table S9) and compared the results from these two inputs.
We relied on CREEDS database which contains 5170 single-drug perturbation-induced gene expression signatures (875 manually curated and 4295 automatically extracted) collected from Gene Expression Omnibus (Wang et al. 2016). Specifically, we screened the CREEDS to identify drug perturbations that could reverse the aging DEGs (i.e., perturbations that up-regulate those repressed aging gene expressions and/or down-regulate those elevated aging gene expressions). We considered three methods to rank a drug-perturbation signature. The first method is a Signed Jaccard Index provided by CREEDS itself (Wang et al. 2016). In this method, the age-associated up- and down-regulated genes are jointly compared with drug-induced up- and down-regulated genes. When there is a perfect match between both up- and down-regulated genes, the Signed Jaccard Index is 1. If genes in the opposite direction match perfectly, the Signed Jaccard Index is −1. For all other situations the score is somewhere between −1 and 1. Since CREEDS only provides Signed Jaccard Index without its associated p-values, we decided to only include the top 50 records based on the Signed Jaccard Index for further consideration. The cutoff at 50 is mainly for practical consideration since in many cases, we can only experimentally follow-up with a limited number of top hits. The second method is based on Gene Set Enrichment Analysis (Subramanian et al. 2005). We first sorted all genes based on their expression fold changes between old and young samples, we then calculated for each drug signature (separating up- and down-regulated genes as two signatures) a normalized enrichment score (NES). A positive NES indicates that a drug perturbation signature is enriched for the top ranked genes which are up-regulated in old samples, while a negative NES indicates an opposite scenario. Similarly, we selected the top 50 records based on NES as more than several hundred records received significant adjusted p-values <0.05. Finally, we used Fisher’s Exact Test to calculate the enrichment between drug perturbation signatures with aging DEGs. To adjust for the potential influence by the gene set size, we calculated a permutation based Z-score to ensure that drug signatures received significant p-values were not due to size-bias, using a method developed by the Enrichr (Chen et al. 2013). We considered all the top significant outputs (FDR < 0.05 or the top 50, whichever is less) by this method. We provide detailed results from each method in the supplementary Table S12. There are in total 232 unique hits by all the three methods. To further narrow down the list, we considered candidates that emerged in the top-ranked lists by at least two methods.
From this analysis, 31 drugs were identified using aging DEGs in the turquoiseold as a query input (Table 2). Among these drugs, 19 of them have at least some supportive evidence suggesting that the compound can improve phenotypes related to metabolism, diabetes, or lifespan (Table 2). These drugs cover a range of distinct mechanisms, suggesting the complexity of the aging process and its interconnection with diseases. It is of note that several type 2 diabetes drugs were identified in this list, including rosiglitazone, pioglitazone, and troglitazone, which are all peroxisome proliferator-activated receptors (PPARs) agonists. In addition to these well-recognized compounds that have already been used to treat type 2 diabetes, several lipids also entered the list such as conjugated linoleic (CLA), oleic acid, and stearic acid.
Table 2.
Top drug identified by querying CREEDS that may reverse the aging DEGs in the turquoiseold module for adipose tissue
| Drug | Direction# | Unique** | Data* | Possible effects | Evidence## (PMID) |
|---|---|---|---|---|---|
| amoxicillin | opposite | Y | M | ||
| ascorbic acid | opposite | Y | M | Reduces blood glucose | + (22242019) (Dakhale et al. 2011); − (24,757,251, 18,160,753, 29,862,091) (Vasudevan and Hirsch 2014; Afkhami-Ardekani and Shojaoddiny-Ardekani 2007; Sharma et al. 2018); Δ (19580823) (Shibamura et al. 2009) |
| Baclofen | opposite | Y | A | Increases brown fat thermogenesis, body weight reduction | + (3,748,315, 20,930,428) (Addae et al. 1986; Arima and Oiso 2010); − (30688773) (Triplett et al. 2019) |
| cis-9, trans-11 CLA (conjugated linoleic) | opposite | A | Reduces body fat mass | + (11110851) (Blankson et al. 2000). | |
| clofibrate | opposite | A | Lipid-lowering, extends C elegans lifespan | Δ(23603800) (Brandstadt et al. 2013) | |
| cyclosporine A | opposite | Y | A | Extends C elegans lifespan | + (24134630) (Ye et al. 2014); − (26787364) (Pottecher et al. 2016) |
| dexamethasone | opposite | M | Anti-inflammatory, extends glucose sensor | + (19885112) (Klueh et al. 2007); − (30927408) (Martin et al. 2019) | |
| diisononyl phthalate | opposite | A | |||
| dioctyl phthalate | opposite | A | |||
| episesamin | opposite | Y | A | Increases mitochondrial activity, antioxidant | + (30,840,309, 31,150,685) (Le et al. 2019; Farbood et al. 2019) |
| equilin | opposite | A | |||
| eribulin | #inconsistent | Y | A | ||
| FA | #inconsistent | Y | A | Oxidative stress resistance | Δ(26546011) (Rathor et al. 2015) |
| GW3965 | #inconsistent | A | Liver X receptor agonist | + (27057732) (Sandoval-Hernandez et al. 2016); − (23,765,184) (Pettersson et al. 2013) | |
| hydrogen peroxide | opposite | Y | M | Extends lifespan | + (8138188) (Ku et al. 1993); − (15298493) (Watt et al. 2004) |
| lactobacillus brevis | opposite | Y | A | ||
| MEK inhibitor U0126 | #inconsistent | Y | A | ||
| oleic acid | opposite | Y | A | Increases insulin sensitivity | + (26055507) (Perdomo et al. 2015) |
| paclitaxel | #inconsistent | Y | A | ||
| parathyroid hormone | opposite | A | Induces lipolysis | + (3598394) (Taniguchi et al. 1987) | |
| perfluorooctanoic acid | opposite | Y | M | ||
| phosgene | opposite | M | |||
| pioglitazone | opposite | Y | M | Treats type 2 diabetes with anti-aging properties | + (17628757) (Jafari et al. 2007) |
| plx4720 | #inconsistent | Y | A | ||
| resveratrol | #inconsistent | M | Extends lifespan | Δ(16461283) (Valenzano et al. 2006) | |
| rosiglitazone | #inconsistent | M | Treats type 2 diabetes, bone loss | + (20971975) (Millar et al. 2011); − (17332064) (Lazarenko et al. 2007) | |
| stearic acid | opposite | A | Can be converted to oleic acid | + (27717956) (Han et al. 2017) | |
| trichloroethylene | opposite | M | |||
| troglitazone | #inconsistent | M | Previously used for diabetes, liver toxicity | + (15793255) (Knowler et al. 2005); − (31294483) (Jia et al. 2019) | |
| ubiquinol | #inconsistent | M | Antioxidant | Δ(19428456) (Yang et al. 2009) | |
| WY-14643 | opposite | A | Accelerates mitochondrial biogenesis | + (16204368) (Bogacka et al. 2005) |
#The direction of compound-induced signature compared with the aging DEG direction: opposite, #inconsistent (drug shows different direction in different methods)
**Compounds identified using turquoise module DEGs but not using all aging DEGs
*M- manually curated drug signatures; A- automatically curated signatures
##Evidence: Δ indicates that drug is in DrugAge build 3; ± indicates the drug has positive/negative evidence, respectively; Representative PubMed IDs are listed for either + or - evidence
From the enrichment analysis using Fisher’s Exact Test, isomers of CLA (Table S13), either cis-9, trans-11 CLA or trans-10, cis-12 CLA, or their mixture up-regulated the expression of multiple genes that were down-regulated in the old-age GTEx adipose tissues. These genes encompass functions such as enzymes in various metabolic pathways (PFKFB3, PGM1, GLUL, MTHFD1, ACO1 and HADH), cofactor and metabolite transporters (SLC19A3 and AQP7), redox homeostasis (FAM213A and PRDX6), PPAR signaling pathway genes (ADIPOQ), cell cycle regulation (CDKN2C), and lipid metabolism (PLIN1 and HADH). The function of these genes is generally consistent with the reported favorable effects of CLA on cancer, atherosclerosis, body weight and fat mass (Pariza 2004).
Another distinct hit is the liver X receptor (LXR) agonist GW3965 (GSE41223) (Pettersson et al. 2013) (Table S13). The canonical role of LXR is regulation of reverse cholesterol transport and cholesterol efflux, but its role also extends to the regulation of glucose and fatty acid metabolism and a wide variety of endocrine processes (Calkin and Tontonoz 2012). The targeted genes that overlap with the aging DEGs are consistent with their function in lipid synthesis (FASN and ACSL1), glucose metabolism (PGD, PC and PGM1), as well as other metabolic pathways (CYB5A, HADH, ACO1, AQP7 and SHMT1).
As another example, episesamin, formed during the refining process of non-roasted sesame seed oil, is a geometrical isomer of sesamin. Sesamin is one of the most abundant lignans (polyphenols) in sesame seed (Kushiro et al. 2002). Both episesamin and sesamin are found to increase the mitochondrial and peroxisomal palmitoyl-CoA oxidation rates and lower the activity and gene expression of hepatic lipogenic enzymes to one-half in rat (Kushiro et al. 2002). Treating 3 T3-L1 preadipocytes with episesamin decreased hormone-induced 3 T3-L1 differentiation as shown by reduced accumulation of intracellular lipid droplets and diminished protein expression of GLUT-4. Episesamin also showed anti-inflammatory activity by counteracting the lipopolysaccharide- and tumor necrosis factor α-induced secretion of interleukin 6 by 3 T3-L1 preadipocytes, suggesting it could be used as a novel potential complementary treatment for obesity (Freise et al. 2013).
To ensure that the drug list output from ANDRU is not random, we estimated how many drug hits we would expect if random aging signatures were provided as input to ANDRU. Specifically, we reshuffled 18,604 protein-coding genes 100 times to generate 100 randomized gene lists, and selected 21 genes from the top and 76 genes from the bottom of each list to form a random aging signature. We then used these random aging signatures as input to ANDRU. From the 100 permutation tests, we only observed 1 or 2 unique drug hits in 4 runs, and no hits in all other runs. Therefore, the p value of obtaining 31 unique drug hits by ANDRU from the real aging signature was very significant (p < 0.01). We listed the drugs identified from the 100 random aging signatures in Table S14. In the list, eribulin and paclitaxel also appeared in our result based on adipose turquoiseold aging signature (Table 2). Except for estradiol, we could not find evidence supporting their function in improving lifespan or metabolic traits.
Comparison of “anti-aging” drug discovery using aging network vs. all age-related DEGs
The general approach of CMap analysis is to use all the DEGs for a disease without considering the underlying network structures. To make a comparison between a general approach with ANDRU, which prioritizes query genes based on the network structures, we compared the results based on query using all the 265 DEGs (Supplement Table S9). Unexpectedly, only 19 drug signatures were found based on the same criterion despite that the input query had a greater number of genes. For these 19 drugs, 11 of them have evidence supporting they may cause improvement in phenotypes related to metabolism, diabetes and lifespan. For the 31 drugs identified using DEGs in turquoiseold, 15 of them were not identified by query using all the DEGs. While using all DEGs, only 2 of the drugs were not found by query using DEGs in turquoiseold, suggesting that DEGs in turquoiseold, although representing fewer genes, are more sensitive in identifying relevant drug perturbation signatures based on our ranking method.
For drugs that were only identified from the turquoiseold module, several of them have evidence to support their protective function in adipose tissue. For example, absortic acid (Vitamin C) was found by querying DEGs in the turquoiseold module but not by using all DEGs. It has been reported that vitamin C can reduce blood glucose and improve glycosylated hemoglobin in type 2 diabetes based on a blinded clinical study (Dakhale et al. 2011).
The prioritized drugs show tissue specificity when ANDRU is applied to another tissue
It is known that aging and ARDs show tissue specificity (Glass et al. 2013; Yang et al. 2015). To evaluate if prioritization of drugs would depend on tissue types, we applied ANDRU to a second tissue type, namely, the artery aorta. The results are summarized in Dataset 2. There were 34 young-age samples (with mean age and standard deviation of 27 ± 4.5) and 33 old-age samples (with mean age and standard deviation of 67 ± 1.1) used by ANDRU. At FDR of 0.05, 459 up-regulated and 325 down-regulated differential protein-coding genes were identified by comparing the young- vs. old-age samples. These DEGs are significantly enriched for glycoprotein and several other categories. The top drug perturbations that reverse the aging DEGs were N-methyl-D-aspartate (NMDA) antagonists. The NMDA receptor has been well-studied for its association with brain aging (Magnusson et al. 2010; Foster et al. 2017). Next to the NMDA antagonists, curcumin was ranked at 4,5,6,8, and 10 in the list. Curcumin is a chemical produced naturally by some plants like ginger root. Curcumin and its metabolite, tetrahydrocurcumin (THC), has been experimentally validated to increase lifespans of nematode roundworm, fruit fly, and mouse (Shen et al. 2013). It is also known that curcumin has anti-oxidative, anti-lipofusinogenesic, and anti-aging effects in the brain (Bala et al. 2006) and might be effective in treating diseases caused by low grade inflammation including cancer and Alzheimer’s disease (Sikora et al. 2010). From network analysis, curcumin was also ranked at the top for aging DEGs in the yellow subnetwork (919 protein coding genes enriched for Zinc finger regions and DNA binding) in old artery aorta tissue (Dataset 2). It is of note that the prioritized drugs are different for the two tissue types we evaluated, implying the underlying aging-related changes and how they interconnect with diseases are tissue specific.
Comparison of “anti-aging” drug discovery using CMap database
While we independently developed this work, Donertas et al. published a closely related study (Donertas et al. 2018). They derived a common aging signature across GTEx brain samples and queried it upon the CMap to infer drugs that may modulate the brain aging signature. Among all the 24 drugs they identified, 7 were known pro-longevity drugs. Donertas et al.’s work suggests that CMap can be valuable resource to work with GTEx-derived aging signatures. To compare this with our work (Donertas et al. 2018), we derived a list of 409 down-regulated and 277 up-regulated age-associated genes common across 13 GTEx brain tissues and queried the CMap (Yang et al. 2015). We were able to identify 8 drugs from CMap whose perturbation signatures significantly correlated with our brain aging signature (FDR <0.05). Among these 8 drugs, 6 of them were consistent with the 18 drugs reported by Donertas et al. by using GTEx data alone (Donertas et al. 2018). The 8 significant drugs are sirolimus, resveratrol, irinotecan, daunorubicin, wortmannin, LY-294002, phenoxybenzamine and chenodeoxycholic acid. The difference between our and Donertas et al.’s results is likely caused by how the aging signatures were generated in the brain tissues. Given the results were largely reproducible, we decided to use CMap to screen the aging signatures derived in adipose tissue and compare with results obtained from CREEDS. Based on querying CMap, rather few hits were returned. Using all aging DEGs, only three drugs were found to be significant (FDR <0.05) (Table S15). Among these, the top one candidate drug was “apigenin” (FDR = 0.017). Apigenin is a natural product belonging to the flavone class. Flavonoid Apigenin (4′,5,7-trihydroxyflavone) has been reported to slow aging by competitive inhibition of NADase activity (Escande et al. 2013) and by inhibition of DNMT activity through reactivation of methylation-silenced genes such as p16INK4A (Fang et al. 2007). It has also been reported that apigenin has properties such as inducing autophagy, anti-inflammatory, antioxidant, anti-carcinogenic and preventing aging signs of skin (Choi et al. 2016). Using DEGs in the turquoiseold, 6 drugs were returned as significant hits (Table S15). The very top compound was pyrvinium, a pinworm anthelmintic drug. It has been found that pyrvinium is a potent Wnt pathway inhibitor that can promote wound repair and post-myocardial infarction cardiac remodeling (Saraswati et al. 2010). Genome Wide Association Studies (GWASs) have suggested a link between Wnt/β-catenin signaling pathway and type 2 diabetes. Wnt/β-catenin signaling pathway is also known to be involved in adipogenesis (Ross et al. 2000), and it is hypothesized that Wnt/β-catenin signaling is crucial for obesity development, and has the potential to be therapeutic target for treatment for obesity (Chen and Wang 2018).
Discussions
In this work, we demonstrated a novel informatics pipeline − ANDRU to construct human aging network and identify interventions that can be geroprotective. We applied ANDRU to GTEx adipose and artery tissues. With aging, adipose tissue undergoes significant changes and contributes to the development of insulin resistance, metabolic dysfunction, inflammation, and impaired regenerative capacity (Palmer and Kirkland 2016). From more than eighty adipose subnetworks, we prioritized and highlighted the turquoiseold for its putative role as the interface between aging and several ARDs. We then queried CREEDs and CMap databases with the aging DEGs within the turquoiseold. Among the top-hits, a large proportion of them received evidence supporting their protective roles in improving metabolic traits and lifespan. As we restricted query genes to the turquoiseold module, which were enriched for genes related to both aging and diseases, we were able to obtain more hits and some of these hits showed clear supportive evidence for their protective functions. Since different subnetworks often have distinct functions and show differential connections to various diseases, it may provide options to select desirable subnetworks to address distinct types of diseases (e.g., the inflammation related diseases vs. metabolic related diseases). Since the diseases of concern can vary among individuals, considering individual’s genetic and genomic profiles to prioritize optimal aging subnetworks could facilitate more personalized geroprotection in future.
The importance and feasibility of performing in silico drug screening for geroprotection has been first demonstrated by Zhavoronkov et al. (Aliper et al. 2016). Our method is conceptual similarity to their method but has a different implementation. For example, we relied on a de novo network construction approach to build aging networks while GeroScope used by Zhavoronkov et al. considered prior knowledge of known aging pathways. While both approaches considered age-related transcriptional changes, the input dataset and methods used to incorporate such information were different. Therefore, our approach represents a significant alternative option for performing in silico geroprotector discovery.
The current implementation of ANDRU and in general the approach based on CMap concept has its limitations and will require future improvement. First, as we relied on existing gene expression signature databases, we are limited to the perturbations collected by these databases and important drugs could be missed if the data was not generated or not included into the database. This is clearly demonstrated that when we used the same aging signature from adipose tissue to query CREEDS and CMap databases, the returned top hits were quite different and CMap only returned a very limited number of significant compounds for the tissue we studied. Although the “incompleteness” of perturbation signatures will likely persist in the near future, perturbation datasets are continuing to grow. For example, the NIH LINCS project is generating high throughput perturbation expression signature data at very large scale (Duan et al. 2016), making it another useful resource for the network pharmacogenomics development. Jointly considering multiple such perturbation pharmacogenomic databases will help to provide more comprehensive coverage. Second, CREEDS contains data collected from multiple species, and the non-human data or data obtained from different tissue types could cause either false positive or false negative hits. This would require additional conformational experiments to ensure perturbation signatures are reproducible in the desirable tissue types. Third, in this pilot work, we considered only a limited number of tissue types to illustrate the pipeline. The top ranked drug candidates were different in each tissue. This suggests that to achieve optimized systemic geroprotection, we may need rely on combining drugs. As GTEx profiles more than 40 tissue types, it is a substantial future work to investigate aging networks and search for geroprotectors at a “whole-body” scale. Fourth, our method contains multiple steps and criteria for which the parameters were empirically determined and therefore arbitrary. In future development, we will explore ways to reduce the complexity of the ANDRU to minimize the number of parameters without sacrificing the overall performance.
In summary, with the accumulation of an unprecedented amount of human omics data, we believe that integrating them to refine the aging networks, and leveraging them to search for the most effective human geroprotectors represents a promising strategy to speed-up the geroscience research.
Electronic supplementary material
(DOCX 593 kb)
(XLSX 11.7 kb)
(XLSX 464 kb)
(XLSX 148 kb)
(XLSX 3.89 mb)
(XLSX 506 kb)
(XLSX 391 kb)
(XLSX 516 kb)
Acknowledgments
JY was supported by a postdoctoral fellowship from Unity Biotechnology. ZT receives financial support from Unity Biotechnology as a consultant. This work is partially supported by NIH/NIA R01-AG055501, U24-CA210993, and a Leducq foundation award to ZT. This work was also supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Authors contributions
ZT conceived the concept of the work. JY, SP and ZT performed the analysis. JY, SP, SH, YS, ZT wrote the manuscript. All authors helped to discuss and improve the work.
Compliance with ethical standards
Competing interests
The authors have declared no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jialiang Yang and Shouneng Peng contributed equally to this work.
References
- Addae JI, Rothwell NJ, Stock MJ, Stone TW. Activation of thermogenesis of brown fat in rats by baclofen. Neuropharmacology. 1986;25:627–631. doi: 10.1016/0028-3908(86)90215-7. [DOI] [PubMed] [Google Scholar]
- Afkhami-Ardekani M, Shojaoddiny-Ardekani A. Effect of vitamin C on blood glucose, serum lipids & serum insulin in type 2 diabetes patients. Indian J Med Res. 2007;126:471–474. [PubMed] [Google Scholar]
- Aliper A, Belikov AV, Garazha A, Jellen L, Artemov A, Suntsova M, Ivanova A, Venkova L, Borisov N, Buzdin A, Mamoshina P, Putin E, Swick AG, Moskalev A, Zhavoronkov A. In search for geroprotectors: in silico screening and in vitro validation of signalome-level mimetics of young healthy state. Aging. 2016;8:2127–2152. doi: 10.18632/aging.101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arima H, Oiso Y. Positive effect of baclofen on body weight reduction in obese subjects: a pilot study. Intern Med. 2010;49:2043–2047. doi: 10.2169/internalmedicine.49.3918. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bala K, Tripathy BC, Sharma D. Neuroprotective and anti-ageing effects of curcumin in aged rat brain regions. Biogerontology. 2006;7:81–89. doi: 10.1007/s10522-006-6495-x. [DOI] [PubMed] [Google Scholar]
- Barardo D, Thornton D, Thoppil H, Walsh M, Sharifi S, Ferreira S, Anžič A, Fernandes M, Monteiro P, Grum T, Cordeiro R, de-Souza EA, Budovsky A, Araujo N, Gruber J, Petrascheck M, Fraifeld VE, Zhavoronkov A, Moskalev A, de Magalhães JP. The DrugAge database of aging-related drugs. Aging Cell. 2017;16:594–597. doi: 10.1111/acel.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankson H, Stakkestad JA, Fagertun H, Thom E, Wadstein J, Gudmundsen O. Conjugated linoleic acid reduces body fat mass in overweight and obese humans. J Nutr. 2000;130:2943–2948. doi: 10.1093/jn/130.12.2943. [DOI] [PubMed] [Google Scholar]
- Bogacka I, Ukropcova B, McNeil M, Gimble JM, Smith SR. Structural and functional consequences of mitochondrial biogenesis in human adipocytes in vitro. J Clin Endocrinol Metab. 2005;90:6650–6656. doi: 10.1210/jc.2005-1024. [DOI] [PubMed] [Google Scholar]
- Brandstadt S, Schmeisser K, Zarse K, Ristow M. Lipid-lowering fibrates extend C. elegans lifespan in a NHR-49/PPARalpha-dependent manner. Aging (Albany NY) 2013;5:270–275. doi: 10.18632/aging.100548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert S, Tacutu R, Sharifi S, Teixeira R, Ghosh P, Magalhães J. A network pharmacology approach reveals new candidate caloric restriction mimetics in C. elegans. Aging Cell. 2016;15:256–266. doi: 10.1111/acel.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Wang J. Wnt/β-catenin signaling and obesity. Front Physiol. 2018;9:792–792. doi: 10.3389/fphys.2018.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Youn J, Kim K, Joo DH, Shin S, Lee J, Lee HK, An IS, Kwon S, Youn HJ, Ahn KJ, An S, Cha HJ. Apigenin inhibits UVA-induced cytotoxicity in vitro and prevents signs of skin aging in vivo. Int J Mol Med. 2016;38:627–634. doi: 10.3892/ijmm.2016.2626. [DOI] [PubMed] [Google Scholar]
- Csermely P, Korcsmáros T, Kiss H, London G, Nussinov R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery a comprehensive review. Pharmacol Ther. 2013;138:333–408. doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhale GN, Chaudhari HV, Shrivastava M. Supplementation of vitamin C reduces blood glucose and improves glycosylated hemoglobin in type 2 diabetes mellitus: a randomized, double-blind study. Adv Pharmacol Sci. 2011;2011:5. doi: 10.1155/2011/195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donertas HM, Fuentealba Valenzuela M, Partridge L, Thornton JM. Gene expression-based drug repurposing to target aging. Aging Cell. 2018;17:e12819. doi: 10.1111/acel.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Reid SP, Clark NR, Wang Z, Fernandez NF, Rouillard AD, Readhead B, Tritsch SR, Hodos R, Hafner M, Niepel M, Sorger PK, Dudley JT, Bavari S, Panchal RG, Ma’ayan A. L1000CDS(2): LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl. 2016;2:16015. doi: 10.1038/npjsba.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant Topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande C, Nin V, Price NL, Capellini V, Gomes AP, Barbosa MT, O'Neil L, White TA, Sinclair DA, Chini EN. Flavonoid Apigenin is an inhibitor of the NAD(+)ase CD38 implications for cellular NAD(+) metabolism, protein acetylation, and treatment of metabolic syndrome. Diabetes. 2013;62:1084–1093. doi: 10.2337/db12-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AV, Hilmer SN, Brand-Miller JC, Jamieson HA, Truswell AS, Sharma AP, Mason RS, Morris BJ, Couteur DGL. Dietary approaches that delay age-related diseases. Clin Interv Aging. 2006;1:11–31. doi: 10.2147/ciia.2006.1.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang MZ, Chen DP, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223s–228s. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- Farbood Y, Ghaderi S, Rashno M, Khoshnam SE, Khorsandi L, Sarkaki A. Sesamin: a promising protective agent against diabetes-associated cognitive decline in rats. Life Sci. 2019;230:169–177. doi: 10.1016/j.lfs.2019.05.071. [DOI] [PubMed] [Google Scholar]
- Fernandes M, Wan C, Tacutu R, Barardo D, Rajput A, Wang JW, Thoppil H, Thornton D, Yang C, Freitas A, de Magalhães JP. Systematic analysis of the gerontome reveals links between aging and age-related diseasesSystematic analysis of the gerontome reveals links between aging and age-related diseases. Hum Mol Genet. 2016;25:4804–4818. doi: 10.1093/hmg/ddw307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T, Kyritsopoulos C, Kumar A. Central role for NMDA receptors in redox mediated impairment of synaptic function during aging and Alzheimer's disease. Behav Brain Res. 2017;322:223–232. doi: 10.1016/j.bbr.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Freise C, Trowitzsch-Kienast W, Erben U, Seehofer D, Kim KY, Zeitz M, Ruehl M, Somasundaram R. (+)-Episesamin inhibits adipogenesis and exerts anti-inflammatory effects in 3T3-L1 (pre)adipocytes by sustained Wnt signaling, down-regulation of PPARγ and induction of iNOS. J Nutr Biochem. 2013;24:550–555. doi: 10.1016/j.jnutbio.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Glass D, Viñuela A, Davies MN, Ramasamy A, Parts L, Knowles D, et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 2013;14:1–12. doi: 10.1186/gb-2013-14-7-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Xu G, Du H, Hu J, Liu Z, Li H, et al. Natural variations in stearoyl-acp desaturase genes affect the conversion of stearic to oleic acid in maize kernel. Theor Appl Genet. 2017;130:151–161. doi: 10.1007/s00122-016-2800-5. [DOI] [PubMed] [Google Scholar]
- Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari M, Khodayari B, Felgner J, Bussel II, Rose MR, Mueller LD. Pioglitazone: an anti-diabetic compound with anti-aging properties. Biogerontology. 2007;8:639–651. doi: 10.1007/s10522-007-9105-7. [DOI] [PubMed] [Google Scholar]
- Jia R, Oda S, Tsuneyama K, Urano Y, Yokoi T (2019). Establishment of a mouse model of troglitazone-induced liver injury and analysis of its hepatotoxic mechanism. J Appl Toxicol [DOI] [PubMed]
- Johnson SC, Dong X, Vijg J, Suh Y. Genetic evidence for common pathways in human age-related diseases. Aging Cell. 2015;14:809–817. doi: 10.1111/acel.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Gonzalez B, Zhang Q, Milholland B, Zhang Z, Suh Y. Network analysis of mitonuclear GWAS reveals functional networks and tissue expression profiles of disease-associated genes. Hum Genet. 2016;136:55–65. doi: 10.1007/s00439-016-1736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klueh U, Kaur M, Montrose DC, Kreutzer DL. Inflammation and glucose sensors: use of dexamethasone to extend glucose sensor function and life span in vivo. J Diabetes Sci Technol. 2007;1:496–504. doi: 10.1177/193229680700100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Hamman RF, Edelstein SL, Barrett-Connor E, Ehrmann DA, Walker EA, Fowler SE, Nathan DM, Kahn SE, Diabetes Prevention Program Research Group Prevention of type 2 diabetes with troglitazone in the diabetes prevention program. Diabetes. 2005;54:1150–1156. doi: 10.2337/diabetes.54.4.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HH, Brunk UT, Sohal RS. Relationship between mitochondrial superoxide and hydrogen peroxide production and longevity of mammalian species. Free Radic Biol Med. 1993;15:621–627. doi: 10.1016/0891-5849(93)90165-q. [DOI] [PubMed] [Google Scholar]
- Kumar S, Lombard DB. Finding Ponce de Leon’s Pill: Challenges in Screening for Anti-Aging Molecules. F1000Research. 2016;5:406. doi: 10.12688/f1000research.7821.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro M, Masaoka T, Hageshita S, Takahashi Y, Ide T, Sugano M. Comparative effect of sesamin and episesamin on the activity and gene expression of enzymes in fatty acid oxidation and synthesis in rat liver. J Nutr Biochem. 2002;13:289–295. doi: 10.1016/s0955-2863(01)00224-8. [DOI] [PubMed] [Google Scholar]
- Lamb J. Innovation - the connectivity map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarenko OP, Rzonca SO, Hogue WR, Swain FL, Suva LJ, Lecka-Czernik B. Rosiglitazone induces decreases in bone mass and strength that are reminiscent of aged bone. Endocrinology. 2007;148:2669–2680. doi: 10.1210/en.2006-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TD, Nakahara Y, Ueda M, Okumura K, Hirai J, Sato Y, et al. Sesamin suppresses aging phenotypes in adult muscular and nervous systems and intestines in a Drosophila senescence-accelerated model. Eur Rev Med Pharmacol Sci. 2019;23:1826–1839. doi: 10.26355/eurrev_201902_17146. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson KR, Brim BL, Das SR. Selective vulnerabilities of N-methyl-D-aspartate (NMDA) receptors during brain aging. Front Aging Neurosci. 2010;2:11. doi: 10.3389/fnagi.2010.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LF, Richardson LS, da Silva MG, Sheller-Miller S, Menon R. Dexamethasone induces primary amnion epithelial cell senescence through telomere-P21 associated pathwaydagger. Biol Reprod. 2019;100:1605–1616. doi: 10.1093/biolre/ioz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie AT, Katsyv I, Song WM, Wang MH, Zhang B. DGCA: a comprehensive R package for differential gene correlation analysis. BMC Syst Biol. 2016;10:106. doi: 10.1186/s12918-016-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JS, Ikewaki K, Bloedon LT, Wolfe ML, Szapary PO, Rader DJ. Effect of rosiglitazone on HDL metabolism in subjects with metabolic syndrome and low HDL. J Lipid Res. 2011;52:136–142. doi: 10.1194/jlr.P008136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza N, Sills GJ, Pirmohamed M, Marson AG. Identifying New Antiepileptic Drugs Through Genomics-Based Drug Repurposing. Hum Mol Genet. 2017;26:527–537. doi: 10.1093/hmg/ddw410. [DOI] [PubMed] [Google Scholar]
- Moskalev A, Chernyagina E, de Magalhães J, Barardo D, Thoppil H, Shaposhnikov M, et al. Geroprotectors.org: a new, structured and curated database of current therapeutic interventions in aging and age-related disease. Aging (Albany NY) 2015;7:616. doi: 10.18632/aging.100799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AK, Kirkland JL. Aging and adipose tissue: potential interventions for diabetes and regenerative medicine. Exp Gerontol. 2016;86:97–105. doi: 10.1016/j.exger.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariza MW. Perspective on the safety and effectiveness of conjugated linoleic acid. Am J Clin Nutr. 2004;79:1132S–1136S. doi: 10.1093/ajcn/79.6.1132S. [DOI] [PubMed] [Google Scholar]
- Perdomo L, Beneit N, Otero YF, Escribano O, Diaz-Castroverde S, Gomez-Hernandez A, et al. Protective role of oleic acid against cardiovascular insulin resistance and in the early and late cellular atherosclerotic process. Cardiovasc Diabetol. 2015;14:75. doi: 10.1186/s12933-015-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson AML, Stenson BM, Lorente-Cebrián S, Andersson DP, Mejhert N, Krätzel J, Åström G, Dahlman I, Chibalin AV, Arner P, Laurencikiene J. LXR is a negative regulator of glucose uptake in human adipocytes. Diabetologia. 2013;56:2044–2054. doi: 10.1007/s00125-013-2954-5. [DOI] [PubMed] [Google Scholar]
- Pottecher J, Kindo M, Chamaraux-Tran TN, Charles AL, Lejay A, Kemmel V, Vogel T, Chakfe N, Zoll J, Diemunsch P, Geny B. Skeletal muscle ischemia-reperfusion injury and cyclosporine a in the aging rat. Fundam Clin Pharmacol. 2016;30:216–225. doi: 10.1111/fcp.12180. [DOI] [PubMed] [Google Scholar]
- Rathor L, Akhoon BA, Pandey S, Srivastava S, Pandey R. Folic acid supplementation at lower doses increases oxidative stress resistance and longevity in Caenorhabditis elegans. Age (Dordr) 2015;37:113. doi: 10.1007/s11357-015-9850-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald O. Inhibition of Adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Sandoval-Hernandez AG, Restrepo A, Cardona-Gomez GP, Arboleda G. LXR activation protects hippocampal microvasculature in very old triple transgenic mouse model of Alzheimer's disease. Neurosci Lett. 2016;621:15–21. doi: 10.1016/j.neulet.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Saraswati S, Alfaro MP, Thorne CA, Atkinson J, Lee E, Young PP. Pyrvinium, a potent small molecule Wnt inhibitor, promotes wound repair and post-MI cardiac remodeling. PLoS One. 2010;5:e15521. doi: 10.1371/journal.pone.0015521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter F, Fauredelanef L, Guenot F, Rouger H, Froguel P, Lesueurginot L, et al. Genetic associations with human longevity at the Apoe and ace loci. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- Sharma E, Resta C, Park P. A case of factitious hyperglycemia in a patient on intravenous ascorbic acid. Case Rep Endocrinol. 2018;2018:7063137. doi: 10.1155/2018/7063137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- Shen LR, Parnell LD, Ordovas JM, Lai CQ. Curcumin and aging. Biofactors. 2013;39:133–140. doi: 10.1002/biof.1086. [DOI] [PubMed] [Google Scholar]
- Shibamura A, Ikeda T, Nishikawa Y. A method for oral administration of hydrophilic substances to Caenorhabditis elegans: effects of oral supplementation with antioxidants on the nematode lifespan. Mech Ageing Dev. 2009;130:652–655. doi: 10.1016/j.mad.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Sikora E, Bielak-Zmijewska A, Mosieniak G, Piwocka K. The promise of slow down ageing may come from curcumin. Curr Pharm Des. 2010;16:884–892. doi: 10.2174/138161210790883507. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhães JP. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–D1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi A, Kataoka K, Kono T, Oseko F, Okuda H, Nagata I, Imura H. Parathyroid hormone-induced lipolysis in human adipose tissue. J Lipid Res. 1987;28:490–494. [PubMed] [Google Scholar]
- Tchkonia T, Morbeck DE, Zglinicki T, Deursen J, Lustgarten J, Scrable H, et al. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JD, Lawn ND, Dunne JW. Baclofen neurotoxicity: a metabolic encephalopathy susceptible to exacerbation by benzodiazepine therapy. J Clin Neurophysiol. 2019;36:209–212. doi: 10.1097/WNP.0000000000000565. [DOI] [PubMed] [Google Scholar]
- Valenzano DR, Terzibasi E, Genade T, Cattaneo A, Domenici L, Cellerino A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr Biol. 2006;16:296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Hirsch IB. Interference of intravenous vitamin C with blood glucose testing. Diabetes Care. 2014;37:e93–e94. doi: 10.2337/dc13-2452. [DOI] [PubMed] [Google Scholar]
- Wagner A, Cohen N, Kelder T, Amit U, Liebman E, Steinberg DM, Radonjic M, Ruppin E. Drugs that reverse disease transcriptomic signatures are more effective in a mouse model of dyslipidemia. Mol Syst Biol. 2015;11:791. doi: 10.15252/msb.20145486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang S, Wang Y, Chen L, Zhang XS. Disease-aging network reveals significant roles of aging genes in connecting genetic diseases. PLoS Comput Biol. 2009;5:e1000521. doi: 10.1371/journal.pcbi.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZC, Monteiro CD, Jagodnik KM, Fernandez NF, Gundersen GW, Rouillard AD, et al. Extraction and analysis of signatures from the gene expression omnibus by the crowd. Nat Commun. 2016;7:12846. doi: 10.1038/ncomms12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt BE, Proudfoot AT, Vale JA. Hydrogen peroxide poisoning. Toxicol Rev. 2004;23:51–57. doi: 10.2165/00139709-200423010-00006. [DOI] [PubMed] [Google Scholar]
- Yang YY, Gangoiti JA, Sedensky MM, Morgan PG. The effect of different ubiquinones on lifespan in Caenorhabditis elegans. Mech Ageing Dev. 2009;130:370–376. doi: 10.1016/j.mad.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, Hill WG, Landi MT, Alonso A, Lettre G, Lin P, Ling H, Lowe W, Mathias RA, Melbye M, Pugh E, Cornelis MC, Weir BS, Goddard ME, Visscher PM. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Huang T, Petralia F, Long Q, Zhang B, Argmann C, et al. Synchronized age-related gene expression changes across multiple tissues in human and the link to complex diseases. Sci Rep. 2015;5:15145. doi: 10.1038/srep15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Huang T, Song WM, Petralia F, Mobbs CV, Zhang B, et al. Discover the network mechanisms underlying the connections between aging and age-related diseases. Sci Rep. 2016;6:32566. doi: 10.1038/srep32566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Linton JM, Schork NJ, Buck LB, Petrascheck M. A pharmacological network for lifespan extension in Caenorhabditis elegans. Aging Cell. 2014;13:206–215. doi: 10.1111/acel.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Nogales-Cadenas R, Lin J-R, Zhang W, Cai Y, Vijg J, et al. Systems-level analysis of human aging genes shed new light on mechanisms of aging. Hum Mol Genet. 2016;25:2934–2947. doi: 10.1093/hmg/ddw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 593 kb)
(XLSX 11.7 kb)
(XLSX 464 kb)
(XLSX 148 kb)
(XLSX 3.89 mb)
(XLSX 506 kb)
(XLSX 391 kb)
(XLSX 516 kb)