Abstract
Optimizing the perception of external cues and regulating physiology accordingly help plants to cope with the constantly changing environmental conditions to which they are exposed. An array of photoreceptors and intricate signaling pathways allow plants to convey the surrounding light information and synchronize an endogenous timekeeping system known as the circadian clock. This biological clock integrates multiple cues to modulate a myriad of downstream responses, timing them to occur at the best moment of the day and the year. Notably, the mechanism underlying entrainment of the light-mediated clock is not clear. This review addresses known interactions between the light signaling and circadian clock networks, focusing on the role of light in clock entrainment and the known molecular players in that process.
Short Summary
Light perception and signaling modulate plant growth and physiology. The circadian clock, an endogenous timekeeping mechanism, allows plants to predict and anticipate daily changes in the external environment. This review focuses on the interaction between the circadian clock and light signaling networks, and the molecular mechanisms underlying the clock entrainment.
Introduction
Plants adjust their growth, physiology and developmental transitions to the ever-changing environmental conditions to which they are exposed. The circadian clock, an internal timekeeping mechanism, helps to integrate endogenous and external cues to ensure that coordinate growth and developmental processes occur at the right time of the day and year. On a rotating planet, light-dark and temperature cycles are highly dynamic, yet predictable, and play crucial roles in setting the endogenous clock on time through a process called entrainment. It has been shown that the resonance between the internal oscillator and the external conditions increases the fitness of Arabidopsis thaliana (Dodd et al., 2005; Michael et al., 2003).
Important features of circadian systems include their ability to be dynamically adjusted and to exhibit robustness towards non-relevant perturbations. Although these two characteristics might seem contradictory, they set the basis for the effective performance of an essential molecular network that influences biological rhythms. For example, dynamic adjustment of gene expression to phase protein activity at different times depending on day length is essential for flowering to occur in the correct season (Song et al., 2015). However, being insensitive to small and random changes in light or temperature protects the system from wrongly setting the time (Gil and Park, 2019). To accomplish this set of complex and comprehensive responses, multiple levels of signal integration are exhibited and numerous pathways converge on particular molecules, such as transcription factors. This multilayer control mechanism also allows plants to achieve perceptual disambiguation, which occurs when a single signal does not provide enough information to univocally specify the external condition (e.g. the same day length can occur in different seasons) and another signal is necessary to resolve the ambiguity (e.g. the memory of winter temperatures complements day length information to define the season) (Casal and Qüesta, 2018).
To ensure synchronicity with environmental conditions, plants must constantly reset; sensing and integrating external cues is fundamental to this process. This review focuses on our current understanding of how light information is integrated into the circadian system of Arabidopsis thaliana, and the phenotypic consequences of disruptions in the interaction between light signaling and the clock network. We also briefly discuss the crosstalk with other signaling pathways, how the molecular clockwork feeds back into light perception, and the lack of a comprehensive model to address the complex molecular output of the circadian system.
The Arabidopsis Core Clock
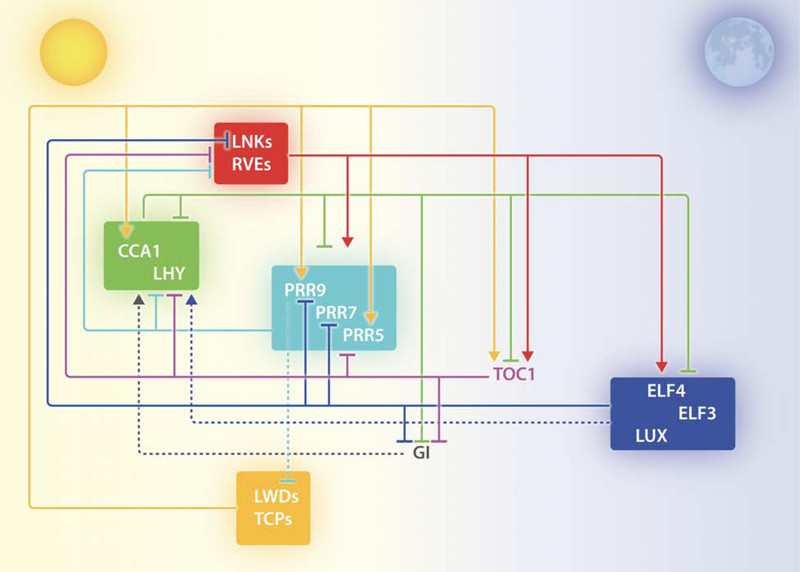
The core of the biological clock is composed of a set of transcriptional-translational feedback loops (TTFL) (Huang and Nusinow, 2016; McClung, 2019) which are modulated by other cellular mechanisms including post-transcriptional regulation, and post-translational and chromatin modifications (Mateos et al., 2018; McClung, 2019; Nohales and Kay, 2016; Yang et al., 2018). Briefly, these TTFL start at dawn with the induction of expression of CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL), which encode for MYB-like transcription factors (Schaffer et al., 1998; Wang and Tobin, 1998). Then, the molecular components of the biological oscillator are expressed in temporal waves. The PRR (PSEUDO-RESPONSE REGULATOR) family of genes is expressed sequentially: the PRR9 transcript peaks after CCA1/LHY, followed by PRR7, PRR5, PRR3, and PRR1 (also known as TOC1 -TIMING OF CAB EXPRESSION 1-) (Makino et al., 2001; Mizuno and Nakamichi, 2005). Finally, a rise in mRNA levels is observed for the evening/night players of the clock: GI (GIGANTEA), ELF3 (EARLY FLOWERING 3), ELF4 and LUX (LUX ARRHYTHMO). The proteins encoded by ELF3, ELF4 and LUX interact to give rise to a transcriptional regulatory complex known as the Evening Complex (EC) (Nusinow et al., 2011). Each of these molecules represses the expression of many other core-clock components (Figure 1). CCA1 and LHY form homo- and heterodimers which bind a promoter motif called the evening element to repress the expression of evening-phased genes, including GI and the members of the EC (Harmer et al., 2000; Kamioka et al., 2016; Lu et al., 2009; Lu et al., 2012; Nagel et al., 2015; Yakir et al., 2009). CCA1/LHY also negatively regulate their own expression, as well as that of the PRR family members (Adams et al., 2015; Alabadí et al., 2001; Schaffer et al., 1998; Wang and Tobin, 1998). In turn, CCA1/LHY transcription can be repressed by PRR9/7/5/1 as well as the EC, closing the central negative feedback loop of the clock (Adams et al., 2015; Helfer et al., 2011; Huang et al., 2012).
Figure 1: Simplified representation of the molecular network underlying the circadian clock of Arabidopsis thaliana.
Clock components are shown from left to right, according their time of expression throughout the day. Functional groups are enclosed in boxes. Lines with blunt ends and arrows indicate repression and activation, respectively. Broken lines represent regulatory steps not proven to be direct.
The positive arms of the clock are represented by three groups of proteins: LWD1 (LIGHT-REGULATED WD 1) and LWD2 in the morning; and LNK1 to 4 (NIGHT LIGHT–INDUCIBLE AND CLOCK-REGULATED 1) together with RVE4 (REVEILLE 4), RVE6 and RVE8 acting at midday (Hsu et al., 2013; Rugnone et al., 2013; Wang et al., 2011). LWD1 is recruited to the DNA by TCP20 (TEOSINTE BRANCHED1-CYCLOIDEA-PCF 20) and TCP22, and activates the transcription of CCA1 (Wu et al., 2016). LWD1/LWD2 also promote the expression of PRR9, PRR5 and TOC1 (Wang et al., 2011). RVE8, a MYB-like transcription factor, associates with LNK1 and LNK2 to bind TOC1 and PRR5 promoters (Pérez-García et al., 2015; Xie et al., 2014). RVE4, RVE6, and RVE8 have been shown to induce the expression of the EC components and PRRs (Farinas and Mas, 2011; Hsu et al., 2013; Rawat et al., 2011; Xie et al., 2014). The latter repress RVE8 expression, and promoters of the LNK genes have been shown to be bound by LUX, closing another loop of the TTFL(Mizuno et al., 2014b; Zhang et al., 2019a). The role of GI within the core-clock seems to be different than other central components: it interacts with the F-box protein ZTL (ZEITLUPE) in a blue-light enhanced manner and helps sustain and modulate ZTL rhythmic accumulation (Kim et al., 2013; Kim et al., 2007). In the dark, ZTL-GI dissociate and ZTL promotes TOC1 and PRR5 proteasomal degradation (Fujiwara et al., 2008; Kim et al., 2007). GI also induces the transcription of CCA1 and LHY (Martin-Tryon et al., 2007). Recently, it was shown that PIFs (PHYTOCHROME-INTERACTING FACTORS) repress CCA1 expression and that this activity is modulated at different levels by GI, suggesting that GI regulation of the CCA1 locus could occur, at least in part, through PIF proteins (Nohales et al., 2019). Finally, the expression of GI is negatively regulated by the EC, TOC1 and CCA1/LHY, thereby closing another loop (Huang et al., 2012; Mizuno et al., 2014a).
The circadian clock is known to regulate a myriad of physiological processes and developmental transitions, and has been proposed to work as an integrator of endogenous and external signals (Sanchez and Kay, 2016). The known molecular pathways connecting the internal timekeeping mechanism and its outputs, as well as their respective feedback to the central oscillator, have been recently reviewed (Creux and Harmer, 2019; Greenham and McClung, 2015; Sanchez and Kay, 2016; Singh and Mas, 2018).
Light Sensing and the Circadian Clock
Light is crucial for plant survival and success, not only because it is a very useful tool to determine the environment in which they are growing, but also because it is their source of energy. Consequently, different and versatile signaling pathways have evolved to correctly sense and translate light information. The first step in this complex molecular cascade of events is light perception itself, achieved through a set of molecules known as photoreceptors which convert detected photons into a biochemical signal capable of being transduced through multiple molecular events (Schäfer and Nagy, 2006).
In Arabidopsis, there are at least 5 families of photoreceptors: 1) phytochromes, 2) cryptochromes, 3) the ZTL family 4) phototropins, and 5) UVR8 (UV Resistance Locus 8). All of these, except UVR8, bind a ligand termed chromophore (light-absorbing molecule). Each of these families present a distinct wavelength absorption spectrum, as well as different biochemical properties and molecular partners. Together, these photoreceptors allow plants to determine incoming irradiance, spectral composition, and light direction and duration (photoperiod) - in other words, to have a thorough and timely outline of the light environment (Briggs, 2006; Rizzini et al., 2011; Schäfer and Nagy, 2006).
All families of photoreceptors (except phototropins) have been described to participate in light entrainment of the circadian clock, as well as contributing to setting its pace (Table 1) (Litthauer et al., 2015). However, it is worth noting that phytochromes, cryptochromes and UVR8 are not considered intrinsic components of the clockwork because none (individually or as a family) are necessary for clock entrainment nor for maintenance of robust circadian oscillations under free-running conditions (Baudry et al., 2010; Devlin and Kay, 2000; Fankhauser and Staiger, 2002; Fehér et al., 2011; Hu et al., 2013; Strasser et al., 2010; Yanovsky et al., 2000b).
Table 1.
Circadian period length phenotype of light signaling mutants.
| Genotype | Phenotype | Continuous Condition | Reference |
|---|---|---|---|
| phyA | Long period | Red and blue light (fluence rate dependent) | (Somers et al., 1998) |
| PhyA OXa | Short period | Red light; darkness | (Anderson et al., 1997; Kolmos et al., 2011) |
| phyB | Long period | Red light (fluence rate dependent) | (Hajdu et al., 2015; Somers et al., 1998) |
| PhyB OXa | Short period | Dark; white light; red light (fluence rate dependent) | (Hajdu et al., 2015; Hall et al., 2002; Kolmos et al., 2011; Somers et al., 1998) |
| phyC | Long period | Red light | (Jones et al., 2015) |
| phyABCDE | Short period | Red light (low-fluence); white light | (Hu et al., 2013; Strasser et al., 2010) |
| Long period | Red light (high-fluence) | (Hu et al., 2013) | |
| cry1 | Long period | Blue and white light; red light (fluence rate dependent) | (Devlin and Kay, 2000; Somers et al., 1998) |
| CRY1 OXa | Short period | Blue and white light | (Somers et al., 1998) |
| cry2 | Short period | Blue light (low-fluence rate); red light (fluence rate dependent) | (Devlin and Kay, 2000; Somers et al., 1998) |
| Long period | Blue light (high-fluence ate) | (Somers et al., 1998) | |
| cry1;cry2 | Long period | Blue light; red light (low-fluence rate) | (Devlin and Kay, 2000) |
| ztl | Long period | Dark; blue and red light | (Somers et al., 2004) |
| ZTL OXa | Arrhythmic | Red light | (Somers et al., 2004) |
| uvr8 | Long period | White light supplemented with UV-B light | (Fehér et al., 2011) |
| pif4;pif5 | Long period | White light | (Nohales et al., 2019) |
| pif1;pif3;pif4 | Long period | White light (with 2% sucrose)b | (Shor et al., 2017) |
| pif1;pif4;pif5 | Long period | White light (with 2% sucrose)b | (Shor et al., 2017) |
| Pif3;pif4;pif5 | Long period | White light (with 2% sucrose)b | (Shor et al., 2017) |
| pifQ | Long period | Red and white light (high-fluence rate) | (Shor et al., 2017) |
| PIF1 OXa | Short period | White light (with 2% sucrose)b | (Shor et al., 2017) |
| PIF3 OXa | Short period | White light (with 2% sucrose)b | (Shor et al., 2017) |
| PIF4 OXa | Short period | White light | (Nohales et al., 2019) |
| PIF5 OXa | Short period | White light | (Nohales et al., 2019; Shor Shor et al., 2017) |
| cop1 | Short Period | Dark; white light | (Millar et al., 1995; Yu et al., 2008) |
| det1 | Short period | Blue, red and white light | (Lau et al., 2011; Millar et al., 1995) |
| hy5 | Short period | Dark; blue, red and white light | (Andronis et al., 2008; Hajdu et al., 2018) |
| Hyh | Short period | Blue light | (Hajdu et al., 2018) |
| hy5;hyh | Short period | Dark; blue, red and white light | (Hajdu et al., 2018) |
| fhy3 | Long period/arrhythmic | Red Light | (Allen et al., 2006) |
| Short period | Blue light | (Allen et al., 2006) | |
| Short period/arrhythmic | White light | (Li et al., 2011a) | |
| far1 | Short/ long period (Assay dependent) | White light | (Li et al., 2011a) |
| hy1 | Long period | Red light | (Millar et al., 1995) |
| cor27 | Long period | White light | (Li et al., 2016) |
| cor28 | Long period | White light | (Li et al., 2016) |
| cor27;cor28 | Long period | White light | (Li et al., 2016) |
OX; Overexpressing line.
Sucrose was specified as an important variable for the phenotype.
In etiolated, temperature-entrained seedlings (i.e. those not exposed to light), the clock controls a smaller set of genes than it does in plants entrained with light-dark cycles and transferred to continuous light (Wenden et al., 2011). Because this observation was made in plants grown in the presence of exogenous sugar, it suggests that the reduction in the number of clock-controlled genes is not associated with starvation. Furthermore, it has been shown that the amplitude of mRNA rhythms of many genes, including core-clock genes, are significantly dampened under constant darkness (Bognár et al., 1999; Covington et al., 2001; Doyle et al., 2002; Millar et al., 1995). A possible explanation is that the drop in photosynthates in dark-grown plants affects transcriptional activity. However, it has been shown that the amplitude of CCR2 and CCA1 oscillations can be restored not only by the addition of exogenous sugar, but also by the expression of a constitutively active form of PhyB (Phytochrome B), a member of the phytochrome family of photoreceptors (see below) (Jones et al., 2015). This evidence suggests that light, instead of merely acting as an entrainment cue, might play an additional role through different photoreceptors to target the core-clock components and be a fundamental variable for sustaining the robustness of biological rhythms.
Additionally, it has been shown that light can acutely induce the expression of multiple core-clock genes (Nohales and Kay, 2016). For example, CCA1 expression is induced upon red light exposure as short as 90 seconds (Wang and Tobin, 1998). Etiolated seedlings irradiated with different wavelength lights for longer times (≤2 hours) also exhibit an acute response, inducing the transcription of CCA1, as well as LHY, PRR9, LNK1, LNK3, ELF4, and TOC1 (Makino et al., 2001; Shikata et al., 2014; Tepperman et al., 2001). This rapid effect of light on the abundance of key clockwork components could impact its resetting and affect the regulation of expression of its downstream targets, which would ultimately induce a rearrangement of the transcriptome.
Phytochromes
Phytochromes have a wide spectrum of light absorption response including red (R) and far red (FR), and to a lesser degree blue (B) wavelengths. The apoprotein of phytochromes is synthetized in the cytoplasm and attached to a linear tetrapyrrole chromophore (phytochromobilin) to give rise to the inactive, red light-absorbing form of phytochromes: Pr. After R illumination, the Pr form converts to the active, far red light-absorbing form (Pfr) and translocates to the nucleus (Casal, 2013). These two photoconvertible states of phytochromes, and the photo-equilibrium between them, are essential for regulation of numerous plant responses, including light input to the clock. In Arabidopsis, this family of photoreceptors has five members - PhyA through PhyE - with PhyA and PhyB playing dominant roles (Franklin and Quail, 2010; Wang and Wang, 2015). The apoprotein of each phytochrome is encoded by a different gene, and each exhibits particular biochemical characteristics, as well as distinct and overlapping biological functions. For example, PhyB is the major phytochrome species in light-grown plants, whereas PhyA is highly abundant in dark-grown seedlings and is rapidly degraded upon exposure to light (Sharrock and Clack, 2002). PhyA is the only type I phytochrome, (known as “light labile”) whereas the other four members of this family are type II (“light stable”) (Franklin and Quail, 2010; Li et al., 2011b; Wang and Wang, 2015). PhyA is considered to be the main FR sensor and has also been shown to participate in the blue light signaling cascade. PhyB, together with PhyC-PhyE, are the dominant regulators of the responses to red light (Neff and Chory, 1998; Reed et al., 1994; Wang and Wang, 2015). PhyA and PhyB were the first and remain the best characterized members of this family of photoreceptors (Schäfer and Nagy, 2006). The existence of mutant alleles of these genes allowed researchers to do an initial phenotypic characterization and determine that both, PhyA and PhyB, participate in light input to the clock.
The free running period of phyB mutant lines has been shown to be longer than that of wild type plants at high-fluence red light, as well as to have an altered phase; phyA mutants have a lengthened period under low-fluence red or blue light (Table 1) (Devlin and Kay, 2000; Salomé et al., 2002; Somers et al., 1998). PhyA and PhyB act additively in red light input to the clock, suggesting the presence of distinct signaling pathways (Devlin and Kay, 2000). Also, these two photoreceptors are important for proper re-entrainment of the clock. PhyA is necessary to re-entrain the oscillator in response to either a different “T cycle” (i.e. a 20-hour day, being 10h blue light/ 10 h darkness) or to a blue light pulse at the end of the night, which shifts the phase of the clock. Although discrepancies were observed in the fluence rate of blue light that affects resetting of the endogenous rhythms in phyA mutants, its role in light input to the clock in response to blue light was consistent (Somers et al., 1998; Yanovsky, 2001). Moreover, it was established that PhyA is necessary for resetting of the clock mediated by far red light, not only in Arabidopsis but also in potato (Solanum tuberosum) (Yanovsky et al., 2000a; Yanovsky, 2001). PhyB was shown to be involved in R light resetting in both Arabidopsis and tomato. This was determined by assessing the ability of modified plants (phyB mutant Arabidopsis and an underexpressor line of PhyB tomato) to adjust the phase of CCA1:LUC (CCA1 promoter fused to the LUCIFERASE gene reporter) and leaf angle respectively, after red light illumination at the end of the night (Hajdu et al., 2015; Yanovsky et al., 2000a). It has been suggested that endogenous levels of PhyB are required and sufficient for the proper resetting of the clock mediated by red wavelengths, as PhyB overexpressing lines do not exhibit an exacerbated response to a red light pulse compared to wild type plants (Hajdu et al., 2015). Nevertheless, the pace of the period correlates with phytochromes dose: phyA and phyB mutants have increased period length and PhyA and PhyB overexpressors have reduced period length (Table 1) (Devlin and Kay, 2000; Hall et al., 2002; Kolmos et al., 2011; Somers et al., 1998).
Gene expression of morning and evening core-clock components has been shown to be decreased and increased respectively, by FR light in a PhyA-dependent manner, suggesting this photoreceptor also has an important role in FR light input to the oscillator (Wenden et al., 2011). Moreover, the analysis of CCA1:LUC expression profile under continuous FR light on gi, toc1 and elf4 mutants suggests these genes contribute to the FR signaling pathway of the clock (Wenden et al., 2011).
Phytochromes C, D and E are less abundant than PhyA and PhyB. PhyD and PhyE display a partially redundant function with PhyB (Franklin and Quail, 2010). The evidence suggests that PhyC and PhyE are not able to homodimerize (a necessary condition for signaling activity in Arabidopsis), but they heterodimerize either with PhyB or PhyD, and are thus hypothesized to modulate the activity of the latter (Clack et al., 2009; Jones et al., 2015). Nonetheless, despite not having a prevailing role in the biological oscillator, PhyC, PhyD and PhyE have been shown to participate in red light input to the clock (Devlin and Kay, 2000; Jones et al., 2015).
In spite of evidence for the function of phytochromes on the entrainment and fine-tuning of the circadian clock, two research groups have shown that plants lacking this whole family of photoreceptors are still able to achieve rhythmicity after light-dark entrainment and to maintain oscillations under free running conditions, indicating that phytochromes are not an intrinsic component of the clockwork itself (Hu et al., 2013; Strasser et al., 2010; Zhong et al., 1998).
Phytochrome Signaling
Phytochrome signaling is extremely complex and divergent, as it controls numerous cellular processes including gene expression, alternative promoter selection, post-transcriptional regulation (alternative splicing), cytosolic mRNA translation and protein localization (Paik et al., 2012; Palágyi et al., 2010; Shikata et al., 2014; Ushijima et al., 2017; Wang and Wang, 2015). Specifically, PhyB has been shown to regulate the transcriptional activity and phase of several core-clock genes (Palágyi et al., 2010). The signaling pathway underlying that regulation and the mechanism responsible for the ability to modulate the period length remain unclear. However, many molecular interactions between central components of light signaling cascades and the circadian clock have been identified, paving the way for unraveling the mechanisms that connect light input with the clockwork (Table 2, Figure 2). One of the earliest interactions to be described was the association between PhyB and ELF3; based on genetic data, ELF3 was proposed to be part of PhyB signaling for at least some physiological responses such as early morphogenesis (Liu et al., 2001). By then, the relevance of ELF3 in light signaling and in the clock network was clearly established as the photoperiod-insensitive flowering of the elf3–1 mutant had suggested its involvement on the circadian clock (Zagotta et al., 1996). Later, it was shown that seedlings carrying elf3 mutant alleles are arrhythmic under constant light, but not under constant dark conditions (Hicks et al., 1996; Reed et al., 2000). Moreover, elf3 mutants kept under constant light but with temperature cycles (i.e. temperature entrainment), partially maintain their rhythmicity, supporting the relevance of ELF3 in light input to the clock (McWatters et al., 2000). Finally, the expression of the CAB2 (CHLOROPHYLL A/B-BINDING PROTEIN 2) gene is known to be acutely induced by a pulse of light, but the magnitude of the induction depends on the time of the day at which the stimulus occurs. This process, known as gating, is controlled by the endogenous oscillator and modulated by ELF3; the acute response of CAB2 expression upon light exposure becomes “time-insensitive” and is enhanced in elf3 mutants (McWatters et al., 2000). Consistently, ELF3 was found to play a role in the resetting of the circadian clock in response to light treatment at different times of the day, and was suggested to antagonize light input during the night (Covington et al., 2001). This evidence, together with the more recent confirmation that ELF3 is a component of the core-clock itself, positioned this gene as a strong candidate for connecting light and circadian networks (Thines and Harmon, 2010).
Table 2.
Protein-protein interactions between core-clock and light signaling network components.
| Core-Clock Protein | Light Signaling Protein | Experimental Technique | Reference |
|---|---|---|---|
| CCA1 | PhyB | Y2H, Co-IP | (Yeom et al., 2014) |
| FHY3 | Y2H, Co-IP, LCI | (Li et al., 2011a) | |
| DET1 | Y2H, in vitro assay, BiFC, LCI | (Lau et al., 2011) | |
| HY5 | Y2H, in vitro assay | (Andronis et al., 2008) | |
| CSU4 | Y2H, BiFC, Co-IP | (Zhao et al., 2018) | |
| LHY | PhyB | Co-IP | (Yeom et al., 2014) |
| FHY3 | Y2H | (Li et al., 2011a) | |
| DET1 | in vitro assay, LCI | (Lau et al., 2011) | |
| PRR9 PRR7 |
PIF3 | Y2H, in vitro assay, BiFC | (Martín et al., 2018; Zhang et al., 2020) |
| PIF4 | Y2H, in vitro assay | (Martín et al., 2018; Zhang et al., 2020) | |
| PIF7 | in vitro assay | (Zhang et al., 2020) | |
| PRR5 | PIF3 | Y2H, BiFC | (Martín et al., 2018) |
| PIF4 | Y2H, BiFC, Co-IP | (Martín et al., 2018; Zhang et al., 2020; Zhu et al., 2016) | |
| PIF7 | in vitro assay, BiFC, Co-IP | (Zhang et al., 2020) | |
| ZTL | Y2H, in vitro assay, Co-IP, | (Fujiwara et al., 2008; Kiba et al., 2007; Yasuhara et al., 2004) | |
| FKF1 | Y2H, in vitro assay, Co-IP | (Baudry et al., 2010) | |
| LKP2 | Y2H, in vitro assay, Co-IP | (Baudry et al., 2010; Yasuhara et al., 2004) | |
| GI | PhyB | Y2H, Co-IP | (Yeom et al., 2014) |
| PIF1 PIF4 |
Y2H, in vitro assay | (Nohales et al., 2019) | |
| PIF3 PIF5 |
Y2H, in vitro assay, Co-IP | (Nohales et al., 2019) | |
| ZTL | Y2H, in vitro assay, Co-IP | (Kim et al., 2007) | |
| FKF1 | Y2H, in vitro assay, Co-IP | (Sawa et al., 2007) | |
| LKP2 | Y2H, in vitro assay | (Kim et al., 2007) | |
| TOC1 | PhyB | Y2H, Co-IP | (Yeom et al., 2014) |
| PIF1 (PIL5) | Y2H | (Yamashino et al., 2003) | |
| PIF2 (PIL1) | Y2H | (Makino, 2002) | |
| PIF3 | Y2H, BiFC, Co-IP | (Makino, 2002; Soy et al., 2016) | |
| PIF4 | Y2H, Co-IP | (Yamashino et al., 2003; Zhu et al., 2016) | |
| PIF5 (PIL6) | Y2H | (Fujimori et al., 2004; Yamashino et al., 2003) | |
| PIF6 (PIL2) | Y2H | (Yamashino et al., 2003) | |
| PIF7 | 2HP, BiFC | (Kidokoro et al., 2009) | |
| ZTL | Y2H, in vitro assay, Co-IP | (Fujiwara et al., 2008; Mas et al., 2003) | |
| FKF1 | Y2H, in vitro assay, Co-IP | (Baudry et al., 2010; Mas et al., 2003) | |
| LKP2 | Y2H, in vitro assay | (Baudry et al., 2010; Mas et al., 2003; Yasuhara et al., 2004) | |
| LUX | PhyB | Co-IP | (Yeom et al., 2014) |
| PCH1 | AP-MS | (Huang et al., 2015; Huang et al., 2016b) | |
| ELF4 | PCH1 | AP-MS | (Huang et al., 2015; Huang et al., 2016b) |
| ELF3 | PhyB | Y2H, in vitro assay, Co-IP, AP-MS | (Huang et al., 2015; Kim and Somers, 2019; Liu et al., 2001; Yeom et al., 2014) |
| PhyA PhyC PhyD PhyE |
AP-MS | (Huang et al., 2015) | |
| PIF4 | Y2H, BiFC, | (Nieto et al., 2015) | |
| PIF7 | AP-MS | (Huang et al., 2015) | |
| COP1 | Y2H, Co-IP, BiFC, AP-MS | (Huang et al., 2015; Yu et al., 2008) | |
| SPAs (SPA1 to SPA3) | AP-MS | (Huang et al., 2015) | |
| PCH1 | AP-MS | (Huang et al., 2015; Huang et al., 2016b) | |
| ZTL | PhyB | Y2H, in vitro assay | (Jarillo et al., 2001) |
| CRY1 | Y2H, in vitro assay | (Jarillo et al., 2001) | |
| FKF1 | Y2H, Co-IP, FRET | (Takase et al., 2011) | |
| LKP2 | Y2H | (Yasuhara et al., 2004) |
Due to its dual function, ZTL was included as a clock protein as well as a member of the light signaling pathway. AP-MS, Affinity Purification and Mass Spectrometry; BiFC, Bimolecular Fluorescence Complementation; Co-IP, Co-Immunoprecipitation; FRET, Förster Resonance Energy Transfer analysis; LCI, firefly Luciferase Complementation Imaging; Y2H, Yeast Two-Hybrid; 2HP, Two-hybrid system in Protoplasts.
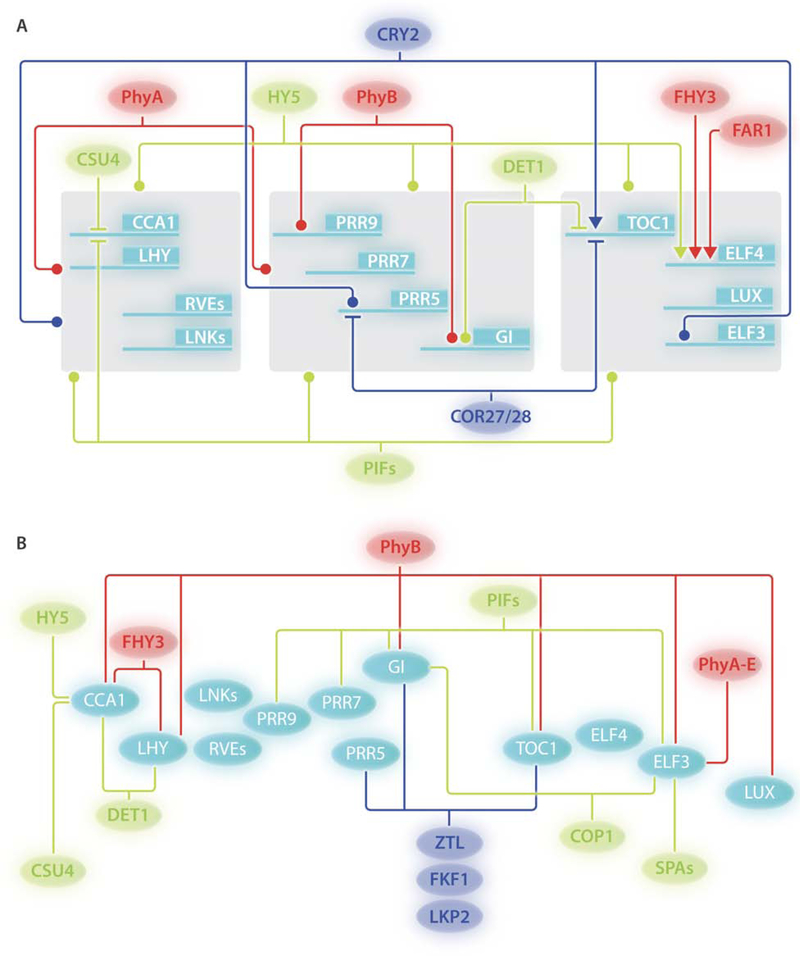
Figure 2: Regulation of core-clock elements by light signaling proteins.
(A) Transcriptional regulation of core-clock genes by photoreceptors or components of light signaling pathways. The presumed regulation is based on various sources of experimental evidence but does not include genetic data. Dot-ended lines represent protein-chromatin associations for which the biological role (i.e. transcriptional regulation) has not been described. Lines with blunt ends and arrows indicate repression and activation, respectively. Cyan lines and boxes indicate clockwork genes. Gray boxes group genes targeted by the same component.
(B) Protein-protein interactions between core-clock players and components involved in light perception or signaling. Cyan ovals indicate clockwork elements. Regulatory elements belonging to different light wavelength signaling cascades are represented with distinct colors: Red elements, red light; blue elements, blue light; green elements, regulatory players shared by different light wavelength perception pathways. Ovals represent proteins.
Further studies confirmed the PhyB-ELF3 interaction. In experiments described in a “Letter to the Editor”, Yeom et al. used yeast two-hybrid to show that PhyB can interact not only with ELF3, but also with GI, TOC1 and CCA1. Moreover, using transiently transfected protoplasts and co-immunoprecipitation assays, the authors confirmed those results and showed that LUX and LHY also associate with PhyB (Yeom et al., 2014). Interestingly, some of those interactions are light quality-dependent, suggesting that PhyB binding might depend on its Pr/Pfr state, and implying a significant biological relevance: it could represent a strategy by which phytochromes convey to the clock both light/dark information and also more subtle and complex information on light status (i.e. shaded conditions or dusk, characterized by a lower red/far red light ratio). Finally, Huang et al. conducted an unbiased experiment to determine partners of ELF3 and ELF4 in vivo. The authors employed transgenic lines in which the level of accumulation of the protein of interest was similar to that of the endogenous and purified protein extracts from plants under normal growth conditions. By using affinity purification followed by mass spectrometry, the researchers discovered that ELF3 associates not only with PhyB but also with the other four phytochromes and with pivotal light signaling components such as COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1), three members of the SPA (SUPRESSOR OF PHYA-105) family of proteins, and PIF7 (PHYTOCHROME INTERACTING FACTOR 7) (see below). Furthermore, it was shown that ELF4 interacts with most of these proteins in an ELF3-dependent manner, reinforcing the notion that ELF3 acts as a hub to link circadian and light signaling networks (Huang et al., 2016a). Although the biological relevance of PhyB-ELF3 interaction is not fully understood, it has been shown that PhyB stabilizes ELF3 (Nieto et al., 2015). Additionally, elf3–14, a hypomorphic allele of ELF3 carrying a single amino acid substitution (A37T) within the known ELF3-PhyB interaction domain, exhibits reduced association with the photoreceptor (Kim and Somers, 2019). This data, together with the developmental phenotypes displayed by the mutant alleles of these genes, suggest that PhyB-ELF3 interaction could be at least somewhat important for PhyB positive signaling through ELF3. The dynamic of this partnership (which has not yet been addressed) is essential for understanding the molecular mechanism of light input to the clock, and should be further studied.
PCH1 (PHOTOPERIODIC CONTROL OF HYPOCOTYL1) directly interacts with the five members of the phytochrome family and is required for the formation of photobodies, the characteristic subnuclear foci formed by PhyB after photoactivation (Huang et al., 2016a; Huang et al., 2019; Huang et al., 2016b). Photobodies were shown to be critical for transducing light signals (Huang et al., 2019). Overexpressing lines of a constitutive active allele of PhyB maintain circadian oscillations after transfer to darkness in a wild type background, but fail to do so in the pch1 mutant genotype94. This demonstrates that PCH1 and, likely the formation of photobodies, are necessary for PhyB-mediated light input to the circadian oscillator (Huang et al., 2019). This finding confirms the essential role of PhyB in light input to the clock. Additionally, it suggests that PhyB is not sufficient to convey light information to the clockwork and that its signaling pathway must also be functional, at least under the conditions tested (i.e. after transfer to darkness).
It is tempting to speculate that the role of phytochromes on light signal integration to the core clock occurs through multiple mechanisms, including by targeting clockwork components directly, as well as regulating molecular hubs (e.g. COP1 and PIFs -see below-) which in turn modulate transcriptional activity and protein function of essential clock players. Additionally, PhyB associates with chromatin and the bound regions exhibit a significant enrichment of G-box motifs. Particularly, this photoreceptor has been shown to bind GI and PRR9 promoters and has been proposed to be a transcriptional repressor (Jung et al., 2016). Although the physiological relevance of these associations has yet to be determined, they suggest that PhyB could directly regulate transcriptional activity of core-clock genes.
PhyA, despite lacking a known DNA binding domain, also has the ability to associate with chromatin and has been found to bind many core-clock gene promoters (Figure 2). The current model proposes that PhyA interacts with its target regions through canonical transcription factors, providing a global mode of action to regulate gene transcription (Chen et al., 2014).
FHY3 (FAR-RED ELONGATED HYPOCOTYL 3) is a transcription factor that has been identified as a component of the PhyA signaling pathway, specifically in the PhyA-mediated high-irradiance response. fhy3 mutant is hyperresponsive to red light during seedling establishment, suggesting it might also have a role downstream of other phytochromes (Whitelam et al., 1993; Yanovsky et al., 2000c). Supporting this hypothesis, FHY3 was shown to regulate different features of the biological timekeeper, almost exclusively in a red-dependent manner. fhy3 mutants exhibit an altered phase and amplitude of gene expression compared to wild type plants, as well as defects in resetting upon red light treatments, suggesting this gene could be involved in red light input to the clock (Allen et al., 2006).
FAR1 (FAR-RED IMPAIRED RESPONSE 1), a paralogue of FHY3, is also part of the PhyA signal transduction and participates in the regulation of seedling development (Lin et al., 2007). FAR1 and FHY3 are essential for the amplitude and rhythmic expression of ELF4, whereas HY5 (Long Hypocotyl 5) and its homolog HYH (HY5 HOMOLOG), two basic leucine zipper domain (bZIP) transcription factors also involved in PhyA signaling, only modulate the amplitude of ELF4 expression (Li et al., 2011a). Although a role for HY5 in light input to the clock has been long established, its molecular mechanism was not understood (Anderson et al., 1997). Recently, FAR1, FHY3 and HY5 have been shown to bind ELF4 promoter in vivo and induce its expression. Moreover, FHY3 is able to interact with CCA1, LHY and HY5. Thus, the proposed model suggests that FAR1, FHY3 and HY5 induce ELF4 expression by direct association with its promoter, and the binding of CCA1 and LHY to that complex on ELF4 locus represses their ability to activate transcription. The activation of ELF4 transcription, mediated by FAR1, FHY3 and HY5, contributes to the circadian profile of expression of this key component of the EC and represents a light input pathway to the clock, as these three transcription factors are regulators of the PhyA signaling (Gangappa and Botto, 2016; Li et al., 2011a). HY5 has been found to have an additional role as part of the blue light input to the clockwork (see details below) which could help to integrate information from multiple wavelengths.
PIFs Signaling
PIFs are a subfamily of basic helix-loop-helix (bHLH) transcription factors, originally described as pivotal components of red/far-red light signaling through characterization of their role in hypocotyl growth. For example, PIF3, the founding member of this protein family, was identified as a direct partner of PhyA and PhyB, and those interactions were shown to be necessary for signal transduction of both photoreceptors and the consequent regulation of seedling deetiolation (Castillon et al., 2007; Kim et al., 2003a; Monte et al., 2004; Ni, 1998). However, it is now clear that PIFs are molecular hubs at the crossroads of multiple cellular pathways such as light (not exclusively through phytochromes) and temperature signaling, biotic and abiotic responses, hormone signaling, sugar metabolism, and the circadian clock (Leivar and Monte, 2014; Paik et al., 2017). Integration of such an assorted internal and external information allows PIFs to shape plant growth and development.
Essentially, upon light exposure, activated phytochromes translocate from the cytoplasm to the nucleus where they promote PIFs turnover through phosphorylation, ubiquitylation and degradation. Hence, a reprogramming of the transcriptional landscape induces physiological and developmental changes (Leivar and Monte, 2014). Nevertheless, the current knowledge broadens that paradigm (Pham et al., 2018a). For example, PIF2 (also known as PIL1) has been shown to be stabilized by PhyB upon red light irradiation (Luo et al., 2014). PIF7, a major regulator of shade response, exhibits a different mode of action in which phosphorylated PIF7 associates with photoactivated PhyB in the nucleus, and upon exposure to shade conditions (low red/far red light ratio), PIF7 becomes rapidly dephosphorylated and binds target promoters (Li et al., 2012). Notably, PIF7 activity is likely regulated by phosphorylation/ dephosphorylation and subcellular localization, instead of ubiquitylation and proteasome-mediated degradation, as is the case for other PIFs (Huang et al., 2018; Li et al., 2012; Pham et al., 2018a). Finally, PIFs are also known to regulate PhyB abundance, which is dependent on their interaction and imposes a mutually-negative feedback-loop (Leivar et al., 2008; Leivar et al., 2012).
The convergence of core-clock components and PIFs to regulate clock outputs has been clearly established and multiple interactions between players of those two signaling pathways have been identified (Table 2). A role for PIF4 and PIF5 linking the circadian clock and red light signal transduction pathways was early proposed (Fujimori et al., 2004; Yamashino et al., 2003). The TOC1-PIF3 module has been established as a diurnal growth pattern regulator, with TOC1 directly interacting with PIFs and repressing their transcriptional activity during the early night (Soy et al., 2016). Furthermore, PRR9, PRR7 and PRR5 bind to PIF3 and PIF4, repress their transcriptional activity and limit the time of action of PIF4 to the end of the night/pre-dawn, when they trigger hypocotyl elongation (Martín et al., 2018). PRR9, PRR7 and PRR5 also bind PIF7 (Zhang et al., 2020). Plant growth regulation mediated by PIFs is also modulated by ELF3 (which represses PIF4 protein activity upon association) and by GI-PIFs interaction (which interferes with PIFs’ stability and DNA binding ability) (Nieto et al., 2015; Nohales et al., 2019). Moreover, the EC has been shown to bind to and regulate PIF4 and PIF5 expression, contributing to the circadian control of hypocotyl growth dynamics (Nozue et al., 2007; Nusinow et al., 2011). TOC1-PIF4 interaction has been associated with the circadian gating of thermoresponsive growth and TOC1-PIF7 are involved in the transcriptional regulation of low-temperature stress-responsive genes (Kidokoro et al., 2009; Zhu et al., 2016).
In view of the numerous interactions between core-clock components and PIF proteins, and considering that these transcription factors bind to DNA (particularly to G-Box elements), that there is an overrepresentation of G-Box motifs on the promoter of clock-controlled genes (including central players of the circadian oscillator) and that PIFs activity is repressed upon interaction with some key clock proteins, it is tempting to speculate that each one of those associations could feed-back into the central oscillator (and other outputs). Thus, besides modulating downstream growth responses, interactions between core-clock components and PIF proteins are one more gear of the oscillator: clock-components modulate PIFs transcriptional activity, which impacts the expression pattern of core-clock genes (Hsu et al., 2013; Martinez-Garcia et al., 2000; Nohales et al., 2019; Zhu et al., 2016). It has been shown that overexpression of PIF4 can destabilize ELF3, suggesting another possible mode of action for signaling into the clock (Nieto et al., 2015). Nevertheless, the role of PIFs in the functioning of the biological timekeeper has been elusive until recently, probably because of their partial functional redundancy and the robustness of the circadian network (Fujimori et al., 2004; Nohales et al., 2019; Nusinow et al., 2011; Viczián et al., 2005).
PIF overexpressing lines have shown distinct clock behavior in different studies, but accumulating evidence suggests that higher levels of PIFs induce a shortening of the free running period (Fujimori et al., 2004; Nohales et al., 2019; Shor et al., 2017; Viczián et al., 2005). In addition, the double mutant pif4; pif5 has no significant period length difference compared to the wild type plant when pTOC1:LUC reporter is assayed, but exhibits a long period if pCCA1:LUC is assessed (Nohales et al., 2019; Nusinow et al., 2011). These discrepancies can be easily explained by multiple factors, including a more direct/indirect effect on different promoters or variations within the experimental set up (e.g. light quality and irradiation); most likely they also represent the complex effect this transcription factor family exerts on the ticking of the molecular clock, particularly on the pace of the oscillator.
Recently, molecular evidence supporting the notion of PIFs signaling into the circadian network and functioning as important pieces of the light and metabolic input was found. PIFs were shown to bind (in vitro and in vivo) to the promoters of core-clock components (Hornitschek et al., 2012; Martinez-Garcia et al., 2000; Nohales et al., 2019; Oh et al., 2012; Pfeiffer et al., 2014; Shor et al., 2017; Zhang et al., 2013). Gene-expression analysis comparing pifQ (pif1;pif3;pif4;pif5) mutant and wild type plants grown under short day conditions, as well as trans-activation assays, showed that PIFs are able to repress CCA1 expression. In addition, assessment of pCCA1:LUC reporter activity in etiolated seedlings transferred to light revealed that wild type plants exhibit a higher light-induced transcriptional activity than PIF overexpressing lines but a lesser level of induction than the pif4;pif5 mutant background. This, together with the hypersensitive phase-shift response to light pulses observed on the pif4;pif5 mutant, establishes that PIFs have a role in the light input to the clock (Nohales et al., 2019). Surprisingly, another study also reported that PIFs are involved in setting the pace of the clock, but as players of the metabolic input (Shor et al., 2017). Shor et al. determined that the long period phenotype of the pifQ mutant is fluence rate dependent. This prompted the authors to question the role of PIFs in light input to the clock and to investigate their role in sugar signaling, the results of which showed the expression level of PIFs to be modulated by sugar. The presence of 3% sucrose enhanced PIFs ability to bind CCA1 and LHY promoters at subjective dawn, which correlated with a peak of expression of those two genes in the wild type background, and a delayed occurrence of such a peak in pifQ mutants. These findings led the authors to suggest that PIFs may be required for sucrose-mediated induction of CCA1 and LHY expression (Shor et al., 2017). This apparently conflicting evidence could be the result of PIFs acting as hubs of different signaling pathways, as has been determined for other physiological responses (Leivar and Monte, 2014; Paik et al., 2017).
Sucrose signaling to the central oscillator depends on PRR7, expression of which is repressed by photosynthetically derived sugars, likely in a PIFs-independent manner (Haydon et al., 2013; Shor et al., 2017). PRR7, a core-clock component and CCA1/LHY transcriptional repressor, associates with chromatin and has been shown to co-bind with PIFs to many dawn-phased genes (Liu et al., 2013; Liu et al., 2016b; Martín et al., 2018). Furthermore, other PRR family members interact with PIFs and gate their activity to gene expression (Martín et al., 2018). It could be hypothesized that low fluence rates of light -and therefore low levels of sucrose- imply high expression of PRR7, which would translate to strong repression of CCA1/LHY. Under such conditions the role of PIFs on the CCA1/LHY repression could be masked due to the reduced levels of those transcripts. In contrast, higher-fluence rates – and higher concentration of sucrose- imply repression of PRR7 and higher expression of CCA1/LHY, which sensitizes the system and exposes the proposed role of PIFs on repressing these two morning genes. Moreover, GI can hinder the binding of PIFs to chromatin (Nohales et al., 2019). It would be valuable to address whether PRR7 has the same ability, as it could also explain the increased binding of PIFs to CCA1/LHY promoters in the presence of sugar, a condition in which PRR7 would be less abundant. This hypothesis locates PRR7 and PIFs as hubs for signaling integration and transduction to the circadian clock, able to modulate the expression of two pivotal core-clock genes depending on two of the most relevant environmental and internal conditions: light and sugar metabolism. Reinforcing the idea of significant crosstalk between PIFs and PRR7 on the regulation of CCA1 expression is the fact that the sucrose-mediated morning induction of CCA1 has been shown to depend on both PIFs and PRR7 (Haydon et al., 2013; Shor et al., 2017). This hypothesis can be tested by assessing the clock behavior of pifQ and pifQ;prr7 mutants under different concentrations of sucrose, as well as the binding of PIFs to CCA1/LHY promoters under different concentrations of sugar in both wild type and prr7 mutant backgrounds. This set of experiments will contribute to better understand the role and interaction of these key players within the system.
Simultaneously, PIFs could be the convergence point for additional endogenous (e.g. hormone pathways) and/or exogenous (e.g. temperature) signals. Thus, PIFs could integrate and convey multiple cues to the central clock, functioning as a calibration system able to fine-tune the biological oscillator by modulating features such as amplitude, pace and resetting capability.
Cryptochromes and Cryptochrome Signaling
Cryptochromes are present in many different organisms, from bacteria to human, and in spite of their shared similarity with a group of light activated DNA repair enzymes known as photolyases, animal and plant cryptochromes have lost DNA-repair activity (Cashmore, 2003; Liu et al., 2016a). In Arabidopsis, the cryptochrome family encompasses three members, CRY1 (CRYPTOCHROME 1), CRY 2 and CRY3, and while the physiological relevance of CRY3 remains to be clarified, CRY1 and CRY2 have been extensively characterized and are the focus of this section (Chaves et al., 2011; Kleine et al., 2003).
CRY1 and CRY2 play a major role as blue light photoreceptors but they can sense a broader spectrum of wavelengths, including UV-A light (~390–480 nm) (Liu et al., 2016a; Yu et al., 2010). CRY2 is rapidly down-regulated by blue light and functions primarily under low-fluence irradiances, whereas CRY1 is more stable and works at higher intensities of light (Lin et al., 1998). Both bind non-covalently to the chromophore flavin adenine dinucleotide (FAD), which is presumed to undergo a redox photocycle and trigger photochemical reactions that induce structural rearrangements in the protein (Ahmad, 2016; Zeugner et al., 2005). Photoexcited cryptochromes are subject to protein modifications and protein-protein interactions that allow them to mediate their biological function (Liu et al., 2016a; Yang et al., 2017; Yu et al., 2010).
CRY1 and CRY2 act as homodimers and through interaction with different partners are able to modulate gene expression, either by transcriptional or post-translational regulation. CRY2 associates with chromatin, likely through canonical transcription factors that positively or negatively regulate transcriptional activity (Pedmale et al., 2016). Thus, these photoreceptors are involved in a variety of blue-light-mediated physiological responses including de-etiolation, photomorphogenesis, flowering, and entrainment of the circadian clock (Yang et al., 2017).
Genetic studies have revealed that both CRY1 and CRY2 play important roles in the proper functioning of the endogenous oscillator and contribute to determining its features. Under continuous blue light, cry1 mutants exhibit a lengthened free-running period compared to wild type plants, but the magnitude of that difference is fluence rate-dependent (Somers et al., 1998). cry2 mutants have a more elusive phenotype and show slightly different results depending on the experimental set-up (Devlin and Kay, 2000; Somers et al., 1998; Yanovsky, 2001). However, under continuous blue light, the double cry1;cry2 mutant exhibits a longer period compared with wild type or single mutant plants, independent of fluence rate, suggesting a partial redundancy in the function of these two proteins on blue light input to the clock (Devlin and Kay, 2000). Interestingly, both single and double cry1;cry2 mutants also present differences on the period length under red light, and cry1;cry2 mutants have also been found to participate in far red light signaling, suggesting a crosstalk between the signaling pathways sensing these different wavelengths (see section below) (Devlin and Kay, 2000; Yanovsky, 2001). Additionally, the resetting ability of the clock in response to different light wavelengths was addressed in multiple genomic backgrounds and the results also suggest that CRY1 and CRY2 are partially functionally redundant.
The mechanism linking the blue light perception to the core of the biological oscillator is still not understood. Recent evidence showing that CRY2 binds several core-clock gene promoters provides a reasonable mode of action, especially considering that such association could be modulated by PIF transcription factors, which were already implicated on the fine-tuning of the oscillator (Pedmale et al., 2016). Besides PIFs, other hub molecules acting at the crossroad of multiple light wavelengths have been found to contribute to convey blue light information (e.g. COP1 -see below-). Additional players regulating the core-clock in response to this particular wavelength have been identified, although whether they are part of the cryptochrome signal transduction or other blue light photoreceptors remain to be elucidated. For example, it was shown that blue light is able to stabilize COR27 (COLD REGULATED GENE 27) and COR28 proteins, which bind to the promoters of PRR5 and TOC1 and repress their transcription (Li et al., 2016). Similarly, blue light induces HY5 and HYH gene expression and protein accumulation. HY5, previously shown to be involved in PhyA signaling and known to regulate ELF4 expression, was found to associate with many core-clock genes in a light quality-dependent manner: binding to CCA1, PRR9, LUX, among other promoters was increased under blue light and in the case of PRR5, LUX and BOA (BROTHER OF LUX ARRHYTHMO), it correlated with changes in the expression level (Hajdu et al., 2018; Li et al., 2011a). Interestingly, the biological relevance of HY5/HYH on the functioning of the oscillator is revealed by the fact that the hy5;hyh double mutant has a shorter period than wild type plants under free running conditions either in dark, or white, blue or red light, but the phenotype is more severe under blue light (Hajdu et al., 2018).
ZTL Family and its Role on Light Input to the Clock
Blue light perception can also be achieved by the ZTL (ZEITLUPE) family of photoreceptors, comprised of three members: ZTL, FKF1 (FLAVIN-BINDING, KELCH REPEAT, F-BOX 1), and LKP2 (LOV KELCH PROTEIN 2). The photosensory domain of these proteins is the LOV (Light, Oxygen or Voltage) domain, which binds non-covalently to the chromophore (flavin mononucleotide), and undergoes structural changes upon blue light exposure that modulate the protein activity. Additionally, these photoreceptors exhibit two other functional domains: a Kelch repeat domain that mediates protein-protein interactions, and an F-box domain that allows these proteins to function as part of the E3 ubiquitin ligase Skp-Cullin-F-box (SCF) complex. This multimeric complex has the ability to trigger proteasomal-mediated protein degradation in a blue light-dependent manner (Ito et al., 2012).
ZTL, LKP2 and FKF1 have been associated with control of the photoperiodic pathway of flowering and the circadian clock by modulating the accumulation of essential proteins in each network (Ito et al., 2012). As shown by single and high-order mutant analysis, these three proteins have partially overlapping functions on the control of the circadian oscillator but ZTL appears to play a predominant role: ztl mutant plants exhibit either a long period or arrhythmic phenotype depending on the wavelength of irradiation, whereas lkp2 and fkf1 mutants do not present significant differences with wild type plants (Baudry et al., 2010; Jarillo et al., 2001). Nevertheless, overexpressing lines of either LKP2 or ZTL present an arrhythmic phenotype, supporting the idea that these loci play an overlapping but important role on the functioning of the circadian clock (Schultz et al., 2001; Somers et al., 2004).
ZTL interacts with GI in a blue-light enhanced manner, and both proteins undergo a reciprocal co-stabilization depending on light (Kim et al., 2013; Kim et al., 2007). GI facilitates folding and maturation of ZTL, likely through the formation of a ternary complex with the chaperone HSP90 (HEAT SHOCK PROTEIN 90), which promotes ZTL stabilization (Cha et al., 2017; Kim et al., 2011). Additionally, GI can recruit either of two deubiquitylases (UBP12 (UBIQUITIN-SPECIFIC PROTEASE 12) or UBP13) to the ZTL-GI complex, contributing to ZTL protein stabilization and accumulation by the end of the light period (Lee et al., 2019). After dusk, the ZTL-GI complexes dissociate and ZTL triggers PRR5 and TOC1 proteasomal-mediated degradation by ubiquitination through the SCF complex (Kiba et al., 2007; Mas et al., 2003). Thus, modulation of PRR5 and TOC1 protein accumulation by ZTL in a light-dependent manner directly impinges on the pace of the biological clock. Similarly, ZTL has been shown to ubiquitylate CHE (CCA1 HIKING EXPEDITION), a TOC1 interacting protein that regulates CCA1 expression, and control its stability in a light- and ubiquitin proteasome system-dependent way (Lee et al., 2018; Pruneda-Paz et al., 2009). Furthermore, ZTL photocycle kinetics play a fundamental role in determining the length of the circadian period. As shown by Pudasaini et al., plants carrying variants of ZTL with altered photocycle kinetics of the LOV domain present a different profile of PRR5 and TOC1 degradation, which correlates with the pace of the circadian clock (Pudasaini et al., 2017). Considering that LOV domains have been proposed to distinguish fluctuations in light intensity, ZTL could be a critical player in the light resetting mechanism of the plant clock, conveying information not only from the light to dark transition, but also contributing to the parametric entrainment, which implies that the clock accelerates as the light fluence increases (Aschoff, 1979; Pudasaini and Zoltowski, 2013).
Ultraviolet B Light Sensing and Signaling
Visible light plays a central role in the physiological and developmental responses of Arabidopsis, with phytochromes and cryptochromes being the major photoreceptors of those wavelengths. Ultraviolet B (UV-B) light (280–315 nm), although representing a minor portion of the sunlight spectrum, has a significant role in plant physiology. UV-B light can represent a stress factor, inducing DNA damage and impacting on development and growth, but at low intensities the same wavelengths can promote photomorphogenesis (Favory et al., 2009; Yin and Ulm, 2017).
UV-B light is sensed by UVR8 (UV Resistance Locus 8), which notably does not require the presence of a chromophore to convert the received photons into a biochemical signal (Rizzini et al., 2011). Instead, the photocycle of UVR8 is accomplished by a large number of aromatic residues within its structure, which induce the homodimerization of the protein and its retention in the cytoplasm. Upon UV-B light irradiation, the charges on the aromatic residues (tryptophan dominated) are re-distributed, promoting the dissociation into active monomers. As a monomer, UVR8 is translocated to the nucleus and triggers regulatory changes in gene expression (Podolec and Ulm, 2018; Yin and Ulm, 2017).
UVR8 physically associates with different partners. It interacts with COP1 in a UV-B dependent manner, promoting UVR8 nuclear accumulation (Favory et al., 2009; Rizzini et al., 2011). The UVR8-COP1 interaction compromises the E3 ubiquitin ligase activity of COP1, which results in the reduction of ubiquitylation and degradation of the transcription factor HY5, and could therefore represent a mechanism to modify gene expression (Huang et al., 2013; Liang et al., 2019; Podolec and Ulm, 2018; Yin and Ulm, 2017). Interestingly, UVR8 and COP1 are both necessary for UV-B light entrainment of the circadian clock, but in a HY5/HYH-dispensable manner, suggesting an alternative signaling pathway to incorporate UV-B light information to the endogenous oscillator (Fehér et al., 2011). Consistently, UV-B light pulses induce transcription of several core-clock genes in an UVR8- and COP1-dependent manner. The level of transcriptional induction depends on the time of day at which the pulse is applied, suggesting that this response is gated by the circadian clock, giving rise to a loop of feedback regulation between UV-B light perception and the circadian clock function (Fehér et al., 2011).
It has been shown that UV-B inhibits PIF4 expression, thereby reducing PIF4 abundance. Despite the known roles of ELF3 and HY5 as transcriptional regulators of PIF4, these two factors were not found to be involved in the UV-B-mediated reduction of PIF4 levels (Delker et al., 2014; Hayes et al., 2017; Nusinow et al., 2011). Moreover, UV-B light triggers degradation of PIF4 and PIF5 and stabilizes the bHLH factor HFR1 (LONG HYPOCOTYL IN FAR RED 1), which can inhibit the ability of PIF4 and PIF5 to bind DNA, thereby repressing their transcriptional activity (Hayes et al., 2017; Hayes et al., 2014; Hornitschek et al., 2009). Considering the recent evidence revealing that PIFs can repress CCA1 expression, it is tempting to speculate that UV-B light information could be conveyed to the clockwork, at least partially, by modulating the abundance of PIFs (Nohales et al., 2019).
Signal Integration and Crosstalk Between Pathways
In nature, day-night cycles provide distinct types of information to plants about the surrounding environment. For example, the length of daylight, or photoperiod, has a decisive role on flowering time of many species. This is a key physiological response for plant survival, as well as of a great interest from an agronomic perspective. The current knowledge of both the role of photoperiod on synchronizing the circadian clock and the role of the biological oscillator on sensing this external variable to integrate it and modulate plant physiology have recently been reviewed (Creux and Harmer, 2019; Oakenfull and Davis, 2017; Webb et al., 2019).
Another important feature of the natural environment is the spectral composition of the incoming light, as it changes along the day and the year, and also depends on the presence of neighbor plants (Schäfer and Nagy, 2006). Therefore, is not surprising that plants have a manifold of strategies to integrate signals from different photoreceptors, each of which senses a particular wavelength. The crosstalk of light signaling cascades occurs at almost every molecular level. Here, we describe just some of those convergence points, particularly those which are known or suspected to regulate the circadian clock.
Phytochromes are the canonical R/FR photoreceptors but they can also serve as blue light sensors. In fact, several pieces of genetic data have shown that they participate in blue light input to the circadian clock: resetting of phyA mutants under low-fluence blue light-dark cycles is impaired, they show a lengthened period under continuous blue light, and they exhibit a reduced phase shift compared with wild type plants under pulses of this wavelength. Additionally, whereas the cry1;cry2 mutant is less sensitive than wild type plants to blue light pulses, the triple mutant phyA;cry1;cry2 is insensitive to that treatment, suggesting these three loci are partially redundant components of blue light input to the clock (Somers et al., 1998; Yanovsky, 2001). Conversely, cry1;cry2 mutants exhibit a long period under continuous red light and CRY1 was early proposed to act downstream of PhyA in white light signaling to the clock (Devlin and Kay, 2000). This evidence shows that although different photoreceptors have a major role in sensing particular wavelengths, they are also able to sense others. Interestingly, PhyB and CRY2, as well as CRY1 and PhyA, are able to directly interact at the protein level, revealing a possible molecular target of convergence and regulation of different wavelengths (Figure 3) (Ahmad et al., 1998; Mas et al., 2000). Moreover, ZTL has also been shown to partner with both, CRY1 and PhyB, increasing the complexity of this network (Jarillo et al., 2001).
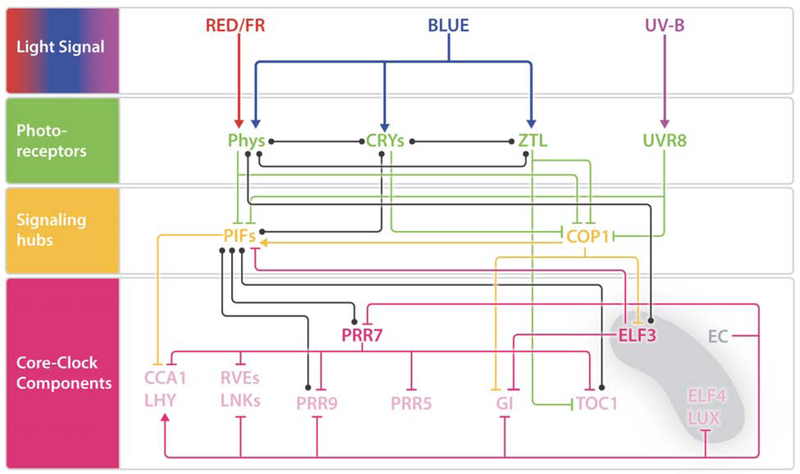
Figure 3: Conceptual framework highlighting crosstalk among light signaling pathways and their integration to the circadian clock network.
Light signal is sensed by different groups of photoreceptors, which convey the environmental information to molecular signaling hubs. Core-clock components are targets of regulation of photoreceptors, signaling hubs, other clock proteins, and additional molecular players not shown in this figure. Multiple points of crosstalk among photoreceptors, signaling hubs, as well as other components of the network, provide plasticity and accuracy to the system, and allow the entrainment of the endogenous oscillator. Lines with blunt ends and arrows indicate repression and activation, respectively. The biological significance of the interaction between components linked by black, dot ended lines has not been described. Green and orange lines represent photoreceptor- and signaling hubs-mediated regulation, respectively; pink lines indicate PRR7 or ELF3/EC regulatory steps (see text for more detail). FR, far-red; UV-B, ultraviolet B; EC, Evening Complex. Red, blue, and purple arrows: Light signal. Green colored elements: Photoreceptors. Orange colored elements: Signaling hubs. Pink colored elements: Core-clock components.
CRY1 and CRY2 interact with PIF4 and PIF5, and CRY2 shares chromatin binding regions with these two PIFs. Phenotypic analysis of different mutant combinations and biochemical assays suggest CRYs regulate the ability of PIF4 and PIF5 to promote growth. Thus, PIFs become a molecular hub at which different photoreceptors converge to modulate at least some developmental responses (Pedmale et al., 2016). Research to address whether PIFs play a role in blue light input to the clock has not yet been reported, but is of great interest as this could be a new link between CRYs and the clockwork, and could become a core integration point for diverse environmental signals.
The convergence of light signaling cascades of distinct photoreceptor families on common components is becoming highly relevant as these points of convergence could be key players in the integration of signals arising from the multiple wavelengths present in “white light” under which plants are growing. These pivotal components would integrate environmental light information and modulate downstream responses. One such element is COP1.
COP1 locus was first described by its role in repressing photomorphogenesis in darkness and it was shown early on that such repressive activity is reversed by light (Deng et al., 1991). A lot of work was dedicated to understanding the molecular function of COP1 and the many partnerships this protein establishes to modulate plant growth and development (Lau and Deng, 2012). COP1 is a RING E3 ubiquitin ligase that targets key regulators for proteasomal-mediated degradation. In vivo, COP1 functional activity depends on interaction with SPA accessory proteins. The SPA family comprises four members (SPA1 to SPA4), which exhibit partially overlapping functions and a differential pattern of expression. COP1 homodimerizes and associates with two SPA proteins. The assembled tetrameric complex, COP1/SPA, is supposed to be the substrate receptor of a bigger, multimeric complex known as the CUL4-DDB1 E3 ligase complex (Lau and Deng, 2012; Podolec and Ulm, 2018).
COP1 is known to be targeted by phytochromes, cryptochromes, UVR8 and FKF1. Upon light exposure, phytochromes and cryptochromes are known to repress COP1 activity by numerous modes of action that promote photomorphogenesis as well as other physiological responses (Podolec and Ulm, 2018). Conversely, the components of these two families of photoreceptors are negatively regulated by COP1/SPA, although not exactly through the same mechanism (Kim et al., 2017). Interestingly, mammalian cryptochromes, which are central components of the circadian clock, are not able to directly interact with the human COP1 ortholog but have retained their ability to negatively regulate its activity by interacting with a different protein of the human CUL4-RING ubiquitin complex (of which COP1 is a subunit). This suggests that the cryptochrome-mediated negative regulation of COP1 activity has been evolutionarily conserved (Rizzini et al., 2019).
As previously mentioned, UVR8 also signals through COP1. Activated UVR8 induces an increase in nuclear COP1 but the COP1/SPA complex gets separated from the CUL4-DDB1 E3 ligase complex, becoming inactive and therefore, some of its targets accumulate and trigger light responses (Podolec and Ulm, 2018). Light-activated FKF1 can interact with and negatively regulate COP1 by inhibiting its homodimerization ability. This molecular regulation is associated with the modulation of flowering time (Lee et al., 2017). It would be interesting to assess whether FKF1 regulation of COP1 activity can also impact the ticking of the circadian clock.
ELF3 physically interacts with COP1 and bridges COP1 and GI association, which triggers degradation of GI. This is likely to occur upon transition from light to dark, when ELF3 and COP1 accumulate in the nucleus and GI has been shown to undergo proteasome-mediated degradation. Thus, the GI accumulation pattern is modulated by ELF3 and COP1 in what could be considered a molecular link between photoreceptors that modulate COP1 activity and the core of the circadian clock, because modulating GI abundance will impact the biological oscillator (Yu et al., 2008). Moreover, upon its interaction with COP1, ELF3 becomes ubiquitylated and degraded by the proteasome, which could also impact the function of the EC and the clockwork itself (Yu et al., 2008). Consistently, it has been shown that cop1 mutants exhibit circadian phenotypes (Millar et al., 1995; Yu et al., 2008).
The co-regulation among PIFs and the COP1/SPA complex provides an additional layer of complexity to the integration of light signals. PIF3 binds to the promoter and likely directly regulates expression of SPA1; the COP1/SPA complex stabilizes PIFs in the dark (Martinez-Garcia et al., 2000; Pham et al., 2018a; Pham et al., 2018b). HY5 is a key transcription factor promoting photomorphogenesis, a known target of the COP1/SPA complex, and a hub where multiple signaling cascades converge. Repression of COP1 activity - mediated by light - allows an increase in HY5 abundance, which triggers photomorphogenesis. Interestingly, hy5 mutant plants exhibit a short period length under continuous light of different wavelengths, but the phenotype is enhanced under blue-light (Hajdu et al., 2018). Moreover, HY5 binds to the promoter of most of the core-clock genes under both red and blue wavelengths (preferably under the latter), which correlates with an induction of HY5 upon exposure to blue light at both transcriptional and post-transcriptional levels. As a molecular hub, HY5 interacts with a myriad of transcription factors to coordinately regulate gene expression. It has been proposed that the light quality-dependent composition of those complexes (i.e. having more or less HY5 availability) could modulate the functioning of the circadian clock as a response to the wavelength composition of the white light under which plants are growing (Hajdu et al., 2018). Additionally, HY5 directly interacts with CCA1, and it has been suggested that CCA1 could contribute to the recruitment of HY5 to certain promoters during the early morning when CCA1 peaks and light-inactivation of COP1 allows the accumulation of HY5 (Andronis et al., 2008).
DET1 (DE-ETIOLATED 1) is part of the CDD complex (COP10-DET1-CCB1) which has a key role on repressing photomorphogenesis. Its action has not been associated with a particular wavelength, but with general light signaling. Specifically, DET1 directly interacts with CCA1 and LHY, which is believed to allow its recruitment to the promoter of TOC1, such that DET1 represses transcription of TOC1. det1 mutant plants exhibit a short period phenotype, suggesting this protein is essential for determining the pace of the oscillator (Lau et al., 2011; Millar et al., 1995). Additionally, DET1 positively regulates PIFs abundance in the dark, although whether that constitutes another input pathway to the oscillator has not been addressed (Dong et al., 2014).
The CSU4 (COP1 SUPPRESSOR 4) locus was identified by its genetic role as a suppressor of the cop1 mutant phenotype. Although the gene encodes a protein with a domain of unknown function, the molecular characterization showed that CSU4 directly binds CCA1 and represses its transcriptional repression activity. Likely as a consequence, csu4 mutants exhibit increased levels of CCA1 and PIF4 (Zhao et al., 2018).
Finally, the family of PPKs (PHOTOREGULATORY PROTEIN KINASES 1 to 4, previously known as MLKs -MUT9-LIKE KINASES-) could also represent a convergence point for the input of multiple light wavelengths to the clock. ppk2, ppk3 and ppk4 single mutants (mlk1, mlk2, mlk3, respectively) exhibit a long period phenotype, which is rescued by a ppk1 (mlk4) mutant allele through an unknown mechanism. Interestingly, PPKs associate in vivo with ELF3 (Huang et al., 2016a). PPKs also interact with PIF3 and PhyB in a light-induced manner and are required for light-mediated phosphorylation and degradation of PIF3 (Ni et al., 2017). PPKs additionally associate with and phosphorylate photoexcited CRY2, which is proposed to activate and destabilize the photoreceptor (Liu et al., 2017). Thus, this recently studied family of kinases could play a central role to integrate and transduce the signal of multiple wavelengths perceived by phytochromes and cryptochromes by affecting PIF3 accumulation and/or through their interaction with ELF3, ultimately impacting circadian clock rhythmicity.
ELF3 and PRR7 as Key Integrators
Different pieces of evidence led to the broadly accepted view that the Arabidopsis clock is set on time mainly by the transition from dark to light (McWatters et al., 2000; Millar and Kay, 1996; Seaton et al., 2018). Data from other organisms suggest that either the night length or both dawn and dusk information synchronize their endogenous oscillators (Hayama et al., 2018; Ramos-Sánchez et al., 2019). Nevertheless, considering that light (through PIFs inactivation) activates the expression of CCA1, that metabolic status (defined by sugar levels) modulates PRR7, and that dusk (through ZTL) induces TOC1 degradation, the more recently proposed hypothesis of a dynamic and continuous entrainment fits best with the observed behavior of the circadian system in Arabidopsis (Frank et al., 2018; Haydon et al., 2013; Mas et al., 2003; Nohales et al., 2019; Seaton et al., 2018; Webb et al., 2019).
The large number of elements comprising the oscillator and their intricate cross-regulation makes the prediction of the circadian network behavior difficult, even through mathematical modeling (Fogelmark and Troein, 2014; Pokhilko et al., 2012). However, a new approach proposes to conceive the clock as a binary system with two states: the morning and evening, characterized by CCA1/LHY and TOC1/PRR5 activity, respectively. In this model there are two rapid switchers that allow the system to move forward and transition from one state to the other: PRR9/PRR7 are the elements pushing the system from the morning to the evening state; the EC does the opposite (Joanito et al., 2018). Interestingly, PRR7 and ELF3 (one of the three components of the EC) both have a set of particular features that support the hypothesis of these two genes as particularly important within the circadian network and perhaps, reliable components to determine the switch of the system from the morning to the night state, and vice versa (Figure 3).
PRR7 plays a partially redundant role with PRR9. However, prr7 single mutants exhibit a longer period than wild type plants and are insensitive to different red fluence rates, whereas the wild type genotype exhibits a shortening of the period as the irradiated light increases (as expected by Aschoff’s rule) (Aschoff, 1979; Farré et al., 2005). This suggests that PRR7 is involved in red light input to the clock. Notably, analogous results are obtained for prr9 mutants under blue light, which raises the question whether these two genes function complementarily for this response under these two wavelengths (Farré et al., 2005). PRR7 is necessary to gate the clock resetting driven by sugar, as prr7 mutants induce the same phase shift to a pulse of exogenous sucrose independently of the time of the day in which the treatment is applied (Haydon et al., 2013). Additionally, the prr7 mutant period length is not adjusted to different concentrations of nicotinamide, a metabolite known to slow down the pace of the clock (Mombaerts et al., 2019). PRR7 is also necessary for the proper response of the circadian clock to different temperature treatments (Salomé and McClung, 2005). In summary, the evidence suggests that PRR7 is essential for red light, sugar, other metabolites (such as nicotinamide) and temperature input to the clock.
ELF3 is required to sustain rhythms under continuous light but not under continuous darkness. Overexpressing ELF3 lines show an increased period length in both constant blue and red light, suggesting ELF3 is associated with input of information from both wavelengths to the clockwork (Covington et al., 2001). ELF3 is also necessary to properly entrain the clock to thermocycles (Thines and Harmon, 2010). Besides its role as a transcriptional regulator, ELF3 is described to act as a hub protein and has been found to associate in vivo with the five members of the phytochrome family, as well as with many other elements of the red and far-red light signaling pathways (Huang et al., 2016a). ELF3 has been shown to interact with both forms of PhyB (Pr and Pfr), suggesting these two proteins could partner either in the nucleus or in the cytoplasm (Liu et al., 2001; Yeom et al., 2014). Finally, under dark conditions, ELF3 bridges COP1 and GI, which triggers COP1-mediated degradation of GI and could indirectly impact ZTL accumulation and ZTL-mediated degradation of TOC1. These steps likely contribute to setting the time by conveying the “dark signal” to the core-clock (Nohales and Kay, 2016). Additionally, the direct association of COP1 with ELF3 links the latter to virtually every light signaling cascade, as COP1 is a common player to all.
Both PRR7 and ELF3 have been shown to partner with PIFs, although the biological relevance of such interactions has not been clarified for PRR7, and has been associated with the control of output responses but not to clock input for ELF3 (Martín et al., 2018; Nieto et al., 2015). PIFs and PRR7 are also targeted by other molecular pathways, conveying to the clock other input signals. This evidence led us to propose that PIFs and COP1 (described above as fundamental components of light signaling pathways) act downstream of the photoreceptors and comprise a first layer of signal integration. Then, PRR7 and ELF3 could represent a second layer of signal integration and at the same time, core-clock components (Figure 3).
Feedback Signaling of the Clock into Light Perception
Light perception and its signaling cascades are modulated by the endogenous clock at different levels. This is not an exclusive feature of light sensing pathways as it is known that the biological oscillator creates signaling feedback loops with several of its entraining cues, as well as in its downstream responses, which both contribute to its fine-tuning (Sanchez and Kay, 2016).
The first clue suggesting that the endogenous clock has the ability to modulate light responses originated from gating experiments in which the induction (or phase shift) of a clock target gene was assessed after plants were treated with light pulses at different times of the day (Millar and Kay, 1996). With the molecular identification of clock components, the loci associated with this response, as well as the wavelength specificity, were also addressed (McWatters et al., 2000). The mechanism underlying this control is not yet completely understood, but some pieces of the puzzle have been fit in place (Oakenfull and Davis, 2017).
Members of the phytochrome family exhibit a circadian profile of expression, peaking at different times of the day (Harmer et al., 2000; Schaffer et al., 2001; Tóth et al., 2001). It has been hypothesized that such patterns of mRNA accumulation could imply a physiological role as PhyB (which encodes a photostable photoreceptor known to mediate responses under high-fluence rate wavelengths) reaches its maximum level of expression around mid-morning, which correlates with high light intensities in natural environments. On the other hand, PhyA (a photolabile molecule associated with low-fluence rate and far-red light responses) shows a peak of mRNA expression at the end of the day, which correlates with low light intensities and enrichment of far-red wavelengths under natural conditions (Tóth et al., 2001). Nevertheless, at the protein level, PhyA exhibit a peak at the end of the night under light-dark conditions, whereas PhyB and PhyE accumulation do not present significant changes in amplitude and PhyC displays only a very weak oscillation under the same growth conditions (Sharrock and Clack, 2002). Consistently, PhyB protein level in tobacco plants has been shown to be fairly stable throughout the day under light-dark conditions (Bognár et al., 1999). Moreover, PhyA, PhyB, PhyC, and PhyE, do not show significant changes in their protein levels under circadian conditions (continuous light) (Sharrock and Clack, 2002), so it remains to be elucidated whether the circadian profile of mRNA expression of phytochromes is biologically relevant. It has been shown, however, that subcellular localization of phytochromes is regulated by different wavelengths and impacts signaling ability (Li et al., 2011b). Additionally, it is conceivable that the circadian clock modulates the signaling ability of PhyB through PCH1 because PCH1 is required for photobodies formation (which is in turn necessary for PhyB signaling and regulation of numerous physiological and developmental processes; see “Phytochrome signaling”) and PCH1 mRNA expression and protein levels exhibit a circadian profile of oscillation, peaking in the evening. Together, these data suggest that the biological clock contributes to time light responses by modulating the PhyB-signaling pathway (Huang et al., 2016b).
PIFs play an important role in the feedback signaling of the clock to light input as they can induce PhyB degradation and regulate PhyA expression and PhyA activity (Leivar et al., 2008; Leivar et al., 2012; Seaton et al., 2018). As explained above, PIFs and core-clock elements are tightly linked: PIFs are direct transcriptional targets of core-clock components and their encoded proteins interact with multiple clockwork elements to coordinate downstream responses. It is possible that PIFs-clock interactions may also modulate the regulatory role of PIFs on phytochromes, although to date no evidence has been found to support this hypothesis.
The cryptochrome profile of expression is also under the control of the circadian clock and resembles that of phytochrome family: CRY1 peaks in the late-morning whereas CRY2 (the protein of which is rapidly down-regulated by light) reaches its maximum at the end of the day, consistent with the light dependent stability and biological function of each photoreceptor (Harmer et al., 2000; Lin et al., 1998; Tóth et al., 2001). As discussed, cryptochromes act in concert with PIF4 and PIF5 to modulate downstream gene expression (Pedmale et al., 2016). Hence, if core-clock components impinge on PIFs activity, this could be an additional way of the endogenous timekeeper modulating blue light responses.
Despite ZTL expression level not exhibiting a circadian profile, ZTL abundance does show a rhythmic pattern of accumulation under light-dark cycles reaching its peak at the end of the day, which correlates with blue-light enrichment in natural conditions and could contribute to its physiological role associated with regulation of TOC1 and PRR5 and CHE stability (Kim et al., 2003b; Nohales and Kay, 2016). UV-B light signaling is also gated by the circadian clock, an example of the feedback regulation of the clockwork (Fehér et al., 2011; Takeuchi et al., 2014). However, the level of UVR8 monomer is relatively constant under diurnal conditions, suggesting the clock and UV-B light signaling networks interact at a different level (Findlay and Jenkins, 2016).
Entrainment of the Clock: A Wider Perspective
Crosstalk between wavelengths and feedback from the core-clock to the light perception shape a convoluted network of interactions (Figure 3). However, this system is more complex still as it includes many additional synchronizing cues. First, light allows plants to produce sugar through photosynthesis. It has been shown that the rhythmic metabolic state driven by sugars can entrain the circadian clock, representing an indirect mechanism for light input to the clock (Frank et al., 2018; Haydon et al., 2013; Sanchez and Kay, 2016). Second, hormone levels are circadian-modulated and can in turn regulate different parameters of the pacemaker (Covington et al., 2008; Hanano et al., 2006; Lee et al., 2016; Legnaioli et al., 2009; Nováková et al., 2005; Sanchez and Kay, 2016; Thain et al., 2004; Zhang et al., 2019b). Because the interaction between light and hormone signaling pathways has been established, it introduces an additional indirect path for light to modulate the biological rhythms (Leivar and Monte, 2014; Paik and Huq, 2019). Third, temperature, together with light, is a fundamental variable for biological clock entrainment (McClung, 2019). Temperature is known to input to the clock at different levels including regulating LUX at the transcriptional level, modulating alternative splicing of several core-clock genes and through PRR7 and PRR9 by an unknown mechanism (Chow et al., 2014; Mateos et al., 2018; Salomé and McClung, 2005). Furthermore, PhyB has the ability to act as a thermosensor, so temperature-mediated regulation of PhyB could contribute to setting the pace of the core-clock (Jung et al., 2016; Legris et al., 2016). Lastly, although not described here, is important to note that other endogenous and exogenous variables, including biotic and abiotic stress and nutrient homeostasis, can fine-tune the functioning of the biological oscillator (Haydon et al., 2015; Seo and Mas, 2015).
Concluding Remarks
The circadian system regulates virtually every single aspect of plant growth and development, and therefore understanding how it is synchronized is of great importance to our ability to predict how changes in environmental stimuli will impact endogenous rhythms and consequently, plant physiology. It has been clearly established that light is one of the key entrainment factors of the internal oscillator, and many efforts have been focused over the last 20 years on describing the mechanism underlying that process. However, despite significant advances made and many pieces uncovered, a comprehensive view and understanding of its functioning is still lacking. Likely, the high degree of crosstalk and feedback between multiple endogenous and external cues that signal back and forth to the core clock, make difficult the unraveling of the entrainment system (Figure 3). System-level approaches, together with mathematical modeling and genome-scale analysis, have contributed to the elucidation of this complex network as they have identified missing pieces and/or connections, which would have been difficult to find by traditional methods (Gould et al., 2013; Joanito et al., 2018; Seaton et al., 2018).
Protein dynamics, including protein-protein and protein-chromatin interactions, as well as the post-translational modifications required for such associations represent essential regulatory steps that have been poorly explored. In fact, specific examples of the importance of such dynamics have been identified. The COP1-ELF3 interaction has been suggested to be dark-dependent (Yu et al., 2008). PhyB appears to associate with different clock components depending on its Pr or Pfr state (Yeom et al., 2014). Remarkably, in vivo experiments have shown the five phytochromes interacting with ELF3, in spite of the fact that these photoreceptors are thought to be mainly active during the daytime, whereas ELF3 tends towards the night (Huang et al., 2016a). Additionally, many interactions are known to be dependent on subcellular localization and post-translational modifications (Nohales and Kay, 2016). Many researchers have deployed strategies to assess context-free interactions (Y2H or in vitro assays; Table 2); great contributions that provide the opportunity to overcome functional redundancy and evaluate direct interaction, but also have limitations. Hence, to answer the important questions of where (within the cell or the plant) and when different proteins associate, as well as which post-translational modifications are required for those interactions to occur, in vivo experiments should be done. The additional information gained from such approaches will likely contribute to comprehension of the biological relevance of those partnerships, and ultimately enrich our understanding of the mechanisms underlying the clock entrainment.
Acknowledgements
The authors apologize for not citing all relevant publications by colleagues because of space limitations. S.E.S. is partially supported by a postdoctoral fellowship from the Pew Latin American Fellows Program. This work is supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award Nos. R01GM067837 and R01GM56006. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams S, Manfield I, Stockley P, and Carré IA (2015). Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 10:e0143943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M (2016). Photocycle and signaling mechanisms of plant cryptochromes. Curr Opin Plant Biol. 33:108–115. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, and Cashmore AA (1998). The CRY1 Blue Light Photoreceptor of Arabidopsis Interacts with Phytochrome A In Vitro. Molecular Cell 1:939–948. [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, and Kay SA (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293:880–883. [DOI] [PubMed] [Google Scholar]
- Allen T, Koustenis A, Theodorou G, Somers DE, Kay SA, Whitelam GC, and Devlin PF (2006). Arabidopsis FHY3 Specifically Gates Phytochrome Signaling to the Circadian Clock. Plant Cell 18:2506–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, and Kay SA (1997). Attenuation of phytochrome A and B signaling pathways by the arabidopsis circadian clock. Plant Cell 9:1721–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronis C, Barak S, Knowles SM, Sugano S, and Tobin EM (2008). The Clock Protein CCA1 and the bZIP Transcription Factor HY5 Physically Interact to Regulate Gene Expression in Arabidopsis. Molecular Plant 1:58–67. [DOI] [PubMed] [Google Scholar]
- Aschoff J (1979). Circadian Rhythms: Influences of Internal and External Factors on the Period Measured in Constant Conditions1. Zeitschrift für Tierpsychologie 49:225–249. [DOI] [PubMed] [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, et al. (2010). F-Box Proteins FKF1 and LKP2 Act in Concert with ZEITLUPE to Control Arabidopsis Clock Progression. Plant Cell 22:606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bognár LK, Hall A, Ádám É, Thain SC, Nagy F, and Millar AJ (1999). The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc. Natl. Acad. Sci. U.S.A 96:14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR (2006). BLUE/UV-A RECEPTORS: HISTORICAL OVERVIEW In: PHOTOMORPHOGENESIS IN PLANTS AND BACTERIA--Schafer E, and Nagy F, eds. Dordrecht: Springer Netherlands; 171–197. [Google Scholar]
- Casal JJ (2013). Photoreceptor Signaling Networks in Plant Responses to Shade. Annu. Rev. Plant Biol 64:403–427. [DOI] [PubMed] [Google Scholar]
- Casal JJ, and Qüesta JI (2018). Light and temperature cues: multitasking receptors and transcriptional integrators. New Phytologist 217:1029–1034. [DOI] [PubMed] [Google Scholar]
- Cashmore AR (2003). Cryptochromes: Enabling Plants and Animals to Determine Circadian Time. Cell 114:537–543. [PubMed] [Google Scholar]
- Castillon A, Shen H, and Huq E (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12:514–521. [DOI] [PubMed] [Google Scholar]
- Cha JY, Kim J, Kim TS, Zeng Q, Wang L, Lee SY, Kim WY, and Somers DE (2017). GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat Commun 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen L-O, Horst G.T.J.v.d., Batschauer A, and Ahmad M (2011). The Cryptochromes: Blue Light Photoreceptors in Plants and Animals. Annu. Rev. Plant Biol 62:335–364. [DOI] [PubMed] [Google Scholar]
- Chen F, Li B, Li G, Charron JB, Dai M, Shi X, and Deng XW (2014). Arabidopsis Phytochrome A Directly Targets Numerous Promoters for Individualized Modulation of Genes in a Wide Range of Pathways. Plant Cell 26:1949–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow Brenda Y., Sanchez Sabrina E., Breton G, Pruneda-Paz Jose L., Krogan Naden T., and Kay Steve A. (2014). Transcriptional Regulation of LUX by CBF1 Mediates Cold Input to the Circadian Clock in Arabidopsis. Curr. Biol 24:1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, and Sharrock RA (2009). Obligate Heterodimerization of Arabidopsis Phytochromes C and E and Interaction with the PIF3 Basic Helix-Loop-Helix Transcription Factor. Plant Cell 21:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M, Maloof J, Straume M, Kay S, and Harmer S (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, and Kay SA (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13:1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creux N, and Harmer S (2019). Circadian Rhythms in Plants. Cold Spring Harbor Perspectives in Biology. Advance Access published May 28, 2019, doi: 10.1101/cshperspect.a034611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James Geo V., Janitza P, Ibañez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J, et al. (2014). The DET1-COP1-HY5 Pathway Constitutes a Multipurpose Signaling Module Regulating Plant Photomorphogenesis and Thermomorphogenesis. Cell Reports 9:1983–1989. [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, and Quail PH (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5:1172–1182. [DOI] [PubMed] [Google Scholar]
- Devlin PF, and Kay SA (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12:2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, and Webb AAR (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309:630–633. [DOI] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, and Chen H (2014). Arabidopsis DE-ETIOLATED1 Represses Photomorphogenesis by Positively Regulating Phytochrome-Interacting Factors in the Dark. Plant Cell 26:3630–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, and Amasino RM (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419:74–77. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, and Staiger D (2002). Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216:1–16. [DOI] [PubMed] [Google Scholar]
- Farinas B, and Mas P (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66:318–329. [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, and Kay SA (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol 15:47–54. [DOI] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. The EMBO Journal 28:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér B, Kozma-Bognár L, Kevei É, Hajdu A, Binkert M, Davis SJ, Schäfer E, Ulm R, and Nagy F (2011). Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 67:37–48. [DOI] [PubMed] [Google Scholar]
- Findlay KM, and Jenkins GI (2016). Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions. Plant, Cell & Environment 39:1706–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelmark K, and Troein C (2014). Rethinking Transcriptional Activation in the Arabidopsis Circadian Clock. PLoS Comput Biol 10:e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE, Wells Newman D, Yochikawa A, Cano-Ramirez DL, Chembath A, et al. (2018). Circadian Entrainment in Arabidopsis by the Sugar-Responsive Transcription Factor bZIP63. Curr. Biol 28:2597–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, and Quail PH (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot 61:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, and Mizuno T (2004). Circadian-Controlled Basic/Helix-Loop-Helix Factor, PIL6, Implicated in Light-Signal Transduction in Arabidopsis thaliana. Plant and Cell Physiology 45:1078–1086. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, and Somers DE (2008). Post-translational Regulation of the Arabidopsis Circadian Clock through Selective Proteolysis and Phosphorylation of Pseudo-response Regulator Proteins. Journal of Biological Chemistry 283:23073–23083. [DOI] [PubMed] [Google Scholar]
- Gangappa Sreeramaiah N., and Botto Javier F. (2016). The Multifaceted Roles of HY5 in Plant Growth and Development. Molecular Plant 9:1353–1365. [DOI] [PubMed] [Google Scholar]
- Gil KE, and Park CM (2019). Thermal adaptation and plasticity of the plant circadian clock. New Phytologist 221:1215–1229. [DOI] [PubMed] [Google Scholar]
- Gould PD, Ugarte N, Domijan M, Costa M, Foreman J, MacGregor D, Rose K, Griffiths J, Millar AJ, Finkenstädt B, et al. (2013). Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Molecular Systems Biology 9:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, and McClung CR (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet 16:598–610. [DOI] [PubMed] [Google Scholar]
- Hajdu A, Ádám É, Sheerin DJ, Dobos O, Bernula P, Hiltbrunner A, Kozma-Bognár L, and Nagy F (2015). High-level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. Plant J. 83:794–805. [DOI] [PubMed] [Google Scholar]
- Hajdu A, Dobos O, Domijan M, Bálint B, Nagy I, Nagy F, and Kozma-Bognár L (2018). ELONGATED HYPOCOTYL 5 mediates blue light signalling to the Arabidopsis circadian clock. Plant J. 96:1242–1254. [DOI] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognár L, Bastow RM, Nagy F, and Millar AJ (2002). Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 32:529–537. [DOI] [PubMed] [Google Scholar]
- Hanano S, Domagalska MA, Nagy F, and Davis SJ (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes to Cells 11:1381–1392. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, and Kay SA (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113. [DOI] [PubMed] [Google Scholar]
- Hayama R, Mizoguchi T, and Coupland G (2018). Differential effects of light-to-dark transitions on phase setting in circadian expression among clock-controlled genes in Pharbitis nil. Plant Signaling and Behavior 13:e1473686:1473681–1473687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, and Webb AAR (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502:689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Román Á, and Arshad W (2015). Nutrient homeostasis within the plant circadian network. Frontiers in Plant Science 6:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Sharma A, Fraser DP, Trevisan M, Cragg-Barber CK, Tavridou E, Fankhauser C, Jenkins GI, and Franklin KA (2017). UV-B Perceived by the UVR8 Photoreceptor Inhibits Plant Thermomorphogenesis. Curr. Biol 27:120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Velanis CN, Jenkins GI, and Franklin KA (2014). UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. U.S.A 111:11894–11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, and Kay SA (2011). LUX ARRHYTHMO Encodes a Nighttime Repressor of Circadian Gene Expression in the Arabidopsis Core Clock. Curr. Biol 21:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, and Kay SA (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274:790–792. [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71:699–711. [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, and Fankhauser C (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. The EMBO Journal 28:3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, and Harmer SL (2013). Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2:e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, and Lagarias JC (2013). Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc. Natl. Acad. Sci. U.S.A 110:1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel RK, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, and Nusinow DA (2015). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Molecular & Cellular Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel RK, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, and Nusinow DA (2016a). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Molecular & Cellular Proteomics 15:201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, McLoughlin KE, Sorkin ML, Burgie ES, Bindbeutel RK, Vierstra RD, and Nusinow DA (2019). PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 116:8603–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, and Nusinow DA (2016). Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends in Genetics 32:674–686. [DOI] [PubMed] [Google Scholar]
- Huang H, Yoo CY, Bindbeutel R, Goldsworthy J, Tielking A, Alvarez S, Naldrett MJ, Evans BS, Chen M, and Nusinow DA (2016b). PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife 5:e13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, and Mas P (2012). Mapping the Core of the Arabidopsis Circadian Clock Defines the Network Structure of the Oscillator. Science 336:75–79. [DOI] [PubMed] [Google Scholar]
- Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, and Deng XW (2013). Conversion from CUL4-based COP1–SPA E3 apparatus to UVR8–COP1–SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. U.S.A 110:16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang Q, Jiang Y, Yang C, Wang Q, and Li L (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14–3-3 proteins in Arabidopsis. eLife 7:e31636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, and Imaizumi T (2012). LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Molecular Plant 5:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang R-H, Yang H-Q, Alonso JM, Ecker JR, and Cashmore AR (2001). An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410:487–490. [DOI] [PubMed] [Google Scholar]
- Joanito I, Chu J-W, Wu S-H, and Hsu C-P (2018). An incoherent feed-forward loop switches the Arabidopsis clock rapidly between two hysteretic states. Scientific Reports 8:13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Hu W, Litthauer S, Lagarias JC, and Harmer S (2015). A Constitutively Active Allele of Phytochrome B Maintains Circadian Robustness in the Absence of Light. Plant Physiol. 169:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354:886–889. [DOI] [PubMed] [Google Scholar]
- Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, and Nakamichi N (2016). Direct Repression of Evening Genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis Circadian Clock. Plant Cell 28:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, and Chua NH (2007). Targeted Degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL Complex Regulates Clock Function and Photomorphogenesis in Arabidopsis thaliana. Plant Cell 19:2516–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, et al. (2009). The Phytochrome-Interacting Factor PIF7 Negatively Regulates DREB1 Expression under Circadian Control in Arabidopsis. Plant Physiol. 151:2046–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Geng R, Gallenstein RA, and Somers DE (2013). The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140:4060–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, and Choi G (2003a). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15:2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song JT, and Seo HS (2017). COP1 regulates plant growth and development in response to light at the post-translational level. J Exp Bot 68:4737–4748. [DOI] [PubMed] [Google Scholar]
- Kim TS, Kim WY, Fujiwara S, Kim J, Cha JY, Park JH, Lee SY, and Somers DE (2011). HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl. Acad. Sci. U.S.A 108:16843–16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, and Somers DE (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449:356–360. [DOI] [PubMed] [Google Scholar]
- Kim WY, Geng R, and Somers DE (2003b). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U.S.A 100:4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, and Somers DE (2019). Luciferase-Based Screen for Post-translational Control Factors in the Regulation of the Pseudo-Response Regulator PRR7. Frontiers in Plant Science 10:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Lockhart P, and Batschauer A (2003). An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 35:93–103. [DOI] [PubMed] [Google Scholar]
- Kolmos E, Herrero E, Bujdoso N, Millar AJ, Tóth R, Gyula P, Nagy F, and Davis SJ (2011). A Reduced-Function Allele Reveals That EARLY FLOWERING3 Repressive Action on the Circadian Clock Is Modulated by Phytochrome Signals in Arabidopsis. Plant Cell 23:3230–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, and Deng XW (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17:584–593. [DOI] [PubMed] [Google Scholar]
- Lau On S., Huang X, Charron JB, Lee JH, Li G, and Deng Xing W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in Mediating Transcriptional Repression in the Plant Circadian Clock. Molecular Cell 43:703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BD, Kim MR, Kang MY, Cha JY, Han SH, Nawkar GM, Sakuraba Y, Lee SY, Imaizumi T, McClung CR, et al. (2017). The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nature Communications 8:2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-M, Feke A, Li M-W, Adamchek C, Webb K, Pruneda-Paz J, Bennett EJ, Kay SA, and Gendron JM (2018). Decoys Untangle Complicated Redundancy and Reveal Targets of Circadian Clock F-Box Proteins. Plant Physiol. 177:1170–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Li MW, Feke A, Liu W, Saffer AM, and Gendron JM (2019). GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nature Communications 10:3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Mas P, and Seo PJ (2016). MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Scientific Reports 6:17754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, and Mas P (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. The EMBO Journal 28:3745–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Costigliolo C, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, and Casal JJ (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354:897–900. [DOI] [PubMed] [Google Scholar]
- Leivar P, and Monte E (2014). PIFs: Systems Integrators in Plant Development. Plant Cell 26:56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, and Quail PH (2008). The Arabidopsis Phytochrome-Interacting Factor PIF7, Together with PIF3 and PIF4, Regulates Responses to Prolonged Red Light by Modulating phyB Levels. Plant Cell 20:337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Cohn MM, and Quail PH (2012). Phytochrome Signaling in Green Arabidopsis Seedlings: Impact Assessment of a Mutually Negative phyB–PIF Feedback Loop. Molecular Plant 5:734–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan XY, Li J, Lau OS, Ouyang X, Dai M, Wan J, et al. (2011a). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol 13:616–622. [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, and Wang Deng X (2011b). Phytochrome signaling mechanisms. In: Arabidopsis Book: American Society of Plant Biologists; e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung H-S, et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ma D, Lu SX, Hu X, Huang R, Liang T, Xu T, Tobin EM, and Liu H (2016). Blue Light- and Low Temperature-Regulated COR27 and COR28 Play Roles in the Arabidopsis Circadian Clock. Plant Cell 28:2755–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Yang Y, and Liu H (2019). Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytologist 221:1247–1252. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, and A, C. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. U.S.A. 95:2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, and Wang H (2007). Transposase-Derived Transcription Factors Regulate Light Signaling in Arabidopsis. Science 318:1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litthauer S, Battle MW, and Jones MA (2015). Phototropins do not alter accumulation of evening-phased circadian transcripts under blue light. Plant Signaling & Behavior 11:e1126029–e1126029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang Z, Gomez A, Liu B, Lin C, and Oka Y (2016a). Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res 129:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Deng W, Wang X, Piao M, Cai D, Li Y, Barshop WD, Yu X, Zhou T, et al. (2017). Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nature Communications 8:15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Carlsson J, Takeuchi T, Newton L, and Farré EM (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76:101–114. [DOI] [PubMed] [Google Scholar]
- Liu TL, Newton L, Liu MJ, Shiu SH, and Farre EM (2016b). A G-Box-Like Motif Is Necessary for Transcriptional Regulation by Circadian Pseudo-Response Regulators in Arabidopsis. Plant Physiol 170:528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, and Wagner DR (2001). ELF3 Encodes a Circadian Clock–Regulated Nuclear Protein That Functions in an Arabidopsis PHYB Signal Transduction Pathway. Plant Cell 13:1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Andronis C, Ong MS, and Tobin EM (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL Function Synergistically in the Circadian Clock of Arabidopsis. Plant Physiol. 150:834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Webb CJ, Knowles SM, Kim SHJ, Wang Z, and Tobin EM (2012). CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in arabidopsis. Plant Physiol. 158:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, and Yang HQ (2014). COP1 and phyB Physically Interact with PIL1 to Regulate Its Stability and Photomorphogenic Development in Arabidopsis. Plant Cell 26:2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Oda Y, and Mizuno T (2001). Light Response of the Circadian Waves of the APRR1/TOC1 Quintet: When Does the Quintet Start Singing Rhythmically in Arabidopsis? Plant and Cell Physiology 42:334–339. [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiology 43:58–69. [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, and Harmer SL (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143:473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM, Boix M, Henriques R, Minguet EG, Alabadí D, et al. (2018). Circadian Waves of Transcriptional Repression Shape PIF-Regulated Photoperiod-Responsive Growth in Arabidopsis. Curr. Biol 28:311–318. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, and Quail PH (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288:859–863. [DOI] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, and Kay SA (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408:207–211. [DOI] [PubMed] [Google Scholar]
- Mas P, Kim W, Somers D, and Kay SA (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426:567–570. [DOI] [PubMed] [Google Scholar]
- Mateos JL, De Leone MJ, Torchio J, Reichel M, and Staiger D (2018). Beyond Transcription: Fine-Tuning of Circadian Timekeeping by Post-Transcriptional Regulation. Genes 9:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR (2019). The Plant Circadian Oscillator. Biology 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, and Millar AJ (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408:716–720. [DOI] [PubMed] [Google Scholar]
- Michael T, Salomé P, Yu H, Spencer T, Sharp E, McPeek M, Alonso J, Ecker J, and McClung C (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302:1049–1053. [DOI] [PubMed] [Google Scholar]
- Millar AJ, and Kay SA (1996). Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci 93:15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, and Kay SA (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267:1163–1166. [DOI] [PubMed] [Google Scholar]
- Mizuno T, and Nakamichi N (2005). Pseudo-Response Regulators (PRRs) or True Oscillator Components (TOCs). Plant and Cell Physiology 46:677–685. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, and Yamashino T (2014a). Ambient Temperature Signal Feeds into the Circadian Clock Transcriptional Circuitry Through the EC Night-Time Repressor in Arabidopsis thaliana. Plant and Cell Physiology 55:958–976. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Takeuchi A, Nomoto Y, Nakamichi N, and Yamashino T (2014b). The LNK1 night light-inducible and clock-regulated gene is induced also in response to warm-night through the circadian clock nighttime repressor in Arabidopsis thaliana. Plant Signaling & Behavior 9:e28505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts L, Carignano A, Robertson FC, Hearn TJ, Junyang J, Hayden D, Rutterford Z, Hotta CT, Hubbard KE, Maria MRC, et al. (2019). Dynamical differential expression (DyDE) reveals the period control mechanisms of the Arabidopsis circadian oscillator. PLoS Computational Biology 15:e1006674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, and Quail PH (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. U.S.A 101:16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, and Kay SA (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 112:E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M, and Chory J (1998). Genetic Interactions between Phytochrome A, Phytochrome B and Cryptochrome 1 during Arabidopsis Development. Plant Physiol. 118:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998). PIF3, a Phytochrome-Interacting Factor Necessary for Normal Photoinduced Signal Transduction, Is a Novel Basic Helix-Loop-Helix Protein. Cell 95:657–667. [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang ZY, and Quail PH (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nature Communications 8:15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière J-M, and Prat S (2015). ELF3-PIF4 Interaction Regulates Plant Growth Independently of the Evening Complex. Curr. Biol 25:187–193. [DOI] [PubMed] [Google Scholar]
- Nohales MA, and Kay SA (2016). Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23:1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Liu W, Duffy T, Nozue K, Sawa M, Pruneda-Paz JL, Maloof JN, Jacobsen SE, and Kay SA (2019). Multi-level Modulation of Light Signaling by GIGANTEA Regulates Both the Output and Pace of the Circadian Clock. Developmental Cell 49:840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková M, Motyka V, Dobrev PI, Malbeck J, Gaudinová A, and Vanková R (2005). Diurnal variation of cytokinin, auxin and abscisic acid levels in tobacco leaves. J Exp Bot 56:2877–2883. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington M, Duek P, Lorrain S, Fankhauser C, Harmer S, and Maloof J (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 19:358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, and Kay SA (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull RJ, and Davis SJ (2017). Shining a light on the Arabidopsis circadian clock. Plant, Cell & Environment 40:2571–2585. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, and Wang Z-Y (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14:802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, and Huq E (2019). Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Seminars in Cell & Developmental Biology 92:114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim JI, and Huq E (2017). Expanding Roles of PIFs in Signal Integration from Multiple Processes. Molecular Plant 10:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, and Choi G (2012). Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. U.S.A 109:1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palágyi A, Terecskei K, Ádám É, Kevei É, Kircher S, Mérai Z, Schäfer E, Nagy F, and Kozma-Bognár L (2010). Functional Analysis of Amino-Terminal Domains of the Photoreceptor Phytochrome B. Plant Physiol. 153:1834–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale Ullas V., Huang, S.-shan C, Zander M, Cole Benjamin J., Hetzel J, Ljung K, Reis Pedro A.B., Sridevi P, Nito K, Nery Joseph R., et al. (2016). Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 164:233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García P, Ma Y, Yanovsky MJ, and Mas P (2015). Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. U.S.A 112:5249–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, and Quail PH (2014). Combinatorial Complexity in a Transcriptionally Centered Signaling Hub in Arabidopsis. Molecular Plant 7:1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, and Huq E (2018a). Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 176:1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Xu X, and Huq E (2018b). Molecular bases for the constitutive photomorphogenic phenotypes in Arabidopsis. Development 145:dev169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R, and Ulm R (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol. 45:18–25. [DOI] [PubMed] [Google Scholar]
- Pokhilko A, Fernandez AP, Edwards KD, Southern MM, Halliday KJ, and Millar AJ (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, and Kay SA (2009). A Functional Genomics Approach Reveals CHE as a Component of the Arabidopsis Circadian Clock. Science 323:1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudasaini A, Shim JS, Song YH, Shi H, Kiba T, Somers DE, Imaizumi T, and Zoltowski BD (2017). Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. eLife 6:e21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudasaini A, and Zoltowski BD (2013). Zeitlupe Senses Blue-Light Fluence To Mediate Circadian Timing in Arabidopsis thaliana. Biochemistry 52:7150–7158. [DOI] [PubMed] [Google Scholar]
- Ramos-Sánchez JM, Triozzi PM, Alique D, Geng F, Gao M, Jaeger KE, Wigge PA, Allona I, and Perales M (2019). LHY2 Integrates Night-Length Information to Determine Timing of Poplar Photoperiodic Growth. Curr. Biol 29:2402–2406. [DOI] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, and Harmer SL (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 Form a Negative Feedback Loop within the Arabidopsis Circadian Clock. PLoS Genet 7:e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, and Chory J (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104:1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, Elumalai RP, and Millar AJ (2000). Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 122:1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L, Favory J-J, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011). Perception of UV-B by the Arabidopsis UVR8 Protein. Science 332:103–106. [DOI] [PubMed] [Google Scholar]
- Rizzini L, Levine DC, Perelis M, Bass J, Peek CB, and Pagano M (2019). Cryptochromes-Mediated Inhibition of the CRL4Cop1-Complex Assembly Defines an Evolutionary Conserved Signaling Mechanism. Curr. Biol 29:1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone ML, Faigón Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P, et al. (2013). LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. U.S.A 110:12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, and McClung CR (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 Are Partially Redundant Genes Essential for the Temperature Responsiveness of the Arabidopsis Circadian Clock. Plant Cell 17:791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Michael TP, Kearns EV, G, F.-N.A., Sharrock R, and McClung RC (2002). The out of phase 1 Mutant Defines a Role for PHYB in Circadian Phase Control in Arabidopsis. Plant Physiol. 129:1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, and Kay SA (2016). The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harbor Perspectives in Biology 8:a027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow D, Kay S, and Imaizumi T (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer E, and Nagy F (2006). Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms. Dordrecht: Springer. [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, and Wisman E (2001). Microarray Analysis of Diurnal and Circadian-Regulated Genes in Arabidopsis. Plant Cell 13:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, and Coupland G (1998). The late elongated hypocotyl Mutation of Arabidopsis Disrupts Circadian Rhythms and the Photoperiodic Control of Flowering. Cell 93:1219–1229. [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, and Kay SA (2001). A Role for LKP2 in the Circadian Clock of Arabidopsis. Plant Cell 13:2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton DD, Toledo-Ortiz G, Ganpudi A, Kubota A, Imaizumi T, and Halliday KJ (2018). Dawn and photoperiod sensing by phytochrome A. Proc. Natl. Acad. Sci. U.S.A 115:10523–10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, and Mas P (2015). STRESSing the role of the plant circadian clock. Trends Plant Sci. 20:230–237. [DOI] [PubMed] [Google Scholar]
- Sharrock RA, and Clack T (2002). Patterns of Expression and Normalized Levels of the Five Arabidopsis Phytochromes. Plant Physiol. 130:442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, and Matsushita T (2014). Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 111:18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Paik I, Kangisser S, Green R, and Huq E (2017). PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytol 215:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, and Mas P (2018). A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes 9:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, and Kay SA (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488–1490. [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, and Geng R (2004). The F-Box Protein ZEITLUPE Confers Dosage-Dependent Control on the Circadian Clock, Photomorphogenesis, and Flowering Time. Plant Cell 16:769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, and Imaizumi T (2015). Photoperiodic Flowering: Time Measurement Mechanisms in Leaves. Annu. Rev. Plant Biol 66:441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, and Monte E (2016). Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A 113:4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Sánchez-Lamas M, Yanovsky MJ, Casal JJ, and Cerdán PD (2010). Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. U.S.A 107:4776–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase T, Nishiyama Y, Tanihigashi H, Ogura Y, Miyazaki Y, Yamada Y, and Kiyosue T (2011). LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 67:608–621. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Newton L, Burkhardt A, Mason S, and Farré EM (2014). Light and the circadian clock mediate time-specific changes in sensitivity to UV-B stress under light/dark cycles. J Exp Bot 65:6003–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman J, Zhu T, Chang H, Wang X, and Quail P (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. U.S.A. 98:9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Vandenbussche F, Laarhoven LJJ, Dowson-Day MJ, Wang Z-Y, Tobin EM, Harren FJM, Millar AJ, and Van Der Straeten D (2004). Circadian Rhythms of Ethylene Emission in Arabidopsis. Plant Physiol. 136:3751–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, and Harmon FG (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 107:3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, Kevei E, Hall A, Millar A, Nagy F, and Kozma-Bognár L (2001). Circadian Clock-Regulated Expression of Phytochrome and Cryptochrome Genes in Arabidopsis. Plant Physiol. 127:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima T, Hanada K, Gotoh E, Yamori W, Kodama Y, Tanaka H, Kusano M, Fukushima A, Tokizawa M, Yamamoto YY, et al. (2017). Light Controls Protein Localization through Phytochrome-Mediated Alternative Promoter Selection. Cell 171:1316–1325. [DOI] [PubMed] [Google Scholar]
- Viczián A, Kircher S, Fejes E, Millar AJ, Schäfer E, Kozma-Bognár L, and Nagy F (2005). Functional Characterization of Phytochrome Interacting Factor 3 for the Arabidopsis thaliana Circadian Clockwork. Plant and Cell Physiology 46:1591–1602. [DOI] [PubMed] [Google Scholar]
- Wang H, and Wang H (2015). Phytochrome Signaling: Time to Tighten up the Loose Ends. Molecular Plant 8:540–551. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu J-F, Nakamichi N, Sakakibara H, Nam H-G, and Wu S-H (2011). LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 Form a Positive Feedback Regulatory Loop in the Arabidopsis Circadian Clock. Plant Cell 23:486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, and Tobin EM (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93:1207–1217. [DOI] [PubMed] [Google Scholar]
- Webb AAR, Seki M, Satake A, and Caldana C (2019). Continuous dynamic adjustment of the plant circadian oscillator. Nature Communications 10:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenden B, Kozma-Bognár L, Edwards KD, Hall AJW, Locke JCW, and Millar AJ (2011). Light inputs shape the Arabidopsis circadian system. Plant J. 66:480–491. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, and Harberd NP (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-F, Tsai H-L, Joanito I, Wu Y-C, Chang C-W, Li Y-H, Wang Y, Hong JC, Chu J-W, Hsu C-P, et al. (2016). LWD–TCP complex activates the morning gene CCA1 in Arabidopsis. Nature Communications 7:13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, et al. (2014). LNK1 and LNK2 Are Transcriptional Coactivators in the Arabidopsis Circadian Oscillator. Plant Cell 26:2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, and Green RM (2009). Posttranslational Regulation of CIRCADIAN CLOCK ASSOCIATED1 in the Circadian Oscillator of Arabidopsis. Plant Physiol. 150:844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Matsushika A, Fujimori T, Sato S, Kato T, Tabata S, and Mizuno T (2003). A Link between Circadian-Controlled bHLH Factors and the APRR1/TOC1 Quintet in Arabidopsis thaliana. Plant and Cell Physiology 44:619–629. [DOI] [PubMed] [Google Scholar]
- Yang P, Wang J, Huang F-Y, Yang S, and Wu K (2018). The Plant Circadian Clock and Chromatin Modifications. Genes 9:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Liu B, Su J, Liao J, Lin C, and Oka Y (2017). Cryptochromes Orchestrate Transcription Regulation of Diverse Blue Light Responses in Plants. Photochemistry and Photobiology 93:112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Izaguirre M, Wagmaister JA, Gatz C, Jackson SD, Thomas B, and Casal JJ (2000a). Phytochrome A resets the circadian clock and delays tuber formation under long days in potato. Plant J. 23:223–232. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazzella MA, and Casal JJ (2000b). A quadruple photoreceptor mutant still keeps track of time. Curr. Biol 10:1013–1015. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazzella MA, Whitelam GC, Casal JJ (2001). Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J. Biol. Rhythms 16:523–530. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Whitelam GC, and Casal JJ (2000c). fhy3–1 retains inductive responses of phytochrome A. Plant Physiol. 123:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, and Kiyosue T (2004). Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot 55:2015–2027. [DOI] [PubMed] [Google Scholar]
- Yeom M, Kim H, Lim J, Shin A-Y, Hong S, Kim J-I, and Nam HG (2014). How Do Phytochromes Transmit the Light Quality Information to the Circadian Clock in Arabidopsis? Molecular Plant 7:1701–1704. [DOI] [PubMed] [Google Scholar]
- Yin R, and Ulm R (2017). How plants cope with UV-B: from perception to response. Curr Opin Plant Biol. 37:42–48. [DOI] [PubMed] [Google Scholar]
- Yu J-W, Rubio V, Lee N-Y, Bai S, Lee S-Y, Kim S-S, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Molecular Cell 32:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, and Lin C (2010). The Cryptochrome Blue Light Receptors. In: Arabidopsis Book: American Society of Plant Biologists; e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, and Meeks-Wagner DR (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10:691–702. [DOI] [PubMed] [Google Scholar]
- Zeugner A, Byrdin M, Bouly J-P, Bakrim N, Giovani B, Brettel K, and Ahmad M (2005). Light-induced Electron Transfer in Arabidopsis Cryptochrome-1 Correlates with in Vivo Function. Journal of Biological Chemistry 280:19437–19440. [DOI] [PubMed] [Google Scholar]
- Zhang C, Gao M, Seitz NC, Angel W, Hallworth A, Wiratan L, Darwish O, Alkharouf N, Dawit T, Lin D, et al. (2019a). LUX ARRHYTHMO mediates crosstalk between the circadian clock and defense in Arabidopsis. Nature Communications 10:2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bo C, and Wang L (2019b). Novel Crosstalks between Circadian Clock and Jasmonic Acid Pathway Finely Coordinate the Tradeoff among Plant Growth, Senescence and Defense. International Journal of Molecular Sciences 20:5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, and Quail PH (2013). A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in Arabidopsis. PLoS Genet 9:e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pfeiffer A, Tepperman JM, Dalton-Roesler J, Leivar P, Gonzalez Grandio E, and Quail PH (2020). Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc. Natl. Acad. Sci. U.S.A, doi: 10.1073/pnas.1918317117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jiang Y, Li J, Huq E, Chen ZJ, Xu D, and Deng XW (2018). COP1 SUPPRESSOR 4 promotes seedling photomorphogenesis by repressing CCA1 and PIF4 expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 115:11631–11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, Painter JE, Salomé PP, Straume M, and McClung R (1998). Imbibition, but not release from stratification, sets the circadian clock in Arabidopsis seedlings. Plant Cell 10:2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-Y, Oh E, Wang T, and Wang Z-Y (2016). TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nature Communications 7:13692. [DOI] [PMC free article] [PubMed] [Google Scholar]