High-throughput sequencing (HTS) of amplified fragments of rRNA genes provides unprecedented insight into the diversity of prokaryotic and eukaryotic microorganisms. Unfortunately, HTS data are prone to quantitative biases, which may lead to an erroneous picture of microbial community composition and thwart efforts to advance its understanding. These concerns motivated us to investigate how accurately HTS data characterize the variability of microbial communities, the relative abundances of specific phylotypes, and their relationships with environmental factors in comparison to an established microscopy-based method. We compared results obtained by HTS and catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) from three independent aquatic time series for both prokaryotic and eukaryotic microorganisms (almost 900 data points, the largest obtained with both methods so far). HTS and CARD-FISH data disagree with regard to relative abundances of bacterial and eukaryotic phylotypes but identify similar environmental drivers shaping bacterial and eukaryotic communities.
KEYWORDS: CARD-FISH, amplicon sequencing, bacterial communities, bacterial community structure, bacterial dynamics, eukaryotic communities, eukaryotic community structure, eukaryotic dynamics, microbial abundance, microbial communities, microbial community structure, microbial dynamics
ABSTRACT
High-throughput sequencing (HTS) of gene amplicons is a preferred method of assessing microbial community composition, because it rapidly provides information from a large number of samples at high taxonomic resolution and low costs. However, mock community studies show that HTS data poorly reflect the actual relative abundances of individual phylotypes, casting doubt on the reliability of subsequent statistical analysis and data interpretation. We investigated how accurately HTS data reflect the variability of bacterial and eukaryotic community composition and their relationship with environmental factors in natural samples. For this, we compared results of HTS from three independent aquatic time series (n = 883) with those from an established, quantitative microscopic method (catalyzed reporter deposition-fluorescence in situ hybridization [CARD-FISH]). Relative abundances obtained by CARD-FISH and HTS disagreed for most bacterial and eukaryotic phylotypes. Nevertheless, the two methods identified the same environmental drivers to shape bacterial and eukaryotic communities. Our results show that amplicon data do provide reliable information for their ecological interpretations. Yet, when studying specific phylogenetic groups, it is advisable to combine HTS with quantification using microscopy and/or the addition of internal standards.
IMPORTANCE High-throughput sequencing (HTS) of amplified fragments of rRNA genes provides unprecedented insight into the diversity of prokaryotic and eukaryotic microorganisms. Unfortunately, HTS data are prone to quantitative biases, which may lead to an erroneous picture of microbial community composition and thwart efforts to advance its understanding. These concerns motivated us to investigate how accurately HTS data characterize the variability of microbial communities, the relative abundances of specific phylotypes, and their relationships with environmental factors in comparison to an established microscopy-based method. We compared results obtained by HTS and catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) from three independent aquatic time series for both prokaryotic and eukaryotic microorganisms (almost 900 data points, the largest obtained with both methods so far). HTS and CARD-FISH data disagree with regard to relative abundances of bacterial and eukaryotic phylotypes but identify similar environmental drivers shaping bacterial and eukaryotic communities.
INTRODUCTION
High-throughput sequencing (HTS) of 16S and 18S rRNA gene amplicons has revolutionized microbiome research in environmental samples (1) because it allows for the unprecedented time- and cost-effective processing of large numbers of samples, providing data at high taxonomic resolution (2). Presently, HTS methods are a commonly used tool to study microbial communities in diverse environments (3–8). They provide data on the abundance of sequence reads affiliated with a particular phylotype relative to the total numbers of reads in the sample. In many studies, such data are treated as actual proportions of the studied microbes in the original habitat, and they are used to generate correlation-based hypotheses on the importance of environmental factors for specific microbial groups (9). Unfortunately, biases introduced during sample processing, such as DNA extraction (10), PCR amplification (11), and uneven coverage of primers across phylogenetic groups (12), result in low quantitative accuracy of amplicon data with respect to translating the relative abundance of a specific phylotype in a sequencing library to its contribution in the samples. This shortcoming has been repeatedly documented using mock communities (13–15) but is largely ignored in the absence of an alternative. It is further aggravated by the uneven distribution of rRNA operons in prokaryotes (1 to 15 copies of rRNA genes) (16) and even more pronounced in eukaryotes (1 to 315,000 copies of rRNA genes) (17, 18). While current pipelines for the analysis of amplicon data minimize errors arising from sequencing and chimera formation (19–22), none of the postsequencing bioinformatic analysis can mitigate the previously listed biases. Thus, statistical analysis based on the relative abundances of individual microbial lineages derived solely from HTS data may hinder, or even misguide, our understanding of microbial community dynamics and functioning.
Catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) provides estimates of relative abundance (percent contribution to total bacterial or eukaryotic numbers) of individual microbial lineages defined based on rRNA gene phylogeny (23, 24), and it is a verified quantitative tool in numerous studies on bacterial and eukaryotic communities (25–30). The accuracy of CARD-FISH may be compromised by imperfect probe coverage and specificity, uneven permeabilization across phylogenetic groups, differences in the presence of endogenous peroxidases between phylogenetic groups and environmental samples, poor detection of low abundance or inactive community members, and difficulties in counting aggregated cells (23, 24). Despite these limitations, relative abundances obtained with CARD-FISH corresponded well to the actual proportions of phylotypes in mock communities (31). The main advantage of CARD-FISH over the HTS methods is that the relative abundance of a particular lineage can be evaluated independent of the other taxa in the samples. Moreover, the CARD-FISH procedure can be separately optimized for each target group (probe), which is not possible for PCR with primers that target many different templates. Finally, CARD-FISH results can be readily combined with results from direct enumeration methods, such as microscopy or flow cytometry, to provide absolute abundance estimates of microbial lineages in the samples. Nevertheless, the considerably less labor-intensive HTS methods have largely replaced CARD-FISH for studies of microbial communities (32).
Regardless of their complexity, mock communities used for an assessment of accuracy of HTS methods are substantially simpler than natural communities. So far, comparative studies of HTS versus microscopic counts from environmental samples have focused on few phylotypes and/or were based on a low number of samples, yielding rather inconsistent results (33–39). Moreover, in the case of eukaryotes, HTS data have usually been compared with abundance data derived from morphological analyses, even though the correspondence between morphotypes and phylotypes is limited by sequence availability in repositories (36, 40).
Despite these limitations, changes in and differences between microbial communities are typically deduced only from proportions of read numbers and so are the proposed external factors potentially affecting them. The latter are often elucidated from statistical multidimensional correlation models based on dissimilarity between the samples (41). Although significant correlations between HTS and CARD-FISH data (33) indicate that the similarity matrixes calculated from both types of data might also agree, to our knowledge this has not been tested on larger data sets so far.
We analyzed three data sets from distinct aquatic habitats that were investigated in parallel by using 454 pyrosequencing with general bacterial or eukaryotic primers (HTS data) and by CARD-FISH, to evaluate the correspondence of these methods in estimating the composition and variability of microbial communities and their relationships with external factors. The eukaryotic data set consisted of 31 samples collected weekly from the Baltic Sea, analyzed by HTS and by CARD-FISH using 11 probes. The bacterial data sets originate from two high-frequency sampling campaigns in contrasting freshwater environments. The first one included 24 samples collected from the humic Jiřická Pond (Czech Republic), analyzed by HTS (V1-V3 region) and by CARD-FISH using 20 probes. The second data set consisted of 24 samples from the oligo-mesotrophic Lake Zurich (Switzerland), analyzed by HTS (V3-V5 region) and by CARD-FISH using 5 probes. All together, this yielded 883 data points (278 for eukaryotes and 605 for bacteria), representing the largest comparative data set from environmental studies available so far.
RESULTS AND DISCUSSION
Direct comparison between HTS and CARD-FISH data (relative abundance/biovolume).
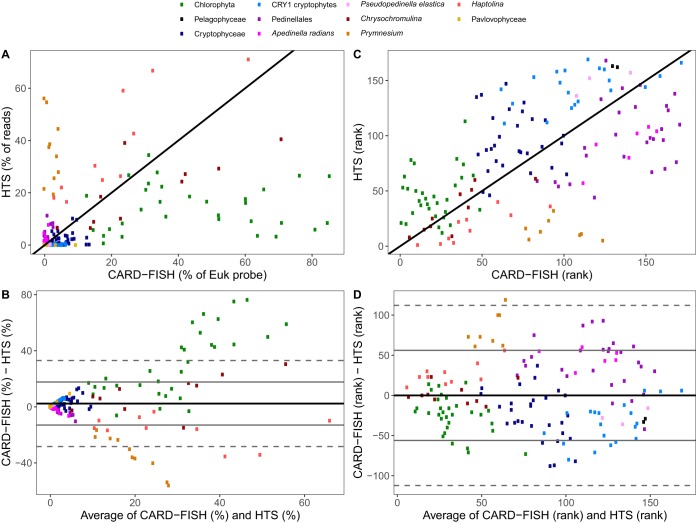
The general agreement between the relative abundances of eukaryotic taxa determined by either CARD-FISH or HTS was poor (Fig. 1A; see Fig. S1 in the supplemental material). Correlations were not significant, and regression slopes differed significantly from the value of 1 for most of the lineages except for the haptophyte genus Haptolina (Table 1). HTS data of freshwater pelagic ciliates correlated better with biomass than with abundance (40), likely due to a higher number of rRNA genes in larger species (18). Unfortunately, of the studied nanoplanktonic groups, this was the case only for cryptophytes (Table 1; Fig. S2 and S3). The agreement between the two approaches was also analyzed by plotting differences between the relative abundances of CARD-FISH and HTS data points against their means (42). The average difference between these two values was greater than zero, both for abundance (Fig. 1B) and biomass (Fig. S2), providing further evidence for rather poor correspondence between HTS and CARD-FISH data (relative abundance or biomass) for specific eukaryotic phylotypes.
FIG 1.
(A) Scatterplot of relative abundances of studied eukaryotic groups by 454 sequencing libraries (HTS) and CARD-FISH. (B) Scatterplot of differences between relative abundances of studied eukaryotic groups estimated by CARD-FISH and HTS against the average of the two values. (C) Scatterplot of ranked relative abundances of studied eukaryotic groups by HTS and CARD-FISH. (D) Scatterplot of differences between ranked relative abundances of studied eukaryotic groups estimated by CARD-FISH and HTS against the average of the two values. Black lines in panels A and C show a 1:1 relationship. Solid black lines in panels B and D show the average difference for the whole data set, solid gray lines show 1 standard deviation, and dashed gray lines show 2 standard deviations. Different eukaryotic groups are color coded. Individual plots for panels A and C are shown in Fig. S1 and S2 in the supplemental material, respectively.
TABLE 1.
Statistics for regressions and Spearman correlations between relative contributions to HTS or CARD-FISH dataa
| Group | Regression |
Spearman correlation |
n | ||||
|---|---|---|---|---|---|---|---|
| Adjusted r2 | Slope | P (r) | P value for slope of 1 | Rho | P (S) | ||
| Eukaryotes—abundance | |||||||
| Chlorophyta | −0.01 | 0.13 | 0.4420 | <0.0001 | 0.10 | 0.5881 | 31 |
| Pedinellales | 0.04 | 0.31 | 0.1608 | 0.0029 | 0.36 | 0.0580 | 28 |
| Cryptophyceae | −0.02 | −0.32 | 0.5409 | 0.0167 | 0.02 | 0.8991 | 30 |
| CRY1 cryptophytes | 0.04 | 0.46 | 0.1869 | 0.1295 | 0.47 | 0.0320 | 21 |
| Chrysochromulina | 0.62 | 0.62 | 0.0042 | 0.0411 | 0.92 | 0.0005 | 10 |
| Haptolina | 0.70 | 0.69 | 0.0015 | 0.0661 | 0.92 | 0.0005 | 10 |
| Prymnesium | −0.04 | −0.13 | 0.7570 | <0.0001 | −0.42 | 0.2696 | 9 |
| Eukaryotes—biomass | |||||||
| Chlorophyta | −0.02 | 0.12 | 0.4980 | <0.0001 | 0.16 | 0.3954 | 31 |
| Pedinellales | 0.11 | 0.42 | 0.0468 | 0.0084 | 0.45 | 0.0181 | 28 |
| Cryptophyceae | 0.23 | 0.95 | 0.0043 | 0.8612 | 0.54 | 0.0024 | 30 |
| CRY1 cryptophytes | 0.08 | 0.46 | 0.1138 | 0.0644 | 0.49 | 0.0258 | 21 |
| Chrysochromulina | 0.53 | 0.58 | 0.0101 | 0.0419 | 0.81 | 0.0082 | 10 |
| Haptolina | 0.53 | 0.62 | 0.0106 | 0.0774 | 0.75 | 0.0184 | 10 |
| Prymnesium | −0.14 | −0.01 | 0.9410 | 0.0005 | −0.23 | 0.5517 | 9 |
| Bacteria—Jiřická Pond | |||||||
| Alphaproteobacteria | −0.01 | −0.38 | 0.4043 | 0.0053 | −0.27 | 0.2094 | 24 |
| Actinobacteria | 0.48 | 0.47 | <0.0001 | <0.0001 | 0.72 | 0.0001 | 24 |
| “Ca. Nanopelagicales” | 0.90 | 0.74 | <0.0001 | <0.0001 | 0.93 | <0.0001 | 24 |
| Luna-2 cluster, Actinobacteria | 0.17 | 0.51 | 0.0279 | 0.0354 | 0.39 | 0.0681 | 23 |
| “Ca. Nanopelagicus” | 0.71 | 0.73 | <0.0001 | 0.0146 | 0.84 | <0.0001 | 23 |
| “Ca. Planktophila versatilis” | 0.20 | 0.83 | 0.0374 | 0.6432 | 0.81 | <0.0001 | 18 |
| Bacteroidetes | 0.23 | 0.52 | 0.0100 | 0.0149 | 0.45 | 0.0277 | 24 |
| Betaproteobacteria | 0.35 | 0.59 | 0.0015 | 0.0187 | 0.56 | 0.0055 | 24 |
| Uncult. lineage GKS98 Betaproteobacteria |
0.46 | 0.71 | 0.0002 | 0.0758 | 0.71 | 0.0002 | 24 |
| Limnohabitans cluster LimA | 0.59 | 0.73 | <0.0001 | 0.0408 | 0.76 | <0.0001 | 24 |
| Limnohabitans cluster LimB | −0.05 | −0.08 | 0.8340 | 0.0078 | 0.59 | 0.0036 | 22 |
| Limnohabitans clusters LimBCD | 0.03 | −0.19 | 0.2100 | <0.0001 | −0.39 | 0.0808 | 21 |
| All Limnohabitans | 0.35 | 0.66 | 0.0013 | 0.0684 | 0.60 | 0.0020 | 24 |
| Methylophilaceae | 0.19 | 0.64 | 0.0190 | 0.1770 | 0.31 | 0.1433 | 24 |
| Polynucleobacter clusters PnecABD | 0.70 | 1.04 | <0.0001 | 0.7979 | 0.89 | <0.0001 | 24 |
| Polynucleobacter cluster PnecC | 0.76 | 0.72 | <0.0001 | 0.0034 | 0.57 | 0.0046 | 24 |
| “Ca. Methylopumilus turicensis” | −0.03 | 0.15 | 0.5310 | 0.0020 | −0.15 | 0.4739 | 24 |
| Opitutae | 0.65 | 0.98 | <0.0001 | 0.8775 | 0.81 | <0.0001 | 24 |
| Verrucomicrobia (excluding Opitutae) | 0.04 | 0.69 | 0.1795 | 0.5489 | 0.01 | 0.9597 | 24 |
| All Verrucomicrobia | 0.67 | 1.28 | <0.0001 | 0.1508 | 0.70 | 0.0002 | 24 |
| Bacteria—Lake Zurich | |||||||
| Bacteroidetes | 0.67 | 0.82 | <0.0001 | 0.1430 | 0.75 | <0.0001 | 24 |
| “Ca. Nanopelagicales” | −0.02 | −0.06 | 0.44 | <0.0001 | −0.17 | 0.4241 | 24 |
| “Ca. Methylopumilus planktonicus” | −0.03 | 0.12 | 0.56 | 0.0002 | 0.02 | 0.9164 | 24 |
| Polynucleobacter cluster PnecB | 0.17 | 0.73 | 0.025 | 0.3874 | 0.52 | 0.0100 | 24 |
Regression statistics include adjusted r2, slope value, and significance level (P [r]) and Spearman correlation statistics include rho and significance level (P [S]) between relative contributions (percentages) to HTS or CARD-FISH data. A P value slope of 1 indicates a significance level against the desired value of 1, while a P value slope of >0.05 indicates that the slope is not significantly different from 1. n, number of data points for each group. Uncult., uncultured.
Individual scatter plots of relative abundances of studied eukaryotic groups by 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S1, PDF file, 0.1 MB (151.2KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Scatterplot of relative biovolumes of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. (B) Scatterplot of ranked relative biovolumes of studied eukaryotic groups from HTS and CARD-FISH. (C) Scatterplot of differences between relative biovolumes of studied eukaryotic groups estimated by CARD-FISH and HTS against the average of the two values. (D) Scatterplot of differences between ranked relative biovolumes of studied eukaryotic groups estimated by CARD-FISH and HTS against the average of the two values. Black lines in panels A and B show a 1:1 relationship. Solid black lines in panels C and D show average differences for the whole data set, solid gray lines show 1 standard deviation, and dashed gray lines show 2 standard deviations. Different eukaryotic groups are color coded. Individual plots for panels A and B are shown in Fig. S3 and S4, respectively. Download FIG S2, PDF file, 0.2 MB (243.5KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of relative biovolumes of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S3, PDF file, 0.1 MB (155.9KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The agreement between HTS and CARD-FISH was better when samples in each data set were ranked from highest to lowest relative abundance (Fig. 1C and D; Fig. S4 and S5). The Spearman rank correlations were strong for most eukaryotic groups and significant for biovolume (Table 1). These findings speak for the use of nonparametric methods for the analysis of HTS data (9).
Individual scatterplots of rank relative biovolumes of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. Samples with zeroes in both CARD-FISH and HTS data were removed, because they were arbitrarily ranked. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S4, PDF file, 0.1 MB (124.5KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of rank relative abundances of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. Samples with zeroes in both CARD-FISH and HTS data were removed, because they were arbitrarily ranked. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S5, PDF file, 0.1 MB (127.3KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
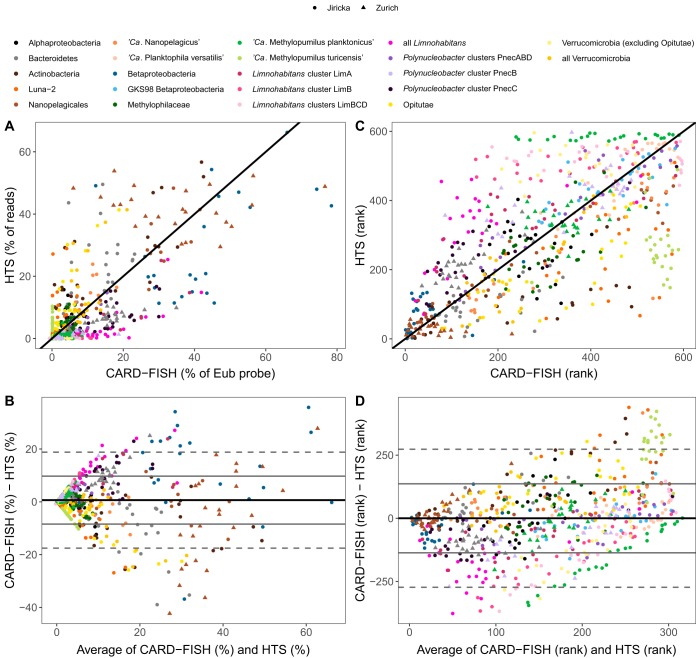
In general, the agreement between the two approaches was better for bacteria than for eukaryotes, for both relative and rank data (Fig. 2; Table 1). Most bacterial groups were underrepresented in HTS data compared to CARD-FISH data, e.g., different lineages of Limnohabitans, “Candidatus Methylopumilus planktonicus,” Polynucleobacter subclusters B and C, “Ca. Planktophila vernalis”, the uncultivated Betaproteobacteria lineage GKS98 and Verrucomicrobia (without Opitutae). In contrast, “Ca. Methylopumilus turicensis.” Opitutae, or the Luna-2 cluster of Actinobacteria were overrepresented in HTS data compared to CARD-FISH data (Table 1; Fig. S6). Such phylotype-dependent agreement between the HTS and CARD-FISH data was also observed in marine mesocosms (43), indicating that a simple interpretation of HTS data in terms of relative abundance should be avoided for bacteria as well.
FIG 2.
(A) Scatterplot of relative abundances of studied bacterial groups (pooled data sets from both lakes) by 454 sequencing libraries (HTS) and CARD-FISH. (B) Scatterplot of differences between relative abundances of studied bacterial groups estimated by CARD-FISH and HTS against the average of the two values. (C) Scatterplot of ranked relative abundances of studied bacterial groups by HTS and CARD-FISH. (D) Scatterplot of differences between ranked relative abundances of studied bacterial groups estimated by CARD-FISH and HTS against the average of the two values. Black lines in panels A and C show a 1:1 relationship. Solid black lines in panels B and D show average differences for the whole data set, solid gray lines show 1 standard deviation, and dashed gray lines show 2 standard deviations. Different bacterial groups are color coded, and lakes of sample collection are indicated by shape. Individual plots for panels A and C are shown in Fig. S6 and S7, respectively.
Individual scatterplots of relative abundances of studied bacterial groups by 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S6, PDF file, 0.3 MB (343.3KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of rank relative abundances of studied bacterial groups from 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S7, PDF file, 0.3 MB (363.4KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
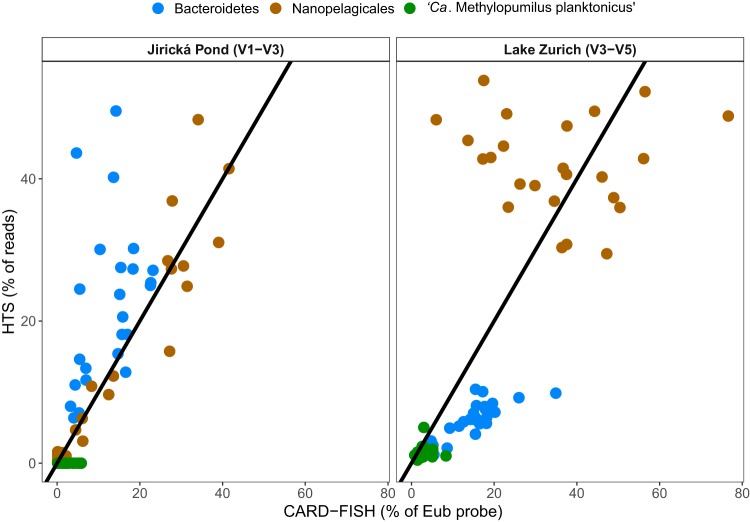
Considerably higher agreement between the two approaches was obtained when rank-transformed relative abundances were used (Fig. 2C and D), especially for “Ca. Nanopelagicus,” Polynucleobacter cluster A, “Ca. Planktophila vernalis,” and the GKS98 lineage (Table 1; Fig. S7). Interestingly, there was a site-specific difference for “Ca. Nanopelagicales” (Actinobacteria), which yielded a good correspondence for samples from Jiřická Pond but a poor one for Lake Zurich (Fig. 3). The opposite was observed for “Ca. Methylopumilus planktonicus,” which was not detected at all by HTS in Jiřická Pond. These incongruities could not be explained by discrepancies in the respective coverage of the different primer sets used for generating the two data sets, as both primer pairs displayed very good in silico coverages of these two bacterial groups (90.9% versus 92.9% for “Ca. Nanopelagicales” and 98.3% versus 90.9% for “Ca. Methylopumilus planktonicus”) (Tables S1 and S2).
FIG 3.
Scatterplots of relative abundances of the same bacterial groups by 454 sequencing libraries (HTS) and CARD-FISH in Jiřická Pond and Lake Zurich. HTS data for each lake were generated with a different primer set (Table S1). Black lines show 1:1 relationship.
List of primers used in this study. Download Table S1, PDF file, 0.3 MB (324KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The overall poor agreement between relative abundances derived from HTS and CARD-FISH could not be explained by different specificities of probes and primers. On the one hand, the 10-fold-higher HTS-derived relative abundance of the haptophyte genus Prymnesium (Fig. S1), detection of which is a major concern in areas where it forms toxic blooms (44), could be attributed to the lower coverage of the group by the CARD-FISH probe than by the primer (70.3% and 87.2%, respectively) (Tables S1 and S2). On the other hand, Pavlovophyceae, whose coverages by probe and primers are very similar (87.2% and 83.0%, respectively) (Tables S1 and S2), were completely undetected by sequencing (Fig. S1). All bacterial lineages that were overrepresented in the HTS data set displayed comparable or even slightly higher in silico coverages of CARD-FISH probes than of HTS primers (“Ca. M. turicensis,” Opitutae, “Ca. Nanopelagicus,” and Actinobacteria of the Luna-2 lineage) (Tables S1 and S2). Likewise, some lineages that were overrepresented by CARD-FISH had a much higher coverage with general PCR primers (e.g., Verrucomicrobia excluding Opitutae, the Betaproteobacteria lineage GKS98), while coverage was very similar in others (e.g., “Ca. Planktophila vernalis,” Polynucleobacter lineage PnecC) (Tables S1 and S2). In fact, agreement between the differences in coverage by HTS primers (80%) and CARD-FISH probes (100%) and of the relative abundances detected by either approach was found only in Polynucleobacter lineage PnecB. In any case, differences in coverage between probes and primers cannot explain phenomena such as the good agreement between HTS and CARD-FISH for “Ca. Nanopelagicales” in Jiřická Pond but not in Lake Zurich (Fig. 3), as the group coverages of both HTS primer sets were very similar (92.2% and 90.9%, respectively) (Table S1) and much higher than that of the probe (68.0%) (Table S2). This indicates the importance of other, unknown biases besides primer and probe coverage.
List of probes, helpers, and competitors used in this study. HB (%), concentration of formamide in the hybridization buffer; T, hybridization temperature (washing temperature was 2°C higher); Probe, coverage (in percent of targeted sequences) of the probe; Primers 1, coverage of primers TAReuk454FWD1 and TAReukREV3 (eukaryotes) or 341F and 907R (bacteria, Lake Zurich); Primers 2, coverage of primers TAReuk454FWD1 and HaptoR1 (eukaryotes) or 27F andUni522R (bacteria, Jiřická Pond). The numbers are based on search in the reference Silva database release 132, conducted on 23 November 2019. Download Table S2, PDF file, 0.6 MB (618KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Limitations connected with PCR biases and imperfect probe and primer coverage and specificity may be potentially overcome by the use of taxonomically profiled metagenomic data to estimate relative abundances of specific groups, but so far tests with mock communities have suggested otherwise (45). Recently, an addition of known amounts of Escherichia coli cells has been proposed as an internal standard for amplicon read normalization in freshwater bacterial communities (31). This approach provided substantially improved estimates for relative changes of phylotype contributions between samples compared to those of nonnormalized reads. It would be valuable to explore its applicability for eukaryotes as well.
Comparison of statistical models.
The above results provide further evidence that the relative abundances of phylotypes as derived from HTS data should not be directly translated into the proportions or biovolumes of cells from these lineages in a sample. However, the generally better agreement of rank data suggests that nonparametric distance-based ordination methods might be appropriate for the analysis of HTS data, e.g., to study differences between microbial communities in different habitats (9). To test this hypothesis, we calculated Bray-Curtis distance matrices for both CARD-FISH and HTS data for all data sets and compared those using two-tailed Mantel tests. For eukaryotes, we found a weak but significant nonparametric correlation between relative abundances derived from HTS and both relative abundances (Spearman’s rho = 0.1533, P < 0.0001) and relative biovolumes determined by CARD-FISH (rho = 0.1083, P = 0.0211). Similar results were obtained for bacteria from Lake Zurich, where relative abundances of HTS and CARD-FISH also correlated weakly but significantly (rho = 0.2025, P = 0.0005). In contrast, very strong and significant correlations of HTS and CARD-FISH data were observed for bacteria from Jiřická Pond (rho = 0.8104, P < 0.0001).
Multivariate methods are often used to generate correlation-based hypotheses about the respective importance of different environmental variables for microbial community dynamics. We analyzed both CARD-FISH and HTS data with distance-based linear models (DistML). The agreement was very good for the eukaryotic data set, as in both cases DistML pointed to soluble reactive phosphorus (SRP) as the only explanatory variable (Table 2). However, this result was largely driven by a single outlier sample, for which we observed elevated SRP concentrations and a massive bloom of the dinoflagellate Heterocapsa triquetra (46). The abundance of nanophytoplankton was substantially lower in this sample, and the sequencing library was dominated by reads from the dinoflagellate. When this sample was excluded from the analysis, a combination of SRP and temperature best explained the variability of CARD-FISH relative abundance data, but there was no significant model for the HTS data. Interestingly, this agreed with the CARD-FISH biovolume data, for which a significant model was not found in both cases (Table 2). This imperfect agreement between the models can be partially attributed to the fact that eukaryotic abundance and biovolume may respond differently to changing conditions (46). In general, it seems that statistical models calculated from HTS and CARD-FISH data may identify the same environmental drivers affecting eukaryotic microbial communities, but it is advisable to combine and calibrate HTS with microscopic methods during experiments aimed to test hypotheses derived from statistical models on observational data.
TABLE 2.
DistML models for the eukaryotic data set calculated from HTS and CARD-FISH data (relative abundance and biovolume)
| Sample(s) | Variablea | DistML model for data calculated from: |
||
|---|---|---|---|---|
| HTS: relative abundance [P value (% explained variation)] |
CARD-FISH |
|||
| Relative abundance [P value (% explained variation)] |
Relative biovolume | |||
| All samples | SRP | 0.0276 (13.5) | 0.0165 (12.8) | No significant model |
| No outlier sample | SRP | No significant model | 0.0015 (14.2) | No significant model |
| Temp | 0.0072 (14.5) | |||
SRP, soluble reactive phosphorus.
Excellent agreement between HTS and CARD-FISH was found in the bacterial data sets. In the case of Lake Zurich, the patterns of relative abundance derived by either approach pointed to temperature and abundance of virus-like particles (VLP) as the best explanatory variables (Table 3). For Jiřická Pond, all models included dissolved organic carbon, total phosphorus, water residence time, and either dissolved nitrogen (HTS) or chlorophyll a (CARD-FISH) (Table 3). This almost perfect correspondence indicates that use of distance-based multivariate analyses for bacterial amplicon HTS data allows for the generation of models and hypotheses similar to those obtained from relative abundance data from CARD-FISH.
TABLE 3.
DistML models for bacterial data sets calculated from HTS and CARD-FISH data
| Sampling site | Variablea | DistML model for data calculated from: |
|
|---|---|---|---|
| HTS: relative abundance [P value (% explained variation)] |
CARD-FISH: relative abundance [P value (% explained variation)] | ||
| Lake Zurich | Temp | 0.0001 (42.9) | 0.0097 (18.9) |
| VLP | 0.0092 (14.1) | 0.05 (11.3) | |
| Jiřická Pond | WRT 0.5m | 0.0001 (29.4) | 0.0004 (10.8) |
| DOC | 0.0001 (18.9) | 0.0001 (37.3) | |
| TP | 0.0002 (14.9) | 0.0009 (15.9) | |
| DN | 0.0061 (6.4) | ||
| Chl-a 0.5m | 0.0231 (5.1) | ||
VLP, abundance of virus-like particles; WRT 0.5m, water residence time at 0.5-m depth; DOC, dissolved organic carbon; TP, total phosphorus; DN, dissolved nitrogen; Chl-a 0.5m, chlorophyll a at 0.5-m depth.
Caveats of the study.
Our HTS data are based on the pyrosequencing 454 method (Roche) that has been replaced by newer platforms that provide sequencing depth orders of magnitude higher, such as Illumina or Oxford Nanopore. However, as the main biases arise from DNA extraction, PCR amplification, and uneven 16S and 18S rRNA gene copy numbers (47), these newer methods will not necessarily improve the quantitative accuracy of the sequencing data, as shown with mock communities sequenced using Illumina (13–15) and Oxford Nanopore (48, 49) platforms. In contrast, the relative abundances of sequences obtained from the same samples by both pyrosequencing and Illumina correlated very strongly (r2 > 0.99) (50). It has been shown that 3,000 reads per sample are sufficient to capture >90% of alpha-diversity in samples from freshwater lakes and to reveal beta-diversity patterns (2). In our study, the lowest number of reads per sample was 1,724 for the eukaryotic data set (average, 4,742), 34,020 for the bacterial data set from Jiřická Pond (average, 68,450), and 3,877 for the bacterial data set from Lake Zurich (average, 11,863). Finally, although it cannot be completely excluded that we missed some reads of phylotypes targeted by the CARD-FISH probes by using 454 pyrosequencing instead of Illumina, rarefaction analysis indicated that most would belong to the rare biosphere (Fig. S8). All of these suggest that sequencing depth was sufficient to capture most of the diversity in our samples. Thus, our conclusions are not considerably affected by lower sequencing depth and likely apply to all nonnormalized PCR-based sequencing methods.
Individual rarefaction curves for samples from the Baltic Sea (eukaryotes) (A), Jiřická Pond (B), and Lake Zurich (C). Download FIG S8, PDF file, 0.2 MB (217.8KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mock community studies have pointed out the importance of PCR conditions for the accurate recovery of bacterial lineages (primer choice, annealing conditions, polymerase type, and number of cycles) (13–15, 47). The PCR conditions used here were standard at the time of the study (51) but have since then been shown to decrease quantitative accuracy and increase chimera formation (52). However, only very few chimeras were detected in our data sets (eukaryotes, 1.2% of operational taxonomic units [OTUs]; bacteria in Jiřická Pond, 9.8% of OTUs; bacteria in Lake Zurich, 5.7% of OTUs). The phylotype-dependent agreement between HTS and CARD-FISH data (Fig. 1 and 3; Fig. S1 to S7) indicates that a template-dependent PCR bias might have dominated in our samples (47). Finally, it has been shown that even optimized PCR conditions do not reproduce original communities with perfect qualitative accuracy (13, 47). All together, although the use of fewer PCR cycles and proofreading polymerase might have arguably improved the agreement (correlations) between the two methods, our main conclusions likely remain unaffected.
Conclusions.
Our study presents the largest data set comparing HTS and CARD-FISH data from natural samples (almost 900 data points) to date. It expands previous observations derived from mock communities, i.e., that the relative abundances of specific phylotypes obtained by HTS may not necessarily correspond to their relative abundances in the original samples. Despite this limitation, we show that nonparametric distance-based multivariate analyses based on HTS and CARD-FISH data often agree and thus seem to allow for reliable ecological interpretations of the relationship between microbial community structure and environmental parameters. This appears to work especially well under conditions that cause substantial changes in community composition, as observed for Jiřická Pond. In summary, it appears that studies focusing on the relationship of whole microbial communities with environmental variables can perhaps rely solely on HTS data. In contrast, we recommend that sequence-based community analysis (optimally using internal standards) be combined with CARD-FISH when aiming at more accurate estimates of abundances or biomass of specific bacterial taxa or when studying eukaryotes.
MATERIALS AND METHODS
Eukaryotes. (i) Sample collection.
Coastal waters of the Gulf of Gdańsk (Baltic Sea) were sampled weekly from 12 April to 7 November 2012. Twenty liters of surface seawater was prefiltered through a 20-μm net and transported to the laboratory within 15 min in a darkened, closed container. Temperature and salinity were measured in situ with an InoLab probe (WTW).
Biomass for amplicon sequencing was collected from 0.8 to 4.6 liters of sampled water filtered onto polyethersulfone filters (0.22-μm pore size, 47-mm diameter; GPWP04700; Millipore-Merck KGaA, Darmstadt, Germany) under aseptic conditions. The filters were stored at –80°C.
For CARD-FISH, a 200-ml subsample was fixed by the Lugol-formalin-sodium thiosulfate method recommended for preservation of fragile protists (53). Fixed samples were stored in the dark at 4°C for 16 h, filtered onto white polycarbonate filters (0.8-μm pore size, 47-mm diameter; Cyclopore; Whatmann, Maidstone, UK) under low pressure (<2 × 104 Pa), rinsed with 50 ml of sterile MilliQ water, air dried, and stored at –20°C.
(ii) Environmental variables.
Concentrations of dissolved inorganic nitrogen (DIN) as well as soluble reactive phosphorus (SRP) and dissolved silicate (DSi) were determined by methods recommended for the Baltic Sea (54). For this purpose, 500 ml of water was collected in an acid-cleaned container, frozen at –20°C, and analyzed within 1 month.
(iii) DNA extraction and sequencing.
DNA was extracted using a PowerWater DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA). Extracted DNA samples were processed by Research and Testing Laboratories (Lubbock, TX, USA). The V-4 fragments of 18S rRNA genes were amplified with TAReuk454FWD1 and TAReukREV3 (see Table S1 in the supplemental material). Amplifications were performed in 25-μl reaction volumes with recombinant Hot Start Taq polymerase (Qiagen HotStarTaq master mix; Qiagen, Inc., Valencia, CA, USA), 1 μl of each 5 μM primer, and 1 μl of template on ABI Veriti thermocyclers (Applied Biosytems, Carlsbad, CA, USA) under the following thermal profile: 95°C for 5 min, followed by 10 cycles of 94°C for 30 s, 57°C for 45 s, and 72°C for 1 min and then 25 additional cycles of 94°C for 30 s, 45°C for 45 s, and 72°C for 1 min, and a final 2-min extension at 72°C (51). As the reverse primer TAReukREV3 poorly targets haptophytes, we additionally sequenced samples with high haptophyte abundance (23 May to 30 July) using the reverse primer HaptoR1 (Table S1) under the following thermal profile: 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min, and a final 2-min extension at 72°C (55). The amplicons were sequenced using the Roche 454 GS FLX Titanium platform with an average sequencing depth of 10,000 raw reads per sample.
(iv) CARD-FISH.
The CARD-FISH procedure was performed with Alexa 488-labeled tyramides (Molecular Probes, Thermo Fisher Scientific, Waltham, MA, USA), as previously described (56), and analyzed manually using 10 to 20 microphotographs randomly taken by epifluorescence microscopy at ×1,000 magnification (AxioVision.M1; Carl Zeiss, Jena, Germany). Biovolume was estimated by multiplying cell abundance by average cell volume, which was calculated based on manual measurements of cell width and length and assuming the cell shape to be prolate spheroid, as described by Piwosz in 2019 (46). The relative abundance of an individual lineage was calculated as the proportion of cells hybridized with the specific probe to that of cells hybridized with the general eukaryotic probe. A full list of applied probes (n = 11) can be found in Table S2.
(v) Bioinformatics analysis.
Sequences were analyzed using a custom-made pipeline as previously described (57). Raw sff flowgrams were denoised using AmpliconNoise (52). The demultiplexed and primer-free reads were quality filtered and trimmed to a length of 250 bp using USEARCH (58) (bases with a Phred score of <30 were trimmed), and chimeric sequences were discarded with UCHIME (59). OTUs were clustered by average linkage at similarity levels of 97% upon the pairwise alignment by the Needleman-Wunsch algorithm. The most closely related sequence for each OTU was identified using pairwise alignment to the curated eukaryotic PR2 reference data (60), and the corresponding taxonomic information, together with the coverage and dissimilarity to the query sequence, was assigned. The final number of reads in samples ranged from 1,707 to 15,233.
Bacteria. (i) Sample collection.
Jiřická Pond is a shallow, humic pond in the southern region of the Czech Republic and is characterized by short-term flooding events, severely shortening its hydraulic retention time, which triggers sudden fluctuations in microbial communities (61, 62). An intensive sampling campaign took place between 5 May and 27 June 2014, with samples taken three times per week. Water samples from a 0.5-m depth were taken with a Friedinger sampler and split into subsamples. Samples for prokaryotic cell counts and CARD-FISH were fixed with formalin (2%, vol/vol). Fixed subsamples for CARD-FISH were filtered onto white polycarbonate filters within 16 h after sampling (0.2-μm pore size, 47-mm diameter; Millipore-Merck KGaA, Darmstadt, Germany) and stored at –20°C. Samples for enumeration of virus-like particles (VLP) were fixed with glutaraldehyde (0.5%, vol/vol) for 10 min, flash-frozen in liquid nitrogen, and stored at –80°C until evaluation via flow cytometry (63). Prokaryotic biomass for amplicon sequencing was collected on polysulfone Sterivex filters (0.22-μm pore size; Millipore-Merck KGaA, Darmstadt, Germany). Additionally, 2 liters of water was taken for chemical analyses. These samples were delivered in a ThermoBox to the laboratory and analyzed within 24 h.
For Lake Zurich, a longitudinal transect of eight sampling stations along Lake Zurich (26) and the connected Upper Lake was sampled in summer 2010 (27 and 28 July). Vertical profiles of temperature, conductivity, turbidity, and concentrations of oxygen and chlorophyll a (differentiating pigments of diatoms and Planktothrix rubescens [64]) were recorded with a YSI multiprobe (model 6600; Yellow Springs Instruments, Yellow Springs, OH, USA) and a bbe FluoroProbe (TS-16-12; bbe Moldaenke GmbH, Schwentinental, Germany), respectively. Water samples from three different depths representing the epilimnion (2 to 5 m), metalimnion (12.5 to 15 m), and hypolimnion (20 m) were taken with a Friedinger sampler for each sampling station and split in subsamples for (i) total counts of prokaryotes, (ii) VLP, (iii) CARD-FISH analyses, (iv) prokaryotic biomass for amplicon sequencing, and (v) chemical analyses. Subsamples (i) were fixed with formalin (2% vol/vol) and stored at 4°C. Subsamples (ii) and (iii) were processed as described above for Jiřická Pond. Subsamples (iv) (600 ml) were filtered on the same day onto polysulfone filters (0.2-μm pore size, 47-mm diameter; Millipore-Merck KGaA, Darmstadt, Germany) and stored at –80°C.
(ii) Environmental variables.
For Jiřická Pond, water temperature, water retention time, and dissolved organic carbon (DOC) and chlorophyll a concentrations at 0.5-m depth were assessed as previously described (61). Concentrations of nitrate, nitrite, and ammonium ions were determined by ion chromatography (IC25; Dionex, USA). Values of total and dissolved phosphorus (TP and DP, respectively) were measured as described by Porcal and Kopáček (62). Dissolved nitrogen (DN) concentrations were obtained using a vario TOC cube (Elementar, Germany).
For Lake Zurich, concentrations of TP, DP, DOC, and different nitrogen species were determined by standard techniques by the Zurich Water Supply Company.
(iii) DNA extraction and sequencing.
For Jiřická Pond, nucleic acid isolation was conducted using phenol-chloroform-isoamyl alcohol extraction according to a previously described protocol (65). The variable regions V1-V3 of the 16S rRNA gene were amplified with primers 27Fand Uni522R (Table S1). A single-step PCR using a HotStarTaq Plus master mix kit (Qiagen, Inc., Valencia, CA, USA) was conducted using the following profile: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, and a final elongation step at 72°C for 5 min. After PCR, all amplicon products were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). The amplicons were sequenced using the Roche 454 GS FLX Titanium platform at MR DNA laboratory (Shallowater, TX, USA) with an overage sequencing depth of 50,000 raw reads per sample.
For Lake Zurich, DNA was isolated with a PowerWater DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA). Extracted DNA samples were processed by Research and Testing Laboratories (Lubbock, TX, USA). V3-V5 fragments of 16S rRNA genes were amplified with primers 341F and 907R (Table S1). Amplifications were performed in 25-μl reaction mixtures with recombinant HotStart Taq polymerase (Qiagen HotStarTaq master mix; Qiagen, Inc., Valencia, CA), 1 μl of each 5 μM primer, and 1 μl of template on ABI Veriti thermocyclers (Applied Biosystems, Carlsbad, CA) under the following thermal profile: 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 54°C for 40 s, and 72°C for 1 min, followed by a final 10-min extension at 72°C. The amplicons were sequenced using the Roche 454 GS FLX Titanium platform with an average sequencing depth of 10,000 reads per sample.
(iv) CARD-FISH.
CARD-FISH for bacteria was carried out as previously described with fluorescein-labeled tyramides (66) and analyzed with a fully automated microscope (AxioImager.Z1; Carl Zeiss) as outlined by Salcher et al. (67). The relative abundance of an individual lineage was calculated as the proportion of cells hybridized with the specific probe to that of cells hybridized with the general bacterial probe. A full list of applied probes (n = 20) is provided in Table S2.
(v) Bioinformatics analysis.
The demultiplexed and primer-free reads were quality filtered and trimmed to a length of 350 bp according to quality report using USEARCH (58). Chimeric sequences were detected and discarded using UCHIIME (59). OTUs were clustered at similarity levels of 97% using the UPARSE-OTU algorithm (68). A taxonomical assignment for representative sequences for each OTU was done with a parallel BLAST search against the SILVA-database SSURef_NR99_132 (69). The final numbers of reads in samples ranged from 34,020 to 104,696 for samples from Jiřická Pond and from 3,877 to 25,031 for samples from Lake Zurich. Data sets were rarefied to the smallest sample prior to statistical analysis.
Statistical analysis.
The read numbers of all OTUs affiliated with lineages that corresponded to those targeted by probes were pooled, and their percent contributions to the total number of reads in each sample were compared with relative abundances (and biovolumes for eukaryotes) estimated by CARD-FISH. Relative abundances and biovolumes of individual eukaryotic or bacterial lineages were calculated as percentages of all hybridized cells (i.e., counts with general eukaryotic [Euk516] or bacterial [EubI-III] probes, respectively). The agreement between the two methods was assessed using graphical techniques, as described by Bland and Altman (42). The same methods were used to compare sample rankings by HTS and CARD-FISH. In addition, linear regressions and Spearman correlations were calculated between relative abundances derived from HTS and CARD-FISH. Multiple null values in data obtained using one of the approaches were pooled, and an average value was calculated for the second approach (i.e., if in HTS data there were three data points with null values, an average value for CARD-FISH data for these three points was calculated). The calculations were performed in the R environment version 3.3.3 (70), and the figures were prepared using functions from the ggplot2 package version 3.2.0 (71) and the ggpubr package version 0.2.1. Mantel tests were performed with XLSTAT 14 (Addinsoft) to determine Spearman correlations of proximity matrices calculated using the Bray-Curtis dissimilarity algorithm.
Correlations with environmental variables.
The relationships between environmental data and the relative abundances of studied bacterial and eukaryotic groups were analyzed by Bray-Curtis dissimilarity distance-based linear models (DistML) (72) in the PERMANOVA+ add-on package of the PRIMER7 software (Primer Ltd., Plymouth, UK). Environmental variables were normalized, and a correlation matrix for the whole set was calculated. From the variables that were strongly correlated (the absolute value of the correlation coefficient was >0.7), only one was chosen for further analysis. Analyses were performed on untransformed relative abundance data using a stepwise selection procedure, and the best model was selected based on the statistical significance (9,999 permutations) and the values of the Akaike’s information criterion (AIC) and the Bayesian information criterion (BIC).
Data availability.
The eukaryotic HTS data obtained with the general primers (TAReuk454FWD1 and TAReukREV3) were deposited in the ENA database under BioProject no. PRJEB23971, and those obtained with primers TAReuk454FWD1 and HaptoR1 were deposited under BioProject no. PRJEB31858. Bacterial HTS data from Jiřická Pond were deposited in NCBI as BioSamples SAMN11974970 to SAMN11974993 as part of BioProject PRJNA547706, and those from Lake Zurich were deposited under BioProject no. PRJNA545726.
ACKNOWLEDGMENTS
We thank Hanna Wróblewska from the National Marine Fisheries Research Institute (Poland) for analyses of nutrients from the Baltic Sea and Jörg Villiger and Eugen Loher from the University of Zurich (Switzerland) for processing the sequence data and help with sampling of Lake Zurich.
The study was supported by the following projects from the Czech Science Foundation: no. 18-14095Y awarded to K.P., no. 20-23718Y awarded to T.S., no. 13-00243S awarded to K.Š., no. 19-00113S awarded to P.P., and no. 19-23469S awarded to M.M.S. The study was also supported by project no. 310030E-160603 awarded to T.P. and project no. 310030_185108 awarded to M.M.S. from the Swiss National Science Foundation.
K.P. designed the study, collected and analyzed samples for the eukaryotic data set, performed statistical analysis except for Mantel tests, analyzed and interpreted the data, and wrote the manuscript. T.S. collected and analyzed samples for the bacterial data sets from Jiřická Pond and Lake Zurich, performed Mantel tests, analyzed and interpreted the data, and revised the manuscript. J.P. contributed to the interpretation of data and revised the manuscript. T.P. collected and analyzed samples for the bacterial data set from Lake Zurich, contributed to interpretation of data, and revised the manuscript. K.Š. collected and analyzed samples for the bacterial data set from Jiřická Pond, contributed to interpretation of data, and revised the manuscript. P.P. collected and analyzed samples for the bacterial data set from Jiřická Pond, contributed to interpretation of data, and revised the manuscript. M.M.S. collected and analyzed samples for the bacterial data sets from Jiřická Pond and Lake Zurich, contributed to interpretation of data, and revised the manuscript.
We declare that there are no competing financial and nonfinancial interests in relation to the work described.
REFERENCES
- 1.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 2.Lundin D, Severin I, Logue JB, Ostman O, Andersson AF, Lindstrom ES. 2012. Which sequencing depth is sufficient to describe patterns in bacterial alpha- and beta-diversity? Environ Microbiol Rep 4:367–372. doi: 10.1111/j.1758-2229.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 3.de Vargas C, Tara Oceans Coordinators, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, Carmichael M, Poulain J, Romac S, Colin S, Aury J-M, Bittner L, Chaffron S, Dunthorn M, Engelen S, Flegontova O, Guidi L, Horák A, Jaillon O, Lima-Mendez G, Lukeš J, Malviya S, Morard R, Mulot M, Scalco E, Siano R, Vincent F, Zingone A, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Acinas SG, Bork P, Bowler C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Raes J, Sieracki ME, Speich S, Stemmann L, Sunagawa S, Weissenbach J, Wincker P, Karsenti E. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 4.Ramirez KS, Knight CG, de Hollander M, Brearley FQ, Constantinides B, Cotton A, Creer S, Crowther TW, Davison J, Delgado-Baquerizo M, Dorrepaal E, Elliott DR, Fox G, Griffiths RI, Hale C, Hartman K, Houlden A, Jones DL, Krab EJ, Maestre FT, McGuire KL, Monteux S, Orr CH, van der Putten WH, Roberts IS, Robinson DA, Rocca JD, Rowntree J, Schlaeppi K, Shepherd M, Singh BK, Straathof AL, Bhatnagar JM, Thion C, van der Heijden MGA, de Vries FT. 2018. Detecting macroecological patterns in bacterial communities across independent studies of global soils. Nat Microbiol 3:189–196. doi: 10.1038/s41564-017-0062-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Xu X, Zhu L. 2017. Structure and function of the microbial consortia of activated sludge in typical municipal wastewater treatment plants in winter. Sci Rep 7:17930. doi: 10.1038/s41598-017-17743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams CL, Dill-McFarland KA, Sparks DL, Kouba AJ, Willard ST, Suen G, Brown AE. 2018. Dietary changes during weaning shape the gut microbiota of red pandas (Ailurus fulgens). Conserv Physiol 6:cox075. doi: 10.1093/conphys/cox075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, D'Amato M, Bonfiglio F, McDonald D, Gonzalez A, McClure EE, Dunklebarger MF, Knight R, Jansson JK. 2017. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirová D, Bárta J, Šimek K, Posch T, Pech J, Stone J, Borovec J, Adamec L, Vrba J. 2018. Hunters or farmers? Microbiome characteristics help elucidate the diet composition in an aquatic carnivorous plant. Microbiome 6:225. doi: 10.1186/s40168-018-0600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, Xia LC, Xu ZZ, Ursell L, Alm EJ, Birmingham A, Cram JA, Fuhrman JA, Raes J, Sun F, Zhou J, Knight R. 2016. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 10:1669–1681. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol 67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen MC, Tolker-Nielsen T, Givskov M, Molin S. 1998. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol 26:141–149. doi: 10.1111/j.1574-6941.1998.tb00500.x. [DOI] [Google Scholar]
- 12.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGovern E, Waters SM, Blackshields G, McCabe MS. 2018. Evaluating established methods for rumen 16S rRNA amplicon sequencing with mock microbial populations. Front Microbiol 9:1365. doi: 10.3389/fmicb.2018.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh YC, Needham DM, Sieradzki ET, Fuhrman JA. 2018. Taxon disappearance from microbiome analysis reinforces the value of mock communities as a standard in every sequencing run. mSystems 3:9. doi: 10.1128/mSystems.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KF, Kohli GS, Murray SA, Rhodes LL. 2017. Assessment of the metabarcoding approach for community analysis of benthic-epiphytic dinoflagellates using mock communities. N Z J Marine Freshwater Res 51:555–576. doi: 10.1080/00288330.2017.1298632. [DOI] [Google Scholar]
- 16.Lee Z-P, Bussema C 3rd, Schmidt TM. 2009. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong J, Dong J, Liu X, Massana R. 2013. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist 164:369–379. doi: 10.1016/j.protis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaechter M. 2015. A brief history of bacterial growth physiology. Front Microbiol 6:289. doi: 10.3389/fmicb.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wendeberg A, Pernthaler J, Amann R. 2004. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms. Mol Microb Ecol Manual 3:711–726. [Google Scholar]
- 24.Amann R, Fuchs BM. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol 6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 25.Eilers H, Pernthaler J, Amann R. 2000. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl Environ Microbiol 66:4634–4640. doi: 10.1128/aem.66.11.4634-4640.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salcher MM, Pernthaler J, Frater N, Posch T. 2011. Vertical and longitudinal distribution patterns of different bacterioplankton populations in a canyon-shaped, deep prealpine lake. Limnol Oceanogr 56:2027–2039. doi: 10.4319/lo.2011.56.6.2027. [DOI] [Google Scholar]
- 27.Shabarova T, Kasalický V, Šimek K, Nedoma J, Znachor P, Posch T, Pernthaler J, Salcher MM. 2017. Distribution and ecological preferences of the freshwater lineage LimA (genus Limnohabitans) revealed by a new double hybridization approach. Environ Microbiol 19:1296–1309. doi: 10.1111/1462-2920.13663. [DOI] [PubMed] [Google Scholar]
- 28.Piwosz K, Kownacka J, Ameryk A, Zalewski M, Pernthaler J. 2016. Phenology of cryptomonads and the CRY1 lineage in a coastal brackish lagoon (Vistula Lagoon, Baltic Sea). J Phycol 52:626–637. doi: 10.1111/jpy.12424. [DOI] [PubMed] [Google Scholar]
- 29.Massana R, Guillou L, Terrado R, Forn I, Pedrós-Alió C. 2006. Growth of uncultured heterotrophic flagellates in unamended seawater incubations. Aquat Microb Ecol 45:171–180. doi: 10.3354/ame045171. [DOI] [Google Scholar]
- 30.Not F, Latasa M, Scharek R, Viprey M, Karleskind P, Balagué V, Ontoria-Oviedo I, Cumino A, Goetze E, Vaulot D, Massana R. 2008. Protistan assemblages across the Indian Ocean, with a specific emphasis on the picoeukaryotes. Deep-Sea Res Part I Oceanogr Res Pap 55:1456–1473. doi: 10.1016/j.dsr.2008.06.007. [DOI] [Google Scholar]
- 31.Piwosz K, Shabarova T, Tomasch J, Šimek K, Kopejtka K, Kahl S, Pieper DH, Koblížek M. 2018. Determining lineage-specific bacterial growth curves with a novel approach based on amplicon reads normalization using internal standard (ARNIS). ISME J 12:2640–2654. doi: 10.1038/s41396-018-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stern R, Kraberg A, Bresnan E, Kooistra W, Lovejoy C, Montresor M, Morán XAG, Not F, Salas R, Siano R, Vaulot D, Amaral-Zettler L, Zingone A, Metfies K. 2018. Molecular analyses of protists in long-term observation programmes—current status and future perspectives. J Plankton Res 40:519–536. doi: 10.1093/plankt/fby035. [DOI] [Google Scholar]
- 33.Giner CR, Forn I, Romac S, Logares R, de Vargas C, Massana R. 2016. Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl Environ Microbiol 82:4757–4766. doi: 10.1128/AEM.00560-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibarbalz FM, Perez MV, Figuerola ELM, Erijman L. 2014. The bias associated with amplicon sequencing does not affect the quantitative assessment of bacterial community dynamics. PLoS One 9:e99722. doi: 10.1371/journal.pone.0099722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monchy S, Grattepanche JD, Breton E, Meloni D, Sanciu G, Chabe M, Delhaes L, Viscogliosi E, Sime-Ngando T, Christaki U. 2012. Microplanktonic community structure in a coastal system relative to a Phaeocystis bloom inferred from morphological and tag pyrosequencing methods. PLoS One 7:e39924. doi: 10.1371/journal.pone.0039924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao W, Chen Z, Li Y, Pan Y, Zhu J, Guo S, Hu L, Huang J. 2018. Bioassessment of a drinking water reservoir using plankton: high throughput sequencing vs. traditional morphological method. Water 10:82. doi: 10.3390/w10010082. [DOI] [Google Scholar]
- 37.Herlemann DPR, Woelk J, Labrenz M, Jürgens K. 2014. Diversity and abundance of “Pelagibacterales” (SAR11) in the Baltic Sea salinity gradient. Syst Appl Microbiol 37:601–604. doi: 10.1016/j.syapm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Bergen B, Herlemann DP, Labrenz M, Jürgens K. 2014. Distribution of the verrucomicrobial clade Spartobacteria along a salinity gradient in the Baltic Sea. Environ Microbiol Rep 6:625–630. doi: 10.1111/1758-2229.12178. [DOI] [PubMed] [Google Scholar]
- 39.Okazaki Y, Fujinaga S, Tanaka A, Kohzu A, Oyagi H, Nakano SI. 2017. Ubiquity and quantitative significance of bacterioplankton lineages inhabiting the oxygenated hypolimnion of deep freshwater lakes. ISME J 11:2279–2293. doi: 10.1038/ismej.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitsch G, Bruni EP, Forster D, Qu Z, Sonntag B, Stoeck T, Posch T. 2019. Seasonality of planktonic freshwater ciliates: are analyses based on V9 regions of the 18S rRNA gene correlated with morphospecies counts?. Front Microbiol 10:248. doi: 10.3389/fmicb.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carr A, Diener C, Baliga NS, Gibbons SM. 2019. Use and abuse of correlation analyses in microbial ecology. ISME J 13:2647–2655. doi: 10.1038/s41396-019-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310. doi: 10.1016/S0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- 43.Bakenhus I, Wemheuer B, Akyol P, Giebel H-A, Dlugosch L, Daniel R, Simon M. 2019. Distinct relationships between fluorescence in situ hybridization and 16S rRNA gene- and amplicon-based sequencing data of bacterioplankton lineages. Syst Appl Microbiol 42:126000. doi: 10.1016/j.syapm.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Graneli E, Edvardsen B, Roelke DL, Hagstrom JA. 2012. The ecophysiology and bloom dynamics of Prymnesium spp. Harmful Algae 14:260–270. doi: 10.1016/j.hal.2011.10.024. [DOI] [Google Scholar]
- 45.Jovel J, Patterson J, Wang W, Hotte N, O'Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK-S. 2016. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7:459–459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piwosz K. 2019. Weekly dynamics of abundance and size structure of specific nanophytoplankton lineages in coastal waters (Baltic Sea). Limnol Oceanogr 64:2172–2186. doi: 10.1002/lno.11177. [DOI] [Google Scholar]
- 47.Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, Gould TJ, Clayton JB, Johnson TJ, Hunter R, Knights D, Beckman KB. 2016. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34:942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 48.Calus ST, Ijaz UZ, Pinto AJ. 2018. NanoAmpli-Seq: a workflow for amplicon sequencing for mixed microbial communities on the nanopore sequencing platform. Gigascience 7:16. doi: 10.1093/gigascience/giy140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acharya K, Khanal S, Pantha K, Amatya N, Davenport RJ, Werner D. 2019. A comparative assessment of conventional and molecular methods, including MinION nanopore sequencing, for surveying water quality. Sci Rep 9:11. doi: 10.1038/s41598-019-51997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT. 2012. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS One 7:e30087. doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner H-W, Richards TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 52.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherr BF, Sherr EB, Fallon RD. 1987. Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol 53:958–965. doi: 10.1128/AEM.53.5.958-965.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grasshoff K, Ehrhardt M, Kremling K. 1976. Methods for sea water analysis. Verlag Chemie: 1–419. [Google Scholar]
- 55.Egge E, Bittner L, Andersen T, Audic S, de Vargas C, Edvardsen B. 2013. 454 Pyrosequencing to describe microbial eukaryotic community composition, diversity and relative abundance: a test for marine haptophytes. PLoS One 8:e74371. doi: 10.1371/journal.pone.0074371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piwosz K, Pernthaler J. 2010. Seasonal population dynamics and trophic role of planktonic nanoflagellates in coastal surface waters of the Southern Baltic Sea. Environ Microbiol 12:364–377. doi: 10.1111/j.1462-2920.2009.02074.x. [DOI] [PubMed] [Google Scholar]
- 57.Shabarova T, Villiger J, Morenkov O, Niggemann J, Dittmar T, Pernthaler J. 2014. Bacterial community structure and dissolved organic matter in repeatedly flooded subsurface karst water pools. FEMS Microbiol Ecol 89:111–126. doi: 10.1111/1574-6941.12339. [DOI] [PubMed] [Google Scholar]
- 58.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 59.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, Boutte C, Burgaud G, de Vargas C, Decelle J, del Campo J, Dolan JR, Dunthorn M, Edvardsen B, Holzmann M, Kooistra W, Lara E, Le Bescot N, Logares R, Mahe F, Massana R, Montresor M, Morard R, Not F, Pawlowski J, Probert I, Sauvadet AL, Siano R, Stoeck T, Vaulot D, Zimmermann P, Christen R. 2013. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res 41:D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gabaldon C, Devetter M, Hejzlar J, Šimek K, Znachor P, Nedoma J, Seďa J. 2017. Repeated flood disturbance enhances rotifer dominance and diversity in a zooplankton community of a small dammed mountain pond. J Limnol 76:292–304. doi: 10.4081/jlimnol.2016.1544. [DOI] [Google Scholar]
- 62.Porcal P, Kopáček J. 2018. Photochemical degradation of dissolved organic matter reduces the availability of phosphorus for aquatic primary producers. Chemosphere 193:1018–1026. doi: 10.1016/j.chemosphere.2017.11.140. [DOI] [PubMed] [Google Scholar]
- 63.Brussaard C. 2004. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol 70:1506–1513. doi: 10.1128/aem.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beutler M, Wiltshire KH, Meyer B, Moldaenke C, Luring C, Meyerhofer M, Hansen UP, Dau H. 2002. A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynth Res 72:39–53. doi: 10.1023/A:1016026607048. [DOI] [PubMed] [Google Scholar]
- 65.Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. 2005. Bacterial populations active in metabolism of C-1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol 71:6885–6899. doi: 10.1128/AEM.71.11.6885-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol 69:2928–2935. doi: 10.1128/aem.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salcher MM, Pernthaler J, Posch T. 2011. Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria ‘that rule the waves’ (LD12). ISME J 5:1242–1252. doi: 10.1038/ismej.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 69.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 71.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 72.Anderson MJ, Legendre P. 1999. An empirical comparison of permutation methods for tests of partial regression coefficients in a linear model. J Stat Comput Simul 62:271–303. doi: 10.1080/00949659908811936. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual scatter plots of relative abundances of studied eukaryotic groups by 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S1, PDF file, 0.1 MB (151.2KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Scatterplot of relative biovolumes of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. (B) Scatterplot of ranked relative biovolumes of studied eukaryotic groups from HTS and CARD-FISH. (C) Scatterplot of differences between relative biovolumes of studied eukaryotic groups estimated by CARD-FISH and HTS against the average of the two values. (D) Scatterplot of differences between ranked relative biovolumes of studied eukaryotic groups estimated by CARD-FISH and HTS against the average of the two values. Black lines in panels A and B show a 1:1 relationship. Solid black lines in panels C and D show average differences for the whole data set, solid gray lines show 1 standard deviation, and dashed gray lines show 2 standard deviations. Different eukaryotic groups are color coded. Individual plots for panels A and B are shown in Fig. S3 and S4, respectively. Download FIG S2, PDF file, 0.2 MB (243.5KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of relative biovolumes of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S3, PDF file, 0.1 MB (155.9KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of rank relative biovolumes of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. Samples with zeroes in both CARD-FISH and HTS data were removed, because they were arbitrarily ranked. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S4, PDF file, 0.1 MB (124.5KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of rank relative abundances of studied eukaryotic groups from 454 sequencing libraries (HTS) and CARD-FISH. Samples with zeroes in both CARD-FISH and HTS data were removed, because they were arbitrarily ranked. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S5, PDF file, 0.1 MB (127.3KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of relative abundances of studied bacterial groups by 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S6, PDF file, 0.3 MB (343.3KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual scatterplots of rank relative abundances of studied bacterial groups from 454 sequencing libraries (HTS) and CARD-FISH. Black lines show a 1:1 relationship. Axis scales differ between the panels. Download FIG S7, PDF file, 0.3 MB (363.4KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of primers used in this study. Download Table S1, PDF file, 0.3 MB (324KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of probes, helpers, and competitors used in this study. HB (%), concentration of formamide in the hybridization buffer; T, hybridization temperature (washing temperature was 2°C higher); Probe, coverage (in percent of targeted sequences) of the probe; Primers 1, coverage of primers TAReuk454FWD1 and TAReukREV3 (eukaryotes) or 341F and 907R (bacteria, Lake Zurich); Primers 2, coverage of primers TAReuk454FWD1 and HaptoR1 (eukaryotes) or 27F andUni522R (bacteria, Jiřická Pond). The numbers are based on search in the reference Silva database release 132, conducted on 23 November 2019. Download Table S2, PDF file, 0.6 MB (618KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Individual rarefaction curves for samples from the Baltic Sea (eukaryotes) (A), Jiřická Pond (B), and Lake Zurich (C). Download FIG S8, PDF file, 0.2 MB (217.8KB, pdf) .
Copyright © 2020 Piwosz et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The eukaryotic HTS data obtained with the general primers (TAReuk454FWD1 and TAReukREV3) were deposited in the ENA database under BioProject no. PRJEB23971, and those obtained with primers TAReuk454FWD1 and HaptoR1 were deposited under BioProject no. PRJEB31858. Bacterial HTS data from Jiřická Pond were deposited in NCBI as BioSamples SAMN11974970 to SAMN11974993 as part of BioProject PRJNA547706, and those from Lake Zurich were deposited under BioProject no. PRJNA545726.