Abstract
Colorectal cancer (CRC) is highly prevalent worldwide. In 2018, there were over 1.8 million new cases. Most sporadic CRC develop from polypoid adenomas and are preceded by intramucosal carcinoma (stage 0), which can progress into more malignant forms. This developmental process is known as the adenoma‐carcinoma sequence. Early detection and endoscopic removal are crucial for CRC management. Accumulating evidence suggests that the gut microbiota is associated with CRC development in humans. Comprehensive characterization of this microbiota is of great importance to assess its potential as a diagnostic marker in the very early stages of CRC. In this review, we summarized recent studies on CRC‐associated bacteria and their carcinogenic mechanisms in animal models, human cell lines and human cohorts. High‐throughput technologies have facilitated the identification of CRC‐associated bacteria in human samples. We have presented our metagenome and metabolome studies on fecal samples collected from a large Japanese cohort that revealed stage‐specific phenotypes of the microbiota in CRC. Furthermore, we have discussed the potential carcinogenic mechanisms of the gut microbiota, from which we can infer whether changes in the gut microbiota are a cause or effect in the multi‐step process of CRC carcinogenesis.
Keywords: adenoma‐carcinoma sequence, colorectal cancer, gut microbiome, metabolome, metagenome
This review summarizes recent studies on colorectal cancer (CRC)‐associated bacteria and their carcinogenic mechanisms in animal models, human cell lines and human cohorts. In addition, we present our most recent metagenome and metabolome studies on fecal samples collected from a large Japanese cohort, which revealed stage‐specific phenotypes of the microbiota in CRC. Finally, we discuss the potential carcinogenic mechanisms of the gut microbiota and whether changes in the gut microbiota are a cause or effect in the multi‐step process of CRC carcinogenesis.

1. INTRODUCTION
In Japan, the mortality rate from colorectal cancer (CRC) has increased significantly over the past 30 years, and in 2014, CRC surpassed gastric cancer to become the most prevalent cancer type in Japan (https://ganjoho.jp/reg_stat/statistics/index.html). Japan has one of the highest CRC incidences in the world (https://www.wcrf.org/dietandcancer/cancer-trends/colorectal-cancer-statistics). Westernized dietary habits are considered to be one of the causal factors, but the underlying mechanisms have not been clarified.1, 2, 3 Most CRC cases are sporadic, and hereditary CRC cases account for <5% of all CRC cases. Most sporadic CRC develop from polypoid adenomas and are preceded by intramucosal carcinomas (stage 0), which can progress into malignant forms.4 This developmental process is known as the adenoma‐carcinoma sequence, which occurs through a multistep mechanism that involves specific mutations.
Approximately 40 trillion bacteria inhabit the human body,5 and disturbances of intestinal bacteria are related to various gastrointestinal diseases, such as inflammatory bowel disease.6 Approximately 2% of CRC are linked to preexisting inflammation, known as colitis‐associated cancer.7 In these cases, enterotoxigenic Bacteroides fragilis toxin and the adherent‐invasive Escherichia coli strain NC101 have been reported to be involved in cancer development (Table 1).8, 9 In 2012, Fusobacterium nucleatum, a periodontal pathogen, was reported to be associated with non–colitis‐associated CRC among cases of sporadic CRC.10, 11 The underlying mechanisms were studied in mouse models and human cell lines, and will be discussed in more detail in the following sections. Later, with the advent of next‐generation sequencing technologies that produce large amounts of metagenome data, more CRC‐associated gut bacteria were identified (Table 1). However, it has not been fully elucidated whether changes in the intestinal microbiota are a cause or a result of carcinogenesis, or whether the microbiota contributes to cancer progression. Some gut bacteria produce genotoxic metabolites: for example, a few species of the genus Clostridium produce deoxycholic acid (DCA)12 and pks + E coli strains produce colibactin.13 Current knowledge on the potential links between diet and intestinal residues as a trophic resource for the gut microbiota and carcinogenesis is only fragmental. To address this issue, we established a large human cohort study in Japan, in which we collected fecal samples and a food frequency questionnaire from the same subjects and conducted multi‐omics analyses to comprehensively study dietary habits and metagenome and metabolome data in relation to CRC.
Table 1.
Association between gut bacteria and colorectal cancer (CRC)
| Bacteria related to colitis‐associated CRC |
| Enterotoxigenic Bacteroides fragilis 8 |
| Adherent‐invasive Escherichia coli 9 |
| Genotoxic compound producing bacteria |
| Pks + Escherichia coli strains (colibactin)13 |
| Clostridium hylemonae (deoxycholic acid)12 |
| Clostridium hiranonis (deoxycholic acid)12 |
| Bacteria related to non–colitis‐associated CRC |
| Fusobacterium nucleatum 10, 11, 14, 15, 16, 19, 20 |
| Parvimonas micra 20, 26 |
| Peptostreptococcus stomatis 19, 20 |
| Peptostreptococcus anaerobius 20, 25 |
| Atopobium parvulum 28 |
| Actinomyces odontolyticus 28, 30 |
2. CARCINOGENIC ROLE OF FUSOBACTERIUM NUCLEATUM
Fusobacterium nucleatum is a gram‐negative, anaerobic oral bacterium that causes periodontitis. Studies in mouse models and human cell lines have revealed that F. nucleatum accelerates CRC via several pathways.14, 15, 16 Kostic et al reported the role of F. nucleatum in colorectal tumor progression via a pathway independent from that involved in colitis‐associated CRC. The introduction of human isolates of F. nucleatum in APC Min/+ mice resulted in tumor multiplicity and recruitment of tumor‐infiltrating myeloid cells, but this bacterium did not induce colitis.14 Han et al reported that F. nucleatum invades endothelial and epithelial cells.17 Later, they performed a structural analysis on the F. nucleatum FadA protein, which revealed that it binds to E‐cadherin,18 and a series of experiments in human cell lines that revealed that FadA‐E‐cadherin binding resulted in the activation of β‐catenin signaling and promoted cell proliferation.15 Gur et al showed that F. nucleatum Fap2 protein inhibits tumor‐killing natural killer cells via the inhibitory receptor TIGIT.16 Collectively, these findings suggested that F. nucleatum is not just associated with CRC but might accelerate/promote CRC, possibly at early‐stage carcinogenesis.
3. IDENTIFICATION OF COLORECTAL CANCER‐ASSOCIATED MICROBIOTA BASED ON HIGH‐THROUGHPUT SEQUENCE DATA FROM HUMAN COHORT STUDIES
16S ribosomal RNA gene and whole shotgun metagenome sequencing technologies have facilitated the quantitative profiling of the intestinal microbiota in human fecal and tissue samples. Large cohort studies have identified several species associated with adenoma and CRC,19, 20, 21, 22, 23, 24 including gram‐positive anaerobic cocci, such as Peptostreptococcus anaerobius, Peptostreptococcus stomatis and Parvimonas micra (Table 1). Subsequent mouse experiments have demonstrated these species' involvement in CRC. Mice treated with azoxymethane had a higher incidence of intestinal dysplasia following exposure to P. anaerobius.25 APC Min/+ mice gavaged with P. micra exhibited higher tumor burden and tumor load.26 However, their carcinogenic mechanisms have not been fully elucidated. In contrast to gastric cancer, which is caused mostly by a single species, Helicobacter pylori, CRC can be affected by various gut bacteria and their coexistence networks. Furthermore, the immune responses to such gut bacteria may affect carcinogenesis.
4. THE INTESTINAL MICROBIOTA AND ITS INVOLVEMENT IN COLORECTAL CANCER IN A JAPANESE POPULATION
Large cohort studies have identified several bacteria and their metabolites that are associated with CRC, mostly in stage I‐IV cases; however, very limited data are available for intramucosal carcinoma (stage 0), the very early stage of CRC. In addition, it has not been clarified whether the reported changes in these bacteria and their metabolites are causative factors in carcinogenesis, or whether they are a result of microbial adaptation to the tumor environment. Nevertheless, CRC‐associated microbiota are currently globally accepted as CRC screening biomarkers that can be used as an alternative for the fecal occult blood test.
To address these challenges, we have established a large Japanese cohort. We collected fecal samples from 631 participants, including patients with multiple polypoid adenomas, intramucosal carcinoma (stage 0), stages I‐IV, and healthy controls, undergoing colonoscopy checks at the National Cancer Center Hospital, Tokyo, Japan.27 From 28 of these patients in stages I‐III, fecal samples were collected before and after surgical tumor resection. We subjected these samples to whole‐genome shotgun metagenomic sequencing and capillary electrophoresis time‐of‐flight mass spectrometry‐based metabolomic studies to assess taxonomic and functional characteristics of the gut microbiota and metabolites (Figure 1).28 In addition, data on lifestyle were obtained from a detailed questionnaire (475 question items, 25 pages) that was developed on the basis of the questionnaire used in the Japan Public Health Center‐based Prospective Study.29 It includes questions on food frequency as well as known CRC risk factors (age, sex, smoking habits and alcohol consumption). Based on a metagenome analysis, we found three distinct patterns of CRC‐related changes in gut microbiota abundance (Figure 2). First, F. nucleatum abundance was significantly (P < .005) elevated continuously from stage 0 to more advanced stages. This suggests that F. nucleatum contributes in the very early stage of the adenoma‐carcinoma sequence, which was in line with the abovementioned findings in mouse models and human cell lines. In contrast, P. anaerobius, P. stomatis and P. micra were predominantly enriched in stage I/II and stage III/IV, and their abundances decreased after tumor resection,28 which implies that these species might not cause carcinogenesis but are adapted to the cancerous environment. Second, we found that Atopobium parvulum and Actinomyces odontolyticus, whose relative abundances were higher than those of other CRC‐associated bacteria, were significantly (P < .005) increased in multiple polypoid adenomas and/or in stage 0 but were not increased as significantly (P > .005) in more advanced stages. Kasai et al reported the presence of A. odontolyticus in fecal samples of patients with carcinoma in adenoma, although the sample size in their study was very small (n = 6).30 They suggested that these bacteria may be one of the factors causing carcinogenesis, but this remains to be validated. Third, butyrate producers (ie, Lachnospira multipara and Eubacterium eligens) and Bifidobacterium longum were depleted in several CRC stages. Comparison of colibactin gene abundances revealed no significant differences across the patient groups (data not shown).
Figure 1.
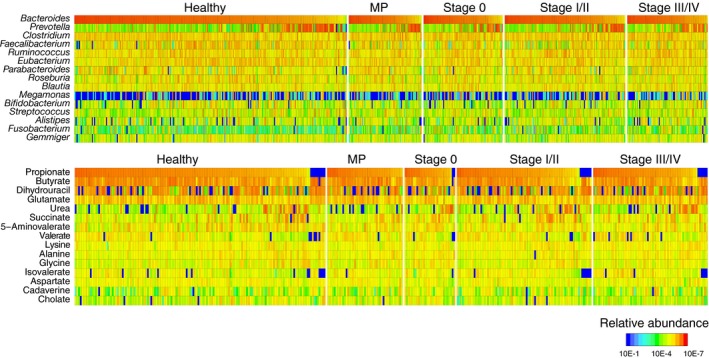
Metagenomic and metabolomic data from our Japanese cohort study. (Upper panel) Relative abundances of the top 15 genera for 576 subjects in the healthy control (N = 251), multiple polypoid adenomas (MP) (N = 67), stage 0 (N = 73), stage I/II (N = 111) and stage III/IV (N = 74) groups. Bifidobacterium and Megamonas are highly abundant, a notable characteristic of the Japanese cohort. Whereas Bifidobacterium is prevalent in the entire population, Megamonas abundance strongly differs among individuals. (Lower panel) Percentage concentrations of top 15 metabolites for 372 subjects in the healthy control (N = 149), MP (N = 45), stage 0 (N = 30), stage I/II (N = 80) and stage III/IV (N = 68) groups
Figure 2.
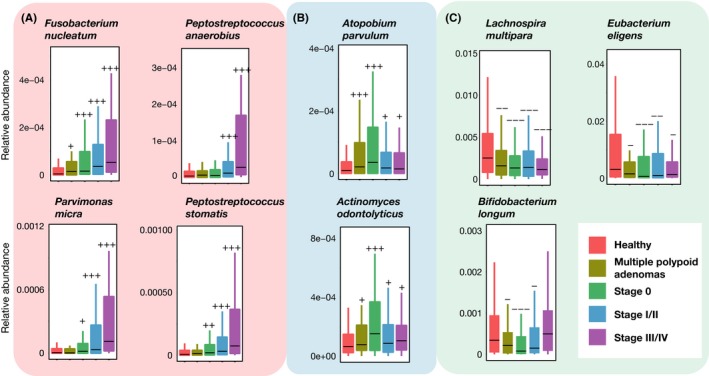
Three types of colorectal cancer (CRC)‐associated taxonomic signatures in our Japanese cohort study. Relative abundances of known CRC‐related species (A), species that are newly associated to multiple polypoid adenomas and stage 0 (B), and species that are depleted in CRC (C), in healthy control, multiple polypoid adenomas, stage 0, stage I/II and stage III/IV groups. The boxes represent the 25th‐75th percentiles; the median is indicated by a black horizontal line, and maximum and minimum values within 1.5 times the interquartile range are indicated by a vertical line. +++, elevation at P < .005; ++, elevation at P < .01; +, elevation at P < .05; −−−, depletion at P < .005; −−, depletion at P < .01; −, depletion at P < .05 (one‐sided Mann‐Whitney U test). P < .005 was considered statistically significant
5. SULFIDOGENIC BACTERIA IMPAIR THE BARRIER FUNCTION OF THE INTESTINAL WALL, CAUSING INFLAMMATION AND TUMORIGENESIS
Atopobium parvulum is a gram‐positive obligate anaerobic bacterium that is frequently isolated from the human oral cavity. It is associated with halitosis arising from malodorous compounds such as volatile sulfur compounds, including hydrogen sulfide (H2S). A. parvulum has been associated with colitis via the production of toxic H2S in IL‐10–/– mice, a mouse model for Crohn's disease.31 It has been shown that a defective mucus layer leads to inflammatory bowel diseases32 and colorectal cancer,33, 34 and the gut bacteria can facilitate this process in various ways. H2S is one of the factors that can cause mucus barrier impairment.34, 35 Using a mouse model of CRC, Grivennikov et al showed that impaired barrier function at tumor sites facilitates the penetration of bacterial products, resulting in the activation of the tumorigenic cytokines IL‐23 and IL‐17 in tumor‐infiltrating cells and “tumor‐elicited inflammation.”7 In our Japanese cohort study, A. parvulum displayed a higher abundance in patients with multiple polypoid adenomas and intramucosal carcinoma (stage 0) (Figure 2), which corroborates the possible role of H2S in intestinal barrier impairment in the very early stage of non–colitis‐associated sporadic CRC development. In regard to diet, sulfidogenic bacteria (eg, Bilophila wadsworthia, Pyramidobacter spp.) have been associated with higher intake of protein and meat.36
6. DEOXYXCHOLIC ACID ACCELERATES COLORECTAL CANCER CARCINOGENESIS
Deoxycholic acid is a secondary bile acid produced by a specific group of gut bacteria. Primary bile acids are generated from cholesterol in the liver and secreted into the intestine. Ninety‐five percent of them are transported back to the liver via a process known as enterohepatic circulation. The remaining 5% escape this process and are metabolized by gut bacteria (eg, Clostridium hylemonae and Clostridium hiranonis), primarily in the colon, into secondary bile acids, including lithocholic acid and DCA.12 A number of animal studies in the past few decades have shown the carcinogenic effect of DCA in gastroenterological organs.37, 38, 39, 40 Studies in mouse models and human cell lines have identified Wnt signaling pathway activation as the tumor‐promoting mechanism. Clinical studies have shown that patients with CRC have higher levels of bile acids in the colonic lumen.41 In addition, bile acids are known causal factors of remnant gastric cancer,42, 43 which is why Billroth II, one of the reconstruction types after gastrectomy, is no longer considered the most appropriate procedure. A metabolome analysis in our Japanese cohort study displayed higher concentrations of DCA in the feces of patients with multiple polypoid adenomas (Figure 3), the precursor of CRC. The findings in the abovementioned studies and ours support that DCA might be involved in very early carcinogenesis.
Figure 3.
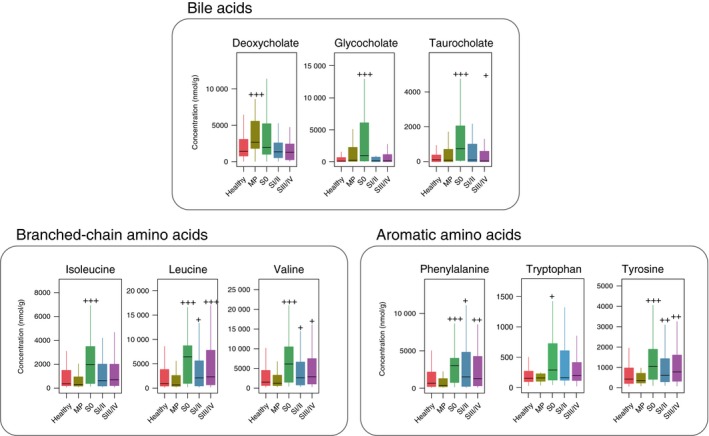
Stage‐specific changes of bile acids and amino acids in our Japanese cohort study. Fecal concentrations of bile acids, branched‐chain amino acids, and aromatic amino acids in patients in healthy control, multiple polypoid adenomas (MP), stage 0 (S0), stage I/II (SI/II) and stage III/IV (SIII/IV) groups. The boxes represent the 25th‐75th percentiles, the median is indicated by a black horizontal line, and maximum and minimum values within 1.5 times the interquartile range are indicated by a vertical line. +++, elevation at P < .005; ++, elevation at P < .01; +, elevation at P < .05; −−−, depletion at P < .005; −−, depletion at P < .01; −, depletion at P < .05 (one‐sided Mann‐Whitney U test)
7. DISTINCT STAGE‐SPECIFIC PHENOTYPES OF THE GUT MICROBIOTA IN COLORECTAL CANCER AND THEIR APPLICATION AS AN EARLY‐STAGE DIAGNOSTIC MARKER TOWARDS CANCER PREVENTION
Our metabolome study also revealed higher concentrations of branched‐chain amino acids (valine, leucine and isoleucine) and aromatic amino acids (phenylalanine, tryptophan and tyrosine) in the feces of patients in different CRC stages than in those of healthy controls (Figure 3). Higher levels of amino acids have also been found in a study of resected CRC tumors, based on which the authors discussed the role of altered metabolism in cancer cells.44 Using metagenome and metabolome data from the same fecal samples, we examined microbial metabolic changes that may play a role in the elevation of branched‐chain and aromatic amino acids. We observed dynamic changes in the amino acid metabolic capacity of the gut microbiota in various CRC stages. In particular, we observed a higher abundance of the cyclohexadienyl dehydratase gene pheC in stage 0, and a gene cluster encoding the aerobic phenylacetate catabolic pathway. A cross‐cohort study by another group also revealed an enrichment of amino acid degradation pathways in CRC metagenomes.23 Collectively, these findings suggest that changes in intestinal bacteria and fecal metabolites may occur from the very early stages of CRC development, and may be useful in elucidating and diagnosing carcinogenic mechanisms.
The abovementioned fecal branched‐chain amino acids, phenylalanine and pheC were identified as the best‐scoring markers to distinguish stage 0 cases from healthy controls in our study,28 suggesting their potential as non–invasive diagnostic markers for early stages of CRC. This relates to a diet‐based cancer prevention strategy in the sense that gut microbial fermentation of dietary amino acids may result in the production of detrimental metabolites such as phenylacetic acid, indole, p‐cresol and N‐nitroso compounds.45 Furthermore, we investigated the relationships between food frequency and CRC stages; we found that multiple polypoid adenomas and CRC were associated with lower intake of total dietary fiber and calcium (Figure 4). This is in agreement with a 10‐year follow‐up survey of approximately 90 000 subjects in the context of the Japan Public Health Center‐based Prospective Study, which revealed a strong correlation between a high‐meat, low‐fiber dietary pattern and CRC incidence,46 and an inverse association between calcium intake and CRC incidence.47 However, how gut microbiota functions contribute to these phenomena requires further analyses. In particular, the potential link between low fiber intake and low abundance of butyrate producers and B. longum needs to be clarified.
Figure 4.
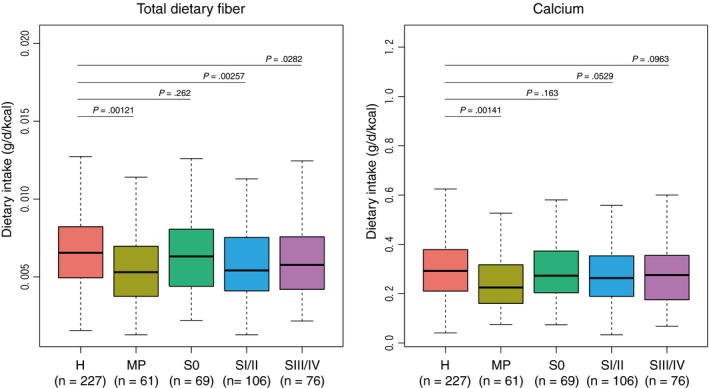
Dietary fiber and calcium intakes in the Japanese cohort study. Dietary intake data were collected from a food frequency questionnaire. Total fiber and calcium intake (g/d) were adjusted to 1 kcal of energy. Data from outliers (energy < 600 kcal or > 3500 kcal) were excluded. The boxes represent the 25th‐75th percentiles, the median is indicated by a black horizontal line, and maximum and minimum values within 1.5 times the interquartile range are indicated by a vertical line. P‐values are obtained from one‐sided Mann‐Whitney U test with the alternative hypothesis that dietary intake in each of the stages is lower than in healthy controls. H, healthy controls; MP, multiple polypoid adenomas; S0, stage 0; SI/II, stage I/II; SIII/IV, stage III/IV
8. CONCLUSION
Previous findings have revealed that intestinal bacteria are involved in the development and progression of CRC through various mechanisms (eg, production of carcinogenic substances, induction of inflammation, and modulation of host signaling and the immune system). In particular, F. nucleatum has been shown to promote cancer by acting on signal transduction pathways involved in epithelial cell proliferation and cancer immune mechanisms. In our large‐scale cohort study, we collected metagenome and metabolome data to evaluate how the intestinal microbial composition and metabolomic profile change as CRC progresses. We showed that CRC‐associated bacteria (eg, F. nucleatum) and microbial metabolites (eg, DCA) are increased at CRC onset. We also revealed that A. parvulum and A. odontolyticus demonstrated significant increases early in carcinogenesis. Based on these findings, we propose a cancer microbiota cascade; that is, a stage‐specific involvement of microbiota in the multi‐step process of CRC carcinogenesis (Figure 5).
Figure 5.
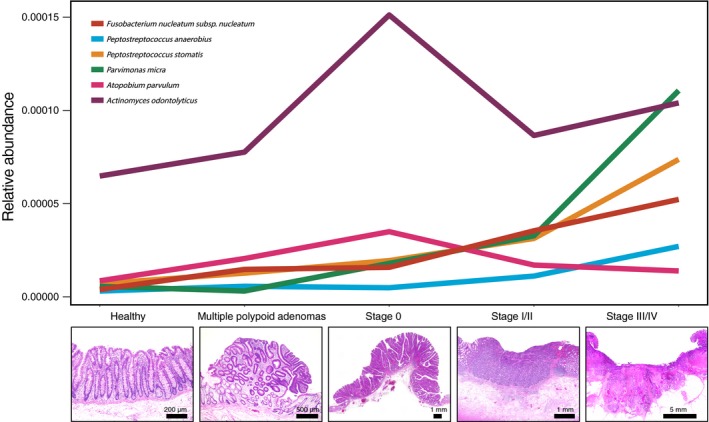
Distinct colorectal cancer (CRC) stage‐specific changes in the gut microbiota. (Upper panel) Bacterial relative abundances in our Japanese cohort study. Values are the median of all samples in each of the five participant groups: healthy control (N = 251), multiple polypoid adenomas (N = 67), stage 0 (N = 73), stage I/II (N = 111) and stage III/IV (N = 74). Three patterns of elevations in abundance can be distinguished. First, Fusobacterium nucleatum abundance starts to increase at stage 0 and continues to increase with cancer progression. Second, Atopobium parvulum and Actinomyces odontolyticus are significantly increased in cases with multiple polypoid adenomas and/or stage 0 but no longer increase in more advanced stages. Third, Peptostreptococcus anaerobius, Peptostreptococcus stomatis,and Parvimonas micra increase in later stages, stage I/II and stage III/IV. (Lower panels) Representative microscopic images of H&E‐stained colorectal tissue sections from each of the participant groups
Prospective studies with a regular follow‐up of healthy individuals and analyses of their intestinal microbiota and metabolites would elucidate the relationship between gut environments and carcinogenesis. Immunological studies have largely elucidated the carcinogenic mechanisms of gut microbiota, whereas biochemical research on their metabolites and the carcinogenic effects thereof is limited. In particular, it remains unclear whether the presence of the CRC‐associated bacteria or the bacterial metabolic products adversely affect the intestinal epithelium. Furthermore, research on how dietary habits are related to the intestinal bacteria that promote cancer development has just begun. Further metagenome and metabolome studies might provide answers to these questions. Genome‐based precision medicine for cancer treatment has recently entered the clinic. We believe that the accumulated knowledge on the gut microbiota will open up new possibilities for microbiota‐based precision medicine in the near future.
DISCLOSURE
Takuji Yamada is a founder of Metabologenomics. The company is focused on the design and control of the gut environment for human health. The company has no control over the interpretation, writing or publication of this work. The terms of these arrangements are being managed by the Tokyo Institute of Technology in accordance with its conflict of interest policies.
ACKNOWLEDGEMENTS
We wish to thank all the patients and their families who participated in this study. We are grateful to Dr Yutaka Saito, Dr Takeshi Nakajima, Dr Taku Sakamoto and Dr Satoshi Shiba (National Cancer Center Hospital) for collecting samples, Dr Shinji Fukuda for metabolome analyses, and Dr Tatsuhiro Shibata for technical advice. This work was supported by grants from: the National Cancer Center Research and Development Fund (25‐A‐4, 28‐A‐4 and 29‐A‐6); the Japan Agency for Medical Research and Development (JP18ek0109187 to SM, TY and SY; JP18jk0210009 to SY; JP19cm0106464 to SY and TY); the Japan Science and Technology Agency‐PRESTO (JPMJPR1507 to TY); JST‐AIP Acceleration Research (JPMJCR19U3 to TY, and SY); the Japan Society for the Promotion of Science (KAKENHI; 16J10135, 142558 and 221S0002 to TY); the Joint Research Project of the Institute of Medical Science, University of Tokyo (to SY); the Takeda Science Foundation (to SY), the Yasuda Medical Foundation (to SY); and the Yakult Bio‐Science Foundation (to SY); and the Princess Takamatsu Cancer Research Fund (to SY).
Mizutani S, Yamada T, Yachida S. Significance of the gut microbiome in multistep colorectal carcinogenesis. Cancer Sci. 2020;111:766–773. 10.1111/cas.14298
REFERENCES
- 1. Kolonel LN, Altshuler D, Henderson BE. The multiethnic cohort study: exploring genes, lifestyle and cancer risk. Nat Rev Cancer. 2004;4:519‐527. [DOI] [PubMed] [Google Scholar]
- 2. Kuriki K, Tajima K. The increasing incidence of colorectal cancer and the preventive strategy in Japan. Asian Pac J Cancer Prev. 2006;7:495‐501. [PubMed] [Google Scholar]
- 3. O'Keefe SJD, Li JV, Lathi L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759‐767. [DOI] [PubMed] [Google Scholar]
- 5. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aggeletopoulou I, Konstantakis C, Assimakopoulos SF, Triantos C. The role of the gut microbiota in the treatment of inflammatory bowel diseases. Microb Pathog. 2019;137:103774. [DOI] [PubMed] [Google Scholar]
- 7. Grivennikov SI, Wang K, Mucida D, et al. Adenoma‐linked barrier defects and microbial products drive IL‐23/IL‐17‐mediated tumour growth. Nature. 2012;491:254‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016‐1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arthur JC, Perez‐Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer‐inducing activity of the microbiota. Science. 2012;338:120‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ridlon JM, Devendran S, Alves JM, et al. The ‘lifestyle’ of bile acid 7α‐dehydroxylating bacteria: comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes. 2019;1‐24. 10.1080/19490976.2019.1618173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson MR, Jiang Y, Villalta PW, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019;363:eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe. 2013;14:207‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E‐cadherin/β‐catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han YW, Shi W, Huang GT, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140‐3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fardini Y, Wang X, Témoin S, et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468‐1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early‐stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non–invasive biomarkers for colorectal cancer. Gut. 2017;66:70‐78. [DOI] [PubMed] [Google Scholar]
- 21. Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun. 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 22. Nakatsu G, Li X, Zhou H, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wirbel J, Py PT, Kartal E, et al. Meta‐analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679‐689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas AM, Manghi P, Asnicar F, et al. Metagenomic analysis of colorectal cancer datasets identifies cross‐cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25:667‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsoi H, Chu ESH, Zhang X, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152:1419–1433.e5. [DOI] [PubMed] [Google Scholar]
- 26. Yu J, Feng W, Wong SH, et al. The role of Parvimonas micra in intestinal tumorigenesis in germ‐free and conventional APCmin/+ mice. J Clin Oncol. 2019;37:531‐531. [Google Scholar]
- 27. Nishimoto Y, Mizutani S, Nakajima T, et al. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut. 2016;65:1574‐1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage‐specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968‐976. [DOI] [PubMed] [Google Scholar]
- 29. Tsugane S, Sawada N. The JPHC study: design and some findings on the typical Japanese diet. Jpn J Clin Oncol. 2014;44:777‐782. [DOI] [PubMed] [Google Scholar]
- 30. Kasai C, Sugimoto K, Moritani I, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next‐generation sequencing analyses. Oncol Rep. 2016;35:325‐333. [DOI] [PubMed] [Google Scholar]
- 31. Mottawea W, Chiang CK, Mühlbauer M, et al. Altered intestinal microbiota–host mitochondria crosstalk in new onset Crohn's disease. Nat Commun. 2016;7:13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johansson MEV, Gustafsson JK, Holmen‐Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726‐1729. [DOI] [PubMed] [Google Scholar]
- 34. Ijssennagger N, Belzer C, Hooiveld GJ, et al. Gut microbiota facilitates dietary heme‐induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc Natl Acad Sci U S A. 2015;112:10038‐10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ijssennagger N, van der Meer R, van Mil SWC. Sulfide as a mucus barrier‐breaker in inflammatory bowel disease? Trends Mol Med. 2016;22:190‐199. [DOI] [PubMed] [Google Scholar]
- 36. Yazici C, Wolf PG, Kim H, et al. Race‐dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66:1983‐1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Payne CM, Weber C, Crowley‐Skillicorn C, et al. Deoxycholate induces mitochondrial oxidative stress and activates NF‐kappaB through multiple mechanisms in HCT‐116 colon epithelial cells. Carcinogenesis. 2007;28:215‐222. [DOI] [PubMed] [Google Scholar]
- 38. Cao H, Luo S, Xu M, et al. The secondary bile acid, deoxycholate accelerates intestinal adenoma–adenocarcinoma sequence in Apc min/+ mice through enhancing Wnt signaling. Fam Cancer. 2014;13:563‐571. [DOI] [PubMed] [Google Scholar]
- 39. Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta‐catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15:2156‐2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshimoto S, Loo TM, Atarashi K, et al. Obesity‐induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97‐101. [DOI] [PubMed] [Google Scholar]
- 41. Bernstein H, Bernstein C, Payne C, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47‐65. [DOI] [PubMed] [Google Scholar]
- 42. Bechi P, Amorosi A, Mazzanti R, Romagnoli P, Tonelli L. Gastric histology and fasting bile reflux after partial gastrectomy. Gastroenterology. 1987;93:335‐343. [DOI] [PubMed] [Google Scholar]
- 43. Păduraru DN, Nica A, Ion D, Handaric M, Andronic O. Considerations on risk factors correlated to the occurrence of gastric stump cancer. J Med Life. 2016;9:130‐136. [PMC free article] [PubMed] [Google Scholar]
- 44. Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time‐of‐flight mass spectrometry. Cancer Res. 2009;69:4918‐4925. [DOI] [PubMed] [Google Scholar]
- 45. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661‐672. [DOI] [PubMed] [Google Scholar]
- 46. Shin S, Saito E, Sawada N, et al. Dietary patterns and colorectal cancer risk in middle‐aged adults: A large population‐based prospective cohort study. Clin Nutr. 2018;37:1019‐1026. [DOI] [PubMed] [Google Scholar]
- 47. Ishihara J, Inoue M, Iwasaki M, et al. Dietary calcium, vitamin D, and the risk of colorectal cancer. Am J Clin Nutr. 2008;88:1576‐1583. [DOI] [PubMed] [Google Scholar]


