Abstract
Objective
The objective of this work was to causatively link biofilm properties of bacterial infection to specific pathogenic mechanisms in wound healing.
Background
Staphylococcus aureus is one of the four most prevalent bacterial species identified in chronic wounds. Causatively linking wound pathology to biofilm properties of bacterial infection is challenging. Thus, isogenic mutant stains of S. aureus with varying degree of biofilm formation ability was studied in an established preclinical porcine model of wound biofilm infection.
Methods
Isogenic mutant strains of S. aureus with varying degree (ΔrexB > USA300 > ΔsarA) of biofilm-forming ability were used to infect full-thickness porcine cutaneous wounds.
Results
Compared with that of ΔsarA infection, wound biofilm burden was significantly higher in response to ΔrexB or USA300 infection. Biofilm infection caused degradation of cutaneous collagen, specifically collagen 1 (Col1), with ΔrexB being most pathogenic in that regard. Biofilm infection of the wound repressed wound-edge miR-143 causing upregulation of its downstream target gene matrix metalloproteinase-2. Pathogenic rise of collagenolytic matrix metalloproteinase-2 in biofilm-infected wound-edge tissue sharply decreased collagen 1/collagen 3 ratio compromising the biomechanical properties of the repaired skin. Tensile strength of the biofilm infected skin was compromised supporting the notion that healed wounds with a history of biofilm infection are likely to recur.
Conclusion
This study provides maiden evidence that chronic S. aureus biofilm infection in wounds results in impaired granulation tissue collagen leading to compromised wound tissue biomechanics. Clinically, such compromise in tissue repair is likely to increase wound recidivism.
Keywords: biofilm, extracellular matrix, wound
Systematic review and meta-analysis of several hundred wound studies reported a 78.2% prevalence of biofilms in chronic wounds.1 A rapidly growing body of evidence establishes biofilm infection as a major cause of delayed wound healing.2–5 Staphylococcus is one of the predominant (65%) causes of persistent infections in chronic wounds.6 Bacteria of the Staphylococcus genus are highly efficient in establishing biofilms resulting in persistent infection.7 Gram-positive Staphylococcus is a nonmotile, and nonspore-forming bacteria that readily adheres to highly proteinaceous surfaces of chronic wounds and forms matrix-encased communities resulting in persistent chronic wound infection that is recalcitrant to antibacterial therapies.8
Despite substantial evidences linking biofilm infection with impaired wound healing, mechanisms underlying impairments in wound healing caused by biofilm infections have only recently started to unfold.4,5 Majority of studies investigating mechanisms underlying the pathogenicity of biofilm utilize in vitro biofilm models.9 While this approach provides the opportunity of elegant traceability, the absence of host factors compromises the relevance of such studies to wound infection in vivo.10 Although of a lesser magnitude, similar concerns apply to the study of biofilm grown on tissue explants lacking immune surveillance. Recent studies demonstrated a clear role of the host immune system in shaping microbial mechanisms relevant to biofilm pathology.11,12 Such mechanisms unfold over time.13,14 Thus, to understand the cascade of mechanisms following biofilm infection of the cutaneous wound, a long-term model involving deliberate infection of clinically isolated bacteria is necessary. It is widely accepted that porcine skin wound healing most closely resembles the human healing process. Furthermore, the human immune system has a higher similarity to the porcine immune system compared with rats or mice, making it a better suited model for studies on the host interactions that are integral to the complexities of the pathological biofilm in wound infections.10 The Wound Healing Society recommends the porcine model as the most relevant preclinical model of skin wound healing.15
Our interest in understanding the mechanism underlying biofilm infection in wound healing led to the development of a preclinical porcine persistent biofilm infection wound model.4,5 Using this model, we provided first evidence that biofilm-inducible microRNAs specifically silence tight junction proteins resulting in impaired barrier function of the neo-epidermis at the wound-site.5 Furthermore, our recent work underscores the significance of the preclinical porcine persistent biofilm infection model to study novel interventions.4 Staphylococcus aureus (SA) is one of the four most prevalent bacterial species identified in chronic wounds.16 While there are numerous studies testing the efficacy of a specific treatment for management of SA biofilm, understanding of molecular mechanisms explaining the pathogenicity of SA biofilm is scantly studied.17 While there is limited information from short-term rodent studies, there is a clear void in time-dependent understanding of molecular mechanisms in the porcine or human systems.
The current work is the first to specifically address the biofilm component of SA pathogenicity by the comparative use of three isogenic mutant strains of SA. Importantly, each of these strains is known to possess varying degrees of biofilm-forming ability. Well-characterized S. aureus USA300LAC (USA300) served as the model strain for wound infection. The biofilm-forming capability of this strain is well documented.18 The isogenic mutant strains USA300::-sarA (ΔsarA) and USA300::rexB (ΔrexB) were used as hypo- and hyperbiofilm-forming mutants, respectively.19,20 Staphylococcal accessory regulator (sarA) is one of the global regulators implicated in biofilm formation.20 Biofilm-forming capacity is compromised in sarA mutants,19,20 The Δrex B is a transposon mutant that was created by disruption of rexB, which encodes for an ATP-dependent helicase/nuclease subunit that is important in DNA repair of double-stranded breaks. We report the hyperbiofilm activity of this strain. This work represents the maiden effort to utilize mutant bacterial strains of graded biofilm-forming ability to interrogate biofilm-dependent mechanisms of action in the healing wound.
METHODS
Ethics Statement
The Ohio State University Institutional Animal care and Use Committee approved all animal experiments.
Porcine Full-thickness Burns and Biofilm Infection
Porcine infections were carried out as previously described.5 In brief, under general anesthesia, reproducible, full-thickness burn wounds were created bilaterally on the dorsolateral trunk. After hair removal with clippers, the dermis was prepped using alternating chlorhexidine and alcohol wipes. A total of six, 2″ × 2″ wounds were generated using a pressure and temperature controlled metallic dice. Wounds were individually covered with Tegaderm™ (3 M) dressing and the entire wound area secured with consecutive bandaging with V.A.C drape (Owens & Minor), VetWrap™, and Elasticon (3 M). After 3 days, the bandages were removed under general anesthesia to allow for bacterial inoculation of the burn wounds. A total of 108 CFU of S. aureus (SA) mutants sarA and rexB and wild-type SA, USA300 was inoculated onto the wounds topically and dispersed across the surface with sterile spatula. Control wounds were inoculated with vehicle only. The wounds were covered individually after bacterial inoculation and bandaged as described above. Full-thickness, excisional wound biopsies (oriented in the sagittal plane) were removed at days 7, 14, and 35 postbacterial inoculation and placed in 10% neutral-buffered formalin for >72 hours. Normal/nonburned skin (~1 cm) from each side of the wound was included in the biopsy. Skin samples were placed on wooden tongue depressors to maintain skin shape prior to fixation.
Histopathology, Immunocytochemistry, and Imaging
Histopathology immunocytochemistry and imaging was performed as described previously.4
Scanning Electron Microscope Imaging
Sample processing and imaging was performed as previously described.4
Secondary-intention Excisional Murine Dermal Wound Model
Young male (8 wk of age) C57BL/6 mice were used. Two 8 × 16-mm full-thickness excisional wounds21 were placed on the dorsal skin, equidistant from the midline and adjacent to the 4 limbs. Each of the 2 wounds was topically infected with isogenic strains of S. aureus USA300, USA300::rexB (ΔrexB) or USA300::sarA (ΔsarA). The animals were euthanized d5 postwound closure, and the wound areas excised for further analyses.
Skin Biomechanics
Tensile strength of the healed murine skin was quantified using tensile testing to failure. Mouse fur was removed from the excised, healed full-thickness skin, being careful to exclude the panniculus. Samples were obtained using a dog bone shaped punch (gauge length of 9.35 mm, gauge width of 3 mm) with the center of the wound positioned in the central region of the punch. Each skin sample was placed into the grips of the tensile tester (TestResources 100R, Shakopee, MN) outfitted with a 2.2lb load cell. Skin was placed in between 2 squares of gauze prior to manual tightening of the grips and subsequently strained at a rate of 2 mm/s until failure. If samples did not fail within the gauge length, data was discarded (note: in this study all samples failed within the gauge length). Maximum load at failure for each sample was extrapolated from the load (N)-position (mm) plot.
RNA Isolation and Quantitative Real-time PCR
Total RNA, including the miRNA, was isolated using mirVana RNA isolation kit followed by quantitative PCR assay as previously reported.22,23
Enzyme-linked Immunosorbent Assay
Levels of matrix metalloproteinase-2 (MMP-2) were measured using commercially available enzyme-linked immunosorbent assay kit following manufacturer’s protocol as previously described.22,23
miR-target 3′—UTR Luciferase Reporter Assay
miRIDIAN mimic-miR-143 were transfected to human Fibroblast cells followed by transfection with pmiR target MMP-2 3’UTR plasmids. Luciferase assays were performed using Gaussian luciferase reporter assay (Genecopia). Normalization was achieved by seap value. Data were presented as ratio of Gaussian luciferase to secreted alkaline phosphatase.
Hydroxyproline Assay
Collagen content of wound edge tissue was assessed by determining the amount of hydroxyproline using a commercially available hydroxy proline assay kit (MAK008 Sigma Aldrich) following manufacturers protocol. In brief, frozen wound tissue was pulverized under liquid nitrogen followed by acid hydrolysis using 12N hydrochloric acid (HCl) and water (1:1) for 3 hrs at 110°C. The hydrolyzed material was dehydrated overnight was at 60°C. For the assay, a mixture of Chloramine T and oxidation buffer was added to reaction and incubated at room temperature for 5 minutes. p-dimethylaminobenzaldehyde, percholic acid and isopropanol mixture was added to each well and incubated at 60°C for 90 minutes. The samples were cooled to room temperature and absorbance was measured at 562 nm, and the amount of hydroxyproline present in samples was determined by comparison to a standard curve.
Zymography
Gelatin zymography was performed. For each sample, 30 μg of total serum protein was loaded in 10% zymogram gels (Novex). Electrophoresis was carried out using the minigel slab apparatus Mini Protean 3 (Biorad) at a constant voltage of 150 V, until the dye reached the bottom of the gel. Following electrophoresis, gels were washed in renaturation buffer (2.5% Triton X-100 in 50 mM Tris–HCl (pH 7.5)) for 1 hour in an orbital shaker. Then the zymograms were incubated for 18 hours at 37°C in incubation buffer (0.15M NaCl, 10mM CaCl2, 0.02% NaN3 in 50mM Tris–HCl (pH 7.5)). Gels were then stained with Coomassie blue and destained with 7% methanol and 5% acetic acid. Areas of enzymatic activity appeared as clear bands over the dark background.
Cell Culture
Human immortalized fibroblasts were grown under standard culture conditions (at 37°C in a humidified atmosphere consisting of 95% air and 5% CO2) in DMEM growth medium supplemented with 10% FBS, 100 IU/mL antimicrobial-antimycotic, 10 mmol/L L-glutamine.
Preparation of Conditioned Media
Mutants and wild-type strains of S. aureus, ΔsarA, ΔrexB and USA300 (108 CFU/mL) bacterial biofilms were grown on a membrane and after 48 hours of growth the membranes were incubated with fibroblast media without antibiotics for 18 hours and filtered through 0.2 μm filter.
Statistics
In vitro data are reported as mean ± SEM of 3 to 8 experiments as indicated in the respective figure legends. Comparisons among multiple groups were tested using analysis of variance. For animal studies, data are reported as mean ± SEM of 3–6 animals as indicated. Student t test (2-tailed) was used to determine significant differences between means. Comparisons among multiple groups were tested using analysis of variance. P < 0.05 was considered statistically significant.
RESULTS
Biofilm-forming Ability of S. aureus (SA) Transposon Mutants in Porcine Burn Wounds
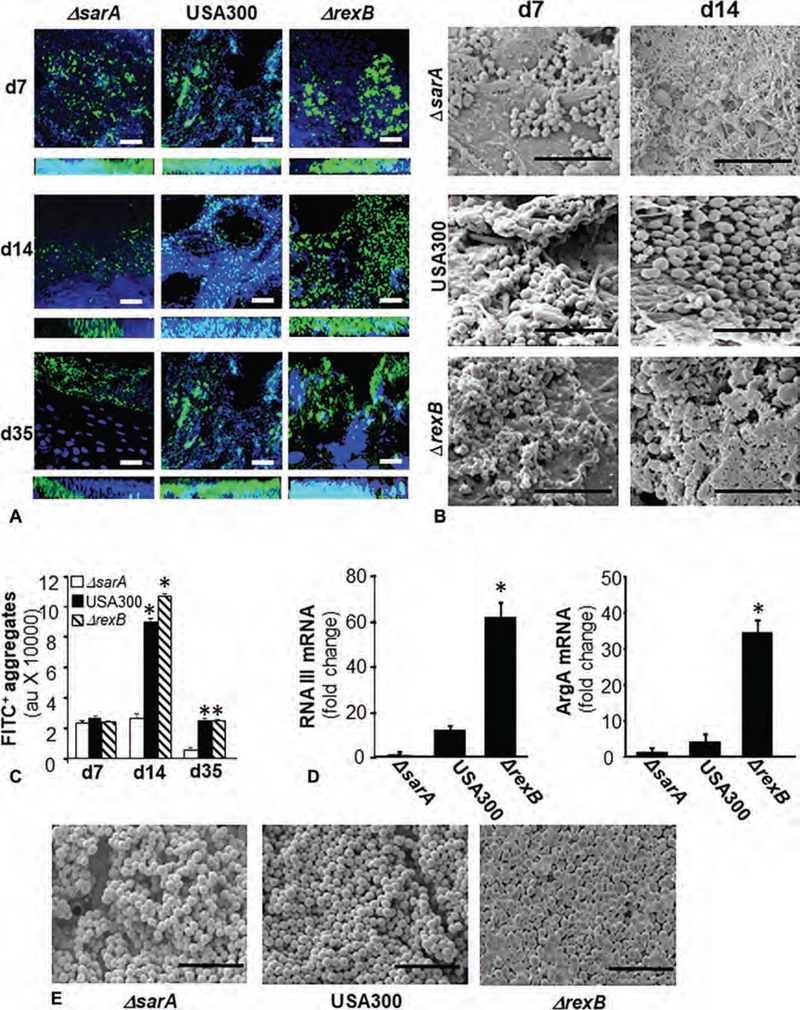
Day 3 porcine burn wounds were infected by isogenic strains of S. aureus USA300, ΔsarA or ΔrexB. The wounds were harvested at specific time points and biofilm formation ability was evaluated using 2 standardized approaches: 1) anti-SA antibody and confocal laser scanning microscopy (CLSM), and 2) Scanning Electron Microscopy (SEM) imaging. CLSM imaging studies revealed thick biofilm (Z section, 20 μm) at day 7 produced by all 3 strains. On day 14 and day 35 postinfection, USA 300 and Δrex B showed thick biofilm and large green SA aggregates. In contrast, biofilm by ΔsarA mutant was diminished at these latter time-points (Fig. 1A, C). Interestingly on day 14, SEM imaging show SA cocci embedded in thick Extracellular Polymeric Substance (EPS) in the case of ΔrexB mutant. Although a large number of cocci persisted in USA300 infected wounds, these cells were not embedded in thick EPS (Fig. 1B). A markedly diminished number of cocci not embedded in EPS were noted in ΔsarA supporting the CLSM data (Fig. 1B).
FIGURE 1.
Evidence of hyperbiofilm formation by S. aureus, transposon mutant USA300::rexB while USA300::sarA mutant is a hypobiofilm-forming strain in porcine burn wound infections. Six 2 × 2 sq inch size burn wounds were created on back of pigs. On day 3 post-burn, the wounds were infected by isogenic strains of S. aureus USA300: USA300, USA300::rexB (ΔrexB) or USA300::sarA (ΔsarA). A, Representative confocal laser scanning microscopy images from burn wounds on days 7, 14, and 35 postinfection showing S. aureus (anti-SA, green) aggregates. Counterstaining was performed using DAPI (blue, nucleus). Z-stack images of tissues sections (20 μm) from burn wound are shown. Scale bar = 50 μm. B, Representative scanning electron microscopy (SEM) images from SA infected burn wounds on days 7 and 14 postinfection. Scale bar = 5 μm. C, Quantifications of S. aureus aggregates for images shown in (A). Data are mean ± SEM (n = 3), *P < 0.05 compared with ΔsarA. D, Quantification of mRNA levels of biofilm/virulence specific genes in biofilm infected burn wound tissue was determined using real-time PCR. The data was normalized against 16 s rRNA. Data are mean ± SEM (n = 6), *P < 0.05 compared with ΔsarA. E, SEM images of in vitro SA biofilms developed on polycarbonate filters. Scale bar = 5 μm.
The gene expressions of RNAIII and agrA in day 35 wound-edge tissue were consistent with observations on magnitude of biofilm as observed by SEM. The expression of RNAIII and agrA genes were highly upregulated in ΔrexB as compared with ΔsarA mutant (Fig. 1D). CLSM and SEM imaging of in vitro SA biofilms developed on polycarbonate membrane supported the in vivo wound observations (Fig. 1E, S1, http://links.lww.com/SLA/B514) establishing ΔrexB as hyperbiofilm forming and ΔsarA as hypobiofilmforming USA300 mutant strains both in vitro and in vivo. Following the characterization of these mutant strains for their biofilm-forming ability, these mutants were used for investigating host wound healing responses.
Compromised Wound Epithelialization and Granulation Tissue Collagen Biosynthesis
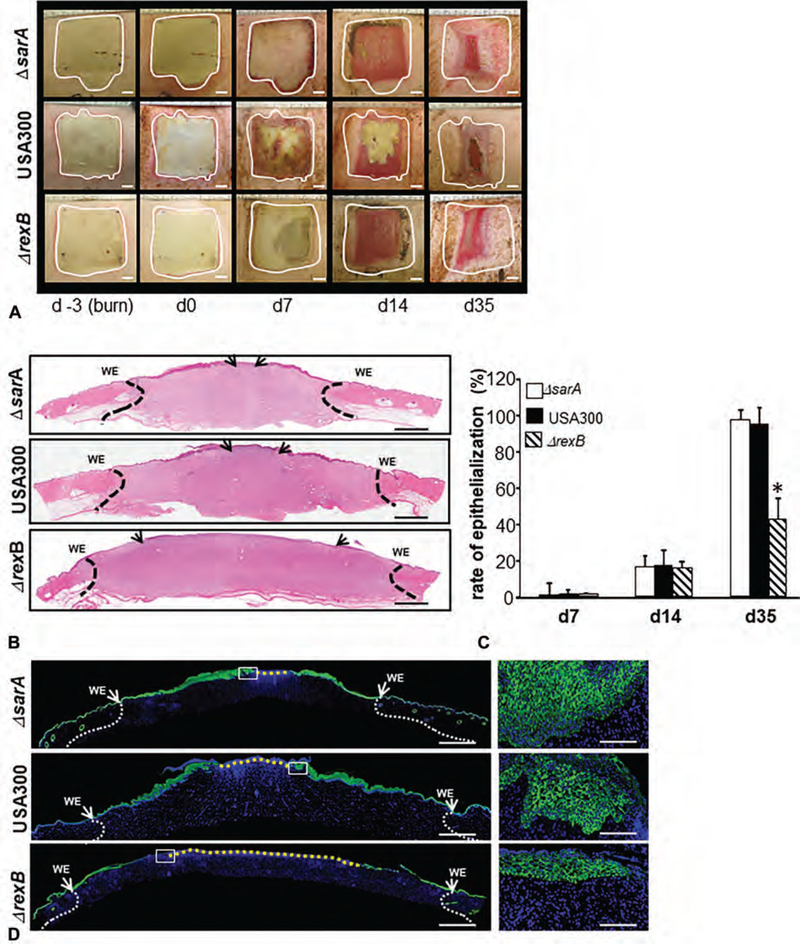
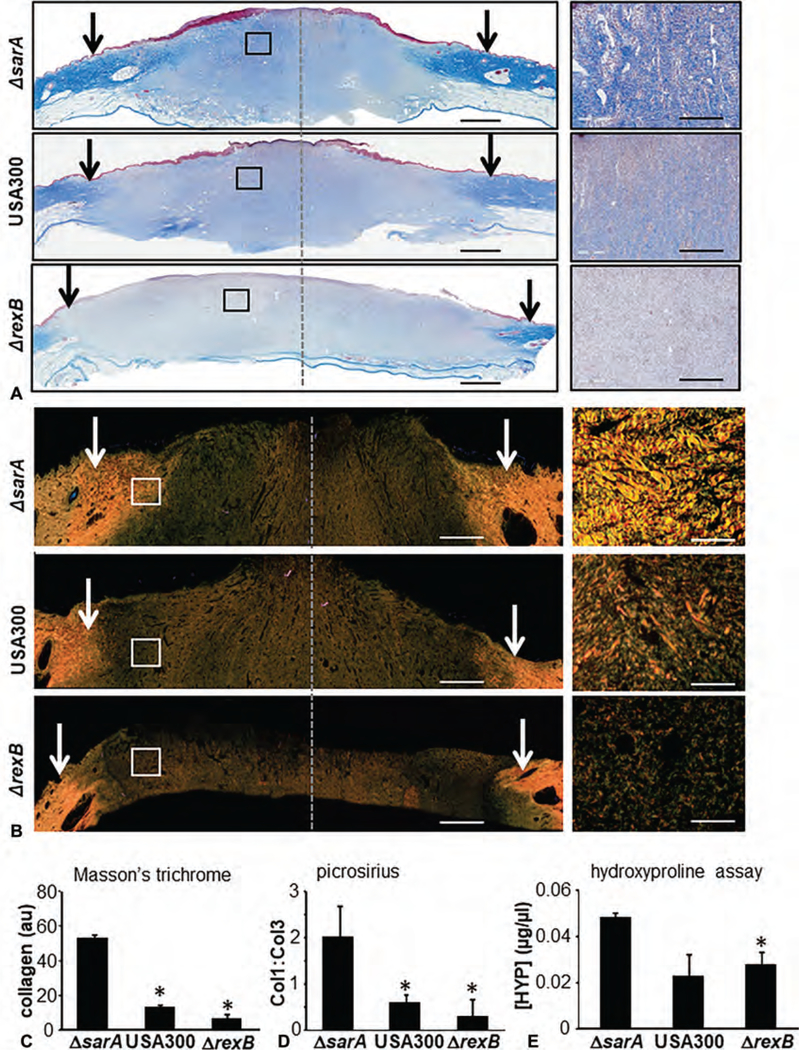
Wound closure, as determined by digital planimetry, was comparable among 3 types of infections studied (Fig. 2A). However, significant attenuation (~50%) of re-epithelialization was noted in whole wound cross-section histological images in hyperbiofilm-forming ΔrexB infected wounds compared with ΔsarA or USA300 infections (Fig. 2B–D). Masson’s trichrome staining revealed a marked reduction in granulation tissue collagen content (blue stain) in burn wounds infected with ΔrexB (hyperbiofilm) or USA300 infected wounds compared with group infected with ΔsarA (hypo-biofilm; Fig. 3A, C). The detection and analysis of collagen fibers in granulation tissue were additionally performed using Picrosirius red staining. This staining, under polarized light, allows the visualization of collagen bundles as green, red, or yellow that are clearly distinct from the black background allowing for quantitative analysis (Fig. 3B).24 Marked reduction in granulation tissue collagen level in ΔrexB (hyperbiofilm) and USA300 infected wounds were noted when compared with ΔsarA (hypo-biofilm) infected burn wounds (Fig. 3B, D). Hydroxyproline assay was performed to quantify collagen levels in wound-edge tissue. Significant loss of collagen in hyperbiofilm infected wounds was noted compared with hypobiofilm-infected SA mutant (Fig. 3E). To test the functional significance of this observation, tensile strength of the healed skin was studied in murine wounds infected with USA300, ΔsarA or ΔrexB. Concomitant with significantly compromised tensile strength following hyperbiofilm infection, the murine model recapitulated hyperbiofilm infection mediated loss of Collagen 1, repression of miR-143 and de-silencing of MMP-2 expression (Fig S3, http://links.lww.com/SLA/B514) as observed in the porcine model.
FIGURE 2.
Effects of S. aureus (SA) biofilm infection on burn wound healing evaluated using 3 strains of USA300 with varying degrees of biofilm-forming capability. On day 3 postburn, the wounds were infected by isogenic strains of S. aureus USA300, USA300::rexB (ΔrexB) or USA300::sarA (ΔsarA). A, Representative digital images of biofilm infected burn wounds on days 0, 7, 14, and 35 postinfection. The wound area on the day of burn has been demarcated with white dashed line. B, hematoxylin and eosin stained whole mount cross sections of infected burn wounds show re-epithelialization. The wound edges have been shown with dash black lines while the tip of migrated epithelium is shown with black arrows. WE indicates wound edge. Scale bar = 200 μm. C, rate of re-epithelialization (%) quantified from H&E stained images shown in (B). D, Immunofluorescence images of anti-K14 (green) stained biofilm infected burn wound tissue sections. Counter staining was performed with DAPI (blue, nucleus). Scale bar = 200 μm. Insets, zoom of the boxed area. Scale bar = 50 μm.
FIGURE 3.
Hyperbiofilm infection by S. aureus USA::ΔrexB in burn wounds severely compromises granulation tissue collagen content. A–C, On day 3 postburn, the porcine wounds were infected by isogenic strains of S. aureus USA300, USA300::rexB (ΔrexB) or USA300::sarA (ΔsarA). A, Representative images of formalin-fixed paraffin-embedded (FFPE) biofilm infected day 35 burn wound biopsy sections (5 μm) were stained using Masson Trichrome (MT). MT staining results in blue-black nuclei, blue collagen, and light red or pink cytoplasm. Epidermal cells appear reddish. Scale bar = 200 μm. The edges of the wound have been shown with black/white arrows. Right panels are the zoom in images of the boxed areas within the images in the left panels. Scale bar, 50 μm. B, Representative images of formalin-fixed paraffin- embedded (FFPE) biofilm infected day 35 burn wound biopsy sections (5 μm) were stained using Picrosirius red (PS) staining. Scale bar = 200 μm. Insets zoom of the box area. Scale bar = 50 μm. C, Bar graph shows quantitation of collagen abundance using MT stains. Data are mean ± SEM (n = 6), *P < 0.05 compared with ΔsarA. D, Bar graph shows quantitation of collagen abundance using PS stains. Data are mean ± SEM (n = 3), *P < 0.05 compared with ΔsarA. E, Granulation (d35 postinfection) tissue collagen content was determined using hydroxyproline assay. Data are mean ± SEM (n = 6), *P < 0.05 compared with ΔsarA.
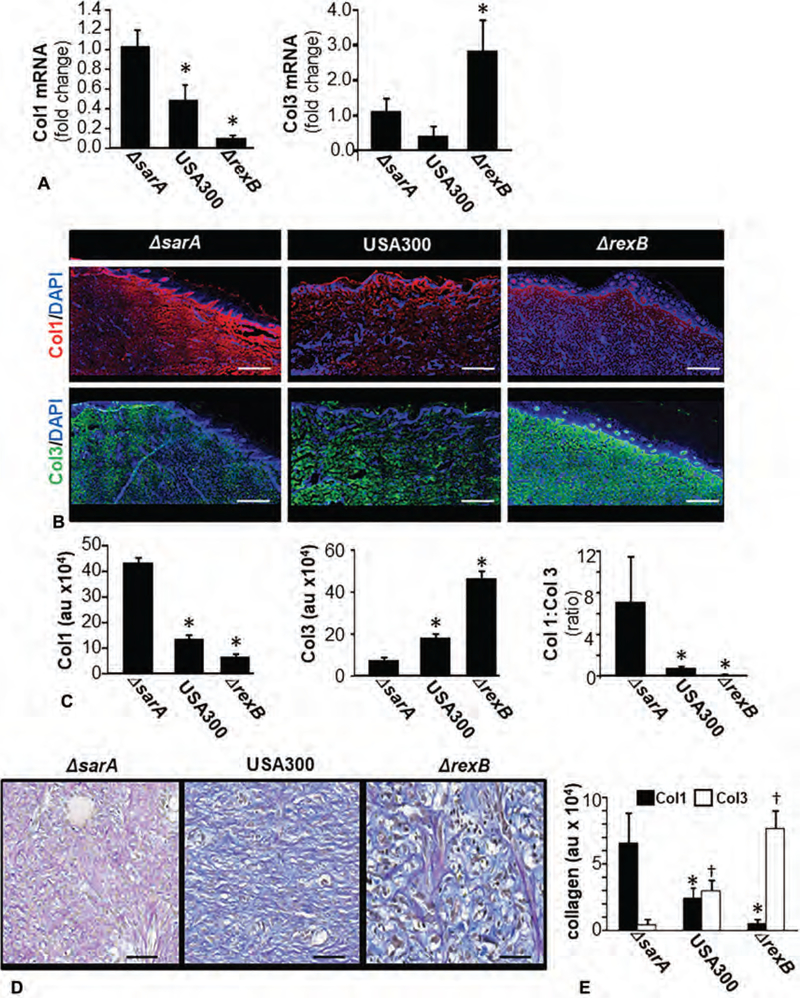
Loss of Collagen Type I (Col1) in Granulation Tissue
Significant reduction in both Col1 mRNA and protein expression were noted in USA300 as well as ΔrexB infected compared with the ΔsarA infected wounds (Fig. 4A–C). Levels of the immature collagen Col3 were higher in the granulation tissue of wounds infected with ΔrexB (hyperbiofilm; Fig. 4A–C). Findings with Col1 and Col3 were further characterized using Herovici staining (Fig. 4D–E). This polychrome staining specifically identifies and quantifies the expression and distribution of types I (purple/red) and III (blue) collagen.25
FIGURE 4.
Dysregulation of Col1 and Col3 at wound-site granulation tissue in S. aureus biofilm-infected burn wounds. On day 3 post-burn, the wounds were infected by isogenic strains of S. aureus USA300, USA300::rexB (ΔrexB) or USA300::sarA (ΔsarA). Wound biopsies were collected at specified time-points after inoculation with S. aureus USA300 isogenic strains: USA300, USA300::rexB (ΔrexB), or USA300::sarA (ΔsarA). A, Expressions of Col1 and Col3 mRNA in wound biopsies collected on day 35 post-inoculation. Data presented as mean ± SEM (n = 4), * P < 0.05 compared with ΔsarA. B, Representative images of Col1 (red, anti-col1) and Col3 (green, anti-Col3) stained sections on days 35 post-inoculation. The sections were counterstained using DAPI (nuclear, blue). Scale bar = 100 μm. C, Bar graphs present quantitation of Col1 and Col3 signal intensity and their ratio in (B). Data are presented as mean ± SEM (n = 4), *P < 0.05 compared with ΔsarA. D, Herovici stained images of biofilm infected burn wound tissue sections from d35 post-infection burn wounds. Herovici stains young collagen reticulum blue (Col3) and mature collagen red (Col1) while providing a yellow cytoplasm counterstain. Nuclei are stained to black with Weigert Hematoxylin. E, Bar graphs present quantitation of Col1 and Col3 signal intensity and their ratio in (D). scale bar = 50 μm. Data are presented as mean ± SEM (n = 4), *P < 0.05 compared with Col1 of ΔsarA +P < 0.05 compared with Col3 of ΔsarA.
miR-143 and MMP-2 Expression and Activity in Wound Fibroblasts Is Regulated by SA Biofilm Infection
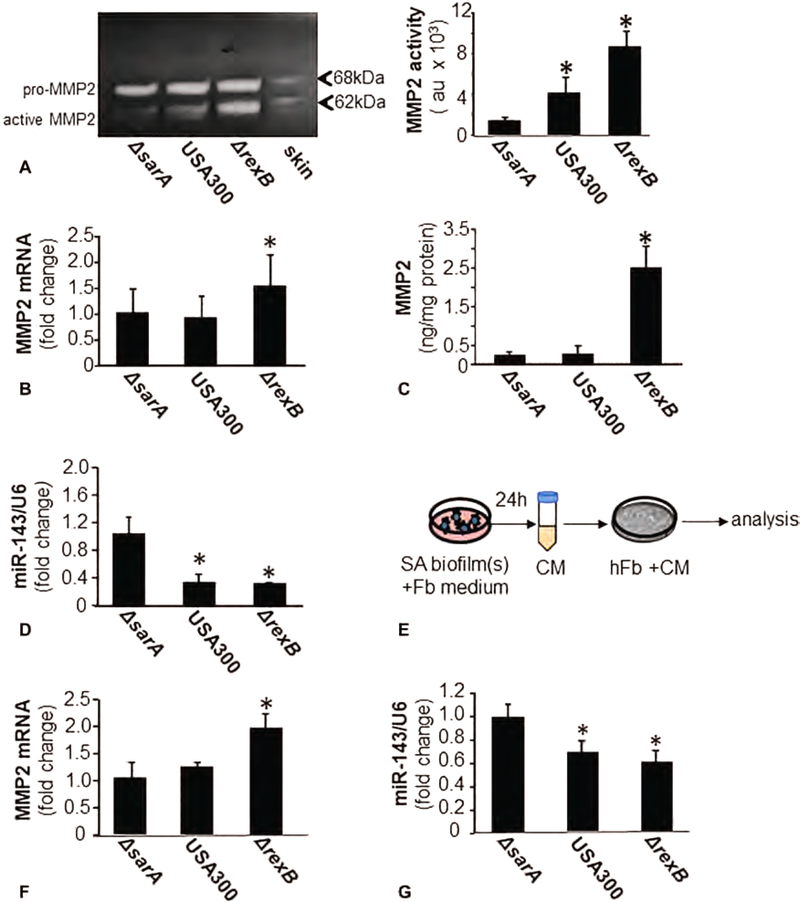
In our search for mechanisms silencing Col1 in the granulation tissue, we hypothesized that biofilm infection induces host and/or bacterial collagenases at the wound-site. Thus, bacterial collagenase activity was measured from SA-biofilm-infected wounds using a bacterial collagenase-specific synthetic peptide FALGPA (N-[3-(2-Furyl)acryloyl]-Leu-Gly-Pro-Ala). Bacterial collagenases, compared with vertebrate collagenases, show broader substrate specificity and make multiple cleavage within the triple helical region.26 No significant change was noted in the bacterial collagenase activity in wounds infected with any of the three isogenic strains of bacteria (Fig S5, http://links.lww.com/SLA/B514). Zymography is routinely employed for measurement of collagenolytic activity of tissue extracts.27 Zymography of denatured Col1, also referred to as gelatin, demonstrated a marked increase in activated MMP-2 expression in ΔrexB and USA 300 infected wounds as compared with ΔsarA (hypobiofilm) infected burn wounds (Fig. 5A). A significant induction in the transcript and protein expression of MMP-2 was observed in hyperbiofilm-infected wounds (Fig. 5B, C). We identified miR-143 as a S. aureus biofilm-sensitive miRNA which when repressed leads to the desilencing of MMP-2 and loss of collagen. miR-143 is abundantly expressed in vascular smooth muscle cells and fibroblasts. Interestingly, ΔrexB and USA300 biofilm infection in burn wounds downregulated miR-143 (Fig. 5D). In wounds, fibroblasts are the primary cell type of the granulation tissue. We used hTERT immortalized cultured human fibroblasts (hFb) and exposed these cells to conditioned media (CM) from in vitro mature biofilms from the three isogenic strains of SA studied in this work (Fig. 5E). Exposure of hFb to CM from hyperbiofilm-forming ΔrexB resulted in increased MMP-2 and loss of miR-143 (Fig. 5F, G). The translational significance of the findings in porcine and murine wounds was tested in human engineered (HE) skin. HE skin was treated with CM from in vitro mature biofilms. Exposure of HE skin to CM from hyperbiofilm-forming ΔrexB suppressed miR-143, desilenced MMP-2 causing loss of Collagen 1 protein (Figs S6, http://links.lww.com/SLA/B514). This observation supports the notion that part of the biofilm-induced loss of Col1 in wounds could be attributed to the effect of biofilm-released soluble factors on wound-site fibroblasts.
FIGURE 5.
miR-143 and MMP-2 expression/activity in wounds fibroblasts is regulated by SA biofilm infection. Wound biopsies were collected at specified time points post-inoculation with S. aureus USA300 isogenic strains: USA300, USA300::rexB (ΔrexB) or USA300::sarA (ΔsarA). A, Representative, gelatin zymogram of d35 porcine infected burn wounds tissue. Based on molecular weight, pro- (70 kDa) and active MMP-2 (62 kDa) bands have been indicated. Quantification of active MMP-2 (62 kDa) band using densitometry. Data are mean ± SEM (n = 4) *P < 0.05 compared with ΔsarA. B, MMP-2 mRNA in d35 post-inoculation porcine infected burn wounds tissues quantified using real-time PCR. Data was normalized against 18S RNA as housekeeping gene. Data are mean ± SEM (n = 6) *P < 0.05 compared with ΔsarA. C, MMP-2 protein in d35 post-inoculation porcine infected burn wounds tissues quantified using ELISA. Data are mean ± SEM (n = 6). *P < 0.05 compared with ΔsarA. D, miR-143 expression in d35 post-inoculation porcine infected burn wounds tissues quantified using real-time PCR. Data was normalized against U6 snRNA as housekeeping genes. Data are mean ± SEM (n = 6) *P < 0.05 compared with ΔsarA. E-G, Human fibroblasts (hFb) were cultured with overnight conditioned media from SA biofilms. Mature biofilms developed on polycarbonate membrane in vitro were placed in cultured dish and incubated with fibroblast (Fb) culture media overnight. The conditioned media was harvested, centrifuged, filtered and then added to hFb. E, Schematic presentation of the experimental model. F, MMP-2 mRNA expression in hTERT immortalized cultured human fibroblasts (hFb) with conditioned media from biofilms cultured in vitro. Data are mean ± SEM (n = 6) *P < 0.05 compared with ΔsarA. G, miR-143 expression in hTERT immortalized cultured human fibroblasts (hFb) with conditioned media from biofilms cultured in vitro. Data are mean ± SEM (n = 3) *P < 0.05 compared with ΔsarA.
MMP-2, a Direct Target of miR-143 in Fibroblasts
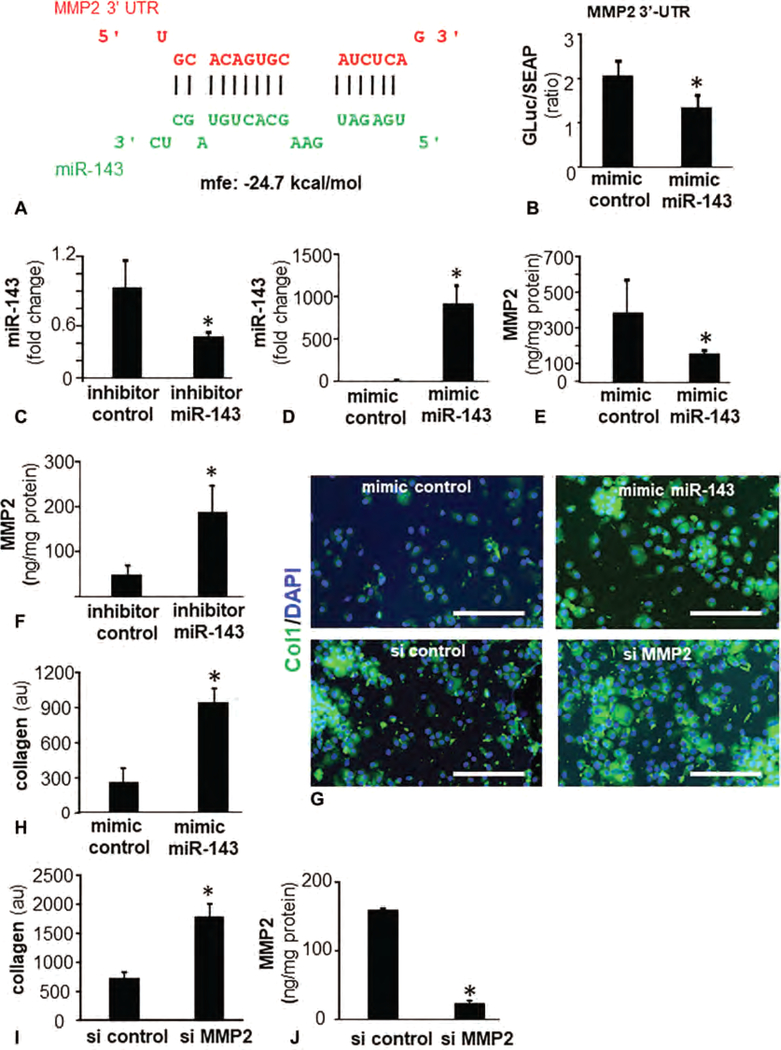
Bioinformatics analyses using RNAHybrid28 recognized miR-143 with potential binding sites on MMP-2 mRNA (Fig. 6A). Using MMP-2 3’-UTR firefly luciferase expression constructs the effect of miR-143 on MMP-2 transcription was tested (Fig. 6B). To directly test the significance of this miR on MMP-2 and collagen expression, studies using miRidian miR-143 mimic and inhibitors (Fig. 6C, D) were performed (Fig. 6C–G). Delivery of miR-143 mimics or their corresponding inhibitors showed that the miRNA was inversely associated with the expression of corresponding MMP-2 protein (Fig. 6E, F). Bolstering miR-143 levels in hFb using mimic resulted in strong induction of cellular collagen expression (Fig. 6G, H). Knockdown of MMP-2 using siRNA significantly increased collagen 1 protein (Fig. 6I, J). Thus, lowering of miR-143 as observed following ΔrexB hyperbiofilm infection desilenced and thus upregulated MMP-2 resulting in lowering of collagen 1 (Fig. 7).
FIGURE 6.
MMP-2 is a direct target of miR-143 and regulates fibroblast collagen levels. A, miR-143 predicted to target MMP-2 3’UTR based on RNA Hybrid algorithm. MMP-2 transcript is NM_004530. Binding position of miR-143 (green) corresponds to position 604–602 of 3′-UTR of MMP-2 (red). B, Luciferase activity in human fibroblasts after transfection with mimic miR-143 and mimic control. Data are mean ± SEM (n = 6) *P < 0.05 compared with mimic control. C and D, Expression of miR-143 in human fibroblasts transfected with (C) miR-143 inhibitor (D) miR-143 mimic. E and F, Expression of MMP-2 in human fibroblasts transfected with (E) miR-143 mimic (F) miR-143 inhibitor. G, Fluorescence microscopy images of collagen 1 (green) Col1 expression in hFb following bolstering of miR143 with mimic miR-143 (upper panel) and knockdown of MMP-2 with siMMP2. Counterstaining was performed using DAPI (blue, nuclear). H, Bar graph presents quantitation of Col1 from (G, upper panel). Data are mean ± SEM (n = 3). *P < 0.05 compared with mimic control. I–J, Knockdown of MMP-2 with siMMP2. Data are mean ± SEM (n = 4). *P < 0.05 compared with si control.
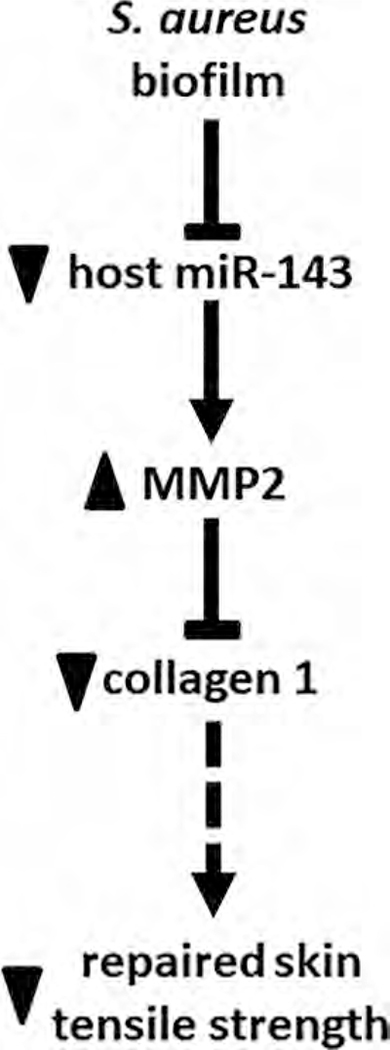
FIGURE 7.
S. aureus biofilm results in loss of Col1 and reduces tensile strength through a miR-143-MMP-2-dependent pathway. Solid lines indicate pathways based on data from this work. Broken lines are based on literature (Eleswarapu SV et al, Tensile properties, collagen content, and crosslinks in connective tissues of the immature knee joint. PLoS One 2011; 6(10):e26178. Lodish H et al., Collagen: The Fibrous Proteins of the Matrix. In: Lodish H, Berk A, Zipursky Sea, eds. Molecular Cell Biology. New York: W. H. Freeman;2000. Roeder BA et al., Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J Biomech Eng - Transactions of the Asme 2002; 124(2):214–222.
DISCUSSION
Collagenopathies, caused by genetic defects in collagen formation, have substantial health impact affecting almost each and every tissue system.29 Collagenopathy of the skin markedly changes the biomechanical properties of the skin such that they are easily torn by the slightest trauma.30 In wound healing, this poses the risk of wound recidivism which represents a major threat to the growing cost of wound care.31 This work compares three isogenic mutant strains of SA with graded biofilm-forming ability to come to the conclusion that wound biofilm infection causes significant reduction in collagen type I in the wound bed compromising tensile strength of the repaired tissue. Such outcomes increase the risk of wound recidivism.32 Given the high incidence of biofilm infection reported in chronic wounds, the current work is of extraordinary significance drawing attention to the quality of closure of biofilm infected wounds. Our previous work has reported that although the effects of chronic biofilm infection on wound closure may be marginal over a period of two months, such infection results in failure of the skin barrier function at the site of healing.4,5 The current work demonstrates that biofilm infection impairs granulation tissue collagen deposition leading to increased risk of wound recurrence as predicted by compromised tensile strength of the repaired tissue. Skin fibroblasts are critical in contributing the extracellular matrix (ECM) to support physiological wound healing. Collagen is one of the most abundant ECM proteins in skin as well as in a healing wound.33 In a healing wound, Col3 appears first as an early collagen in granulation tissue which is then gradually replaced by Col1, a central protein that is responsible for the wound tensile strength.34 The counter balance of ECM synthesis is provided by matrix MMPs. On one hand, MMPs are essential for physiological wound healing. MMPs break down ECM, thus allowing migration of cells and remodeling of the injured tissue. On the other hand, excessive MMPs in the wound milieu could be detrimental, resulting in nonhealing chronic wounds. High levels of MMPs are a common characteristic property of chronic wounds.35
This study utilizes a unique approach of in vivo graded biofilm infection in a preclinical porcine burn wounds to establish a direct cause and effect relationship between methicillin-resistant S. aureus (MRSA) biofilm infected wound and defective collagen levels via a novel miRNA-MMP axis. S. aureus is one of the most common causes of hospital/community acquired wound infections. In our efforts to establish an in vivo preclinical burn chronic biofilm infection model where the host responses to SA biofilm infection may be causatively linked to specific host wound healing response, we utilized USA300 and isogenic mutant strains ΔsarA and ΔrexB, with varying biofilm-forming capabilities. In 1959, with the introduction of semisynthetic penicillin, methicillin, the wave of resistance emerged with MRSA that gained considerable attention due to invasive nature of the infection.36 MRSA is capable of forming robust biofilm and persists in host contributing to relapsing chronic infections in wound tissue.3 USA300, the model organism used in the current study is a specific MRSA lineage that was first reported by McDougal et al37 in 2003. USA300 is a predominant strain in US hospitals.38
Fibroblasts from chronic wounds have been reported to produce defective collagen.39 Consistently, we noted that in hyper-biofilm-infected wounds there was a marked reduction in both Col1 transcript as well as protein indicative of blocked Col1 synthesis. Col1 is a predominant collagen in mature dermis as well as in late phase of wound healing.34 In the wound granulation tissue, fibroblasts differentiate to form myofibroblasts which deposit collagen.40 Soluble factors released from SA biofilm are known to limit migration and differentiation of fibroblasts.41 In this work, we observed that exposure of isolated fibroblasts to secreted products from SA biofilms blunted Col1 expression. Collagen biosynthesis involves: 1) expression of polypeptides and posttranslational modification, 2) folding into a triple helix, and 3) secretion and maturation of procollagen to collagen.42 Post-transcriptional control plays a pivotal role in the synthesis and production of Col1 polypeptides.43 miRNA are about 18 to 24 nucleotide long, single-stranded RNA molecules that have emerged as key post-transcriptional regulators of gene expression.44 A number of miRNA, including miR-29 and 143/145 clusters, regulate collagen expression.45,46 While miR-29 was nonresponsive to biofilm-infection in burn wounds, this study reports potent inhibition of miR-143 expression in response to hyperbiofilm infection. miR-143/145 is abundantly expressed in cells of mesenchymal origin.47 A major role of this cluster in intestinal wound repair is evident.48 Under conditions of hypertrophic scarring, miR-143–3p attenuates ECM production-associated proteins including the expression of Col1 and 3. This is achieved by targeting CTGF via the Akt/mTOR pathway.49 In fibroblasts, direct inverse regulation of Col3 by miR-143 has been reported.46 Increased Col3 expression in hyper-biofilm infected wounds may be explained by the loss of miR-143 in wound granulation tissue fibroblasts. Tight control of collagen levels in physiological wound healing is maintained by a fine balance of biosynthesis and breakdown. In the wound microenvironment, MMPs contribute to the breakdown of collagen.35 This work recognizes S. aureus MRSA biofilm infection as a potent inducer of MMP-2 activity. Such regulation is indirect and caused by biofilm-dependent depletion of miR-143. miR-143 is directly implicated in posttranscriptional silencing of MMP-2. Thus, this work provides a mechanistic explanation to the common observation that chronic wounds are rich in MMP-2.50
Taken together, this study utilized SA strains with graded biofilm-forming ability to specifically dissect the biofilm-dependent pathogenic mechanisms in the healing wound. Long-term outcomes studied in an established porcine model of biofilm infection, show striking collagen 1 deficiency in the wound granulation tissue. Such deficiency compromised wound re-epithelialization. Biofilm inhibited collagen synthesis and induced collagenolytic MMP-2 by lowering miR-143. Low collagen 1, a major structural protein of ECM, in wounds culminated in poor wound tensile strength of the repaired skin. Clinical studies looking for higher wound recidivism of closed wounds with a history of biofilm infection are warranted.
Supplementary Material
ACKNOWLEDGMENTS
The following reagent was provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH: Staphylococcus aureus subsp. aureus, Strain JE2, Transposon Mutant NE1012 (SAUSA300_0869), NR-47555, NE1193 (SAUSA300_0605), NR-47736. Technical help from Dr Kasturi Ganesh Barki is acknowledged for pig experiments and tissue collection.
This work was partly supported by National Institute of Health NR015676, NR013898 and Diacomp, NIDDK (Augusta University). In addition, it benefited from the following National Institutes of Health awards: GM077185, GM069589, GM108014, DK076566, DK114718, AI097511, and NS42617.
Footnotes
The authors report no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.annalsofsurgery.com).
REFERENCES
- 1.Malone M, Bjarnsholt T, McBain AJ, et al. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care. 2017;26:20–25. [DOI] [PubMed] [Google Scholar]
- 2.Wolcott R Disrupting the biofilm matrix improves wound healing outcomes. J Wound Care. 2015;24:366–371. [DOI] [PubMed] [Google Scholar]
- 3.Percival SL, McCarty SM, Lipsky B. Biofilms and wounds: an overview of the evidence. Adv Wound Care (New Rochelle). 2015;4:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barki KG, Das A, Dixith S, et al. Electric field based dressing disrupts mixed-species bacterial biofilm infection and restores functional wound healing. Ann Surg. 2017. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy S, Elgharably H, Sinha M, et al. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J Pathol. 2014;233:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. [DOI] [PubMed] [Google Scholar]
- 7.Götz F Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–1378. [DOI] [PubMed] [Google Scholar]
- 8.Kowalewska-Grochowska K, Richards R, Moysa GL, et al. Guidewire catheter change in central venous catheter biofilm formation in a burn population. Chest. 1991;100:1090–1095. [DOI] [PubMed] [Google Scholar]
- 9.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. [DOI] [PubMed] [Google Scholar]
- 10.Ganesh K, Sinha M, Mathew-Steiner SS, et al. Chronic wound biofilm model. Adv Wound Care (New Rochelle). 2015;4:382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanke ML, Kielian T. Deciphering mechanisms of staphylococcal biofilm evasion of host immunity. Front Cell Infect Microbiol. 2012;2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulcahy LR, Isabella VM, Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb Ecol. 2014;68:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielen KS, Jongers B, Boddaert J, et al. Biofilm-induced type 2 innate immunity in a cystic fibrosis model of pseudomonas aeruginosa. Front Cell Infect Microbiol. 2017;7:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omar A, Wright JB, Schultz G, et al. Microbial biofilms and chronic wounds. Microorganisms. 2017;5 E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordillo GM, Bernatchez SF, Diegelmann R, et al. Preclinical models of wound healing: is man the model? proceedings of the wound healing society symposium. Adv Wound Care (New Rochelle). 2013;2:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schierle CF, De la Garza M, Mustoe TA, et al. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 2009;17:354–359. [DOI] [PubMed] [Google Scholar]
- 18.Hidron AI, Low CE, Honig EG, et al. Emergence of community-acquired meticillin-resistant Staphylococcus aureus strain USA300 as a cause of necrotising community-onset pneumonia. Lancet Infect Dis. 2009;9:384–392. [DOI] [PubMed] [Google Scholar]
- 19.Abdelhady W, Bayer AS, Seidl K, et al. Reduced vancomycin susceptibility in an in vitro catheter-related biofilm model correlates with poor therapeutic outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57:1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archer NK, Mazaitis MJ, Costerton JW, et al. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy S, Khanna S, Nallu K, et al. Dermal wound healing is subject to redox control. Mol Ther. 2006;13:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das A, Ghatak S, Sinha M, et al. Correction of MFG-E8 resolves inflammation and promotes cutaneous wound healing in diabetes. J Immunol. 2016;196: 5089–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das A, Ganesh K, Khanna S, et al. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol. 2014;192:1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lattouf R, Younes R, Lutomski D, et al. Picrosirius red staining: a useful tool to appraise collagen networks in normal and pathological tissues. J Histochem Cytochem. 2014;62:751–758. [DOI] [PubMed] [Google Scholar]
- 25.Turner NJ, Pezzone MA, Brown BN, et al. Quantitative multispectral imaging of Herovici’s polychrome for the assessment of collagen content and tissue remodelling. J Tissue Eng Regen Med. 2013;7:139–148. [DOI] [PubMed] [Google Scholar]
- 26.Mookhtiar KA, Van Wart HE. Clostridium histolyticum collagenases: a new look at some old enzymes. Matrix Suppl. 1992;1:116–126. [PubMed] [Google Scholar]
- 27.Tyagi SC, Matsubara L, Weber KT. Direct extraction and estimation of collagenase(s) activity by zymography in microquantities of rat myocardium and uterus. Clin Biochem. 1993;26:191–198. [DOI] [PubMed] [Google Scholar]
- 28.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison B, Sanniec K, Janis JE. Collagenopathies-implications for abdominal wall reconstruction: a systematic review. Plast Reconstr Surg Glob Open. 2016;4:e1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellini MH, Caldini ET, Scapinelli MP, et al. Increased elastic microfibrils and thickening of fibroblastic nuclear lamina in canine cutaneous asthenia. Vet Dermatol. 2009;20:139–143. [DOI] [PubMed] [Google Scholar]
- 31.Sen CK, Gordillo GM, Roy S, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doillon CJ, Dunn MG, Bender E, et al. Collagen fiber formation in repair tissue: development of strength and toughness. Coll Relat Res. 1985;5:481–492. [DOI] [PubMed] [Google Scholar]
- 33.Albaugh VL, Mukherjee K, Barbul A. Proline precursors and collagen synthesis: biochemical challenges of nutrient supplementation and wound healing. J Nutr. 2017;147:2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clore JN, Cohen IK, Diegelmann RF. Quantitation of collagen types I and III during wound healing in rat skin. Proc Soc Exp Biol Med. 1979;161:337–340. [DOI] [PubMed] [Google Scholar]
- 35.Trengove NJ, Stacey MC, MacAuley S, et al. Analysis of the acute and chronic wound environments: the role of proteases and their inhibitors. Wound Repair Regen. 1999;7:442–452. [DOI] [PubMed] [Google Scholar]
- 36.Carrel M, Perencevich EN, David MZ. USA300 Methicillin-Resistant Staphylococcus aureus, United States, 2000–2013. Emerg Infect Dis. 2015;21: 1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDougal LK, Steward CD, Killgore GE, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diekema DJ, Richter SS, Heilmann KP, et al. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol. 2014;35:285–292. [DOI] [PubMed] [Google Scholar]
- 39.Cook H, Davies KJ, Harding KG, et al. Defective extracellular matrix reorganization by chronic wound fibroblasts is associated with alterations in TIMP-1, TIMP-2, and MMP-2 activity. J Invest Dermatol. 2000;115:225–233. [DOI] [PubMed] [Google Scholar]
- 40.Gabbiani G The myofibroblast in wound healing and fibrocontractive diseases. J Pathol. 2003;200:500–503. [DOI] [PubMed] [Google Scholar]
- 41.Kshetri P, Brennan RE, Vaughan MB. The effects of Staphylococcus aureus biofilm conditioned medium on fibroblast tension generation and migration. FASEB J. 2016;30(suppl 1). 1034.9. [Google Scholar]
- 42.Kivirikko KI. Collagen biosynthesis: a mini-review cluster. Matrix Biol. 1998;16:355–356. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Stefanovic B. Akt mediated phosphorylation of LARP6; critical step in biosynthesis of type I collagen. Sci Rep. 2016;6:22597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. [DOI] [PubMed] [Google Scholar]
- 45.Wang B, Komers R, Carew R, et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naito Y, Sakamoto N, Oue N, et al. MicroRNA-143 regulates collagen type III expression in stromal fibroblasts of scirrhous type gastric cancer. Cancer Sci. 2014;105:228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent OA, McCall MN, Cornish TC, et al. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014;42:7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chivukula RR, Shi G, Acharya A, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu S, Kang B, Zeng W, et al. MicroRNA-143–3p inhibits hyperplastic scar formation by targeting connective tissue growth factor CTGF/CCN2 via the Akt/mTOR pathway. Mol Cell Biochem. 2016;416:99–108. [DOI] [PubMed] [Google Scholar]
- 50.Krejner A, Grzela T. Modulation of matrix metalloproteinases MMP-2 and MMP-9 activity by hydrofiber-foam hybrid dressing—relevant support in the treatment of chronic wounds. Cent Eur J Immunol. 2015;40:391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.