Abstract
Fosmidomycin inhibits IspC (1-deoxy-d-xylulose 5-phosphate reductoisomerase), the first committed enzyme in the methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis. The MEP pathway of isoprenoid biosynthesis is essential to the causative agent of the plague, Yersinia pestis, and is entirely distinct from the corresponding mammalian pathway. To further drug development, we established structure–activity relationships of fosmidomycin analogues by assessing a suite of 17 α-phenyl-substituted reverse derivatives of fosmidomycin against Y. pestis IspC. Several of these compounds showed increased potency over fosmidomycin with IC50 values in the nanomolar range. Additionally, we performed antimicrobial susceptibility testing with Y. pestis A1122 (YpA1122). The bacteria were susceptible to several compounds with minimal inhibitory concentration (MIC) values ranging from 128 to 512 μg/mL; a correlation between the IC50 and MIC values was observed.
Introduction
The plague, also known as the “Black Death,” is caused by the bacterium Yersinia pestis.1 The discovery of antibiotics, beginning with the introduction of penicillin in the 1940s,2 was anticipated to be the end of bacterial diseases.3 However, the evolution of antibiotic resistance has since resolved this notion. In fact, the increase in drug resistance has coincided with a decline in antibiotic discovery.3 Compounding the issue of antibiotic resistance is the threat of bioterrorism, which became a reality with the 2001 anthrax attacks.3,4
In 2000, the US Centers for Disease Control and Prevention (CDC) released a response plan for bioterrorism, wherein they categorized several infectious agents according to their likelihood to be used as biothreats.5 According to the CDC, Y. pestis is presently classified as a “category A” bioterrorism agent, that is, a high-priority agent.5 High-priority agents are classified as such by their ease of transmission, high mortality rate, and likelihood to require a specialized public health response.5
Antibiotic treatment is effective against plague bacteria;6 however, a 2017 plague outbreak in Madagascar resulted in approximately 2417 cases and 209 deaths within a period of 4 months.7 The plague takes two main clinical forms: bubonic and pneumonic.6 Thirty to sixty percent of cases of bubonic plague result in fatality,6 and, if left untreated, pneumonic plague is always fatal.1 Given the nature of antibiotic resistance, the need for preventative biothreat countermeasures, and the recent plague epidemics in Madagscar,7 continued development of antibiotics is necessary for the prevention of widespread outbreaks and deaths.
Isoprenoids are one of the largest and most diverse group of natural products, enumerating over 30,000 known products.8,9 They are fundamental biomolecules involved in vital biological functions such as electron transport and peptidoglycan biosynthesis in bacteria.8,10,11 Bacteria synthesize isoprenoids via the methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis.12,13 The MEP pathway is entirely distinct from the corresponding mammalian pathway, the mevalonic acid pathway, making it an attractive target for antibiotic development.12,13
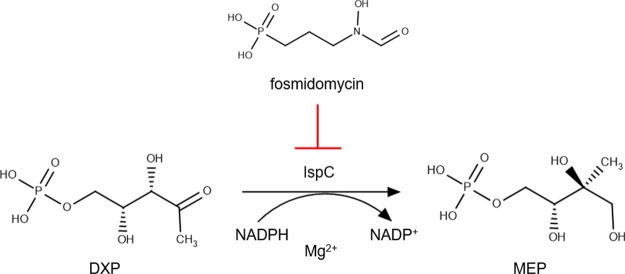
Previously, we cloned, expressed, and characterized the first committed enzyme of the MEP pathway, 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR/IspC) from Y. pestis.14 IspC catalyzes the reduction and isomerization of 1-deoxy-d-xylulose 5-phosphate (DXP) to yield 2-C-methylerythritol 4-phosphate (MEP) (Figure 1).15 Furthermore, we demonstrated the effectiveness of inhibiting both the purified Y. pestis enzyme (YpIspC) and liquid cultures of Y. pestis using the known IspC inhibitor, fosmidomycin (Figure 1).14
Figure 1.
Inhibition of IspC by fosmidomycin.
Previous studies have shown that α-phenyl substitutions of reverse derivatives of fosmidomycin are efficacious.16−18 Furthermore, crystal structures of IspC- and α-phenyl-substituted reverse derivatives of fosmidomycin have been resolved.19−21 However, no crystal structures of YpIspC in the presence of fosmidomycin or one of its analogues have been resolved. Therefore, to elucidate the structure–activity relationships (SARs) of α-phenyl-substituted reverse derivatives of fosmidomycin, we systematically assessed the potency of 17 compounds against YpIspC and performed antimicrobial susceptibility assays with YpA1122. Specifically, we assessed the effect of methylene, oxygen, or sulfur at the β-position, the electron-donating or -withdrawing effects of the substituents on the α-phenyl ring, and the substitution pattern of the α-phenyl ring (Figure 2).
Figure 2.
α-Phenyl-substituted β-carba, thia, oxa reverse fosmidomycin analogues. The β position, X, is either carbon (β-carba), sulfur (β-thia), or oxygen (β-oxa). R, R1, and R2 are either CH3, OCH3, Cl, or F.
Results and Discussion
Biological Evaluation of IspC Inhibitors
All assayed compounds were synthesized according to previously described procedures.19−24 The addition of differently substituted phenyl rings at the α-position of fosmidomycin analogues can increase their potency against IspC enzymes from bacteria and the protozoan, Plasmodium falciparum.17,19−21,24 However, to our knowledge, no inhibitors from this class have been tested against YpIspC. Furthermore, no crystal structures of YpIspC bound to fosmidomycin or one of its analogues have been resolved. To expand upon our understanding of SARs between YpIspC and fosmidomycin analogues, we screened a comprehensive library of our α-phenyl substituted reverse fosmidomycin analogues (Table 1). The determined half-maximal inhibitory concentrations (IC50 values) and the determined minimal inhibitory concentrations (MICs) for each compound are presented in Table 1. YpA1122 is indicated as being either resistant (R), or susceptible (S), to each compound.
Table 1. Inhibition of Y. pestis A1122 and Y. pestis IspC by α-Phenyl-Substituted Reverse Fosmidomycin Analogues.
| compound | X | R | YpIspC IC50 (μM)a | 95% confidence interval of YpIspC IC50 (μM) | YpA1122 MIC (μg/mL)b |
|---|---|---|---|---|---|
| 1a | CH2 | Ph | 1.06 | 0.88–1.27 | 512 (S) |
| 1b | S | Ph | 0.39 | 0.29–0.51 | 128 (S) |
| 1c | O | Ph | 4.03 | 2.84–5.74 | >512 (R) |
| 2a | CH2 | 4-CH3-Ph | 3.42 | 2.43–4.81 | >512 (R) |
| 2b | S | 4-CH3-Ph | 0.72 | 0.54–0.97 | 256 (S) |
| 2c | O | 4-CH3-Ph | 6.21 | 3.93–9.80 | >512 (R) |
| 2d | CH2 | 4-OCH3-Ph | 2.38 | 1.47–3.86 | >512 (R) |
| 3a | CH2 | 3,4-Cl-Ph | 1.36 | 1.06–1.74 | >512 (R) |
| 3b | S | 3,4-Cl-Ph | 0.19 | 0.15–0.23 | 128 (S) |
| 3c | O | 3,4-Cl-Ph | 1.80 | 1.25–2.61 | >512 (R) |
| 3d | CH2 | 3,4-F-Ph | 0.30 | 0.24–0.38 | 128 (S) |
| 3e | S | 3,4-F-Ph | 0.25 | 0.18–0.36 | 128 (S) |
| 3f | O | 3,4-F-Ph | 1.90 | 1.34–2.70 | >512 (R) |
| 3g | CH2 | 3,4-OCH3-Ph | 12.75 | 9.75–16.66 | >512 (R) |
| 4a | S | 3,5-OCH3-Ph | 3.82 | 2.22–6.55 | >512 (R) |
| 4b | S | 3,5-F-Ph | 0.32 | 0.23–0.46 | 256 (S) |
| 5 | O | 2,4-F-Ph | 6.24 | 3.94–9.89 | >512 (R) |
| fosmidomycin | 0.71c | 128 (S) |
YpIspC = IspC from Y. pestis, IC50 = half-maximal inhibitory concentration. All assays were performed in duplicate.
YpA1122 = Y. pestis strain A1122, MIC = minimal inhibitory concentration, R = resistant, S = susceptible. Six replicates were performed for each compound.
The most potent compounds in Table 1 (β-thia and β-carba analogues 3b, 3d, 3e, and 4b) are twice as potent as fosmidomycin, with IC50 values in the nanomolar range. To assess the effect of replacing the β-methylene group with either a sulfur or oxygen atom, the potency of four isosteric sets of β-carba, β-thia, and β-oxa analogues (1a–1c, 2a–2c, 3a–3c, and 3d–3f) was determined. For each set of isosteres, the β-thia analogues showed increased inhibition over their β-carba and β-oxa counterparts, with the β-oxa analogues showing the lowest levels of inhibition among the three. This pattern of inhibition was observed for both the enzymes and the bacteria. These results are consistent with our previous findings with IspC orthologs from Escherichia coli and Mycobacterium tuberculosis.20 Taken together, these results further establish that replacement of the β-methylene group with a sulfur atom is an effective strategy for the development of reverse fosmidomycin analogues for IspC proteins of bacterial origins.
Concomitantly, the electron-donating or -withdrawing effects of the substituents on the α-phenyl ring were assessed. The compounds contained either electron-donating groups (CH3, OCH3, compounds 2a–d, 3g, 4a) or electron-withdrawing groups (Cl, F, compounds 3a–f, 4b, 5). Compounds without substituents on the α-phenyl ring were assessed for comparison (compounds 1a–c). Compounds containing electron-donating groups showed decreased potency over analogues without substitutions on the α-phenyl ring. Compound 1a is three times more potent than its methylated analog, 2a, and 12 times more potent than its 3,4-dimethoxyphenyl analog 3g. Furthermore, compounds 2a and 3g have no activity against the bacteria at 512 μg/mL. Similarly, YpA1122 is susceptible to 1b at 128 μg/mL, whereas YpA1122 is susceptible to its methylated analog, 2b, at 256 μg/mL, and resistant to its 3,5-dimethoxyphenyl analog, 4a, at 512 μg/mL.
Conversely, compounds containing electron-withdrawing groups showed increased potency over analogues without substitutions or electron-donating groups on the α-phenyl ring. 3,4-Difluorophenyl analog, 3d, is three times more potent than 1a, and 42 times more potent than 3g. Similarly, 3,4-dichlorophenyl analog, 3b, is 20 times more potent than 3,5-dimethoxyphenyl analog, 4a. Four compounds were further examined to probe the effect of the substitution pattern of the α-phenyl ring. 3,4-Difluorophenyl compound, 3e, was more potent than its 3,5-difluorophenyl isostere, 4b. This correlation can also be observed with the Y. pestis bacterium. YpA1122 is susceptible to 3e at 128 μg/mL, whereas YpA1122 is susceptible to 4b at 256 μg/mL. Additionally, 3,4-difluorophenyl compound, 3f, is three times more potent than its 2,4-difluorophenyl isostere, 5. Altogether, these results demonstrate that compounds containing electron-withdrawing groups (Cl, F) show promise for inhibitor design and suggest that 3,4-substitution patterns are more promising than 3,5- or 2,4-substitution patterns. Further study is warranted to confirm the SARs of the substitution pattern conclusively.
In E. coli, fosmidomycin is known to enter bacterial cells via the glycerol-3-phosphate transporter (GlpT).25−27 Conversely, fosmidomycin uptake is limited in other organisms, such as M. tuberculosis, by the lack of a GlpT.26 Previously, using a BLAST search with the E. coli K12 GlpT sequence (accession no. P08194), we identified a homologous transport protein (accession no. YP_002347496) in the Y. pestis CO92 proteome.14 A BLAST search with this transport protein (accession no. YP_002347496) identifies a homologous transporter (accession no. AEL73320, 100% identity) in the Y. pestis A1122 proteome (taxonomy ID: 1035377).
It is possible that uptake of fosmidomycin and/or fosmidomycin analogues 1a, 1b, 2b, 3b, 3d, 3e, and 4b is fully or partially dependent on the Y. pestis A1122 transporter. Previous studies have also shown that uptake of fosmidomycin analog, FR900098, is only partially dependent on GlpT, and that uptake of lipophilic phosphonate prodrugs of fosmidomycin analogues is not dependent on GlpT.28 In these studies, the more hydrophobic nature of these compounds was attributed as a source of their partial-dependence or independence on GlpT.28 Future study is warranted to confirm the mechanism of uptake of these analogues and to assess their dependence on a transporter protein to enter the cell. Nonetheless, the enzyme SARs offer insight for the direction of future synthesis and possibly a lipophilic phosphonate prodrug strategy may yield better bacterial growth inhibition.
Conclusions
In summary, we report kinetic data and antimicrobial susceptibility data to establish SARs of a suite of 17 α-phenyl substituted reverse derivatives of fosmidomycin for YpIspC. These compounds varied by substitution of the β-methylene with oxygen or sulfur, addition of electron-donating or -withdrawing substituents on the α-phenyl ring, and by the substitution pattern of the α-phenyl ring. Our results showed that replacement of the β-methylene group with a sulfur atom is a useful strategy for developing reverse fosmidomycin analogues for YpIspC. This result is consistent with other IspC proteins of bacterial origins. We also found that attaching electron-donating substituents to the α-phenyl ring is not a useful strategy for developing reverse fosmidomycin analogues for YpIspC. Lastly, our results show that 3,4-difluorophenyl substitutions are more potent than 3,5- or 2,4-difluorophenyl substitutions; however, further study is needed to unequivocally establish the relationship between potency and the substitution pattern. These results provide useful information for the future development of novel IspC inhibitors.
Experimental Section
Bacterial Cell Culture
Recombinant proteins were expressed in E. coli BL21 CodonPlus (DE3)-RIL cells (Stratagene, La Jolla, CA). E. coli was cultured at 37 °C in Luria–Bertani (LB) media supplemented with 100 μg/mL ampicillin and 50 μg/mL chloramphenicol with constant shaking at 250 rpm. Agar (1.5% wt/vol) was added to prepare solid media. Y. pestis strain A1122 was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH. The YpA1122 bacterial isolates were subcultured on blood agar plates (tryptic soy agar with 5% sheep blood) 48 h prior to antimicrobial susceptibility testing.
Expression and Purification of Y. pestis IspC
Y. pestis IspC was cloned, expressed, and purified as described previously.14 The Y. pestis ispC gene was synthesized (GenScript USA Inc, Piscataway, NJ) and cloned into a pET101/D-TOPO vector to facilitate the expression of a C-terminal His6-tagged protein. The plasmid was transformed into chemically competent E. coli BL21 CodonPlus (DE3)-RIL cells (Stratagene, La Jolla, CA) for protein expression.
To express the His6-tagged protein, 1 L of LB media was inoculated with a 10 mL overnight seed culture and incubated with shaking at 37 °C and 250 rpm. At an OD600 of 1.8, protein expression was induced by addition of isopropyl b-d-thiogalactopyranoside to 0.5 mM. After protein induction, the culture was incubated with shaking at 37 °C and 250 rpm for an additional 18 h. Cells were harvested via centrifugation (4648g, 20 min, 4 °C) and stored at −80 °C. Protein was subsequently isolated and purified from the cells via chemical lysis and affinity chromatography.
Cells were lysed with lysis buffer 1 (100 mM Tris pH 8.0, 0.032% lysozyme, 3 mL per gram cell pellet), followed by lysis buffer 2 (0.1 M CaCl2, 0.1 M MgCl2, 0.1 M NaCl, 0.020% DNase, 0.3 mL per gram cell pellet). The clarified cell lysate was collected after centrifugation (48,000g, 20 min, 4 °C) and passed through a TALON immobilized metal affinity column (Clontech Laboratories, Mountain View, CA).
The column was washed with 20 column volumes of equilibrium buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 300 mM NaCl), 10 column volumes of wash buffer 1 (50 mM HEPES pH 7.5, 300 mM NaCl, 10 mM imidazole), and 15 column volumes of wash buffer 2 (100 mM HEPES pH 7.5, 600 mM NaCl, 20 mM imidazole). The protein was eluted with 5 column volumes of elution buffer (150 mM imidazole pH 7.0, 300 mM NaCl), and then exchanged into storage buffer (0.1 M Tris pH 7.5, 1 mM NaCl, 5 mM dithiothreitol) during concentration by ultrafiltration. The protein concentration was determined using Advanced Protein Assay Reagent (Cytoskeleton, Denver CO) with γ-globulins (Sigma-Aldrich) as the standard. Purified protein was visualized via Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The yield of YpIspC averaged 30 mg per 1 L of culture.
Enzyme Assays
Y. pestis IspC activity was assayed at 37 °C by spectrophotometrically monitoring the enzyme catalyzed oxidation of NADPH upon addition of 1-deoxy-d-xylulose 5-phosphate (DXP, Echelon Biosciences, Salt Lake City, UT) to the assay mixture, as described previously.29 The oxidation of NADPH was monitored at 340 nm using an Agilent 8453 UV–visible Spectrophotometer equipped with a temperature-regulated cuvette holder. All assays were performed in duplicate.
Half-maximal inhibition (IC50) of enzyme activity was determined using nonlinear regression of a plot of fractional enzyme activity as a function of inhibitor concentration (sigmoidal dose–response curve) using GraphPad Prism 5.0, wherein the top plateau of the curve was set to 100% residual enzyme activity. As fosmidomycin is a slow, tight-binding inhibitor,15 the fosmidomycin analogues were preincubated with the enzyme at 37 °C for 10 min prior to the addition of the substrate, DXP.
Antimicrobial Susceptibility Assays
The MIC of each compound was determined using the broth microdilution method outlined by the Clinical and Laboratory Standards Institute (CLSI).30 Briefly, the compounds were twofold diluted in cation-adjusted Mueller–Hinton Broth (CAMHB) in 96-well round-bottom, polystyrene microtiter plates to final concentrations ranging from 1 to 512 μg/mL. Wells containing broth only served as growth and sterility controls. YpA1122 was cultured on blood agar plates (tryptic soy agar with 5% sheep blood) at 37 °C for 48 h. After 48 h, the colonies were directly suspended in phosphate buffered saline and diluted in CAMHB before inoculation into the wells of the 96-well plate to yield a final starting inoculum concentration of approximately 5 × 105 colony-forming units (CFUs)/mL. The plates were incubated at 37 °C for 24 h. The MIC was recorded as the lowest concentration of compound that inhibited visible bacterial growth after 24 h of incubation (i.e., no turbidity is observed and optical density at 600 nm is zero). Six replicates were performed for each compound.
Synthesis of Compounds
The synthesis of the carba analog 1a was performed according to Behrendt et al., 2010 and the carba analogues 2a, 2d, 3a, and 3d according to Behrendt et al., 2011.19,23 The thia analogues 1b, 2b, 3b–c were synthesized following the same procedures described by Kunfermann et al., 2013 and the analogues 4a–b according to Lienau et al., 2019.20,24 The oxa analogues 1c, 2c, 3c, 3f, and 5 were synthesized according to Brücher et al., 2012 and the 3,4 dimethoxy analog, 3g, was synthesized according to Konzuch et al., 2014.21,22
Acknowledgments
This work was generously supported by the George Mason University Department of Chemistry and Biochemistry, the U.S. Army MRMC (W81XWH-17-C-0066 to R.D.C.), the Military Infectious Diseases Research Program (W0161_15_WR to S.M.N.), and by an appointment to the Student Research Participation Program at the Walter Reed Army Institute of Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRMC.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b04171.
The authors declare no competing financial interest.
Supplementary Material
References
- Pechous R. D.; Sivaraman V.; Stasulli N. M.; Goldman W. E. Pneumonic Plague: The Darker Side of Yersinia Pestis. Trends Microbiol. 2016, 24, 190–197. 10.1016/j.tim.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Lobanovska M.; Pilla G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future?. Yale J. Biol. Med. 2017, 90, 135–145. [PMC free article] [PubMed] [Google Scholar]
- Davies J. Where Have All the Antibiotics Gone?. Can. J. Infect Dis. Med. Microbiol. 2006, 17, 287–290. 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Moayeri M.; Leppla S. H. Anthrax Lethal and Edema Toxins in Anthrax Pathogenesis. Trends Microbiol 2014, 22, 317–325. 10.1016/j.tim.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biological and Chemical Terrorism: Strategic Plan for Preparedness and Response. Recommendations of the CDC Strategic Planning Workgroup. Morbidity and Mortality Weekly Report, 2000; Vol. 49 (RR-4), pp 1–14. [PubMed]
- Plague https://www.who.int/news-room/fact-sheets/detail/plague (accessed Jan 23, 2019).
- Nguyen V. K.; Parra-Rojas C.; Hernandez-Vargas E. A. The 2017 Plague Outbreak in Madagascar: Data Descriptions and Epidemic Modelling. Epidemics 2018, 25, 20–25. 10.1016/j.epidem.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Lange B. M.; Rujan T.; Martin W.; Croteau R. Isoprenoid Biosynthesis: The Evolution of Two Ancient and Distinct Pathways across Genomes. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 13172–13177. 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard J.; Moreira D. Origins and Early Evolution of the Mevalonate Pathway of Isoprenoid Biosynthesis in the Three Domains of Life. Mol. Biol. Evol. 2011, 28, 87–99. 10.1093/molbev/msq177. [DOI] [PubMed] [Google Scholar]
- Jawaid S.; Seidle H.; Zhou W.; Abdirahman H.; Abadeer M.; Hix J. H.; van Hoek M. L.; Couch R. D. Kinetic Characterization and Phosphoregulation of the Francisella Tularensis 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (MEP Synthase). PLoS One 2009, 4, e8288 10.1371/journal.pone.0008288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuston S.; Begley M.; Gahan C. G. M.; Hill C. Isoprenoid Biosynthesis in Bacterial Pathogens. Microbiology 2012, 158, 1389–1401. 10.1099/mic.0.051599-0. [DOI] [PubMed] [Google Scholar]
- Rohdich F.; Bacher A.; Eisenreich W. Isoprenoid Biosynthetic Pathways as Anti-Infective Drug Targets. Biochem. Soc. Trans. 2005, 33, 785–791. 10.1042/bst0330785. [DOI] [PubMed] [Google Scholar]
- Singh N.; Cheve G.; Avery M.; McCurdy C. Targeting the Methyl Erythritol Phosphate (MEP) Pathway for Novel Antimalarial, Antibacterial and Herbicidal Drug Discovery: Inhibition of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase (DXR) Enzyme. Curr. Pharm. Des. 2007, 13, 1161–1177. 10.2174/138161207780618939. [DOI] [PubMed] [Google Scholar]
- Haymond A.; Johny C.; Dowdy T.; Schweibenz B.; Villarroel K.; Young R.; Mantooth C. J.; Patel T.; Bases J.; Jose G. S.; et al. Kinetic Characterization and Allosteric Inhibition of the Yersinia Pestis 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase (MEP Synthase). PLoS One 2014, 9, e106243 10.1371/journal.pone.0106243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppisch A. T.; Fox D. T.; Blagg B. S. J.; Poulter C. D. E. coli MEP Synthase: Steady-State Kinetic Analysis and Substrate Binding. Biochemistry 2002, 41, 236–243. 10.1021/bi0118207. [DOI] [PubMed] [Google Scholar]
- Andaloussi M.; Henriksson L. M.; Wiȩckowska A.; Lindh M.; Björkelid C.; Larsson A. M.; Suresh S.; Iyer H.; Srinivasa B. R.; Bergfors T.; et al. Design, Synthesis, and X-ray Crystallographic Studies of α-Aryl Substituted Fosmidomycin Analogues as Inhibitors ofMycobacterium tuberculosis1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase. J. Med. Chem. 2011, 54, 4964–4976. 10.1021/jm2000085. [DOI] [PubMed] [Google Scholar]
- Haemers T.; Wiesner J.; Poecke S. V.; Goeman J.; Henschker D.; Beck E.; Jomaa H.; Calenbergh S. V. Synthesis of α-substituted fosmidomycin analogues as highly potent Plasmodium falciparum growth inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 1888–1891. 10.1016/j.bmcl.2005.12.082. [DOI] [PubMed] [Google Scholar]
- Jansson A. M.; Więckowska A.; Björkelid C.; Yahiaoui S.; Sooriyaarachchi S.; Lindh M.; Bergfors T.; Dharavath S.; Desroses M.; Suresh S.; et al. DXR Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues. J. Med. Chem. 2013, 56, 6190–6199. 10.1021/jm4006498. [DOI] [PubMed] [Google Scholar]
- Behrendt C. T.; Kunfermann A.; Illarionova V.; Matheeussen A.; Pein M. K.; Gräwert T.; Kaiser J.; Bacher A.; Eisenreich W.; Illarionov B.; et al. Reverse Fosmidomycin Derivatives against the Antimalarial Drug Target IspC (Dxr). J. Med. Chem. 2011, 54, 6796–6802. 10.1021/jm200694q. [DOI] [PubMed] [Google Scholar]
- Kunfermann A.; Lienau C.; Illarionov B.; Held J.; Gräwert T.; Behrendt C. T.; Werner P.; Hähn S.; Eisenreich W.; Riederer U.; et al. IspC as Target for Antiinfective Drug Discovery: Synthesis, Enantiomeric Separation, and Structural Biology of Fosmidomycin Thia Isosters. J. Med. Chem. 2013, 56, 8151–8162. 10.1021/jm4012559. [DOI] [PubMed] [Google Scholar]
- Konzuch S.; Umeda T.; Held J.; Hähn S.; Brücher K.; Lienau C.; Behrendt C. T.; Gräwert T.; Bacher A.; Illarionov B.; et al. Binding Modes of Reverse Fosmidomycin Analogues toward the Antimalarial Target IspC. J. Med. Chem. 2014, 57, 8827–8838. 10.1021/jm500850y. [DOI] [PubMed] [Google Scholar]
- Brücher K.; Illarionov B.; Held J.; Tschan S.; Kunfermann A.; Pein M. K.; Bacher A.; Gräwert T.; Maes L.; Mordmüller B.; et al. α-Substituted β-Oxa Isosteres of Fosmidomycin: Synthesis and Biological Evaluation. J. Med. Chem. 2012, 55, 6566–6575. 10.1021/jm300652f. [DOI] [PubMed] [Google Scholar]
- Behrendt C. T.; Kunfermann A.; Illarionova V.; Matheeussen A.; Gräwert T.; Groll M.; Rohdich F.; Bacher A.; Eisenreich W.; Fischer M.; et al. Synthesis and Antiplasmodial Activity of Highly Active Reverse Analogues of the Antimalarial Drug Candidate Fosmidomycin. ChemMedChem 2010, 5, 1673–1676. 10.1002/cmdc.201000276. [DOI] [PubMed] [Google Scholar]
- Lienau C.; Gräwert T.; Alves Avelar L. A.; Illarionov B.; Held J.; Knaab T. C.; Lungerich B.; van Geelen L.; Meier D.; Geissler S.; et al. Novel Reverse Thia-Analogues of Fosmidomycin: Synthesis and Antiplasmodial Activity. Eur. J. Med. Chem. 2019, 181, 111555. 10.1016/j.ejmech.2019.07.058. [DOI] [PubMed] [Google Scholar]
- Armstrong C. M.; Meyers D. J.; Imlay L. S.; Meyers C. F.; Odom A. R. Resistance to the Antimicrobial Agent Fosmidomycin and an FR900098 Prodrug through Mutations in the Deoxyxylulose Phosphate Reductoisomerase Gene (Dxr). Antimicrob. Agents Chemother. 2015, 59, 5511–5519. 10.1128/aac.00602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. C.; Parish T. Dxr Is Essential in Mycobacterium Tuberculosis and Fosmidomycin Resistance Is Due to a Lack of Uptake. BMC Microbiol. 2008, 8, 78. 10.1186/1471-2180-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y.; Furukawa S.; Ogihara H.; Yamasaki M. Fosmidomycin Resistance in Adenylate Cyclase Deficient (cya) Mutants ofEscherichia coli. Biosci., Biotechnol., Biochem. 2003, 67, 2030–2033. 10.1271/bbb.67.2030. [DOI] [PubMed] [Google Scholar]
- McKenney E. S.; Sargent M.; Khan H.; Uh E.; Jackson E. R.; Jose G. S.; Couch R. D.; Dowd C. S.; van Hoek M. L. Lipophilic Prodrugs of FR900098 Are Antimicrobial against Francisella Novicida In Vivo and In Vitro and Show GlpT Independent Efficacy. PLoS One 2012, 7, e38167 10.1371/journal.pone.0038167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S.; Kuzuyama T.; Watanabe H.; Seto H. A 1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase Catalyzing the Formation of 2-C-Methyl-D-Erythritol 4-Phosphate in an Alternative Nonmevalonate Pathway for Terpenoid Biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 9879–9884. 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, 2015; Vol. 3. CLSI supplement M45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.