Abstract
The epidermis has an essential function in creating a barrier against the external environment to retain proper fluid balance and block the entry of pathogens. When damage occurs to this barrier, the wound must quickly be sealed to avoid fluid loss, cleared of invading pathogens, and then keratinocytes must re-form an intact barrier. This requires complex integration of temporally and spatially distinct signals to execute orderly closure of the wound, and failure of this process can lead to chronic ulceration. Transcription factors serve as a key integration point for the myriad of information coming from the external environment, allowing for an orderly process of re-epithelialization. Importantly, transcription factors engage with and alter the chromatin structure around key target genes through association with different chromatin-modifying complexes. In this review, we will discuss the current understanding of how transcription is regulated during the initiation of re-epithelialization, and the exciting technological advances that will allow for a more refined mechanistic understanding of the re-epithelialization process.
Keywords: Wound healing, transcription, skin, keratinocytes, migration, re-epithelialization
Introduction
The skin is the largest organ in the human body and is responsible for preventing fluid loss, blocking pathogens, and providing a physical barrier against the external environment. Efficient and rapid wound healing following skin injury is therefore critical to maintain proper homeostasis. This requires coordinated action between the many cell types that make up the skin (Figure 1a). Cutaneous wound healing can be divided into three major stages, the Inflammatory Phase, the Proliferative Phase, and the Matrix Remodeling Phase [1-4]. Immediately following wounding, vasoconstriction and recruitment of platelets quickly occurs, resulting in the formation of a fibrin clot that effectively stops hemorrhage. In addition to re-establishing hemostasis, platelets secrete cytokines such as Platelet-Derived Growth Factor (PDGF), which trigger the inflammatory response by attracting immune cells to clear wound bed from pathogenic agents and cell debris [5]. Release of cytokines and growth factors from infiltrating immune cells then help to initiate the Proliferative Phase, where keratinocytes at the wound edge break down their hemidesmosomes, severing their secure attachment to the basement membrane and adopt an activated state, characterized by a change in cytoskeletal network, cell surface receptors, the expression of integrins αVβ5, αVβ6, and α5β1, and of cytokeratins KRT6, KRT16, and KRT17 [4,6]. These leading edge keratinocytes switch to a non-proliferative, migratory mode and move to fill in the damaged epidermis while keratinocytes further away from the wound proliferate to replace the migrating cells [3,4]. When the keratinocytes have covered the wound area, they re-form a stratified epidermis, while coordinated action of macrophages, fibroblasts, and endothelial cells results in the formation of the granulation tissue beneath the wound [3,4].
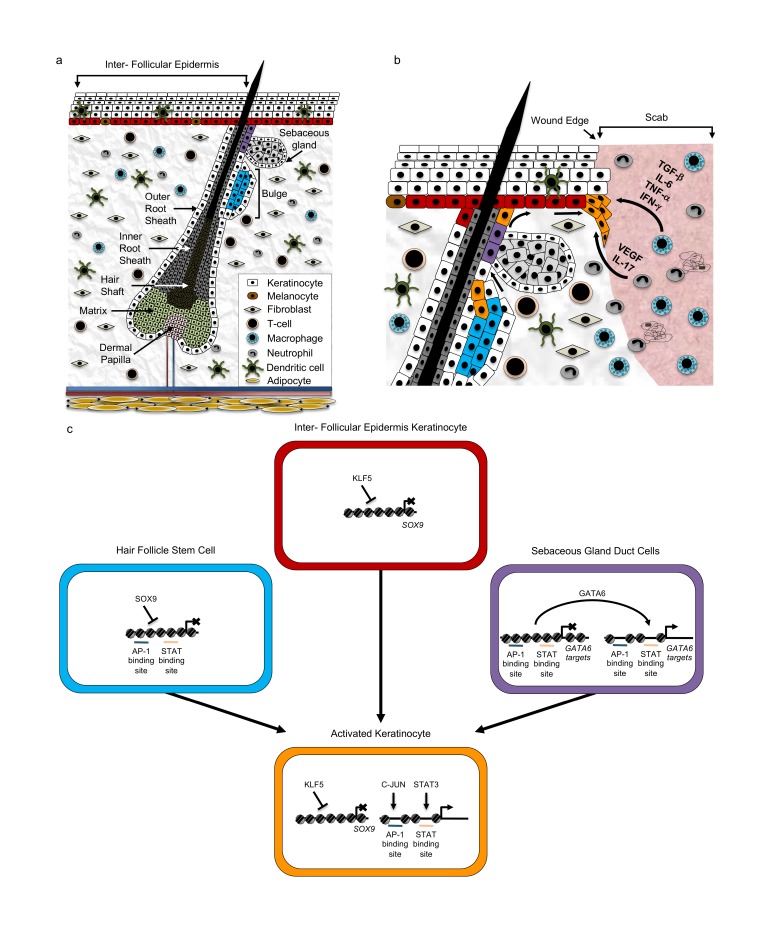
Figure 1.
Changes in the skin microenvironment during cutaneous wound healing. (a) Diagram of major structures and cell types in the skin. Hair Follicle Stem Cells (HF-SC) are indicated in blue, Sebaceous Gland Duct Cells are indicated in purple, and basal Interfollicular Epidermis (IFE) cells are indicated in red. (b) Movement of key keratinocyte stem cell populations in response to skin wounding. Cytokines released from wound-infiltrating macrophages and neutrophils induce activation of keratinocytes from the Interfollicular Epidermis (red), Sebaceous Gland Ducts (purple), and Hair Follicles (blue). These populations acquire a common transcriptional program of activated wound keratinocytes (orange) and begin migration toward the wound edge. Direction of migration is indicated by black arrows. (c) Key transcriptional programs in keratinocyte stem cells converge on a common activated keratinocyte transcriptional program following wounding. In HF-SCs (blue), Sox9 facilitates maintenance of closed chromatin, blocking access to AP-1 and STAT binding sites and keeping key wound healing genes turned off. In contrast, IFE cells (Red) express the transcription factor KLF5, which represses SOX9 transcription. Although it is associated with Sebaceous Gland lineage commitment, GATA6 can also as a pioneer factor in Sebaceous Gland Duct cells (purple) to open up the chromatin around AP-1 and STAT binding sites. After wounding, all these cells express KLF5, which represses SOX9, and open up chromatin regions around AP-1 and STAT binding sites, to help initiate the wound repair process.
As humans age, the ability to heal wounds decreases [7], and improper control of wound healing can lead to persistent, chronic ulceration and significant morbidity, which is the largest driver of skin disease costs at ~$15 billion per year [8]. One population at particularly high risk is patients with diabetes, as non-healing ulcers led to lower limb amputations in approximately 73,000 adults in 2010[9], a number that rose to 108,000 in 2014 [10], correlating with the rise in incidence of diabetes [9,10]. A variety of therapeutic strategies have been developed seeking to improve wound healing and prevent ulceration. New methods of debridement and novel wound dressings incorporating matrix components [11,12] allow for induction of the natural wound response and protection from the external environment, but do not address the underlying pathology of chronic ulceration. Alternatively, many studies have sought to modulate the wound microenvironment with the addition of cytokines and growth factors, an approach that has shown promise in pre-clinical models [13-16]. Unfortunately, only one growth factor based drug (rPDGF-BB, Ortho-McNeil) has been FDA approved for non-healing ulcers, and the clinical benefit of this treatment is modest [17-19]. In addition, post-marketing retrospective studies found increased cancer incidence from systemic treatment, resulting in a FDA black box warning [20], suggesting that new therapeutic approaches are needed. While the mixture of cytokines and growth factors in the wound bed is complex, all the information they convey is integrated by a set of transcription factors to elicit a transcriptional response that allows keratinocytes to migrate across the wound bed. Thus, understanding the regulation of the transcriptional network controlling re-epithelialization may uncover new therapeutic targets that can promote re-epithelialization and proper wound healing.
Transcriptional Plasticity of Epidermal Stem Cells in Wound Healing
Sequence-specific transcription factors serve as integration hubs for various cellular signaling pathways, allowing for ordered changes in sets of transcripts required for different physiological functions. Transcription factors often work together as regulatory modules [21], and the regulatory regions for genes that contain binding sites for multiple different transcription factors are concentrated into small stretches of DNA with high information content [22]. During wound healing, there is coordinate regulation of target genes by multiple transcription factors, which changes over time [23-25] allowing for integration of external signals to generate the appropriate physiological output. The ability of many transcription factors to bind to these regulatory regions in different cell types [26], or in response to stimuli [27] is often determined by pre-existing genome-wide chromatin accessibility. Transcriptional networks are remarkably cell-type specific [22], and even in different cell types with similarly accessible chromatin, a subset of transcription factors have sequence specific binding discrimination [28].
Keratinocyte migration into the wound bed is the earliest event of re-epithelialization, and starts about 24 to 48 hours after injury [11]. The numerous paracrine factors released by immune cells present in the wound bed triggers profound phenotypic changes in keratinocytes. In particular, Epidermal Growth Factor (EGF), Tumor Necrosis Factor-α (TNF-α, Interleukin-6 (IL-6), and Transforming Growth Factor-β (TGF-β) are important to fine tune the balance between keratinocyte migration and proliferation [29] (Figure 1b). Recent work has revealed the presence of at least two distinct populations of keratinocytes around the wound site: cells that are proximal to the wound edge that do not proliferate and actively express pro-migratory genes, and cells distal to the wound edge which proliferate, therefore supplying a new pool of cells that can in turn migrate into the wound [23]. These cells can come from a variety of stem and progenitor cell populations found in the hair follicle (HF-SC) [24,30-34], the interfollicular epidermis (IFE) [24,34-36], and the upper isthmus [32]. Importantly, spatial confinement of the stem cell populations is lost during wound healing, allowing for their recruitment to the injury site and their participation in epidermal regeneration [25] (Figure 1b). Genetic labeling experiments have shown that stem cell lineage plasticity is important for proper re-epithelialization [37]. When activated upon injury, HF-SCs transiently express the IFE-specific transcription factor KLF5, which suppresses the HF-SC transcription factor SOX9, establishing an IFE-like transcriptional state. This effect is mediated by the increased accessibility of binding sites for the transcription factors AP1 (JUN/FOS) and STAT3, which have increased expression around the wound [37]. This plasticity can also be seen in terminally differentiated cells, such as GATA6+ cells from the sebaceous gland duct, which can migrate into the wound bed and acquire IFE cell characteristics. In this case, GATA6 was proposed to act as a pioneer factor, opening up chromatin and revealing binding sites for AP1 and STAT transcription factors [38]. Thus, as stem cells exit their different niches, they converge on common transcriptional programs, which allows for the ordered repair of the epidermis (Figure 1c).
Inflammatory Signaling Drives Early Transcriptional Responses
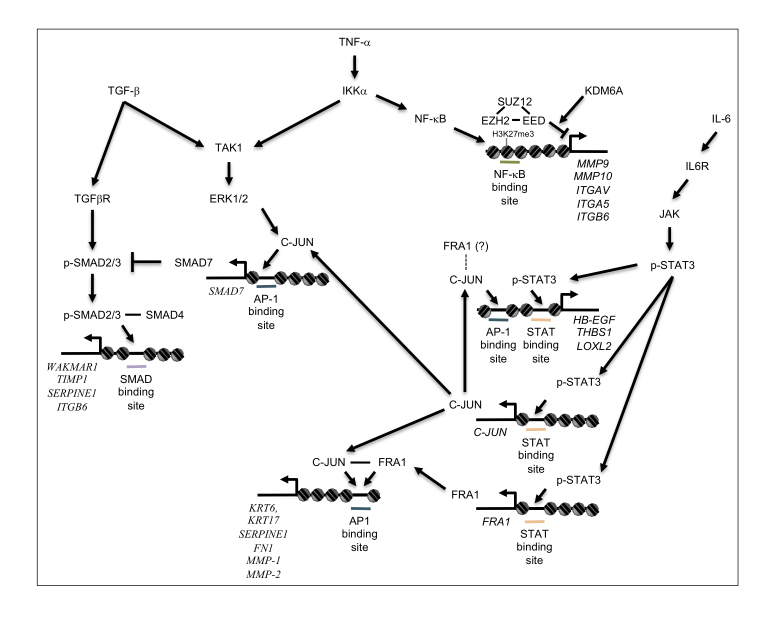
Inflammation is one of the earliest events in wound healing and is critical not only to prevent infections but also to trigger keratinocyte activation and re-epithelialization. Platelets and leukocytes present in the wound release numerous chemical signals, including IL-1α, IL-1β, IL-6, IL-8, TNF-α, PDGF, and TGF-β [39]. Besides their role as a feedback loop regulating immune cell infiltration, activity and inflammation, these cytokines have a profound impact on keratinocytes. TNF-α can activate NF-κB in keratinocytes (Figure 2), which increases the expression of genes involved in integrin signaling, cell adhesion, and motility (ITGB6, ITGAV, ITGA5, NINJ1, RANBP9, NEF3, MMP9, MMP10, MMP13) [40], while suppressing cell proliferation (reduced CDC25, MCM3) [41].
Figure 2.
Transcriptional network regulating initiation of re-epithelialization. Schematic diagram of signaling pathways activated by cytokines and growth factors present in the wound bed, leading to initiation of re-epithelialization. TGF-β, TNF-α, and IL-6 released into the wound environment activate interconnected pathways, which collaborate with AP-1 transcription factors expressed in wound edge keratinocytes to activate transcription of target genes (Italics) required for matrix re-modeling and initiation of keratinocyte migration.
Cytokines and growth factors released into the early wound environment such as EGF and IL-6 can activate another key signaling pathway, the JAK-STAT pathway (Figure 2) [42,43]. Interestingly, increased accessibility of STAT binding sites in cells at the edge of the wound [37,38] is consistent with previous studies implicating STAT3 in wound healing. Keratinocyte-specific deletion of Stat3 in mice causes a severely reduced rate of wound closure in vivo, and reduced mitogen-dependent migration in vitro [44], but has little effect on proliferation. STAT3 can also directly activate a variety of genes involved in migration and matrix remodeling, such as THBS1, TIAM1, SERPINE2, and LOXL2 [45]. In addition, IL-6-mediated activation of STAT3 induces direct up-regulation of Skint2/3 genes resulting in recruitment of dendritic epidermal T-cells [16], which can improve re-epithelialization in part by secreting Insulin Like Growth Factor 1 (IGF-1) and Keratinocyte Growth Factor (KGF) [46]. Increased STAT3 activity also serves to re-enforce early chromatin changes [37,38] and promote AP-1 dependent transcription by repressing SOX9 and inducing transcription of GATA6, FRA1, JUNB, and JUN (Figure 1c) [45].
Transcriptional Control of Keratinocyte Migration
Re-epithelialization requires migration of keratinocytes, and there are many potential ways cells can move through tissues. One of the most studied mechanisms leading to individual cell migration is Epithelial-to-Mesenchymal Transition (EMT), in which polarized epithelial cells lose their adhesion to basement membranes and to each other and acquire mesenchymal cell properties. On the opposite end of the spectrum is collective cell migration, in which cells maintain most of their epithelial identity genes and move as a tightly connected sheet of cells [47,48]. While many studies have worked to elucidate what controls the EMT switch, there is an emerging appreciation that EMT is not an “either/or” process, but is a continuum [29,47-49].
One of the major pathways regulating migration in wound healing is the TGF-β pathway (Figure 2). When bound to ligands, TGF-β superfamily receptors phosphorylate and activate receptor-regulated SMAD proteins (SMAD2, SMAD3, SMAD5, SMAD8), which in turn bind and activate SMAD4. The heterodimer then translocates to the nucleus where it regulates the expression of genes involved in cell motility and EMT. This pathway is also negatively regulated by SMAD7, which inhibits TGF-β induced signaling through receptor-regulated SMAD protein degradation or inhibitory interaction with TGF-β receptors [50].
The role of the TGF-β pathway in wound repair has been somewhat confusing, likely due to differing effects of TGF-β signaling on the various cell populations in the wound area. Transgenic mice overexpressing the Tgf-β1 ligand under control of a Keratin 14 promoter display a greater rate of re-epithelialization in partial thickness ear wounds, where only the epidermis is injured [51]. In contrast, full thickness back skin wounds had slower re-epithelialization [51], consistent with independent results seen with burn-induced skin wounds [52]. One potential explanation is that TGF-β1 increases keratinocyte migration, providing an advantage in partial thickness wounds, but impairs inflammation and dermal repair. This is supported by data from Smad3-/- keratinocytes, which have reduced migration in response to Tgf-β1. Interestingly, full thickness wounds in Smad3-/- mice actually heal faster than in Smad3+/+ mice, an effect attributed to reduced monocyte infiltration in granulation tissue [53].
The TGF-β pathway inhibitor Smad7 also plays a role in cutaneous wound healing, where expression gradually increases and peaks approximately halfway through the healing process before returning to baseline levels [54]. Smad7 expression is stimulated by pro-inflammatory cytokines, such as IL-1, Interferon γ (IFN γ) and TNF-α which are present in the wound environment [55], and is transcriptionally activated by AP-1 transcription factors [56]. Interestingly, transforming growth factor-β-activated kinase 1 (TAK1) stimulates NF-κB activity [57], which can directly activate expression of SMAD7, resulting in negative feedback and attenuation of TGF-β pathway signaling [58]. Overexpression of Smad7 in Keratin 14-expressing cells accelerated wound healing and was associated with increasing proliferation and migration. Intriguingly, migrating cells with Smad7 overexpression also expressed Keratin 14 and had activation of Erk signaling, and increased migration was dependent on MAPK pathway activation [54].
While activation of the TGF-β pathway can induce EMT in some situations, such as cancer metastasis, the role of EMT in wound healing is less clear [59]. In epidermal wounds, leading edge cells appear to maintain the epithelial markers Keratin 5 and 14 during wound closure [23,60,61], and recent data suggesting an extending shield re-epithelialization mechanism [61] supports a role for collective migration. TGF-β has recently been shown to promote collective cell migration in cells, which is correlated with high activity of Erk1/2 [62]. Importantly, Erk1/2 activity is required for early migratory events induced by TGF-β1 [63,64], and potentiates TGF-β pathway signaling by enhancing Smad2-dependent transcriptional activity [65]. In addition, transcription factors downstream of Erk1/2 such as JunB [66] can modulate the transcriptional effects of TGF-β1. These data highlight the importance of signal integration in modifying the TGF-β pathway response to promote a collective migration phenotype during cutaneous wound healing.
AP-1 Family Transcription Factors in Early Re-epithelization
A key step in re-epithelialization is the activation of AP-1 family transcription factors, some of which (c-Jun, JunB, JunD, and c-Fos) are induced around 8 hours after wounding in the leading edge of migrating keratinocytes [67]. Jun family transcription factors (c-Jun, JunB, JunD) can form both heterodimers and homodimers, while Fos family proteins (Fos, FosB, Fra-1, and Fra-2) form heterodimers with Jun family proteins [68]. While this mix-and-match feature of AP-1 transcription factors has presented challenges in dissecting the role of individual Jun and Fos family proteins, knock-out experiments have clearly demonstrated roles for c-Jun and JunB in wound healing. Mice lacking c-Jun in either Keratin 14 [69] or Keratin 5 [70] expressing cells show a reduced basal proliferative rate in the skin, and severely reduced proliferation in vitro, which was attributed to decreased Epidermal Growth Factor (EGFR) signaling due to reduced expression of EGFR ligands such as HB-EGF. Interestingly, while there was slightly reduced wound healing with c-Jun loss in Keratin 14 expressing cells [69], this was not seen with c-Jun loss in Keratin 5-expressing cells [70]. It is important to note that this discrepancy may be due to the fact that the reported effect on wound healing was modest [69], and unlike human skin, wound healing in mice occurs largely by contraction, which may obscure effects on migration if murine models do not employ splinting [71,72]. Another potential confounding factor in these genetic experiments is that c-Jun expression is bi-phasic in the leading edge, which has little c-Jun protein at early timepoints (before 8 hours), and increases later during migration [67], while expression remains unchanged away from the leading edge. This is consistent with the opening of chromatin around AP-1 binding sites at early timepoints, allowing for later AP-1 regulated transcription [37,38]. In vitro experiments show more support for the role of c-Jun in migration, which is significantly reduced following scratch assay with loss of c-Jun, in conjunction with reduced EGF-dependent phosphorylation of Focal Adhesion Kinase (FAK) [69]. Loss of c-Jun causes an increase in stable focal contacts [73], consistent with the role of FAK in promoting turnover of focal adhesions. In addition, c-Jun directly activates transcription of Src, which along with FAK, also promotes turnover of focal adhesions and promotes migration [73]. These data point to a key role for c-Jun in promoting migration early in the process of re-epithelialization.
JunB has also been shown to contribute to epidermal wound healing using mice lacking JunB in keratinocytes and fibroblasts [74]. Mice lacking JunB in the skin demonstrate reduced keratinocyte migration as well as epidermal hyper proliferation, sustained inflammation and increased granulation tissue after wounding [74]. Interestingly, paracrine signaling by JunB-/- fibroblasts may cause some of these phenotypes, such as epidermal hyperproliferation [75]. In addition, these differential effects of JunB loss compared to c-Jun loss may be simply due to expression patterns [67,76], as genetic knock-in of JunB to the c-Jun locus can rescue many of the phenotypes seen in c-Jun-/- mice [77].
Beyond c-Jun and JunB, data on the functions of AP-1 family members in normal keratinocytes during wound healing is generally lacking. While it is likely that Fos family members form heterodimers with Jun family proteins during wound healing, this is yet to be rigorously tested in vivo. Intriguingly, Fra1 is activated by JAK-STAT signaling [37,38,45], and it was shown to Fra1 can activate transcription of the pro-migratory genes KRT6, KRT17, FN1, SERPINE1, MMP-1, MMP-2, and MMP-12, when co-expressed with c-Jun. Fra1 overexpression can also accelerate migration of immortalized HaCAT keratinocytes in scratch assays [78], making this an important transcription factor for further study in the context of re-epithelialization.
The Hypoxic Response to Tissue Injury
Tissue injury causes vascular disruption and vasoconstriction, and coupled with high oxygen consumption by cells, the resulting wound environment becomes hypoxic [79]. The Hypoxia-Inducible Factor (HIF) transcription factors play a central role in the adaptation to low oxygen levels. In normoxic conditions, HIF proteins are hydroxylated by the HIF Prolyl-hydroxylase EGLN1, targeting it to the pVHL (Von Hippel-Lindau tumor suppressor protein) E3 ubiquitin ligase, ultimately leading to its degradation via the proteasome [80-82]. In hypoxic condition, EGLN1 is inhibited resulting in HIF protein accumulation and activation of its transcriptional program [83]. In cutaneous wounds, HIF1α protein is increased at the wound margin, as well as in the leading edge of migrating keratinocytes in vitro [84]. This is consistent with observations that hypoxia causes an increase in keratinocyte migration [85]. Keratinocyte specific deletion of Hif1α significantly impairs closure of in vivo punch wounds, an effect primarily attributed to a lack of migration, as proliferation, apoptosis, and inflammation are similar to controls. In addition, knock-down of HIF1α reduced migration of keratinocytes in vitro as assessed by scratch assays [84,86]. This phenotype is at least in part due to activation of SERPINE1, LAMA3, LAMB3, LAMC2, ITGA6, and ITGB1 by HIF1α, which promote migration [84,86-89]. In addition, secreted factors such as VEGFA, which is activated by HIF1α, are likely to contribute to proper wound healing by promoting angiogenesis [90]. Further support for a key role for HIF1α in promoting wound healing comes from the finding that mice lacking Egln1 in the skin, resulting in stabilized Hif1α, have accelerated in vivo wound healing and in vitro migration of keratinocytes [91]. The HIF2α protein is regulated by oxygen in a similar manner to HIF1α, but has non-redundant functions in wound healing [84-86]. Interestingly, loss of Hif2α in keratinocytes results in accelerated wound healing [85], and combined Hif2α/Vhl loss leading to stabilized HIF1α also have accelerated wound healing, consistent with previous data suggesting opposing roles for HIF1α and HIF2α [92].
Epigenetic Control of Re-epithelialization
Transcription factors do not act in isolation, but form complexes with histone-modifying enzymes, which facilitate changes in chromatin accessibility to mediate activation or repression of transcriptional programs. It is now widely appreciated that histone proteins that serve as a scaffold for DNA are not passive bystanders in gene regulation, but active players [93-95]. The nucleosome core consists of a dimer of the tetrameric unit containing histones H2A, H2B, H3, and H4 that directly interacts with 146bp of DNA that is wrapped around it [96]. The post-translational modification of histone proteins on their exposed tails facilitates the opening or compaction of the chromatin allowing for the activation or repression of transcription. The modification of histones by acetylation, methylation, and ubiquitylation has been well studied, giving insight into the chromatin structures associated with different transcriptional states [95,97]. In general, acetylation of Histone H3 at lysines 9 and 27, as well as methylation at lysines 4, 36, and 79 have been strongly associated with genes that are active. Conversely, tri-methylation of Histone H3 at lysines 9 and 27, mono-methylation of Histone H4 at lysine 20, and monoubiquitylation of H2A lysine 119 are all associated with the repressive heterochromatin state [98-102]. Interestingly, a subset of genes that are not actively transcribed sit in a “poised” state and exhibit both active (H3K4me3) and repressive (H3K27me3) marks, allowing for developmentally appropriate activation during lineage specification and differentiation [103,104].
While a variety of chromatin modifiers have been shown to regulate physiological processes important for wound healing, such as proliferation, migration, and differentiation [103,105-107], relatively little is known about epigenetic regulation in the specific context of re-epithelialization. The regulation of H3K27me3 is the most well studied and is controlled by the dynamic balance between the PRC2 complex and KDM6A/KDM6B (Figure 2). The core Polycomb Repressive Complex 2 (PRC2) subunits EZH2, EED, and SUZ12 have significantly reduced protein expression in the leading edge of migrating keratinocytes, correlating with reduced H3K27me3. Functionally, EZH2 has been found to repress the Ink4a/Arf locus [108], and upregulation of p16Ink4a is associated with increased migration [109]. Somewhat paradoxically, loss of EZH1 and EZH2 results in delayed closure of split-thickness wounds, although single knock-outs were not examined [108]. This might be explained by the differential expression of EZH2 in leading edge cells, where expression is reduced, and the proliferative hub cells, which still express EZH2 [110]. Alternatively, loss of EZH2 may cause a shift in lineage or increase differentiation, similar to what is seen in during pre-natal epidermal development [111].
The H3K27me3 histone demethylase KDM6B is a crucial NFκB coactivator in keratinocytes. Its expression is increased in the wound edge where it helps activate the transcription of matrix metallo-proteinases, which promote matrix remodeling and help keratinocytes carve their way through the wound bed [112]. In addition, the KDM6B-NFκB axis is necessary for the expression of various cytokines by keratinocytes, including IL-6 and TNF-α, which may promote a positive feedback loop and install a molecular crosstalk between keratinocytes and other cell types within the wound. In contrast to PRC2 proteins, KDM6A/B expression is increased early in wounding, and then reduced at late stages [110,113]. Loss of KDM6B has been shown to reduce migration and is required for optimal induction of NF-kB target genes (IL-1β, IL-6, IL-18, TNF-α, MMP-3, MMP-9, MMP-14, HB-EGF) [112]. KDM6B also activates NOTCH1 transcription and induction of NOTCH1 target genes (RHOU and PLAU) promoting loss of focal adhesion and migration [113]. PRC2-mediated repression is also countered by the activity of the Histone Methyltransferase ASH1L, which methylates H3K36 in the bodies of actively transcribed genes [100]. ASH1L inactivation leads to increased proliferation and delayed re-epithelialization during wounding [114], but the mechanism remains unclear.
Methylation is not restricted to histone proteins, and indeed a major epigenetic regulator of gene expression is DNA methylation. Early during embryogenesis, de novo methylation of DNA is facilitated by DNA methyltransferases DNMT3A and DNMT3B [115]. After establishment of methylation patterns, DNMT1 functions to maintain DNA methylation patterns following replication by binding to hemi-methylated CpG regions, and adding a methyl group to the newly synthesized strand [116]. DNA methylation at promoter CpG islands can promote the formation of heterochromatin and transcriptional repression [117]. DNA methylation facilitated by DNMT proteins is opposed by the activity of the Tet Methylcytosine Dioxygenase protein family (TET1, TET2, and TET3), which convert 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC), then 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC), ultimately leading to the removal of cytosine methylation [118]. Interestingly, many transcription factors implicated in wound healing (JUN, JUNB, E2F1, E2F4, ETS2, MYC) have altered affinity when the CpG regions of their binding sites are methylated [119-121], suggesting that DNA methylation changes are important for the wound healing response. Somewhat paradoxically, both proliferation and migration were reduced following DNMT1 knockdown, but it is unclear if the effects on migration are due to specific regulation of migration, or are secondary effects of reduced cell division in the proliferative hub causing a lack of new cells to fill in behind the leading edge cells as they migrate [122]. Further evidence for the potential role of DNA methylation in regulating wound healing comes from a recent study on the skin-specific long non-coding RNA (lncRNA) WAKMAR1, which is expressed in leading edge keratinocytes and promotes migration [123]. LncRNAs can regulate transcription in many ways, such as physically interacting with chromatin-modifying complexes like PRC2 [124]. In human keratinocytes, WAKMAR1 was found to bind to DNMT1, and block inactivating methylation of the E2F promoter [123]. While this seems to contradict the results seen with DNMT1 knock-down [122], lncRNAs have been proposed to target DNMT1 to specific loci [125], and DNMT1 may have gene-specific effects during wound healing. It is also possible that DNMT1 is required only in the proliferative hub, and suppression of DNMT1 activity is required for migration in leading edge cells, which do not proliferate. Given that WAKMAR1 is the first lncRNA shown to be involved in the regulation of wound healing, it is likely that future studies will uncover a much more robust network of non-coding RNAs that play significant roles in transcriptional control during re-epithelialization.
Conclusions and Outlook
A major challenge in understanding transcriptional regulation in wound healing remains the complex spatial and temporal changes that occur during the re-epithelialization. Key cell populations, such as the leading-edge cells that interface directly with the wound, are too small to capture using bulk population analysis, leaving the spatio-temporal dynamics of transcription unclear. Recently however, advances in single-cell analysis including RNA profiling (scRNA-seq) [126], DNA binding protein mapping (ChIP-seq) [127,128], methylation profiling (snmC-seq2) [129], chromatin accessibility (scATAC-seq) [130-132], and spatial positioning [133] along with new computation methods of integrating this complex data [134,135] have opened up new ways to explore the different transcriptional states of individual cells during the wound healing process. A key feature of these emerging technologies is the ability to integrate large-scale information from many unique cell populations that change over time. This will allow for the development of a high-resolution map of the movement and transcriptional changes of all cells involved in the repair of wounds and facilitate dissection of the functional interplay between the many cell types that contribute to wound healing. Indeed, these approaches are already being applied to wound healing model systems to examine the differential contributions of hair follicle bulge stem cells (Lgr5+) and Interfollicular Epidermis (IFE) cells (Lgr6+) stem cells. Lineage tracing experiments combined with scRNA-seq identified at least eight distinct cell states associated with unique transcriptional profiles during the course of wound regeneration, which were previously unappreciated. Interestingly, Lgr5+ cells converge on an IFE-like state [24], suggesting that different stem cell populations converge on a common transcriptional program that is necessary with proper repair. This is consistent with data suggesting lineage plasticity in hair follicle stem cells during wound healing, where transient co-expression of the transcription factors Sox9, Klf5, Tcf3, and AP2γ occurs in early phases of repair, then become lineage-restricted again after completion of repair [37]. Thus, single-cell analysis of transcriptional changes in Lgr5+ and Lgr6+ cells has both confirmed previous studies and identified new transcriptional states in cells that contribute to the re-epithelialization process.
A thorough understanding of the changes that occur in each cell during the wound healing process is essential for determining the key events that initiate re-epithelialization and identifying how these events are altered in chronic wounding. Currently, most wound healing studies assess the essential functions of genes in wound healing employing whole-body knock-outs or more restricted tissue specific inducible alleles [25]. The approach of using germline knock-out mice has the major drawback of affecting multiple different cell types, some of which may have opposing functions, thus obscuring major effects. Another confounding factor in many of these studies is that the proliferative hub cells have a very different function than migrating cells, yet both populations are often targeted in knockout mice, even when using tissue restricted Cre transgenes. The ability to analyze experiments at the single cell level may allow for dissection of the effects of gene disruption on these different populations. In addition, a more refined understanding of gene expression could allow for more precise targeting to cells of interest. Moving forward, these new technologies are likely to dramatically improve our understanding of the mechanisms of transcriptional control regulating the many complex processes that occur in multiple cell types that contribute to proper cutaneous wound healing.
The ageing population and increased occurrence of metabolic diseases worldwide will fuel a continuous rise of the chronic wound epidemic, and finding better therapies is urgent. Current therapies include surgical or enzymatic debridement of necrotic tissue, specific dressings aimed at preventing infections while maintaining adequate moist levels, autologous skin grafts, extracellular matrix protein, and growth factors treatment such as PDGF [11,19]. While these treatment advances have made meaningful differences in the lives of patients, therapies that effectively correct the underlying causes of non-healing and chronic wounds remain elusive. Transcription factor complexes integrate the myriad signals emanating from the wound to control proper re-epithelialization, and as such represent promising therapeutic targets. By dissecting the mechanisms of transcriptional regulation underlying the normal wound healing process, a more complete picture of pathologic wound healing can emerge. Importantly, drugs inhibiting epigenetic regulators, such as EZH2, have been developed [136], and given the reductions in EZH2 activity in leading edge cells [110], inhibition of EZH2 activity could potentially induce keratinocyte migration and re-epithelialization. The targeting of the enzymatic activity of transcriptional complexes in pathological states such as cancer shows great promise [137,138] and such approaches could offer new ways to more precisely promote the process of re-epithelialization in normal and pathological wound healing.
Acknowledgments
RB is funded by a SunPharma/SID Innovation Research Fellowship. This work is also funded by a Harvard Stem Cell Institute seed grant (DP-0156-16-00) and Brigham Research Institute Fund to Sustain Research Excellence.
Glossary
- PDGF
Platelet-Derived Growth Factor
- EGF
Epidermal Growth Factor
- TNF-α
Tumor Necrosis Factor-α
- IL-6
Interleukin-6
- TGF-β
Transforming Growth Factor-β
- IGF-1
Insulin Like Growth Factor 1
- IFN γ
Interferon γ
- KGF
Keratinocyte Growth Factor
- HF-SC
hair follicle stem cell
- IFE
interfollicular epidermis
- EMT
Epithelial-to-Mesenchymal Transition
- PRC2
Polycomb Repressive Complex 2
- lncRNA
long non-coding RNA
- FAK
Focal Adhesion Kinase
- TAK1
transforming growth factor-β-activated kinase 1
- HIF
Hypoxia-Inducible Factor
- KLF5
Kruppel Like Factor 5
- SOX9
SRY-Box Transcription Factor 9
- NF-κB
Nuclear Factor Kappa B
- STAT3
Signal Transducer and Activator of Transcription 3
- JAK
Janus Kinase
- ERK
Extracellular Signal-Regulated Kinase
- MAPK
Mitogen-Activated Protein Kinase
- C-Jun
Jun Proto-Oncogene
- EGLN1
Egl-9 Family Hypoxia Inducible Factor 1
- KDM6B
Lysine Demethylase 6B
- WAKMAR1
Wound and Keratinocyte Migration-Associated LncRNA
- TET
Tet-eleven translocation Methylcytosine Dioxygenase
- DNMT
DNA Methyltransferase
- 5-hmC
5-hydroxymethylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- scRNA-seq
Single cell Ribonucleic Acid sequencing
- ChIP-seq
Chromatin Immunoprecipitation-sequencing
- snmC-seq
Single-cell DNA methylome sequencing
- ATAC-seq
Assay for Transposase-Accessible Chromatin using sequencing
Author Contributions
Both MRR and RB wrote and edited the manuscript and figures.
References
- Demidova-Rice TN, Hamblin MR, Herman IM. Acute and impaired wound healing: pathophysiology and current methods for drug delivery, part 1: normal and chronic wounds: biology, causes, and approaches to care. Adv Skin Wound Care. 2012;25(7):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falanga V. Wound healing and its impairment in the diabetic foot. Lancet. 2005;366(9498):1736–43. [DOI] [PubMed] [Google Scholar]
- Hopkinson SB, Hamill KJ, Wu Y, Eisenberg JL, Hiroyasu S, Jones JC. Focal Contact and Hemidesmosomal Proteins in Keratinocyte Migration and Wound Repair. Adv Wound Care (New Rochelle). 2014;3(3):247–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I, Stojadinovic O, Yin NC, Ramirez H, Nusbaum AG, Sawaya A, et al. Epithelialization in Wound Healing: A Comprehensive Review. Adv Wound Care (New Rochelle). 2014;3(7):445–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16(5):585–601. [DOI] [PubMed] [Google Scholar]
- Coulombe PA. Towards a molecular definition of keratinocyte activation after acute injury to stratified epithelia. Biochem Biophys Res Commun. 1997;236(2):231–8. [DOI] [PubMed] [Google Scholar]
- Gould L, Abadir P, Brem H, Carter M, Conner-Kerr T, Davidson J, et al. Chronic wound repair and healing in older adults: current status and future research. Wound Repair Regen. 2015;23(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HW, Collins SA, Resneck JS, Jr, Bolognia J, Hodge JA, Rohrer TA, et al. A risk adjustment approach to estimating the burden of skin disease in the United States. J Am Acad Dermatol. 2018;78(1):129–40. [DOI] [PubMed] [Google Scholar]
- Prevention. CfDCa. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014:2014.
- Prevention. CfDCa. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2017:2017. Available from: https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf
- Rousselle P, Braye F, Dayan G. Re-epithelialization of adult skin wounds: cellular mechanisms and therapeutic strategies. Adv Drug Deliv Rev. 2018: 10.1016/j.addr.2018.06.019 [DOI] [PubMed] [Google Scholar]
- Rousselle P, Montmasson M, Garnier C. Extracellular matrix contribution to skin wound re-epithelialization. Matrix Biol. 2018: 10.1016/j.matbio.2018.01.002 [DOI] [PubMed] [Google Scholar]
- Pierce GF, Mustoe TA, Lingelbach J, Masakowski VR, Griffin GL, Senior RM, et al. Platelet-derived growth factor and transforming growth factor-beta enhance tissue repair activities by unique mechanisms. J Cell Biol. 1989;109(1):429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh DG, Sprugel KH, Murray MJ, Ross R. PDGF and FGF stimulate wound healing in the genetically diabetic mouse. Am J Pathol. 1990;136(6):1235–46. [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Liu PH, Pietramaggiori G, Ibrahim SI, Hechtman HB, Orgill DP. Effect of recombinant platelet-derived growth factor (Regranex) on wound closure in genetically diabetic mice. J Burn Care Res. 2006;27(2):202–5. [DOI] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, et al. Impaired Epidermal to Dendritic T Cell Signaling Slows Wound Repair in Aged Skin. Cell. 2016;167(5):1323-38 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees RS, Robson MC, Smiell JM, Perry BH. Becaplermin gel in the treatment of pressure ulcers: a phase II randomized, double-blind, placebo-controlled study. Wound Repair Regen. 1999;7(3):141–7. [DOI] [PubMed] [Google Scholar]
- Smiell JM, Wieman TJ, Steed DL, Perry BH, Sampson AR, Schwab BH. Efficacy and safety of becaplermin (recombinant human platelet-derived growth factor-BB) in patients with nonhealing, lower extremity diabetic ulcers: a combined analysis of four randomized studies. Wound Repair Regen. 1999;7(5):335–46. [DOI] [PubMed] [Google Scholar]
- Whittam AJ, Maan ZN, Duscher D, Wong VW, Barrera JA, Januszyk M, et al. Challenges and Opportunities in Drug Delivery for Wound Healing. Adv Wound Care (New Rochelle). 2016;5(2):79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Administration USFD. Postmarket Drug Safety Information for Patients and Providers, REGRANEX (becaplermin) 2008. Epub 05/2008.
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47(5):810–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489(7414):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Dekoninck S, Rulands S, Lenglez S, Mascre G, Simons BD, et al. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat Commun. 2017;8:14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S, Jacob T, Sun X, Annusver K, La Manno G, Sur I, et al. Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell Rep. 2018;25(3):585-97 e7. [DOI] [PubMed] [Google Scholar]
- Dekoninck S, Blanpain C. Stem cell dynamics, migration and plasticity during wound healing. Nat Cell Biol. 2019;21(1):18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21(3):456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvey A, Agius P, Noble WS, Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012;22(9):1723–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haensel D, Dai X. Epithelial-to-mesenchymal transition in cutaneous wound healing: where we are and where we are heading. Dev Dyn. 2018;247(3):473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11(12):1351–4. [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 2007;21(7):1358–66. [DOI] [PubMed] [Google Scholar]
- Page ME, Lombard P, Ng F, Gottgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell. 2013;13(4):471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnozzi AN, Reiter JF, Wong SY. Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration. Cell Cycle. 2015;14(21):3408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascre G, Dekoninck S, Drogat B, Youssef KK, Brohee S, Sotiropoulou PA, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489(7415):257–62. [DOI] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, et al. Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol. 2016;18(6):619–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell. 2017;169(4):636-50 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Rognoni E, Hiratsuka T, Liakath-Ali K, Hoste E, Kar G, et al. Wounding induces dedifferentiation of epidermal Gata6(+) cells and acquisition of stem cell properties. Nat Cell Biol. 2017;19(6):603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Liang H, Clarke E, Jackson C, Xue M. Inflammation in Chronic Wounds. Int J Mol Sci. 2016;17(12): 10.3390/ijms17122085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno T, Gazel A, Blumenberg M. Pathway-specific profiling identifies the NF-kappa B-dependent tumor necrosis factor alpha-regulated genes in epidermal keratinocytes. J Biol Chem. 2005;280(19):18973–80. [DOI] [PubMed] [Google Scholar]
- Banno T, Gazel A, Blumenberg M. Effects of tumor necrosis factor-alpha (TNF alpha) in epidermal keratinocytes revealed using global transcriptional profiling. J Biol Chem. 2004;279(31):32633–42. [DOI] [PubMed] [Google Scholar]
- Gallucci RM, Sloan DK, Heck JM, Murray AR, O’Dell SJ. Interleukin 6 indirectly induces keratinocyte migration. J Invest Dermatol. 2004;122(3):764–72. [DOI] [PubMed] [Google Scholar]
- Cha D, O’Brien P, O’Toole EA, Woodley DT, Hudson LG. Enhanced modulation of keratinocyte motility by transforming growth factor-alpha (TGF-alpha) relative to epidermal growth factor (EGF). J Invest Dermatol. 1996;106(4):590–7. Epub 1996 Apr 01. [DOI] [PubMed] [Google Scholar]
- Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18(17):4657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24(21):3397–408. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Y, Zhou L, Liu M, Liang G, Yan R, et al. Vgamma4 T Cells Inhibit the Pro-healing Functions of Dendritic Epidermal T Cells to Delay Skin Wound Closure Through IL-17A. Front Immunol. 2018;9:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–83. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K, Casanova J. A common framework for EMT and collective cell migration. Development. 2016;143(23):4291–300. [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Miyazono K. Regulation of TGF-beta Family Signaling by Inhibitory Smads. Cold Spring Harb Perspect Biol. 2017;9(3): 10.1101/cshperspect.a022095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tredget EB, Demare J, Chandran G, Tredget EE, Yang L, Ghahary A. Transforming growth factor-beta and its effect on reepithelialization of partial-thickness ear wounds in transgenic mice. Wound Repair Regen. 2005;13(1):61–7. [DOI] [PubMed] [Google Scholar]
- Yang L, Chan T, Demare J, Iwashina T, Ghahary A, Scott PG, et al. Healing of burn wounds in transgenic mice overexpressing transforming growth factor-beta 1 in the epidermis. Am J Pathol. 2001;159(6):2147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft GS, Yang X, Glick AB, Weinstein M, Letterio JL, Mizel DE, et al. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol. 1999;1(5):260–6. [DOI] [PubMed] [Google Scholar]
- Han G, Li F, Ten Dijke P, Wang XJ. Temporal smad7 transgene induction in mouse epidermis accelerates skin wound healing. Am J Pathol. 2011;179(4):1768–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH. Fine tuning and cross-talking of TGF-beta signal by inhibitory Smads. J Biochem Mol Biol. 2005;38(1):9–16. [DOI] [PubMed] [Google Scholar]
- Brodin G, Ahgren A, ten Dijke P, Heldin CH, Heuchel R. Efficient TGF-beta induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J Biol Chem. 2000;275(37):29023–30. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270(5244):2008–11. [DOI] [PubMed] [Google Scholar]
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, et al. A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev. 2000;14(2):187–97. [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–74. [DOI] [PubMed] [Google Scholar]
- Iglesias-Bartolome R, Uchiyama A, Molinolo AA, Abusleme L, Brooks SR, Callejas-Valera JL, et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Sci Transl Med. 2018;10(451): Epub 2018 Jul 27. 10.1126/scitranslmed.aap8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safferling K, Sutterlin T, Westphal K, Ernst C, Breuhahn K, James M, et al. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. J Cell Biol. 2013;203(4):691–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapnick DA, Liu X. Leader cell positioning drives wound-directed collective migration in TGFbeta-stimulated epithelial sheets. Mol Biol Cell. 2014;25(10):1586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J, Tsou R, Wallace K, Gibran N, Isik F. Early gene expression profile of human skin to injury using high-density cDNA microarrays. Wound Repair Regen. 2001;9(5):360–70. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98(12):6686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funaba M, Zimmerman CM, Mathews LS. Modulation of Smad2-mediated signaling by extracellular signal-regulated kinase. J Biol Chem. 2002;277(44):41361–8. [DOI] [PubMed] [Google Scholar]
- Gervasi M, Bianchi-Smiraglia A, Cummings M, Zheng Q, Wang D, Liu S, et al. JunB contributes to Id2 repression and the epithelial-mesenchymal transition in response to transforming growth factor-beta. J Cell Biol. 2012;196(5):589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neub A, Houdek P, Ohnemus U, Moll I, Brandner JM. Biphasic regulation of AP-1 subunits during human epidermal wound healing. J Invest Dermatol. 2007;127(10):2453–62. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117(Pt 25):5965–73. Epub 2004 Nov 27. [DOI] [PubMed] [Google Scholar]
- Li G, Gustafson-Brown C, Hanks SK, Nason K, Arbeit JM, Pogliano K, et al. c-Jun is essential for organization of the epidermal leading edge. Dev Cell. 2003;4(6):865–77. [DOI] [PubMed] [Google Scholar]
- Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4(6):879–89. [DOI] [PubMed] [Google Scholar]
- Ansell DM, Holden KA, Hardman MJ. Animal models of wound repair: are they cutting it? Exp Dermatol. 2012;21(8):581–5. [DOI] [PubMed] [Google Scholar]
- Nunan R, Harding KG, Martin P. Clinical challenges of chronic wounds: searching for an optimal animal model to recapitulate their complexity. Dis Model Mech. 2014;7(11):1205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Katiyar S, Liu M, Mueller SC, Lisanti MP, Li A, et al. Disruption of c-Jun reduces cellular migration and invasion through inhibition of c-Src and hyperactivation of ROCK II kinase. Mol Biol Cell. 2008;19(4):1378–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Knebel J, Zigrino P, Vonderstrass B, Mauch C, Schorpp-Kistner M, et al. Delayed wound healing and epidermal hyperproliferation in mice lacking JunB in the skin. J Invest Dermatol. 2006;126(4):902–11. [DOI] [PubMed] [Google Scholar]
- Szabowski A, Maas-Szabowski N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig NE, et al. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell. 2000;103(5):745–55. [DOI] [PubMed] [Google Scholar]
- Mehic D, Bakiri L, Ghannadan M, Wagner EF, Tschachler E. Fos and jun proteins are specifically expressed during differentiation of human keratinocytes. J Invest Dermatol. 2005;124(1):212–20. [DOI] [PubMed] [Google Scholar]
- Passegue E, Jochum W, Behrens A, Ricci R, Wagner EF. JunB can substitute for Jun in mouse development and cell proliferation. Nat Genet. 2002;30(2):158–66. [DOI] [PubMed] [Google Scholar]
- Ma S, Rao L, Freedberg IM, Blumenberg M. Transcriptional control of K5, K6, K14, and K17 keratin genes by AP-1 and NF-kappaB family members. Gene Expr. 1997;6(6):361–70. [PMC free article] [PubMed] [Google Scholar]
- Schreml S, Szeimies RM, Prantl L, Karrer S, Landthaler M, Babilas P. Oxygen in acute and chronic wound healing. Br J Dermatol. 2010;163(2):257–68. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–8. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92(12):5510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–72. [DOI] [PubMed] [Google Scholar]
- To KK, Huang LE. Suppression of hypoxia-inducible factor 1alpha (HIF-1alpha) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J Biol Chem. 2005;280(45):38102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitsialos G, Bourget I, Augier S, Ginouves A, Rezzonico R, Odorisio T, et al. HIF1 transcription factor regulates laminin-332 expression and keratinocyte migration. J Cell Sci. 2008;121(Pt 18):2992–3001. [DOI] [PubMed] [Google Scholar]
- Cowburn AS, Alexander LE, Southwood M, Nizet V, Chilvers ER, Johnson RS. Epidermal deletion of HIF-2alpha stimulates wound closure. J Invest Dermatol. 2014;134(3):801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani HR, Ali N, Serrano-Sanchez M, Dubus P, Varon C, Ged C, et al. Loss of epidermal hypoxia-inducible factor-1alpha accelerates epidermal aging and affects re-epithelialization in human and mouse. J Cell Sci. 2011;124(Pt 24):4172–83. [DOI] [PubMed] [Google Scholar]
- Keely S, Glover LE, MacManus CF, Campbell EL, Scully MM, Furuta GT, et al. Selective induction of integrin beta1 by hypoxia-inducible factor: implications for wound healing. FASEB J. 2009;23(5):1338–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Providence KM, Higgins SP, Mullen A, Battista A, Samarakoon R, Higgins CE, et al. SERPINE1 (PAI-1) is deposited into keratinocyte migration “trails” and required for optimal monolayer wound repair. Arch Dermatol Res. 2008;300(6):303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson DA, Thurston G, Huang LE, Ginzinger DG, McDonald DM, Johnson RS, et al. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1alpha. Genes Dev. 2001;15(19):2520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka J, Ettinger A, Franke K, Mamlouk S, Singh RP, Farhat K, et al. Loss of epithelial hypoxia-inducible factor prolyl hydroxylase 2 accelerates skin wound healing in mice. Mol Cell Biol. 2013;33(17):3426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A, Jozkowicz A, Dulak J. HIF-1 versus HIF-2—is one more important than the other? Vascul Pharmacol. 2012;56(5-6):245–51. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. Epub 2009 Nov 06. [DOI] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. Epub 2009 Apr 10. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20(6):845–54. [DOI] [PubMed] [Google Scholar]
- Huang C, Zhu B. Roles of H3K36-specific histone methyltransferases in transcription: antagonizing silencing and safeguarding transcription fidelity. Biophys Rep. 2018;4(4):170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442(7100):307–11. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242 Epub 2008 Nov 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. J Cell Sci. 2010;123(Pt 13):2207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138(4):660–72. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25(5):485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan E, Omobono JD, 2nd, Guo Z, Hopkinson S, Lazar AJ, Brenn T, et al. A keratinocyte hypermotility/growth-arrest response involving laminin 5 and p16INK4A activated in wound healing and senescence. Am J Pathol. 2006;168(6):1821–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10(8):881–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the Roles of Polycomb Repressive Complex 2 Subunits in the Control of Skin Development. J Invest Dermatol. 2016;136(8):1647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J, Lee K, Na W, Shin JY, Lee MJ, Yune TY, et al. Histone H3K27 Demethylase JMJD3 in Cooperation with NF-kappaB Regulates Keratinocyte Wound Healing. J Invest Dermatol. 2016;136(4):847–58. [DOI] [PubMed] [Google Scholar]
- Na J, Shin JY, Jeong H, Lee JY, Kim BJ, Kim WS, et al. JMJD3 and NF-kappaB-dependent activation of Notch1 gene is required for keratinocyte migration during skin wound healing. Sci Rep. 2017;7(1):6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ye Z, Shi C, Sun L, Han M, Zhuang Y, et al. The Histone Methyltransferase Ash1l is Required for Epidermal Homeostasis in Mice. Sci Rep. 2017;7:45401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295(5850):620–2. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988;53(1):3–4. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337): 10.1126/science.aaj2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251(4990):186–9. [DOI] [PubMed] [Google Scholar]
- Gustems M, Woellmer A, Rothbauer U, Eck SH, Wieland T, Lutter D, et al. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 2014;42(5):3059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Jing X, Yang S, Peng D, Dong J, Li L, et al. DNA Methylation Regulates Corneal Epithelial Wound Healing by Targeting miR-200a and CDKN2B. Invest Ophthalmol Vis Sci. 2019;60(2):650–60. [DOI] [PubMed] [Google Scholar]
- Li D, Kular L, Vij M, Herter EK, Li X, Wang A, et al. Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration. Proc Natl Acad Sci USA. 2019;116(19):9443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. [DOI] [PubMed] [Google Scholar]
- Ai S, Xiong H, Li CC, Luo Y, Shi Q, Liu Y, et al. Profiling chromatin states using single-cell itChIP-seq. Nat Cell Biol. 2019;21(9):1164–72. Epub 2019 Sep 05. [DOI] [PubMed] [Google Scholar]
- Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, et al. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet. 2019;51(6):1060–6. Epub 2019 Jun 04. [DOI] [PubMed] [Google Scholar]
- Luo C, Rivkin A, Zhou J, Sandoval JP, Kurihara L, Lucero J, et al. Robust single-cell DNA methylome profiling with snmC-seq2. Nat Commun. 2018;9(1):3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun. 2018;9(1):3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci. 2018;21(3):432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell. 2019;176(1-2):361-76 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362(6416): 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888-902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell. 2019;177(7):1873-87 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sermer D, Pasqualucci L, Wendel HG, Melnick A, Younes A. Emerging epigenetic-modulating therapies in lymphoma. Nat Rev Clin Oncol. 2019;16(8):494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–41. Epub 2016 Sep 16. [DOI] [PubMed] [Google Scholar]
- Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25(3):403–18. Epub 2019 Mar 08. [DOI] [PubMed] [Google Scholar]