Abstract
The pancreatic peptide hormone, amylin, plays a critical role in the control of appetite, and synergizes with other key metabolic hormones such as glucagon-like peptide 1 (GLP-1). There is opportunity to develop potent and long-acting analogues of amylin or hybrids between these and GLP-1 mimetics for treating obesity. To achieve this, interrogation of how the 37 amino acid amylin peptide engages with its complex receptor system is required. We synthesized an extensive library of peptides to profile the human amylin sequence, determining the role of its disulfide loop, amidated C-terminus and receptor “capture” and “activation” regions in receptor signaling. We profiled four signaling pathways with different ligands at multiple receptor subtypes, in addition to exploring selectivity determinants between related receptors. Distinct roles for peptide subregions in receptor binding and activation were identified, resulting in peptides with greater activity than the native sequence. Enhanced peptide activity was preserved in the brainstem, the major biological target for amylin. Interpretation of our data using full-length active receptor models supported by molecular dynamics, metadynamics, and supervised molecular dynamics simulations guided the synthesis of a potent dual agonist of GLP-1 and amylin receptors. The data offer new insights into the function of peptide amidation, how allostery drives peptide–receptor interactions, and provide a valuable resource for the development of novel amylin agonists for treating diabetes and obesity.
Keywords: amylin, CGRP, calcitonin receptor, GPCR, IAPP, RAMP
Appetite control involves an intricate multifaceted system of hedonic and homeostatic mechanisms influenced by genetic and environmental factors. Multiple hormones, molecules, and neurotransmitters interact via the gut–brain axis to elicit both short- and long-term effects on energy balance.1,2 A multitude of neuroendocrine hormones play various roles in orexigenic or satiation signaling. These hormones are released by a variety of tissues, thus insulin is released from the pancreas, glucagon-like peptide-1 (GLP-1) from the gut, and leptin from adipose tissue to confer peripheral and centrally mediated metabolic effects.3
Amylin is a key part of this axis, being a pancreatic hormone that mediates widespread effects on energy homeostasis via brain centers that affect feeding behavior, resulting in suppressed food intake and reductions in body weight and adiposity.4,5 These effects complement those of GLP-1, and a combination of both amylin and GLP-1 agonists may have superior metabolic effects.6 Amylin also acts as a leptin-sensitizer, with combinations of both hormones showing remarkable metabolic benefits.7,8 An amylin-mimetic drug, pramlintide, which differs in amino acid sequence from human amylin by only three amino acids, is approved for use in humans as a treatment for diabetes, in conjunction with insulin. Pramlintide has also shown proof-of-concept clinical efficacy for the treatment of obesity.9 However, pramlintide is short-acting, requires subcutaneous injection at meal times and cannot be coformulated with insulin.4 There is tremendous scope for developing novel amylin-mimetics with increased potency, half-life, and improved physicochemical properties, or generating combinations with other metabolic peptides, such as GLP-1. The lack of information on how amylin engages its receptor binding site to trigger signaling is hampering these developments.
Amylin receptors reside in the small class B G protein-coupled receptor (GPCR) grouping. Recent structures highlight how class B peptide ligands bind to their receptors in an extended conformation, with conformational changes likely propagated through multiresidue contacts between the peptide N-terminus and the upper portion of the receptor transmembrane bundle and extracellular loops (ECL), known as the juxtamembrane region.10,11 Though valuable, this body of data cannot easily be applied to amylin because high affinity amylin binding requires the presence of a second protein to form a heterodimeric receptor complex.12 Amylin activates the calcitonin receptor (CTR), which is also a receptor for the osteogenic calcitonin peptide. The association of a single transmembrane-spanning receptor activity-modifying protein (RAMP) with the CTR alters its pharmacology, resulting in receptors with higher affinity for amylin. This mechanism creates multiple amylin receptor (AMY) subtypes (hAMY1, hAMY2, and hAMY3) from CTR with RAMP1, 2, or 3, respectively (Figure 1a).13−15 To add further complexity, RAMPs can alter G protein-coupling, receptor trafficking, and downstream signaling of an increasing number of GPCRs, including the calcitonin receptor-like receptor (CLR) to form calcitonin gene-related peptide (CGRP) and adrenomedullin receptors (AM1 and AM2), with the different RAMPs.16 Drug discovery efforts at RAMP-coupled receptors including amylin, CGRP, and adrenomedullin receptors cannot be truly effective unless RAMP contribution to ligand interactions can be defined and the key molecular drivers of selectivity between closely related peptides and receptors can be identified.
Figure 1.
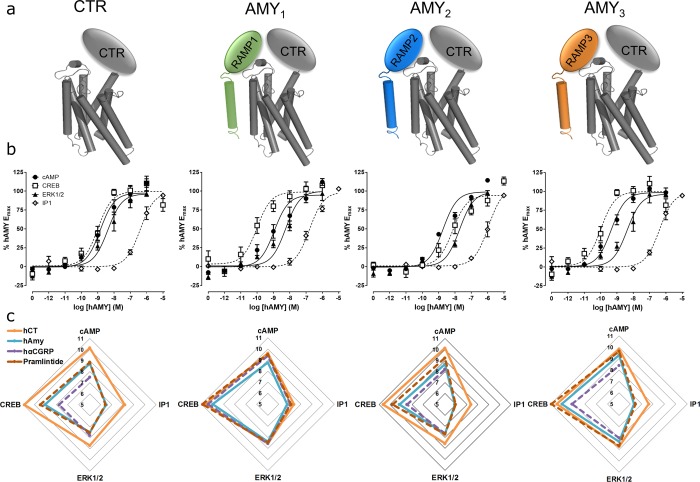
(a) Receptor subunit composition, (b) activation of signaling pathways at the corresponding receptors by human amylin (hAMY), (c) activation of signaling pathways at the corresponding receptors by all peptides. In panel b, the concentration response curves are the combined mean data from four or five independent experiments (cAMP, pCREB n = 5, IP1, pERK1/2 n = 4). In panel c, potency data are summarized in radial plots showing mean pEC50 values from between three and five individual experiments. Exact experimental n is provided in Tables SB1–4. All errors are s.e.m. pERK1/2 data is the 15 min time point.
Using peptide synthesis and determination of peptide activity at multiple receptors we report key drivers for amylin–receptor interactions and identify distinct roles for its two post-translational modifications in affinity, activity, and selectivity. The mechanism of selectivity between receptors is remarkably subtle and not clearly linked to sequence changes between related peptides, strengthening the notion that selectivity is principally driven via an allosteric effect of the RAMP to augment the peptide binding site within CTR. Our data identify a key region of the amylin peptide that provides an area of focus to generate higher potency amylin mimetics. We use our mechanistic data and dynamic molecular models to develop a potent dual agonist of amylin and GLP-1 receptors.
Results and Discussion
Pramlintide, a first-in-class amylin mimetic, is available for the treatment of insulin-requiring diabetes. However, further improvements are required to develop a novel amylin receptor-directed drug for metabolic disease or other conditions, including Alzheimer’s disease.4,17,18 This potential improvement is constrained because little is currently known about the mechanisms of amylin receptor binding and activation. Extensive study into the structure and function of the related glucoregulatory hormone, GLP-1, has guided the production of the most promising antiobesity and antidiabetes class of therapy currently available, such as the drugs liraglutide and semaglutide.19 Scrutinizing the properties of amylin is critical for progressing drug discovery efforts. This is especially important given the complex heterodimeric assembly of the different amylin receptor subtypes (Figure 1a), and the proposed bimodal (two stage) receptor binding mechanism involving the N and C termini of the peptide.11
Amylin Receptors Display Similar Pharmacological Profiles Irrespective of Signaling Pathway
GPCRs are well-known for pleiotropic intracellular signaling, giving substantial scope for “functional selectivity” or “biased signaling”, in which different ligands preferentially activate particular signaling pathways at one receptor, via unique receptor or G protein conformations, or linked to the kinetics of ligand binding and unbinding.20−22 CTR and CTR/RAMP complexes have multiple potential ligands and are reported to couple to Gαs, resulting in the downstream activation of adenylyl cyclase and cAMP production, to Gαq or to Gαi and to promote other downstream signaling events, such as ERK1/2 phosphorylation. How much impact the presence of RAMPs/different ligands has on signaling is not well-defined.23 We therefore profiled multiple signaling pathways in cells transfected with CTR alone or CTR with different RAMPs using multiple ligands. We compared human calcitonin as the cognate endogenous ligand of CTR to the drug pramlintide and human amylin as amylin receptor agonists and hαCGRP as a second high affinity ligand of the CTR/RAMP1 complex.24
Human amylin, human calcitonin, hαCGRP and pramlintide were all capable of inducing cAMP responses at all receptors (Figure 1b,c). Similar results were obtained for downstream CREB phosphorylation, although potency in general was higher at this pathway (Figure 1b,c). Human amylin, human calcitonin, and pramlintide were all capable of inducing IP1 accumulation at the different receptors, although hαCGRP was only able to elicit a measurable IP1 response at the hAMY1 receptor. All peptides produced ERK1/2 phosphorylation at two time-points at all four receptors. Figure 1 shows the 15 min data. Although IP1 and pERK1/2 were more weakly activated than cAMP or pCREB, the relative potencies of ligands were similar across all pathways (Figure 1b,c). Concentration–response curves for all peptides at all pathways are shown in Figure SB1 and corresponding potencies and Emax data are presented in Tables SB1–4. These data suggest the effect of RAMP on CTR pharmacology is largely independent of signaling pathway, at least with respect to the pathways measured in this study.
The Amylin C-Terminus Contains RAMP-Dependent Drivers of Affinity
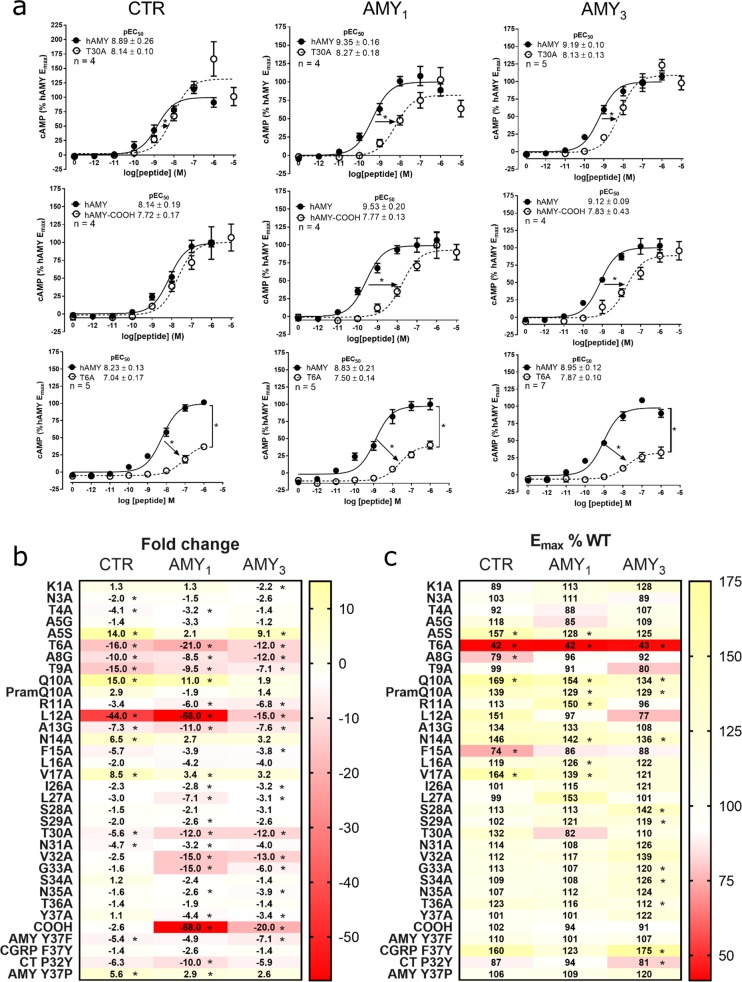
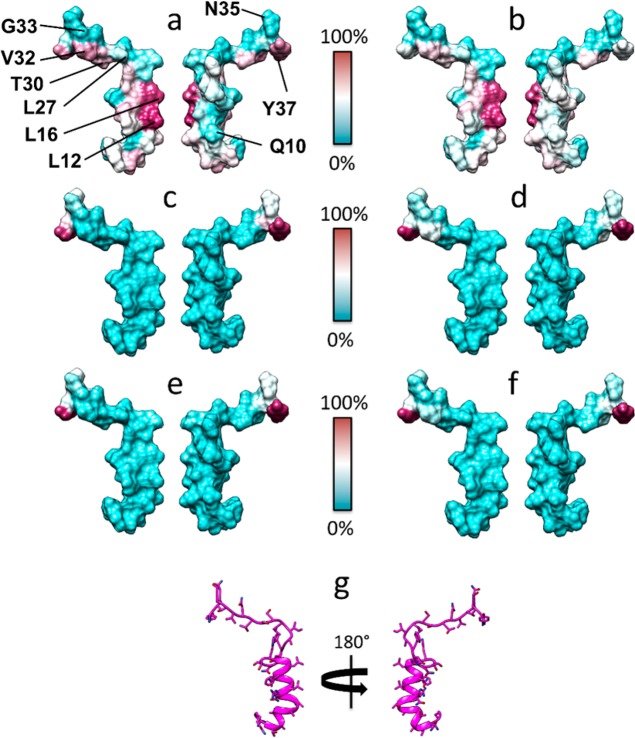
To understand the mechanisms through which human amylin triggers receptor signaling, we proceeded to explore the role of different regions of this peptide, using cAMP as our functional readout because amylin potently activates this pathway at CTR alone or in complex with RAMP. Structural data from related CGRP and AM receptors suggests that the amylin C-terminus will contain key residues for high affinity binding and potentially selectivity determinants between receptors.25 This region is also highly conserved across multiple species (Figure SB2). Therefore, we synthesized a series of alanine substituted peptides (Figure 2 and Figure SC1), testing their bioactivity at CTR, AMY1, and AMY3 receptors (see Supplementary Biology). We excluded the AMY2 receptor from these analyses because of its weaker induction of amylin phenotype in Cos 7 cells.23 Analysis of the C-terminal 12 amino acids of human amylin (residues 26–37) revealed that a single C-terminal residue (T30) was important with and without RAMP coexpression (Figure 3a–c, Figures SB3–5, Tables SB5–7). Substitution of T30 with alanine resulted in reductions in amylin potency at all three receptors, although the presence of RAMP influenced the magnitude of the reduction (∼5-fold at CTR, ∼10-fold at AMY1, and AMY3). An additional two C-terminal residues (V32, G33) had a 6–15-fold reduction in amylin potency but only in the presence of RAMP1 and RAMP3, not at CTR alone. Figure 3 panels b and c summarize the potency and Emax data, respectively; full data and statistical information can be found in the accompanying Supplementary Biology file. Functional data were supported by binding data at AMY1, where reductions in affinity generally mirrored changes in potency (Figure SB6). Hence, the C-terminal sequence of human amylin contains amino acids that are more important for receptor binding in the presence of RAMP. We speculated that this could be because the extreme amylin C-terminus docks less effectively into CTR in the absence of RAMP. To examine this and to provide mechanistic insight into our data, we developed the first active models of full-length amylin receptors with amylin bound (Figure 4). The starting model, along with key peptide residues, taken from 750 ns of molecular dynamics (MD) simulations is shown in Figure 4 and Figure SM1.
Figure 2.
Amino acid sequences of peptides used during the study. Further details are provided in Table SC1. Blue shading illustrates alanine or glycine substitution (or serine substitution in the C2S,C7S peptide), green shading shows a modification, red shading shows the three substitutions in pramlintide, purple shading shows incorporation of a CGRP residue into amylin, gray shading shows incorporation of an amylin residue into another peptide, and orange shading illustrates incorporation of a calcitonin residue.
Figure 3.
(a) cAMP production at three receptors by three selected amylin analogues. (b,c) Heat maps for all peptide analogues showing effect on potency, as a fold-change from control (b) and Emax as a percentage of control (c), in cAMP production at three receptors. Concentration response curves are the combined mean data from between four and seven independent experiments, with exact experimental n shown on each graph. All errors are s.e.m. (∗) P < 0.05 by unpaired t-test for pEC50 or where 95% confidence intervals did not include 100 for Emax. Experimental n for all data in the heat map is provided in Tables SB5–7.
Figure 4.
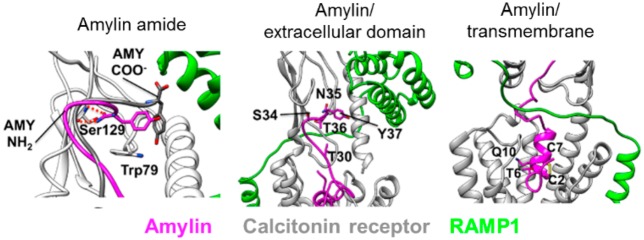
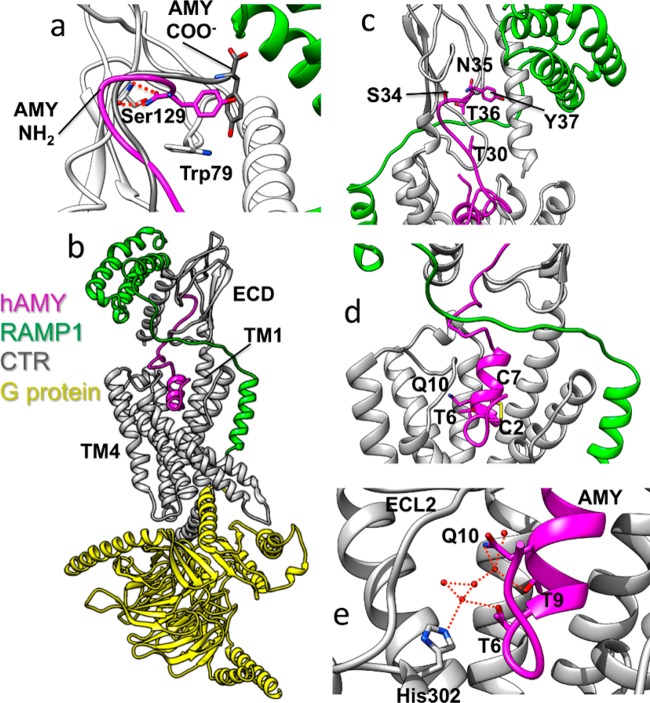
(a) Superposition of the equilibrated CTR:amylin–amide (AMY NH2, magenta) and the equilibrated CTR:amylin-COO– (AMY COO–, gray) complexes. Both peptides are shown in the presence of the CTR ECD (white) and RAMP1 (green) taken from the CTR:amylin–amide simulation. The peptide amide group hydrogen bonds with the backbone of Ser129, while the carboxylate forces the C-terminus to adopt a different orientation. (b) The AMY1 receptor model. (c) The ECD of the AMY1 receptor model. (d) The peptide N-terminal region of the AMY1 receptor model. (e) T6–His302 interactions. Besides the contact between T6 of amylin (magenta) and His302 of CTR (gray), a water-mediated interaction occurs during MD simulations. This is part of a more extended water network, stabilized at the interface between the peptide and the ECL2. Dashed red lines represent hydrogen bonds between donor and acceptor atoms.
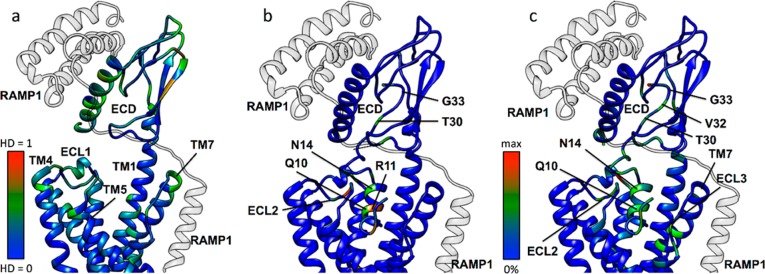
We compared the conformations of the CTR during MD simulations in the presence and absence of RAMP1 by evaluating the difference in the distribution of the backbone torsional angles using the Hellinger distance (HD).26Figure 5a shows that, at the extracellular vestibule, these differences are mainly localized at the CTR ECD, ECL1, ECL2, and TM1, 2, 3, 4, and 7. We also evaluated differences in the number of residue–residue contacts, both van der Waals contacts and hydrogen bonds. The predicted number of peptide–receptor residue–residue hydrogen bonds and contacts in the CTR ECD, ECLs and extracellular region of the helices is dependent on whether RAMP1 is present or absent (Figure 5b,c). The bound simulations suggest that T30 and V32 make different contacts in the presence of RAMP1 (Figure 5b,c), supporting the experimental data showing a RAMP-dependent difference in the effect of mutation of these residues. While G33 does not show differential contacts along the binding pathway, analysis of the Ramachandran plot (Figure SM2) shows that in the absence of RAMP1 the distribution of G33 backbone angles is similar regardless of whether the peptide is already bound to or is approaching the receptor (Figure SM2b,d); the presence of RAMP1, instead, has a deep influence on the torsional angle distribution, with notable differences when the peptide is approaching the ECD compared to the bound state (Figure SM2a,c).
Figure 5.
(a) Hellinger distance (HD) analysis of the ϕ,ψ protein backbone angles for CTR in the presence and absence of RAMP1 (transparent gray ribbon). For each residue, the higher HD value between ϕ and ψ is displayed on a model of the CTR structure (peptide omitted for clarity), with small values shown in blue and large values (indicating significant conformational differences) in red. (b) The difference in CTR and amylin (both ribbons) intermolecular hydrogen bond contacts in the presence and absence of RAMP1, with small values shown in blue and large values in red. (c) The difference in CTR and amylin (both ribbons) intermolecular contacts in the presence and absence of RAMP1, with small values shown in blue and large values in red.
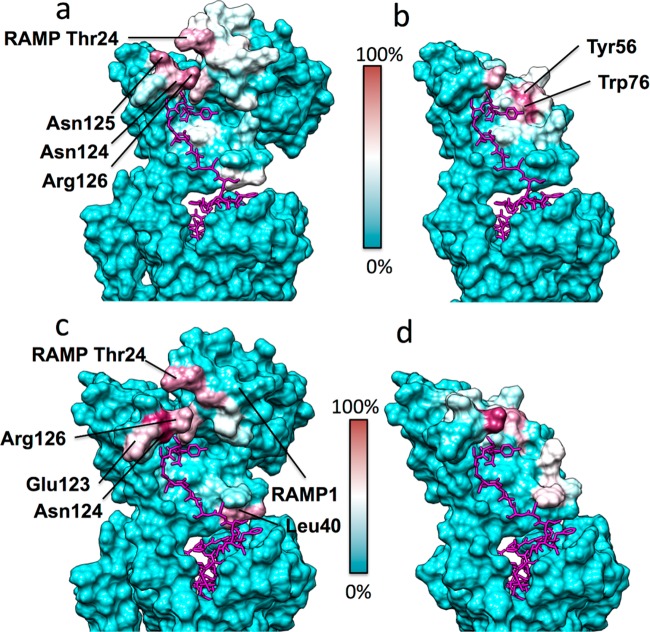
Figure 6a,b shows significant differences in the early recognition events on the binding pathway in the presence and absence of RAMP1, as determined by the supervised MD (SuMD) simulations (Movies 1 and 2); most notably the presence of RAMP1 increases the predicted interactions with Asn124, Asn125, and Arg126 during amylin approach to the ECD (Figure 6a and Movie 1), although for Asn125 this could be affected by glycosylation which we did not test here.27 On the other hand, in absence of RAMP1 the peptide makes contacts with residues inaccessible when RAMP1 is present, such as Tyr56 and Trp76 (Figure 6b and Movie 2).
Figure 6.
CTR/AMY1–amylin-amide (a, b, magenta) and CTR/AMY1–amylin-carboxylate form (c, d, magenta) contacts identified during SuMD simulations, plotted on the CTR/AMY1 molecular surface. The CTR/AMY1 residues least engaged by amylin (0% contact) are colored cyan, while residues most engaged by amylin (100% contact) are colored purple. (a) SuMD simulations of amylin–amide binding to AMY1 receptor ECD. (b) SuMD simulations of amylin–amide binding to the CTR ECD. (c) SuMD simulations of amylin–carboxylate form binding to AMY1 receptor ECD. (d) SuMD simulations of amylin–carboxylate form binding to the CTR ECD.
Amylin C-Terminal Amide Is Critical for High Affinity Binding to CTR/RAMP Complexes
The amidated C-terminus is strictly conserved among all known amylin sequences even though amidation enhances the in vitro propensity to aggregate and form amyloid.28 Intriguingly, this could be substituted in amylin with carboxylate with no loss of peptide activity at CTR alone. However, the human amylin-COOH peptide lost potency by 20 and 58-fold in the presence of RAMP3 and 1, respectively, which was the largest effect for any C-terminal analogue (Figure 3a–c, Figures SB3–5, Tables SB5–7). Binding affinity was also substantially reduced (Figure SB6). In the CTR ECD crystal structure, the salmon calcitonin proline amide makes critical contacts with the Ser129 backbone, supported by Asp77, Lys110, and Tyr131, plus hydrophobic interactions with Trp79.29 We propose that the C-terminal amide is critical for receptor selectivity.
Our models suggest that this could be driven by conformational and electrostatic differences in the CTR ECD as a consequence of RAMP interaction. The electrostatic potential of the receptor ECD in the vicinity of Y37 is much more negative in the presence of RAMP1 than in its absence (Figure SM3), ensuring that the C-terminal–COO– group experiences more repulsive interactions than the usual C-terminal amide (−CONH2) group; this may underlie the very large experimental reduction in binding seen for the −COOH C-terminal analogue in the presence of RAMP1 (Figure SB6). This is also coupled with the reduction in hydrogen-bonding complementarity between the carboxylate and the backbone of Ser129 (Figure 4a). As a consequence, the terminal −COO– group may no longer be able to form the same tight binding interactions as amylin is predicted to with Ser129, but the Y37 side chain could still make hydrophobic interactions with Trp79, as indicated in Figure 4a. RAMP3 has a similar effect, but the electrostatic potential is not as negative, and this may explain why the effect of RAMP3 is less marked than that of RAMP1 (Figure 3, Figure SM3c). As highlighted by SuMD simulations, the electrostatic repulsion elicited by RAMP1 affects the amylin binding pathway toward the ECD of CTR (Figure 6 and Figure SM4). The models and simulations suggest that when approaching the receptor, the C-terminal amide form of the peptide makes contacts with a higher number of residues on the surface of RAMP1 (Figure 6a and Figure SM4), compared to the C-terminal carboxylate form (Figure 6c and Figure SM5), indicating a higher number of stabilizing interactions during the early stages of the intermolecular recognition.
C-terminal amidation occurs in many different bioactive peptides, including other GPCR peptide ligands such as gastrin, vasointestinal peptide, and GLP-1.30 This post-translational modification is known to be crucial for bioactivity in many peptides.30 That the C-terminal amide was a critical determinant of high affinity binding and activity at RAMP-associated amylin receptors but not at the CTR alone suggests that the C-terminal amidation in amylin is not necessarily universally important but depends on the context. The structural availability or conformation in which this C-terminal post-translational modification is presented to the receptor(s) may act to modulate the activity of amylin and related peptides at their receptors. Our modeling suggests that this could occur by the amide affecting the binding pathway of a peptide when engaging its receptor.
The Extreme C-Terminal Amino Acid of Amylin Does Not Exclusively Dictate Receptor Selectivity
The X-ray crystal structures of CGRP and AM bound to the CLR ECD indicate minimum interactions between the peptide and the RAMP, which primarily involve the C-terminal residue side-chain, namely F37 of CGRP and Y52 of AM.25 This C-terminal residue is conserved as tyrosine in amylin (Figure SB2) and the minimal RAMP interactions naturally carry over into the CTR-RAMP models from the template through the homology modeling process. However, alanine substitution had only a small effect at AMY1 and AMY3 (Figure 3b,c). To further experimentally interrogate the role of this position as a possible RAMP contact in CTR/RAMP complexes and as a potential selectivity determinant between CTR and CLR-based receptors we synthesized full-length human amylin and human αCGRP with their C-terminal residue exchanged (Figure 2; that is, Amylin Y37F and CGRP F37Y). Incorporation of the CGRP C-terminal phenylalanine to replace the amylin tyrosine did not increase potency at the CLR/RAMP1 CGRP receptor. Instead, this peptide had universally decreased potency at all six receptors tested (CTR, AMY1, AMY3, CGRP, AM1, AM2) (Figure SB7). The reciprocal substitution in CGRP had an increase in potency at the AM1 receptor and an increase in Emax at the AMY3 receptor but little effect at the other receptors (Figure SB8, Table SB8). These data suggest that the nature of the C-terminal residue in amylin does not exclusively or clearly drive selectivity between CLR or CTR-based receptors, consistent with the lack of persistent hydrogen bonding between Y37 and RAMP1 in MD simulations (Movie 3). We extended this work to explore exchange of Pro/Tyr between amylin and calcitonin, generating amylin Y37P and human calcitonin P32Y (Figure SB9, Table SB8). P32Y calcitonin had a small reduction in potency at all receptors but no change in affinity at the AMY1 receptor (Figure SB9). Y37P amylin had a small increase in potency, mirrored by an increase in affinity at the AMY1 receptor (Figure SB9). This is consistent with data for an amylin antagonist fragment.31 Thus, the C-terminal amino acid in the calcitonin peptide family is not a clear and exclusive signature for selectivity or affinity.
The Amylin N-Terminus Contains RAMP-Independent Drivers of Amylin Affinity and Efficacy
A recent low resolution cryoelectron microscopy structure of CTR suggests that the calcitonin N-terminal loop formed by a disulfide bond and adjacent α-helix may make several contacts with the upper transmembrane bundle and ECLs of CTR.10 To determine whether this may be a conserved mechanism for amylin and CTR/RAMP complexes, we synthesized human amylin-derived peptides that explored different elements of the binding and activation mechanism. We synthesized three linearized full-length human amylin peptides lacking the C2–C7 disulfide bond, and we divided human amylin into two segments (Figure 2). Omission of the cysteine to cystine oxidation step in the human amylin synthesis enabled generation of a linear peptide. However, this cysteine-containing peptide spontaneously oxidized in our assay conditions (data not shown). Therefore, we synthesized two other full-length variants to probe the role of the disulfide-containing loop; these were a double serine variant, replacing the two native cysteines with serines and also a CAM variant, where the two cysteines were attached to carboxyamidomethyl blocking groups (Figure 2). See Supplementary Chemistry for details. Both peptides displayed a decrease in potency, which was greater for the double serine variant. This was accompanied by a decrease in Emax for this peptide (Figures SB10–12, Tables SB5–7). An intact ring structure in amylin is evidently necessary for full bioactivity. Linearized variants of CGRP, human, and salmon calcitonin have been previously been synthesized. These show a range of activities, depending on the peptide and assay but there is a precedent for peptides within this family to have some activity in the absence of an intact N-terminal ring.32−35
We further explored the role of the N-terminal ring using the amylin8–37 fragment, which was expected to antagonize the receptors and lack efficacy.36 Instead we observed weak partial agonism from this peptide which was almost identical to the profile of the double-serine peptide (Figures SB10–12, Tables SB5–7). This suggests that the 8–37 sequence contains molecular determinants of efficacy, as well as affinity. However, this peptide was of lower purity than all of the other peptides and therefore we confirmed this result with a second synthesis of amylin8–37 (amylin8–37(DR)) from our collaborating laboratory (Figures SB10–12). For unknown reasons, this second synthesis was also difficult to purify to >90%. Interestingly, N-terminal acetylation afforded a >10-fold gain in potency, compared to amylin8–37 (Figures SB10–12). Given that this acetylated peptide was 97% pure, it suggests that the prior results with the lower purity amylin8–37 peptides were not artifacts of any impurity. N-terminal acetylation removes the positive charge of the N-terminus and helps to partially deconvolute the effects due to removal of residues 1 to 7 and the disulfide loop from any effects caused by introduction of a positive charge. Removal of the charge and the addition of an N-capping acetyl group is also expected to increase the helical propensity of the peptide. This could explain the improved potency of this analogue, compared to that of amylin8–37.
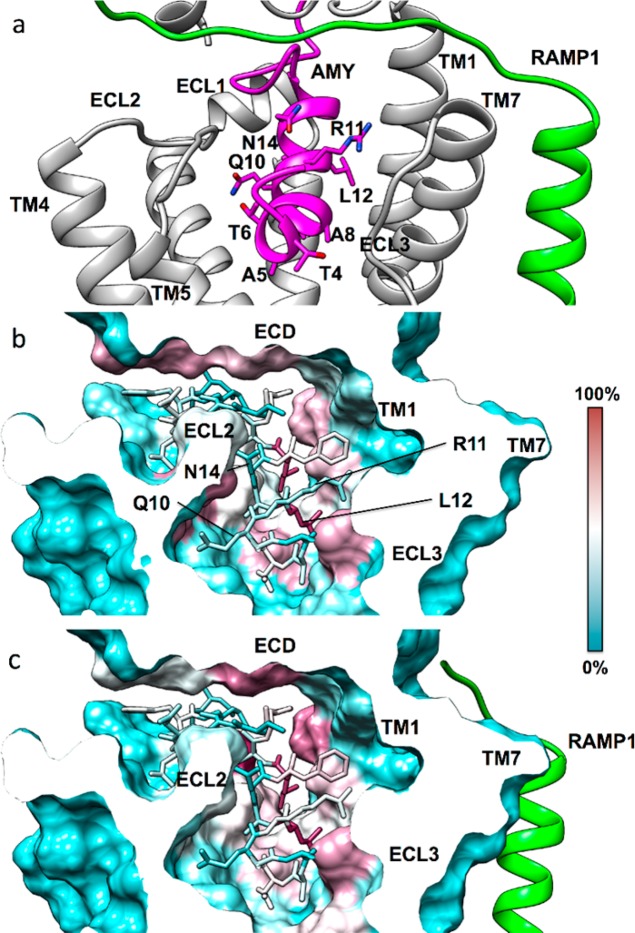
We next made a peptide that retained only the loop and the predicted α-helix, based on the solution structure of human amylin in SDS micelles (PDB code 2KB8(37)); amylin1–17. This peptide was also a partial agonist, suggesting that the 7–17 helix contains important residues for receptor activation (Figures SB10–12, Tables SB5–7). This is supported by the data in Figure 3b, whereby apart from C2 and C7, T6 is the only significant residue missing from amylin8–37. The amylin1–17 peptide could provide a useful lead for the future development of shorter peptides, such as have been achieved in the near wild-type 11mer GLP-1 analogue,38 which activates the class B GLP-1 receptor.
We synthesized alanine-substituted peptides from position 1 to 17 within full length amylin, excluding the cysteines, to determine the role of individual amylin amino acids within this region (see Supplementary Chemistry). Where a native residue was alanine, we replaced this with glycine. Figure 3b summarizes these results. Seven analogues exhibited decreased potency (T4A, T6A, A8G, T9A, R11A, L12A, A13G) at two or more receptors, which was generally accompanied by a similar reduction in affinity (Figure 3b, Tables SB5–7, Figures SB13–16). T6A and L12A had large effects, with decreased potency for both and a substantial decrease in Emax for T6A (Figure 3a–c). The large effect on potency upon mutating L12 (Figure 3b) probably arises through its predicted hydrophobic interactions to TM1 (Figure 7), similar to the interactions shown or inferred in the class B GPCR cryoelectron microscopy structures.10,39 The importance of this residue is underlined by its high conservation as hydrophobic in class B GPCR peptide hormones and its complete conservation within amylin sequences (Figure SB2).40 T6 lies within the critical N-terminal region that is absent in weak agonists such as amylin8–37 and is probably one of the main drivers of activation. The main interaction of T6, either directly or via bridged water molecules (Figures 4, 7), is predicted to be with His302 on TM5 of CTR; TM5 is known to play a key role in activation. The adjacent residue at position 5 is alanine in human amylin, where glycine substitution had no effect. This residue is serine in calcitonin and is also predicted to interact with His302.10 We hypothesized that the sequence divergence at this position may underlie pharmacological differences between CTR and CTR/RAMP complexes and synthesized human amylin with serine at position 5 in place of the native alanine. This peptide had increased activity at CTR, AMY1 and AMY3 receptors, with the greatest increase at CTR (Figure 3b, Figure SB17, Tables SB5–7). Thus, position 5 has the potential to be an important driver of activity, as has also been observed in CGRP.41
Figure 7.
(a) The modeled amylin (magenta) N-terminus binding mode inside the CTR (gray) transmembrane domain (RAMP1 is green). The hydrophobic residue L12 orients toward TM1, while the opposite side of the peptide is characterized by more hydrophilic amino acids (T6, Q10, N14). Overall contacts established by the amylin N-terminus (stick representation) inside the CTR transmembrane domain, plotted on the receptor molecular surface. (b) Intermolecular contacts identified during MD simulations of amylin bound to CTR, plotted on the CTR molecular surface, (c) Intermolecular contacts identified during MD simulations of amylin bound to the AMY1 receptor (RAMP1 in green). The CTR residues least engaged by amylin (0% contact) are colored cyan, while residues most engaged by amylin (100% contact) are colored purple.
The predicted interactions of T4–N14 with multiple residues in the juxtamembrane region of CTR are shown in Figure 7 and Movies 4 and 5. These suggest how the mutation of each of these residues has effects on peptide activity. Interestingly. the experimental effect of mutations in this region is not hugely dependent on the presence or absence of the RAMP and indeed, the significant predicted interactions observed are, bar L12, generally similar regardless of whether the RAMP is present or not (Figure 7). Metadynamics simulations show the partial unbinding of the amylin N-terminus under the input of energy, but the initial part of the simulation also justifies the bound simulations as the peptide does not explore novel interactions as a result of this energy. Figure 7 shows that the center of gravity of the peptide N-terminus interactions within the juxtamembrane region is predicted to shift away from ECL2 and toward the hydrophobic surface on TM1/TM2/TM7 when RAMP1 is present.
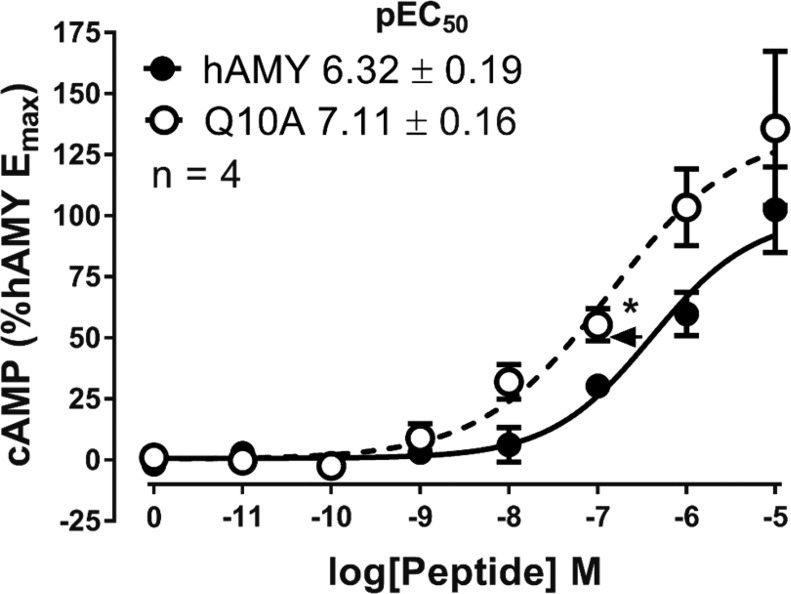
Other experimentally important positions for regulating peptide activity were Q10, N14, and V17. Alanine substitution in human amylin at each of these positions unexpectedly increased activity, particularly at CTR and AMY1 (Figure 3b,c, Figures SB13–16, Tables SB5–7), suggesting that alteration of the side-chains of these residues yields modified peptide–receptor interactions. In our models, Q10 is predicted to interact with Trp290 but in Q10A, this interaction is lost. In our simulations, Glu294 flips into the resulting hydrated cavity, thereby modifying the ECL2 conformation (Movie 6) and interacts with K1 of amylin; either of these effects may contribute to the increased activity of Q10A.
Q10, N14, and V17 lie on the opposite side of the amylin helix to L12 and face toward ECL2. Hence we hypothesized that alanine substitution of any of these residues could alter the helical propensity of unbound amylin, affecting receptor interactions and potency. The AGADIR algorithm suggests that two variants are predicted to have a lower helical propensity (Q10A and N14A) than human amylin, whereas V17A is predicted to have a higher helical propensity than the unmodified peptide (Figure SB18). Thus, there is no clear correlation between receptor activity and predicted helical propensity for these variants. Future studies could consider the effect of these substitutions on receptor binding kinetics, given the known importance to kinetics of the helical region in salmon calcitonin.42
The RAMP-dependent behavior of many analogues, together with our modeling indicates that the principal mechanism for how RAMPs alter the binding pocket of the receptor is allosteric, in line with other data.31,40,43 This contrasts with CLR, where direct RAMP-peptide interactions have been demonstrated. The allosteric mechanism RAMPs employ to modulate CTR pharmacology and signaling may have broader implications for other GPCRs. RAMPs may sculpt the peptide-binding pocket to differentially expose receptor residues that can associate with the C-terminal amide or other residues, affecting affinity and downstream activation and signaling. Our modeling suggests that this can be achieved in a number of different ways, including steric effects, electrostatic effects, and effects on the binding pathway. The repertoire of RAMP-associating GPCRs identified spans the GPCR superfamily.16 The ubiquitous expression of RAMPs and their coevolution with GPCRs suggest that they will have many more receptor partners than is currently appreciated, emphasizing the need for greater understanding of their effects on GPCRs.44 It may be fruitful to explore the correlation between the presence or absence of a C-terminal amide on peptide ligands with phenotypic effects of RAMPs on given GPCRs.
Amylin Analogue Signaling
Amylin analogues with increased activity could be valuable drug leads, particularly if the peptide is shorter than human amylin. It was therefore important to determine whether increased activity of some analogues translated into a system that endogenously expresses amylin receptors. The major target site for amylin is the brainstem,45 which has abundant high affinity amylin binding sites.46 We prepared primary rat brainstem cultures from the medulla; this includes the area postrema and the nucleus of the solitary tract, as well as other nuclei that are reported to express CTR and other amylin receptor subunits.47−50 We confirmed that CTR was present with two different antibodies (Figure SB19), although we were unable to confirm colocalization of RAMPs in our cultures due to a lack of suitable antibodies. Nevertheless, we tested the activity of human amylin and Q10A amylin at increasing cAMP production in these cultures. Increased activity of this analogue was retained in this physiologically relevant system (Figure 9). However, it is not clear from these data whether amylin is acting via CTR alone or an amylin receptor. Amylin potency is relatively low and this could suggest it is acting via CTR alone, but this could be a consequence of this particular mixed nuclei culture. Future studies should examine amylin action in a more defined culture from discrete brainstem nuclei.
Figure 9.
cAMP production in rat brainstem cultures by human amylin or Q10A human amylin. Concentration–response curves are the combined mean data from four independent experiments. All errors are s.e.m. (∗) P < 0.05 by unpaired t-test.
We questioned whether increased activity would also occur if we substituted Q10 in the approved drug pramlintide with alanine. Q10A pramlintide showed small increases Emax at AMY1 and AMY3, when compared to unmodified pramlintide (Figure 3b, Tables SB5–7).
Any alteration to a peptide can affect how it engages its receptors and trigger signaling, with the potential for signal bias.51 Therefore, we tested a selection of peptide analogues for their ability to affect other pathways, namely CREB or ERK1/2 phosphorylation and IP1 accumulation. We chose T6A, which had decreased cAMP activity and a smaller reduction in affinity, along with Q10A and V17A both of which had increased activity with respect to cAMP production and distinct effects on affinity and predicted helical propensity. Relative to amylin, all analogue peptides exhibited similar signaling profiles at all receptors for each pathway (Tables SB9–12, Figures SB20,21). T6A displayed lower relative efficacy, calculated as Δlog(τ/KA), when compared to amylin. Q10A and V17A displayed higher or equivalent Δlog(τ/KA) to amylin (Figure SB21a). When compared to a reference pathway (cAMP) to account for differences in the relative efficacy between the different analogue peptides, no significant signaling bias, calculated as ΔΔlog(τ/KA), was observed (Figure SB21b). Thus, despite differences in the relative efficacy, these analogue peptides appear to have relatively balanced signaling for the measured pathways.
Retention of Potent Dual Receptor Activity by a Peptide Combining Pramlintide and Exendin-4
Our data and models provide a valuable resource for the design of novel amylin-based peptides. Metabolic disease results from the dysregulation of a multitude of hormones and thus, combination hormone therapies containing amylin could be a valuable approach for successful treatment.8 We chose to exploit the synergistic behavior exhibited by amylin agonists and GLP-1 agonists4 to produce a novel dual agonist of GLP-1 and amylin receptors, coining the phrase “DAGAR”. Such molecules could be used as pharmacological probes to further explore this intriguing biology. Previous attempts to create agonists with dual amylin and GLP-1 receptor activity have resulted in reduced activity compared to the single parent peptides. For example, CTR was used to probe amylin-like activity with a marked reduction in agonism of approximately 25-fold.52,53 This reduced activity could be a consequence of joining the peptides via the amylin analogue N-terminus, whereby the modified peptide could no longer fit effectively within the transmembrane domain, or because the method of conjugation was suboptimal for peptide-receptor activity.52,53
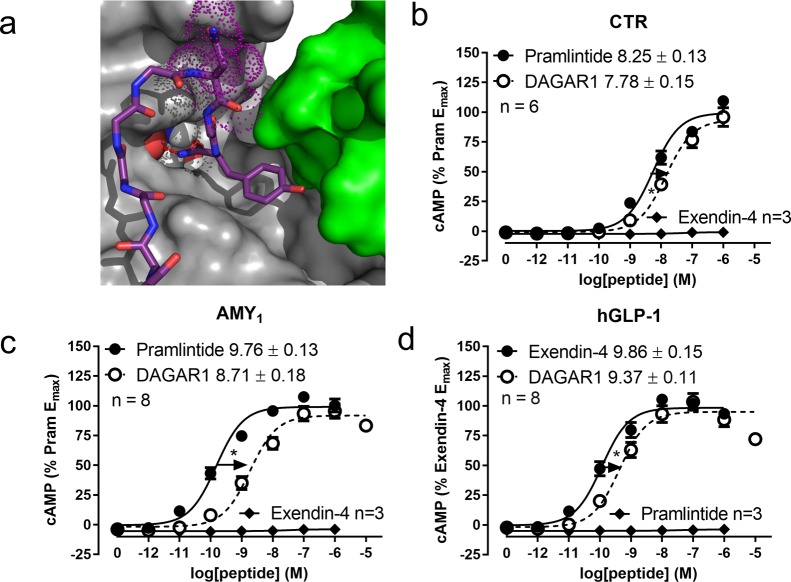
Here we developed a new approach using Cu(I)-catalyzed alkyne azide cycloaddition to join the C-terminus of the GLP-1 receptor agonist drug exendin-4 to position 35 toward the C-terminus of pramlintide (Figure SC3), which our amylin MD simulations (Figure 8, 10a, Movie 3) and existing data on exendin-454,55 suggested would be well-tolerated. We also selected position 35 in pramlintide because we have previously reported that this position appears to tolerate glycosylation, even with large sugars.56 Exendin-4 was therefore modified with an azido-lysine tag at the C-terminus and pramlintide was modified with a complementary acetylene-containing propargylglycine residue at position 35. Subsequent “click chemistry” smoothly afforded the novel triazole-linked hybrid peptide termed DAGAR1. Pleasingly, DAGAR1 retained equivalent Emax to pramlintide and had only a small reduction in potency compared to pramlintide at CTR, demonstrating an improvement over prior efforts (Figure 10b). At the AMY1 receptor, this peptide had equivalent Emax to pramlintide and retained nanomolar potency but had an ∼10-fold reduction in potency and binding affinity compared to pramlintide (Figure 10c, Figure SB22). This greater reduction in the presence of RAMP1 could be a consequence of interference with the allosteric mechanism. At the GLP-1 receptor DAGAR1 retained subnanomolar potency, which was only 3.8-fold lower than that of unmodified exendin-4 (Figure 10d). The Emax of DAGAR1 was equivalent to that of Exendin-4 at the GLP-1 receptor. The potent activity of this bifunctional peptide highlights the value of our structure–function driven strategy and the power of click chemistry for site-specific conjugation of long peptides that contain only minor modifications compared to the native sequence. This approach illustrates how long peptides can be efficiently joined together, creating opportunities for pairing diverse combinations of peptides.
Figure 8.
Predicted CTR/AMY1 - amylin contacts identified during MD simulations, plotted on the amylin molecular surface. The amylin residues least engaged by the receptor (0% contact) are colored cyan, while residues most engaged by the receptor (100% contact) are colored purple. (a) MD simulations of amylin bound to the AMY1 receptor. (b) MD simulations of amylin bound to CTR. (c) Supervised MD (SuMD) simulations of amylin binding to the AMY1 ECD. (d) SuMD simulations of amylin binding to the CTR ECD. (e) MD simulations of amylin after the SuMD simulations performed on the AMY1 receptor. (f) MD simulations of amylin after the SuMD performed on CTR. For the SuMD results, the data normalization is heavily weighted by the high number of contacts made by T37, so other contacts may not be shown very strongly. (g) Amylin (magenta, two different views), is reported as the reference structure.
Figure 10.
Receptor activity of a dual amylin and GLP-1 receptor agonist (DAGAR1). (a) Position N35 (dots) in amylin (magenta) when bound to the AMY1 receptor ECD, (b,c,d) cAMP production at human CTR (b) AMY1 (c) and GLP-1 (d) receptors. Concentration response curves from transfected Cos-7 cells are the combined mean data from six to eight independent experiments. All errors are s.e.m. (∗) P < 0.05 by unpaired t-test.
Conclusion
Our extensive characterization of the human amylin sequence distinguished discrete residues and structural features that were important for receptor binding and activation. The data, combined with extensive molecular modeling offer new insights into the function of peptide amidation and how allostery may drive peptide–receptor interactions. The data also provide a valuable resource for the development of novel amylin agonists for treating diabetes and obesity.
Methods
Cell Culture and Transfection
Receptors were expressed in mammalian cells via transient transfection. Cos-7 cells were used because these lack endogenous expression of RAMPs, CLR, and CTR, allowing careful control of the receptor that is expressed.57 The cells were cultured as previously described.57 Briefly, cells were maintained in Dulbecco’s modified eagle media (DMEM) supplemented with 8% heat-inactivated fetal bovine serum (FBS) in a 37 °C humidified incubator with 5% CO2. Cells were seeded into 96 well or 48 well plates at a density of 20 000 or 50 000 cells per well (determined using a Countess Counter, Thermo Fisher, New Zealand) and left for 24 h prior to transfecting. Transfections were performed using polyethylenamine as previously described.57 All DNA constructs were human receptors inserted in the mammalian expression vectors pcDNA3 or pcDNA3.1. Multiple splice variants of CTR have been reported, with the majority of research focusing on the CT(a) receptor isoform, which is conserved across mammals.58 In this manuscript, CTR is the CT(a) receptor splice variant, according to International Union of Basic and Clinical Pharmacology guidelines, generating AMY1(a), AMY2(a) and AMY3(a) subtypes with RAMP1, RAMP2, and RAMP3, respectively.12,15 In the main manuscript, CTR, AMY1–3 are used for simplicity. Specifically, constructs were the Leu447 polymorphic variant of hemeagglutinin (HA)-tagged hCT(a) receptor (kindly provided by Prof Patrick Sexton, Monash Institute of Pharmaceutical Sciences, Australia), hGLP-1 receptor (also from Prof Patrick Sexton), FLAG-tagged hRAMP2,59 myc-tagged hRAMP1 and untagged hRAMP3 (Kindly provided by Steven Foord, GlaxoSmithKline, UK). HA-CLR was also used in some experiments. All N-terminally tagged constructs have been characterized and previously reported to not affect receptor function.35,59−61
Peptides
All peptides were synthesized by Fmoc solid phase peptide synthesis (Fmoc SPPS), using different conditions, depending on the peptide sequence. These are outlined below and in Supplementary Chemistry. Table SC1 provides a summary of conditions.
Peptide Synthesis: General Procedure
Human amylin is notoriously difficult to synthesize, can be insoluble, and forms fibrils under some conditions.62 We have previously reported the successful synthesis and bioactivity of human amylin, with no evidence of cell death under our assay conditions.63,64 SPPS was carried out on-resin using the Fmoc/tBu strategy (Figure SC1). Briefly, in order to afford a C-terminal amide for peptide analogues a 4-[(R,S)-α-[1-(9H-floren-9-yl)]methoxycarbonylamino]- 2,4-dimethoxy]phenoxyacetic acid (Fmoc Rink amide) was attached to aminomethyl Chemmatrix (AM-CM) resin or aminomethyl polystyrene (AM-PS) resin. To obtain a C-terminal acid for human amylin (−COOH) Fmoc-O-tert-butyl-l-tyrosine attached to a 3-(4-hydroxymethylphenoxy)propionic acid (Fmoc-Tyr(tBu)-HMPP) was coupled to AM-CM as previously described.65 The peptide was elongated using either a microwave-assisted Biotage initiator + alstra (Biotage, Uppsala, Sweden) or Liberty (CEM, Matthews, NC, USA) or a room temperature Tribute or PS3 (Gyros Protein Technologies, Tucson, AZ, USA) peptide synthesizers (see Supplementary Chemistry for further details). Cleavage from the resin with simultaneous side-chain deprotection was achieved using trifluoroacetic acid/triisopropylsilane/water/3,6-dioxa-1,8-octane-dithiol (TFA/iPr3SiH/H2O/DODT, 94/1/2.5/2.5, v/v/v/v) for 2–3 h, precipitated with cold diethyl ether, isolated by centrifugation, dissolved in 50% aqueous acetonitrile containing 0.1% TFA and lyophilized. For formation of the disulfide bond, the crude peptides were dissolved in DMSO (10 mg/mL), and a solution of 2,2′-dithiobis(5-nitropyridine) (DTNP, 0.5 equiv) in DMSO (20 mg/mL) was added and the mixture shaken for 20 min. The mixture was diluted with H2O containing 0.1% TFA to a concentration of 1 mg/mL and immediately purified by semipreparative reversed phase high-performance liquid chromatography (RP-HPLC). Figure 2 shows the amino acid sequences of all peptides used in this study. Further details, including information on peptide purity are provided in the Supplementary Chemistry file.
Dual Agonist Synthesis
Nα-Fmoc-Nε-azide-l-Lysine [Fmoc-Lys(N3)–OH] and Nα-Fmoc-l-propargylglycine (Fmoc-Pra–OH)66 building blocks were used for incorporation of azide- and alkyne-handles during Fmoc SPPS of [Lys(N3)]40-exenatide and [Pra]35-pramlintide analogues, respectively, required for subsequent “click chemistry”.67,68 For each, the C-terminal amide was installed by the use of the Rink amide linker covalently bonded to AM-CM resin using conditions specified in Supplementary Chemistry. For synthesis of crude [K(N3)]40-exenatide, Nα-Fmoc deprotection was initially carried out using 20% piperidine in DMF for 2 × 5 min, followed by coupling of Fmoc-Lys(N3)–OH using O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU), and 2,4,6-collidine at room temperature for 1 h. The remaining sequence was elongated using a Biotage inititator + Alstra peptide synthesizer. Synthesis of crude [Pra]35-pramlintide was performed using the PS3 peptide synthesizer (see Supplementary Chemistry for further details). Both peptides were individually cleaved from the resin, isolated and lyophilized using conditions described in the peptide synthesis general protocol. The crude [K(N3)]40-exenatide (1.15 mg, 2.7 × 10–4 mmol) and [Pra]35-pramlintide (1.06 mg, 2.7 × 10–4 mmol) were dissolved in 40 μL of DMSO (degassed, N2). 0.25 M CuSO4·5H2O (4 μL, 1 × 10–6 mol) and 0.1 M Na ascorbate (10 μL, 1 × 10–6 mol) were added, and the resulting mixture was shaken for 5 min at 80 °C. The crude product was diluted (H2O, 500 μL), and purified by RP-HPLC.
Experimental Design
For signaling pathways apart from cAMP, time-course experiments were first conducted with a saturating concentration of peptide to determine the optimal time to conduct concentration–response experiments (data not shown). Concentration–response experiments were then conducted with the same experimental design for all pathways, including cAMP assays. For signaling assays, the relevant control peptide was included in each independent experiment and on each assay plate. For radioligand binding studies a control peptide curve was included for each experimental day but not for each plate due to the difference in plate size (48 well plates). In all cases, duplicate or triplicate technical replicates were included for each biological replicate (independent experiment), and each experiment was repeated at least three times. This minimum sample size was chosen based on prior extensive experimentation using this experimental design.40,43,60,69,70 In some cases, n is larger where peptides were resynthesized and retested. Each biological replicate involved plating cells from a distinct passage, separate transient transfection, and separate peptide dilutions, constituting experimental n. In the case of primary brainstem cultures, where the receptors are endogenously expressed, experimental n relates to separate preparations of cultures from individual litters and separate peptide dilutions. The sample size for primary cultures was estimated from prior cAMP data in primary trigeminal ganglia neurons.71 Blinding was not conducted but peptides were randomized between assay plates or within assay plates to ensure that there was no bias from plate position.
Cell Signaling Assays–cAMP
cAMP assays were performed using the LANCE cAMP detection kit (PerkinElmer Life and Analytical Sciences, Waltham, MA, USA) as previously described with minor modifications.72 All cAMP assays were performed in the presence of 1 mM 3-isobutyl-1-methylxanthine (IBMX) (Sigma-Aldrich, St. Louis, MO, USA) and contained 0.1% DMSO. Briefly, Cos-7 cells were serum starved in cAMP assay media (DMEM + 0.1% BSA + 1 mM IBMX) for 30 min at 37 °C prior to peptide stimulation. Peptides were serially diluted in cAMP assay media and cells incubated with assay media alone or each concentration of peptide at 37 °C for 15 min. Media was then aspirated, and the reaction was stopped by incubating with 50 μL of ice-cold ethanol for 10 min at −20 °C. Ethanol was evaporated off the samples in a fume hood, and cAMP was extracted in 50 μL (brainstem cultures 20 μL) of cAMP detection buffer (0.35% Triton X-100, 50 mM HEPES and 10 mM calcium chloride in ddH2O, pH 7.4) and shaken at room temperature for 10 min. Five microliters of cell lysates were transferred to a 384-well optiplate and cAMP measured. Five microliters of antibody mix (1:200 Alexafluor 647 anti-cAMP in detection buffer) was added and incubated at room temperature for 30 min. Ten microliters of detection mix (1:4500 Europium-W8044 labeled streptavidin and 1:1500 biotin-cAMP in detection buffer) was added and incubated for 4 h at room temperature. Plates were read on an Envision plate reader (PerkinElmer Life and Analytical Sciences, Waltham, MA, USA). The concentration of cAMP in each sample was determined from a standard curve that was generated in each assay.
Cell Signaling Assays–IP1
IP1 assays were performed using the IP-One Tb kit (Cisbio, Bedford, MA, USA) with minor modifications from the manufacturer’s protocol. Briefly, Cos-7 cells were serum starved in assay media (DMEM + 0.1% BSA + 0.1% DMSO) for 30 min at 37 °C prior to peptide stimulation in the presence of 50 mM LiCl to prevent IP1 degradation. Cells were incubated with assay media containing 50 mM LiCl alone or containing each concentration of peptide at 37 °C for 90 min. Media was aspirated and detection mix was added (14 μL of buffer, 3 μL of IP1-coupled d2 fluorophore, and 3 μL of Eu-cryptate conjugated anti-IP1 monoclonal antibody). Samples were then incubated at room temperature for 1 h on a shaker before 15 μL was transferred to a white 384-well optiplate and measured on an Envision plate reader (PerkinElmer). The concentration of IP1 in each sample was determined from a standard curve that was generated in each assay.
Cell Signaling Assays—ERK1/2 and CREB phosphorylation
Phosphorylated (p) extracellular signal-regulated kinase 1/2 (ERK1/2) and CREB were detected using the AlphaLISA SureFire Ultra pERK1/2 (Thr202/Tyr204) or the AlphaLISA SureFire Ultra pCREB (Ser133) assay kits (PerkinElmer Life and Analytical Sciences, Waltham, MA, USA) as per the manufacturer’s protocol. These assays are well-characterized and are a sensitive method for the detection of phosphoproteins, displaying equivalent or greater sensitivity than Western blotting and other methodologies.72−75 Briefly, Cos-7 cells were serum starved in assay media (DMEM + 0.1% BSA + 0.1% DMSO) for 4 h at 37 °C/5% CO2 prior to peptide stimulation. Peptides were serially diluted in assay media, and cells were incubated with assay media alone or each concentration of peptide for 7 or 15 min for pERK1/2 detection or 15 min for pCREB detection. FBS (50%) in pERK1/2 or 50 μM forskolin in pCREB assays were used as a positive controls. Media was then aspirated, and the cells were lysed in 25 μL of the kit lysis buffer, followed by shaking for 10–15 min at room temperature. Ten microliters of cell lysate was transferred to a white 384-well optiplate. Five microliters of acceptor beads coated with a Captsure tag immobilizing an ERK1/2 or CREB-specific antibody was added and incubated at room temperature in the dark for 1 h. Five microliters of donor beads coated with streptavidin, which captures a biotinylated antibody specific for the phosphorylated protein, was added and incubated in the dark at room temperature for 1 h. Plates were read on an Envision plate reader (PerkinElmer). In these assays, the signal is directly proportional, and so no standard curve was used.
Data Analysis—Signaling Assays
All data are the mean ± the standard error of the mean (s.e.m.), combined from n independent experiments. Most data were analyzed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). For each individual experiment, concentration–response curves were fitted using three-parameter nonlinear regression to determine the pEC50, after first determining that the Hill slope was not significantly different from one via four parameter nonlinear regression and F-test. Individual pEC50 values were combined to generate mean data. Due to day-to-day variability because of transient transfection (Figure SB23), the Emax in each experiment was normalized such that the data are expressed as a percentage of the Emax for the control curve performed in parallel. The percentage Emax values were then combined to generate mean data. To determine the effect of a peptide analogue compared to control, statistical significance was accepted at (∗) P < 0.05 using unpaired two-tailed t-test for pEC50 or where 95% confidence intervals did not include 100 for Emax. Ligand bias was quantified in GraphPad Prism by analyzing the concentration–response curves using the operational model of agonism, as described previously.76 The system maximum was defined as the highest Emax determined for each signaling pathway at a particular receptor using individual three parameter concentration–response curves. This analysis was conducted on data normalized to the maximal amylin response to estimate Δlog(τ/KA) and ΔΔlog(τ/KA) values. These values were then combined and compared to the control (amylin) by one-way ANOVA with a posthoc Dunnet’s test. Statistical significance was defined at (∗) P < 0.05.
Radioligand Binding Assays
Competition binding assays were used to determine comparative affinities between control and test peptide at AMY1 receptors, using radiolabeled I[125]-CGRP.43,77 Control experiments were completed to ensure that the probe was functional and behaved as expected with binding to the CGRP and hAMY1 receptors (Figure SB24). Following transfection, plates were removed from the incubator, old media was removed, and the wells were washed once in binding buffer (37 °C) composed of DMEM and 0.1% BSA (250 μL/well). Binding buffer was aspirated, and 100 μL of binding buffer was added per well followed by 50 μL of radiolabeled I[125]-hαCGRP (PerkinElmer) at 30 000 cpm/well and finally 50 μL of competitor peptide at a range of concentrations. Total binding was obtained from four wells per plate with radioligand alone, and nonspecific binding was obtained from two wells per plate. To define nonspecific binding, we used 3 μM human amylin. The plates were incubated at room temperature for 1 h. After which, plates were aspirated and 250 μL of ice-cold PBS was added per well. The PBS was aspirated and 0.2 M NaOH (200 μL) was added to each well to lyse the cells. The lysates were then transferred to 1.2 mL of microdilution tubes and read on a Wizard2 gamma counter (PerkinElmer).
Data Analysis—Radioligand Binding
Mean nonspecific binding was subtracted from the raw data to obtain specific binding, which was then expressed as a percentage of the total binding to obtain % specific binding in each experiment. Curves were fitted to these data using a nonlinear regression three-parameter logistic equation to obtain pIC50 values in GraphPad Prism 7.02. These were combined and compared by two-tailed unpaired t-test with statistical significance defined at (∗) P < 0.05.
Brainstem Cultures–Isolation and cAMP Signaling
All procedures involving the use of animals were conducted in accordance with the New Zealand animal welfare act (1999) and approved by the University of Auckland Animal Ethics Committee. The isolation and culture of brainstem medulla cells was performed based on previously described methods.71 For each experiment, four 5 day-old postnatal Wistar rat pups (male and female) were euthanized by decapitation, and the medulla was collected in ice-cold Hank’s balanced salt solution (HBSS) containing HEPES (25 mM), pH 7.2–7.4. Medulla were dissociated by incubation in the same buffer with added Dispase II (10 mg/mL) for 30 min at 37 °C. Cells were pelleted by centrifugation at 500g for 3 min, resuspended in the HBSS/HEPES buffer without Dispase, and triturated with a 1 mL pipet 15 times. Cells were pelleted again by centrifugation at 500g and resuspended in L15 media containing HEPES (25 mM), pH 7.2–7.4. Cells were then enriched by differential centrifugation through a BSA gradient. The medulla cell pellet was resuspended in Neurobasal A containing B27 and diluted penicillin, streptomycin, and l-glutamine mix (Thermo Fisher, New Zealand) and preplated for 1 h at 37 °C. Cells were then plated into 96 well poly-d-lysine-coated cell culture plates. Cultures were maintained at 37 °C in a humidified incubator for 5 days. During this time the media was replaced twice (24 and 96 h). cAMP assays were then performed as described previously.71 cAMP was measured using the LANCE ultra cAMP detection kit (PerkinElmer Life and Analytical Sciences, Waltham, MA, USA).
Brainstem Cultures—Immunofluorescence
For immunofluorescence, cells that were prepared as per the above procedures were fixed in 96 well cell culture plates with 4% paraformaldehyde for 10 min, washed twice with PBS, and stored in PBS at 4 °C before processing. Cells were blocked with 10% goat serum in PBS for 1 h at room temperature. Cells were then incubated at 4 °C overnight with anti-CTR primary antibody (pAb 188/10 1:500 or mAb 9B4 1:100; Welcome receptor antibodies Pty Ltd., Melbourne, Australia) in PBS containing 1% goat serum. These antibodies were selected as they are well characterized to recognize CTR.50,78 Additional controls for 188/10 are provided in Figure SB19. Cells were washed with PBS and incubated with secondary antibody (Alexa Fluor 568 goat antirabbit IgG, 1:200, A11011, lot no. 1778025 or Alexa Fluor 594 goat antimouse, 1:200, A11032; Thermofisher, New Zealand) at room temperature for 1 h in the dark. Cells were washed with PBS and then counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Thermofisher) for 5 min. The DAPI was replaced with PBS, and the cells were imaged using an Operetta high content screening system (PerkinElmer Life and Analytical Sciences). Images were collected using the Harmony and Columbus software packages (PerkinElmer Life and Analytical Sciences). Three independent cultures were prepared, and representative images are shown.
Modeling
Models of the human CTR:amylin or CTR:RAMP:amylin complexes were generated from the cryoelectron microscopy structure of CTR (PDB code 5UZ7),10 the X-ray structure of the CTR extracellular domain (ECD) (PDB code 5II0)29 and the X-ray structure of the CLR-RAMP1 ECD complex (PDB code 4RWG),25 combined using Modeler,79 in line with approaches described elsewhere.40,80 MD simulations of the complex embedded in a POPC bilayer were carried out using ACEMD81 as for previous work,51,80 but additional SuMD simulations were carried out to investigate putative mechanisms of amylin C-terminus binding to the receptor ECD. The enhanced sampling of SuMD, an adaptive sampling method,82 means that ligand binding83−86 and peptide binding87 can be studied within the nanosecond (ns) rather than microsecond (μs) time scale without the introduction of any energy bias by monitoring the distance between the centers of masses of the ligand and the binding site during short classical MD simulations. In addition, metadynamics simulations88,89 were performed, primarily to check amylin bound states as predicted by the modeling and MD simulations, but also to model contacts with the receptor along the initial stages of the dissociation pathway. The CHARMM36 force field90 was used for all MD simulations. Electrostatic potential calculations were carried out using APBSmem,91 as described elsewhere.40 The HD and related metrics were used to compare the CTR structure in the presence and absence of RAMP1. Full details are given in the Supplementary Modeling file.
Calculation of Predicted Helical Propensity
The AGADIR algorithm92 was used to predict the helical propensity of selected peptides at 5 and 25 °C, at a pH of 7.4 and an ionic strength of 0.14 M, to reflect conditions similar to PBS.
Data Availability
Most data generated or analyzed in this study are included in the published article (or Supporting Information). Modeling data sets are available from the url provided in Supplementary Modeling. Raw data are available from corresponding authors on reasonable request.
Acknowledgments
This work was supported by the Maurice Wilkins Centre for Molecular Biodiscovery, Marsden Fund (Royal Society of New Zealand), Lottery Health (New Zealand), the BBSRC (UK) to CAR (BB/M006883/1) and a Wellcome Trust senior investigator award 107927/Z/15/Z to D.P.R. A.A.P. is supported by NIH Grant R01GM104251. C.A.R. acknowledges receipt of a Royal Society Industrial Fellowship, and D.L.H. acknowledges receipt of a James Cook Research Fellowship from the Royal Society of New Zealand. C.S.W. acknowledges receipt of a Sir Charles Hercus Fellowship from the Health Research Council (New Zealand), and E.R.H. acknowledges receipt of a Ph.D. scholarship from the Auckland Medical Research Foundation. Z.R. was supported in part by a GAANN fellowship from the U.S. Department of Education. We wish to acknowledge Patrick Sexton for providing the CTR coordinates and David Poyner for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsptsci.8b00002.
Author Contributions
∇ R.L.B. and L.Y. contributed equally to this work.
The authors declare no competing financial interest.
This article is made available for a limited time sponsored by ACS under the ACS Free to Read License, which permits copying and redistribution of the article for non-commercial scholarly purposes.
Supplementary Material
References
- Field B. C.; Chaudhri O. B.; Bloom S. R. (2010) Bowels control brain: gut hormones and obesity. Nat. Rev. Endocrinol. 6, 444–453. 10.1038/nrendo.2010.93. [DOI] [PubMed] [Google Scholar]
- Richard D. (2015) Cognitive and autonomic determinants of energy homeostasis in obesity. Nat. Rev. Endocrinol. 11, 489–501. 10.1038/nrendo.2015.103. [DOI] [PubMed] [Google Scholar]
- Murphy K. G.; Bloom S. R. (2006) Gut hormones and the regulation of energy homeostasis. Nature 444, 854–859. 10.1038/nature05484. [DOI] [PubMed] [Google Scholar]
- Hay D. L.; Chen S.; Lutz T. A.; Parkes D. G.; Roth J. D. (2015) Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol. Rev. 67, 564–600. 10.1124/pr.115.010629. [DOI] [PubMed] [Google Scholar]
- Lutz T. A. (2010) The role of amylin in the control of energy homeostasis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1475–1484. 10.1152/ajpregu.00703.2009. [DOI] [PubMed] [Google Scholar]
- Bello N. T.; Kemm M. H.; Ofeldt E. M.; Moran T. H. (2010) Dose combinations of exendin-4 and salmon calcitonin produce additive and synergistic reductions in food intake in nonhuman primates. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R945–952. 10.1152/ajpregu.00275.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. E.; Lutz T. A. (2017) Amylin and Leptin: Co-Regulators of Energy Homeostasis and Neuronal Development. Trends Endocrinol. Metab. 28, 153–164. 10.1016/j.tem.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Roth J. D.; Roland B. L.; Cole R. L.; Trevaskis J. L.; Weyer C.; Koda J. E.; Anderson C. M.; Parkes D. G.; Baron A. D. (2008) Leptin responsiveness restored by amylin agonism in diet-induced obesity: evidence from nonclinical and clinical studies. Proc. Natl. Acad. Sci. U. S. A. 105, 7257–7262. 10.1073/pnas.0706473105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. R.; Aronne L. J.; Burns C. M.; Kesty N. C.; Halseth A. E.; Weyer C. (2008) Sustained Weight Loss Following 12-Month Pramlintide Treatment as an Adjunct to Lifestyle Intervention in Obesity. Diabetes Care 31, 1816–1823. 10.2337/dc08-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. L.; Khoshouei M.; Radjainia M.; Zhang Y.; Glukhova A.; Tarrasch J.; Thal D. M.; Furness S. G. B.; Christopoulos G.; Coudrat T.; Danev R.; Baumeister W.; Miller L. J.; Christopoulos A.; Kobilka B. K.; Wootten D.; Skiniotis G.; Sexton P. M. (2017) Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123. 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf C.; Song G.; Cao C.; Zhao Q.; Wang M. W.; Wu B.; Stevens R. C. (2017) Extending the Structural View of Class B GPCRs. Trends Biochem. Sci. 42, 946–960. 10.1016/j.tibs.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Hay D. L.; Garelja M. L.; Poyner D. R.; Walker C. S. (2018) Update on the pharmacology of calcitonin/CGRP family of peptides: IUPHAR Review 25. Br. J. Pharmacol. 175, 3–17. 10.1111/bph.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff R.; Buhlmann N.; Fischer J. A.; Born W. (1999) An amylin receptor is revealed following co-transfection of a calcitonin receptor with receptor activity modifying proteins-1 or −3. Endocrinology 140, 2924–2927. 10.1210/endo.140.6.6930. [DOI] [PubMed] [Google Scholar]
- Christopoulos G.; Perry K. J.; Morfis M.; Tilakaratne N.; Gao Y.; Fraser N. J.; Main M. J.; Foord S. M.; Sexton P. M. (1999) Multiple Amylin Receptors Arise from Receptor Activity-Modifying Protein Interaction with the Calcitonin Receptor Gene Product. Mol. Pharmacol. 56, 235–242. 10.1124/mol.56.1.235. [DOI] [PubMed] [Google Scholar]
- Poyner D. R.; Sexton P. M.; Marshall I.; Smith D. M.; Quirion R.; Born W.; Muff R.; Fischer J. A.; Foord S. M. (2002) International Union of Pharmacology. XXXII. The Mammalian Calcitonin Gene-Related Peptides, Adrenomedullin, Amylin, and Calcitonin Receptors. Pharmacol. Rev. 54, 233–246. 10.1124/pr.54.2.233. [DOI] [PubMed] [Google Scholar]
- Hay D. L.; Pioszak A. A. (2016) Receptor Activity-Modifying Proteins (RAMPs): New Insights and Roles. Annu. Rev. Pharmacol. Toxicol. 56, 469–487. 10.1146/annurev-pharmtox-010715-103120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W.; Patel A.; Kimura R.; Soudy R.; Jhamandas J. H. (2017) Amylin Receptor: A Potential Therapeutic Target for Alzheimer’s Disease. Trends Mol. Med. 23, 709–720. 10.1016/j.molmed.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Qiu W. Q. (2017) Amylin and its G-protein-coupled receptor: A probable pathological process and drug target for Alzheimer’s disease. Neuroscience 356, 44–51. 10.1016/j.neuroscience.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly D. (2012) The structure and function of the glucagon-like peptide-1 receptor and its ligands,. Br. J. Pharmacol. 166, 27–41. 10.1111/j.1476-5381.2011.01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Herenbrink C.; Sykes D. A.; Donthamsetti P.; Canals M.; Coudrat T.; Shonberg J.; Scammells P. J.; Capuano B.; Sexton P. M.; Charlton S. J.; Javitch J. A.; Christopoulos A.; Lane J. R. (2016) The role of kinetic context in apparent biased agonism at GPCRs. Nat. Commun. 7, 10842. 10.1038/ncomms10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T.; Christopoulos A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discovery 12, 205–216. 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- Furness S. G. B.; Liang Y. L.; Nowell C. J.; Halls M. L.; Wookey P. J.; Dal Maso E.; Inoue A.; Christopoulos A.; Wootten D.; Sexton P. M. (2016) Ligand-Dependent Modulation of G Protein Conformation Alters Drug Efficacy. Cell 167, 739–749.e11. 10.1016/j.cell.2016.09.021. [DOI] [PubMed] [Google Scholar]
- Morfis M.; Tilakaratne N.; Furness S. G.; Christopoulos G.; Werry T. D.; Christopoulos A.; Sexton P. M. (2008) Receptor activity-modifying proteins differentially modulate the G protein-coupling efficiency of amylin receptors. Endocrinology 149, 5423–5431. 10.1210/en.2007-1735. [DOI] [PubMed] [Google Scholar]
- Hay D. L.; Walker C. S. (2017) CGRP and its receptors. Headache 57, 625–636. 10.1111/head.13064. [DOI] [PubMed] [Google Scholar]
- Booe J. M.; Walker C. S.; Barwell J.; Kuteyi G.; Simms J.; Jamaluddin M. A.; Warner M. L.; Bill R. M.; Harris P. W.; Brimble M. A.; Poyner D. R.; Hay D. L.; Pioszak A. A. (2015) Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor. Mol. Cell 58, 1040–1052. 10.1016/j.molcel.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton G., and Cook I. (2008) A Dictionary of Statistics, Hellinger distance, Oxford University Press. [Google Scholar]
- Lee S. M.; Booe J. M.; Gingell J. J.; Sjoelund V.; Hay D. L.; Pioszak A. A. (2017) N-Glycosylation of Asparagine 130 in the Extracellular Domain of the Human Calcitonin Receptor Significantly Increases Peptide Hormone Affinity. Biochemistry 56, 3380–3393. 10.1021/acs.biochem.7b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L. H.; Serrano A. L.; Zanni M. T.; Raleigh D. P. (2014) Mutational analysis of preamyloid intermediates: the role of his-tyr interactions in islet amyloid formation. Biophys. J. 106, 1520–1527. 10.1016/j.bpj.2013.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E.; Hansen J. L.; Hansen A. M.; Shaw A. C.; Becker P.; Schaffer L.; Reedtz-Runge S. (2016) Type II Turn of Receptor-bound Salmon Calcitonin Revealed by X-ray Crystallography. J. Biol. Chem. 291, 13689–13698. 10.1074/jbc.M116.726034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper B. A.; Stoffers D. A.; Mains R. E. (1992) The biosynthesis of neuropeptides: peptide alpha-amidation. Annu. Rev. Neurosci. 15, 57–85. 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- Lee S.-M.; Hay D. L.; Pioszak A. A. (2016) Calcitonin and Amylin Receptor Peptide Interaction Mechanisms: insights into peptide-binding modes and allosteric modulation of the calcitonin receptor by receptor activity-modifying proteins. J. Biol. Chem. 291, 8686–8700. 10.1074/jbc.M115.713628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski R. C.; Epand R. M.; Stafford A. R. (1987) Biologically potent analogues of salmon calcitonin which do not contain an N-terminal disulfide-bridged ring structure. Eur. J. Biochem. 162, 399–402. 10.1111/j.1432-1033.1987.tb10615.x. [DOI] [PubMed] [Google Scholar]
- Dennis T.; Fournier A.; St Pierre S.; Quirion R. (1989) Structure-activity profile of calcitonin gene-related peptide in peripheral and brain tissues. Evidence for receptor multiplicity. J. Pharmacol. Exp. Ther. 251, 718–725. [PubMed] [Google Scholar]
- Dumont Y.; Fournier A.; St. Pierre S.; Quirion R. (1997) A potent and selective CGRP2 agonist, [Cys(Et)2,7]hCGRPa: comparison in prototypical CGRP1 and CGRP2 in vitro bioassays. Can. J. Physiol. Pharmacol. 75, 671–676. 10.1139/y97-079. [DOI] [PubMed] [Google Scholar]
- Hay D. L.; Christopoulos G.; Christopoulos A.; Poyner D. R.; Sexton P. M. (2005) Pharmacological Discrimination of Calcitonin Receptor: Receptor Activity-Modifying Protein Complexes. Mol. Pharmacol. 67, 1655–1665. 10.1124/mol.104.008615. [DOI] [PubMed] [Google Scholar]
- Bower R. L.; Hay D. L. (2016) Amylin structure–function relationships and receptor pharmacology: implications for amylin mimetic drug development. Br. J. Pharmacol. 173, 1883–1898. 10.1111/bph.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanga R. P.; Brender J. R.; Vivekanandan S.; Ramamoorthy A. (2011) Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment. Biochim. Biophys. Acta, Biomembr. 1808, 2337–2342. 10.1016/j.bbamem.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang H. N.; Song K.; Hill T. A.; Derksen D. R.; Edmonds D. J.; Kok W. M.; Limberakis C.; Liras S.; Loria P. M.; Mascitti V.; Mathiowetz A. M.; Mitchell J. M.; Piotrowski D. W.; Price D. A.; Stanton R. V.; Suen J. Y.; Withka J. M.; Griffith D. A.; Fairlie D. P. (2015) Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists. J. Med. Chem. 58, 4080–4085. 10.1021/acs.jmedchem.5b00166. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Sun B.; Feng D.; Hu H.; Chu M.; Qu Q.; Tarrasch J. T.; Li S.; Sun Kobilka T.; Kobilka B. K.; Skiniotis G. (2017) Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253. 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins H. A.; Chakravarthy M.; Abhayawardana R. S.; Gingell J. J.; Garelja M.; Pardamwar M.; McElhinney J. M. W. R.; Lathbridge A.; Constantine A.; Harris P. W. R.; Yuen T.-Y.; Brimble M. A.; Barwell J.; Poyner D. R.; Woolley M. J.; Conner A. C.; Pioszak A. A.; Reynolds C. A.; Hay D. L. (2016) Receptor Activity-modifying Proteins 2 and 3 Generate Adrenomedullin Receptor Subtypes with Distinct Molecular Properties. J. Biol. Chem. 291, 11657–11675. 10.1074/jbc.M115.688218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D. L.; Harris P. W.; Kowalczyk R.; Brimble M. A.; Rathbone D. L.; Barwell J.; Conner A. C.; Poyner D. R. (2014) Structure-activity relationships of the N-terminus of calcitonin gene-related peptide: key roles of alanine-5 and threonine-6 in receptor activation. Br. J. Pharmacol. 171, 415–426. 10.1111/bph.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton J. M.; Dowton M.; Houssami S.; Sexton P. M. (2000) Identification of key components in the irreversibility of salmon calcitonin binding to calcitonin receptors. J. Endocrinol. 166, 213–226. 10.1677/joe.0.1660213. [DOI] [PubMed] [Google Scholar]
- Gingell J. J.; Simms J.; Barwell J.; Poyner D. R.; Watkins H. A.; Pioszak A. A.; Sexton P. M.; Hay D. L. (2016) An allosteric role for receptor activity-modifying proteins in defining GPCR pharmacology. Cell Discovery 2, 16012. 10.1038/celldisc.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash S.; Lorenzen E.; Persson T.; Huber T.; Sakmar T. P. (2017) GPCRs globally coevolved with receptor activity-modifying proteins, RAMPs. Proc. Natl. Acad. Sci. U. S. A. 114, 12015–12020. 10.1073/pnas.1713074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potes C. S.; Lutz T. A. (2010) Brainstem mechanisms of amylin-induced anorexia. Physiol. Behav. 100, 511–518. 10.1016/j.physbeh.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Sexton P. M.; Paxinos G.; Kenney M. A.; Wookey P. J.; Beaumont K. (1994) In vitro autoradiographic localization of amylin binding sites in rat brain. Neuroscience 62, 553–567. 10.1016/0306-4522(94)90388-3. [DOI] [PubMed] [Google Scholar]
- Liberini C. G.; Boyle C. N.; Cifani C.; Venniro M.; Hope B. T.; Lutz T. A. (2016) Amylin receptor components and the leptin receptor are co-expressed in single rat area postrema neurons. Eur. J. Neurosci. 43, 653–661. 10.1111/ejn.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrikse E. R.; Bower R. L.; Hay D. L.; Walker C. S. (2018) Molecular studies of CGRP and the CGRP family of peptides in the central nervous system. Cephalalgia 765787. 10.1177/0333102418765787. [DOI] [PubMed] [Google Scholar]
- Bower R. L.; Eftekhari S.; Waldvogel H. J.; Faull R. L.; Tajti J.; Edvinsson L.; Hay D. L.; Walker C. S. (2016) Mapping the calcitonin receptor in human brain stem. Am. J. Physiol. Regul. Integr. Comp. Physiol. 310, R788–793. 10.1152/ajpregu.00539.2015. [DOI] [PubMed] [Google Scholar]
- Becskei C.; Riediger T.; Zund D.; Wookey P.; Lutz T. A. (2004) Immunohistochemical mapping of calcitonin receptors in the adult rat brain. Brain Res. 1030, 221–233. 10.1016/j.brainres.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Wootten D.; Reynolds C. A.; Smith K. J.; Mobarec J. C.; Koole C.; Savage E. E.; Pabreja K.; Simms J.; Sridhar R.; Furness S. G.; Liu M.; Thompson P. E.; Miller L. J.; Christopoulos A.; Sexton P. M. (2016) The Extracellular Surface of the GLP-1 Receptor Is a Molecular Trigger for Biased Agonism. Cell 165, 1632–1643. 10.1016/j.cell.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.; Trevaskis J. L.; Jodka C. M.; Neravetla S.; Griffin P.; Xu K.; Wang Y.; Parkes D. G.; Forood B.; Ghosh S. S. (2013) Bifunctional PEGylated exenatide-amylinomimetic hybrids to treat metabolic disorders: an example of long-acting dual hormonal therapeutics. J. Med. Chem. 56, 9328–9341. 10.1021/jm401418s. [DOI] [PubMed] [Google Scholar]
- Trevaskis J. L.; Mack C. M.; Sun C.; Soares C. J.; D’Souza L. J.; Levy O. E.; Lewis D. Y.; Jodka C. M.; Tatarkiewicz K.; Gedulin B.; Gupta S.; Wittmer C.; Hanley M.; Forood B.; Parkes D. G.; Ghosh S. S. (2013) Improved glucose control and reduced body weight in rodents with dual mechanism of action peptide hybrids. PLoS One 8, e78154. 10.1371/journal.pone.0078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood C. R.; Garibay P.; Knudsen L. B.; Hastrup S.; Peters G. H.; Rudolph R.; Reedtz-Runge S. (2010) Crystal structure of glucagon-like peptide-1 in complex with the extracellular domain of the glucagon-like peptide-1 receptor. J. Biol. Chem. 285, 723–730. 10.1074/jbc.M109.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge S.; Thogersen H.; Madsen K.; Lau J.; Rudolph R. (2008) Crystal structure of the ligand-bound glucagon-like peptide-1 receptor extracellular domain. J. Biol. Chem. 283, 11340–11347. 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- Kowalczyk R.; Brimble M. A.; Tomabechi Y.; Fairbanks A. J.; Fletcher M.; Hay D. L. (2014) Convergent chemoenzymatic synthesis of a library of glycosylated analogues of pramlintide: structure-activity relationships for amylin receptor agonism. Org. Biomol. Chem. 12, 8142–8151. 10.1039/C4OB01208A. [DOI] [PubMed] [Google Scholar]
- Bailey R. J.; Hay D. L. (2006) Pharmacology of the human CGRP1 receptor in Cos 7 cells. Peptides 27, 1367–1375. 10.1016/j.peptides.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Furness S. G. B.; Wootten D.; Christopoulos A.; Sexton P. M. (2012) Consequences of splice variation on Secretin family G protein-coupled receptor function. Br. J. Pharmacol. 166, 98–109. 10.1111/j.1476-5381.2011.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T.; Dong M.; Watkins H. A.; Wootten D.; Miller L. J.; Hay D. L. (2013) Receptor activity-modifying protein-dependent impairment of calcitonin receptor splice variant Δ(1–47)hCT(a) function. Br. J. Pharmacol. 168, 644–657. 10.1111/j.1476-5381.2012.02197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner A. C.; Hay D. L.; Simms J.; Howitt S. G.; Schindler M.; Smith D. M.; Wheatley M.; Poyner D. R. (2006) A Key Role for Transmembrane Prolines in Calcitonin Receptor-Like Receptor Agonist Binding and Signalling: Implications for Family B G-Protein-Coupled Receptors. J. Biol. Chem. 67, 20–31. 10.1074/jbc.M510064200. [DOI] [PubMed] [Google Scholar]
- McLatchie L. M.; Fraser N. J.; Main M. J.; Wise A.; Brown J.; Thompson N.; Solari R.; Lee M. G.; Foord S. M. (1998) RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393, 333–339. 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- Cao P.; Abedini A.; Raleigh D. P. (2013) Aggregation of islet amyloid polypeptide: from physical chemistry to cell biology. Curr. Opin. Struct. Biol. 23, 82–89. 10.1016/j.sbi.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingell J. J.; Burns E. R.; Hay D. L. (2014) Activity of Pramlintide, Rat and Human Amylin but not Abeta1–42 at Human Amylin Receptors. Endocrinology 155, 21–26. 10.1210/en.2013-1658. [DOI] [PubMed] [Google Scholar]
- Harris P. W. R.; Kowalczyk R.; Hay D. L.; Brimble M. A. (2013) A Single Pseudoproline and Microwave Solid Phase Peptide Synthesis Facilitates an Efficient Synthesis of Human Amylin 1–37. Int. J. Pept. Res. Ther. 19, 147–155. 10.1007/s10989-012-9325-9. [DOI] [Google Scholar]
- Harris P. W. R.; Yang S. H.; Brimble M. A. (2011) An improved procedure for the preparation of aminomethyl polystyrene resin and its use in solid phase (peptide) synthesis. Tetrahedron Lett. 52, 6024–6026. 10.1016/j.tetlet.2011.09.010. [DOI] [Google Scholar]
- Jensen K. J.; Meldal M.; Bock K. (1993) Glycosylation of phenols: preparation of 1,2-cis and 1,2-trans glycosylated tyrosine derivatives to be used in solid-phase glycopeptide synthesis. J. Chem. Soc., Perkin Trans. 1 1, 2119–2129. 10.1039/p19930002119. [DOI] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. (2002) A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ″ligation″ of azides and terminal alkynes. Angew. Chem., Int. Ed. 41, 2596–2599. . [DOI] [PubMed] [Google Scholar]
- Tornoe C. W.; Christensen C.; Meldal M. (2002) Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 67, 3057–3064. 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Watkins H. A.; Walker C. S.; Ly K. N.; Bailey R. J.; Barwell J.; Poyner D. R.; Hay D. L. (2014) Receptor activity-modifying protein-dependent effects of mutations in the calcitonin receptor-like receptor: implications for adrenomedullin and calcitonin gene-related peptide pharmacology. Br. J. Pharmacol. 171, 772–788. 10.1111/bph.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley M. J.; Watkins H. A.; Taddese B.; Karakullukcu Z. G.; Barwell J.; Smith K. J.; Hay D. L.; Poyner D. R.; Reynolds C. A.; Conner A. C. (2013) The role of ECL2 in CGRP receptor activation: a combined modelling and experimental approach,. J. R. Soc., Interface 10, 20130589. 10.1098/rsif.2013.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. S.; Sundrum T.; Hay D. L. (2014) PACAP receptor pharmacology and agonist bias: analysis in primary neurons and glia from the trigeminal ganglia and transfected cells. Br. J. Pharmacol. 171, 1521–1533. 10.1111/bph.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. S.; Raddant A. C.; Woolley M. J.; Russo A. F.; Hay D. L. (2018) CGRP receptor antagonist activity of olcegepant depends on the signalling pathway measured. Cephalalgia 38, 437–451. 10.1177/0333102417691762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch M. F.; Osmond R. I. (2008) New strategies in drug discovery for GPCRs: high throughput detection of cellular ERK phosphorylation. Comb. Chem. High Throughput Screening 11, 344–356. 10.2174/138620708784534806. [DOI] [PubMed] [Google Scholar]
- Osmond R. I.; Sheehan A.; Borowicz R.; Barnett E.; Harvey G.; Turner C.; Brown A.; Crouch M. F.; Dyer A. R. (2005) GPCR screening via ERK 1/2: a novel platform for screening G protein-coupled receptors. J. Biomol. Screening 10, 730–737. 10.1177/1087057105277968. [DOI] [PubMed] [Google Scholar]
- van der Westhuizen E. T.; Werry T. D.; Sexton P. M.; Summers R. J. (2007) The relaxin family peptide receptor 3 activates extracellular signal-regulated kinase 1/2 through a protein kinase C-dependent mechanism. Mol. Pharmacol. 71, 1618–1629. 10.1124/mol.106.032763. [DOI] [PubMed] [Google Scholar]
- van der Westhuizen E. T.; Breton B.; Christopoulos A.; Bouvier M. (2014) Quantification of ligand bias for clinically relevant beta2-adrenergic receptor ligands: implications for drug taxonomy. Mol. Pharmacol. 85, 492–509. 10.1124/mol.113.088880. [DOI] [PubMed] [Google Scholar]
- Kuwasako K.; Cao Y.-N.; Nagoshi Y.; Tsuruda T.; Kitamura K.; Eto T. (2004) Characterization of the Human Calcitonin Gene-Related Peptide Receptor Subtypes Associated with Receptor Activity-Modifying Proteins. Mol. Pharmacol. 65, 207–213. 10.1124/mol.65.1.207. [DOI] [PubMed] [Google Scholar]
- Walker C. S.; Eftekhari S.; Bower R. L.; Wilderman A.; Insel P. A.; Edvinsson L.; Waldvogel H. J.; Jamaluddin M. A.; Russo A. F.; Hay D. L. (2015) A second trigeminal CGRP receptor: function and expression of the AMY1 receptor. Ann. Clin. Transl. Neurol. 2, 595–608. 10.1002/acn3.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N.; Webb B.; Marti-Renom M. A.; Madhusudhan M. S.; Eramian D.; Shen M. Y.; Pieper U.; Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 15, 6. 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C.; Winfield I.; Harris M.; Hodgson R.; Shah A.; Dowell S. J.; Mobarec J. C.; Woodlock D. A.; Reynolds C. A.; Poyner D. R.; Watkins H. A.; Ladds G. (2016) Receptor Activity-modifying Protein-directed G Protein Signaling Specificity for the Calcitonin Gene-related Peptide Family of Receptors. J. Biol. Chem. 291, 21925–21944. 10.1074/jbc.M116.751362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M. J.; Giupponi G.; Fabritiis G. D. (2009) ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J. Chem. Theory Comput. 5, 1632–1639. 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- Deganutti G.; Moro S. (2017) Estimation of kinetic and thermodynamic ligand-binding parameters using computational strategies. Future Med. Chem. 9, 507–523. 10.4155/fmc-2016-0224. [DOI] [PubMed] [Google Scholar]
- Sabbadin D.; Moro S. (2014) Supervised molecular dynamics (SuMD) as a helpful tool to depict GPCR-ligand recognition pathway in a nanosecond time scale. J. Chem. Inf. Model. 54, 372–376. 10.1021/ci400766b. [DOI] [PubMed] [Google Scholar]
- Deganutti G.; Welihinda A.; Moro S. (2017) Comparison of the Human A2A Adenosine Receptor Recognition by Adenosine and Inosine: New Insight from Supervised Molecular Dynamics Simulations. ChemMedChem 12, 1319–1326. 10.1002/cmdc.201700200. [DOI] [PubMed] [Google Scholar]
- Cuzzolin A.; Sturlese M.; Deganutti G.; Salmaso V.; Sabbadin D.; Ciancetta A.; Moro S. (2016) Deciphering the Complexity of Ligand-Protein Recognition Pathways Using Supervised Molecular Dynamics (SuMD) Simulations. J. Chem. Inf. Model. 56, 687–705. 10.1021/acs.jcim.5b00702. [DOI] [PubMed] [Google Scholar]
- Deganutti G.; Cuzzolin A.; Ciancetta A.; Moro S. (2015) Understanding allosteric interactions in G protein-coupled receptors using Supervised Molecular Dynamics: A prototype study analysing the human A3 adenosine receptor positive allosteric modulator LUF6000. Bioorg. Med. Chem. 23, 4065–4071. 10.1016/j.bmc.2015.03.039. [DOI] [PubMed] [Google Scholar]
- Salmaso V.; Sturlese M.; Cuzzolin A.; Moro S. (2017) Exploring Protein-Peptide Recognition Pathways Using a Supervised Molecular Dynamics Approach. Structure 25, 655. 10.1016/j.str.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Valsson O.; Tiwary P.; Parrinello M. (2016) Enhancing Important Fluctuations: Rare Events and Metadynamics from a Conceptual Viewpoint. Annu. Rev. Phys. Chem. 67, 159–184. 10.1146/annurev-physchem-040215-112229. [DOI] [PubMed] [Google Scholar]
- Barducci A.; Bonomi M.; Parrinello M. (2011) Metadynamics. Wiley Interdisciplinary Reviews: Computational Molecular Science 1, 826–843. 10.1002/wcms.31. [DOI] [Google Scholar]
- Huang J.; MacKerell A. D. Jr (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145. 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callenberg K. M.; Choudhary O. P.; de Forest G. L.; Gohara D. W.; Baker N. A.; Grabe M. (2010) APBSmem: a graphical interface for electrostatic calculations at the membrane. PLoS One 5, e12722. 10.1371/journal.pone.0012722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz V.; Serrano L. (1994) Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Mol. Biol. 1, 399. 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Most data generated or analyzed in this study are included in the published article (or Supporting Information). Modeling data sets are available from the url provided in Supplementary Modeling. Raw data are available from corresponding authors on reasonable request.