SKP1 coordinates chromosome synapsis and meiotic recombination with progression through the first meiotic cell division in mouse.
Abstract
The meiotic prophase I to metaphase I (PI/MI) transition requires chromosome desynapsis and metaphase competence acquisition. However, control of these major meiotic events is poorly understood. Here, we identify an essential role for SKP1, a core subunit of the SKP1–Cullin–F-box (SCF) ubiquitin E3 ligase, in the PI/MI transition. SKP1 localizes to synapsed chromosome axes and evicts HORMAD proteins from these regions in meiotic spermatocytes. SKP1-deficient spermatocytes display premature desynapsis, precocious pachytene exit, loss of PLK1 and BUB1 at centromeres, but persistence of HORMAD, γH2AX, RPA2, and MLH1 in diplonema. Strikingly, SKP1-deficient spermatocytes show sharply reduced MPF activity and fail to enter MI despite treatment with okadaic acid. SKP1-deficient oocytes exhibit desynapsis, chromosome misalignment, and progressive postnatal loss. Therefore, SKP1 maintains synapsis in meiosis of both sexes. Furthermore, our results support a model where SKP1 functions as the long-sought intrinsic metaphase competence factor to orchestrate MI entry during male meiosis.
INTRODUCTION
During the prophase I (PI) of meiosis, homologous chromosomes undergo pairing, synapsis, and meiotic recombination (1, 2). The lengthy pachytene stage is characterized by full synapsis of homologs and completion of meiotic recombination, which are interdependent in many species including mouse. Chromosome synapsis requires the assembly of the synaptonemal complex (SC), a tripartite protein structure, along chromosome axes. Meiotic recombination involves repair of several hundred programmed DNA double-strand breaks (DSBs) into a finite number of crossovers. Coordinated execution of these meiotic events promotes proper segregation of homologous chromosomes during meiosis I, whereas meiotic abnormality can cause pregnancy loss, infertility, and birth defects.
The transition from PI to metaphase I (PI/MI) in meiosis involves chromosome desynapsis, chromatin condensation, and compaction of MI chromosomes. The meiotic PI/MI transition largely corresponds to the G2-M transition in the mitotic cell cycle. Okadaic acid (OA), an inhibitor of protein phosphatases PP1 and PP2A, induces premature PI/MI progression in pachytene spermatocytes (pachynema) (3). MI competence is acquired in mid-to-late, but not early, pachynema, because treatment of mid-to-late, but not early, pachytene spermatocytes with OA causes desynapsis, chromatin condensation, and chromosome compaction (4). The relatively long duration of the pachytene stage in male meiosis (6 days in mouse) is enigmatic. In females, the arrest of oocytes at the diplotene stage provides ample time to develop MI competence. Therefore, it is possible that the long time is necessary for pachynema in males to gain MI competence. However, the molecular nature of the meiotic MI competence acquisition is unknown.
SCF (SKP1–Cullin–F-box) is a family of ubiquitin E3 ligases and consists of four subunits: SKP1, CUL1, the RING finger protein RBX1, and an F-box protein. The cullin subunit CUL1 functions as a scaffold that interacts with both RBX1 and SKP1. RBX1 recruits an ubiquitin E2 enzyme. SKP1 (S-phase kinase-associated protein 1) associates with different F-box proteins to form distinct E3 ligase complexes to target specific proteins for proteasome-mediated degradation, because the F-box proteins provide target specificity (5). SCF has emerged as a major E3 ligase crucial for the ubiquitin-mediated degradation of a host of cellular proteins, including cell cycle regulators and transcription factors.
Many species including yeast, mouse, and human have only one Skp1 gene. Other species have multiple Skp1 homologs due to gene duplications. For example, Caenorhabditis elegans has 21 Skp1-related (Skr) genes, which are involved in cell proliferation, development, and meiosis (6). Arabidopsis also has 21 Skp1-related genes (7). Mouse SKP1 was previously identified as one of the meiotic chromatin–associated proteins in testis (8). Here, we find that SKP1 localizes to the SC in meiotic germ cells and plays critical roles in chromosome synapsis, meiotic progression, and oocyte survival. Strikingly, SKP1 is essential for the PI/MI phase transition during male meiosis.
RESULTS
SKP1 associates with meiotic chromatin and localizes to the lateral elements of the SC
The SKP1 protein abundance increased sharply in testes at postnatal day 14, when pachytene spermatocytes first appeared (Fig. 1A). Immunofluorescence showed that SKP1 was detected in spermatogonia and preleptotene through zygotene spermatocytes, but at much higher levels in pachytene to MI spermatocytes and postmeiotic round spermatids (Fig. 1B). SKP1 was predominantly cytoplasmic (Fig. 1B). Strikingly, analysis of spermatocyte nuclear spread showed that SKP1 localized to the SC exclusively in the synapsed regions of meiotic chromosomes from zygotene, pachytene, and diplotene spermatocytes but not in unsynapsed regions, including the unsynapsed regions of X-Y chromosomes (Fig. 1C). We next examined the localization of SKP1 in meiotic chromosomes in oocytes and found a consistent localization pattern: localization to the SC in the synapsed, but not unsynapsed, regions in zygotene, pachytene, and diplotene stages (fig. S1). These results suggest a conserved function of SKP1 in both male and female meiosis.
Fig. 1. Localization of SKP1 to SC LEs coincides with chromosome synapsis.
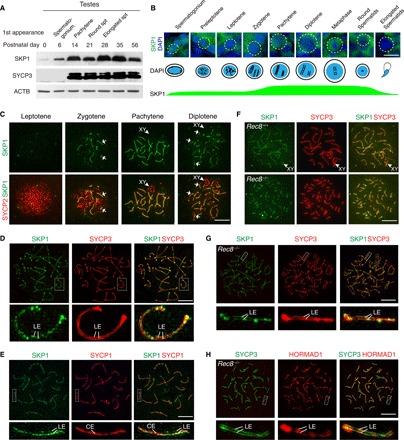
(A) Western blot analysis of SKP1 in developing mouse testes. Timing of the first appearance of spermatogonium, pachytene spermatocytes, round spermatids (spt), and elongated spermatids in developing testes is shown. SYCP3, meiosis-specific protein control; ACTB, loading control. (B) Spatiotemporal expression pattern of SKP1 in postnatal male germ cells. DAPI, 4′,6-diamidino-2-phenylindole. (C) Localization of SKP1 to synapsed regions of the SC (indicated by arrows) in wild-type (WT) spermatocytes. (D) Super-resolution localization of SKP1 and SYCP3 in pachytene spermatocytes. (E) Super-resolution localization of SKP1 and SYCP1 in pachytene spermatocytes. SYCP1 is a component of both CE and transverse filaments. (F) Localization of SKP1 to the SC between sister chromatids in Rec8−/− spermatocytes. (G) Super-resolution localization of SKP1 in Rec8−/− spermatocytes. (H) Super-resolution localization of HORMAD1 in Rec8−/− spermatocytes. Scale bars, 10 μm.
The SC is composed of two lateral elements (LEs) and one central element (CE) at the pachytene stage. Super-resolution imaging revealed that SKP1 localized to the LEs in the SC, marked by SYCP3, and that SKP1 localization appeared as both filaments and foci (Fig. 1D). As expected, the SKP1 filaments flanked the SC central element, marked by SYCP1 (Fig. 1E). We next sought to investigate whether SKP1 localizes to the synapsed sister chromatids using the Rec8 mouse model. REC8 is a meiosis-specific cohesin required for sister chromatid cohesion. Rec8−/− germ cells exhibit synapsis between sister chromatids rather than homologous chromosomes (9). We found that SKP1 localized to the synapsed sister chromatids and specifically to the LEs in Rec8−/− spermatocytes (Fig. 1, F and G, and fig. S2). These data suggest that SKP1 may play a critical role in chromosomal synapsis during meiosis.
SKP1 is essential for viability and male meiosis
To elucidate the function of Skp1 in meiosis, we generated Skp1f/f mice, in which exons 3 to 5 are flanked by loxP sites (fig. S3A). Skp1f/f mice were fertile. Germ cell–specific inactivation of Skp1 using the constitutively expressed Ddx4-Cre transgene caused a complete loss of germ cells in Skp1f/− Ddx4-Cre males after postnatal day 6, showing that Skp1 is essential for survival of mitotic germ cells including spermatogonia (fig. S3B). Interbreeding of Skp1f/− mice failed to produce Skp1−/− offspring, showing that Skp1 is required for embryogenesis.
To circumvent the premeiotic germ cell loss in Skp1f/− Ddx4-Cre males, we carried out tamoxifen-induced inactivation of Skp1 specifically in germ cells using Ddx4-CreERT2 (Fig. 2A). Intraperitoneal injection of adult Skp1f/− Ddx4-CreERT2 (referred to as Skp1cKO) males with tamoxifen resulted in progressive decrease in testis weight over time (Fig. 2B). Histological analyses of Skp1cKO testes showed severe defects in spermatogenesis (Fig. 2, C to J). Notably, metaphase spermatocytes were absent in Skp1cKO testes at 4 days post-tamoxifen (dpt) and beyond (Fig. 2, E to J). Multinucleated giant cells were frequently observed in the mutant seminiferous tubules, indicative of spermatogenic defects (Fig. 2, H and I). Skp1cKO testes at 10 dpt and beyond contained diplotene spermatocytes but lacked postmeiotic spermatids (Fig. 2, G to J), demonstrating that SKP1 is essential for meiotic progression to MI in spermatogenesis.
Fig. 2. Premature chromosome desynapsis and precocious pachytene exit in Skp1-deficient spermatocytes.
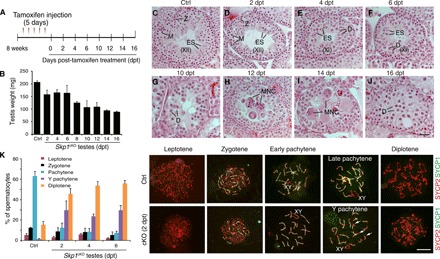
(A) Tamoxifen treatment regimen in 8-week-old Skp1f/− Ddx4-CreERT2 males. (B) Testis weight of adult Skp1cKO males. (C to J) Histological analysis of control (C) and Skp1cKO (D to J) adult testes. Stage XII (C and D) and stage XI (E and F) tubules are shown. Please note that early-stage (I to VII) tubules from control and Skp1cKO testes at 2, 4, and 6 dpt contained round spermatids. Scale bar, 25 μm. Z, zygonema; D, diplonema; M, metaphase; ES, elongated spermatid; MNC, multinucleated giant cells. (K) Surface nuclear spread analysis of control and Skp1cKO spermatocytes. The plot shows the percentage of each type of spermatocytes. Scale bar, 10 μm.
Precocious chromosomal desynapsis and pachytene exit in Skp1-deficient spermatocytes
To probe the nature of meiotic block in Skp1cKO testes, we examined chromosomal synapsis by nuclear spread analysis. In untreated adult testes, pachynema and diplonema accounted for ~60% and ~18% of spermatocytes, respectively. In contrast, in Skp1cKO testes, pachynema decreased to less than 40% at 4 and 6 dpt, and diplonema increased to more than 50% (Fig. 2K). SKP1 was absent from SCs in Skp1cKO pachynema, showing that the localization signal to synapsed chromosomal axes is specific (fig. S3C). Notably, in Skp1cKO testes, late pachynema were absent, and most pachynema contained unsynapsed chromosomal ends (termed Y pachynema), whereas the few pachynema had normal synapsis and were clearly at the early pachytene stage (Fig. 2K). Histone H1t is absent in early pachynema, begins to express in mid-pachynema, and is strongly expressed in late pachynema and diplonema (fig. S4A) (4). Most Y pachynema were H1t-negative and thus were early pachynema (Y1 type), while the remaining were weakly H1t-positive (Y2 type) and thus were mid-pachynema (fig. S4B). In Y pachynema, only one end was unsynapsed and corresponded to the centromeric end (CREST-positive; fig. S4, C and D), showing that SKP1 is required for chromosomal synapsis only at the pericentromeric regions in early pachynema and a fraction of mid-pachynema. The Y pachynema could arise from early pachynema that underwent desynapsis at centromeric ends and/or from zygonema that failed to synapse at centromeric ends but still progressed to early pachytene stage. The lack of late pachynema and the concurrent increase of diplonema in Skp1cKO testes suggest that loss of SKP1 causes premature chromosome desynapsis and precocious pachytene exit in spermatocytes.
Loss of SKP1 leads to accumulation of HORMAD proteins through TRIP13 reduction in spermatocytes
HORMAD proteins are associated with unsynapsed or desynapsed chromosome axes (Fig. 3A) (10–12). HORMAD1 promotes DSB formation through interaction with IHO1 (13). In contrast with its exclusive localization to XY axes in wild-type (WT) pachynema, HORMAD1 was highly enriched on both synapsed and unsynapsed regions of all chromosomes in Skp1-deficient pachynema (Fig. 3A). In addition, this enrichment of HORMAD1 on the SC persisted in Skp1-deficient diplonema (Fig. 3A). Two categories of diplonema in Skp1cKO testes were observed: high HORMAD1 and low HORMAD1. The level of HORMAD1 in the low category of mutant diplonema was still higher than the WT. A similar pattern of enrichment in Skp1 mutant spermatocytes was observed for HORMAD2 (Fig. 3B). HORMAD2 localized only to XY axes in WT pachynema and diplonema, but in Skp1cKO spermatocytes, it localized strongly on unsynapsed centromeric regions and weakly on synapsed regions in pachynema and persisted in most diplonema (Fig. 3B). HORMAD2 was only present on the XY axes in the remaining Skp1cKO (~30%) diplonema, as in the WT. We proposed that HORMAD-high and HORMAD-low diplonema were derived from spermatocytes, in which SKP1 was depleted in the late pachynema and diplonema, respectively. Super-resolution imaging revealed that HORMAD1 localized to the LEs in Skp1cKO pachynema (Fig. 3C) and in Rec8−/− spermatocytes (Fig. 1H).
Fig. 3. SKP1 removes HORMAD1 and HORMAD2 from SCs in pachytene spermatocytes.
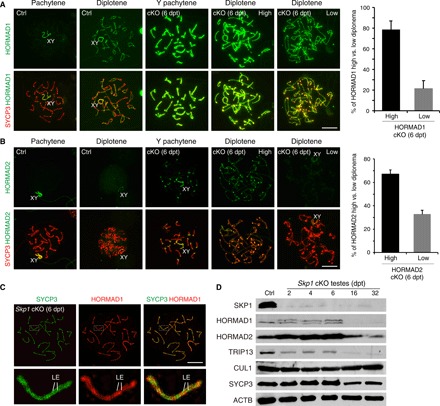
(A) Localization of HORMAD1 in pachynema and diplonema. The plot shows the count of HORMAD1-high versus HORMAD1-low Skp1cKO diplonema. (B) Localization of HORMAD2 in pachynema and diplonema. The count of HORMAD2-high versus HORMAD2-low Skp1cKO diplonema is shown in the plot. (C) Super-resolution localization of HORMAD1 to the SC LEs in Skp1cKO pachynema. Scale bars, 10 μm. (D) Western blot analysis of SKP1, HORMAD1, HORMAD2, TRIP13, CUL1, and SYCP3 in Skp1cKO testes at 2, 4, 6, 16, and 32 dpt.
SKP1 was markedly depleted in Skp1cKO testes (Fig. 3D). In contrast, the abundance of CUL1, also a core component of SCF, remained comparable between control and Skp1cKO testes (Fig. 3D). Western blot analyses confirmed the increased abundance of both HORMAD1 and HORMAD2 in Skp1cKO testes at 2, 4, and 6 dpt, in contrast with the marked depletion of the SKP1 protein (Fig. 3D). A slower migrating band of HORMAD1 appeared in Skp1cKO testes and could be due to phosphorylation (Fig. 3D). These results demonstrate that SKP1 is required for removal of HORMAD proteins from synapsed chromosome axes in pachynema and reduces (HORMAD1) or eliminates (HORMAD2) their return to autosomal axes in diplonema. The TRIP13 protein is required for meiotic recombination but not synapsis (14). Like SKP1, TRIP13 is essential for excluding HORMAD1/2 from synapsed chromosome axes in pachynema, although it is not known to localize to SC (10). However, we found that SKP1 still localized to synapsed chromosome axes in Trip13Gt/Gt pachynema, suggesting that SKP1 may exclude HORMAD proteins from SC in WT pachynema by stabilizing TRIP13 (fig. S5). We tested this possibility by Western blotting analysis and found that TRIP13 protein abundance decreased in Skp1cKO testes at 2, 4, and 6 dpt (Fig. 3D). These data demonstrate that loss of SKP1 leads to reduction in TRIP13 abundance, which, in turn, may cause accumulation of HORMADs on SCs in Skp1cKO pachynema and diplonema.
Impaired meiotic recombination in Skp1cKO spermatocytes
We monitored the progression of meiotic recombination in Skp1cKO spermatocytes. In WT, γH2AX was abundant in leptonema/zygonema, reduced in early pachynema, and confined to the XY body in mid-late pachynema and diplonema (Fig. 4A). In Skp1cKO spermatocytes, γH2AX persisted on autosomes in all Y pachytene spermatocytes and ~50% of diplotene spermatocytes; in addition, the γH2AX-positive domain (XY body) was much larger than the WT XY body and occupied some autosomes (Fig. 4A). Replication protein A (RPA) is a single-strand DNA binding heterotrimeric complex of RPA1, RPA2, and RPA3. RPA is essential for meiotic recombination (15). We examined the localization of RPA2 as a representative of the RPA complex. RPA2 localizes as foci on DSBs in leptonema through early pachynema in WT (Fig. 4B). In contrast, in Skp1cKO testes, RPA2 foci abnormally persisted in Y pachynema and a substantial fraction of diplonema (Fig. 4B). The persistence of γH2AX on autosomes in Y pachytene spermatocytes were most likely due to unrepaired DSBs as marked by RPA2 foci. The abundant γH2AX signal in mutant diplotene spermatocytes could be due to a combination of unrepaired DSBs (RPA2 foci) and MSUC (meiotic silencing of unpaired chromatin) response (16). MLH1 foci, markers of future crossovers, were present in mid-late pachynema but not in diplonema in WT; however, MLH1 foci were present in 25% of Skp1cKO diplotene spermatocytes (Fig. 4C). MLH1-positive diplotene cells were most likely derived from premature desynapsis of chromosomes in mid-late pachytene spermatocytes. The fact that the mutant diplotene spermatocytes are MLH1-positive is consistent with premature chromosomal desynapsis in Skp1-deficient pachytene spermatocytes. The ubiquitin-proteasome system regulates meiotic recombination in mouse and budding yeast (17, 18). SKP1 functions in resolution of meiotic recombination intermediates in fission yeast (19). Therefore, in addition to its requirement for maintenance of chromosome synapsis, SKP1 plays an evolutionarily conserved role in meiotic recombination in mouse.
Fig. 4. Depletion of SKP1 impairs meiotic recombination in spermatocytes.
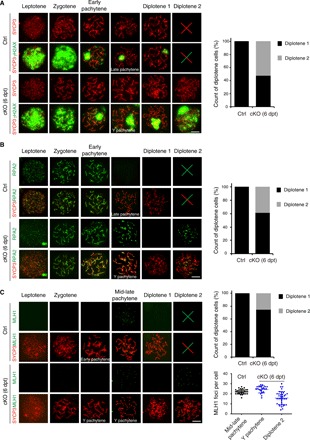
Surface nuclear spread analysis of WT and Skp1cKO (6 dpt) spermatocytes was performed by immunostaining of SYCP3 and γH2AX (A), RPA2 (B), and MLH1 (C). The percentage of different types of diplotene spermatocytes is shown as graphs. The count of MLH1 foci in MLH1-positive pachytene and diplotene spermatocytes is shown in a dot plot (C). Scale bars, 10 μm.
SKP1 maintains chromosomal synapsis and regulates meiotic recombination in oocytes
Western blot analysis showed that SKP1 was expressed in oocytes and preimplantation embryos (Fig. 5A). To examine the function of Skp1 in female fertility, Ddx4-Cre (constitutively active) was used to inactivate Skp1 in female germ cells in Skp1fl/− Ddx4-Cre mice. Ddx4-Cre is expressed between embryonic day 15 (E15) and E18 (20). SKP1 localizes to the synapsed regions of meiotic chromosomes in both sexes (Fig. 1C and fig. S1). We next analyzed the impact of loss of SKP1 on chromosome synapsis in oocytes from E18.5 ovaries (fig. S6A). At E18.5, most oocytes (~80%) from WT ovaries were at the pachytene stage and ~20% were at the diplotene stage. However, <30% of oocytes from Skp1cKO ovaries were at the pachytene stage but >70% were at the diplotene stage. In addition, we monitored repair of meiotic DSBs by examining RPA2 in oocytes. In WT oocytes, RPA2 foci were present at the pachytene stage but absent at the diplotene stage due to repair of DSBs; however, RPA2 foci were present in Skp1cKO diplotene oocytes (fig. S6B). Collectively, inactivation of SKP1 causes premature chromosomal desynapsis of pachytene-stage germ cells and impairs meiotic recombination in both males and females.
Fig. 5. SKP1 is essential for maintenance of postnatal oocytes.
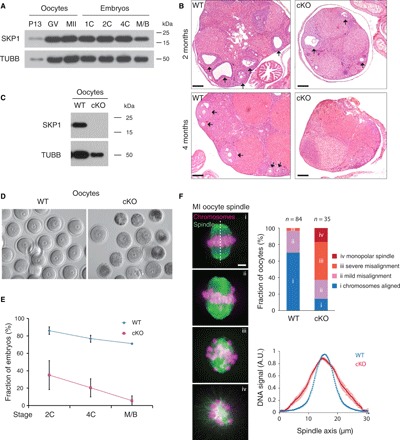
All the Skp1cKO females used here were Skp1f/− Ddx4-Cre females. The Ddx4-Cre was constitutively active in oocytes (20). (A) Western blot analysis of SKP1 in oocytes and preimplantation embryos. P13, postnatal day 13; GV, germinal vesicle stage; MII, metaphase II; 1C, one-cell embryos; 2C, two-cell embryos; 4C, four-cell embryos; M/B, morula/blastocyst. (B) Histological analysis of WT and Skp1cKO ovaries at 2 and 4 months of age. Arrows indicate follicles. Scale bars, 200 μm. (C) Western blot analysis of WT and Skp1cKO GV oocytes. (D) Morphology of GV oocytes collected from 2-month-old WT and Skp1cKO mice. (E) Failure of preimplantation embryo development in Skp1cKO females. IVF was performed with superovulated oocytes from 2-month-old females. One-cell embryos were cultured until morula/blastocyst stage and counted. WT, 228 one-cell embryos (5 females); Skp1cKO, 40 one-cell embryos (13 females). (F) Analysis of chromosome alignment in MI oocytes. MI oocytes from WT and Skp1cKO mice were fixed and stained for tubulin and DNA. Oocytes were divided into four categories (i to iv) based on chromosome alignment and spindle defects (χ2 = 198, P = 0). Bottom right graph shows quantification of chromosome alignment in WT and Skp1-deficient MI oocytes (n = 25) as DNA density distribution along the spindle axes. Data are combined results from three independent experiments. Scale bar, 5 μm. A.U., arbitrary units.
SKP1 is required for survival of postnatal oocytes and proper chromosome alignment
Histological analysis revealed that oocytes were present in the ovaries from Skp1cKO females at 2 months of age but were depleted in the mutant ovaries at 4 months, showing that SKP1 is essential for maintenance of postnatal oocytes (Fig. 5B). Consistently, Western blotting analysis demonstrated that the SKP1 protein was absent in postnatal oocytes from Skp1cKO females (Fig. 5C).
Mating tests showed that Skp1cKO females were sterile. Thirteen 2-month-old Skp1cKO females were mated with WT males for 2 months: Only one dead pup from one Skp1cKO female was observed, whereas no pups were found from other Skp1cKO females. To further probe the cause of sterility of 2-month-old Skp1cKO females, we examined the morphology of superovulated oocytes. While WT oocytes were uniform in size at the germinal vesicle (GV) stage, oocytes from Skp1cKO females varied in size and appearance: Some were smaller and appeared darker (Fig. 5D). We next performed in vitro fertilization (IVF) to examine the developmental potential of resulting embryos at two-cell, four-cell, and morula/blastocyst stages. The percentage of two-cell or four-cell embryos derived from Skp1cKO oocytes was much lower compared to the WT and rarely reached the morula/blastocyst stage (Fig. 5E). These results show that Skp1-deficient oocytes fail to sustain preimplantation embryonic development.
To further investigate the defects in Skp1-deficient oocytes, we examined chromosome alignment at MI. In WT MI oocytes, all chromosomes were aligned at the metaphase plate, as expected (Fig. 5F, i). However, >80% of Skp1-deficient MI oocytes exhibited various degrees of chromosome misalignment: mild misalignment, severe misalignment, and monopolar spindle (Fig. 5F, ii to iv). Consistently, quantification of chromosomal signal across the spindle axes showed that chromosomes were frequently located out of the metaphase plate in the Skp1-deficient MI oocytes compared to WT oocytes (Fig. 5F). Together with the centromere protein localization defects observed in Skp1-deficient spermatocytes described below, this result suggests that SKP1 is essential for normal centromere behavior in MI oocytes.
Loss or reduction of centromere proteins in Skp1-deficient spermatocytes
Polo-like kinase 1 (PLK1) is implicated in SC disassembly (21). We found that PLK1 localized to the centromeric ends of SC in pachynema and diplonema in WT spermatocytes (fig. S7A). In Skp1cKO spermatocytes, PLK1 was still present at the SC end in pachynema but absent in diplonema (fig. S7A). We next investigated whether other centromere components were affected in the absence of SKP1. In yeast, SKP1 is a core kinetochore component and recruits BUB1, a component of the spindle checkpoint, to the centromere (22). Immunolocalization analysis revealed that BUB1 localized prominently to centromeric regions in WT diplotene spermatocytes (fig. S7B). However, 89 of 92 Skp1cKO diplotene spermatocytes lacked BUB1 at centromeres, and only three had weak BUB1 signal, showing that SKP1 is required for BUB1 localization at centromeres in diplotene spermatocytes (fig. S7B). Likewise, we analyzed CENP-C, a constitutive centromere protein. CENP-C signal was strong at the centromeres in WT diplotene spermatocytes but was sharply reduced at the centromeres in Skp1cKO diplotene spermatocytes (fig. S7C), implying defects in assembly of the constitutive centromere-associated network (CCAN) of proteins (23). Collectively, our results demonstrate that SKP1 is required for localization of signaling molecules such as PLK1 and BUB1 to the centromeres and is also important for CCAN integrity in spermatocytes.
SKP1 is essential for MPF activity and MI entry in spermatocytes
To investigate the lack of MI spermatocytes in Skp1cKO testes, we examined phosphorylation of histone H3 at Ser10 (pHH3), a marker of chromatin condensation during the diplotene to MI transition (24). pHH3-positive MI spermatocytes were frequently observed in WT tubules (stage XII) but not in Skp1cKO testes (Fig. 6A). Consistently, Western blotting showed that pHH3 was absent in Skp1cKO testes (Fig. 6B). Early meiotic inhibitor 2 (EMI2), an inhibitor of the anaphase-promoting complex APC/C, was also absent in Skp1cKO testes (Fig. 6B) (25). OA induces chromosomal desynapsis in pachynema and entry into MI in WT spermatocytes (Fig. 6C) (24). However, Skp1cKO spermatocytes were resistant to OA induction and failed to enter MI, further supporting the idea that Skp1cKO spermatocytes were incompetent for MI entry (Fig. 6C). Collectively, multiple lines of evidence have shown that SKP1 is required for PI to MI transition in male meiosis.
Fig. 6. SKP1 is required for MPF activity and PI-MI transition during male meiosis.
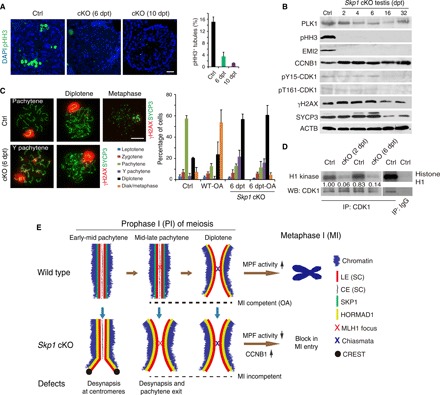
(A) Count of metaphase spermatocytes (pHH3-positive) in seminiferous tubules from WT and Skp1cKO testes. Scale bar, 25 μm. (B) Western blot (WB) analysis of cell cycle regulators in Skp1cKO testes. (C) Skp1-deficient spermatocytes are incompetent for MI entry with OA induction. Scale bar, 10 μm. (D) Histone H1 kinase assay for the MPF activity in WT (control) and Skp1cKO testes at 2 and 6 dpt. Immunoglobulin G (IgG) serves as a negative control. IP, immunoprecipitation. (E) Key functions of SKP1 during male meiosis. This diagram summarizes the major meiotic defects in Skp1cKO testes. Note the accumulation of HORMAD1 on LEs in Y pachynema and its increased abundance on LEs in diplonema in the absence of SKP1. For illustration purpose, SKP1 and HORMAD1 are shown next to the LEs instead of overlaying.
To probe the failure in MI entry of Skp1cKO spermatocytes, we examined the metaphase-promoting factor (MPF), which consists of cyclin-dependent kinase 1 (CDK1) and cyclin B1 (CCNB1). The MPF kinase activity is regulated by CCNB1 binding/dissociation and reversible phosphorylation of CDK1. In Skp1cKO testes (2 to 6 dpt), pY15-CDK1 (inhibitory phosphorylation) and pT161-CDK1 (activating phosphorylation) remained relatively constant; however, the abundance of CCNB1 increased significantly (Fig. 6B). CCNB1 was predominantly cytoplasmic in zygonema and pachynema in both WT and Skp1cKO males (fig. S7D). While CCNB1 was still cytoplasmic in WT diplonema, it accumulated in the nucleus of Skp1cKO diplonema (fig. S7D). We next tested the histone H1 kinase activity of immunoprecipitated MPF from testes and found that the MPF activity was sharply reduced in Skp1cKO testes at 2 and 6 dpt (Fig. 6D). This result provided further support for the lack of MI entry in Skp1cKO spermatocytes.
DISCUSSION
There is a wealth of information on the genetic requirement of chromosome synapsis (1). For instance, the SC physically holds together the homologous chromosomes and thus is required for chromosome synapsis (26, 27). However, little is known about the regulation of chromosome desynapsis. Homologs must undergo desynapsis at the diplotene stage in preparation for the first meiotic cell division. Mouse HSPA2 and PLK1 localize to the SC and have been postulated to promote chromosome desynapsis in pachynema (21, 28). Inactivation of HSPA2 causes arrest at the pachytene stage in mouse testis (28). The function of PLK1 in mouse meiosis has not been examined genetically (29). In budding yeast, Cdc5 (PLK1 homolog) is required for SC disassembly and pachytene exit (30). In contrast, inactivation of Skp1 causes premature chromosome desynapsis in both pachytene spermatocytes and oocytes. The Skp1 mutant reported here is the only mouse mutant known to display premature chromosome desynapsis.
Chromosome desynapsis is a prerequisite for the PI/MI transition in meiosis (Fig. 6E) (24). Spermatocytes lacking CCNA1 or EMI2 exhibit meiotic arrest at the diplotene stage (25, 31). However, pachytene spermatocytes from neither of these mutant (Ccna1 or Emi2) mice undergo premature chromosome desynapsis, suggesting that these factors regulate PI/MI transition but not MI competence acquisition. Ccna1-deficient spermatocytes can be induced to enter MI by OA treatment and thus are MI competent (32). Trip13Gt/Gt spermatocytes can also be induced to the MI phase by OA treatment (14). However, the Trip13Gt mutant allele, a gene trap allele, appears to be hypomorphic rather than null (14). Mid-late, but not early, pachytene spermatocytes are competent for OA-induced MI entry (4). SKP1-deficient spermatocytes fail to progress to MI and are resistant to OA-induced MI entry, demonstrating that SKP1 constitutes an intrinsic competence factor for MI entry in spermatocytes. In addition, only mid-late pachytene Skp1-deficient spermatocytes undergo extensive premature chromosome desynapsis, implying that synapsis maintenance and MI competence acquisition might be linked at least in spermatocytes. Therefore, we propose that SKP1 maintains chromosome synapsis at the prolonged pachytene stage (6 days in male mouse) to promote MI competence acquisition until the pachytene exit in spermatocytes (Fig. 6E). SKP1 is required for localization of PLK1 and BUB1 to the centromeres in diplotene spermatocytes. In addition, SKP1 is important for CENP-C localization to centromeres and thus CCAN integrity. It is possible that SKP1 might ensure MI licensing through centromeres in spermatocytes.
The Skp1 conditional deletion mutants exhibit sexual dimorphism in meiotic progression, as in other meiotic mutants (33). While Skp1cKO spermatocytes fail to progress to the MI stage, Skp1cKO oocytes can proceed to the MI stage and complete meiosis. One possible explanation for the sex difference is that SKP1 is required for meiotic PI-MI phase transition in males but not females. Sexual dimorphism of infertility is also present in mice lacking CCNA1, EMI2, or HSPA2, in which only males are infertile (25, 28, 31). The duration of the meiotic PI including the diplotene (dictyate) stage is much longer in females than in males, as the oocytes are arrested at the dictyate stage at birth until ovulation. Different conditional knockout strategies were used for males and females and thus could, although less likely, contribute to the sexual dimorphism of the mutant phenotypes. We propose that the extremely long duration of PI allows the oocytes to acquire MI competence independent of SKP1. Despite this difference, our results show that Skp1-deficient oocytes are defective, display various degrees of chromosome misalignment, fail to support early embryogenesis, and are completely lost by 4 months of age, suggesting additional functions of SKP1. Furthermore, centromere defects were observed in Skp1-deficient diplotene spermatocytes (fig. S7, A to C). Similar centromere defects may underlie the chromosome misalignment phenotype in MI oocytes (Fig. 5F).
Genetic studies of SKP1 and SKP1-related homologs have been reported in yeast, C. elegans, and Arabidopsis. Like mammals, yeast has only one Skp1 gene. The temperature-sensitive Skp1 fission yeast mutant exhibits abnormal spindle bending in meiosis I. It was proposed that yeast SKP1 functions in resolution of meiotic recombination intermediates, persistence of which results in chromosome entanglement and spindle bending (19). Here, we find that mouse SKP1 is involved in meiotic recombination, evidenced by the abnormal persistence of meiotic recombination proteins such as γH2AX, RPA, and MLH1 in Skp1cKO diplotene spermatocytes (Fig. 4). A systematic study of the 21 Skr genes in C. elegans by RNA interference (RNAi) reveals critical functions of Skr genes in cell proliferation, morphogenesis, and meiosis (6). Notably, zygotic RNAi of Skr1-1/2 (two closely related homologs) results in an arrest at the pachytene stage (6). Among the 21 Arabidopsis Skp1-like (Ask) genes, Ask1 is essential for male meiosis (7, 34–37). ASK1 is required for chromosome synapsis (37), nuclear reorganization and release from nuclear membrane (35, 36), and homologous chromosome separation (34). Therefore, together with the mouse study described here, the requirement of SKP1 in meiosis is evolutionarily conserved in yeast, C. elegans, Arabidopsis, mouse, and likely other sexually reproducing organisms.
HORMAD proteins localize to unsynapsed and desynapsed chromosome axes (10–12). In Skp1-deficient spermatocytes, HORMAD proteins accumulate on both synapsed and unsynapsed regions of SC at the pachytene stage and persist on SC until the diplotene stage (Fig. 3). These results indicate that SKP1 is required for removal of HORMAD proteins from SC. Trip13Gt/Gt is also known to display the similar phenotype in terms of accumulation of HORMAD proteins on SC (10). It has been hypothesized that TRIP13 removes HORMAD proteins from SCs (10). Our results suggest that SKP1 may exclude HORMAD proteins from SCs by stabilizing the TRIP13 protein. In addition, both Skp1-deficient (Fig. 2K) and Trip13Gt/Gt spermatocytes exhibit desynapsis at pericentromeric regions at the pachytene stage (38), implying that SKP1 and TRIP13 may function in the same molecular pathway. SKP1 binds to different F-box proteins to form distinct E3 ligase complexes. F-box proteins provide target specificity for proteasome-mediated degradation (5). Therefore, it would be important to biochemically identify the F-box proteins for the SCF/SKP1 ubiquitin E3 ligase in meiotic germ cells in future studies.
MATERIALS AND METHODS
Targeted inactivation of the Skp1 gene
In the Skp1-targeting construct, a 3.1-kb genomic DNA segment harboring exons 3 to 5 was flanked by loxP sites (fig. S3A). The two homologous arms (2.6 and 2.4 kb) were amplified from a Skp1-containing bacterial artificial chromosome (BAC) clone (RP23-223A11) by polymerase chain reaction (PCR) with high-fidelity DNA polymerase. The HyTK selection cassette was cloned before the right arm. V6.5 embryonic stem (ES) cells were electroporated with linearized targeting construct, cultured in the presence of hygromycin B (120 μg/ml; Invitrogen), and screened by long-distance PCR for homologously targeted Skp13lox clones. Two Skp13lox ES cell lines were electroporated with the pOG231 plasmid that expresses Cre recombinase. ES cells were subjected to selection with ganciclovir (2 μM; Sigma) for removal of the HyTK cassette. ES cell colonies were screened by PCR. Recombination between the immediate HyTK-flanking loxP sites resulted in the Skp1f allele (fig. S3A). Two Skp1f/+ ES cell lines were injected into B6C3F1 (Taconic) blastocysts that were subsequently transferred to the uteri of pseudopregnant ICR females. The Skp1f allele was transmitted through the germ line from chimeric males. To generate germ cell–specific Skp1 knockout mice, we used two strategies. First, to study the function of Skp1 in germ cells, we generated Skp1f/− Ddx4-Cre mice, in which the constitutively active Cre is expressed specifically in germ cells, around E15 in both males and females (20). Second, to study the function of Skp1 at any specific stage during germ cell development, we generated Skp1f/− Ddx4-CreERT2 mice. Ddx4-CreERT2 is tamoxifen-inducible Cre (39). Tamoxifen injection induces Cre-mediated deletion of the floxed exons. Briefly, tamoxifen (Sigma, catalog number T5648) was resuspended with corn oil (Sigma, catalog number C8267) to a final concentration of 20 mg/ml and injected intraperitoneally into the 8-week-old Skp1f/− Ddx4-CreERT2 males at a dose of 2 mg/30 g body weight for five consecutive days. Untreated littermates with the same genotype were used as controls, because the same tamoxifen treatment of Skp1f/− or Skp1f/+ adult males or corn oil treatment of Skp1f/− Ddx4-CreERT2 adult males did not adversely affect meiosis when evaluated by testis histology and meiotic nuclear spread analysis. Tamoxifen treatment in combination with Ddx4-CreERT2 in WT males does not cause defects in meiosis as previously reported (40). Testes were collected 2, 4, 6, 8, 10, 12, 14, 16, and 32 days post-treatment after the last tamoxifen injection.
Genotyping for Skp1, Ddx4-Cre, and Ddx4-CreERT2 alleles was performed separately by PCR on genomic DNA isolated from tails. Skp1 floxed and WT alleles [537 and 349 base pairs (bp)] were assayed with primers CCTGAGGAGATTCGTAAAAC and GCACATTATGCCTTTGTATCA. The Skp1− (knockout) allele (320 bp) was assayed by PCR with primers TTGGCTCATTTGTGGGTTGG and GCACATTATGCCTTTGTATCA. The Ddx4-Cre allele (240 bp) was genotyped with primers CACGTGCAGCCGTTTAAGCCGCGT and TTCCCATTCTAAACAACACCCTGAA. The Ddx4-CreERT2 allele (205 bp) was assayed with primers ATACCGGAGATCATGCAAGC and GGCCAGGCTGTTCTTCTTAG. Mice were maintained and used for experimentation according to the guidelines of the Institutional Care and Use Committee of the University of Pennsylvania.
Targeted inactivation of the Rec8 gene
The Rec8 deletion was generated by the CRISPR-Cas9–mediated genome editing method as described before (41). Briefly, two single-guide RNAs (sgRNAs) that target the first and last intron of the mouse Rec8 gene were designed to generate about 6.5-kb DNA deletion (fig. S2). The two sgRNA sequences are as follows: CGCGTTGTGCAGACCTTTTT and ACAGCACACTCTAGATACTG. For each sgRNA, the two oligos were phosphorylated, annealed, and cloned to PX330 plasmid (Addgene). After in vitro transcription with the MEGAshortscript T7 Kit (AM1354, Invitrogen) and purification with the MEGAclear Transcription Clean-Up Kit (AM1908, Invitrogen), a mixture of 1 μl of Cas9 mRNA (500 ng/μl; Trilink, catalog number L-7206) + 0.5 μl of each sgRNA (500 ng/μl) was injected into zygotes. The injected zygotes were cultured in KSOM (potassium simplex optimization medium) medium at 37°C in a 5% CO2 incubator until the two-cell stage. The two-cell embryos were transferred into oviducts of 0.5-day post-coitum pseudopregnant ICR foster mothers. Founder mice were bred to WT mice to produce Rec8+/− mice. The Rec8− allele was sequenced to confirm the deletion. The WT allele (426 bp) was assayed by primers AGCAGAGTCGAAGAAGGCCTCTTG and CAGATGGTGGCGAAGCAGCCTGT. The knockout (Rec8−) allele (212 bp) was assayed by primers AGCAGAGTCGAAGAAGGCCTCTTG and TTGCTCAGGGGAATTTGGGTC.
PLK1 antibody production
Mouse PLK1 (amino acids 414 to 565) was expressed as a 6×His-PLK1 fusion protein in Escherichia coli using the pQE-30 vector and affinity-purified with Ni–nitrilotriacetic acid (NTA) agarose. Two rabbits were immunized with the recombinant protein, yielding antisera UP2456 and UP2457 (Cocalico Biologicals Inc.). Antiserum UP2456 was used for immunofluorescence analysis in this study.
Histological, immunofluorescence, and surface nuclear spread analyses
For histological analysis, testes or ovaries were fixed in Bouin’s solution at room temperature overnight, embedded with paraffin, and sectioned. Sections were stained with hematoxylin and eosin. In terms of immunofluorescence analysis, testes were fixed in 4% paraformaldehyde [in 1× phosphate-buffered saline (PBS)] for 6 hours at 4°C, dehydrated in 30% sucrose (in 1× PBS) overnight, and sectioned. For surface nuclear spread analysis (42), spermatocytes from 6-dpt testes were used unless noted otherwise, and oocytes from E18.5 ovaries were used. Testicular tubules or oocytes were extracted in hypotonic treatment buffer [30 mM tris, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. Cells were suspended in 100 mM sucrose and were then spread on a thin layer of paraformaldehyde solution containing Triton X-100. The following primary antibodies were used for immunofluorescence analyses: rabbit anti-SYCP1 (1:100, catalog number ab15090, Abcam), mouse anti-SYCP1 (1:200; a gift from C. Hoog), guinea pig anti-SYCP2 (43), mouse anti-SYCP3 (1:200; catalog number ab97672, Abcam), rabbit anti-SYCP3 (1:200; 23024-1-AP), rabbit anti-SKP1 (1:50; catalog number ab10546, Abcam), rabbit anti-HORMAD1 (12), rabbit anti-HORMAD1 (1:200; 13917-1-AP, Proteintech Group), rabbit anti-HORMAD2 (10), mouse anti-γH2AX (1:500; catalog number 16-202A, clone JBW301, Millipore), guinea pig anti-H1T (1:500; a gift from M. A. Handel) (4), rabbit anti-RPA2 (1:200; UP2436) (15), mouse anti-MLH1 (1:50; catalog number 550838, clone G168-15, BD Biosciences), human anti-CREST (1:100; 15-234, Antibodies Incorporated), mouse anti-CCNB1 (1:200; ab72, Abcam), rabbit anti-pHH3 (1:300; 9701S, Cell Signaling Technology), rabbit anti-PLK1 (1:50; UP2456; this study), mouse anti-BUB1 (1:50; a gift from Y. Watanabe) (44, 45), and rabbit anti–CENP-C (1:500; a gift from Y. Watanabe) (46).
Imaging analysis
Histological images were captured on a Leica DM5500B microscope with a DFC450 digital camera (Leica Microsystems). Most immunolabeled chromosome spread images were taken on a Leica DM5500B microscope with an ORCA Flash4.0 digital monochrome camera (Hamamatsu Photonics). Super-resolution immunolabeled chromosome spread images were taken on an Olympus BX51 microscope with an XM10 monochrome camera at the Penn Vet Imaging Core. Images were processed using Photoshop (Adobe) and ImageJ v1.44 software packages. Super-resolution imaging microscopy analysis was performed using DeltaVision OMX SR imaging system and softWoRx processing software.
Western blot analysis
Adult testes were homogenized in four volumes of protein extraction buffer [62.5 mM tris-HCl (pH 6.8), 3% SDS, 10% glycerol, and 5% 2-mercaptoethanol]. Samples were boiled at 95°C for 10 to 15 min to obtain soluble testicular protein extracts. Protein was quantified with spectrophotometer (Eppendorf), and 20 μg of protein samples was resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes using iBlot (Invitrogen) and immunoblotted with indicated antibodies. The following primary antibodies were used for Western blotting analysis: rabbit anti-SKP1 (1:1000; ab80586, Abcam), rabbit anti-HORMAD1 (1:1000; a gift from A. Rajkovic) (12), rabbit anti-HORMAD2 (1:2000; a gift from A. Toth) (10), mouse anti-SYCP3 (1:3000; ab97672, Abcam), rabbit anti-CCNB1 (1:500; 12231S, Cell Signaling Technology), mouse anti-γH2AX (1:500; clone JBW301, Millipore), rabbit anti-TRIP13 (1:500; 19602-1-AP, Proteintech Group), rabbit anti-CUL1 (1:1000; ab75817, Abcam), rabbit anti-pHH3 (1:500; 9701S, Cell Signaling Technology), rabbit anti-EMI2 (1:500; 55176-1-AP, Proteintech Group), mouse anti-CCNB1 (1:500; ab72, Abcam), rabbit anti–pY15-CDK1 (1:500; 4539S, Cell Signaling Technology), rabbit anti–pT161-CDK1 (1:500; ab194874, Abcam), mouse anti-PLK1 (1:500; SC-17783, Santa Cruz Biotechnology), mouse anti-ACTB (1:10,000; A5441, Sigma), and mouse anti-TUBB (T4026, Sigma).
Culture of spermatocytes and OA treatment
Short-term culture of testicular cells was carried out as reported previously with modifications (47). Freshly dissected testes of 2- to 3-month-old male mice were decapsulated and placed into Krebs buffer (120 mM NaCl, 4.8 mM KCl, 25.2 mM NaHCO3, 1.2 mM KH2PO4, 1.2 mM MgSO4, 1.3 mM CaCl2, and 11.1 mM dextrose) containing collagenase (1.6 mg/ml). Testicular tubules were digested at 32°C for 10 min and were transferred into Krebs buffer containing collagenase (1.6 mg/ml) and 0.01% trypsin. Cells were incubated at 32°C for another 10 min with repeated pipetting to become single-cell suspension. Then, the cells were passed through a 100-μm nylon mesh. About 2 × 106 testicular cells were washed twice with Krebs buffer and cultured overnight in 1 ml of MEMα (minimal essential media with alpha modifications) containing 5% streptomycin, 7.5% penicillin, 0.29% dl-lactic acid sodium salt, 0.59% Hepes, and 5% fetal bovine serum in each well of a six-well plate at 32°C. In the following morning, a small number of cells (2%) were used to measure cell viability with trypan blue, and the rest of the cells were treated with OA (5934S, Cell Signaling Technology) at a final concentration of 4 μM. Cells were harvested 5 hours after the addition of OA and used for nuclear spread analysis.
H1 kinase assay
H1 kinase assay was carried out by following the procedures described before (32). In brief, adult testes (200 mg) were homogenized in 1 ml of radioimmunoprecipitation assay (RIPA) buffer [10 mM tris (pH 8.0), 140 mM NaCl, 1% Trion X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 1 mM EDTA] with 1 mM PMSF. MPF complexes were immunoprecipitated from 50 μg of cellular protein lysate using CDK1 antibody (ab194874, Abcam). The beads were washed three times with RIPA buffer and equilibrated with EB kinase buffer [50 mM Hepes (pH 7.5), 10 mM MgCl2, 80 mM β-glycerophosphate, 20 mM EGTA, 1 mM DTT]. Immunoprecipitates were incubated in 20 μl of kinase buffer supplemented with 10 μM adenosine triphosphate (ATP), 5 μM adenosine 3′,5′-monophosphate (cAMP)–dependent protein kinase inhibitor, calf thymus histone H1 (50 μg/ml; 14-155, Sigma-Aldrich), and 4 μCi of [γ-32P]ATP for 20 min at 30°C. The reaction was terminated by adding 20 μl of 2 × SDS sample buffer and boiling for 10 min. The eluted samples were separated by 12% SDS-PAGE gels, and gels were dried and exposed to autoradiography overnight. Quantification of the bands was analyzed by ImageJ software.
Oocyte collection and culture
WT and Skp1cKO (Skp1f/− Ddx4-Cre) female mice (3 to 4 weeks old) were hormonally primed with 5 U of PMSG (pregnant mare’s serum gonadotropin) (catalog number 367222, Calbiochem) 48 hours before oocyte collection. GV-intact oocytes were collected in bicarbonate-free minimal essential medium with polyvinylpyrrolidone and Hepes (MEM-PVP) (48), denuded from cumulus cells, and cultured in Chatot-Ziomek-Bavister (CZB) medium (49) covered with mineral oil (catalog number M5310, Sigma) in a humidified atmosphere of 5% CO2 at 37°C. Meiotic resumption was inhibited by addition of 2.5 μM milrinone. Milrinone was subsequently washed out to allow meiotic resumption 1.5 hours after collection. Oocytes were checked for nuclear envelope breakdown (GVBD) 1.5 hours after washout, and those that did not enter GVBD were removed from the culture. Confocal images were collected with a microscope (DMI4000 B, Leica) equipped with a 63× 1.3 numerical aperture (NA) glycerol immersion objective lens, an xy piezo Z stage (Applied Scientific Instrumentation), a spinning disc confocal scanner (Yokogawa Electric Corporation), an electron multiplier charge-coupled device camera (ImageEM C9100-13; Hamamatsu Photonics), and an LMM5 laser merge module with 488- and 593-nm diode lasers (Spectral Applied Research) controlled by MetaMorph software (Molecular Devices).
Oocyte immunocytochemistry, spindle morphology, and chromosome alignment analysis
MI oocytes were fixed 7 to 7.5 hours after GVBD, in freshly prepared 2% paraformaldehyde in PBS with 0.1% Triton X-100 (pH 7.4), for 25 min at room temperature, placed in blocking solution [PBS, 0.3% bovine serum albumin (BSA), and 0.01% Tween-20] overnight at 4°C, then permeabilized in PBS with 0.3% BSA and 0.1% Triton X-100 for 20 min, and washed in blocking solution for 20 min before primary antibody staining. Rabbit anti–β-tubulin (9F3) monoclonal antibody conjugated to Alexa Fluor 488 (1:75 dilution; Cell Signaling Technology) was used to visualize MI spindles. Cells were washed three times for 15 min in blocking buffer and mounted in Vectashield with bisbenzimide (Hoechst 33342, Sigma-Aldrich) to visualize chromosomes.
Oocyte images were collected as a series of 0.4-μm z-stacks covering the whole spindle. To analyze chromosome alignment along the spindle axis, maximum contrast total z-projection images of MI spindles parallel to the coverslip were selected into four distinct categories: (i) chromosome aligned (bipolar spindles with all chromosomes aligned at the metaphase plate), (ii) mild misalignment (bipolar spindles with less than three chromosomes misaligned), (iii) severe misalignment (bipolar spindles with more than three chromosomes misaligned; frequently >10 chromosomes were misaligned), and (iv) monopolar spindles.
To quantify chromosome alignment, DNA signal intensity along the spindle axis was measured by line scan (line thickness = 100) on maximal intensity z-projection images (n = 25 WT and Skp1cKO oocytes). After background subtraction, each DNA line scan was normalized to its highest value and aligned to each other based on the middle value, and all line scans were then averaged. Oocyte data analyzed in this part are combined from three independent experiments.
Supplementary Material
Acknowledgments
We thank A. Rajkovic for HORMAD1 antibody, A. Toth for HORMAD2 antibody, M. A. Handel for H1t antibody, C. Hoog for SYCP1 antibody, Y. Watanabe and T. Akera for BUB1 and CENP-C antibodies, and R. Guo, J. Chotiner, and F. Yang for critical reading of the manuscript. Funding: This work was supported by NIH/NIGMS grant R35GM118052 (to P.J.W.); National Key Research and Development Program of China 2018YFC1003400, National Natural Science Foundation of China 31771588, and the Thousand Youth Talents Plan (to M.L.); NIH/NICHD grant R01HD082568 (to J.C.S.); NIH/NICHD T32HD057854 (to J.C.B.); and NIH/NIGMS grant R35GM122475 (to M.A.L.). Author contributions: P.J.W., Y.G., and M.L. conceptualized the study and designed the experiments. Y.G. carried out most of the experiments and generated the Rec8 knockout mice. M.L. observed the localization of SKP1 to SC, made the Skp1 floxed mice, and produced the PLK1 antibody. N.A.L. did the blastocyst injection of Skp1 ES cells and zygote injection of Rec8 gRNAs/Cas9. J.C.B. and J.C.S. contributed to the Trip13 data. J.M., L.C., and M.A.L. contributed to the oocyte data. G.R. contributed to the super-resolution microscopy. P.J.W. supervised all aspects of the work and wrote the manuscript. Y.G. and L.C. contributed to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/13/eaaz2129/DC1
Fig. S1. Localization of SKP1 to the SC in oocytes.
Fig. S2. Inactivation of the Rec8 gene by CRISPR-Cas9–mediated genome editing.
Fig. S3. Targeted inactivation of the Skp1 gene.
Fig. S4. H1t expression and CREST localization in WT and Skp1cKO spermatocytes.
Fig. S5. Localization of SKP1 in WT and Trip13Gt/Gt pachytene spermatocytes.
Fig. S6. Precocious chromosome desynapsis and persistence of RPA2 foci in E18.5 Skp1cKO oocytes.
Fig. S7. Expression and localization of PLK1, BUB1, CENP-C, and CCNB1 in spermatocytes.
REFERENCES AND NOTES
- 1.Handel M. A., Schimenti J. C., Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124–136 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Zickler D., Kleckner N., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiltshire T., Park C., Caldwell K. A., Handel M. A., Induced premature G2/M-phase transition in pachytene spermatocytes includes events unique to meiosis. Dev. Biol. 169, 557–567 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Cobb J., Cargile B., Handel M. A., Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev. Biol. 205, 49–64 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Cardozo T., Pagano M., The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Nayak S., Santiago F. E., Jin H., Lin D., Schedl T., Kipreos E. T., The Caenorhabditis elegans Skp1-related gene family: Diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12, 277–287 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Zhao D., Ni W., Feng B., Han T., Petrasek M. G., Ma H., Members of the Arabidopsis-SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol. 133, 203–217 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo M., Yang F., Leu N. A., Landaiche J., Handel M. A., Benavente R., La Salle S., Wang P. J., MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat. Commun. 4, 2788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H., Beasley M. D., Warren W. D., van der Horst G. T., McKay M. J., Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C. R., Cooke H. J., Jasin M., Keeney S., McKay M. J., Toth A., Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLOS Genet. 5, e1000702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda T., Daniel K., Wojtasz L., Toth A., Höög C., A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp. Cell Res. 316, 158–171 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Shin Y.-H., Choi Y., Erdin S. U., Yatsenko S. A., Kloc M., Yang F., Wang P. J., Meistrich M. L., Rajkovic A., Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLOS Genet. 6, e1001190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanzione M., Baumann M., Papanikos F., Dereli I., Lange J., Ramlal A., Tränkner D., Shibuya H., de Massy B., Watanabe Y., Jasin M., Keeney S., Tóth A., Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X. C., Schimenti J. C., Mouse pachytene checkpoint 2 (Trip13) is required for completing meiotic recombination but not synapsis. PLOS Genet. 3, e130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi B., Xue J., Yin H., Guo R., Luo M., Ye L., Shi Q., Huang X., Liu M., Sha J., Wang P. J., Dual functions for the ssDNA-binding protein RPA in meiotic recombination. PLOS Genet. 15, e1007952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner J. M. A., Meiotic silencing in mammals. Annu. Rev. Genet. 49, 395–412 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Ahuja J. S., Sandhu R., Mainpal R., Lawson C., Henley H., Hunt P. A., Yanowitz J. L., Börner G. V., Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science 355, 408–411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao H. B., Qiao H., Bhatt S. K., Bailey L. R., Tran H. D., Bourne S. L., Qiu W., Deshpande A., Sharma A. N., Beebout C. J., Pezza R. J., Hunter N., A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto S. Y., Sato M., Toda T., Yamamoto M., SCF ensures meiotic chromosome segregation through a resolution of meiotic recombination intermediates. PLOS ONE 7, e30622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallardo T., Shirley L., John G. B., Castrillon D. H., Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan P. W., Karppinen J., Handel M. A., Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J. Cell Sci. 125, 5061–5072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagawa K., Abdulle R., Bansal P. K., Cagney G., Fields S., Hieter P., Requirement of Skp1-Bub1 interaction for kinetochore-mediated activation of the spindle checkpoint. Mol. Cell 11, 1201–1213 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Klare K., Weir J. R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., Musacchio A., CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun F., Handel M. A., Regulation of the meiotic prophase I to metaphase I transition in mouse spermatocytes. Chromosoma 117, 471–485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinathan L., Szmyd R., Low D., Diril M. K., Chang H. Y., Coppola V., Liu K., Tessarollo L., Guccione E., van Pelt A. M. M., Kaldis P., Emi2 is essential for mouse spermatogenesis. Cell Rep. 20, 697–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F., Wang P. J., The mammalian synaptonemal complex: A scaffold and beyond. Genome Dyn. 5, 69–80 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Cahoon C. K., Hawley R. S., Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23, 369–377 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Dix D. J., Allen J. W., Collins B. W., Poorman-Allen P., Mori C., Blizard D. R., Brown P. R., Goulding E. H., Strong B. D., Eddy E. M., HSP70-2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development 124, 4595–4603 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Wachowicz P., Fernández-Miranda G., Marugan C., Escobar B., de Cárcer G., Genetic depletion of polo-like kinase 1 leads to embryonic lethality due to mitotic aberrancies. Bioessays 38 (suppl. 1), S96–S106 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Sourirajan A., Lichten M., Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 22, 2627–2632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu D., Matzuk M. M., Sung W. K., Guo Q., Wang P., Wolgemuth D. J., Cyclin A1 is required for meiosis in the male mouse. Nat. Genet. 20, 377–380 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Liu D., Liao C., Wolgemuth D. J., A role for cyclin A1 in the activation of MPF and G2–M transition during meiosis of male germ cells in mice. Dev. Biol. 224, 388–400 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Hunt P. A., Hassold T. J., Sex matters in meiosis. Science 296, 2181–2183 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Yang M., Hu Y., Lodhi M., McCombie W. R., Ma H., The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. U.S.A. 96, 11416–11421 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao D., Yang X., Quan L., Timofejeva L., Rigel N. W., Ma H., Makaroff C. A., ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol. Biol. 62, 99–110 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Timofejeva L., Ma H., Makaroff C. A., The Arabidopsis SKP1 homolog ASK1 controls meiotic chromosome remodeling and release of chromatin from the nuclear membrane and nucleolus. J. Cell Sci. 119, 3754–3763 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Wu H., Liang G., Yang M., Defects in nucleolar migration and synapsis in male prophase I in the ask1-1 mutant of Arabidopsis. Sex. Plant Reprod. 16, 273–282 (2004). [Google Scholar]
- 38.Roig I., Dowdle J. A., Toth A., de Rooij D. G., Jasin M., Keeney S., Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLOS Genet. 6, e1001062 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John G. B., Gallardo T. D., Shirley L. J., Castrillon D. H., Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev. Biol. 321, 197–204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe H., Alavattam K. G., Kato Y., Castrillon D. H., Pang Q., Andreassen P. R., Namekawa S. H., CHEK1 coordinates DNA damage signaling and meiotic progression in the male germline of mice. Hum. Mol. Genet. 27, 1136–1149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H., Wang H., Jaenisch R., Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 9, 1956–1968 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Peters A. H., Plug A. W., van Vugt M. J., de Boer P., A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Yang F., De La Fuente R., Leu N. A., Baumann C., McLaughlin K. J., Wang P. J., Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 173, 497–507 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawashima S. A., Yamagishi Y., Honda T., Ishiguro K.-i., Watanabe Y., Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Akera T., Trimm E., Lampson M. A., Molecular strategies of meiotic cheating by selfish centromeres. Cell 178, 1132–1144.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J., Ishiguro K.-i., Nambu A., Akiyoshi B., Yokobayashi S., Kagami A., Ishiguro T., Pendas A. M., Takeda N., Sakakibara Y., Kitajima T. S., Tanno Y., Sakuno T., Watanabe Y., Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 517, 466–471 (2015). [DOI] [PubMed] [Google Scholar]
- 47.La Salle S., Sun F., Handel M. A., Isolation and short-term culture of mouse spermatocytes for analysis of meiosis. Methods Mol. Biol. 558, 279–297 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Stein P., Schindler K., Mouse oocyte microinjection, maturation and ploidy assessment. J. Vis. Exp. 2011, e2851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatot C. L., Ziomek C. A., Bavister B. D., Lewis J. L., Torres I., An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 86, 679–688 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/13/eaaz2129/DC1
Fig. S1. Localization of SKP1 to the SC in oocytes.
Fig. S2. Inactivation of the Rec8 gene by CRISPR-Cas9–mediated genome editing.
Fig. S3. Targeted inactivation of the Skp1 gene.
Fig. S4. H1t expression and CREST localization in WT and Skp1cKO spermatocytes.
Fig. S5. Localization of SKP1 in WT and Trip13Gt/Gt pachytene spermatocytes.
Fig. S6. Precocious chromosome desynapsis and persistence of RPA2 foci in E18.5 Skp1cKO oocytes.
Fig. S7. Expression and localization of PLK1, BUB1, CENP-C, and CCNB1 in spermatocytes.


