Key Points
Viral reproduction involves not only replication but also interactions with host defences. Although various viral proteins can take part in counteracting innate and adaptive immunity, many viruses possess a subset of proteins that are specifically dedicated to counter-defensive activities. These proteins are sometimes referred to as 'virulence factors', but here we argue that the term 'security proteins' is preferable, for several reasons.
The concept of security proteins of RNA-containing viruses can be considered using the leader (L and L*) and 2A proteins of picornaviruses as examples. The picornaviruses are a large group of human and animal viruses that include important pathogens such as poliovirus, hepatitis A virus and foot-and-mouth disease virus. The genomes of different picornaviruses have a similar organization, in which the genes for L and 2A occupy fixed positions upstream and downstream of the capsid genes, respectively.
Both L and 2A are extremely heterogeneous with respect to size, sequence and biochemical properties. The similarly named proteins can be completely unrelated to each other in different viral genera, and the variation can be striking even among members of the same genus. A subset of picornaviruses lacks L altogether.
The properties and functions of L and 2A of many picornaviruses are unknown, but in those viruses that have been investigated sufficiently it has been found that these proteins can switch off various aspects of host macromolecular synthesis and specifically suppress mechanisms involved in innate immunity. Thus, notwithstanding their unrelatedness, the security proteins carry out similar biological functions. It is proposed that other picornavirus L and 2A proteins that have not yet been investigated should also be primarily involved in security activities.
The L, L* and 2A proteins are dispensable for viral reproduction, but their elimination or inactivation usually renders the viruses less pathogenic. The phenotypic changes associated with inactivation of security proteins are much less pronounced in cells or organisms that have innate immunity deficiencies. In several examples, the decreased fitness of a virus in which a security protein has been inactivated could be rescued by the experimental introduction of an unrelated security protein.
It can be argued that L and 2A were acquired by different picornaviruses independently, and possibly by exploiting different mechanisms, late in the evolution of this viral family.
It is proposed that the concept of security proteins is of general relevance and can be applied to viruses other than picornaviruses. The hallmarks of security proteins are: structural and biochemical unrelatedness in related viruses or even absence in some of them; dispensability of the entire protein or its functional domains for viral viability; and, for mutated versions of the proteins, fewer detrimental effects on viral reproduction in immune-compromised hosts than in immune-competent hosts.
Supplementary information
The online version of this article (doi:10.1038/nrmicro2452) contains supplementary material, which is available to authorized users.
Subject terms: Virus-host interactions, Viral proteins, Viral infection
Viral security proteins are structurally and biochemically unrelated proteins that function to counteract host defences. Here, Agol and Gmyl consider the impact of the picornavirus security proteins on viral reproduction, pathogenicity and evolution.
Supplementary information
The online version of this article (doi:10.1038/nrmicro2452) contains supplementary material, which is available to authorized users.
Abstract
Interactions with host defences are key aspects of viral infection. Various viral proteins perform counter-defensive functions, but a distinct class, called security proteins, is dedicated specifically to counteracting host defences. Here, the properties of the picornavirus security proteins L and 2A are discussed. These proteins have well-defined positions in the viral polyprotein, flanking the capsid precursor, but they are structurally and biochemically unrelated. Here, we consider the impact of these two proteins, as well as that of a third security protein, L*, on viral reproduction, pathogenicity and evolution. The concept of security proteins could serve as a paradigm for the dedicated counter-defensive proteins of other viruses.
Supplementary information
The online version of this article (doi:10.1038/nrmicro2452) contains supplementary material, which is available to authorized users.
Main
All bona fide RNA viruses encode at least two proteins, a capsid protein and an RNA-dependent RNA polymerase (RdRP). In this article, we do not consider 'abnormal' RNA viruses such as hepatitis delta virus, which does not possess its own replication machinery and borrows a capsid from another virus1, or the capsid-less narnaviruses, which encode only an RdRP2; hepatitis delta virus and narnaviruses are closer to viroids and plasmids, respectively, than to fully fledged RNA viruses. RNA viruses encoding only a capsid and an RdRP are known to infect protists such as Giardia spp.3. Some RNA phages and fungal RNA viruses express only three proteins. However, most RNA viruses do not rely on such a scant protein repertoire. The proteomes of the majority of RNA viruses comprise up to 12 different proteins, with the genomes of some coronaviruses encoding nearly 30 proteins.
To infect, viruses must reach an appropriate intracellular environment and, if necessary, adapt this environment to their own requirements. Thus, viral infection requires not only replication but also interactions with host defences. To carry out these tasks, RNA viruses have only a few proteins at their disposal, with the available protein arsenal being limited by the genome size. As the proteomes of these viruses are strikingly small and specific viral functions generally require more than one protein, most proteins encoded by RNA viruses are multifunctional. At the same time, however, there is a certain division of labour between viral proteins. We consider some aspects of this problem using, as an example, the picornaviruses, a large family of small animal viruses with a medium-sized, single-stranded, positive-sense RNA genome (Box 1). Although they share a common genome organization, these viruses exhibit sufficient genetic variation to be separated into at least 12 distinct genera (Box 2). The family includes important human and animal pathogens that cause a range of disorders, including poliomyelitis, foot-and-mouth disease, the common cold, gastroenteritis, hepatitis, meningitis, myocarditis and uveitis. In addition to acute diseases, these viruses can also cause chronic persistent disorders.
Of the 12 'mature' (fully processed) picornaviral proteins (Box 1), the most ancient group includes the capsid proteins and a set of conserved proteins considered to be picornavirus signature proteins4, comprising the RdRP (3Dpol), the primer for RNA synthesis (VPg), a protease (3Cpro) and an ATPase (2CATPase). The capsid and signature proteins are indispensable for viral viability. Two less conserved proteins, 2B and 3A, are also essential for viability but are less directly involved in viral reproduction. The main functions of these two proteins are to target replicative proteins to the correct destinations and aid in the creation of a suitable replicative niche.
Finally, two other non-structural proteins that flank the capsid precursor in the polyprotein molecule, the leader (L) and 2A proteins, constitute a distinct group, although they have no common structural or biochemical features. They are the most variable among the picornavirus proteins, and some viruses even lack L altogether (Fig. 1; Tables 1, 2). This group also includes the so-called L* protein, which is encoded in a different reading frame of the RNA of certain cardioviruses. Functionally, these proteins (L, L* and 2A) have been characterized in some detail for only a few picornaviruses, and in these cases their major function has been shown to be counteracting host defences, thereby ensuring optimal conditions for viral reproduction. In this Review we argue that picornaviral L and 2A proteins that have yet to be characterized are likely to fulfil the same biological role.
Figure 1. Leader and 2A proteins of picornaviruses.
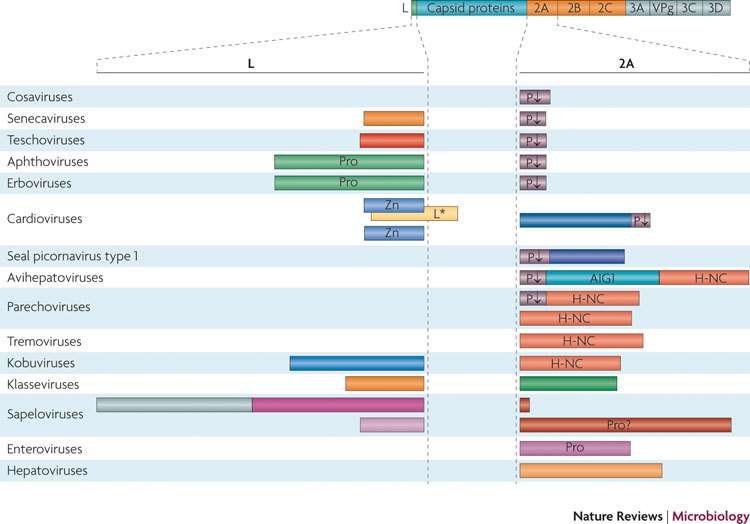
The organization of an 'idealized' picornaviral polyprotein is shown, with specific viral leader (L) and 2A proteins given below; protein sizes are not to scale but give approximate relative lengths. There is great variability in L and 2A proteins. Multiple L and 2A proteins are known for some viral genera. Several picornaviruses do not possess L but have large 2A proteins. Cosaviruses contain no L and only a very short 2A. Other viruses (for example, some sapeloviruses) possess an unusually long L (we propose that in this case it might correspond to at least two separate polypeptides; see Supplementary information S1 (figure)) and a very short 2A (if there is a 2A peptide at all). Remarkably, other sapeloviruses possess a long 2A and a short L. Notable differences in the organization of the L and 2A proteins can occur among representatives of the same genus, such as in cardioviruses and parechoviruses. Well-defined amino acid motifs are indicated. See main text for details about the H-NC and AIG1 domains. L*, alternative leader protein encoded by an alternative reading frame beginning in the L-encoding sequence; P↓, the NPG(P) motif, which interrupts translation at the proline residue; Pro, protease; VPg, primer for RNA synthesis; Zn, zinc finger.
Table 1.
Properties of the leader proteins of picornaviruses*
| Genus | Protein | Size (residues)‡ | Isoelectric point§ | Identifiable domains | Number of species | Interspecies sequence conservation|| |
|---|---|---|---|---|---|---|
| Senecavirus | SVV L | 79 | 4.74 | None | 1 | NA |
| Teschovirus | PTV L | 86 | 7.67 | None | 1 | NA |
| Aphthovirus | FMDV Lab | 199–204 | 4.91–5.13 | Peptidase_C28¶ | 3 | 32% (51%) |
| FMDV Lb | 170–175 | 4.50–4.71 | ||||
| ERAV Lab | 209 | 8.74 | ||||
| ERAV Lb | 188 | 8.29 | ||||
| BRV Lab | 207 | 4.86 | ||||
| BRV Lb | 183 | 4.37 | ||||
| Erbovirus | ERBV L | 218–219 | 5.70 | Peptidase_C28¶ | 1 | NA |
| Cardiovirus | EMCV L | 67 | 3.80 | Zn finger# | 2 | 47% (59%) |
| TMEV L | 76 | 3.64 | ||||
| TMEV L* | 156 | 10.39 | ||||
| Sapelovirus | ASV L1 | 215 | 8.66 | None | 3 | ND for all 3 species; 15% (34%) for SSV and PSV |
| ASV L2 | 236 | 8.54 | ||||
| SSV | 88 | 8.95 | ||||
| PSV | 84 | 8.46 | ||||
| Kobuvirus | AiV L | 170 | 5.92 | None | 3 | 25% (35%) |
| BKV L | 187 | 5.51 | ||||
| PKV L | 195 | 5.38 | ||||
| Klassevirus | HKV L | 111–114 | 10.35 | None | 1 | NA |
| AiV, Aichi virus; ASV, avian sapelovirus; BKV, bovine kobuvirus; BRV, bovine rhinitis B virus; EMCV, encephalomyocarditis virus; ERAV, equine rhinitis A virus; ERBV, equine rhinitis B virus; FMDV, foot-and-mouth disease virus; HKV, human klassevirus; NA, not applicable; ND, not determined; PKV, porcine kobuvirus; PSV, porcine sapelovirus; PTV, porcine teschovirus; SSV, simian sapelovirus; SVV, Seneca Valley virus; TMEV, Theiler's murine encephalomyelitis virus. | ||||||
| *For aphthoviruses, there are two leader (L) proteins, Lab and Lb. For avian sapelovirus, we propose the existence of two leader proteins, L1 and L2 (see Supplementary information S1 (figure)), rather than one L protein. For TMEV, an alternative L protein, L*, is encoded in an alternative reading frame that starts within the L coding sequence. | ||||||
| ‡According to the data in GenBank. | ||||||
| §Values were calculated using ProtParam132. | ||||||
| ||For the viral genera with more than one species, the level of interspecies amino acid identity (and similarity, in parenthesis) was calculated with the aid of CLUSTAL_X2 alignments, using utilities implemented in BioEdit and the BLOSUM62 similarity matrix133. To focus on the core protein sequences, the internal insertions of >15 amino acid residues, as well as terminal insertions, were not taken into account. | ||||||
| ¶The peptidase_C28 motif was revealed by BLAST searches in the NCBI Conserved Domain Database134. | ||||||
| #The conserved non-canonical zinc (Zn) finger domain (with a CHCC motif) was found to be present. | ||||||
Table 2.
Properties of the 2A proteins and peptides of picornaviruses
| Virus | Size (residues)‡ | Isoelectric point§ | Identifiable domains/motifs | Number of species | Interspecies sequence conservation|| | |
|---|---|---|---|---|---|---|
| Genus | Species* | |||||
| Cosavirus | All 5 species | 30–37 | ND | NPG(P) | 5 | ND |
| Senecavirus | SVV | 9 | ND | NPG(P) | 1 | NA |
| Teschovirus | PTV | 21 | ND | NPG(P) | 1 | NA |
| Erbovirus | ERBV | 16 | ND | NPG(P) | 1 | NA |
| Aphthovirus | FMDV | 18 | ND | NPG(P) | 3 | ND |
| ERAV | 18 | ND | ||||
| BRV | 19 | ND | ||||
| Cardiovirus | EMCV | 143 | 9.67 | NPG(P) | 2 | 24% (42%) |
| TMEV | 133 | 8.89 | ||||
| Sapelovirus | ASV | 12 | ND | None | 3 | ND for all 3 species; 27% (39%) for SSV and PSV |
| SSV | 292–302 | 5.00 | Protease¶ | |||
| PSV | 226 | 5,55 | Protease¶ | |||
| Kobuvirus | AiV | 111 | 5.92 | H-NC¶ | 3 | 54% (66%) |
| BKV | 134 | 5.51 | ||||
| PKV | 136 | 5.38 | ||||
| Parechovirus | LV 2A1 | 20 | ND | NPG(P) | 2 | 44% (64%) for 2A2 of LV and 2A of HPeV |
| LV 2A2 | 135 | 6.60 | H-NC | |||
| HPeV | 160 | 5.28 | H-NC | |||
| Avihepatovirus | DHV 2A1 | 20 | ND | NPG(P) | 1 | NA |
| DHV 2A2+2A3 | 285 | 8.65 | AIG1 and H-NC | |||
| Hepatovirus | HAV | 189 | 8.78 | None | 1 | NA |
| Tremovirus | AEV | 163 | 8.22 | H-NC | 1 | NA |
| Enterovirus | All 10 species | 142–150 | 5.22–6.30 | Pico_P2A | 10 | 35% (51%) |
| Klassevirus | HKV | 126 | 4.80 | None | 1 | NA |
| Unclassified | SePV-1 2A1 | 29 | ND | NPG(P)# | 1 | NA |
| SePV-1 2A2 | 100 | 7.92 | None | 1 | NA | |
| AEV, avian encephalomyelitis virus; AiV, Aichi virus; ASV, avian sapelovirus; BKV, bovine kobuvirus; BRV, bovine rhinitis B virus; DHV, duck hepatitis A virus; EMCV, encephalomyocarditis virus; ERAV, equine rhinitis A virus; ERBV, equine rhinitis B virus; FMDV, foot-and-mouth disease virus; HAV, hepatitis A virus; HKV, human klassevirus; HPeV, human parechovirus; LV, Ljungan virus; NA, not applicable; ND, not determined; PKV, porcine kobuvirus; PSV, porcine sapelovirus; PTV, porcine teschovirus; SePV-1, seal picornavirus type 1; SSV, simian sapelovirus; SVV, Seneca Valley virus; TMEV, Theiler's murine encephalomyelitis virus. | ||||||
| *For those viruses with more than one 2A protein, the specific protein analysed is indicated. | ||||||
| ‡According to the data in GenBank. | ||||||
| §Values were calculated only for peptides with >40 residues using ProtParam132. | ||||||
| ||For the viral genera with more than one species, the level of interspecies amino acid identity (and similarity, in parentheses) was calculated with the aid of CLUSTAL_X2 alignments, using utilities implemented in BioEdit and the BLOSUM62 similarity matrix134. To focus on the core protein sequences, the internal insertions of >15 amino acid residues, as well as terminal insertions, were not taken into account. | ||||||
| ¶These are manually recognizable motifs. | ||||||
| #One of the strains is reported to harbour an NPR(P) motif instead of NPG(P). | ||||||
We have proposed that this group of proteins should be called security proteins5. They are sometimes also referred to as virulence factors6; such a designation, although it may be more familiar, is somewhat less preferable to us for the reasons explained below. Viruses have no special 'desire' to be virulent, that is, to harm, even less to kill, their hosts. The pathogenic properties of a virus are not a prerequisite for viral fitness. In fact, the most severe harm in a viral infection comes not from viral reproduction but from the (sometimes miscalculated) host defence response. Host damaging or even suicidal defensive reactions include RNA degradation, inhibition of translation, and the induction of endoplasmic reticulum stress, apoptosis, autophagy and inflammation, and these reactions are aimed at limiting viral reproduction and spread. As a natural response, viruses have evolved tools directed not only at overcoming the specific innate and adaptive immune responses but also at inhibiting the general host metabolic functions on which these specific defences are based — that is, processes such as transcription and translation, cell signalling and intracellular trafficking. Inhibiting these processes is the major function of the security proteins. Accordingly, the capacity to withstand host defences, rather than virulence (the ability to harm the host) per se, is the property that is selected for during viral evolution. The long-term co-evolution of a virus and its host will probably result in their mutual adaptation accompanied by a decrease in viral virulence. Thus, in our opinion, the term 'security protein' is a better reflection of the evolutionary origin of the relevant proteins than the term 'virulence factor'. Moreover, the possession of efficient security proteins does not necessarily make a virus particularly virulent.
The aim of this Review is to consider the properties and biological significance of picornavirus security proteins and, on the basis of this knowledge, to put forward a general view of security proteins as dedicated counter-defensive proteins that evolved primarily to overcome various mechanisms of host resistance.
Properties of picornavirus security proteins
Both L and 2A are extremely heterogeneous with respect to size, sequence and biochemical properties (Fig. 1; Tables 1, 2). The length of L varies from ∼70 amino acid residues in cardioviruses to ∼450 residues in some sapeloviruses, and the variation is striking even in a single genus; for example, in sapeloviruses, the length varies from ∼80 residues (one L protein) to ∼450 residues (two L proteins). Although certain sapeloviruses seem to express two L proteins (Supplementary information S1 (figure)), approximately half the picornavirus genera lack L altogether. The length of 2A varies ∼30-fold (from 9 residues in senecaviruses to ∼300 residues in avihepatoviruses and some sapeloviruses), and it can comprise one to three separate polypeptides. The L proteins of cardioviruses and klasseviruses are strongly acidic and markedly basic, respectively, whereas the 2A proteins of these viruses exhibit the reverse ratio of acidity. The overall charge of the proteins will influence the choice of their interaction partners. The primary structures of L and 2A are, in most cases, totally unrelated when compared across different genera, and even intra-genus conservation among seemingly homologous proteins can be low, as is the case, for example, with the L proteins of kobuviruses and 2A proteins of cardioviruses and sapeloviruses.
With regard to their biochemical activities, only a few security proteins have been characterized in any detail. 2A of enteroviruses (2Apro) is a protease7 with a chymotrypsin-like fold in which the catalytic serine residue has been replaced by cysteine8. 2Apro performs an essential function in viral reproduction through its involvement in the so-called 'primary' co-translational cis-cleavage of the polyprotein between the amino-terminal amino acid of 2Apro and the carboxy-terminal residue of the capsid protein VP1. Aphthovirus L is also a protease (Lpro) with a papain-like fold9. Translation of aphthovirus RNA can start at two in-frame AUG codons, generating distinct L proteins, Lab (∼200 residues) and Lb (∼170 residues)10, with Lb predominating in vivo. Lpro cleaves the bond at the Lpro–VP4 boundary11,12. Erbovirus Lpro exhibits similar enzymatic activity13. Owing to the presence of a characteristic motif, the 2A proteins of some sapeloviruses have also been proposed to be proteases14.
2A of aphthoviruses is a short peptide that promotes co-translational interruption of polyprotein synthesis just downstream of its own coding sequence (and before the 2B moiety); cleavage at the aphthovirus VP1–2A boundary is accomplished by 3Cpro. Interruption of polyprotein synthesis (also known as ribosome skipping) is proposed to be caused by an interaction between the 2A peptide and the exit tunnel of the ribosome, preventing the interaction of the ribosome with Pro-tRNA (proline is the amino-terminal residue of aphthovirus 2B)15,16. To achieve this, a short peptide harbouring an NPG(P) motif (the parentheses mark the interruption site) is sufficient. Short 2A peptides from erboviruses17, teschoviruses18, senecaviruses19 and cosaviruses20 also possess NPG(P) motifs and seem to be involved in polyprotein processing at the 2A–2B border. Separation of these peptides from the VP1 capsid protein has been assumed but not yet clearly demonstrated. As these peptides possess no other known activities, their assignment as security proteins is not warranted; we refer to them here as 2Asp (2A short peptide).
The ribosome-skipping NPG(P) motif is also present in some 'composite' 2A proteins, such as those of Ljungan virus21 (a parechovirus), avihepatoviruses22 and seal picornavirus type 1 (Ref. 23), in which they seem to ensure self-separation from the downstream 2A moieties. In cardiovirus 2A, the NPG(P) motif is located at the C terminus and interrupts polyprotein synthesis at the 2A–2B boundary24, which corresponds to the polyprotein 'primary' cleavage site in this genus25.
Apart from the NPG(P)-containing peptides, 2A proteins of cardioviruses, avihepatoviruses, Ljungan virus and seal picornavirus type 1 contain distinct but poorly characterized sequences. The cardiovirus encephalomyocarditis virus (EMCV) encodes a 2A protein with RNA-binding affinity26. The bipartite 2A of Ljungan virus, the tripartite 2A of avihepatoviruses and the NPG(P)-lacking 2A proteins of kobuviruses, tremoviruses and some parechoviruses all have a ∼140–150-residue H-NC domain, which contains a histidine with a downstream asparagine-cysteine dipeptide and a putative transmembrane domain21,27. The observation that a similar H-NC domain is present in certain cellular tumour suppressors21 suggests that these viral proteins are also involved in the control of host activities. Between the NPG(P) and H-NC motifs, 2A of the avihepatovirus duck hepatitis A virus possesses an additional moiety that contains the so-called AIG1 domain22, which is also found in representatives of the Ras-like GTPase superfamily.
Cardiovirus L proteins28, which are devoid of any enzymatic activity and exhibit noticeable intra-genus variability (Table 1), contain a non-classical but functional zinc finger (Cys-His-Cys-Cys) motif29,30 and a downstream acidic motif31 that, in some of these L proteins, contains potential phosphorylation sites31,32. Certain strains of Theiler's murine encephalomyelitis virus (TMEV) are unique among picornaviruses in expressing a functional protein, L*, that is encoded in an alternative translation frame33,34 that starts within the L coding sequence, goes through the VP4 coding sequences and terminates in the VP2 coding sequence of the main reading frame. The 2A and L proteins of other picornaviruses neither contain easily identifiable amino acid motifs nor have known specific biochemical activities.
Effects on general host metabolism
Security proteins contribute substantially to the shut-off of host macromolecular synthesis that occurs in response to infection with many picornaviruses. One could perhaps argue that such effects of security proteins contradict the key proposal of this Review that these proteins are dedicated to counter-defensive functions. However, as already mentioned, inflicting harm on their hosts does not bring viruses any benefits per se. The only reason (or at least, the main reason) why viruses evolved the ability to damage infected cells is their need to incapacitate the cellular defensive machinery. This machinery includes several specific mechanisms (such as innate and adaptive immunity), the implementation of which requires general cellular functions such as translation, transcription and controllable nucleocytoplasmic trafficking. Therefore, virus-induced impairment of these all-purpose metabolic functions can be regarded as a component of the viral counter-defensive strategy.
The effect of security proteins on cap-dependent translation of cellular mRNA is particularly important. 2Apro from diverse enteroviruses35,36,37,38 and Lpro from aphthoviruses12,39 cleave eukaryotic translation initiation factor eIF4G. Interestingly, erbovirus Lpro does not seem to cleave eIF4G and does not trigger translational shut-off13. Poly(A)-binding protein 1 (PABP1; also known as cytoplasmic PABP), a host protein that is also involved in translational control, is another target of enterovirus 2Apro (Refs 40, 41).
Security proteins can inhibit host translation by mechanisms other than proteolysis. Mutations of cardiovirus 2A (which is not a protease) alleviate virus-induced translational shut-off42,43. It is thought that association of cardiovirus 2A with ribosomes44,45, which seems to take place in the nucleolus46,47 and possibly occurs through the RNA-binding activity of 2A26, might contribute to preferential use of the internal ribosome entry site (IRES)-dependent viral templates. Cardiovirus L was also reported to mediate translational shut-off48. However, this effect could largely be due to L-mediated inhibition of nuclear export of mRNA rather than to inhibition of translation per se49,50. On the basis of ectopic expression experiments, hepatitis A virus 2A has also been implicated in inhibition of cap-dependent translation51, but the relevance of this observation is uncertain, as hepatitis A virus does not exert translational shut-off.
The effects of security proteins on cellular transcription are less well studied, although synthesis of the mRNA for cytokines and chemokines is inhibited in certain cases (see below). Individually expressed poliovirus 2Apro was reported to cleave the general transcription factors TATA-box-binding protein52 and cyclic AMP-responsive element-binding protein 1 (Ref. 38), but the biological significance of these effects is debatable52,53. Poliovirus 2Apro cleaves GEMIN3 (also known as DDX20), a protein that is involved in the formation of spliceosomes54. Cardiovirus 2A has also been implicated in virus-triggered inhibition of host transcription47, but this effect was not investigated further. Foot-and-mouth disease virus (FMDV) Lpro (Refs 55, 56) and human parechovirus 2A57 accumulate in the nucleus during the course of infection and as a result of ectopic expression, respectively, but their nuclear effects are unknown. Enterovirus 2Apro cleaves some cytoskeletal proteins, such as cytokeratin 8 (Ref. 58) and dystrophin, a protein that connects the cytoskeleton to the plasma membrane59.
The security proteins of several picornaviruses profoundly affect nucleocytoplasmic transport in infected cells. The targets of enterovirus 2Apro include nucleoporins, which are nuclear pore components that control nucleocytoplasmic exchange60,61. As a result, bidirectional passive diffusion through the nuclear envelope is facilitated. Another manifestation of the perturbation of intracellular trafficking is the suppression of nuclear export of mRNAs, ribosomal RNAs and U spliceosomal small nuclear RNAs62. Passive nucleocytoplasmic diffusion of proteins is also facilitated by the cardiovirus L protein63,64, and this effect seems to be caused by L-triggered phosphorylation of nucleoporins50,65,66.
Effects on innate immunity and viral pathogenicity
One major consequence of the effects of security proteins on host metabolism is the downregulation of innate immunity. FMDV Lpro suppresses interferon production56,67 and action68,69, largely through the inhibition of nuclear factor-κB-dependent transcription that is caused by the degradation of the p65 subunit of this transcription factor55,56. The transcription of genes encoding various cytokines and chemokines (including tumour necrosis factor (TNF; also known as TNFa), T cell-specific protein RANTES (also known as CCL5), myxovirus resistance protein 1 and interferon regulatory factor 7) is also suppressed by FMDV Lpro(Ref. 56).
Poliovirus reproduction, which is largely resistant to the effects of interferon, becomes interferon sensitive in 2Apro mutants. Introduction of the poliovirus 2Apro gene into interferon-sensitive EMCV facilitated its replication in cells pretreated with interferon70. The ability of rhinovirus 2Apro to cleave mitochondrial antiviral-signalling protein (MAVS; also known as VISA, CARDIF and IPS1), an intermediate in the interferon generation pathway, might contribute to the insensitivity of these viruses to interferons71. 2Apro also cleaves the catalytic subunit of DNA-dependent protein kinase, which, among other activities, is involved in the induction of pro-inflammatory cytokines72. Cardiovirus L also suppresses interferon production by affecting the activation of interferon regulatory factor 3, but the exact mechanism of this interference has not yet been identified32,50,73,74. TMEV L* was proposed to suppress the antiviral cytotoxic T cell response in TMEV-infected mice75,76.
In line with these observations, the functions of security proteins are less crucial for viral 'well-being' in hosts with innate immunity defects. Mutations in cardiovirus L48,77,78 or 2A79 or in FMDV Lpro(Ref. 56) decrease viral reproductive potential, but such mutations are less detrimental for viral growth in BHK-21 cells, which are deficient in interferon production, than in immune-competent cells. Similarly, cardiovirus mutants lacking L grow better in mice that have a deficient interferon system than in wild-type mice50,74. For the L*-expressing TMEV strains, functional L* is important for the ability of the virus to infect macrophages or microglia80,81.
One of the components of the innate immune response is apoptosis, which can potentially limit viral reproduction and spread, although certain viruses can subvert the apoptotic machinery to their benefit. Conflicting results have been reported on the relationship between the security proteins and the apoptotic machinery. Ectopic expression of enterovirus 2Apro triggers an apoptotic response38,82,83 and enhances the sensitivity of cells to the apoptogenic activity of tumour necrosis factor84. However, in the context of the whole genome, 2Apro possesses anti-apoptotic activity85,86. Similarly, expression of TMEV L in macrophage-like cells induces apoptosis, as occurs during TMEV infection of these cells87, whereas EMCV and mengovirus L are anti-apoptotic in the context of the whole virus (at least in HeLa cells, in which the virus itself elicits necrotic death)5. Whether this apparent discrepancy is a result of the use of different assays or hosts or of the intrinsic peculiarities of cardiovirus L proteins remains unknown. TMEV L* has also been reported to exhibit anti-apoptotic activity88.
Several illuminating examples strongly suggest that security proteins can markedly modulate viral pathogenicity. FMDV lacking L is highly attenuated89, and both L73,90 and L* (Refs 33, 81, 88) of TMEV strains BeAn 8386 and DA have been implicated in persistence in the central nervous system and in demyelinating disease. 2Apro of human coxsackievirus B4, which can cleave dystrophin in the cardiac muscle, seems to be involved in the pathogenesis of human acquired dilated cardiomyopathy59. Moreover, a major virulence determinant of swine vesicular disease virus, an enterovirus, has been mapped to 2Apro(Ref. 91).
Exchangeability and dispensability
Notwithstanding the low level of conservation of L proteins between EMCV and TMEV (Table 1), these proteins can be functionally exchanged with respect to their ability to inhibit interferon formation and to 'open' nuclear pores90. The replacement of L in the full-length cardiovirus genome with FMDV Lpro generates a virus that can overcome host defences more efficiently than its leaderless counterpart, although it has lower fitness92,93. Such interchangeability of structurally and biochemically distinct proteins attests to the similarity of their biological functions.
It is hardly by chance that viruses that encode long or multiple Ls tend to have a short 2A or lack 2A altogether (assuming that 2Asp is not a security protein), and vice versa (Fig. 1). For example, simian and porcine sapeloviruses harbour a long 2A and a short L, whereas avian sapelovirus encodes the longest L identified to date and the predicted 2A ORF is very short, if it encodes a protein at all. This tendency is consistent with the notion that L and 2A have similar roles in virus–host interactions. As already mentioned, some picornaviruses, for example, cosaviruses, encode no security proteins. Even viruses that do encode security proteins can, under certain circumstances, survive and replicate after these proteins have been inactivated or eliminated. Notably, cardiovirus L31,48,77,78 and L* (Refs 34, 81, 94) and FMDV Lpro(Ref. 95) are not essential for viral viability, and extended deletions in 2A of cardioviruses42,79 and hepatitis A virus96,97 do not kill these viruses. Furthermore, although some data suggest that poliovirus 2Apro has an essential replicative function98, recent experiments have demonstrated its dispensability for viral viability86.
Division of labour in picornavirus proteins
Although this Review focuses on the counter-defensive functions of security proteins, it should be kept in mind that other picornavirus proteins are also often engaged in similar functions. The targets of 3Cpro (or its proteolytically active precursors) might include proteins that are involved in innate immunity99,100,101,102. 2B and 3A are also important players in the virus–host struggle, being involved in the rearrangement of cytoplasmic membranes and in suppression of trafficking, secretion and antigen presentation on cellular plasma membranes84,103,104,105. 2C can also participate in some of these activities103. Even specialized proteins such as VPg106 and capsid proteins107 can sometimes assist in overcoming host defences. In all these cases, however, the 'security' functions are neither the main nor the conserved roles of these proteins.
Conversely, as well as representing a dedicated counter-defensive system, security proteins can be directly involved in viral reproduction. The products of the cleavage of eIF4G, and possibly of some other host proteins, by FMDV Lpro and enterovirus 2Apro can stimulate IRES-dependent cell-free translation108,109; the effect of 2Apro is partly the result of stabilization of the viral RNA110. The physiological relevance of these phenomena is unclear, however. The poliovirus IRES dysfunction that is caused by some mutations can be compensated for by mutations in 2A in a host-dependent manner111, suggesting the participation of host proteins. Cardiovirus L was also implicated in control of IRES activity, because EMCV RNA with deletions in the L-coding sequence exhibited a decrease in cell-free translatability31.
Poliovirus 2Apro was reported to stimulate strand initiation of negative-strand RNA, and this was seemingly independent of its effects on RNA stability and translation110. Deletion of a carboxy-terminal sequence from this protein does not substantially affect its protease activity but does inhibit RNA replication112. Deletion of the amino-terminal region notably, but incompletely, suppresses replication of the relevant replicon113. Similarly, deletions in 2A of Aichi virus (a kobuvirus) inhibit viral RNA replication114. However, the mechanisms responsible for these effects have not been elucidated, casting doubts on whether these 2A proteins affect viral replication directly or by modulating host cell activities.
A role for hepatitis A virus 2A in virion assembly and maturation has been established97,115. The primary co-translational cleavage of the viral polyprotein takes place between 2A and 2B and is accomplished by the viral proteinase 3Cpro (or its precursor), leaving the 2A sequence fused to the carboxyl terminus of capsid protein VP1 (Refs 116, 117). In immature virions, VP1 retains this 2A extension, which is eventually cleaved off by an unidentified enzyme118. The involvement of 2A in the maturation of some other picornaviruses cannot be excluded therefore. Notably, the primary co-translational scission of the cardiovirus polyprotein generates a fusion between 2A and the capsid protein precursor25.
Origins and evolution of security proteins
Obviously, it is not possible to construct a genealogical tree of either L or 2A, because they are unrelated. These proteins are not considered at all in the hypothesis on the origin of picorna-like viruses4, and only the short, translation-interrupting 2A peptides (that is, the 2Asp peptides) are briefly mentioned in the proposal on picornavirus classification119. There is no correlation between the nature (or even the presence) of security proteins and either the type of IRES that lies upstream of the L protein or the RdRP lineage (which is generally accepted as the most reliable indicator of viral relatedness) (Fig. 2).
Figure 2. Relationships between the presence of distinct security proteins and other evolutionary hallmarks of picornaviruses.
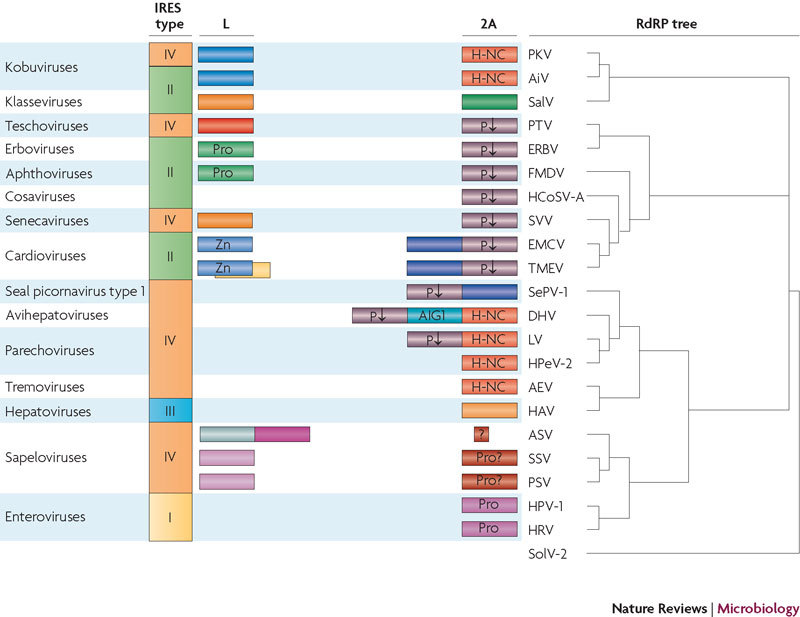
The distribution of security proteins among different viruses is not congruent with either the type of internal ribosome entry site (IRES; the key cis-acting element responsible for cap-independent translation of picornavirus RNAs) or the topology of the RNA-dependent RNA polymerase (RdRP) tree. For example, viruses harbouring type II IRESs can possess different L proteins (aphthoviruses and cardioviruses) or be devoid of this protein (cosaviruses). A similar situation occurs with viruses that use type IV IRESs. Conversely, different kobuviruses can possess unrelated IRESs. 2A proteins with the H-NC motif are present in viruses of distant RdRP lineages (kobuviruses on the one hand and avihepatoviruses, parechoviruses and tremoviruses on the other), but this motif is not shared by more closely related viruses (for example, it is present in tremoviruses but absent in hepatoviruses). The same pattern is characteristic of the 2A proteins that contain the NPG(P) motif (which interrupts translation at the indicated proline residue; shown by P↓). The closely related seal picornavirus type 1 (SePV-1), duck hepatitis A virus (DHV), Ljungan virus (LV) and human parechovirus type 2 (HPeV-2) each harbour a distinct 2A protein. Well-defined amino acid motifs are indicated. Viral RdRP protein sequences were taken from GenBank. Multiple alignments of protein sequences were constructed using CLUSTAL-X2. The RdRP tree was constructed by using MrBayes with default parameters. Solenopsis invicta virus 2 (SolV-2) was used as the outgroup. AEV, avian encephalomyelitis virus; AiV, Aichi virus; ASV, avian sapelovirus; EMCV, encephalomyocarditis virus; ERBV, equine rhinitis B virus; FMDV, foot-and-mouth disease virus; HAV, hepatitis A virus; HCoSV-A, human cosavirus A; HPV-1, human poliovirus type 1 str. Mahoney; HRV, human rhinovirus A101; PKV, porcine kobuvirus; Pro, protease; PSV, porcine sapelovirus; PTV, porcine teschovirus; SalV, Salivirus NG-J1; SSV, simian sapelovirus; SVV, Seneca Valley virus; TMEV, Theiler's murine encephalomyelitis virus; Zn, zinc finger.
The diversity of the security proteins and their absence from many picornaviruses suggest that they are independent and late evolutionary acquisitions5,120. It can be speculated that the most ancient of the 2A molecules are the 2Asp peptides, as hinted by their presence in most picornavirus genera, including those belonging to different lineages (Fig. 2). Interruption of polyprotein synthesis after translation of the capsid proteins might be advantageous for viral reproduction. It might, for example, facilitate proper protein folding, ensure optimal kinetics of protein synthesis or control the ratios of structural to non-structural proteins, which could theoretically be achieved by incomplete translational re-initiation at the second proline residue of the NPG(P) motif. It is worth noting that there is a difference in the translation factor requirements for translation of the EMCV polyprotein upstream and downstream of the 2A–2B boundary121.
Strikingly, 2Asp peptides — or more accurately, the DXEXNPG(P) motifs (where X is any amino acid) that are characteristic of these peptides — are found in some other picorna-like and unrelated viruses, where they seem to serve the same function122,123,124. Assuming that the acquisition of NPG(P) was an early event, this motif might have been lost by some parechoviruses (and other picornaviruses) at a certain step of evolution21. In contrast to picornaviruses, the acquisition of 2Asp by members of some other families of RNA viruses has been proposed to have occurred at a late stage122. The idea that these peptides might have originated in picornaviruses is an attractive hypothesis. The NPG(P) motif can also be found in several cellular proteins, but the current data do not allow researchers to determine whether the recoding ability of this motif was a viral or cellular invention.
The non-NPG(P)-containing regions of the 2A proteins are unrelated acquisitions. Cardiovirus 2A possesses these additional moieties in its amino-terminal region, whereas other viruses of this subset (parechoviruses, avihepatoviruses and seal picornavirus type 1) contain these moieties in their carboxy-terminal regions (Fig. 1). This may suggest that these moieties were acquired after the NPG(P) motif. In avihepatoviruses and some parechoviruses, these newly acquired sequences harbour the H-NC motif, which is also present in the NPG(P)-lacking 2A proteins of other parechoviruses, kobuviruses and tremoviruses (that is, viruses of different RdRP lineages) (Fig. 2). A plausible hypothesis is that H-NC-containing proteins from cellular organisms were hijacked by certain picornaviruses on several independent occasions, but the possibility that 2Asp peptides were formed by deletions from larger 2A proteins120 cannot be ruled out.
Taking into account the chymotrypsin-like fold that is found in enterovirus 2Apro, it is reasonable to assume that this protein was derived from picornavirus 3Cpro or a cellular protease125. A similar assumption could perhaps be made for the putative sapelovirus 2A protease. There are no obvious clues to the possible origins of the 2A proteins of hepatoviruses and klasseviruses or of the non-NPG(P) moieties of the 2A proteins of cardioviruses and seal picornavirus type 1.
With regard to the picornavirus L proteins, one hypothesis is that the papain-like Lpro proteases of aphthoviruses and erboviruses originated from cellular enzymes. No obvious relatives of other L proteins can currently be identified among cellular or viral proteins. Only the origin of cardiovirus L* seems certain: the first L* probably came into being accidentally, by translation of an alternative reading frame, and was then shaped by mutations preserving the functional integrity of L, VP4 and VP2. It is worth noting that recently identified human TMEV-like cardioviruses lack this alternative reading frame126.
Theoretically, several mechanisms could underlie the acquisition of security proteins. For example, one possibility is that viral genes were duplicated and subsequently substantially modified (such a scenario can be imagined for the origin of enterovirus 2Apro(Ref. 125)). Other scenarios are: recombination with viral or cellular RNAs encoding related proteins such as proteases; conversion of non-coding RNA sequences into coding sequences (which could occur through different mechanisms, such as interspecies recombination127 (V.I.A., A.P.G., E. V. Khitrina and W. J. Melchers, unpublished observations), or introduction or activation of an upstream in-frame AUG codon); frame shifting120 (or double frame shifting, in the case of 2A proteins); and using an alternative reading frame. Unfortunately, we can only pinpoint a definite mechanism for the case of L*, for which the last mechanism has obviously been operative.
The acquisition of security proteins, by whatever mechanism, required that the new 'additions' did not interfere with the function of the adjacent viral proteins — VP4 (or VP0) in the case of L, and VP1 and 2B in the case of 2A. The new proteins should either be separated from these neighbours (by self-proteolysis, the action of other viral proteases or translation interruption) or, if they remain fused, they should not impair the functions of these neighbours (as is the case with the hepatitis A virus VP1–2A fusion).
A separate issue is the evolution of the security proteins themselves. It was proposed that the L proteins of TMEV and EMCV diverged during evolution to adapt to the different replication fitnesses of these viruses90. The data are too scarce, however, for any generalizations at this point.
Conclusions
L and 2A constitute a distinct and remarkable set of picornavirus proteins. They exhibit striking structural and biochemical diversity but (with the exception of the 2Asp peptides) accomplish similar biological functions by counteracting host defensive reactions. However, their abilities to solve similar problems might involve fundamentally different molecular mechanisms. This is illustrated in Fig. 3, which summarizes the properties of the three best studied security proteins. The biological functions of enterovirus 2Apro and cardiovirus L are strikingly similar: inhibition of host macromolecular synthesis, permeabilization of the nuclear envelope and inhibition of active nucleocytoplasmic transport, suppression of specific innate immunity mechanisms and control of the apoptotic machinery of the host cells. No less striking is the difference in the mechanisms by which the two proteins achieve these goals. Conversely, aphthovirus Lpro can solve only a subset of these problems. An intriguing question is how, and whether, the diverse security functions exhibited by a given protein (Fig. 3) are related to each other. The fact that mutations inactivating one function usually also impair other functions suggests the existence of a common upstream target.
Figure 3. Major biological functions of the best studied but unrelated security proteins.
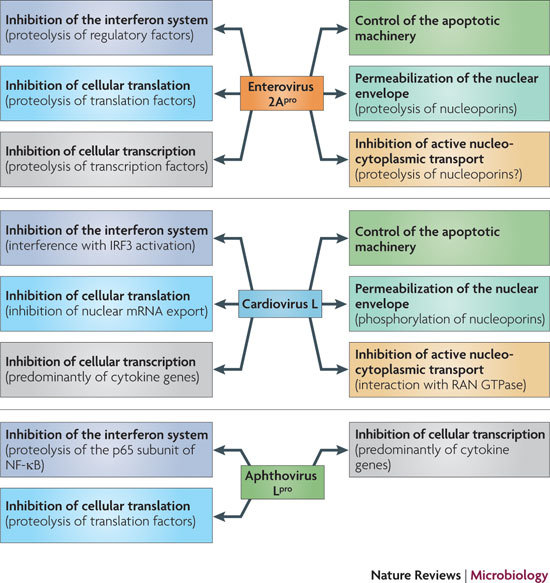
There is a striking similarity between the functional activities of the enterovirus 2A protease (2Apro) and the cardiovirus leader protein (L), but the underlying mechanisms by which these functions are carried out are fundamentally different. By contrast, the known functional activities of aphthovirus L, which is a protease (Lpro), seem to be more limited. IRF3, interferon regulatory factor 3; NF-κB, nuclear factor-κB.
Most of the picornavirus L and 2A proteins still await researchers' attention. Nevertheless, the data discussed here allow us to provisionally assign security functions even to those L and 2A proteins that have not yet been characterized. Indeed, L and 2A do not definitely belong to the set of essential reproductive proteins that ensure translation and replication of the viral genome (although 2A of hepatitis A virus assists virion maturation).
We propose that the concept of security proteins is of general relevance and can be applied to viruses other than picornaviruses. The hallmarks of these proteins are as follows: structural and biochemical unrelatedness or even absence in related viruses; the dispensability of the entire protein or its functional domains for viral viability; and, for mutated versions of the proteins, fewer detrimental effects on viral reproduction in immune-compromised hosts than in immune-competent hosts. Possessing one of these features would make a viral protein a good candidate security protein, whereas a combination of these features would probably confirm this designation.
Viruses with large DNA genomes possess impressive arsenals of security proteins. The complement of security proteins is much more limited in RNA viruses but is sufficient for their evolutionary success. There are also tentative examples of security proteins in non-picornavirus RNA viruses. Coronaviruses have several so-called accessory proteins, which are neither conserved nor essential and exhibit the capacity to suppress host innate immunity by a range of mechanisms128. Some, but not all, flaviviruses use ribosome frame shifting to express a non-essential non-structural protein, NS1′, mutational inactivation of which results in viral attenuation129. The NSS protein of the Rift Valley fever phlebovirus suppresses the interferon system, and its inactivation does not kill the virus but attenuates its pathogenicity130. This list can readily be extended.
The spectrum of picornavirus-induced diseases is extraordinarily broad. The reasons for this variability are poorly understood, although receptor compatibility and effects on viral protein and RNA synthesis that are caused by differences in the availability of host factors are surely important contributors. However, the interaction between host defences and viral counter-defence is certainly one of the key factors underlying the pathogenicity of picornaviruses and other viruses. This interaction cannot be fully understood without elucidation of the roles of the security proteins. Treatment and prevention of viral diseases may also markedly benefit from such elucidation. The study of security proteins is therefore an underdeveloped but highly promising research area.
Box 1 | Picornavirus genes and proteins.
Picornaviruses possess a single-stranded positive-sense RNA genome (see the figure) comprising approximately 7,000–9,000 nucleotides. The RNA is packaged into an icosahedral capsid usually composed of four distinct proteins (VP1–VP4), but in some viruses VP4 and VP2 are fused (forming VP0). After productive contacts with specific cellular receptors, which differ between viruses, the genome is uncoated and enters the cytoplasm, where the main steps of viral reproduction occur. The viral RNA, which contains a single ORF (with one exception described in the main text), is translated in a cap-independent manner into a 2,200–2,500 amino acid polypeptide, which is eventually processed by limited proteolysis into a dozen 'mature' proteins. These proteins include: capsid proteins; an RNA-dependent RNA polymerase (3Dpol); a protein (VPg, or 3B) that serves as a primer for the initiation of RNA synthesis; an ATPase with a conserved superfamily 3 helicase motif (2CATPase) and an essential but poorly defined role in viral RNA replication; a chymotrypsin-like protease (3Cpro), which, as the mature protein or as a precursor, is a major factor in polyprotein processing; two hydrophobic membrane-binding proteins (2B and 3A) that participate in the generation of a virus-friendly environment; and, flanking the precursor of the capsid proteins, one or two highly variable proteins (L and 2A), the structure and functions of which are the subject of this Review. The entire coding region of the RNA is surrounded with untranslated regions (UTRs) harbouring regulatory elements that control viral translation (for example, the internal ribosome entry site (IRES)) and replication (the origins oriL and oriR), and this entire RNA sequence is flanked by the covalently linked viral protein VPg and a poly(A) tract at the 5′ and 3′ termini, respectively. There are four structurally and functionally distinct types of IRES131 and many types of oriL and oriR.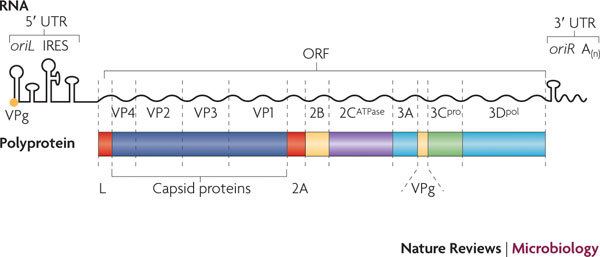
Box 2 | Picornavirus taxonomy.
Members of the Picornaviridae family are small (∼30 nm), non-enveloped animal viruses with similar genome organizations and mechanisms of reproduction (Box 1). This family contains important human and animal pathogens, studies of which have made crucial contributions to our understanding of the molecular biology of viruses and the mechanisms of virus–cell interactions. The first animal virus (foot-and-mouth disease virus; FMDV) and the first human virus (poliovirus) to be discovered belong to this family.
The current official classification of picornaviruses (see the International Committee on Taxonomy of Viruses) includes 12 genera: Aphthovirus (for example, FMDV), Avihepatovirus (for example, duck hepatitis A virus), Cardiovirus (including important model viruses such as encephalomyocarditis virus, mengovirus and Theiler's murine encephalomyelitis virus, as well as human and animal pathogenic viruses such as the newly recognized Saffold virus), Enterovirus (a large genus that includes poliovirus and many other human and animal pathogenic viruses, such as coxsackieviruses, echoviruses and other 'numbered' enteroviruses, as well as rhinoviruses, the causative agents of the common cold), Erbovirus (equine rhinitis B virus), Hepatovirus (hepatitis A virus), Kobuvirus(for example, Aichi virus, which induces gastroenteritis in humans, as well as some animal pathogens), Parechovirus (the causative agents of various human diseases, particularly in children; also isolated from rodents), Sapelovirus (including an avian, a porcine and a simian pathogen), Senecavirus (Seneca Valley virus, a candidate oncolytic agent), Teschovirus (a porcine pathogen) and Tremovirus (avian encephalomyelitis virus). In addition, there are two candidate genera, not yet officially approved, that are associated with human diarrhoea: Cosavirus and Klassevirus. Finally, seal picornavirus type 1 is a distinct virus not assigned to any of the above genera. Several new picornavirus genera have been identified recently, and it seems plausible that this number will grow, particularly with the breakthroughs in sequencing techniques and the achievements of metagenomics.
Supplementary information
(PDF 171 kb)
Acknowledgements
We thank F. van Kuppeveld for critical reading of this manuscript. Recent research in the authors' laboratory was supported by grants from the Russian Foundation for Basic Research (RFBR), the Scientific School Support Program and The Netherlands Organization for Scientific Research–RFBR (NWO–RFBR).
Glossary
- Cap-dependent translation
The mode of translation for which initiation is dependent on the presence of the so-called cap structure (m7GpppN) at the 5′ end of an mRNA. Specific cap-binding proteins (translation initiation factors) recruit the ribosome to the 5′ end of the mRNA, and the ribosome scans the template until it encounters the initiation codon. This translation mode is exploited by most mRNAs in eukaryotic cells.
- Poly(A)-binding protein 1
A protein that binds to the 3′ poly(A) tail of eukaryotic mRNAs on the one hand and to cap-dependent translation initiation factors on the other hand. This dual binding results in a non-covalent circularization of the mRNA template, which is accompanied by a significant increase in translation efficiency.
- Spliceosome
A complex that is formed of several small nuclear ribonucleoproteins and is involved in splicing.
- U spliceosomal small nuclear RNA
An RNA component of a spliceosomal small nuclear ribonucleoprotein.
Biographies
Vadim I. Agol graduated from the First Moscow Institute of Medicine, USSR, in 1951. He earned a Cand.Sc. (equivalent to Ph.D.) in biochemistry in 1954 for the work done while he was an undergraduate and a D.Sc. in virology in 1967. He joined the Laboratory of Biochemistry of the Institute for Poliomyelitis Research (now the M.P. Chumakov Institute of Poliomyelitis and Viral Encephalitides of the Russian Academy of Medical Sciences, Moscow, Russia) in 1956 as a junior researcher and became head of this laboratory in 1961 (until 2009). He is now a chief research scientist at the same institute. In 1963, he participated in the organization of the Chair of Virology at the Moscow State University, where he started teaching as a dozent and then became a professor (until present). In 1965 he established the Department of Virus-Cell Interactions at what is now known as the A.N.Belozersky Institute of Physico-Chemical Biology, Moscow State University. His main scientific interests are focused on the molecular biology of RNA viruses, in particular picornaviruses. He has published numerous papers on the translation, replication, recombination, virulence, molecular epidemiology and evolution of these viruses as well as on different aspects of virus–host cell interactions.
Anatoly P. Gmyl joined the Laboratory of Biochemistry (headed by V. Agol) of the M.P. Chumakov Institute of Poliomyelitis and Viral Encephalitides in 1986 as an undergraduate of the Moscow Institute of Physics and Technology, and he continues to work there today. He earned his M.S. and Ph.D. in 1989 and 1996, respectively. For several years following this, he participated in projects studying various aspects of cap-independent translation of picornavirus RNAs. Since then, his major interests have been focused on replication and evolution of RNA-containing viruses as well as on virus–host cell interactions. In particular, he has made substantial contributions to the problem of RNA recombination. Since 2009, he has been Head of the Laboratory of Biochemistry.
Related links
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
References
- 1.Lai MM. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 2005;79:7951–7958. doi: 10.1128/JVI.79.13.7951-7958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solorzano A, Rodríguez-Cousiño N, Esteban R, Fujimura T. Persistent yeast single-stranded RNA viruses exist in vivo as genomic RNA·RNA polymerase complexes in 1:1 stoichiometry. J. Biol. Chem. 2000;275:26428–26435. doi: 10.1074/jbc.M002281200. [DOI] [PubMed] [Google Scholar]
- 3.Wang AL, Yang HM, Shen KA, Wang CC. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc. Natl Acad. Sci. USA. 1993;90:8595–8599. doi: 10.1073/pnas.90.18.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV, Wolf YI, Nagasaki K, Dolja VV. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nature Rev. Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 5.Romanova LI, et al. Antiapoptotic activity of the cardiovirus leader protein, a viral “security” protein. J. Virol. 2009;83:7273–7284. doi: 10.1128/JVI.00467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grubman MJ, Moraes MP, Diaz-San Segundo F, Pena L, de los Santos T. Evading the host immune response: how foot-and-mouth disease virus has become an effective pathogen. FEMS Immunol. Med. Microbiol. 2008;53:8–17. doi: 10.1111/j.1574-695X.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 7.Toyoda H, et al. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 8.Bazan JF, Fletterick RJ. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl Acad. Sci. USA. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Koonin EV, Lai MM. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck E, Forss S, Strebel K, Cattaneo R, Feil G. Structure of the FMDV translation initiation site and of the structural proteins. Nucleic Acids Res. 1983;11:7873–7885. doi: 10.1093/nar/11.22.7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strebel K, Beck E. A second protease of foot-and-mouth disease virus. J. Virol. 1986;58:893–899. doi: 10.1128/jvi.58.3.893-899.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medina M, Domingo E, Brangwyn JK, Belsham GJ. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 13.Hinton TM, Ross-Smith N, Warner S, Belsham GJ, Crabb BS. Conservation of L and 3C proteinase activities across distantly related aphthoviruses. J. Gen. Virol. 2002;83:3111–3121. doi: 10.1099/0022-1317-83-12-3111. [DOI] [PubMed] [Google Scholar]
- 14.Oberste MS, Maher K, Pallansch MA. Genomic evidence that simian virus 2 and six other simian picornaviruses represent a new genus in Picornaviridae. Virology. 2003;314:283–293. doi: 10.1016/S0042-6822(03)00420-3. [DOI] [PubMed] [Google Scholar]
- 15.Ryan MD, King AMQ, Thomas GP. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 1991;72:2727–2732. doi: 10.1099/0022-1317-72-11-2727. [DOI] [PubMed] [Google Scholar]
- 16.Doronina VA, et al. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wutz G, et al. Equine rhinovirus serotypes 1 and 2: relationship to each other and to aphthoviruses and cardioviruses. J. Gen. Virol. 1996;77:1719–1730. doi: 10.1099/0022-1317-77-8-1719. [DOI] [PubMed] [Google Scholar]
- 18.Doherty M, Todd D, McFerran N, Hoey EM. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J. Gen. Virol. 1999;80:1929–1941. doi: 10.1099/0022-1317-80-8-1929. [DOI] [PubMed] [Google Scholar]
- 19.Hales LM, et al. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J. Gen. Virol. 2008;89:1265–1275. doi: 10.1099/vir.0.83570-0. [DOI] [PubMed] [Google Scholar]
- 20.Kapoor A, et al. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc. Natl Acad. Sci. USA. 2008;105:20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson S, Niklasson B, Maizel J, Gorbalenya AE, Lindberg AM. Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J. Virol. 2002;76:8920–8930. doi: 10.1128/JVI.76.17.8920-8930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding C, Zhang D. Molecular analysis of duck hepatitis virus type 1. Virology. 2007;361:9–17. doi: 10.1016/j.virol.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Kapoor A, et al. A highly divergent picornavirus in a marine mammal. J. Virol. 2008;82:311–320. doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly MLL, Gani D, Flint M, Monoghan S, Ryan MD. The cleavage activity of aphtho- and cardiovirus 2A proteins. J. Gen. Virol. 1997;78:13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]
- 25.Kazachkov YA, Svitkin YV, Agol VI. The position of polypeptide G on the encephalomyocarditis virus polyprotein cleavage map. FEBS Lett. 1983;154:161–165. doi: 10.1016/0014-5793(83)80895-3. [DOI] [PubMed] [Google Scholar]
- 26.Gorbalenya AE, Chumakov KM, Agol VI. RNA-binding properties of nonstructural polypeptide G of encephalomyocarditis virus. Virology. 1978;88:183–185. doi: 10.1016/0042-6822(78)90122-8. [DOI] [PubMed] [Google Scholar]
- 27.Hughes PJ, Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- 28.Kazachkov YA, et al. Leader polypeptides encoded in the 5′-region of the encephalomyocarditis virus genome. FEBS Lett. 1982;141:153–156. doi: 10.1016/0014-5793(82)80035-5. [DOI] [PubMed] [Google Scholar]
- 29.Chen HH, Kong WP, Roos RP. The leader peptide of Theiler's murine encephalomyelitis virus is a zinc-binding protein. J. Virol. 1995;69:8076–8078. doi: 10.1128/jvi.69.12.8076-8078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornilescu CC, Porter FW, Zhao KQ, Palmenberg AC, Markley JL. NMR structure of the mengovirus Leader protein zinc-finger domain. FEBS Lett. 2008;582:896–900. doi: 10.1016/j.febslet.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 31.Dvorak CM, et al. Leader protein of encephalomyocarditis virus binds zinc, is phosphorylated during viral infection, and affects the efficiency of genome translation. Virology. 2001;290:261–271. doi: 10.1006/viro.2001.1193. [DOI] [PubMed] [Google Scholar]
- 32.Zoll J, Melchers WJ, Galama JM, van Kuppeveld FJ. The mengovirus leader protein suppresses α/β interferon production by inhibition of the iron/ferritin-mediated activation of NF-κB. J. Virol. 2002;76:9664–9672. doi: 10.1128/JVI.76.19.9664-9672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong WP, Roos RP. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J. Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen HH, Kong WP, Zhang L, Ward PL, Roos RP. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nature Med. 1995;1:927–931. doi: 10.1038/nm0995-927. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein HD, Sonenberg N, Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol. Cell. Biol. 1985;5:2913–2923. doi: 10.1128/MCB.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kräusslich HG, Nicklin MJ, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liebig H, et al. Purification of two picornaviral 2A proteinases: interaction with eIF4γ and influence on translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- 38.Chau DH, et al. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis. 2007;12:513–524. doi: 10.1007/s10495-006-0013-0. [DOI] [PubMed] [Google Scholar]
- 39.Devaney MA, Vakharia VN, Lloyd RE, Ehrenfeld E, Grubman MJ. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 1988;62:4407–4409. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joachims M, van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerekatte V, et al. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoll J, van Kuppeveld FJ, Galama JM, Melchers WJ. Genetic analysis of mengovirus protein 2A: its function in polyprotein processing and virus reproduction. J. Gen. Virol. 1998;79:17–25. doi: 10.1099/0022-1317-79-1-17. [DOI] [PubMed] [Google Scholar]
- 43.Svitkin YV, Hahn H, Gingras AC, Palmenberg AC, Sonenberg N. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J. Virol. 1998;72:5811–5819. doi: 10.1128/jvi.72.7.5811-5819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medvedkina OA, Scarlat IV, Kalinina NO, Agol VI. Virus-specific proteins associated with ribosomes of Krebs II cells infected with encephalomyocarditis virus. FEBS Lett. 1974;39:4–8. doi: 10.1016/0014-5793(74)80003-7. [DOI] [PubMed] [Google Scholar]
- 45.Groppo R, Palmenberg AC. Cardiovirus 2A protein associates with 40S but not 80S ribosome subunits during infection. J. Virol. 2007;81:13067–13074. doi: 10.1128/JVI.00185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aminev AG, Amineva SP, Palmenberg AC. Encephalomyocarditis viral protein 2A localizes to nucleoli and inhibits cap-dependent mRNA translation. Virus Res. 2003;95:45–57. doi: 10.1016/S0168-1702(03)00162-X. [DOI] [PubMed] [Google Scholar]
- 47.Aminev AG, Amineva. SP, Palmenberg AC. Encephalomyocarditis virus (EMCV) proteins 2A and 3BCD localize to nuclei and inhibit cellular mRNA transcription but not rRNA transcription. Virus Res. 2003;95:59–73. doi: 10.1016/S0168-1702(03)00163-1. [DOI] [PubMed] [Google Scholar]
- 48.Zoll J, Galama JM, van Kuppeveld FJ, Melchers WJ. Mengovirus leader is involved in the inhibition of host cell protein synthesis. J. Virol. 1996;70:4948–4952. doi: 10.1128/jvi.70.8.4948-4952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porter FW, Bochkov YA, Albee AJ, Wiese C, Palmenberg AC. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl Acad. Sci. USA. 2006;103:12417–12422. doi: 10.1073/pnas.0605375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ricour C, et al. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein. J. Gen. Virol. 2009;90:177–186. doi: 10.1099/vir.0.005678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maltese E, et al. Inhibition of cap-dependent gene expression induced by protein 2A of hepatitis A virus. J. Gen. Virol. 2000;81:1373–1381. doi: 10.1099/0022-1317-81-5-1373. [DOI] [PubMed] [Google Scholar]
- 52.Yalamanchili P, Banerjee R, Dasgupta A. Poliovirus-encoded protease 2APro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J. Virol. 1997;71:6881–6886. doi: 10.1128/jvi.71.9.6881-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies MV, Pelletier J, Meerovitch K, Sonenberg N, Kaufman RJ. The effect of poliovirus proteinase 2Apro expression on cellular metabolism. Inhibition of DNA replication, RNA polymerase II transcription, and translation. J. Biol. Chem. 1991;266:14714–14720. [PubMed] [Google Scholar]
- 54.Almstead LL, Sarnow P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes Dev. 2007;21:1086–1097. doi: 10.1101/gad.1535607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de los Santos T, Diaz-San Segundo F, Grubman MJ. Degradation of nuclear factor kappa B during foot-and-mouth disease virus infection. J. Virol. 2007;81:12803–12815. doi: 10.1128/JVI.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de los Santos T, et al. A conserved domain in the leader proteinase of foot-and-mouth disease virus is required for proper subcellular localization and function. J. Virol. 2009;83:1800–1810. doi: 10.1128/JVI.02112-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krogerus C, Samuilova O, Poyry T, Jokitalo E, Hyypia T. Intracellular localization and effects of individually expressed human parechovirus 1 non-structural proteins. J. Gen. Virol. 2007;88:831–841. doi: 10.1099/vir.0.82201-0. [DOI] [PubMed] [Google Scholar]
- 58.Seipelt J, Liebig HD, Sommergruber W, Gerner C, Kuechler E. 2A proteinase of human rhinovirus cleaves cytokeratin 8 in infected HeLa cells. J. Biol. Chem. 2000;275:20084–20089. doi: 10.1074/jbc.275.26.20084. [DOI] [PubMed] [Google Scholar]
- 59.Badorff C, et al. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nature Med. 1999;5:320–326. doi: 10.1038/6543. [DOI] [PubMed] [Google Scholar]
- 60.Belov GA, et al. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 2004;78:10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park N, Katikaneni P, Skern T, Gustin KE. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 2008;82:1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castelló A, Izquierdo JM, Welnowska E, Carrasco L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J. Cell Sci. 2009;122:3799–3809. doi: 10.1242/jcs.055988. [DOI] [PubMed] [Google Scholar]
- 63.Delhaye S, van Pesch V, Michiels T. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 2004;78:4357–4362. doi: 10.1128/JVI.78.8.4357-4362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lidsky PV, et al. Nucleo-cytoplasmic traffic disorder induced by cardioviruses. J. Virol. 2006;80:2705–2717. doi: 10.1128/JVI.80.6.2705-2717.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bardina MV, et al. Mengovirus-induced rearrangement of the nuclear pore complex: hijacking cellular phosphorylation machinery. J. Virol. 2009;83:3150–3161. doi: 10.1128/JVI.01456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Porter FW, Palmenberg AC. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 2009;83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chinsangaram J, Piccone ME, Grubman MJ. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 1999;73:9891–9898. doi: 10.1128/jvi.73.12.9891-9898.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chinsangaram J, Koster M, Grubman MJ. Inhibition of L-deleted foot-and-mouth disease virus replication by α/β interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 2001;75:5498–5503. doi: 10.1128/JVI.75.12.5498-5503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de los Santos T, de Avila Botton S, Weiblen R, Grubman MJ. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 2006;80:1906–1914. doi: 10.1128/JVI.80.4.1906-1914.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morrison JM, Racaniello VR. Proteinase 2Apro is essential for enterovirus replication in type I interferon-treated cells. J. Virol. 2009;83:4412–4422. doi: 10.1128/JVI.02177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drahos J, Racaniello VR. Cleavage of IPS-1 in cells infected with human rhinovirus. J. Virol. 2009;83:11581–11587. doi: 10.1128/JVI.01490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graham KL, et al. Proteolytic cleavage of the catalytic subunit of DNA-dependent protein kinase during poliovirus infection. J. Virol. 2004;78:6313–6321. doi: 10.1128/JVI.78.12.6313-6321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Pesch V, van Eyll O, Michiels T. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 2001;75:7811–7817. doi: 10.1128/JVI.75.17.7811-7817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hato SV, et al. The mengovirus leader protein blocks interferon-a/b gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 2007;9:2921–2930. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- 75.Lin X, Roos RP, Pease LR, Wettstein P, Rodriguez M. A Theiler's virus alternatively initiated protein inhibits the generation of H-2K-restricted virus-specific cytotoxicity. J. Immunol. 1999;162:17–24. [PubMed] [Google Scholar]
- 76.Lin X, Ma X, Rodriguez M, Roos RP. CD4 T cells are important for clearance of the DA strain of TMEV from the central nervous system of SJL/J mice. Int. Immunol. 2004;16:1237–1240. doi: 10.1093/intimm/dxh125. [DOI] [PubMed] [Google Scholar]
- 77.Kong WP, Ghadge GD, Roos RP. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc. Natl Acad. Sci. USA. 1994;91:1796–1800. doi: 10.1073/pnas.91.5.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calenoff MA, Badshah CS, Dal Canto MC, Lipton HL, Rundell MK. The leader polypeptide of Theiler's virus is essential for neurovirulence but not for virus growth in BHK cells. J. Virol. 1995;69:5544–5549. doi: 10.1128/jvi.69.9.5544-5549.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michiels T, Dejong V, Rodrigus R, Shaw-Jackson C. Protein 2A is not required for Theiler's virus replication. J. Virol. 1997;71:9549–9556. doi: 10.1128/jvi.71.12.9549-9556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takata H, et al. L* protein of the DA strain of Theiler's murine encephalomyelitis virus is important for virus growth in a murine macrophage-like cell line. J. Virol. 1998;72:4950–4955. doi: 10.1128/jvi.72.6.4950-4955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Eyll O, Michiels T. Influence of the Theiler's virus L* protein on macrophage infection, viral persistence, and neurovirulence. J. Virol. 2000;74:9071–9077. doi: 10.1128/JVI.74.19.9071-9077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goldstaub D, et al. Poliovirus 2A protease induces apoptotic cell death. Mol. Cell. Biol. 2000;20:1271–1277. doi: 10.1128/MCB.20.4.1271-1277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuo RL, Kung SH, Hsu YY, Liu WT. Infection with enterovirus 71 or expression of its 2A protease induces apoptotic cell death. J. Gen. Virol. 2002;83:1367–1376. doi: 10.1099/0022-1317-83-6-1367. [DOI] [PubMed] [Google Scholar]
- 84.Neznanov N, et al. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 2001;75:10409–10420. doi: 10.1128/JVI.75.21.10409-10420.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burgon TB, Jenkins JA, Deitz SB, Spagnolo JF, Kirkegaard K. Bypass suppression of small-plaque phenotypes by mutation in poliovirus 2A that enhances apoptosis. J. Virol. 2009;83:10129–10139. doi: 10.1128/JVI.00642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Igarashi H, et al. 2Aprotease is not a prerequisite for poliovirus replication. J. Virol. 2010;84:5947–5957. doi: 10.1128/JVI.02575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fan J, Son KN, Arslan SY, Liang Z, Lipton HL. Theiler's murine encephalomyelitis virus leader protein is the only nonstructural protein tested that induces apoptosis when transfected into mammalian cells. J. Virol. 2009;83:6546–6553. doi: 10.1128/JVI.00353-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghadge GD, Ma L, Sato S, Kim J, Roos RP. A protein critical for a Theiler's virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J. Virol. 1998;72:8605–8612. doi: 10.1128/jvi.72.11.8605-8612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown CC, Piccone ME, Mason PW, McKenna TS, Grubman MJ. Pathogenesis of wild-type and leaderless foot-and-mouth disease virus in cattle. J. Virol. 1996;70:5638–5641. doi: 10.1128/jvi.70.8.5638-5641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paul S, Michiels T. Cardiovirus leader proteins are functionally interchangeable and have evolved to adapt to virus replication fitness. J. Gen. Virol. 2006;87:1237–1246. doi: 10.1099/vir.0.81642-0. [DOI] [PubMed] [Google Scholar]
- 91.Sakoda Y, Ross-Smith N, Inoue T, Belsham GJ. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 2001;75:10643–10650. doi: 10.1128/JVI.75.22.10643-10650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Piccone ME, Chen HH, Roos RP, Grubman MJ. Construction of a chimeric Theiler's murine encephalomyelitis virus containing the leader gene of foot-and-mouth disease virus. Virology. 1996;226:135–139. doi: 10.1006/viro.1996.0637. [DOI] [PubMed] [Google Scholar]
- 93.Hato SV, et al. Differential IFN-α/β production suppressing capacities of the leader proteins of mengovirus and foot-and-mouth disease virus. Cell. Microbiol. 2010;12:310–317. doi: 10.1111/j.1462-5822.2009.01395.x. [DOI] [PubMed] [Google Scholar]
- 94.Stavrou S, et al. Theiler's murine encephalomyelitis virus (TMEV) L* amino acid position 93 is important for virus persistence and virus-induced demyelination. J. Virol. 2010;84:1348–1354. doi: 10.1128/JVI.01585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piccone ME, Rieder E, Mason PW, Grubman MJ. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. J. Virol. 1995;69:5376–5382. doi: 10.1128/jvi.69.9.5376-5382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harmon S, Emerson SU, Huang YK, Summers DF, Ehrenfeld E. Hepatitis A viruses with deletions in the 2A gene are infectious in cultured cells and marmosets. J. Virol. 1995;69:5576–5581. doi: 10.1128/jvi.69.9.5576-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cohen L, Bénichou D, Martin A. Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis. J. Virol. 2002;76:7495–7505. doi: 10.1128/JVI.76.15.7495-7505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Molla A, Paul AV, Schmid M, Jang SK, Wimmer E. Studies on dicistronic polioviruses implicate viral proteinase 2Apro in RNA replication. Virology. 1993;196:739–747. doi: 10.1006/viro.1993.1531. [DOI] [PubMed] [Google Scholar]
- 99.Dasgupta A, et al. Molecular Biology of Picornaviruses. 2002. pp. 321–333. [Google Scholar]
- 100.Neznanov N, et al. Proteolytic cleavage of the p65-RelA subunit of NF-κB during poliovirus infection. J. Biol. Chem. 2005;280:24153–24158. doi: 10.1074/jbc.M502303200. [DOI] [PubMed] [Google Scholar]
- 101.Yang Y, et al. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl Acad. Sci. USA. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barral PM, Sarkar D, Fisher PB, Racaniello VR. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Egger D, Gossert R, Bienz K. Molecular Biology of Picornaviruses. 2002. [Google Scholar]
- 104.Doedens JR, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kemball CC, et al. Coxsackievirus B3 inhibits antigen presentation in vivo, exerting a profound and selective effect on the MHC class I pathway. PLoS Pathog. 2009;5:e1000618. doi: 10.1371/journal.ppat.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pacheco JM, Henry TM, O'Donnell VK, Gregory JB, Mason PW. Role of nonstructural proteins 3A and 3B in host range and pathogenicity of foot-and-mouth disease virus. J. Virol. 2003;77:13017–13027. doi: 10.1128/JVI.77.24.13017-13027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Autret A, et al. Early phosphatidylinositol 3-kinase/Akt pathway activation limits poliovirus-induced JNK-mediated cell death. J. Virol. 2008;82:3796–3802. doi: 10.1128/JVI.02020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ziegler E, Borman AM, Kirchweger R, Skern T, Kean KM. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ohlmann T, Rau M, Pain VM, Morley SJ. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. doi: 10.1002/j.1460-2075.1996.tb00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jurgens CK, et al. 2Apro is a multifunctional protein that regulates the stability, translation and replication of poliovirus RNA. Virology. 2006;345:346–357. doi: 10.1016/j.virol.2005.09.067. [DOI] [PubMed] [Google Scholar]
- 111.Rowe A, Ferguson GL, Minor PD, Macadam AJ. Coding changes in the poliovirus protease 2A compensate for 5′NCR domain V disruptions in a cell-specific manner. Virology. 2000;269:284–293. doi: 10.1006/viro.2000.0244. [DOI] [PubMed] [Google Scholar]
- 112.Li X, Lu HH, Mueller S, Wimmer E. The C-terminal residues of poliovirus proteinase 2Apro are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. J. Gen. Virol. 2001;82:397–408. doi: 10.1099/0022-1317-82-2-397. [DOI] [PubMed] [Google Scholar]
- 113.Collis PS, O'Donnell BJ, Barton DJ, Rogers JA, Flanegan JB. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 1992;66:6480–6488. doi: 10.1128/jvi.66.11.6480-6488.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sasaki J, Taniguchi K. Aichi virus 2A protein is involved in viral RNA replication. J. Virol. 2008;82:9765–9769. doi: 10.1128/JVI.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Probst C, Jecht M, Gauss-Müller V. Intrinsic signals for the assembly of hepatitis A virus particles. Role of structural proteins VP4 and 2A. J. Biol. Chem. 1999;274:4527–4531. doi: 10.1074/jbc.274.8.4527. [DOI] [PubMed] [Google Scholar]
- 116.Anderson DA, Ross B. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J. Virol. 1990;64:5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martin A, et al. Identification and site-directed mutagenesis of the primary (2A/2B) cleavage site of the hepatitis A virus polyprotein: functional impact on the infectivity of HAV RNA transcripts. Virology. 1995;213:213–222. doi: 10.1006/viro.1995.1561. [DOI] [PubMed] [Google Scholar]
- 118.Morace G, Kusov Y, Dzagurov G, Beneduce F, Gauss-Muller V. The unique role of domain 2A of the hepatitis A virus precursor polypeptide P1–2A in viral morphogenesis. BMB Rep. 2008;41:678–683. doi: 10.5483/BMBRep.2008.41.9.678. [DOI] [PubMed] [Google Scholar]
- 119.Le Gall O, et al. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008;153:715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 120.Gromeier M, Wimmer E, Gorbalenya AE. Origin and Evolution of Viruses. 1999. [Google Scholar]
- 121.Svitkin YV, Agol VI. Translational barrier in central region of encephalomyocarditis virus genome. Modulation by elongation factor 2 (eEF-2) Eur. J. Biochem. 1983;133:145–154. doi: 10.1111/j.1432-1033.1983.tb07440.x. [DOI] [PubMed] [Google Scholar]
- 122.Luke GA, et al. Occurrence, function and evolutionary origins of '2A-like' sequences in virus genomes. J. Gen. Virol. 2008;89:1036–1042. doi: 10.1099/vir.0.83428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Atkins JF, et al. A case for ''StopGo'': reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go) RNA. 2007;13:803–810. doi: 10.1261/rna.487907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zeddam JL, et al. Euprosterna elaeasa virus genome sequence and evolution of the Tetraviridae family: Emergence of bipartite genomes and conservation of the VPg signal with the dsRNA Birnaviridae family. Virology. 2010;397:145–154. doi: 10.1016/j.virol.2009.10.042. [DOI] [PubMed] [Google Scholar]
- 125.Gorbalenya AE. Molecular Basis of Viral Evolution. 1995. pp. 49–66. [Google Scholar]
- 126.Drexler JF, et al. Genomic features and evolutionary constraints in Saffold-like cardioviruses. J. Gen. Virol. 2010;91:1418–1427. doi: 10.1099/vir.0.018887-0. [DOI] [PubMed] [Google Scholar]
- 127.Agol VI. The Picornaviruses. 2010. pp. 239–252. [Google Scholar]
- 128.Frieman M, Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. Rev. 2008;72:672–685. doi: 10.1128/MMBR.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Melian EB, et al. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J. Virol. 2010;84:1641–1647. doi: 10.1128/JVI.01979-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Habjan M, et al. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 2009;83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Belsham GJ. Divergent picornavirus IRES elements. Virus Res. 2009;139:183–192. doi: 10.1016/j.virusres.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 132.Gasteiger E, et al. The Proteomics Protocols Handbook. 2005. pp. 571–607. [Google Scholar]
- 133.Larkin MA. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 134.Marchler-Bauer A. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 171 kb)


