Key Points
The rapid evolution of many pathogens, particularly RNA viruses, means that their evolution and ecology occur on the same timescale, and therefore must be studied jointly to be fully understood.
The rapid growth in gene sequence data and the development of new analysis techniques has enabled researchers to study the evolutionary dynamics of important human pathogens such as HIV, influenza, hepatitis C and dengue virus. The term phylodynamics has come to be associated with such studies.
Phylodynamic questions arise in a number of practical contexts, including epidemic surveillance, outbreak control, forensics and clinical medicine.
Evolutionary analysis methods can be applied to the investigation of viral dynamics at different organizational scales, from global studies of pathogen dissemination among continents, to the dynamics of infection within the tissues of individual infected hosts.
Viral genomes are an important and independent source of information about epidemiological processes, thereby supporting and corroborating epidemiological results obtained using standard surveillance methods.
The introduction of next-generation sequencing technologies will greatly increase the amount of viral genetic data available for analysis. Substantial changes and improvements to analysis methodologies will be necessary to deal with this exciting change.
The rapid evolution of many important pathogens, particularly RNA viruses, means that their ecological and evolutionary dynamics occur on the same timescale. This Review discusses the insights into the transmission and epidemiology of viruses that have been provided by analyses of their evolutionary dynamics across a wide range of biological scales.
Abstract
Many organisms that cause infectious diseases, particularly RNA viruses, mutate so rapidly that their evolutionary and ecological behaviours are inextricably linked. Consequently, aspects of the transmission and epidemiology of these pathogens are imprinted on the genetic diversity of their genomes. Large-scale empirical analyses of the evolutionary dynamics of important pathogens are now feasible owing to the increasing availability of pathogen sequence data and the development of new computational and statistical methods of analysis. In this Review, we outline the questions that can be answered using viral evolutionary analysis across a wide range of biological scales.
Main
Rapidly evolving pathogens are unique in that their ecological and evolutionary dynamics occur on the same timescale and can therefore potentially interact. For example, the exceptionally high nucleotide mutation rate of a typical RNA virus1 — a million times greater than that of vertebrates — allows these viruses to generate mutations and adaptations de novo during environmental change, whereas other organisms must rely on pre-existing variation maintained by population structure or balancing selection. In addition, many viruses frequently recombine, further increasing the opportunity for genetic novelty. Consequently, populations of fast-evolving pathogens can accumulate detectable genetic differences in just a few days and can adapt brutally swiftly, even when the adapted genotype would have been strongly deleterious in a previous environment. The interaction between evolution and epidemiology is reciprocal: the maintenance of onward transmission may be crucially dependent on continuous viral adaptation, just as the fate of a viral mutant may be decided by its hosts' position in a transmission network.
The term phylodynamics has been coined2 to describe infectious disease behaviour that arises from a combination of evolutionary and ecological processes, and we adopt the term in this Review as a convenient shorthand for the existence and investigation of such behaviour. We focus on studies that infer viral transmission dynamics from genetic data; these are typically based on concepts from phylogenetics and population genetics, but they also link pathogen evolution to the dynamics of infection and transmission. In the last decade, such studies have matured from theoretical and qualitative investigations (for example, Refs 3,4) to global genomic investigations of key human pathogens (for example, Refs 5, 6, 7). Understandably, most studies have focused on important human RNA viruses such as influenza virus, HIV, dengue virus and hepatitis C virus (HCV); therefore, this Review concentrates on these infections. However, the range of pathogens and hosts to which phylodynamic methods are applied is expanding, and we also discuss infectious diseases of wildlife, crops and livestock.
The field of viral evolutionary analysis has greatly benefited from three developments: the increasing availability and quality of viral genome sequences; the growth in computer processing power; and the development of sophisticated statistical methods. Although the explosion in viral genomic data is outpacing our ability to develop methods that fully exploit the potential of these data, we provide an overview of the key biological questions that can be tackled using current evolutionary analysis methods (Box 1). For example, when did a newly emergent epidemic begin, and from which population or reservoir species did it originate? Can genetic data resolve the order and timing of transmission events during an outbreak? How swiftly do pathogen strains move between continents, regions and epidemiological risk groups, or even between different tissues in a single infected host? Perhaps the most recognizable achievements of viral evolutionary analysis to date are the reconstruction of the origin and worldwide dissemination of HIV-1 (Refs 8, 9, 10, 11, 12, 13, 14, 15, 16), and the explanation of influenza A epidemics through the combined effects of natural selection and global migration5,6,17,18,19,20,21,22,23,24.
We describe the range of empirical questions that phylodynamic studies can address by outlining the findings of important studies, most of which have been published in the last few years. Our Review also highlights the variety of practical contexts in which such questions arise, including epidemic management and control, understanding variation in clinical disease, the design of effective vaccines, and criminal trials in which negligent transmission has been alleged. To emphasize the general applicability of the phylodynamic approach, we consider the various organizational scales at which analyses are undertaken, from the global evolutionary behaviour of pathogens to evolution in a single infected host. It is clear that, even for the same pathogen, evolutionary and ecological processes combine in different ways at different scales2 (Box 2). For example, influenza A virus displays strong genetic evidence of antigenic selection when studied over many years, but seems to be dominated by stochastic processes when only a single epidemic in one location is considered22. We also discuss aspects of data collection, pathogen biology and analysis methodology that may promote or hinder the generation of reliable conclusions.
Methods to analyse viral evolutionary dynamics
Investigating the joint evolutionary and ecological dynamics of infectious disease requires a common frame of reference within which models and data from different fields can be integrated. As we illustrate, this is often achieved by reconstructing evolutionary change on a natural timescale of months or years, enabling researchers to date epidemiologically important events such as zoonotic transmissions. A real timescale also allows pathogen evolution to be directly compared with known surveillance or time series data, perhaps revealing the time period during which a pathogen existed in a population before its discovery, or indicating the impact of public health interventions on viral genetic diversity.
Phylodynamic analyses commonly use molecular clock models to represent the relationship between genetic distance and time (Box 1). Early simplistic models that assume a constant rate of virus evolution have been superseded by those that explicitly incorporate rate variation, either between strains or through time (for example, Ref. 25).
A second, and increasingly popular, common frame of reference is provided by the geographic or spatial distribution of disease isolates (Box 1). Combined spatial and genetic analyses not only reveal the location of origin of emerging infections, but can also discern the route of transmission and the rate of geographic spread. In addition, statistical models based on coalescent theory are used to directly link patterns of genetic diversity to ecological processes, such as changing population size and population structure (Box 1). Using these models, it becomes possible to infer the characteristics of pathogen populations, such as their rate of growth, from a small sample of genomes. The resolution and scope of phylodynamic methods depends on the rate of pathogen evolution relative to that of ecological or spatial change — epidemics that fluctuate faster than mutations accumulate among pathogens will not leave an imprint in genetic diversity, although longer-term dynamic trends will.
Dynamics on a global scale
The broadest perspective on the evolutionary dynamics of a pathogen is obtained by sampling its worldwide genetic diversity over a suitable period of time. Not all viruses are geographically widespread — some might be limited by the range and dispersal of their hosts — but for those that are, it is essential to understand the geographic structure of viral genetic diversity. For example, HCV shows genotype-specific responses to antiviral drugs, and the clinical severity of dengue virus infection may depend on previous exposure to genetically distinct strains. Genetic data also reveal the rate and route of global spread, which have been most effectively studied for highly infectious airborne viruses such as severe acute respiratory syndrome (SARS) coronavirus and influenza viruses.
Influence of human movement. Humans are an atypical host species as urban population densities and international transport provide opportunities for pathogen transmission that would be otherwise absent. The role of contemporary human migration in determining global viral dynamics has been most comprehensively studied for the influenza A virus by the systematic collection, sequencing and analysis of thousands of viral isolates. Historically, influenza has caused intense bursts of human mortality, most notably associated with the reassortment of human and non-human influenza viruses, which creates strains for which humans have no acquired immunity. Evolutionary analysis of the antigenic haemagglutinin gene (HA) of the dominant H3N2 strain has shown that the influenza A virus evolves rapidly through time, yet viruses sampled concurrently from different continents exhibit limited diversity and are typically descended from a common ancestor only a few years earlier5,22. Recent evolutionary studies have revealed that the virus re-emerges each year from a persistent Southeast Asian 'source' and follows global aviation networks to temperate 'sink' regions, seeding new winter epidemics there that die out over summer5,6 (Fig. 1). The global restriction on the diversity of influenza A virus is caused by selective sweeps driven by the host's acquired immunity, which generates rapid antigenic evolution24 and corresponding high rates of amino acid change at HA antigenic sites19. Evolution of influenza A virus is even more dynamically complex when the whole genome is considered — reassortment between genome segments modulates the action of selection, so that some selective sweeps are genome-wide, whereas others only restrict the diversity of HA5.
Figure 1. The global dynamics of influenza A virus.
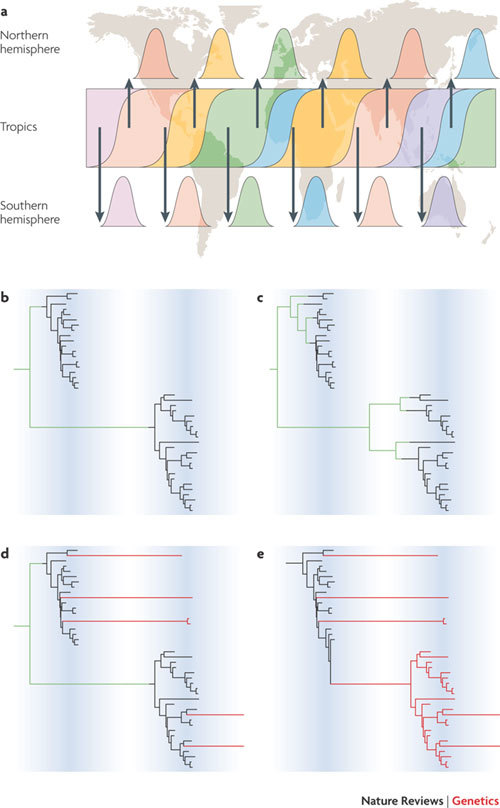
Human influenza A virus exhibits a complex pattern of global seasonal dynamics, with epidemics in temperate areas occurring during the winter and year-round sporadic outbreaks in the tropics. Recent analyses indicate that these dynamics are best described by a source–sink model of viral population structure, with a persistent reservoir in South-East Asia driving viral diversity worldwide5,6. a | Complete genome sequences sampled from New York State, USA, and from Australia and New Zealand have provided a high-resolution snapshot of diversity in these locales over successive seasons5,22. Continuous transmission of influenza in the reservoir populations allows natural selection for antigenic diversity, whereas the sink populations with seasonal dynamics will tend to be a representative sample of this diversity. b–d | Different patterns of global gene flow will be reflected in the phylogenies of influenza isolates sampled from sequential epidemics in one location. b | The entire diversity of the second season is descended from a single lineage originating from the global reservoir (lineages representing this global reservoir are in green). c | As part b, but with multiple lineages from the global reservoir seeding each season. d | As part b, but with a few lineages persisting locally (red) from one season to the next. e | The entire second season is descended from local lineages, implying that transmission persists from season to season in this location. Part a is modified, with permission, from Ref. 5 © Nature (2008) Macmillan Publishers Ltd, all rights reserved.
Reconstructing histories of epidemics. Influenza A dynamics are clearly the result of intricate and ongoing interactions between evolutionary and ecological processes. However, not all pathogens with a worldwide distribution show such complex behaviour at this scale. Although the HIV-1 pandemic is truly international, it is the result of simpler ecological processes that are less strongly coupled to viral evolution. Evolutionary analysis has proven successful in reconstructing the global epidemic history of HIV-1. Viral sequences sampled at various times since the discovery of HIV in 1983 have been used to date the origin of the pandemic to the first half of the twentieth century10,15 and to pinpoint west-central Africa as its geographic source14. These results have been validated and refined by the recovery of genomic fragments from older isolates, notably two 50-year old preserved tissue samples from Kinshasa, Democratic Republic of Congo15,16, which indicate that considerable HIV diversity had accrued there by 1960.
The worldwide dissemination of HIV-1 from its central African source over several decades was propelled by multiple 'founder events', whereby individual HIV-1 lineages moved to new regions and established epidemics, sometimes recombining in the process, thus generating an array of circulating recombinant forms. The nature and timing of both founder and recombination events have been estimated by evolutionary analysis8,13,26. In contrast to influenza A, the absence of protective immunity against HIV means that viral adaptation probably played little part in shaping the current geographical distribution of HIV-1 subtypes, although there is evidence that the virus acquired specific mutations after zoonosis to enable efficient transmission among humans27 and that HIV-1 is now adapting to the diversity of human leukocyte antigen class I molecules28,29.
Simple epidemic dynamics also explain the global dissemination of HCV, which has infected humans for at least several centuries30. A handful of endemic HCV strains, originally from Asia and Africa, exploded in prevalence worldwide during the twentieth century owing to their chance association with new routes of transmission, such as transfused blood31.
Emerging insights. Although few pathogens have been sampled as comprehensively as influenza A virus or HIV-1, new insights are being gained as large data sets are compiled for other viruses. For example, recent studies of echovirus 30, a transmissible human enterovirus that causes periodic outbreaks of meningitis, have revealed a fascinating picture of evolutionary forces that vary among viral genes32,33. Echoviral capsid genes diverge continuously and rapidly, show rapid global transmission, but exhibit limited concurrent variation. This is analogous to the immune-driven turnover of influenza A HA lineages, but there is substantially less genetic evidence of positive selection for immunologically novel echoviral variants32,33. By contrast, echovirus 30 polymerase gene lineages are geographically structured, diverse, and coexist on a global scale. Frequent recombination between the capsid and polymerase genes generates transient recombinant forms that are estimated to persist for approximately 5 years33. This modular nature of echovirus 30 evolution is all the more remarkable given that it takes place in an unsegmented linear genome that is less than 8 kb long.
Human metapneumovirus, a recently discovered and common cause of childhood respiratory illness, exhibits complex behaviour that is less fully understood. The virus forms several lineages, each of which contains little genetic diversity — suggesting that genetic bottlenecks are common, but only partial or local in effect34.
Evolutionary analysis has helped track the global spread of the H5N1 highly pathogenic avian influenza (HPAI). Because the virus has been continuously sampled since its emergence in China in 1996, phylogenies can provide accurate reconstructions of its movements, both internationally35 and locally36. Molecular clock results indicate that HPAI lineages typically reside at a location for several months before their official detection37. HPAI strains in Asia also undergo frequent reassortment, which may be facilitated by the dense and interconnected duck and poultry populations in the region36,37.
As more pathogens are studied on a global scale, we should remember that conclusions drawn from small and local samples will underestimate dynamic complexity. Indeed, our understanding of both HIV-1 and influenza virus population dynamics changed appreciably after comprehensive surveys of viral diversity were published6,14. If we extrapolate from the examples of echovirus 30 and influenza A virus, then it seems that the most complex global behaviour occurs in highly transmissible viruses that cause acute infections and short-lived epidemics, possibly because their dynamics arise from a three-way interplay between transmission, host herd immunity and viral adaptation. Human viruses that might show such behaviour — when sampled on a sufficiently large scale — include enteroviruses, rhinoviruses, caliciviruses and paramyxoviruses.
Regionally or genetically defined epidemics
A large proportion of evolutionary analyses of pathogens consider individual lineages, strains or subtypes circulating in a specific location, which may be a whole continent or just one town or district. Such outbreaks frequently correspond to a single epidemic, as defined by surveillance organizations, and may involve a single lineage or cluster of infections, as defined by phylogenetic analysis. Evolutionary analysis at this scale can determine the source and time of origin of an epidemic, reveal its genetic composition, and is often used to estimate the rate of viral transmission and spatial spread in the affected region.
Locating the source of an epidemic. Studies on a regionally or genetically defined scale often begin by seeking the source of the new strain, which could be either a zoonotic reservoir or an epidemiologically distinct or distant human population. The origin of an epidemic is typically inferred by finding the most genetically similar non-epidemic strain. This is a simple procedure but is greatly dependent on previous sampling. For example, the SARS coronavirus was highly distinct with no close relatives when initially characterized in April 2003 (Ref. 38). The discovery in October 2003 of related viruses in civet cats from animal markets39 suggested that SARS originated from a zoonotic source, but further sampling has shown that bats are the primary reservoir of these viruses40. Molecular clock analysis of bat coronaviruses indicates that the cross-species transfer to civet cats occurred only 4 years before the onset of the human epidemic41.
Epidemic origins are also hard to locate if the source is geographically or temporally remote; West Nile virus strains sampled from the Mediterranean in 1998 were quickly identified as the source of the 1999 North American epidemic42, whereas the discovery of the probable zoonotic source of pandemic HIV-1 — Pan troglodytes troglodytes chimpanzees in south-eastern Cameroon — was the culmination of many years of research9.
In some instances, genetic analysis can reveal hidden multiple origins for epidemics that initially seemed homogenous. The 1980s HIV epidemic in the UK among men who have sex with men and the 1990s outbreak of HCV in a subset of the same population are both comprised of at least five distinct strains, each with similar epidemiological behaviours43,44. Similarly, phylodynamic analysis of whole viral genomes indicates that the 2005 Singapore dengue virus epidemic comprised multiple viral lineages of different geographical origins45. The existence of hidden genetic heterogeneity within an epidemic implies that rapid movement of lineages at a higher geographic scale is likely.
Spatial dynamics. Viral isolates sampled from regional epidemics can contain valuable information about the spatial dynamics of infection. For example, Biek et al.46 estimated the spread of raccoon rabies across the north-eastern United States from sequences sampled over three decades. Viral movement was initially rapid but slowed considerably after a few years as individual lineages became established in different locales, and ecological data on outbreak size closely matched the estimates obtained using coalescent methods (see next section). A similar process of invasion and establishment was also reported for dengue virus in the Americas47. Interestingly, dengue virus diversity was maintained across epidemic cycles by the metapopulation structure built up during the invasion phase (Fig. 2). If both the location and sampling date of viral sequences is specified it is possible to estimate the distance pathogens move per year solely from genetic data, as demonstrated by reconstructions of Ebola virus spread in central Africa48 and feline immunodeficiency virus infection of Rocky Mountain cougars49.
Figure 2. A spatially and temporally defined epidemic.
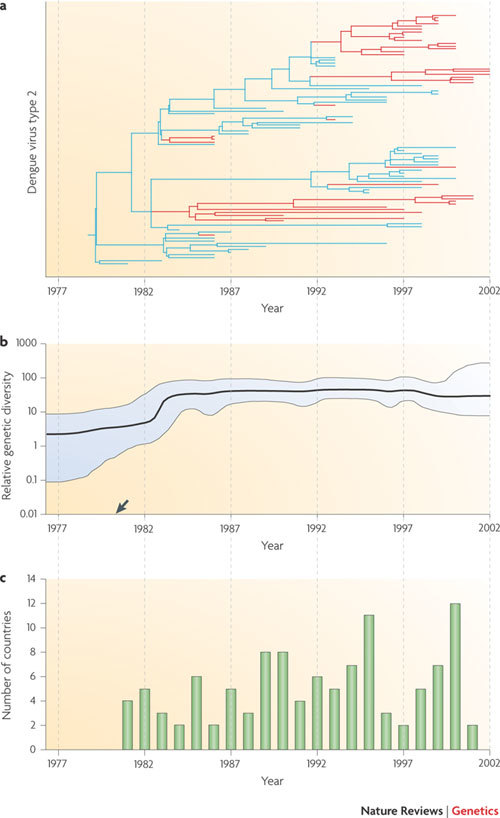
a | A molecular clock phylogeny that illustrates the history of dengue virus genotype 2 infection in the Caribbean and in Central and South America47. A simple parsimony approach has been used to reconstruct the likely location of each phylogenetic branch (blue, Caribbean islands; red, mainland Central America and mainland South America). By combining phylogenetic and geographic information, the phylogeny indicates that the outbreak began in the Caribbean before repeatedly and independently invading mainland locations some years later. b | An estimate of the relative genetic diversity of the same dengue virus epidemic, which shows an initial increase before stabilizing (95% confidence limits shown in blue). This stabilization does not match the varying number of reported dengue outbreaks (shown in part c), probably because spatial population structure maintains viral diversity across epidemic peaks and troughs. More generally, when the sampled population exhibits strong positive selection or population structure then the y-axis cannot be reliably interpreted as proportional to effective population size. The estimated common ancestor of the sampled sequences (arrow) is dated slightly earlier than the first reported outbreak in the region (see part c). c | Shows the number of countries affected by dengue virus genotype 2 infection per year. Figure is modified, with permission, from Ref. 47 © (2005) American Society for Microbiology.
Coalescent theory analysis. Regionally or genetically defined outbreaks most closely represent the typical 'epidemic' that is described by models of mathematical epidemiology. In some cases this representation can be formalized using population genetic models based on coalescent theory, which directly link phylogenetic structure with ecological processes (Box 1). This approach is typically used to infer past rates of epidemic growth from sampled viral sequences3 but can, in some circumstances, be used to directly estimate the fundamental epidemiological parameter(R0) from such data30,46,50. Coalescent-based methods have been successfully applied to HCV and HIV-1. This success is partly because of the chronic nature of infection and the absence of cross-immunity for these viruses, which result in comparatively slow changes in prevalence that leave clear footprints in the patterns of viral diversity. Analysis of HCV genomes indicates that, during the twentieth century, strains varied significantly in their rates of growth according to the transmission route by which each strain was spread30,51. The reliability of coalescent-based methods — which make a number of limiting assumptions — was tested in an analysis of HCV in Egypt: here, the methods correctly reconstruct a mid-twentieth century explosion in transmission that was caused by widespread unsafe injection during campaigns against schistosomiasis52. Comparable phylodynamic studies of HIV-1 subtypes also show agreement between genetic and epidemiological reconstructions53,54, even though commonly used coalescent methods ignore the presence of HIV recombination.
As well as describing the origin and spread of many individual outbreaks, analyses of regional epidemics have helped reveal conceptual connections between the different fields of epidemiology, population genetics and phylogenetics, and have validated methods of statistical inference. Despite the choice of examples above, analysis at this scale is not limited to human and animal pathogens. For example, Fargette et al.55 linked the timescale of the emergence of rice yellow mottle virus to the nineteenth century expansion of rice culture in Africa, and Almeida et al.56 used similar methods to conclude that the human transport of contaminated plants disseminated banana bunchy top virus among Hawaiian islands after it was introduced to the islands in 1989.
Infection clusters and transmission chains
If an outbreak or infection cluster occurs on a small enough scale then we can realistically expect to sample viruses from all or most of the individuals involved. Studies of such outbreaks tend to fall into two categories: those for which the transmission history (that is, who infected whom, and when) is mostly or wholly known, and those for which it is unknown. Examples in which the transmission history is known are highly informative, as the specified infection history allows evolutionary processes to be investigated with a greater degree of certainty. When the transmission chain is unknown, the primary goal may be the reconstruction of the chain or the identification of its source, timescale or transmission route.
Known transmission histories. Naturally occurring outbreaks for which the transmission event details are known are understandably rare; the majority of those with known details are HIV outbreaks. Known chains of transmission have been used to measure the rate of HIV evolution57 (Box 2) and the magnitude of the bottleneck in virus diversity generated at transmission58. The Irish anti-D cohort — a well-studied group of HCV-infected women who were accidentally infected with almost identical strains at the same time — has also provided valuable information about variation in viral evolution, host immune selection and disease outcome between patients59,60. Using a different HCV transmission cluster, Wrobel et al.61 demonstrated that molecular clock methods can reliably estimate the date that a patient was infected. Transmission chains can also resolve whether the same viral adaptation arises in different hosts (convergent evolution)62.
Known transmission chains have been used to test whether sequence-based phylogenies match the true history of transmission among epidemiologically connected infections. Although several studies of HIV clusters have reported close agreement62,63,64, it is often not appreciated that there are good reasons to expect occasional mismatches between the phylogeny and the true transmission history of a cluster. When one 'donor' infection transmits the virus to multiple recipients, the common ancestors of viral lineages sampled from the recipients will exist in the donor. If the amount of viral diversity in the donor is comparatively high, then the relative order of phylogenetic splitting events (one for each common ancestor) may differ from the order of infection events (Fig. 3). The branching order of transmission for genetically diverse infections is therefore best analysed using metapopulation models that integrate the process of transmission with that of lineage coalescence65. This issue is not only restricted to specialized phylogenetic studies — evolutionary analyses of transmission chains are presented in criminal proceedings in which individuals are accused of intentional or negligent transmission66.
Figure 3. Reconstruction of a known HIV-1 transmission chain.
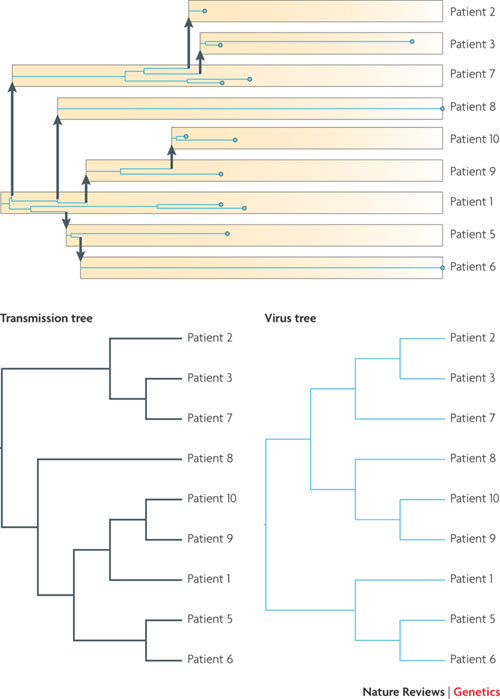
A phylogeny of 13 HIV-1 viral particles (blue circles) sampled at different times (horizontal axis) from 9 different patients for whom the times and direction of viral transmission are known. The virus phylogeny (blue lines) can be mapped within the transmission tree (yellow boxes and arrows), analogous to the mapping of a gene genealogy within a species tree. We can trace all the viruses sampled from one patient back to the time of transmission. Whether more than one lineage is transmitted at this time from the donor will depend on the size of the genetic bottleneck at transmission. Even in the presence of a tight bottleneck, a diverse population in the donor can result in lineage sorting, with the result that the topology of the virus phylogenetic tree does not exactly match the transmission tree.
Reconstructing transmission histories. Anew and interesting approach to the analysis of transmission chains is presented in recent studies of UK outbreaks of foot and mouth disease virus (FMDV). These studies describe the infection process at the level of individual farms, with transmission between farms mainly caused by the transport of infected livestock. Cottam et al.67 developed dynamic models that provide a probability distribution for the date of infection of a particular infected farm and its likely period of 'infectiousness' before FMDV diagnosis and culling of the animals. This temporal information was then combined with the genome sequences of viruses that were sampled from the infected herds to identify the most likely chains of transmission linking the farms in time and space. A joint analysis was particularly suitable because FMDV spread is so rapid that comparatively few genetic changes accrue between inter-farm transmissions.
Not all studies of infection clusters focus on the pathways of transmission; sometimes the initiation date of an outbreak is of most interest68 and at other times the precise epidemic source is sought69. However, coalescent-based estimates of population processes are not suitable for infection clusters because this approach requires that the sequences analysed represent a small fraction of the sampled population. Despite this restriction, transmission chain phylogenies can still provide important information about populations, such as the minimum time between transmission events70. Furthermore, modern sequencing technology is fast enough for genetic analysis to assist contact tracing and control as an epidemic unfolds. For example, phylogenies confirmed epidemiological suspicions that the 2007 Italian chikungunya outbreak originated from an Indian index case71. Considered together, the studies discussed in this section highlight the relevance of transmission chain analyses to applied problems in clinical medicine, forensics and public health. The microevolutionary dynamics of infection events will become a major focus of infectious disease research as high-resolution longitudinal studies will be made possible by the application of next-generation sequencing.
Within-host dynamics
The exceptionally rapid rate of evolution of RNA viruses means that viral evolution in a single host can be studied for the duration of an infection. Dynamics at this scale are fundamental as within-host evolution is the ultimate source of all viral genetic diversity, and therefore it must be understood before models that link different evolutionary scales can be properly developed (Box 2). Additionally, within-host analyses can reveal the evolutionary processes that underlie some aspects of clinical disease. In practice, such analyses have so far been limited to viruses that establish chronic infections lasting months or years, and for which measurable amounts of genetic change occur between viral samples; this is particularly the case for HIV infection and, to a lesser extent, for HCV and hepatitis B virus72 infection.
Strong natural selection is clearly the dominant force determining HIV evolutionary dynamics in hosts: HIV phylogenies display a high turnover of short-lived lineages that is driven by host immune selection, analogous to the pattern observed for influenza A virus at the global scale2 (Box 2). Correspondingly, HIV genetic diversity at any particular time is low but slowly increases over the course of chronic infection73. Numerous analyses have quantified HIV adaptation and evolution using gene sequences, particularly for the viral envelope gene. These studies have found that these processes correlate with the rate of progression to clinical AIDS74,75,76 and the rate at which HIV evades neutralizing antibody responses77. Equivalent studies of HCV infection have found that viral adaptation predicts the outcome of acute infection78,79 and that HCV diversity correlates with levels of liver damage80. Perhaps the most important outcome of HIV within-host evolution is the generation of T cell escape mutants that can elude host cytotoxic T lymphocyte responses81 — this is a major barrier to the development of effective HIV vaccines. Although much of the work on T cell escape is not explicitly phylogenetic, there has been a trend away from cross-sectional surveys of viral variation (for example, Ref. 82) towards longitudinal and evolutionary studies at all organizational scales, from the level of the pandemic83 to that of small transmission chains81 and in individual hosts84. The rate at which HIV evolves during an infection depends not only on viral adaptation but also on the replication rate of the virus and its population size: these factors combine to generate measurable variation in viral evolutionary rate both within and between hosts. As a result, evolutionary rates estimated from sequence data may be crucially dependent on the scale of analysis (Box 2).
Spatial dynamics at the cellular level. Phylodynamic methods have detected and measured the compartmentalization of viral lineages into specific tissues during chronic infection, which creates within-host subpopulations (so-called virodemes), which are analogous to the location-specific clusters of infection seen at higher scales. Highly distinct strains of HIV are found in the brains of patients with neurological illness85,86, suggesting that virus movement across the blood–brain barrier is not common and might be unidirectional. Finer genetic structure is apparent even among viruses from different brain regions, which seem to evolve at different rates87. HIV subpopulations in other tissues have been proposed, including in the cervix88 and seminal fluid89, as has compartmentalization in livers with chronic HCV infection90.
Integrating levels of phylodynamic processes
The evolutionary and ecological dynamics of viral pathogens take place in a hierarchy of organizational scales, from within-host processes to the global dynamics of pandemics, but it is not obvious how dynamics at lower scales combine to generate higher-order behaviour. Such hierarchical processes can be studied from the perspective of both populations genetics65 and mathematical epidemiology91. Multiscale interactions are of great public health importance as well as being of theoretical interest; for example, the success of antiviral drug treatment campaigns will depend on the degree to which drug resistance mutations that arise in treated hosts can accumulate at the epidemic level92.
There are intriguing parallels between processes in hosts and those at the epidemic or global level2. First, within-host studies reconstruct the dynamics of large viral populations from small samples, hence techniques commonly applied to large-scale epidemics (particularly coalescent models) can be re-employed with an appropriate change in perspective — each sequence represents an infected cell or virion, rather than an infected host. Secondly, within-host evolution is closely intertwined with ecological processes, such as the turnover of virions, host cells and components of the host immune response. These dynamics are studied using virus kinetics models93, which were directly inspired by related models developed by mathematical epidemiologists. As at higher scales, within-host studies have attempted to integrate evolutionary and ecological processes94,95,96; for example, in vivo HIV cell-to-cell generation times can be accurately estimated by coalescent analysis of sampled virus sequences97,98. There is great potential for further development of models that combine the abundant longitudinal data on infection kinetics with those on viral evolution.
Conclusions
The field of infectious disease evolutionary dynamics is currently seeing a revolution in all three of the technologies on which it relies: genomic sequencing, statistical methodology and high-performance computing. This confluence has produced a burgeoning interest in the evolutionary and epidemiological processes that leave their imprint on pathogen genomes, as reflected in the empirical studies and analysis techniques reviewed here. However, it is our opinion that many investigations still fail to fully appreciate or utilize the rich source of epidemiological information contained in viral genome sequences. Genetic data can independently corroborate surveillance data during an epidemic and can shed light on events before the initial report of the outbreak. Furthermore, evolutionary and surveillance data provide alternative perspectives on the same underlying phylodynamic process and can therefore be validated against one another. The practicality of this approach was demonstrated during the H1N1 'swine flu' epidemic, first detected in April 2009. Tens of viral sequences were made publically available within days of discovery of the virus, and evolutionary analysis was incorporated into initial assessments of the pandemic potential of the new strain50.
Large-scale sampling and sequencing could also revolutionize our understanding of medically important RNA viruses, such as caliciviruses, rotaviruses and enteroviruses, the genetics of which are currently comparatively neglected. DNA viruses with small genomes that evolve at similar rates to RNA viruses1 will be equally suitable for phylodynamic analysis. When applied to slower-evolving DNA viruses, bacteria and protozoa, evolutionary analyses similar to those introduced here can help elucidate longer-term processes, such as host–pathogen co-divergence and pathogen speciation99,100,101.
In the near future, the greatest impact on viral evolutionary analysis will come from the increasing accessibility of new high-throughput sequencing technologies102. For RNA viruses, which have genomes that are on average only 15,000 nucleotides long, it is likely that hundreds or thousands of complete genomes sampled from both viral epidemics and infected hosts can be routinely subjected to molecular epidemiological analysis. Ensuring that computational and statistical developments keep pace with this revolution in data acquisition will be a great challenge. One promising solution is to harness the power of 'multi-core' or massively parallel computing technologies in evolutionary analysis103. The coming genomic era will also allow us to determine how much information can be inferred from gene sequences alone — only those ecological processes that occur on the same timescale as genetic change will leave their mark on genetic data, and robust evolutionary inferences carry a statistical uncertainty that should be accurately estimated and reported.
Therefore, a clear goal for the future is to further develop analytic methods that combine genetic and epidemiological data to reconstruct epidemic history and to predict future trends, a task to which Bayesian inference methods of statistical inference are well suited. Further development of analysis methods is required in three key areas: the quantification of viral adaptation by natural selection; the explicit integration of evolutionary and spatial information; and the measurement of rates of viral reassortment or recombination. Advances in these areas could raise new questions for phylodynamic analysis. For example, do lineages differ in their rates of spatial diffusion? And are bursts of viral adaptation associated with recombination events? However, such analytical finesse is of little use if basic epidemiological information, such as the date and location of sampling, is unavailable, and we implore researchers generating viral sequences to attach as much sample information to each sequence as ethical constraints permit.
Box 1 | Phylodynamic techniques.
Rooted molecular phylogenies can be estimated from viral gene sequences (see the figure, part a). Depending on the scale of the analysis undertaken, the sampled sequences (red circles) may represent infected individuals, infected cells, virions or higher-level units such as villages. The phylogeny branching order shows the shared ancestry of the sequences, which usually — but not always — reflects the history of pathogen transmission between these units (discussed in main text). This phylogeny has no timescale, so the branch lengths represent the genetic divergence from the ancestor (black circle). If the sequences of interest undergo recombination, then a single phylogenetic tree may not adequately describe evolutionary history and alternative methods can be applied (for example, Ref. 104).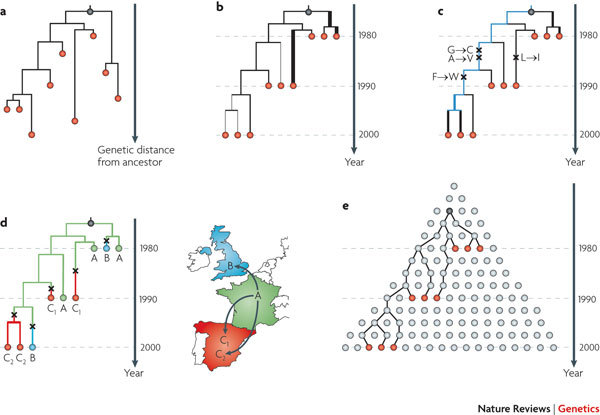
The same phylogeny can also be reconstructed using a molecular clock model (see the figure, part b), which defines a relationship between genetic distance and time. The pathogen sequences have been sampled at known time points and the phylogeny branches have lengths in units of years. This approach estimates the ages of branching events, including that of the common ancestor. The simplest, 'strict' clock model assumes that all lineages evolve at the same rate. More complex, 'relaxed' models allow evolutionary rates to vary through time or among lineages, resulting in variation around an average rate25. In this phylogeny, unusually fast or slow evolving lineages are shown as thick or thin lines, respectively. The relationships among genetic distance, evolutionary rate and time can be understood by comparing the branch lengths in part a and part b.
Phylodynamic data can also highlight the evolution through time of mutations that may reflect viral adaptations (see the figure, part c). Observed amino acid changes (crosses) are shown mapped onto specific phylogeny branches. Amino acid sites under positive selectioncan be identified using dn/ds methods, which compare the rate of replacement substitutions (that change the amino acid) with the rate of silent substitutions (that do not change the amino acid)18,105. Such methods are most powerful when detecting diversifying selection, making them appropriate for the analysis of infectious disease, but the results obtained using these methods require careful interpretation106. Of particular interest are the replacement mutations that are found on the persisting phylogenetic 'backbone' that represents the ancestor of future virus populations (blue branches), as opposed to those occurring on branches that die out (black branches).
The data can also be analysed using temporal phylogeography (see the figure, part d). The nine sequences were sampled from France (green, A), the United Kingdom (blue, B) and two locations in Spain (red, C1 and C2). Statistical methods can be used to reconstruct the history of pathogen spread, so that each branch is labelled with its estimated geographic position. Current reconstruction methods mostly use simple parsimony approaches107 that reconstruct a minimum set of migration events consistent with the observed phylogeny. Lineage movement events are marked on the phylogeny with crosses. Combining the spatial and temporal information provides further insights — this hypothetical pathogen spread to location C1 years before independently arriving at location C2. Such analyses are not limited to hypotheses concerning physical geography, as the labels A, B, C can stand for any trait of interest, for example, host species, cell tropism during infection, host risk factors or clinical outcome.
The principles of coalescent analyses, which incorporate an explicit model of the sampled pathogen population, are illustrated in figure, part e. Each circle represents an infection, and circles on the same row occur during the same period of time. The increasing width of each row therefore reflects the growth of the epidemic through time. Starting from the sampled infections (red), the sampled lineages (black lines) can be traced back through unsampled infections (grey) to the common ancestor (black circle). The rate at which the sampled lineages merge or coalesce depends on population processes such as population dynamics, population structure, selection and recombination (only change in population size is represented here). Coalescent methods are used to infer these processes from randomly sampled pathogen sequences.
Box 2 | Linking evolutionary scales: HIV as an example.
To illustrate the challenges involved in understanding dynamics at multiple levels jointly, we consider here the well-characterized rate of HIV-1 genome evolution at the within-host and between-host scales. The divergence rates of a series of infections can be plotted against time (see the figure, parts a–d). Each infection is represented by a differently coloured cone of divergence — the gradient of each cone equals the mean rate of within-host virus evolution and the width of each cone represents the variance of this rate. The long-term accumulation of virus divergence at the epidemic level (dashed lines) depends on three factors: the variation in evolutionary rate among strains within a host; whether the average viral rate varies over the course of infection; and whether the strain transmitted to the next host is selected randomly with respect to its evolutionary rate. Empirical analyses indicate a high variance in evolutionary rate among lineages within a host75, which is caused, at least in part, by latent non-replicative infection of cells108. Provided that the lineages are transmitted to subsequent hosts randomly (see the figure, part a), the long-term virus evolutionary rate will, on average, equal the average within-host evolutionary rate, even when these average rates differ between patients (P) (see the figure, part e).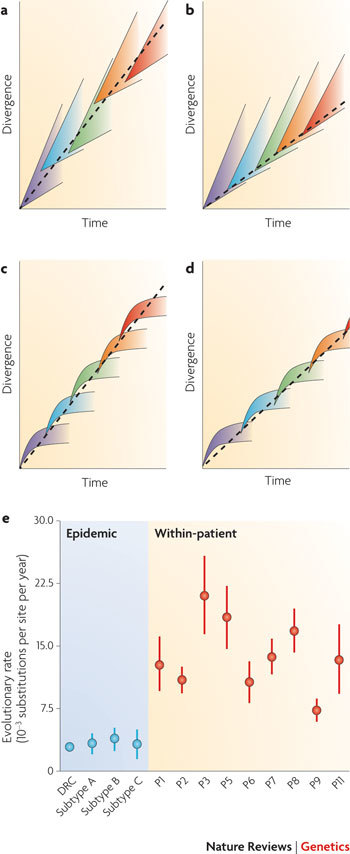
Discrepancy between within- and between-host rates
In contrast to the above, it seems that HIV-1 evolutionary rates are slower when measured at the epidemic level (see the figure, part e; DRC, Democratic Republic of Congo) than when measured at the within-host level109 (see the figure, part e; P1–P9 and P11). One explanation for this difference is that transmission is nonrandom, such that slower-evolving lineages are more likely to successfully generate the next infection than faster ones, with the result that the long-term rate is less than the average within-host rate (see the figure, part b). Indeed, the short-sighted action of natural selection will tend to favour those strains with higher within-host fitness, even at the cost of lowered transmissibility. Thus, transmitted viruses could be preferentially drawn from lineages that have accumulated fewer mutations, such as those that have spent a greater proportion of time in a latent state. This effect may be enhanced by the existence of a genetically distinct HIV subpopulation in genital mucosa88,89.
The discrepancy between within- and between-host rates can also be explained if viral evolutionary rates decrease over the course of infection (see the figure, parts c,d). Several processes could cause such a decrease: the rate of viral replication declines as the disease progresses75,110; selection for viral immune escape variants weakens later in infection76,105; and adaptation of the viral population is fastest early in infection, soon after its transmission to a new host environment. As yet, the possible effect of recombination on HIV evolutionary rates at different scales is unknown.
Whatever the underlying cause, if average evolutionary rates vary during infection then the long-term rate of evolution becomes dependent on when transmission occurs. If within-host rates decline during infection then more rapid transmission will result in a faster long-term rate of evolution (see the figure, part c) than slower transmission (see the figure, part d). This has been shown for the human T cell lymphotropic virus type II, a leukaemia-causing relative of HIV, which seems to evolve many times faster in rapidly transmitting drug users than in populations that are vertically infected during breastfeeding4. Conversely, it has been argued that within-host rates increase over the first weeks of infection, owing to the activation of the immune response that drives viral adaptation, hence fast early transmission could alternatively lead to slower long-term rates111.
Acknowledgements
We would like to thank E. Holmes and three referees for commenting on the manuscript and improving it immeasurably. We thank A. Drummond and P. Lemey for providing rates of HIV-1 evolution for FIG. 4. Finally we gratefully acknowledge The Royal Society of London, which supports both authors.
Glossary
- Balancing selection
Any form of natural selection that results in the maintenance of genetic polymorphisms in a population, as opposed to their loss through fixation or elimination.
- Diversifying selection
Any form of natural selection that generates high levels of genetic diversity; for example, recurrent positive selection or balancing selection.
- Parsimony approach
A principle of evolutionary inference, based on the assumption that the best-supported evolutionary history for a characteristic is the one that requires the fewest number of changes in that characteristic.
- Molecular clock
A statistical model that describes the relationship between time and the genetic distances among nucleotide sequences. In contrast to older molecular clock models, contemporary models no longer require the assumption that the rates of nucleotide change are constant through time.
- Coalescent theory
A theory that describes the shape and size of genealogies that represent the shared ancestry of sampled genes. It describes how the statistical distribution of branch lengths in genealogies depends on population processes such as size change and structure.
- Reassortment
A form of genome recombination occasionally exhibited by viruses, such as influenza, which have a genome composed of multiple RNA molecules (genomic segments). The resulting virus produced by reassortment possesses a mixture of genomic segments from two or more parental viruses.
- Selective sweep
The rapid increase in frequency of a mutation owing to positive selection for that mutation.
- Zoonosis
An infectious disease transmitted from animals to humans, or the event of cross-species transmission.
- Positive selection
Also known as directional selection. A form of natural selection that results from an increase in the relative frequency of one genetic variant compared with other variants. It often results in the fixation of the selected variant in the population.
- Herd immunity
The protection of susceptible members of a population from infection owing to the sufficiently high prevalence of immune individuals.
- Metapopulation structure
A metapopulation is composed of multiple subpopulations, among which there is gene flow. Subpopulations also arise and become extinct dynamically through time.
- Fundamental epidemiological parameter
(R0). The basic reproductive number of an infectious disease, from which many epidemiological predictions can be made. It is equal to the number of secondary infections caused by a single infection in a wholly susceptible host population.
- Index case
The first infection in an epidemic or outbreak, from which all subsequent infections are ultimately descended.
- Cross-sectional survey
An investigation that samples a population at a specific point in time. A longitudinal survey, by contrast, samples a population at several different times.
- Bayesian inference
A method of statistical inference that uses Bayes' theorem to calculate the probability of a hypothesis. Such methods combine prior information with new observations or data.
Biographies
Oliver Pybus is a Royal Society University Research Fellow at the Department of Zoology, University of Oxford, UK, where he received his DPhil in 2000, and is Tutor in Biology at New College, University of Oxford, UK. He studies the evolutionary dynamics of infectious disease, particularly human pathogenic viruses, and develops statistical methods for the analysis of gene sequence data. He is still searching for the meaning of Silbury Hill.
Andrew Rambaut is a Royal Society University Research Fellow at the Institute of Evolutionary Biology, University of Edinburgh, UK, where he also studied as an undergraduate. He received his DPhil from the University of Oxford, UK, in 1997. He is interested in the evolution of all creatures great and small but particularly the microscopic and ugly, and is a leading developer of phylogenetic software, including BEAST.
Related links
FURTHER INFORMATION
Contributor Information
Oliver G. Pybus, Email: oliver.pybus@zoo.ox.ac.uk
Andrew Rambaut, Email: a.rambaut@ed.ac.uk.
References
- 1.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nature Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 2.Grenfell BT, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 3.Holmes EC, Nee S, Rambaut A, Garnett GP, Harvey PH. Revealing the history of infectious disease epidemics through phylogenetic trees. Philos. Trans. R. Soc. Lond. B. 1995;349:33–40. doi: 10.1098/rstb.1995.0088. [DOI] [PubMed] [Google Scholar]
- 4.Salemi M, et al. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc. Natl Acad. Sci. USA. 1999;96:13253–13258. doi: 10.1073/pnas.96.23.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rambaut A, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell CA, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 7.Bird BH, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J. Virol. 2007;81:2805–2816. doi: 10.1128/JVI.02095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert MTP, et al. The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl Acad. Sci. USA. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korber B, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 11.Lemey P, et al. The molecular population genetics of HIV-1 group O. Genetics. 2004;167:1059–1068. doi: 10.1534/genetics.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemey P, et al. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl Acad. Sci. USA. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. Human immunodeficiency virus — phylogeny and the origin of HIV-1. Nature. 2001;410:1047–1048. doi: 10.1038/35074179. [DOI] [PubMed] [Google Scholar]
- 14.Vidal N, et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J. Virol. 2000;74:10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worobey M, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu TF, et al. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 17.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. Predicting the evolution of human influenza A. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 18.Fitch WM, Bush RM, Bender CA, Cox NJ. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl Acad. Sci. USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitch WM, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl Acad. Sci. USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes EC, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson MI, et al. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 2008;4:e1000133. doi: 10.1371/journal.ppat.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 2007;3:1220–1228. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson MI, et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008;4:e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 25.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PloS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tee KK, et al. Estimating the date of origin of an HIV-1 circulating recombinant form. Virology. 2009;387:229–234. doi: 10.1016/j.virol.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Wain LV, et al. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 2007;24:1853–1860. doi: 10.1093/molbev/msm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawashima Y, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosakovsky Pond SL, et al. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pybus OG, et al. The epidemic behavior of the hepatitis C virus. Science. 2001;292:2323–2325. doi: 10.1126/science.1058321. [DOI] [PubMed] [Google Scholar]
- 31.Pybus OG, et al. Genetic history of hepatitis C virus in East Asia. J. Virol. 2009;83:1071–1082. doi: 10.1128/JVI.01501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailly JL, et al. Phylogeography of circulating populations of human echovirus 30 over 50 years: nucleotide polymorphism and signature of purifying selection in the VP1 capsid protein gene. Infect. Genet. Evol. 2009;9:699–708. doi: 10.1016/j.meegid.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 33.McWilliam Leitch EC, et al. Transmission networks and population turnover of echovirus 30. J. Virol. 2009;83:2109–2118. doi: 10.1128/JVI.02109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Graaf M, Osterhaus ADME, Fouchier RAM, Holmes EC. Evolutionary dynamics of human and avian metapneumoviruses. J. Gen. Virol. 2008;89:2933–2942. doi: 10.1099/vir.0.2008/006957-0. [DOI] [PubMed] [Google Scholar]
- 35.Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proc. Natl Acad. Sci. USA. 2007;104:4473–4478. doi: 10.1073/pnas.0700435104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam TT, et al. Evolutionary and transmission dynamics of reassortant H5N1 influenza virus in Indonesia. PLoS Pathog. 2008;4:e1000130. doi: 10.1371/journal.ppat.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vijaykrishna D, et al. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PloS Pathog. 2008;4:e1000161. doi: 10.1371/journal.ppat.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peiris JSM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan Y, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 40.Li WD, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 41.Hon CC, et al. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanciotti RS, et al. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 43.Danta M, et al. Recent epidemic of acute hepatitis C virus in HIV-positive men who have sex with men linked to high-risk sexual behaviours. Aids. 2007;21:983–991. doi: 10.1097/QAD.0b013e3281053a0c. [DOI] [PubMed] [Google Scholar]
- 44.Hue S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc. Natl Acad. Sci. USA. 2005;102:4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber MJ, et al. Genomic epidemiology of a dengue virus epidemic in urban Singapore. J. Virol. 2009;83:4163–4173. doi: 10.1128/JVI.02445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc. Natl Acad. Sci. USA. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrington CVF, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J. Virol. 2005;79:14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh PD, Biek R, Real LA. Wave-like spread of Ebola Zaire. PloS Biol. 2005;3:e371. doi: 10.1371/journal.pbio.0030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biek R, et al. Epidemiology, genetic diversity, and evolution of endemic feline immunodeficiency virus in a population of wild cougars. J. Virol. 2003;77:9578–9589. doi: 10.1128/JVI.77.17.9578-9589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fraser C., Donnelly C. A., Cauchemez S., Hanage W. P., Van Kerkhove M. D., Hollingsworth T. D., Griffin J., Baggaley R. F., Jenkins H. E., Lyons E. J., Jombart T., Hinsley W. R., Grassly N. C., Balloux F., Ghani A. C., Ferguson N. M., Rambaut A., Pybus O. G., Lopez-Gatell H., Alpuche-Aranda C. M., Chapela I. B., Zavala E. P., Guevara D. Ma. E., Checchi F., Garcia E., Hugonnet S., Roth C. Pandemic Potential of a Strain of Influenza A (H1N1): Early Findings. Science. 2009;324(5934):1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka Y, et al. A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc. Natl Acad. Sci. USA. 2002;99:15584–15589. doi: 10.1073/pnas.242608099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pybus OG, Drummond AJ, Nakano T, Robertson BH, Rambaut A. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol. Biol. Evol. 2003;20:381–387. doi: 10.1093/molbev/msg043. [DOI] [PubMed] [Google Scholar]
- 53.Deng X, Liu H, Shao Y, Rayner S, Yang R. The epidemic origin and molecular properties of B′: a founder strain of the HIV-1 transmission in Asia. AIDS. 2008;22:1851–1858. doi: 10.1097/QAD.0b013e32830f4c62. [DOI] [PubMed] [Google Scholar]
- 54.Paraskevis D, et al. Increasing prevalence of HIV-1 subtype A in Greece: estimating epidemic history and origin. J. Infect. Dis. 2007;196:1167–1176. doi: 10.1086/521677. [DOI] [PubMed] [Google Scholar]
- 55.Fargette D, et al. Diversification of rice yellow mottle virus and related viruses spans the history of agriculture from the Neolithic to the present. PloS Pathog. 2008;4:e1000125. doi: 10.1371/journal.ppat.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Almeida RP, Bennett GM, Anhalt MD, Tsai CW, O'Grady P. Spread of an introduced vector-borne banana virus in Hawaii. Mol. Ecol. 2009;18:136–146. doi: 10.1111/j.1365-294X.2008.04009.x. [DOI] [PubMed] [Google Scholar]
- 57.Leitner T, Albert J. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl Acad. Sci. USA. 1999;96:10752–10757. doi: 10.1073/pnas.96.19.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edwards CTT, et al. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol. Biol. 2006;6:28. doi: 10.1186/1471-2148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McAllister J, et al. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J. Virol. 1998;72:4893–4905. doi: 10.1128/jvi.72.6.4893-4905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N. Engl. J. Med. 1999;340:1228–1233. doi: 10.1056/NEJM199904223401602. [DOI] [PubMed] [Google Scholar]
- 61.Wrobel B, et al. Analysis of the overdispersed clock in the short-term evolution of hepatitis C virus: using the E1/E2 gene sequences to infer infection dates in a single source outbreak. Mol. Biol. Evol. 2006;23:1242–1253. doi: 10.1093/molbev/msk012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lemey P, et al. Molecular footprint of drug-selective pressure in a human immunodeficiency virus transmission chain. J. Virol. 2005;79:11981–11989. doi: 10.1128/JVI.79.18.11981-11989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leitner T, Escanilla D, Franzen C, Uhlen M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc. Natl Acad. Sci. USA. 1996;93:10864–10869. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paraskevis D, et al. Phylogenetic reconstruction of a known HIV-1 CRF04_cpx transmission network using maximum likelihood and Bayesian methods. J. Mol. Evol. 2004;59:709–717. doi: 10.1007/s00239-004-2651-6. [DOI] [PubMed] [Google Scholar]
- 65.Wilson DJ, Falush D, McVean G. Germs, genomes and genealogies. Trends Ecol. Evol. 2005;20:39–45. doi: 10.1016/j.tree.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 66.Pillay D, Rambaut A, Geretti AM, Brown AJL. HIV phylogenetics — criminal convictions relying solely on this to establish transmission are unsafe. BMJ. 2007;335:460–461. doi: 10.1136/bmj.39315.398843.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cottam EM, et al. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc. R. Soc. Lond. B. 2008;275:887–895. doi: 10.1098/rspb.2007.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Oliveira T, et al. Molecular epidemiology — HIV-1 and HCV sequences from Libyan outbreak. Nature. 2006;444:836–837. doi: 10.1038/444836a. [DOI] [PubMed] [Google Scholar]
- 69.Guan Y, et al. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Brown AJL. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PloS Med. 2008;5:392–402. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rezza G, et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 72.Lim SG, et al. Viral quasi-species evolution during hepatitis Be antigen seroconversion. Gastroenterology. 2007;133:951–958. doi: 10.1053/j.gastro.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 73.Shankarappa R, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolinsky SM, et al. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 75.Lemey P, et al. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 2007;3:282–292. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol. Biol. Evol. 2003;20:1318–1325. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- 77.Frost SD, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 2005;79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farci P, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 79.Sheridan I, Pybus OG, Holmes EC, Klenerman P. High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression. J. Virol. 2004;78:3447–3454. doi: 10.1128/JVI.78.7.3447-3454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farci P, et al. Evolution of hepatitis C viral quasispecies and hepatic injury in perinatally infected children followed prospectively. Proc. Natl Acad. Sci. USA. 2006;103:8475–8480. doi: 10.1073/pnas.0602546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leslie AJ, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nature Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 82.Moore CB, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 83.Bhattacharya T, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 84.Asquith B, Edwards CT, Lipsitch M, McLean AR. Inefficient cytotoxic T lymphocyte-mediated killing of HIV-1-infected cells in vivo. PLoS Biol. 2006;4:e90. doi: 10.1371/journal.pbio.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong JK, et al. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J. Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Korber BT, et al. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salemi M, et al. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J. Virol. 2005;79:11343–11352. doi: 10.1128/JVI.79.17.11343-11352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Iversen AKN, et al. Preferential detection of HIV subtype C′ over subtype A in cervical cells from a dually infected woman. AIDS. 2005;19:990–993. doi: 10.1097/01.aids.0000171418.91786.ad. [DOI] [PubMed] [Google Scholar]
- 89.Pillai SK, et al. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J. Virol. 2005;79:1734–1742. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sobesky R, et al. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology. 2007;46:1704–1712. doi: 10.1002/hep.21898. [DOI] [PubMed] [Google Scholar]
- 91.Mideo N, Alizon S, Day T. Linking within- and between-host dynamics in the evolutionary epidemiology of infectious diseases. Trends Ecol. Evol. 2008;23:511–517. doi: 10.1016/j.tree.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 92.Wensing AM, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J. Infect. Dis. 2005;192:958–966. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar]
- 93.Nowak MA, May RM. Virus Dynamics. 2000. [Google Scholar]
- 94.Nickle DC, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J. Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kelly JK, Williamson S, Orive ME, Smith MS, Holt RD. Linking dynamical and population genetic models of persistent viral infection. Am. Nat. 2003;162:14–28. doi: 10.1086/375543. [DOI] [PubMed] [Google Scholar]
- 96.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc. Natl Acad. Sci. USA. 2007;104:17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodrigo AG, et al. Coalescent estimates of HIV-1 generation time in vivo. Proc. Natl Acad. Sci. USA. 1999;96:2187–2191. doi: 10.1073/pnas.96.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Achaz G, et al. A robust measure of HIV-1 population turnover within chronically infected individuals. Mol. Biol. Evol. 2004;21:1902–1912. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- 99.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 100.Ehlers B, et al. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. J. Virol. 2008;82:3509–3516. doi: 10.1128/JVI.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Joy DA, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- 102.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 105.Bonhoeffer S, Holmes EC, Nowak MA. Causes of HIV diversity. Nature. 1995;376:125. doi: 10.1038/376125a0. [DOI] [PubMed] [Google Scholar]
- 106.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 2008;8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 108.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 109.Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- 110.Lee HY, Perelson AS, Park SC, Leitner T. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput. Biol. 2008;4:e1000240. doi: 10.1371/journal.pcbi.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maljkovic Berry I, et al. Unequal evolutionary rates in the human immunodeficiency virus type 1 (HIV-1) pandemic: the evolutionary rate of HIV-1 slows down when the epidemic rate increases. J. Virol. 2007;81:10625–10635. doi: 10.1128/JVI.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


