Abstract
We describe the isolation, biological and genetic characterization of a host-range variant of bovine coronavirus (BCoV) detected in water buffalo (Bubalus bubalis). By conventional and real-time RT-PCR assays, the virus was demonstrated in the intestinal contents of two 20-day-old buffalo calves dead of a severe form of enteritis and in the feces of additional 17 buffalo calves with diarrhea. Virus isolation, hemagglutination and receptor-destroying enzyme activity showed that the buffalo coronavirus (BuCoV) is closely related to BCoV but possesses some different biological properties. Sequence and phylogenetic analyses of the 3′ end (9.6 kb) of the BuCoV RNA revealed a genomic organization typical of group 2 coronaviruses. Moreover, the genetic distance between BuCoV and BCoV was proven to be the same or even higher than the distance between other ruminant coronaviruses and BCoV. In conclusion, our data support the existence of a host-range variant of BCoV associated with enteritis in buffaloes.
Keywords: Buffalo, Enteritis, Bubaline coronavirus, Biological characterization, Molecular characterization
Introduction
Coronaviruses (CoVs) (order Nidovirales, family Coronaviridae) are enveloped, positive-sense single-stranded RNA particles that are responsible for enteric and/or respiratory disease in mammals and birds. Their ability to evolve through genetic recombination and/or point mutation is well-known so that they give rise to new viral genotypes or mutants with different tissue or host tropism. Currently, CoVs are organized into three antigenic groups with group 2 including bovine-like (subgroup 2a) and SARS-like (subgroup 2b) viruses. Bovine coronavirus (BCoV) belongs to subgroup 2a together with mouse hepatitis virus, sialodacryadenitis virus, porcine hemagglutinating encephalomyelitis virus, human coronavirus (HCoV) OC43, human enteric coronavirus (HECV) 4408 (Enjuanes et al., 2000) and the newly recognized equine coronavirus (ECoV) (Guy et al., 2000), HCoV-HKU1 (Woo et al., 2005), and canine respiratory coronavirus (CRCoV) (Decaro et al., 2007a, Erles et al., 2003). There are multiple genetic and antigenic evidences that several subgroup 2a CoVs, such as HCoV-OC43, HECV-4408, PHEV and CRCoV, have arisen as a consequence of trans-species infections caused by BCoV (Zhang et al., 1994, Vijgen et al., 2005, Vijgen et al., 2006, Erles et al., 2007). BCoV can cause severe diarrhea in newborn calves (Snodgrass et al., 1986), “winter dysentery” in adult cows (Cho et al., 2000, Saif et al., 1991) and respiratory tract illness in calves and cows (Lathrop et al., 2000, Storz et al., 2000). The same virus strain could be responsible for simultaneous appearance of enteric and respiratory disease in the same animals (Chouljenko et al., 2001) as well as in both calves and cows (Tråvén et al., 2001). BCoV infection is responsible for severe economic losses in cattle herds due to decreased milk production in dairy cows and reduced weight gain in beef calves (Saif et al., 1998), with generally low mortality rates.
Water buffalo (Bubalus bubalis) represents a ruminant species important in the economy of several countries, including Brazil, India, Vietnam and some regions of southern Italy, where the breed Mediterranean Italian buffalo produces high quality milk employed for production of the buffalo “mozzarella cheese” (Romano et al., 2001). Recently, bovine-like CoVs have been identified in wild or domesticated ruminants, including several species of deer, waterbuck antelope (Tsunemitsu et al., 1995), giraffe (Giraffa camelopardalis) (Hasoksuz et al., 2007), alpaca (Lama pacos) (Jin et al., 2007) and sable antelope (Hippotragus niger) (Spiro D., R. Halpin, S. Wang, M. Hasoksuz, X. Zhang, K. Alekseev, A. Vlasova, D. Janies, E. Ghedin, and L. Saif, unpublished). However, CoVs have never been isolated from buffaloes, although there is a single report on the detection of BCoV antibodies in Bulgarian buffaloes by hemagglutination inhibition and virus neutralization tests (Muniiappa et al., 1985). In the same report, hemagglutinating activity which could be suppressed by a BCoV-specific bovine serum was also demonstrated in some bubaline fecal samples. In this paper, we describe the isolation and biological and genomic characterization of a bovine-like coronavirus from a buffalo herd affected by severe diarrhea and calf mortality.
Results
Clinical outbreak
The outbreak occurred between October 2006 and April 2007 in a herd of Mediterranean Italian buffalo (B. bubalis) in Campania (southern Italy). No cattle, sheep or goat farms were present in the vicinity of the outbreak. At that time the herd consisted of 460 buffaloes, including 215 lactating cows, all vaccinated against colibacillosis, clostridiosis and salmonellosis. Buffalo calves were removed from their dams shortly after their birth and placed in separate hutches according to the gender where they were hand-fed fresh colustrum for 5 days. Neonatal mortality was firstly observed in October 2006 in 30 5- to 20-day-old calves (out of 40 newborns) that displayed severe diarrhea and died despite treatment with antibiotics (oxytetracyclin and amoxicillin). Simultaneously, gastroenteric disease was also observed in older calves (1–3 months of age). On April 2007, neonatal mortality and enteric signs in calves persisted and two carcasses of 20-day-old dead calves together with fecal samples from additional 17 diseased calves were submitted to our laboratory for routine analysis.
Identification of a bovine-like CoV in buffalo calves
At necropsy, the carcasses of the two dead buffalo calves showed severe gastroenteritis, with enlargement of the mesenteric lymph nodes and gall bladder. By conventional RT-PCR (Erles et al., 2003), bovine-like CoV RNA was detected in the intestinal content of the dead animals as well as in all 17 fecal samples from calves with diarrhea. Sequence analysis of the S-gene fragments showed a 100% nucleotide identity among the CoV strains detected in different animals.
Gastrointestinal parasites of ruminants, including Cryptosporidium spp., were not detected by zinc sulfate flotation or Ziehl Nielsen staining, whereas by bacteriological investigations only common bacteria with poor pathogenic significance (E. coli, Streptococcus spp., Staphylococcus spp., Corynebacterium spp.) were isolated from the intestinal contents or fecal samples. Using real-time RT-PCR, bovine-like CoV RNA was detected at low titers in the intestinal contents of the dead animals, whereas higher viral loads were found in the fecal samples of calves with diarrhea, with a peak of 5.23 × 107 RNA copies/μl of template in calf 179/07-11 (Table 1 ).
Table 1.
Bovine-like CoV RNA titers detected by real-time RT-PCR in buffalo calves with diarrhea
| Prot. no. | Age of the animals | Sample type | RNA copies/μl of template |
|---|---|---|---|
| 179/07-A | 20 days | Intestinal content | 2.13 × 103 |
| 179/07-B | 20 days | Intestinal content | 9.41 × 102 |
| 179/07-1 | 8 days | Feces | 1.25 × 103 |
| 179/07-2 | 2 months | Feces | 4.10 × 104 |
| 179/07-3 | 2 months | Feces | 5.98 × 106 |
| 179/07-4 | 5 days | Feces | 4.55 × 104 |
| 179/07-5 | 1 month | Feces | 8.14 × 104 |
| 179/07-6 | 2 months | Feces | 3.27 × 105 |
| 179/07-7 | 5 days | Feces | 2.90 × 102 |
| 179/07-8 | 12 days | Feces | 6.71 × 103 |
| 179/07-9 | 2 months | Feces | 2.76 × 103 |
| 179/07-10 | 2 months | Feces | 7.18 × 102 |
| 179/07-11 | 3 months | Feces | 5.23 × 107 |
| 179/07-12 | 1 month | Feces | 2.04 × 102 |
| 179/07-13 | 1 month | Feces | 1.77 × 105 |
| 179/07-14 | 2 months | Feces | 3.34 × 104 |
| 179/07-15 | 1.5 months | Feces | 4.29 × 102 |
| 179/07-16 | 2 months | Feces | 6.83 × 103 |
| 179/07-17 | 2 months | Feces | 2.69 × 104 |
Biological characterization of the bubaline CoV
The fecal sample of calf 179/07-11 was used for virus isolation attempts on human rectal tumor (HRT-18) and Madin Darby bovine kidney (MDBK) cells. Cytopathic effect (CPE) consisting of syncytia formation and subsequent cell lysis was evident at the 2nd passage on HRT-18 cells (Fig. 1A), where the growth of a bovine-like CoV was confirmed by the cytoplasmic fluorescence detected by the IF assay using a BCoV-specific serum (Fig. 1B). At the 5th passage on HRT cells the viral titer reached values of 106.25 TCID50/50 μl of viral suspension and 1.97 × 108 RNA copies/ml of template, as calculated by the Karber's method and real-time RT-PCR, respectively. Replication of strain 179/07-11 in MDBK cells was not as efficient as in HRT-18 cells, considering that CPE was not observed on MDBK monolayers and only few positive cells were detected by the IF assay at the 1st passage, with viral titers (as calculated by real-time RT-PCR) decreasing progressively in the subsequent passages until definitive loss of the replication ability. MDBK cells were confirmed to be poorly permissive to 179/07-11 replication even when the HRT-18 cell-cultured virus was inoculated.
Fig. 1.
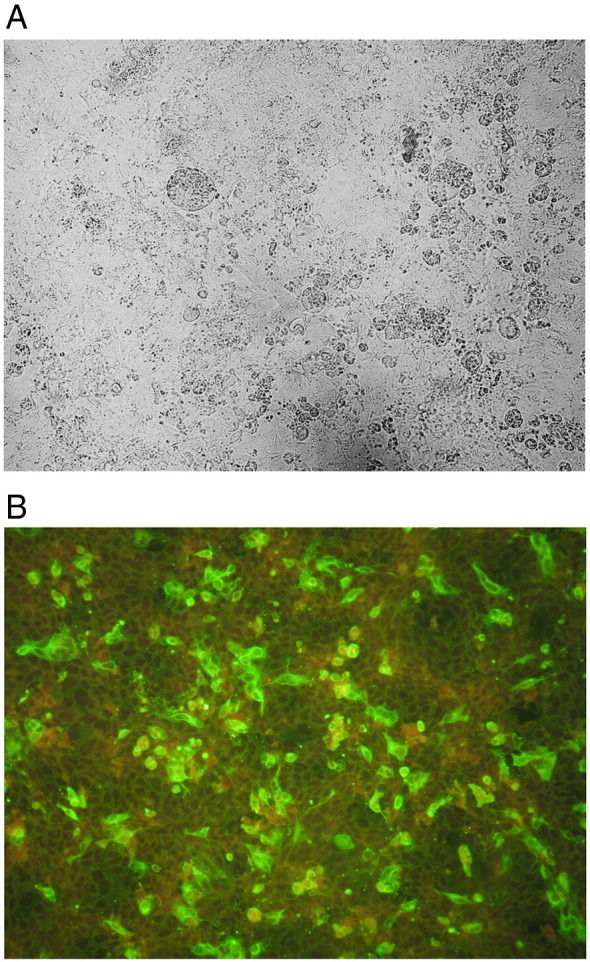
Virus isolation of the bubaline CoV on human rectal tumor cells. (A) Cytopathic effect (syncytia) caused by CoV strain 179/07-11 (72 h postinoculation). (B) Cytoplasmic fluorescence detected by the immunofluorescence assay using a BCoV-specific serum (24 h postinoculation).
The isolated strain was evaluated by assessment of hemagglutination (HA) and receptor-destroying enzyme (RDE) activities in comparison with the BCoV isolates 339/06 (Decaro et al., 2007b), 438/06-2 (Decaro et al., in press) and 9WBL7 (kindly provided by Dr Paolo Cordioli, Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna, Brescia, Italy). All isolates agglutinated mouse erythrocytes at titers higher at 4 °C than at 37 °C. The bovine isolates caused HA with chicken erythrocytes at low titers at both 4 °C and 37 °C, whereas the bubaline strain 179/07-11 did not hemagglutinate those cells despite the high HA titers obtained with mouse red blood cells (Table 2 ). Using mouse erythrocytes, RDE activity (loss of the HA pattern formed at 4 °C) was observed for the bovine and bubaline isolates (Table 2).
Table 2.
Hemagglutination (HA) and receptor-destroying enzyme (RDE) titers of bubaline CoV 179/07-11 and BCoV strains
| CoV strain | Origin | HA titera |
RDE titera |
||||
|---|---|---|---|---|---|---|---|
| 4 °C |
37 °C |
||||||
| Mouse | Chicken | Mouse | Chicken | Mouse | Chicken | ||
| BuCoV-179/07-11 | Feces | 512 | − | 128 | − | 128 | − |
| BCoV-339/06 | Feces | 128 | 64 | 64 | 32 | 64 | 16 |
| BCoV-438/06-2 | Nasal swab | 512 | 256 | 128 | 16 | 128 | 64 |
| BCoV-9WBL7 | Unknown | 1024 | 256 | 1024 | 64 | 256 | − |
HA and RDE titers per 50 μl of fecal suspension are expressed as reciprocals of the highest dilutions producing HA and resulting in complete disappearance of HA, respectively. Titers < 2 are indicated as negative (−).
Genetic characterization of the bubaline CoV
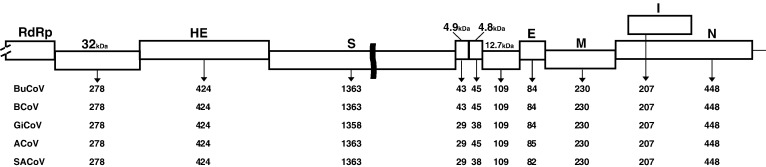
A 9.6-kb region encompassing the entire 3′ end of the viral genome (from the 32-kDa to nucleocapsid protein genes) was determined through PCR amplification and subsequent sequencing of overlapping fragments. At the 3′ end of viral RNA, bubaline CoV strain 179/07-11 had the same genomic organization of other ruminant CoVs (Fig. 2 ). Nine ORFs were identified by both ORF Finder program and sequence comparison with reference ruminant CoV sequences. All predicted ORFs but the 4.8-kDa gene were preceded by a repeated intergenic sequence, CUAAAC or CCAAAC, which is predicted to interact with the viral polymerase during the discontinuous transcription of the negative strand subgenomic RNA of the Nidovirales members (Table 3 ).
Fig. 2.
Schematic comparison of the genomes (3′ end) of different ruminant CoVs. RdRp, RNA dependent-RNA polymerase. Below the diagram, the length in amino acids is reported for the encoded proteins of bubaline coronavirus 179/07-11 (BuCoV), bovine coronavirus Mebus (BCoV), giraffe coronavirus US/OH3/2003 (GiCoV), alpaca coronavirus (ACoV) and sable antelope coronavirus US/OH1/2003 (SACoV).
Table 3.
Coding potential and putative transcription regulatory sequences of the 3′ end of the BuCoV genome
| Putative gene |
Putative TRS |
||
|---|---|---|---|
| Gene segment | Coding sequence | Start nt position | TRS sequence |
| 32 kDa | 200–1036 | 187 | CUAAAC |
| HE | 1048–2322 | 1033 | CUAAAC |
| S | 2337–6428 | 2330 | CUAAAC |
| 4.9 kDa | 6418–6549 | 6095 | CCAAAC |
| 4.8 kDa | 6585–6722 | – | Not detected |
| 12.7 kDa | 6806–7135 | 6725 | CCAAAC |
| E | 7122–7376 | 6993 | CCAAAC |
| M | 7390–8082 | 7381 | CCAAAC |
| N | 8092–9438 | 8079 | CUAAAC |
| I | 8153–8776 | 8079 | CUAAAC |
Table 4 shows the unique aa changes encountered in the encoded proteins of strain 179/07-11 with respect to other ruminant CoVs. The predicted 32-kDa nonstructural protein (nsp) had the same length (278 aa) as most other bovine-like CoVs, including ruminant CoVs giraffe coronavirus (GiCoV), alpaca coronavirus (ACoV) and sable antelope coronavirus (SACoV), as well as CRCoV and HCoV-OC43. Three aa changes were found to be unique to strain 179/07-11. The highest aa identity (98.9%) of the 32-kDa protein of strain 179/07-11 was found to enteric and respiratory BCoVs, as well as to ACoV. The hemagglutinin-esterase (HE) protein (424 aa) showed three unique aa mutations in comparison with other ruminant CoVs and it was more closely related (99% aa identity) to BCoVs DB2 and ENT and SACoV. By analysis with the NetNglyc server, the HE of strain 179/07-11 was found to contain nine potential N-glycosylation sites, analogously to BCoV reference strains. Moreover, the predicted site for neuraminidate-O-acetylesterase activity, FGDS, was detected at the N-terminus. The spike (S) protein was long 1363 aa and matched the best aa identity (98.1%) to BCoV-E-AH65 and SACoV-US/OH1/03. Twenty-one potential N-glycosylation sites were identified throughout the protein, one more than other ruminant CoVs, due to the presence of an additional N-glycosylation site, NLS, at position 1260. No deletions or insertions were observed in strain 179/07-11 compared with bovine-like CoVs. Also, the aa stretch KRRSRR, responsible for the proteolytic cleavage of the S protein at residue 768 into subunits S1 and S2, was conserved. In comparison to ruminant CoV reference strains, 16 unique residues were identified in the S protein of strain 179/07-11. Nsp 4.9 kDa was 43-aa long as for BCoVs Mebus, Quebec and DB2 and HECV-4408, whereas in other ruminant CoVs it is truncated in the C-terminus (only 29 aa in length). The length of nsp 4.8 kDa was 45 aa as for HECV-4408 and ruminant CoVs with the exception of GiCoV and SACoV (38 aa). Nsp 12.7 kDa showed the same size as for HECV-4408 and ruminant CoVs (109 aa), although BCoV Quebec exhibits a truncate form of the protein (87 aa). In the three nonstructural proteins, strain 179/07-11 displayed the highest identity to HECV-4408, with three, seven and one unique residues in nsp 4.9 kDa, 4.8 kDa and 12.7 kDa, respectively. The envelope (E) protein was 84-aa long exactly as BCoV and GiCoV but 1-aa shorter and 2-aa longer than the analogous protein of ACoV and SACoV, respectively. No residue unique to the bubaline strain was detected at this level. A 100% aa identity in the E protein was found between the bubaline strain and GiCoV and most BCoV strains, with the exception of BCoV-Mebus (98.8%), whereas the bubaline isolate was slightly less related to ACoV and SACoV. The membrane (M) protein was 220-aa long and contained two unique residues, with the I58L change shared with HECV-4408. In this protein the highest identity (99.5%) was matched to HECV-4408. One potential N-glycosylation and four potential O-glycosylation sites were detected at the N-terminus of the M protein, as for all bovine-like CoVs including the closely related HECV-4408, with which the bubaline isolates shared an aa mutation with respect to other ruminant CoVs. However, both programs indicated that M gene sequence of the ruminant CoVs might not contain a signal peptide. Proteins without signal peptides are unlikely to be exposed to the O-glycosylation and N-glycosylation machineries and thus may not be glycosylated in vivo even though they contain potential motifs. The nucleocapsid (N) protein of strain 179/07-11 had a length of 448 aa and was closely related (99.3% of aa identity) to BCoV-DB2. Two unique aa changes were found in comparison to other bovine-like viruses. The I protein (207 aa) encoded by the N-gene internal ORF showed four unique residues and the best aa identity (96.1%) to BCoV reference strains Mebus, Quebec and DB2. When the 9.6-kb sequence corresponding to the 3′ end of CoV genome was analyzed, the bubaline strain showed over 97.6% nt identity to BCoV strains and other ruminant CoVs, whereas it was less genetically related to other bovine-like CoVs, including CRCoV (96.5% nt identity), HCoV-OC43 (91.4%), HECV-4408 (85.1%) and PHEV-VW572 (83.1%).
Table 4.
Unique amino acid residues observed in structural and nonstructural proteins encoded by the 3′ end of the bubaline CoV genome in comparison with other ruminant CoVs
| CoV strain | 32 kDaa |
HE |
S |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 205 | 217 | 275 | 108 | 158 | 210 | 143 | 147 | 151 | 157 | 257 | 318 | 328 | 525 | 608 | 785 | 805 | 828 | 888 | 909 | 927 | 1260 | |
| BuCoV-179/07-11 | K | N | F | H | P | N | Y | F | I | V | N | T | F | Y | G | K | D | T | K | R | A | N |
| BCoV-Mebus | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | D |
| BCoV-ENT | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | D |
| BCoV-Quebec | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | D |
| BCoV-DB2 | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | H |
| GiCoV-US/OH3/2003 | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | D |
| ACoV | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | D |
| SACoV-US/OH1/2003 | M | K | V | Y | S | K | H | L | L | I | T | I | L | H | D | N | E | A | N | K | S | D |
| CRCoV-4182 | M | K | V | Y | S | K | H | L | L | I | T | I | L | Y | G | N | E | V | N | K | S | D |
| HECV-4408 | NA | NA | NA | Y | S | K | H | L | L | I | S | I | L | H | G | N | E | A | N | R | A | D |
| HCoV-OC43 | M | K | V | Y | A | R | R | S | V | V | T | I | L | D | D | N | E | A | N | K | S | D |
| PHEV-VW572 | M | NP | NP | Y | P | K | H | I | L | I | S | V | L | T | D | N | E | T | N | R | S | D |
| CoV strain | 4.9 kDab |
4.8 kDab |
12.7 kDa |
M |
N |
I |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 | 32 | 40 | 2 | 9 | 16 | 28 | 29 | 40 | 41 | 70 | 58 | 158 | 152 | 354 | 56 | 116 | 122 | 132 | |
| BuCoV-179/07-11 | P | A | T | Q | D | K | G | F | N | W | F | L | S | H | S | K | I | S | M |
| BCoV-Mebus | L | E | M | P | G | M | C | S | T | C | S | I | G | Q | N | R | T | L | R |
| BCoV-ENT | L | NP | NP | P | G | M | V | S | T | C | S | I | G | Q | N | R | T | L | R |
| BCoV-Quebec | L | E | M | NP | G | M | C | S | T | C | S | I | G | Q | N | R | T | L | R |
| BCoV-DB2 | L | E | M | P | G | M | V | S | T | C | S | I | G | Q | N | R | T | L | R |
| GiCoV-US/OH3/2003 | L | NP | NP | P | V | M | V | S | NP | NP | S | I | G | Q | N | R | T | L | R |
| ACoV | L | NP | NP | P | V | M | V | S | T | S | S | I | G | Q | N | R | T | L | R |
| SACoV-US/OH1/2003 | L | NP | NP | P | V | M | V | S | NP | NP | S | I | G | Q | N | R | T | L | R |
| CRCoV-4182 | NP | NP | NP | NP | NP | NP | NP | NP | NP | NP | S | I | G | Q | N | R | T | L | R |
| HECV-4408 | L | E | T | Q | D | M | G | S | N | W | S | L | G | Q | N | R | T | L | R |
| HCoV-OC43 | NP | NP | NP | NP | NP | NP | NP | NP | NP | NP | S | I | G | Q | N | NP | T | L | R |
| PHEV-VW572 | F | NP | NP | P | NP | NP | NP | NP | NP | NP | S | I | G | Q | N | R | T | L | R |
NA, not available; NP, not present.
Nsp 32 kDa of HECV-4408 is not available in the GenBank database, whereas it is truncated in PHEV-VW572.
Nsp 4.9 kDa and 4.8 kDa are not present in the CRCoV-4182 and HCoV-OC43 genomes. Nsp 4.9 kDa is truncated in PHEV-VW572.
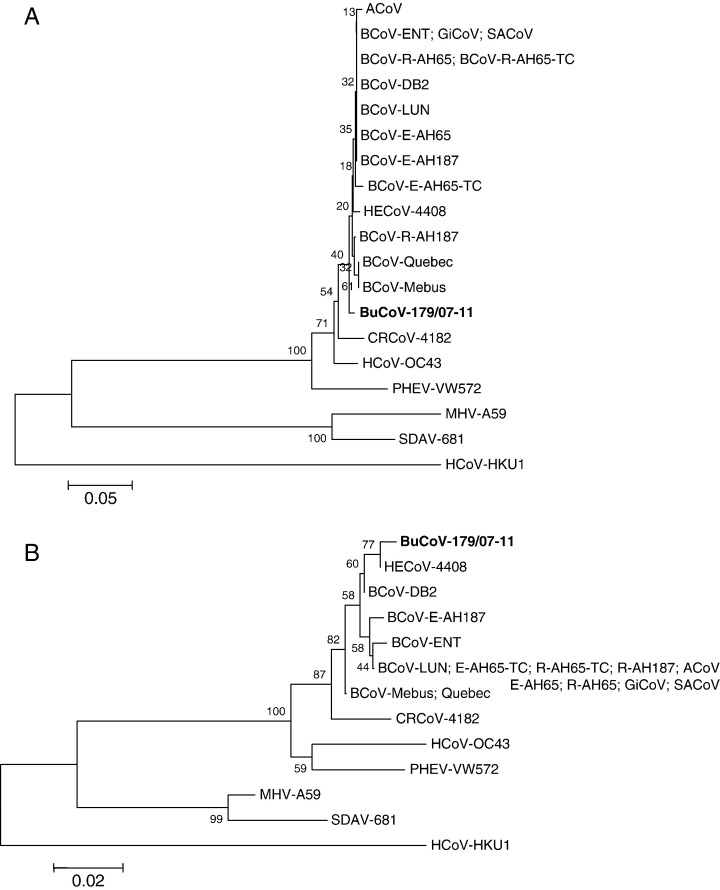
Phylogenetic analysis with the structural protein HE showed that the bubaline strain 179/07-11 clusters together with the bovine-like CoVs, being more related to ruminant viruses (Fig. 3 ). Similar phylogenetic trees were generated for the other proteins (data not shown) but the M protein, where strain 179/07-11 forms a separate cluster with HECV-4408 into the bovine subgroup (Fig. 3).
Fig. 3.
Neighbor-joining trees based on the hemagglutinin-esterase (A) and membrane (B) proteins of group 2 CoVs. For phylogenetic tree construction, the following CoV strains were used (GenBank accession numbers are reported in parentheses): BCoV-Mebus (U00735), Quebec (AF220295), DB2 (DQ811784), ENT (AF391541), LUN (AF391542), E-AH65 (EF424615), R-AH65 (EF424617), E-AH65-TC (EF424616), R-AH65-TC (EF424618), E-AH187 (EF424619), R-AH187 (EF424620); GiCoV-US/OH3/2003 (EF424623); ACoV (DQ915164); SACoV-US/OH1/2003 (EF424621); CRCoV-4182 (DQ682406); HCoV-OC43 (NC_005147); HECV-4408 (HE, L07747; M, AY316299); PHEV-VW572 (DQ011855); MHV-A59 (AY700211); SDAV (AF207551); HCoV-HKU1 (NC_006577). A statistical support was provided by bootstrapping over 1000 replicates. The scale bars indicate the estimated numbers of amino acid substitutions per site.
Evaluation of the genetic distance among ruminant CoVs
In order to better define the taxonomic position of the bubaline CoV 179/07-11, a sequence comparison between BCoV reference strain ENT and ruminant CoVs GiCoV, ACoV, SACoV and the bubaline strain was conduced in the nonstructural and structural proteins (Table 5 ). For such a comparison, strain ENT was chosen among the available BCoV sequences as it is an enteric strain isolated very recently (Chouljenko et al., 2001). The results showed that strain 179/07-11 is less related to BCoV-ENT than to other ruminant CoVs in all major structural and nonstructural proteins, whereas closer relationship (98.8% aa identity) to BCoV-ENT with respect to SACoV (97.6% aa identity) was found only in the E protein. Analysis of the nt sequence corresponding to the entire 3′ end of CoV genome confirmed that the bubaline strain has a lower genetic relationship (97.8% nt identity) to the reference BCoV than other recently isolated ruminant CoVs (99.1–99.3% nt identity). Confirmation of the conclusions drawn from Table 5 was derived with the use of SimPlot analysis.
Table 5.
Sequence identity (%) of ruminant CoVs to reference BCoV-ENT in nonstructural and structural proteins and in the full-length sequence of the genomic RNA 3′ end (9.6 kb)
| Gene segment | CoV strain |
|||
|---|---|---|---|---|
| BuCoV 179/07-11 | GiCoV US/OH3/2003 | ACoV | SACoV US/OH1/2003 | |
| 32 kDaa | 98.2 | 98.5 | 99.2 | 98.5 |
| HEa | 99.0 | 100 | 99.7 | 100 |
| Sa | 98.0 | 98.8 | 99.1 | 99.3 |
| 4.9 kDaa | 55.8 | 100 | 100 | 100 |
| 4.8 kDaa | 75.5 | 82.2 | 93.3 | 82.2 |
| 12.7 kDaa | 98.1 | 99.0 | 99.0 | 98.1 |
| Ea | 98.8 | 100 | 98.8 | 97.6 |
| Ma | 98.2 | 99.5 | 99.5 | 99.5 |
| Na | 98.4 | 99.5 | 99.7 | 99.5 |
| Ia | 95.1 | 98.0 | 99.0 | 98.5 |
| 9.6-kb 3′ endb | 97.8 | 99.1 | 99.3 | 99.2 |
Amino acid identity.
Nucleotide identity.
Discussion
In the present report, a bovine-like CoV in water buffalo (B. bubalis) has been described, along with its biological and genetic characterization. The bubaline strain was detected in the feces of buffalo calves with severe gastroenteritis, causing the death of at least 30 animals. A virus (strain 179/07-11) antigenically related to BCoV was isolated on HRT-18 cells, producing CPE typical of BCoV and being recognized by a BCoV-specific serum in an IF assay. Analysis of the biological properties of the bubaline strain showed that, unlike classical BCoV strains (Fukutomi et al., 1999), the virus was not able to replicate efficiently on MDBK cells or cause hemagglutination with chicken erythrocytes. However, Benfield and Saif (1990) demonstrated that winter dysentery strains of BCoV from adult cattle were also unable to replicate in MDBK cells. Among wild ruminant CoVs, only few strains have been characterized at a biological level. GiCoV displayed HA activity also with chicken erythrocytes at a temperature of 4 °C, whereas no data are available about its replication efficiency on MDBK cells (Hasoksuz et al., 2007). Using chicken erythrocytes, other ruminant CoVs, including the waterbuck, sambar deer and white-tailed deer isolates and some BCoV strains, exhibited very low HA titers at 4 °C and no HA activity at 37 °C. Thus, the loss of ability to replicate on MDBK cells and to agglutinate chicken erythrocytes appears as characteristic of the bubaline CoV with respect to the other ruminant CoVs.
Sequence analysis of the 3′ end of the viral genome showed that strain 179/07-11 has a genomic organization similar to BCoV, including the presence of three ORFs encoding for small nonstructural proteins between the S and E genes. Moreover, the virus has a close genetic relatedness to the bovine CoV subgroup in the major structural and nonstructural proteins. However, as shown by sequence and SimPlot analyses, the genetic distance between the bubaline virus and reference BCoV strain ENT was approximately the same or even higher than observed between the BCoV strain and other ruminant CoVs. Unique aa substitutions were detected in all proteins of the bubaline virus but they were mainly within the S protein (Table 4). As observed in BCoV (Chouljenko et al., 1998), GiCoV (Hasoksuz et al., 2007) and ACoV (Jin et al., 2007), these changes occurred mostly in the S1 subunit which has been proven to be involved in receptor binding and host specificity in the well-characterized MHV (de Haan et al., 2006). Certain residues in the S protein had been associated to enteric or respiratory tropism of BCoV, including residues 531, 769 and 1026 (Chouljenko et al., 1998). The bubaline strain displayed the same aa residues encountered in enteric BCoVs in only one position (D531). The remaining two aa residues (S769, G1026) suggest a potential respiratory tropism, which is in contrast with the exclusive enteric clinical pattern observed in water buffalo. However, more recent analysis of two enteric and respiratory BCoV pairs failed to identify specific aa residue determinants in the S proteins as well as in other structural and nonstructural proteins, except for a T22I mutation in nsp 4.8 kDa (Zhang et al., 2007). Interestingly, strain 179/07-11 showed the same aa determinant T22 as the enteric BCoV strains. Other authors have identified a truncated form of nsp 4.9 kDa (29 instead of 43 aa) as a genetic marker for the respiratory tropism (Gelinas et al., 2001, Vijgen et al., 2006). The bubaline strain presented an intact nsp 4.9 kDa as enteric BCoVs, but even this aa deletion has been recently shown not to be involved in tissue tropism (Zhang et al., 1994).
Based on the unique biological properties and the more distant relatedness of strain 179/07-11 to BCoV with respect to other ruminant CoVs, we propose to designate this strain as prototype of a host-range variant of BCoV, namely bubaline CoV (BuCoV). With regard to the its possible origin, BuCoV may have arisen through interspecies transmission of a BCoV strain from cattle to water buffaloes, although no recent contacts with cattle or other ruminants were reported. This hypothesis is supported by the high genetic relatedness between BuCoV and BCoV. Moreover, it should be considered that a bovine origin has been strongly suggested for other group 2 CoVs less genetically related to BCoV, such as HCoV-OC43 (Vijgen et al., 2005), PHEV (Vijgen et al., 2006) and CRCoV (Erles et al., 2007).
Accumulation of point mutations, as well as small insertions and deletions in coding and non-coding sequences, are the dominant forces in the microevolution of plus-stranded (+) RNA viruses, resulting in proliferation of virus strains, serotypes and subtypes (Dolja and Carrington, 1992). Coronaviruses are thought to mutate at high frequency like most RNA viruses as a consequence of high error rates of the RNA polymerase that are predicted to accumulate several base substitutions per round of replication (Jarvis and Kirkegaard, 1991, Lai and Holmes, 2001). Changes in tissue tropisms and/or interspecies transmission of CoVs occur through genetic variations in structural and/or nonstructural proteins (Laude et al., 1993, Vennema et al., 1998, Guan et al., 2003, Rottier et al., 2005, Song et al., 2005, Vijgen et al., 2005, Decaro et al., 2007c). Genetic determinants that may have caused the cattle-to-buffalo interspecies transmission have not been identified so far. Although this study has detected a CoV strain in buffalo calves with severe diarrhea, the pathogenicity of this virus and its etiologic role in enteric disease of water buffalo still have to be determined more definitively. In addition, epidemiological studies could assess whether BuCoV is widespread among water buffalo herds as well as whether cross-species transmission between buffalo and cattle occurs mainly in areas where both closely related ruminant species are raised intensively.
Materials and methods
(RT-)PCR assay for screening for bovine viral pathogens
Nucleic acids for (RT-)PCR assays were purified using the commercial kits DNeasy Tissue Kit (QIAGEN S.p.A., Milan, Italy) and QIAamp® Viral RNA Mini Kit (Qiagen S.p.A., Milan, Italy) from the fecal samples and intestinal contents and QIAamp® RNeasy Mini Kit (Qiagen S.p.A.) from the tissue samples.
RNA extracts were used for detection of BCoV (Erles et al., 2003), toroviruses (Hoet et al., 2002), rotaviruses (Gouvea et al., 1994), caliciviruses (Jiang et al., 1999), bovine viral diarrhea virus (BVDV) (Sullivan and Akkina, 1995) and bovine respiratory syncytial virus (Valarcher et al., 1999). Detection of bovine herpesvirus types 1 (BoHV-1) (Vilcek, 1993) and 4 (BoHV-4) (Boerner et al., 1999) was carried out on the DNA templates. RT-PCR and PCR assays were performed using SuperScript™ One-Step RT-PCR for Long Templates (Life Technologies) and LA PCR Kit Ver. 2.1 (TaKaRa Bio Inc., Shiga, Japan), respectively.
Bacteriological and parasitological investigations
The samples were examined for bacterial and parasitic pathogens by standardized methods. For bacteriological investigations, the fecal samples were plated out on 5% sheep blood agar and cultured aerobically at 37 °C for 24 h to exclude the presence of aerobic pathogens. One hundred milligrams of fecal samples was resuspended in 900 μl of fluid thioglycolate medium (FTG). Ten-fold dilutions (10− 2 to 10− 8) were subsequently plated onto 5% sheep blood agar and egg yolk agar with d-cycloserine 400 μg/ml. Bacteria were allowed to grow overnight at 37 °C in anaerobic condition. Detection of the most common enteric parasites was achieved using zinc sulfate flotation. The Ziehl Nielsen staining was also performed on fecal samples or intestinal sections for detection of Cryptosporidium spp.
Real-time RT-PCR for quantification of bovine-like CoVs
A real-time RT-PCR assay based on TaqMan technology (G. Elia et al., manuscript in preparation) was used to quantify the viral load in samples tested positive for bovine-like CoVs by conventional RT-PCR (Erles et al., 2003). The TaqMan assay had been proven to be able to detect BCoV as well as the closely related CRCoV (Elia et al., manuscript in preparation). BCoV-like RNA copy numbers were calculated on the basis of the standard curves generated by 10-fold dilutions of a synthetic RNA obtained by in vitro transcription of a plasmid containing the M gene of BCoV strain 339/07 (Decaro et al., 2007b). Reverse transcription was carried out using GeneAmp® RNA PCR (Applied Biosystems, Applera Italia, Monza, Italy), following the manufacturer's recommendations. The quantitative assay targeting the M gene was conducted in a 50-μl reaction mixture containing 25 μl of IQ™ Supermix (Bio-Rad Laboratories Srl), 600 nM of primers BCoV-F (CCTTCATATCTATACACATCAAGTTGTT) and BCoV-R (ACCAGCCATTTTAAATCCTTCA), 200 nM of probe BCoV-Pb (6FAM− CCTTCATATCTATACACATCAAGTTGTT-BHQ1) and 20 μl of c-DNA. The thermal profile consisted of activation of iTaq DNA polymerase at 95 °C for 10 min followed by 45 cycles of denaturation at 95 °C for 15 s and annealing extension at 60 °C for 1 min.
Virus isolation
Virus isolation was carried out on the buffalo fecal sample containing the highest RNA titer of BCoV-like coronavirus (strain 179/07-11), as determined by real-time RT-PCR. Feces were homogenized in Dulbecco's minimal essential medium (D-MEM) containing antibiotics (penicillin 5000 IU/ml, streptomycin 2500 μg/ml, amphotericin B 10 μg/ml). After centrifugation at 3000×g for 15 min, the supernatant was used to inoculate confluent monolayers of HRT-18 and MDBK cells in the presence of trypsin (5 μg/ml). Viral growth was monitored by an immunofluorescence (IF) assay using a BCoV-positive bovine serum and a rabbit anti-bovine IgG conjugated with fluorescein isothiocyanate (Sigma Aldrich srl, Milan, Italy).
HA and RDE activity
Two-fold dilutions (starting from dilution 1:2) of the supernatant of the CoV-infected HRT-18 cells tested positive by the IF assay were made in phosphate buffered saline (PBS, pH 7.2) using 96-well V-plates. Mouse (0.8%) or chicken (0.4%) erythrocytes were added to each dilution. Results were read after 1 h at 4 °C or 37 °C and expressed as the reciprocal of the highest dilution producing HA. The plates were then incubated for additional 4 h at 37 °C to assess RDE activity, which was expressed as the reciprocal of the highest dilution of virus resulting in complete disappearance of HA (Hasoksuz et al., 1999).
PCR amplifications of the 3′ end of the BCoV-like virus
The 3′ end of the genome of strain 179/07-11 was amplified from the original fecal sample using SuperScript™ One-Step RT-PCR for Long Templates (Life Technologies, Invitrogen. Milan, Italy), according to the manufacturer's instructions. Thirteen partially overlapping fragments encompassing from the very 3′ end of ORF1b to the 5′ end of the untranslated region (UTR) were amplified using primer pairs designed on conserved regions among BCoV-like group 2 coronaviruses (Table 6 ).
Table 6.
Primers used for RT-PCR amplification and sequence analysis
| Primer | Sequence 5′ to 3′ | Sense | Positiona | Amplicon size (bp) |
|---|---|---|---|---|
| BCV-22001F | TAGACTTGAAATAGTTAAGCTTGGTG | + | 22,001–22,026 | 893 |
| BCV-22894R | AAATTAGCTTCACGAGCTATATATGC | − | 22,868–22,893 | |
| BCV-22769F | TATCGCAGCCTTACTTTTGTTAATG | + | 22,769–22,793 | 527 |
| BCV-23295R | CGAAAATAACAGTACGGGGGTTGACA | − | 23,270–23,295 | |
| BCV-23112F | TACCCTCTGGTAATTATTTAGCCATTTCA | + | 23,112–23,140 | 776 |
| BCV-23887R | TTCCCTTCAGTGCCATATTACGATATGT | − | 23,860–23,887 | |
| BCV-23510F | TATGATCCGCTACCAATTATTTTGCTTGGCA | + | 23,510–23,540 | 817 |
| BCV-24326R | ACAACACCAGTGTCTGTAAAATATGCA | − | 24,300–24,326 | |
| BCV-24182F | TAGAACTATGGCATTGGGATACAGGTGTTG | + | 24,182–24,211 | 1254 |
| BCV-25435R | TACACCTATCCCCTTGTAAACAAGAGTA | − | 25,409–25,435 | |
| BCV-25301F | ACTTAGTTGGCATAGGTGAGCACTGTTC | + | 25,301–25,328 | 851 |
| BCV-26151R | ACATGCTACATAATCACCACAGACAA | − | 26,126–26,151 | |
| BCV-26028F | TTTGTATGAAATTCAAATACCTTCAGAG | + | 26,028–26,055 | 877 |
| BCV-26904R | GTCTATCTGAGCTTGCGCTTCAAGAGCA | − | 26,877–26,904 | |
| BCV-26760F | TAAAATTCAAGCTGTTGTTAATGCAAAT | + | 26,760–26,787 | 1059 |
| BCV-27818R | GCTCGACCTAAATGGGTCTTATAATTAGA | − | 27,791–27,818 | |
| BCV-27667F | GGTGGTTGTTGTGATGATTATACTGGACA | + | 27,667–27,695 | 853 |
| BCV-28519R | ACTACAACTATTATAACCAATAAACACAT | − | 28,491–28,519 | |
| BCV-28380F | TCTTAGCTGTTGACTTTATTACCTGG | + | 28,380–28,405 | 919 |
| BCV-29298R | ACATAAACAGCAAAACCACTAGTATCGCC | − | 29,270–29,298 | |
| BCV-29160F | AAGGTATAAAACTAGGTACTGGCTAT | + | 29,160–29,185 | 876 |
| BCV-30035R | TGCGCGATCCTGCACTAGAGGCTCTAC | − | 30,009–30,035 | |
| BCV-29900F | GAGGCTATTCCGACTAGGTTTCCGCCT | + | 29,900–29,926 | 827 |
| BCV-30726R | GTGTCTTCAGTAAAGGGCTCATCCATC | − | 30,700–30,726 | |
| BCV-30610F | TGATAATATAAGTGTTGCAGCGCCCAAA | + | 30,610–30,637 | 419 |
| BCV-31028R | GTGATTCTTCCAATTGGCCATAATT | − | 31,004–31,028 | |
Primers position is referred to the sequence of BCoV strain ENT (accession number AF391541).
Sequence analysis and phylogeny
The PCR-amplified products were sequenced by Genome Express (Meylan, France) and the obtained sequences were assembled and analyzed using the BioEdit software package (Hall, 1999) and the NCBI's (htttp://www.ncbi.nlm.nih.gov) and EMBL's (http://www.ebi.ac.uk) analysis tools. Additional RT-PCR assays and sequencing attempts were performed to close gaps between assembled contigs using strain-specific primers. ORFs contained in the amplified genomic region were determined either with the ORF Finder tool of NCBI or on the basis of the similarity to known coronavirus proteins. The ORFs identified in this manner were translated and the predicted amino acid (aa) sequences were saved as individual files for further analyses. Putative N-glycosylation sites were predicted using the NetNglyc 1.0 Server program (http://www.cbs.dtu.dk/services/NetNGlyc/), whereas the NetOGlyc 3.1 Server program (http://www.cbs.dtu.dk/services/NetOGlyc/) was used for prediction of O-linked glycosylation sites. Phylogenetic and molecular evolutionary analyses were conducted using Mega3 (Kumar et al., 2004). Phylogenetic trees, based on the nonstructural 32-kDa protein, on the structural spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins and on the genomic RNA 3′ end (9.6 kb) of CoV strain 179/07-11, were elaborated using both parsimony and neighbor-joining methods, supplying a statistical support with bootstrapping over 1000 replicates.
SimPlot analysis
The SimPlot program (version 3.2) was used to analyze the genetic distance of the 3′ end of the genomes of ruminant CoVs, including the bubaline strain, in reference to the same region of BCoV strain ENT and this genetic distance was plotted versus nucleotide (nt) positions (Lole et al., 1999).
Nucleotide sequence accession number
The nt sequence of the bubaline strain 179/07-11 has been deposited in GenBank under accession number EU019216.
Acknowledgments
This work was supported by grants from University of Bari, Italy: project ex 60% 2007 “Messa a punto di un sistema real-time RT-PCR per la identificazione e la quantificazione dell'RNA del coronavirus bovino”.
References
- Benfield D.A., Saif L.J. Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J. Clin. Microbiol. 1990;28:1454–1457. doi: 10.1128/jcm.28.6.1454-1457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner B., Weigelt W., Buhk H.J., Castrucci G., Ludwig H. A sensitive and specific PCR/Southern blot assay for detection of bovine herpesvirus 4 in calves infected experimentally. J. Virol. Methods. 1999;83:169–180. doi: 10.1016/s0166-0934(99)00117-2. [DOI] [PubMed] [Google Scholar]
- Cho K.O., Halbur P.G., Bruna J.D., Sorden S.D., Yoon K.J., Janke B.H., Chang K.O., Saif L.J. Detection and isolation of coronavirus from feces of three herds of feedlot cattle during outbreaks of winter dysentery-like disease. J. Am. Vet. Med. Assoc. 2000;217:1191–1194. doi: 10.2460/javma.2000.217.1191. [DOI] [PubMed] [Google Scholar]
- Chouljenko V.N., Kousoulas K.G., Lin X., Storz J. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes. 1998;17:3–42. doi: 10.1023/A:1008048916808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouljenko V.N., Lin X.Q., Storz J., Kousoulas K.G., Gorbalenya A.E. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 2001;82:2927–2933. doi: 10.1099/0022-1317-82-12-2927. [DOI] [PubMed] [Google Scholar]
- de Haan C.A., Te Lintelo E., Li Z., Raaben M., Wurdinger T., Bosch B.J., Rottier P.J. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J. Virol. 2006;80:10909–10918. doi: 10.1128/JVI.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Elia G., Mari V., Lucente M.S., Cordioli P., Colaianni M.L., Martella V., Buonavoglia C. Serological and molecular evidence that canine respiratory coronavirus is circulating in Italy. Vet. Microbiol. 2007;121:225–230. doi: 10.1016/j.vetmic.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Desario C., Campolo M., Elia G., Martella V., Greco G., Cirone F., Colaianni M.L., Cordioli P., Buonavoglia C. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet. Microbiol. 2007 doi: 10.1016/j.vetmic.2007.06.024. Electronic publication ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Elia G., Campolo M., Desario C., Cirone F., Tempesta M., Buonavoglia C. Molecular characterisation of the virulent canine coronavirus CB/05 strain. Virus Res. 2007;125:4–60. doi: 10.1016/j.virusres.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro, N., Campolo, M., Desario, C., Cirone, F., D'abramo, M., Lorusso, E., Greco, G., Mari, V., Colaianni, M.L., Elia, G., Martella, V., Buonavoglia C., in press. Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy. J. Vet. Diagn. Invest. [DOI] [PubMed]
- Dolja V.V., Carrington J.C. Evolution of positive-strand RNA viruses. Semin. Virol. 1992;3:315–326. [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S., Spaan W.J.M., Taguchi F., Talbot P. Family Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy, Classification and Nomenclature of Viruses. Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310:216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Shiu K.B., Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 2007;124:78–87. doi: 10.1016/j.virusres.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutomi T., Tsunemitsu H., Akashi H. Detection of bovine coronaviruses from adult cows with epizootic diarrhea and their antigenic and biological diversities. Arch. Virol. 1999;144:997–1006. doi: 10.1007/s007050050562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas A.M., Boutin M., Sasseville A.M., Dea S. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res. 2001;76:43–57. doi: 10.1016/S0168-1702(01)00243-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V., Santos N., Mdo C. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guy J.S., Breslin J.J., Breuhaus B., Vivrette S., Smith L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hasoksuz M., Lathrop S.L., Gadfield K.L., Saif L.J. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 1999;60:1227–1233. [PubMed] [Google Scholar]
- Hasoksuz M., Alekseev K., Vlasova A., Zhang X., Spiro D., Halpin R., Wang S., Ghedin E., Saif L.J. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 2007;81:4981–4990. doi: 10.1128/JVI.02361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet A.E., Cho K.O., Chang K.O., Loerch S.C., Wittum T.E., Saif L.J. Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 2002;63:342–348. doi: 10.2460/ajvr.2002.63.342. [DOI] [PubMed] [Google Scholar]
- Jarvis T.C., Kirkegaard K. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 1991;7:186–191. doi: 10.1016/0168-9525(91)90434-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Huang P.W., Zhong W.M., Farkas T., Cubitt D.W., Matson D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods. 1999;83:145–154. doi: 10.1016/s0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- Jin L., Cebra C.K., Baker R.J., Mattson D.E., Cohen S.A., Alvarado D.E., Rohrmann F. Analysis of the genome sequence of an alpaca coronavirus. Virology. 2007;365:198–203. doi: 10.1016/j.virol.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lai M.M.C., Holmes K.V. Coronaviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 4th edition. Lippincott Williams and Wilkins; Philadelphia, PA: 2001. pp. 1163–1185. [Google Scholar]
- Lathrop S.L., Wittum T.E., Brock K.V., Loerch S.C., Perino L.J., Bingham H.R., McCollum F.T., Saif L.J. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am. J. Vet. Res. 2000;61:1062–1066. doi: 10.2460/ajvr.2000.61.1062. [DOI] [PubMed] [Google Scholar]
- Laude H., Van Reeth K., Pensaert M. Porcine respiratory coronavirus: molecular features and virus–host interactions. Vet. Res. 1993;24:125–150. [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R., Gadkari S., Kulkarni D., Novak S.S., Ingersoll N.G., Sheppard R., Ray H.W. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniiappa L., Mitov B.K., Kharalambiev Kh.E. Demonstration of coronavirus infection in buffaloes. Vet. Med. Nauki. 1985;22:27–32. [PubMed] [Google Scholar]
- Romano P., Ricciardi A., Salzano G., Suzzi G. Yeasts from water buffalo mozzarella, a traditional cheese of the Mediterranean area. Int. J. Food Microbiol. 2001;69:45–51. doi: 10.1016/s0168-1605(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Rottier P.J., Nakamura K., Schellen P., Volders H., Haijema B.J. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005;79:14122–14130. doi: 10.1128/JVI.79.22.14122-14130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J., Brock K.V., Redman D.R., Kohler E.M. Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet. Rec. 1991;128:447–449. doi: 10.1136/vr.128.19.447. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Redman D.R., Brock K.V., Kohler E.M., Heckert R.A. Winter dysentery in adult dairy cattle: detection of coronavirus in the faeces. Vet. Rec. 1998;123:300–301. doi: 10.1136/vr.123.11.300. [DOI] [PubMed] [Google Scholar]
- Snodgrass D.R., Terzolo H.R., Sherwood D., Campbell I., Menzies J.D., Synge B.A. Aetiology of diarrhoea in young calves. Vet. Rec. 1986;119:31–34. doi: 10.1136/vr.119.2.31. [DOI] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu H.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J., Purdy C.W., Lin X., Burrell M., Truax R.E., Briggs R.E., Frank G.H., Loan R.W. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp from cattle involved in two natural outbreaks of shipping fever. J. Am. Vet. Med. Assoc. 2000;216:1599–1604. doi: 10.2460/javma.2000.216.1599. [DOI] [PubMed] [Google Scholar]
- Sullivan D.G., Akkina R.K. A nested polymerase chain reaction assay to differentiate pestiviruses. Virus Res. 1995;38:231–239. doi: 10.1016/0168-1702(95)00065-x. [DOI] [PubMed] [Google Scholar]
- Tråvén M., Naslund K., Linde N., Linde B., Silvan A., Fossum C., Hedlund K.O., Larsson B. Experimental reproduction of winter dysentery in lactating cows using BCV—comparison with BCV infection in milk-fed calves. Vet. Microbiol. 2001;81:127–151. doi: 10.1016/S0378-1135(01)00337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemitsu H., el-Kanawati Z.R., Smith D.R., Reed H.H., Saif L.J. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J. Clin. Microbiol. 1995;33:3264–3269. doi: 10.1128/jcm.33.12.3264-3269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarcher J.F., Bourhy H., Gelfi J., Schelcher F. Evaluation of a nested reverse transcription-PCR assay based on the nucleoprotein gene for diagnosis of spontaneous and experimental bovine respiratory syncytial virus infections. J. Clin. Microbiol. 1999;37:1858–1862. doi: 10.1128/jcm.37.6.1858-1862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Thoelen I., Wollants E., Lemey P., Vandamme A.M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek S. Detection of the bovine herpesvirus-1 (BHV-1) genome by PCR. J. Virol. Methods. 1993;41:245–247. doi: 10.1016/0166-0934(93)90132-b. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H, Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hasoksuz M., Spiro D., Halpin R., Wang S., Vlasova A., Janies D., Jones L.R., Ghedin E., Saif L.J. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology. 2007;363:1–10. doi: 10.1016/j.virol.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]