Abstract
The discovery of novel viruses is of great importance to human health—both in the setting of emerging infectious disease outbreaks and in disease syndromes of unknown etiology. Despite the recent proliferation of many efficient virus discovery methods, careful selection of a combination of methods is important to demonstrate a novel virus, its clinical associations, and its relevance in a timely manner. The identification of a patient or an outbreak with distinctive clinical features and negative routine microbiological workup is often the starting point for virus hunting. This review appraises the roles of culture, electron microscopy, and nucleic acid detection–based methods in optimizing virus discovery. Cell culture is generally slow but may yield viable virus. Although the choice of cell line often involves trial and error, it may be guided by the clinical syndrome. Electron microscopy is insensitive but fast, and may provide morphological clues to choice of cell line or consensus primers for nucleic acid detection. Consensus primer PCR can be used to detect viruses that are closely related to known virus families. Random primer amplification and high-throughput sequencing can catch any virus genome but cannot yield an infectious virion for testing Koch postulates. A systematic approach that incorporates carefully chosen combinations of virus detection techniques is required for successful virus discovery.
CME Accreditation Statement: This activity (“JMD 2015 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2015 CME Program in Molecular Diagnostics”) for a maximum of 36 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
The Importance of Virus Discovery
Novel viruses are important causes of emerging infectious diseases, as illustrated by multinational outbreaks of severe respiratory illness due to two novel coronaviruses in recent times: severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus. Such outbreaks entail considerable public health concern, requiring investigators to discover the infectious agent in a timely manner by efficiently using virus diagnostic tools.
Even outside outbreak settings, many clinical syndromes encountered by clinicians on a daily basis, such as coryza, pneumonia, diarrhea, meningoencephalitis, hepatitis, epididymo-orchitis, and sialadenitis, have no identifiable infectious etiology, raising the possibility of infections by as-yet undiscovered pathogens. At the present time, >200 viruses are known to cause human disease; extrapolation of recent trends anticipates that pathogenic virus discovery is likely to continue unabated in the near future.1 Given the importance of microbial stimuli in determining short- and long-term immunological stimulation, it is conceivable that some of these undiscovered viruses might even contribute to autoimmune, degenerative, and neoplastic conditions of currently unknown etiology.2 Novel viruses may also play a role in the etiology of common bacterial infections by impairing local and systemic immune mechanisms—a phenomenon that is frequently noted in viral infections, such as measles.3
The importance of discovering viruses that play an etiological role in infectious disease outbreaks and clinical syndromes transcends academic interest. If direct causality between the novel virus and the disease syndrome is established by clinical associations and animal models, then further study of the novel virus is indicated for defining specific diagnostic, therapeutic, and preventative measures. This was illustrated by the discovery of the SARS-CoV, which paved the way to an understanding of the origins of the outbreak, specific control measures, diagnostics, and therapeutic initiatives. Discovery of novel viruses may also carry important implications for blood product, organ transplant, bioterrorism, and laboratory safety. Furthermore, the experience with smallpox has shown that viruses and the diseases they cause may be eradicated by global health initiatives. Therefore, rigorous investigation for novel viral agents is of considerable importance to human health.
The Steps in Virus Discovery
Case Identification and Selection
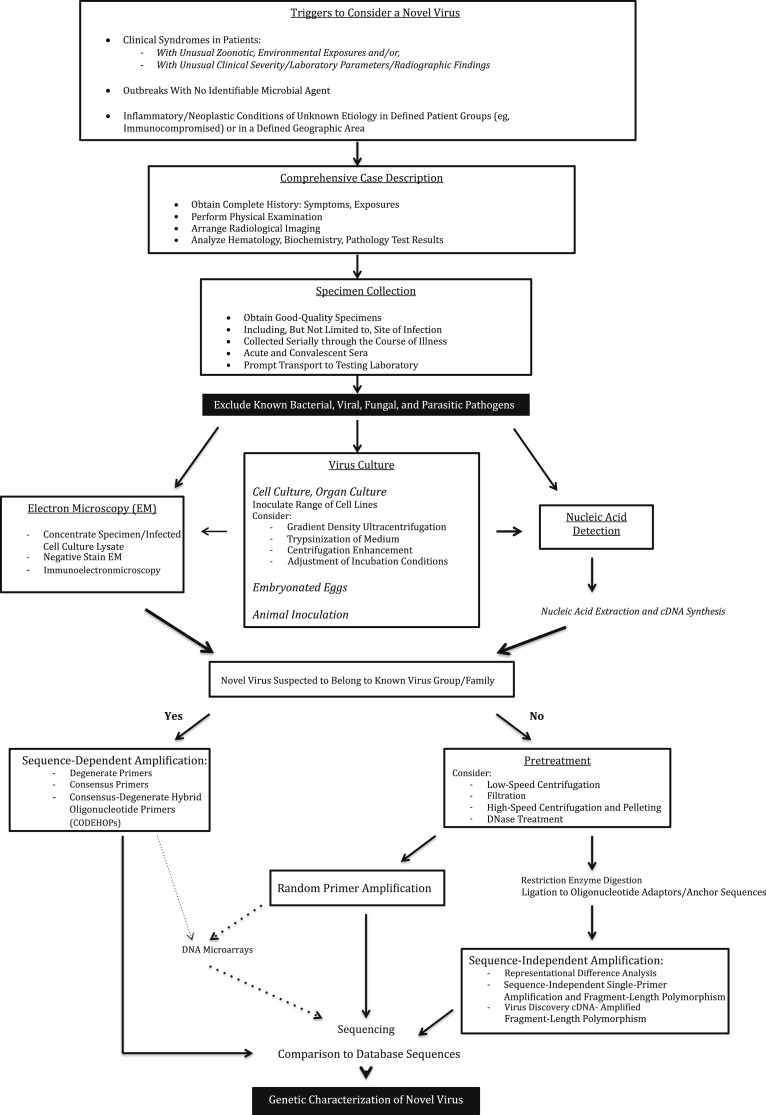
The process of virus discovery begins with case selection (namely, the identification of a patient or a group of patients with distinctive clinical features that could represent infection by a novel virus in a population with little herd immunity) (Figure 1 ). Such distinctive features may include unusually severe clinical presentations, an increased incidence of the disease syndrome above the baseline, uncommonly seen radiological features, and/or idiosyncratic laboratory test results. Patients may report unusual dietary, environmental, zoonotic, sexual, or travel-related exposures that alert the clinician to the possibility of a rare or new pathogen. The emerging novel virus may also manifest as an outbreak of illness with no identifiable microbial cause. A thorough history, physical examination, and basic laboratory results will enable the formulation of a precise case definition, and facilitate case finding and outbreak investigation. Looking for potentially affected cases among close contacts (household, occupational, nosocomial, and sexual contacts) must always be considered because it provides evidence for transmissibility of the disease, indicating an infectious etiology. In addition to infectious disease syndromes, viruses play a direct carcinogenic role in many neoplasias; infectious etiologies should be considered in neoplasias occurring in immunocompromised patients4 (eg, HIV-positive or post-organ transplant patients) or in defined geographical areas.
Figure 1.
Algorithm for novel virus discovery.
Scenario 1: Discovery of SARS-CoV
In 2002, rumors of an outbreak of fatal pneumonia of unknown cause were reported in southern China by the mass media and later confirmed by government officials. Affected individuals were often linked in time and nosocomial transmission chains, with some early cases reporting an occupational exposure to wild caged animals in markets and restaurants. Case definitions and outbreak information widely disseminated over the World Wide Web enabled the efficient identification of SARS patients by health care workers. Research efforts culminated in the cell culture isolation (discussed in detail later) of the novel SARS-CoV from two such patients presenting with severe pneumonia.5
Scenario 2: Discovery of Hepatitis E Virus
A major epidemic of waterborne icteric illness occurred in the Kashmir valley of India from 1978 to 1979. In a resource-poor setting with minimal access to serological confirmation, the epidemic was initially attributed to hepatitis A. However, the unusual severity of the illness in pregnant women, a feature not associated with hepatitis A infection, prompted investigators to perform extensive clinical and serological surveillance of affected patients. This led to the recognition that patients in the epidemic did not have any serological evidence of recent hepatitis A infection. Case definitions and clinical description of the novel non-A, non-B hepatitis entity enabled the recognition of similar patients globally, leading to the discovery of the hepatitis E virus from the stool of a human volunteer (discussed in detail later) through immunoelectron microscopy.6
Specimen Collection and Processing
Careful specimen collection from patients suspected to harbor novel viruses is crucial to their successful recovery. Clinical specimens must be of good quality and obtained serially throughout the course of illness to capture the novel virus at the time of peak viral load. Viral load dynamics vary, but in general, specimens that are collected early in the course of the illness just before the illness nadir are likely to contain a high viral load. The clinical syndrome often dictates the sites of specimen collection. For example, lower respiratory tract specimens are particularly valuable in patients presenting with pneumonia. However, specimen collection must not be limited to these sites alone because the detection of the novel virus at body sites distant from the focus of infection may illustrate important points regarding tissue tropism and routes of transmission of the new virus, as was illustrated by the high fecal viral loads of SARS-CoV. In general, respiratory tract aspirates, throat swabs, urine, feces, buffy coat, plasma, tissue biopsy specimens, vesicle fluids, and cerebrospinal fluid may all be considered for virus detection in appropriate clinical settings. In addition, acute and convalescent sera should always be stored to retrospectively identify an antibody response to the newly detected virus. Discovery of a novel virus in specimens does not necessarily mean causality, but the detection of an increasing antibody response in temporal relation to the illness is good evidence in support of an infectious role for the novel agent, especially in situations where Koch postulates cannot be fulfilled.
Freshly collected specimens must be properly labeled and speedily transported to the laboratory in leak-proof containers holding virus transport medium. Preliminary processing includes inoculation of the specimen into cell cultures and nucleic acid extraction. Clinical material may also be inoculated into embryonated eggs or suckling mice, depending on availability. The specimen may be filtered and centrifuged before RNA and DNA extraction to remove as much cell and bacterial debris as possible. The specimens should be refrigerated at −80°C with cryopreservative if any delay in processing is anticipated. However, when facing a novel virus with unknown stability outside living systems, such delays may adversely affect recovery and should be avoided as far as possible.
While subjecting clinical specimens to tests for novel virus detection, it is important to concomitantly undertake exhaustive specific assays for known bacterial, viral, fungal, and parasitic pathogens that could also account for the clinical scenario. The detection of a known pathogen not only has a significant impact on patient management, but also weakens the etiological link between the disease syndrome and any new virus that may be discovered. However, the detection of a known agent may not necessarily exclude the necessity of searching for a new virus, especially if the patient shows clinical features that are atypical. Polymicrobial or coinfections are always possible; this is especially true of bacterial respiratory tract infections, which are often preceded or accompanied by viral infections.7
Discovery of coronavirus HKU1 was performed as follows. An elderly patient presented with acute community-acquired pneumonia. Chest radiography showed patchy infiltrates over the left lower zone. Blood and sputum cultures were negative for recognized bacterial pathogens. Immunofluorescence staining of nasopharyngeal aspirates tested negative for influenza, parainfluenza, respiratory syncytial virus, and adenovirus. RT-PCR results for SARS-CoV, rhinovirus, and metapneumovirus were negative as well. The patient did not mount an antibody response against Legionella pneumophila, Mycoplasma pneumoniae, or Chlamydia species. A novel agent was suspected, and the patient's nasopharyngeal aspirate was subjected to consensus primer RT-PCR for the coronavirus pol gene, which revealed a compatible gene product consistent with a novel coronavirus on sequencing.8
Laboratory Tests for Detecting New Viruses
Before embarking on investigations to detect the novel virus, the safety of laboratory workers should be carefully considered. When dealing with novel viruses that cause severe clinical infection, laboratory acquisition of infection is a concern. The severity of the patients' clinical manifestation and suspected route of transmission serves as a guide to the level of physical containment required.
Each virus detection method has relative merits and demerits (Table 1 ). These must be carefully considered before the method is selected, especially when specimen quantity is limited.
Table 1.
Merits and Demerits of Virus Detection Strategies
| Detection method | Merits | Demerits |
|---|---|---|
| Culture | ||
| Inoculation of clinical specimens into the following: cell lines, organ cultures, embryonated eggs, and small mammals | Isolation of viable virus and provides information on cell tropism and pathogenesis of novel virus | Available cell lines may not be susceptible to novel virus, time consuming in outbreak settings, adaptation of novel virus to culture environment to produce quasispecies that may have different characteristics to the virus in vivo |
| Electron microscopy | Unbiased screening method and morphological appearance can guide choice of consensus PCR primers | Poor sensitivity and labor intensive |
| DNA microarrays | Parallel comparison of novel viral sequence to sequences from many virus families | Non-specific hybridization and divergent novel viruses may be missed |
| Sequence-dependent amplification using degenerate primers, consensus primers, and consensus-degenerate hybrid oligonucleotide primers | Fast, suitable screening method for outbreak settings | Divergent novel viruses may be missed and challenging primer design |
| Sequence-independent amplification using DNase-SISPA, VIDISCA, and random primer PCR | No prior knowledge of viral genome required and convenient and efficient detection of viral genomes in combination with high-throughput sequencing | Effect of exonuclease pretreatment on recovery of fragile enveloped viruses uncertain, unique virus sequences may still be missed, and expensive, not readily available in resource-limited settings |
Virus Culture
The culture of novel viruses should always be attempted; this is the only means to obtain viable virus for enabling study of phenotypic and growth characteristics of the novel agent while also facilitating the design of diagnostic assays, animal models, and vaccines. Despite its advantages, virus culture often entails a hit-and-miss approach, because the growth requirements of the novel virus are unknown. It must also be remembered that the culture technique may select out quasispecies of the novel virus that are most adapted to the culture medium used, especially on repeated passage, as exemplified by the case of influenza virus. Such culture adaptation may result in different receptor specificity and growth characteristics to the virus in vivo.9
The culture of novel viruses can be attempted using cell line, organ culture, small mammal, or embryonated egg inoculation.
Cell Culture
Although the clinical syndrome may guide the choice of cell line for inoculation, the specimen should be inoculated into as wide range of cell lines as possible. This proved to be the case with SARS-CoV when initial culture in conventional cell lines, such as Madin Darby canine kidney, LLC-Mk2, HEp-2, MRC-5, and RDE cells, proved unsuccessful, only for the virus to produce cytopathic effect (CPE) within 2 to 4 days of inoculation in fetal rhesus kidney cells.5
However, even if a susceptible cell line with appropriate receptors and coreceptors for the virus is fortuitously chosen, the isolation of a novel virus may still be limited by numerous other physicochemical factors.10 Viable virus may fail to bind to cell surface receptors in vitro due to suboptimal electrostatic interactions between the virus and cell surface. This may be overcome by pretreatment of the inoculum with cationic liposomes and polymers, such as diethylaminoethyl-dextran, polybrene, and poly-l-lysine. This has been shown to boost infectivity, CPE, and viral plaque sizes.11, 12 This enhancement may occur by reducing the electrostatic repulsion between the virus particle and cell surface and by increasing the sedimentation rate of flocculated virus particles onto the cell surface.
Gradient density ultracentrifugation can be used to reduce impurities in clinical specimens and culture suspensions before inoculation or passage into cell lines. Sucrose gradients are most commonly used and are suitable for enveloped viruses. Cesium chloride gradients are easier to prepare, but generally require longer centrifugation times and are unsuitable for enveloped viruses. Iodixanol density gradients offer the advantage of being isosmotic and noncytotoxic to cell lines, thereby preserving the infectivity of fragile enveloped viruses.13
Trypsinization of the medium should always be considered. Trypsin treated with protease inhibitor tosyl phenylalanyl chloromethyl ketone results in proteolytic cleavage and activation of virus surface glycoproteins, which is a prerequisite for cell binding and fusion for some RNA viruses and dramatically increases their infectivity in vitro. The biological niche of the novel virus must always be considered in deciding additives to culture media. For example, replication of porcine calicivirus associated with epidemic diarrhea (in pigs) is increased in the presence of intestinal fluid filtrate and bile acids.14
Low-speed centrifugation of infected cells at 1000 to 2000 × g in shell vials before incubation has been established to increase the recovery of many viruses that may be slow growing in conventional culture.10 Cultures are usually maintained at 35°C to 37°C, but incubation at lower temperatures may be rewarding if the natural site of infection is at a cooler area, such as the upper respiratory tract, as is the case with rhinoviruses.
Culture in Embryonated Eggs
Embryonated eggs may be used concomitantly with cell cultures. Because the cellular tropism of the putative novel virus is uncertain, inoculation into the amniotic cavity is always advisable because this route offers a greater variety of tissue types for virus propagation due to direct ingestion and inhalation by the chick embryo. Ten- to eleven-day-old embryos are commonly used for this purpose. If novel poxviruses are suspected, inoculation into the chorioallantoic membrane will enable detection of pocks. Novel viruses isolated by these techniques may then be adapted to grow in the allantoic entoderm to increase yield for further study.
Culture in Small Mammals
If a small mammal facility is available, inoculation of newborn suckling mice with suspensions of novel virus should also be considered. The animals should be observed for clinical symptoms over a fixed period of time. After sacrifice, harvested tissues are subjected to histological/electron microscopy (EM) examination or nucleic acid amplification/detection strategies. Homogenized organ tissues containing amplified novel virus may also be used for cell line inoculation or further passage in suckling mice after freeze thawing. Small mammal inoculation enables the study of the clinical and histological effects of experimental inoculation in a healthy living organism. Furthermore, tissue suspensions from inoculated mice provide a source of whole virus or virus antigen for many applications, such as designing serological assays. Finally, successful virus passage in laboratory animals demonstrates that the novel virus produces transmissible infection in an experimental host and forms the basis for developing animal models of the novel virus infection. Intracerebral, intraperitoneal, subcutaneous, and intranasal routes of suckling mice inoculation may all be used.
Inoculated cell cultures, embryonated eggs, or suckling mice should be monitored daily for evidence of CPE or disease, as appropriate (Figure 2 A). Cell or tissue lysates should be subjected to blind passage in the absence of CPE. EM and nucleic acid detection should be performed on cell lysates to look for evidence of the novel virus.
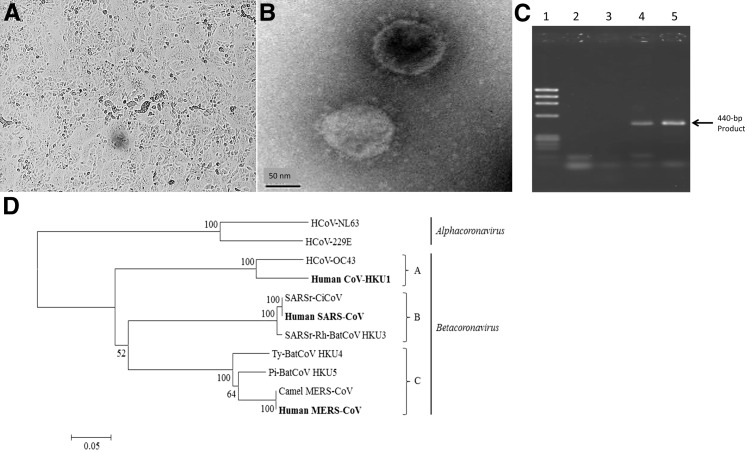
Figure 2.
Methods of virus detection applied to novel coronavirus discovery. A: Vero kidney epithelial cells infected by Middle East respiratory syndrome–coronavirus (MERS-CoV) showing cytopathic effect in the form of cell shrinkage and syncytium formation. B: Electron micrograph of severe acute respiratory syndrome coronavirus (SARS-CoV) showing characteristic enveloped virus particles with protruding spikes. C: Gel electrophoresis image of DNA amplified by coronavirus pol gene-derived consensus primer RT-PCR. Lane 1, bacteriophage ΦX174 DNA HaeIII digest marker; 2, negative sample; 3, negative control; 4, positive control; 5, nasopharyngeal aspirate sample with human coronavirus HKU1. D: Phylogenetic analysis of selected human and animal coronaviruses on the basis of the pol gene by the neighbor joining method. Lineage A betacoronavirus (A), lineage B betacoronavirus (B), and lineage C betacoronavirus (C).
Electron Microscopy
EM offers an unbiased method for visualization of virus-like particles in clinical specimens, cell culture fluid, and tissue sections (Figure 2B). Yield is not affected by the viability of the virus, and detection is possible as long as the virus is present in sufficient concentrations in the specimen. However, this method lacks sensitivity, especially when used directly on clinical specimens, because successful visualization requires a virus concentration of 105 to 106 particles/mL of specimen. Such concentrations are found in few clinical specimens other than stool; however, the high viral loads in sera of hepatitis B, ebola, early phase of parvovirus B19 virus infections and vesicular fluid containing poxvirus and herpes viruses are notable exceptions. Therefore, EM is more commonly used for visualization of putative novel viruses producing CPE in culture systems and facilitating choice of consensus primers on the basis of morphological appearance.
There are several methods that may improve the sensitivity of EM screening. Some form of specimen concentration (eg, ultracentrifugation, ultrafiltration, or agar diffusion) before EM screening is recommended. Glow discharging to render grid carbon support films hydrophilic for improved adsorption of biological material is widely available. Another useful technique for virus concentration is immunoelectron microscopy, in which clinical specimens suspected to harbor a novel virus are incubated with the patient's convalescent serum before visualization. Demonstration of aggregation of virus-like particles after treatment with convalescent sera was critical for the discovery of hepatitis A, hepatitis E, and diarrheal viruses.5, 15, 16 The advantages of immunoelectron microscopy are twofold—through aggregation, it enables the identification of small particles with indistinct surface features as viruses; by comparison of virus-like particle–antibody coating density between acute and convalescent sera, seroconversion can be indirectly demonstrated. This provides support for a specific host response to the agent, thus strengthening the causative link between the new virus and the disease syndrome.
Nucleic Acid Detection
Sensitive methods for the amplification, detection, sequencing, and analysis of viral nucleic acids have revolutionized the process of virus discovery (Table 2 ). When faced with a multitude of methods and protocols, it is important for investigators to ask themselves whether the suspected novel virus belongs to a known virus family or, alternatively, represents the first member of a distinct taxonomic archipelago. This can be inferred with varying degrees of certainty from factors such as the clinical disease syndrome, culture isolation pattern, and EM appearance (if available). Such information guides the choice of method for the detection of novel viral nucleic acid when no knowledge of the novel viral sequence is available. One distinct advantage of direct sequencing from clinical specimens is the avoidance of virus mutations, which is inevitable during adaptation of the virus in cell culture systems.
Table 2.
Examples of Novel Human and Animal Viruses Discovered by Various Nucleic Acid Detection Strategies
| Discovery method | Human viruses | Animal viruses |
|---|---|---|
| Virus discovery microarrays | Novel human cardiovirus17 | Beluga whale coronavirus18 |
| Sequence-dependent amplification techniques | Coronavirus HKU1,7 Bundibugyo ebola virus,19 novel Hanta virus,20 TTV-like minivirus,21 and rhinovirus C22 | Bat sapovirus23; bovine astrovirus24; avian, bat, camel, rabbit, and dolphin coronaviruses25, 26, 27, 28, 29, 30, 31; canine and murine noroviruses32, 33; porcine, bat, feline, and murine paramyxoviruses34, 35, 36, 37; feline, canine, bat, and avian picornaviruses38, 39, 40, 41; feline and canine bocavirus42; and porcine and bovine parvovirus43 |
| Sequence-independent amplification techniques | ||
| Representational difference analysis/digital transcriptome subtraction | Human herpesvirus 8,4 Torque teno virus,44 and Merkel cell polyomavirus2 | Chimpanzee GB virus45 |
| Virus discovery cDNA amplified fragment-length polymorphism, with or without high-throughput sequencing | Novel genotype of torque teno minivirus,46 novel parechovirus,47 and novel HIV-1 variant48 | Novel bat parvoviruses49 |
| DNase sequence-independent single-primer amplification | Novel picornavirus,50 novel parvovirus,51 and novel adenovirus genotype52 | Bovine parvoviruses53 |
| Unbiased high-throughput sequencing of random primer PCR amplification products | Transplant-associated arenavirus,54 Cosavirus, Klassevirus/Salivirus,55, 56 novel human bocavirus,57 novel picobirnavirus,58 Saffold virus,59 New Jersey polyomavirus,60 novel astrovirus,61 and novel papillomavirus62 | Novel bat papillomavirus63 and camel hepatitis E virus genotype64 |
Methods to Identify Novel Viruses Suspected to Belong to a Known Family
Hybridization-Based Assays and Microarrays
An early example of this method was the discovery of human papillomavirus 16 DNA in cervical cancer specimens,65 which was achieved by the demonstration of novel papillomaviral DNA in cancer specimens by using human papillomavirus 11 probe hybridization under low-stringency conditions. Specific probes developed using the novel DNA were found to hybridize with cervical cancer DNA from several patients, even under high-stringency conditions.
Probe hybridization using panviral DNA microarrays that incorporate many 70-mer oligonucleotides derived from highly conserved sequences of several pathogenic virus families can enable virus discovery.17, 18 The long probes hybridize to novel viral sequences even if sequence complementarity is less than perfect. Novel viruses that are related to represented viruses may produce a unique hybridization pattern, despite not being explicitly represented in the microarray. This provides an efficient, unbiased, massively parallel screening strategy for viral nucleic acid in clinical specimens after amplification by random PCR. However, the use of long-nucleotide probes in virus discovery arrays increases the risk of hybridization with non-specific random PCR products. Microarray analysis was used in parallel with consensus primer amplification-sequencing by a group of investigators to identify coronavirus sequences in patients with SARS.66
Amplification-Based Assays
If the novel virus is strongly suspected to be a member of a known group of viruses, then pools of degenerate primers that encompass all possible nucleotide differences in a given sequence may be used to amplify gene segments in the clinical specimen or infected cell culture extract. Such degenerate primers are derived from amino acid motifs of highly conserved proteins of the virus group of interest. Conserved blocks (usually pentapeptides) are selected from the amino acid sequences of these proteins that preferably contain amino acids encoded by fewer codons (eg, methionine) so as to limit the level of degeneracy of the primer pool to <128-fold. A higher degeneracy would lead to a decrease in sensitivity of the assay. Degeneracy at the 3′ end of the primer should be especially avoided because this compromises primer extension.
An alternative approach is to derive consensus primers directly from the nucleotide sequences of conserved proteins of a particular virus family. Consensus primer PCR uses primer pairs constituted from the most common nucleotide variant at each position of the primer sequence and relies on the ability of this primer pair to amplify closely related target DNA. However, the sensitivity of this approach is entirely dependent on the degree of complementarity of the target DNA sequence to the consensus primers. Consensus primers derived by multiple alignment of known coronavirus pol gene nucleotide sequences enabled the discovery of human coronavirus HKU1 and several other novel animal coronaviruses8, 25, 26, 27, 28, 29, 30, 31 (Figure 2, C and D).
A different strategy combining the strengths of degenerate and consensus primers is the use of consensus-degenerate hybrid oligonucleotide primers (CODEHOPs).67 CODEHOPs contain a degenerate 3′ core region and a conserved 5′ clamp sequence. The limited level of degeneracy in the 3′ core maintains the relative lack of bias of degenerate primers, whereas the conserved 5′ clamp sequence stabilizes hybridization of the 3′ core and allows higher annealing temperatures. Interactive software programs for designing CODEHOPs are now available.68
A recent approach to the discovery of novel equine hepaciviruses is selection of horse serum specimens that are immunoreactive to highly conserved hepacivirus antigens using a luciferase immunoprecipitation assay.69 Positive sera were then tested with hepacivirus consensus primers, followed by sequencing of amplification products. When a large specimen pool is available, this approach enables efficient selection of specimens for screening by consensus primer PCR or high-throughput sequencing. However, antibody responses are often associated with decreasing viral loads for many acute viral infections. Therefore, this strategy could lack sensitivity for the discovery of novel viruses that do not establish chronicity.
Methods to Identify Novel Viruses of Unknown Sequence Suspected to Belong to a New Taxon
The utility of consensus primer PCR is limited if the sequence of the novel virus is highly divergent from that of known virus families. This scenario requires unbiased screening of the specimen for viral nucleic acid. However, the quantity of putative viral nucleic acid is usually scarce compared to host DNA, making such screening a tedious exercise. Overcoming this obstacle generally requires targeted amplification of nucleic acid extracted from nuclease-pretreated specimens, followed by selective detection and sequencing of viral nucleic acid.
Generation and Analysis of cDNA Libraries
The discovery of the hepatitis C virus is an early example of amplification and selective detection of viral nucleic acid.70 cDNA phage libraries obtained from infected chimpanzees were screened for a clone that failed to hybridize with human or chimpanzee DNA, but hybridized selectively with nucleic acid from infected liver tissues. Analysis of these cDNA clones enabled the design of specific primers and serological assays that could be used to directly screen for hepatitis C infection in different patient populations. This technique is time consuming and labor intensive. It also relies on the amplification of the virus in a biological system, which is unlikely to be available for most novel viral infections.
Representational Difference Analysis
Representational difference analysis was first developed for defining the difference between tester DNA, believed to contain the target nucleic acid, and driver DNA, which represents the wild type.71 Oligonucleotide adaptor ligation and adaptor-specific PCR amplify both cDNA populations. Hybridization of tester with adaptor-ligated driver DNA follows. Sequences common to both populations are subtracted while adaptor-specific primers amplify sequences unique to the tester population for subsequent sequencing. The discovery of human herpesvirus 8 and TT virus was made using this method.4, 44 However, this method is laborious. Furthermore, obtaining noninfected tissue for driver amplicon extraction from the same patient is often not possible.
Digital Transcriptome Subtraction
The availability of high-quality human genome sequences enables subtraction of high-fidelity sequences derived from serial analysis of gene expression libraries to be performed in silico by comparison with reference human sequence databases. Developed for the purpose of screening for viral nucleic acid in tumor tissues, this method has been used for the discovery of polyomavirus sequences in patients with Merkel cell carcinoma.2
SISPA
DNase-sequence–independent single-primer amplification (SISPA) is based on the nonselective amplification of template DNA by a single primer.53 The filtered specimen is pretreated with DNase to selectively degrade host DNA (capsid-associated viral DNA is relatively protected from enzymatic degradation) before nucleic acid extraction. Extracted cDNA is then digested with a restriction enzyme (eg, CspI). DNA fragments are ligated to primer-binding oligonucleotide adaptors. Amplification is performed using a single primer specific to the adaptor sequence. Analysis of the SISPA products is achieved by gel electrophoresis, followed by sequencing of bands of interest. This method has proved useful for the discovery of novel viruses from human specimens.50, 51 DNase SISPA was also used to characterize a novel adenovirus genotype isolated from patients in an outbreak of gastroenteritis.52
VIDISCA
Virus discovery cDNA amplified fragment length polymorphism (VIDISCA) was originally described for the detection of polymorphisms in a DNA population.72 As in SISPA, specimens are treated with DNase to degrade host genomic DNA before nucleic acid extraction. Extracted cDNA is digested and ligated to anchor sequences at restriction sites. Two rounds of PCR amplification then follow: the first round uses primers that anneal to the anchor sequences; the second round uses identical primers as the first round, extended by one nucleotide at the 3′ end, outside the anchor sequence and within the putative viral nucleic acid. Because the additional nucleotide may be any one of the nucleotides (A, T, G, or C), it provides 16 possible primer combinations for analysis of the infected material. The PCR products are analyzed by gel electrophoresis to identify unique bands in infected material, which are sequenced. VIDISCA was used to characterize a novel HIV-1 subtype and a torque teno virus variant.46, 48 The VIDISCA procedure has been combined with next-generation sequencing (VIDISCA 454) for the analysis of PCR products and has been applied to tissue, serum, stool, respiratory samples, and cell culture specimens. A procedure for pretreatment of respiratory and stool specimens with convalescent serum for further enrichment of virus particles before VIDISCA 454 amplification and sequencing has been described.73
RCA
Rolling circle amplification (RCA) protocols for the detection of novel viruses use multiple random hexamer primers that bind to different points of the circular genome. Most RCA applications use phi29 DNA polymerase, which possesses strand displacement and 3′ to 5′ exonuclease activities, to produce linear concatamerized double-stranded DNA copies of the novel viral genome. PCR products are then digested with a restriction enzyme that is likely to only have a single recognition site in the viral genome, which is then followed by gel electrophoresis, sequencing, and phylogenetic analysis. This method has been used for the discovery of several novel viruses that possess a circular DNA genome, such as papillomaviruses, polyomaviruses, and anneloviruses.74, 75
Random Primer PCR and High-Throughput Sequencing
Random primer PCR and high-throughput sequencing have gained popularity among investigators undertaking metagenomic analysis of specimens from patients with viral infection syndromes of unknown etiology. The specimen is pretreated with DNase. Extracted cDNA is then used as a template for amplification using random primers. The amplification products are separated by gel electrophoresis. Unique fragments not found in controls are sequenced and analyzed. Random primer PCR and high-throughput sequencing have facilitated the discovery of human bocavirus, SARS-CoV, novel picobirnavirus, arenavirus, and picornaviruses.5, 54, 55, 56, 57, 58
Unbiased high-throughput sequencing of randomly amplified DNA is a powerful method for identification of novel pathogens and has been used for the identification of several novel viruses using a metagenomic approach.54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64 The reads generated by high-throughput sequencing are trimmed to remove primer, eukaryotic, bacterial, and highly repetitive sequences. They are then assembled to form nucleotide sequences from which amino acid sequences can be inferred. Bioinformatic analysis then follows to assess for homology with known viral nucleotide and protein sequences and even de novo virus genome assembly. These sequences are then used to design specific PCR primers that enable subsequent case finding and further genomic characterization. With increasingly sophisticated automation and software support, unbiased high-throughput sequencing of randomly amplified PCR products from pretreated clinical specimens is likely to be an increasingly important method for novel virus screening in clinical specimens and environmental samples. However when compared with other nucleic acid amplification approaches, most of the discovered viruses using the DNase-SISPA approach are nonenveloped virus (Table 2),50, 51, 52, 53 because the presequencing enrichment step may damage the delicate virus envelope.
Conclusions: Beyond Virus Discovery
Virus discovery is the first step in a long process of establishing a clinical association and a causative link between the virus and the disease entity, which rely on case-control studies and demonstration of seroconversion in patients and infected animals. Demonstration of viral antigen and nucleic acid in tissue specimens using immunohistochemical stains and specific primers for the novel agent serves as another link in the chain of causation. Causation is definitively established by the fulfillment of traditional Koch postulates in animal models, if available. Once causation is established, the newly found phenotypic and genotypic characteristics of the novel virus enable the development of diagnostic tests, vaccines, and specific antiviral therapy. In addition, the origins of the novel virus may be inferred by field studies of vectors and nonhuman hosts, as in the case of SARS-CoV. Even when the novel virus cannot be conclusively associated with human disease at the time of discovery, the availability of its genetic sequences in public databases is important because the novel virus or a close relative may evolve into an emerging human infection in the future. Rapid recognition of new diseases is possible with pre-existing knowledge of the virus, as illustrated by the relationship between Middle East respiratory syndrome coronavirus and the previously identified bat coronaviruses HKU4 and HKU5.27 The discovery of novel viruses continues to play an important role in the control of infectious diseases.
With the established trend of increasing technological sophistication involved in uncovering novel viruses, virus discovery sometimes falls outside the capabilities of clinical virology laboratories and requires facilities in research or reference laboratories. Increasingly, discovery requires a collaborative effort from various parties (ie, frontline clinicians, clinical virologists, laboratory technicians, and basic scientists). Such collaborations will increasingly become multinational because clinical specimens from novel disease outbreaks in resource-poor settings can be rapidly transported to overseas laboratories for timely virus discovery using a battery of methods that are not available in the field.
The detection of novel viruses cannot depend on any one method. Rather, discovery often relies on a combination of methods after taking into account the merits and demerits of each method. The exact combination will depend on specific circumstances, as was the case with SARS-CoV. Although culture-, EM-, and consensus primer PCR-based methods will continue to play important roles, molecular methods based on unbiased next-generation sequencing offer unique opportunities for efficient and systematic virus discovery.
Acknowledgment
We thank Alan Tsang for designing the phylogenetic tree.
Footnotes
Supported in part by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the Hong Kong Special Administrative Region (HKSAR) Department of Health and the HKSAR Research Fund for the Control of Infectious Diseases commissioned grant.
Disclosures: None declared.
References
- 1.Woolhouse M.E., Howey R., Gaunt E., Reilly L., Chase-Topping M., Savill N. Temporal trends in the discovery of human viruses. Proc Biol Sci. 2008;275:2111–2115. doi: 10.1098/rspb.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffin D.E. Measles virus-induced suppression of immune responses. Immunol Rev. 2010;236:176–189. doi: 10.1111/j.1600-065X.2010.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 5.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y., SARS Study Group Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balayan M.S., Andjaparidze A.G., Savinskaya S.S., Ketiladze E.S., Braginsky D.M., Savinov A.P., Poleschuk V.F. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology. 1983;20:23–31. doi: 10.1159/000149370. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs S.E., Lamson D.M., St. George K., Walsh T.J. Human rhinoviruses. Clin Microbiol Rev. 2013;26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roedig J.V., Rapp E., Höper D., Genzel Y., Reichl U. Impact of host cell line adaptation on quasispecies composition and glycosylation of influenza A virus hemagglutinin. PLoS One. 2011;6:e27989. doi: 10.1371/journal.pone.0027989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes J.H. Physical and chemical methods for enhancing rapid detection of viruses and other agents. Clin Microbiol Rev. 1993;6:150–175. doi: 10.1128/cmr.6.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konopka K. Enhancement of retroviral transduction by cationic liposomes. Methods Enzymol. 2003;373:493–506. doi: 10.1016/S0076-6879(03)73031-6. [DOI] [PubMed] [Google Scholar]
- 12.Pagano J.S., Vaheri A. Enhancement of infectivity of poliovirus RNA with diethylaminoethyl-dextran (DEAE-D) Arch Gesamte Virusforsch. 1965;17:456–464. doi: 10.1007/BF01241201. [DOI] [PubMed] [Google Scholar]
- 13.Gias E., Neilsen S.U., Morgan L.A.F., Toms G.L. Purification of human respiratory syncytial virus by ultracentrifugation in iodixanol density gradient. J Virol Methods. 2008;147:328–332. doi: 10.1016/j.jviromet.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang K.O., Sosnovtsev S.V., Belliot G., Kim Y., Saif L.J., Green K.Y. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc Natl Acad Sci U S A. 2004;101:8733–8738. doi: 10.1073/pnas.0401126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinstone S.M., Kapikian A.Z., Purcell R.H. Hepatitis A: detection by immune electron microscopy of a virus like antigen associated with acute illness. Science. 1973;182:1026–1028. doi: 10.1126/science.182.4116.1026. [DOI] [PubMed] [Google Scholar]
- 16.Kapikian A.Z., Wyatt R.G., Dolin R., Thornhill T.S., Kalica A.R., Chanock R.M. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. 1972;10:1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu C.Y., Greninger A.L., Kanada K., Kwok T., Fischer K.F., Runckel C., Louie J.K., Glaser C.A., Yagi S., Schurr D.P., Haggerty T.D., Parsonnet J., Ganem D., DeRisi J.L. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A. 2008;105:14124–14129. doi: 10.1073/pnas.0805968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mihindukulasuriya K.A., Wu G., St Leger J., Nordhausen R.W., Wang D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towner J.S., Sealy T.K., Khristova M.L., Albariño C.G., Conlan S., Reeder S.A., Quan P.L., Lipkin W.I., Downing R., Tappero J.W., Okware S., Lutwama J., Bakamutumaho B., Kayiwa J., Comer J.A., Rollin P.E., Ksiazek T.G., Nichol S.T. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nichol S.T., Spiropoulou C.F., Morzunov S., Rollin P.E., Ksiazek T.G., Feldmann H., Sanchez A., Childs J., Zaki S., Peters C.J. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi K., Iwasa Y., Hijikata M., Mishiro S. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch Virol. 2000;145:979–993. doi: 10.1007/s007050050689. [DOI] [PubMed] [Google Scholar]
- 22.Lamson D., Renwick N., Kapoor V., Liu Z., Palacios G., Ju J., Dean A., St George K., Briese T., Lipkin W.I. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse H., Chan W.M., Li K.S., Lau S.K., Woo P.C., Yuen K.Y. Discovery and genomic characterization of a novel bat sapovirus with unusual genomic features and phylogenetic position. PLoS One. 2012;7:e34987. doi: 10.1371/journal.pone.0034987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tse H., Chan W.M., Tsoi H.W., Fan R.Y., Lau C.C., Lau S.K., Woo P.C., Yuen K.Y. Rediscovery and genomic characterization of bovine astroviruses. J Gen Virol. 2011;92:1888–1898. doi: 10.1099/vir.0.030817-0. [DOI] [PubMed] [Google Scholar]
- 25.Woo P.C., Lau S.K., Li K.S., Poon R.W., Wong B.H., Tsoi H.W., Yip B.C., Huang Y., Chan K.H., Yuen K.Y. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo P.C., Lau S.K., Lam C.S., Tsang A.K., Hui S.W., Fan R.Y., Martelli P., Yuen K.Y. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J Virol. 2014;88:1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo P.C., Lau S.K., Wernery U., Wong E.Y., Tsang A.K., Johnson B., Yip C.C., Lau C.C., Sivakumar S., Cai J.P., Fan R.Y., Chan K.H., Mareena R., Yuen K.Y. Novel betacoronaviruses in dromedaries of the Middle East, 2013. Emerg Infect Dis. 2014;20:560–572. doi: 10.3201/eid2004.131769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau S.K., Woo P.C., Yip C.C., Fan R.Y., Huang Y., Wang M., Guo R., Lam C.S., Tsang A.K., Lai K.K., Chan K.H., Che X.Y., Zheng B.J., Yuen K.Y. Isolation and characterization of a novel betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol. 2012;86:5481–5496. doi: 10.1128/JVI.06927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tse H., Chan W.M., Lam C.S., Lau S.K., Woo P.C., Yuen K.Y. Complete genome sequences of novel rat noroviruses in Hong Kong. J Virol. 2012;86:12435–12436. doi: 10.1128/JVI.01976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tse H., Lau S.K., Chan W.M., Choi G.K., Woo P.C., Yuen K.Y. Complete genome sequences of novel canine noroviruses in Hong Kong. J Virol. 2012;86:9531–9532. doi: 10.1128/JVI.01312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo P.C., Lau S.K., Wong B.H., Fan R.Y., Wong A.Y., Zhang A.J., Wu Y., Choi G.K., Li K.S., Hui J., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in cats. Proc Natl Acad Sci U S A. 2012;109:5435–5440. doi: 10.1073/pnas.1119972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau S.K., Woo P.C., Wu Y., Wong A.Y., Wong B.H., Lau C.C., Fan R.Y., Cai J.P., Tsoi H.W., Chan K.H., Yuen K.Y. Identification and characterization of a novel paramyxovirus, porcine parainfluenza virus 1, from deceased pigs. J Gen Virol. 2013;94:2184–2190. doi: 10.1099/vir.0.052985-0. [DOI] [PubMed] [Google Scholar]
- 36.Lau S.K., Woo P.C., Wong B.H., Wong A.Y., Tsoi H.W., Wang M., Lee P., Xu H., Poon R.W., Guo R., Li K.S., Chan K.H., Zheng B.J., Yuen K.Y. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology. 2010;404:106–116. doi: 10.1016/j.virol.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo P.C., Lau S.K., Wong B.H., Wong A.Y., Poon R.W., Yuen K.Y. Complete genome sequence of a novel paramyxovirus, Tailam virus, discovered in Sikkim rats. J Virol. 2011;85:13473–13474. doi: 10.1128/JVI.06356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau S.K., Woo P.C., Yip C.C., Choi G.K., Wu Y., Bai R., Fan R.Y., Lai K.K., Chan K.H., Yuen K.Y. Identification of a novel feline picornavirus from the domestic cat. J Virol. 2012;86:395–405. doi: 10.1128/JVI.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo P.C., Lau S.K., Choi G.K., Yip C.C., Huang Y., Tsoi H.W., Yuen K.Y. Complete genome sequence of a novel picornavirus, canine picornavirus, discovered in dogs. J Virol. 2012;86:3402–3403. doi: 10.1128/JVI.07228-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau S.K., Woo P.C., Lai K.K., Huang Y., Yip C.C., Shek C.T., Lee P., Lam C.S., Chan K.H., Yuen K.Y. Complete genome analysis of three novel picornaviruses from diverse bat species. J Virol. 2011;85:8819–8828. doi: 10.1128/JVI.02364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo P.C., Lau S.K., Huang Y., Lam C.S., Poon R.W., Tsoi H.W., Lee P., Tse H., Chan A.S., Luk G., Chan K.H., Yuen K.Y. Comparative analysis of six genome sequences of three novel picornaviruses, turdiviruses 1, 2 and 3, in dead wild birds, and proposal of two novel genera, Orthoturdivirus and Paraturdivirus, in the family Picornaviridae. J Gen Virol. 2010;91:2433–2448. doi: 10.1099/vir.0.021717-0. [DOI] [PubMed] [Google Scholar]
- 42.Lau S.K., Woo P.C., Yeung H.C., Teng J.L., Wu Y., Bai R., Fan R.Y., Chan K.H., Yuen K.Y. Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J Gen Virol. 2012;93:1573–1582. doi: 10.1099/vir.0.042531-0. [DOI] [PubMed] [Google Scholar]
- 43.Lau S.K., Woo P.C., Tse H., Fu C.T., Au W.K., Chen X.C., Tsoi H.W., Tsang T.H., Chan J.S., Tsang D.N., Li K.S., Tse C.W., Ng T.K., Tsang O.T., Zheng B.J., Tam S., Chan K.H., Zhou B., Yuen K.Y. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J Gen Virol. 2008;89:1840–1848. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- 44.Nishizawa T., Okamoto H., Konishi K., Yoshizawa H., Miyakawa Y., Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 45.Birkenmeyer L.G., Desai S.M., Muerhoff A.S., Leary T.P., Simons J.N., Montes C.C., Mushahwar I.K. Isolation of a GB virus-related genome from a chimpanzee. J Med Virol. 1998;56:44–51. doi: 10.1002/(sici)1096-9071(199809)56:1<44::aid-jmv8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 46.Jazaeri Farsani S.M., Jebbink M.F., Deijs M., Canuti M., van Dort K.A., Bakker M., Grady B.P., Prins M., van Hemert F.J., Kootstra N.A., van der Hoek L. Identification of a new genotype of Torque Teno Mini virus. Virol J. 2013;10:323. doi: 10.1186/1743-422X-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Hoek L., Pollakis G., Lukashov V.V., Jebbink M.F., Jeeninga R.E., Bakker M., Dukers N., Jurriaans S., Paxton W.A., Back N.K., Berkhout B. Characterization of an HIV-1 group M variant that is distinct from the known subtypes. AIDS Res Hum Retroviruses. 2007;23:466–470. doi: 10.1089/aid.2006.0184. [DOI] [PubMed] [Google Scholar]
- 48.de Souza Luna L.K., Baumgarte S., Grywna K., Panning M., Drexler J.F., Drosten C. Identification of a contemporary human parechovirus type 1 by VIDISCA and characterization of its full genome. Virol J. 2008;5:26. doi: 10.1186/1743-422X-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canuti M., Eis-Huebinger A.M., Deijs M., de Vries M., Drexler J.F., Oppong S.K., Müller M.A., Klose S.M., Wellinghausen N., Cottontail V.M., Kalko E.K., Drosten C., van der Hoek L. Two novel parvoviruses in frugivorous New and Old World bats. PLoS One. 2011;6:e29140. doi: 10.1371/journal.pone.0029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones M.S., Lukashov V.V., Ganac R.D., Schnurr D.P. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J Clin Microbiol. 2007;45:2144–2150. doi: 10.1128/JCM.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones M.S., Kapoor A., Lukashov V.V., Simmonds P., Hecht F., Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones M.S., 2nd, Harrach B., Ganac R.D., Gozum M.M., Dela Cruz W.P., Riedel B., Pan C., Delwart E.L., Schnurr D.P. New adenovirus species found in a patient presenting with gastroenteritis. J Virol. 2007;81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A. 2001;98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P.L., Hui J., Marshall J., Simons J.F., Egholm M., Paddock C.D., Shieh W.J., Goldsmith C.S., Zaki S.R., Catton M., Lipkin W.I. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 55.Kapoor A., Victoria J., Simmonds P., Slikas E., Chieochansin T., Naeem A., Shaukat S., Sharif S., Alam M.M., Angez M., Wang C., Shafer R.W., Zaidi S., Delwart E. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A. 2008;105:20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Victoria J.G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapoor A., Slikas E., Simmonds P., Chieochansin T., Naeem A., Shaukat S., Alan M.M., Sharif S., Angez M., Zaidi S., Delwart E. A newly identified bocavirus species in human stool. J Infect Dis. 2009;199:196–200. doi: 10.1086/595831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Leeuwen M., Williams M.M., Koraka P., Simon J.H., Smits S.L., Osterhaus A.D. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol. 2010;48:1787–1794. doi: 10.1128/JCM.02452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zoll J., Erkens Hulshof S., Lanke S., Verduyn Lunel F., Melchers W.J., Schoondermark-van de Ven E., Roivanen M., Galama J.M., van Kuppeveld F.J. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 2009;5:e1000416. doi: 10.1371/journal.ppat.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishra N., Pereira M., Rhodes R.H., An P., Pipas J.M., Jain K., Kapoor A., Briese T., Faust P.L., Lipkin W.I. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis. 2014;210:1595–1599. doi: 10.1093/infdis/jiu250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finkbeiner S.R., Li Y., Ruone S., Conrardy C., Gregoricus N., Toney D., Virgin H.W., Anderson L.J., Vinjé J., Wang D., Tong S. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol. 2009;83:10836–10839. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mokili J.L., Dutilh B.E., Lim Y.W., Schenider B.S., Taylor T., Haynes M.R., Metzqar D., Myers C.A., Blair P.J., Nosrat B., Wolfe N.D., Rohwer P. Identification of a novel human papillomavirus by metagenomic analysis of samples from patients with febrile respiratory illness. PLoS One. 2013;8:e58404. doi: 10.1371/journal.pone.0058404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tse H., Tsang A.K., Tsoi H.W., Leung A.S., Ho C.C., Lau S.K., Woo P.C., Yuen K.Y. Identification of a novel bat papillomavirus by metagenomics. PLoS One. 2012;7:e43986. doi: 10.1371/journal.pone.0043986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woo P.C., Lau S.K., Teng J.L., Tsang A.K., Joseph M., Wong E.Y., Tang Y., Sivakumar S., Xie J., Bai R., Wernery R., Wernery U., Yuen K.Y. New hepatitis E virus genotype in camels, the Middle East. Emerg Infect Dis. 2014;20:1044–1048. doi: 10.3201/eid2006.140140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dürst M., Gissmann L., Ikenberg H., zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 67.Rose T.M., Schultz E.R., Henikoff J.G., Pietrokovski S., McCallum C.M., Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acid Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boyce R., Chilana P., Rose T.M. iCODEHOP: a new interactive program for designing COnsensus-DEgenerate Hybrid Oligonucleotide Primers from multiply aligned protein sequences. Nucleic Acids Res. 2009;37:W222–W228. doi: 10.1093/nar/gkp379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burbelo P.D., Dubovi E.J., Simmonds P. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012;86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 71.Lisitsyn N., Lisitsyn N., Wigler M. Cloning the difference between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 72.Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Detection of new viruses by VIDISCA: virus discovery based on cDNA-amplified fragment length polymorphism. Methods Mol Biol. 2008;454:73–89. doi: 10.1007/978-1-59745-181-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oude Munnink B.B., Jazaeri Farsani S.M., Deijs M., Jonkers J., Verhoeven J.T., Ieven M., Goosens H., de Jong M.D., Berkhout B., Loens K., Kellam P., Bakker M., Canuti M., Cotten M., van der Hoek L. Autologous antibody capture to enrich immunogenic viruses for viral discovery. PLoS One. 2013;8:e78454. doi: 10.1371/journal.pone.0078454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niel C., Diniz-Mendes L., Devalle S. Rolling-circle amplification of Torque teno virus (TTV) complete genomes from human and swine sera and identification of a novel swine TTV genogroup. J Gen Virol. 2005;86:1343–1347. doi: 10.1099/vir.0.80794-0. [DOI] [PubMed] [Google Scholar]
- 75.Li J., Pan Y., Deng Q., Cai H., Ke Y. Identification and characterization of eleven novel human gamma-papillomavirus isolates from healthy skin, found at a low frequency in a normal population. PLoS One. 2013;8:e77116. doi: 10.1371/journal.pone.0077116. [DOI] [PMC free article] [PubMed] [Google Scholar]